Abstract
Drug-related kidney stones are a diagnostic problem, since they contain a large matrix (protein) fraction and are frequently incorrectly identified as matrix stones. A urine proteomics study patient produced a guaifenesin stone during her participation, allowing us to both correctly diagnose her disease and identify proteins critical to this drug stone-forming process. The patient provided three random midday urine samples for proteomics studies; one of which contained stone-like sediment with two distinct fractions. These solids were characterized with optical microscopy and Fourier transform infrared spectroscopy. Immunoblotting and quantitative mass spectrometry were used to quantitatively identify the proteins in urine and stone matrix. Infrared spectroscopy showed that the sediment was 60 % protein and 40 % guaifenesin and its metabolite guaiacol. Of the 156 distinct proteins identified in the proteomic studies, 49 were identified in the two stone-components with approximately 50 % of those proteins also found in this patient’s urine. Many proteins observed in this drug-related stone have also been reported in proteomic matrix studies of uric acid and calcium containing stones.
More importantly, nine proteins were highly enriched and highly abundant in the stone matrix and 8 were reciprocally depleted in urine, suggesting a critical role for these proteins in guaifenesin stone formation. Accurate stone analysis is critical to proper diagnosis and treatment of kidney stones. Many matrix proteins were common to all stone types, but likely not related to disease mechanism. This protocol defined a small set of proteins that were likely critical to guaifenesin stone formation based on their high enrichment and high abundance in stone matrix, and it should be applied to all stone types.
Keywords: Guaifenesin, Kidney stones, Proteomics, Stone analysis, Urinary proteins, Stone matrix
Introduction
Kidney stones affect about 10 % of the general population and are typically composed of crystalline components and organic matrix (largely protein with small amounts of other organic materials and cellular debris) [1, 2]. Characterization of the crystalline components (typically calcium oxalate, calcium phosphate, uric acid, or admixtures of these components) [3] remains critical to the proper diagnosis and treatment of affected individuals in current clinical practice. However, a recent survey of stone analysis from a selection of commercial laboratories was surprisingly inaccurate, even for typical stone crystals [4]. Stone characterization is further complicated by the observation of relatively rare matrix stones that are almost exclusively composed of macromolecular components [5, 6]. Drug-induced kidney stones often suffer the same diagnostic fate as matrix stones as they are both poorly imaged by standard radiologic methods and often misidentified by stone analysis laboratories with inaccurate or incomplete reference libraries. Since drug-induced stones frequently contain a large fraction of matrix, they are frequently assigned to the matrix stone category.
We and others have hypothesized that the protein components of stone matrix play a critical role in stone formation. Proteins can act as molecular templates or inhibitors for crystal nucleation, growth and aggregation, and they can affect crystal attachment to cells and tissues [2]. Earlier reports demonstrated that proteins comprise the majority of matrix [7, 8]. More recently, the matrix proteome has been characterized using mass spectrometry-based techniques, with more than one hundred individual proteins reported in association with various stone crystals [9–11]. Unfortunately, the large number of proteins observed obscures their roles in stone formation, but suggests that many proteins are likely adventitiously included in matrix. A critical need remains to find methods to not only identify proteins that regulate stone formation, but also to define the mechanism by which they regulate this process.
We present here a detailed analysis of the urine and matrix proteomics of samples from a guaifenesin-associated stone. Since the first report of guaifenesin renal stones in 1999 [12] other publications have further characterized the drug and its metabolite guaiacol in nephrolithiasis (Fig. 1), but they remained focused on the crystal-forming components [13, 14]. In this study, we had the “stone” and the urine from which it formed, as well as the same patient’s normal baseline urine. We were therefore able to identify proteins critical to the guaifenesin stone formation based on their selective enrichment in stone matrix and reciprocal depletion in urine. The proteomic comparison of a patient’s urine with their stone matrix outlines a new protocol for stone matrix characterization that will help identify proteins critical to stone formation and reinvigorate this aspect of stone research.
Fig. 1.
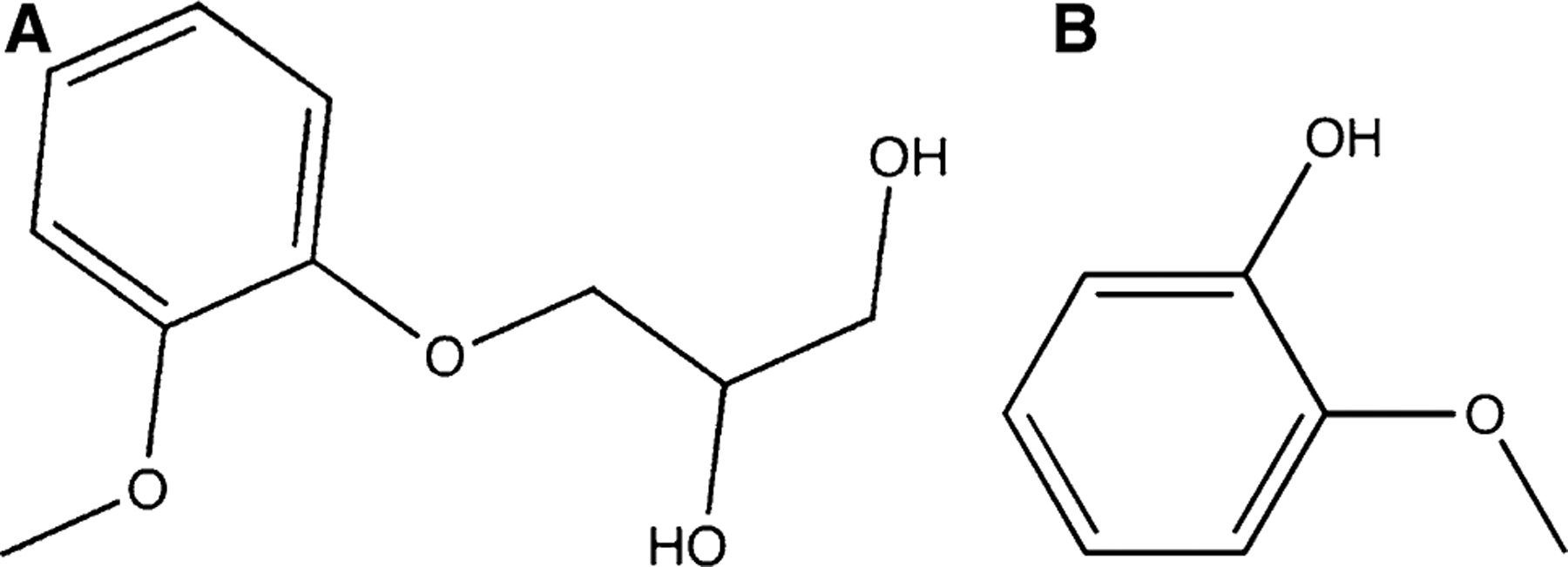
Drug chemical structure. A Guaifenesin and B guaiacol a metabolite of guaifenesin
Materials and methods
Patient history
All samples were obtained from a 30-year-old Caucasian woman who presented for the evaluation and treatment of recurrent matrix stones, and she was recruited to our IRB-approved study of urine proteomics (VA-IRB protocol: 9305-01P). Her initial diagnosis of stone disease occurred 3 years earlier, when she presented with lower abdominal pain, dysuria, and inability to void. Abdominal CT scan showed bilateral obstructing stones (7 mm right and 8 mm left) and serum studies showed acute kidney injury (creatinine = 5.3 mg/dl). Bilateral stent placement resolved her symptoms and restored normal renal function. Stone analysis from an outside clinical laboratory identified the sediment as 100 % mucin. During the next 3 years she continued to have intermittent attacks of abdominal pain and dysuria with resolution of symptoms upon passing gravel-like material.
Her other medical history was significant only for amenorrhea and a thin build (BMI = 21.6). She admitted smoking a half pack of cigarettes per day for 15 years, but denied the use of alcohol or illicit drugs. Laboratory investigations showed normal serum electrolytes and normal renal function. A urine toxicology screen was positive for methamphetamine and amphetamine. Her prescription medications included potassium citrate and allopurinol for her stones, along with over the counter multivitamins and a daily salicylate plus methenamine tablet for bladder pain relief. She reported occasional allergy symptoms, but only admitted later to using guaifenesin, ibuprofen, and loratadine to treat these symptoms. Two 24-h urine collections (UroRisk® kit) are summarized in Table 1.
Table 1.
24-h urine data (URO-RISK® kit)
| Test | Reference range | 2010 | 2013 |
|---|---|---|---|
| Volume | >2.00 l | 1.59 | 2.17 |
| pH | 5.5–7.0 | 5.5 | 6.4 |
| Calcium | <250 mg | 33 | 339 |
| Oxalate | <45 mg | 59 | 75 |
| Citrate | >320 mg | 272 | 669 |
| Uric acid | <700 mg | 534 | 757 |
| Sodium | <200 mEq | 322 | 280 |
| Phosphorous | <1100 mg | 1146 | 1200 |
| Magnesium | >60 mg | 190 | 171 |
| Potassium | 19–135 mEq | 85 | 75 |
| Creatinine | 600–1800 mg | 1474 | 2110 |
| Calcium oxalate URO-RISK | <2.00 | 3.58 | |
| Brushite URO-RISK | <2.00 | 3.34 | |
| Sodium urate URO-RISK | <2.00 | 3.49 | |
| Uric acid URO-RISK | <2.00 | 0.64 |
Study urine collections
Freshly voided random, midday urine samples were collected on three separate occasions (samples a, b, and c corresponding to 0, 3, and 10 weeks from recruitment). Sample c was immediately preceded by renal colic, and only this sample contained an orange gravel-like material (GF) with a small matrix strand (MX). Urine sample c, GF, and MX fractions were separated physically by decanting the supernatant urine (sample c) and separating MX from GF using a pipette to remove the viscous liquid-like MX substance from GF. Each was frozen separately (−20 °C) until analyzed. The urine samples were also stored frozen (−20 °C) with protease inhibitors for later characterization [15]. Urine ion chemistries were measured using ion chromatography (Table 2) and were consistent with her 24-h urine data from the clinical laboratory (Table 1) and not unusual.
Table 2.
Random urine characteristics
| Urine samples | a | b | c | |
|---|---|---|---|---|
| Volume | 95 ml | 110 ml | 120 ml | |
| Notes | Cloudy | None | Orange sediment & matrix strand (MX) | |
| Urine dipstick | ||||
| SG | 1.020 | 1.015 | 1.030 | |
| pH | 5.5 | 6.5 | 5.0 | |
| Notes | Bright orange bilirubin | Trace protein | Bright orange bilirubin | |
| Urine pellet fraction post-low-speed centrifugation | ||||
| Pellet fraction | Yes, small | Yes, trace | Sediment | MX |
| FTIR analysis | 20 % COD/struvite; 80 % protein | Not conducted | 40 % GC 60 % Prot. | Protein with trace GCa |
| Urine analytes, mg/dL | Reference values [20] | |||
| Cations | ||||
| Sodium | 470 | 390 | 280 | 220 (450) |
| Ammonium | 40 | 45 | 160 | 55 (125) |
| Potassium | 430 | 280 | 330 | 160 (300) |
| Magnesium | 5.4 | 5.2 | 14.2 | 9 (15) |
| Calcium | 0.2 | 5.8 | 26 | 11 (26) |
| Anions | ||||
| Chloride | 730 | 600 | 520 | - |
| Sulfate | 90 | 150 | 150 | 115 (190) |
| Phosphate | 280 | 300 | 200 | 155 (265) |
| Oxalate | 6.9 | 4.9 | 9 | 1.1 (1.8) |
| Creatinine | 274 | 233 | 312 | 90 (250) |
| Protein, mg/L | 112 | 176 | 97 | - |
| P/C, mg/g | 41 | 76 | 31 | <150 |
GC guaifenesin/guaiacol crystal mixture
Based on microscopic examination. Urine reference values are based on 94 normal subjects 24-h urine data and shown as set mean (set high value) [20]
Urine analysis
Filtered urine samples were analyzed for both anion and cation concentrations using a Dionex ICS3000 dual pump ion chromatography system (ThermoFisher-Dionex, Bannockburn, IL) equipped with ion suppression and conductivity detection, using their Chromeleon software for system control and data analysis. Concentrations were based on peak areas from duplicate analysis (at minimum). Cations were separated on a CS12 column using a methylsulfonic acid cartridge for eluent generation. Creatinine was determined using ultraviolet detection (210 nm) from the CS12 elution. The system was calibrated with cation and creatinine standards (TECO, Anaheim, CA). Anions were separated on an AS12 column (carbonate–bicarbonate buffer) calibrated with a Dionex anion standard mixture. Oxalate standards were made from sodium oxalate (Sigma-Aldrich, Milwaukee, WI). Urine total protein concentrations were determined using a pyrogallol red assay with bovine serum albumin as a standard.
Urine sample handling
Urine samples were defrosted in a warm water bath (37 °C) and clarified by low-speed centrifugation (1,000g × 10 min) prior to ultrafiltration (Amicon Ultra 10 kDa mwco, Millipore) against 10 mM NaCl to obtain the urinary macromolecules (UM). Pellet fractions were evaluated by polarized light microscopy and FTIR.
Matrix protein isolation
The proteins from the solid GF “stone” were isolated by exploiting the guaifenesin and guaiacol solubility with subsequent protein ultrafiltration (>10 kDa cutoff) [16]. The MX “stone” fraction was solubilized for protein characterization in 200 μl 0.25 M Tris-base, 1.92 M glycine, 1 % SDS with subsequent addition of 200 μl 0.5 M dithiothreitol (60 °C water bath, 1 h), and then desalted by ultrafiltration.
Crystal identification-fourier transform infrared spectroscopy (FTIR)
All solid pellet fractions were air-dried and analyzed for composition in the Mandel International Stone and Molecular Analysis Center (MIS.MAC, Clement J. Zablocki Veterans Affairs Medical Center, Milwaukee, WI), using attenuated total reflectance data collection on a Thermo Nicolet Nexus 870 FTIR spectrometer. Spectra were collected at room temperature with 32 scans per data collection between 700 and 3500 cm−1. The spectral data were compared with a locally constructed reference library using a correlation algorithm [17].
Gel electrophoresis
Urine macromolecules and stone matrix fractions were characterized using stand gels and blotting protocols as described earlier [15, 18]. Primary antibodies for Tamm–Horsfall glycoprotein or uromodulin (UROM) [18], osteopontin (OSTP) [15], transferrin (TRFE), albumin (ALBU), zinc-α-2 glycoprotein (ZA2G), IgKappa (IGKC) and Histone (HIS) were used with details given in supplementary data (S1. Antibodies). Images were collected using a 4 mega-pixel imaging system and accompanying software (IS4000R; MI software; CareStream Health, Rochester, NY USA).
Mass spectrometry
Proteomic studies were performed at the MCW Innovation Center, Milwaukee, WI. Equivalent samples (20 μg) of macromolecules from each urine (a, b and c), GF-associated proteins, and matrix strand (MX) were lyophilized, reconstituted, and then in-gel trypsin digested [19] prior to loading on the ThermoFinnigan LTQ Ion Trap LC-MS/MS Instrument (linear ion trap with MSn capability) with a Thermo nanoelectrospray ionization source. The accompanying nano-HPLC system included a ThermoFinnigan Surveyor quaternary pump plus Surveyor autosampler and capillary columns (10 cm × 75 μm) packed with 3 μm Magic C18AQ particles (Michrom-Bruker, Auburn, CA). Protein peaks were determined using established criteria and matched to the human Uniprot database following previously established procedures [19]. The analysis was based on a summation of at least two column injections. Proteomic datasets were reviewed excluding keratins (due to their variable origin), redundant proteins, proteins with a peptide probability <0.85, and proteins with less than 2 peptides identified. The resulting mass spectrometry data yielded spectral counts (number of times that any peptide associated with a given protein is detected). The relative abundance was defined as % spectral counts (% SC; percentage of all assigned spectral counts that were attributed to this protein).
Results
The primary observations for the three urine samples (samples a, b, and c) are shown in Table 2. Sample c contained two distinct solid phases or stones prior to centrifugation: GF and MX fractions. Her normal urine, exemplified by samples a and b, did not contain a visible solid phase. Microscopic examination of the two solid phases revealed many spherical, birefringent crystals and traces of proteinaceous material in GF (Fig. 2A, B), while occasional spherical crystals and an abundance of proteinaceous fibers were observed in MX (Fig. 2C, D). These observations were confirmed using FTIR, finding 60 % protein, 16 % guaifenesin and 24 % guaiacol in the GF sample and predominantly protein with trace drug/metabolite in the MX sample (S2. FTIR data). Gravimetric analysis of the drug and metabolite crystal mixture suggested that the initial drug concentration was about 6 mg/ml in the voided urine. Urine ion analysis was remarkable for variable calcium and consistently high oxalate and creatinine excretion, while protein/creatinine ratios for urine samples a, b and c were normal (Table 2). All other urine ion data were near the boundaries of normal urine values [20].
Fig. 2.
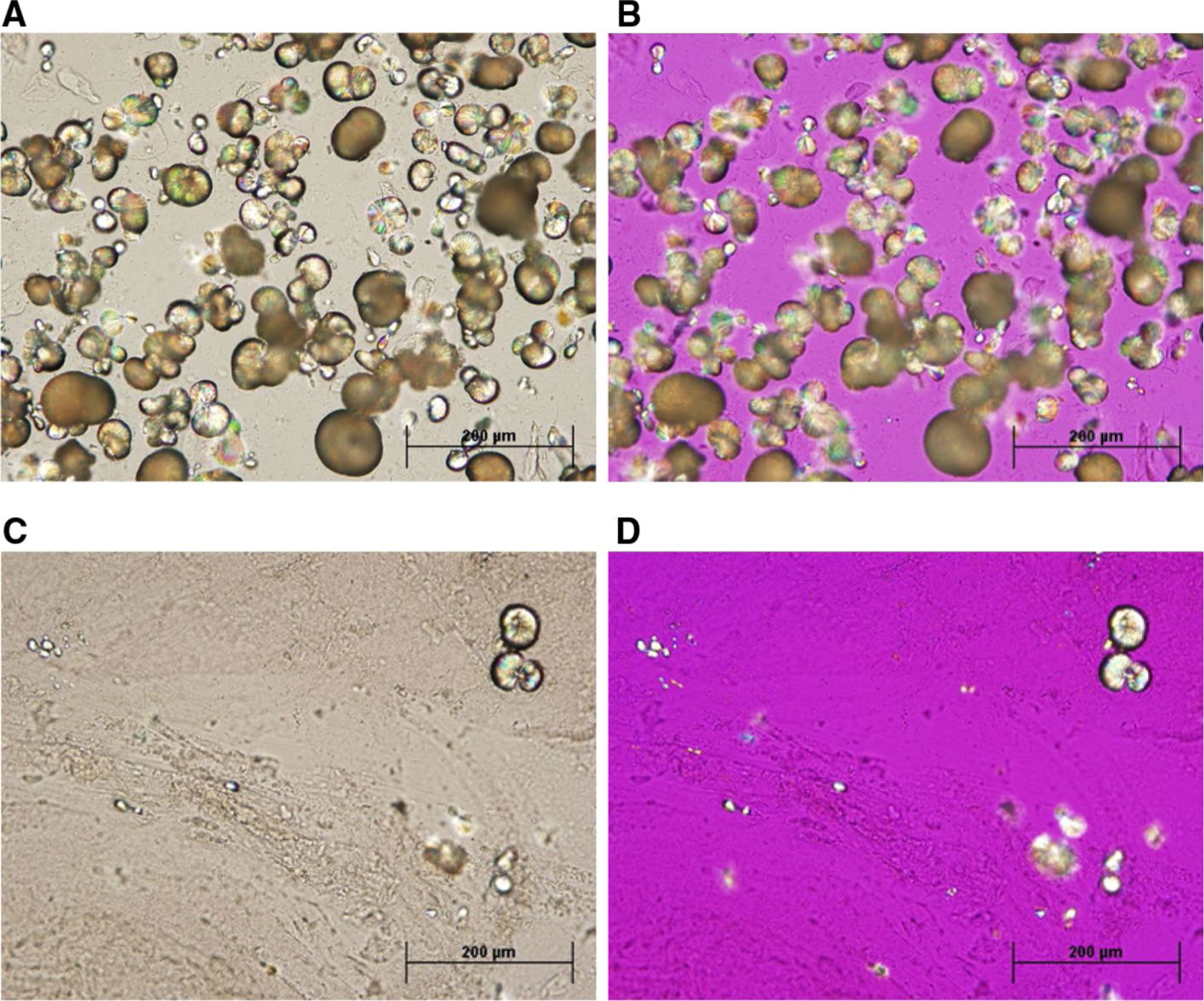
Optical microscopy of urine sample-c sediment and matrix strand. Top row: sample c orange, stone-like sediment (A bright field, B polarized). Bright field microscopic examination of the sediment (top left) revealed spherical crystals that were birefringent under polarized light (top right) with small traces of protein strands evident. Bottom row: sample c mucus strand (C bright field, D polarized). Examination of the mucus strand showed long polymeric protein fibrils with an occasional spherical crystal (bottom left), which were birefringent under polarized light (bottom right). All images were at 20× magnification
Immunoblot results from equal total protein loadings are shown in Fig. 3 for her normal UM (samples a and b), the depleted stone UM (sample c), and the protein isolate from GF. For these 7 proteins, Histone (HIS) was only detected in GF, while transferrin (TRFE) was almost undetectable in GF and c samples. Uromodulin (UROM), osteopontin (OSTP) and possibly Ig Kappa (IGKC) showed increased abundance in GF with reduced abundance in UM c, compared to UM samples a and b, suggesting preferential adsorption of these proteins to the stone crystals/matrix. Conversely, albumin (ALBU) was reduced in GF, suggesting only adventitial inclusion in GF. Furthermore, ALBU was increased in UM c, suggesting that it was left behind in solution during stone formation. Zinc alpha 2 glycoprotein (ZA2G) was not detected in GF. Data from MX protein immunoblots (data not shown) showed that UROM and ALBU were present at comparable quantities to normal UM samples, while HIS and IGKC were increased in MX. Conversely, OSTP and ZA2G were not detected and TRFE was only detected at trace levels in MX.
Fig. 3.
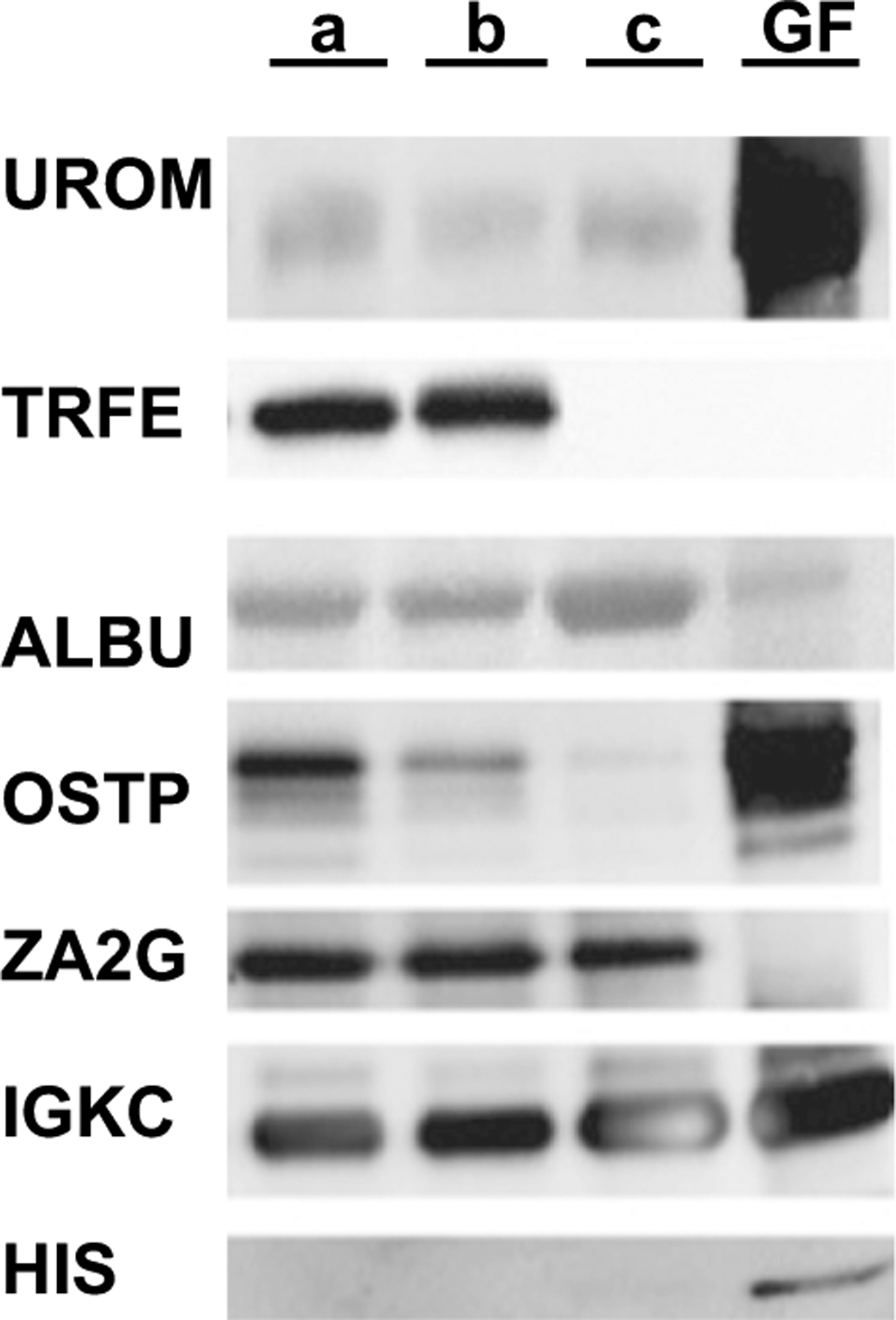
Immunoblot analysis of equivalent UM loadings (10 μg). Specific antibodies for UROM (Tamm–Horsfall glycoprotein), TRFE (transferrin), ALBU (albumin), OSTP (osteopontin), ZA2G (zinc α 2 glycoprotein), and IGKC (Ig Kappa C chain) and HIS (histone) were used in staining. Lanes (left to right) UM samples a, b, c and GF fraction. MX fraction is not shown
Mass spectrometry experiments identified a total of 156 proteins from urine (a, b and c), GF, and MX samples (131, 27 and 29 proteins, respectively), with complete data summary given in Supplementary Data (S3). We compared relative abundance data from mass spectrometry (% SC) to quantitative immunoblots for three highly abundant urine proteins; UROM, TRFE, and ALBU (Fig. 4), confirming the validity of using % SC as a quantitative proteomics measure [19, 21]. Urine contained 107 proteins that were not associated with either GF or MX. The GF sample had a strong overlap with the urine proteome (>90 % of GF protein mass), while >50 % of MX protein mass was unique to that fraction. The 49 proteins that comprise the “guaifenesin stone matrix proteome” (GF and MX samples) are enumerated in Table 3. These proteins were largely overlapping with stone matrix proteins from various stone types (uric acid, calcium oxalate, calcium phosphate, and matrix stone). Overlap was indicated by checkmarks in the appropriate columns, since relative abundances were not report in these earlier publications [9–11, 22]. It is evident from the list of tissue location in column 2 that the stone matrix proteins common with urine were predominantly extracellular proteins with limited inclusions of cytosolic, nuclear, and plasma membrane proteins. The urine proteins overlapping between the guaifenesin stone matrix and more conventional stone types were largely extracellular with some immune and complement proteins, while the proteins unique to guaifenesin stone matrix were largely cell membrane and cytosolic proteins.
Fig. 4.
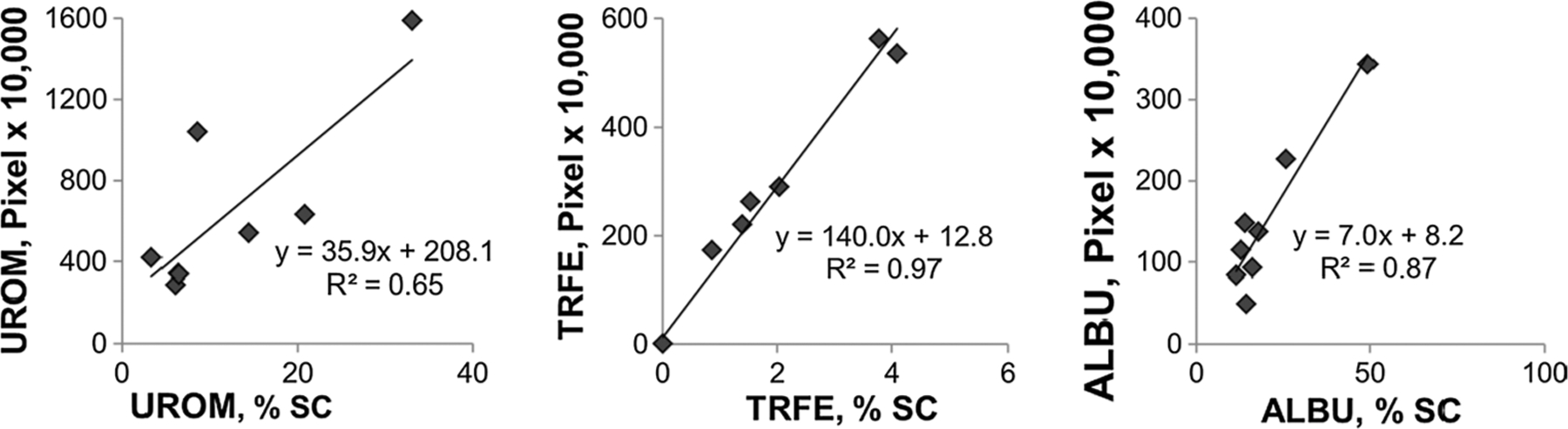
Relative urine protein abundance: proteomics vs. immunoblot. Antibody signal from immunoblots (Fig. 3 and additional samples of known protein content) compared to the mass spectroscopy proteomic data for representative proteins (UROM = Tamm–Horsfall Protein, TRFE = transferrin and ALBU = albumin). Immnunoblot data shown as pixels ×10,000 per given area. Mass spectrometry-based proteomic data given as % of total spectral count (% SC) for a given sample
Table 3.
Proteomic data for patient urine samples (a, b and c), GF and matrix strand (MX)
| Net work | Tissue location | Protein | % Spectral counts | UA stone | Ca stone | Matrix stone | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| a | b | c | GF | MX | |||||||
| Common urine and stone matrix proteins | |||||||||||
| 1 | Extracellular | ALBU | Albumin | 17.8 | 25.8 | 49.1 | 14.2 | 22.3 | ✓ | ✓ | ✓ |
| 1 | Extracellular | UROM | Uromodulin (THP) | 6.3 | 6.0 | 6.4 | 33.0 | 2.3 | ✓ | ✓ | ✓ |
| 1 | Extracellular | TRFE | Transferrin | 3.8 | 4.1 | 0.0 | 0.3 | 1.3 | ✓ | ✓ | ✓ |
| 1 | Extracellular | IGKC | Ig Kappa | 0.1 | 1.6 | 2.7 | 1.8 | 1.0 | ✓ | ✓ | ✓ |
| 1 | Extracellular | IGHG1 | Ig Gamma | 1.9 | 2.3 | 0.1 | 0.6 | 2.2 | ✓ | ✓ | ✓ |
| 1 | Extracellular | A1AT | α-1-Antitrypsin | 1.8 | 1.7 | 0.1 | 0.8 | 2.4 | ✓ | ✓ | ✓ |
| 1 | Cytosol | HBB | Hemoglobin β | 0.6 | 0.5 | 0.0 | 0.2 | 5.6 | ✓ | ✓ | ✓ |
| 1 | Extracellular | APOD | Apolipoprotein D | 3.5 | 2.9 | 0.0 | 7.9 | 0.0 | ✓ | ✓ | |
| 1 | Extracellular | KNG1 | Kininogen 1 | 2.5 | 1.7 | 0.0 | 5.0 | 0.0 | ✓ | ✓ | |
| 1 | Cytosol | IGLC | Ig Lambda | 2.4 | 3.1 | 1.5 | 2.6 | 0.0 | ✓ | ✓ | ✓ |
| 2 | Extracellular | OSTP | Osteopontin | 2.8 | 1.5 | 0.3 | 2.5 | 0.0 | ✓ | ✓ | |
| 1 | Extracellular | KV309 | Ig Kappa chain V-III | 0.0 | 0.0 | 0.0 | 2.5 | 0.0 | ✓ | ✓ | ✓ |
| 1 | Extracellular | A1BG | α-1-β Glycoprotein | 3.2 | 1.3 | 0.0 | 1.2 | 0.0 | ✓ | ✓ | |
| 1 | Cytosol | S10A8 | Calgranulin | 0.0 | 0.0 | 0.1 | 0.4 | 0.0 | ✓ | ✓ | ✓ |
| 1 | Extracellular | CO3 | Complement C3 | 0.0 | 0.0 | 0.0 | 0.0 | 15.3 | ✓ | ✓ | ✓ |
| 1 | Extracellular | HPT | Haptoglobin | 1.9 | 1.1 | 0.0 | 0.0 | 5.2 | ✓ | ✓ | |
| 1 | Extracellular | FIBG | Fibrinogen γ | 0.0 | 0.0 | 0.0 | 0.0 | 3.4 | ✓ | ✓ | |
| 1 | Extracellular | FIBB | Fibrinogen β | 0.0 | 0.0 | 0.0 | 0.0 | 2.5 | ✓ | ✓ | |
| 1 | Nuclear | H4 | Histone H4(HIS) | 0.0 | 0.0 | 0.0 | 0.0 | 1.0 | ✓ | ✓ | ✓ |
| 1 | Extracellular | FIBA | Fibrinogen α | 0.0 | 0.0 | 0.0 | 0.0 | 0.9 | ✓ | ✓ | |
| 2 | Cytosol | CAH1 | Carbonic anhydrase | 0.0 | 0.0 | 0.0 | 0.0 | 0.7 | ✓ | ✓ | |
| 1 | Extracellular | CO5 | Complement C5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.6 | ✓ | ✓ | |
| 1 | Extracellular | CO9 | Complement C9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | ✓ | ✓ | |
| 1 | Extracellular | AMBP | Bikunin | 8.5 | 7.6 | 5.5 | 5.8 | 0.0 | ✓ | ||
| 1 | Extracellular | ITIH4 | Inter-α-trypsin inh (C4) | 1.0 | 0.7 | 0.0 | 0.7 | 0.0 | ✓ | ||
| 2 | Cell membrane | GELS | Gelsolin | 2.2 | 1.4 | 0.0 | 0.7 | 0.0 | ✓ | ||
| 2 | Extracellular | A2MG | α-2-Macroglobulin | 0.0 | 0.0 | 0.0 | 0.0 | 7.9 | ✓ | ||
| 1 | Extracellular | ITIH1 | Inter-α-trypsin inh (C1) | 0.0 | 0.0 | 0.0 | 0.0 | 2.8 | ✓ | ||
| 1 | Other | HBD | Hemoglobin δ | 0.0 | 0.0 | 0.0 | 0.0 | 2.2 | ✓ | ||
| 1 | Cell membrane | MUC | Ig Mu | 0.0 | 0.0 | 0.0 | 0.0 | 1.5 | ✓ | ||
| 1 | Extracellular | ITIH2 | Inter-α-trypsin inh (C2) | 0.0 | 0.0 | 0.0 | 0.0 | 2.8 | ✓ | ||
| 1 | Extracellular | CFAB | Complement factor B | 0.0 | 0.0 | 0.0 | 0.0 | 0.4 | ✓ | ||
| Proteins uniquely GF or MX associated | |||||||||||
| 2 | Cell membrane | PDC15 | Protocadherin 15 | 8.5 | 2.9 | 0.6 | 12.8 | 0.0 | |||
| 3 | Cytosol | SPRR3 | Small Pro-rich protein 3 | 0.0 | 0.0 | 0.7 | 2.7 | 0.0 | |||
| 3 | Extracellular | A2GL | α-2-Glycoprotein | 3.6 | 2.9 | 0.0 | 0.7 | 0.0 | |||
| 1 | Extracellular | PGBM | BM Proteoglycan | 0.0 | 0.0 | 0.1 | 0.6 | 0.0 | |||
| 2 | Cell membrane | CAD13 | Cadherin 13 | 0.2 | 0.0 | 0.0 | 0.5 | 0.0 | |||
| 3 | Cytosol | PSMD6 | 26S non-ATPase | 0.0 | 0.1 | 0.0 | 0.5 | 0.0 | |||
| 2 | Cell membrane | CNTN3 | Contactin-3 | 0.0 | 0.0 | 0.1 | 0.4 | 0.0 | |||
| 2 | Cell membrane | AT2B3 | PM Ca-ATPase 3 | 0.0 | 0.0 | 0.0 | 0.4 | 0.0 | |||
| 3 | Cell Membrane | ARFG3 | GTPase activating protein | 0.0 | 0.0 | 0.0 | 0.2 | 0.0 | |||
| 2 | Cytosol | PCLO | Protein piccolo | 0.0 | 0.0 | 0.0 | 0.2 | 0.0 | |||
| 3 | Other | MUC5B | Mucin-5B | 0.0 | 0.0 | 0.0 | 0.0 | 10.2 | |||
| 3 | Extracellular | PZP | A2MG-like protein | 0.0 | 0.0 | 0.0 | 0.0 | 3.3 | |||
| 3 | Cell membrane | DESP | Desmoplakin | 0.0 | 0.0 | 0.0 | 0.0 | 1.3 | |||
| 3 | Cell membrane | DMBT1 | Glycoprotein 340 | 0.0 | 0.0 | 0.0 | 0.0 | 0.8 | |||
| 3 | Extracellular | TENA | Tenascin | 0.0 | 0.0 | 0.0 | 0.0 | 0.7 | |||
| 3 | Cell membrane | LDLR | LDL receptor | 0.0 | 0.0 | 0.0 | 0.0 | 0.4 | |||
| 2 | Cell membrane | EZRI | Ezrin (Villin) | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 | |||
Protein relative abundances are given for each matrix associated protein in urine samples (a, b and c), GF, and MX, and compared to the list of proteins reported in uric acid stone matrix (UA) [10], calcium oxalate/calcium phosphate stone matrix (Ca) [9, 11], and a matrix stone [22], with overlap denoted by a ✓ in the appropriate column. Approximately 80 % of the protein mass from GF/MX samples was common with uric acid stones. Metabolomics network analysis (www.ingenuity.com) identified three physiologic networks (column 1) and five tissue locations (column 2). Network 1 is a metabolic disease response network rich in immune and complement factors. Network 2 is a cell–cell signaling and development network. Network 3 is related to hereditary disorders and other disease states
A far more interesting picture emerges in comparing the protein relative abundance data between normal urine, depleted urine, and stone matrix. Bar graphs in Fig. 5 show protein inclusion patterns observed in mass spectrometry data, mirroring the same findings observed in immunoblotting data (Fig. 3). Most proteins demonstrating enrichment in either stone matrix sample were depleted in UM sample c compared to the baseline urine samples (a and b). Conversely, UROM (Fig. 5A) remained equally abundant in the depleted urine sample (c), as compared with normal urine (a and b), despite demonstrating a strong preference for the GF matrix. The relatively small size of the GF and MX fractions compared to the total urine volume in sample c may explain the lack of UROM depletion. On the other hand, ALBU (Fig. 5B) was included in decreased abundance in the guaifenesin stone matrix. ALBU was increased in sample c consistent with a relative depletion of other proteins in the supernatant urine due to selective adsorption. The similarity of total protein content in the three urine samples argues against the possibility that this sample had higher ALBU content than normal. OSTP (Fig. 5C) and A1AT (Fig. 5D) demonstrated patterns of selective adsorption in stone matrix (with OSTP in GF and A1AT in MX) and depletion in UM sample c.
Fig. 5.
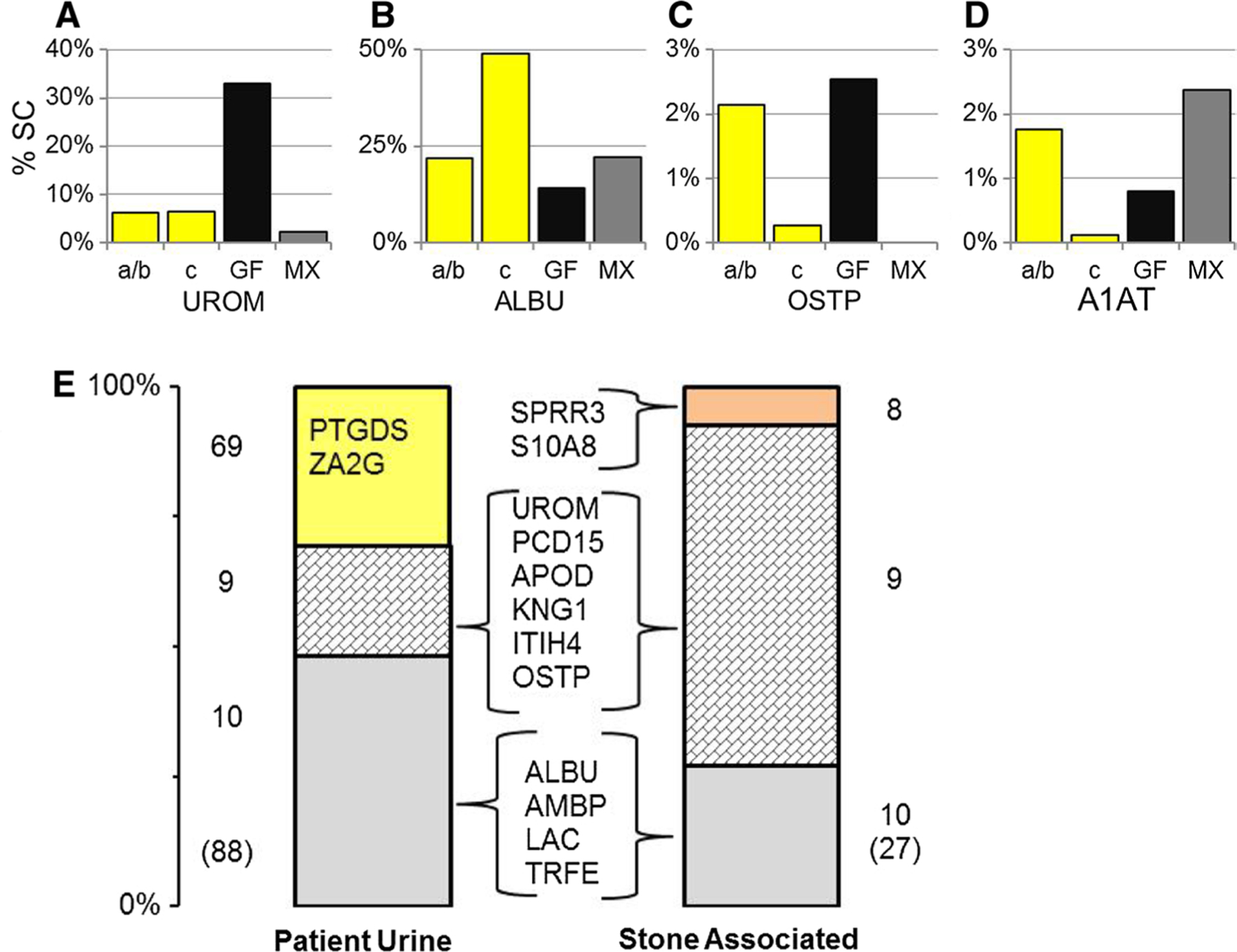
Selective protein inclusion in either the GF-MX proteome. Relative protein abundance data are shown for four individual proteins (A UROM, B ALBU, C OSTP, D A1AT) illustrating different patterns of enrichment comparing normal baseline urine (samples a/b, yellow bar) to depleted urine (sample c, yellow bar) and the two different matrix phases, GF (black bar) and MX (gray bar). E Cumulative abundances for various protein subsets in normal urine (a/b) were compared to similarly defined groupings in GF matrix. In the normal urine bar 69 proteins were found in urine only (yellow, approximately 30 % of total protein mass) dominated by PTGDS and ZA2G (prostaglandin D isomerase and zinc-α-2 glycoprotein). In the GF matrix bar 8 proteins were uniquely found in this fraction dominated by SPRR3 and S10A8 (light orange, approximately 5 % of total protein mass). Overlapping between the two proteome distributions were 9 urine proteins that were enriched in GF matrix (block tiled gray, illustrated by OSTP and UROM (C, A) Approximately 20 % of total urine protein mass, but 75 % of stone matrix protein mass) and 10 abundant urine proteins that were detected in lesser abundance in GF matrix (light gray, illustrated by ALBU and A1AT (B, D). (Approximately 50 % of total urine protein mass, but only 20 % of stone matrix protein mass.) All abbreviations are defined in Table 3 unless noted
The overall proteomic shift in the guaifenesin stone matrix is shown in an integrative way in Fig. 5E [23]. In this representation, each bar graph represents the total protein mass of the sample; normal baseline urine samples (a and b) in the left hand bar and GF proteins in the right hand bar. Each bar has been divided into three groups of proteins based on their comparative protein abundance. The majority of protein mass included proteins common to both phases shown in two groups with nine proteins present in GF at increased abundance (block tiled gray) and ten proteins present in GF at decreased abundance (solid gray section). The nine proteins common to both samples that were increased in GF likely drove the stone-formation process, as they accounted for almost 75 % of GF protein mass, notably including UROM, OSTP, and APOD. On the other hand, the ten proteins common to both samples that were decreased in GF included most other high-abundance urine proteins, such as ALBU, AMBP, and TRFE. The remaining section of each bar shows proteins unique to that phase with 69 proteins found only in urine (yellow bar) and 8 proteins found only in GF (orange bar). The unique urine proteins were generally low abundance, though they totaled almost 30 % of UM protein mass, while the 8 proteins unique to GF accounted for only about 5 % of GF protein mass. The number of proteins found to be unique in any phase would likely decrease if more sensitive mass spectrometry methods were applied, such that lower abundance proteins could be detected; however, expanding the list of proteins detected would not change the identity or relative contribution of the abundant proteins reported. Interestingly, UM sample c contained 43 proteins that were not observed in normal urine (a and b), which may indicate an altered physiologic response during stone formation.
Discussion
This case illustrates the critical importance of accurate stone analysis, as our patient’s true clinical diagnosis and definitive treatment were dependent on this information. The 24-h urine data predicted risk for stone types (calcium oxalate and uric acid in this case) that were not relevant to this patient’s disease process, as is frequently observed clinically. The discovery of guaifenesin/guaiacol crystals in this patient’s stone sample completely redirected her clinical care. Only when presented with this result did she admit to long-term extensive use of over the counter medications containing guaifenesin (including the timeframe of her first stone episodes). The estimated guaifenesin concentration of 6 mg/ml could be readily achieved with cold medicine products on the market containing 200–600 mg of guaifenesin per dose. While it is not a primary focus of this study, commercial stone analysis laboratories have been shown to perform poorly and misdiagnosis as this patient example suggests is very common [4]. The urologic community will clearly need to push for higher quality stone analysis to fight the trend toward lowest bidder contracts for pathology services.
Several apparently unrelated findings are consistent with her use of guaifenesin. Her positive tests for methamphetamine and amphetamines in urine screenings could easily have resulted from ingesting cold remedies containing ephedrine, a common companion drug, which is known to cause false-positive results in drug screening [24]. Guaifenesin is a uricosuric agent [25], which likely created urine conditions that prompted allopurinol and citrate prescriptions from her initial presentation. Finally, in our ion chromatography-based creatinine determinations, we noted significant overlap in the peaks for creatinine and guaifenesin/guaiacol which caused an overestimation of creatinine content by about 20 %. Consequently, the surprisingly large creatinine contents reported in her commercial laboratory 24-h urine data may well be explained by the presence of guaifenesin/guaiacol in these samples, rather than the initially suspected “over-collection.” Aberrantly high creatinine determinations would skew the urine protein to creatinine ratios low, though her values were still normal after correction.
At a superficial level, this drug containing stone seems quite different from calcium stones, where the matrix is only 2–3 % of the stone by weight [1] compared to the 60 % matrix content reported here. However, substantial overlap was noted in Table 3 between the proteomes from the guaifenesin stones and both uric acid and calcium-based stones [9–11], implying similarity in mechanism. Even the reported proteome from a single matrix stone shared many of the same proteins [22]. While GF and MX contained somewhat different proteins, they showed the greatest commonality with uric acid stone matrix reports, particularly including some abundant urine proteins, such as bikunin (AMBP), inter-α-trypsin inhibitor (ITIH4), gelsolin (GELS), and α-2-macroglobulin (A2MG). The uricosuric effect of guaifenesin may explain this similarity due to a physiologic response to elevated uric acid in the urine [25]. The fact that many highly abundant urine proteins were found in all stone samples suggests that the mechanisms underlying protein inclusion in matrix may involve protein properties that are widely shared.
The data summarized in Fig. 5 and Table 3 demonstrated clear evidence that a small subset (9) of proteins with moderate-to-high urinary abundance were highly enriched in stone matrix and ultimately accounted for 75 % of the total matrix mass. Certainly, these nine proteins must dominate the guaifenesin stone-forming process based on their overwhelming presence in matrix, through whatever combination of protein–crystal and protein–protein interactions constitute this process. The remaining 40 proteins likely have no more than a minor influence on the process based on their low relative abundance, even though some of these proteins were also highly enriched in stone matrix. While these nine proteins demonstrated increased abundance in GF matrix compared to the patient’s normal urine (samples a and b), the depletion of these components in sample c urine further strengthens support for selective inclusion of these proteins in matrix. We do note that UROM and OSTP were prominent in this subset, and they have been implicated in calcium stone disease also [2, 15]. Of these, UROM has been shown previously to be prone to aggregation, possibly related to diminished glycosylation levels, which could trigger crystal aggregation regardless of crystal type [18]. Unfortunately, we are unable to compare relative enrichment data between stone types to know if the same proteins might be enriched in different stone types, because earlier studies did not adequately report relative abundance data [9–11].
More interesting perhaps for their apparent lack of contribution are the 10 moderate-to-high abundance urinary proteins observed in GF matrix, but at lower abundance than they were found in urine, including proteins such as ALBU, AMBP, and TRFE that were also reported in typical stones. Their relatively low abundance in GF stone matrix suggests that their inclusion was the result of weak, nonspecific interactions with either the crystals or more likely the matrix, analogous to that described in studies of desialylated UROM with other urinary proteins [18]. The desialylated UROM was prone to self-aggregation and subsequent aggregation with many other urine proteins, including ALBU. While we cannot say that these urine proteins would be inert in typical stone formation, the clear implication from this study is that many proteins reported in stone matrix may have no significant contribution to the generation of a stone, and simply appeared as innocent bystanders in the stone, because of non-specific interactions.
Clearly, the eight proteins unique to the GF sample must also be counted as highly enriched, though their relatively low abundance (~5 %) in GF matrix argues against them playing a major role in stone formation. These eight proteins may have been simply below our detection limit in normal urine, because we used tight criteria for protein identification by mass spectrometry, but we cannot exclude the possibility that they were excreted in response to the stone-forming process. Kidney damage has been reported with increased exposure to over the counter headache medicines, characterized by increased excretion of UROM, AMBP, zinc-α-2 glycoprotein (ZA2G), ITIH4, and IGKC [26]. ZA2G, a marker of kidney injury in diabetes, was enriched in all three urine samples compared to literature reports [27], though it was not found in the GF fraction. HIS, a very basic nuclear protein associated with kidney injury, was found in the stone samples, though not in any of the urine samples. IGKC and other immunoglobulins were also found in these samples, supporting separately a role for immune response [28].
Some drug-induced calcium stones are clearly related to supersaturation effects, such as the association of calcium oxalate stones with loop diuretics or calcium phosphate stones with the carbonic anhydrase inhibitors [29]. Proteomic shifts observed in the current study are likely a response to drug crystal interactions with tubular epithelial surfaces, but no data have been published for guaifenesin drug or crystal toxicity in renal cell culture. Both calcium oxalate and uric acid crystals are known to be toxic to renal epithelial cells [30, 31]. In the current study, heavy drug intake likely contributed to high drug supersaturation and crystalluria as previously reported [32]. Cell culture studies of lung cells exposed to guaifenesin drug concentrations have shown decreased excretion of mucin [33], a result contrary to this report; however, the presence of numerous unique proteins in UM sample c (complement factors C3, C5 and C9, factor B, α-2-macroglobulin, and cell surface and plasma membrane-related proteins) argues for altered physiology at the time of stone formation. In this regard, both the GF and MX proteins correlated to network 3, which is associated with other disease states (i.e., cancer, neurologic disorders, and hepatologic disorders).
Conclusion
This protocol of comparing a patient’s urine proteome to their stone matrix proteome demonstrated that only a small fraction of proteins were both highly enriched and highly abundant in stone matrix, and therefore likely controlled the stone-formation process. Conversely, many abundant urine proteins were shown to be present in stone matrix at reduced abundance levels, consistent with their inclusion being a secondary phenomenon, involving non-selective processes. The similarities in matrix proteins between guaifenesin and more typical stones suggest that the inclusion of many proteins in stone matrix is a universal property regardless of stone type; however, the specific protein inclusion pattern may differ when compared on the basis of both enrichment and abundance. Identifying the proteins that regulate stone formation in this manner should help elucidate their mechanism of action, which remains a critical need in understanding stone formation.
Supplementary Material
Acknowledgments
We gratefully acknowledge the primary financial support provided in part with resources and the use of facilities at the Clement J. Zablocki Department of Veterans Affairs Medical Center, Milwaukee, WI, and in part by the National Institutes of Health/National Institute for Diabetes, Digestive, and Kidney Diseases (DK 82550) (JAW). Additional financial support was provided by the Medical College of Wisconsin and in part by the National Institutes of Health/National Institute for Diabetes, Digestive, and Kidney Diseases (DK 74741) (JGK). We also gratefully acknowledge the technical support MIS.MAC (Mandel International Stone and Molecular Analysis Center), Milwaukee, WI, for stone analysis and the technical support of Brian Halligan, PhD, for proteomic analysis.
Funding
This study was primarily funded with resources and the use of facilities at the Clement J. Zablocki Department of Veterans Affairs Medical Center, Milwaukee, WI, and in part by a grant from the National Institutes of Health (NIDDK, DK 82550-JAW). Additional financial support was provided by the Medical College of Wisconsin and in part by the National Institutes of Health (NIDDK, DK 74741-JGK).
Abbreviations
- BMI
Body mass index
- FTIR
Fourier transform infrared spectroscopy
- GC
Guaifenesin.guaiacol crystal mixture
- GF
Orange stone sediment containing guaifenesin and guaiacol crystals
- IRB
Institutional Review Board
- LC–MS
Liquid chromatography coupled with mass spectrometry
- MCW
Medical College of WISCONSIN
- MIS.MAC
Mandel International Stone and Molecular Analysis Center
- MS
Mass spectrometry
- MX
Matrix strand stone sediment containing few drug crystals
- P/C
Protein/creatinine ratio in urine (mg/g)
- SG
Specific gravity
- UA
Uric acid stone matrix
- UM
Urine macromolecules
- VA
US Department of Veterans Affairs
- % SC
-
Percentage of total assigned scan counts attributable to an individual or group of proteins
All protein abbreviations are defined in Table 3
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00240-016-0907-4) contains supplementary material, which is available to authorized users.
The contents do not represent the views of the US Department of Veterans Affairs or the United States Government.
Conflict of interest None of the authors has any conflicts of interest to report.
Human studies The participating patient in this study was recruited with informed consent to an established study under VA IRB approval (VA-IRB protocol: 9305-01P). All procedures performed in these studies were in accordance with the ethical standards of the institutional and national research committee and with the 1964 Helsinki declaration and its later amendments.
References
- 1.Boyce Garvey FK (1956) The amount and nature of the organic matrix in urinary calculi: a review. J Urol 76:213–227 [DOI] [PubMed] [Google Scholar]
- 2.Aggarwal KP, Narula S, Kakkar M, Tandon C (2013) Nephrolithiasis: molecular mechanism of renal stone formation and the critical role played by modulators. Biomed res int. doi: 10.1155/2013/292953 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 3.Mandel NS, Mandel GS (1989) Urinary tract stone disease in the united states veteran population. I. Geographical frequency of occurrence. J Urol 142:1513–1515 [DOI] [PubMed] [Google Scholar]
- 4.Krambeck AE, Lingeman JE, McAteer JA, Williams JC Jr (2010) Analysis of mixed stones is prone to error: a study with US laboratories using micro CT for verification of sample content. Urol Res 38:469–475. doi: 10.1007/s00240-010-0317-y [DOI] [PubMed] [Google Scholar]
- 5.Gage H, Beal HW (1908) V. fibrinous calculi in the kidney. Ann Surg 48:378–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bani-Hani AH, Segura JW, Leroy AJ (2005) Urinary matrix calculi: our experience at a single institution. J Urol 173:120–123 [DOI] [PubMed] [Google Scholar]
- 7.Dussol B, Geider S, Lilova A, Leonetti F, Dupuy P, Daudon M, Berland Y, Dagorn JC, Verdier JM (1995) Analysis of the soluble organic matrix of five morphologically different kidney stones. evidence for a specific role of albumin in the constitution of the stone protein matrix. Urol Res 23:45–51 [DOI] [PubMed] [Google Scholar]
- 8.Khan SR, Atmani F, Glenton P, Hou Z, Talham DR, Khurshid M (1996) Lipids and membranes in the organic matrix of urinary calcific crystals and stones. Calcif Tissue Int 59:357–365 [DOI] [PubMed] [Google Scholar]
- 9.Canales BK, Anderson L, Higgins L, Ensrud-Bowlin K, Roberts KP, Wu B, Kim IW, Monga M (2010) Proteome of human calcium kidney stones. Urology 76:1017.e13–1017.e20. doi: 10.1016/j.urology.2010.05.005 [DOI] [PubMed] [Google Scholar]
- 10.Jou YC, Fang CY, Chen SY, Chen FH, Cheng MC, Shen CH, Liao LW, Tsai YS (2012) Proteomic study of renal uric acid stone. Urology 80:260–266. doi: 10.1016/j.urology.2012.02.019 [DOI] [PubMed] [Google Scholar]
- 11.Kaneko K, Kobayashi R, Yasuda M, Izumi Y, Yamanobe T, Shimizu T (2012) Comparison of matrix proteins in different types of urinary stone by proteomic analysis using liquid chromatography-tandem mass spectrometry. Int J Urol 19:765–772. doi: 10.1111/j.1442-2042.2012.03005.x [DOI] [PubMed] [Google Scholar]
- 12.Assimos DG, Langenstroer P, Leinbach RF, Mandel NS, Stern JM, Holmes RP (1999) Guaifenesin and ephedrine induced stones. J Endourol 13:665–667 [DOI] [PubMed] [Google Scholar]
- 13.Bennett S, Hoffman N, Monga M (2004) Ephedrine- and guaifenesin-induced nephrolithiasis. J Altern Complement Med 10:967–969 [DOI] [PubMed] [Google Scholar]
- 14.Pickens CL, Milliron AR, Fussner AL, Dversdall BC, Langenstroer P, Ferguson S, Fu X, Schmitz FJ, Poole EC (1999) Abuse of guaifenesin-containing medications generates an excess of a carboxylate salt of beta-(2-methoxyphenoxy)-lactic acid, a guaifenesin metabolite, and results in urolithiasis. Urology 54:23–27 [DOI] [PubMed] [Google Scholar]
- 15.Kolbach AM, Afzal O, Halligan B, Sorokina E, Kleinman JG, Wesson JA (2012) Relative deficiency of acidic isoforms of osteopontin from stone former urine. Urol Res 40:447–454. doi: 10.1007/s00240-012-0459-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mani N, Jun HW, Beach JW, Nerurkar J (2003) Solubility of guaifenesin in the presence of common pharmaceutical additives. Pharm Dev Technol 8:385–396 [DOI] [PubMed] [Google Scholar]
- 17.Mandel IC, Mandel NS (2007) Chapter 5. Structural and compositional analysis of kidney stones. In: Stoller ML, Meng MV (eds) Urinary stone disease: the practical guide to medical and surgical management. Humana Press, USA, pp 69–81 [Google Scholar]
- 18.Viswanathan P, Rimer JD, Kolbach AM, Ward MD, Kleinman JG, Wesson JA (2011) Calcium oxalate monohydrate aggregation induced by aggregation of desialylated tamm–horsfall protein. Urol Res 39:269–282. doi: 10.1007/s00240-010-0353-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu H, Wakim B, Li M, Halligan B, Tint GS, Patel SB (2007) Quantifying raft proteins in neonatal mouse brain by ‘tube-gel’ protein digestion label-free shotgun proteomics. Proteome Sci 5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemann J Jr, Pleuss JA, Worcester EM, Hornick L, Schrab D, Hoffmann RG (1996) Urinary oxalate excretion increases with body size and decreases with increasing dietary calcium intake among healthy adults. Kidney Int 49:200–208 [DOI] [PubMed] [Google Scholar]
- 21.Li M, Gray W, Zhang H, Chung CH, Billheimer D, Yarbrough WG, Liebler DC, Shyr Y, Slebos RJ (2010) Comparative shotgun proteomics using spectral count data and quasi-likelihood modeling. J Proteome Res 9:4295–4305. doi: 10.1021/pr100527g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Canales BK, Anderson L, Higgins L, Frethem C, Ressler A, Kim IW, Monga M (2009) Proteomic analysis of a matrix stone: a case report. Urol Res 37:323–329. doi: 10.1007/s00240-009-0213-5 [DOI] [PubMed] [Google Scholar]
- 23.Okumura N, Tsujihata M, Momhara C, Yoshioka I, Suto K, Nonomura N, Okuyama A, Toshifumi T (2013) Diversity in protein profiles of individual calcium oxalate kidney stones. PLoS One 8(7):e68624. doi: 10.1371/journal.pone.0068624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brahm NC, Yeager LL, Fox MD, Farmer KC, Palmer TA (2010) Commonly prescribed medications and potential false-positive urine drug screens. Am J Health Syst Pharm 67:1344–1350. doi: 10.2146/ajhp090477 [DOI] [PubMed] [Google Scholar]
- 25.Ramsdell CM, Postlethwaite AE, Kelley WN (1974) Uricosuric effect of glyceryl guaiacolate. J Rheumatol 1:114–116 [PubMed] [Google Scholar]
- 26.Bellei E, Cuoghi A, Monari E, Bergamini S, Fantoni LI, Zappaterra M, Guerzoni S, Bazzocchi A, Tomasi A, Pini LA (2012) Proteomic analysis of urine in medication-overuse headache patients: possible relation with renal damages. J Headache Pain 13:45–52. doi: 10.1007/s10194-011-0390-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riaz S, Alam SS, Srai SK, Skinner V, Riaz A, Akhtar MW (2010) Proteomic identification of human urinary biomarkers in diabetes mellitus type 2. Diabetes Technol Ther 12:979–988. doi: 10.1089/dia.2010.0078 [DOI] [PubMed] [Google Scholar]
- 28.Merchant ML, Cummins TD, Wilkey DW, Salyer SA, Powell DW, Klein JB, Lederer ED (2008) Proteomic analysis of renal calculi indicates an important role for inflammatory processes in calcium stone formation. Am J Physiol Renal Physiol 295:F1254–F1258. doi: 10.1152/ajprenal.00134.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matlaga BR, Shah OD, Assimos DG (2003) Drug-induced urinary calculi. Rev urol 5:227–231 [PMC free article] [PubMed] [Google Scholar]
- 30.Riese RJ, Kleinman JG, Wiessner JH, Mandel GS, Mandel NS (1990) Uric acid crystal binding to renal inner medullary collecting duct cells in primary culture. J Am Soc Nephrol 1:187–192 [DOI] [PubMed] [Google Scholar]
- 31.Riese RJ, Mandel NS, Wiessner JH, Mandel GS, Becker CG, Kleinman JG (1992) Cell polarity and calcium oxalate crystal adherence to cultured collecting duct cells. Am J Physiol (Renal Fluid Electrolyte Physiol) 262(31):F117–F184 [DOI] [PubMed] [Google Scholar]
- 32.Cockerill PA, de Cogain MR, Krambeck AE (2013) Acute bilateral ureteral obstruction secondary to guaifenesin toxicity. Can J Urol 20:6971–6973 [PubMed] [Google Scholar]
- 33.Seagrave J, Albrecht H, Park YS, Rubin B, Solomon G, Kim KC (2011) Effect of guaifenesin on mucin production, rheology, and mucociliary transport in differentiated human airway epithelial cells. Exp Lung Res 37:606–614. doi: 10.3109/01902148.2011.623116 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


