Abstract
Oral squamous cell carcinoma (OSCC) occurs as a malignancy of the oral cavity. RANK ligand (RANKL) is essential for osteoclast formation/bone resorption. Recently, we showed autoregulation of receptor activator of nuclear factor-κB ligand (RANKL) stimulates OSCC cell proliferation. OSCC cells show resistance to tumor necrosis factor related apoptosis inducing ligand (TRAIL) treatment. Therefore, we hypothesize that RANKL promotes resistance for TRAIL induction of OSCC apoptotic cell death. In this study, SCC14A and SCC74A cells cultured with TRAIL revealed high-level expression of RANKL which increased resistance to TRAIL inhibition of tumor cell proliferation. RANKL stimulation inhibited terminal deoxynucleotidyl transferase dUTP nick end labeling positive staining in TRAIL-treated cells. CRISPR/Cas-9 knockout of RANKL (RANKL-KO) increased caspase-9, caspase-3 activity and cytochrome c release in OSCC cells. RANKL inhibited proapoptotic proteins BAD and BAX expression. TRAIL treatment suppressed the SQSTM1/p62 and RANKL restored the expression. Interestingly, RANKL alone significantly increased proteasome activity. RANKL-KO in OSCC cells inhibited autophagic activity as evidenced by decreased light chain 3B-II and beclin-1 expression. Thus, RANKL stimulation of OSCC tumor cells triggered resistance for TRAIL-induced OSCC cell death. Taken together, blockade of RANKL may inhibit OSCC tumor progression and enhance the potential of TRAIL induced OSCC tumor cell apoptosis.
Keywords: apoptosis, autophagy, bone, oral squamous cell carcinoma, RANKL, TRAIL
1 |. INTRODUCTION
Oral cancer incidence is approximately 50,000 cases annually causing about 10,000 deaths in a year (Cronin et al., 2018). Oral squamous cell carcinoma (OSCC) commonly occurs in the oral cavity, oropharynx, mucoginigval tissues or tongue and accounts for the majority of oral malignancies (Attar et al., 2010). OSCC etiology includes genetic predisposition, exposure to carcinogens such as tobacco, papilloma virus and chronic inflammation (Choi & Myers, 2008). In the US, oral cancer incidence has increased by 0.7% each year over the last 10 years (Cronin et al., 2018). OSCC has been shown to invade into the alveolar bone, causing metastasis (Jimi et al., 2011). Stromal pre-osteoblastic cells and several cancer cells secrete receptor activator of nuclear factor-κB ligand (RANKL) (Renema, Navet, Heymann, Lezot, & Heymann, 2016). RANKL binds to RANK surface receptor expressed in pre-osteoclast cells, which is necessary for osteoclast formation/bone resorption (Boyle, Simonet, & Lacey, 2003). In addition to functioning as a critical osteoclastogenic cytokine, RANKL plays an important role in cancer cell migration (Jones et al., 2006). Autonomous to RANKL derived from stromal pre-osteoblastic cells lining the bone, RANKL expressed in cancer cells contributes to bone metastasis (Zhang et al., 2001). Tumor cell expression of RANKL enhances the local osteoclast activity, which mediates the osteolysis and bone invasion (Dougall, Holen, & Gonzalez Suarez, 2014).
Earlier, we established an in vivo model to study invasion of bone, which involves subcutaneous injection of OSCC cells over calvaria in NCr-nu/nu athymic nude mice (Pandruvada et al., 2010). Expression of RANKL in OSCC cells has been shown to play a vital role in tumor osteolysis (Sambandam et al., 2013; Yuvaraj et al., 2009). Most recently, we demonstrated that RANKL expression is autoregulated in OSCC cells, which promotes tumor cell proliferation and osteoclast formation (Sambandam et al., 2018).
TRAIL (tumor necrosis factor related apoptosis-inducing ligand) mediates cell death in several cancer cells, but not in normal cell types (Wiley et al., 1995). Normal cells show low levels of TRAIL expression. Further, TRAIL-deficient mice have shown no change in bone phenotype (Sedger et al., 2002). TRAIL modulates osteoclast activity in physiologic and pathologic conditions (Brunetti et al., 2011), which is mediated through the TRAF6 signaling pathway (Yen, Hsu, Liao, Lee, & Tsai, 2012). Tumor osteolysis associated with multiple myeloma cells is correlated with high levels of TRAIL expression (Kawano et al., 2012). Interestingly, OSCC is relatively resistant for TRAIL induction of apoptosis (Younes & Kadin, 2003). However, underlying mechanisms for regulation of apoptosis in OSCC cells is unclear. Therefore, we investigated the hypothesis that RANKL promotes resistance to TRAIL-induced apoptotic cell death in OSCC cells.
2 |. MATERIALS AND METHODS
2.1 |. Culture of OSCC cells
Human SCC14A, SCC74A, and SCC14A-KO cells derived from OSCC tumors (Brenner et al., 2010) were cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum and 100 units/ml penicillin and streptomycin at 37°C and 5% CO2. mRANKL and hTRAIL recombinant proteins were obtained from the R&D System Inc., MN.
2.2 |. OSCC cell growth assay
SCC14A and SCC74A cells were seeded in a 96-well plate (1 × 104) at 37°C with TRAIL (100 ng/ml), RANKL (80 ng/ml) and in combination for 24 hr. The cells were supplemented with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reagent (5 mg/ml) and incubated for 4 hr. The formazan crystals formed were dissolved in dimethyl sulfoxide and absorbance was monitored at 595 nm using a microplate reader (PerkinElmer, Waltham, MA) (Ethiraj, Veerappan, Samuel, & Sivapatham, 2016).
2.3 |. Generation of stable OSCC cells with CRISPR/Cas-9 knockout of RANKL expression
SCC14A cells were transfected with CRISPR/Cas-9 double nickase construct (Santa Cruz Biotech, Inc., Dallas, TX) to knockout (KO) RANKL expression. hRANKL plasmid pair contained D10A mutated Cas-9 with a specific guide RNA (20 nucleotides). The plasmids contained the puromycin gene for resistance and green fluorescent protein (GFP) to validate the efficiency of transfection. Lipofectamine reagent (Invitrogen, Carlsbad, CA) was used for transfection, and after 24 hr transfection efficiency (>80%) was confirmed by fluorescence microscopy for GFP expression. Stable cell line (SCC14A-KO) was established by selection for puromycin (5 mg/ml) resistance (Santa Cruz Biotech., Inc., Dallas, TX).
2.4 |. Immunoblotting
OSCC cells (5 × 105/well) were seeded in six-well plates in the presence of TRAIL (100 ng/ml), RANKL (80 ng/ml), and in combination. Cell lysates were obtained using radioimmunoprecipitation assay buffer containing protease inhibitors. The samples were assayed for protein levels using Bradford reagent (Bio-Rad, Hercules, CA). Protein samples (100 μg) were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis using 4–15% gels and transferred onto polyvinylidene difluoride membrane. Western blot analysis was done with anti-RANKL, anti-caspase-9 and -3, anti-cytochrome c, anti-BAX, anti-BAD, anti-SQSTM1/p62, anti-beclin-1 (BECN1), anti-PCNA (Santa Cruz Biotech., Dallas, TX), anti-light chain 3B (LC3B) (Cell Signaling Tech., Danvers, MA), and anti-β-actin (Sigma Aldrich, St. Louis, MO) antibodies. Enhanced chemiluminescence substrate was used to identify the bands and quantified by the NIH ImageJ densitometry program (Ethiraj, Veerappan, Doraisami, & Sivapatham, 2014).
2.5 |. Real-time reverse-transcription polymerase chain reaction
SCC14A and SCC74A cells seeded in 6-well plate (5 × 105/well) were cultured with TRAIL (100 ng/ml) for 24 hr. Messenger RNA (mRNA) expression in OSCC cells were determined by quantitative polymerase chain reaction (Yuvaraj et al., 2009) performed with IQTM SYBR Green in an iCycler (Bio-Rad, Hercules, CA) and specific primers to hRANKL 5′-ACCAGCATCAAAATCCCAAG-3′ (sense), 5′-TAAGGAGTTGGAGACCT-3′ (antisense); hDR4 5′-GTTTGCTTGCCTCCCATGTA-3′ (sense), 5′-GGAAAGTTCCTGGTTTGCAC-3′ (antisense); hDR5 5′-TGCATCTCCTGCAAATATGG-3′ (sense), 5′-GTGGTGCAGGGACTTAGCTC-3′ (antisense); hDcR1 5′-AGTGGGGAAGTCCAAGTCAG-3′ (sense), 5′-CTTCAGCAGCTGGGGTTTC-3′ (antisense); hDcR2 5′-ATTTGCCTTCTTGCCTGCTA-3′ (sense), 5′-CCTTTTTCACACTGACACACG-3′ (antisense); hGA PDH 5′-CCTACCCCCAATGTATCCGTTGTG-3′ (sense), and 5′-GGAGGAATGGGAGTTGCTGTTGAA-3′ (antisense). PCR conditions used were: 3 min at 94°C, 35 cycles for 30 s at 94°C, 1 min at 58°C, 2 min at 72°C, and a final extension for 10 min at 72°C followed by melt curve analysis from 59°C to 95°C with 0.5°C for gene specific peaks. mRNA expression was normalized with GAPDH.
2.6 |. Terminal deoxynucleotidyl transferase dUTP nick end labeling assay
OSCC cells cultured in eight-well chamber slide overnight were treated with TRAIL (100 ng/ml) and in a combination of TRAIL and RANKL (80 ng/ml) for 24 hr followed by 4% paraformaldehyde fixation for 1 hr at room temperature. Cells were incubated with Triton X-100 (0.1%) and sodium citrate (0.1%). Terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay was done immunocytohemically with anti-fluorescent antibody conjugated with the alkaline phosphatase reporter enzyme (Roche Applied Science, IN). TUNEL-positive and- negative cells were digitally imaged in bright field at ×400 on a Zeiss Axioskop 2 plus and fluorescence microscope.
2.7 |. Proteasome activity assay
SCC14A and SCC74A cells in 96-well plates (1 × 104/well) were cultured with TRAIL (100 ng/ml), RANKL (80 ng/ml), and in combination for 24 hr. Then, proteasome activity was monitored in cell extracts using the fluorometric assay kit (Biovision, Milpitas, CA).
2.8 |. Acridine orange staining
OSCC cells (5 × 105/well) seeded in six-well plates were cultured with TRAIL (100 ng/ml), in the presence of RANKL (80 ng/ml) and in combination for 24 hr. Cells were added with acridine orange (5 μg/ml) reagent (Invitrogen, Grand Island, NY) at 37°C for 30 min. Cells were washed with saline and incubated at 37°C for 10 min and analyzed with a confocal microscope (excitation: 450–490 nm and emission: 515–565 nm or 600 nm) (Ethiraj et al., 2018).
2.9 |. Statistics
The results were expressed as mean ± standard deviation for triplicate studies and assessed by Student’s t test for statistical significance (*p < .05).
3 |. RESULTS
3.1 |. TRAIL stimulates RANKL expression in OSCC cells
We showed that OSCC cells expressed elevated levels of RANKL, a critical factor involved in bone resorbing osteoclast differentiation (Yuvaraj et al., 2009). It has been reported that TRAIL inhibits the human tumor xenografts growth with no toxicity (Dong, Hu, Zhao, Xu, & Liu, 2007; Ishii et al., 2007). However, TRAIL resistance to cell death in various cancer types, including OSCC cells, has been reported (Younes & Kadin, 2003). Under steady-state conditions, OSCC cells produce very low levels of TRAIL. Interestingly, western blot analysis of OSCC cells cultured with TRAIL revealed elevated RANKL expression (Figure 1a). In addition, real-time polymerase chain reaction identified increased levels of RANKL expression in OSCC cells treated with TRAIL (Figure 1b). Further, we observed high-level mRNA of TRAIL receptors DR4, DR5, and moderate levels of DcR1, DcR2 decoy receptors expression (Figure 1c). Thus, our results suggest that TRAIL signaling modulates RANKL expression in OSCC cells.
FIGURE 1.
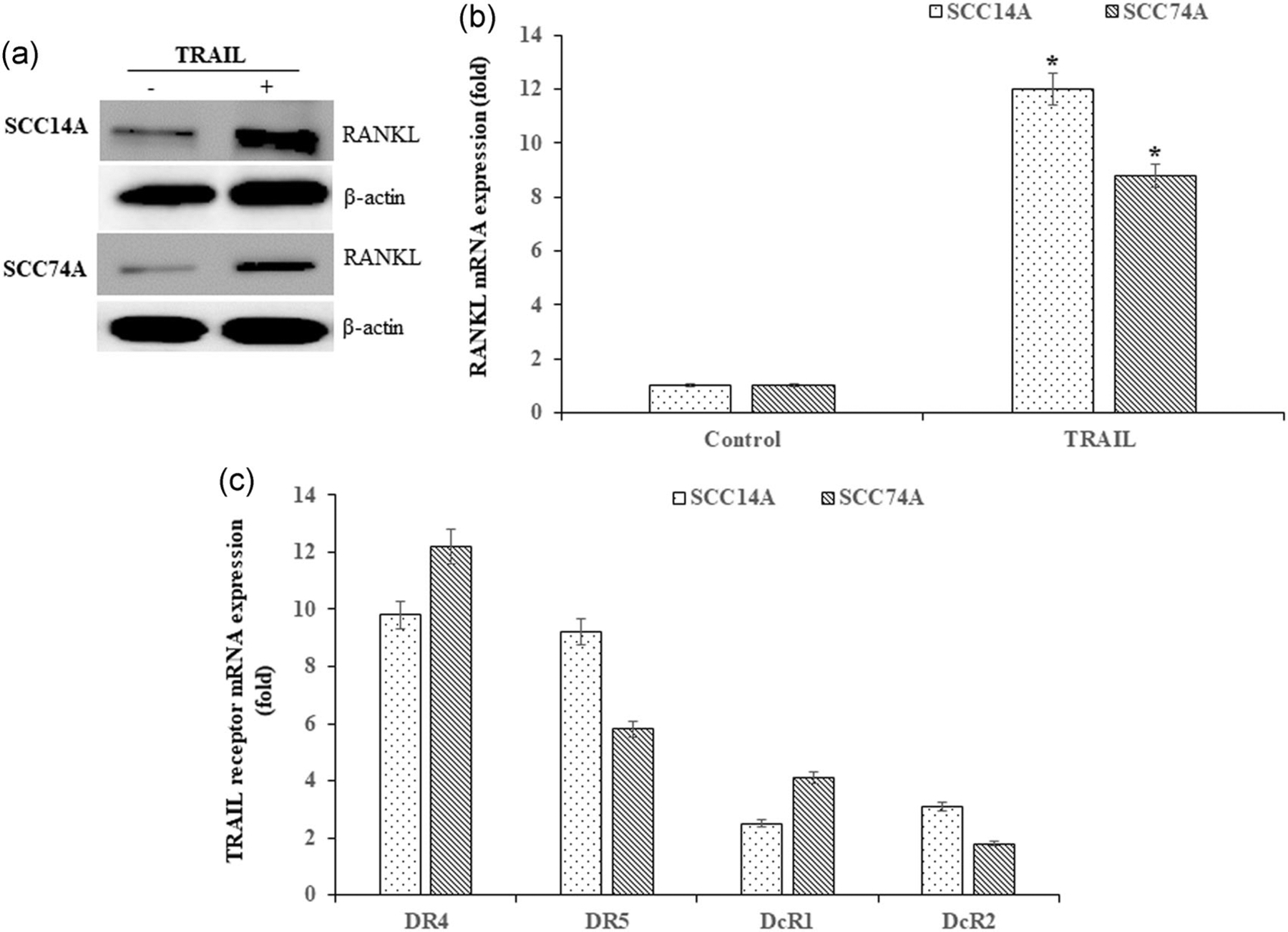
Expression of RANKL and TRAIL receptors in OSCC cells. Immunoblotting for RANKL expression in SCC14A and SCC74A cells cultured with TRAIL (100 ng/ml) for 24 hr. β-actin served as a loading control (a). Quantitative PCR analysis for RANKL expression in tumor cells treated with TRAIL for 24 hr (b). Real-time RT-PCR analysis of TRAIL receptors DR4, DR5, and decoy receptors DcR1 and DcR2 expression in OSCC cells. The mRNA amplification was normalized for GAPDH expression in all the samples analyzed (c). Values represent mean ± SD (*p < 0.05). mRNA, messenger RNA; OSCC, oral squamous cell carcinoma; PCR, polymerase chain reaction; RANKL, receptor activator of nuclear factor-κB ligand; SD, standard deviation; TRAIL, tumor necrosis factor related apoptosis inducing ligand
3.2 |. RANKL suppress TRAIL inhibition of OSCC cell proliferation and apoptosis
RANKL stimulation of OSCC cells induces a dose-dependent increase in tumor cell proliferation (Sambandam et al., 2018). SCC14A and SCC74A cell cultures stimulated with TRAIL decreased cell proliferation, which is more profound at 24 hr compared to 12 hr treatment period. Addition of RANKL to OSCC cultures stimulated with TRAIL markedly suppressed TRAIL inhibition of cell proliferation. RANKL suppression of TRAIL-mediated inhibitory effects on OSCC cell proliferation was time-dependent (Figure 2a,b). Further, western blot analysis for proliferative cell nuclear antigen (PCNA) expression confirms the inhibition of OSCC cell proliferation upon RANKL and TRAIL treatment (Figure 2c). These results demonstrate that RANKL stimulation suppressed TRAIL inhibition of OSCC cell proliferation.
FIGURE 2.
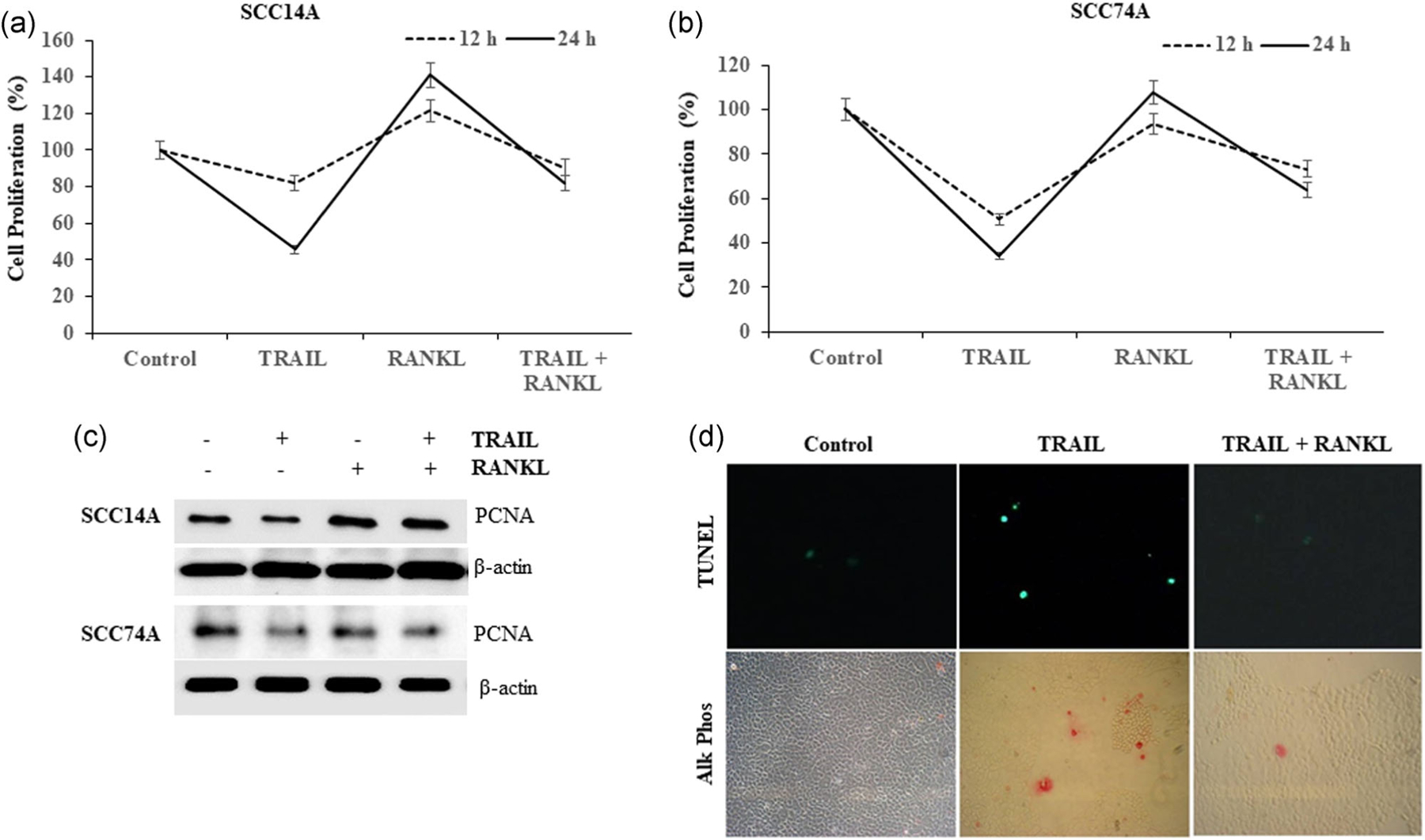
RANKL triggers resistance to TRAIL inhibition of OSCC cell proliferation and cell death. SCC14A and SCC74A cells were treated with TRAIL (100 ng/ml), stimulated with and without RANKL (80 ng/ml) for 24 hr. MTT assay was performed as described in methods to assess the cell proliferation (a,b). RANKL enhances PCNA (proliferative cell nuclear antigen) expression in OSCC tumor cells. SCC14A and SCC74A cells treated with TRAIL (100 ng/ml) and stimulated with and without RANKL (80 ng/ml) for 24 hr. Cell lysates obtained were analyzed by immunoblotting for PCNA expression. The levels of β-actin served as a loading control (c). SCC14A cells were cultured in the 8-well chamber slide overnight. Then, cells were treated with TRAIL (100 ng/ml) and in combination with RANKL (80 ng/ml) for 24 hr. Cells were washed with PBS, then fixed with 4% paraformaldehyde and performed TUNEL assay, immunocytochemically using the alkaline phosphatase detection kit. TUNEL-positive and- negative cells digitally imaged in a fluorescence microscope (upper panel) and bright field at ×400 on a Zeiss Axioskop 2 plus showing DNA damage by alkaline phosphatase reporter activity staining (lower panel) (d). MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; OSCC, oral squamous cell carcinoma; RANKL, receptor activator of nuclear factor-κB ligand; TRAIL, tumor necrosis factor related apoptosis inducing ligand; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling
TRAIL-treated (100 ng/ml) SCC14A cells demonstrated ~10% cell death of OSCC cells. Cells stimulated with combination of RANKL and TRAIL as shown by TUNEL assay, induced resistance to TRAIL-mediated apoptosis (Figure 2d). As shown in the upper panel, the fluorescent microscopy demonstrated RANKL inhibition of TRAIL-treated TUNEL-positive cells (green), as validated by alkaline phosphatase activity staining with fast red chromogen (red), is shown in the bottom panel. These data reveal that RANKL stimulation suppressed TRAIL-induced OSCC cell death.
3.3 |. RANKL-KO in OSCC cells promote TRAIL-induced caspase activation
We further determined the functional role of RANKL on TRAIL induction of apoptotic cell death in OSCC cells. SCC14A cells, which endogenously express high levels of RANKL, were used to generate the stable RANKL-KO cells by CRISPR/Cas-9 as described in the methods. Fluorescence microscopy analysis identified transfection efficiency >80% as evident from the GFP content in tumor cells (Figure 3a). We also confirmed the knockout of RANKL in transfected cells by western blot analysis (Figure 3b).
FIGURE 3.
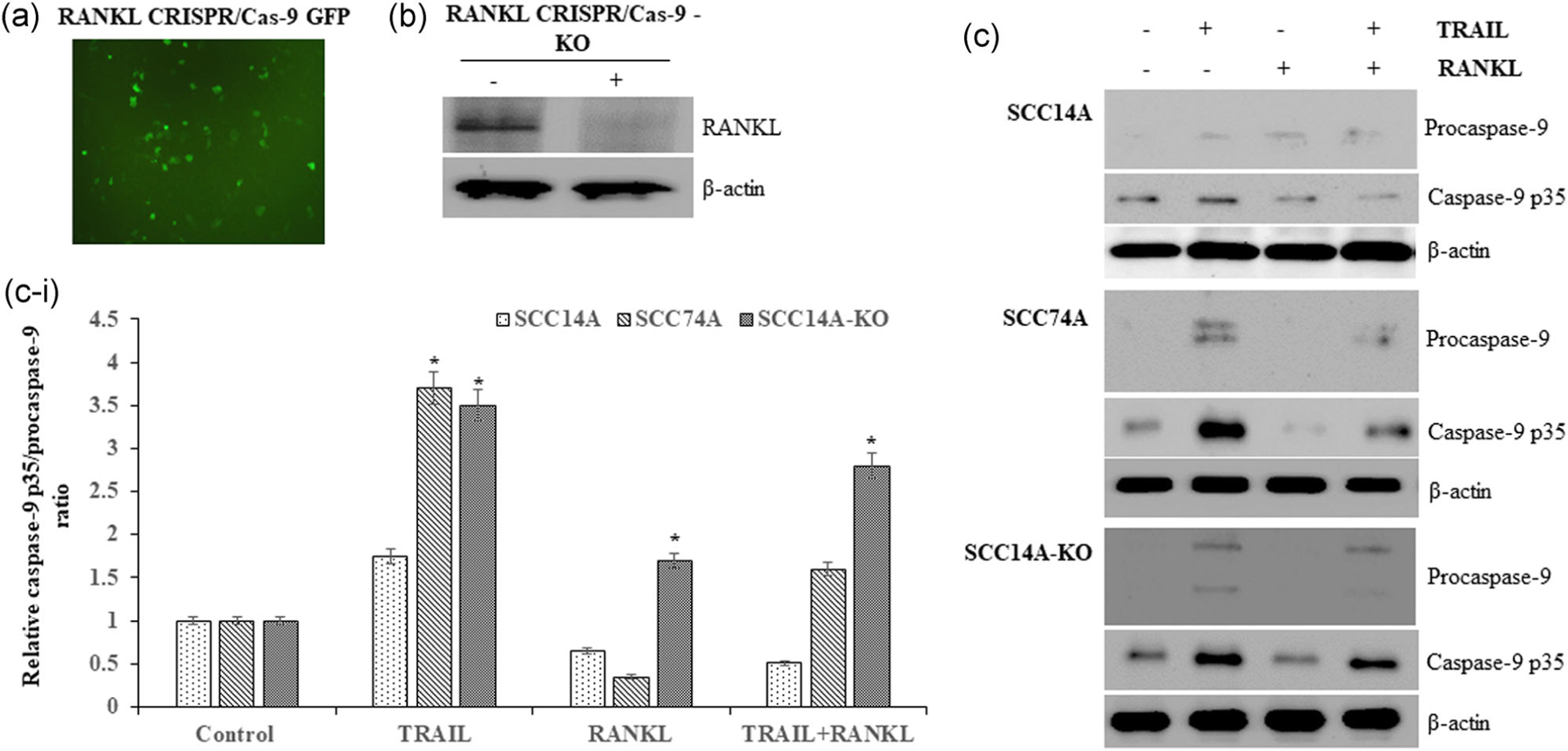
RANKL knockout (KO) in OSCC cells promotes TRAIL-induced cell death. RANKL CRISPR/Cas-9 plasmid transfected into SCC14A cells as visualized under fluorescence microscopy for GFP (a). Immunoblotting for RANKL knockout (RANKL-KO) in SCC14A transfected cells (b). SCC14A, SCC74A, and SCC14A-KO cells were treated with TRAIL (100 ng/ml) with and without RANKL (80 ng/ml) for 24 hr. Cell lysates collected were analyzed by western blot for procaspase-9 and active caspase-9 p35 expression. The levels of β-actin served as a loading control (c). Densitometric analysis of relative levels of caspse-9 p35/procaspase-9 ratio (c-i). OSCC, oral squamous cell carcinoma; RANKL, receptor activator of nuclear factor-κB ligand; TRAIL, tumor necrosis factor related apoptosis inducing ligand
SCC14A, SCC74A, and SCC14A-KO cells were cultured with TRAIL in presence and absence of RANKL stimulation. Cell lysates obtained were analyzed by immunoblotting to assess RANKL effects on TRAIL-induced OSCC apoptosis. TRAIL stimulation alone upregulated caspase-9 p35 activation in SCC14A and SCC74A cells, which was blunted by RANKL costimulation (Figure 3c). Knockout of RANKL in SCC14A-KO cells showed increased levels of caspase-9 p35 activation when treated with TRAIL compared to wild-type (WT) SCC14A cells (Figure 3c-i).
Effector caspase-3, which is downstream of initiator caspase-9 (Dillon & Green, 2016), was assessed to delineate the effect of RANKL on TRAIL induction of OSCC cell death. RANKL stimulated SCC14A cells cultured with TRAIL showed no change in levels of caspase-3 p17 activation, whereas investigation in SCC74A cells revealed that RANKL stimulation inhibited TRAIL prompted caspase-3 p17 activation (Figure 4a). Consistent with increased caspase-9 p35 activation found in TRAIL-stimulated SCC14A-KO cells, SCC14A-KO cells showed an elevated level of caspase-3 p17 activation in response to TRAIL (Figure 4a,a-i). TRAIL treatment in the presence of RANKL stimulation also induced high-level caspase-9 p35 and caspase-3 p17 activation in SCC14A-KO cells. Taken together, these data provide novel mechanistic insights suggesting that RANKL modulated TRAIL-induced apoptosis via intrinsic caspase pathway in OSCC cells.
FIGURE 4.
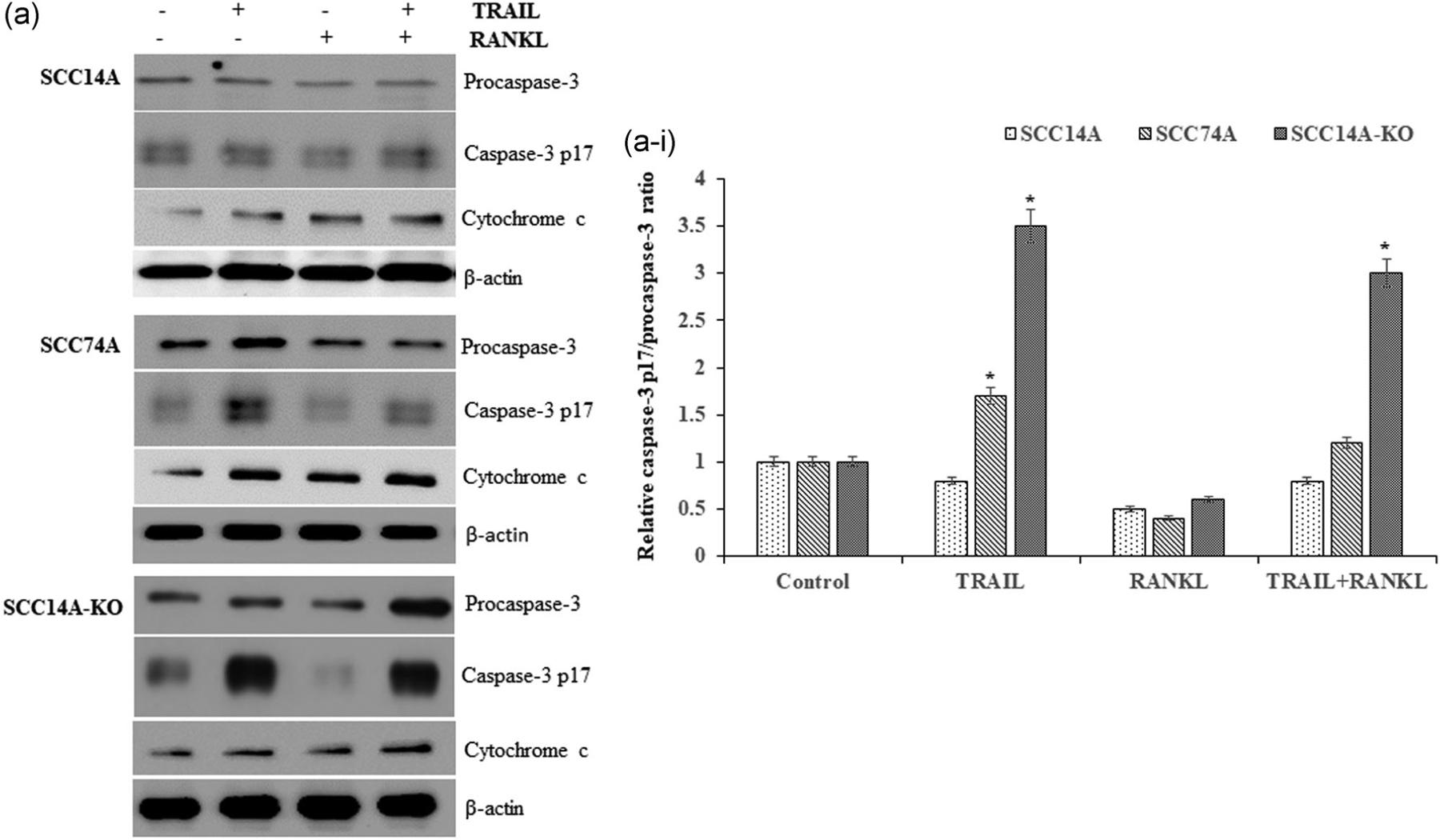
RANKL knockout (KO) enhances caspase-3 activation and cytochrome c release. Western blot analysis for procaspase-3 and active caspase-3 p17 expression and cytochrome c release from SCC14A, SCC74A, and SCC14A-KO cells in response to TRAIL and RANKL stimulation (a). Densitometric analysis of relative levels of caspase-3 p17/procaspase-3 ratio (a-i). The levels of β-actin served as a loading control. RANKL, receptor activator of nuclear factor-κB ligand; TRAIL, tumor necrosis factor related apoptosis inducing ligand
Cytochrome c released from mitochondria activates caspase-9 (Dillon & Green, 2016), which was also assessed via western blot analysis in SCC14A, SCC74A, and SCC14A-KO cells. These mechanistic studies detected an increased level of cytochrome c expression in RANKL-KO cells when cultured with TRAIL in the presence of RANKL compared to those in WT SCC14A cells (Figure 4a-i). Thus, our results further support that RANKL autoregulation inhibits TRAIL-mediated apoptosis in OSCC cells.
3.4 |. TRAIL-induced cell death in RANKL-KO OSCC cells occurs through caspase-dependent mitochondrial pathway
Prior studies have demonstrated that TRAIL death receptors (DR4/5) are associated with caspase-mediated cell death in tumor cells (Chan et al., 2012). BAD and Bcl-xL dimerization result in the dislocation of BAX from the Bcl-xL-BAX complex to recover from BAX-mediated apoptosis (Tait & Green, 2010). BAD and BAX proapoptotic protein expressions were analyzed by immunoblotting. SCC14A, SCC74A, and SCC14A-KO cells cultured with TRAIL were stimulated with and without RANKL. SCC14A and SCC74A cells treated with TRAIL showed elevated expression of BAD/BAX, whereas cells stimulated with RANKL alone and in combination with TRAIL showed inhibition of BAD/BAX expression compared to untreated control cells. Interestingly, BAD/BAX expression levels were elevated in SCC14A-KO cells following treatment with TRAIL alone, as well as in combination of RANKL and TRAIL when compared to WT SCC14A cells (Figure 5a,a-i,a-ii). These results indicated that RANKL suppression in tumor cells increases TRAIL-induced apoptotic cell death by caspase-dependent mitochondrial pathway.
FIGURE 5.
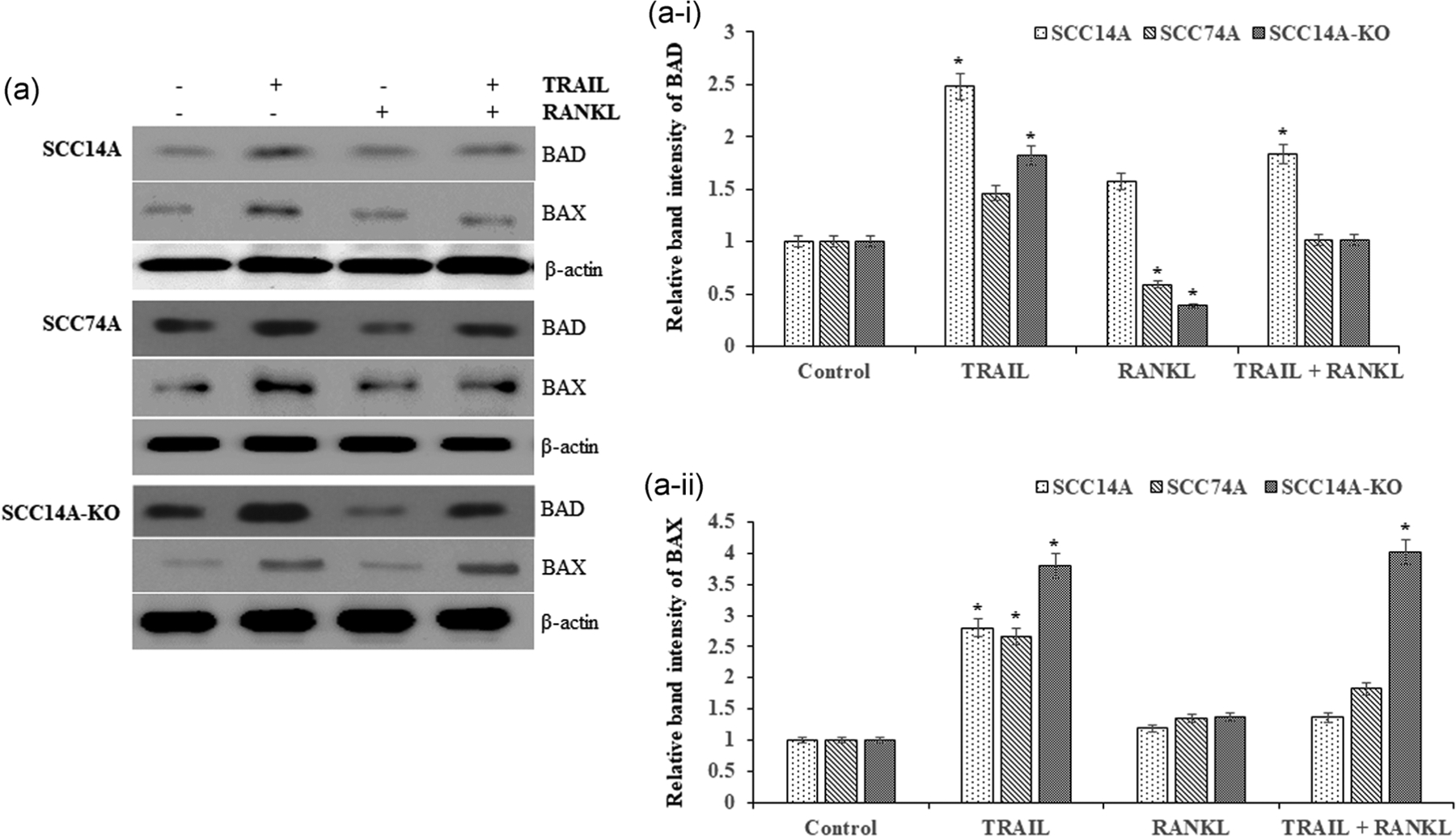
Proapoptotic marker proteins BAD/BAX in OSCC tumor cells treated with TRAIL and RANKL. SCC14A, SCC74A, and SCC14A-KO cells were treated with TRAIL (100 ng/ml) stimulated with and without RANKL (80 ng/ml) for 24 hr. Cell lysates collected were analyzed by western blot for BAD and BAX (a). Densitometric analysis of relative levels of BAD (a-i) and BAX expression (a-ii). The levels of β-actin served as a loading control. OSCC, oral squamous cell carcinoma; RANKL, receptor activator of nuclear factor-κB ligand; TRAIL, tumor necrosis factor related apoptosis inducing ligand
3.5 |. RANKL reverses TRAIL inhibition of proteasome activity in OSCC cells
SQSTM1/p62 has been implicated in RANK signaling through binding to TRAF6 adaptor protein (McManus & Roux, 2012). SCC14A and SCC74A cells cultured with TRAIL demonstrated inhibition of p62 expression. RANKL treatment alone induced expression of p62. In contrast, OSCC cells treated with TRAIL in the presence of RANKL showed no change in the level of p62 (Figure 6a). Furthermore, TRAIL treatment markedly inhibited proteasome activity in OSCC cells. RANKL stimulation alone increased the proteasome activity by 1.5-fold in these cells compared to basal activity. Notably, RANKL stimulation significantly increased proteasome activity in OSCC cells cultured with TRAIL (Figure 6b). These results revealed RANKL reversal of TRAIL inhibition of proteasome activity in OSCC cells.
FIGURE 6.
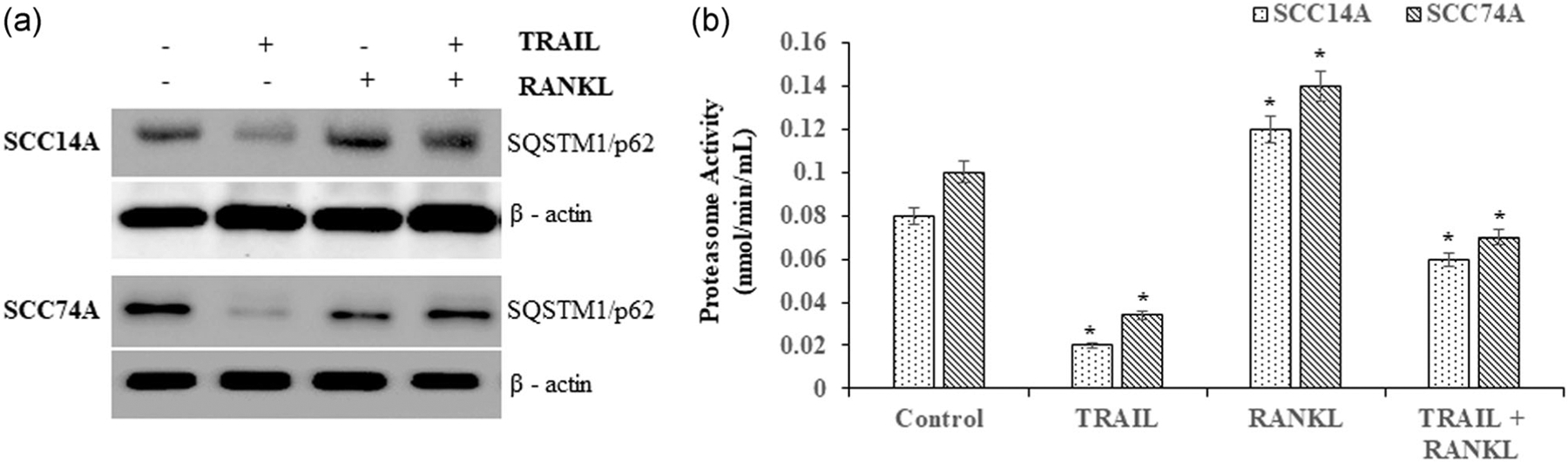
RANKL restore TRAIL inhibition of SQSTM1/p62 expression and modulates autophagy in OSCC cells. SCC14A and SCC74A cells were treated with TRAIL (100 ng/ml) stimulated with and without RANKL (80 ng/ml) for 24 hr. Cell lysates obtained were analyzed by western blot for SQSTM1/p62. The levels of β-actin served as a loading control. (a). Proteasome activity was assayed as described in methods (b). Values represent mean ± SD (*p < .05). OSCC, oral squamous cell carcinoma; RANKL, receptor activator of nuclear factor-κB ligand; SD, standard deviation; TRAIL, tumor necrosis factor related apoptosis inducing ligand
3.6 |. RANKL modulates the autophagic activity in OSCC cells treated with TRAIL
Autophagy is a cellular process for the recycling of nutrients through autophago-lysosomal degradation of cytoplasmic contents (Reggiori & Klionsky, 2002). OSCC cells were cultured with TRAIL in the presence and absence of RANKL for 24 hr. Acridine orange staining of SCC14A and SCC74A cells demonstrated low autophagic activity in these cells treated with TRAIL or RANKL alone. Intriguingly, cells stimulated with RANKL and treated with TRAIL showed increased autophagic activity. Furthermore, knockout of RANKL in SCC14A-KO cells demonstrated inhibition of autophagic activity (Figure 7a).
FIGURE 7.
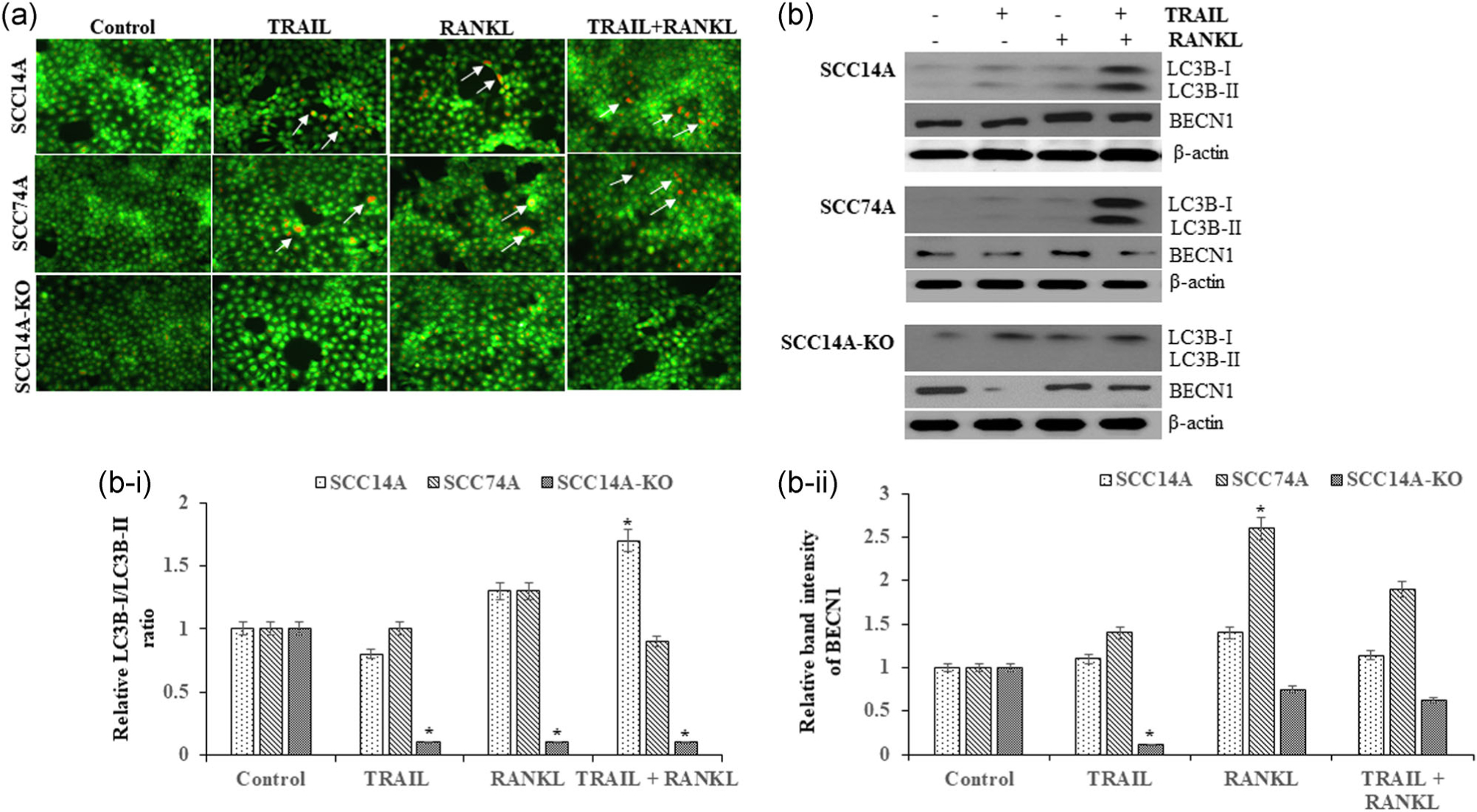
RANKL modulates autophagic marker proteins. SCC14A, SCC74A, and SCC14A-KO cells were treated with TRAIL in the presence and absence of RANKL for 24 hr. Representative confocal images of OSCC cells stained with acridine orange were shown and arrows point the autophagic activity (a). OSCC cells stimulated with RANKL and TRAIL for 24 hr. Cell lysates obtained were analyzed by western blot for LC3B-I and II isoforms and BECN1. The levels of β-actin served as a loading control (b). Densitometric analysis for LC3B-I/LC3B-II ratio (b-i) and BECN1 (b-ii) in OSCC cells. Values represent mean ± SD (*p < .05). BECN1, beclin-1; LC3, light chain 3; OSCC, oral squamous cell carcinoma; RANKL, receptor activator of nuclear factor-κB ligand; SD, standard deviation; TRAIL, tumor necrosis factor related apoptosis inducing ligand
The expression of microtubule-associated protein 1 light chain 3 (LC3) and BECN1 involved in autophagosome formation were analyzed by immunoblot to further elucidate the role of RANKL and TRAIL in autophagic activity in OSCC cells. RANKL and TRAIL alone moderately increased LC3B-II isoform expression in SCC14A and SCC74A cells, and RANKL stimulation elevated the LC3B-II isoform expression in these cells treated with TRAIL. While RANKL alone increased BECN1 expression in SCC14A and SCC74A cells, TRAIL treatment alone and in combination with RANKL did not change BECN1 expression. RANKL knockout in SCC14A-KO cells displayed the expression of LC3B-I isoform but abrogated the expression of LC3B-II active form. SCC14A-KO cells treated with TRAIL alone suppressed BECN1, whereas cells stimulated with RANKL and in combination with TRAIL demonstrated BECN1 expression similar to control cultures (Figure 7b). Densitometry analysis of LC3B-I/LC3B-II ratio suggested that RANKL knockout in SCC14A-KO cells significantly inhibits LC3B-II expression compared to WT SCC14A cells (Figure 7b,b-i) and TRAIL treatment suppress BECN1 expression in SCC14A-KO cells (Figure 7b,b-ii). These results indicate that RANKL modulates the autophagic activity in OSCC cells, and RANKL-KO may promote TRAIL function in inducing apoptosis.
4 |. DISCUSSION
RANK receptor and RANKL expression has been demonstrated in breast, prostate, myeloma, and osteosarcoma malignancies (Li et al., 2014; Marley, Bracha, & Seguin, 2015; Yoneda, Tanaka, & Hata, 2013). Importantly, RANK/RANKL molecular signaling axis is involved in all stages of tumorigenesis (Renema et al., 2016). We recently showed RANKL/RANK expression in primary human OSCC tumor sample specimens and that RANKL stimulation increases OSCC cell proliferation. We also have shown that RANKL autoregulation facilitates OSCC tumor growth and the osteolytic process (Sambandam et al., 2018). Therefore, RANKL autoregulation in OSCC plays a functional role in chemotherapeutic resistance. We previously reported that TRAIL induces expression of RANKL through STAT-6 dependent transcriptional mechanism in bone marrow stromal and preosteoblasts (Sundaram et al., 2015). However, this study delineates inhibitory effects of RANKL on TRAIL induction of OSCC cell death. Our results demonstrated that OSCC cells expressed death receptors essential for TRAIL signaling associated cell death. Further, we showed that TRAIL induces the expression of RANKL in tumor cells. Hence, our study indicates that TRAIL-induced RANKL in OSCC cells plays a vital role against TRAIL-induced apoptotic cell death and tumor progression. RANKL inhibition of TRAIL-induced TUNEL labeling in SCC14A cells discern that RANKL stimulation suppresses the TRAIL induction of cell death in OSCC cells.
Bcl-2 family members include pro/antiapoptotic proteins, which play a pivotal role in cell death and determining mitochondrial membrane integrity (Luna-Vargas & Chipuk, 2016). We evaluated the proapoptotic intrinsic caspase pathway, inducing cytochrome c, caspase-3, and caspase-9. Proapoptotic signaling leads to release of mitochondrial cytochrome c into cytosol. Cytochrome c activates caspase-9 resulting in activation of caspase-3 (Dillon & Green, 2016). We observed no change in caspase levels in WT SCC14A cells upon TRAIL treatment, suggesting that these cells are highly resistant to TRAIL induction of cell death. Furthermore, our results demonstrated that RANKL knockout in OSCC cells (SCC14A-KO) increased caspase-9 and caspase-3 activation consistent with an elevation in cytochrome c release. Therefore, these results suggest that TRAIL-mediated cell death in SCC14A-KO was associated with caspase-dependent mitochondrial pathway. Knockout of RANKL in OSCC cells increased TRAIL-induced BAD/BAX expression, which initiated apoptosis by mitochondrial cytochrome c release into the cytoplasm, activating proapoptotic caspase cascades (Tait & Green, 2010). BAD and Bcl-xL dimerization resulted in dislocation of BAX from Bcl-xL-BAX complex, causing BAX related apoptosis. Our results that RANKL inhibited caspase-9 and caspase-3 activation in OSCC cells treated with TRAIL suggest that RANKL induced tumor cell resistance against TRAIL-induced cell death. Furthermore, proapoptotic markers BAD and BAX expression were restored in cells upon costimulation with RANKL and TRAIL. These results further confirm that RANKL inhibited TRAIL-mediated cell death in OSCC cells.
Proteasomes play a critical role in degradation of ubiquitinylated proteins (Castro, Bernis, Vigneron, Labbe, & Lorca, 2005; Orlowski, 1999). SQSTM1/p62 is involved with the transport of ubiquitynylated proteins to the proteasome system (Liu et al., 2016). We have shown that TRAIL suppressed p62 expression in OSCC cells. RANKL stimulation restored the TRAIL inhibition of p62 expression. Our results indicated that RANKL modulation of p62 levels and proteasome activity are associated with enhanced resistance against TRAIL induction of tumor cell growth. We also confirmed that TRAIL significantly inhibited the proteasome activity, whereas RANKL stimulation alone increased proteasome activity in OSCC cells. However, RANKL stimulation significantly increased proteasome activity in TRAIL-treated OSCC cells. Thus, our results reveal that p62 had a functional link between TRAIL and RANKL associated resistance, which played a significant role in transporting ubiquitinated proteins for degradation by proteasome or autophagy pathways.
Evidence suggests that p62 is associated with autophagy processes and proteasomes for protein degradation (Liu et al., 2016). The autophagic markers BECN1 and LC3 play an essential role in tumor progression (Hamurcu et al., 2018). To understand the functional role of RANKL in LC3B-II isoform activation and to analyse BECN1 expression, we performed RANKL knockout in OSCC cells. We demonstrated RANKL stimulation in the presence of TRAIL restores BECN1 expression and LC3B-II isoform in OSCC cells, which indicates a functional role of RANKL in autophagy for resistance to TRAIL related OSCC cell death. This study revealed that caspase activity, induction of proapoptotic proteins had a functional role in suppression of RANKL on TRAIL induction of cell death. In contrast, RANKL induced proteasome and autophagic activity played a role in OSCC cell growth. Earlier, we reported that RANKL modulates autophagic activity in OSCC cells and autophagy inhibitor suppress RANK/RANKL signaling (Sambandam et al., 2014; Sambandam, Sakamuri, Balasubramanian, & Haque, 2016).
Targeting RANK/RANKL signaling axis has clinical advantages in the management of cancer invasion and metastases to bone (Renema et al., 2016). Hence, this study revealed that RANKL signaling suppressed the efficacy of TRAIL-associated OSCC cell death, which may implicate a functional role in tumor progression and bone destruction. Our findings in SCC14A and SCC74A cells and the knockout of RANKL in SCC14A-KO cells provide novel insight suggesting that RANKL autoregulation modulates TRAIL related cell death in OSCC. Schematic illustration displayed represents the molecular mechanism of RANKL inhibitory effect on TRAIL induction of apoptotic OSCC cell death (Figure 8). Thus, our results highlight the blockade of RANKL in tumor cells enhanced the therapeutic efficacy of TRAIL in controlling OSCC tumor progression.
FIGURE 8.
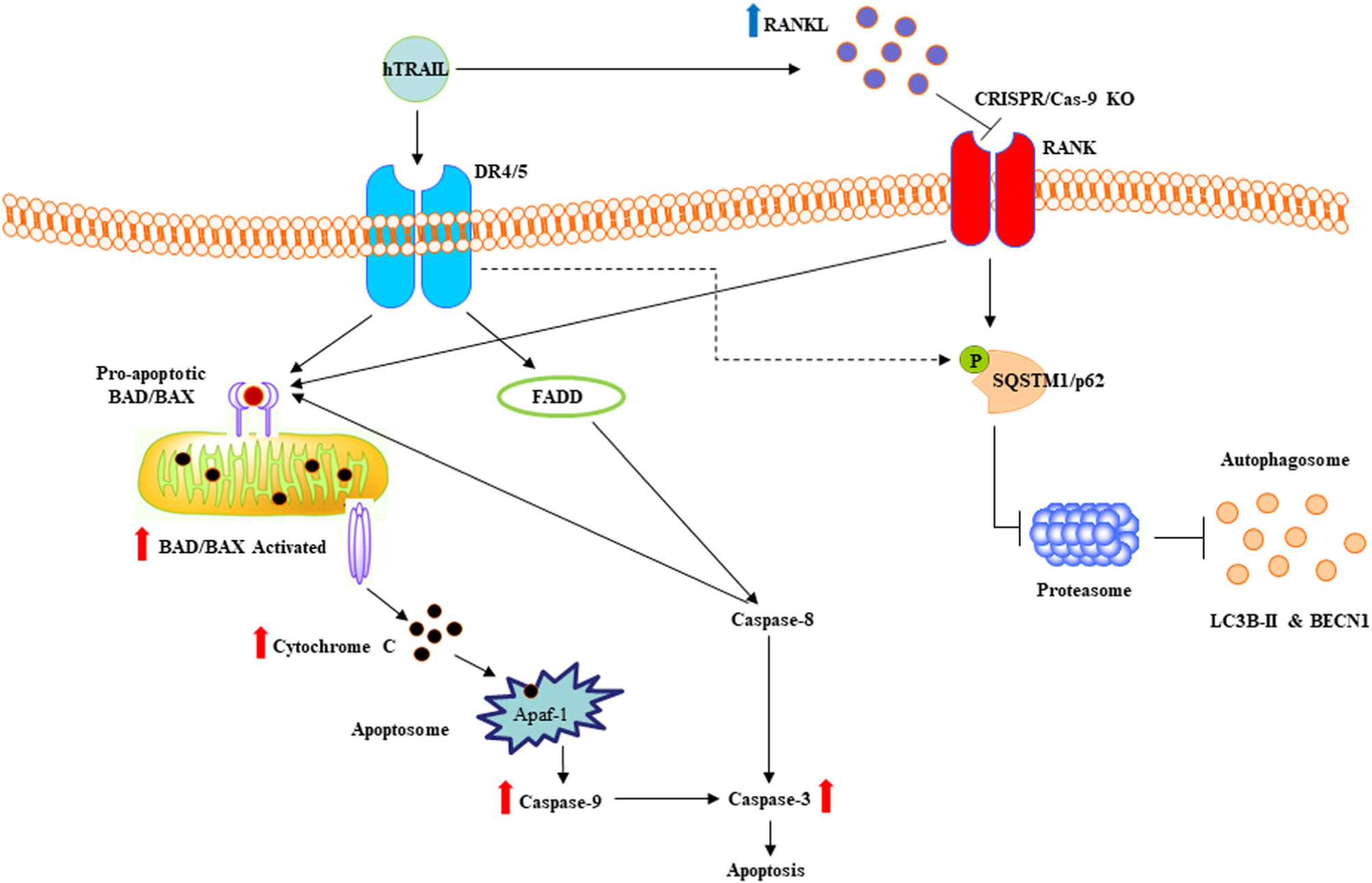
Schematic illustration of RANKL inhibitory effect on TRAIL-induced cell death in OSCC cells. TRAIL increase RANKL expression in OSCC cells is represented by upright blue arrow. CRISPR/Cas-9 knockout of RANKL in tumor cells increases activation of apoptotic markers shown by upright red arrows. TRAIL treatment inhibits SQSTM1/p62 expression is shown by a dotted arrow. OSCC, oral squamous cell carcinoma; RANKL, receptor activator of nuclear factor-κB ligand; TRAIL, tumor necrosis factor related apoptosis inducing ligand
ACKNOWLEDGMENT
We thank Thomas E. Carey, University of Michigan for providing OSCC cells.
Footnotes
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
REFERENCES
- Attar E, Dey S, Hablas A, Seifeldin IA, Ramadan M, Rozek LS, Soliman AS (2010). Head and neck cancer in a developing country: A population-based perspective across 8 years. Oral Oncology, 46(8), 591–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle WJ, Simonet WS, & Lacey DL (2003). Osteoclast differentiation and activation. Nature, 423(6937), 337–342. [DOI] [PubMed] [Google Scholar]
- Brenner JC, Graham MP, Kumar B, Saunders LM, Kupfer R, Lyons RH, … Carey TE (2010). Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head and Neck, 32(4), 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti G, Oranger A, Mori G, Sardone F, Pignataro P, Coricciati M, … Colucci S (2011). TRAIL effect on osteoclast formation in physiological and pathological conditions. Frontiers in Bioscience, 3, 1154–1161. [DOI] [PubMed] [Google Scholar]
- Castro A, Bernis C, Vigneron S, Labbe JC, & Lorca T (2005). The anaphase-promoting complex: A key factor in the regulation of cell cycle. Oncogene, 24(3), 314–325. [DOI] [PubMed] [Google Scholar]
- Chan LP, Chou TH, Ding HY, Chen PR, Chiang FY, Kuo PL, & Liang CH (2012). Apigenin induces apoptosis via tumor necrosis factor receptor- and Bcl-2-mediated pathway and enhances susceptibility of head and neck squamous cell carcinoma to 5-fluorouracil and cisplatin. Biochimica et Biophysica Acta, 1820(7), 1081–1091. [DOI] [PubMed] [Google Scholar]
- Choi S, & Myers JN (2008). Molecular pathogenesis of oral squamous cell carcinoma: Implications for therapy. Journal of Dental Research, 87(1), 14–32. [DOI] [PubMed] [Google Scholar]
- Cronin KA, Lake AJ, Scott S, Sherman RL, Noone AM, Howlader N, … Jemal A (2018). Annual report to the nation on the status of cancer, part I: National cancer statistics. Cancer, 124(13), 2785–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon CP, & Green DR (2016). Molecular Cell Biology of Apoptosis and Necroptosis in Cancer. Advances in Experimental Medicine and Biology, 930, 1–23. [DOI] [PubMed] [Google Scholar]
- Dong A, Hu J, Zhao L, Xu H, & Liu X (2007). Regulation and pharmacokinetics of inducible recombinant TRAIL expression. Cancer Biology & Therapy, 6(12), 1978–1985. [DOI] [PubMed] [Google Scholar]
- Dougall WC, Holen I, & Gonzalez Suarez E (2014). Targeting RANKL in metastasis. BoneKEy Reports, 3, 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ethiraj P, Link JR, Sinkway JM, Brown GD, Parler WA, & Reddy SV (2018). Microgravity modulation of syncytin-A expression enhance osteoclast formation. Journal of Cellular Biochemistry, 119(7), 5696–5703. [DOI] [PubMed] [Google Scholar]
- Ethiraj P, Veerappan K, Doraisami B, & Sivapatham S (2014). Synergistic anti-carcinogenic effect of interferon-beta with cisplatin on human breast adenocarcinoma MDA MB231 cells. International Immunopharmacology, 23(1), 222–228. [DOI] [PubMed] [Google Scholar]
- Ethiraj P, Veerappan K, Samuel S, & Sivapatham S (2016). Interferon beta improves the efficacy of low dose cisplatin by inhibiting NF-kappaB/p-Akt signaling on HeLa cells. Biomedicine and Pharmacotherapy, 82, 124–132. [DOI] [PubMed] [Google Scholar]
- Hamurcu Z, Delibasi N, Gecene S, Sener EF, Donmez-Altuntas H, Ozkul Y, … Ozpolat B (2018). Targeting LC3 and Beclin-1 autophagy genes suppresses proliferation, survival, migration and invasion by inhibition of Cyclin-D1 and uPAR/Integrin beta1/Src signaling in triple negative breast cancer cells. Journal of Cancer Research and Clinical Oncology, 144(3), 415–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii M, Iwai M, Harada Y, Kishida T, Asada H, Shin-Ya M, … Mazda O (2007). Soluble TRAIL gene and actinomycin D synergistically suppressed multiple metastasis of TRAIL-resistant colon cancer in the liver. Cancer Letters, 245(1–2), 134–143. [DOI] [PubMed] [Google Scholar]
- Jimi E, Furuta H, Matsuo K, Tominaga K, Takahashi T, & Nakanishi O (2011). The cellular and molecular mechanisms of bone invasion by oral squamous cell carcinoma. Oral Diseases, 17(5), 462–468. [DOI] [PubMed] [Google Scholar]
- Jones DH, Nakashima T, Sanchez OH, Kozieradzki I, Komarova SV, Sarosi I, … Penninger JM (2006). Regulation of cancer cell migration and bone metastasis by RANKL. Nature, 440(7084), 692–696. [DOI] [PubMed] [Google Scholar]
- Kawano Y, Ueno S, Abe M, Kikukawa Y, Yuki H, Iyama K, … Hata H (2012). TRAIL produced from multiple myeloma cells is associated with osteolytic markers. Oncology Reports, 27(1), 39–44. [DOI] [PubMed] [Google Scholar]
- Li X, Liu Y, Wu B, Dong Z, Wang Y, Lu J, … Wang Z (2014). Potential role of the OPG/RANK/RANKL axis in prostate cancer invasion and bone metastasis. Oncology Reports, 32(6), 2605–2611. [DOI] [PubMed] [Google Scholar]
- Liu WJ, Ye L, Huang WF, Guo LJ, Xu ZG, Wu HL, … Liu HF (2016). p62 links the autophagy pathway and the ubiqutin-proteasome system upon ubiquitinated protein degradation. Cellular and Molecular Biology Letters, 21(29), 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Vargas MPA, & Chipuk JE (2016). Physiological and Pharmacological Control of BAK, BAX, and Beyond. Trends in Cell Biology, 26(12), 906–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marley K, Bracha S, & Seguin B (2015). Osteoprotegerin activates osteosarcoma cells that co-express RANK and RANKL. Experimental Cell Research, 338(1), 32–38. [DOI] [PubMed] [Google Scholar]
- McManus S, & Roux S (2012). The adaptor protein p62/SQSTM1 in osteoclast signaling pathways. Journal of Molecular Signaling, 7, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlowski RZ (1999). The role of the ubiquitin-proteasome pathway in apoptosis. Cell Death and Differentiation, 6(4), 303–313. [DOI] [PubMed] [Google Scholar]
- Pandruvada SN, Yuvaraj S, Liu X, Sundaram K, Shanmugarajan S, Ries WL, … Reddy SV (2010). Role of CXC chemokine ligand 13 in oral squamous cell carcinoma associated osteolysis in athymic mice. International Journal of Cancer, 126(10), 2319–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori F, & Klionsky DJ (2002). Autophagy in the eukaryotic cell. Eukaryotic Cell, 1(1), 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renema N, Navet B, Heymann MF, Lezot F, & Heymann D (2016). RANK-RANKL signalling in cancer. Bioscience Reports, 36(4), e00366–e00366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandam Y, Ethiraj P, Hathaway-Schrader JD, Novince CM, Panneerselvam E, Sundaram K, Reddy SV (2018). Autoregulation of RANK ligand in oral squamous cell carcinoma tumor cells. Journal of Cellular Physiology, 233(8), 6125–6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandam Y, Sakamuri S, Balasubramanian S, & Haque A (2016). RANK ligand modulation of autophagy in oral squamous cell carcinoma tumor cells. Journal of Cellular Biochemistry, 117(1), 118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandam Y, Sundaram K, Liu A, Kirkwood KL, Ries WL, & Reddy SV (2013). CXCL13 activation of c-Myc induces RANK ligand expression in stromal/preosteoblast cells in the oral squamous cell carcinoma tumor-bone microenvironment. Oncogene, 32(1), 97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandam Y, Townsend MT, Pierce JJ, Lipman CM, Haque A, Bateman TA, Reddy SV (2014). Microgravity control of autophagy modulates osteoclastogenesis. Bone, 61, 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedger LM, Glaccum MB, Schuh JC, Kanaly ST, Williamson E, Kayagaki N, … Gliniak B (2002). Characterization of the in vivo function of TNF-alpha-related apoptosis-inducing ligand, TRAIL/Apo2L, using TRAIL/Apo2L gene-deficient mice. European Journal of Immunology, 32(8), 2246–2254. [DOI] [PubMed] [Google Scholar]
- Sundaram K, Sambandam Y, Balasubramanian S, Pillai B, Voelkel-Johnson C, Ries WL, Reddy SV (2015). STAT-6 mediates TRAIL induced RANK ligand expression in stromal/preosteoblast cells. Bone, 71, 137–144. [DOI] [PubMed] [Google Scholar]
- Tait SW, & Green DR (2010). Mitochondria and cell death: Outer membrane permeabilization and beyond. Nature Reviews. Molecular Cell Biology, 11(9), 621–632. [DOI] [PubMed] [Google Scholar]
- Wiley SR, Schooley K, Smolak PJ, Din WS, Huang CP, Nicholl JK, … Goodwin RG (1995). Identification and characterization of a new member of the TNF family that induces apoptosis. Immunity, 3(6), 673–682. [DOI] [PubMed] [Google Scholar]
- Yen ML, Hsu PN, Liao HJ, Lee BH, & Tsai HF (2012). TRAF6 dependent signaling pathway is essential for TNF-related apoptosis-inducing ligand (TRAIL) induces osteoclast differentiation. PLoS One, 7(6), e38048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda T, Tanaka S, & Hata K (2013). Role of RANKL/RANK in primary and secondary breast cancer. World Journal of Orthopedics, 4(4), 178–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younes A, & Kadin ME (2003). Emerging applications of the tumor necrosis factor family of ligands and receptors in cancer therapy. Journal of Clinical Oncology, 21(18), 3526–3534. [DOI] [PubMed] [Google Scholar]
- Yuvaraj S, Griffin AC, Sundaram K, Kirkwood KL, Norris JS, & Reddy SV (2009). A novel function of CXCL13 to stimulate RANK ligand expression in oral squamous cell carcinoma cells. Molecular Cancer Research, 7(8), 1399–1407. [DOI] [PubMed] [Google Scholar]
- Zhang J, Dai J, Qi Y, Lin DL, Smith P, Strayhorn C, … Keller ET (2001). Osteoprotegerin inhibits prostate cancer-induced osteoclastogenesis and prevents prostate tumor growth in the bone. Journal of Clinical Investigation, 107(10), 1235–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]


