Abstract
Interleukin-17a (IL-17a) has been highly conserved during evolution of the vertebrate immune system and widely studied in contexts of infection and autoimmunity. Studies suggest that IL-17a promotes behavioral changes in experimental models of autism and aggregation behavior in worms. Here, through a cellular and molecular characterization of meningeal γδ17 T cells, we defined the nearest central nervous system associated source of IL-17a under homeostasis. Meningeal γδ T cells express high levels of the chemokine receptor CXCR6 and seed meninges shortly after birth. Physiological release of IL-17a by these cells was correlated with anxiety-like behavior in mice and was partially dependent on T cell receptor engagement and commensal-derived signals. IL-17a receptor was expressed in cortical glutamatergic neurons under steady state and its genetic deletion decreased anxiety-like behavior in mice. Our findings suggest that IL-17a production by meningeal γδ17 T cells represents an evolutionary bridge between this conserved anti-pathogen molecule and survival behavioral traits in vertebrates.
Infectious pathogens are among the strongest selective forces in mammalian evolution, allowing resistance alleles to emerge and spread over time. The host inflammatory response to such pathogens is orchestrated predominantly by cytokines, which may have evolved to shunt energy resources into fighting infectious agents and altering host behavior to avoid death from pathogen exposure3. Recently, a rich diversity of immune cells in the healthy mouse meninges has been described, where they are ideally positioned for immune surveillance of the central nervous system (CNS) and its borders4. Meningeal immune cells, and their derived cytokines, have also been shown to affect brain functions, including sociability and spatial learning5,6. Here, we sought to identify and explore additional meningeal immune populations, with potential to impact brain functions. We uncovered the molecular mechanisms by which physiological levels of IL-17a derived from dural-associated γδ17 T cells control anxiety-like behavior in mice through neuronal IL-17Ra signaling. Our findings provide new insights into the neuroimmune interactions at the meningeal-brain interface and support further research into new therapies for neuropsychiatric conditions.
Results
Steady state meninges harbor a CNS-associated source of IL-17a.
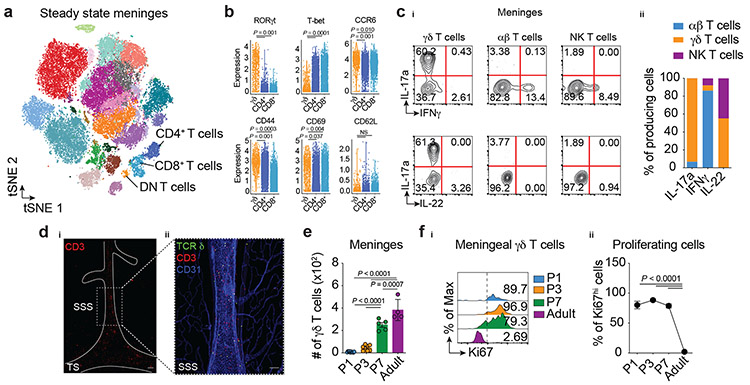
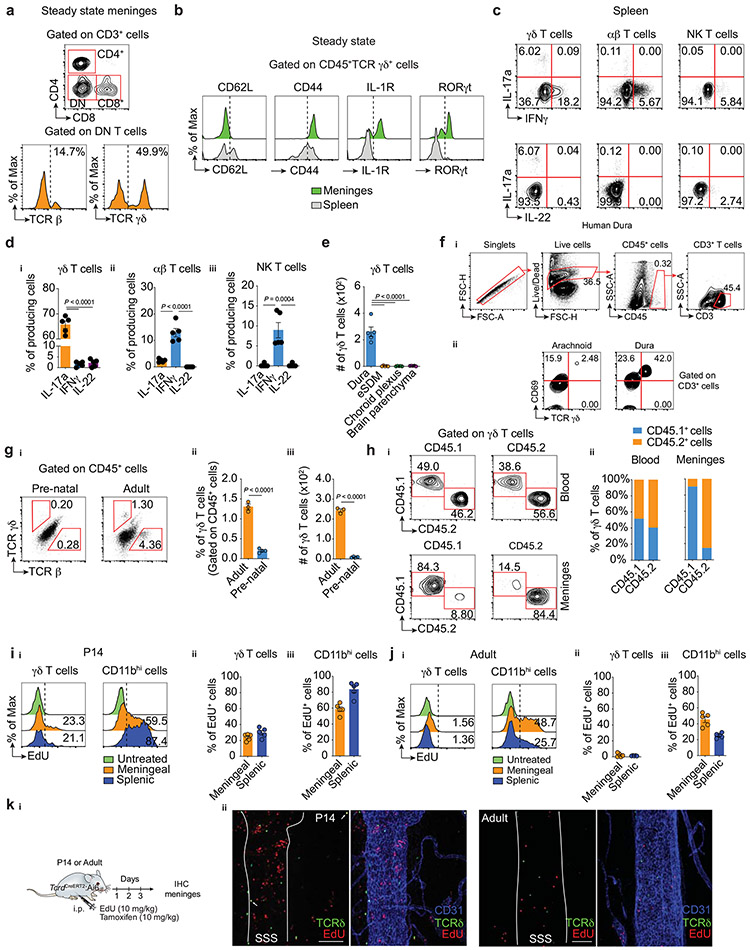
Using mass cytometry (CyTOF) to conduct a broad immunophenotyping analysis of the dural meningeal immune compartment, we identified a cluster of CD3+ T cells lacking expression of the conventional T-cell coreceptors CD4 and CD8 (Fig. 1a). Flow cytometric analysis revealed a high representation of γδ-T cell receptor (TCR)-expressing cells in the double-negative (DN) T-cell fraction (Extended Data Fig. 1a), a recently reported population in the meninges7,8. High dimensional analysis by CyTOF revealed a high proportion of γδ-TCR-positive cells expressing chemokine receptor CCR6, activation markers CD44 and CD69 and the lineage-defining transcription factor of IL-17a-producing γδ T cells (γδ17 T cells) RORγt, (Fig. 1b). Similar results were obtained by flow cytometry staining (Extended Data Fig. 1b). Mirroring their RORγt expression, the dural meninges demonstrated enrichment in γδ T cells expressing IL-17a upon ex vivo stimulation, which comprised more than 95% of the potential cytokine source in the meningeal space (Fig. 1c, Extended Data Fig. 1c-d).
Figure 1. ∣. Dural meninges harbor IL-17 producing (γδ17) T cells.
a, Mass cytometry analysis of immune cells isolated from meninges of naïve animals. Meningeal T cells (CD3+Thy1.2+) were clustered as CD4+, CD8+ or double negative (DN). b, Expression of T cell markers in meningeal γδ T cells, CD4+ and CD8+ αβ T cells analyzed by mass cytometry (n = 8 pooled mice; each dot represents one cell). Wilcoxon rank sum test with Benjamini Hochberg post hoc adjustments. Data represent one single experiment. NS, not significant. ci, Expression of IL-17a, IFNγ and IL-22 in meningeal γδ T cells, conventional αβ T cells (TCR β+NK1.1neg) and NK T lymphocytes (TCR β+NK1.1+) by flow cytometric analysis. cii, Contribution of each isolated population in total IL-17a, IFNγ and IL-22 expression in steady state meninges using backgating analysis. d, Meningeal whole-mounts stained for CD3 (red) and CD31 (blue) showing the presence of γδ T cells (CD3+TCR δ (ZsGreen)+ cells, in yellow). TS, transverse sinus, SSS, superior sagittal sinus. di, Scale bar, 200 μm and dii, 100 μm. Representative image of 3 independent experiments with similar results (n = 3 in each experiment). e, Absolute numbers of meningeal γδ T cells isolated from postnatal animals at P1, P3, P7 and adult (8-week-old) analyzed by flow cytometry. P1 (n = 10); P3 (n = 7); P7 (n = 5); and Adult (n = 5). One-way ANOVA followed by Tukey's multiple comparisons test fi, Ki67 expression in meningeal γδ T cells depicted in e. fii, Percentage of Ki67hi γδ T cells isolated from postnatal mice. One-way ANOVA followed by Bonferroni's multiple comparisons test. Data pooled from two independent experiments with similar results. Data are shown as mean ± s.e.m. and each dot symbol represents individual mouse.
To assess γδ T cell distribution in additional CNS-associated barriers, we micro-dissected border regions and performed flow cytometry analysis. γδ T cells were highly prevalent in the dura mater when compared to samples obtained from subdural meninges (leptomeninges, comprising of pia and arachnoid), choroid plexus, or the brain parenchyma (Extended Data Fig. 1e). Similar γδ T-cell populations were also detected in fresh autopsy samples of human dura, with fewer cells present in the arachnoid mater (Extended Data Fig. 1f).
To visualize the spatial localization of these intradural γδ17 T cells, we relied on whole-mount immunostaining of healthy mouse meninges. Since γδ T cell-receptor staining has proven to be challenging by commercial antibodies, we generated a tamoxifen-inducible TcrdCreERT2:Ai6 γδ T cell reporter line and through co-staining with anti-CD3, observed a high concentration of these cells restricted to regions along the dural sinuses (Fig. 1d). Collectively, these findings indicate that γδ17 T cells are situated in the dural meninges and constitute the nearest CNS-related potential source of IL-17a under steady state conditions.
γδ17 T cells populate meninges shortly after birth and are maintained by slow self-renewal.
γδ T cells are the first T cells generated during embryonic development and rapidly seed peripheral tissues where specialized subsets are maintained for life as tissue-resident cells. To determine the migratory kinetics of meningeal γδ17 T cells to the dura mater, we obtained tissues from prenatal, neonatal, and adult mice. Flow cytometry analysis revealed that γδ T cells were virtually absent in prenatal dural meninges (Extended Data Fig. 1g) but displayed a progressive seeding from postnatal day three with substantial numbers present by day seven (Fig. 1e).
In both humans and mice, γδ T cells represent a minor part of the circulating T cell compartment, however, certain subsets are present in much higher proportions in barrier tissues such as the skin, gut, and reproductive tract9. To determine whether meningeal (dural) γδ17 T cells are constantly replenished from the periphery or represent a long-lived population, we performed parabiotic experiments using congenic CD45.1 and CD45.2 mice. Unlike blood circulating γδ T cells, which reached near-complete chimerism after 4 weeks, meningeal γδ17 T cells were found to originate almost exclusively from the host mice, indicating a tissue-resident phenotype (Extended Data Fig. 1h).
Given the early post-natal seeding and the low chimerism between blood and meninges in adulthood, we hypothesized that highly active γδ T cells in neonatal meninges undergo rapid local expansion to fill the niche and reach homeostatic numbers in adult mice. In support of this idea, Ki67 staining revealed that meningeal γδ17 T cells exhibited high homeostatic proliferation at early postnatal periods (P1-7) yet maintained very low proliferation rates in adult mice (Fig. 1f). Corroborating the Ki67 staining, meningeal γδ17 T cells isolated from neonates displayed higher levels of in vivo EdU incorporation when compared to adult mice (Extended Data Fig. 1i-k). Taken together, these data indicate that γδ T cells populate the meninges at perinatal stages and may be maintained by low-rate self-renewal.
Molecular diversity and transcriptional landscape of meningeal γδ17 T cells.
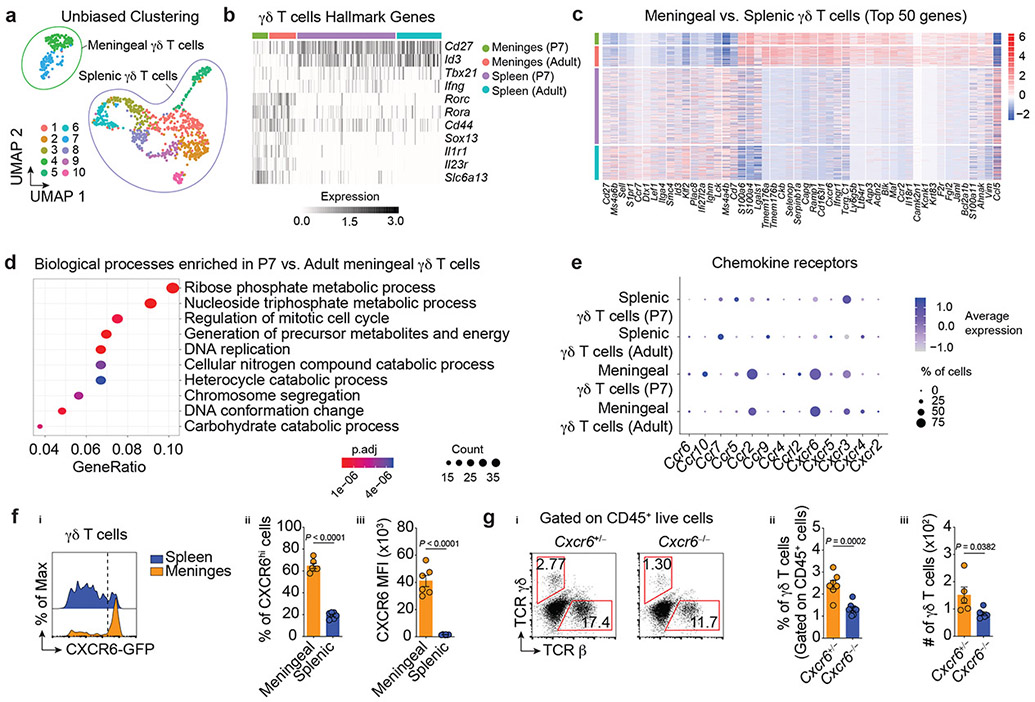
To gain further insight into the molecular diversity and transcriptional landscape of the meningeal γδ17 T cells, sorted γδ T cells from the dural meninges and spleen of P7 or 8-week-old adult mice were profiled using single-cell RNA-sequencing (scRNA-Seq). Uniform manifold approximation and projection (UMAP) for dimensionality reduction revealed 10 clusters, demonstrating heterogeneity within the γδ T-cell lineage (Fig. 2a). Nonetheless, this cluster distribution was clearly partitioned into two main subsets, based on meningeal or splenic origin (Fig. 2a), thereby supporting maximal similarity between meningeal γδ T cells regardless of the age of the mice.
Figure 2. ∣. Functional and molecular characterization of meningeal γδ17 T cells.
a, UMAP visualization of 1,041 γδ T cells isolated from adult and neonates (P7) colored by each cluster. b, Log normalized expression per cell of hallmark γδ T cell markers, with cells ordered by sample. c, Mean centered, log normalized expression per cell of the top fifty significantly differentially expressed genes (adj. P < 0.05) between adult meningeal and splenic γδ T cells ranked by absolute log fold change. d, Top ten enriched biological processes for the set of significantly differentially expressed genes (adj. P < 0.05) between adult and P7 meningeal γδ T cells using two-sided Fisher’s Exact test with Benjamini Hochberg corrections (FDR < 0.05). e, Expression of chemokine receptors in meningeal and splenic γδ T cells isolated from adult or P7 mice. fi, CXCR6-GFP expression on γδ T cells from adult mice by flow cytometry. fii, Percentage of CXCR6hi meningeal or splenic γδ T cells. fiii, Analysis of CXCR6-GFP MFI in γδ T cells. Meningeal (n = 6); Splenic (n = 6). Data is representative of 5 independent experiments. gi, Frequency of TCR γδ- or αβ-expressing cells isolated from CXCR6-sufficient or deficient mice. gii, Graph bars for the frequency of γδ T cells. Cxcr6+/− (n = 7); Cxcr6−/− (n = 8). Data are pooled from two independent experiments with similar results. giii, Absolute numbers. Cxcr6+/− (n = 5); Cxcr6−/− (n = 6). Data are pooled from two independent experiments with similar results. f-g, Data are shown as mean ± s.e.m. and each dot symbol represents one mouse. Unpaired two-tailed t test.
We then investigated the expression of known hallmark regulators of γδ T-cell subsets. In splenic populations the genes Cd27, Id3 and Tbx21, all characteristic of IFNγ-producing γδ T cells, were overrepresented (Fig. 2b). In contrast to splenic γδ T cells and in agreement with previous findings, we observed higher representations of transcripts for Rorc, Rora, Cd44, Sox13, Il1r1 and Il23r in the meningeal γδ17 T cells (Fig. 2b). Meningeal γδ17 T cells were also enriched for Tcrgv6 transcripts encoding the TCR-Vγ4 chain (Garman nomenclature, Extended Data Fig. 2a), which characterizes a subset of fetal thymus-derived γδ17 T cells9. Further assessment of the TCR-Vγ chain representation in the meningeal compartment using three commercially available flow antibodies (Vγ1, Vγ2, Vγ3) supported these findings (Extended Data Fig. 2b-c).
To identify unique meningeal γδ17 T cell-related transcripts, we assessed differentially expressed genes between the meningeal and splenic subsets. While meningeal γδ17 T cells shared many functional genes (such as Maf, Blk and Cd163l1) with their IL-17a-producing counterparts residing in other tissues, the functional roles of a sizeable number of genes remains unexplored (Fig. 2c). Notably, we noticed a substantial expression of Slc6a13 transcripts in meningeal γδ17 T cells (Fig. 2b, Extended Data Fig. 2d), a solute carrier responsible for the gamma-aminobutyric acid (GABA) and taurine transporter (GAT2)10, a function of which may relate to their location and should be further explored.
We next performed gene set enrichment analysis for gene ontology biological processes in the set of transcripts differentially expressed between meningeal γδ17 T cells isolated from P7 and adult mice. Our results demonstrated enrichment of biological processes that participate in cell cycle and DNA replication in the P7 meningeal γδ17 T cells (Fig. 2d). In summary, these findings reveal a transcriptional signature for meningeal γδ17 T cells and corroborate their higher activation status during early postnatal stages.
CXCR6 is highly expressed in meningeal γδ17 T cells.
Immune cell migration is mediated predominantly by the expression of chemokines and their receptors. Analysis of chemokine receptor expression in each γδ T cell population revealed a significant enrichment of CXCR6 (a chemokine receptor highly expressed in activated/memory T cells11) on meningeal γδ17 T cells when compared to the spleen (Fig. 2e). Using publicly available scRNA-Seq data8, we found that Cxcl16 (the ligand for CXCR6) was highly expressed in dural-resident myeloid cells (Extended Data Fig. 2e), indicating a potential receptor-ligand axis for its dural recruitment, retention and/or activation. In line with these scRNA-Seq results, we observed high CXCR6 expression on meningeal T cells (Extended Data Fig. 2f), with more than 60% of γδ17 T cells positive using flow cytometric analysis (Fig. 2f). To examine the functional role of CXCR6-CXCL16 axis for meningeal γδ17 T cell trafficking, we isolated the dural meninges from CXCR6–deficient or –sufficient mice. Significantly fewer meningeal γδ17 T cells were present in mice lacking CXCR6 in both frequency and absolute number (Fig. 2g). Interestingly, the number of conventional αβ T cells, was also decreased by CXCR6 deficiency (Extended Data Fig. 2g). In addition to CXCR6, ~20% of meningeal γδ17 T cells also expressed CCR2 (Fig. 2e, Extended Data Fig. 2h), however, Ccr2−/− mice displayed elevated T cell frequencies and numbers in the meningeal compartment (Extended Data Fig. 2i-j). Collectively, these findings suggest that CXCR6-CXCL16 axis plays a role in γδ17 T cell recruitment, retention and/or activation in the dural meninges. Further studies will address its cell-autonomous role, particularly during perinatal stages.
Meningeal γδ17 T cells control anxiety-like behaviors.
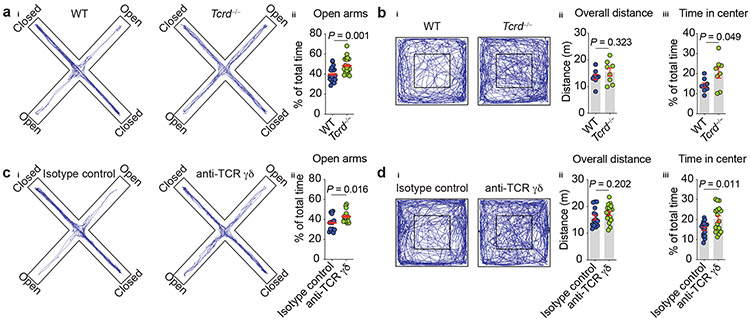
Given that aspects of meningeal immunity reportedly affect CNS function in both health and disease4,12,13, we investigated whether mice deficient in γδ T cells (Tcrd−/−) would exhibit any behavioral alterations. Profiling of Tcrd−/− mice in a battery of behavioral tests revealed no deficit in spatial memory task, social preference, or foraging behavior (Extended Data Fig. 2k-n). However, γδ T cell-deficient mice exhibited significantly increased exploration time in the open arms of the elevated plus maze test when compared to their wild type (WT) littermates (Fig. 3a). Typically, mice tested in the elevated plus maze naturally avoid unprotected open areas, favoring the darker and safer enclosed arms. This conflict between approach-avoidance paradigms capitalizes on the innate, conflicting drives of rodents to explore novelty and to avoid environmental threats – behaviors that are typically thought to reflect the anxiety state of the organism14.
Figure 3. ∣. Meningeal γδ17 T cells regulate anxiety-like behavior.
ai, Cumulative movement of wild type (WT) and γδ T cell-deficient (Tcrd−/−) mice in the elevated plus maze. aii, Percentage time spent in the open arms of the maze. WT (n = 18); Tcrd−/− (n = 20). Pool of two independent experiments. bi, Cumulated movement of WT and Tcrd−/− animals in the open field test. bii, Total ambulatory distance in the arena. biii, Percentage time spent in the center of the arena. WT (n = 8); Tcrd−/− (n = 8). c, WT mice were injected (i.c.m.) with 2.5 μg of anti-TCR γδ or isotype control and three days later were assessed in the elevated plus maze. ci, Cumulated movement of isotype control– or anti-TCR γδ–injected mice. cii, Percentage time spent in the open arms of the maze. Isotype control (n = 15); anti-TCR γδ (n = 16). Results pooled from two independent experiments. d, WT mice were injected anti-TCR γδ or isotype control and three days later were assessed in the open field task. di, Cumulative movement in the open field arena. dii, Total distance traveled. diii, Percentage time spent in the center of the arena. Isotype control (n = 16); anti-TCR γδ (n = 17). Data were pooled of two independent experiment. a-d, Data are expressed as mean ± s.e.m. and each dot symbol represents individual mouse. Unpaired two-tailed t test.
As another approach to measure anxiety-like behavior we assessed γδ T cell-deficient mice in the open field test, for which the typical tendency is to spend more time exploring the periphery and avoid the center of the arena15. In agreement with the elevated plus maze findings, the Tcrd−/− mice spent more time in the center of the arena than their WT counterparts (Fig. 3b), supporting the notion that mice lacking γδ T cells show decreased basal levels of vigilance, which we interpreted as decreased anxiety-behavior.
Given that Tcrd−/− mice lack γδ T cells in all tissues from birth, this model cannot be used to determine the specific contribution of the meningeal subset to the observed phenotype. Further, it could be argued that the lack of γδ T cells during ontogenesis may have pleiotropic effects, complicating our interpretation of the behavior phenotype. To rule out these possibilities and to target meningeal γδ17 T cells more specifically, we injected anti-TCR γδ antibodies into the cerebrospinal fluid (intra-cisterna magna, i.c.m.) of WT mice, and three days later evaluated their performance in the elevated plus maze and open field tests. Of note, this is currently the best available approach to primarily target meningeal immune cells and it has been previously demonstrated that antibodies and tracers injected into the CSF reach meningeal spaces16. While the complete pathway remains unknown, CSF macromolecules are likely reaching the dura by traversing the arachnoid layer underlying the dural sinuses17. Supporting the meningeal contribution to the observed behavioral phenotype, anti-TCR γδ treatment decreased anxiety levels of animals in both paradigms (Fig. 3c-d). Altogether, these findings suggested that meningeal γδ17 T cells might act as homeostatic regulators of anxiety-like behaviors.
Homeostatic activation of meningeal γδ17 T regulates anxiety-like behavior.
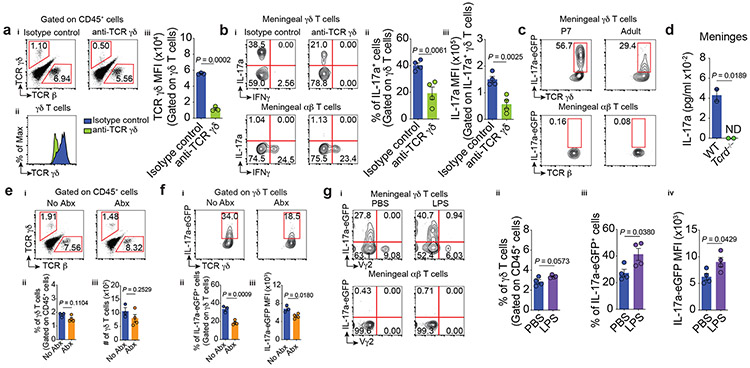
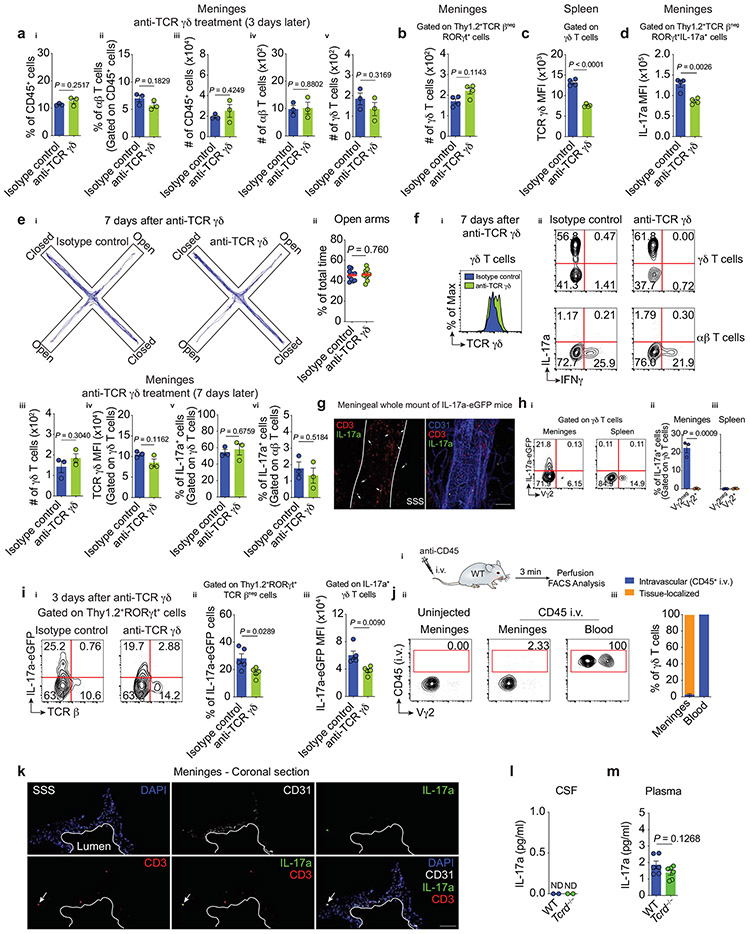
Having shown that the use of anti-TCR γδ antibodies decreased anxiety-like behaviors in mice, we next sought to determine how meningeal γδ17 T cells regulate this process. In vivo administration of anti-TCR γδ monoclonal antibodies was associated with internalization of TCR γδ, without actual loss of the meningeal γδ T cells (Fig. 4a, Extended Data Fig. 3a-b), similar to what has been previously shown in the intestine18. It is important to note that in addition to its primarily meningeal specificity, an off-target effect of anti-TCR γδ treatment was also observed in splenic γδ T cells (Extended Data Fig. 3c).
Figure 4. ∣. Steady state expression of IL-17a by meningeal γδ T cells is partially regulated by TCR engagement and commensal-derived signals.
a-b, WT mice were injected (i.c.m.) with 2.5 μg of anti-TCR γδ or isotype control and their meningeal immunity was analyzed three days later by flow cytometry.ai, Representative dot plots of meningeal TCR β- or TCR γδ-expressing cells. aii, Histogram of TCR γδ levels on meningeal γδ T cells. aiii, MFI of TCR γδ. Isotype control (n = 3); anti-TCR γδ (n = 3). bi, Representative contour plots of the percentage of IL-17a– and IFNγ–producing T cells. bii, Frequency of IL-17a+ meningeal γδ T cells. biii, MFI of IL-17a in IL-17a+ meningeal γδ T cells. Isotype control (n = 4); anti-TCR γδ (n = 4). c, Representative contour plots of IL-17a-eGFP expression in fresh meningeal T cell subsets by FACS. P7 (n = 3); Adult (n = 3). d, Steady state concentration of IL-17a in meningeal whole tissue lysates obtained from WT and Tcrd−/− mice by MSD assay. Technical replicate of meninges pooled from WT or Tcrd−/− mice (n = 10 per group); ND, not detected. ei, Percentage of conventional αβ and γδ T cells obtained from meninges of untreated (No Abx) or antibiotics-treated (Abx) animals. eii, Frequency and eiii, absolute number of meningeal γδ T lymphocytes. No Abx (n = 3); Abx (n = 4). fi, Percentage of IL-17a-eGFP+ meningeal γδ T cells from mice treated with Abx. fii, Frequency of IL-17a-eGFP + γδ T cells and fiii, IL-17a-eGFP MFI in IL-17a+ γδ T cells. No Abx (n = 3); Abx (n = 4). gi, Frequency of IL-17a–eGFP+ meningeal T cells four hours after i.p. injection with PBS or LPS (1 mg/kg). gii, Frequency of total meningeal γδ T lymphocytes, giii, IL-17a–eGFP+ innate-like γδ T cells (Vγ2neg). giv, IL-17a–eGFP MFI. PBS (n = 4); LPS (n = 4). Data are expressed as mean ± s.e.m. and each dot symbol represents individual mouse. Unpaired two-tailed t test.
We further hypothesized that meningeal γδ17 T cells with lower TCR levels on their cell surfaces would become poor responders to TCR-specific stimulation and hence would down-regulate the production of IL-17a. To this end, we repeated the i.c.m. injections of anti-TCR γδ and evaluated the meningeal T-cell subsets by flow cytometry. Administration of anti-TCR γδ decreased the percentage of meningeal IL-17a-producing γδ T cells as well as the cell-based production of IL-17a (Fig. 4b, Extended Data Fig. 3d). Importantly, TCR γδ expression, IL-17a production, and anxiety levels returned to normal one week later, which indicates γδ T-cell receptor recycling, antigen engagement, and reciprocal regulation of anxiety-like behaviors (Extended Data Fig. 3e-f). Collectively, these observations demonstrate that meningeal γδ17 T cells are crucial modulators of anxiety-like behaviors.
The aforementioned findings relied on ex vivo restimulation as a readout for cytokine production. However, this approach detects the potential, but not the true extent, of cytokine production in vivo. We utilized IL-17a-eGFP reporter mice to directly assess IL-17a transcription in vivo. Consistent with its steady-state expression, we observed a CD3+IL-17a-eGFP+ population by immunofluorescence, mainly confined to regions of the dural sinuses (Extended Data Fig. 3g), which mirrored the γδ T cell distribution (Fig. 1d). Flow cytometry analysis revealed restricted expression of IL-17a-eGFP in freshly isolated meningeal γδ17 T cells with virtual absence in their splenic counterparts (Fig. 4c, Extended Data Fig. 3h). Notably, this baseline expression was downregulated following anti-TCR γδ treatment (Extended Data Fig. 3i). Taken together, these findings demonstrate that meningeal γδ17 T cells are actively expressing IL-17a under steady state and that TCR engagement partially regulates this process.
Meningeal γδ17 cells were distributed along the dural sinuses, which drain blood from the brain into the internal jugular veins19. To determine whether the γδ17 T cells are tissue-localized and thus constantly release IL-17a within the meningeal stroma, we injected intravenously fluorescently labeled anti-CD45 antibodies and assessed γδ T cells for labeling. More than 99% of blood-circulating γδ T cells were stained with the i.v. anti-CD45, while only 1-2% of meningeal γδ17 were labelled (Extended Data Fig. 3j), suggesting their localization to dural parenchyma. Immunostaining of coronal sections of the dura mater obtained from IL-17a-eGFP mice confirmed this parenchymal localization (Extended Data Fig. 3k). IL-17a detection in dural meningeal homogenates from WT but not Tcrd−/− mice further suggests that IL-17a is likely released by γδ17 T cells in the meningeal stroma (Fig. 4d). It is noteworthy that CSF levels of IL-17a were below the technical limits of detection (Extended Data Fig. 3l), however its plasma levels were comparable between groups (Extended Data Fig. 4m). Collectively, these results demonstrate that meningeal γδ17 T cells are bona fide tissue-localized cells and the nearest CNS-associated source of IL-17a under homeostasis.
Molecular mediators regulating IL-17a production by meningeal γδ17 T cells.
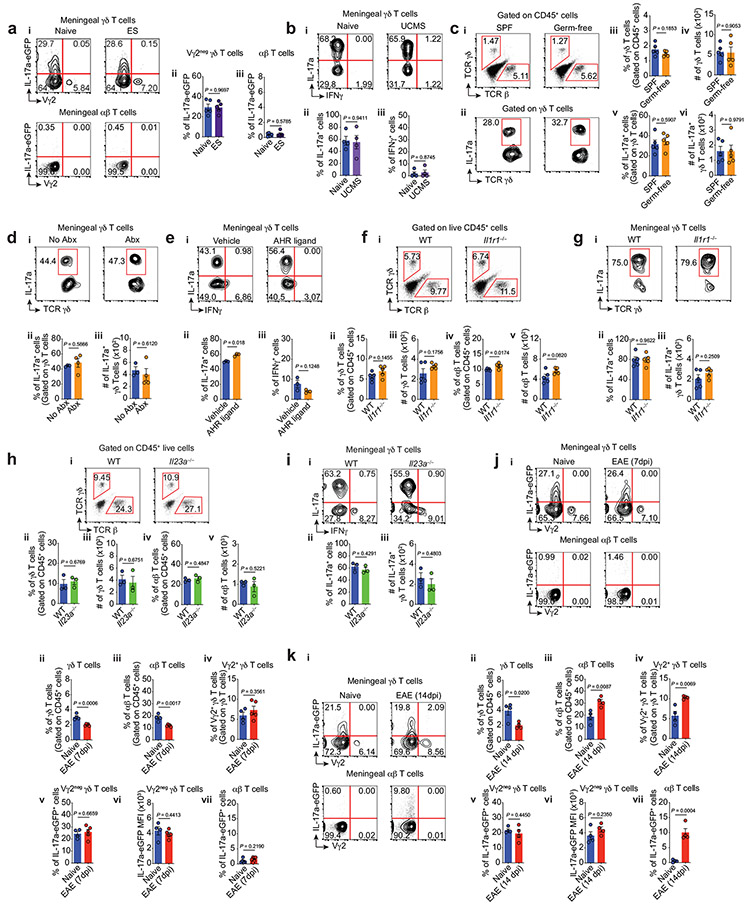
γδ T cells express many receptors that regulate their responsiveness to the environment9. As such, we next investigated the ability of several endogenous and exogenous stimuli to modulate IL-17a production in meningeal γδ17 T cells. Physical and mental stress have been frequently correlated with an increased risk to develop anxiety disorders15. However, neither seven consecutive days of electrical foot shock (ES) nor unpredictable chronic mild stress (UCMS) over 8 weeks modulated IL-17a expression in meningeal γδ T cells (Extended Data Fig. 4a-b).
While the identity of TCR ligands for γδ T cells have been obscure, some reports suggest that commensal bacteria are necessary for their expansion and activation, via direct or indirect pathways20. Abnormal behaviors have also been reported in the absence of microbiota, including decreased anxiety-like behavior in the elevated plus maze21. We next tested whether commensals may, in part, modulate IL-17a production by meningeal γδ17 T cells. Although the disruption of the commensal microbiota with broad-spectrum antibiotics and the use of germ-free mice did not affect meningeal γδ17 T cell numbers, we found a mild, but significant, decrease of IL-17a transcription by these cells (Fig. 4e-f, Extended Data Fig. 4c-d). Moreover, acute peripheral LPS injection, a pathogen-associated molecular pattern (PAMP), and stimulation of the aryl hydrocarbon receptor increased IL-17a expression in meningeal γδ17 T cells (Fig. 4g and Extended Data Fig. 4e, respectively). Deficiency of IL-1R or IL-23, known to promote IL-17a production in T cells, did not alter the number of γδ17 T cells or their IL-17a production following ex vivo stimulation (Extended Data Fig. 4f-i). Likewise, a chronic neuroinflammation characteristic of experimental autoimmune encephalomyelitis (EAE) had no effect on IL-17a expression by γδ17 (Extended Data Fig. 4j-k). Taken together, these findings suggest that meningeal γδ17 T cells are pre-programmed to quickly respond independently of microbiota and inflammatory signals (IL-1β and IL-23), but pathogen/commensal-derived signals affect their basal transcription of IL-17a.
IL-17a signaling through cortical neurons regulates anxiety-like behavior.
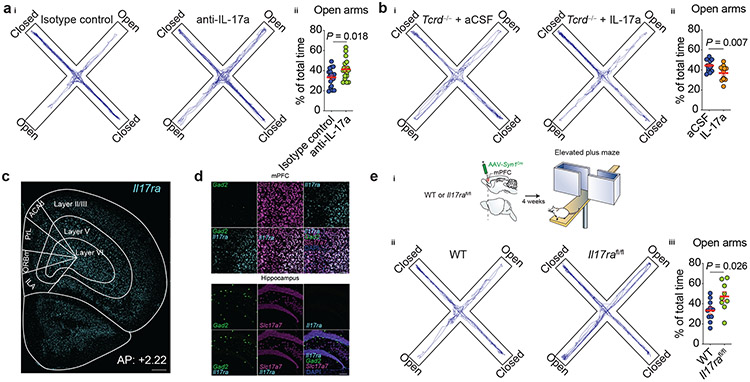
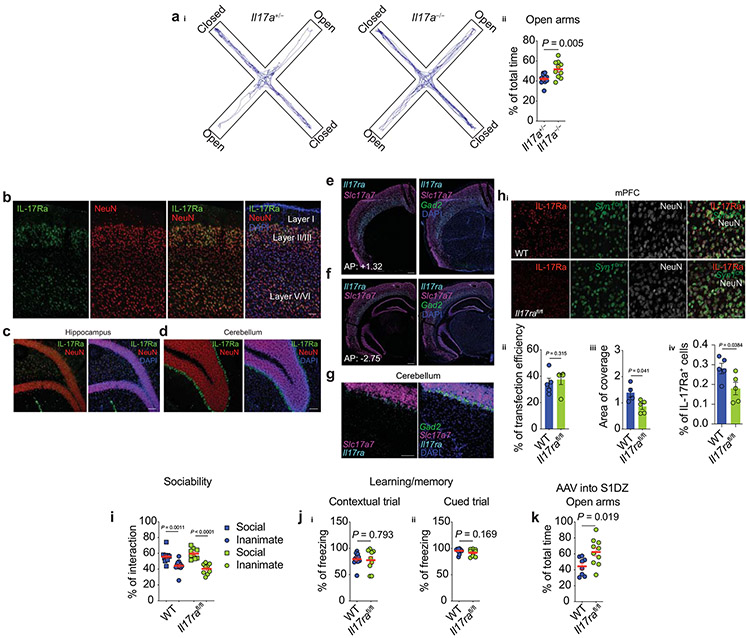
On the basis of our aforementioned finding of TCR-dependent downregulation of IL-17a by anti-TCR γδ-treatment in mice, we next sought to determine whether IL-17a deficiency would recapitulate the behavioral abnormalities observed after the “functional depletion” of meningeal γδ17 T cells. We first tested behavior in global IL-17a-deficient mice. In agreement with our previous observations, Il17a−/− mice spent significantly more time in the open arms than their heterozygous littermate controls (Extended Data Fig. 5a). Consistent with a meningeal-CSF route, injection of neutralizing antibodies for IL-17a into the cerebrospinal fluid (CSF) of WT mice decreased anxiety-like behavior (Fig. 5a). A single injection of recombinant IL-17a into the CSF of Tcrd−/− mice was sufficient to increase anxiety levels when tested in the elevated plus maze three hours after injection (Fig. 5b). Collectively, these findings further support the reciprocal link between meningeal γδ17 T cells, IL-17a production by these cells, and homeostatic brain function.
Figure 5. ∣. IL-17a signaling through cortical neurons regulates anxiety-like behavior.
ai, Cumulated movement of isotype control– or anti-IL-17a–injected mice in the elevated plus maze 14 h after treatment. aii, Percentage time spent in the open arms of the plus maze. Isotype control (n = 18) and anti-IL-17a (n = 18). Data were pooled of two independent experiments. b, Tcrd−/− animals were given 25 ng of recombinant IL-17a or artificial CSF (aCSF) via cisterna magna (i.c.m) and after three hours were tested in the elevated plus maze. bi, Total movement in the maze. bii, Percentage of total time spent in the open arms. aCSF (n = 14) and IL-17a (n = 12). Data were pooled of two independent experiments with similar results. c, Expression of Il17ra (Cyan) within the prefrontal cortex regions of naive mice according to cortical layers. ILA, infralimbic cortex; ORBm, medial orbifrontal cortex; PrL, prelimbic cortex; ACAd, dorsal anterior cingulate cortex. Scale bar, 500 μm. Representative image of 5 independent experiments (n = 3 in each experiment). d, Il17ra (Cyan) expression on GABAergic (Gad2, green) and glutamatergic neurons (Slc17a7, magenta) in the mouse mPFC (top) or hippocampus (bottom). DAPI (blue). Scale bar, 100 μm. Representative image of 3 independent experiments with similar results (n = 3 in each experiment). ei, Experimental approach used to ablate the IL-17a signaling in mPFC neurons. AAV Syn1Cre was bilaterally injected into the mPFC of WT or Il17rafl/fl mice. Mice were tested in the elevated plus maze four weeks later. eii, Total movement in the maze. eiii, Percentage of total time spent in the open arms of the maze. WT: AAV Syn1Cre (n = 10) and Il17rafl/fl: AAV Syn1Cre (n = 9). Data are expressed as mean ± s.e.m. and each dot symbol represents individual mouse. Unpaired two-tailed t test.
To assess a situation as threatening and render an anxiety-like response, an individual must first detect environmental stimuli through sensory systems and identify them as aversive or potentially dangerous14. An integral structure for threat interpretations is the medial prefrontal cortex (mPFC), which regulates subcortical responses to threatening stimuli14. IL-17a receptor subunit a (IL-17Ra) has been observed in cortical neurons in a mouse model of maternal immune activation (MIA)23. We therefore asked whether IL-17Ra is also expressed in mPFC neurons under steady state. In situ hybridization revealed high expression of Il17ra throughout all cortical layers of the mPFC, predominantly in excitatory glutamatergic neurons (Fig. 5c-d). Importantly, at steady state, IL-17Ra expression was found in multiple other brain areas (Extended Data Fig. 5b-g), but low to none expression was detected in the hippocampus (Fig. 5d).
Next we investigated whether the loss of neuronal IL-17a signaling could recapitulate the alleviated anxiety-like behavior. To this end, we injected virally expressed Cre recombinase under the synapsin I promoter (AAV Syn1Cre) into the mPFC of either WT or Il17rafl/fl mice, resulting in a significant reduction of IL-17Ra expressing neurons in Il17rafl/fl mice (Extended Data Fig. 5h). After four weeks the mice were tested in the elevated plus maze, and in agreement with our previous results, attenuation of neuronal IL-17a signaling sufficed to decrease anxiety-like behavior in mice (Fig. 5e), with no alterations in tests that assess sociability or memory (Extended Data Fig. 5i-j). Loss of IL-17Ra in the somatosensory cortex (S1DZ), previously implicated in neuronal IL-17a signaling to mediate social abnormalities1,23, also decreased anxiety levels of animals (Extended Data Fig. 5k), indicating that the behavioral phenotype observed in the absence of neuronal IL-17Ra is complex and is not limited to prefrontal cortex.
IL-17a signaling shapes the transcriptional landscape of mPFC neurons.
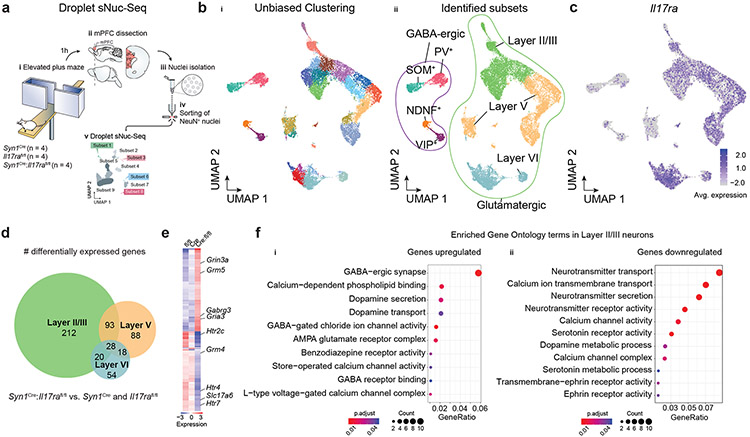
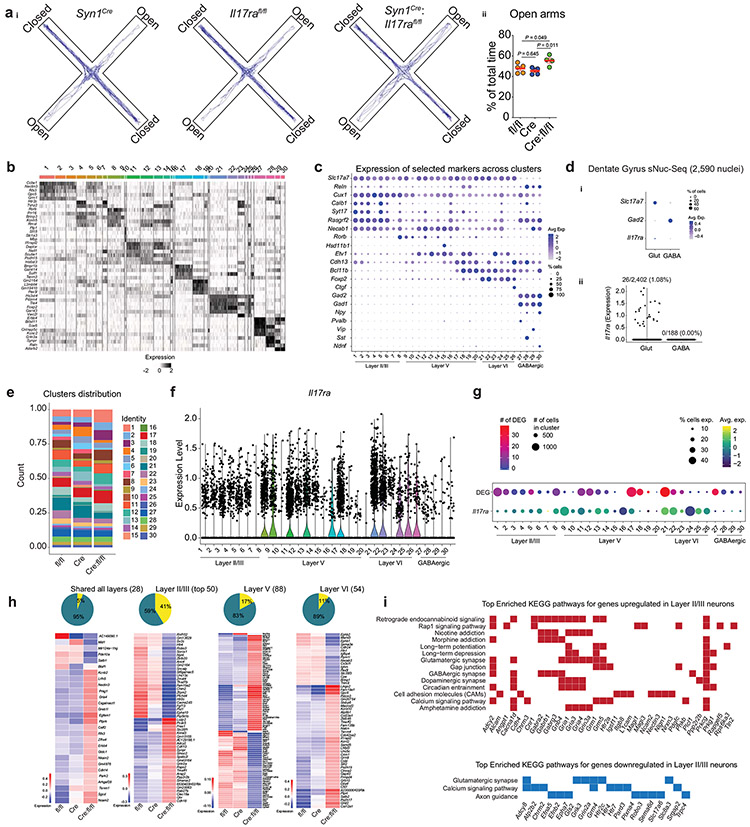
To assess the transcriptional landscape of neurons displaying a homeostatic loss of IL-17a signaling, we performed single-nuclei RNA-sequencing (sNuc-Seq) of NeuN+ nuclei from mPFC samples dissected from IL-17Ra- sufficient or deficient mice (Syn1Cre, Il17rafl/fl and Syn1Cre:Il17rafl/fl) one hour after they had been submitted to the elevated plus maze task (Fig. 6a, Extended Data Fig. 6a). Unbiased clustering analysis identified 30 cell clusters, corresponding to known neuronal subtypes (GABAergic and glutamatergic neurons) and from anatomically distinct cortical regions (II/III, V and VI) (Fig. 6b, Extended Data Fig. 6b-c). We found high similarity between Il17ra distribution in the sNuc-Seq (Fig. 6c) and our in situ hybridization studies (Fig. 5c-d). sNuc-Seq performed from samples isolated from the mouse hippocampus revealed very low to none neuronal Il17ra expression (Extended Data Fig. 6d), supporting the preferential IL-17Ra expression in cortical glutamatergic neurons.
Figure 6. ∣. Neuronal IL-17a signaling shapes the transcriptional landscape of mPFC neurons.
a, Schematic of the experimental approach used to perform single-nuclei RNA-Seq from mPFC neurons in the absence of neuronal IL-17a signaling. bi, UMAP projection of nuclei isolated from the mPFC colored by cluster membership. bii, UMAP projection indicating subpopulations of neurons based on expression of cortical layer, excitatory, and inhibitory genetic markers. c, UMAP projection colored by the expression of Il17ra. d, Overlap among cortical layers in significantly differentially expressed genes (DEG) found between both Syn1Cre:Il17rafl/fl vs. Syn1Cre and Syn1Cre:Il17rafl/fl vs. Il17rafl/fl based on the layers identified in bii. e, Heatmap of DEG between both Syn1Cre:Il17rafl/fl vs. Syn1Cre and Syn1Cre:Il17rafl/fl vs. Il17rafl/fl in Layer II /III. f, Significantly enriched gene ontology (GO) terms for the set of significantly up (i)- or down- (ii) regulated genes in Syn1Cre:Il17rafl/fl compared to both Syn1Cre and Il17rafl/fl in Layer II/III. Color indicating the Benjamini-Hochberg adjusted P value and dot size indicating the number of genes contributing to the enrichment of the term.
To better understand the transcriptomic changes associated with a lack of IL-17a signaling in mPFC neurons, we further investigated the differentially expressed genes (DEG) between groups. Overall, we did not find any striking difference in the representation of individual clusters (Extended Data Fig. 6e), but we observed over 300 DEG, in which the great majority were enriched in neurons annotated for the cortical layers II/III (Fig. 6d, Extended Data Fig. 6f-g). Notably, a significant proportion of these genes are also dysregulated in anxiety and depression disorders, including Htr2c, Htr4, Htr7, and Grm5 (Fig. 6e, Extended Data Fig. 6h). Gene ontology and KEGG pathway analysis performed with upregulated genes in the absence of neuronal IL-17a signaling revealed several enriched processes involved in γ-aminobutyric acid (GABA)-benzodiazepine activity (Fig. 6f, Extended Data Fig. 6i), the prototypical target system for anxiolytic drugs15. Altogether, these observations demonstrate that homeostatic neuronal IL-17a signaling shapes the transcriptional landscape of mPFC neurons and might be correlated with the behavioral changes in the anxiety state of the organism.
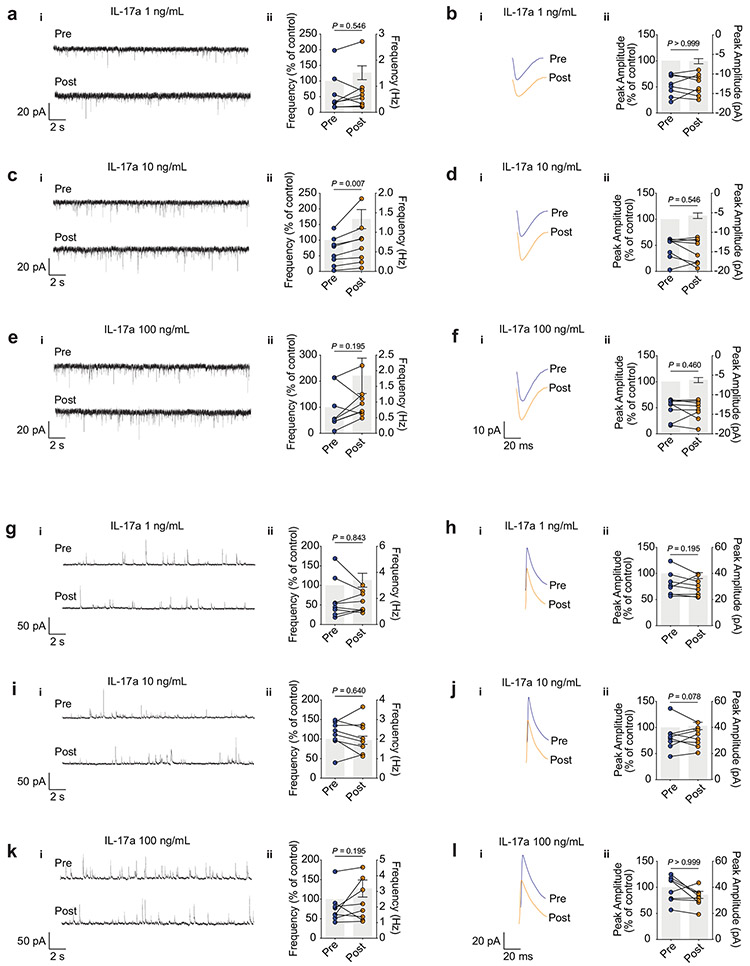
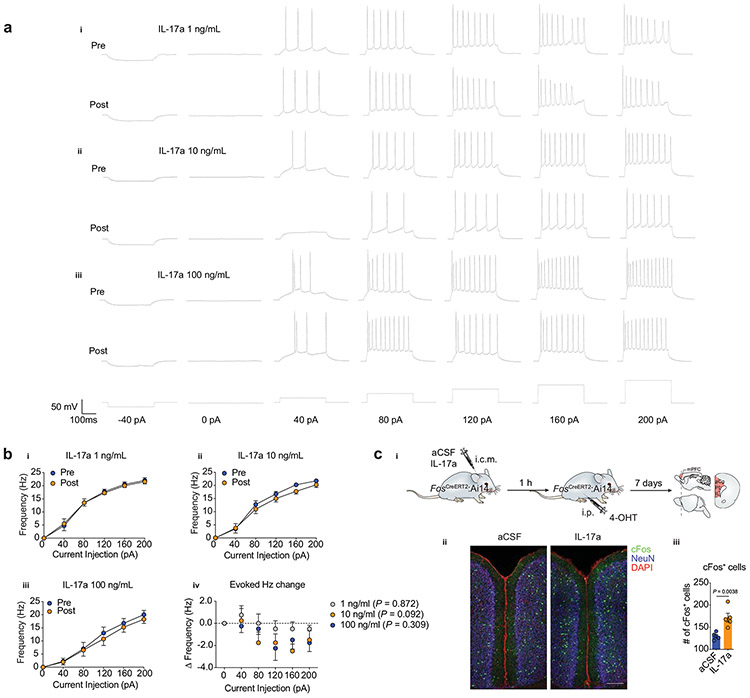
To explore whether these transcriptional changes occur in parallel with changes in synaptic transmission, we used whole-cell patch-clamp recordings from layer II/III mPFC neurons to measure the spontaneous excitatory or inhibitory post-synaptic currents (sEPSCs and sIPSCs respectively; Extended Data Fig. 7). Consistent with changes in the properties of neurotransmitter release, we observed a significant increase in the frequency, but not amplitude, of sEPSCs in neurons exposed to 10 ng/ml of IL-17a (Extended Data Fig. 7c and Extended Data Fig. 7d, respectively). No significant changes were found in any parameter evaluated for the sIPSCs (Extended Data Fig. 7g-l). We also saw no changes in the evoked action potentials (Extended Data Fig. 8a-b), which suggests that IL-17a signaling does not contribute to the intrinsic neuronal excitability. Additionally, IL-17a injection directly into the mouse CSF increased neuronal activation in the mPFC, as seen by the quantification of c-Fos+ cells, a marker of early neuronal activation (Extended Data Fig. 8c). These results suggest that the basal neuronal IL-17a signaling contributes to proper neurotransmitter release from excitatory presynaptic terminals of mPFC neurons, thus regulating anxiety-like traits in mouse.
Discussion
Here we provide molecular and functional insights into how a population of γδ17 T cells seeds the meningeal spaces and controls anxiety-like behavior in mice through neuronal IL-17Ra signaling.
Previously described non-immune properties of IL-17 included (i) the regulation of social behavior in MIA models1,23, (ii) body temperature24,25, and (iii) neuromodulation in Caenorhabditis elegans2. More recently, meningeal γδ T cells were linked to short-term memory via IL-17a signaling in glial cells7. In our hands, using another hippocampal-dependent task, γδ T cells knockout mice did not exhibit learning deficits, and IL-17Ra expression was highly concentrated to cortical neurons, with low to no expression in the hippocampus.
Injection of antibodies (anti-IL-17a and anti-TCR γδ) into the cisterna magna led to decreased anxiety-like behavior and suggested that the physiological release of IL-17a in the meningeal-brain interface contributes to the anxiety state of the organism. However, a peripheral leak of these antibodies does not rule out the contribution of blood-derived IL-17a in our behavioral readouts. Genetic deletion of neuronal IL-17Ra from the mouse mPFC, an integral structure for threat interpretations, recapitulated the behavioral changes observed in γδ T cell-deficient mice and supported a direct effect of IL-17a in neurons. Loss of endogenous IL-17a signaling revealed more than 300 regulated genes in mPFC neurons by sNuc-Seq, many of them enriched in processes involving GABA-benzodiazepine activity, the prototypical pathway for anxiolytic drugs. Further studies are necessary to fully address whether these transcriptional changes may have a causal connection with the behavioral readouts, and which are the downstream pathways following the basal IL-17a signaling in neurons.
Unexpectedly, ablation of neuronal IL-17a signaling in the primary somatosensory cortex (S1DZ) also led to similar results, possibly suggesting that anxiety readout is part of a complex behavioral phenotype observed in the absence of neuronal IL-17Ra, encompassing brain areas beyond prefrontal cortex. The complex role of IL-17Ra signaling in the brain has been previously appreciated in mouse models of autism. While embryonic IL-17Ra signaling on S1DZ results in social impairment1, same signaling during adulthood promotes sociability23.
Alleviated anxiety-like behavior has been previously linked to the lack of microbiota21. Interestingly, we found that commensal-derived signals regulate the transcriptional levels of IL-17a by meningeal γδ17 cells. This is in disagreement with the previously published work7, which may have missed the commensal participation by assessing IL-17a expression by ex vivo stimulation. Similar caveats should be noted with regard to our studies using IL-23- and IL-1R-deficient mice. It remains unclear how signals derived from commensals activate meningeal γδ17 T cells and whether such activation depends on TCR signaling. Further studies are needed to address how TCR specificity and diversity could modulate meningeal immunity and host behavior.
IL-17a is highly conserved across the evolution of the vertebrate immune system, probably owing to its key role in fighting infections at barrier sites. Anxiety is a normal component of the emotional repertoire, aiding survival by increasing awareness while also enabling rapid responses to environmental threats14. In the highly pathogenic ancestral environments, evolutionary conservation of tissue sentinels such as meningeal γδ17 T cells may have allowed organisms to respond rapidly to environmental stresses while also adapting to additional physiological processes, such as host alertness and exposure to external threats including predators and pathogens. In the modern world, given the growing interest in the involvement of IL-17a in neuropsychiatric conditions such as anxiety, depression and autism, our findings may shed new light on the understanding of these mechanisms and support further research on the development of new therapeutic targets.
Methods
Mice.
All mice (C57/BL/6J, TcrdCreERT2, Ai6, CD45.1, Cxcr6GFP, Tcrd−/−, Il17a−/−, Il1r1−/−, FosCreERT2, Ai14, Il17rafl/fl, Syn1Cre) were either bred in-house or purchased from the Jackson Laboratory. The Il17aeGFP and Il23a−/− (Genentech) mice were kindly donated by Dr. Alban Gaultier and Dr. Morgan Salmon respectively. Dr. Alex Kuan kindly donated the Ccr2CreERT2: Ai6 and Ccr2RFP/RFP strains. For the microbiota experiments, germ-free (GF) and specific-pathogen free (SPF) C57/BL6/J animals were purchased from Taconic and used immediately after arrival. When purchased from JAX, animals were maintained for at least 1 week to habituate before experimentation. All experiments were approved by the Institutional Animal Care and Use Committee of the University of Virginia and adhered to ethical consideration in animal research.
Human Samples.
Autopsy specimens of adult human dura and arachnoid (n = 2) were obtained from the Department of Pathology at the University of Virginia (UVA). All samples were from consenting patients that gave no restriction to the use of their body for research and teaching (through an UVA’s Institutional Review Board for Health Sciences Research). Patients had no previous record of neurological diseases and samples were processed as soon as four hours after patient death. Samples were physically minced before digested and processed as described in the flow cytometry section. Cell surface staining was performed at 4°C for 30 min. The list of antibodies is shown in Supplementary Information Table 1 and antibodies were diluted according to the manufacture’ instructions.
Antibiotic Treatment.
For ablation of commensal bacteria, an antibiotic cocktail of 1 g/L each of Ampicillin sodium salt (Sigma Aldrich), Neomycin trisulfate salt hydrate (Sigma Aldrich), Metronidazole (Sigma Aldrich), and 0.5 g/L Vancomycin hydrochloride (Alvogen) was used during 14 days, as previously described26. Antibiotics were replaced into the drinking water every other day. Antibiotic activity was confirmed at the day of the experiment by changes in cecum size due to bacterial death.
Intra-cisterna magna (i.c.m.) injections.
Mice were anaesthetized by intraperitoneal (i.p.) injection of a mixed solution of ketamine (100 mg kg−1) and xylazine (10 mg kg−1) in saline. The skin of the neck was shaved and cleaned with iodine and 70% ethanol, ophthalmic solution placed on the eyes to prevent drying and the head of the mouse was secured in a stereotaxic frame. After making a skin incision, the muscle layers were retracted, and the cisterna magna exposed. Using a Hamilton syringe (coupled to a 33-gauge needle), 5 μl of artificial CSF (Harvard Apparatus) containing cytokine or antibody was injected into the CSF-filled cisterna magna compartment. For meningeal γδ T cell “functional depletion”, it was injected 2.5 μg of anti-TCR γδ (UC7-13D5, BioXCell) or isotype control (polyclonal Armenian hamster IgG, BioXCell). For IL-17a blockade, it was used 2.5 μg of anti-IL-17a (17F3, BioXCell) or isotype control (MOPC-21, BioXCell). For IL-17a injections into Tcrd−/− animals, mice were deeply anesthetized with isoflurane and 25 ng of IL-17a (eBioscience) or aCSF were injected three hours before the behavior task. For the aryl hydrocarbon receptor (AHR) stimulation, 5 μl of the FICZ (10 ng, Tocris, #Cat 5304) was injected 24 h before euthanasia. After injections, the neck skin was then sutured, and mice were subcutaneously injected with ketoprofen (2 mg kg−1) and allowed to recover on a heat pad until fully awake.
Parabiosis.
Mice were anesthetized to full muscle relaxation with xylazine and ketamine by intraperitoneal injection. The corresponding lateral aspects of each mouse was shaved, and an incision was made from the olecranon to the knee joint of each mouse, bluntly dissecting the subcutaneous fascia to create 0.5 cm of free skin on each side of the incision. The olecranon and knee joints were attached by an absorbable 5-0 Vicryl suture. The dermis of the parabiotic partners were pushed together (excluding epidermal layers in the junction of the dermis) and closed with sutures. Post-surgery, systemic analgesics (Ketoprofen 2-5 mg kg−1 every 24h) were used along the intraperitoneal injection of antibiotics (Baytril, Bayer). All parabiotic pairs were sacrificed after four weeks, disjoined, and separately perfused.
Prefrontal cortex (PFC) AAV delivery.
Il17rafl/fl or C57BL/6 (WT) mice were anaesthetized by i.p. injection of ketamine and xylazine in saline and the head was secured in a stereotaxic frame. An incision was made in the skin to expose the skull and a hole was drilled bilateral at 2.22 mm in the anterior–posterior axis and 0.4 mm in the medial–lateral axis relative to bregma. Then, using a Hamilton syringe (coupled to a 33-gauge needle) placed at 2.5 mm in the dorsal–ventral axis (relative to bregma), 0.5 μl of artificial CSF of AAV9-hSyn1-eGFP-Cre (purchased from Addgene, Watertown, MA, USA) was injected at a rate of 0.5 μl min−1 into the brain parenchyma. The scalp skin was sutured, after which the mice were subcutaneously injected with ketoprofen (2 mg kg−1) and allowed to recover on a heat pad until further experiments.
Behavior.
Experimental groups were blinded and randomly assigned before the start of experimentation and remained blinded until all data were collected. Mice were housed under standard 12 h light/dark cycle conditions in rooms equipped with control for temperature and humidity. Most of our behavior studies were performed on male mice, and our key findings were also tested in female mice. We never mix the results from different genders. Unless stated otherwise, mice were tested at 8-10 weeks of age. Sample sizes were chosen on the basis of a power analysis using estimates from previously published experiments. For cohorts tested with multiple behavioral assays, the elevated plus maze was performed first and then followed by the open field before any other test. Before all experiments, mice were transported to the behavior room and given at least 30 min to habituate. All behavioral testing was conducted during daylight hours. More details regarding the behavior experiments are described in Supplementary Information Table 2.
Elevated plus maze.
Briefly, mice were transported to the testing room and habituated for at least 30 min. The room was maintained in dim light and a white noise generator used to mitigate any unforeseen noises. The plus maze consisted of four arms (two open without walls and two enclosed by 20 cm high walls) 35 cm long and 6 cm wide (Med associates, Inc.). Each arm of the maze was elevated 75 cm off of the floor. For the behavior analysis, mice were placed into the center hub and allowed to explore the plus maze for 5 min. Video tracking software (TopScan, CleverSys, Inc. and Noldus Ethovision) were used to quantify time spent in the open arms.
Open field.
Animals were previously habituated for at least 30 min in the testing room and then placed into the open field (35 cm x 35 cm) to explore for 15 min. Total distance and time spent in the center (23 cm x 23 cm) were quantified using video tracking software (TopScan, CleverSys, Inc. and Noldus Ethovision).
Novel location recognition (NLR) test.
The novel location recognition test was performed as described in27. Mice were firstly habituated to the apparatus for 15 min. After the habituation phase, two plastic objects (with different colors and shapes) were placed in the arena. Mice were then placed in the arena again and allowed to explore for 10 min. In the next day, one of the objects had switched location (novel) and the mice were allowed to explore the arena for other 10 min. The time spent exploring the objects in the familiar and novel locations was measured using a video tracking software (TopScan, CleverSys, Inc.). The object location preference (percentage of time with object) was calculated as the exploration time of the objects in the familiar or in the novel location/total exploration time.
Contextual Fear Conditioning (CFC) test.
CFC was performed as previously described with minor modifications27. Briefly, on day 1, mice were placed in the conditioning chamber and allowed to habituate for 3 min. Then, mice received three pairs of cue aversive stimuli, consisting of tone (18 s, 5 kHz, 75 dB)–shock (2 s, 0.5 mA) pairings, separated by an interval of 40 s (total of 3 min). On day 2, mice were tested and scored for conditioned fear to the training context for 3 min (context trial), in the absence of the cue stimulus. After 2 h, animals were presented to a novel context and allowed to habituate for 3 min. After the habituation phase, they received a continuous cue stimulus (tone) for an additional 3 min (cued trial). Mice behavior was recorded by a digital video camera mounted above the conditioning chamber and freezing was manually scored by a blinded experimenter using the Etholog v.2.2 software. Parameters analyzed included the percentage of time freezing during the 3 min of the context test and the last 3 min of the cued test.
Three-chamber sociability assay.
The three-chamber sociability test was conducted as previously published with minor changes6. Test mice were placed in the center chamber and allowed to explore for 10 min (habituation phase). After habituation, animals were returned to the center chamber. A C57BL/6J mouse (8- to 10-week-old) was placed under one cup and an object placed under the other. Tested mice were allowed to explore additional 15 min (social phase) and video tracking (TopScan, CleverSys, Inc.) was used to quantify the time spent around each target. The percentage of interaction was calculated as the exploration time in the mouse (social) or object (inanimate) chamber divided by the total exploration time.
Foraging behavior.
The foraging behavior was conducted as previously described with minor changes28. Briefly, mice were first habituated with sand (Jurassic play sand, Jurassic Sand) and seeds (Whole millet, Living Whole Foods) for two days in their home cage. For testing, mice were habituated into the behavior room and placed into the testing-cage attached to the arena 15 minutes prior to testing. At the start of testing, the testing-cage was attached to the arena via the tunnel, the mouse had access to the arena and video recording started for the Exploration phase. Mouse behavior was recorded continuously during the 30 min Exploration phase trial. After completing the Exploration phase, the mouse was transferred to a holding cage and four hours later tested for the Foraging phase. Video recording was also performed for additional 30 minutes. Between each Exploration and Foraging phase trial, the entire arena, including walls, platform, tunnel and steel pots, were wiped clean with 70% ethanol.
Unpredictable chronic mild stress (UCMS).
Briefly, mice were given daily unpredictable acute and overnight stresses for 8 weeks. Physical restraint, loud white noise, crowded housing, and strobe light were used as acute stressors. Mice were additionally submitted to overnight lights on during the dark cycle, wet bedding, cage tilting, frequent cage changing.
Acute stress.
Acute stress was induced by the electric shock model. Mice were individually placed in a chamber with a grid floor connected to a shock generator. The mice were exposed to a 3 s foot shock (0.6 mA) for 5 times during 120 s randomly for 7 consecutive days. For the control group, mice were placed in the chamber at the same time without foot shock.
EAE induction.
For active induction of EAE, mice were immunized by subcutaneous injection of 200 μg MOG35-55 (CSBio, Menlo Park, CA) in Complete Freund’s Adjuvant (CFA; Sigma Aldrich) and received 200 ng of pertussis toxin (List Biological Laboratories, Inc) intraperitoneally on days 0 and 2.
Meninges dissection and immunohistochemistry (IHC).
Mice were given a lethal dose of anesthetics by i.p. euthasol (10% v/v) and transcardially perfused with ice-cold 0.025% (w/v) heparin in PBS. Mice were decapitated immediately posterior to the occipital bone, and overlying skin and muscle was removed from the skull. The mandibles and skull rostral to maxillae was removed and the remaining skull was drop fixed in 4% paraformaldehyde (PFA) at 4 °C for 24 hours. The skull cap was then removed with fine surgical scissors by clockwise incisions, beginning and ending at the occipital bone, and was stored in 1X PBS 0.02% azide. The brain was removed, placed in 4% PFA for an additional 24 hours, then transferred to 30% sucrose in PBS until brains had completely sunk (24-48 hours). Brains were embedded in O.C.T. (Fisher Healthcare), rapidly frozen over dry ice and stored at −20 °C. Coronal cryosections (40 μm) were cut using a cryostat (Leica) and free-floating sections were stored in PBS with 0.02% azide until use. Meningeal whole mounts were prepared by careful peeling from the skull cap using fine surgical forceps as previously described19 and stored in 1X PBS 0.02% azide. Free floating brain sections and meningeal whole mounts were blocked and permeabilized for one hour at room temperature in 24-well plates with constant agitation using block/stain buffer (PBS with 0.2% Triton X-100 and 2% chicken serum). Sections were then incubated with primary antibodies in block/stain buffer at 4 °C for 24 hours with agitation, washed three times in PBS 0.2% triton (PBS-T), and incubated with secondary antibodies (1:500 dilution) for two hours at room temperature with gentle agitation. Sections were washed once in PBS-T and incubated with DAPI (1 μg/mL) in PBS-T. Sections were mounted on Superfrost Plus slides (Fisher Scientific), and coversliped with Aqua-Mount (Lerner). The list of antibodies used and their catalog numbers are presented in Supplementary Information Table 1.
In situ hybridization.
Brains were extracted and embedded in optimal cutting temperature (OCT) compound on dry ice and sections were cut at 16-μm thickness on a cryostat. In situ hybridizations were performed using the RNAscope Multiplex Fluorescent V2 Assay accordingly to the manufacturer’s recommendations (323100, Advanced Cell Diagnostics). For Il17ra staining we used the probe Mm-Il17ra-C1 (403741, Advanced Cell Diagnostics). The probe DapB (310043, Advanced Cell Diagnostics) was used as a negative control. For detection of glutamatergic and GABAergic populations we used the probes Mm-Slc17a7-C2 (416631) and Mm-Gad2-C3 (439371), respectively (both from Advanced Cell Diagnostics).
Confocal microscopy.
Meningeal whole mounts or brain slices were acquired with a Leica TCS SP8 confocal system (Leica Microsystems) using the LAS AF Software. Quantitative analysis of c-Fos imaging measurements was performed blinded with the FIJI package for ImageJ, using the cell counter. For the AAV-delivered experiments, IL-17Ra-positive cells were quantified in FIJI. Cells were segmented in max-projected images using the StarDist plugin29. Protein punctae were segmented with the ‘Analyze Particles’ function after 3D difference-of-Gaussian filtering with the CLIJ2 plugin30 and thresholding. The number of puncta was counted for each segmented cell, and further processing of the data was performed in R. For all measurements, 5 sites per brain/meninges were collected, and results were averaged to generate the value utilized for a single mouse.
c-Fos reporter activation.
4-hydroxytamoxifen (Sigma Aldrich, H6278) was dissolved at 20 mg/mL in ethanol by shaking at 37 °C for 15 min and was then aliquoted and stored at −20 °C for up to several weeks. Before use, 4-OHT was re-dissolved in ethanol by shaking at 37 °C for 15 min. Chen oil (1:4 mixture of castor oil (Both from Sigma Aldrich) was added to give a final concentration of 10 mg/mL 4-OHT, and the ethanol was evaporated by vacuum under centrifugation. To determine the number of “TRAPed” cells, Fos-CreERT2:Ai14 mice were injected i.c.m. with 25 ng of IL-17a (eBioscience) and one hour later were given an intraperitoneal injection of 10 mg/kg 4-OHT. Mice were killed and perfused seven days after injection for immunohistochemistry analysis.
Flow cytometry.
Mice were given a lethal dose of 10% (v/v) euthasol in saline (i.p.) and transcardially perfused with ice-cold 0.025% (w/v) heparin in PBS. Meninges were dissected as previously described and digested for 15 min at 37 °C with 1.4 U/mL of Collagenase VIII (Sigma Aldrich) and 35 U/mL of DNAse I (Sigma Aldrich) in IMDM (Sigma Aldrich) media. Following the digestion step, the tissue was gently pressed through 70 μm nylon mesh cell strainers. Cells were then centrifuged at 450g at 4°C for 4 min. The cell pellets were resuspended in ice-cold FACS buffer (2 mM EDTA, 25 mM HEPES, 1% BSA in 1X PBS) and stained for extracellular markers at 1:300 dilution. The list of flow antibodies used is presented in Supplementary Information Table 1. Samples were run on a flow cytometer Gallios (Beckman Coulter) then analyzed using FlowJo software v10 (Treestar).
Intracellular staining.
Intracellular staining was performed as described31. Briefly, single-cell isolates from meninges were stimulated 4 h in IMDM (supplemented with 1X non-essential amino acids, 50 U/ml penicillin, 50 μg/ml streptomycin, 50 μM β-mercaptoethanol, 1 mM sodium pyruvate, and 10% FBS, all for Gibco) with PMA/ionomycin (Cell Stimulation Cocktail – eBioscience) and 1X brefeldin A (eBioscience) at 37°C before extracellular staining as stated above. Cells were then permeabilized with Foxp3/Transcription Factor Staining Buffer Set (eBioscience) and stained for 30 min at 4 °C. All antibodies used were diluted at 1:250.
Labelling of the vascular compartment.
To assess the abluminal localization of the sinusal γδ T cells, mice were injected i.v. with 2.5 μg of eFluor 450-conjugated anti-CD45 antibody (eBioscience, clone 30-F11) 3 min before euthanasia.
In vivo cell labelling and EdU detection.
Mice received daily i.p. injections of 10mg/kg EdU (Carbosinth; #Cat 61135-33-9) during three consecutive days. EdU detection was performed using the Click-iT™ Plus EdU Alexa Fluor™ 647 Flow Cytometry Assay Kit and Click-iT™ Plus EdU Cell Proliferation Kit for Imaging, Alexa Fluor™ 647 dye, following the manufacturer’s instructions.
Mass cytometry.
Single cell suspensions of dural meninges were stained with metal-conjugated antibodies and reagents summarized in Supplementary Information Table 3. Each antibody was validated on meningeal tissue, titrated and optimal concentrations used in the panel. If not otherwise stated, cells were washed and stained in MaxPar (MP) buffers (Fluidigm). The entire meninges were transferred to a 96 well plate and incubated with 5 μM Cisplatin (Cell-ID; Fluidigm; diluted in MP PBS) for 5 minutes at room temperature. After two washes with MP Cell Staining Buffer (MP CSB), cells were pre-incubated with anti-mouse CD16/32 to block Fc receptors for 15 min on ice. An equal volume of a cocktail of fixation sensitive markers and fluorescent antibodies was added and the samples incubated on ice for an additional 25 minutes. Following two washes with MP CSB, cells were fixed in 1.6% EM-grade paraformaldehyde (Electron Microscopy) in MP PBS for 10 min at room temperature. Individual samples were barcoded with the Cell-ID 20-plex barcoding kit according to the manufacturer’s instructions (Fluidigm), combined and multiplexed samples stained with the remaining surface markers of the panel for 30 minutes on ice. Intracellular staining of markers within the nucleus were performed using the MP Nuclear Antigen Staining Buffer Set (Fluidigm) for 30 minutes on ice. Following two washes, cells were incubated in 1:2000 dilution of Ir intercalator (in MP Fix/perm buffer) overnight at 4°C. Prior to acquisition on a Helios mass cytometer, samples were washed with MP fix/perm, MP water and MP Cell acquisition solution. The stained and intercalated cell pellet was resuspended to a concentration of ~106 cells per ml in MP Cell acquisition solution and a five-element bead standard added.
Pre-processing of mass cytometry data.
Samples were debarcoded using the Zunder lab single-cell debarcoder (https://github.com/zunderlab/single-cell-debarcoder) in MATLAB and files uploaded in Cytobank. Raw data were manually gated to exclude debris, doublets, dead cells, normalization beads and live single CD45 cell events from Cytobank.
Automated population identification in high-dimensional data analysis.
Gated FCS files were read into R, and expression values for each marker were transformed using the arcsinh transformation. Cells were further analyzed as previously described in the Robinson workflow (https://bioconductor.org/packages/release/bioc/vignettes/cytofkit/inst/doc/cytofkit_example.html), using the Rphenograph clustering algorithm with all markers from the panel as input features32. Expert-guided definition of cell clusters was done based on heatmaps of median expression values of the initial RPhenograph nodes. Differential expression of specific markers between the identified clusters of CD4+ CD8+ and γδ T cells was tested using a pairwise Wilcoxon rank sum test with Benjamini Hochberg post hoc adjustments.
Cell sorting and scRNA-Seq analysis.
Seven days old (P7) or adult (8-week-old) C57BL/6 mice were given a lethal dose of 10% (v/v) euthasol in saline (i.p.) and transcardially perfused with ice-cold 0.025% (w/v) heparin in PBS. Meninges and spleen were harvested and physically minced before digested as previously described. A single-cell suspension was obtained and immunostained with Live/Dead Fixable Aqua Dead (eBioscience), anti-CD45 APC (BD Bioscience), anti-TCR γδ PE (Biolegend) and anti-TCR β FITC (Biolegend) using the BD Influx Cell Sorter (BD Biosciences) and sorted as CD45+TCR γδ+TCR βneg live cells. Cells were encapsulated in one lane of a 10x Chromium instrument, and libraries were constructed with a Single Cell 3′ Reagent Kit (V2 chemistry) following the manufacturer instructions. Libraries were then sequenced on the NextSeq 500 platform.
Nuclei isolation and sorting.
WT mice (n = 3) were sacrificed and transcardially perfused with ice-cold aCSF. The brains were collected, and 400 uM coronal sections were prepared using the vibratome. The dentate gyrus (DG) was dissected from the coronal sections and were placed in 1x HBSS. For the medial prefrontal cortex (mPFC) isolation, Syn1Cre (n = 4), Il17rafl/fl (n = 4) and Syn1Cre : Il17rafl/fl (n = 4) mice were firstly tested in the elevated plus maze task and one hour later sacrificed and transcardially perfused with ice-cold PBS-Heparin. The brains were collected, and the medial prefrontal cortex (mPFC) was isolated as prescribed by33. Briefly, coronal sections were made using a brain slicer matrix and a sharp razor blade. Sections were made until the anterior commissure is visible, and subsequent sections were prepared, which contain the darker area in the middle that represents the mPFC. The mPFC was then dissected from these sections and were placed in 1x HBSS on ice. The DG and mPFC sections were transferred in the nuclei isolation medium (0.25 M sucrose, 25 mM KCl, 5 mM MgCl2, 10 mM Tris-Cl (pH 7.4), 100 uM DTT, 1X Protease inhibitor (Roche) in PBS) and were triturated using a wide-bore pipette. The triturated tissue was gently homogenized using a Dounce tissue grinder. The homogenized tissue was filtered through a 70-μM nylon-mesh cell strainer and centrifuged at 1000 xg for 8 minutes at 4°C. The supernatant was removed carefully. The isolated nuclei were washed and resuspended in 1% BSA in PBS. They were subsequently stained with Hoechst 33342 (1: 1000) and anti-NeuN AlexaFluor® 647 (1: 500). The stained nuclei were washed with 1% BSA, pelleted, and resuspended in 1% BSA in PBS. They were sorted using the Influx Cell Sorter (Beckman Dickinson) available at the University of Virginia Flow Cytometry Core Facility. Intact nuclei were gated on the basis of positive Hoechst 33342 staining. From that population, singlets were isolated following gating on forward scatter height vs. forward scatter width and on forward scatter height vs. side scatter height. The NeuN+ nuclei were then sorted from that population into 1.5 mL LoBind tubes containing 400 uL of 0.04% non-acetylated BSA in PBS. After sorting, the nuclei were pelleted and were resuspended in 10-20 uL of 0.04% non-acetylated BSA in PBS for preparation of single-nuclei libraries using the 10X genomics platform.
Single cell data preprocessing.
Base call files were converted to Cellranger compatible fastq files using the Illumina bcl2fastq2 software. Reads were then aligned to the mm10 transcriptome using the Cellranger software pipeline (version 3.0.2) provided by 10x genomics, specifically the count function with the pre-mrna reference used for nuclei. The resulting raw gene by cell matrices of UMI counts for each sample, adult meningeal γδ T cells, P7 meningeal γδ T cells, adult splenic γδ T cells, and P7 splenic γδ T cells, were read into R using the read10xCounts function from the Droplet Utils package and filtered to remove barcodes which did not show significant deviations from the ambient RNA profile using the EmptyDrops method 34,35. The count-filtered matrices were then merged by gene symbol to create one matrix with all γδ T cells. Similarly, neuronal nuclei isolated from the prefrontal cortex were analyzed together, and neuronal nuclei isolated from the Dentate Gyrus comprised a third separate analysis. For each dataset, filtering was applied in order to remove low quality cells by excluding those with low UMI counts, low numbers of unique genes, or high levels of mitochondrial gene expression. Expression values for the remaining cells were then normalized using the scran and scater packages and the resulting log2 values were transformed to the natural log scale for compatibility with the Seurat (v3.0) pipeline36-38.
Dimensionality reduction and clustering of γδ T cells.
The filtered and normalized matrix was used as input to the Seurat pipeline and after a preliminary exploration of the dataset, one population of macrophages was identified based on the expression of H2.Aa, H2.Ab1, Ly6d, and Cd79a. After exclusion of this population, expression values for the remaining 1,041 cells were scaled across each gene and highly variable genes were identified using the FindVariableFeatures function with the variance stabilizing transformation method. The top 2000 highly variable genes were filtered to remove ribosomal and Riken genes and the remaining genes were used for Principal Components Analysis. Statistical significance for the first twenty components was calculated using the jackstraw test and the first eight components were used for tSNE analysis, UMAP, and clustering. Shared Nearest Neighbor (SNN) clustering optimized with the Louvain algorithm, as implemented by the Seurat FindNeighbors and FindClusters functions, resulted in 10 clusters and a modularity score of 0.8313.
Analysis of Neuronal Nuclei.
The filtered and normalized matrices of normalized counts were used to create Seurat Objects for each region (mPFC and DG). Each dataset was then scaled across all features to remove the effects of sequencing depth and mitochondrial gene expression. Variable genes for each dataset were identified using the variance stabilizing transformation method and the first 50 principal components were computed for each dataset using the top 2,000 highly variable genes. The number of principal components used for SNN clustering and for projection into two dimensions with the Uniform Manifold Approximation and Projection algorithm were determined based on significance as evaluated with the jackstraw test as well as based on the percentage of variance in the dataset explained by each component. Clusters were then identified as GABAergic or Glutamatergic based on their expression of canonical marker genes such as Gad1, Gad2, Slc17a7, and Slc17a6. In the dentate gyrus, three clusters of either Aqp4 or Olig2 expressing cells were removed to prevent potential contamination by glial populations and the remaining cells were reanalyzed using the same process as described. In the prefrontal cortex, glutamatergic clusters were further categorized into cortical layers using genes previously described in the literature, such as Cux1, Rasgrf2, Etv1, Cdh13, and Bcl11b. Descriptive markers for each cluster were identified using the FindAllMarkers with the Wilcoxon test and Bonferroni corrections to test genes with average log fold changes of at least 0.5 in the cluster of interest compared to all other clusters. For testing for differences between samples (Syn1Cre, Il17rafl/fl, and Syn1Cre:Il17rafl/fl) the log fold change threshold for testing was set to 0.1 and only genes expressed in a minimum of 10% cells were tested. For functional enrichment the clusterProfiler package was used to enrich the sets of genes that were significantly up or down regulated in both the comparison of Syn1Cre:Il17rafl/fl to the Syn1Cre control and the comparison of Syn1Cre:Il17rafl/fl to the Il17rafl/fl control for Gene Ontology (GO) terms or Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways.
Differential expression.
For all differential expression testing, the Wilcoxon rank sum test was used with the Bonferroni correction using the FindMarkers function. Unless otherwise stated previously, only genes showing a minimum log fold change of 0.1 between groups were tested for significance.
Pathway enrichment.
Fisher’s Exact test was used to determine significantly enriched GO biological processes, cellular components, and molecular for the sets of significantly differentially expressed genes (adjusted p < 0.05) for each comparison using the enrichGO function from the ClusterProfiler package 39,40. For each enrichment analysis the gene set size was set to a range of 10-500 genes and the Benjamini Hochberg adjustment was used to correct for false discovery rates. The enrichment results for meningeal γδ T cells (adult vs. pup) were simplified using the simplify function from the ClusterProfiler package to reduce redundant terms before filtering the top ten terms by significance for visualization. Enrichment results were visualized using the DOSE heatplot and dotplot functions. For enriched pathways visualized in the mPFC neuronal nuclei dataset, terms were chosen based on descriptions containing the following words or phrases: serotonin, kynurenine, ephrin, dopamine, calcium ion transport, calcium channel, neurotransmitter, NMDA, AMPA, GABA, benzodiazepine, postsynaptic density, postsynaptic membrane, axon, and potassium.
Whole-patch clamp recording.
Studies were conducted in collaboration with AfaSci (Redwood City, CA). C57BL/6 mice (Charles River, P9 to P58 days) were used in the experiments. Recombinant IL-17a (eBioscience) was prepared in PBS at a stock concentration of 0.1 mg/ml. Aliquots of the stock solutions were frozen at −20 °C until use. Artificial cerebrospinal fluid (aCSF) was prepared and contained (in mM): NaCl (124), KCl (2.5), CaCl2 (2.4), KH2PO4 (1.2), NaHCO3 (26), glucose (10), and MgSO4 (1.3). aCSF was bubbled with 5 % CO2 and 95 % O2 for at least 15 minutes prior to use and continuously throughout use. For brain slice preparation, aCSF was stored at −20 °C for 30 minutes until becoming a partially frozen slush. Mice were deeply anesthetized with isoflurane and then decapitated. The brains were quickly excised and transferred to the aCSF slush preparation and were allowed to chill for 1 minute. The brains were then transferred to a filter paper soaked with ice cold aCSF where the caudal region of the brain was removed by a coronal cut in order to attach, using super glue, the remainder of the brain flat on the specimen holding plate of the vibrating blade microtome (Leica VT1000s). The brains were bathed in ice cold/slush aCSF while preparing the slices. Coronal slices of the rostral brains were cut with a thickness of 250 μm and were transferred to an elevated mesh platform inside a glass beaker filled with continuously bubbled aCSF at a temperature of 20 to 25 °C. Slices were cut until the appearance of the genu corpus callosum connecting both hemispheres of the brain. Experiments were performed 2 to 4 layers prior to the final slice made in the mPFC. The slices were allowed to rest for at least 1 hour prior to performing electrophysiological recordings. Whole-cell responses were recorded using a MultiClamp 700B (Molecular Devices) amplifier and head stage and low-pass filtered at 10 kHz before digitization using a Digidata 1440 data acquisition system (Molecular Devices). Data was stored on a PC running pClamp software (version 10.4, Molecular Devices). Patch-pipettes were fabricated from 1.5 mm OD borosilicate capillary glass (Warner Instruments) using a micropipette puller (Sutter Instrument, Model P-87) and had tip resistances of 4-6 MΩ. The series resistance for all recordings ranged from 10 to 30 MΩ. Capacitance transients and series resistance errors were compensated for (70 %) using the amplifier circuitry for voltage clamp recordings. For spontaneous action potential, evoked action potential, and spontaneous excitatory post-synaptic current (sEPSC) recordings, the intracellular solution contained (in mM): K-gluconate (140), HEPES (10), EGTA (11), CaCl2 (0.5), MgCl2 (0.5), NaGTP (0.25), and MgATP (2). The internal solution was brought to pH 7.4 and had a liquid junction potential of 14.5 mV in the bathing solution. For inhibitory post-synaptic current (sIPSC) recordings, the intracellular solution contained (in mM): Cs-gluconate (140), HEPES (10), EGTA (11), CaCl2 (1), and MgCl2 (1). The internal solution was brought to pH 7.4 and had a liquid junction potential of 14.7 in the bathing solution. Brain slices in the recording chamber that was mounted on a phase contrast microscope (Olympus BX51WI) were perfused with aCSF (1 to 2 ml/min) continuously bubbled with 5 % CO2 and 95 % O2. Cells were identified using bright field phase contrast microscopy under the surface of the slices. Cells were patched in the whole-cell configuration and were held at −65 mV or −80 mV for spontaneous or evoked action potential recordings, respectively, in the current-clamp configuration, as well as −90 mV or −15 mV for excitatory or inhibitory post-synaptic current recordings, respectively, in the voltage-clamp configuration. Spontaneous EPSCs were observed as downward spikes that could be blocked by the addition of 30 μM DNQX. Spontaneous IPSCs were observed as upward spikes that could be blocked by the addition of 10 μM bicuculline. Data reduction for sampling intervals of 200 μs as well as low pass filtering was used to reduce noise in raw traces of sEPSC and sIPSC recordings. Spontaneous EPSC and IPSC events were measured using Clampfit’s ‘Template search’ program. Templates were created by selecting 5 to 10 events. EPSC and IPSC events were scored and manually verified. For each set of data, i.e. control and treatment at a particular concentration in the same cell, the same template and matching threshold was used for analysis. For tests with IL-17a, working concentrations (1, 10, and 100 ng/ml) were freshly made up in aCSF. Control spontaneous action potentials, sEPSCs, and sIPSCs were recorded for 5 to 10 minutes and drug solutions were applied for 20 to 25 minutes. The final 5 minutes of control application and 5 minutes of IL-17a application after a minimum of 15 minutes application was analyzed for spontaneous action potential, sEPSC, and sIPSC parameters. For evoked action potentials, a current injection step protocol (−40, 0, 40, 80, 120, 160, 200 pA; 500 ms) was run before and after 20 minute IL-17a application. Analysis was performed using Clampfit 10.4 and Clampfit 10.7.
Statistical analysis.
One-way ANOVA with appropriate multiple-comparison tests were used to compare three independent groups. Two-group comparisons were made using two-tailed unpaired Student’s t test. For comparisons of multiple factors, two-way ANOVA with appropriate multiple-comparisons tests were used. Statistical analysis (data are always presented as mean ± s.e.m.) was performed using Prism 8.0 (GraphPad Software, Inc.).
Extended Data
Extended Data Fig. 1. Meningeal γδ17 T cells are long-lived cells with low proliferative capacity in steady state.
a, Representative contour plot and histogram of the frequency of meningeal TCR β– or TCR γδ– expressing cells on the double negative (DN, CD4negCD8neg) T cell population under steady state by flow cytometry. b, Representative histograms of CD62L, CD44, IL-1R and RORγt expression in meningeal and splenic γδ T cells from naïve mice by flow cytometry. c, Representative contour plots of the intracellular staining for IL-17a, IFNγ and IL-22 in γδ T cells (TCR γδ+), conventional αβ T cells (TCR β+NK1.1neg) and NK T lymphocytes (TCR β+NK1.1+) isolated from spleen. d, Graph bars for the frequency of IL-17a, IFNγ and IL-22 for each isolated meningeal T cell subset. di, Meningeal γδ T cells; dii, conventional αβ T cells and diii, NK T lymphocytes (n = 5 per group). One-way ANOVA followed by Bonferroni's multiple comparisons test. e, Absolute number of γδ T cells isolated from dura, enriched subdural meninges (eSDM), choroid plexus or brain parenchyma under homeostasis (n = 5 per group). One-way ANOVA followed by Bonferroni's multiple comparisons test. fi, Gating strategy for the flow analysis of fresh dura sample isolated from humans. Single live cells were gated as CD45+ and the γδ T cell frequency was determined in the CD3+. fii, Contour plots of the frequency of γδ T cells isolated from arachnoid or dura mater by flow cytometry (n = 2). gi, Representative dot plots of the percentage of TCR γδ– or αβ– expressing cells obtained from meninges during prenatal stages (around E18.5) or at 8-weeks of age (Adult). gii, Graph bars of the percentage and giii, absolute number of γδ T cells. Each pre-natal sample represents a pool of three embryos. Unpaired two-tailed t test. Data are from one single experiment. hi, Representative flow cytometry analysis of γδ T lymphocytes in the blood (top), and meninges (bottom) of CD45.1+ and CD45.2+ congenic parabiotic pairs joined for 4 weeks (n = 4 per group). hii, Mean frequency of either CD45.1+ or CD45.2+ γδ T cells in each parabiotic pair in the blood and meninges. i, WT mice at post-natal day 14 (P14) or j, adult (8 week-old) were injected daily with EdU (i.p., 10 mg/kg) and three days later the proliferating EdU+ cells were determined by flow cytometry. ii, Histograms showing the EdU frequency in γδ T cells and CD11bhi myeloid cells isolated from meninges and spleen of P14 mice. iii, Graph bars of the percentage of γδ T cells or iiii, CD11bhi myeloid cells co–expressing EdU. P14 (n = 5 per group). ji, Representative histograms of γδ T lymphocytes and CD11bhi myeloid cells expressing EdU in meninges and spleen of adult mice. jii, Graph bars of the percentage of γδ T cells and jiii, CD11bhi myeloid cells co-expressing EdU. Adult (n = 5 per group). ki, Scheme of the experimental details used to assess the proliferation capacity of meningeal γδ T cells by immunohistochemistry. P14 or adult TcrdCreERT2:Ai6 mice were injected daily with EdU and tamoxifen (both i.p., 10 mg/kg) and three days later the proliferation of γδ T cells was determined in the whole-mount meninges by confocal microscopy. kii, Representative images of meningeal whole-mounts isolated from P14 (left) and Adult (right) mice and stained for CD31 (blue) and EdU (red). Arrows indicate γδ T cells proliferating in P14 meninges (TCR δ (ZsGreen)+EdU+). SSS, superior sagittal sinus. Scale bar, 100 μm. Representative image of one single experiment (n = 5). Data are shown as mean ± s.e.m. and each dot symbol represents individual mouse.
Extended Data Fig. 2. Meningeal γδ T cells expression profile and additional behavior paradigms using γδ T cell-deficient mice.
a, Normalized expression of Tcrgv6 transcripts (Vγ4 chain, Garman nomenclature) in meningeal and splenic γδ T cells obtained by single cell RNA-Seq. b, Graph bars of the TCR Vγ-usage in γδ T cells. bi, Meningeal and bii, splenic γδ T cells. biii, Overview of Vγ-chain usage (Vγ1, Vγ2, Vγ3 or others (Vγ4 or Vγ5)). ci, Representative contour plots of the intracellular staining for IL-17a and IFNγ in αβ T cells (TCR β+NK1.1neg) and γδ T lymphocytes (Vγ1, Vγ2, Vγ3–negative or positive fractions) isolated from steady state meninges. cii-v, Graph bars of the percentage of IL-17a– or IFNγ– producing cells in these different meningeal T cell subsets. One-way ANOVA followed by Bonferroni's multiple comparisons test. d, UMAP projection of γδ T cells colored by the scaled expression of Slc6a13 (GAT2). e, Average expression of chemokines in CD45+ cells isolated from steady state dura mater obtained from public database1. f, Representative histogram of the expression of CXCR6-GFP in meningeal T cell subsets. g, Absolute numbers of conventional αβ T cells isolated from CXCR6–sufficient and –deficient mice. Unpaired two-tailed t test. Data represent one independent experiment. hi, Adult Ccr2CreERT2:Ai6 mice were injected daily with tamoxifen (i.p., 10 mg/kg) and three days later meninges, spleen and blood were harvested for FACS analysis. Representative histograms of CCR2-ZsGreen expression in γδ T cells and conventional αβ T lymphocytes by flow cytometry. hii, Graph bars of the percentage of γδ T cell– and hiii, αβ T cell– expressing CCR2-ZsGreen in spleen, blood and meninges. hiv, Histograms of the expression of CCR2-ZsGreen in meningeal γδ and αβ T lymphocytes. hv, Frequency of CCR2–expressing cells. One-way ANOVA followed by Bonferroni's multiple comparisons test. ii, Representative dot plots of the percentage of meningeal TCR γδ– and TCR β– expressing cells isolated from WT or Ccr2−/− (Ccr2RFP/ RFP) mice. iii, Percentage and iiii, absolute number of meningeal γδ T cells. iiv, Percentage and iv absolute number of meningeal αβ T cells. Unpaired two-tailed t test. ji, Dot plots of the frequency of inflammatory monocytes (CD11bhiLy6Chi) isolated from WT or CCR2-deficient mice. jii, Percentage and jiii, absolute number of meningeal inflammatory monocytes. Data are pooled from two independent experiments with similar results. Unpaired two-tailed t test. k, Social interaction of wild-type and Tcrd−/− mice was assessed in the three-chamber test and the percentage of time investigating either a mouse (social) or an object (inanimate) was calculated. WT (n = 9); Tcrd−/− (n = 11). Two-way ANOVA followed by Bonferroni's multiple comparisons test. li, For the spatial memory task, WT and γδ T cell-deficient mice were assessed in the training and lii, novel location tasks of the novel location recognition (NLR) test and the time spent with either a familiar or novel object was determined. WT (n = 8); Tcrd−/− (n = 8). Two-way ANOVA followed by Bonferroni's multiple comparisons test. m, Foraging behavior of wild-type and Tcrd−/− mice assessed in the exploratory and n, foraging phase of the foraging paradigm. Data is pooled from three independent cohorts. WT (n = 18); Tcrd−/− (n = 18); Unpaired two-tailed t test. Data are shown as mean ± s.e.m. and each dot symbol represents individual mouse.
Extended Data Fig. 3. γδ17 T cells are localized in the meningeal stroma and actively transcribe IL-17a in vivo.
a, Flow cytometry analysis of meninges three days after treatment with 2.5 μg of isotype control or anti-TCR γδ (i.c.m.). ai, Percentage of CD45+ live cells. aii, Frequency of αβ T lymphocytes gated on the CD45+ fraction. aiii, Absolute number of CD45+ leukocytes, aiv, αβ T cells and av, γδ T cells in the meninges. Isotype control (n = 3); anti-TCR γδ (n = 3). b, Absolute number of γδ T cells of mice treated as described in a. To bypass their TCR staining, γδ T cells were gated as Thy1.2+TCR βnegRORγt+ cells. Isotype control (n = 4); anti-TCR γδ (n = 4). c, Levels of TCR γδ expression in splenic γδ T cells three days after treatment with 2.5 μg of isotype control or anti-TCR γδ (i.c.m.). Isotype control (n = 4); anti-TCR γδ (n = 4). d, Expression per cell of IL-17a in Thy1.2+TCR βnegRORγt+IL-17a+ T cells (“visible” and “invisible” γδ T cells). Isotype control (n = 4); anti-TCR γδ (n = 4). ei, WT mice were injected with 2.5 μg of anti-TCR γδ or isotype control (i.c.m.) and seven days later were assessed in the elevated plus maze. Representative track plots of the cumulated movement. eii, Percentage time spent in the open arms of the maze. Isotype control (n = 10); anti-TCR γδ (n = 10). fi, Representative histogram of the levels of TCR γδ expression in meningeal γδ17 T cells seven days after injection with isotype control or anti-TCR γδ. fii, Contour plots of the percentage of IL-17a– and IFNγ–producing meningeal T lymphocytes. fiii, Absolute number of meningeal γδ T cells. fiv, Analysis of the TCR γδ MFI (gated on γδ T cells). fv, Graph bars for the percentage of IL-17a–producing meningeal γδ T cells or fvi, αβ T cells. Isotype control (n = 3); anti-TCR γδ (n = 3). g, Homeostatic IL-17a-eGFP expression in meningeal T cell by immunohistochemistry using the IL-17a reporter mice (Il17aeGFP/eGFP). Insets of the superior sagittal sinus (SSS) of meningeal dural whole-mounts showing CD3 (red), IL-17a (eGFP, green) and CD31 (blue) staining in adult mice. White arrows highlight γδ T cells (defined by the CD3 and IL-17a double-staining). Scale bar, 100 μm. Representative image of at least three independent experiments with similar results (n = 3 per experiment). hi, Representative FACS plot of the IL-17a-eGFP expression of meningeal and splenic γδ T cells under steady state. hii, Graph bars of IL-17a–expressing cells in the meninges or hiii, spleen (n = 3 per group). ii, Representative dot plots of IL-17a-eGFP+ meningeal T cells three days after treatment with 2.5 μg of anti-TCR γδ treatment (i.c.m.). To bypass their TCR staining, “visible” and “invisible” γδ T cells were gated as Thy1.2+RORγt+ cells. iii, Frequency of IL-17a-eGFP+ meningeal γδ17 T cells (defined as Thy1.2+RORγt+TCR βneg cells). iiii, IL-17a expression per a cell basis on IL-17a-eGFP–expressing γδ T cells. Isotype control (n = 5); anti-TCR γδ (n = 5). ji, Scheme of the experimental approach used to label intravascular and tissue-localized cells in steady state meninges. WT mice were given 0.25 μg of anti-CD45 (i.v.) and 3 minutes later blood was collected. Animals were perfused, and meninges were harvested for flow cytometry analysis. jii, Representative dot plots and jiii, Graph bars of either intravascular (i.v. CD45+) or tissue-localized (i.v. CD45neg) γδ T cells (n = 4 per group). k, Representative meningeal coronal section of IL-17a-eGFP reporter mice stained for DAPI (blue), CD31 (white), and CD3 (red). Arrows indicate a T cell expressing IL-17a-eGFP in meningeal stroma. Scale bar, 100 μm. Representative image of one single experiment (n = 5). l, MSD assay of CSF obtained from WT and Tcrd−/− mice. Technical replicate of meninges pooled from WT or Tcrd−/− mice (n = 10 per group). ND, not detected. m, Steady state concentration of plasmatic IL-17a obtained from WT and Tcrd−/− mice by MSD assay (n = 6 per group). Data are shown as mean ± s.e.m. and each dot symbol represents individual mouse. a-f, h-i, and m, Unpaired two-tailed t test.
Extended Data Fig. 4. Pathways and stimuli involved in the activation of meningeal γδ17 T cells.
a, Stress was induced in IL-17a-eGFP mice by the exposure to electronical foot shock (ES) for seven consecutive days. ai, Representative dot plots of the IL-17a-eGFP expression by meningeal T cells by flow cytometry. aii, Frequency of IL-17a-eGFP+ γδ17 T cells or aiii, conventional αβ T cells. Data are from one single experiment. Naive (n = 5); ES (n = 5). b, Unpredictable chronic mild stress (UCMS) was induced for over eight weeks in WT mice. bi, Representative expression of IL-17a and IFNγ assessed by the intracellular staining following ex vivo stimulation. bii, Percentage of IL-17a– or biii, IFNγ– producing γδ T cells. Naive (n = 4); UCMS (n = 5). Data are from one single experiment. ci, Percentage of conventional αβ and γδ T cells isolated from meninges of specific pathogen free (SPF) and germ-free mice. cii, Representative contour plots of the percentage of IL-17a–positive meningeal γδ cells. ciii, Graph bars of the frequency and civ, absolute number of meningeal γδ T lymphocytes in SPF versus germ-free animals. cv, Analysis of the frequency and cvi, absolute number of meningeal γδ T cell-producing IL-17a. SPF (n = 5); Germ-free (n = 5). di, Contour plots of the percentage of IL-17a–positive meningeal γδ cells obtained from meninges of untreated (No Abx) or broad-spectrum antibiotics-treated (Abx) animals and assessed by ex vivo stimulation. dii, Graph bars of the frequency and diii, absolute number of IL-17a–producing meningeal γδ T lymphocytes. No Abx (n = 3); Abx (n = 4). ei, Representative contour plots of IL-17a and IFNγ expression by meningeal γδ T cells 24 h after aryl hydrocarbon receptor (AHR) activation by the endogenous ligand 6-Formylindolo[3,2-b]carbazole (FICZ, 10 ng, i.c.m.). eii, Analysis of the frequency of IL-17a– and eiii, IFNγ– producing cells assessed by ex vivo stimulation. N = 3 per group. fi, Representative dot plots of the percentage of TCR γδ- or αβ-expressing cells obtained from WT or Il1r1−/− mice. fii, Analysis of the percentage and fiii, absolute number of meningeal γδ T cells. fiv, Graph bars of the frequency and fv, absolute number of meningeal αβ T lymphocytes. WT (n = 5); Il1r1−/− (n = 5). gi, Contour plots of the IL-17a expression by meningeal γδ T cells following the ex vivo stimulation by PMA/Ionomycin. gii, Analysis of the percentage and giii, absolute number of IL-17a–producing γδ T cells. WT (n = 5); Il1r1−/− (n = 5). hi, Representative dot plots of the percentage of TCR γδ- or αβ-expressing cells obtained from WT or Il23a−/− mice. hii, Analysis of the frequency and hiii, absolute number of meningeal γδ T cells. hiv, Graph bars of the percentage and hv, absolute number of meningeal αβ T lymphocytes. WT (n = 3); Il23ra−/− (n = 3). ii, Contour plots of the IL-17a and IFNγ expression by meningeal γδ T cells following ex vivo stimulation. iii, Analysis of the percentage and iiii, absolute number of IL-17a–producing γδ T cells. WT (n = 3); Il23ra−/− (n = 3). ji, Representative contour plots of the percentage of IL-17a–eGFP+ meningeal T cells seven days after active EAE induction (EAE 7dpi). jii, Graph bars of the frequency of meningeal γδ T lymphocytes (total TCR γδ+ cells), jiii, conventional αβ T cells (TCR β+ cells) and jiv, Vγ2+ γδ T cells (gated on total γδ T cells). jv, Percentage of IL-17a–eGFP+ cells gated on innate-like γδ T cells (Vγ2neg) and jvi, expression per cell of IL-17a. jvii, Frequency of IL-17a–eGFP+ αβ T cells. Naive (n = 4); EAE (n = 5). ki, Representative contour plots of the percentage of IL-17a–eGFP+ meningeal T cells 14 days post EAE induction (EAE 14dpi). kii, Graph bars of the frequency of meningeal γδ T lymphocytes (TCR γδ+ cells), kiii, αβ T cells and kiv, Vγ2+ γδ T cells (gated on γδ T cells). kv, Percentage of IL-17a–eGFP+ cells gated on innate-like γδ T cells (Vγ2neg) and kvi, expression per cell of IL-17a–eGFP. kvii, Frequency of IL-17a–eGFP+ cells in αβ T cells. Naive (n = 4); EAE (n = 4). Data are shown as mean ± s.e.m. and each dot symbol represents individual mouse. a-k, Unpaired two-tailed t test.
Extended Data Fig. 5. Lack IL-17a signaling does not affect social or learning behaviors.
ai, Representative track plots of the cumulative total distance of Il17a+/− and Il17a−/− mice in the elevated plus maze test. aii, Percentage of total time spent in the open arms. Il17a+/− (n = 11) and Il17a−/− (n = 10). Unpaired two-tailed t test. b, Representative images of IL-17Ra (green), NeuN (red) and DAPI (blue) staining in the PFC, c, hippocampus and d, cerebellum of WT mice. b-d, Representative image of at least three independent experiments with similar results (n = 3 per experiment). Scale bar, 100 μm. e, Expression of Il17ra (Cyan) at anterior–posterior (AP) +1.32 mm, f, AP: −2.75 and g, in the cerebellum by in situ hybridization. e-f, Scale bar, 500 μm. g, Scale bar, 100 μm. Il17ra (Cyan) was co-labelled with GABAergic (Gad2, green), glutamatergic markers (Slc17a7, magenta), and DAPI (blue). e-g, Representative image of 5 independent experiments with similar results (n = 3 per experiment).hi, AAV Syn1Cre was bilaterally injected into the mPFC of WT or Il17rafl/fl mice. Four weeks later, mPFC slices were prepared for immunohistochemistry analysis. hii Representative images of IL-17Ra expression in the mPFC of WT or Il17rafl/fl. hii Analysis of the efficiency transfection. hiii Coverage of IL-17Ra staining and hiv Quantification of IL-17Ra-positive cells as a fraction of total NeuN+ cells. Scale bar, 30 μm. WT (n = 5) and Il17rafl/fl (n = 5), with 20 slices per mouse. Unpaired two-tailed t test. i, Sociability assay was performed in the three-chamber test and the percentage of time investigating either a mouse (social) or an object (inanimate) was calculated. WT (n = 9) and Il17rafl/fl (n = 11). Two-way ANOVA followed by Bonferroni's multiple comparisons test. ji, In the fear conditioning paradigm, the percentage of time freezing was evaluated in the context and jii, cued trial. WT: AAV Syn1Cre (n = 10) and Il17rafl/fl: AAV Syn1Cre (n = 9). k, AAV Syn1Cre was bilaterally injected into the S1DZ region of WT or Il17rafl/fl mice. Mice were tested in the elevated plus maze four weeks later. Percentage of total time spent in the open arms of the maze is presented. WT (n = 8) and Il17rafl/fl (n = 10). Unpaired two-tailed t test. Data represents one single experiment. Data is presented as mean ± s.e.m. and each dot symbol represents individual mouse.
Extended Data Fig. 6. Neuronal loss of IL-17a signaling changes the transcriptional landscape of mPFC neurons.
ai, Representative track plots of the cumulative total distance of Syn1Cre, Il17rafl/fl and Syn1Cre:Il17rafl/fl tested in the elevated plus maze test just before nuclei isolation for the scRNA-Seq experiment. aii, Percentage of total time spent in the open arms. Il17rafl/fl (n = 5); Syn1Cre (n = 5); and Syn1Cre:Il17rafl/fl (n = 4). Data represents one single experiment. Il17rafl/fl vs. Syn1Cre:Il17rafl/fl, P = 0.049; Syn1Cre vs. Syn1Cre:Il17rafl/fl; P = 0.011; One-way analysis of variance (ANOVA) followed by Bonferroni's multiple comparisons test. b, Heatmap showing cluster signatures across all mPFC nuclei. Each line represents the scaled expression of the corresponding gene within each nucleus, with more highly expressed genes shown in black. The top five most highly expressed genes from each cluster (compared against all other clusters) are shown. c, Dot plot showing the expression of canonical neuronal subclass markers across each of our Seurat identified clusters. Color represents the scaled average expression and size indicates the percentage of cells within each cluster expressing the gene. di, Dot plot showing the expression of Slc17a7, Gad2 and Il17ra among 2,590 nuclei isolated from the mouse hippocampus (dentate gyrus). Color represents the scaled average expression and size indicates the percentage of cells within each cluster expressing the gene. dii, violins show the normalized expression of Il17ra within each cell across clusters. e, Bar graph showing the proportion of cells from each sample belonging to each cluster across all mPFC nuclei. f, Violins show the normalized expression of Il17ra within each cell across clusters. g, Dot plot showing information about each cluster. In the top row, dot size indicates the total number of cells belonging to the cluster and color indicates the number of differentially expressed genes (DEG) overlapping between Syn1Cre:Il17rafl/fl vs. Syn1Cre and Syn1Cre:Il17rafl/fl vs. Il17rafl/fl (adjusted p value < 0.05, absolute log fold change > 0.1). In the second row, the size of the dot shows the percentage of cells within the cluster expressing Il17ra while the color represents the scaled average expression of Il17ra across cells in the cluster. h, Heatmaps showing overlapping differentially expressed genes between Syn1Cre:Il17rafl/fl vs. Syn1Cre and Syn1Cre:Il17rafl/fl vs. Il17rafl/fl that are significantly changed in all three layer subsets (far left), the top 50 (based on absolute average log fold change) differentially expressed genes that changed uniquely in Layer II/III (left), and the significantly differentially expressed genes that were uniquely changed in Layer V (right) and Layer VI (far right) with the loss of functional Il17ra. Above each heatmap is a pie chart showing the percentage of significantly differentially expressed genes combined across all layers, that belong to each category described (shared, unique to Layer II/III, unique to Layer V, or unique to Layer VI). i, Plots showing the most significantly enriched KEGG pathways based on the set of significantly up- (top) or down- (bottom) regulated genes shared between Syn1Cre:Il17rafl/fl vs. Syn1Cre and Syn1Cre:Il17rafl/fl vs. Il17rafl/fl with colored squares indicating the contribution of each gene to the corresponding enriched pathway.
Extended Data Fig. 7. IL-17a effects on electrophysiological parameters of mPFC neurons.
a-f, Spontaneous excitatory postsynaptic action potentials (sEPSCs) in the mouse mPFC before and after IL-17a application via bath-application for 20 to 25 minutes (n = 8 neurons). The data points represent sEPSCs frequency or amplitude on a given neuron pre- and post- IL-17a treatment (blue and orange dots respectively – plotted on the right y-axis). Light grey bars represent the frequency or peak amplitude (pA) as % of control in the given neuron (mean and s.e.m., plotted on the left y-axis). ai, Recordings of spontaneous action potentials before (pre) and after (post) IL-17a application (1 ng/mL). aii, Frequency of spontaneous action potentials pre and post treatment of 1 ng/mL of IL-17a. bi, Representative amplitude of the spontaneous excitatory postsynaptic action potentials in the mouse mPFC before and after application of IL-17a (1 ng/ml) as described before. bii, Analysis of the peak amplitude. ci, Recordings of spontaneous action potentials before and after IL-17a application (10 ng/mL). cii, Frequency of spontaneous action potentials pre and post treatment of 10 ng/mL of IL-17a. di, Representative amplitude of the spontaneous excitatory postsynaptic action potentials before and after application of IL-17a (10 ng/ml). dii, Analysis of the peak amplitude. ei, Recordings of spontaneous action potentials before and after IL-17a application (100 ng/mL). eii, Frequency of spontaneous action potentials pre and post treatment of 100 ng/mL of IL-17a. fi, Representative amplitude of the spontaneous excitatory postsynaptic action potentials before and after application of IL-17a (100 ng/ml). fii, Analysis of the peak amplitude. g-l, Spontaneous inhibitory postsynaptic action potentials (sIPSCs) in the mouse mPFC before and after IL-17a application via bath-application for 20 to 25 minutes (n = 8 neurons). gi, Recordings of spontaneous action potentials before and after IL-17a application (1 ng/mL). gii, Frequency of spontaneous action potentials pre and post treatment of 1 ng/mL of IL-17a. hi, Representative amplitude of the spontaneous inhibitory postsynaptic action potentials before and after application of IL-17a (1 ng/ml). hii, Analysis of the peak amplitude. ii, Recordings of spontaneous action potentials before and after IL-17a application (10 ng/mL). iii, Frequency of spontaneous action potentials pre and post treatment of 10 ng/mL of IL-17a. ji, Representative amplitude of the spontaneous inhibitory postsynaptic action potentials before and after application of IL-17a (10 ng/ml). jii, Analysis of the peak amplitude. ki, Recordings of spontaneous action potentials before and after IL-17a application (100 ng/mL). kii, Frequency of spontaneous action potentials pre and post treatment of 100 ng/mL of IL-17a. li, Representative amplitude of the spontaneous inhibitory postsynaptic action potentials before and after application of IL-17a (100 ng/ml). lii, Analysis of the peak amplitude. a-l, Wilcoxon matched-pairs signed rank test.
Extended Data Fig. 8. IL-17a effects on the evoked action potentials before and after IL-17a treatment.
ai-iii, Recordings of evoked action potentials in a neuron in the mouse mPFC before (pre) and after (post) IL-17a application (n = 8 neurons). The current injection protocol (−40, 0, 40, 80, 120, 160, 200 pA; 500 ms) is shown in gray underneath the traces. bi-iii, Graphs show the frequency of action potentials (y-axis) in response to different levels of current injection (x-axis) before and after IL-17a bath-application for 20 to 25 minutes. biv, Graph summarizes the change in frequency pre and post IL-17a treatment. Two-way ANOVA followed by Sidak's multiple comparisons test. Data points and error bars represent the mean and s.e.m., respectively, of n = 8 sets (Pre vs. Post IL-17a treatment).ci, Scheme of the experimental details to target IL-17a–responding cells. FosCreERT2:Ai14 mice were treated with 25 ng of IL-17a or saline (i.c.m.) and then given an injection of 4-hydroxytamoxifen (4-OHT, i.p.) one hour later. After seven days, animals were perfused and the mPFC was collected for immunohistochemistry staining and analysis. cii, Representative photomicrographs from the mPFC of mice injected with IL-17a or saline and stained for a neuronal marker, NeuN (blue), DAPI (red) and showing c-Fos expression (green). ciii, Quantification of c-Fos+ cells in the mPFC of stimulated mice. Scale bar, 200 μm. aCSF (n = 5); IL-17a (n = 5). Unpaired two-tailed t test. Representative image of two independent experiments with similar results (n = 5 per experiment). Data is presented as mean ± s.e.m. and each dot symbol represents individual mouse.
Supplementary Material
Acknowledgments
We would like to thank S. Smith for editing the manuscript and Jennifer Sokolowski for the help with the human samples’ collection. We thank all the members of the Kipnis lab and the members of the Center for Brain Immunology and Glia (BIG) for their valuable comments during multiple discussions of this work. We also thank UVA Flow Cytometry Core for help with cell sorting, the Genome Analysis and Technology Core for help with library preparation and sequencing, and Jacob Graham from Virginia Commonwealth University (VCU) School of Nursing, for performing the MSD assay. This work was supported by grants from the National Institutes of Health (MH108156, AT010416 and AG034113,) to J.K.
Footnotes
Data availability
Fastq files and quantified gene counts for single cell sequencing are available at the Gene Expression Omnibus (GEO) under accession number GSE147262.
Competing interests
J.K. is an Advisor to PureTech Health, Inc.
References
- 1.Choi GB et al. The maternal interleukin-17a pathway in mice promotes autism-like phenotypes in offspring. Science 351, 933–939 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen C et al. IL-17 is a neuromodulator of Caenorhabditis elegans sensory responses. Nature 542, 43–48 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller AH & Raison CL The role of inflammation in depression: from evolutionary imperative to modern treatment target. Nat. Rev. Immunol 16, 22–34 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alves de Lima K, Rustenhoven J & Kipnis J Meningeal Immunity and Its Function in Maintenance of the Central Nervous System in Health and Disease. Annu. Rev. Immunol 38, 597–620 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Derecki NC et al. Regulation of learning and memory by meningeal immunity: a key role for IL-4. J. Exp. Med 207, 1067–1080 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filiano AJ et al. Unexpected role of interferon-γ in regulating neuronal connectivity and social behaviour. Nature 535, 425–429 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ribeiro M et al. Meningeal γδ T cell–derived IL-17 controls synaptic plasticity and short-term memory. Sci. Immunol 4, eaay5199 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Hove H et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat. Neurosci 22, 1021–1035 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Vantourout P & Hayday A Six-of-the-best: unique contributions of γδ T cells to immunology. Nat. Rev. Immunol 13, 88–100 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Y et al. Deletion of the γ-Aminobutyric Acid Transporter 2 (GAT2 and SLC6A13) Gene in Mice Leads to Changes in Liver and Brain Taurine Contents. J. Biol. Chem 287, 35733–35746 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Unutmaz D et al. The Primate Lentiviral Receptor Bonzo/STRL33 Is Coordinately Regulated with CCR5 and Its Expression Pattern Is Conserved Between Human and Mouse. J. Immunol 165, 3284–3292 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Kipnis J Multifaceted interactions between adaptive immunity and the central nervous system. Science 353, 766–771 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Filiano AJ, Gadani SP & Kipnis J How and why do T cells and their derived cytokines affect the injured and healthy brain? Nat. Rev. Neurosci 18, 375–384 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Calhoon GG & Tye KM Resolving the neural circuits of anxiety. Nat. Neurosci 18, 1394–1404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griebel G & Holmes A 50 years of hurdles and hope in anxiolytic drug discovery. Nat. Rev. Drug Discov 12, 667–687 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Louveau A et al. CNS lymphatic drainage and neuroinflammation are regulated by meningeal lymphatic vasculature. Nat. Neurosci 21, 1380–1391 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ringstad G & Eide PK Cerebrospinal fluid tracer efflux to parasagittal dura in humans. Nat. Commun. 11, 354 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koenecke C et al. In vivo application of mAb directed against the γδ TCR does not deplete but generates “invisible” γδ T cells. Eur. J. Immunol 39, 372–379 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Louveau A et al. Structural and functional features of central nervous system lymphatic vessels. Nature 523, 337–341 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nielsen MM, Witherden DA & Havran WL γδ T cells in homeostasis and host defence of epithelial barrier tissues. Nat. Rev. Immunol 17, 733–745 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heijtz RD et al. Normal gut microbiota modulates brain development and behavior. Proc. Natl. Acad. Sci 108, 3047–3052 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adhikari A, Topiwala MA & Gordon JA Synchronized Activity between the Ventral Hippocampus and the Medial Prefrontal Cortex during Anxiety. Neuron 65, 257–269 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reed MD et al. IL-17a promotes sociability in mouse models of neurodevelopmental disorders. Nature 577, 249–253 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohlgruber AC et al. γδ T cells producing interleukin-17A regulate adipose regulatory T cell homeostasis and thermogenesis. Nat. Immunol 19, 464–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu B et al. γδ T cells and adipocyte IL-17RC control fat innervation and thermogenesis. Nature 578, 610–614 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References
- 26.Ivanov II et al. Specific Microbiota Direct the Differentiation of IL-17-Producing T-Helper Cells in the Mucosa of the Small Intestine. Cell Host Microbe 4, 337–349 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Da Mesquita S et al. Functional aspects of meningeal lymphatics in ageing and Alzheimer’s disease. Nature 560, 185–191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stacher Hörndli CN et al. Complex Economic Behavior Patterns Are Constructed from Finite, Genetically Controlled Modules of Behavior. Cell Rep. 28, 1814–1829.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmidt U, Weigert M, Broaddus C & Myers G Cell Detection with Star-Convex Polygons. in Medical Image Computing and Computer Assisted Intervention – MICCAI 2018 (eds. Frangi AF, Schnabel JA, Davatzikos C, Alberola-López C & Fichtinger G) vol. 11071 265–273 (Springer International Publishing, 2018). [Google Scholar]
- 30.Haase R et al. CLIJ: GPU-accelerated image processing for everyone. Nat. Methods 17, 5–6 (2020). [DOI] [PubMed] [Google Scholar]
- 31.de Lima KA et al. TGFβ1 signaling sustains aryl hydrocarbon receptor (AHR) expression and restrains the pathogenic potential of TH17 cells by an AHR-independent mechanism. Cell Death Dis. 9, 1130 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nowicka M et al. CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. F1000Research 6, 748 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spijker S, Faliagkas L & Rao-Ruiz P Dissection of Rodent Brain Regions: Guided Free-Hand Slicing and Dissection of Frozen Tissue. In Neuroproteomics (ed. Li KW) vol. 146 7–19 (Springer; New York, 2019). [Google Scholar]
- 34.Lun ATL et al. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffiths JA, Richard AC, Bach K, Lun ATL & Marioni JC Detection and removal of barcode swapping in single-cell RNA-seq data. Nat. Commun 9, 2667 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lun ATL, McCarthy DJ & Marioni JC A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Research 5, 2122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCarthy DJ, Campbell KR, Lun ATL & Wills QF Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics btw777 (2017) doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu G, Wang L-G, Han Y & He Q-Y clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol 16, 284–287 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu G, Wang L-G, Yan G-R & He Q-Y DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics 31, 608–609 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.