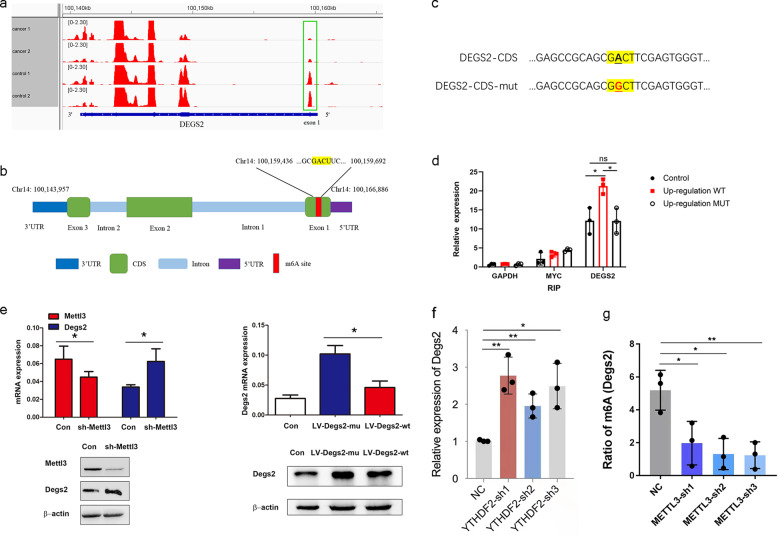
Fig. 4. m6A methylation of the CDS regulates translation of DEGS2.
a m6A peaks were enriched in the CDS and 3′ UTR of the DEGS2 according to the m6A-RIP-seq data. Squares indicate marked decreases in m6A peaks in CRC tissues compared to control tissues; b m6A-RIP assay confirmed the exact m6A location in the CDS region of DEGS2 mRNA. GAPDH mRNA was used as a nontarget control, and MYC mRNA was used as a positive control. ns, not significant; *p < 0.05. c Schematic representation of the position of the m6A motifs within DEGS2 mRNA. d Schematic representation of the mutation (GACT to GGCT) in the CDS to investigate the roles of m6A in DEGS2 expression. e sh-METTL3, LV-DEGS2-mut, and LV-DEGS2-WT were transfected into HCT116 cells for 24 h. Protein expression was measured by WB (bottom panel) and quantitatively analyzed (upper panel). f knockdown of YTHDF2 promoted the DEGS2 mRNA level. g knockdown of METTL3 reduced m6A methylation of DEGS2.