Abstract
Interferons (IFNs) are among the first vertebrate immune pathways activated upon viral infection and are crucial for control of viral replication and dissemination, especially at mucosal surfaces as key locations for host exposure to pathogens. Inhibition of viral establishment and spread at and from these mucosal sites is paramount for preventing severe disease, while concomitantly limiting putative detrimental effects of inflammation. Here, we compare the roles of type I, II, and III IFNs in regulating three archetypal viruses – norovirus, herpes simplex virus, and severe acute respiratory virus coronavirus 2 (SARS-CoV-2) – which infect distinct mammalian mucosal tissues. Emerging paradigms include highly specific roles for IFNs in limiting local versus systemic infection, synergistic activities, and a spectrum of protective versus detrimental effects of IFNs during the infection response.
Keywords: mucosal viruses, interferons, IFN-alpha/beta, IFN-gamma, IFN-lambda, norovirus, herpesvirus, SARS-CoV-2, interferon-stimulated genes, viral antagonism
Interferons at the front line
Mucosal (see Glossary) surfaces are the first line of defense against invasion by most pathogens. The human gastrointestinal tract alone possesses a surface area of at least 30 m2 and, together with other mucosal tissues, including the respiratory and reproductive tracts, represents the majority of potential exposure sites to outside threats. Thus, these tissues must constantly defend against infiltration by a variety of microbes. This response is multifaceted and complex, but one key initial defense against a plethora of pathogens is innate immune interferon (IFN) signaling. IFNs have long been recognized for their role in antagonizing viral pathogens [1] as well as fungal and bacterial infections and are crucial for limiting both local infection and systemic spread in and from mucosal tissues. Recent studies have revealed numerous shared modalities across body sites in the IFN responses to different viruses in humans and rodent models. While all IFNs share effectors, the role of each IFN type is distinct, requiring a careful examination of the regulation and effects of different IFN types in mammalian hosts.
Here, we compare and contrast the signaling pathways and effects of the three types of IFNs, describing their key protective and detrimental roles in the context of viral infections. While IFNs are essential at numerous other sites including the skin, we focus on the mechanisms by which IFNs are regulated and control infection at three key mucosal sites – gastrointestinal, respiratory, and female reproductive tracts, using well-studied ‘model’ pathogens to detail the complex interplay of the different IFNs. Though IFNs have been studied for over 60 years, our understanding of their individual and combinatorial effects continues to evolve, driven by new discoveries of the distinct activities of each IFN type in protecting mucosal sites, a front line against viral infection.
Types I, II, and III IFN signaling and induction
Initially named after their function in ‘interfering’ with viral replication [1], IFNs are a class of cytokines responsible for initiating a cascade of immune responses against pathogens [2]. Subsequently grouped into three families (type I, II, and III; or IFN-I, -II, and -III), IFNs work together in a synergistic manner to induce antiviral activities in mammalian host cells such as epithelial cells and macrophages. In general, pathogen-associated molecular patterns (PAMPs) are sensed by pattern recognition receptors (PRRs), leading to induction and secretion of IFNs by infected immune or epithelial cells. IFNs bind IFN receptors on the surface of neighboring and/or immune cells, triggering a signaling cascade to induce a suite of IFN-stimulated genes (ISGs) that directly mediate the antipathogenic effects of IFNs (Figure 1 ) [3]. There are specificities to each IFN type, such as different receptor utilization, expression patterns, and distinct downstream genes, which are key for their divergent roles (Table 1 ).
Figure 1.
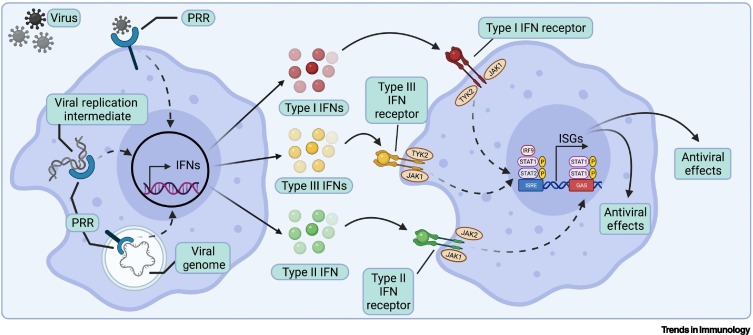
Overview of interferon signaling pathways in vertebrate hosts.
Production of interferons (IFNs) begins with the binding of viral molecules, such as genomic nucleic acids, to either cell surface or intracellular pattern recognition receptors (PRRs) [18,19]. The resulting signaling cascade activates transcription and secretion of IFNs, which then bind to their associated IFN receptor on the same and nearby cells. Binding of IFNs to their receptors activates a signal cascade by Janus tyrosine kinases (JAK) and tyrosine kinase (TYK) that leads to the phosphorylation of STAT1 and/or STAT2 [9,15]. For type I and III IFNs, STAT1 and STAT2 complex with IRF9 and bind to IFN-stimulated response elements (ISREs) to express IFN-stimulated genes (ISGs). For type II IFNs, phosphorylated STAT1 dimers bind to gamma-activated site (GAS) elements for ISG production [9,15]. In turn, ISGs mediate antiviral effects directly within infected cells, or further induce innate and adaptive immune responses [3,120]. Figure created using BioRender (https://biorender.com/). Abbreviation: ISG, IFN-stimulated gene.
Table 1.
Key molecular characteristics contributing to differential roles of type I, II, and III IFNsa
| IFN type | Type I | Type II | Type III |
|---|---|---|---|
| Receptor | IFNAR1, IFNAR2 [9] | IFNGR1, IFNGR2 [125] | IL-28Rα/IFNLR1, IL-10R2 [16] |
| Receptor expression | Ubiquitous [126] | Diverse, especially hematopoietic cells [127] | Epithelial cells, peripheral blood lymphocytes, neutrophils, plasmacytoid DCs [16,17,102] |
| Members in mice and humans | Mice: 14 IFN-α IFN-β IFN-ε IFN-κ IFN-ζ Humans: 13 IFN-α IFN-β IFN-ε IFN-ω IFN-κ [5] |
Mice: IFN-γ Humans: IFN-γ |
Mice: IFN-λ2, IFN-λ3 [128,129] Humans: IFN-λ1, IFN-λ2, IFN-λ3 IFN-λ4 (variably pseudogenic) [130,131] |
| Member expression | IFN-α/IFN-β: Plasmacytoid DCs Macrophages Fibroblasts Epithelial cells [7] Others: Female reproductive tract (IFN-ε) [77] Keratinocytes (IFN-κ) [132] |
NK cells T cells Type I innate lymphoid cells [11] |
Epithelial cells of intestine and lung [133,134] Plasmacytoid DCs [135] |
| Downstream signaling complex | ISGF3 complex (STAT1/STAT2/IRF9) [125] | STAT1 homodimers [125] | ISGF3 complex (STAT1/STAT2/IRF9) [16] |
| Key activators | IRF3, IRF5, IRF7, IRF8, various STAT TFs [2] | AP-1, CREB/ATF, NFAT, T-bet, Eomes, various STAT TFs [2] | IRF1, IRF3, IRF7, NF-κB. various STAT TFs [2] |
Abbreviations: DCs, dendritic cells; TFs, transcription factors.
Type I IFNs are the most diverse IFN family, with prototypical members IFN-α and IFN-β expressed widely in a variety of cell types [4]. There are a variety of additional type I IFNs with distinct expression patterns and functions, with some additional subtypes present in restricted mammalian lineages, such as IFN-τ and IFN-ζ, which are only found in ruminants and mice, respectively [5]. Type I IFNs are also broadly produced by most cells in response to infection, though plasmacytoid dendritic cells (pDCs) are a particularly important source of type I IFNs [6,7]. The binding of type I IFNs to their receptor, IFNAR, leads to the phosphorylation of transcription factors (TFs) STAT1 and STAT2, which bind IFN regulatory factor (IRF) 9 to form a complex called the ISG factor 3 (ISGF3) [8]. ISGF3 translocates to the nucleus, binding IFN-stimulated response elements to induce ISGs [9].
Unlike the diverse suite of type I IFNs, there is only a single type II IFN, IFN-γ [10,11]. Despite its initial association with antiviral responses [12], IFN-γ is now more generally considered for its important roles in immunity against fungi and bacteria [11]. IFN-γ provides crucial support for the function of type I IFNs and is particularly involved in regulating cell-mediated immune responses, such as promotion of macrophage activation and enhancement of antigen presentation [9]. Indeed, type I IFN can subsequently activate natural killer (NK) cells to produce IFN-γ [13]. Unlike the heteromeric ISGF3 signaling complex in type I IFN signaling, type II IFN signals through STAT1 homodimers to induce ISG transcription [14].
Type III IFNs (IFN-λ) – the most recently identified type of IFNs – share many characteristics with type I IFNs, albeit with a more limited set of cells in which they and their receptor are expressed [15]. Like type I IFNs, type III IFNs signal through the ISGF3 complex, making its component STAT1 an essential TF for all three IFN types [8]. However, due to the restricted expression of receptor protein IL28Rα/IFNLR1, responses to IFN-λ in mice and humans are generally confined to epithelial cells, peripheral blood lymphocytes, neutrophils, and plasmacytoid DCs, with most type III IFN responses occurring at epithelial surfaces [15., 16., 17.].
While the intricacies of how the host senses invasion by viral pathogens are outside of the scope of this review, other recent reviews dissect this topic in greater detail [18]. In short, a variety of widely expressed PRRs sense common motifs, PAMPs, present in the context of a viral pathogen, such as double-stranded RNA viral genomes or replication intermediates [19,20] (Figure 1). Additional patterns, such as modified bases, certain DNA motifs, or, rarely, specific viral proteins, may also activate PRRs [21,22]. Ultimately, pathways that sense more ubiquitous motifs are preferred by the host to facilitate responses to both common and unknown pathogens.
PRRs trigger signaling cascades that result in the upregulation of one or more IFN types to induce an antiviral response. These sensing pathways converge on a handful of common mediators of signaling, such as MAVS, which is activated by the PRRs RIG-I and MDA5 to respond to distinct RNA PAMPs [19]. This bottleneck of diverse signaling pathways activating the same suite of mediators allows a variety of PAMPs to induce similar IFN responses. However, not all PRRs or stimulants result in equal induction of all IFNs, likely due to differences in transcriptional regulation of these IFNs. Type I and III IFNs are primarily activated by the IRF TFs, while the type II IFN promoter instead bears a diversity of binding sites for TFs such as the Th1-regulatory protein T-bet [11]. Individual IRFs also have unique proclivities for activation of certain IFN types. For example, IRF1 is essential for activation of type III, but not type I, IFN expression in human cells, whereas the AP-1 TF is required for type I, but not type III, expression [23]. Individual TFs also have unique ranges of expression in diverse cell types; IRF5, for example, is expressed in higher amounts in murine hematopoietic cells and, thus, specifically induces protective IFN responses in immune cells [24]. Thus, in summary, differential requirements for certain TFs to activate IFNs result in specific expression patterns for each IFN. This, combined with the unique patterns of IFN receptor expression on specific cell types as discussed earlier and further detailed in Table 1, contributes to differential roles for each type of IFN in antiviral defense at mucosal surfaces. Additionally, these unique regulatory pathways also contribute to distinct temporal expression for each IFN, with type I IFNs typically induced more rapidly than type III IFNs upon exposure to viral infection or PAMPs [25,26]. Type III IFNs may also be expressed for longer periods of time than type I IFNs based on findings from diverse human cell lines and primary cell culture models [25,26]. These differential kinetics of expression add further complexity to the unique roles played by each IFN.
Furthermore, despite their unique roles, IFNs are usually not activated in isolation. Indeed, individual IFNs can serve to induce other IFN types or intermediate signal transducers as well [26,27]. For example, as mentioned earlier, type I IFN can stimulate type II IFN production by other cell types [13], and mice deficient in Ifnar1 (Ifnar1 –/–) exhibit dampened type III IFN induction in response to influenza A virus (IAV) infection [28]. Treatment of mice with type III IFN can potentiate type II IFN serum concentrations following vaginal viral infection [29]. Thus, IFNs can coordinately promote cross-activation of synergistic antiviral pathways to facilitate broad and efficacious antiviral responses.
Three ‘model’ mucosal viruses
IFNs have been implicated as key cytokines in the control of mucosal viruses. Indeed, mice lacking receptors for IFNs (e.g., Ifnar1 –/–or Ifnlr1 –/–) or the shared downstream TF STAT1 consistently exhibit more severe and/or disseminated viral infections [30., 31., 32.]. Many of these experimental observations are corroborated by the identification of debilitating susceptibility to infection with IAV, herpesviruses, or severe acute respiratory virus coronavirus 2 (SARS-CoV-2), in human patients with deficiencies in IFN signaling molecules, such as mutations in IFNAR1 or IRF9, or harboring autoantibodies against IFNs [33., 34., 35., 36., 37., 38.]. Here, we briefly introduce these three model viruses that infect distinct mucosal sites and subsequently describe the insights they have yielded into IFN-mediated regulation of viral infections.
Norovirus – gastrointestinal tract
Noroviruses (NoVs) belong to the family Caliciviridae and are single-stranded, positive-sense RNA viruses. NoVs infect a wide range of mammals, including humans, mice, pigs, and canines, though individual genotypes exhibit limited cross-species tropism. Human NoV (HNoV) is the leading cause of viral gastroenteritis, causing an estimated 700 million infections annually [39]. Although human infections are usually self-limiting, in the young, the immunocompromised, and the elderly, they can be severe, leading to ~200 000 deaths each year [39]. Murine NoV (MNoV), which infects the small intestine and colon after oral administration, is a robust small animal model for experimental NoV studies [31]. Numerous MNoV strains with varying degrees of lethality and persistence have been characterized, and the MNoV reverse genetics system has allowed for key insights into the contributions of viral and host genetics to infection outcomes [40., 41., 42.]. There are data to support a cellular tropism for intestinal epithelial [43., 44., 45.] and/or immune cells [46., 47., 48., 49.] for HNoV and MNoV that may be strain-specific. Recent insights, which will be subsequently defined, have demonstrated crucial aspects of MNoV–IFN interactions in regulating virulence as well as cellular tropism of this prominent pathogen [50., 51., 52.].
Herpes simplex virus – female reproductive tract
Herpes simplex viruses (HSVs) are herpesviruses with long double-stranded DNA genomes stretching into hundreds of kilobases [53]. HSVs are among the most prominent viral pathogens worldwide and the most prevalent sexually transmitted infections, with up to two-thirds of humans believed to be infected with HSV-1 or HSV-2, which can cause cutaneous infections around the mouth, genitals, or eyes [54,55]. HSVs infect diverse cell types, with the initial infection usually occurring in the epithelium, then progressing to neurons [56]. In severe cases, particularly in immunocompromised humans, HSV can progress to infect the central nervous system (CNS) to cause encephalitis [34,57]. Due to the plethora of genes present in their extremely large genomes, HSVs are masters of immune evasion, by directly interfering with immune sensors and effectors (Box 1 ) and indirectly by entering into latency, wherein they are maintained within the host nucleus [58]. HSVs have provided key insights into viral immunology, such as the protective effects of type II IFN and T cell responses versus the detrimental effects of type I IFNs during infection in mice [59,60]. HSVs remain among the best-studied viruses in laboratory genital infection mouse models.
Box 1. Viral antagonism of IFN signaling.
While hosts mount a myriad of mechanisms to provide immunity to pathogens, pathogens develop counter-immune mechanisms to evade these responses. IFN responses can be inhibited at nearly any stage in the pathway, and many viruses utilize multiple distinct mechanisms to effectively prevent these responses. Murine norovirus (MNoV) possesses at least three separate proteins that antagonize various IFN pathways, while herpes simplex viruses (HSVs) and severe acute respiratory virus coronavirus 2 (SARS-CoV-2), with their large genomes, each carry an arsenal of proteins involved in evasion of IFN responses, described below. For SARS-CoV-2, nearly half of its products are reported to inhibit IFNs, with in vitro studies identifying at least eight distinct viral proteins that inhibit type I IFN responses [135]. Below are some of the known mechanisms utilized by these mucosal viruses to highlight potential targets for viral antagonism.
Some viral proteins involved in immune evasion prevent IFN induction, inhibiting the activity of PRRs or intermediate signaling molecules. Some of these activities remain to be fully elucidated, such as how the MNoV protein VF1 prevents induction of type I IFNs [136]. An additional MNoV nonstructural protein, NS1, is an essential determinant of the epithelial tropism of MNoV by inhibiting IFN-λ signaling through a yet-unknown mechanism [50,51]. Like MNoV, HSV prevents IFN induction, with viral proteins ICP0 and VP24 preventing activation or nuclear translocation of IFN-inducing TF IRF3 in human cell lines [137,138]. Additionally, in human cell lines, another HSV protein, UL36, inactivates PRR signaling mediator TRAF3 via its deubiquitinase activity to prevent IFN induction [139]. Disruption of signaling pathways upstream of IFN production provides for widespread inhibition of IFN signaling.
To further interfere with IFN signaling, viruses also inhibit signaling downstream of the IFN receptors. HSV is particularly effective in this antagonism, with HSV ICP0 promoting degradation of the host deubiquitinase BRCC36, resulting in downregulation of IFNAR1 in a variety of human and mouse cell lines in vitro [140]. HSV infection also represses IFNGR1 expression, limiting IFN-γ activity in human primary dendritic cells [141]. Downstream of the receptors, HSV UL36 prevents IFNAR signaling by blocking its interaction with the JAK1 kinase, which is essential for IFN signal transduction in human cell cultures [142]. Using a different but complementary activity to prevent ISG induction, SARS-CoV-2 nonstructural proteins NSP14 and NSP1 inhibit synthesis of host proteins, including ISGs in a variety of human and primate cell lines [143., 144., 145.].
Finally, some viruses directly inhibit specific ISGs. NS7, the MNoV polymerase, directly antagonizes a specific type II IFN-induced ISG, GBP2, to block its anti-MNoV activity in mice [146]. SARS-CoV-2 nonstructural protein ORF7A similarly antagonizes a specific ISG, BST2, in human epithelial cells in vitro [147]. These direct interactions with ISGs allow for potent, but focused, inhibition of antiviral activities and may aid in identifying ISGs with special importance for the viruses that block them.
Alt-text: Box 1
SARS-CoV-2 – respiratory tract
SARS-CoV-2 is the causative agent of COVID-19, which, within a year of its emergence in November 2019 from an animal reservoir into humans, has caused a catastrophic pandemic with millions of deaths worldwide. Coronaviruses are single-stranded, positive-sense RNA viruses which have among the largest genomes of known RNA viruses at nearly 30 kbp [61]. Although many coronaviruses cause mild colds, some coronaviruses such as MERS-CoV, SARS-CoV-1, and SARS-CoV-2 infect the lungs, causing severe viral pneumonia in humans [62]. SARS-CoV-2 uses the host receptor ACE2 to infect human respiratory and intestinal epithelial cells [63,64]. SARS-CoV-2 may also infect a range of hosts including cats, dogs, mink, ferrets, and bats [62]. Though the literature surrounding SARS-CoV-2 continues to evolve, numerous studies have already demonstrated the importance of IFNs in its control, both clinically and in rodent models, as discussed further in the next section.
Antiviral activities of IFNs
Type I IFNs restrict dissemination of mucosal viruses
While there are overlapping roles for all three IFN types in restricting mucosal viral infections and specific nuances are observed for any specific virus or host site, some common and consistent trends can be identified (Figure 2, Key figure). Type I IFNs are critically important for preventing morbidity and mortality in mice from infection by diverse viruses including IAV, West Nile virus (WNV), and Nipah virus, beyond those detailed here [65., 66., 67.]. The protective effects of type I IFNs often involve restricting systemic viral infection and dissemination from the initial mucosal site of infection.
Figure 2.
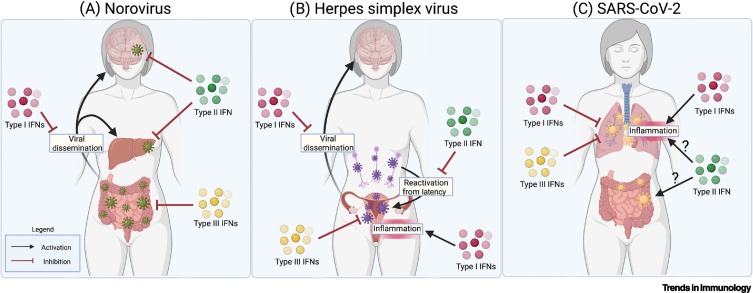
Key figure. Key roles for type I, II, and III interferons in mucosal antiviral responses in mice and humans.
(A) Norovirus initiates infection within the gastrointestinal tract, wherein type III interferons (IFNs) are key for restricting viral growth [30]. Type I IFNs restrict dissemination of the virus from the gut to distal sites such as the liver and brain, wherein type II IFN further controls replication [69,89]. (B) Herpes simplex virus infections initiated in the female reproductive tract are controlled at the site of infection by type III IFNs, while type I IFNs drive inflammation [29,60]. Dissemination from this mucosal tissue to the central nervous system (CNS) is restricted by type I IFNs [32,73]. Herpesviruses undergo latency, remaining mostly transcriptionally silent within neurons, with reactivation of the virus from latency restricted by type II IFN [92]. (C) SARS-CoV-2 is restricted within the respiratory tract by type I and III IFNs [83,84,113]. Conversely, type I IFNs may be drivers of inflammation in the respiratory tract during SARS-CoV-2 infection [28,83,86]. The role of type II IFN is currently less understood, but it may be proviral in some sites by inducing expression of the viral receptor and may also drive inflammation [100]. Figure created using BioRender (https://biorender.com/). Abbreviation: SARS-CoV-2, severe acute respiratory virus coronavirus 2.
Specifically, after oral infection of the gastrointestinal tract, type I IFNs serve to limit the spread of viruses such as MNoV beyond the intestine. Specific MNoV strains induce type I IFNs in vitro in murine dendritic cells (DCs) and in vivo in mouse mesenteric lymph nodes [30,68]. Mice lacking Ifnar1, either globally (Ifnar1 –/–) or specifically in myeloid or DCs (LysM-Cre;Ifnar1 f/f and Cd11c-Cre;Ifnar1 f/f, respectively), are susceptible to the extraintestinal spread of virulent strains of MNoV to the spleen, liver, and brain and exhibit persistent systemic infection or death relative to wild-type controls [31,40,68., 69., 70.]. Although tractable means of studying HNoV in the laboratory have only recently become available with the development of human B cell and intestinal enteroid models [43,71], application of exogenous type I IFNs to human intestinal enteroids has now been shown to restrict growth of HNoV in a viral strain-dependent manner [72]. This finding suggests that the IFN-mediated regulation identified in murine models of NoV may be applicable to humans, although further studies are warranted. In summary, both MNoV and HNoV exhibit strain-specific induction of and restriction by type I IFN, emphasizing the importance of viral genotype in the ability of IFNs to control infection [30,40,72].
In HSV-2 genital tract infections, loss of type I IFN signaling in mice results in increased local viral replication and systemic spread to the CNS [32,73]. As in MNoV infection, DCs are involved in limiting systemic replication and infection of HSV, with pDCs serving as essential producers of type I IFNs [74]. Patients deficient in IFNAR1 or related signaling molecules often present with HSV encephalitis [34,75,76]. Of interest, HSV-2 is restricted by IFN-ε – a recently described type I IFN that is constitutively expressed in a hormonally regulated manner within the female reproductive tract in humans and mice [77,78]. Mice lacking IFN-ε (Ifne –/–) exhibit decreased baseline expression of many ISGs in the reproductive tract, as well as increased replication of HSV-2 within the reproductive tract and increased spread of the virus into the spinal cord and brain stem, relative to wild-type animals [78]. IFN-κ, another understudied type I IFN that is selectively expressed in keratinocytes, has also been proposed to restrict HSV-1 replication in a human keratinocyte cell culture model [79], suggesting that there may be important tissue-restricted roles for some type I IFNs during HSV infection. Although crucial for preventing viral spread, type I IFNs also drive pathology in murine genital HSV infection in a viral strain-specific manner via their interactions with neutrophils [60], demonstrating the potentially detrimental effects of IFN signaling.
As with the protective and detrimental roles of type I IFNs in HSV infections, there appear to be dual roles for these IFNs in SARS-CoV-2 infections. In vitro, type I IFNs control SARS-CoV-2 replication [80., 81., 82.], and mutations in genes including IFNAR1, IRF7, and TLR3 involved in the response to type I IFNs are associated with severe COVID-19 in humans [35], supporting the importance of type I IFNs in the modulation of SARS-CoV-2 infection. Additionally, autoantibodies against type I IFNs have been identified at higher rates in patients with severe COVID-19 than in controls [36,37]. However, while in mouse and hamster models disruption of type I IFN signaling, such as in Ifnar1 –/– or Stat2 –/– animals, does indeed lead to increased viral dissemination, animals are also protected from disease, with decreased pneumonia and lung inflammation [83,84]. We posit that this might hold true in humans as well, as a higher proportion of type III IFN expression relative to type I IFN expression has correlated with viral clearance and improved clinical outcomes in COVID-19 patients, raising the possibility that there may be detrimental effects that are specific to type I IFNs associated with this disease [85]. However, further studies are warranted to fully validate these findings, as other recent reports have implicated both type I and III IFNs in COVID-19-associated morbidity in human patients, as well as in IAV-mediated lung pathology in mice [28,86]. All in all, the SARS-CoV-2 pandemic has emphasized the importance of human genetic variation in determining clinical outcomes and has also highlighted the potential double-edged nature of type I IFN responses in disease; indeed, these IFNs can aid in clearing viral infection, but must also be carefully controlled to prevent deleterious unwanted effects, such as the damaging overinduction of the immune system known as the ‘cytokine storm’ [87].
Type II IFN synergizes with type I IFNs to restrict mucosal viruses
Type II IFN is widely appreciated for its role in controlling bacterial infections but is relatively understudied in the context of viral infections. While the importance of type I and III IFNs in viral restriction is immediately evident by their induction of an antiviral state within infected cells [9], the antipathogenic roles of type II IFNs are often mediated through additional immune cell types. These phenotypes are more difficult to detect in in vitro experiments, limiting the characterization of this cytokine as antiviral. Despite this, important roles of IFN-γ in viral infections have been described [88], with varying mechanisms detailed here.
In contrast to the importance of type I IFNs in preventing systemic infection, the role of type II IFN in controlling MNoV infection is somewhat more subtle, emerging primarily when type I IFN signaling is also disrupted, as evidenced from the enhanced morbidity in Ifnar1 –/– Ifngr1 –/– mice compared with Ifnar1 –/– mice [89]. However, IFN-γ can directly restrict MNoV replication in myeloid cells lines in vitro [89]. Additionally, in mice, IFN-γ acts on myeloid cells to promote assembly of a protein complex that includes autophagy protein Atg16L1, to restrict viral replication and limit pathogenesis [90], along with IFN-γ-inducible GTPases serving to disrupt viral replication complexes [91]. This role appears to be independent of autophagy itself, as pharmacological inhibition of autophagy does not alter IFN-γ-mediated inhibition of MNoV replication [90]. These data provide hints of a nonautophagic role for essential autophagy proteins in IFN-γ-mediated antiviral defense.
IFN-γ is a crucial innate immune regulator of HSV-1 and -2 infections, particularly in the context of disseminated infection beyond the epithelium and in control of viral latency [92]. In multiple mammalian cell culture models, including African green monkey Vero cells and human dorsal root ganglia explants, exogenous type II IFNs exhibit an ability to control HSV-1 replication directly, although this activity is most potent in synergy with type I IFN activity [93,94]. The responses of infected cells to type I or II IFNs differ somewhat, with IFN-γ treatment leading to a more widespread neuronal antiviral response than type I IFN treatment. However, both types of IFNs prevent the transport of HSV virions within neurons to limit viral spread in primary murine neuron cultures [95]. CD8+ T cell-derived IFN-γ also impedes the emergence of HSV-1 from latency in ex vivo murine trigeminal ganglia cultures, thus preventing recurring infections [92,96]. Type II IFN production by NK cells and CD4+ and CD8+ T cells during murine vaginal infection is key to preventing HSV-1, but not HSV-2, neuronal infection, which may be important for better understanding the distinct rates of recurrent genital infections seen with these two viruses [59].
The role of type II IFN in COVID-19 is less clear, although IFN-γ does exhibit antiviral activity in vitro in primary human differentiated bronchial epithelial cells [80]. Some studies have proposed a potentially proviral role for IFN-γ in SARS-CoV-2 infection, as the ACE2 receptor is an ISG that is induced in primary human bronchial and colon epithelial cells upon IFN-γ treatment [80,97]. Clinical data are scattered, with some reports describing lower serum concentrations of IFN-γ in COVID-19 patients presenting with fibrosis than COVID-19 patients without fibrosis [98]. Other studies have shown that individuals who succumb to COVID-19 may have higher concentrations of circulating IFN-γ, suggesting that type II IFN may exacerbate disease, or might even be considered as a putative biomarker for severe disease [99]. This observation is supported by the recent elucidation of a combinatorial role for IFN-γ and TNF-α in the cytokine storm; indeed, increased PANoptosis and tissue inflammation and, ultimately, mortality in SARS-CoV-2-infected mice has been noted, and these outcomes have been rescued in Stat1 –/– or Ripk3 –/– Casp8 –/– mice lacking IFN or apoptosis/necroptosis signaling, respectively, suggesting a relevant role for type II IFN in this process [100]. Further rigorous studies will be necessary to fully determine the role of type II IFN in COVID-19, given the complicated network of cytokines activated during infection. In summary, limited data to date suggest that type II IFN can play a combinatorial role with type I IFN in limiting viral dissemination from mucosal sites, but similar to type I IFN, it can also mediate detrimental effects, including tissue damage during and after certain infections.
Type III IFNs restrict viral replication at mucosal sites
Due to the restricted expression of their receptor primarily to epithelial cells and limited immune cell types in mice and humans, type III IFNs exhibit more localized activity than type I IFNs at mucosal sites [15]. Thus, in mucosal infections, type III IFNs serve as a key means of controlling infection, especially in early stages, without inducing widespread inflammatory processes.
Type III IFNs are crucial for regulating intestinal MNoV replication. Mice lacking either Ifnlr1, globally (Ifnlr1 –/–) or in intestinal epithelial cells (Villin-Cre;Ifnlr1 f/f), or lacking the IFN-λ cytokines (Ifnl2 –/– Ifnl3 –/– ) exhibit increased fecal shedding and intestinal tissue titers of persistent MNoV relative to wild-type mice [30,101,102]. Further, IFN-λ treatment can prevent or cure MNoV infection in wild-type mice [30,102]. HNoV also induces and is restricted by type III IFNs in a human enteroid model, suggesting potential cross-genogroup importance of this cytokine in regulating NoV infection [72]. Of note, type III IFNs are also key regulators of more complex trans-kingdom interactions by which the gut microbiota regulate MNoV infection. Specifically, antibiotic-treated mice lacking gut bacteria are resistant to MNoV, but viral dependence on the microbiota is entirely eliminated in Ifnlr1 –/– mice, indicating that IFN-λ can be important in regulating these complex interactions [103]. Further, microbiota-mediated induction of type III IFN signaling in the proximal small intestine via bile acid regulation limits acute MNoV infection [47]. In the context of excessive type III IFN signaling, either driven by viral interference during chronic murine astrovirus infection [104] or dysregulated autophagy in Epg5 –/– mice [105], the animals are rendered resistant to MNoV infection. Therefore, these observations emphasize the relevance of type III IFNs in restricting enteric viral infections and highlight the concept that at mucosal surfaces, numerous environmental stimuli, including bacterial and viral elements of the microbiota, can converge to modulate these cytokines.
Type III IFNs were initially identified as key antiviral cytokines in the context of murine genital HSV-2 infection, with IFN-λ treatment blocking virus replication in the vaginal mucosa [29]. These findings have since been confirmed in vitro in simian Vero cells as well as primary human astrocytes and neurons, thus indicating that type III IFNs exhibit potent restriction of HSV replication in mucosal epithelial cells as well as in CNS cells [106,107]. Of interest, type III IFNs have been implicated in the maintenance of the blood–brain barrier during WNV and rabies virus infections in mice, with Ifnlr1 –/– mice exhibiting enhanced spread to the brain and IFN-λ treatment in them helping to maintain tight junction and barrier integrity [108,109]. Whether type III IFNs are protective against HSV encephalitis is not yet clear. In the female reproductive tract, IFN-λ also controls replication of Zika virus, as enhanced vaginal viral titers have been observed in Ifnlr1 –/–mice while IFN-λ treatment limits viral replication in Ifnlr1-sufficient mice; this in turn supports the concept that this cytokine is likely active against a variety of pathogens at this mucosal site [110].
Early during the COVID-19 pandemic, human and murine studies quickly sought to understand the importance of type III IFNs in controlling SARS-CoV-2 infection. In vitro studies in immortalized human Calu-3 and simian Vero lines as well as primary human airway and intestinal epithelial cells demonstrated robust IFN-λ-mediated antiviral activity, limiting SARS-CoV-2 replication [82,111,112]. Murine studies leveraging a mouse-adapted strain of SARS-CoV-2 demonstrated that exogenous treatment with type III IFNs could efficiently control lung viral replication and disease [113]. This finding was supported by the observation of accelerated SARS-CoV-2 clearance in patients with mild to moderate COVID-19 who were administered pegylated IFN-λ treatment (Box 2 ) [114]. Additionally, recent work reported that type III IFN-driven expression of ISGs in humans with COVID-19 was presumably protective in the upper airways of individuals with mild cases of the disease, while critical patients exhibited dominant type I IFN responses and weaker ISG responses [115]. However, as discussed earlier, type III IFNs may also coordinate with type I IFNs to cause certain detrimental effects during severe COVID-19 in human patients [28,86]. Patients with severe COVID-19 exhibit increased plasma concentrations of type III IFNs during the first week of symptoms which remain at high amounts in later phases of COVID-19 infection compared with patients with moderate disease [116]. Further, type III IFNs have been implicated as a potential part of the ‘cytokine storm’ associated with SARS-CoV-2-induced morbidity and mortality in human ACE2 transgenic mice [87,117,118]. The relative balance between protective and detrimental effects for type III IFNs during SARS-CoV-2 infection thus needs to be fully defined. Overall, despite the varying characteristics of the viral pathogens and mucosal sites in which these viruses routinely attack, individual IFN types exhibit similar antiviral effects, although in some cases, they may also exhibit detrimental inflammatory responses.
Box 2. Potential roles for IFNs in the treatment of emergent viral diseases.
IFNs have been used therapeutically for a variety of diseases of infectious and noninfectious etiology, such as hepatitis and some cancers. In the wake of the current COVID-19 pandemic, interest in IFN therapies has skyrocketed due to the need to mitigate severe effects of SARS-CoV-2. Much of the current research focus has been on type I IFN, which is used clinically for other diseases and restricts SARS-CoV-2 [148]. Indeed, preliminary phase 2 studies investigating the efficacy of type I IFN in preventing or ameliorating COVID-19 have been promising: data from randomized controlled trials support this type of therapy, including results with IFN-β, which resulted in a shorter length of positive infection or a shorter time to clinical improvement in COVID-19 patients, relative to therapy devoid of IFN-β (NCT04276688i, NCT04343768ii) [149,150]. An additional phase 2 randomized controlled study has demonstrated that inhaled, nebulized IFN-β1a contributes to an improvement in breathlessness for hospitalized patients (NCT04385095iii) [151]. IFN-γ has not been thoroughly assessed for its therapeutic effects against SARS-CoV-2 and may have limited utility for treatment.
Type III IFNs are equally promising in their potential therapeutic effects against SARS-CoV-2, as they exhibit more localized activity and have robust safety and tolerance profiles [152,153]. IFN-λ, coupled with type I IFN, upregulates ISGs in primary human epithelial cell culture models, reducing SARS-CoV-2 replication [112]. A randomized controlled phase 2 trial has shown that a single subcutaneous injection of pegylated IFN-λ accelerated the decrease in SARS-CoV-2 viral load in humans, with the effects being most apparent in patients with higher SARS-CoV-2 viral load (NCT04354259iv) [114]. However, an alternate randomized, single-blind, placebo-controlled phase 2 clinical trial found that while pegylated IFN-λ was well tolerated, a single dose had no effect on the duration of SARS-CoV-2 viral shedding or symptoms in patients with moderate COVID-19 (NCT04331899v) [154]. Because type I and III IFNs induce kinetically distinct ISG responses, with slower and more sustained responses induced by type III than type I IFNs, the application of type I and/or III IFNs as putative therapeutics for COVID-19 requires careful consideration of proper timing for administration [25,26,155].
Better characterization of ways to prevent the damaging effects of IFNs, while promoting their antiviral activities, can greatly expand the potential therapeutic use of these cytokines. Due to their broad antiviral activity, IFNs are a potential therapeutic for essentially any new or emerging viral epidemic, and further exploration of IFN administration to seriously ill patients may help mitigate the impact of future pandemics.
Alt-text: Box 2
Concluding remarks
IFNs have long been recognized for their key roles in restricting viral pathogenesis at the organismal level, but recent studies have revealed an increasingly nuanced set of interactions between IFNs. Here, we have focused on the emerging understanding that there appear to be highly specific roles for different IFNs in regulating local viral replication versus dissemination, with type III IFNs exhibiting a predominant role in initial barrier control at mucosal surfaces, while type I and II IFNs may play more dominant roles in limiting viral dissemination. Synergistic activities of the IFNs are increasingly appreciated, especially for type I and II IFNs, both in mediating protective and also potentially detrimental effects when excessively activated. While we have focused here on three specific viruses that infect at mucosal sites, the regulatory patterns observed might likely extend widely to viral pathogens invading these surfaces, although this remains to be investigated.
Moreover, numerous areas related to IFN biology are still unexplored (see Outstanding questions). As our understanding of the roles of IFNs in limiting infection has grown, research has begun to focus on the tissue- and cell-specific levels at which IFNs exert these effects and how these effects are mediated. Great strides have been made in this arena, but careful consideration should be taken to test not just how these IFNs act individually or in vitro, but more importantly, how they may act in combination in vivo; we argue that this will be key for understanding their bona fide roles in complex diseases. Recent findings such as those demonstrating the importance of cooperative roles of type I and III IFNs in maintaining the intestinal epithelial barrier in chemically induced colitis in mice, coupled to the revelation that type II IFNs restrict MNoV infection only when type I IFNs are absent, support this supposition [89,119]. These studies, as well as those leveraging cell-specific conditional disruption of IFN receptors [69,102], will also continue to provide insights into the compartmental specificity of IFN responses. Further investigation of the relative importance of different inducers, receptor expression, and other regulatory factors is also required to determine the specific effects of each type of IFN.
Outstanding questions.
What are the bona fide sets of genes induced in vivo by each type of IFN, and how do these vary depending on the cell and organ? A better understanding of the distinct effects of IFNs can aid in their safe and efficacious therapeutic use.
How do PRRs differentially regulate multiple types of IFNs? A nuanced understanding of why IFNs are regulated in distinct manners, spatially and temporally, can help clarify their protective versus detrimental effects.
How do IFNs interact with each other to synergistically or antagonistically control mucosal viral infections? How do context and timing of induction of individual IFNs contribute to these combinatorial effects on viruses? Cross-type IFN interactions are likely to extend the impact of immune deficiencies and/or therapies in the context of infection.
What are the mechanisms underlying the individual or combined antiviral activities of ISGs? How do viruses subvert ‘proviral’ IFN-stimulated genes for infection? A clearer definition of the specific ISGs that limit viral infections is likely to offer important insights into viral life cycles.
What is the breadth of mechanisms used by viruses to limit induction of or responses to IFNs, or to directly antagonize ISG activity? Defining how viruses antagonize IFN activity may help identify specific host factors that are particularly important for restricting a given virus.
How can therapies directed against IFNs be targeted to reduce the potential detrimental effects of the immune response? What are key ISGs that may serve as more specific therapeutic targets? Antiviral therapies targeted toward specific ISGs in lieu of IFNs themselves might potentially have fewer side effects.
Alt-text: Outstanding questions
Beyond the need for improved understanding of the factors contributing to IFN responses, our knowledge of the expression and specific antiviral effects of ISGs is quite limited [120]. Many reports delineating which ISGs are induced by individual IFNs rely on tissue culture models, which likely do not recapitulate the complexity of ISG induction in vivo. The recent increase in the use of organoids to interrogate the antiviral functions and mechanisms of IFNs may be key to providing more physiologically relevant insights into IFN and ISG biology [72,112]. Given that IFN-γ exhibits many cell-extrinsic effects, systems leveraging different combinations of primary cells – especially those combining nonimmune and immune cell types – may be essential for yielding insights into IFN-γ roles.
In addition to defining the specific regulons for IFNs and to which extent their profiles overlap, further insight into host–virus interactions should be achieved, and for this, it will be crucial to understand the mechanisms and activities of individual ISGs. As screens across species and cell types identifying ISGs and antiviral effectors continue to reveal broad or specific anti- or proviral effects of individual ISGs [3,121], extensive confirmatory experiments examining their specific roles and mechanisms are essential. Currently, IFN-directed therapies against viral infections typically rely on the administration of these IFNs themselves, which can come with limited therapeutic windows due to the induction of possible pathological side effects. As such, administration or induction of individual ISGs may be a more attractive target in the future, as these proteins exhibit more specific activities than IFNs themselves. Additionally, some ISGs have been recently reported to promote viral infection through various mechanisms, with IAV repurposing of ISGs LY6E and IFIT2 used to facilitate viral entry and viral mRNA translation, respectively, and a herpesvirus co-opting viperin to achieve enhanced infectivity [122., 123., 124.]. In addition, fully characterizing how counterintuitive proviral effects of IFNs might contribute to viral pathogenesis in specific scenarios might allow for a more holistic understanding of the many and varied interactions between viral pathogens and IFNs.
In summary, IFNs provide key protective roles for the vertebrate host during mucosal viral infections. However, recent work has also demonstrated the more complex detrimental effects that IFNs can have, as well as ways in which they synergize to respond to viral infections. Only by carefully characterizing the roles of each IFN alone and in conjunction, as well as by dissecting the mechanisms of action of ISGs, can we fully understand and potentially augment the host response in the context of disease.
Acknowledgments
Acknowledgments
We acknowledge all members of the Baldridge laboratory for helpful discussions, especially Elizabeth A. Kennedy. F.C.W. was supported by National Institutes of Health (NIH) grant T32 GM007067. M.T.B. was supported by NIH grants R01 AI127552, R01 AI139314, and R01 AI141478, the Pew Biomedical Scholars Program of the Pew Charitable Trusts, the Mathers Foundation, and the Burroughs Wellcome Fund. The funders had no role in decision to publish or preparation of the manuscript.
Author contributions
F.C.W., P.S., and M.T.B. wrote and edited the manuscript.
Declaration of interests
No interests are declared.
Glossary
- Autoantibodies
target host proteins, potentially causing detrimental effects.
- Autophagy
process by which unwanted intracellular components, such as pathogens or organelles, are degraded.
- Deubiquitinase
enzyme that removes post-translational modification ubiquitin from a protein, with potential effects, including altering degradation or activity of the target protein.
- Encephalitis
inflammation of the brain.
- Enteroid
type of organoid derived from intestinal stem cells, which contains many of the cell types found within the mature intestine.
- IFN regulatory factor
family of 10 transcription factors that are key interferon-stimulated genes; mediate the initial expression and amplification of interferons.
- Latency
state in which a viral genome is still present in a cell, but most or all viral genes are inactive.
- MAVS
mitochondrial antiviral signaling protein; activated by pattern recognition receptor sensing of pathogens and, in turn, activates transcription factors to initiate the interferon response.
- MDA5
melanoma differentiation-associated protein 5; cytosolic pattern recognition receptor recognizing long double-stranded RNA fragments.
- Mucosal
inner surface of tissues such as the gastrointestinal, respiratory, and reproductive tracts that are covered in mucus.
- Murine astrovirus
single-stranded, positive-sense RNA virus of the Astroviridae family; has a tropism that includes intestinal epithelial cells.
- Organoid
3D tissue culture model, derived from stem cells; used to broadly recapitulate the cell types and organization of the tissue from which they are derived.
- PANoptosis (pyroptosis, apoptosis, necroptosis)
interconnected cell death process that restricts a wide range of pathogens, including bacteria, viruses, and fungi.
- Pathogen-associated molecular patterns
pathogen-derived molecules, for example, nucleic acids or proteins; sensed by the host to activate the initial steps of the innate immune response.
- Pattern recognition receptors
host proteins such as Toll-like receptors or RIG-I; sense various PAMPs to induce the innate immune response, including interferons.
- Plasmacytoid dendritic cells
circulating myeloid cells specialized in sensing viral pathogens; produce large amounts of type I and III IFNs to combat infection.
- Positive-sense RNA virus
virus with single-stranded genome that can function as an mRNA and be directly translated.
- RIG-I
retinoic acid-inducible gene I; cytosolic pattern recognition receptor; recognizes short double-stranded RNA fragments.
- Th1
T helper cells; can secrete IFN-γ and induce cellular immunity.
- Tropism
range of hosts, tissues, or cells which a pathogen can infect.
Resources
ihttps://clinicaltrials.gov/ct2/show/NCT04276688iihttps://clinicaltrials.gov/ct2/show/NCT04343768iiihttps://clinicaltrials.gov/ct2/show/NCT04385095ivhttps://clinicaltrials.gov/ct2/show/NCT04354259vhttps://clinicaltrials.gov/ct2/show/NCT04331899References
- 1.Isaacs A., Lindenmann J. Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 1957;147:258–267. [PubMed] [Google Scholar]
- 2.Negishi H., et al. The interferon (IFN) class of cytokines and the IFN regulatory factor (IRF) transcription factor family. Cold Spring Harb. Perspect. Biol. 2018;10 doi: 10.1101/cshperspect.a028423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schoggins J.W., et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011;472:481–485. doi: 10.1038/nature09907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McNab F., et al. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015;15:87–103. doi: 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li S.-F., et al. Type I interferons: distinct biological activities and current applications for viral infection. Cell. Physiol. Biochem. 2018;51:2377–2396. doi: 10.1159/000495897. [DOI] [PubMed] [Google Scholar]
- 6.Cella M., et al. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat. Med. 1999;5:919–923. doi: 10.1038/11360. [DOI] [PubMed] [Google Scholar]
- 7.Ali S., et al. Sources of type I interferons in infectious immunity: plasmacytoid dendritic cells not always in the driver's seat. Front. Immunol. 2019;10:778. doi: 10.3389/fimmu.2019.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stanifer M.L., et al. Differential regulation of type I and type III interferon signaling. Int. J. Mol. Sci. 2019;20:1445. doi: 10.3390/ijms20061445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee A.J., Ashkar A.A. The dual nature of type I and type II interferons. Front. Immunol. 2018;9:2061. doi: 10.3389/fimmu.2018.02061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wheelock E.F. Interferon-like virus-inhibitor induced in human leukocytes by phytohemagglutinin. Science. 1965;149:310–311. [PubMed] [Google Scholar]
- 11.Fenimore J., Howard A.Y. Regulation of IFN-gamma expression. Adv. Exp. Med. Biol. 2016;941:1–19. doi: 10.1007/978-94-024-0921-5_1. [DOI] [PubMed] [Google Scholar]
- 12.Glasgow L.A. Leukocytes and interferon in the host response to viral infections. II. Enhanced interferon response of leukocytes from immune animals. J. Bacteriol. 1966;91:2185–2191. doi: 10.1128/jb.91.6.2185-2191.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zanoni I., et al. IL-15 cis presentation is required for optimal NK cell activation in lipopolysaccharide-mediated inflammatory conditions. Cell Rep. 2013;4:1235–1249. doi: 10.1016/j.celrep.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 14.Green D.S., et al. Current prospects of type II interferon gamma signaling and autoimmunity. J. Biol. Chem. 2017;292:13925–13933. doi: 10.1074/jbc.R116.774745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wells A.I., Coyne C.B. Type III interferons in antiviral defenses at barrier surfaces. Trends Immunol. 2018;39:848–858. doi: 10.1016/j.it.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou J.-H., et al. Type III interferons in viral infection and antiviral immunity. Cell. Physiol. Biochem. 2018;51:173–185. doi: 10.1159/000495172. [DOI] [PubMed] [Google Scholar]
- 17.Broggi A., et al. IFN-lambda suppresses intestinal inflammation by non-translational regulation of neutrophil function. Nat. Immunol. 2017;18:1084–1093. doi: 10.1038/ni.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen N., et al. RNA sensors of the innate immune system and their detection of pathogens. IUBMB Life. 2017;69:297–304. doi: 10.1002/iub.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Broquet A.H., et al. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J. Immunol. 2011;186:1618–1626. doi: 10.4049/jimmunol.1002862. [DOI] [PubMed] [Google Scholar]
- 20.McCartney S.A., et al. MDA-5 recognition of a murine norovirus. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiu Y.-H., et al. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boehme K.W., et al. Human cytomegalovirus envelope glycoproteins B and H are necessary for TLR2 activation in permissive cells. J. Immunol. 2006;177:7094–7102. doi: 10.4049/jimmunol.177.10.7094. [DOI] [PubMed] [Google Scholar]
- 23.Odendall C., et al. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat. Immunol. 2014;15:717–726. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yanai H., et al. Role of IFN regulatory factor 5 transcription factor in antiviral immunity and tumor suppression. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3402–3407. doi: 10.1073/pnas.0611559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolen C.R., et al. Dynamic expression profiling of type I and type III interferon-stimulated hepatocytes reveals a stable hierarchy of gene expression. Hepatology. 2014;59:1262–1272. doi: 10.1002/hep.26657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pervolaraki K., et al. Differential induction of interferon stimulated genes between type I and type III interferons is independent of interferon receptor abundance. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dickensheets H., et al. Interferon-lambda (IFN-lambda) induces signal transduction and gene expression in human hepatocytes, but not in lymphocytes or monocytes. J. Leukoc. Biol. 2013;93:377–385. doi: 10.1189/jlb.0812395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Major J., et al. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science. 2020;369:712–717. doi: 10.1126/science.abc2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ank N., et al. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J. Virol. 2006;80:4501–4509. doi: 10.1128/JVI.80.9.4501-4509.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nice T.J., et al. Interferon-lambda cures persistent murine norovirus infection in the absence of adaptive immunity. Science. 2015;347:269–273. doi: 10.1126/science.1258100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karst S.M., et al. STAT1-dependent innate immunity to a Norwalk-like virus. Science. 2003;299:1575–1578. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- 32.Iversen M.B., et al. An innate antiviral pathway acting before interferons at epithelial surfaces. Nat. Immunol. 2016;17:150–158. doi: 10.1038/ni.3319. [DOI] [PubMed] [Google Scholar]
- 33.Hoyos-Bachiloglu R., et al. A digenic human immunodeficiency characterized by IFNAR1 and IFNGR2 mutations. J. Clin. Invest. 2017;127:4415–4420. doi: 10.1172/JCI93486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bastard P., et al. Herpes simplex encephalitis in a patient with a distinctive form of inherited IFNAR1 deficiency. J. Clin. Invest. 2021;131 doi: 10.1172/JCI139980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Q., et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bastard P., et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370 doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bastard P., et al. Autoantibodies neutralizing type I IFNs are present in ~4% of uninfected individuals over 70 years old and account for ~20% of COVID-19 deaths. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abl4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Credle J.J., et al. Neutralizing IFNL3 autoantibodies in severe COVID-19 identified using molecular indexing of proteins by self-assembly. bioRxiv. 2021 doi: 10.1101/2021.03.02.432977. Published online March 3, 2021. [DOI] [Google Scholar]
- 39.Patel M.M., et al. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg. Infect. Dis. 2008;14:1224–1231. doi: 10.3201/eid1408.071114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strong D.W., et al. Protruding domain of capsid protein is necessary and sufficient to determine murine norovirus replication and pathogenesis in vivo. J. Virol. 2012;86:2950–2958. doi: 10.1128/JVI.07038-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nice T.J., et al. A single-amino-acid change in murine norovirus NS1/2 is sufficient for colonic tropism and persistence. J. Virol. 2013;87:327–334. doi: 10.1128/JVI.01864-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thackray L.B., et al. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J. Virol. 2007;81:10460–10473. doi: 10.1128/JVI.00783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ettayebi K., et al. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Green K.Y., et al. Human norovirus targets enteroendocrine epithelial cells in the small intestine. Nat. Commun. 2020;11:2759. doi: 10.1038/s41467-020-16491-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilen C.B., et al. Tropism for tuft cells determines immune promotion of norovirus pathogenesis. Science. 2018;360:204–208. doi: 10.1126/science.aar3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grau K.R., et al. The major targets of acute norovirus infection are immune cells in the gut-associated lymphoid tissue. Nat. Microbiol. 2017;2:1586–1591. doi: 10.1038/s41564-017-0057-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grau K.R., et al. The intestinal regionalization of acute norovirus infection is regulated by the microbiota via bile acid-mediated priming of type III interferon. Nat. Microbiol. 2020;5:84–92. doi: 10.1038/s41564-019-0602-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karandikar U.C., et al. Detection of human norovirus in intestinal biopsies from immunocompromised transplant patients. J. Gen. Virol. 2016;97:2291–2300. doi: 10.1099/jgv.0.000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jones M.K., et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science. 2014;346:755–759. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee S., et al. Norovirus cell tropism is determined by combinatorial action of a viral non-structural protein and host cytokine. Cell Host Microbe. 2017;22:449–459.e4. doi: 10.1016/j.chom.2017.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee S., et al. A secreted viral nonstructural protein determines intestinal norovirus pathogenesis. Cell Host Microbe. 2019;25:845–857.e5. doi: 10.1016/j.chom.2019.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walker F.C., et al. Norovirus evolution in immunodeficient mice reveals potentiated pathogenicity via a single nucleotide change in the viral capsid. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gonzalez-Perez A.C., et al. One step ahead: herpesviruses light the way to understanding interferon-stimulated genes (ISGs) Front. Microbiol. 2020;11:124. doi: 10.3389/fmicb.2020.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McQuillan G., et al. Prevalence of herpes simplex virus type 1 and type 2 in persons aged 14-49: United States, 2015-2016. NCHS Data Brief. 2018:1–8. [PubMed] [Google Scholar]
- 55.Looker K.J., et al. Global and regional estimates of prevalent and incident herpes simplex virus type 1 infections in 2012. PLoS One. 2015;10 doi: 10.1371/journal.pone.0140765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cohen J.I. Herpesvirus latency. J. Clin. Invest. 2020;130:3361–3369. doi: 10.1172/JCI136225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bradshaw M.J., Venkatesan A. Herpes simplex virus-1 encephalitis in adults: pathophysiology, diagnosis, and management. Neurotherapeutics. 2016;13:493–508. doi: 10.1007/s13311-016-0433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu H., Zheng C. The race between host antiviral innate immunity and the immune evasion strategies of herpes simplex virus 1. Microbiol. Mol. Biol. Rev. 2020;84 doi: 10.1128/MMBR.00099-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lee A.G., et al. T cell response kinetics determines neuroinfection outcomes during murine HSV infection. JCI Insight. 2020;5 doi: 10.1172/jci.insight.134258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lebratti T., et al. A sustained type I IFN-neutrophil-IL-18 axis drives pathology during mucosal viral infection. eLife. 2021;10 doi: 10.7554/eLife.65762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.V'Kovski P., et al. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hossain M.G., et al. SARS-CoV-2 host diversity: an update of natural infections and experimental evidence. J. Microbiol. Immunol. Infect. 2021;54:175–181. doi: 10.1016/j.jmii.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu B., et al. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zang R., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arimori Y., et al. Type I interferon limits influenza virus-induced acute lung injury by regulation of excessive inflammation in mice. Antivir. Res. 2013;99:230–237. doi: 10.1016/j.antiviral.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 66.Samuel M.A., Diamond M.S. Alpha/beta interferon protects against lethal West Nile virus infection by restricting cellular tropism and enhancing neuronal survival. J. Virol. 2005;79:13350–13361. doi: 10.1128/JVI.79.21.13350-13361.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dhondt K.P., et al. Type I interferon signaling protects mice from lethal henipavirus infection. J. Infect. Dis. 2013;207:142–151. doi: 10.1093/infdis/jis653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.MacDuff D.A., et al. HOIL1 is essential for the induction of type I and III interferons by MDA5 and regulates persistent murine norovirus infection. J. Virol. 2018;92 doi: 10.1128/JVI.01368-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nice T.J., et al. Type I interferon receptor deficiency in dendritic cells facilitates systemic murine norovirus persistence despite enhanced adaptive immunity. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thackray L.B., et al. Critical role for interferon regulatory factor 3 (IRF-3) and IRF-7 in type I interferon-mediated control of murine norovirus replication. J. Virol. 2012;86:13515–13523. doi: 10.1128/JVI.01824-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jones M.K., et al. Human norovirus culture in B cells. Nat. Protoc. 2015;10:1939–1947. doi: 10.1038/nprot.2015.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin S.-C., et al. Human norovirus exhibits strain-specific sensitivity to host interferon pathways in human intestinal enteroids. Proc. Natl. Acad. Sci. U. S. A. 2020;117:23782–23793. doi: 10.1073/pnas.2010834117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Conrady C.D., et al. Loss of the type I interferon pathway increases vulnerability of mice to genital herpes simplex virus 2 infection. J. Virol. 2011;85:1625–1633. doi: 10.1128/JVI.01715-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Swiecki M., et al. Plasmacytoid dendritic cells contribute to systemic but not local antiviral responses to HSV infections. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Andersen L.L., et al. Functional IRF3 deficiency in a patient with herpes simplex encephalitis. J. Exp. Med. 2015;212:1371–1379. doi: 10.1084/jem.20142274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jouanguy E., et al. Human primary immunodeficiencies of type I interferons. Biochimie. 2007;89:878–883. doi: 10.1016/j.biochi.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 77.Nickodem C., et al. Interferon epsilon in the reproductive tract of healthy and genital herpes simplex virus-infected pregnant women: results of a pilot study. Am. J. Reprod. Immunol. 2018;80 doi: 10.1111/aji.12995. [DOI] [PubMed] [Google Scholar]
- 78.Fung K.Y., et al. Interferon-epsilon protects the female reproductive tract from viral and bacterial infection. Science. 2013;339:1088–1092. doi: 10.1126/science.1233321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li Y., et al. Interferon kappa is important for keratinocyte host defense against herpes simplex virus-1. J. Immunol. Res. 2020;2020:5084682. doi: 10.1155/2020/5084682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Busnadiego I., et al. Antiviral activity of type I, II, and III interferons counterbalances ACE2 inducibility and restricts SARS-CoV-2. mBio. 2020;11 doi: 10.1128/mBio.01928-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lokugamage K.G., et al. Type I interferon susceptibility distinguishes SARS-CoV-2 from SARS-CoV. J. Virol. 2020;94 doi: 10.1128/JVI.01410-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stanifer M.L., et al. Critical role of type III interferon in controlling SARS-CoV-2 infection in human intestinal epithelial cells. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Israelow B., et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J. Exp. Med. 2020;217 doi: 10.1084/jem.20201241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boudewijns R., et al. STAT2 signaling restricts viral dissemination but drives severe pneumonia in SARS-CoV-2 infected hamsters. Nat. Commun. 2020;11:5838. doi: 10.1038/s41467-020-19684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Galani I.-E., et al. Untuned antiviral immunity in COVID-19 revealed by temporal type I/III interferon patterns and flu comparison. Nat. Immunol. 2021;22:32–40. doi: 10.1038/s41590-020-00840-x. [DOI] [PubMed] [Google Scholar]
- 86.Broggi A., et al. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. 2020;369:706–712. doi: 10.1126/science.abc3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Karki R., Kanneganti T.-D. The ‘cytokine storm’: molecular mechanisms and therapeutic prospects. Trends Immunol. 2021;42:681–705. doi: 10.1016/j.it.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kang S., et al. Direct antiviral mechanisms of interferon-gamma. Immune Netw. 2018;18 doi: 10.4110/in.2018.18.e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maloney N.S., et al. Essential cell-autonomous role for interferon (IFN) regulatory factor 1 in IFN-gamma-mediated inhibition of norovirus replication in macrophages. J. Virol. 2012;86:12655–12664. doi: 10.1128/JVI.01564-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hwang S., et al. Nondegradative role of Atg5-Atg12/ Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe. 2012;11:397–409. doi: 10.1016/j.chom.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Biering S.B., et al. Viral replication complexes are targeted by LC3-guided interferon-inducible GTPases. Cell Host Microbe. 2017;22:74–85.e7. doi: 10.1016/j.chom.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu T., et al. Gamma interferon can prevent herpes simplex virus type 1 reactivation from latency in sensory neurons. J. Virol. 2001;75:11178–11184. doi: 10.1128/JVI.75.22.11178-11184.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mikloska Z., Cunningham A.L. Alpha and gamma interferons inhibit herpes simplex virus type 1 infection and spread in epidermal cells after axonal transmission. J. Virol. 2001;75:11821–11826. doi: 10.1128/JVI.75.23.11821-11826.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pierce A.T., et al. Beta interferon and gamma interferon synergize to block viral DNA and virion synthesis in herpes simplex virus-infected cells. J. Gen. Virol. 2005;86:2421–2432. doi: 10.1099/vir.0.80979-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Song R., et al. Two modes of the axonal interferon response limit alphaherpesvirus neuroinvasion. mBio. 2016;7 doi: 10.1128/mBio.02145-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Decman V., et al. Gamma interferon can block herpes simplex virus type 1 reactivation from latency, even in the presence of late gene expression. J. Virol. 2005;79:10339–10347. doi: 10.1128/JVI.79.16.10339-10347.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Heuberger J., et al. Epithelial response to IFN-gamma promotes SARS-CoV-2 infection. EMBO Mol. Med. 2021;13 doi: 10.15252/emmm.202013191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hu Z.-J., et al. Lower circulating interferon-gamma is a risk factor for lung fibrosis in COVID-19 patients. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.585647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gadotti A.C., et al. IFN-gamma is an independent risk factor associated with mortality in patients with moderate and severe COVID-19 infection. Virus Res. 2020;289 doi: 10.1016/j.virusres.2020.198171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Karki R., et al. Synergism of TNF-alpha and IFN-gamma triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell. 2021;184:149–168.e17. doi: 10.1016/j.cell.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Peterson S.T., et al. Disruption of type III interferon genes Ifnl2 and Ifnl3 recapitulates loss of the type III IFN receptor in the mucosal antiviral response. J. Virol. 2019;93 doi: 10.1128/JVI.01073-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Baldridge M.T., et al. Expression of Ifnlr1 on intestinal epithelial cells is critical to the antiviral effects of IFN-lambda against norovirus and reovirus. J. Virol. 2017;91 doi: 10.1128/JVI.02079-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Baldridge M.T., et al. Commensal microbes and interferon-lambda determine persistence of enteric murine norovirus infection. Science. 2015;347:266–269. doi: 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ingle H., et al. Viral complementation of immunodeficiency confers protection against enteric pathogens via interferon-lambda. Nat. Microbiol. 2019;4:1120–1128. doi: 10.1038/s41564-019-0416-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee S., et al. Intestinal antiviral signaling is controlled by autophagy gene Epg5 independent of the microbiota. Autophagy. 2021 doi: 10.1080/15548627.2021.1968607. Published online September 14, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lopušná K., et al. Interferon lambda induces antiviral response to herpes simplex virus 1 infection. Acta Virol. 2014;58:325–332. doi: 10.4149/av_2014_03_325. [DOI] [PubMed] [Google Scholar]
- 107.Li J., et al. Interferon lambda inhibits herpes simplex virus type I infection of human astrocytes and neurons. Glia. 2011;59:58–67. doi: 10.1002/glia.21076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lazear H.M., et al. Interferon-lambda restricts West Nile virus neuroinvasion by tightening the blood-brain barrier. Sci. Transl. Med. 2015;7 doi: 10.1126/scitranslmed.aaa4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Li Y., et al. Interferon-lambda attenuates rabies virus infection by inducing interferon-stimulated genes and alleviating neurological inflammation. Viruses. 2020;12:405. doi: 10.3390/v12040405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Caine E.A., et al. Interferon lambda protects the female reproductive tract against Zika virus infection. Nat. Commun. 2019;10:280. doi: 10.1038/s41467-018-07993-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Felgenhauer U., et al. Inhibition of SARS-CoV-2 by type I and type III interferons. J. Biol. Chem. 2020;295:13958–13964. doi: 10.1074/jbc.AC120.013788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vanderheiden A., et al. Type I and type III interferons restrict SARS-CoV-2 infection of human airway epithelial cultures. J. Virol. 2020;94 doi: 10.1128/JVI.00985-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dinnon K.H., 3rd, et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature. 2020;586:560–566. doi: 10.1038/s41586-020-2708-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Feld J.J., et al. Peginterferon lambda for the treatment of outpatients with COVID-19: a phase 2, placebo-controlled randomised trial. Lancet Respir. Med. 2021;9:498–510. doi: 10.1016/S2213-2600(20)30566-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sposito B., et al. The interferon landscape along the respiratory tract impacts the severity of COVID-19. Cell. 2021;184:4953–4968.e16. doi: 10.1016/j.cell.2021.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lucas C., et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Oladunni F.S., et al. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat. Commun. 2020;11:6122. doi: 10.1038/s41467-020-19891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Winkler E.S., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.McElrath C., et al. Critical role of interferons in gastrointestinal injury repair. Nat. Commun. 2021;12:2624. doi: 10.1038/s41467-021-22928-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schoggins J.W. Interferon-stimulated genes: what do they all do? Annu. Rev. Virol. 2019;6:567–584. doi: 10.1146/annurev-virology-092818-015756. [DOI] [PubMed] [Google Scholar]
- 121.Schoggins J.W., et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature. 2014;505:691–695. doi: 10.1038/nature12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tran V., et al. Influenza virus repurposes the antiviral protein IFIT2 to promote translation of viral mRNAs. Nat. Microbiol. 2020;5:1490–1503. doi: 10.1038/s41564-020-0778-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Seo J.-Y., et al. Human cytomegalovirus directly induces the antiviral protein viperin to enhance infectivity. Science. 2011;332:1093–1097. doi: 10.1126/science.1202007. [DOI] [PubMed] [Google Scholar]
- 124.Mar K.B., et al. LY6E mediates an evolutionarily conserved enhancement of virus infection by targeting a late entry step. Nat. Commun. 2018;9:3603. doi: 10.1038/s41467-018-06000-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Platanias L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 126.Stifter S.A., et al. Visualizing the selectivity and dynamics of interferon signaling in vivo. Cell Rep. 2019;29:3539–3550.e4. doi: 10.1016/j.celrep.2019.11.021. [DOI] [PubMed] [Google Scholar]
- 127.Valente G., et al. Distribution of interferon-gamma receptor in human tissues. Eur. J. Immunol. 1992;22:2403–2412. doi: 10.1002/eji.1830220933. [DOI] [PubMed] [Google Scholar]
- 128.Bartlett N.W., et al. Murine interferon lambdas (type III interferons) exhibit potent antiviral activity in vivo in a poxvirus infection model. J. Gen. Virol. 2005;86:1589–1596. doi: 10.1099/vir.0.80904-0. [DOI] [PubMed] [Google Scholar]
- 129.Lasfar A., et al. Characterization of the mouse IFN-lambda ligand-receptor system: IFN-lambdas exhibit antitumor activity against B16 melanoma. Cancer Res. 2006;66:4468–4477. doi: 10.1158/0008-5472.CAN-05-3653. [DOI] [PubMed] [Google Scholar]
- 130.Donnelly R.P., Kotenko S.V. Interferon-lambda: a new addition to an old family. J. Interf. Cytokine Res. 2010;30:555–564. doi: 10.1089/jir.2010.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Prokunina-Olsson L., et al. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 2013;45:164–171. doi: 10.1038/ng.2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.LaFleur D.W., et al. Interferon-kappa, a novel type I interferon expressed in human keratinocytes. J. Biol. Chem. 2001;276:39765–39771. doi: 10.1074/jbc.M102502200. [DOI] [PubMed] [Google Scholar]
- 133.Ingle H., et al. Murine astrovirus tropism for goblet cells and enterocytes facilitates an IFN-lambda response in vivo and in enteroid cultures. Mucosal Immunol. 2021;14:751–761. doi: 10.1038/s41385-021-00387-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Galani I.E., et al. Interferon-lambda mediates non-redundant front-line antiviral protection against influenza virus infection without compromising host fitness. Immunity. 2017;46:875–890.e6. doi: 10.1016/j.immuni.2017.04.025. [DOI] [PubMed] [Google Scholar]
- 135.Yin Z., et al. Type III IFNs are produced by and stimulate human plasmacytoid dendritic cells. J. Immunol. 2012;189:2735–2745. doi: 10.4049/jimmunol.1102038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.McFadden N., et al. Norovirus regulation of the innate immune response and apoptosis occurs via the product of the alternative open reading frame 4. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Melroe G.T., et al. Herpes simplex virus 1 has multiple mechanisms for blocking virus-induced interferon production. J. Virol. 2004;78:8411–8420. doi: 10.1128/JVI.78.16.8411-8420.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhang D., et al. Herpes simplex virus 1 serine protease VP24 blocks the DNA-sensing signal pathway by abrogating activation of interferon regulatory factor 3. J. Virol. 2016;90:5824–5829. doi: 10.1128/JVI.00186-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wang S., et al. Herpes simplex virus 1 ubiquitin-specific protease UL36 inhibits beta interferon production by deubiquitinating TRAF3. J. Virol. 2013;87:11851–11860. doi: 10.1128/JVI.01211-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang L., et al. HSV-1-encoded ICP0 degrades the host deubiquitinase BRCC36 to antagonize interferon antiviral response. Mol. Immunol. 2021;135:28–35. doi: 10.1016/j.molimm.2021.03.027. [DOI] [PubMed] [Google Scholar]
- 141.Eisemann J., et al. Infection of mature dendritic cells with herpes simplex virus type 1 interferes with the interferon signaling pathway. Immunobiology. 2007;212:877–886. doi: 10.1016/j.imbio.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 142.Yuan H., et al. Herpes simplex virus 1 UL36USP antagonizes type I interferon-mediated antiviral innate immunity. J. Virol. 2018;92 doi: 10.1128/JVI.01161-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hsu J.C.-C., et al. Translational shutdown and evasion of the innate immune response by SARS-CoV-2 NSP14 protein. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2101161118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Finkel Y., et al. SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis. Nature. 2021;594:240–245. doi: 10.1038/s41586-021-03610-3. [DOI] [PubMed] [Google Scholar]
- 145.Thoms M., et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369:1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yu P., et al. Guanylate-binding protein 2 orchestrates innate immune responses against murine norovirus and is antagonized by the viral protein NS7. J. Biol. Chem. 2020;295:8036–8047. doi: 10.1074/jbc.RA120.013544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Martin-Sancho L., et al. Functional landscape of SARS-CoV-2 cellular restriction. Mol. Cell. 2021;81:2656–2668.e8. doi: 10.1016/j.molcel.2021.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.King C., Sprent J. Dual nature of type I interferons in SARS-CoV-2-induced inflammation. Trends Immunol. 2021;42:312–322. doi: 10.1016/j.it.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hung I.F.-N., et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395:1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Alavi Darazam I., et al. Role of interferon therapy in severe COVID-19: the COVIFERON randomized controlled trial. Sci. Rep. 2021;11:8059. doi: 10.1038/s41598-021-86859-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Monk P.D., et al. Safety and efficacy of inhaled nebulised interferon beta-1a (SNG001) for treatment of SARS-CoV-2 infection: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Respir. Med. 2021;9:196–206. doi: 10.1016/S2213-2600(20)30511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Muir A.J., et al. Phase 1b study of pegylated interferon lambda 1 with or without ribavirin in patients with chronic genotype 1 hepatitis C virus infection. Hepatology. 2010;52:822–832. doi: 10.1002/hep.23743. [DOI] [PubMed] [Google Scholar]
- 153.Phillips S., et al. Peg-interferon lambda treatment induces robust innate and adaptive immunity in chronic hepatitis B patients. Front. Immunol. 2017;8:621. doi: 10.3389/fimmu.2017.00621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Jagannathan P., et al. Peginterferon Lambda-1a for treatment of outpatients with uncomplicated COVID-19: a randomized placebo-controlled trial. Nat. Commun. 2021;12:1967. doi: 10.1038/s41467-021-22177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Marcello T., et al. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131:1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]