Abstract
Background
The treatment coverage of control programs providing benzimidazole (BZ) drugs to eliminate the morbidity caused by soil-transmitted helminths (STHs) is unprecedently high. This high drug pressure may result in the development of BZ resistance in STHs and so there is an urgent need for surveillance systems detecting molecular markers associated with BZ resistance. A critical prerequisite to develop such systems is an understanding of the gene family encoding β-tubulin proteins, the principal targets of BZ drugs.
Methodology and principal findings
First, the β-tubulin gene families of Ascaris lumbricoides and Ascaris suum were characterized through the analysis of published genomes. Second, RNA-seq and RT-PCR analyses on cDNA were applied to determine the transcription profiles of the different gene family members. The results revealed that Ascaris species have at least seven different β-tubulin genes of which two are highly expressed during the entire lifecycle. Third, deep amplicon sequencing was performed on these two genes in more than 200 adult A. lumbricoides (Ethiopia and Tanzania) and A. suum (Belgium) worms, to investigate the intra- and inter-species genetic diversity and the presence of single nucleotide polymorphisms (SNPs) that are associated with BZ resistance in other helminth species; F167Y (TTC>TAC or TTT>TAT), E198A (GAA>GCA or GAG>GCG), E198L (GAA>TTA) and F200Y (TTC>TAC or TTT>TAT). These particular SNPs were absent in the two investigated genes in all three Ascaris populations.
Significance
This study demonstrated the presence of at least seven β-tubulin genes in Ascaris worms. A new nomenclature was proposed and prioritization of genes for future BZ resistance research was discussed. This is the first comprehensive description of the β-tubulin gene family in Ascaris and provides a framework to investigate the prevalence and potential role of β-tubulin sequence polymorphisms in BZ resistance in a more systematic manner than previously possible.
Author summary
Benzimidazole (BZ) drugs remain the standard of treatment in large-scale deworming programs that aim to control the morbidity caused by intestinal worms. As these deworming programs are expanding world-wide, there is an increasing risk of worms becoming resistant to BZ drugs, highlighting the necessity for tools to detect gene mutations associated with drug resistance. However, the development of such tools is impeded by a lack of insights into the genes that are coding for β-tubulin proteins, which are the principal targets of BZ drugs. The aim of this study was to comprehensively characterize these genes in the worm species Ascaris lumbricoides and Ascaris suum. The findings highlight that these species have at least seven β-tubulin genes. Only two genes are highly expressed throughout the different life stages of the worm, and hence are more likely to be involved in the development of BZ resistance. No mutations that have previously been associated with BZ resistance in other intestinal worms were found. This study provides a baseline towards more efficient and accurate monitoring of drug resistance in large-scale deworming programs.
Introduction
The latest global reports on control programs for soil-transmitted helminths (STHs; Ascaris lumbricoides, Trichuris trichiura, Necator americanus and Ancylostoma duodenale) show that drug coverage continues to rise. In 2019, the benzimidazole (BZ) drugs albendazole (ALB) and mebendazole (MEB) were administered to 777.5 million people worldwide, which covered 58.64% of all (pre-)school-aged children ((pre-)SAC) in need of treatment (in 2010 this was only 30.94%) [1,2]. It is anticipated that this number will continue to increase since the target of the World Health Organization (WHO) is to reduce moderate-to-heavy intensity infection prevalence to less than 2% by 2030 [3]. As demonstrated repeatedly in animal STHs, the world-wide upscale in drug distribution increases the risk for the development of anthelmintic resistance, highlighting the necessity for tools to detect mutations in the genes that are encoding for β-tubulin proteins, the principal targets of BZ drugs [4,5]. A number of single nucleotide polymorphisms (SNPs) in β-tubulin genes (F167Y (TTC>TAC or TTT>TAT), E198A (GAA>GCA or GAG>GCG), E198L (GAA>TTA) and F200Y (TTC>TAC or TTT>TAT)) are associated with BZ resistance in a variety of animal STHs (e.g. Haemonchus contortus, Teladorsagia circumcincta and Ancylostoma caninum [6–24]) and conferred phenotypic resistance in the transgenic model organism Caenorhabditis elegans [25]. However, the number of β-tubulin genes and their relationships vary between nematode species, and so care needs to be taken when extrapolating information between species, particularly those that are more distantly related [15,26,27]. For H. contortus, which has four β-tubulin genes, the F167Y (TTC>TAC or TTT>TAT), E198A (GAA>GCA or GAG>GCG) and F200Y (TTC>TAC or TTT>TAT) resistance mutations have been detected in one β-tubulin gene (Hco-tbb-iso-1), often at high frequency in BZ resistant populations [10,17,19–24,28]. However, there is some evidence that also a second β-tubulin gene (Hco-tbb-iso-2) has a role in BZ resistance, although possibly less important [24,29]. The two remaining H. contortus β-tubulin genes (Hco-tbb-iso-3 and Hco-tbb-iso-4) are expressed at extremely low levels and in a restricted spatial manner and so are unlikely to have a role in resistance [27]. Experimental data for C. elegans, which has six β-tubulin genes, has shown that all BZ resistance mutations generated by chemical mutagenesis map to a single gene, namely Cel-ben-1 [27,30]. But there is evidence that BZ resistance in natural strains of C. elegans is not only associated with the loss of one β-tubulin gene and that there are multiple mechanisms underlying BZ resistance, involving multiple loci [31,32]. Although H. contortus and C. elegans are relatively closely related, both belonging to Clade V in the nematode phylogeny [33], the genes involved in BZ resistance do not share one-to-one orthology [27]. The complex β-tubulin phylogeny thereby not only complicates comparative analysis into the potential involvement of genes in BZ resistance but also makes a common nomenclature for the β-tubulin genes across species difficult. Today, little is known about the composition of the β-tubulin gene family in human STH species. Krücken and colleagues investigated four β-tubulin genes in A. lumbricoides [34], but remaining studies have focused on a single gene to explore the presence of SNPs that may be potentially associated with BZ resistance in human A. lumbricoides [35–41] and porcine Ascaris suum [42] populations. In A. lumbricoides, a reduced efficacy to BZ has been reported but no SNPs were found in the four investigated β-tubulin genes [34]. Other studies detected the SNP TTC>TAC in codon 167 but the mutation was not associated with reduced efficacy [35,37]. In A. suum, to date none of the BZ resistance associated SNPs have been described in the single β-tubulin gene that has been examined [42]. Consequently, for both species, the current evidence for these SNPs as a marker for BZ resistance is not yet well-defined [4,43]. Furthermore, because of the poor knowledge of the β-tubulin gene family composition in the Ascaris species, it is difficult to interpret the relevance of the absence of potential resistance conferring SNPs in a particular β-tubulin gene without knowledge of other β-tubulin loci that may be relevant.
The overall aim of the present study was to comprehensively characterize the β-tubulin gene family in A. lumbricoides and A. suum, both in Clade III of the nematode phylogeny, with the goal of prioritizing the most relevant genes in the context of BZ resistance. The porcine parasite A. suum is phylogenetically very closely related to A. lumbricoides; they have recently been shown to be an interbreeding species complex [44]. For A. suum a high-quality genome assembly and transcriptome is available and worm material is accessible through experimental infection of pigs. Moreover, BZ drugs have already been used for decades to combat Ascaris infections in the pig industry, further highlighting the potential of A. suum as an excellent experimental model with respect to BZ resistance research.
Methods
Ethics statement
A. lumbricoides worms were collected during two worm expulsion studies as part of the Starworms project. Ethical approval to conduct the expulsion studies was obtained from the Institutional Review Board (IRB) of the Faculty of Medicine and Health Sciences (Ghent University) and Ghent University Hospital (Belgium; reference number: B670201837418), The Zanzibar Medical Research and Ethics Committee (Tanzania; reference number: ZAHREC/03/DEC/2018) and Jimma University (Ethiopia; reference number: IHRPG/269/2018). Parent(s)/guardian(s) of participants signed an informed consent document indicating that they understood the purpose and procedures of the study, and that they allowed their child to participate. Children younger than 12 years of age had to orally assent in order to participate, participants of 12 years of age or older were only included if they gave written consent.
Research workflow
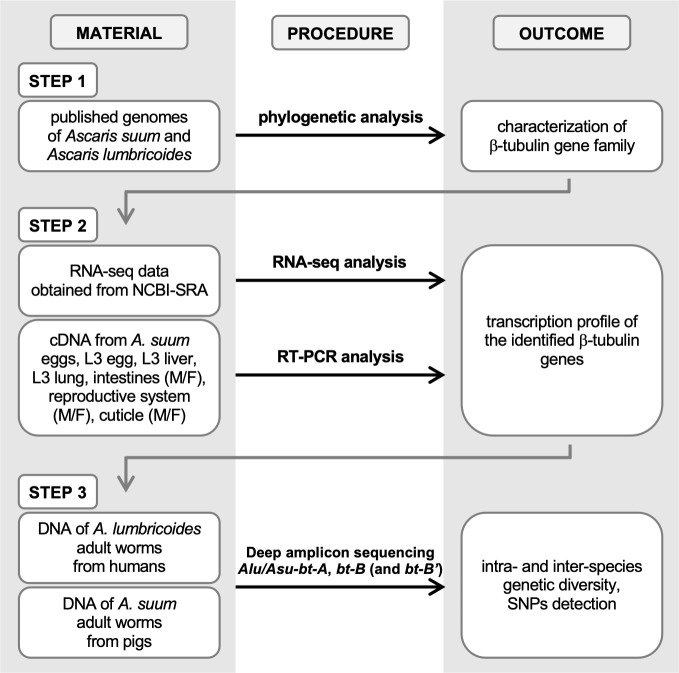
We performed a series of experiments that were organized in three consecutive steps (summarized in Fig 1). In a first step, we characterized and compared the β-tubulin gene families of A. lumbricoides and A. suum. In a second step, we analyzed the transcription profile of the different β-tubulin genes during the life cycle of A. suum. In a third step, we assessed the intra- and inter-species genetic diversity and presence of known BZ resistance associated SNPs in β-tubulin genes in both human and porcine BZ-drug-exposed worm populations applying a deep amplicon sequencing approach. This final step was applied on strategically selected genes based on the outcome of steps 1 and 2.
Fig 1. Schematic representation of the research workflow.
RNA-Seq: RNA sequencing, NCBI-SRA: NCBI Sequence Read Archive, cDNA: complementary DNA, L3: third larval stage, M: male, F: female, RT-PCR: reverse transcription PCR, SNPs: single nucleotide polymorphisms.
Ascaris material
Messenger RNA from A. suum eggs, infectious stage 3 larvae (L3) from eggs, migrating L3 from liver and lungs, as well as cuticle, intestinal, and reproductive tissues from adult male and female A. suum worms was purified by Wang and colleagues [45]. Complementary DNA (cDNA) was synthesized using the iScript cDNA synthesis kit (Bio-rad) following the manufacturer’s instructions.
Genomic DNA was extracted from adult Ascaris worms for which the origin and method of collection are summarized in Table 1. Human A. lumbricoides worms were collected during two worm expulsion studies. The human study population consisted of school children of 7 to 14 years of age, living in two endemic regions, i.e. Pemba Island (Tanzania) and Jimma Town (Ethiopia). A single dose of 400 mg ALB (GlaxoSmithKline) was orally administered to the children for one day (Tanzania) or three consecutive days (Ethiopia). Expelled worms were collected from stool from the past 24 hours for seven consecutive days. As indicated in Table 1, a total of 106 A. lumbricoides worms were analyzed, including 29 worms from 7 Ethiopian children and 77 worms from 53 Tanzanian children. The 109 A. suum worms were collected from porcine intestines obtained at different slaughterhouses in Flanders. The random collection of both male and female worms was done at three different time points within a six-month period. This time interval assured a set of worms coming from different farms.
Table 1. Overview of origin of the adult Ascaris worms used for deep amplicon sequencing.
| Origin | Study site | Study design | Number of worms | Number of subjects | Median (range) of worms per subject |
|---|---|---|---|---|---|
| Human | Ethiopia (Jimma Town) | Expulsion within 7 days after treatment with 400 mg ALB for 3 consecutive days | 29 | 7 | 3 (1–7) |
| Human | Tanzania (Pemba Island) | Expulsion within 7 days after single treatment with 400 mg ALB | 77 | 53 | 1 (1–7) |
| Porcine | Belgium (Flanders) | Collection from intestines after slaughter | 109 | NA | NA |
Anterior sections (0.5–1 cm) of both A. lumbricoides and A. suum adult worms were individually lysed in 300 μL Buffer ATL (Qiagen) and 50 μL Proteinase K (Qiagen) for 24h to 48h at 55°C under gentle agitation (300 rpm). One volume of phenol-chloroform (1:1) (Sigma) was added, followed by centrifugation for 10 min at 10,000 g. The supernatant was recovered and after addition of 3 M sodium acetate (1:10) (Sigma) and isopropanol (1:1), single worm DNA was precipitated by centrifugation for 10 min at 16,000 g. The pellet was washed twice with 80% ethanol and eluted in 50 μL molecular-grade water. DNA concentration was measured with the Nanodrop 2000 (ThermoFisher Scientific).
Characterization and comparison of the β-tubulin gene families
Protein sequences available at WormBase ParaSite from A. lumbricoides (PRJEB4950) and A. suum (PRJNA80881) were used as query sequences in a BLASTP search against a database of C. elegans proteins (PRJNA13758) [46–48]. Ascaris proteins whose top hit was a protein from one of the six β-tubulin genes from C. elegans (B0272.1, C36E8.5, C54C6.2, K01G5.7, T04H1.9, ZK154.3), were considered candidate β-tubulin genes. The alignments were manually inspected to ensure that the Ascaris proteins were at least 400 amino acids in length. All candidate proteins were searched against the Pfam database to ensure that they contain the ‘Tubulin/FtsZ family, GTPase’ domain (PF00091) and the ‘Tubulin C-terminal’ domain (PF03953) [49]. This revealed that the protein with accession GS_10401 contained the N terminus of the ‘Tubulin GTPase’ domain, while the protein with accession GS_22804 contained the rest of that domain and the ‘Tubulin C-terminal’ domain. As their genes are adjacent in the genome assembly, we considered this a mistake in the original annotation and decided to merge the two to give one protein sequence.
The β-tubulin genes identified in Ascaris species (Clade III in nematode phylogeny) were aligned to those of the other three human STH species (T. trichiura (PRJEB535) belonging to Clade I, N. americanus (PRJNA72135) and Ancylostoma ceylanicum (PRJNA231479) belonging to Clade V), and to the well characterized β-tubulin families of H. contortus (PRJEB506) and C. elegans (PRJNA13758) (both Clade V) [30,50]. Additionally, two other members of Clade III, and thus more closely related to Ascaris, were included in the analysis: Parascaris equorum (equines) and Ascaridia galli (poultry) for which sequences published by Tyden and colleagues were used [26]. Gene ID and Transcript ID of all protein sequences covered in the analysis are provided in S1 Info, including accession numbers of source worm genomes. The genome assembly and annotation of A. ceylanicum were used as close representatives of A. duodenale, since the latter genome assembly has low quality metrics [46,47]. β-tubulin gene names were adopted from published literature and genes that have not been named yet were indicated by their accession number.
Protein sequences were aligned using MUSCLE (maximum number of iterations: 8; other parameters: default) [51]. The alignment was viewed in Geneious (v10.2.6; https://geneious.com) and alignment positions with greater than 10% gaps from the phylogenetic reconstructions were masked. Two sequences, TBB-6 from C. elegans and Ttr-TTRE_0000019101 from T. trichiura, were noticeably divergent from other sequences in the alignment and in the preliminary phylogenetic reconstructions confounded the topology, likely due to long-branch attraction, so were removed from subsequent analyses [52]. For phylogenetics, three β-tubulins from Drosophila melanogaster were included to serve as an outgroup. RAxML (version 8) was used to generate maximum likelihood-based trees (protein model: GAMMA LG; algorithm: rapid bootstrapping; replicates: 1,000) [53]. MRBAYES (version 3.2.6) was used to reconstruct a Bayesian phylogeny (rate matrix: poisson; rate variation: gamma; chain length: 1,100,000; subsampling frequency: 200; heated chains: 4; burn-in length: 100,000; heated chain temp: 0.2) [54]. The phylogenetic figures were viewed and further annotated in iTOL (version 5.7) [55] and Adobe Illustrator (https://adobe.com/products/illustrator).
Previously investigated β-tubulin genes from eight papers studying these genes in A. lumbricoides [15,34,35,37–41] and three papers in A. suum [15,34,42] were BLAST searched against the Ascaris genes identified in this study to determine which β-tubulin gene(s) were examined in each of the previous studies [48].
Transcription profiles of the β-tubulin genes during the Ascaris life cycle
Gene specific primers were designed using Primer3 software (http://bioinfo.ut.ee/primer3/), based on the coding sequences of the β-tubulin genes, retrieved from the A. suum published genome (PRJNA62057) available at WormBase ParaSite [46,47]. The full list of forward and reverse primers can be found in S2 Info. The actin gene was used as a control gene (CB039781). Actin primers were adopted from Vlaminck et al., 2011 [56]. To check gene specificity, all primers were searched using BLAST against the published A. suum and A. lumbricoides genomes (PRJNA62057, PRJNA80881 and PRJEB4950) [46,47]. Reverse transcription (RT)-PCR reactions were performed on cDNA from worm eggs, L3 larvae, and cuticle, intestinal, and reproductive tissues from adult male and female A. suum worms under the following conditions: 1X Green GoTaq Flexi Buffer (Promega), 2 mM MgCl2, 0.2 mM dNTPs, 0.25 μM gene specific primer forward, 0.25 μM gene specific primer reverse, 1.25 U GoTaq G2 DNA Polymerase (Promega) and 250–500 ng of cDNA. The thermocycling parameters were 95°C for 2 min, followed by 35 cycles of 95°C for 30 s, 54°C for 30 s, 72°C for 1 min, and a final extension of 72°C for 10 min. Aliquots (5 μL) of individual RT-PCR products were run electrophoretically on 1.5% agarose gels stained with ethidium bromide (0.4 μg/ml) (Sigma-Aldrich). Results were visualized with the Bio-Rad Gel Doc EZ Imager (Bio-Rad Laboratories nv.) by ultraviolet trans-illumination, and fragment sizes determined by comparison with a 100 bp DNA ladder (Promega).
A. suum RNA-seq reads were downloaded from the NCBI Sequence Read Archive (SRA) from SRA studies SRP013573 [57], SRP013609 [57], SRP005511 [58] and SRP010159 [59] (S3 Info). A. suum transcripts were downloaded from WormBase ParaSite (PRJNA80881) [46,47] and the transcripts for the β-tubulins were renamed and joined to match the identified β-tubulin genes. RNA-seq reads were then pseudo-mapped using kallisto v0.46.2 [60] to the modified A. suum transcripts to obtain read counts and transcripts per million (TPM) expression values for every transcript. For each sample type, the mean TPM of each β-tubulin was calculated and the proportion of total β-tubulin expression represented by each β-tubulin gene in each sample type was plotted.
Intra- and inter-species genetic diversity and presence of candidate BZ resistance associated SNPs in drug-exposed adult A. lumbricoides and A. suum worm populations
To amplify a 519 bp fragment of Alu/Asu-bt-A (ALUE_0000927201, GS_23993), a single forward primer (BtA-For) and a single reverse primer (BtA-Rev) were designed for both A. lumbricoides and A. suum. For a 689 bp fragment of Alu/Asu-bt-B (ALUE_0000986501, GS_01240) a single forward primer (BtB-For), but two different reverse primers (BtB-Rev1 and BtB-Rev2) were created to ensure amplification of Asu-bt-B in A. suum, and both Alu-bt-B and Alu-bt-B’ (ALUE_0001827701) in A. lumbricoides for which it was not clear whether these were allelic variants or separate genes. All primers were designed using Geneious (v10.2.6; https://geneious.com). The locus specific primers were adapted for Illumina deep sequencing as described by Avramenko et al. [61]. A complete list of all adapted primers can be found in S4 Info. In general, the primer design was as follows: 5’–Illumina adapter sequence– 0 to 3 random nucleotides–locus specific primer– 3’. Four forward adapter primers, and four or eight reverse adapter primers were mixed in equal concentrations and used for the first round of PCR under the following conditions: 1X KAPA HiFi HotStart Fidelity Buffer (KAPA Biosystems), 0.3 μM forward primer mix, 0.3 μM reverse primer mix, 0.2 mM dNTPs, 0.5 U KAPA HiFi HotStart Polymerase (KAPA Biosystems), 2 μg bovine serum albumin (BSA) and a 1:250 dilution of single worm DNA (variable concentration). The thermocycling parameters were 95°C for 3 min, followed by 35 cycles of 98°C for 20 s, 61°C for 15 s, 72°C for 30 s, and a final extension of 72°C for 2 min. PCR products were purified with AMPure XP Magnetic Beads (1X) (Beckman Coulter Inc.) following the manufacturer’s recommended protocol.
Illumina barcode indices and P5/P7 sequencing regions were added to the amplicons by limited cycle PCR with primers of the Nextera XT Index Kit v2 set (Illumina Inc.). All primer sequences are provided in S4 Info. The PCR was performed as described by Avramenko et al. [61] with 5 μL of first-round clean PCR product as template. Amplicons were purified with AMPure XP Magnetic Beads (1X) (Beckman Coulter Inc.) following the manufacturer’s recommended protocol.
The concentration of the second-round clean PCR product was measured using the Implen NanoPhotometer NP80 and 50 ng of each sample was pooled to produce a master sequencing library. The final concentration of this pooled library was assessed with the KAPA qPCR Library Quantification Kit (KAPA Biosystems), following the manufacturer’s recommended protocol. The prepared pooled library was run on an Illumina MiSeq Desktop Sequencer using a 600-cycle MiSeq Reagent Kit v3 (Illumina Inc., MS-102-3003) at a concentration of 15 pM with the addition of 20% PhiX Control v3 (Illumina Inc., FC-110-3001). The MiSeq was set to generate only FASTQ files with no post-run analysis. Based on their supplied index combinations, samples were automatically demultiplexed by the MiSeq. All protocols were carried out per Illumina’s standard MiSeq operating protocol (Illumina Inc.).
Raw FASTQ files generated were analyzed with the DADA2 v.1.11.5 bioinformatic software package to ascertain the number of unique amplicon sequencing variants (ASV) contained in each sample [62]. This workflow was adapted from the DADA2 analysis described at www.nemabiome.ca, with modifications to accommodate the β-tubulin amplicons analyzed in this paper. Briefly, FASTQ files were prefiltered with the ‘FilterAndTrim’ function to remove any ‘N’s contained in the sequences. Cutadapt was used to remove forward and reverse primer sequences in the amplicons [63]. After primer removal, reads were filtered again with ‘FilterAndTrim’ with no N’s allowed, maxEE = 6, truncQ = 2, a minimum length of 200 bp for each forward and reverse read, and the removal of phiX if identified. As the Alu/Asu-bt-B amplicon is >600 bp, the paired-end reads do not have any overlap; resultantly the reads were trimmed to a length of 278 bp, to ensure all amplicons are merged at a consistent length. The error profile of the reads was assessed and reads denoised accordingly using ‘learnErrors’ and ‘derepFastq’ respectively. Overlapping reads were merged with the mergePairs function, while amplicons without paired read overlap (i.e Alu/Asu-bt-B) were merged with ‘justConcatenate = TRUE’ and ‘NNNNNNNNNN’ placed between the paired reads to denote the forced merger. A sequence table was constructed with ‘makeSequenceTable’ to display all ASVs present in the dataset. Chimeras were removed with ‘removeBimeraDenovo’. This provides a read count of each ASV present in each sample. A fasta file was also generated with the ‘getUniques’ and ‘uniquesToFasta’ commands to provide a list of all ASVs identified and their corresponding nucleotide sequence. Each ASV was blasted against a reference sequence to ensure that the ASV correctly matched the intended amplicon. Any off-target amplicons were subsequently removed from analysis. Furthermore, 21 ASVs with extremely low reads (below 40) and only detected in one or two worms were manually deleted from the Alu/Asu-bt-B dataset since these were most likely PCR artifacts. The resulting sequence list was then screened for the presence of any canonical resistance conferring single nucleotide polymorphisms (SNPs).
Worm genotypes were defined by applying a read count threshold of 1,000 reads. ASVs below threshold level were excluded from further population genetic analysis. As amplicons were generated from single adult worms, a maximum of two ASVs was expected from each sample. There was one A. lumbricoides worm with only reads below threshold and five A. suum worms with reads above threshold for more than two ASVs, these six worms were not included in the genotype data used. Basic population genetic analysis was conducted in R Studio (v1.3.1093) [64] using the package PopGenReport [65,66]. Allele counts and allele richness by locus and by population were calculated. Likewise, pairwise FSTs were estimated according to Nei [67].
Results
Characterization and comparison of the β-tubulin gene families
Seven β-tubulin genes and an eighth putative β-tubulin encoding gene were identified for both A. lumbricoides and A. suum. Table 2 gives an overview of the proposed and previously used nomenclature for the different genes of both species. Regarding the nomenclature used in this analysis; for A. lumbricoides genes (PRJEB4950) the prefix ‘Alu’ is used, while A. suum genes (PRJNA62057) start with ‘Asu’. β-tubulin is abbreviated by ‘bt’. Genes closely related in both species are named correspondingly (e.g. Alu-bt-A and Asu-bt-A). The use of Latin alphabet characters over numbers was preferred as the use of numbers as indexes may lead to erroneous assumptions regarding orthology with genes of other species. For example Cel-tbb-1 and Hco-tbb-iso-1 are not orthologous.
Table 2. Alignment of the proposed with the previously used nomenclature for β-tubulins of Ascaris species.
| Accession number | Nomenclature used in this analysis | Previous nomenclature | References |
|---|---|---|---|
| Ascaris lumbricoides | |||
| ALUE_0000927201 | Alu-bt-A | β-tubulin | Diawara et al., 2009 [41] Diawara et al., 2013a [35] Diawara et al., 2013b [36] Zuccherato et al., 2018 [39] Matamoros et al., 2019 [40] |
| Alutbb-1 | Demeler et al., 2013 [15] | ||
| tbb-1.2 | Krücken et al., 2017 [34] | ||
| β-tubulin isotype 1 | Rashwan et al., 2017 [37] Furtado et al., 2019 [38] |
||
| ALUE_0000986501 | Alu-bt-B | tbb-2 | Krücken et al., 2017 [34] |
| ALUE_0001827701 | Alu-bt-B’ | - | - |
| ALUE_0000494801 | Alu-bt-C | tbb-1 | Krücken et al., 2017 [34] |
| ALUE_0001031701 | Alu-bt-D | - | - |
| ALUE_0000949301 | Alu-bt-E | tbb-4 | Krücken et al., 2017 [34] |
| ALUE_0000949201 | Alu-bt-F | - | - |
| ALUE_0001294101 | Alu-bt-G | - | - |
| Ascaris suum | |||
| GS_23993 | Asu-bt-A | tbb-1.2 | Krücken et al., 2017 [34] |
| β-tubulin | Palma et al., 2020 [42] | ||
| GS_01240 | Asu-bt-B | Asutbb-2 (Asutbb-3) | Demeler et al., 2013 [15] |
| - * | Asu-bt-B’ * | - | - |
| GS_11145 | Asu-bt-C | Asutbb-1 | Demeler et al., 2013 [15] |
| GS_13691 | Asu-bt-D | - | - |
| GS_05353 | Asu-bt-E | Asutbb-4 | Demeler et al., 2013 [15] |
| GS_11773 | Asu-bt-F | - | - |
| GS_10401 & GS_22804 | Asu-bt-G | - | - |
For Ascaris lumbricoides genes (PRJEB4950) the prefix ‘Alu’ is used, while Ascaris suum genes (PRJNA62057) start with ‘Asu’. β-tubulin is abbreviated by ‘bt’. Genes closely related in both species are named correspondingly. *: The amplicon sequencing results from this study suggested that bt-B’ is present in both A. lumbricoides and A. suum, even though it was only identified in the former species in the public datasets. Further research is needed to clarify if Alu/Asu-bt-B’ should be considered as a separate β-tubulin gene or as an allelic variant of Alu/Asu-bt-B.
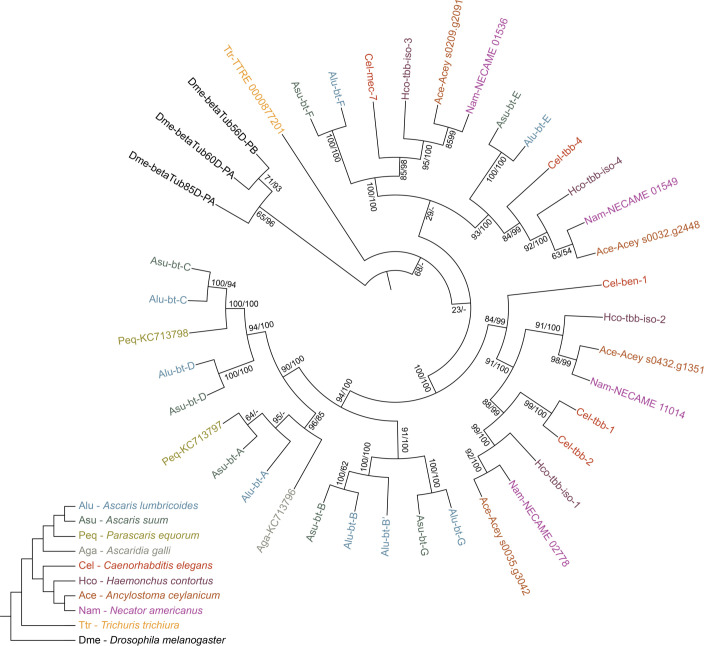
Phylogenetic analysis based on amino acid sequences (Fig 2) showed that the β-tubulin gene family is highly conserved in the two species (β-tubulin (bt) A, B, C, D, E, F and G). Additionally, an eighth putative β-tubulin encoding gene was observed in the published genome for A. lumbricoides but was absent in A. suum. However, due to its close relationship with Alu-bt-B it was not clear if this sequence had to be considered as an extra gene or as an allelic variant of Alu-bt-B. The sequence was therefore named Alu-bt-B’. Furthermore, the amplicon sequencing results from this study contained an ASV of Alu/Asu-bt-B (btB-ASV26) also generated from A. suum samples which showed only one nucleotide difference with Alu-bt-B’. The results therefore suggest that Alu/Asu-bt-B’ is present in both A. lumbricoides and A. suum. Alignment of complete amino acid sequences of the β-tubulins from the published genomes is provided in S5 Info.
Fig 2. Phylogenetic tree of β-tubulin proteins of Ascaris species, other parasitic nematodes, and C. elegans.
Phylogenies were reconstructed with RAxML and MRBAYES. The topology shown is from RaxML. The node support values are percent bootstraps / posterior probabilities. The ‘-‘ indicates that a node was not present in the MRBAYES tree. The branches are equal length to show topology clearly; a tree with branch lengths showing divergence is available in supplementary materials (S14 Info).
To understand the evolutionary relationship of β-tubulins from both human and animal STHs, a protein alignment was generated, including A. suum, A. lumbricoides, P. equorum and A. galli (belonging to Clade III in the nematode phylogeny); T. trichiura (Clade I); H. contortus, A. ceylanicum and N. americanus (Clade V), and the free-living C. elegans (Clade V) (S1 Info). Both maximum likelihood and Bayesian phylogenetic trees returned similar topologies that closely followed the species phylogeny (Fig 2). A. lumbricoides and A. suum have, similar to H. contortus, A. ceylanicum and N. americanus, genes that are orthologues to C. elegans mec-7 and tbb-4 (Alu/Asu-bt-E and bt-F). The relationship to the other C. elegans genes is more complex. H. contortus, A. ceylanicum and N. americanus have two sets of clear one-to-one orthologues, but inclusion of C. elegans suggests a pattern of gene duplication and loss. Resolution of these relationships requires analyses with more species, which are currently being performed. The Ascaris species show a lineage specific expansion, with at least six gene duplication events, one each for Alu/Asu-bt-A, bt-B, bt-C, bt-D, bt-G and then another giving Alu-bt-B’. The included β-tubulins of P. equorum and A. galli, both members of Clade III, show a close phylogenetic relationship with the genes of Ascaris. It has to be noted that for these two species only the sequences published by Tyden et al. [26] were included and no further genomic or transcriptomic data was investigated for other potential β-tubulin genes orthologues to C. elegans mec-7 and tbb-4.
Table 3 gives an overview of the three codons of particular interest for the detection of BZ resistance associated SNPs (167, 198 and 200). BZ susceptible genotypes were present at codons 167 and 198 in all the A. lumbricoides and A. suum β-tubulin gene family members; Phe (TTC) at codon 167 and Glu (GAA or GAG) at codon 198. In contrast, at codon 200, the BZ susceptible genotypes Phe (TTC or TTT) were present in all the genes except for Alu/Asu-bt-D, which has the BZ resistance genotype Tyr (TAT).
Table 3. Genotype of codons 167, 198 and 200 in the identified Ascaris β-tubulin genes.
| Codon | Genotype | Amino acid | Ascaris β-tubulin genes |
|---|---|---|---|
| 167 | TTC | Phe | All identified Ascaris β-tubulin genes |
| 198 | GAA | Glu |
Alu-bt-A and Asu-bt-A Alu-bt-B and Asu-bt-B Alu-bt-B’ and Asu-bt-B’* Alu-bt-C Alu-bt-D and Asu-bt-D Alu-bt-F and Asu-bt-F Alu-bt-G and Asu-bt-G |
| GAG | Glu |
Asu-bt-C Alu-bt-E and Asu-bt-E |
|
| 200 | TTC | Phe |
Alu-bt-A and Asu-bt-A Alu-bt-B and Asu-bt-B ** Alu-bt-B’ and Asu-bt-B’* Alu-bt-C Alu-bt-E and Asu-bt-E Alu-bt-F and Asu-bt-F Alu-bt-G and Asu-bt-G |
| TTT | Phe |
Alu-bt-B and Asu-bt-B ** Asu-bt-C |
|
| TAT | Tyr | Alu-bt-D and Asu-bt-D |
*: Asu-bt-B’ was sequenced from A. suum worms in this study and showed the same genotypes as Alu-bt-B’. **: A synonymous sequence polymorphism was observed at codon 200 of Alu/Asu-bt-B in both A. lumbricoides and A. suum sequenced in this study (btB-ASV01, btB-ASV03 and btB-ASV13). This polymorphism was not concluded from the published genomes, in which TTC was present in Asu-bt-B and TTT in Alu-bt-B.
Transcription profiles of the β-tubulin genes during the Ascaris life cycle
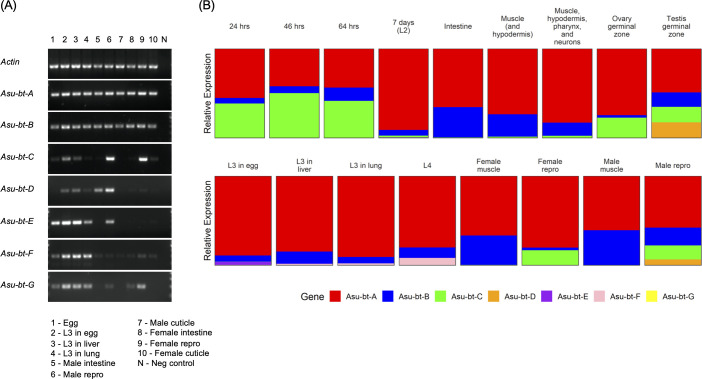
RT-PCR was performed for each of the seven identified β-tubulin genes for multiple life cycle stages and tissues of A. suum (Fig 3, panel A) and the expression of these genes was also examined from the available RNA-seq data (Fig 3, panel B). Both the analyses indicated that only two of the seven identified β-tubulin genes are highly and widely expressed. The gene Asu-bt-A amplified by RT-PCR from cDNA from all life stages and tissues of the parasite and had consistently high RNA-seq expression levels. Also the gene Asu-bt-B is expressed throughout the entire life cycle of the parasite albeit at lower levels than Asu-bt-A. All other β-tubulin genes were transcribed in specific life stages or tissues, suggesting more specialized functions (Fig 3). For some genes, although RT-PCR amplification confirmed the expression in certain tissues, the RNA-seq data clearly indicated low transcription levels. Based on these results, Alu/Asu-bt-A and Alu/Asu-bt-B were selected as the most interesting targets for BZ mode of action and subsequently the most relevant for screening for potential resistance mutations. Consequently, these two genes were selected for deep amplicon sequencing to assess intra- and inter-species genetic diversity and the presence of sequence polymorphisms at codons 167, 198 and 200.
Fig 3. Transcription profiles of the β-tubulins of Ascaris suum.
Panel A. Reverse transcription (RT)-PCR analyses were performed on cDNA samples with gene specific primer sets. The agarose gels show the transcription profiles of the β-tubulins in four developmental stages (egg, third larval stage (L3) in egg, L3 in liver, L3 in lung) and three tissue types of adult male and female worms (intestinal, reproductive, and cuticle). Panel B. RNA-seq analyses were performed using RNA-seq data obtained from the NCBI Sequence Read Archive. The stacked bar charts show the expression of each β-tubulin (relative to total β-tubulin expression) in different developmental stages (24h embryos to L4 stage larvae) and different tissue types of A. suum.
Intra- and inter-species genetic diversity and presence of candidate BZ resistance associated SNPs in drug-exposed A. lumbricoides and A. suum populations
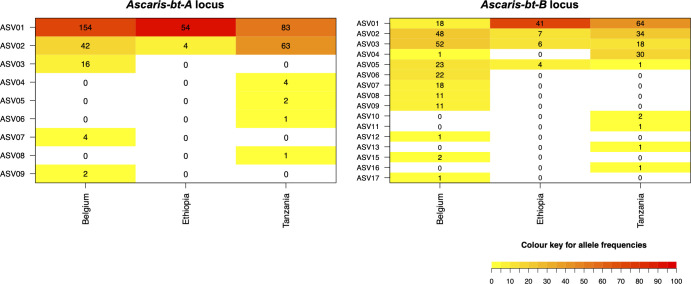
Deep amplicon sequencing was successfully performed on Alu/Asu-bt-A and Alu/Asu-bt-B for 215 and 209 adult Ascaris worms respectively, applying a read threshold of 1,000 reads. The amplicon sequencing data for Alu/Asu-bt-A clearly suggested the primers used target a single locus since there was a maximum of two β-tubulin ASVs amplified from each single worm (S6 Info). The data from Alu/Asu-bt-B was somewhat more complex. While the majority of single worms had a maximum of two ASVs with high read counts (consistent with a diploid genotype), 47.7% (52/109) of A. suum worms and 15.1% (16/106) of A. lumbricoides worms had several additional β-tubulin ASVs at very low read depths (S11 Info). Although additional low frequency ASVs can be generated from samples due to bar-code hopping, there was no pattern related to particular index combinations and so it is more likely to be due to “off target” amplification from a second copy of Alu/Asu-bt-B and other β-tubulin genes. For example, ASVs corresponding to the Alu/Asu-bt-A locus (btA-ASV01, btA-ASV02, btA-ASV08) were also detected in the Alu/Asu-bt-B Illumina samples, but only two worms showed read counts above threshold level (1,000 reads). Both loci appeared to be relatively conserved, with Alu/Asu-bt-A being less diverse than Alu/Asu-bt-B. Allelic richness by locus and population is displayed in Table 4. Overall, the Tanzanian and Belgian Ascaris populations show more diversity than the Ethiopian population. The intra- and inter-population genetic diversity of both loci is graphically presented in Fig 4 as heatmaps.
Table 4. Allelic richness of Alu/Asu-bt-A and Alu/Asu-bt-B in the investigated human and porcine adult Ascaris worm populations.
| Allelic richness | Ascaris suum | Ascaris lumbricoides | |
|---|---|---|---|
| (n = 109) | Ethiopia (n = 29) | Tanzania (n = 77) | |
| Alu/Asu-bt-A | 4.112 | 1.997 | 4.067 |
| Alu/Asu-bt-B | 9.156 | 3.997 | 5.956 |
| Mean | 6.634 | 2.997 | 5.011 |
| total | 13.268 | 5.994 | 10.022 |
Fig 4. Allele counts for Alu/Asu-bt-A and Alu/Asu-bt-B in three adult Ascaris worm populations.
Ascaris suum worms were collected from the intestines of pigs slaughtered in Belgium. Ascaris lumbricoides worms were collected during two expulsion studies in Ethiopia and Tanzania. The total number of each amplicon sequence variant (ASV) per population is displayed (allele count) and the color indicates the proportion within the population (allele frequency). Heatmaps were created using the R package PopGenReport [65,66].
For the Alu/Asu-bt-A locus, only nine ASVs were identified across the three populations (S6 Info). These were highly similar with a percentage of identity being > 99% for all pair-wise comparisons (S7 Info). All variation in the sequenced region was located within the intron. No F167Y (TTC>TAC or TTT>TAT), E198A (GAA>GCA or GAG>GCG), E198L (GAA>TTA) and F200Y (TTC>TAC or TTT>TAT) BZ resistance associated SNPs were found in any of the ASVs (S8 Info). Two ASVs (btA-ASV01 and btA-ASV02) were highly frequent in all three Ascaris populations with a total coverage of 93.0% (400/430) of all sequenced fragments (S9 Info). The most common worm genotypes were the same within the human and porcine Ascaris populations.
For the Alu/Asu-bt-B locus, a total of 21 ASVs were detected in the complete dataset, of which only 16 were included in the population genetic analysis based on the minimum threshold of 1,000 reads (S11 Info). Among the 21 ASVs, the percentage identity ranged from 94.6% to 98.5% and nucleotide variation in this fragment was located in both intron and exon (S12 Info and S13 Info). Two ASVs (btB-ASV23 and btB-ASV26) having only low read counts (< 1,000) in a total of seven worms contained SNPs resulting in amino-acid substitutions (P173H (CCT>CAT), Q191E (CAG>GAG), V193I (GTT>ATT) and D197N (GAT>AAT)). All other polymorphisms were synonymous. Considering the SNPs that could be potentially associated with BZ resistance, as for Alu/Asu-bt-A, no F167Y (TTC>TAC or TTT>TAT), E198A (GAA>GCA or GAG>GCG), E198L (GAA>TTA) or F200Y (TTC>TAC or TTT>TAT) mutations were detected. Although, a synonymous sequence polymorphism was present at codon 200 in three ASVs (btB-ASV01, 03 and 13) having the Phe (TTT) instead of the Phe (TTC) genotype (S13 Info). More detailed data regarding genotype counts and frequencies can be found in supplementary materials (S10 Info).
Based on the two β-tubulin loci, Nei’s pairwise FST values between all pairs of populations were calculated; Belgium-Ethiopia: 0.0710, Belgium-Tanzania: 0.0532, Ethiopia-Tanzania: 0.0713 [67]. These results reveal low levels of population differentiation and suggest that among the investigated populations, the A. suum population collected from pigs is not the most divergent.
Discussion
At least seven β-tubulin genes were identified for both A. lumbricoides and A. suum
This study demonstrated the presence of at least seven β-tubulin genes in Ascaris species. All the identified β-tubulin gene family members, except Alu/Asu-bt-D, have the susceptible genotype at codons 167, 198 and 200. Of the possible candidates, most do not have one-to-one orthologous relationships with C. elegans and H. contortus β-tubulin genes. Thus, prioritizing the genes most likely to play a potential role in BZ resistance by simple extrapolation of information from those species is difficult. The only two exceptions to this are Alu/Asu-bt-E and bt-F which appear orthologous to Cel-tbb-4 and Cel-mec-7 respectively. These C. elegans genes have very specialized functions and are only expressed in a small subset of cells including specific sensory neurons and do not appear to be involved in BZ resistance [68,69]. Consistent with this, Alu/Asu-bt-E and bt-F are only expressed at very low levels and so it seems reasonable to assume these are not likely to be involved in BZ resistance. In the phylogenetic analysis, we noted high rates of variation in the three pairs Alu/Asu-bt-C, bt-D, and bt-G, which is indicative that these genes are under different selective pressures from Alu/Asu-bt-A and bt-B. It might suggest that these three more rapidly evolving genes are adapting to a specific cell or tissue in Ascaris, analogous to Cel-mec-7 and Cel-tbb-4 in C. elegans, and Hco-tbb-iso-3 in H. contortus [27,68,70]. Only two of the β-tubulin genes, namely Alu/Asu-bt-A and Alu/Asu-bt-B, are widely expressed in all stages of the parasite’s lifecycle and all tissues of adult worms. Consequently, we considered these the most likely relevant BZ targets.
Regarding the place of Alu/Asu-bt-B’ in the Ascaris β-tubulin gene family, the results of the deep amplicon sequencing of Alu/Asu-bt-B showed two ASVs almost identical to the Alu-bt-B’ reference sequence (btB-ASV23 and btB-ASV26). These ASVs only appeared at low read counts in nine worms in addition to two highly frequent Alu/Asu-bt-B ASVs. Four SNPs present in Alu-bt-B’, btB-ASV23 and btB-ASV26 result in amino-acid substitutions (P173H (CCT>CAT), Q191E (CAG>GAG), V193I (GTT>ATT) and D197N (GAT>AAT)). Further research is needed to clarify if this sequence should be considered as an allelic variant of Alu/Asu-bt-B or as a separate β-tubulin gene, although the results of the present study suggest the latter since both observations would be unusual for alleles of the same gene.
There is a need for a new nomenclature for β-tubulins in nematodes
Similar to previous studies, the presented data suggested that diversification of β-tubulins occurred independently in different nematode lineages. Hence, β-tubulin families are more conserved between species of the same nematode clade. For Ascaris species in particular, this complex phylogeny has the consequence that the currently used nomenclature is confusing. More specifically, the term Ascaris β-tubulin isotype 1, used for the most frequently studied gene, falsely suggests homology with Hco-tbb-iso-1 of H. contortus or Cel-tbb-1 of C. elegans. To avoid such incorrect assumptions regarding phylogenetic relationships and potential orthology, we have used letters as indexes to name the β-tubulins identified in the present study. In relation to this, a comprehensive nomenclature of β-tubulin genes in parasitic nematodes that considers the complex phylogeny and the continuous expansion of knowledge is needed.
Alu/Asu-bt-A has less allelic diversity than Alu/Asu-bt-B
The genetic diversity of the Alu/Asu-bt-A and bt-B genes was investigated in adult worms collected from porcine and human populations using deep amplicon sequencing. The sequence diversity in the Alu/Asu-bt-A locus within and between worm populations was markedly lower than for Alu/Asu-bt-B. For Alu/Asu-bt-A one haplotype (btA-ASV01) was by far the most frequent in all three populations (adult A. suum worms from Belgium and adult A. lumbricoides worms from Ethiopia and Tanzania). This is consistent with the very close phylogenetic relationship between A. lumbricoides and A. suum, and probably represents ancestral polymorphism. The higher diversity in the porcine population for both Alu/Asu-bt-A and bt-B may be explained by the fact that A. suum worms were randomly collected from the intestines of pigs slaughtered in different slaughterhouses in Belgium. Although there is no information about the farms where the pigs were raised, a diverse origin and a broad distribution among a number of herds is assumed, each with a different management and treatment history. In contrast, human Ascaris worms were collected only from two schools. The reason for the higher genetic diversity of both Alu/Asu-bt-A and bt-B loci in the Tanzanian versus the Ethiopian population is unclear and might suggest a larger effective population size of the parasite in Tanzania, e.g. due to higher infection intensities [71]. Yet, it is also possible that the observed difference is a consequence of the smaller worm population size, sampled from a smaller number of children in the Ethiopian school.
Candidate BZ resistance SNPs are absent in Alu/Asu-bt-A and Alu/Asu-bt-B in adult A. lumbricoides and A. suum worms
All the individual adult A. lumbricoides worms from Ethiopia and Tanzania that were sequenced had the BZ susceptible genotype at codons 167, 198 and 200 since the F167Y (TTC>TAC or TTT>TAT), E198A (GAA>GCA or GAG>GCG), E198L (GAA>TTA) and F200Y (TTC>TAC or TTT>TAT) polymorphisms were not observed in either of the two targeted genes. Diawara and colleagues were previously able to identify the mutation F167Y (TTC>TAC) in the gene Alu/Asu-bt-A in A. lumbricoides worm eggs collected both before and after treatment in Haiti, Kenya and Panama [35]. Furtado and colleagues were the first to report the mutation TTC>TAC at codon 200 of Alu/Asu-bt-A in A. lumbricoides eggs collected in seven Brazilian states, but only at a very low frequency of 0.5% (4/854) [38]. For the set of A. lumbricoides worms investigated in this study, the presence of the potentially BZ resistance associated SNPs was not expected since the worms were collected after expulsion by treatment with BZ drugs and so were susceptible to the drug. Based on the reported national coverage of drug administration for the last 5 years, the site in Ethiopia was considered to have experienced a low drug pressure with MDA administered to SAC since 2015 [72]. The school in Tanzania has a history of MDA since 1994 and was therefore considered as high drug pressure region [70]. Recently, Vlaminck et al. reported an efficacy of ALB against A. lumbricoides of 99% in Ethiopia and 96.8% in Tanzania [71].
Similarly, the F167Y (TTC>TAC or TTT>TAT), E198A (GAA>GCA or GAG>GCG), E198L (GAA>TTA) and F200Y (TTC>TAC or TTT>TAT) polymorphisms were not observed in either of the two targeted genes in any of the A. suum adult worms sequenced from Belgium. Regarding A. suum, there are no reports from the pig sector describing declined efficacy of BZs to date, even though the drugs have been widely used for decades to control infections. However, the fact that Ascaris has minimal acute clinical signs, may result in a lack of recognition of treatment failure in the field. The result of 100% wild-type alleles is in agreement with the study of Palma et al. (2020) likewise unable to demonstrate the presence of SNPs associated with BZ resistance in A. suum collected from pigs [42].
Conclusion
Accurate and reliable detection of molecular markers of BZ resistance in STHs will be critical in the upcoming years, anticipating the continuing increase in number of drug treatments to reach the WHO target in all STH-endemic countries. The characterization of the β-tubulin family in Ascaris species provides a framework to investigate the prevalence and potential role of β-tubulin sequence polymorphisms in BZ resistance in a more systematic manner than previously possible. The work has revealed that Alu/Asu-bt-A and Alu/Asu-bt-B are the obvious β-tubulin genes to prioritize in this context. Nevertheless, further research into the associations between the frequency of SNPs, the drug efficacy assessed by egg counts and the history of drug pressure on investigated worm populations will allow substantiation of the role of the different β-tubulin gene family members in BZ resistance and validation of particular SNPs as molecular markers for BZ resistance.
Supporting information
WormBase ParaSite or NCBI Gene ID and Transcript ID of all β-tubulin proteins included in the phylogenetic analyses and trees. Caenorhabditis elegans and Haemonchus contortus β-tubulin protein sequences were obtained from published genomes [50,73]. The corresponding names were adopted from published literature [27,30]. For Trichuris trichiura, Necator americanus and Ancylostoma ceylanicum keyword search for ‘tubulin beta’ and BLASTP search with the Ascaris β-tubulin genes against the published genomes [74–76] resulted in two sequences for T. trichiura and four sequences for both hookworms. The genome assembly and annotation of A. ceylanicum were used as close representative of A. duodenale, since this genome assembly has low quality metrics. For Parascaris equorum and Ascaridia galli, the sequences published by Tyden et al. (2013) were used [26]. Three β-tubulins from Drosophila melanogaster are included in the list to be used as outgroups.
(XLSX)
β-tubulin gene specific primers used in RT-PCR analysis. All β-tubulin primers are based on coding sequences. The actin gene is used as household gene and primers are adopted from Vlaminck et al., 2011 [56].
(PDF)
Overview of all RNA-seq datasets (NCBI Sequence Read Archive (SRA)) used in the A. suum β-tubulin RNA-seq analysis.
(XLSX)
Alu/Asu-bt-A and Alu/Asu-bt-B (and bt-B’) primers with Illumina Adapters. Locus specific primer sequence bolded, N’s underlined. Illumina adaptor oligonucleotide sequences were obtained from the Illumina Adapter Sequences document dated March 2020 (Illumina Inc.). Forward and Reverse barcoded sequencing primers. Index sequence bolded. Sequences were obtained from the Illumina Adapter Sequences document dated March 2020 (Illumina Inc.).
(PDF)
Alignment of amino acid sequences of the identified β-tubulins of both A. suum and A. lumbricoides. For aesthetic reasons, all sequences were trimmed to a uniform length of 427 amino acids. (N-terminus trimmed: Asu-bt-B, Asu-bt-E; C-terminus trimmed: all).
(PDF)
Excel spreadsheet of amplicon sequencing results for Alu/Asu-bt-A following analysis with the DADA2 v.1.11.5 bioinformatic software package and manual assessment for chimeras, off-target amplicons and PCR artefacts. The first column of the spreadsheet contains the single worm identification. In the adjacent columns the read number for each unique amplicon sequence variant (ASV) is given. Corresponding genotypes were defined by applying a threshold of 1,000 reads. ASVs below threshold level were excluded from further population genetic analyses and are indicated in red in the table. There were no worms with reads above the threshold for more than two ASVs.
(XLSX)
Percentage of bases/residues which are identical (% Identity), and number of bases/residues which are identical (# Identities) and not identical (# Differences). Results for the nine identified amplicon sequence variants (ASVs) of Alu/Asu-bt-A. All pairwise distances were computed using Geneious v10.2.6.
(XLSX)
Nucleotide alignment of the nine identified amplicon sequence variants (ASVs) of Alu/Asu-bt-A. Multiple alignment was performed using Geneious v10.2.6. The three codons of interest in the context of benzimidazole resistance are indicated by their number at the base of the consensus sequence.
(PDF)
The total number of each amplicon sequence variant (ASV) (allele count) and the proportion within the (sub)populations (allele frequency) for both Alu/Asu-bt-A and Alu/Asu-bt-B. A. suum worms were collected from the intestines of pigs slaughtered in Belgium. A. lumbricoides worms were collected during two expulsion studies in Ethiopia and Tanzania.
(XLSX)
The total number of each defined genotype (genotype count) and the proportion within the (sub)populations (genotype frequency) for both Alu/Asu-bt-A and Alu/Asu-bt-B. Genotype per worm was defined by applying a threshold of 1,000 reads. A. suum worms were collected from the intestines of pigs slaughtered in Belgium. A. lumbricoides worms were collected during two expulsion studies in Ethiopia and Tanzania.
(XLSX)
Excel spreadsheet of amplicon sequencing results for Alu/Asu-bt-B following analysis with the DADA2 v.1.11.5 bioinformatic software package and manual assessment for chimeras, off-target amplicons and PCR artefacts. The first column of the spreadsheet contains the single worm identification. In the adjacent columns the read number for each unique amplicon sequence variant (ASV) is given. From the dataset, 21 ASVs with extremely low reads (below 40) and only detected in 1 or 2 worms were manually deleted since these were seen as PCR artefacts. Furthermore, due to a lack of complete specificity of the primers, the dataset contained 10 ASVs corresponding to the Alu/Asu-bt-A locus, which were also manually deleted. The genotype per worm was defined by applying a threshold of 1,000 reads. ASVs below threshold level were excluded from further population genetic analyses and are indicated in red in the table. There was one A. lumbricoides worm with only reads below threshold and five A. suum worms with reads above threshold for more than two ASVs, these six worms were not included in the genotype data used for population genetic analysis.
(XLSX)
Percentage of bases/residues which are identical (% Identity), and number of bases/residues which are identical (# Identities) and not identical (# Differences). Results for the 21 identified amplicon sequence variants (ASVs) of Alu/Asu-bt-B. All pairwise distances were computed using Geneious v10.2.6.
(XLSX)
Nucleotide alignment of the 21 identified amplicon sequence variants (ASVs) of Alu/Asu-bt-B. Multiple alignment was performed using Geneious v10.2.6. The three codons of interest in the context of benzimidazole resistance are indicated by their number at the base of the consensus sequence.
(PDF)
Phylogenies were reconstructed with RAxML and MRBAYES. The topology shown is from RAxML. The node support values are percent bootstraps / posterior probabilities. The ‘-‘ indicates that a node was not present in the MRBAYES tree. The branch lengths show divergence.
(PDF)
Acknowledgments
The authors would like to thank all children, their parents, the schoolteachers and principals that participated in the expulsion trials. Furthermore, we want to acknowledge all the people that provided the necessary laboratory and logistic support during the worm collection studies in Ethiopia, Tanzania and Belgium.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
SR was supported through a Fundamental Research Fellowship (1154819N) of the Research Foundation Flanders (www.fwo.be). The two expulsion studies were a part of the Starworms study, supported through a grant from the Bill and Melinda Gates Foundation (OPP1120972, PI is BL). Part of the sequencing work was supported by a travel award of the FWO for SR (K204820N) and a grant from the Bill and Melinda Gates Foundation (OPP1172974, PI is JG). SMJP is funded by a Natural Sciences and Engineering Research Council of Canada (NSERC) PGS-D award (411306230) and Eyes High Recruitment award from UCalgary. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.World Health Organization. Schistosomiasis and soil-transmitted helminthiases: numbers of people treated in 2019. Weekly Epidemiological Record. 2020;95(50):629–40.
- 2.World Health Organization. Soil-transmitted helminthiases: number of children treated in 2010. Weekly Epidemiological Record. 2012;87(23):225–32. [PubMed]
- 3.World Health Organization (2020). 2030 targets for soil-transmitted helminthiases control programmes. World Health Organization, Geneva. [Google Scholar]
- 4.Vercruysse J, Albonico M, Behnke JM, Kotze AC, Prichard RK, McCarthy JS, et al. Is anthelmintic resistance a concern for the control of human soil-transmitted helminths? International Journal for Parasitology: Drugs and Drug Resistance. 2011;1(1):14–27. doi: 10.1016/j.ijpddr.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lacey E. Mode of action of benzimidazoles. Parasitol Today. 1990;6(4):112–5. doi: 10.1016/0169-4758(90)90227-u . [DOI] [PubMed] [Google Scholar]
- 6.Kwa MS, Veenstra JG, Roos MH. Benzimidazole resistance in Haemonchus contortus is correlated with a conserved mutation at amino acid 200 in beta-tubulin isotype 1. Mol Biochem Parasitol. 1994;63(2):299–303. Epub 1994/02/01. doi: 10.1016/0166-6851(94)90066-3 . [DOI] [PubMed] [Google Scholar]
- 7.Elard L, Cabaret J, Humbert JF. PCR diagnosis of benzimidazole-susceptibility or -resistance in natural populations of the small ruminant parasite, Teladorsagia circumcincta. Veterinary parasitology. 1999;80(3):231–7. doi: 10.1016/s0304-4017(98)00214-3 . [DOI] [PubMed] [Google Scholar]
- 8.Silvestre A, Cabaret J. Mutation in position 167 of isotype 1 beta-tubulin gene of Trichostrongylid nematodes: role in benzimidazole resistance? Mol Biochem Parasitol. 2002;120(2):297–300. doi: 10.1016/s0166-6851(01)00455-8 . [DOI] [PubMed] [Google Scholar]
- 9.Alvarez-Sanchez MA, Perez-Garcia J, Cruz-Rojo MA, Rojo-Vazquez FA. Real time PCR for the diagnosis of benzimidazole resistance in trichostrongylids of sheep. Vet Parasitol. 2005;129(3–4):291–8. Epub 2005/04/23. doi: 10.1016/j.vetpar.2005.02.004 . [DOI] [PubMed] [Google Scholar]
- 10.Ghisi M, Kaminsky R, Maser P. Phenotyping and genotyping of Haemonchus contortus isolates reveals a new putative candidate mutation for benzimidazole resistance in nematodes. Vet Parasitol. 2007;144(3–4):313–20. Epub 2006/11/15. doi: 10.1016/j.vetpar.2006.10.003 . [DOI] [PubMed] [Google Scholar]
- 11.Njue AI, Prichard RK. Cloning two full-length beta-tubulin isotype cDNAs from Cooperia oncophora, and screening for benzimidazole resistance-associated mutations in two isolates. Parasitology. 2003;127(Pt 6):579–88. Epub 2004/01/01. doi: 10.1017/s0031182003004086 . [DOI] [PubMed] [Google Scholar]
- 12.Von Samson-Himmelstjerna G, Blackhall WJ, McCarthy JS, Skuce PJ. Single nucleotide polymorphism (SNP) markers for benzimidazole resistance in veterinary nematodes. Parasitology. 2007;134(Pt 8):1077–86. doi: 10.1017/S0031182007000054 . [DOI] [PubMed] [Google Scholar]
- 13.Pape M, Posedi J, Failing K, Schnieder T, von Samson-Himmelstjerna G. Analysis of the beta-tubulin codon 200 genotype distribution in a benzimidazole-susceptible and -resistant cyathostome population. Parasitology. 2003;127(Pt 1):53–9. Epub 2003/07/30. doi: 10.1017/s0031182003003317 . [DOI] [PubMed] [Google Scholar]
- 14.Drogemuller M, Schnieder T, von Samson-Himmelstjerna G. Beta-tubulin complementary DNA sequence variations observed between cyathostomins from benzimidazole-susceptible and -resistant populations. J Parasitol. 2004;90(4):868–70. Epub 2004/09/11. doi: 10.1645/GE3305RN . [DOI] [PubMed] [Google Scholar]
- 15.Demeler J, Krüger N, Krücken J, von der Heyden VC, Ramünke S, Küttler U, et al. Phylogenetic characterization of β-tubulins and development of pyrosequencing assays for benzimidazole resistance in cattle nematodes. PloS one. 2013;8(8):e70212–e. doi: 10.1371/journal.pone.0070212 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jimenez Castro PD, Howell SB, Schaefer JJ, Avramenko RW, Gilleard JS, Kaplan RM. Multiple drug resistance in the canine hookworm Ancylostoma caninum: an emerging threat? Parasit Vectors. 2019;12(1):576. Epub 2019/12/11. doi: 10.1186/s13071-019-3828-6 ; PubMed Central PMCID: PMC6902405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Redman E, Whitelaw F, Tait A, Burgess C, Bartley Y, Skuce PJ, et al. The emergence of resistance to the benzimidazole anthlemintics in parasitic nematodes of livestock is characterised by multiple independent hard and soft selective sweeps. PLoS Negl Trop Dis. 2015;9(2):e0003494. Epub 2015/02/07. doi: 10.1371/journal.pntd.0003494 ; PubMed Central PMCID: PMC4319741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avramenko RW, Redman EM, Melville L, Bartley Y, Wit J, Queiroz C, et al. Deep amplicon sequencing as a powerful new tool to screen for sequence polymorphisms associated with anthelmintic resistance in parasitic nematode populations. Int J Parasitol. 2019;49(1):13–26. Epub 2018/11/25. doi: 10.1016/j.ijpara.2018.10.005 . [DOI] [PubMed] [Google Scholar]
- 19.Mottier MdL Prichard RK. Genetic analysis of a relationship between macrocyclic lactone and benzimidazole anthelmintic selection on Haemonchus contortus. Pharmacogenetics and Genomics. 2008;18(2). [DOI] [PubMed] [Google Scholar]
- 20.Barrère V, Alvarez L, Suarez G, Ceballos L, Moreno L, Lanusse C, et al. Relationship between increased albendazole systemic exposure and changes in single nucleotide polymorphisms on the β-tubulin isotype 1 encoding gene in Haemonchus contortus. Veterinary Parasitology. 2012;186(3):344–9. doi: 10.1016/j.vetpar.2011.11.068 [DOI] [PubMed] [Google Scholar]
- 21.Brasil BSAF Nunes RL, Bastianetto E Drummond MG, Carvalho DC Leite RC, et al. Genetic diversity patterns of Haemonchus placei and Haemonchus contortus populations isolated from domestic ruminants in Brazil. International Journal for Parasitology. 2012;42(5):469–79. doi: 10.1016/j.ijpara.2012.03.003 [DOI] [PubMed] [Google Scholar]
- 22.Höglund J, Gustafsson K, Ljungström B-L, Engström A, Donnan A, Skuce P. Anthelmintic resistance in Swedish sheep flocks based on a comparison of the results from the faecal egg count reduction test and resistant allele frequencies of the β-tubulin gene. Veterinary Parasitology. 2009;161(1):60–8. doi: 10.1016/j.vetpar.2008.12.001 [DOI] [PubMed] [Google Scholar]
- 23.Kotze AC, Cowling K, Bagnall NH, Hines BM, Ruffell AP, Hunt PW, et al. Relative level of thiabendazole resistance associated with the E198A and F200Y SNPs in larvae of a multi-drug resistant isolate of Haemonchus contortus. International Journal for Parasitology: Drugs and Drug Resistance. 2012;2:92–7. doi: 10.1016/j.ijpddr.2012.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rufener L, Kaminsky R, Mäser P. In vitro selection of Haemonchus contortus for benzimidazole resistance reveals a mutation at amino acid 198 of β-tubulin. Mol Biochem Parasitol. 2009;168(1):120–2. doi: 10.1016/j.molbiopara.2009.07.002 [DOI] [PubMed] [Google Scholar]
- 25.Dilks CM, Hahnel SR, Sheng Q, Long L, McGrath PT, Andersen EC. Quantitative benzimidazole resistance and fitness effects of parasitic nematode beta-tubulin alleles. Int J Parasitol Drugs Drug Resist. 2020;14:28–36. Epub 2020/08/29. doi: 10.1016/j.ijpddr.2020.08.003 ; PubMed Central PMCID: PMC7473882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tydén E, Engström A, Morrison DA, Höglund J. Sequencing of the β-tubulin genes in the ascarid nematodes Parascaris equorum and Ascaridia galli. Mol Biochem Parasitol. 2013;190(1):38–43. Epub 2013/05/21. doi: 10.1016/j.molbiopara.2013.05.003 . [DOI] [PubMed] [Google Scholar]
- 27.Saunders GI, Wasmuth JD, Beech R, Laing R, Hunt M, Naghra H, et al. Characterization and comparative analysis of the complete Haemonchus contortus beta-tubulin gene family and implications for benzimidazole resistance in strongylid nematodes. Int J Parasitol. 2013;43(6):465–75. Epub 2013/02/19. doi: 10.1016/j.ijpara.2012.12.011 . [DOI] [PubMed] [Google Scholar]
- 28.Silvestre A, Humbert JF. Diversity of benzimidazole-resistance alleles in populations of small ruminant parasites. International Journal for Parasitology. 2002;32(7):921–8. doi: 10.1016/s0020-7519(02)00032-2 [DOI] [PubMed] [Google Scholar]
- 29.Kwa MSG, Kooyman FNJ, Boersema JH, Roos MH. Effect of Selection for Benzimidazole Resistance in Haemonchus contortus on β-Tubulin Isotype 1 and Isotype 2 Genes. Biochemical and Biophysical Research Communications. 1993;191(2):413–9. doi: 10.1006/bbrc.1993.1233 [DOI] [PubMed] [Google Scholar]
- 30.Driscoll M, Dean E, Reilly E, Bergholz E, Chalfie M. Genetic and molecular analysis of a Caenorhabditis elegans beta-tubulin that conveys benzimidazole sensitivity. J Cell Biol. 1989;109(6 Pt 1):2993–3003. doi: 10.1083/jcb.109.6.2993 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hahnel SR, Zdraljevic S, Rodriguez BC, Zhao Y, McGrath PT, Andersen EC. Extreme allelic heterogeneity at a Caenorhabditis elegans beta-tubulin locus explains natural resistance to benzimidazoles. PLOS Pathogens. 2018;14(10):e1007226. doi: 10.1371/journal.ppat.1007226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zamanian M, Cook DE, Zdraljevic S, Brady SC, Lee D, Lee J, et al. Discovery of genomic intervals that underlie nematode responses to benzimidazoles. PLoS Negl Trop Dis. 2018;12(3):e0006368. Epub 2018/03/31. doi: 10.1371/journal.pntd.0006368 ; PubMed Central PMCID: PMC5895046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392(6671):71–5. Epub 1998/03/24. doi: 10.1038/32160 . [DOI] [PubMed] [Google Scholar]
- 34.Krücken J, Fraundorfer K, Mugisha JC, Ramünke S, Sifft KC, Geus D, et al. Reduced efficacy of albendazole against Ascaris lumbricoides in Rwandan schoolchildren. International Journal for Parasitology: Drugs and Drug Resistance. 2017;7(3):262–71. doi: 10.1016/j.ijpddr.2017.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diawara A, Halpenny CM, Churcher TS, Mwandawiro C, Kihara J, Kaplan RM, et al. Association between response to albendazole treatment and beta-tubulin genotype frequencies in soil-transmitted helminths. PLoS Negl Trop Dis. 2013;7(5):e2247. Epub 2013/06/06. doi: 10.1371/journal.pntd.0002247 ; PubMed Central PMCID: PMC3667785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Diawara A, Schwenkenbecher JM, Kaplan RM, Prichard RK. Molecular and biological diagnostic tests for monitoring benzimidazole resistance in human soil-transmitted helminths. Am J Trop Med Hyg. 2013;88(6):1052–61. Epub 2013/03/06. doi: 10.4269/ajtmh.12-0484 ; PubMed Central PMCID: PMC3752802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rashwan N, Scott M, Prichard R. Rapid Genotyping of beta-tubulin Polymorphisms in Trichuris trichiura and Ascaris lumbricoides. PLoS Negl Trop Dis. 2017;11(1):e0005205. Epub 2017/01/13. doi: 10.1371/journal.pntd.0005205 ; PubMed Central PMCID: PMC5230752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Furtado LFV, Medeiros CdS, Zuccherato LW, Alves WP, de Oliveira VNGM, da Silva VJ, et al. First identification of the benzimidazole resistance-associated F200Y SNP in the beta-tubulin gene in Ascaris lumbricoides. PloS one. 2019;14(10):e0224108–e. doi: 10.1371/journal.pone.0224108 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuccherato LW, Furtado LF, Medeiros CdS, Pinheiro CdS, Rabelo ÉM. PCR-RFLP screening of polymorphisms associated with benzimidazole resistance in Necator americanus and Ascaris lumbricoides from different geographical regions in Brazil. Plos Neglect Trop D. 2018;12(9):e0006766–e. doi: 10.1371/journal.pntd.0006766 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matamoros G, Rueda MM, Rodríguez C, Gabrie JA, Canales M, Fontecha G, et al. High Endemicity of Soil-Transmitted Helminths in a Population Frequently Exposed to Albendazole but No Evidence of Antiparasitic Resistance. Trop Med Infect Dis. 2019;4(2):73. doi: 10.3390/tropicalmed4020073 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Diawara A, Drake LJ, Suswillo RR, Kihara J, Bundy DA, Scott ME, et al. Assays to detect beta-tubulin codon 200 polymorphism in Trichuris trichiura and Ascaris lumbricoides. PLoS Negl Trop Dis. 2009;3(3):e397. Epub 2009/03/25. doi: 10.1371/journal.pntd.0000397 ; PubMed Central PMCID: PMC2654341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palma A, Matamoros G, Escobar D, Sánchez AL, Fontecha G. Absence of mutations associated with resistance to benzimidazole in the beta-tubulin gene of Ascaris suum. Rev Soc Bras Med Trop. 2020;53:e20190155. Epub 2020/03/19. doi: 10.1590/0037-8682-0155-2019 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gandasegui J, Martínez-Valladares M, Grau-Pujol B, Krolewiecki AJ, Balaña-Fouce R, Gelaye W, et al. Role of DNA-detection–based tools for monitoring the soil-transmitted helminth treatment response in drug-efficacy trials. Plos Neglect Trop D. 2020;14(2):e0007931. doi: 10.1371/journal.pntd.0007931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Easton A, Gao S, Lawton SP, Bennuru S, Khan A, Dahlstrom E, et al. Molecular evidence of hybridization between pig and human Ascaris indicates an interbred species complex infecting humans. Elife. 2020;9. Epub 2020/11/07. doi: 10.7554/eLife.61562 ; PubMed Central PMCID: PMC7647404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang T, Van Steendam K, Dhaenens M, Vlaminck J, Deforce D, Jex AR, et al. Proteomic analysis of the excretory-secretory products from larval stages of Ascaris suum reveals high abundance of glycosyl hydrolases. PLoS Negl Trop Dis. 2013;7(10):e2467. Epub 2013/10/08. doi: 10.1371/journal.pntd.0002467 ; PubMed Central PMCID: PMC3789772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Howe KL, Bolt BJ, Shafie M, Kersey P, Berriman M. WormBase ParaSite—a comprehensive resource for helminth genomics. Mol Biochem Parasitol. 2017;215:2–10. Epub 2016/12/03. doi: 10.1016/j.molbiopara.2016.11.005 ; PubMed Central PMCID: PMC5486357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Howe KL, Bolt BJ, Cain S, Chan J, Chen WJ, Davis P, et al. WormBase 2016: expanding to enable helminth genomic research. Nucleic Acids Res. 2016;44(D1):D774–80. Epub 2015/11/19. doi: 10.1093/nar/gkv1217 ; PubMed Central PMCID: PMC4702863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. Epub 1997/09/01. doi: 10.1093/nar/25.17.3389 ; PubMed Central PMCID: PMC146917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar Gustavo A, Sonnhammer ELL, et al. Pfam: The protein families database in 2021. Nucleic Acids Research. 2021;49(D1):D412–D9. doi: 10.1093/nar/gkaa913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doyle SR, Tracey A, Laing R, Holroyd N, Bartley D, Bazant W, et al. Genomic and transcriptomic variation defines the chromosome-scale assembly of Haemonchus contortus, a model gastrointestinal worm. Communications Biology. 2020;3(1):656. doi: 10.1038/s42003-020-01377-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32(5):1792–7. doi: 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Felsenstein J. Cases in which Parsimony or Compatibility Methods will be Positively Misleading. Systematic Biology. 1978;27(4):401–10. doi: 10.1093/sysbio/27.4.401 [DOI] [Google Scholar]
- 53.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–3. doi: 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–5. doi: 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- 55.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Research. 2019;47(W1):W256–W9. doi: 10.1093/nar/gkz239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vlaminck J, Martinez-Valladares M, Dewilde S, Moens L, Tilleman K, Deforce D, et al. Immunizing pigs with Ascaris suum haemoglobin increases the inflammatory response in the liver but fails to induce a protective immunity. Parasite Immunol. 2011;33(4):250–4. Epub 2011/01/06. doi: 10.1111/j.1365-3024.2010.01274.x . [DOI] [PubMed] [Google Scholar]
- 57.Wang J, Mitreva M, Berriman M, Thorne A, Magrini V, Koutsovoulos G, et al. Silencing of Germline-Expressed Genes by DNA Elimination in Somatic Cells. Developmental Cell. 2012;23(5):1072–80. doi: 10.1016/j.devcel.2012.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, Czech B, Crunk A, Wallace A, Mitreva M, Hannon GJ, et al. Deep small RNA sequencing from the nematode Ascaris reveals conservation, functional diversification, and novel developmental profiles. Genome Res. 2011;21(9):1462–77. Epub 2011/06/21. doi: 10.1101/gr.121426.111 ; PubMed Central PMCID: PMC3166831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jex AR, Liu S, Li B, Young ND, Hall RS, Li Y, et al. Ascaris suum draft genome. Nature. 2011;479(7374):529–33. Epub 2011/10/28. doi: 10.1038/nature10553 . [DOI] [PubMed] [Google Scholar]
- 60.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nature Biotechnology. 2016;34(5):525–7. doi: 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- 61.Avramenko RW, Redman EM, Lewis R, Yazwinski TA, Wasmuth JD, Gilleard JS. Exploring the Gastrointestinal "Nemabiome": Deep Amplicon Sequencing to Quantify the Species Composition of Parasitic Nematode Communities. PLoS One. 2015;10(12):e0143559. Epub 2015/12/03. doi: 10.1371/journal.pone.0143559 ; PubMed Central PMCID: PMC4668017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods. 2016;13(7):581–3. doi: 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011. 2011;17(1):3. Epub 2011-08-02. doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 64.R Core Team (2019). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available from: http://R-project.org. [Google Scholar]
- 65.Adamack AT, Gruber B. PopGenReport: simplifying basic population genetic analyses in R. Methods in Ecology and Evolution. 2014;5(4):384–7. doi: 10.1111/2041-210X.12158 [DOI] [Google Scholar]
- 66.Gruber B, Adamack AT. landgenreport: a new r function to simplify landscape genetic analysis using resistance surface layers. Molecular Ecology Resources. 2015;15(5):1172–8. doi: 10.1111/1755-0998.12381 [DOI] [PubMed] [Google Scholar]
- 67.Nei M. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci U S A. 1973;70(12):3321–3. Epub 1973/12/01. doi: 10.1073/pnas.70.12.3321 ; PubMed Central PMCID: PMC427228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hurd DD, Miller RM, Nunez L, Portman DS. Specific alpha- and beta-tubulin isotypes optimize the functions of sensory Cilia in Caenorhabditis elegans. Genetics. 2010;185(3):883–96. Epub 2010/04/28. doi: 10.1534/genetics.110.116996 ; PubMed Central PMCID: PMC2907207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Savage C, Hamelin M, Culotti JG, Coulson A, Albertson DG, Chalfie M. mec-7 is a beta-tubulin gene required for the production of 15-protofilament microtubules in Caenorhabditis elegans. Genes Dev. 1989;3(6):870–81. Epub 1989/06/01. doi: 10.1101/gad.3.6.870 . [DOI] [PubMed] [Google Scholar]
- 70.Savage C, Xue Y, Mitani S, Hall D, Zakhary R, Chalfie M. Mutations in the Caenorhabditis elegans beta-tubulin gene mec-7: effects on microtubule assembly and stability and on tubulin autoregulation. J Cell Sci. 1994;107 (Pt 8):2165–75. Epub 1994/08/01. . [DOI] [PubMed] [Google Scholar]
- 71.Vlaminck J, Cools P, Albonico M, Ame S, Ayana M, Cringoli G, et al. Therapeutic efficacy of albendazole against soil-transmitted helminthiasis in children measured by five diagnostic methods. PLoS Negl Trop Dis. 2019;13(8):e0007471. Epub 2019/08/02. doi: 10.1371/journal.pntd.0007471 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vlaminck J, Cools P, Albonico M, Ame S, Ayana M, Bethony J, et al. Comprehensive evaluation of stool-based diagnostic methods and benzimidazole resistance markers to assess drug efficacy and detect the emergence of anthelmintic resistance: A Starworms study protocol. Plos Neglect Trop D. 2018;12(11):e0006912. doi: 10.1371/journal.pntd.0006912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282(5396):2012–8. Epub 1998/12/16. doi: 10.1126/science.282.5396.2012 . [DOI] [PubMed] [Google Scholar]
- 74.Foth BJ, Tsai IJ, Reid AJ, Bancroft AJ, Nichol S, Tracey A, et al. Whipworm genome and dual-species transcriptome analyses provide molecular insights into an intimate host-parasite interaction. Nat Genet. 2014;46(7):693–700. doi: 10.1038/ng.3010 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tang YT, Gao X, Rosa BA, Abubucker S, Hallsworth-Pepin K, Martin J, et al. Genome of the human hookworm Necator americanus. Nat Genet. 2014;46(3):261–9. doi: 10.1038/ng.2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.International Helminth Genomes Consortium. Comparative genomics of the major parasitic worms. Nat Genet. 2019;51(1):163–74. doi: 10.1038/s41588-018-0262-1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
WormBase ParaSite or NCBI Gene ID and Transcript ID of all β-tubulin proteins included in the phylogenetic analyses and trees. Caenorhabditis elegans and Haemonchus contortus β-tubulin protein sequences were obtained from published genomes [50,73]. The corresponding names were adopted from published literature [27,30]. For Trichuris trichiura, Necator americanus and Ancylostoma ceylanicum keyword search for ‘tubulin beta’ and BLASTP search with the Ascaris β-tubulin genes against the published genomes [74–76] resulted in two sequences for T. trichiura and four sequences for both hookworms. The genome assembly and annotation of A. ceylanicum were used as close representative of A. duodenale, since this genome assembly has low quality metrics. For Parascaris equorum and Ascaridia galli, the sequences published by Tyden et al. (2013) were used [26]. Three β-tubulins from Drosophila melanogaster are included in the list to be used as outgroups.
(XLSX)
β-tubulin gene specific primers used in RT-PCR analysis. All β-tubulin primers are based on coding sequences. The actin gene is used as household gene and primers are adopted from Vlaminck et al., 2011 [56].
(PDF)
Overview of all RNA-seq datasets (NCBI Sequence Read Archive (SRA)) used in the A. suum β-tubulin RNA-seq analysis.
(XLSX)
Alu/Asu-bt-A and Alu/Asu-bt-B (and bt-B’) primers with Illumina Adapters. Locus specific primer sequence bolded, N’s underlined. Illumina adaptor oligonucleotide sequences were obtained from the Illumina Adapter Sequences document dated March 2020 (Illumina Inc.). Forward and Reverse barcoded sequencing primers. Index sequence bolded. Sequences were obtained from the Illumina Adapter Sequences document dated March 2020 (Illumina Inc.).
(PDF)
Alignment of amino acid sequences of the identified β-tubulins of both A. suum and A. lumbricoides. For aesthetic reasons, all sequences were trimmed to a uniform length of 427 amino acids. (N-terminus trimmed: Asu-bt-B, Asu-bt-E; C-terminus trimmed: all).
(PDF)
Excel spreadsheet of amplicon sequencing results for Alu/Asu-bt-A following analysis with the DADA2 v.1.11.5 bioinformatic software package and manual assessment for chimeras, off-target amplicons and PCR artefacts. The first column of the spreadsheet contains the single worm identification. In the adjacent columns the read number for each unique amplicon sequence variant (ASV) is given. Corresponding genotypes were defined by applying a threshold of 1,000 reads. ASVs below threshold level were excluded from further population genetic analyses and are indicated in red in the table. There were no worms with reads above the threshold for more than two ASVs.
(XLSX)
Percentage of bases/residues which are identical (% Identity), and number of bases/residues which are identical (# Identities) and not identical (# Differences). Results for the nine identified amplicon sequence variants (ASVs) of Alu/Asu-bt-A. All pairwise distances were computed using Geneious v10.2.6.
(XLSX)
Nucleotide alignment of the nine identified amplicon sequence variants (ASVs) of Alu/Asu-bt-A. Multiple alignment was performed using Geneious v10.2.6. The three codons of interest in the context of benzimidazole resistance are indicated by their number at the base of the consensus sequence.
(PDF)
The total number of each amplicon sequence variant (ASV) (allele count) and the proportion within the (sub)populations (allele frequency) for both Alu/Asu-bt-A and Alu/Asu-bt-B. A. suum worms were collected from the intestines of pigs slaughtered in Belgium. A. lumbricoides worms were collected during two expulsion studies in Ethiopia and Tanzania.
(XLSX)
The total number of each defined genotype (genotype count) and the proportion within the (sub)populations (genotype frequency) for both Alu/Asu-bt-A and Alu/Asu-bt-B. Genotype per worm was defined by applying a threshold of 1,000 reads. A. suum worms were collected from the intestines of pigs slaughtered in Belgium. A. lumbricoides worms were collected during two expulsion studies in Ethiopia and Tanzania.
(XLSX)
Excel spreadsheet of amplicon sequencing results for Alu/Asu-bt-B following analysis with the DADA2 v.1.11.5 bioinformatic software package and manual assessment for chimeras, off-target amplicons and PCR artefacts. The first column of the spreadsheet contains the single worm identification. In the adjacent columns the read number for each unique amplicon sequence variant (ASV) is given. From the dataset, 21 ASVs with extremely low reads (below 40) and only detected in 1 or 2 worms were manually deleted since these were seen as PCR artefacts. Furthermore, due to a lack of complete specificity of the primers, the dataset contained 10 ASVs corresponding to the Alu/Asu-bt-A locus, which were also manually deleted. The genotype per worm was defined by applying a threshold of 1,000 reads. ASVs below threshold level were excluded from further population genetic analyses and are indicated in red in the table. There was one A. lumbricoides worm with only reads below threshold and five A. suum worms with reads above threshold for more than two ASVs, these six worms were not included in the genotype data used for population genetic analysis.
(XLSX)
Percentage of bases/residues which are identical (% Identity), and number of bases/residues which are identical (# Identities) and not identical (# Differences). Results for the 21 identified amplicon sequence variants (ASVs) of Alu/Asu-bt-B. All pairwise distances were computed using Geneious v10.2.6.
(XLSX)
Nucleotide alignment of the 21 identified amplicon sequence variants (ASVs) of Alu/Asu-bt-B. Multiple alignment was performed using Geneious v10.2.6. The three codons of interest in the context of benzimidazole resistance are indicated by their number at the base of the consensus sequence.
(PDF)
Phylogenies were reconstructed with RAxML and MRBAYES. The topology shown is from RAxML. The node support values are percent bootstraps / posterior probabilities. The ‘-‘ indicates that a node was not present in the MRBAYES tree. The branch lengths show divergence.
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.