Abstract
Electronic health records (EHR) provide an unprecedented opportunity to conduct large, cost-efficient, population-based studies. However, the studies of heterogeneous diseases, such as chronic obstructive pulmonary disease (COPD), often require labor-intensive clinical review and testing, limiting widespread use of these important resources. To develop a generalizable and efficient method for accurate identification of large COPD cohorts in EHRs, a COPD datamart was developed from 3420 participants meeting inclusion criteria in the Mass General Brigham Biobank. Training and test sets were selected and labeled with gold-standard COPD classifications obtained from chart review by pulmonologists. Multiple classes of algorithms were built utilizing both structured (e.g. ICD codes) and unstructured (e.g. medical notes) data via elastic net regression. Models explicitly including and excluding spirometry features were compared. External validation of the final algorithm was conducted in an independent biobank with a different EHR system. The final COPD classification model demonstrated excellent positive predictive value (PPV; 91.7%), sensitivity (71.7%), and specificity (94.4%). This algorithm performed well not only within the MGBB, but also demonstrated similar or improved classification performance in an independent biobank (PPV 93.5%, sensitivity 61.4%, specificity 90%). Ancillary comparisons showed that the classification model built including a binary feature for FEV1/FVC produced substantially higher sensitivity than those excluding. This study fills a gap in COPD research involving population-based EHRs, providing an important resource for the rapid, automated classification of COPD cases that is both cost-efficient and requires minimal information from unstructured medical records.
Subject terms: High-throughput screening, Machine learning, Predictive medicine, Epidemiology, Population screening, Medical research
Introduction
The estimated world prevalence of COPD ranges from 4–10%, and it is projected to be the third leading cause of mortality by 20301,2. Diagnosis of COPD can be confirmed through spirometry demonstrating a forced expiratory volume in 1 s (FEV1) to forced vital capacity (FVC) ratio of < 0.7 that persists after administration of inhaled bronchodilators3. However, relying solely on FEV1/FVC to diagnose patients in clinical settings is marked by both under- and misdiagnoses4–6, as it comprises only one parameter of the criteria necessary to establish COPD. In most epidemiological studies of COPD, ascertainment of case populations thus frequently requires labor-intensive, direct medical review in addition to testing (e.g. spirometry) by trained clinicians. This process is a significant rate-limiting factor in large-scale COPD studies, especially in the development of new cohorts.
Electronic health record (EHR) databases and linked biorepositories have become increasingly common7–9, and present a major opportunity for disease research. Numerous studies have demonstrated the utility of leveraging EHRs and International Classification of Diseases (ICD) codes for the accurate identification of complex, heterogeneous conditions such as asthma, rheumatoid arthritis, bipolar disorder, and Alzheimer’s disease10–14. The development of EHR algorithms can facilitate rapid, low-cost, and large population-based explorations of disease that can be employed across a variety of studies, such as pharmacologic surveillance15–17, personalized medicine18,19, and genetic association9,10,20.
Prior EHR-based classification heuristics and algorithms for COPD have reported low positive predictive value (PPV) estimates ranging from 36.7% to 80.7%21–23. Therefore, our aim was to develop a high-performing, portable COPD classification algorithm using EHR data in the Mass General Brigham Biobank (MGBB). This diagnostic classification algorithm facilitates rapid identification (i.e., without requiring labor-intensive manual chart reviews by clinicians) of COPD cases with high specificity. It has been independently validated, and has been successfully implemented across a larger national consortium of biobanks called the Electronic Medical Records and Genomics (eMERGE) network.
Methods
Study participants were drawn from the Mass General Brigham (MGB; Boston, MA; formerly Partners Healthcare Systems) Biobank, which is a subset of the MGB Research Patient Data Registry (RPDR). The RPDR is a data warehouse that gathers data from multiple hospital electronic record systems within MGB and includes over 4.6 million patients from MGB hospitals, 227 million encounters, and 2.4 billion distinct, coded clinical facts dating back to 1986 including demographic data, diagnostic codes, procedures, pharmacy data (e.g. RxNorm), inpatient and outpatient encounter information, laboratory data, imaging/pathology data. The MGBB is a collection of DNA, serum, and plasma samples from participants (recruitment ongoing) from MGB hospitals who provided informed consent for a) broad use for genomics, biomarker, epidemiology research, and b) linkage with RPDR EHRs and survey data. At the time of analysis, the MGBB comprised approximately 77,000 consented participants. Demographics of the MGBB are broadly representative of demographic composition of Eastern MA. Minority enrollment reflects population demographics (13% non-White).
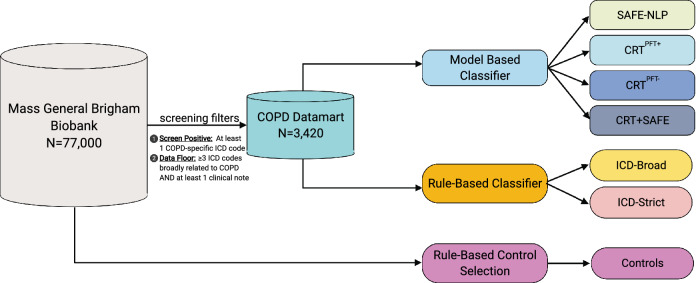
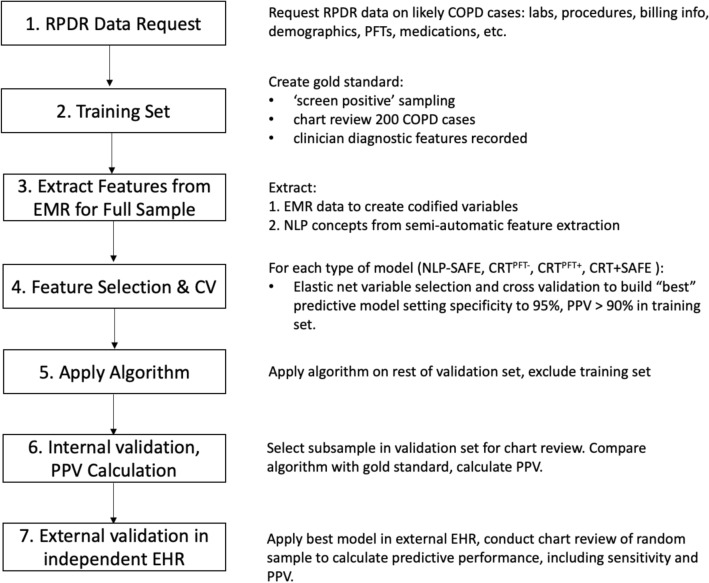
Patients with one International Classification of Diseases (ICD) code specific to COPD (ICD9: 491.2 obstructive chronic bronchitis, 493.2 chronic obstructive asthma, 496.* chronic airway obstruction, not elsewhere classified ; ICD10: J43* emphysema, J44* other chronic obstructive pulmonary disease) were selected to generate a “screen positive” COPD datamart. Next, we implemented a data floor threshold whereby subjects with 3 ICD codes broadly associated with COPD (ICD9: 491.* chronic bronchitis, 492.* emphysema, 493.2 chronic obstructive asthma, 496.* chronic obstructive pulmonary disease, not elsewhere classfiied; ICD10: J40* bronchitis, not specified as acute or chronic, J41* simple and mucopurulent chronic bronchitis, J42* unspecified chronic bronchitis, J43* emphysema, J44* other chronic obstructive pulmonary disease) occurring on distinct dates, and at least one unstructured medical note (i.e., narrative text from patient health records regarding reason for visit, discharge, operation, labs, etc.) in their records, were selected from the COPD datamart to create our patient chart review pool. This was to ensure that study subjects had sufficient EHR and medical record histories to be informative in the algorithm development process. An overview of these filtering steps can be seen in Fig. 1, and an overview of general model development can be seen in Fig. 2. The protocol for this study was approved by the Brigham and Women’s Hospital and the Mass General Brigham institutional review boards, #2015P000983. All methods were performed in accordance with relevant guidelines and regulations.
Figure 1.
Overview of COPD datamart selection and developed algorithms.
Figure 2.
Broad overview of steps in phenotyping algorithm development.
Clinician chart reviews
A random sample of 200 participants was selected from the COPD screen positive set, among whom 182 met the data floor selection criteria for chart review. Chart reviews were performed by two senior pulmonologists (M.H.C. and E.W.) to establish a gold standard training set using an internally developed chart review protocol (Figure S2). To ensure acceptable interrater reliability, 25 charts were reviewed by both pulmonologists and consistency was assessed using Cohen’s kappa statistic. Charts with available pulmonary function tests (PFTs) such as spirometry reports within the past 10 years were reviewed for the presence of values of FEV1/FVC < 0.7 to classify ‘definite’ COPD cases. Charts without available PFT reports were reviewed for clinical COPD criteria based on ≥ 3 of the following: (1) ever smoking, (2) ≥ 2 notes confirming a clinical COPD diagnosis, (3) moderate or severe centrilobular or panacinar emphysema on clinical chest computed tomography (CT) scan, (4) COPD-specific medications or (5) presence of pulmonologist-diagnosed COPD. Reviewers classified patients meeting these criteria but lacking confirmatory spirometry as ‘clinical’ COPD cases. Finally, for internal validation, 100 participants with charts were randomly selected and reviewed from the screen-positive set to comprise a gold-standard test set.
Rule-based algorithms
Algorithms relying strictly on ICD codes to identify COPD cases have previously been proposed and applied22. Therefore, as a performance benchmark for our algorithm, we employed rule-based heuristics to classify COPD cases based only on ICD codes among the screen positive set: (1) ICD-strict: 3 COPD-specific ICD codes, and (2) ICD-broad: 2 COPD-specific codes, where each code was required to have occurred on distinct dates in the EHR for a given patient. A rule-based algorithm was also applied to identify controls (i.e. subjects with no history of COPD-related codes), requiring 0 COPD-specific codes and 2 encounters in the EHR.
Model-based algorithms
To develop model-based algorithms, we first constructed a comprehensive feature space from which predictive variables of COPD case status could be selected. Variables were derived from (1) structured data such as ICD, current procedural terminology, and prescription codes, and (2) unstructured data, such as narrative text present in clinical visit and discharge notes or radiology reports. Coded features derived from the structured data were obtained based on COPD-related risk factors partly curated by the two chart reviewers or drawn from prior literature21. These included derived variables relating to age at first COPD diagnosis, smoking history, medical utilization, PFTs, visit history to pulmonary clinics, ICD codes, COPD and asthma medications, and radiography. Values for FEV1, FVC, and smoking status were obtained from spirometry reports using NLP algorithms in R (Supplementary Information 5 and 6), and all other NLP variables were constructed using surrogate assisted feature extraction (SAFE)24, which has been described in detail previously25. Briefly, the implementation of the SAFE procedure proceeds as follows: (1) extraction of United Medical Language System (UMLS) concepts related to COPD from publicly available databases including Medscape, PubMed, and Wikipedia via named entity recognition, (2) application of Narrative Information Linear Extraction25, to scan EHR clinical narratives for positive mentions of the UMLS terms, which are then totaled to obtain patient-level counts, (3) feature selection based on majority voting (concept present in the majority of databases scanned), frequency control (concept present in > 5% medical notes mentioning COPD), and surrogate selection criteria (concept selection based on predictiveness of COPD ICD counts and primary NLP terms).
After creating the feature space, adaptive elastic net regularization was applied to build logistic regression classifiers of COPD status using the R package glmnet. Tuning parameters, for the regularization penalty and for the mixing parameter between ridge () and lasso () regression, for the elastic net were selected through fivefold cross validation using the package caret in R26.
COPD status from chart reviews was classified as a binary variable combining ‘definite’ and ‘clinical’ into COPD versus non-COPD. Classification models were built using several different feature space combinations: (1) expert and literature curated features excluding spirometry derived variables (CRTPFT-), (2) expert and literature curated features including spirometry derived variables (CRTPFT+), (3) SAFE-extracted features only (SAFE-NLP), and (4) combined CRTPFT+ and SAFE features (CRT + SAFE). For each model, the set of features most predictive of COPD in the gold-standard training set were identified, and the relative weights (beta coefficients) of the features were extracted. After feature selection, classification thresholds were selected holding the specificity level at ~ 95%, where those above the cutoff were assigned case status, and those below were assigned non-case status. The model with the best performance in terms of 1) total area under the curve (AUC) of the receiver operator characteristic curve and 2) estimated sensitivity, positive predictive value (PPV), and negative predictive value (NPV) at the 95% specificity threshold was taken forward for algorithm validation. Confidence intervals for performance estimates were obtained via bootstrap. F1 and F0.5 metrics of performance were also calculated for the final model. An overview of the different criteria for all model and rule-based algorithms can be seen in Table 1.
Table 1.
Algorithms for classifying chronic obstructive pulmonary disease.
| Classification method | Classifier description | Minimum selection criteria | ||
|---|---|---|---|---|
| ICD9/10 Diagnosis criteria |
Visit criteria | Other criteria | ||
| Rule-based | ||||
| ICD-stricta | 3 COPD-specific codes | 3 or more COPD-specific codes | None | |
| ICD-broadb | 2 COPD-specific codes | 2 or more COPD-specific codes | None | |
| Control selection | 0 COPD-specific codes | Subjects with no history of COPD related codes | 2 encounters in MGB Biobank | |
| Model-basedc | ||||
| Automatic extraction NLP features | ||||
| SAFE-NLP | Model selected from surrogate assisted feature extraction with natural language processing of unstructured EHR data (narrative text from clinic notes) | At least 1 COPD-specific code and at least 3 broad COPD codes | 1 visit with electronic clinical note in the EHR | Selected by classifier |
| Curated (CRT) features | ||||
| CRTPFT- | Model selected from literature-based and expert-curated feature inputs primarily derived from structured data, excluding measures of spirometric FEV1/FVC performance | At least 1 COPD-specific code and at least 3 broad COPD codes | 1 visit with electronic clinical note in the EHR | Selected by classifier |
| CRTPFT+ | Model selected from the feature space of CRTPFT-, but inclusive of measures of spirometric FEV1/FVC performance | At least 1 COPD-specific code and at least 3 broad COPD codes | 1 visit with electronic clinical note in the EHR | Selected by classifier |
| Mixed (automatic + curated) features | ||||
| CRT + SAFE | Model based on combining the full feature space for CRTPFT+ and SAFE | At least 1 COPD-specific code and at least 3 broad COPD codes | 1 visit with electronic clinical note in the EHR | Selected by classifier |
aCOPD-specific codes include: 1) ICD9: 491.2, 493.2, and 496.*; 2) ICD10: J43.* or J44.*.
bBroad COPD codes include any codes with the following base numbers: 1) ICD9: 491.*, 492.*, 493.2*, and 496.*; 2) ICD10: J40.*, J41.*, J42.*, J43.*, J44.*.
cAll model-based algorithms were developed using probability-based thresholding via logistic regression models selected using a threshold for specificity at 95%.
Internal and external validation
Internal validation to confirm algorithm performance was conducted in the gold standard test set in the MGBB. To assess the portability of the selected MGBB algorithm, external validation was performed at an independent site with a different EHR system: Marshfield Clinic Health System (MCHS; Marshfield, WI). MCHS is an integrated health care delivery system that provides the majority of healthcare services to 1.5 million patients residing in more than 50 locations in northern, central, and western Wisconsin. The MCHS has coded diagnoses dating back to the early 1960s, and employs a modern integrated, internally developed EHR and data warehouse beginning in the 1990s.
Using identical filtering procedures as in the MGBB, a COPD datamart was constructed in the Marshfield EHR. Among eligible subjects meeting the datamart screening criteria, a random set of 100 subject charts were selected for chart review using the clinical heuristic algorithm developed in the MGBB. After applying the MGBB COPD algorithm, sensitivity, specificity, PPV, NPV, and F1 and F0.5 metrics were calculated to assess algorithm performance in the Marshfield validation sample.
Ethical approval
The eMERGE study protocol was approved by the institutional review board at Mass General Brigham (formerly Partners Healthcare System) and Marshfield Clinic Research Institute. All authors read and approved the manuscript.
Results
The demographic characteristics of the randomly sampled gold standard training and test sets, as well as the full COPD datamart, can be seen in Supplementary Table 1. In total, 77 patients were classified with COPD, and 105 were defined as non-cases in the full gold-standard training set (N = 182), and 46 and 54 patients were classified as cases and non-cases in the gold standard test set (N = 100). Spirometric FEV1/FVC results were available for 129 (71%) patients in the training set, and 80 (80%) patients in the test set. Within a random set of 25 subject charts reviewed by both pulmonologists, high interrater reliability of COPD diagnosis was observed with an estimated Cohen’s kappa of 0.867 (p = 6.6 × 10–9).
We identified and derived 56 variables for consideration in the curated (CRT) feature space (Supplementary File: Code Book for Structured Data Features). Medications associated with pulmonary diseases were classified by specificity for COPD management (Supplementary Table 2). In total, 53 NLP features were identified by SAFE for use in model training to construct the SAFE-NLP and CRT + SAFE classifiers (Supplementary Table 3).
The two rule-based algorithms using the naïve ICD-based thresholding approach to classify COPD status performed similarly in the gold standard training set. The ICD-Strict algorithm had 98.1% sensitivity, 11.7% specificity, 60.2% PPV, and 81.8% NPV, with F1 of 0.746 and F0.5 of 0.652, and the ICD-Broad algorithm demonstrated 100% sensitivity, 3.9% specificity, 58.7% PPV and 100% NPV, with F1 = 0.740 and F0.5 = 0.877. As the specificity for both rule-based algorithms were extremely low, neither was taken forward for assessment in the MGBB test set (Table 2).
Table 2.
Comparison of performance characteristics between different electronic medical record COPD classification algorithms within Mass General Brigham Biobank training set (N = 182).
| Algorithm | Counts (N) | Algorithm Performance (95% CI)* | ||||||
|---|---|---|---|---|---|---|---|---|
| True positive | True negative | False positive | False negative | Sensitivity | Specificity | PPV | NPV | |
| Rule-based | ||||||||
| ICD-Strict | 103 | 9 | 68 | 2 | 0.981 | 0.117 | 0.602 | 0.818 |
| ICD-Broad | 105 | 3 | 74 | 0 | 1 | 0.039 | 0.587 | 1 |
| Automatic NLP features | ||||||||
| SAFE-NLP | 38 | 73 | 4 | 66 |
0.365 (0.270–0.462) |
0.948 (0.896–0.987) |
0.905 (0.816–0.977) |
0.525 (0.490–0.567) |
| Curated features | ||||||||
| CRTPFT- | 42 | 72 | 5 | 63 |
0.400 (0.314–0.495) |
0.935 (0.883–0.987) |
0.894 (0.808–0.976) |
0.533 (0.493–0.578) |
| CRTPFT+ | 62 | 73 | 4 | 43 |
0.590 (0.495–0.676) |
0.948 (0.896–0.987) |
0.939 (0.878–0.987) |
0.629 (0.577–0.688) |
| Mixed features | ||||||||
| CRT + SAFE | 47 | 73 | 4 | 62 |
0.404 (0.317–0.490) |
0.948 (0.896–0.987) |
0.913 (0.830–0.979) |
0.541 (0.503–0.583) |
aCOPD-specific codes include: 1) ICD9: 491.2, 493.2, and 496; 2) ICD10: J43 or J44.
bBroad COPD codes include any codes with the following base numbers: 1) ICD9: 491, 492, 493.2, and 496; 2) ICD10: J40, J41, J42, J43, J44.
*All probabilistic algorithms were assessed at their corresponding thresholds specifying 95% specificity.
SAFE-NLP: Model based on surrogate assisted feature extraction with natural language processing of unstructured EHR data (free text); CRTPFT-: Model based on literature and expert-curated feature inputs primarily derived from structured data, excluding feature weights for spirometric FEV1/FVC performance; CRTPFT+: Model based on feature space of CRTPFT-, but inclusive of feature weights for spirometric FEV1/FVC performance; CRT + SAFE: Model based on combining the full feature space for CRTPFT+ and SAFE.
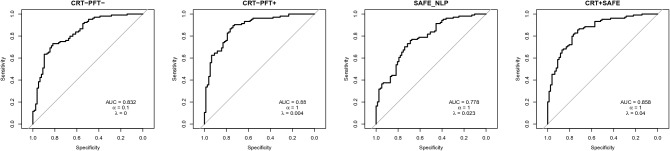
As COPD is not an uncommon disease, we prioritized optimizing specificity (i.e. reducing the false positive rate) over sensitivity (i.e. detecting all true positives) in the model-based algorithms. Among the four model-based algorithms, CRTPFT+ produced the highest AUC of 0.879. AUCs for CRTPFT-, SAFE-NLP, and CRT + SAFE were 0.834, 0.778, and 0.865, respectively (Fig. 3).
Figure 3.
Receiver-operator characteristic curves to assess classification performance of model-based algorithms.
Performance characteristics of the CRTPFT+ model with specificity held at 95% yielded a sensitivity of 59.0%, PPV of 93.9%, an NPV of 62.9%, F1 of 0.725, and F0.5 of 0.840 in the gold standard training set (Table 2). The performance of the CRTPFT- algorithm excluding spirometric results for FEV1/FVC revealed a lower sensitivity of 40%, a PPV of 89.4% and NPV of 53.3%, F1 of 0.553 and F0.5 of 0.450. The SAFE-NLP algorithm had 36.5% sensitivity, 90.5% PPV, 52.5% NPV, 0.520 F1, and 0.414 F0.5, while the CRT + SAFE algorithm had 40.4% sensitivity, 91.3% PPV, 54.1% NPV, F1 of 0.560, and F0.5 of 0.455. The model demonstrating the greatest overall AUC, as well as the highest PPV and sensitivity, was the CRTPFT+ model.
In the MGBB gold standard test set (N = 100), the CRTPFT+ algorithm had sensitivity of 71.7%, specificity of 94.4%, PPV of 91.7%, and NPV of 79.7% (Table 3). The CRTPFT- algorithm had lower sensitivity more consistent with performance in the training set at 43.5%, 94.4% specificity, 87% PPV, and 66.2% NPV. The CRT + SAFE algorithm demonstrated 43.5% sensitivity, 98.1% specificity, 95.2% PPV, and 67.1%NPV. The SAFE-NLP algorithm had the lowest sensitivity of 37%, specificity of 94.4%, PPV of 85% and NPV of 63.8%. As the CRTPFT+ algorithm demonstrated comparably high PPV and higher sensitivity in the test set relative to its performance in the gold standard training set, the CRTPFT+ algorithm was taken forward for external validation in the Marshfield Clinic EHR (Table 3). Among the Marshfield screen positive set, chart reviewers classified 70 cases and 30 non-cases. Applying the CRTPFT+ model revealed independent validation performance of sensitivity 61.4%, specificity of 90%, PPV of 93.5%, NPV of 50% (Table 3) in the Marshfield validation sample.
Table 3.
Comparison of performance characteristics of probabilistic electronic medical record COPD classification algorithms within Mass General Brigham Biobank validation set (N = 100) and external, independent validation of final algorithm in the Marshfield Clinic (N = 100).
| Algorithm | Counts (N) | Algorithm Performance* | ||||||
|---|---|---|---|---|---|---|---|---|
| True positive | True negative | False positive | False negative | Sensitivity | Specificity | PPV | NPV | |
| MGBB Validation (N = 100) | ||||||||
| Automatic NLP features | ||||||||
| SAFE-NLP | 17 | 51 | 3 | 28 | 0.370 | 0.944 | 0.850 | 0.638 |
| Curated features | ||||||||
| CRTPFT- | 20 | 51 | 3 | 26 | 0.435 | 0.944 | 0.870 | 0.662 |
| CRTPFT+ | 33 | 51 | 3 | 13 | 0.717 | 0.944 | 0.917 | 0.797 |
| Mixed features | ||||||||
| CRT + SAFE | 20 | 53 | 1 | 26 | 0.435 | 0.981 | 0.952 | 0.671 |
| External validation (N = 100) | ||||||||
| Curated features | ||||||||
| CRTPFT+ | 43 | 27 | 3 | 27 | 0.614 | 0.900 | 0.935 | 0.500 |
*All probabilistic algorithms were assessed at their corresponding thresholds specifying 95% specificity.
SAFE-NLP Model based on surrogate assisted feature extraction with natural language processing of unstructured EHR data (free text), CRTPFT- Model based on literature and expert-curated feature inputs primarily derived from structured data, excluding feature weights for spirometric FEV1/FVC performance, CRTPFT+ Model based on feature space of CRTPFT-, but inclusive of feature weights for spirometric FEV1/FVC performance, CRT + SAFE Model based on combining the full feature space for CRTPFT+ and SAFE.
Feature weight specifications for the CRTPFT+ algorithm are available in Table 4, and the alternative model weights are available in the supplement (Supplementary Table 4).
Table 4.
Patient medical history features and weights used in the final Mass General Brigham Biobank CRTPFT+ algorithm for classification of COPD.
| Model feature | Model weight | Variable type | Description |
|---|---|---|---|
| Intercept | − 1.871 | ||
| everPFTlt70 | 1.750 | NLP | Ever had a pulmonary function test with spirometry indicating pre-bronchodilator FEV1/FVC ratio < 0.7 OR post-bronchodilator FEV1/FVC ratio < 0.7 |
| nCOPDGTE3_365 | 0.465 | Coded | Ever diagnosed with 3 or more COPD-related ICD codes within any rolling time window of 365 days |
| everTiotropium | 0.334 | Coded | Ever been prescribed tiotropium |
| iNotWhite | − 0.239 | Coded | Race category denoting whether subject is White or Not White |
| smkEver | 0.175 | NLP | Any current/former history of smoking |
| everdxAtPulmClinic | 0.056 | Coded | Ever diagnosed with a COPD-related ICD code at a pulmonary clinic |
| everCOPDmed | 0.048 | Coded | Ever been prescribed a medication used to treat COPD? |
| nmedLAMA | 0.017 | Coded | Total count of distinct prescription codes for long acting muscarinic antagonists in participant medical record for treatment of lung diseases |
| pftCount | − 0.016 | Coded | Total count of any kind of pulmonary function test |
| ageCOPDt1Specific | 0.013 | Coded | Age (in years) at first ICD code specific to COPD |
| nCOPD_ICD | 0.013 | Coded |
The COPD feature count of distinct dates on which a subject has a code from this feature ICD10: J40–J44 ICD9: 491, 492, 493.2, 496 |
| nBronchitis | − 0.009 | Coded |
The count of distinct dates on which a subject has a Bronchitis ICD code ICD10: J40, J41, J42 ICD9: 490, 491 |
| nBronchiectasis | − 0.008 | Coded |
The count of distinct dates on which a subject has a Bronchiectasis ICD code ICD10: J47 ICD9: 494 |
| patient_dxenct | − 0.001 | Coded | Total number of encounters (visits) per subject with a coded diagnosis (any diagnosis not limited to COPD) |
The case assignment threshold for this model, holding specificity at 95%, was 0.754. For subjects who were missing PFT results, the everPFTlt70 variable was classified as ‘No’ in this model.
Discussion
In this study, we developed and compared several phenotyping algorithms for the classification of COPD in large EHR databases. We prioritized specificity and PPV for case selection in our algorithms, as the unbiased detection and estimation of associations between exposures and COPD is conditional on correct case classification. Among the models, we found that curated model CRTPFT+ performed with the greatest AUC, and also yielded the highest sensitivity and PPV while maintaining specificity of ~ 95%. The feature weight for everPFTlt70 was the largest weight in CRTPFT+, with several other clinical variables weighted highly including smoking history, having been diagnosed with a COPD-specific code 3 times in a rolling 365 day period, and having ever been prescribed tiotropium.
We also developed the CRTPFT- model without inclusion of FEV1/FVC results from spirometry, as not all patients receive spirometric confirmation of COPD at diagnosis, particularly in primary care settings. This model demonstrated 19% reduced sensitivity and 4.5% reduced PPV relative to the CRTPFT+ model in the training set, and lower sensitivity by 28.2% and PPV by 4.7% in the test set (model weights available in Supplementary Table 4). Taken together, our findings are in line with what has been previously observed with respect to under- and mis-diagnosis trends for COPD in practice, where spirometry and clinical symptoms are both important features for appropriate diagnosis of COPD. Importantly, these findings demonstrate the inadequacy of relying solely on structured data features (e.g. counts of diagnostic codes) to identify high-confidence COPD cases in the EHR.
Strengths of this study include the large variety of features that were considered in the model building process, as the most essential step in developing prediction models is the curation of the feature space. We explored semi-automatic feature extraction approaches, in addition to manual development. In our study, we found that the use of SAFE did not perform as well as our expert curated models, and that combining the curated and SAFE-based feature spaces for model training did not appreciably improve the performance of the model, while making the algorithm significantly more laborious to apply. Therefore, we selected the CRTPFT+ model, which was largely based on derived variables from structured data in patient EHRs, with only two NLP-based variables for PFT and smoking (an overview of the steps for extracting the specific NLP variables in this model can be seen in Supplementary Information 5 and 6), to make the model more portable across institutions. More importantly, portability of the model was confirmed through external validation at the Marshfield Clinic which uses an entirely different home-grown EHR management system, with higher sensitivity (61.4%) reported than in our gold standard training set and an excellent PPV of 93.5%.
The advantage of employing an algorithm to classify COPD cases over manual chart review in a large EHR context is tremendous. Completing each chart reviews took an average of 15 min per patient in this study; manual chart review of the full MGBB COPD datamart (N = 3420) would have required 8 h/day of clinician labor for 107 days. Once the model terms are extracted from the EHR, our approach can be applied almost instantaneously and it is easily re-applied as new participants are enrolled or as EHR records update over time.
Aside from case identification for cohort development or population-based epidemiological research both within and across different institutions/biobanks, there may be more immediate potential clinical applications for the algorithms developed in this work. For example, the CRTPFT- algorithm could be used to encourage providers to order PFTs among high-scoring patients. If employed as a pre-screening step, we note that probabilistic threshold indicating putative COPD could be relaxed to screen more subjects and thus improve overall capture of COPD cases, especially outside of specialty pulmonology clinics.
Our study is not without limitations. First, all of our models were developed using a COPD datamart built with a screening paradigm (i.e. multiple instances of COPD-related ICD codes, rather than multiple instances of any ICD codes) tailored for distinguishing between patients with COPD-like symptoms vs true COPD, which may have resulted in a less sensitive algorithm. However, an advantage of training on a more stringently screened sample is that the specificity for COPD is likely higher when applied to the general population, where we expect that distinguishing between COPD cases vs general controls and non-cases is less challenging than in our training set. In addition, the use of a screening step enhances PPV by increasing the prior probability of true cases in the sample to which the algorithm is applied. While none of the models performed outstandingly well with respect to NPV, our goal was to successfully make the more challenging distinction between COPD cases vs non-cases in the COPD datamart, rather than COPD cases vs controls in the general EHR; high confidence case ascertainment is essential for improving the power to detect even small associations between a given exposure and COPD (e.g. in molecular epidemiologic contexts). Thus, control selection was conceived as a rule-based approach requiring no history of any COPD-specific ICD codes, as has been successfully implemented previously11,12.
Second, our models are subject to the same vulnerabilities as any other predictive model in that that their development, and therefore their performance, depends significantly on the quality and source of the data inputs on which they are trained. In particular, while the demographic distribution of the MGB datamart was predominantly white, the randomly selected chart review sample from the external validation at Marshfield Clinic was entirely so; in both cases, the sampled participants were reflective of the demographic composition of the populations from which they were drawn. However, the consequence was that the predictive weight assigned to the indicator for White vs Not White was not applied in the execution of the algorithm at Marshfield and was not technically validated. Indeed, the weights assigned to this indicator variable in our curated models may reflect racial disparities in COPD diagnosis27,28 rather than a reliable predictor of true clinical COPD case status, and should thus be considered with appropriate caution in the absence of evidence to the contrary.
Finally, to apply our algorithm requires additional computational time to extract terms that are typically not reliably captured in structured EHR and are more often found in test results, lab reports, and clinical notes; however, we note that our final algorithm limits this work by restricting such extractions to just two features. On balance, the time saved by obviating more traditional, manual clinical review should still considerably outweigh the time required to extract even more extensive lists of NLP-based terms than the two proposed in our model.
In summary, we provide a high-performing, externally validated, and generalizable algorithm for the rapid classification of COPD using EHRs obviating the need for laborious manual chart review. Our algorithm relies primarily on structured data features, while minimizing dependency on unstructured records to just two features: smoking history and spirometry. In settings where support for text-mining of unstructured records is limited, we supply an alternative algorithm with high PPV as a viable alternative, albeit with lower sensitivity. While prior studies have used a variety of ICD-based strategies for identifying COPD patients, none have previously examined the impact of spirometry on classification performance. This study provides strong supportive evidence for the inclusion of spirometric features, in tandem with clinical features derived from structured records, in EHR investigations requiring classification of COPD.
Supplementary Information
Acknowledgements
We thank Tianrun Cai and Kumar Dahal from the Verity Bioinformatics Core for their assistance with applying the SAFE algorithm in the Mass General Brigham Biobank.
Author contributions
S.H.C., J.L.-S., S.T.W., and E.K. conceived of the study. S.H.C. designed the algorithm, conducted statistical analyses, and wrote the manuscript. E.S.W. and M.H.C. helped develop the algorithm, generated data, and reviewed cases. S.G., V.G., S.M. assisted in data acquisition, generation, and processing. J.L., E.J.S., and S.J.H. assisted in data acquisition, analysis, and validation at the Marshfield Clinic Research Institute. E.K., J.W.S., J.L.-S., and S.T.W. provided critical methodological guidance and support. All authors reviewed and approved the final manuscript.
Funding
This work was supported by the National Human Genome Research Institute of the National Institutes of Health [Grant numbers U01HG008685 and U01HG008701], the National Institute on Aging at the National Institute of Health [Grant number R01 AR049880], the National Heart, Lung, and Blood Institute [Grant numbers R01 HL123915, R01 HL141826, and R01 HL135142, R01 HL089856 and K01HL153941], and the Department of Veterans Affairs [Grant number IK2RX002165].
Data availability
Data are available to investigators whose proposed use of the data are approved through an institutional review committee of Mass General Brigham.
Competing interests
STW is an author for UpToDate. MHC has received grant support from GSK and Bayer, speaking fees from Illumina, and consulting fees from AstraZeneca. STW is an author for UpToDate. MHC has received grant support from GSK and Bayer, speaking fees from Illumina, and consulting fees from AstraZeneca. The remaining authors have no conflicts of interest to declare.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Su H. Chu, Email: su.chu@channing.harvard.edu
Elizabeth Karlson, Email: ekarlson@bwh.harvard.edu.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-98719-w.
References
- 1.Mathers CD, Loncar D. Projections of global mortality and burden of disease from 2002 to 2030. PLoS Med. 2006;3(11):e442. doi: 10.1371/journal.pmed.0030442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soriano JB, Abajobir AA, Abate KH, et al. Global, regional, and national deaths, prevalence, disability-adjusted life years, and years lived with disability for chronic obstructive pulmonary disease and asthma, 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet Respir. Med. 2017;5(9):691–706. doi: 10.1016/S2213-2600(17)30293-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogelmeier CF, Criner GJ, Martinez FJ, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017 report. GOLD executive summary. Am. J. Respir. Crit. Care Med. 2017;195(5):557–582. doi: 10.1164/rccm.201701-0218PP. [DOI] [PubMed] [Google Scholar]
- 4.Hill K, Goldstein RS, Guyatt GH, et al. Prevalence and underdiagnosis of chronic obstructive pulmonary disease among patients at risk in primary care. CMAJ. 2010;182(7):673–678. doi: 10.1503/cmaj.091784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamprecht B, Soriano JB, Studnicka M, et al. Determinants of underdiagnosis of COPD in national and international surveys. Chest. 2015;148(4):971–985. doi: 10.1378/chest.14-2535. [DOI] [PubMed] [Google Scholar]
- 6.Diab N, Gershon AS, Sin DD, et al. Underdiagnosis and overdiagnosis of chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2018;198(9):1130–1139. doi: 10.1164/rccm.201804-0621CI. [DOI] [PubMed] [Google Scholar]
- 7.Gottesman O, Kuivaniemi H, Tromp G, et al. The electronic medical records and genomics (eMERGE) network: Past, present, and future. Genet. Med. 2013;15(10):761–771. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bycroft C, Freeman C, Petkova D, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dewey FE, Murray MF, Overton JD, et al. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science. 2016;354:6319. doi: 10.1126/science.aaf6814. [DOI] [PubMed] [Google Scholar]
- 10.Almoguera B, Vazquez L, Mentch F, et al. Identification of four novel loci in asthma in European American and African American populations. Am. J. Respir. Crit. Care Med. 2017;195(4):456–463. doi: 10.1164/rccm.201604-0861OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carroll RJ, Thompson WK, Eyler AE, et al. Portability of an algorithm to identify rheumatoid arthritis in electronic health records. J. Am. Med. Inform. Assoc. 2012;19(e1):e162–169. doi: 10.1136/amiajnl-2011-000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castro VM, Minnier J, Murphy SN, et al. Validation of electronic health record phenotyping of bipolar disorder cases and controls. Am. J. Psychiatry. 2015;172(4):363–372. doi: 10.1176/appi.ajp.2014.14030423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei W-Q, Teixeira PL, Mo H, Cronin RM, Warner JL, Denny JC. Combining billing codes, clinical notes, and medications from electronic health records provides superior phenotyping performance. J. Am. Med. Inform. Assoc. JAMIA. 2016;23(e1):e20–27. doi: 10.1093/jamia/ocv130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pacheco JA, Avila PC, Thompson JA, et al. A highly specific algorithm for identifying asthma cases and controls for genome-wide association studies. AMIA Annual Symposium proceedings AMIA Symposium. 2009;2009:497–501. [PMC free article] [PubMed] [Google Scholar]
- 15.Tatonetti NP, Denny JC, Murphy SN, et al. Detecting drug interactions from adverse-event reports: Interaction between paroxetine and pravastatin increases blood glucose levels. Clin. Pharmacol. Ther. 2011;90(1):133–142. doi: 10.1038/clpt.2011.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallagher PJ, Castro V, Fava M, et al. Antidepressant response in patients with major depression exposed to NSAIDs: A pharmacovigilance study. Am. J. Psychiatry. 2012;169(10):1065–1072. doi: 10.1176/appi.ajp.2012.11091325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brownstein JS, Murphy SN, Goldfine AB, et al. Rapid identification of myocardial infarction risk associated with diabetes medications using electronic medical records. Diabetes Care. 2010;33(3):526–531. doi: 10.2337/dc09-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuppa AF, Benitez GR, Zane NR, et al. Morphine dose optimization in critically ill pediatric patients with acute respiratory failure: A population pharmacokinetic-pharmacogenomic study. Crit. Care Med. 2019;47(6):e485–e494. doi: 10.1097/CCM.0000000000003741. [DOI] [PubMed] [Google Scholar]
- 19.Zuppa AF, Conrado DJ, Zane NR, et al. Midazolam dose optimization in critically ill pediatric patients with acute respiratory failure: A population pharmacokinetic-pharmacogenomic study. Crit. Care Med. 2019;47(4):e301–e309. doi: 10.1097/CCM.0000000000003638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khawaja AP, Cooke Bailey JN, Wareham NJ, et al. Genome-wide analyses identify 68 new loci associated with intraocular pressure and improve risk prediction for primary open-angle glaucoma. Nat. Genet. 2018;50(6):778–782. doi: 10.1038/s41588-018-0126-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee TM, Tu K, Wing LL, Gershon AS. Identifying individuals with physician-diagnosed chronic obstructive pulmonary disease in primary care electronic medical records: a retrospective chart abstraction study. NPJ Primary Care Respir. Med. 2017;27(1):34. doi: 10.1038/s41533-017-0035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooke CR, Joo MJ, Anderson SM, et al. The validity of using ICD-9 codes and pharmacy records to identify patients with chronic obstructive pulmonary disease. BMC Health Serv. Res. 2011;11:37. doi: 10.1186/1472-6963-11-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birtwhistle R, Keshavjee K, Lambert-Lanning A, et al. Building a pan-Canadian primary care sentinel surveillance network: Initial development and moving forward. J. Am. Board Fam. Med. 2009;22(4):412–422. doi: 10.3122/jabfm.2009.04.090081. [DOI] [PubMed] [Google Scholar]
- 24.Yu S, Chakrabortty A, Liao KP, et al. Surrogate-assisted feature extraction for high-throughput phenotyping. J. Am. Med. Inform. Assoc. 2017;24(e1):e143–e149. doi: 10.1093/jamia/ocw135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu S, Cai T, Cai T. NILE: Fast Natural Language Processing for Electronic Health Records. arXiv e-prints. 2019.
- 26.R: A language and environment for statistical computing [computer program]. Vienna, Austria: R Foundation for Statistical Computing; 2019.
- 27.Fuller-Thomson E, Chisholm RS, Brennenstuhl S. COPD in a population-based sample of never-smokers: Interactions among sex, gender, and race. Int. J. Chronic. Dis. 2016;2016:5862026. doi: 10.1155/2016/5862026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mamary AJ, Stewart JI, Kinney GL, et al. Race and gender disparities are evident in COPD underdiagnoses across all severities of measured airflow obstruction. Chronic Obstr. Pulm. Dis. 2018;5(3):177–184. doi: 10.15326/jcopdf.5.3.2017.0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available to investigators whose proposed use of the data are approved through an institutional review committee of Mass General Brigham.