Abstract
Supply shortage for the development and production of preventive, therapeutic, and diagnosis tools during the COVID-19 pandemic is an important issue affecting the wealthy and poor nations alike. Antibodies and antigens are especially needed for the production of immunological-based testing tools such as point-of-care tests. Here, we propose a simple and quick magnetic nanoparticle (MNP)–based separation/isolation approach for the repurposing of infected human samples to produce specific antibodies and antigen cocktails. Initially, an antibody cocktail was purified from serums via precipitation and immunoaffinity chromatography. Purified antibodies were conjugated onto MNPs and used as an affinity matrix to separate antigens. The characterization process was performed by ELISA, SDS-PAGE, electrochemistry, isothermal titration calorimetry, and LC-Q-TOF-MS/MS analyses. The MNP-separated peptides can be used for mass spectrometry–based as well as paper-based lateral flow assay diagnostic. The exploitation of the current workflow for the development of efficient diagnostic tools, specific treatments, and fundamental research can significantly impact the present or eventual pandemic. This workflow can be considered as a two birds, one stone–like strategy.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s00216-021-03654-4.
Keywords: SARS-CoV-2, Antigen/antibody purification, Magnetic nanoparticle (MNP)–based separation, Mass spectrometry, Lateral flow assay
Introduction
Since the arrival of the SARS-CoV-2 pandemic, research has been working to provide tools for the fight against the virus. These efforts consisted of developing diagnostic tools and the production of treatments and vaccines. The majority of the reports published during this period have been based on affinity interactions using antibodies. Simple and rapid diagnosis has a significant impact on the screening of individuals to provide treatment and preventive measures. Point-of-care (POC) diagnosis kits have seen more popularity due to their portability, ease of use, and low cost. Immunological assays based on antigens and antibody detection have shown great potential in large-group screening. Nevertheless, the preparation of highly specific antigens and antibodies is a time- and labor-consuming process in addition to the expensiveness related to monoclonal or recombinant antibody production [1].
Monoclonal antibodies (mAbs) demonstrate various attractive features that make them the obvious choice in biomedicine applications compared to polyclonal antibodies. The homogeneity of mAbs permits the precise detection of single specific targets, which allows them to be considered a single entity [2]. Yet, with the urgency needed for the current pandemic, the similarities between the SARS-CoV-2, its variants, and the previous strains made the limitations of mAbs more apparent. Additionally, they are expensive and labor-intensive, and might show low efficacy in vivo. On the other hand, polyclonal Abs can target various epitopes that can cover the whole antigen. They have high avidity and can tolerate slight changes such as glycosylation and minor denaturation compared to mAb [3]. With these concerns, the preparation of antibody cocktails recognizing various epitopes of different targets can potentially relieve diagnostics and treatment development. Indeed, it has been shown in a recent report that the use of an antibody cocktail (either monoclonal or serum-derived polyclonal antibodies) can neutralize the COVID-19 virus and resist SARS-CoV-2 escape [4]. The concept of a “cocktail” that allows the recognition of various epitope targets proves more attractive for diagnostics and treatment compared with single-target approaches.
Another issue faced during the current pandemic is explicitly seen in the middle- and low-income countries. They are still struggling to obtain diagnostic tools and supplies that they urgently require to mitigate the disease. Undeniably, even the wealthiest nations have been hindered by resource shortage which further accentuates the need for a simple approach to produce raw materials such as antibodies and antigens to develop diagnostics locally for urgent times [5].
The present study proposes a simple and quick approach to repurpose human SARS-CoV-2 samples to produce antibody and antigen cocktails that can be used for various applications such as diagnostics, therapy, or specific analysis of protein structures. The purification is based on immunoaffinity procedures using protein A and functionalized magnetic nanoparticles (MNPs) to purify antibodies and antigens (Fig. 1). Furthermore, the purified proteins were characterized using various analytical methods such as LC-Q-TOF-MS/MS, electrochemistry, isothermal titration calorimetry, and electrophoresis to demonstrate the capabilities of the approach in producing highly purified proteins. Given the versatility of the antibody cocktail, the MNP-based purification procedure was shown to produce antibodies able to properly separate and detect antigens from wild-type SARS-CoV-2 and the English B.1.1.7 variant. As a proof of concept, the antigens and antibodies were used for the development of a paper-based lateral flow assay (LFA) system used for the diagnosis of SARS-CoV-2.
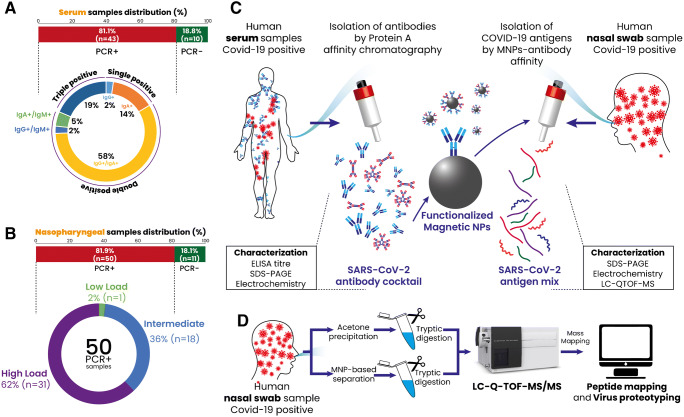
Fig. 1.
Schematic illustration of the current study design. Samples were obtained and analyzed via PCR to determine the presence of SARS-CoV-2 infection. A Serum samples were analyzed with specific COVID-19 anti-IgG, anti-IgA, and anti-IgM. B Nasopharyngeal swab samples were divided into high, intermediate, and low viral load according to the Ct scores obtained from RT-PCR analysis. C The samples (serums and swabs) were selected, pooled, and used to purify SARS-CoV-2-specific antibodies and antigens, respectively. The purified proteins were characterized and identified using different techniques using SDS-PAGE, electrochemistry, and LC-Q-TOF-MS/MS. D Proteotyping of the obtained peptide and swab samples (acetone precipitated) using LC-Q-TOF-MS/MS
Experimental section
The study on infected human samples was performed in an SBL-2+ level laboratory following the highest standard of biosafety guidelines, and all samples were handled by experimented staff. The samples were also inactivated using standard heat treatment for safety. The proteomic analyses and sample handling comply with the minimum information about a proteomics experiment (MIAPE) [6].
Chemicals and reagents
An affinity column (protein A) was obtained from Thermo Fisher. Amine-functionalized iron oxide magnetic nanoparticles (30 nm) were purchased from Sigma-Aldrich (Germany). Anti-COVID IgG, anti-COVID IgA, and anti-COVID IgM ELISA immunoassays were obtained from EUROIMMUN (Luebeck, Germany). Recombinant SARS-CoV spike proteins (S1 and S2) were purchased from RayBiotech (GA, USA). All other chemicals are of analytical grade and were purchased from Sigma-Aldrich.
Human samples and selection
Human samples (serums: n = 53 and nasopharyngeal swabs: n = 61) were provided by Ege University Hospital (Izmir, Turkey) between December 2020 and April 2021. Patients were rigorously diagnosed for COVID-19 using RT-PCR analysis in addition to clinical and radiological observations following national and international guidelines. Samples were stored at − 80 °C until subsequent use. The study on human samples was approved by Ege University Clinical Research Ethics Committee (20-8T/28).
SARS-CoV-2 specific antibody purification
Human serum samples with COVID-19 infection were tested for the presence of anti-COVID-19 IgG, IgA, and IgM using ELISA kits. Samples demonstrating high COVID-19-specific IgG, IgA, and IgM levels were selected and pooled together for the antibody purification process (Fig. 1A, C). After the serums were pooled, 45% ammonium sulfate was applied to precipitate the immunoglobulins. The precipitated sample was dialyzed and applied into an immunoaffinity spin column (protein A column) to purify the immunoglobulins. The eluted fractions were combined and concentrated using a 10-kDa MWCO (molecular weight cut off) membrane. The obtained antibody cocktail solution was stored at − 20 °C in aliquots until used. Protein concentrations were determined via UV measurement at 280 nm with a BSA calibration curve. Purified antibodies were run through gel electrophoresis using a standard protocol [7]. The specificity of the purified antibodies for COVID-19 was determined using commercial anti-COVID-19 IgG, IgA, and IgM ELISA kits and calculating the antibody titer.
Preparation of MNP-Abs and SARS-CoV-2 antigen purification
Antigen purification was performed by affinity-based interaction using antibody-functionalized magnetic nanoparticles (MNPs) according to the protocol described by Denison et al. [8] with modification. The procedure for functionalization was performed according to our previous work [9]. Briefly, purified antibodies were activated via EDC/NHS chemistry to bind on the surface of MNPs. For this purpose, a mixture of 4.0 mg EDC and 9.0 mg NHS was added over the antibody solution and allowed to react for 30 min at room temperature with gentle shaking. The activated proteins were added to the solution containing MNPs with a ratio of 1.5:100 (Ab:MNP) and allowed to react at room temperature for 16 h.
Swab samples with high viral load identified through RT-PCR were pooled (2.1 ± 0.2 mg protein/mL) and mixed with the Ab-functionalized MNPs (200 mg/mL) (Fig. 1B, C). The mixture was left at room temperature for 30 min and then washed three times using PBS to eliminate non-bound substances. The elution process of antigens was performed using an elution buffer containing formic acid/acetonitrile (10%/3.0%, v/v). Purified antigens were collected, aliquoted, and stored at − 20 °C till further use. Each step of the purification procedure was characterized by scanning electron microscopy (SEM) (Thermo Fischer Scientific Apreo S LoVac, OR, USA).
The process of antigen purification via the antibody-conjugated MNPs was optimized for different parameters (binding time, binding capacity, binding buffer pH, and elution buffer composition). The selection of the binding buffer pH was performed using various buffers at different pH levels: sodium citrate pH 5.0, sodium phosphate buffer pH 7.0, ammonium bicarbonate buffer pH 9.0, and sodium carbonate buffer pH 11. The optimization of the elution buffer was realized using three different buffers: formic acid/acetonitrile (10%/3.0%, v/v), glycine buffer (0.1 M, pH 3.0), and ammonium bicarbonate (0.2 M, pH 9.0). The next factor is the antigen-binding time which was estimated through a period of 1.0 h (5, 15, 30, 45, and 60 min). Lastly, the loading capacity was measured by applying several sample protein concentrations over the MNP-Ab immunoaffinity system (30, 40, 50, 60, 80, and 100 μg protein). Purified antigens were analyzed by standard sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) [7]. The reusability of the MNP-Ab for repeatable purifications was explored by applying successive purifications (n = 3) over the same MNP-Ab material.
Electrochemical analysis was carried out to confirm the antibody-antigen interaction using a screen-printed electrode (SPE) with differential pulse voltammetry (DPV) measurement to exhibit the signal responses with a potential range from − 0.4 to + 0.8 V. A neodymium magnet was placed in the center backside of the SPE’s working electrode. After measuring the DPV signal of bare SPE, 10 μL of MNP-Ab suspension was deposited onto the SPE surface, and the DPV signal was measured again. After the SPE was washed, 60 μL of the sample (purified antigen and positive and negative nasopharyngeal swabs) was deposited over the SPE/MNPs-Abs and allowed to incubate for 60 min at room temperature, after which the DPV signal of MNP-Ab-Ag was measured. The biosensor responses were calculated using the difference in DPV signals before and after the addition of the sample, which gives the difference between current values (ΔμA). In another similar experiment set, pure COVID-19 spike proteins (S1 and S2) were attached to MNPs. They were reacted with the purified antibody to confirm the specificity of the MNP-based system in purifying COVID-19 antigens. Isothermal titration calorimetry (ITC) was used to measure the physical binding interaction of the purified antigens with the antibodies. Samples (antigens or antibodies) were deposited in their respective cells, and the interaction was followed through the thermodynamic parameters generated.
Antigen analysis via mass spectrometry
Sample preparation
Protein bands were excised from the SDS-PAGE gel and destained using 100 mM (NH4)HCO3/50% MeOH, 25 mM (NH4)HCO3/50% acetonitrile, and 100% acetonitrile [10, 11]. Destained and dried gel samples were re-suspended in 25 mM ammonium bicarbonate and subjected to the same trypsin digestion process overnight. Peptides were extracted using formic acid/acetonitrile (0.1%/70%, v/v) for 10 min under vigorous shaking. The supernatant was concentrated and used for mass spectrometry analyses.
To identify the isolated antigens after gel electrophoresis, an in-gel digest protocol was used. Briefly, samples (acetone-precipitated swabs or isolated antigens) were dried under N2 gas and re-suspended in 100 μL ammonium bicarbonate buffer (50 mM, pH 8.5). The sample solutions were incubated with 6.0 μL trypsin (20 mg/mL) for 90 min at 37 °C. The reaction was stopped by formic acid addition (final concentration 1.0%) over ice for 10 min [12]. The samples were centrifuged at 4000×g, and the obtained supernatants were taken for mass spectrometric analysis.
LC-Q-TOF-MS/MS analysis
Liquid chromatography quadrupole time-of-flight mass spectrometry (LC-Q-TOF-MS) analysis was performed using an Agilent Technologies 1260 infinity series liquid chromatography system coupled with an Agilent Technologies G6550A Q-TOF LC/MS device (with a dual AJS-ESI ion source). Agilent MassHunter Workstation software was used for both instrument control and data acquisition. The experimental conditions for measurement were performed according to the work of Kotecka-Majchrzak et al. [12].
The SARS-CoV-2 peptides and proteins were identified in the tested samples using the online Mascot search engine (http://www.matrixscience.com/). LC-Q-TOF-MS/MS raw data were first converted to peptide mass value by using Agilent Bioconfirm software. The search parameters were set as follows: both contaminants and hSARS-CoV-2 databases were selected, trypsin enzyme with up to 2 missed cleavages was allowed, peptide mass tolerance was set at ± 1.2 Da, MS/MS tolerance ± 0.6 Da, smaller than seven amino acid length peptides were filtered, taxonomy all entries, formyl (protein N-term) as variable modification, and instrument ESI-QUAD-TOF. Protein sequences were identified using the UniProtKB/Swiss-Prot database, and proteins with scores greater than 38 were considered significant and accepted for protein identification. MassProfiler software (version B.08.01) was used to compare data before and after MNP-based separation.
Development of a paper-based colorimetric lateral flow assay for the detection of SARS-CoV-2 antigens and antibodies
As a proof of concept in the utility of the purified antibodies and antigens in diagnosis application, two paper-based colorimetric lateral flow assays (LFA) were developed: one sandwich-type LFA for the detection of antigens in the nasopharyngeal samples using a competitive-type immunoreaction. The second is for the detection of antibodies in serum samples. The platform was constructed using a nitrocellulose membrane containing two test lines: a control line (C-line) containing rabbit anti-human IgG used as a control for the successful function of the system and a test line (T-line) containing the purified antibodies or antigens which produces an antigen diagnosis LFA or antibody detection LFA, respectively. The color signal was provided by synthesizing a dye-loaded polymersome that was bioconjugated with the purified antibody cocktail and loaded with Coomassie blue dye. The detailed protocol for the biosensor’s design, preparation, and application can be found in our recent work [13].
Statistical analysis
All data were obtained in triplicates, and results are presented as mean ± SD. Statistical significance was measured using the ANOVA test (GraphPad Prism, version 8.0.1). p values lower than 0.05 were considered statistically significant.
Results and discussion
The current study aims to provide a simple approach for the selective purification of antibodies and antigens by repurposing infected human samples (serums and nasopharyngeal swabs) to provide a pool of raw materials that are ready to use for different applications, including diagnosis. The isolation of proteins using magnetic nanoparticle (MNP)–based immunoaffinity could be seen as a quick approach for producing raw materials that can be prioritized for the development of performant diagnostic, therapeutic, and many other applications.
Purification of a SARS-CoV-2-specific antibody cocktail
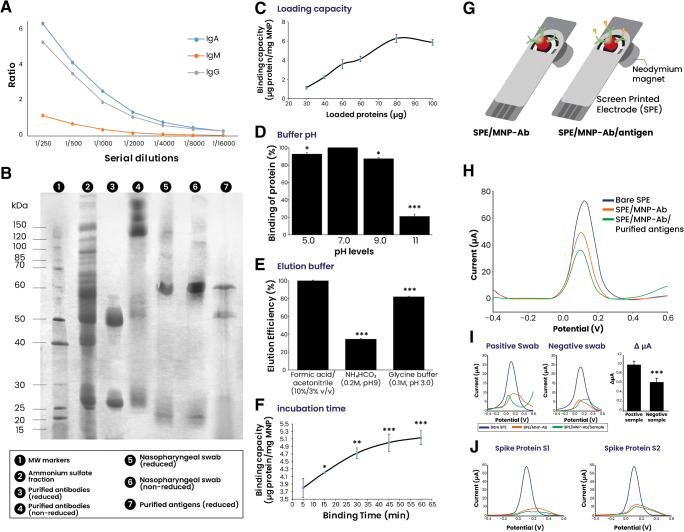
The first step in the present work aimed to purify a SARS-CoV-2-specific antibody cocktail using commercial protein A–based affinity chromatography. The results demonstrated that simple ammonium sulfate precipitation followed by chromatography allows the successful purification of an IgG/IgA/IgM cocktail (yield = 42 ± 5%) that is majorly comprised of COVID-19 specific antibodies. The selection of patient samples with high immunoglobulin levels provided substantial amounts of purified antibodies as shown in the antibody titer (1:2000 for IgG and IgA; 1:250 for IgM) (Fig. 2A). In addition to the ELISA testing, electrophoresis analysis of the purified antibodies under reducing and non-reducing conditions demonstrated bands around 50 and 25 kDa, typical for the heavy and light chains that make the antibody structure. Heavy chains from other antibodies are known to have variations in their molecular weights being 60 kDa for IgA and 75 kDa for IgM. A band of ~ 57 kDa is observed and can be attributed to the IgA heavy chain. There were no observable IgM heavy chains due to the low concentration of IgM in the purified antibody cocktail, as shown in the low antibody titer.
Fig. 2.
Purification and characterization of antibody and antigen cocktails. Serum samples from COVID-19-positive patients were taken and analyzed via ELISA for IgG and IgA levels, after which the samples with the highest amounts were subjected to simple purification via protein A affinity. A Purified antibody cocktail titer analysis using COVID-19-specific ELISA kits. B Electrophoresis analysis of both purified antibodies and antigen cocktails under reduced and non-reduced conditions. C–F Optimization of the various parameters implicated in the functioning of MNP-based immunoaffinity antigen purification. G Illustration of the working principle of the electrochemical-based analysis. H Electrochemical confirmation of the antibody/antigen attachment. I Confirmation of the antigen presence via testing a positive and negative swab sample and calculating ΔμA. J Confirmation of the antibody specificity using MNPs functionalized with pure SARS-CoV-2 spike proteins (S1 and S2). *p < 0.05, **p < 0.01, and ***p < 0.001 vs the optimal condition marker
On the other hand, bands of 150 kDa or higher are representative of native antibody structures in monomer, dimer (IgA), and pentamer forms (IgM) (Fig. 2B). The specificity of the purified antibodies was further confirmed using electrochemistry analyses and isothermal titration calorimetry (Fig. 2G–J, Fig. S2). Pure spike proteins (S1 and S2) were functionalized over the MNP surface. After adding the antibodies, the attachment of the antibodies induced a difference in the signal measurement compared to negative samples. Many studies described that a high antibody titer is often related to severe-infection cases [14, 15]. In addition, it has been shown that antibody cocktails have more potential for the precise recognition of the virus antigens than a single monoclonal antibody due to their capacity to recognize multiple epitope targets of the virus [4, 16]. Polyclonal antibodies have a high affinity to their targets, making them a good choice for application in immunoassays. Furthermore, they are easy to store and can handle minor deviations in epitopes (conformational changes, polymorphism, or denaturation) [17].
Antibody conjugation to MNPs and antigen separation
In the aim to prepare an immunoaffinity system for the quick purification of SARS-CoV-2 antigens, the antibodies purified earlier were conjugated to MNPs. The separation and purification process of proteins from other unneeded components as well as possible impurities in the medium of complex matrices is an essential step in sample preparation and analysis. As such, the use of novel strategies based on nanomaterials has become popular due to their usefulness in bypassing the limitations observed with orthodox extraction methods (high volume consumption, extended time, etc.) [18]. The current work aimed to explore MNPs for the purification process. The functionalization of the MNPs demonstrated a 12.5-μg Ab bound/mg MNP.
The magnetic features of MNPs are of great interest due to facilitating and hastening the proteins’ separation process. The general procedure involves mixing Ab-functionalized MNPs (MNP-Ab) with the target matrix solution (serums, nasopharyngeal liquid, etc.). After the target proteins are captured, MNPs are immobilized via strong magnets, and then the unattached molecules are eliminated. Lastly, the target proteins are eluted (by changing solvents) and stocked for further use after being washed [18]. In the current work, the preparation of the MNP-based system for the separation and purification of COVID-19 antigens was characterized via SEM analysis. The attachment of antibodies onto the MNP surface showed a higher density of the material and a slight change in the surface color (Fig. S1).
To optimize the purification system, various factors (binding time, buffer pH, loading capacity, and the elution buffer type) have been explored to determine the optimal conditions (Fig. 2C–F). Results showed that the binding capacity of the MNP-based system was defined as 6.0 μg protein/mg MNP (yield = 49 ± 2%). The increase of sample amount showed exponential antigen binding reaching a maximum (6.0 μg protein/mg MNP) at 80 μg of the loaded sample. Higher amounts of the loaded sample had no changes suggesting saturation of all binding sites (Fig. 2C). Physiological pH levels (pH 7.0) showed the highest protein binding compared to other pH levels. A highly basic pH (pH 11) showed poor binding that can be related to protein damage or environment electrons and proton perturbation (Fig. 2D). The type of buffer employed is another factor that may affect the purification process. Data demonstrated that a mixture of formic acid/acetonitrile (10%/3%, v/v) produced the most optimal elution process compared with other types of buffers (Fig. 2E). The last factor to optimize was the binding time where it was time-dependent. Although the increase continued till 60 min of incubation, it had no statistical significance compared with 30 min (Fig. 2F). As such, the current system showed a maximum binding capacity of 6.0 μg antigens/mg MNP at pH 7.0 with an extraction time of 30 min and elution with a mixture of formic acid/acetonitrile (10%/3%, v/v). There exist many commercially available MNPs functionalized with a wide variety of functionalities such as protein A/G, amine groups, biotin/streptavidin, etc., which further expand their potential applications. Generally, MNP-based separation methods take around 15 min to 120 min for the purification, depending on the protein type [18]. Furthermore, the main advantage of such an MNP-based system is their reusability after simple washing processes, which further underline their economic potential in parallel to their effectiveness and easy-to-use operations.
A COVID-19-positive patient’s nasopharyngeal samples were selected (with high viral load determined from PCR data) and applied through the MNP-based separation system to purify antigens. The eluted samples were characterized via electrophoresis (Fig. 2B) and electrochemical analysis compared with commercially obtained pure S1 and S2 spike proteins (Fig. 2G–J). The use of S1 and S2 proteins demonstrated that the purified antibody cocktail at least contains spike protein–recognizing antibodies. In a similar logic, the antigen mixtures can also be said to possess at least spike proteins in their content. The electrophoretic analysis of the purified antigens demonstrated two major bands of a molecular weight around 48 and 60 kDa and a less obvious band around 20–25 kDa. The composition of the SARS-CoV-2 proteins has been extensively analyzed, and the major proteins were found to be at 47–55 kDa for N protein, 60–80 kDa for S1 and S2 separately (130–150 kDa for complete monomeric spike protein), and 15 kDa for membrane protein [19]. Our findings align with the statements above, demonstrating protein bands of the same interval molecular weights.
Additionally, the purified antigens were characterized using electrochemical analysis using screen-printed electrodes (SPE) and the earlier prepared MNPs-Ab. The SPE-based electrochemical analysis is a simple tool that has excellent biosensing potential for analyte detection. It consists of trapping the MNP-Ab on the SPE surface using magnets, after which the sample is added over the surface (Fig. 2G). The addition of purified antigens or purified spike proteins induces a change in the current measured via differential pulse voltammetry (DPV) (Fig. 2H–J). The difference in signals between SPE-Ab and SPE-Ab/antigen highlights the attachment of the purified antigens over the functionalized MNPs.
Furthermore, positive and negative swab samples were tested, and the obtained differences between the signal further demonstrate the recognition of COVID-19 antigens by the MNP-Ab. The use of MNPs for the specific detection of SARS-CoV-2 antigens has been reported in many researches [20] and showed excellent sensitivity in different matrices such as saliva [21]. To further confirm the specificity of the purified antigens and antibodies, isothermal titration calorimetry was employed to measure the affinity between them (Fig. S2). The obtained results underline the recognition and affinity between the two proteins, which further prove the efficiency of the proposed system for the repurposing of human samples in the purification of antigens and antibodies. The capacity of the MNP-Ab utilization for several successive purifications was explored and showed 100%, 97%, and 90% from the first to the third application, respectively (Fig. S3). The capacity of the MNP-Ab was tested after 2 months from preparation and showed a capacity of 50% (data not shown).
Antigen sequence identification and analysis
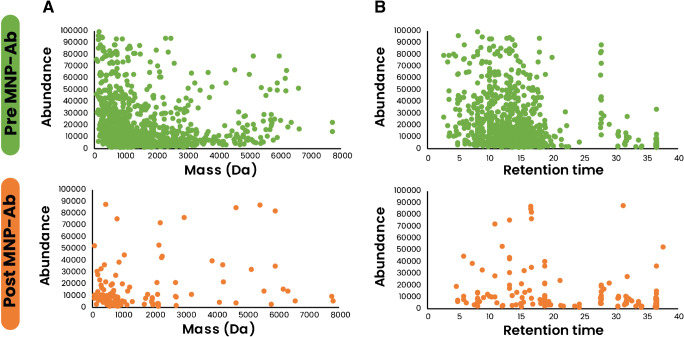
The use of MNPs has a significant role in developing biosensors by taking advantage of magnetic separation. The latter can facilitate the cleaning and enrichment of target molecules, enabling the purified biomolecules’ easy exploitation for different applications [22]. In this sense, the characterization demonstrated earlier using the repurposed human samples for the purification of antibodies and antigens presents a great potential for the development of an electrochemical biosensor for the diagnosis of SARS-CoV-2 taking advantage of the multiple epitope target–recognizing antibody cocktails or the antigen mix that can be immobilized over the MNPs for the detection of antigen or antibody presence, respectively. Furthermore, the MNP-based separation can be seen as a sample preparatory method to clean, enrich, or explore the antigen protein sequences. Indeed, the current study used mass spectroscopy to analyze nasopharyngeal antigens pre- and post-passage through the MNP system. The use of MNP-based separation allowed the selective purification of COVID-19-specific antigens and eliminating unnecessary proteins going from 946 peptides pre-MNP-Ab to 66 peptides post-MNP-Ab separation (Table S1, Fig. 3).
Fig. 3.
Comparison between the peptide content in nasopharyngeal swabs and the purified antigens after passing through the MNP-based system. Comparison according to A mass data and B retention time
Identifying the separated proteins in the pooled nasopharyngeal samples and the eluted samples further demonstrated the specificity of the separation. Proteins and peptides identified through LC-Q-TOF-MS in the pooled swabs and the purified antigens showed an important number of SARS-CoV-2 proteins and peptides that were preserved after purification (Table S1). The peptide sequences identified were further filtered, and only peptides of 7 amino acids or higher were considered. These peptides were mainly represented by the spike protein and nucleoprotein. It should be mentioned that other proteins (membrane, envelope, and ORF) were also identified but showed lower scores than S and N proteins.
In-gel digestion of the purified peptides observed after SDS-PAGE further confirmed our initial estimation that the bands corresponded to S, N, M, E, and other structural proteins (Fig. 2, Table 1). These potential peptides were again checked for their positions over the entire sequence of each respective protein to determine if key regions or domains were identified. The raw data of the analysis with all possible identified peptides were also taken into consideration, and only peptides with a higher score were selected and presented (Table S2). It has been reported that COVID-19 patient antibodies mainly react to peptides derived from S and N proteins followed by ORF1. However, ORF1 was also detected by antibodies from patients infected with other coronaviruses. This suggests that the recognition of ORF1 is a result of cross-reactivity due to prior infections. There was also a low detection of peptides derived from M, ORF3, and ORF9 from SARS-CoV-2 patients’ antibodies [16]. These results corroborate our findings, especially with the low peptide hits found in proteins other than S and N proteins. In the process of infection, the spike protein is cut into S1 and S2 subunits. The S1 protein consists of the N-terminal domain (NTD) and the receptor-binding domain (RBD), responsible for the interaction with ACE2. Besides, the NTD function is still not fully understood. It has been seen that in other coronaviruses, the NTD interacts with glycated moieties during attachment and thus has a potential role in the transition between prefusion and postfusion states of the S protein during infection. Furthermore, the NTD of the S protein was shown to be a key target for neutralizing antibodies in MERS-CoV [23].
Table 1.
Peptide identification from the in-gel digestion of the purified antigen
| Protein name/ accession number |
Molecular weight (Da) | Position | Peptide sequence | Domain | Sequence coverage (%) | Expected value | Protein score | Retention time (min) | Mass |
|---|---|---|---|---|---|---|---|---|---|
|
Nucleoprotein/ NCAP_SARS2 |
45,598 |
33–38 62–68 94–100 101–107 144–149 196–203 250–256 294–299 362–369 |
SGARSK EDLKFPR IRGGDGK MKDLSPR SAAEASK NSTPGSSR QGTDYK DHIGTR TFPPTEPK |
NTD NTD NTD NTD NTD SR-rich linker SR-rich linker CTD CTD |
14 | 2.8e−05 | 71 |
1.748 10.009 8.99 14.535 11.876 3.106 8.141 4.463 17.195 |
605.3003 905.3679 702.3656 846.4745 663.2919 805.4151 711.3777 699.3434 916.4878 |
|
Spike Glycoprotein/ SPIKE_SARS2 |
141,088 |
35–41 196–202 238–246 305–310 409–417 458–466 530–535 568–577 815–825 987–995 1020–1028 1039–1045 |
GVYYPDK NIDGYFK FQTLLALHR SFTVEK QIAPGQTGK KSNLKPFER STNLVK DIADTTDAVR RSFIEDLLFNK VEAEVQIDR ASANLAATK RVDFCGK |
NTD NTD NTD NTD RBD RBD SD1 SD1 FP – HR HR |
7 | 3.5e−05 | 70 |
15.271 14.761 11.536 8.141 7.293 7.293 5.765 8.537 8.537 8.537 14.535 16.515 |
840.3916 855.4207 1099.5145 711.3777 898.4402 559.2666 660.3676 538.2224 1381.5842 1059.4718 846.4745 824.4274 |
|
Membrane/ VME1_SARS2 |
25,130 |
43–50 151–158 175–180 181–186 |
NRFLYIIK IAGHHLGR TLSYYK LGASQR |
– | 12 | 0.0031 | 51 |
19.514 15.836 7.01 10.065 |
1065.5635 859.4678 774.3603 632.3762 |
|
ORF3a/ AP3A_SARS2 |
31,103 |
22–30 62–67 69–75 193–198 |
DATPSDFVR IITLKK WQLALSK WESGVK |
– | 10 | 0.0067 | 47 |
8.537 7.575 14.535 6.387 |
1007.4616 716.3357 846.4745 704.3564 |
|
ORF8/ NS8_SARS2 |
13,822 |
45–52 95–101 116–121 |
WYIRVGAR LGSLVVR VVLDFI |
– | 17 | 0.011 | 45 |
11.536 10.009 6.387 |
1021.46 371.655 704.3564 |
|
Envelope/ VEMP_SARS2 |
8360 |
62–69 64–69 70–75 |
VKNLNSSR NLNSSR VPDLLV |
CTD | 18 | 0.012 | 38 |
7.575 14.705 2.653 |
916.4513 689.3618 655.3016 |
Placeholder TextPeptides with 7 amino acids or higher are considered as plausible sequences for further analyses
The use of an antibody cocktail has great potential in enhancing the recognition and enrichment properties of the proposed MNP-based system. It has been demonstrated that from a population point of view, IgG and IgA have a similar recognition frequency of antigens. Nevertheless, at an individual level, there have been various discrepancies showing that some epitopes are recognized by only IgG, only IgA, or both antibodies. This suggests that patients generate distinct antibody responses to SARS-CoV-2. As for IgM, it is suggested that the low presence is probably due to a monoclonal response, and given some signals, IgM clones transform into IgA or IgG antibodies [16].
Application potential of the current study
Identification capabilities using mass spectrometry
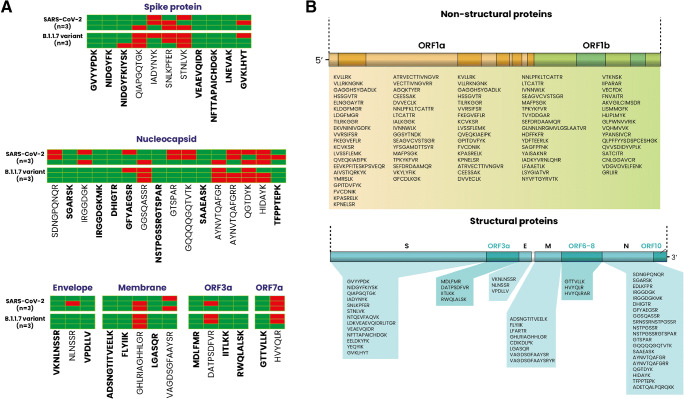
The collection of proposed therapeutic antibodies and vaccines were initially designed based on the first described spike protein. However, with the emergence of the various variants, many preliminary studies have suggested that these antibodies’ neutralization/detection efficiency might be reduced against these variants containing mutations [4]. From this logic, some of the described peptides risk being different on the proteins of the virus variants. Our approach consisted of examining nasopharyngeal samples taken from confirmed cases of the B.1.1.7 (English variant). The data showed that some of the found peptide sequences in the earlier SARS-CoV-2 samples were not found in the B.1.1.7 variant suggesting that the proposed peptides might not have a great potential for large spectrum diagnostic, therapy, or vaccine purposes (Fig. 4A). Results showed various sequences from both structural (spike protein, nucleoprotein, envelope, membrane) and non-structural (ORF3a and ORF7) proteins that are common between the wild type and the English variant. This demonstrates that the purified antibodies could potentially be used in diagnostic applications with a wide range of recognition that can detect other variants given the various target epitopes it can recognize. This consequently provides a group of target peptides that can be used as an identification base in LC-Q-TOF-MS/MS or other methods for diagnostics. Indeed, the use of mass spectrometry analysis for diagnostics can provide a high level of accuracy due to the detection of a myriad of peptides that can give a general idea of the protein content and allow for a diagnosis verdict compared with other methods that target only one protein or a specific region. Our experimental data was able to detect peptides from structural and non-structural proteins (Fig. 4B). The peptide sequences can be used as a base for diagnosis or explored to develop different immunological-based diagnosis tools.
Fig. 4.
A Peptide identification and analysis of SARS-CoV-2 samples and its B.1.1.7 variant via LC-Q-TOF-MS/MS (n = 6). Bold peptides are sequences found in both SARS-CoV-2 and B.1.1.7 variants (n = 3 each). B Ensemble of the sequences that could be detected through LC-Q-TOF-MS/MS and their positioning over the SARS-CoV-2 genome. Non-structural proteins are represented mainly by replicase 1a (from ORF1a) and replicase 1ab (from ORF1a and b). Red and green colors represent the absence or presence of the peptide sequence in the tested samples
Lateral flow assay development for diagnostics
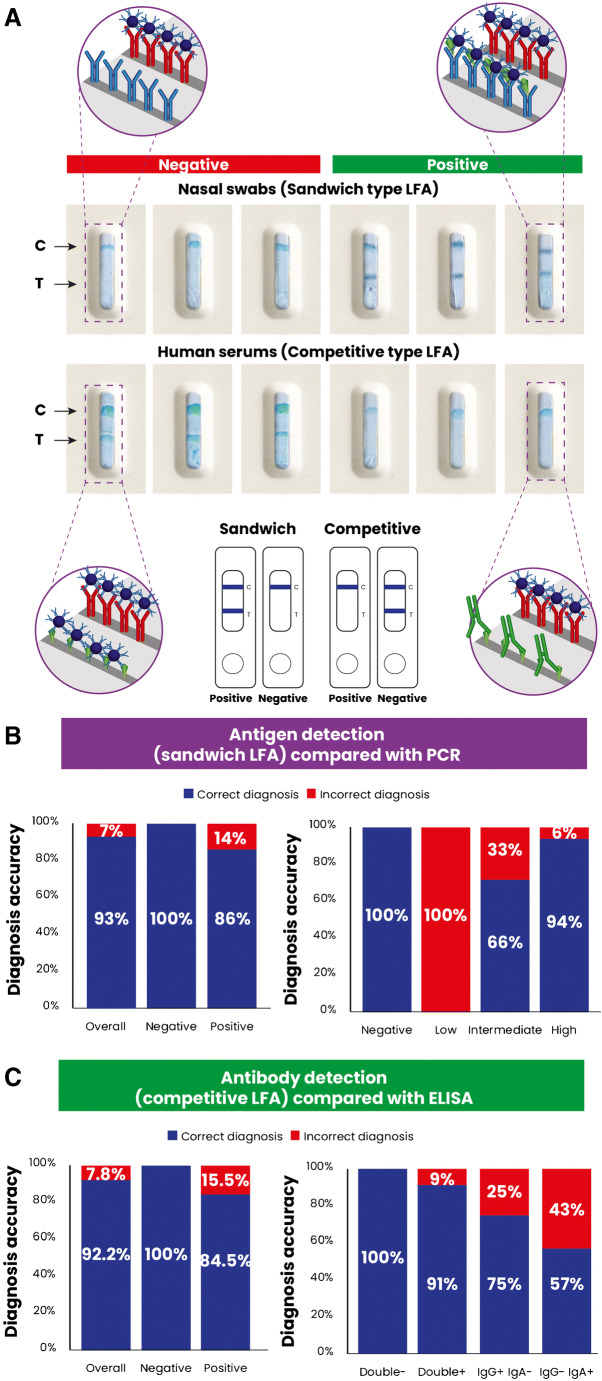
As a proof of concept of the feasibility of the purified antigen and antibody application in developing highly sensitive diagnostic tools, two types of lateral flow assays (LFA) were designed to detect antigens or antibodies in nasopharyngeal or serum samples, respectively (Fig. 5A). The human samples were tested compared to RT-PCR or commercial ELISA results. The obtained data demonstrated a 93% correlation with the RT-PCR results for the antigen detection capacity. On the other hand, 92.2% agreement was seen between our antibody test and the commercial ELISA (Fig. 5B, C). It has been shown that the specificity and sensitivity of biosensors towards antigen detection are closely related to Ct scores. We have also explored the sensitivity of the proposed platform according to Ct values and divided the samples into high-, middle-, and low-viral-load groups. The results showed 94% correct diagnosis for the high-viral-load groups followed by the middle and low groups. On the one hand, this decrease in sensitivity can be attributed to the limited number of antigens to be detected in the samples but also the limited number of samples used for the study (Fig. 5B, C). The antibody detection produced a 91% correct detection when samples contain SARS-CoV-2-specific IgG and IgA. This value also decreases when samples are either IgG+ or IgA+ alone. The rationale is that the development of COVID-19 antibody production happens in different steps where IgM is generally associated with infection within the first days, followed by IgG production, and then IgA [24]. The detection of antibodies is important for epidemiological studies and can help predict the date of the infection onset. Additionally, it helps predict immune response patterns that are necessary for developing prevention methods. A more thorough study of the application of the current purified antigens and antibodies has been reported by our research group in our recent publication [13].
Fig. 5.
Development of a lateral flow assay (LFA) using the purified antigens and antibodies to diagnose SARS-CoV-2. A Schematic representation and visual observations of the sandwich-type and competitive-type LFAs. B, C Analytical assessment of the LFA platform and estimation of the accuracy compared with RT-PCR. Reprinted with permission from [13]. Copyright 2021 American Chemical Society
Limitations, perspectives, and conclusions
The current study describes a methodology for a quick and simple workflow in repurposing SARS-CoV-2 samples to produce precise and accurate targeting and generate a low-cost pool of raw materials (antibodies and antigens) used for critical research related to diagnostics, therapy, or vaccine development. The advantage resides in the fact that the human samples are destined to be eliminated after PCR or ELISA analysis, thus making the source free of recruiting patients or volunteers. In addition, while it is understandable that outside pandemic times, finding enough patients is difficult, the current report proposes this approach for urgent times such as pandemics where the supply chain cannot be predicted. It should be noted that biosafety is an important factor in this situation and needs to be handled carefully using proper lab safety measures and experimented staff. The use of MNP-based separation methods to isolate specific proteins and peptides could be adapted to a broad range of proteomics-based applications. The current study demonstrated that with simple sample preparatory techniques, highly purified materials could be produced. The purified antibodies show great potential as a tool to be explored in diagnostic procedures such as ELISA, lateral flow assay, biosensors, etc. We have explored the potential of the purified antigens and antibodies in the current study by developing a colorimetric paper-based LFA where the system efficiently recognized positive and negative samples with a sensitivity of 93% compared with RT-PCR results [13]. These materials can also be exploited in combination with mass spectrometry to explore the antigen sequences for identification. Some interesting research has used mass spectrometry techniques to identify and diagnose SARS-CoV-2, providing a fast turnaround reaching as low as 3 min [25–27]. Here, it is indeed difficult to draw strong conclusions about the feasibility of the identified peptide sequences for such applications. However, it is indeed a promising approach for bulk production of material supplies in urgent times such as pandemics or the case of low-financial power. The produced antibodies are designed for a wide-range screening. However, additional studies to check cross-reactivity against other microorganisms should be cleared. Moreover, performing deeper profiling of the antigens in larger cohorts with different demographic factors (age, race, and sex) and other virus variants will provide a better understanding of the epitope landscape which can pave a stepping stone for the isolation and creation of efficient tools that can standardize the approaches to fight the current or any eventual pandemic.
Supplementary information
(DOCX 482 kb)
(XLSX 147 kb)
(XLSX 23.8 kb)
(DOCX 14.1 kb)
Acknowledgements
We acknowledge the staff of EGE University Hospital who are on the front line fighting against the COVID-19 pandemic. The research project was supported by TUBITAK (Project number: 120R021). The Republic of Turkey Ministry of Development provided the support for the infrastructure of EGE MATAL (Ege University/Izmir) via 2010K120810/2020K12150700 and 2016K121190 grants. The authors thank the SEM A.S Company for the technical support and the help provided during the mass spectrometry data analysis.
Author contribution
The manuscript was written through the contributions of all the authors. All of the authors approved the final version of the manuscript.
Funding
The study was supported by TUBITAK (Project number: 120R021). The Republic of Turkey Ministry of Development provided the support for the infrastructure of EGE MATAL (Ege University/Izmir) via 2010K120810/2020K12150700 and 2016K121190 grants.
Data availability
All data obtained in the current study are provided as Online Resources.
Declarations
Ethics approval
The study was approved by the Ege University Clinical Research Ethics Committee (20-8T/28).
Consent to participate
All samples were taken after the obtention of signed consent from the patients.
Consent for publication
All authors have read and approved the manuscript for publication.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kerem Tok and Hichem Moulahoum contributed equally to this work.
Contributor Information
Hichem Moulahoum, Email: Hic_moul@hotmail.com.
Figen Zihnioglu, Email: figen.zihnioglu@ege.edu.tr.
References
- 1.Orooji Y, Sohrabi H, Hemmat N, Oroojalian F, Baradaran B, Mokhtarzadeh A, et al. An overview on SARS-CoV-2 (COVID-19) and other human coronaviruses and their detection capability via amplification assay, chemical sensing, biosensing, immunosensing, and clinical assays. Nano Lett. 2021;13(1):18. doi: 10.1007/s40820-020-00533-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Read AJ, Gauci CG, Lightowlers MW. Purification of polyclonal anti-conformational antibodies for use in affinity selection from random peptide phage display libraries: a study using the hydatid vaccine EG95. J Chromatogr B Anal Technol Biomed Life Sci. 2009;877(14–15):1516–1522. doi: 10.1016/j.jchromb.2009.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuravsky ML, Schmalhausen EV, Pozdnyakova NV, Muronetz VI. Isolation of antibodies against different protein conformations using immunoaffinity chromatography. Anal Biochem. 2012;426(1):47–53. doi: 10.1016/j.ab.2012.03.026. [DOI] [PubMed] [Google Scholar]
- 4.Chen RE, Zhang X, Case JB, Winkler ES, Liu Y, VanBlargan LA, et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonalantibodies. Nat Med 27 (4):717–26.10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed]
- 5.McMahon DE, Peters GA, Ivers LC, Freeman EE. Global resource shortages during COVID-19: bad news for low-income countries. PLoS Negl Trop Dis. 2020;14(7):e0008412. doi: 10.1371/journal.pntd.0008412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor CF, Paton NW, Lilley KS, Binz PA, Julian RK, Jr, Jones AR, et al. The minimum information about a proteomics experiment (MIAPE) Nat Biotechnol. 2007;25(8):887–893. doi: 10.1038/nbt1329. [DOI] [PubMed] [Google Scholar]
- 7.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 8.Denison MIJ, Raman S, Duraisamy N, Thangavelu RM, Riyaz SUM, Gunasekaran D, et al. Preparation, characterization and application of antibody-conjugated magnetic nanoparticles in the purification of begomovirus. RSC Adv. 2015;5(121):99820–99831. doi: 10.1039/C5RA17982C. [DOI] [Google Scholar]
- 9.Sanli S, Ghorbani-Zamani F, Moulahoum H, Gumus ZP, Coskunol H, Odaci Demirkol D, et al. Application of biofunctionalized magnetic nanoparticles based-sensing in abused drugs diagnostics. Anal Chem. 2020;92(1):1033–1040. doi: 10.1021/acs.analchem.9b04025. [DOI] [PubMed] [Google Scholar]
- 10.Glatter T, Ludwig C, Ahrne E, Aebersold R, Heck AJ, Schmidt A. Large-scale quantitative assessment of different in-solution protein digestion protocols reveals superior cleavage efficiency of tandem Lys-C/trypsin proteolysis over trypsin digestion. J Proteome Res. 2012;11(11):5145–5156. doi: 10.1021/pr300273g. [DOI] [PubMed] [Google Scholar]
- 11.Jimenez CR, Huang L, Qiu Y, Burlingame AL. In-gel digestion of proteins for MALDI-MS fingerprint mapping. Curr Protoc Protein Sci. 2001; Chapter 16(1):Unit 16 4. [DOI] [PubMed]
- 12.Kotecka-Majchrzak K, Sumara A, Fornal E, Montowska M. Proteomic analysis of oilseed cake: a comparative study of species-specific proteins and peptides extracted from ten seed species. J Sci Food Agric. 2021;101(1):297–306. doi: 10.1002/jsfa.10643. [DOI] [PubMed] [Google Scholar]
- 13.Ghorbanizamani F, Tok K, Moulahoum H, Harmanci D, Hanoglu SB, Durmus C, et al. Dye-loaded polymersome-based lateral flow assay: rational design of a COVID-19 testing platform by repurposing SARS-CoV-2 antibody cocktail and antigens obtained from positive human samples. ACS Sens 6 (8):2988–97. 10.1021/acssensors.1c00854. [DOI] [PubMed]
- 14.Liu L, To KK, Chan KH, Wong YC, Zhou R, Kwan KY, et al. High neutralizing antibody titer in intensive care unit patients with COVID-19. Emerg Microbes Infect. 2020;9(1):1664–1670. doi: 10.1080/22221751.2020.1791738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salazar E, Kuchipudi SV, Christensen PA, Eagar TN, Yi X, Zhao P, et al. Relationship between anti-spike protein antibody titers and SARS-CoV-2 in vitro virus neutralization in convalescent plasma. bioRxiv. 2020:2020.06.08.138990.
- 16.Shrock E, Fujimura E, Kula T, Timms RT, Lee IH, Leng Y, et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370(6520):eabd4250. doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lipman NS, Jackson LR, Trudel LJ, Weis-Garcia F. Monoclonal versus polyclonal antibodies: distinguishing characteristics, applications, and information resources. ILAR J. 2005;46(3):258–268. doi: 10.1093/ilar.46.3.258. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez-Garcia E, Marina ML, Garcia MC. Nanomaterials in protein sample preparation. Sep Purif Rev. 2020;49(3):229–264. doi: 10.1080/15422119.2019.1581216. [DOI] [Google Scholar]
- 19.Bates TA, Weinstein JB, Farley S, Leier HC, Messer WB, Tafesse FG. Cross-reactivity of SARS-CoV structural protein antibodies against SARS-CoV-2. Cell Rep 2021;34 (7):108737. 10.1016/j.celrep.2021.108737. [DOI] [PMC free article] [PubMed]
- 20.Pishva P, Yuce M. Nanomaterials to tackle the COVID-19 pandemic. Emergent Mater. 2021;4(1):1–19. doi: 10.1007/s42247-021-00184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fabiani L, Saroglia M, Galata G, De Santis R, Fillo S, Luca V, et al. Magnetic beads combined with carbon black-based screen-printed electrodes for COVID-19: a reliable and miniaturized electrochemical immunosensor for SARS-CoV-2 detection in saliva. Biosens Bioelectron. 2021;171:112686. doi: 10.1016/j.bios.2020.112686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yildiz I. Applications of magnetic nanoparticles in biomedical separation and purification. Nanotechnol Rev. 2016;5(3):331–340. [Google Scholar]
- 23.Chi X, Yan R, Zhang J, Zhang G, Zhang Y, Hao M, et al. A neutralizing human antibody binds to the N-terminal domain of the spike protein of SARS-CoV-2. Science. 2020;369(6504):650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu L, Zhang H, Zhan M, Jiang J, Yin H, Dauphars DJ, et al. Antibody response and therapy in COVID-19 patients: what can be learned for vaccine development? Sci China Life Sci. 2020;63(12):1833–1849. doi: 10.1007/s11427-020-1859-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gouveia D, Miotello G, Gallais F, Gaillard JC, Debroas S, Bellanger L, et al. Proteotyping SARS-CoV-2 virus from nasopharyngeal swabs: a proof-of-concept focused on a 3 min mass spectrometry window. J Proteome Res. 2020;19(11):4407–4416. doi: 10.1021/acs.jproteome.0c00535. [DOI] [PubMed] [Google Scholar]
- 26.Ihling C, Tanzler D, Hagemann S, Kehlen A, Huttelmaier S, Arlt C, et al. Mass spectrometric identification of SARS-CoV-2 proteins from gargle solution samples of COVID-19 patients. J Proteome Res. 2020;19(11):4389–4392. doi: 10.1021/acs.jproteome.0c00280. [DOI] [PubMed] [Google Scholar]
- 27.Nikolaev EN, Indeykina MI, Brzhozovskiy AG, Bugrova AE, Kononikhin AS, Starodubtseva NL, et al. Mass-spectrometric detection of SARS-CoV-2 virus in scrapings of the epithelium of the nasopharynx of infected patients via nucleocapsid N protein. J Proteome Res. 2020;19(11):4393–4397. doi: 10.1021/acs.jproteome.0c00412. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 482 kb)
(XLSX 147 kb)
(XLSX 23.8 kb)
(DOCX 14.1 kb)
Data Availability Statement
All data obtained in the current study are provided as Online Resources.