Abstract
DNA, a genetic material, has been employed in different scientific directions for various biological applications as driven by DNA nanotechnology in the past decades, including tissue regeneration, disease prevention, inflammation inhibition, bioimaging, biosensing, diagnosis, antitumor drug delivery, and therapeutics. With the rapid progress in DNA nanotechnology, multitudinous DNA nanomaterials have been designed with different shape and size based on the classic Watson–Crick base-pairing for molecular self-assembly. Some DNA materials could functionally change cell biological behaviors, such as cell migration, cell proliferation, cell differentiation, autophagy, and anti-inflammatory effects. Some single-stranded DNAs (ssDNAs) or RNAs with secondary structures via self-pairing, named aptamer, possess the ability of targeting, which are selected by systematic evolution of ligands by exponential enrichment (SELEX) and applied for tumor targeted diagnosis and treatment. Some DNA nanomaterials with three-dimensional (3D) nanostructures and stable structures are investigated as drug carrier systems to delivery multiple antitumor medicine or gene therapeutic agents. While the functional DNA nanostructures have promoted the development of the DNA nanotechnology with innovative designs and preparation strategies, and also proved with great potential in the biological and medical use, there is still a long way to go for the eventual application of DNA materials in real life. Here in this review, we conducted a comprehensive survey of the structural development history of various DNA nanomaterials, introduced the principles of different DNA nanomaterials, summarized their biological applications in different fields, and discussed the current challenges and further directions that could help to achieve their applications in the future.
Subject terms: Nanobiotechnology, Biomaterials
Introduction
DNA was used for the storage of genetic information as a common biomolecule and fundamental hereditary material in different creatures.1,2 DNA materials possess the potential for nanofabrication due to its characteristic nature, i.e., high precision of Watson–Crick base-pairing and high controllability.1,3–5 In 1980s, Seeman et al.4 proposed the rules for the synthesis of DNA nanostructures for the first time, which triggered a booming years of DNA nanotechnology development.6–10 In the following decades, the technology of DNA nanostructures developed and improved rapidly, and various DNA nanostructures have been designed and widely applied as generic polymeric materials to form complex nanoparticles for specific usages in the fields of chemistry and biomaterial.11–18
DNA, the carrier and transmitter of genetic information in all living systems, consists of four different deoxynucleotide monomers.5 Each monomer is formed with a phosphate group, a deoxyribose, and one of four nitrogen-involving nucleobases, while the nucleobases include thymine (T), adenine (A), guanine (G), and cytosine (C).1,19 Between the monomers, the phosphate group and the deoxyribose can form the phosphodiester bond, which links a certain number of monomers into a DNA chain, named single-stranded DNA (ssDNA).1,19 Based on the principle of Watson–Crick base-pairing, the continuous ssDNA can pair with another one in antiparallel directions to form a double-helix DNA structure, that is double-stranded DNA (dsDNA). Between the two polymeric chains, the nucleobases of C and G can form three hydrogen bonds, and A and T can form two hydrogen bonds, resulting in the base pairs with the certain complementary ssDNA inside the dsDNA.20–24
Despite that DNA molecule is composed of simple units, the variety of dsDNA and the programmability of the structure can be realized by accurate design and freely defined constitutions. In artificially synthesized dsDNAs, the quantity and order of the four monomers are accurately arranged as designed, which determines the diversity of dsDNAs. The freely defined constitutions of DNA are based on sequence-specific base attacking interactions, which are determined by both the external conditions and the sequences of DNA duplexes.25–28 With the rapid development of nanotechnologies and availability to manage the constructions of DNA nanostructures, a variety of designable principles and plentiful DNA-based rigid titles have been used to prepare various dimensional architectures into rationally assembling nanoparticle templates or biomolecular scaffolds,29–34 such as tile assembly,30,35 origami structure,36–39 and dynamic nanomechanical system.40–45 Owing to the inherent nature of biocompatibility and biodegradability of DNA material, different kinds of structural and functional materials based on DNA have been reported with promising potential for many biological usages in different fields.46–49 Subsequently, DNA nanotechnology and nanomaterials kept gaining increasing interests in DNA material science and engineering, especially with the development of DNA nanotechnology triggering by the growing demands in biological, chemical, or medical-related usages.50–56
DNA nanomaterials were developed with a wide variety of structures, including two- and three-dimension (2D and 3D) constructions. Furthermore, according to the various molecular building-styles of functional DNA materials, the constructions of artificially synthesized DNA nanostructures contain single-layer and multilayer nanostructures, which also can be sorted into linear, circular, and branched form.57 The DNA origami technique makes it easy for preparation of DNA, which also promotes the explosive development of structural DNA nanotechnology. The advanced development of DNA nanotechnology then breeds various novel DNA nanomaterials that are applied widely, including tissue engineering, immune engineering, drug delivering, disease diagnosis, and as tools for molecular biology or as biosensors.54–56,58–67 However, for therapeutic applications, some multifunctional polymeric DNA nanomaterials still maintain drawbacks, including being easily-metabolized, lack of targeting, and instability.68–70 Therefore, the combinations of DNA nanomaterials and other nanoparticles and hyper-polymeric compounds, such as gold nanoparticle (AuNPs),69,71,72 polyethylenimine (PEI),73–77 chitosan,78–80 and poly-l-lysine (PLL),81–83 are also developed to overcome those limitations.
While the structural DNA nanotechnology has developed quickly and proved important, the studies on the application of those DNA nanostructures are actually more practical and critical.29,84–87 In such efforts, some previous studies demonstrated that some DNA scaffolds with specific structures or other materials could affect cell biological behaviors, such as cell viability,68,88 proliferation,62,63,65,66 migration,89 differentiation,90,91 morphology,92 and autophagy.67 Owing to the special nature, DNA-based materials could be employed in tissue regeneration engineering and potential treatments for some diseases.81,93–95 In this aspect, many DNA nanostructures were also combined with other materials, such as nucleic acid aptamer (DNA and RNA),96–100 polypeptide,101–103 protein,87,104–106 and some chemical drugs,107–110 in which the DNA nanostructures were designed as delivery systems. When combined with nucleic acid aptamer, the complex DNA-based nanostructures could enhance the ability of targeting,68,88,100 inhibition of malignant cells,100 and antitumor effects.88,100,111 More complex designs of DNA origami enable more advanced applications in other fields, such as in photonics,112–114 electronics,115–117 reaction networks of artificial enzymes,118,119 and etc. This review will summarize the structural design and self-assembly of DNA nanostructures and their biological applications, and discuss the current challenges and future directions.
Fundamentals of DNA origami design
The structural DNA nanotechnology was pioneered by Ned Seeman and colleagues from 1982.6 There arouse an interest in DNA structures, which developed tremendously thereafter.120–125 The DNA formed stabilized branch junctions, which can be further modified via sticky ends to form higher-order structures and lattices, is the vital foundation of structural DNA nanotechnology including DNA origami.6 As for the intrinsic internal sequence symmetry, the branched DNA structures are naturally mobile,126 and the sequence symmetry should be minimized to form stable branched junction.6,126 Seeman et al. suggested to possibly use the stale junction to synthesis 3D frameworks of nucleic acids via legitimately designed sticky-end associations. They assembled different multi-arm junctions, including 4-, 5-, 6-, 8-, and 12-arm junctions, demonstrating that DNA junctions could be precisely branched.127–129 However, considering the structural flexibility and the unpredictable conformations, multi-arm junctions are not suitable to act as the basic structural elements, which are needed to assemble higher-order periodic lattice structures. To solve this problem, Seeman and colleagues constructed a DNA double-crossover molecule by creating a structural motif with two 4-way junctions, constructing a rigid DNA molecule by two crossovers.130,131 In 1990s, a periodic 2D crystalline, the first higher-order DNA lattice nanostructure, was successfully made from crossover molecule.132 In 2003, LaBean and colleagues133 added crossover molecule into a 4 × 4 DNA title with four different direction arms. The 4 × 4 DNA title has a long central strand to connect four Holliday junctions together into a rigid, branched, and fourfold symmetry, similaring to the crossover molecule with a central strand to link two Holliday junctions by end to end.133 The 4 × 4 title reduced the probability of stacking interactions between each arm by placing T4 loops at each of the four concerns of the central strand. Nanoribbon or 2D grid-like lattice was successfully synthesized via supporting 4 × 4 title with appropriate sticky end association sites. For the inspiration of the 4 × 4 title and crossover molecule, some DNA basic units, such as three- and six-point star title, and six-helix bundle, were successfully assembled.134–136 These DNA basic units can further self-assemble to construct 2D arrays with various patterns.137 The 3D macroscopic crystal could also be successful synthesized by the assembly of a rationally designed tensegrity triangle motif.138
In addition to synthesizing periodic DNA nanostructures by titles, LaBan et al. also investigated a long scaffold to guide the nucleation of DNA titles into barcode DNA lattices with complicated aperiodic nanostructures.139 The long DNA scaffold consisted of shorter synthetic oligos, for adding binding sites to shorter strands and forming larger individual complex with barcode DNA nanostructures, which could be further constructed into latticed nanostructures if different sticky ends provided. Shih et al.140 constructed an octahedron using a ssDNA with 1.7 kilobases. Six four-way junctions at each vertex and five double-crossover struts were embedded into the octahedron structure. The synthesis of the octahedron structure includes two steps as follows: (1) five staples with 40 nt bind to the 1699-nt scaffold, assembling a giant branched molecule with bulges at different arms; (2) each arm associating with its partner by the paranemic cohesions interactions, forming the final octahedron-shaped nanostructure.140
The fundamental concepts of DNA nanotechnology and previous explorations established a solid foundation, facilitating greatly the development and the design of DNA nanostructures to be more convenient, complex, and diverse.
Various designs and constructions of DNA nanostructures
Single-layered DNA nanostructure designs
The DNA origami technology was firstly reported by Paul Rothumend in 2006.38 The synthesis of some single-layered, complanate structures, whose structures ranged from triangular, simple rectangular and five-point star shape to intricate smiley faces, each with its unique size, ranges roughly 100 nm in diameter.38 The procedures related to the design and self-assembly process have been described in detail by Paul Rothumend et al.38 Since then, other studies were inspired and successfully synthesized 2D planar patterns, including a depiction of dolphins and a map of China.141,142 Then, the creation of complex curvatures within 2D planar patterns and 3D planar nanostructures were reported by Han et al.143 The distances of the consecutive crossovers, which can concatenate adjacent helices, are usually constant along the parallel alignment of DNA helices. Nevertheless, if the distance of the crossovers along the inner helices is less than that of outer helices, the tension could make the outer helices bend. This method was successfully adopted to incorporate DNA into concentric ring or square with rounded angels.144
Some previous studies reported the applications of two general strategies to investigate 3D nanostructures, and the 3D nanostructures were based on the single-layered design of double-crossover. The first strategy depends on a second-step folding to build 2D planar DNA nanostructures. However, some 2D planar DNA nanostructures are not completely paired with staples. The 2D planar DNA nanostructure is intentionally left as a single-strand at the certain parts so that some 2D planar DNA nanostructures could form some well-cut pieces, such as squares and triangles. When 2D shapes join in with the matched well-cut pieces, the 2D nanostructures could fold and transform into 3D shapes. For instance, a boxlike DNA nanostructure with the external size of 36 × 36 × 42 nm3 had a lid which opened and closed like a box.145 A 3D DNA box-shaped structure with a switchable lid was also reported, and the DNA boxlike nanostructure had a unique electron microscopy, which could potentially be used widely including delivering drugs, controlling the function of some single molecules, and molecule computing.146–148 Other studies developed a stepwise folding mechanism that was self-assembled by a set of staples, to form a DNA prime and a cuboidal boxlike nanostructure.148,149
The second strategy applying to develop 3D DNA nanostructure was used to introduce crossovers and to use plane linkages. The parallel DNA nanostructures did not form the single plane, such as DNA nanotubes.150 By using the approach with the in-plane curvature strategy, some tanglesome nanostructures including the 3D sphere, nanosized flask, and football were developed.151 The double-crossovers DNA nanostructures are relatively rigid structural motifs. However, when compared to duplex DNA at the large size (such as ~100 nm), the structures of single-layered DNA nanostructure are still flexible.
Wireframe single-layered designs
Given that the designs were based on the double-crossover motifs, the wireframe single-layered DNA nanostructures that were conducted with different design strategies were formed by aligning DNA helices in the parallel and compact way. If some new motifs are added to the DNA nanostructure, new nanostructures will be achieved, such as multi-arm junctions allowing for the mash style of desired patterns. A square junction was used for creating branched nanostructures, which was connected with the stem of the branch region to only one of the base helices of the desired feature, and allowed to the connecting helix to remain at the right angle.152 The first wireframe DNA nanostructure with a gridiron pattern was created by Han et al.153 The gridiron pattern consisted of four 4-arm junctions, and they joined in each other to form a square frame.153 Connecting some gridiron patterns together with a consecutive strand could result in a series of 2D lattice DNA nanostructures.153 Subsequently, Zhang et al.154 studied another strategy to make much more intricate wireframe nanostructures with a number of multi-arm junctions. Based on the multi-arm junction, the design algorithm named DAEDALUS was created to fully automate the scaffold routing and staple assignment for some specific target DNA nanostructures.155
In addition, Benson et al.156 created a new DNA nanostructure with a more open conformation with one helix per edge. The structure is therefore stable under the ionic conditions that were always applied for biological assay. The structure was conducted arbitrary target objects to the triangulated meshes to create DNA nanostructures in a wireframe style by using an alternative way.156,157 A single DNA helix rather than two is the majority of the edge designs, which requires the DNA scaffold traveling through all of the edges at once, while a few edges require the DNA scaffold to travel twice. The novel multi-arm DNA structures combined with the DNA origami method for the synthesis of 2D DNA scaffolds with folding into 3D by connection strands on the 2D nanostructures, and the novel multi-arm DNA structures were designed with a scaffold routing whose fundamental structures were the A-trails to form the center of the nanostructures through a computational algorithm.149,158 The availability of this method was further demonstrated by means of the successful preparation of a series of meshed 3D nanostructures, such as ball, bottle and stanford bunny. The same design concept was also used to synthesize triangulated DNA origami trusses.159
Based on the design concept once adopted, many different approaches can be used to create single-layered DNA nanostructures. In spite of successful formation and high yield, several limitations remain when the DNA structures were used in practical applications. For instance, single-layered DNA nanostructures have structural heterogeneity and conformation, which might restrict the applications in precise addressability.160 Owing to the vertex discontinuity of the DNA chains, it is difficult to avoid the structural flexibility in the single-layered wireframe design. In addition, the technologies of single-layered DNA nanostructures also shows the potential of biochemical reactions at the single-molecule level.161
Multilayered designs
Single-layered DNA nanostructures could be aggregated to form multilayered 3D nanostructures with the increase of rigidity, which could overcome some limitations and improve the design methods. In the first study on designing 3D solid DNA nanostructures, the DNA nanostructures were based on a six-helix bundle as the fundamental unit.162 Douglas et al.162 demonstrated the design and self-assembly process of DNA nanostructures with six different shapes, including monolith, railed bridge, square nut, stacked cross, genie bottle, and slotted cross, and the dimensions with precise controlling ranged from 10 to 100 nm. Douglas et al. also reported the validity of the design method by assembling a monolith, square net, and slotted cross by using a honeycomb alignment. In another study, Ke et al.163 reported a new approach to design a multilayer DNA structure with a square lattice, which was based on a four-helix bundle. In this design, the square lattice could be folded into the nanostructures of designed dimensions by one-step annealing process, in spite of the intensive density of DNA helices.163 The three-helix bundle was regarded as the basic unit in a triangular lattice.164 In this study, Ke et al. reported the successful folding of a multilayer DNA structure with helices arranged on the close-packed hexagonal lattice. This study also presented that hybrid DNA structures could incorporate three different shapes all in one design, including square lattice, honeycomb-lattice, and hexagonal-lattice packing of helices.164
More work reported that the multilayered DNA nanostructures could be designed to incorporate more various packaging styles, which possess the programmable curvatures and twists.165 In this research, Dietz et al. systematically demonstrated that the strands of DNA with programable self-assembly could form a custom-shaped bundle of tightly cross-linked double helices, and they also created several different shapes of intricate DNA nanostructures via combining multiple curved elements together. Another method combined single-strand DNA flexibility and multilayered nanostructure rigidity to build the tensegrity structures.166 Tensegrity, also named tensional integrity, is a structural principle related to the usage of isolated component, indicating a reliable balance between components in pure compression or pure tension for stability.166 Liedl et al. successfully synthesized a tensegrity prism that was formed with three multilayered DNA bundle nanostructures as compression components and nine ssDNAs regions as tension cables.166
Ke et al.167 introduced that the multilayered DNA nanostructures had much better rigidity than that of the single-layered DNA nanostructures. Nevertheless, the longer folding course need to avoid the kinetic traps, and the buffer solution should contain a high concentration of magnesium (Mg2 + ), which could suppress the electrostatic repulsion of the negatively charged phosphate backbones between adjacent helices. Furthermore, due to the lower accessibility and shorter nucleation region of folding DNA strands, the yield of the single-layered DNA nanostructures is usually higher than that of the compact multilayered DNA nanostructures. The careful design of the scaffold procedure and staple break positions could boost the folding yield of DNA nanostructures.167 The parallel alignments of DNA nanostructures in a layer-by-layer fashion are not necessarily required for the 3D DNA nanostructures. Hong et al.168 reported a strategy to design a layered 3D wireframe DNA nanostructure by employing crossover pairs, which could connect neighboring layers of DNA double strands together. The layered crossovers could make the scaffold or helper DNA strands go through different layers and get command of the relative direction of DNA strands within the neighboring layers. In this research, the authors successfully created a nine-layered wireframe DNA nanostructure and reported that the neighboring helices could be controlled within well-defined interlayer angles. The 3D latticed nanostructures could also be constructed by applying 3D point-star junctions. However, it would be a challenge to availably control its relative direction in the interspace of 3D.
The design methodologies of DNA nanostructures depend on the usage of DNA molecules rendering objects. An object can be generated by scientists via using of DNA origami or a 3D modeling software. With the adoptions of DNA origami and 3D modeling software, objects can be folded from the 2D plane, visualized as outlined meshes, or filled with layered lines. The preparation procedures of various DNA nanostructures are conducted under a good framework now: (1) composing an object nanostructure, (2) creating the scaffold style layout and defining the DNA sequence by using a software program,169,170 (3) designing the oligoes of the scaffold and staple, (4) enabling DNA self-assembling under a buffer solution with the ramp of temperature, (5) purifying and functionalizing the DNA nanostructure when needed, (6) analyzing and visualizing the DNA nanostructure, and then (7) expanding and exploring the various applications of the DNA nanostructure.171–173 Various methods have been created to improve the synthesis processes of DNA nanostructures, such as increasing the production efficiency of DNA nanostructures,174–178 packaging the DNA nanostructures in a Mg2 + -free buffer solution with an isothermal environment,179–181 and purifying the functionalized DNA nanostructures to wipe off any misfolding nanostructures or polymers.182–186 Some other studies have also developed a few methods to apply DNA nanostructures into different experimental conditions, such as the application of photoinduced crosslinking with 8-methoxypsoralen (8-MOP), which could improve the heat resistance of the DNA nanostructures;187,188 the application of external signal triggers such as magnetic or electric fields, chemical, light, pH, and temperature, which could control the formation of DNA nanostructures;189–193 and the application of imaging and folding DNA nanostructures in hydrated glycholine under isothermal conditions.194
Biological applications of various DNA nanostructures
The final objects of various DNA nanomaterials are the application for different field, such as biomedicine, chemistry, materials, and etc. However, the structural development is way ahead of the applications of DNA nanomaterials. In this part, we will summarize the biological applications of different DNA and DNA-based materials, and discuss the challenges of them. The contents of this part are mainly devided into six small parts shown as follows.
Tissue engineering
Tissue engineering is a cutting-edge field that study seeded cells, suitable physical and biochemical factors, and biocompatible materials, as well as combinations thereof, to promote the creation of tissue-like structures.195,196 Tissue engineering mainly involves cartilage and bone construction, vascular tissue engineering,197 nerve tissue engineering, skin tissue engineering,198,199 oral tissue engineering, tendon and ligament tissue engineering, corneal tissue engineering, and tissue engineering of some other important organs (e.g., liver, pancreas, kidney, and lung).200,201 The therapeutical strategies adopted mainly include seeding cells,202–205 synthesizing biological materials,206–209 constructing tissues and organs with advanced methods and techniques,210–213 and clinical applications.197,214,215 At present, there are mainly three ways of tissue repairing commonly used in clinical practice: autologous tissue transplantation, allogeneic tissue transplantation, and artificial substitutes. For tissue regeneration engineering including cell seeding, proliferation, orientation, and differentiation of the stem or progenitor cells on the injured sites, various bio-mimicked materials or medicines with the use of nanotechnology were studied to provide the adhesive surfaces.216–220 The nanotechnology can be used to develop and investigate different nanomaterials and medicines, which can provide different substrates for cell seeding, adhesion and growth, to aid the tissue regeneration on the defects.63,64,221–224 The most important purpose of regenerative nanomaterials and medicines is to rebuild or create functional tissue replacements to repair the function and morphology of defected tissues.225 In this part of the review, we will summarize the applications of DNA nanostructures, discuss the limitations of different studies, and present the gap between the studies and clinical applications.
With the rapid development of DNA nanotechnology, the applications of DNA nanostructures in tissue regeneration engineering are embodied in altering biological behaviors of cells, such as morphology, viability, growth, and migration.226,227 The potential applications of DNA nanostructures in tissue regeneration include the regeneration of bone and cartilage (Fig. 1g),65,228,229 nervous system (Fig. 1a, f),63,64,230 and integumentary system (Fig. 1b),231–236 as well as changing the stress response of tissue, such as anti-inflammatory effects,237,238 anti-aging effects,239 and anti-apoptosis (Fig. 1).240 Owing to the interesting feature of DNA with the ability to intercalate synthetic or natural molecules, such as daunorubicin, amsacrine, and adriamycin, the combinations of DNA nanostructures and other molecules are regarded as complex drug delivery systems (DDSs) for many active and photosensitive compounds (Table 1).241–245 Therefore, the DNA-based nanostructures or nanomedicines are exerting their effects on tissue regeneration engineering.246–248
Fig. 1.
The biological applications of various DNA-based and DNA-encoding nanomaterials in tissue engineering. a, f The biological applications of different DNA-based nanomaterials in neural tissue (e.g., TDNs, NPs loading various pDNA, polycation loading various pDNA, and some nanofibrous loading pDNA). b The biological applications of different DNA-based nanomaterials in skin tissue (e.g., TDNs, polycation loading various pDNA, and some basement materials loading pDNA). c, e The biological applications of different DNA-based nanomaterials in skeletal and cardiac muscle engineering (e.g., skeletal tissue: NIR-DA, NPs loading genomic DNA, and various polycation loading pDNA; cardiac muscle: nanofibrous loading pDNA and TDNs). d The biological applications of different DNA-based nanomaterials in corneal tissue (e.g., TDNs, NPs loading various pDNA, and polycation loading various pDNA). g The biological applications of different DNA-based nanomaterials in bone tissue engineering (e.g., TDNs, LNPs loading pDNA, polypeptides loading various pDNA, and polycation loading various pDNA)
Table 1.
The biological applications of various DNA-based and DNA-encoding nanomaterials in tissue engineering
| Tissue regeneration engineering | Material format | Composition | DNA nanotructure | Biological function | References |
|---|---|---|---|---|---|
| Bone tissue regeneration | DNA/chitosan nanoparticles |
1. Poly(d, l-lactic-co-glycolic acid)/hydroxylapatite (PLGA/HAp) 2. pDNA/BMP-2 |
pDNA/BMP-2 | PLGA/HAp composite scaffolds could deliver the pDNA/BMP-2 into human marrow stem cells (hMSCs), which showed the higher cell attachment, higher cell viablity and desirable transfection efficiency of DNA. | 275–279 |
| pBMP-2/Polyethylenimine nanoparticles |
1. PEI 2. Human pDNA/BMP-2 |
Human pDNA/BMP-2 | Bone formation. | 282,283 | |
| DNA-loaded calcium phosphate nanoparticles |
1. Nano-calcium phosphate paste 2. pDNA/BMP-7 3. pDNA/VEGF-A |
1. pDNA/BMP-7 2 pDNA/VEGF-A |
Enhanced regeneration of bone volume and a significantly faster healing. | 255,284 | |
| Chitosan-gold nanoparticles mediated gene delivery of c-myb |
1. Chitosan-gold nanoparticles 2. pDNA/c-myb |
pDNA/c-myb | Suppressing osteoclastogenesis and promoting osteogenesis of dental implant even in osteoporotic condition | 256,285 | |
| CS/CSn(pDNA/BMP2)-GP |
1. chitosan (CS)-based hydrogel with a,b-glycerophosphate (a,b-GP) 2. pDNA/BMP-2 |
pDNA/BMP-2 | Enhancing cell proliferation, maintaining great potential in gene delivery system and tissue regeneration of periodontium | 286–290 | |
| Chitosan/β-glycerophosphate (CS/β-GP) hydrogel as a VEGF-sustained release system |
1. Chitosan/β-glycerophosphate (CS/β-GP) hydrogel 2. VEGF gene |
VEGF gene | Delivering and releasing VEGF, and promoting cell proliferation and differentiation of dental pulp stem cells (DPSCs). | 291 | |
| Lipopolysaccharide-amine nanopolymersomes (LNPs) modified with Noggin small interfering (si)RNA (siNoggin) and pDNA/BMP-2 |
1. Lipopolysaccharide-amine nanopolymersomes 2. siNoggin 3. pDNA/BMP-2 |
1. siNoggin 2. pDNA/BMP-2 |
Enhancing osteogenic differentiation. | 257 | |
| Star-shaped poly(l-lysine) polypeptides/genomic DNA |
1. Star-shaped poly(l-lysine) polypeptides 2. pDNA/VEGF and pDNA/BMP-2 |
1. pDNA/VEGF 2. pDNA/BMP-2 |
Irritating mesenchymal stem cells (MSCs) to differentiate into bone tissue. | 292 | |
| DNA/protamine complex scaffold |
1. Protamine 2. pDNA/FGF-2 or pDNA/BMP-2 |
1. pDNA/FGF-2 2. pDNA/BMP-2 |
Enhancement of bone regeneration, and gene delivery. | 293,295 | |
| Tetrahedral DNA nanostructures (TDNs) | TDNs | TDNs |
Enhancing the cell growth and motility of different cells, such as adipose stem cells (ASCs), human periodontal ligament stem cells (PDLSCs), MSCs, and chondrocytes. TDNs could act the anti-inflammatory effects on the periodontist model, and could promote osteogenic differentiation |
65,226,296–299,301 | |
| TDNs/Wogonin |
1. TDNs 2. Wogonin |
TDNs | Enhancing expression of chondrogenic markers, downregulating matrix metalloproteinases and inflammatory mediators, and promoting the expression of tissue inhibitor of metalloproteinase 1 and B-cell lymphoma 2. | 302 | |
| Neural tissue engineering | Nanofibrous modified with pDNA |
1. A nanofibrous micro cell carrier 2. A self-assembled nano-sized polymer 3. pDNA/NR4A1 |
pDNA/NR4A1 | Promoting intervertebral disc (IVD) regeneration and suppress the fibrosis. | 94 |
| A novel DNA-based chemical approach with the synthetic DNA-binding inhibitor | Pyrrole-imidazole polyamides (PIPs) | N/A | Targeting cell differentiation controlling genes of human induced pluripotent stem cells (hiPSCs), and the DNA-binding inhibitor could target the SOX2 gene. | 341 | |
| DNA aptamer | yly12 aptamer | yly12 aptamer | The aptamer yly12 could bind neural cell adhesion molecule L1 (L1CAM), which was expressed in normal neural tissue as surface antigen. | 342 | |
| Poly(lactide-co-glycolide) (PLG) delivery DNA complexes |
1. PLG 2. pDNA |
pDNA | Promoting the transgene expression and the regeneration of complicated neural system. | 332 | |
| Nanoparticles loading pDNA and RNA |
1. Nanoparticles 2. pDNA or RNA (e.g., siRNA) |
1. pDNA 2. RNA (e.g., siRNA) |
Delivering genetic materials into neural stem cells (NSCs) and guiding cell differentiation. | 343–347 | |
| TDNs | TDNs | TDNs |
1. Enhancing cell proliferation, migration, and neuronal differentiation of NSCs. 2. Improving the recovery of motor function and the neural tissue regeneration in the injured site of spine cord. 3. Potential neuroprotective effects on the cell models of Alzheimer’s Disease (AD) and Parkinson’s Disease (PD). |
63,64,229,239,329,348 | |
| Skeletal and cardiac muscle engineering | Near-infrared light-activated DNA agonist (NIR-DA) nanodevice |
1. Nongenetic manipulation 2. DNA agonist |
DNA agonist | Delivering the cell signaling and regulating the receptor tyrosine kinase (RTK) signaling, cell migration, and myogenesis of skeletal muscle satellite cells. | 89 |
| Iron-oxide nanoparticles loading genomic DNA |
1. Magnetic nanoparticle 2. Genomic DNA |
Genomic DNA | Capturing some chemotherapy agents from human serum, including cisplatin, epirubicin, and doxorubicin (DOX), and protecting cultured cardiac myoblasts from the lethal levels of chemotherapy agents | 363 | |
| TDNs | TDNs | TDNs | Cardio-protection effects from myocardial ischemia-reperfusion injury (MIRI) by taking the advantage of the anti-inflammatory and antioxidative potential. | 365 | |
| Skin tissue engineering | N,N,N-trimethyl chitosan chloride (TMC)/pDNA-VEGF complexes |
1. TMC 2. pDNA-VEGF |
pDNA-VEGF | Promoting the expression of VEGF gene and wound healing. | 388 |
| Collagen-chitosan scaffold/silicone membrane bilayer dermal equivalent (BDE)/ pDNA-VEGF |
1. BDE 2. pDNA-VEGF |
pDNA-VEGF | Effective transfection abilities to promote the expression of VEGF gene. | 390,391 | |
| Hydrogel embedded with resveratrol (Res) and pDNA-VEGF (Gel-Res/pDNA-VEGF) |
1. Gel-Res 2. pDNA-VEGF |
pDNA-VEGF | Promoting the healing of splinted excisional burn wounds. | 392 | |
| Nanofibers loading pDNA-bFGF and pDNA of angiopoietin (pDNA-ANG) |
1. PLLA/POSS 2. pDNA-bFGF 3. pDNA-ANG |
1. pDNA-bFGF 2. pDNA-ANG |
Promoting revascularization of the deep skin defect healing. | 231 | |
| Electrospun core-sheath fibers loaded with pDNA-bFGF |
1. Electrospun core-sheath fibers 2. pDNA-bFGF |
pDNA-bFGF | Promoting skin regeneration in diabetic rats. | 393 | |
| TDNs | TDNs | TDNs |
1. TDNs (sizes ≤ 75 nm) could effectively penetrate the skin of mice and human, and reach dermis layer, which could load and deliver DOX to subcutaneous tumor site. 2. Promoting fibroblast (HSF cell line) and keratinocyte (HaCaT cell line) growth and migrating. |
230,398 | |
| Other tissue engineering | N, N, N-trimethyl chitosan chloride (TMC) modified with pDNA-VEGF |
1. TMC 2. pDNA-VEGF |
pDNA-VEGF | Enhancing the angiogenesis. | 400 |
| Scaffolds containing both pDNA-VEGF and pDNA-FGF2 |
1. Scaffolds (e.g., collagen-heparin scaffolds) 2. pDNA-VEGF and pDNA-FGF2 |
pDNA-VEGF and pDNA-FGF2 | Enhancing the angiogenesis. | 401,402 | |
| Aptamer-conjugated hydroxyapatite (Apt-HA) | Apt-HA | Apt-HA | Promoting angiogenesis and bone regeneration. | 399 | |
| TDNs | TDNs | TDNs |
1. Promoting angiogenesis in vitro and the model of the jaw bisphosphonate-related osteonecrosis. 2. Facilitating the corneal wound healing, promoting the re-epithelialization of wounds, and improving the corneal transparency. 3. Preventing retina ischemia-reperfusion injury from the oxidative. 4. TDNs could induce the immune tolerance and prevent the onset of Type 1 diabetes. |
62,403–406,411 | |
| PEI-DNA nanoparticles |
1. PEI 2. Genomic DNA |
Genomic DNA | Delivering corneal gene for the corneal gene therapy. | 408 | |
| DNA-based carrier systems | DNA nanoparticles | DNA nanoparticles | Ophthalmic drug delivery. | 407 | |
| AuNPs-encoding pDNA |
1. AuNPs 2. pDNA |
pDNA | Transfecting human retinal pigment epithelium cells. | 409 |
Bone tissue engineering
The loss and dysfunction of bone tissue, which are mainly caused by aging, injury, inflammation, such as rheumatoid arthritis (RA) or osteoarthritis (OA), and tumors, are associated with individual susceptibility, and can result in severe morbidity and socio-economic issues.249,250 In the skeletal tissue, the DNA nanostructures are mainly used to provide the adhesive scaffolds or delivery some biological agents to the injured articulations, and the DNA nanostructures as the delivery system could overcome some drawbacks of other biological agents, such as poor penetration level and low stability.
At the first decades, the main application of DNA nanostructures in bone tissue was gene therapy by using plasmid DNA (pDNA).215,226,251–253 In previous studies, the pDNA or other DNA nanomaterials were usually designed as the delivery systems to transport some growth factors and genes, including fibroblast growth factor (FGF), bone morphogenetic protein (BMP), and vascular endothelial growth factor (VEGF),254–258 which was regarded as viral gene delivery (Fig. 1g).259 The combinations with different materials and seed cells were also conducted in a vast of studies, and the purpose was to promote the seed cell proliferation and differentiation into the functional target tissues including bone and cartilage.260–264 The treatments based on different recombinant human growth factors have been investigated and the therapeutic effects were encouraging. Owing to the short half-lives of recombinant human growth factors and the needed for clinical effectiveness, supraphysiological dosages were used, and their production was expensive.265,266 The supraphysiological dosages of recombinant human growth factors could also cause several side effects, such as ectopic formation of bone tissue and soft tissue swelling.267 Therefore, the non-viral gene therapy then became a promising alternative strategy for bone tissue repair.268–270
In studies within recent decade, the non-viral gene vectors were developed by combining DNA with other scaffolds.259,271–275 The complex of DNA and poly(lactic-co-glycolic acid) (PLGA)/HAp composite scaffolds was constructed to delivery pDNA-BMP-2 into bone marrow stem cells (BMSCs) and enhance the DNA transfection efficiency, which was quite encouraging and promising for bone regeneration.276–280 Previous studies reported that the delivery of FGF-2 and BMP-2 genes could upregulate the transcription of Runx2 and osteocalcin,281,282 and Salem et al. reported that non-viral gene delivery of FGF-2 and BMP-2 via pDNA could attenuate the harmful effects of diabetes mellitus on bone regeneration at the defect sites.254 PLGA was used to encapsulate the novel nanoparticles that consisted of polyethylenimine (PEI) and human pDNA-BMP-2 to create pBMP-2/PEI nanoparticles for bone formation.283,284 The BMP-7 and VEGF-A genes were encoded into the injectable DNA-loaded nano-calcium phosphate paste, and the nanocomplex was regarded as bioactive bone substitution material.256 The DNA-loaded bone paste encoding with BMP-7 and VEGF-A showed enhanced regeneration of bone volume and significantly faster healing in critical site of bone loss at the early stages.256 The DNA-loaded nanoparticles could release DNA, resulting in cell transfection, and then promote the expression of BMP and VEGF in bone cells.285 Takanche et al. found that pDNA-c-myb conjugated with chitosan-gold nanoparticles (Ch-GNPs/c-myb) could suppress osteoclastogenesis and promote osteogenesis of dental implant even in osteoporotic condition, making an applicable material to sustain dental implant integration and therapy in some degenerative bone diseases.257,286 Some studies loaded BMP-2 gens (pDNA-BMP-2) into a chitosan (CS)-based hydrogel with a,b-glycerophosphate (a,b-GP), termed CS/CSn(pDNA-BMP2)-GP. The CS/CSn(pDNA-BMP2)-GP then possessed excellent capability in enhancing cell proliferation, maintaining great potential in gene delivery system and tissue regeneration of periodontium.287–291 Chitosan/β-glycerophosphate (CS/β-GP) hydrogel as a VEGF-sustained release system could delivery and release VEGF, and promote cell proliferation and differentiation of dental pulp stem cells (DPSCs).292 Huang et al.258 applied lipopolysaccharide-amine nanopolymersomes (LNPs) to delivery Noggin small interfering (si)RNA (siNoggin) and pDNA-BMP-2 to transfect cells, respectively. This study demonstrated that LNPs/siNoggin and LNPs/pBMP-2 could both enhance the osteogenic differentiation, and the osteogenic differentiation of LNPs/siNoggin was better than that in LNPs/pBMP-2. Moreover, different structural variations of star-shaped poly(l-lysine) polypeptides were used as promising nano-viral gene delivery systems for mesenchymal stem cells (MSCs), which could irritate MSCs to differentiate into bone tissue by delivering pDNA-VEGF and pDNA-BMP-2.293 Novel DNA/protamine complex scaffold was developed for a number of clinical usages, such as enhancement of bone regeneration,294,295 and pasting and delivery the genes of pDNA-FGF-2 or pDNA-BMP-2.294,296
Beyond the application of pDNA in bone tissue regeneration, some other DNA nanostructures were also developed for bone regeneration with their own features in recent decades. Some functional DNA materials were synthesized to promote the reconstruction of bone tissue, and some DNA materials could deliver drug monomers to exert anti-osteoarthritis effects. Tetrahedral DNA nanostructures (TDNs) have the specific framework and can be internalized by different cells, in which they can change the cell biological behaviors (Fig. 1g). Lin et al. reported that TDNs could enhance the cell growth and motility of different cells, such as adipose stem cells (ASCs), human periodontal ligament stem cells (PDLSCs), MSCs, and chondrocytes.65,227,297–300 TDNs could also induce stem cells toward osteogenic differentiation.65,297 Zhou et al.301 also reported that the TDNs could act the anti-inflammatory effects on the periodeontitis model, and could promote osteogenic differentiation. In this study, the TDNs acted the effects of anti-inflammations and regeneration on bone tissue, and the results suggested that DNA nanomaterials might be applied for some inflammatory diseases. In a recent study, TDNs were employed as the delivery loaded with Wogonin to form the TDNs/Wogonin complexes.302 The TDNs/Wogonin complexes were verified as a potential nanomedicine for osteoarthritis by executing different biological effects, including enhancing expression of chondrogenic markers, downregulating matrix metalloproteinases and inflammatory mediators, and promoting the expression of tissue inhibitor of metalloproteinase 1 and B-cell lymphoma 2. The TDNs/Wogonin thus proved itself with great potential as an injectable nanomedicine in the treatment of osteoarthritis.
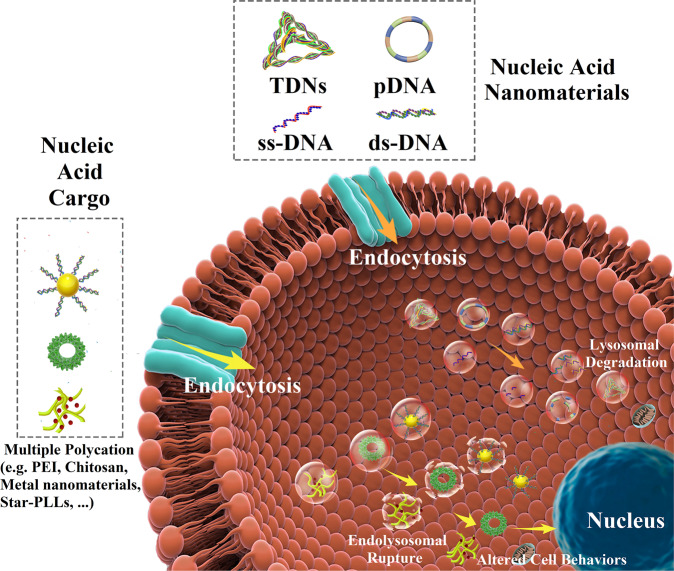
The DNA materials applied for bone regeneration were mainly the combinations of pDNA and other materials for enhancing the gene transfection efficiency to turn gene on or off, and the pDNA used was usually adenovirus plasmid.303,304 In general, for the easy laboratory manipulation, considering the features of porosity, adjustable degradation rate, and good mechanical property of natural and synthetic polymers, vast polymers were used to modify DNA materials for bone regeneration, such as alginate, chitosan, proteins (eg. fibrin, silk, and collagen), glycosaminoglycans, polylactides (PLA), polyglycolides (PGA), co-polymers PLGA, polycaprolactone (PCL), and polyethylene-glycol (PEG).305–308 When these complex materials were used in cells or organs, cellular uptake via endocytosis is the first challenge, since the efficiency and process of cellular uptake depend on the cell types and the physicochemical properties of the complex materials, such as the surfaces of cell and materials, and the shape and size of materials.309,310 The mechanisms of endocytosis are diverse, including both caveolae and clathrin dependent, micropinocytosis, and independent mechanisms.311 For DNA nanomaterials without any modification, when they are internalized by cells, the first existing form within cells is early endocytic vesicles, which can be rapidly acidified and thereby form late endosomes, and then DNA nanomaterials will be degraded by lysosomes (Fig. 2).312 Those nano-viral vectors mentioned above have some excellent properties, such as being non-immunogenic, high cellular uptake, outstanding endosomal escape, and rapid gene release.313–316 However, these studies mainly focused on the gene delivery and release, the investigations of the biological application in bone regeneration were limited in cytology experiments and small animal experiments, primarily the pre-clinical trials. The gene release processes of some delivery systems were even unclearly conducted and demonstrated. More importantly, the underlying mechanisms and the biosafety of these DNA materials combined with other scaffolds in bone regeneration remain poorly understood, and the effects on bone tissue regeneration of these nanomaterial complexes should be elucidated in further investigations.
Fig. 2.
The fate of DNA materials following delivery into cells. Nucleic acid nanomaterials (e.g., TDNs, pDNA, ssDNA, and dsDNA) with the excellent ability to enter cells, can be internalized by cells and degraded by lysosomal. Nucleic acid cargos combine the DNA materials and different delivery systems. These vectors can protect the DNA materials from degradation and promote cellular internalization. When inside the cells, these vector-DNA material complexes are embedded in an endosome, and these vectors can help DNA materials to escape lysosomal degradation
Neural tissue engineering
Neural tissue is considered as the vital system for all lives.317 For human being, neurodegenerative disorders and damages to neural tissue can result in severe and permanent disabilities,317,318 bringing heavy burdens for individuals, families, and socity.319 Owing to the lack of regeneration ability and complicated connections between different physiology systems, it is a great challenge to recuperate injured neural system, and make it work normally.317,320–322 Therefore, neural tissue engineering has emerged as a potential remedy with great capacity in repairing and regenerating nerve tissue injuries.323 In recent decades, biocompatible materials have been largely employed to achieve the regeneration and functional recover of injured neural tissue. Some successfully synthesized nanomaterials can provide mechanical support for neurite growing and inhibit the formation of scar tissue, affect biological cues to steer the growth of axons, promote regeneration, and incite the integration with the resident healthy tissue.321,324 Currently, the natural (e.g., collagen, gelatin, hyaluronic acid, and alginate) and synthetic nanomaterials (e.g., PLA, PGA, PLGA, and PEG) are considered as neural scaffolds for their biocompatible and biodegradable properties.325–329 Those nanomaterials could also combine with some specific DNA materials to obtain other positive effects for gene delivery or cell activation in neural tissue regeneration (Fig. 1a, f).240,330–333
The applications of gene therapy with DNA materials in neural tissue regeneration are also significant, and the pDNA is the most common material used.331,332,334 Feng et al.94 reported an injectable two-stage gene delivery system, which consisted of a nanofibrous micro cell carrier and a self-assembled nano-sized polymer, and found that the combination of the two-stage pDNA delivery system could promote intervertebral disc (IVD) regeneration and suppress the fibrosis (Fig. 1f). In this study, the pDNA was loaded with orphan nuclear receptor 4A1 (NR4A1). The NR4A1 was demonstrated as a safe and efficient gene agent with pleiotropic regulatory function to restrain pathogenic fibrosis, which was associated with vascular homeostasis,335,336 inflammatory responses,337,338 and metabolism of lipid and glucose.339–341 Taniguchi et al.342 reported a synthetic DNA-binding inhibitor with the ability of targeting vital cell differentiation controlling genes, and demonstrated that the DNA-binding inhibitor could target the SOX2 gene. The downregulation of SOX2 gene could trigger pluripotent stem cells differentiate into the mesoderm, which could form the notochords to wrap neural tubes. Wang et al.343 developed a DNA aptamer, named yly12, which could strongly target neurites. The aptamer yly12 could bind neural cell adhesion molecule L1 (L1CAM), which was expressed in normal neural tissue as surface antigen. Therefore, the aptamer yly12 could be used as a potential molecular probe to investigate the development of nervous system and the diagnosis of neurological diseases. Shea et al. used the poly(lactide-co-glycolide) (PLG) delivery DNA complexes to achieve gene delivery.333 In this study, the neurons and accessory cells were co-cultured together to measure the outgrowth progress of neurites when the PLG-DNA substrates were applied. Shea et al. found that surface immobilization of PLG-DNA substrates, which were modified with pDNA as the gene carrier and deliver systems, could promote the transgene expression and the regeneration of complicated neural system. In other studies, some nanoparticles (e.g., magnetic nanoparticles (MNPs), magnetic core-shell nanoparticles (MCNPs), and gold nanoparticles (AuNPs)) loading pDNA or RNA (e.g., short interfering RNA (siRNA)) were used to deliver genetic materials into neural stem cells (NSCs) and guide cell differentiation.344–348 Till now, although the above-mentioned work has presented numerous nanomaterials modified with different DNA materials, their applications in neural tissue engineering are still greatly limited, becasue the work is at the initial stage and the results are prelimilary. The mechanisms of different DNA-based and DNA-modified nanomaterials studied in neural tissue regeneration remains unclear. However, these studies intiated the step by providing the design principles for DNA-based and DNA-modified nanomaterials that were used in in vitro and in vivo nerve regeneration.
Recently, TDNs were reported as promising DNA nanomaterials for neural tissue regeneration (Fig. 1f).63,64,230 In these studies, it was demonstrated that NSCs treated with TDNs could exhibit enhanced cell proliferation, migration, and neuronal differentiation.63,64 In the in vivo experiment, a concomitant approach of NSCs and TDNs was applied in the animal model of spine cord injury (SCI).230 The study found that the TDNs possessed the capacity to increase the cell survival of the transplanted NSCs, and the concomitant approach of NSCs and TDNs could improve the recovery of motor function and the neural tissue regeneration in the injured site of spine cord. These results provided an interesting treatment strategy for neural tissue regeneration. Furthermore, Lin et al. also reported potential neuroprotective effects of TDNs on the cell models of Alzheimer’s Disease (AD) and Parkinson’s Disease (PD).240,330,349 Although these results were prelimilary, they might provide a promising synergistic agent for treating nerve damage and degenerative diseases. For futher studies, we would believe that TDNs and other DNA nanomaterials might be used to deliver some functional drugs and DNA/RNA sequences to the neural tissue for disease therapy.
Skeletal and cardiac muscle engineering
The obligations of skeletal and cardiac muscle are to support the body movements and blood circulation, where the chemical energy of nutrients are turned into kinetic and heat energy.350 Although the skeletal and cardiac muscle are both differentiated from the mesoderm, physiology and morphology properties of them are dissimilar.351 Lack of regeneration ability, the adult mammalian cardiac muscle could not self-renew when some injuries happen, such as heart attack.352–354 In contrast, skeletal muscle tissue can naturally repair minor injuries by self-regenerating, thanks to the property instigated by the activation of residual stem cells named satellite cells.355–357 However, severe damage and some myopathies can result in irreversible loss of muscle and functional disorder.206,358,359 Current therapies of broken skeletal muscle and some myopathies include injection of in vitro cultivated muscle cells and autologous skeletal muscle transplantation, but with many limitations in clinical applications of treating patients.206,360–362 In some previous studies, it was reported that some biomaterials could stimulate skeletal muscle regeneration via supplying chemical and physical cues to muscle cells, which could mimic the natural cascade of muscle tissue regeneration.206,212,359,363 The possibility of skeletal muscle regeneration is of great promise in curing the movement disability of bones and muscles,89 and novel medicines/biomaterials with some positive effects on cardiac muscle, such as protective effects from drug toxicity and hypoxia injury, are also of great significance in treating heart diseases.364
In recent studies, DNA-based nanomaterials were used for regenerating skeletal muscle and exerting protective effects from hypoxia injury of cardiac muscle, respectively.89,365 Wang et al.89 creatively developed a fancy near-infrared light-activated DNA agonist (NIR-DA) nanodevice (Fig. 1c). The NIR-DA system could deliver the cell signaling by nongenetic manipulation, and change the tissue phenotype of deep sites by activating the receptor tyrosine kinase (RTK), which enables the control of cell polarization, cytoskeletal remodeling, and directional migration. The study demonstrated that NIR-DA system could be employed in vivo to regulate RTK signaling, cell migration, and myogenesis of skeletal muscle satellite cells. Therefore, the NIR-DA system was regarded as a promising regeneration medicine for skeletal muscle injury.
Blumenfeld et al.364 reported a new-style covalent attachment of genomic DNA to iron-oxide nanoparticles. These magnetic nanoparticle complexes could capture some chemotherapy agents from human serum, including cisplatin, epirubicin, and doxorubicin (DOX). It was found that the DNA-coated nanoparticle complexes could protect cultured cardiac myoblasts from the lethal levels of chemotherapy agents, which could be developed as substrates for medicine capturing. Zhang et al. found the cardio-protection effects from myocardial ischemia-reperfusion injury (MIRI) by taking the advantage of the anti-inflammatory and antioxidative potential of TDNs (Fig. 1e), which could suppress the expression of reactive oxygen species (ROS) and then weaken the oxidative damage and apoptosis of cardiomyocytes.366 These studies offered some new ideas of using DNA-modified nanomaterials for treating cardiovascular and cerebrovascular diseases.367,368 Although the applications of DNA-based nanomaterials are few in this aspect, an innovative research design idea was provided by these studies for the skeletal and cardiac muscle engineering.
Skin tissue engineering
Skin, the biggest organ covering the entire body, consists of the epidermis and the dermis, with a complex blood supply and nerve innervation.369,370 For the resident stem cells in skin, the functions include hair regeneration, epidermal homeostasis, barrier resistance, and wound repair.371,372 Therefore, the health of skin organ is a significant element for human.373–375 The injuries of skin commonly happen with causes including burn, surgical operation, traumatism, chronic skin disease (e.g., parapsoriasis guttata and vitiligo), aging, systemic diseases (e.g., chronic kidney disease (CKD), hypertension, diabetes, and rheumatologic or inflammatory disease), infection (bacterial or fungal) and dermatitis.376–381 Nowadays, promoting the wound healing in skin is hot in clinical research, which can be affected by the interaction of several cells, growth factors, and cytokines.382–384 Some biomaterials have the abilities of the biomedical functions and tissue regeneration engineering, including implantable devices and some drug and gene delivery.385–387 Furthermore, some biomaterials were combined with DNA nanomaterials to overcome some shortcomings and enhance positive effects of these materials in biological application.388,389 DNA biomaterials applied for skin regeneration have also been widely studied in different skin diseases (Fig. 1b).232,388–390
For the therapy of full-thickness burn wounds, Gao et al.389 used the pDNA encoded VEGF-165/N,N,N-trimethyl chitosan chloride (TMC) complexes to load a bilayer porous collagen-chitosan/silicone membrane dermal equivalents (BDEs). The cells treated with TMC/pDNA-VEGF complexes could highly express VEGF gene and quickly promote wound healing. For diabetic chronic wounds, Lou et al.391 put the gene of pDNA-VEGF into a collagen-chitosan scaffold/silicone membrane bilayer dermal equivalent (BDE), and the DNA complexes showed the effective transfection abilities to promote the expression of VEGF gene, which could then promote the angiogenesis and change the immunomodulation.392 This study showed that the gene-activated bilayer dermal equivalents (Ga-BDEs) possessed the versatile potentials for accelerating wound healing of diabetic chronic wounds.391 Furthermore, Li et al. used hydrogel embedded with resveratrol (Res) and pDNA-VEGF (Gel-Res/pDNA-VEGF) to promote the healing of splinted excisional burn wounds. The Gel-Res/pDNA-VEGF was proved to possess the abilities to enhance the formation of microvascular and suppress the inflammation response.393 Additionally, the pDNA of basic fibroblast growth factor (bFGF; pDNA-bFGF) was also widely studied.232,390,394–396 Li et al.232 applied nanofibers (poly(l-lactic acid)/polyhedral oligomeric silsesquioxane (PLLA/POSS)) to load pDNA-bFGF and pDNA of angiopoietin (pDNA-ANG). The PLLA/POSS/pANG/pbFGF (Fab) composite nanofibers presented the capacities to promote revascularization of the deep skin defect healing. As reported, the electrospun core-sheath fibers loaded with pDNA-bFGF could promote skin regeneration in diabetic rats.394 Furthermore, dual delivery systems of the pDNA-VEGF, pDNA-bFGF, or pDNA-ANG were applied to promote regeneration of different skin defects.395–398
Fan et al. reported that TDNs (sizes ≤ 75 nm) could effectively penetrate the skin of mice and human, and reach dermis layer, which could load and deliver DOX to subcutaneous tumor site.399 This study inspired that TDNs could deliver other drugs to treat more subcutaneous tumors. Zhu et al.231 demonstrated that TDNs could promote fibroblast (HSF cell line) and keratinocyte (HaCaT cell line) growth and migrating. In addition, TDNs could promote the secretion of VEGF and bFGF and enhance the wound healing. Therefore, TDNs might be an novel DNA materials with the potential in skin tissue regeneration and drug delivery for subcutaneous tumors.
Other tissue engineering
Beyond above-mentioned tissue engineering applications, the DNA materials also have been used in vascular tissue and ocular tissue.62,400–404 Angiogenesis is important for other tissue regeneration, such as bone, muscle, and neural tissue. Abundant blood supply can enhance the nutrition supply for different tissue regeneration. For angiogenesis, materials loading pDNA or DNA aptamer are widely studied. Mao et al.401 used N,N,N-trimethyl chitosan chloride (TMC) that was modified with pDNA-VEGF to promote VEGF gene expression and thereby effectively enhance the angiogenesis. The increased angiogenesis could also be promoted by some scaffolds containing both pDNA-VEGF and pDNA-FGF2.402,403 Son et al.400 developed aptamer-conjugated hydroxyapatite (Apt-HA) to promote angiogenesis and bone regeneration. In recent years, TDNs were employed to promote angiogenesis in vitro and the model of the jaw bisphosphonate-related osteonecrosis.405–407 Although the DNA-based complexes applied for angiogenesis are in their early stages, DNA-based materials might be developed as regeneration medicine to promote angiogenesis in different tissues.
As for the research on ocular tissue, the number of studies keeps increasing in recent years, indicating growing interests and great potential of applications for ocular tissue regeneration and ophthalmic drug delivery (Fig. 1d). Heremann et al. combined DNA and nanoparticles as DNA-based complexes to treat eye infections, which was the first report describing the usage of DNA-based carrier systems in ophthalmic drug delivery.408 Then, PEI-DNA nanoparticles were employed to deliver corneal gene, with great efficiency in gene delivery for the corneal gene therapy.409 AuNPs-encoding pDNA were used to transfect human retinal pigment epithelium cells.410 Furthermore, TDNs were used to facilitate the corneal wound healing, promote the re-epithelialization of wounds, and improve the corneal transparency.62 Cai et al. reported that TDNs could prevent retina ischemia-reperfusion injury from the oxidative by reducing the ROS.404 Moreover, Gao et al.411 proven that TDNs could induce the immine tolerance and prevent the onset of Type 1 diabetes (T1D). For T1D occurring with T cell-mediated autommune, inhibition of autoreactive T cell and inducing regulatory Tregs to rebuild the immune tolerance are the two promising strategies. In this study, the researchers reported that the TDNs could control the level of glycemis, protect β-cell, and prevent the onsed of T1D by regulating the immunity. Therefore, some DNA nanomaterials might possess the abilities of immunomodulation for some immune diseases. The regeneration and protection of the ocular tissue are vital for human healthcare, and DNA-based nanomaterials will maintain attractive in this field in the following years.
Drug delivery and tumor therapy
In recent decades, DNA nanotechnology has also made amazing progress in biomedical applications in drug delivery and tumor therapy. In this part of the review, the development trajectories and latest advances of pristine DNA-based nanostructures will be highlighted.
Compared with conventional nanoscale drug carriers, such as gold nanoparticles and liposomes, DNA-based nanostructures are more suitable candidates characterized by high biocompatibility, structural diversity, low cytotoxicity, high accessibility, and capability of penetrating cell membranes without transfection.412–414 Owing to their nucleotide-composed backbones and flexible joints, DNA-based nanostructures possess high editability and modifiability. They can be built into arbitrary constructions like stacking building blocks.38,415 According to their intrinsic natures, multifunctional ligands can be integrated into frameworks through different ways, including conjugation, intercalation, encapsulation and loading.416,417 Thus, numerous assemblies of DNA nanoparticles were developed and further applied to multiple biomedical fields, especially in drug delivery (Fig. 3).30,418 In the following, diverse DNA nanoparticles applied in biomedicine are listed and illuminated (Table 2).
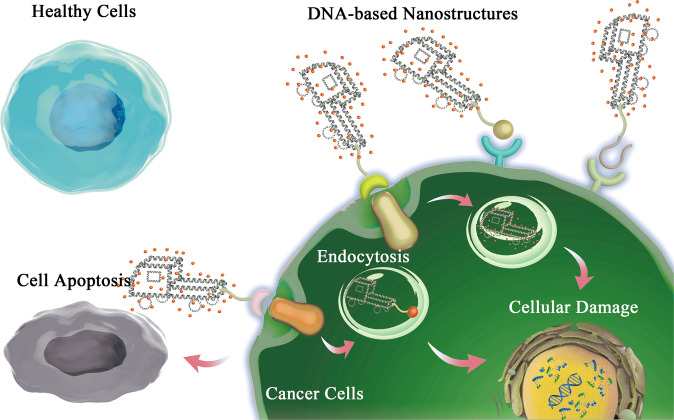
Fig. 3.
DNA-based nanostructures on tumor therapy by modification with drug, aptamer, or other functional ligands act various biological functions, which can bind to the target protein of cells. DNA-based nanostructures could distinguish cancer cells via ligand-receptor binding and permeate the membranes through endocytosis. The corresponding release of therapeutical agents would cause cellular damage and lead to apoptosis of cancer cells
Table 2.
DNA-based nanostructures on tumor therapy and cell imaging
| Number | DNA-based nanostructures | Proteins or aptamers /target | Modification type | Payloads | Loading way | Application | References |
|---|---|---|---|---|---|---|---|
| 1 | Straight and twisted DNA tubes, DNA triangle | N/A | N/A | DOX | Intercalation | Cancer therapy | 424,425 |
| 2 | TDNs | FA/folate receptor | Click chemistry | DOX | Intercalation | Cancer therapy | 426 |
| 3 | DNA tube | AS1411/NCL | Linkage at the ends | Thrombin | conjugating through cross-linkers | NCL-overexpressed cancer therapy | 427 |
| 4 | DNA icosahedron | MUC1 aptamer /MUC1 | Chemical bond | DOX | Intercalation | MUC-positive cancer therapy | 429 |
| 5 | DNA dendrimer | AS1411/NCL + MUC1 aptamer /MUC1 + ATP aptamer/ ATP | Chemical bonds | EPI | Intercalation | Cancer therapy | 430 |
| 6 | TDNs | HApt /HER2 | Chemical bond | N/A | N/A | HER2-positive cancer therapy | 439 |
| 7 | TDNs | Affibody/HER2 | Chemical bond | DOX | Intercalation | HER2-positive cancer therapy | 437 |
| 8 | TDNs | AS1411 /NCL | Chemical bond | 5-FU | Chemical bond | NCL-overexpressed cancer therapy | 88 |
| 9 | TDNs | SL28 /VEGF-165 + FA /folate receptor | click chemistry | DOX | intercalation | Colorectal cancer therapy | 438 |
| 10 | TDNs | MUC1 aptamer /MUC1 + AS1411 /NCL | Extend from vertex + overhang on vertex | DOX | Intercalation | Cancer therapy and cell imaging | 449 |
| 11 | TDNs | AS1411 /NCL + GS24 /TRF | Chemical bond | TMZ | Load | Glioblastoma therapy | 450 |
| 12 | TDNs | AS1411/NCL + MUC1 aptamer/MUC1 | chemical bond | Ir | Load | Glioma therapy | 452 |
| 13 | TDNs | N/A | N/A | MB | Interaction | Cancer therapy | 454 |
| 14 | DNA triangle | N/A | N/A | BMEPC | Interaction | Cancer therapy | 455 |
| 15 | TDNs | Nuclear localization peptide /nucleus | Chemical modification | ASOs | Disulfide linkage | Gene c-raf silence | 433 |
| 16 | X-Y-shaped DNA | Sgc8/ CCRF-CEM cancer cells | Linked with Y-shaped DNA strand | DOX | Intercalation | T cell acute lymphoblastic leukemia therapy and MDR | 462 |
| 17 | DNA nanotrain | Sgc8/ CCRF-CEM cancer cells or AS1411/NCL | Chimeric aptamer-trigger | DOX, EPI or daunorubicin | Intercalation | Targeted cancer theranostic | 463 |
| 18 | NFs | Sgc8c /PTK7 | Integrated with template sequence | DOX | Intercalation | PTK7-overexpressed cancer therapy | 464 |
| 19 | DNA nanocircuit | Aptamer/cancer cells | Linked with an overhung catalyst sequence | Chlorin e6 | Modified on ssDNA | Cancer therapy | 465 |
| 20 | DNA nanocentipede | Zy1/ hepatoma SMC-7721 cells | Streptavidin-based linkage | DOX | Intercalation | Hepatoma cells SMC-7721 targeted delivery | 466 |
| 21 | DNA nanorod | N/A | N/A | DOX | Intercalation | MDR | 108 |
| 22 | NFs | N/A | N/A | DOX | Intercalation | MDR | 484 |
| 23 | TDNs | N/A | N/A | DOX | Intercalation | MDR | 109 |
| 24 | DNA triangle and tube | N/A | N/A | DOX | Intercalation | MDR | 482 |
| 25 | TDNs | N/A | N/A | PTX | load | MDR | 107 |
| 26 | TDNs, square, pentagon-based pyramid and prism | PfLDH aptamer /PfLDH | Linked with ssDNA | N/A | N/A | Malaria diagnosis | 501 |
| 27 | TDNs | ANG /LPR-1 | Click chemistry | N/A | N/A | Brain tumor imaging | 502 |
| 28 | TDNs | FAM and HE | Amido bond | N/A | N/A | H + and O2- detection | 504 |
DNA polyhedron and origami
Since the first closed DNA polyhedron (poly-DNA) developed by Seeman et al., multifarious DNA-based predetermined motifs (tubes, buckyballs and prisms) were created.419–421 By means of DNA origami technique, the long scaffold ssDNA coupled with short and flexible staples provide more conceivements for different DNA motifs. For further developments and applications, poly-DNA was served as a nanocage to accommodate pharmaceutical molecules owing to its inherent spatiality.416–418 Extended sticky ends can also be designed for the attachment of additional aptamers, biotins or valuable peptides thanks to their modifiable external surface.422 With the modification of available ligands, it holds great promise in tumor targeted therapy.423
It is common to utilize DOX to form a drug-DNA nanocomplex. For instance, Hogberg et al. employed two types of DNA tubes (straight and twisted with different numbers) for carrying DOX and compared their carrying performance.424 Triangular DNA and tubular DNA of different forms were constructed as a vehicle to deliver DOX into cancer cells for effective cancer therapy.425 Folic acid (FA) coupled with DOX were also linked to TDNs,426 which indicating the excellent delivery abilities of TDNs.
A DNA rectangle bent into a hollow DNA tube, and the butt seam of which was fastened with AS1411 (a DNA aptamer).427 Its open state depended on the affinity of aptamer with the nucleolin targeting. With the structural separation, inner thrombin was exposed selectively to induce blood coagulation and cause cell death by vascular occlusion. Biocca et al. reported that truncated DNA octahedron (octa-DNA) could be recognized by low-density lipoprotein receptor-1 (LOX-1) because of its similar property with oxidized low-density lipoprotein (ox-LDL).428 It could thereby invade into LOX-1 related cells via substrate-receptor mechanism, which indicated the inherent targeting property of octa-DNA. Huang et al. proposed a unique DNA icosahedron (icosa-DNA) that was composed of six-pointed star skeleton.429 MUC1 aptamer was appended to its primary synthetic unit, and DOX was simultaneously embedded into double-stranded docking sites.416 This MUC1-DOX modified icosa-DNA could distinguish the MUC1-positive cells and achieve controllable release of DOX intracellularly.
As the non-stop development, multiple-bundle DNA-based nanostructures for different purposes appeared. An aptamer-Epirubicin (EPI) DNA dendrimer was designed by Khalil et al.430 Distinctively, more than two aptamers (AS1411, MUC1 and ATP aptamers) coupled with Epi were embraced in this bracket, equipping it with multitargetness and potent cytotoxicity.423
Applications of TDNs in drug delivery
Among multitude architectures, TDNs is of the greatest interest.412,413,431–434 Owing to its tripod-shaped framework, excellent structural stability was conferred so that it could resist deformation under enzymatic digestion. As for the synthesis of TDNs, several synthetic schemes were put forward, and TDNs of different sizes were successfully produced.169,435,436 These products were examined with potential to remedy many disorders. In previous studies, aptamers (such as AS1411, anti-HER2 aptamer (HApt), and SL2B), antibodies and proteins (affibody, FA) were conjugated with TDNs for cancer cell recognition and inhibition.437–439 Coupled with the loading of therapeutical agents like 5-fluoroutacil (5-FU), DOX, paclitaxel (PTX), the products were supposed to selectively enter cancer cells and exert potent antitumor effects.88,107,426
As a proof of concept, HApt was linked with TDNs to anchor HER2-overexpressed cancer cells.439 HER2 is a member of transmembrane proteins, and a hub for signal transduction pathways among cancer cells. Its overexpression is associated with malignancy and the poor prognosis of breast cancer.440 According to previous work, HApt could be specifically chosen to bond with HER2 and supress its expression.441,442 With this in mind, HApt-TDNs was prepared to specifically combine with HER2 and then transfer it into lysosome for the following degradation. This behavior consequently contributed to cell apoptosis and inhibit tumorigenic growth. Compared to simple HApt, the stability of this nanocomplex was considerably improved and response time was prolonged. It might stand for a new perspective in breast cancer therapy. In addition, a tailored affibody molecule was built to mimic the structure of monoclonal antibody, which was confirmed with specificity to target HER2.437 With the additional intercalation of DOX, this DNA-affibody-drug chimera showed excellent specificity and inhibition to HER2-overexpressed cancer cells.
With regards to aptamers, G-quadruplex aptamer (AS1411) was the representative one that was systermatically investigated. It was equipped with both targetness and pleiotropic antiproliferation through its combination with targeting receptor-nucleolin (NCL).443,444 It could be directly attached to the vertice of poly-DNA or linked with the ssDNA to form an overhang at the edge of 3D structure.445 These AS1411-modified poly-DNA could act as an effective drug carrier or a nanomedicine with targeting ability for therapy.100 Studies have also been concentrating on the structural upgrading of TDNs.446–448 AS1411 and 5-FU were simultaneously conjugated to TDNs to establish a splendid nanomedicine that specifically inhibit the cell viability of NCL-overexpressed cancers.88
SL28 is a VEGF-165-related DNA aptamer. Its dimerization with VEGF-165 plays an important role in angiogenesis in vivo. Zhao et al. presented a dual-targeting nanosystem for SL28 and FA, the cooperation of which could obviously improve targeting ability of nanosystem towards colon cancer cells.438 With a follow-up loading of DOX, the synergic antitumor effect was obtained. Liu et al.449 united MUC1-probe and AS1411 with TDNs, as well as the payload of DOX. The selective MUC1 aptamer-protein binding induced an unwinding of quenched sequence and fluorescence recovery to differentiate MUC1-positive cancer cells. The follow-up AS1411-nucleolin bound caused its translocation into nucleus and DOX release intranuclearly. This superior combination carved a path to cancer theranostics. Furthermore, temozolomide (TMZ) loaded TDNs along with two DNA aptamers (AS1411 and GS24) for glioblastoma treatment was reported.450 GS24 was reported to be a transferrin (TRF) aptamer of 64 nt in length.451 Owing to its unique spatial structure, TRF on cerebrovascular endothelial cells of BALB/c nude mice can seize and combine with GS24, and thereby pass through cerebrovascular barrier. Specifically, after injection with this nanointegrate, the function route was as follows: firstly, it moved across the blood-brain barrier (BBB) of mouse and permeated into brain parenchyma via receptor-mediated transcytosis; secondly, the assistance of AS1411 enabled it to enter the cell nucleus, and then offloaded TMZ for its lethality towards tumor cells, which could alleviate the resistance and myelosuppression of TMZ at the same time.
Other elements including metal complex and photosensitizer were also added as adjuvants to substantially enhance the medical effects of TDNs. Tian et al.452 employed a dual aptamer-tethered TDNs for carrying metal compounds. A representative platinum agent-[Ir(ppy)2phen]+PF6 (Ir) was loaded onto MUC1- and AS1411-modified TDNs. It anchored glioma cells to destroy their vascular mimicry processes and cause cell death through mitochondrial and ROS pathways, which contributes significantly to glioma therapy.
In addition to routine drug treatment, photodynamic therapy (PDT) is a minimally invasive means, which is frequently used in tumor therapy. Photosensitizers including porphyrinoid and cyanines can be activated under irradiation, thereby produced excess ROS and ultimately drove cancer cells to necrose.453 When they are attached to or loaded on TDNs, their disadvantages such as low stability, poor solubility and tissue penetration were greatly minimized. Methylene blue (MB)454 and BMEPC,455 which are commonly used as imagine probes and methemoglobinemia agents, can also interact with TDNs and DNA origami. Subsequently, when the preprocessed tumor tissues were exposed to light, a satisfying cytotoxicity against tumor cells would be observed.
Other than chemical agents, the delivery of antisense oligonucleotides (ASOs), proteins and RNA are also spotlighted. Dual-bundle TDNs with targeting peptides and ASOs were reported to effectively deliver ASOs into nucleus of tumor cells.433 The increased release of ASOs could lead to an enhanced cytotoxicity and the knockdown of proto-oncogene c-raf. Studies regarding small interfering RNA (siRNA) were also reported.431,456,457 Similar DNA nanoparticles with cancer-targeting peptides were utilized to accurately transport siRNA into tumor cells and trigger related genes silencing. The low expression of particular genes is the main inducement of cell inhibition.
In addition to the above-mentioned, TDNs were also proved to affect various pathological courses. Lin et al. reported that TDNs could modulate the M1 polarization of macrophages into an appropriate level, avoiding an exaggerated inflammatory response.458 It also exhibited antioxidative and anti-inflammatory potentials through suppressing mitogen-activated protein kinases (MAPK, including JNK1/2/3, ERK1/2, and p38 family) and upregulating the expression level of HO-1, which is associated with anti-inflammation, anti-apoptosis and antioxidation.
In the microbiological researches, TDNs have sparkled as a component of innovative antimicrobial agents. With the advent of superbugs, bacterial resistance caused by antibiotics abuse has become so acute that effective measures should be immediately taken to prevent the deterioration of this condition. An antisense peptide nucleotide (asPNA) with high stability and affinity was proposed to inhibit the transcription of an intended bacterial gene (ftsZ).459 Its encoded protein (FtsZ) was confirmed to be involved in bacterial cell division and has been used as a validated target in bacterial prevention. Lin et al. incorporated asPNA with TDNs to deliver it into methicillin-resistant Staphylococcus aureus (MRSA), interfere the expression of ftsZ, and subsequently result in bacterial growth inhibition. Although humans are collective organisms with microbes, the colonization of bacterial biofilm is a decisive factor of chronic inflammation, which is the main culprit of critical illnesses in essential organs. Microbial communities form compact biofilms, along with exopolysaccharides (EPS) and other extracellular matrix, to protect themselves from antibiotics. Therefore, the most fundamental method to diminish the vitality of bacteria is to prevent the initial formation of bacterial biofilms. The same study group designed an ASOs on the basis of VicK protein binding domains to regulate the secretion of EPS for early prevention against biofilm formation.460 It could specifically target gtfBCD (from glucosyltransferase gene family), gbpB, and ftf genes, which encoded a series of Streptococcus mutans biofilm formation proteins, a glucan-binding and an adhesion-associated protein, respectively. Similarly, ASOs were carried by TDNs to degrade the expression of targeted genes and availably block EPS production, which were both prerequisites for initial stage of biofilm.
DNA intelligence nanodevice
Along with the progress in DNA nanotechnology, deoxynucleotides were concatenated, folded, and assembled into a more intricate, multifunctional and intelligent nanodevice.461 Primitively, Tan et al. created X-Y-shaped DNA nano-assembly462 and aptamer modified DNA nano-train.463 For the former, three branches of Y-shaped DNA units were developed to link or intercalate with aptamers, drugs or oligonucleotides, and the chimera then jointed in the X-shaped central bonder through predesigned supplementary arms. After compact crosslinking of these X-Y-shaped building blocks, a simple nano-assembly with targeting and multiple drug resistance (MDR) reversion ability could be obtained. The latter one was composed of two hairpin-patterned DNA sequences (M1, M2) and an aptamer trigger. The hybridization of M1 and M2 was initiated by the mixture of chimeric trigger, resulting in a nano-train with a chimeric aptamer locomotive and many chambers for drug loading. The same group intensively studied a noncanonical DNA nanoflower (NFs) without following Watson–Crick.464 It was self-assembled from DNA concatemers generated by rolling circle replication and the building template could be tailored with the attachment of functional moieties for biotherapy and bioimaging, etc. DOX and aptamer sgc8c were embedded and incorporated into NFs to recognize PTK-overexpressed cells and exert therapeutic effects. The optical labels could be similarly applied for fluorescent imaging. Moreover, several novel DNA nanocircuits were also established.465 The trigger sequence linked with the aptamer could precisely activate a series of related cascade reactions in target cells to eventually produce therapeutic effects.
Xu et al. fabricated a DNA nanocomplex with a trunk and several legs, resembling a centipede.466 Two hairpin sequences constituted a DNA scaffold through hybridization chain reaction (HCR) to form a long DNA trunk, providing abundant docking sites for pharmaceutical molecules. Aptamers such as Zy1 were combined with the trunk-like couples of legs via streptavidin-biotin affinity. It could grasp hepatoma cells SMC-7721 and release the payload to influence the cell activity. Theoretically, this nanocentipede could be further used in multivalent drug delivery.
Moreover, smarter 3D architectonics designed for drug containers like DNA boxes were set up with movable switches.145 Their hollow constructions provide sufficient spaces for functional cargoes. Contents could be reversibly exposed or manageably released under genetic, optical or many other biochemical triggers.467 Additionally, more sophisticated lock-and-key systems were fabricated to control the accessibility of cargoes or for controlled reaction rate as required.468 Their structures were also refined to the greatest extent so that they could readily pass through the biological barriers in vivo.
For DNA nanodevices illustrated above, the appended molecules were mostly replaceable, revealing that they were superior to drug carriers with broad feasibility. However, the microstructures of the combination between DNA nanomaterials and different drugs cannot the clear observed.
DNA nanohydrogels
As for particle size, apart from individual nanoscale structures, micron-sized DNA polymers were constructed via liquid phase crystallization and dense packing of dsDNAs instead of Watson–Crick base-pairing.469,470 Thus, they exhibit excellent monodispersity and biocompatibility. Tan et al. proposed a multi-module DNA nanohydrogel with one linker and two functional monomers.471 The size could be adjusted by controlling the ratio of building units. Elements including aptamers and genes could be incorporated to build an efficient nanomedicine. Hitherto, not just linear as nanowires, other devisable and rigorous nanostructures have been obtained. They were applied extensively in both biosensing and biomedicine, and would be further utilized in biological nanotechnology.
Switchable DNA nanomachine
In order to meet the growing demands, mechanically controlled DNA nanomachines with operational strands that could be switched on and off by active domains emerged.472–474 For instance, complementary fuel strands were designed to initiate DNA tweezers.475 Their hybridization and removal enable these tweezer-like machines to open and close reversibly. In addition, the structural transformation could be triggered by following fuses: changes of pH, light excitation, binding with specific sequences or incorporating with proteins, aptamers or other functional ligands. A pH-dependent DNA origami plier with two levers connected at a fulcrum was observed to transform according to pH value.476 Its X-shaped open state could turn into parallel or antiparallel closed form in response to acidic conditions.
At present, a new concept-DNA nanorobot is emerging. It refers to an array of highly intellectual nanodevices. They could be equipped with multiple functions through precision assembly. The mode of action was controlled by organized switching system, which includes but was not limited to simple on-off setting, and the logic gates (AND, NOT, OR, etc.) could also be set up.477,478 Thubagere et al.43 devised an autonomous DNA robot for cargo sorting. Unlike previous ones that walked following a prescribed route, it could walk towards different directions randomly without energy supply for automatic cargo detection and classification. Three building blocks were designedly composable to select various cargoes at uncertain locations and classify them into two groups. This sorting task could be carried out on the surface of a 2D DNA origami sheet. In more sophisticated scenarios, several robots could be assigned to work cooperatively. When it comes to other molecules, such as antibodies, proteins or nanoparticles, promising applications including molecular computation and biomedical diagnosis can be explored.
Multi-drug resistance (MDR) reversion
DNA-based nanostructures can not only achieve effective delivery of chemical agents to tumor sites, but also reverse multi-drug resistance (MDR) in cancer cells. MDR, either inherent or acquired, has always been a tough challenge in drug development and utilization. To overcome it, multifarious drug combination strategies are proposed and different drugs are formulated, but their effects are not that satisfactory.479 The causes of MDR are extremely complex, including DNA repair, decreased drug flow and detoxification. According to previous studies, the most common reason for this is the excessive efflux pumps, which can eject drug and result in ineffective.479,480 Taken together, the fundamental way to bypass this efflux-pump mediated MDR is to improve drug concentration in the pathological areas.
DNA-based targeted drug delivery system has become a focus worldwide because of its ability to increase drug accumulation specifically in cancer cells via enhanced permeability and retention effects (EPR).481 It proved that DNA nano-systems could enter cells through caveolin- and clathrin-mediated endocytosis, independent of efflux pump pathway.413 Therefore, their high delivery efficiency can be expected to reverse obstinate MDR observed in most drug tests.
Conventional agents such as DOX, PTX, 5-FU have been confirmed to elicit MDR in tumor cells during their long-term use. Investigators have been striving for a more optimized program-various types of DNA nanoparticles that were applied for DOX resistance.481–483 As a consequence, DNA origami was reported to show better performance in delivering effective levels of DOX than poly-DNA owing to its higher density of DOX in more abundant docking sites. DOX loaded rod-like,108 X-Y-shaped DNA origami and an apt-NFs nano-assembly484 were fabricated for human leukemia/MDR cells, respectively. A DOX loaded TDNs was found to inhibit MCF-7/MDR cells activities.109 MDR reversion in these cases were achieved through efflux evasion, which directly enhanced local drug concentration, and thus, extended effective action time and reduced the general toxic effects. Additionally, Ding et al. constructed 2D (DNA triangle) and 3D (DNA tube)482 nanostructures to carry DOX. Both of them could improve drug sensitivity of DOX/MDR human breast cancer cells through efflux pathway. They were verified to be able to prompt drug molecules to disperse into active sites through elevating lysosomal pH.
PTX loaded TDNs (PTX-TDNs) presented by Lin’s group was surprisingly valuable for treatment of PTX/MDR non-small cell lung cancer.107 It exerted cytotoxicity towards PTX/MDR cancer cells through downregulating the expression of P-glycoprotein (P-gp), a membranal transporter associated with drug efflux. In other words, PTX-TDNs could increase intracellular excretion of drugs in MDR cells, resulting in potent microtubule polymerization and cell apoptosis.
Immunostimulatory effect
Similar to aptamers and drugs, ligands with immunostimulatory (e.g., CpG), or catalytic activities (e.g., enzyme) can be appended to DNA frameworks.485 Since the basic component of DNA nanoparticles is deoxyribonucleic acid, they exhibit low immunogenicity and their internalization would not intensify immunoreaction. Unmethylated CpG oligodeoxynucleotides can be recognized by mammalian immune system because of its existence in bacterial or viral genome.486,487 It can specifically combine with Toll-like receptor 9 (TLR9), and activate it allosterically, which subsequently induce cascade reactions and promote the secretion of remarkable cytokines and chemokines, including interleukin-12 (IL-12) and tumor necrosis factor-α (TNF-α).488,489 Thereout, DNA-CpG nanosubstance was conceived an immunomodulating nanomaterial (Fig. 4).
Fig. 4.
DNA-based nanostructures on immunostimulatory reactions. CpG-modified DNA-based nanostructures could enter immune cells, including macrophages, mast, dendritic and monocyte cells through integrating with TLR9, which subsequently induce cascade reactions and promote the secretion of remarkable cytokines and chemokines
DNA nanotubes490,491 of different dimensions and DNA dendrimers492,493 with CpG were reported in previous studies. Fan’s team attached CpG to four ssDNA building units and prepared a CpG-bearing TDNs.494 Variform DNA nanoribbons based on rolling circle amplification (RCA) technique were proposed by the same group as well.495
In general, macrophages along with dendritic, monocytes and mast cells are of great significance in maintaining immune homeostasis and play an important role not only in autoimmunity, but also in inflammatory diseases, metabolic disorders, neoplasia, anaphylaxis and wound healing. CpG-modified DNA-based nanostructures could enter immune cells, including macrophage RAW264.7 cells noninvasively and efficiently, and keep intact in their endosomes, whereupon trigger enhanced immune responses.496 Through this activation process, they were expected an alternative to surmount tumor-associated immunodepression and regulate essential pathological reactions. Afterwards, CpG was respectively linked to ssDNA, dsDNA, Y-shaped DNA (Y-DNA), and three types of dendritic DNA (DD-DNA).492,497 In the literature, Y-DNA with three dendritic branches served as a single element in DD-DNA’s building-up process. One Y-DNA was ligated to three other duplicates to obtain a primary DD-DNA (D1). One D1 was thereby ligated to another six Y-DNA to form a secondary DD-DNA (D2). By that analogy, tertiary DD-DNA (D3) was synthesized. It was noteworthy that individual Y-DNA, D1, D2 and D3 without CpG were substantiated to have intrinsic immune activation properties, and there was an increase with the growth of branch number, maybe partially due to their increased uptake by macrophages. Then, CpG were positioned on the outermost layer of each structures. In other words, D1 (CpG), D2 (CpG) and D3 (CpG) were decorated with 6, 12 and 24 CpG sequences, respectively. Detailed experiments revealed that the secretion levels of TNF-α and IL-6 were strikingly increased in DD-DNA (CpG) and Y-DNA (CpG), compared to ssDNA (CpG), dsDNA (CpG) and simple mixture of several CpG sequences. However, the increasing trends were not proportional but much higher than the enhanced immunoreaction triggered by non-CpG structures. It was indicated that the immunostimulation of CpG motifs could be greatly amplified by the structural characteristics of Y-shaped DNA dendrimer. A similar profile was detected in the comparison of Y-DNA modified with 1 and 3 CpG motifs. Thus, more CpG motifs on one architecture might stimulate stronger immune response. This finding was then extensively applied to inhibit the growth of tumor cells. After adding Y-DNA (CpG)-treated RAW264.7 cells, a significant decrease was observed on the number of murine melanoma cells. To clarify the relationship between structural features and biological properties, the same team established some other polypod DNA (p-DNA).498 They used 3 to 12 oligodeoxynucleotides of different lengths to generate tri-, tetra- to dodecapods DNA of different sizes, and CpG was attached in a similar way. Through rigorous examinations, it was discovered that p-DNA with more pods entered RAW264.7 cells more easily. The release levels of TNF-α and IL-6 showed the same trend, and an increasing order was found in tri-, tetra-, penta-, hexa- and octapods DNA, which symbolized a stronger activation of immune response. These results provided definite evidence that the growing number of pods had an impact on activating the level of immunostimulation.
To the best of our knowledge, the invasion of foreign substances would cause varying degree of host immune defense. In general, it manifests as an excess secretion and accumulation of inflammatory cytokines, which is the origin of systemic complications. To explore the immunological role of TDNs, previous studies established lipopolysaccharides (LPS)-induced inflammatory cell model to act RAW264.7 cell line.458 After retreatment with TDNs, the production and the expression of nitric oxide (NO) and inducible NO synthase (iNOS, closely related to NO production) were decreased and downregulated to regulate NO within a normal concentration. Herein, it could be inferred that TDNs is an outstanding biomaterial without immune rejection. Based on this, Chang et al. utilized TDNs as an antigen platform for vaccine.432 As CpG conventionally serves as an adjuvant in vaccine for immunotherapy, Chang et al. assembled an antigen-streptavidin (STV), CpG and TDNs into a vaccine complex (SCT). Both in vitro and in vivo studies showed that STV and CpG could be more easily internalized by antigen-presenting cells due to the application of TDNs delivery system. Then, more T cells were activated, facilitating the differentiation of STV-related B cells and the production of relevant antibodies. Importantly, the 3D architecture of SCT could render a close proximity of adjuvant and antigen, which highly augmented immunogenicity of the vaccine. Given results above, DNA-based nanosystems maintained an impressive immunostimulation potential that might be further utilitized in immunotherapy and vaccine development.
Biosensing, biodetection, and diagnosis
Early diagnosis of diseases is of great importance in developing timely and effective treatment stragety to prevent deterioration and cure illness.499,500 The traces of intracellular molecules, proteins or nucleic acids have great benefits in disease prevention, diagnosis and therapy. Conventionally, experimental approaches such as western blotting to assay their expressions are time-consuming and laborious. Micromolecular probes that can achieve real-time detection and dynamic imaging are preferable. A robust probe delivery vehicle is on demand to conquer the instability of simple probes. More recently, DNA-based nanostructures show extraordinary promise in molecule detection and biomedical imaging (Fig. 5).413
Fig. 5.
DNA-based nanostructures on bioimaging and diagnosis. Targeted ligands (e.g., affibody, peptides and aptamers) and cargoes (e.g., drugs, fluorescein and radioisotopes) were simultaneously attached to DNA-based nanostructures. After specific combination with biomarkers, the integrates entered target cells and released fluorescent labels to sensitively detect the diseased regions
Generally, aptamer-based DNA diagnostic nanodevices were widely explored. TDNs, square, pentagon-based pyramids and prisms were incorporated with Plasmodium falciparum lactate dehydrogenase (PfLDH) aptamer and fluorescent labels to distinguish malaria biomarker- PfLDH from human lactate dehydrogenase.501 Recent progress has been made that angiopep-2 (ANG) linked TDNs (ANG-TDNs) was a potential imaging medium for brain tumors. Owing to the high affinity of ANG with low-density lipoprotein receptor protein-1 (LPR-1) on brain endothelial and glioma cells, ANG-TDNs was able to cross BBB and recognize cancer cells,502 and combined ues of luminescent dyes could specifically label brain tumor regions.
In Argonaute2 (Ago2) detection, Zhang et al.503 designed a switchable nanoprobe using TDNs as a scaffold. An RNA-hairpin sequence was linked to the terminal of one strand, and a photoinduced electron transfer pair (PET) was attached to the terminal of the complementary strand. The vicinity of them caused a fluorescent quenching, while the emergency of Ago2 and its corresponding siRNA could restore its fluorescence. Under conditions of inflammation, neurodegeneration and most diseases, abnormalities of hydrogen ions (H+ ) and superoxide anion (O2-) can be discovered. Li et al.504 incorporated hydroethidine (HE) and fluorescein (FAM) with the vertex of TDNs, the intensity of fluorescence would change in response to pH and O2−. Hence, a sensitive nanoprobe monitoring microenvironmental changes emerged for future disease diagnosis. The reason why TDNs was frequently employed as a bioimaging vehicle is that its rigid skeleton renders a fixed distance between fluorescent labels. It can prevent attached probes from self-quenching, and control their fluorescence recovery. In some cases, aptamer strand acts as not only a molecular recognition probe but also an activatable initiator. An electrochemical TDNs without additional reagent operated as follows:505 Once the target was captured by strand a and bounded with it, strand b on another vertex could pull them down through hybridization; the consequential collision of the terminal probe with electrode surface was induced, and eventually caused electron transfer. This linkage mechanism can be achieved in living animals, and thus expected to be applied in point-of care testing.
In practice, bioimaging-related agents (e.g., 99mTc) are all candidates that could be integrated with TDNs. Indeed, multiple probes can be simultaneously functionalized for a more sensitive imaging nanodevice. Jiang et al.506 incorporated FA, radioisotope and ordinary label on TDNs with the linkage of multiple arms. A multimodality probe that can be observable under both near-infrared (NIR) and single-photon emission computed tomography (SPECT) was achieved.
Beyond that, metallics and fluorophores were also commonly used in bioimaging and biosensing. A novel DNA nanoprobe containing AuNP, three Cyanine-5 (Cy5) and custom telomerase strand primer (TSP) was integrated for telomerase monitoring. Its fluorescence was quenched by fluorescence resonance energy transfer (FRET)-induced AuNP at an original state, which was regarded as “off”. After catching the signal of telomerase, hairpin structural TSP linked with AuNP would unfold, leading to the fluorescence recovery of Cy5 (“on” state). The sensitive detection of telomerase in cancer cells was a referable forebode in early diagnosis.507
In clinical, analysis of lymphonodi is essential in estimating oncology progress. Sentinel lymph nodes, the primary ones which malignant cells invade, become the emphasis of auxiliary examinations. However, case investigation reveals that patients with aggressive tumors like melanoma are susceptible to metastases after sentinel lymph nodes biopsy. Alternative noninvasive means are necessary. Dye-labeled TDNs with high in vivo stability, long circulation and great visualization was prepared, not only for lymph node imaging but also for tissues evaluation.434
Cell surface engineering
The cell surface membrane is the primary mediator for interactions of the cell-cell and the cell-microenvironment.508 Recently, the DNA nanomaterials were also investigated to modify the cell surface membrane for different aims, such as targeting, drug delivery, and enhanced interactions of cell-cell and cell-microenvironment.509 Gartner et al.510 have demonstrated that hybridization of complementary DNA sequences enabled the assembly of multicellular structures with defined cell-cell contacts. Akbari et al.511 also reported a DNA nanoplatform that could act as a membrane-bound breadboard (MBB) with nano-scale. This work developed a foundation to apply DNA nanomaterials as membrane engineering technologies, which could mimic and direct complex biomedical processes on the cell surface of different cells in tissues. Some DNA nanomaterials would be investigated for the cell surface engineering with some targeted aims, such as drug delivery and gene transduction. Furthermore, some DNA nanomaterials could combine with the proteins of cell membrane, and then could be internalizated by cells.Some studies have been deomenstrated that some DNA nanomaterials could transfer the cell membrane into the biomimic materials, which could act some functions in the palsma of the living cells.
In the future studies, the membrane-bound DNA origami structures could enable programed assembly within the membrane to mimic functional biomolecular complexes, such as an immune synapse, especially given the wide use of DNA origami to template or organize proteins.512
Summary and perspective
In recent decades, DNA-based nanostructures have become one of the most attractive focuses because of their structural diversity and scientific values. As a basic component of living entities, DNA is endowed with excellent biocompatibility. Its 3D double-helix structure and small molecular weight confer variability and nanosized magnitude. As mentioned above, DNA nanoparticles can be assembled into arbitrary shapes including 2D triangle, rectangle, 3D polyhedron, liquid nanohydrogel, smarter nanocontainer and biomimetic constructions. More fancy structures might be available in the future.
More importantly, DNA nanoparticles have showed influences on cell biological activities, especially on cell proliferation. Accordingly, their roles in tissue regeneration were broadly investigated in tissues including bone, cartilage, blood vessel, nerve, skeletal and cardiac muscles, skin and corneal systems. They also attract tremendous attentions when they serve as nanocarriers. Compared with traditional nanoparticles, they are more variform for the following reasons: (1) flexible joints because every strand of DNA-based nanostructures can be concatenated or linked with an extended arm; (2) abundant binding sites, as the assembly of DNA frameworks provides a hollow internal space for drug molecules; (3) negative charge, so that positive-charged substances can be integrated with them under electrostatic attraction. In this context, studies started applying them as nanoscale vehicles to transport various drugs into cellular focal sites for a desired efficacy. Therapeutical moieties with distinctive natures were chosen to incorporate with DNA nanoparticles. Aptamers (AS1411, MUC1, HApt, etc.) and antineoplastics (DOX, PTX, 5-FU, etc.) modified nanostructures were the most attractive. Their capabilities of antitumor effect, MDR reversion and photodynamic promotion were discovered. Then, the notion of multi-modality nanosystems were put forward to enhance their targeting performance. In addition, through the combination with immune agents (e.g., CpG) and fluorescent labels (e.g., FAM), DNA nanoparticles were also of great significance in immunostimulating, biosensing and bioimaging. More advanced nanotools with logic gates or structural switches were developed for further accurate administration.
Although the biological applications of various DNA nanostructures were comprehensive and extensive, there remain some limitations to overcome, such as, the interaction mechanisms between DNA nanomaterials and cells, the short blood circulation time, and poor aggregation in target tissues. The cellular internalization processes of various DNA nanomaterials are the prerequisites for biological applications. For example, the TDNs have been widely investigated in bioimaging, drug delivery, targeted delivery of functional DNA or RNA sequence, and biocatalysis.502,513–515 Unlike ssDNA with poor cellular uptake properties, previous studies reported that TDNs could be internalized by cells without the assistance of transfection agents.516–518 In the recent study, researcher combined drug affinity responsive target stability (DARTS) with liquid chromatography/tandem mass spectrometry (LC-MS/MS) techniques to explore the endocytosis process of TDNs. The study reported that the endocytosis of TDNs were related to caveolin-1 (CAV1) and micropinocytosis-related protein sorting nexin5 (SNX5). However, the cellular internalization processes of other DNA nanomaterials were not clear still now, which need more explorations.
For the nature of DNA nanomaterials, different DNA nanomaterials encounter the challenges of poor blood circulation time and poor aggregation in target tissues. Some previous studies have demonstrated that the DNA nanomaterials (TDNs, HApt-TDNs, and DNA aptamer, etc) could circulate in body no >1 h, which were quickly eliminated by different cells and organs (such as macrophages, liver, and kidney) before DNA nanomaterials arriving at target tissues.519 Therefore, some other bionic materials were investigated to prolong the blood circulation time, such as liposomes and cell membranes.520 Some previous studies have reported various DNA-coated liposomes for drug delivery.520–522 In a recent report, Han et al.526 found that tannic acid (TA) could mediate the co-assembly of branched-DNA/RNA and A549 lung cancer cell membrane-camouflaged to form nanocomplex (nanocomplex@A549m), which could strengthen the affinity between TA and nucleic acids/membrane proteins. The objectives of nanocomplex@A549m with the application of A549 lung cancer cell membrane-camouflaged were to prolong the blood circulation and reduce macrophage clearance. Moreover, the nanocomplex@A549m could effectively bind to target cells. From this study, we think that the cell membrane-camouflaged method (stem cells, cancer cells, blood cells, and exosomes, etc) might inspire other researchers to develop more DNA-based nanocomplex@cell-membrance materials.520,523 In further studies, DNA nanomaterials might be combine with other materials for more applications, such as hydrogel,524 nanozyme,525 and bimimic cell membrane nanomaterials.526
In conclusion, due to their structural flexibility, multiform customized DNA-based nanostructures were created. Some innovative creations discussed in this review provided valuable directions for the development of DNA-based nanostructures in biomedicine. Regarding the biological performance, acquired nanostructures were explored in tissue engineering and regeneration. As a high-efficient drug delivery system, they could be used as assembly templates to generate agent-modified nanodevices that execute rationally designed behaviors, including tumor therapy, MDR reversion, immunotherapy and biosensing. While studies and analyses were mostly conducted in vitro and the biological effects in different aspects are encouraging, in vivo studies are critical and needed to confirm the observed effects, assess the side effects and make the comprehensive evaluation before achieving the ultimate clinical application of DNA-based nanostructures.
Acknowledgements
This work was supported in part by National Key R&D Program of China (Grant Nos. 2019YFA0110600) and National Natural Science Foundation of China (Grant Nos. 81970916 and 81671031).
Author contributions
All authors designed and supervised the review. W.M. and Y.Z. wrote the manuscript. Y.Z., C.M., and X.X. provided the document retrieval. Y.L. provided administrative and technical support. All authors reviewed and provided feedback on the review.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Wenjuan Ma, Yuxi Zhan
References
- 1.Dong Y, et al. DNA functional materials assembled from branched DNA: design, synthesis, and applications. Chem. Rev. 2020;120:9420–9481. doi: 10.1021/acs.chemrev.0c00294. [DOI] [PubMed] [Google Scholar]
- 2.Broker TR, Lehman IR. Branched DNA molecules: intermediates in T4 recombination. J. Mol. Biol. 1971;60:131–149. doi: 10.1016/0022-2836(71)90453-0. [DOI] [PubMed] [Google Scholar]
- 3.Seeman NC. DNA nanotechnology: from the pub to information-based chemistry. Methods Mol. Biol. 2018;1811:1–9. doi: 10.1007/978-1-4939-8582-1_1. [DOI] [PubMed] [Google Scholar]
- 4.Seeman NC. DNA nanotechnology: novel DNA constructions. Annu. Rev. Biophys. Biomol. Struct. 1998;27:225–248. doi: 10.1146/annurev.biophys.27.1.225. [DOI] [PubMed] [Google Scholar]
- 5.Seeman NC, Sleiman HF. DNA nanotechnology. Nat. Rev. Mater. 2018;3:17068. doi: 10.1038/natrevmats.2017.68. [DOI] [Google Scholar]
- 6.Seeman NC. Nucleic-acid junctions and lattices. J. Theor. Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 7.Seeman NC. DNA engineering and its application to nanotechnology. Trends Biotechnol. 1999;17:437–443. doi: 10.1016/S0167-7799(99)01360-8. [DOI] [PubMed] [Google Scholar]
- 8.Seeman NC, et al. Two dimensions and two States in DNA nanotechnology. J. Biomol. Struct. Dyn. 2000;17:253–262. doi: 10.1080/07391102.2000.10506629. [DOI] [PubMed] [Google Scholar]
- 9.Seeman NC. Key experimental approaches in DNA nanotechnology. Curr. Protoc. Nucleic Acid Chem. 2002;Chapter 12:11. doi: 10.1002/0471142700.nc1201s09. [DOI] [PubMed] [Google Scholar]
- 10.Fan S, et al. Create nanoscale patterns with DNA origami. Small. 2019;15:e1805554. doi: 10.1002/smll.201805554. [DOI] [PubMed] [Google Scholar]
- 11.Jones MR, Seeman NC, Mirkin CA. Nanomaterials. programmable materials and the nature of the DNA bond. Science. 2015;347:1260901. doi: 10.1126/science.1260901. [DOI] [PubMed] [Google Scholar]
- 12.Hu Y, Niemeyer CM. From DNA nanotechnology to material systems engineering. Adv. Mater. 2019;31:e1806294. doi: 10.1002/adma.201806294. [DOI] [PubMed] [Google Scholar]
- 13.Chao J, et al. Solving mazes with single-molecule DNA navigators. Nat. Mater. 2019;18:273–279. doi: 10.1038/s41563-018-0205-3. [DOI] [PubMed] [Google Scholar]
- 14.Harroun SG, et al. Programmable DNA switches and their applications. Nanoscale. 2018;10:4607–4641. doi: 10.1039/C7NR07348H. [DOI] [PubMed] [Google Scholar]
- 15.Hu Y, Cecconello A, Idili A, Ricci F, Willner I. Triplex DNA nanostructures: from basic properties to applications. Angew. Chem. Int. Ed. Engl. 2017;56:15210–15233. doi: 10.1002/anie.201701868. [DOI] [PubMed] [Google Scholar]
- 16.Seeman NC. DNA in a material world. Nature. 2003;421:427–431. doi: 10.1038/nature01406. [DOI] [PubMed] [Google Scholar]
- 17.Jia S, et al. Programming DNA origami patterning with non-canonical DNA-based metallization reactions. Nat. Commun. 2019;10:5597. doi: 10.1038/s41467-019-13507-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gopinath A, Miyazono E, Faraon A, Rothemund PW. Engineering and mapping nanocavity emission via precision placement of DNA origami. Nature. 2016;535:401–405. doi: 10.1038/nature18287. [DOI] [PubMed] [Google Scholar]
- 19.Wing R, et al. Crystal structure analysis of a complete turn of B-DNA. Nature. 1980;287:755–758. doi: 10.1038/287755a0. [DOI] [PubMed] [Google Scholar]
- 20.Franklin RE, Gosling RG. Molecular structure of nucleic acids. Molecular configuration in sodium thymonucleate. 1953. Ann. N. Y. Acad. Sci. 1995;758:16–17. [PubMed] [Google Scholar]
- 21.Franklin RE, Gosling RG. Molecular configuration in sodium thymonucleate. Nature. 1953;171:740–741. doi: 10.1038/171740a0. [DOI] [PubMed] [Google Scholar]
- 22.Wilkins MH, Stokes AR, Wilson HR. Molecular structure of nucleic acids. Molecular structure of deoxypentose nucleic acids. 1953. Ann. N. Y. Acad. Sci. 1995;758:13–16. doi: 10.1111/j.1749-6632.1995.tb24824.x. [DOI] [PubMed] [Google Scholar]
- 23.Wilkins MH, Stokes AR, Wilson HR. Molecular structure of deoxypentose nucleic acids. Nature. 1953;171:738–740. doi: 10.1038/171738a0. [DOI] [PubMed] [Google Scholar]
- 24.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 25.Mergny JL, Sen D. DNA quadruple helices in nanotechnology. Chem. Rev. 2019;119:6290–6325. doi: 10.1021/acs.chemrev.8b00629. [DOI] [PubMed] [Google Scholar]
- 26.Leslie AG, Arnott S, Chandrasekaran R, Ratliff RL. Polymorphism of DNA double helices. J. Mol. Biol. 1980;143:49–72. doi: 10.1016/0022-2836(80)90124-2. [DOI] [PubMed] [Google Scholar]
- 27.Basu HS, Feuerstein BG, Zarling DA, Shafer RH, Marton LJ. Recognition of Z-RNA and Z-DNA determinants by polyamines in solution: experimental and theoretical studies. J. Biomol. Struct. Dyn. 1988;6:299–309. doi: 10.1080/07391102.1988.10507714. [DOI] [PubMed] [Google Scholar]
- 28.Irobalieva RN, et al. Structural diversity of supercoiled DNA. Nat. Commun. 2015;6:8440. doi: 10.1038/ncomms9440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang F, Nangreave J, Liu Y, Yan H. Structural DNA nanotechnology: state of the art and future perspective. J. Am. Chem. Soc. 2014;136:11198–11211. doi: 10.1021/ja505101a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu J, Liu M, Liu Y, Yan H. Spatially-interactive biomolecular networks organized by nucleic acid nanostructures. Acc. Chem. Res. 2012;45:1215–1226. doi: 10.1021/ar200295q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alivisatos AP, et al. Organization of ‘nanocrystal molecules’ using DNA. Nature. 1996;382:609–611. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]
- 32.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 33.Julin S, Nummelin S, Kostiainen MA, Linko V. DNA nanostructure-directed assembly of metal nanoparticle superlattices. J. Nanopart. Res.: Interdiscip. Forum Nanoscale Sci. Technol. 2018;20:119. doi: 10.1007/s11051-018-4225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aldaye FA, Palmer AL, Sleiman HF. Assembling materials with DNA as the guide. Science. 2008;321:1795–1799. doi: 10.1126/science.1154533. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, et al. Paranemic crossover DNA: there and back again. Chem. Rev. 2019;119:6273–6289. doi: 10.1021/acs.chemrev.8b00207. [DOI] [PubMed] [Google Scholar]
- 36.Loescher S, Groeer S, Walther A. 3D DNA origami nanoparticles: from basic design principles to emerging applications in soft matter and (bio-)nanosciences. Angew. Chem. Int. Ed. Engl. 2018;57:10436–10448. doi: 10.1002/anie.201801700. [DOI] [PubMed] [Google Scholar]
- 37.Hong F, Zhang F, Liu Y, Yan H. DNA origami: scaffolds for creating higher order structures. Chem. Rev. 2017;117:12584–12640. doi: 10.1021/acs.chemrev.6b00825. [DOI] [PubMed] [Google Scholar]
- 38.Rothemund PW, Folding DNA. to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 39.Torring T, Voigt NV, Nangreave J, Yan H, Gothelf KV. DNA origami: a quantum leap for self-assembly of complex structures. Chem. Soc. Rev. 2011;40:5636–5646. doi: 10.1039/c1cs15057j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petersen P, Tikhomirov G, Qian L. Information-based autonomous reconfiguration in systems of interacting DNA nanostructures. Nat. Commun. 2018;9:5362. doi: 10.1038/s41467-018-07805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang SS, Ellington AD. Pattern generation with nucleic acid chemical reaction networks. Chem. Rev. 2019;119:6370–6383. doi: 10.1021/acs.chemrev.8b00625. [DOI] [PubMed] [Google Scholar]
- 42.Simmel FC, Yurke B, Singh HR. Principles and applications of nucleic acid strand displacement reactions. Chem. Rev. 2019;119:6326–6369. doi: 10.1021/acs.chemrev.8b00580. [DOI] [PubMed] [Google Scholar]
- 43.Thubagere AJ, et al. A cargo-sorting DNA robot. Science. 2017;357:eaan6558. doi: 10.1126/science.aan6558. [DOI] [PubMed] [Google Scholar]
- 44.Zhang DY, Seelig G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 2011;3:103–113. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- 45.Kopperger E, et al. A self-assembled nanoscale robotic arm controlled by electric fields. Science. 2018;359:296–301. doi: 10.1126/science.aao4284. [DOI] [PubMed] [Google Scholar]
- 46.Hu Q, Li H, Wang L, Gu H, Fan C. DNA nanotechnology-enabled drug delivery systems. Chem. Rev. 2019;119:6459–6506. doi: 10.1021/acs.chemrev.7b00663. [DOI] [PubMed] [Google Scholar]
- 47.Kohman RE, Kunjapur AM, Hysolli E, Wang Y, Church GM. From designing the molecules of life to designing life: future applications derived from advances in DNA technologies. Angew. Chem. Int. Ed. Engl. 2018;57:4313–4328. doi: 10.1002/anie.201707976. [DOI] [PubMed] [Google Scholar]
- 48.Li J, Green AA, Yan H, Fan C. Engineering nucleic acid structures for programmable molecular circuitry and intracellular biocomputation. Nat. Chem. 2017;9:1056–1067. doi: 10.1038/nchem.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang D, et al. DNA materials: bridging nanotechnology and biotechnology. Acc. Chem. Res. 2014;47:1902–1911. doi: 10.1021/ar5001082. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y, et al. Programmable and multifunctional DNA-based materials for biomedical applications. Adv. Mater. 2018;30:e1703658. doi: 10.1002/adma.201703658. [DOI] [PubMed] [Google Scholar]
- 51.Zhou L, et al. Functional DNA-based hydrogel intelligent materials for biomedical applications. J. Mater. Chem. B. 2020;8:1991–2009. doi: 10.1039/C9TB02716E. [DOI] [PubMed] [Google Scholar]
- 52.Jiang Q, et al. Rationally designed DNA-based nanocarriers. Adv. Drug Deliv. Rev. 2019;147:2–21. doi: 10.1016/j.addr.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 53.Elfwing A, et al. DNA based hybrid material for interface engineering in polymer solar cells. ACS Appl. Mater. Interfaces. 2018;10:9579–9586. doi: 10.1021/acsami.7b17807. [DOI] [PubMed] [Google Scholar]
- 54.Li J, et al. Functional nucleic acid-based hydrogels for bioanalytical and biomedical applications. Chem. Soc. Rev. 2016;45:1410–1431. doi: 10.1039/C5CS00586H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maeda M, Kojima T, Song Y, Takayama S. DNA-based biomaterials for immunoengineering. Adv. Healthc. Mater. 2019;8:e1801243. doi: 10.1002/adhm.201801243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chandrasekaran AR, et al. DNA nanotechnology approaches for microRNA detection and diagnosis. Nucleic Acids Res. 2019;47:10489–10505. doi: 10.1093/nar/gkz580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Roh YH, Ruiz RC, Peng S, Lee JB, Luo D. Engineering DNA-based functional materials. Chem. Soc. Rev. 2011;40:5730–5744. doi: 10.1039/c1cs15162b. [DOI] [PubMed] [Google Scholar]
- 58.Khajouei S, Ravan H, Ebrahimi A. DNA hydrogel-empowered biosensing. Adv. Colloid Interface Sci. 2020;275:102060. doi: 10.1016/j.cis.2019.102060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li M, et al. Engineering multifunctional DNA hybrid nanospheres through coordination-driven self-assembly. Angew. Chem. Int. Ed. Engl. 2019;58:1350–1354. doi: 10.1002/anie.201810735. [DOI] [PubMed] [Google Scholar]
- 60.Liu S, Jiang Q, Wang Y, Ding B. Biomedical applications of DNA-based molecular devices. Adv. Healthc. Mater. 2019;8:e1801658. doi: 10.1002/adhm.201801658. [DOI] [PubMed] [Google Scholar]
- 61.Samanta A, Medintz IL. Nanoparticles and DNA-a powerful and growing functional combination in bionanotechnology. Nanoscale. 2016;8:9037–9095. doi: 10.1039/C5NR08465B. [DOI] [PubMed] [Google Scholar]
- 62.Liu N, et al. Tetrahedral framework nucleic acids promote corneal epithelial wound healing in vitro and in vivo. Small. 2019;15:e1901907. doi: 10.1002/smll.201901907. [DOI] [PubMed] [Google Scholar]
- 63.Ma W, et al. Self-assembled tetrahedral DNA nanostructures promote neural stem cell proliferation and neuronal differentiation. ACS Appl Mater. Interfaces. 2018;10:7892–7900. doi: 10.1021/acsami.8b00833. [DOI] [PubMed] [Google Scholar]
- 64.Ma WJ, et al. Tetrahedral DNA nanostructures facilitate neural stem cell migration via activating RHOA/ROCK2 signalling pathway. Cell Prolif. 2018;51:e12503. doi: 10.1111/cpr.12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shao X, et al. Tetrahedral DNA nanostructure: a potential promoter for cartilage tissue regeneration via regulating chondrocyte phenotype and proliferation. Small. 2017;13:1602770. doi: 10.1002/smll.201602770. [DOI] [PubMed] [Google Scholar]
- 66.Shi S, et al. Self-assembled tetrahedral DNA nanostructures promote adipose-derived stem cell migration via lncRNA XLOC 010623 and RHOA/ROCK2 signal pathway. ACS Appl. Mater. Interfaces. 2016;8:19353–19363. doi: 10.1021/acsami.6b06528. [DOI] [PubMed] [Google Scholar]
- 67.Shi SR, et al. Effects of tetrahedral DNA nanostructures on autophagy in chondrocytes. Chem. Commun. 2018;54:1327–1330. doi: 10.1039/C7CC09397G. [DOI] [PubMed] [Google Scholar]
- 68.Ma W, et al. An intelligent DNA nanorobot with in vitro enhanced protein lysosomal degradation of HER2. Nano Lett. 2019;19:4505–4517. doi: 10.1021/acs.nanolett.9b01320. [DOI] [PubMed] [Google Scholar]
- 69.Lee H, Dam DH, Ha JW, Yue J, Odom TW. Enhanced human epidermal growth factor receptor 2 degradation in breast cancer cells by lysosome-targeting gold nanoconstructs. ACS Nano. 2015;9:9859–9867. doi: 10.1021/acsnano.5b05138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Petrizzo A, et al. Functional characterization of biodegradable nanoparticles as antigen delivery system. J. Exp. Clin. Cancer Res. 2015;34:114. doi: 10.1186/s13046-015-0231-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu B, et al. DNA-origami-based assembly of anisotropic plasmonic gold nanostructures. Small. 2017;13:1603991. doi: 10.1002/smll.201603991. [DOI] [PubMed] [Google Scholar]
- 72.Edwardson TG, Lau KL, Bousmail D, Serpell CJ, Sleiman HF. Transfer of molecular recognition information from DNA nanostructures to gold nanoparticles. Nat. Chem. 2016;8:162–170. doi: 10.1038/nchem.2420. [DOI] [PubMed] [Google Scholar]
- 73.Utsuno K, Uludag H. Thermodynamics of polyethylenimine-DNA binding and DNA condensation. Biophys. J. 2010;99:201–207. doi: 10.1016/j.bpj.2010.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang Y, et al. Ionic-crosslinked polysaccharide/PEI/DNA nanoparticles for stabilized gene delivery. Carbohydr. Polym. 2018;201:246–256. doi: 10.1016/j.carbpol.2018.08.063. [DOI] [PubMed] [Google Scholar]
- 75.Xie L, Ding X, Budry R, Mao G. Layer-by-layer DNA films incorporating highly transfecting bioreducible poly(amido amine) and polyethylenimine for sequential gene delivery. Int. J. Nanomed. 2018;13:4943–4960. doi: 10.2147/IJN.S162353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ewe A, et al. Storage stability of optimal liposome-polyethylenimine complexes (lipopolyplexes) for DNA or siRNA delivery. Acta Biomaterialia. 2014;10:2663–2673. doi: 10.1016/j.actbio.2014.02.037. [DOI] [PubMed] [Google Scholar]
- 77.Tian TR, et al. Synthesis of an ethyleneimine/tetrahedral DNA nanostructure complex and its potential application as a multi-functional delivery vehicle. Nanoscale. 2017;9:18402–18412. doi: 10.1039/C7NR07130B. [DOI] [PubMed] [Google Scholar]
- 78.Liu W, et al. An investigation on the physicochemical properties of chitosan/DNA polyelectrolyte complexes. Biomaterials. 2005;26:2705–2711. doi: 10.1016/j.biomaterials.2004.07.038. [DOI] [PubMed] [Google Scholar]
- 79.Gu T, Wang J, Xia H, Wang S, Yu X. Direct electrochemistry and electrocatalysis of horseradish peroxidase immobilized in a DNA/chitosan-Fe(3)O(4) magnetic nanoparticle bio-complex film. Materials. 2014;7:1069–1083. doi: 10.3390/ma7021069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kumar S, et al. Enhanced chitosan-DNA interaction by 2-acrylamido-2-methylpropane coupling for an efficient transfection in cancer cells. J. Mater. Chem. B. 2015;3:3465–3475. doi: 10.1039/C4TB02070G. [DOI] [PubMed] [Google Scholar]
- 81.Walsh DP, et al. Bioinspired star-shaped poly(l-lysine) polypeptides: efficient polymeric nanocarriers for the delivery of DNA to mesenchymal stem cells. Mol. Pharm. 2018;15:1878–1891. doi: 10.1021/acs.molpharmaceut.8b00044. [DOI] [PubMed] [Google Scholar]
- 82.Boylan NJ, et al. Enhancement of airway gene transfer by DNA nanoparticles using a pH-responsive block copolymer of polyethylene glycol and poly-L-lysine. Biomaterials. 2012;33:2361–2371. doi: 10.1016/j.biomaterials.2011.11.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.von Erlach T, et al. Formation and characterization of DNA-polymer-condensates based on poly(2-methyl-2-oxazoline) grafted poly(L-lysine) for non-viral delivery of therapeutic DNA. Biomaterials. 2011;32:5291–5303. doi: 10.1016/j.biomaterials.2011.03.080. [DOI] [PubMed] [Google Scholar]
- 84.Chen T, Romesberg FE. Enzymatic synthesis, amplification, and application of dna with a functionalized backbone. Angew. Chem. Int. Ed. Engl. 2017;56:14046–14051. doi: 10.1002/anie.201707367. [DOI] [PubMed] [Google Scholar]
- 85.Zhan Y, et al. Diversity of DNA nanostructures and applications in oncotherapy. Biotechnol. J. 2020;15:e1900094. doi: 10.1002/biot.201900094. [DOI] [PubMed] [Google Scholar]
- 86.Schnitzbauer J, Strauss MT, Schlichthaerle T, Schueder F, Jungmann R. Super-resolution microscopy with DNA-PAINT. Nat. Protoc. 2017;12:1198–1228. doi: 10.1038/nprot.2017.024. [DOI] [PubMed] [Google Scholar]
- 87.Ngo TA, et al. Protein adaptors assemble functional proteins on DNA scaffolds. Chem. Commun. (Camb.) 2019;55:12428–12446. doi: 10.1039/C9CC04661E. [DOI] [PubMed] [Google Scholar]
- 88.Zhan Y, et al. DNA-Based nanomedicine with targeting and enhancement of therapeutic efficacy of breast cancer cells. ACS Appl. Mater. Interfaces. 2019;11:15354–15365. doi: 10.1021/acsami.9b03449. [DOI] [PubMed] [Google Scholar]
- 89.Wang M, et al. Near-infrared light-activated DNA-agonist nanodevice for nongenetically and remotely controlled cellular signaling and behaviors in live animals. Nano Lett. 2019;19:2603–2613. doi: 10.1021/acs.nanolett.9b00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Taniguchi J, et al. A synthetic DNA-binding inhibitor of SOX2 guides human induced pluripotent stem cells to differentiate into mesoderm. Nucleic Acids Res. 2017;45:9219–9228. doi: 10.1093/nar/gkx693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hendrikson WJ, et al. Biological and tribological assessment of poly(ethylene oxide terephthalate)/poly(butylene terephthalate), polycaprolactone, and poly (L\DL) lactic acid plotted scaffolds for skeletal tissue regeneration. Adv. Healthc. Mater. 2016;5:232–243. doi: 10.1002/adhm.201500067. [DOI] [PubMed] [Google Scholar]
- 92.Jayme CC, et al. DNA polymeric films as a support for cell growth as a new material for regenerative medicine: compatibility and applicability. Exp. Cell Res. 2017;360:404–412. doi: 10.1016/j.yexcr.2017.09.033. [DOI] [PubMed] [Google Scholar]
- 93.Basu S, et al. Harnessing the noncovalent interactions of DNA backbone with 2D silicate nanodisks to fabricate injectable therapeutic hydrogels. ACS Nano. 2018;12:9866–9880. doi: 10.1021/acsnano.8b02434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Feng G, et al. Injectable nanofibrous spongy microspheres for NR4A1 plasmid DNA transfection to reverse fibrotic degeneration and support disc regeneration. Biomaterials. 2017;131:86–97. doi: 10.1016/j.biomaterials.2017.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang Y, et al. Nucleic acids and analogs for bone regeneration. Bone Res. 2018;6:37. doi: 10.1038/s41413-018-0042-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Mahlknecht G, Sela M, Yarden Y. Aptamer targeting the ERBB2 receptor tyrosine kinase for applications in tumor therapy. Methods Mol. Biol. 2015;1317:3–15. doi: 10.1007/978-1-4939-2727-2_1. [DOI] [PubMed] [Google Scholar]
- 97.Yu X, et al. Targeting EGFR/HER2/HER3 with a three-in-one aptamer-siRNA chimera confers superior activity against HER2( +) breast cancer. Mol. Ther. Nucleic Acids. 2018;10:317–330. doi: 10.1016/j.omtn.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bai S, et al. High-discrimination factor nanosensor based on tetrahedral dna nanostructures and gold nanoparticles for detection of MiRNA-21 in live cells. Theranostics. 2018;8:2424–2434. doi: 10.7150/thno.23852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zeng D, et al. DNA tetrahedral nanostructure-based electrochemical miRNA biosensor for simultaneous detection of multiple miRNAs in pancreatic carcinoma. ACS Appl. Mater. Interfaces. 2017;9:24118–24125. doi: 10.1021/acsami.7b05981. [DOI] [PubMed] [Google Scholar]
- 100.Li Q, et al. Aptamer-modified tetrahedral DNA nanostructure for tumor-targeted drug delivery. ACS Appl. Mater. Interfaces. 2017;9:36695–36701. doi: 10.1021/acsami.7b13328. [DOI] [PubMed] [Google Scholar]
- 101.Wang L, Arrabito G. Hybrid, multiplexed, functional DNA nanotechnology for bioanalysis. Analyst. 2015;140:5821–5848. doi: 10.1039/C5AN00861A. [DOI] [PubMed] [Google Scholar]
- 102.Li C, et al. A writable polypeptide-DNA hydrogel with rationally designed multi-modification sites. Small. 2015;11:1138–1143. doi: 10.1002/smll.201401906. [DOI] [PubMed] [Google Scholar]
- 103.Estrich NA, Hernandez-Garcia A, de Vries R, LaBean TH. Engineered diblock polypeptides improve DNA and gold solubility during molecular assembly. ACS Nano. 2017;11:831–842. doi: 10.1021/acsnano.6b07291. [DOI] [PubMed] [Google Scholar]
- 104.Nakata E, Dinh H, Nguyen TM, Morii T. DNA binding adaptors to assemble proteins of interest on DNA scaffold. Methods Enzymol. 2019;617:287–322. doi: 10.1016/bs.mie.2018.12.014. [DOI] [PubMed] [Google Scholar]
- 105.Chandrasekaran AR. Programmable DNA scaffolds for spatially-ordered protein assembly. Nanoscale. 2016;8:4436–4446. doi: 10.1039/C5NR08685J. [DOI] [PubMed] [Google Scholar]
- 106.Chandler M, et al. Breaking and joining single-stranded DNA: the HUH endonuclease superfamily. Nat. Rev. Microbiol. 2013;11:525–538. doi: 10.1038/nrmicro3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xie X, et al. Overcoming drug-resistant lung cancer by paclitaxel loaded tetrahedral DNA nanostructures. Nanoscale. 2018;10:5457–5465. doi: 10.1039/C7NR09692E. [DOI] [PubMed] [Google Scholar]
- 108.Halley PD, et al. Daunorubicin-loaded DNA origami nanostructures circumvent drug-resistance mechanisms in a leukemia model. Small. 2016;12:308–320. doi: 10.1002/smll.201502118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim KR, et al. Drug delivery by a self-assembled DNA tetrahedron for overcoming drug resistance in breast cancer cells. Chem. Commun. (Camb.) 2013;49:2010–2012. doi: 10.1039/c3cc38693g. [DOI] [PubMed] [Google Scholar]
- 110.Liu M, et al. Aptamer-targeted DNA nanostructures with doxorubicin to treat protein tyrosine kinase 7-positive tumours. Cell Prolif. 2018;52:e12511. doi: 10.1111/cpr.12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Meng L, et al. Tetrahedral DNA nanostructure-delivered DNAzyme for gene silencing to suppress cell growth. ACS Appl. Mater. Interfaces. 2019;11:6850–6857. doi: 10.1021/acsami.8b22444. [DOI] [PubMed] [Google Scholar]
- 112.Samanta A, Banerjee S, Liu Y. DNA nanotechnology for nanophotonic applications. Nanoscale. 2015;7:2210–2220. doi: 10.1039/C4NR06283C. [DOI] [PubMed] [Google Scholar]
- 113.Wang ZG, Song C, Ding B. Functional DNA nanostructures for photonic and biomedical applications. Small. 2013;9:2210–2222. doi: 10.1002/smll.201300141. [DOI] [PubMed] [Google Scholar]
- 114.Shen B, Kostiainen MA, Linko V. DNA origami nanophotonics and plasmonics at interfaces. Langmuir. 2018;34:14911–14920. doi: 10.1021/acs.langmuir.8b01843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Maune HT, et al. Self-assembly of carbon nanotubes into two-dimensional geometries using DNA origami templates. Nat. Nanotechnol. 2010;5:61–66. doi: 10.1038/nnano.2009.311. [DOI] [PubMed] [Google Scholar]
- 116.Amodio A, Del Grosso E, Troina A, Placidi E, Ricci F. Remote electronic control of DNA-based reactions and nanostructure assembly. Nano Lett. 2018;18:2918–2923. doi: 10.1021/acs.nanolett.8b00179. [DOI] [PubMed] [Google Scholar]
- 117.Gates EP, Dearden AM, Woolley AT. DNA-templated lithography and nanofabrication for the fabrication of nanoscale electronic circuitry. Crit. Rev. Anal. Chem. 2014;44:354–370. doi: 10.1080/10408347.2014.910636. [DOI] [PubMed] [Google Scholar]
- 118.Yang YR, Liu Y, Yan H. DNA nanostructures as programmable biomolecular scaffolds. Bioconjug. Chem. 2015;26:1381–1395. doi: 10.1021/acs.bioconjchem.5b00194. [DOI] [PubMed] [Google Scholar]
- 119.Jaekel A, Stegemann P, Sacca B. Manipulating enzymes properties with DNA nanostructures. Molecules. 2019;24:3694. doi: 10.3390/molecules24203694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Winfree E, Liu FR, Wenzler LA, Seeman NC. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 121.Yan H, Zhang XP, Shen ZY, Seeman NC. A robust DNA mechanical device controlled by hybridization topology. Nature. 2002;415:62–65. doi: 10.1038/415062a. [DOI] [PubMed] [Google Scholar]
- 122.Chen JH, Seeman NC. Synthesis from DNA of a molecule with the connectivity of a cube. Nature. 1991;350:631–633. doi: 10.1038/350631a0. [DOI] [PubMed] [Google Scholar]
- 123.Liu WY, Zhong H, Wang RS, Seeman NC. Crystalline two-dimensional DNA-origami arrays. Angew. Chem. Int Ed. 2011;50:264–267. doi: 10.1002/anie.201005911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Le JD, et al. DNA-templated self-assembly of metallic nanocomponent arrays on a surface. Nano Lett. 2004;4:2343–2347. doi: 10.1021/nl048635+. [DOI] [Google Scholar]
- 125.Selmi DN, et al. DNA-templated protein arrays for single-molecule imaging. Nano Lett. 2011;11:657–660. doi: 10.1021/nl1037769. [DOI] [PubMed] [Google Scholar]
- 126.Holthausen JT, Wyman C, Kanaar R. Regulation of DNA strand exchange in homologous recombination. DNA Repair. 2010;9:1264–1272. doi: 10.1016/j.dnarep.2010.09.014. [DOI] [PubMed] [Google Scholar]
- 127.Wang X, Seeman NC. Assembly and characterization of 8-arm and 12-arm DNA branched junctions. J. Am. Chem. Soc. 2007;129:8169–8176. doi: 10.1021/ja0693441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang YL, Mueller JE, Kemper B, Seeman NC. Assembly and characterization of five-arm and six-arm DNA branched junctions. Biochemistry. 1991;30:5667–5674. doi: 10.1021/bi00237a005. [DOI] [PubMed] [Google Scholar]
- 129.Kallenbach NR, Ma RI, Seeman NC. An immobile nucleic-acid junction constructed from oligonucleotides. Nature. 1983;305:829–831. doi: 10.1038/305829a0. [DOI] [Google Scholar]
- 130.Sa-Ardyen P, Vologodskii AV, Seeman NC. The flexibility of DNA double crossover molecules. Biophys. J. 2003;84:3829–3837. doi: 10.1016/S0006-3495(03)75110-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fu TJ, Seeman NC. DNA double-crossover molecules. Biochemistry. 1993;32:3211–3220. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 132.Winfree E, Liu F, Wenzler LA, Seeman NC. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 133.Yan H, Park SH, Finkelstein G, Reif JH, LaBean TH. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science. 2003;301:1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- 134.He Y, Chen Y, Liu H, Ribbe AE, Mao C. Self-assembly of hexagonal DNA two-dimensional (2D) arrays. J. Am. Chem. Soc. 2005;127:12202–12203. doi: 10.1021/ja0541938. [DOI] [PubMed] [Google Scholar]
- 135.Mathieu F, et al. Six-helix bundles designed from DNA. Nano Lett. 2005;5:661–665. doi: 10.1021/nl050084f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.He Y, Tian Y, Ribbe AE, Mao C. Highly connected two-dimensional crystals of DNA six-point-stars. J. Am. Chem. Soc. 2006;128:15978–15979. doi: 10.1021/ja0665141. [DOI] [PubMed] [Google Scholar]
- 137.Lin C, Liu Y, Rinker S, Yan H. DNA tile based self-assembly: building complex nanoarchitectures. Chemphyschem. 2006;7:1641–1647. doi: 10.1002/cphc.200600260. [DOI] [PubMed] [Google Scholar]
- 138.Zheng J, et al. From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature. 2009;461:74–77. doi: 10.1038/nature08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yan H, LaBean TH, Feng LP, Reif JH. Directed nucleation assembly of DNA tile complexes for barcode-patterned lattices. Proc. Natl Acad. Sci. USA. 2003;100:8103–8108. doi: 10.1073/pnas.1032954100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shih WM, Quispe JD, Joyce GF. A 1.7-kilobase single-stranded DNA that folds into a nanoscale octahedron. Nature. 2004;427:618–621. doi: 10.1038/nature02307. [DOI] [PubMed] [Google Scholar]
- 141.Qian L, et al. Analogic China map constructed by DNA. Chin. Sci. Bull. 2006;51:2973–2976. doi: 10.1007/s11434-006-2223-9. [DOI] [Google Scholar]
- 142.Andersen ES, et al. DNA origami design of dolphin-shaped structures with flexible tails. ACS Nano. 2008;2:1213–1218. doi: 10.1021/nn800215j. [DOI] [PubMed] [Google Scholar]
- 143.Han DR, et al. DNA origami with complex curvatures in three-dimensional space. Science. 2011;332:342–346. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 144.Yan H, LaBean TH, Feng L, Reif JH. Directed nucleation assembly of DNA tile complexes for barcode-patterned lattices. Proc. Natl Acad. Sci. USA. 2003;100:8103–8108. doi: 10.1073/pnas.1032954100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Andersen ES, et al. Self-assembly of a nanoscale DNA box with a controllable lid. Nature. 2009;459:73–76. doi: 10.1038/nature07971. [DOI] [PubMed] [Google Scholar]
- 146.Zadegan RM, et al. Construction of a 4 zeptoliters switchable 3D DNA box origami. ACS Nano. 2012;6:10050–10053. doi: 10.1021/nn303767b. [DOI] [PubMed] [Google Scholar]
- 147.Zadegan RM, Jepsen MDE, Hildebrandt LL, Birkedal V, Kjems J. Construction of a fuzzy and boolean logic gates based on DNA. Small. 2015;11:1811–1817. doi: 10.1002/smll.201402755. [DOI] [PubMed] [Google Scholar]
- 148.Kuzuya A, Komiyama M. Design and construction of a box-shaped 3D-DNA origami. Chem. Commun. (Camb.) 2009;28:4182–4184. doi: 10.1039/b907800b. [DOI] [PubMed] [Google Scholar]
- 149.Endo M, Hidaka K, Kato T, Namba K, Sugiyama H. DNA prism structures constructed by folding of multiple rectangular arms. J. Am. Chem. Soc. 2009;131:15570–15571. doi: 10.1021/ja904252e. [DOI] [PubMed] [Google Scholar]
- 150.Douglas SM, Chou JJ, Shih WM. DNA-nanotube-induced alignment of membrane proteins for NMR structure determination. Proc. Natl Acad. Sci. USA. 2007;104:6644–6648. doi: 10.1073/pnas.0700930104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Han D, et al. DNA origami with complex curvatures in three-dimensional space. Science. 2011;332:342–346. doi: 10.1126/science.1202998. [DOI] [PubMed] [Google Scholar]
- 152.Pound E, Ashton JR, Becerril HA, Woolley AT. Polymerase chain reaction based scaffold preparation for the production of thin, branched DNA origami nanostructures of arbitrary sizes. Nano Lett. 2009;9:4302–4305. doi: 10.1021/nl902535q. [DOI] [PubMed] [Google Scholar]
- 153.Han D, et al. DNA gridiron nanostructures based on four-arm junctions. Science. 2013;339:1412–1415. doi: 10.1126/science.1232252. [DOI] [PubMed] [Google Scholar]
- 154.Zhang F, et al. Complex wireframe DNA origami nanostructures with multi-arm junction vertices. Nat. Nanotechnol. 2015;10:779-+. doi: 10.1038/nnano.2015.162. [DOI] [PubMed] [Google Scholar]
- 155.Veneziano R, et al. DNA NANOTECHNOLOGY Designer nanoscale DNA assemblies programmed from the top down. Science. 2016;352:1534. doi: 10.1126/science.aaf4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Benson E, et al. DNA rendering of polyhedral meshes at the nanoscale. Nature. 2015;523:441–444. doi: 10.1038/nature14586. [DOI] [PubMed] [Google Scholar]
- 157.Benson E, et al. Computer-aided production of scaffolded DNA nanostructures from flat sheet meshes. Angew. Chem. Int. Ed. Engl. 2016;55:8869–8872. doi: 10.1002/anie.201602446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ellis-Monaghan JA, McDowell A, Moffatt I, Pangborn G. DNA origami and the complexity of Eulerian circuits with turning costs. Nat. Comput. 2015;14:491–503. doi: 10.1007/s11047-014-9457-2. [DOI] [Google Scholar]
- 159.Matthies M, Agarwal NP, Schmidt TL. Design and synthesis of triangulated DNA origami trusses. Nano Lett. 2016;16:2108–2113. doi: 10.1021/acs.nanolett.6b00381. [DOI] [PubMed] [Google Scholar]
- 160.Mallik L, et al. Electron microscopic visualization of protein assemblies on flattened DNA origami. ACS Nano. 2015;9:7133–7141. doi: 10.1021/acsnano.5b01841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wu N, et al. Molecular threading and tunable molecular recognition on DNA origami nanostructures. J. Am. Chem. Soc. 2013;135:12172–12175. doi: 10.1021/ja403863a. [DOI] [PubMed] [Google Scholar]
- 162.Douglas SM, et al. Self-assembly of DNA into nanoscale three-dimensional shapes (vol 459, pg 414, 2009) Nature. 2009;459:1154–1154. doi: 10.1038/nature08165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ke YG, et al. Multilayer DNA origami packed on a square lattice. J. Am. Chem. Soc. 2009;131:15903–15908. doi: 10.1021/ja906381y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ke YG, Voigt NV, Gothelf KV, Shih WM. Multilayer DNA origami packed on hexagonal and hybrid lattices. J. Am. Chem. Soc. 2012;134:1770–1774. doi: 10.1021/ja209719k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Dietz H, Douglas SM, Shih WM. Folding DNA into twisted and curved nanoscale shapes. Science. 2009;325:725–730. doi: 10.1126/science.1174251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liedl T, Hogberg B, Tytell J, Ingber DE, Shih WM. Self-assembly of three-dimensional prestressed tensegrity structures from DNA. Nat. Nanotechnol. 2010;5:520–524. doi: 10.1038/nnano.2010.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Ke Y, Bellot G, Voigt NV, Fradkov E, Shih WM. Two design strategies for enhancement of multilayer-DNA-origami folding: underwinding for specific intercalator rescue and staple-break positioning. Chem. Sci. 2012;3:2587–2597. doi: 10.1039/c2sc20446k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Hong F, Jiang S, Wang T, Liu Y, Yan H. 3D framework DNA origami with layered crossovers. Angew. Chem. Int. Ed. Engl. 2016;55:12832–12835. doi: 10.1002/anie.201607050. [DOI] [PubMed] [Google Scholar]
- 169.Douglas SM, et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 2009;37:5001–5006. doi: 10.1093/nar/gkp436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Catherinot V, Labesse G. ViTO: tool for refinement of protein sequence-structure alignments. Bioinformatics. 2004;20:3694–3696. doi: 10.1093/bioinformatics/bth429. [DOI] [PubMed] [Google Scholar]
- 171.Bai XC, Martin TG, Scheres SH, Dietz H. Cryo-EM structure of a 3D DNA-origami object. Proc. Natl Acad. Sci. USA. 2012;109:20012–20017. doi: 10.1073/pnas.1215713109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Fischer S, et al. Shape and interhelical spacing of dna origami nanostructures studied by small-angle x-ray scattering. Nano Lett. 2016;16:4282–4287. doi: 10.1021/acs.nanolett.6b01335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wagenbauer KF, Wachauf CH, Dietz H. Quantifying quality in DNA self-assembly. Nat. Commun. 2014;5:3691. doi: 10.1038/ncomms4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Brown S, et al. An easy-to-prepare mini-scaffold for DNA origami. Nanoscale. 2015;7:16621–16624. doi: 10.1039/C5NR04921K. [DOI] [PubMed] [Google Scholar]
- 175.Nickels PC, et al. DNA origami structures directly assembled from intact bacteriophages. Small. 2014;10:1765–1769. doi: 10.1002/smll.201303442. [DOI] [PubMed] [Google Scholar]
- 176.Kick B, Praetorius F, Dietz H, Weuster-Botz D. Efficient production of single-stranded phage DNA as scaffolds for DNA origami. Nano Lett. 2015;15:4672–4676. doi: 10.1021/acs.nanolett.5b01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kick B, Hensler S, Praetorius F, Dietz H, Weuster-Botz D. Specific growth rate and multiplicity of infection affect high-cell-density fermentation with bacteriophage M13 for ssDNA production. Biotechnol. Bioeng. 2017;114:777–784. doi: 10.1002/bit.26200. [DOI] [PubMed] [Google Scholar]
- 178.Said H, et al. M1.3–a small scaffold for DNA origami. Nanoscale. 2013;5:284–290. doi: 10.1039/C2NR32393A. [DOI] [PubMed] [Google Scholar]
- 179.Martin TG, Dietz H. Magnesium-free self-assembly of multi-layer DNA objects. Nat. Commun. 2012;3:1103. doi: 10.1038/ncomms2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zhang Z, Song J, Besenbacher F, Dong M, Gothelf KV. Self-assembly of DNA origami and single-stranded tile structures at room temperature. Angew. Chem. Int. Ed. Engl. 2013;52:9219–9223. doi: 10.1002/anie.201303611. [DOI] [PubMed] [Google Scholar]
- 181.Kopielski A, Schneider A, Csaki A, Fritzsche W. Isothermal DNA origami folding: avoiding denaturing conditions for one-pot, hybrid-component annealing. Nanoscale. 2015;7:2102–2106. doi: 10.1039/C4NR04176C. [DOI] [PubMed] [Google Scholar]
- 182.Timm C, Niemeyer CM. Assembly and purification of enzyme-functionalized DNA origami structures. Angew. Chem. Int. Ed. Engl. 2015;54:6745–6750. doi: 10.1002/anie.201500175. [DOI] [PubMed] [Google Scholar]
- 183.Shaw A, Benson E, Hogberg B. Purification of functionalized DNA origami nanostructures. ACS Nano. 2015;9:4968–4975. doi: 10.1021/nn507035g. [DOI] [PubMed] [Google Scholar]
- 184.Stahl E, Martin TG, Praetorius F, Dietz H. Facile and scalable preparation of pure and dense DNA origami solutions. Angew. Chem. Int. Ed. Engl. 2014;53:12735–12740. doi: 10.1002/anie.201405991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bellot G, McClintock MA, Lin C, Shih WM. Recovery of intact DNA nanostructures after agarose gel-based separation. Nat. Methods. 2011;8:192–194. doi: 10.1038/nmeth0311-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Lin C, Perrault SD, Kwak M, Graf F, Shih WM. Purification of DNA-origami nanostructures by rate-zonal centrifugation. Nucleic Acids Res. 2013;41:e40. doi: 10.1093/nar/gks1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Rajendran A, Endo M, Katsuda Y, Hidaka K, Sugiyama H. Photo-cross-linking-assisted thermal stability of DNA origami structures and its application for higher-temperature self-assembly. J. Am. Chem. Soc. 2011;133:14488–14491. doi: 10.1021/ja204546h. [DOI] [PubMed] [Google Scholar]
- 188.Hofr C, Brabec V. Thermal and thermodynamic properties of duplex DNA containing site-specific interstrand cross-link of antitumor cisplatin or its clinically ineffective trans isomer. J. Biol. Chem. 2001;276:9655–9661. doi: 10.1074/jbc.M010205200. [DOI] [PubMed] [Google Scholar]
- 189.Eskelinen AP, Rosilo H, Kuzyk A, Torma P, Kostiainen MA. Controlling the formation of DNA origami structures with external signals. Small. 2012;8:2016–2020. doi: 10.1002/smll.201102697. [DOI] [PubMed] [Google Scholar]
- 190.Browne WR, Feringa BL. Making molecular machines work. Nat. Nanotechnol. 2006;1:25–35. doi: 10.1038/nnano.2006.45. [DOI] [PubMed] [Google Scholar]
- 191.Li SL, et al. New light on the ring-chain equilibrium of a hydrogen-bonded supramolecular polymer based on a photochromic dithienylethene unit and its energy-transfer properties as a storage material. Chem.-Eur. J. 2011;17:10716–10723. doi: 10.1002/chem.201100691. [DOI] [PubMed] [Google Scholar]
- 192.ten Brinke G, Ruokolainen J, Ikkala O. Supramolecular materials based on hydrogen-bonded polymers. Adv. Polym. Sci. 2007;207:113–177. doi: 10.1007/12_2006_111. [DOI] [Google Scholar]
- 193.Stuart MA, et al. Emerging applications of stimuli-responsive polymer materials. Nat. Mater. 2010;9:101–113. doi: 10.1038/nmat2614. [DOI] [PubMed] [Google Scholar]
- 194.Gallego I, Grover MA, Hud NV. Folding and imaging of DNA nanostructures in anhydrous and hydrated deep-eutectic solvents. Angew. Chem. Int. Ed. Engl. 2015;54:6765–6769. doi: 10.1002/anie.201412354. [DOI] [PubMed] [Google Scholar]
- 195.Berthiaume F, Maguire TJ, Yarmush ML. Tissue engineering and regenerative medicine: history, progress, and challenges. Annu. Rev. Chem. Biomolecular Eng. 2011;2:403–430. doi: 10.1146/annurev-chembioeng-061010-114257. [DOI] [PubMed] [Google Scholar]
- 196.Orlando G, et al. Regenerative medicine and organ transplantation: past, present, and future. Transplantation. 2011;91:1310–1317. doi: 10.1097/TP.0b013e318219ebb5. [DOI] [PubMed] [Google Scholar]
- 197.Song HG, Rumma RT, Ozaki CK, Edelman ER, Chen CS. Vascular tissue engineering: progress, challenges, and clinical promise. Cell Stem Cell. 2018;22:340–354. doi: 10.1016/j.stem.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Tarassoli SP, et al. Skin tissue engineering using 3D bioprinting: an evolving research field. J. Plast. Reconstr. Aesthet. Surg. 2018;71:615–623. doi: 10.1016/j.bjps.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 199.Yu JR, et al. Current and future perspectives on skin tissue engineering: key features of biomedical research, translational assessment, and clinical application. Adv. Healthc. Mater. 2019;8:e1801471. doi: 10.1002/adhm.201801471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Jana S, Levengood SK, Zhang M. Anisotropic materials for skeletal-muscle-tissue engineering. Adv. Mater. 2016;28:10588–10612. doi: 10.1002/adma.201600240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hassanzadeh P, Atyabi F, Dinarvand R. Tissue engineering: Still facing a long way ahead. J. Control. Release. 2018;279:181–197. doi: 10.1016/j.jconrel.2018.04.024. [DOI] [PubMed] [Google Scholar]
- 202.Polacheck WJ, Kutys ML, Tefft JB, Chen CS. Microfabricated blood vessels for modeling the vascular transport barrier. Nat. Protoc. 2019;14:1425–1454. doi: 10.1038/s41596-019-0144-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wang Y, et al. Engineering vascular tissue with functional smooth muscle cells derived from human iPS cells and nanofibrous scaffolds. Biomaterials. 2014;35:8960–8969. doi: 10.1016/j.biomaterials.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Dufrane D. Impact of age on human adipose stem cells for bone tissue engineering. Cell Transplant. 2017;26:1496–1504. doi: 10.1177/0963689717721203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Ho-Shui-Ling A, et al. Bone regeneration strategies: engineered scaffolds, bioactive molecules and stem cells current stage and future perspectives. Biomaterials. 2018;180:143–162. doi: 10.1016/j.biomaterials.2018.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Kwee BJ, Mooney DJ. Biomaterials for skeletal muscle tissue engineering. Curr. Opin. Biotechnol. 2017;47:16–22. doi: 10.1016/j.copbio.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Aamodt JM, Grainger DW. Extracellular matrix-based biomaterial scaffolds and the host response. Biomaterials. 2016;86:68–82. doi: 10.1016/j.biomaterials.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Shih YV, Varghese S. Tissue engineered bone mimetics to study bone disorders ex vivo: role of bioinspired materials. Biomaterials. 2019;198:107–121. doi: 10.1016/j.biomaterials.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Saldin LT, Cramer MC, Velankar SS, White LJ, Badylak SF. Extracellular matrix hydrogels from decellularized tissues: Structure and function. Acta Biomaterialia. 2017;49:1–15. doi: 10.1016/j.actbio.2016.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Keane TJ, Swinehart IT, Badylak SF. Methods of tissue decellularization used for preparation of biologic scaffolds and in vivo relevance. Methods. 2015;84:25–34. doi: 10.1016/j.ymeth.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 211.Lancaster MA, et al. Guided self-organization and cortical plate formation in human brain organoids. Nat. Biotechnol. 2017;35:659–666. doi: 10.1038/nbt.3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kang HW, et al. A 3D bioprinting system to produce human-scale tissue constructs with structural integrity. Nat. Biotechnol. 2016;34:312–319. doi: 10.1038/nbt.3413. [DOI] [PubMed] [Google Scholar]
- 213.Takebe T, Zhang B, Radisic M. Synergistic engineering: organoids meet organs-on-a-chip. Cell Stem Cell. 2017;21:297–300. doi: 10.1016/j.stem.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 214.Wubneh A, Tsekoura EK, Ayranci C, Uludag H. Current state of fabrication technologies and materials for bone tissue engineering. Acta Biomaterialia. 2018;80:1–30. doi: 10.1016/j.actbio.2018.09.031. [DOI] [PubMed] [Google Scholar]
- 215.Atasoy-Zeybek A, Kose GT. Gene therapy strategies in bone tissue engineering and current clinical applications. Adv. Exp. Med. Biol. 2018;1119:85–101. doi: 10.1007/5584_2018_253. [DOI] [PubMed] [Google Scholar]
- 216.Langer R, Vacanti JP. Tissue engineering. Science. 1993;260:920–926. doi: 10.1126/science.8493529. [DOI] [PubMed] [Google Scholar]
- 217.Salvay DM, Shea LD. Inductive tissue engineering with protein and DNA-releasing scaffolds. Mol. Biosyst. 2006;2:36–48. doi: 10.1039/B514174P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Wobus AM, Boheler KR. Embryonic stem cells: prospects for developmental biology and cell therapy. Physiol. Rev. 2005;85:635–678. doi: 10.1152/physrev.00054.2003. [DOI] [PubMed] [Google Scholar]
- 219.Guan K, Schmidt MM, Ding Q, Chang H, Wobus AM. Embryonic stem cells in vitro-prospects for cell and developmental biology, embryotoxicology and cell therapy. Altex. 1999;16:135–141. [PubMed] [Google Scholar]
- 220.Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- 221.Hersel U, Dahmen C, Kessler H. RGD modified polymers: biomaterials for stimulated cell adhesion and beyond. Biomaterials. 2003;24:4385–4415. doi: 10.1016/S0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 222.Arnold MS, Guler MO, Hersam MC, Stupp SI. Encapsulation of carbon nanotubes by self-assembling peptide amphiphiles. Langmuir. 2005;21:4705–4709. doi: 10.1021/la0469452. [DOI] [PubMed] [Google Scholar]
- 223.Zhang S, et al. Self-complementary oligopeptide matrices support mammalian cell attachment. Biomaterials. 1995;16:1385–1393. doi: 10.1016/0142-9612(95)96874-Y. [DOI] [PubMed] [Google Scholar]
- 224.Vasita R, Katti DS. Nanofibers and their applications in tissue engineering. Int. J. Nanomed. 2006;1:15–30. doi: 10.2147/nano.2006.1.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Murphy WL, Mooney DJ. Controlled delivery of inductive proteins, plasmid DNA and cells from tissue engineering matrices. J. Periodontal Res. 1999;34:413–419. doi: 10.1111/j.1600-0765.1999.tb02275.x. [DOI] [PubMed] [Google Scholar]
- 226.Jang JH, Bengali Z, Houchin TL, Shea LD. Surface adsorption of DNA to tissue engineering scaffolds for efficient gene delivery. J. Biomed. Mater. Res. Part A. 2006;77:50–58. doi: 10.1002/jbm.a.30643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Zhou M, et al. Effect of tetrahedral DNA nanostructures on proliferation and osteo/odontogenic differentiation of dental pulp stem cells via activation of the notch signaling pathway. Nanomedicine. 2018;14:1227–1236. doi: 10.1016/j.nano.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 228.Gonzalez-Fernandez T, et al. Pore-forming bioinks to enable spatio-temporally defined gene delivery in bioprinted tissues. J. Control. Release. 2019;301:13–27. doi: 10.1016/j.jconrel.2019.03.006. [DOI] [PubMed] [Google Scholar]
- 229.Walsh DP, et al. Transfection of autologous host cells in vivo using gene activated collagen scaffolds incorporating star-polypeptides. J. Control. Release. 2019;304:191–203. doi: 10.1016/j.jconrel.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 230.Ma W, et al. Enhanced neural regeneration with a concomitant treatment of framework nucleic acid and stem cells in spinal cord injury. ACS Appl. Mater. Interfaces. 2020;12:2095–2106. doi: 10.1021/acsami.9b19079. [DOI] [PubMed] [Google Scholar]
- 231.Zhu J, et al. Tetrahedral framework nucleic acids promote scarless healing of cutaneous wounds via the AKT-signaling pathway. Signal Transduct. Target. Ther. 2020;5:120. doi: 10.1038/s41392-020-0173-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Li W, et al. A gene-activating skin substitute comprising PLLA/POSS nanofibers and plasmid DNA encoding ANG and bFGF promotes in vivo revascularization and epidermalization. J. Mater. Chem. B. 2018;6:6977–6992. doi: 10.1039/C8TB02006J. [DOI] [PubMed] [Google Scholar]
- 233.Duman N, et al. Topical folinic acid enhances wound healing in rat model. Adv. Med. Sci. 2018;63:347–352. doi: 10.1016/j.advms.2018.04.011. [DOI] [PubMed] [Google Scholar]
- 234.Eke G, Mangir N, Hasirci N, MacNeil S, Hasirci V. Development of a UV crosslinked biodegradable hydrogel containing adipose derived stem cells to promote vascularization for skin wounds and tissue engineering. Biomaterials. 2017;129:188–198. doi: 10.1016/j.biomaterials.2017.03.021. [DOI] [PubMed] [Google Scholar]
- 235.Milan PB, et al. Accelerated wound healing in a diabetic rat model using decellularized dermal matrix and human umbilical cord perivascular cells. Acta Biomaterialia. 2016;45:234–246. doi: 10.1016/j.actbio.2016.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Bhowmick S, Scharnweber D, Koul V. Co-cultivation of keratinocyte-human mesenchymal stem cell (hMSC) on sericin loaded electrospun nanofibrous composite scaffold (cationic gelatin/hyaluronan/chondroitin sulfate) stimulates epithelial differentiation in hMSCs: In vitro study. Biomaterials. 2016;88:83–96. doi: 10.1016/j.biomaterials.2016.02.034. [DOI] [PubMed] [Google Scholar]
- 237.Keil TWM, Baldassi D, Merkel OM. T-cell targeted pulmonary siRNA delivery for the treatment of asthma. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020;12:e1634. doi: 10.1002/wnan.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Liang H, et al. Topical nanoparticles interfering with the DNA-LL37 complex to alleviate psoriatic inflammation in mice and monkeys. Sci. Adv. 2020;6:eabb5274. doi: 10.1126/sciadv.abb5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Mao C, et al. The clearance effect of tetrahedral DNA nanostructures on senescent human dermal fibroblasts. ACS Appl. Mater. Interfaces. 2019;11:1942–1950. doi: 10.1021/acsami.8b20530. [DOI] [PubMed] [Google Scholar]
- 240.Shao XR, et al. Neuroprotective effect of tetrahedral DNA nanostructures in a cell model of Alzheimer’s disease. ACS Appl. Mater. Interfaces. 2018;10:23682–23692. doi: 10.1021/acsami.8b07827. [DOI] [PubMed] [Google Scholar]
- 241.Whittlesey KJ, Shea LD. Delivery systems for small molecule drugs, proteins, and DNA: the neuroscience/biomaterial interface. Exp. Neurol. 2004;190:1–16. doi: 10.1016/j.expneurol.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 242.Pannier AK, Shea LD. Controlled release systems for DNA delivery. Mol. Ther. 2004;10:19–26. doi: 10.1016/j.ymthe.2004.03.020. [DOI] [PubMed] [Google Scholar]
- 243.Amani A, Kabiri T, Shafiee S, Hamidi A. Preparation and characterization of PLA-PEG-PLA/PEI/DNA nanoparticles for improvement of transfection efficiency and controlled release of DNA in gene delivery systems. Iran. J. Pharm. Res.: IJPR. 2019;18:125–141. [PMC free article] [PubMed] [Google Scholar]
- 244.Gajbhiye KR, Chaudhari BP, Pokharkar VB, Pawar A, Gajbhiye V. Stimuli-responsive biodegradable polyurethane nano-constructs as a potential triggered drug delivery vehicle for cancer therapy. Int. J. Pharm. 2020;588:119781. doi: 10.1016/j.ijpharm.2020.119781. [DOI] [PubMed] [Google Scholar]
- 245.Hashemi M, Shamshiri A, Saeedi M, Tayebi L, Yazdian-Robati R. Aptamer-conjugated PLGA nanoparticles for delivery and imaging of cancer therapeutic drugs. Arch. Biochem. Biophys. 2020;691:108485. doi: 10.1016/j.abb.2020.108485. [DOI] [PubMed] [Google Scholar]
- 246.Wolski P, Nieszporek K, Panczyk AT. Carbon nanotubes and short cytosine-rich telomeric DNA oligomeres as platforms for controlled release of doxorubicin-a molecular dynamics study. Int. J. Mol. Sci. 2020;21:3619. doi: 10.3390/ijms21103619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Tan X, Jia F, Wang P, Zhang K. Nucleic acid-based drug delivery strategies. J. Control. Release. 2020;323:240–252. doi: 10.1016/j.jconrel.2020.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Peng Q, et al. Understanding the biomedical effects of the self-assembled tetrahedral DNA nanostructure on living cells. ACS Appl. Mater. Interfaces. 2016;8:12733–12739. doi: 10.1021/acsami.6b03786. [DOI] [PubMed] [Google Scholar]
- 249.Faingold A, Cohen SR, Reznikov N, Wagner HD. Osteonal lamellae elementary units: lamellar microstructure, curvature and mechanical properties. Acta Biomaterialia. 2013;9:5956–5962. doi: 10.1016/j.actbio.2012.11.032. [DOI] [PubMed] [Google Scholar]
- 250.Raftery RM, et al. Delivering nucleic-acid based nanomedicines on biomaterial scaffolds for orthopedic tissue repair: challenges, progress and future perspectives. Adv. Mater. 2016;28:5447–5469. doi: 10.1002/adma.201505088. [DOI] [PubMed] [Google Scholar]
- 251.Betz VM, et al. Recent advances in gene-enhanced bone tissue engineering. J. Gene Med. 2018;20:e3018. doi: 10.1002/jgm.3018. [DOI] [PubMed] [Google Scholar]
- 252.Balmayor ER, Evans CH. RNA therapeutics for tissue engineering. Tissue Eng. Part A. 2019;25:9–11. doi: 10.1089/ten.tea.2018.0315. [DOI] [PubMed] [Google Scholar]
- 253.Shim MS, Kwon YJ. Controlled cytoplasmic and nuclear localization of plasmid DNA and siRNA by differentially tailored polyethylenimine. J. Control. Release. 2009;133:206–213. doi: 10.1016/j.jconrel.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 254.Khorsand B, et al. Regeneration of bone using nanoplex delivery of FGF-2 and BMP-2 genes in diaphyseal long bone radial defects in a diabetic rabbit model. J. Control. Release. 2017;248:53–59. doi: 10.1016/j.jconrel.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Kim TH, Singh RK, Kang MS, Kim JH, Kim HW. Gene delivery nanocarriers of bioactive glass with unique potential to load BMP2 plasmid DNA and to internalize into mesenchymal stem cells for osteogenesis and bone regeneration. Nanoscale. 2016;8:8300–8311. doi: 10.1039/C5NR07933K. [DOI] [PubMed] [Google Scholar]
- 256.Schlickewei C, et al. A bioactive nano-calcium phosphate paste for in-situ transfection of BMP-7 and VEGF-A in a rabbit critical-size bone defect: results of an in vivo study. J. Mater. Sci. Mater. Med. 2019;30:15. doi: 10.1007/s10856-019-6217-y. [DOI] [PubMed] [Google Scholar]
- 257.Takanche JS, et al. Chitosan-gold nanoparticles mediated gene delivery of c-myb facilitates osseointegration of dental implants in ovariectomized rat. Artif. Cells Nanomed. Biotechnol. 2018;46:S807–S817. doi: 10.1080/21691401.2018.1513940. [DOI] [PubMed] [Google Scholar]
- 258.Huang M, et al. Comparison of osteogenic differentiation induced by siNoggin and pBMP-2 delivered by lipopolysaccharide-amine nanopolymersomes and underlying molecular mechanisms. Int. J. Nanomed. 2019;14:4229–4245. doi: 10.2147/IJN.S203540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Liang D, et al. In vitro non-viral gene delivery with nanofibrous scaffolds. Nucleic Acids Res. 2005;33:e170. doi: 10.1093/nar/gni171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Li Y, Liu C. Nanomaterial-based bone regeneration. Nanoscale. 2017;9:4862–4874. doi: 10.1039/C7NR00835J. [DOI] [PubMed] [Google Scholar]
- 261.Wang SJ, et al. Biomimetic nanosilica-collagen scaffolds for in situ bone regeneration: toward a cell-free, one-step surgery. Adv. Mater. 2019;31:e1904341. doi: 10.1002/adma.201904341. [DOI] [PubMed] [Google Scholar]
- 262.Li G, Zhou T, Lin S, Shi S, Lin Y. Nanomaterials for craniofacial and dental tissue engineering. J. Dent. Res. 2017;96:725–732. doi: 10.1177/0022034517706678. [DOI] [PubMed] [Google Scholar]
- 263.Mohammadi M, et al. Micro and nanotechnologies for bone regeneration: Recent advances and emerging designs. J. Control. Release. 2018;274:35–55. doi: 10.1016/j.jconrel.2018.01.032. [DOI] [PubMed] [Google Scholar]
- 264.Lv L, et al. The nanoscale geometry of TiO2 nanotubes influences the osteogenic differentiation of human adipose-derived stem cells by modulating H3K4 trimethylation. Biomaterials. 2015;39:193–205. doi: 10.1016/j.biomaterials.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 265.D’Mello S, et al. Bone regeneration using gene-activated matrices. AAPS J. 2017;19:43–53. doi: 10.1208/s12248-016-9982-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Tierney EG, Duffy GP, Hibbitts AJ, Cryan SA, O’Brien FJ. The development of non-viral gene-activated matrices for bone regeneration using polyethyleneimine (PEI) and collagen-based scaffolds. J. Control. Release. 2012;158:304–311. doi: 10.1016/j.jconrel.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 267.Shimer AL, Oner FC, Vaccaro AR. Spinal reconstruction and bone morphogenetic proteins: open questions. Injury. 2009;40:S32–S38. doi: 10.1016/S0020-1383(09)70009-9. [DOI] [PubMed] [Google Scholar]
- 268.Winn SR, Hu Y, Sfeir C, Hollinger JO. Gene therapy approaches for modulating bone regeneration. Adv. Drug Deliv. Rev. 2000;42:121–138. doi: 10.1016/S0169-409X(00)00057-0. [DOI] [PubMed] [Google Scholar]
- 269.Chan HL, McCauley LK. Parathyroid hormone applications in the craniofacial skeleton. J. Dent. Res. 2013;92:18–25. doi: 10.1177/0022034512464779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Elangovan S, et al. Chemically modified RNA activated matrices enhance bone regeneration. J. Control. Release. 2015;218:22–28. doi: 10.1016/j.jconrel.2015.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Higashi T, et al. Novel lipidated sorbitol-based molecular transporters for non-viral gene delivery. J. Control. Release. 2009;136:140–147. doi: 10.1016/j.jconrel.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 272.des Rieux A, Shikanov A, Shea LD. Fibrin hydrogels for non-viral vector delivery in vitro. J. Control. Release. 2009;136:148–154. doi: 10.1016/j.jconrel.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Lei Y, Rahim M, Ng Q, Segura T. Hyaluronic acid and fibrin hydrogels with concentrated DNA/PEI polyplexes for local gene delivery. J. Control. Release. 2011;153:255–261. doi: 10.1016/j.jconrel.2011.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Togashi R, et al. A hepatic pDNA delivery system based on an intracellular environment sensitive vitamin E-scaffold lipid-like material with the aid of an anti-inflammatory drug. J. Control. Release. 2018;279:262–270. doi: 10.1016/j.jconrel.2018.04.022. [DOI] [PubMed] [Google Scholar]
- 275.Siegman S, Truong NF, Segura T. Encapsulation of PEGylated low-molecular-weight PEI polyplexes in hyaluronic acid hydrogels reduces aggregation. Acta Biomaterialia. 2015;28:45–54. doi: 10.1016/j.actbio.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Nie H, Soh BW, Fu YC, Wang CH. Three-dimensional fibrous PLGA/HAp composite scaffold for BMP-2 delivery. Biotechnol. Bioeng. 2008;99:223–234. doi: 10.1002/bit.21517. [DOI] [PubMed] [Google Scholar]
- 277.Nie H, Wang CH. Fabrication and characterization of PLGA/HAp composite scaffolds for delivery of BMP-2 plasmid DNA. J. Control. Release. 2007;120:111–121. doi: 10.1016/j.jconrel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 278.Fu YC, Nie H, Ho ML, Wang CK, Wang CH. Optimized bone regeneration based on sustained release from three-dimensional fibrous PLGA/HAp composite scaffolds loaded with BMP-2. Biotechnol. Bioeng. 2008;99:996–1006. doi: 10.1002/bit.21648. [DOI] [PubMed] [Google Scholar]
- 279.Nie H, Ho ML, Wang CK, Wang CH, Fu YC. BMP-2 plasmid loaded PLGA/HAp composite scaffolds for treatment of bone defects in nude mice. Biomaterials. 2009;30:892–901. doi: 10.1016/j.biomaterials.2008.10.029. [DOI] [PubMed] [Google Scholar]
- 280.Keeney M, et al. Scaffold-mediated BMP-2 minicircle DNA delivery accelerated bone repair in a mouse critical-size calvarial defect model. J. Biomed. Mater. Res. Part A. 2016;104:2099–2107. doi: 10.1002/jbm.a.35735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Atluri K, Seabold D, Hong L, Elangovan S, Salem AK. Nanoplex-mediated codelivery of fibroblast growth factor and bone morphogenetic protein genes promotes osteogenesis in human adipocyte-derived mesenchymal stem cells. Mol. Pharm. 2015;12:3032–3042. doi: 10.1021/acs.molpharmaceut.5b00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Elangovan S, et al. The enhancement of bone regeneration by gene activated matrix encoding for platelet derived growth factor. Biomaterials. 2014;35:737–747. doi: 10.1016/j.biomaterials.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Qiao C, et al. Using poly(lactic-co-glycolic acid) microspheres to encapsulate plasmid of bone morphogenetic protein 2/polyethylenimine nanoparticles to promote bone formation in vitro and in vivo. Int. J. Nanomed. 2013;8:2985–2995. doi: 10.2147/IJN.S45184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Qiao C, et al. Sustained release poly (lactic-co-glycolic acid) microspheres of bone morphogenetic protein 2 plasmid/calcium phosphate to promote in vitro bone formation and in vivo ectopic osteogenesis. Am. J. Transl. Res. 2015;7:2561–2572. [PMC free article] [PubMed] [Google Scholar]
- 285.Hadjicharalambous C, Kozlova D, Sokolova V, Epple M, Chatzinikolaidou M. Calcium phosphate nanoparticles carrying BMP-7 plasmid DNA induce an osteogenic response in MC3T3-E1 pre-osteoblasts. J. Biomed. Mater. Res. Part A. 2015;103:3834–3842. doi: 10.1002/jbm.a.35527. [DOI] [PubMed] [Google Scholar]
- 286.Bhattarai G, Lee YH, Lee MH, Yi HK. Gene delivery of c-myb increases bone formation surrounding oral implants. J. Dent. Res. 2013;92:840–845. doi: 10.1177/0022034513497753. [DOI] [PubMed] [Google Scholar]
- 287.Li DD, et al. Characterization and cytocompatibility of thermosensitive hydrogel embedded with chitosan nanoparticles for delivery of bone morphogenetic protein-2 plasmid DNA. J. Mater. Sci. Mater. Med. 2016;27:134. doi: 10.1007/s10856-016-5743-0. [DOI] [PubMed] [Google Scholar]
- 288.Li H, et al. Accelerated bony defect healing based on chitosan thermosensitive hydrogel scaffolds embedded with chitosan nanoparticles for the delivery of BMP2 plasmid DNA. J. Biomed. Mater. Res. Part A. 2017;105:265–273. doi: 10.1002/jbm.a.35900. [DOI] [PubMed] [Google Scholar]
- 289.Supper S, et al. Thermosensitive chitosan/glycerophosphate-based hydrogel and its derivatives in pharmaceutical and biomedical applications. Expert Opin. drug Deliv. 2014;11:249–267. doi: 10.1517/17425247.2014.867326. [DOI] [PubMed] [Google Scholar]
- 290.Carballo-Pedrares N, Fuentes-Boquete I, Diaz-Prado S, Rey-Rico A. hydrogel-based localized nonviral gene delivery in regenerative medicine approaches-an overview. Pharmaceutics. 2020;12:752. doi: 10.3390/pharmaceutics12080752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Lauritano D, Limongelli L, Moreo G, Favia G, Carinci F. Nanomaterials for periodontal tissue engineering: chitosan-based scaffolds. a systematic review. Nanomaterials. 2020;10:605. doi: 10.3390/nano10040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Wu S, et al. Evaluation of chitosan hydrogel for sustained delivery of VEGF for odontogenic differentiation of dental pulp stem cells. Stem Cells Int. 2019;2019:1515040. doi: 10.1155/2019/1515040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Walsh DP, et al. Bioinspired star-shaped poly(L-lysine) polypeptides: efficient polymeric nanocarriers for the delivery of dna to mesenchymal stem cells. Mol. Pharm. 2018;15:1878–1891. doi: 10.1021/acs.molpharmaceut.8b00044. [DOI] [PubMed] [Google Scholar]
- 294.Shinozaki Y, et al. Evaluation of bone formation guided by DNA/protamine complex with FGF-2 in an adult rat calvarial defect model. J. Biomed. Mater. Res. Part B, Appl. Biomater. 2014;102:1669–1676. doi: 10.1002/jbm.b.33143. [DOI] [PubMed] [Google Scholar]
- 295.Toda M, Ohno J, Shinozaki Y, Ozaki M, Fukushima T. Osteogenic potential for replacing cells in rat cranial defects implanted with a DNA/protamine complex paste. Bone. 2014;67:237–245. doi: 10.1016/j.bone.2014.07.018. [DOI] [PubMed] [Google Scholar]
- 296.Charles LF, et al. Effects of low dose FGF-2 and BMP-2 on healing of calvarial defects in old mice. Exp. Gerontol. 2015;64:62–69. doi: 10.1016/j.exger.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Shi S, et al. Modulation of chondrocyte motility by tetrahedral DNA nanostructures. Cell Prolif. 2017;50:e12368. doi: 10.1111/cpr.12368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Shi S, et al. Effects of tetrahedral DNA nanostructures on autophagy in chondrocytes. Chem. Commun. (Camb.) 2018;54:1327–1330. doi: 10.1039/C7CC09397G. [DOI] [PubMed] [Google Scholar]
- 299.Shao XR, et al. Effect of tetrahedral DNA nanostructures on osteogenic differentiation of mesenchymal stem cells via activation of the Wnt/beta-catenin signaling pathway. Nanomedicine. 2017;13:1809–1819. doi: 10.1016/j.nano.2017.02.011. [DOI] [PubMed] [Google Scholar]
- 300.Zhou M, et al. Effect of tetrahedral DNA nanostructures on proliferation and osteogenic differentiation of human periodontal ligament stem cells. Cell Prolif. 2019;52:e12566. doi: 10.1111/cpr.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Zhou M, et al. The protective effect of tetrahedral framework nucleic acids on periodontium under inflammatory conditions. Bioact. Mater. 2021;6:1676–1688. doi: 10.1016/j.bioactmat.2020.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Sirong S, et al. Effects of tetrahedral framework nucleic acid/wogonin complexes on osteoarthritis. Bone Res. 2020;8:6. doi: 10.1038/s41413-019-0077-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Wang G, et al. Evaluation of the protective effect of a prime-boost strategy with plasmid DNA followed by recombinant adenovirus expressing BmAMA1 as vaccines against Babesia microti infection in hamster. Acta Parasitol. 2018;63:368–374. doi: 10.1515/ap-2018-0042. [DOI] [PubMed] [Google Scholar]
- 304.Burger C, Chu B. Functional nanofibrous scaffolds for bone reconstruction. Colloids Surf. B Biointerfaces. 2007;56:134–141. doi: 10.1016/j.colsurfb.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 305.Gunatillake PA, Adhikari R. Biodegradable synthetic polymers for tissue engineering. Eur. Cells Mater. 2003;5:1–16. doi: 10.22203/eCM.v005a01. [DOI] [PubMed] [Google Scholar]
- 306.Wang MO, et al. Evaluating 3D-printed biomaterials as scaffolds for vascularized bone tissue engineering. Adv. Mater. 2015;27:138–144. doi: 10.1002/adma.201403943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.O’Brien FJ. Biomaterials & scaffolds for tissue engineering. Mater. Today. 2011;14:88–95. doi: 10.1016/S1369-7021(11)70058-X. [DOI] [Google Scholar]
- 308.Meinel L, Kaplan DL. Silk constructs for delivery of musculoskeletal therapeutics. Adv. Drug Deliv. Rev. 2012;64:1111–1122. doi: 10.1016/j.addr.2012.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Raftery RM, et al. Delivering nucleic-acid based nanomedicines on biomaterial scaffolds for orthopedic tissue repair: challenges, progress and future. Perspectives. 2016;28:5447–5469. doi: 10.1002/adma.201505088. [DOI] [PubMed] [Google Scholar]
- 310.Chirico G, Pallavicini P, Collini M. Gold nanostars for superficial diseases: a promising tool for localized hyperthermia? Nanomedicine (Lond.) 2014;9:1–3. doi: 10.2217/nnm.13.186. [DOI] [PubMed] [Google Scholar]
- 311.Vilella A, et al. Endocytosis of nanomedicines: the case of glycopeptide engineered PLGA nanoparticles. Pharmaceutics. 2015;7:74–89. doi: 10.3390/pharmaceutics7020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Katav T, et al. Modified pectin-based carrier for gene delivery: cellular barriers in gene delivery course. J. Control. Release. 2008;130:183–191. doi: 10.1016/j.jconrel.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 313.Jeong GW, Nah JW. Evaluation of disulfide bond-conjugated LMWSC-g-bPEI as non-viral vector for low cytotoxicity and efficient gene delivery. Carbohydr. Polym. 2017;178:322–330. doi: 10.1016/j.carbpol.2017.09.048. [DOI] [PubMed] [Google Scholar]
- 314.Singh J, Michel D, Chitanda JM, Verrall RE, Badea I. Evaluation of cellular uptake and intracellular trafficking as determining factors of gene expression for amino acid-substituted gemini surfactant-based DNA nanoparticles. J. Nanobiotechnol. 2012;10:7. doi: 10.1186/1477-3155-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Zhang B, Mallapragada S. The mechanism of selective transfection mediated by pentablock copolymers; part II: nuclear entry and endosomal escape. Acta Biomaterialia. 2011;7:1580–1587. doi: 10.1016/j.actbio.2010.11.033. [DOI] [PubMed] [Google Scholar]
- 316.Wang J, Dou B, Bao Y. Efficient targeted pDNA/siRNA delivery with folate-low-molecular-weight polyethyleneimine-modified pullulan as non-viral carrier. Mater. Sci. Eng. C. Mater. Biol. Appl. 2014;34:98–109. doi: 10.1016/j.msec.2013.08.035. [DOI] [PubMed] [Google Scholar]
- 317.Gu X, Ding F, Williams DF. Neural tissue engineering options for peripheral nerve regeneration. Biomaterials. 2014;35:6143–6156. doi: 10.1016/j.biomaterials.2014.04.064. [DOI] [PubMed] [Google Scholar]
- 318.Chooi WH, Chew SY. Modulation of cell-cell interactions for neural tissue engineering: Potential therapeutic applications of cell adhesion molecules in nerve regeneration. Biomaterials. 2019;197:327–344. doi: 10.1016/j.biomaterials.2019.01.030. [DOI] [PubMed] [Google Scholar]
- 319.Madigan NN, McMahon S, O’Brien T, Yaszemski MJ, Windebank AJ. Current tissue engineering and novel therapeutic approaches to axonal regeneration following spinal cord injury using polymer scaffolds. Respir. Physiol. Neurobiol. 2009;169:183–199. doi: 10.1016/j.resp.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Editorial. Engineering approaches to enhance neural tissue regeneration. Exp. Neurol. 319, 112873 (2019). [DOI] [PubMed]
- 321.Koss KM, Unsworth LD. Neural tissue engineering: bioresponsive nanoscaffolds using engineered self-assembling peptides. Acta Biomaterialia. 2016;44:2–15. doi: 10.1016/j.actbio.2016.08.026. [DOI] [PubMed] [Google Scholar]
- 322.Tang X, et al. Bridging peripheral nerve defects with a tissue engineered nerve graft composed of an in vitro cultured nerve equivalent and a silk fibroin-based scaffold. Biomaterials. 2012;33:3860–3867. doi: 10.1016/j.biomaterials.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 323.Yang S, et al. Self-assembling peptide hydrogels functionalized with LN- and BDNF- mimicking epitopes synergistically enhance peripheral nerve regeneration. Theranostics. 2020;10:8227–8249. doi: 10.7150/thno.44276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Willerth SM, Sakiyama-Elbert SE. Approaches to neural tissue engineering using scaffolds for drug delivery. Adv. Drug Deliv. Rev. 2007;59:325–338. doi: 10.1016/j.addr.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Yao Y, et al. Efect of longitudinally oriented collagen conduit combined with nerve growth factor on nerve regeneration after dog sciatic nerve injury. J. Biomed. Mater. Res. Part B, Appl. Biomater. 2018;106:2131–2139. doi: 10.1002/jbm.b.34020. [DOI] [PubMed] [Google Scholar]
- 326.Baiguera S, et al. Electrospun gelatin scaffolds incorporating rat decellularized brain extracellular matrix for neural tissue engineering. Biomaterials. 2014;35:1205–1214. doi: 10.1016/j.biomaterials.2013.10.060. [DOI] [PubMed] [Google Scholar]
- 327.Liang Y, Walczak P, Bulte JW. The survival of engrafted neural stem cells within hyaluronic acid hydrogels. Biomaterials. 2013;34:5521–5529. doi: 10.1016/j.biomaterials.2013.03.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Sufan W, et al. Sciatic nerve regeneration through alginate with tubulation or nontubulation repair in cat. J. Neurotrauma. 2001;18:329–338. doi: 10.1089/08977150151070991. [DOI] [PubMed] [Google Scholar]
- 329.Chong EJ, et al. Evaluation of electrospun PCL/gelatin nanofibrous scaffold for wound healing and layered dermal reconstitution. Acta Biomaterialia. 2007;3:321–330. doi: 10.1016/j.actbio.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 330.Cui W, et al. Neuroprotective and neurotherapeutic effects of tetrahedral framework nucleic acids on Parkinson’s disease in vitro. ACS Appl. Mater. Interfaces. 2019;11:32787–32797. doi: 10.1021/acsami.9b10308. [DOI] [PubMed] [Google Scholar]
- 331.Li J, et al. Ternary polyplex micelles with PEG shells and intermediate barrier to complexed DNA cores for efficient systemic gene delivery. J. Control. Release. 2015;209:77–87. doi: 10.1016/j.jconrel.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 332.Feng G, et al. Gene therapy for nucleus pulposus regeneration by heme oxygenase-1 plasmid DNA carried by mixed polyplex micelles with thermo-responsive heterogeneous coronas. Biomaterials. 2015;52:1–13. doi: 10.1016/j.biomaterials.2015.02.024. [DOI] [PubMed] [Google Scholar]
- 333.Houchin-Ray T, Swift LA, Jang JH, Shea LD. Patterned PLG substrates for localized DNA delivery and directed neurite extension. Biomaterials. 2007;28:2603–2611. doi: 10.1016/j.biomaterials.2007.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Li J, Zha Z, Ge Z. Thermo-responsive polyplex micelles with PEG shells and PNIPAM layer to protect DNA cores for systemic gene therapy. Methods Mol. Biol. 2016;1445:269–276. doi: 10.1007/978-1-4939-3718-9_17. [DOI] [PubMed] [Google Scholar]
- 335.Arkenbout EK, et al. Protective function of transcription factor TR3 orphan receptor in atherogenesis: decreased lesion formation in carotid artery ligation model in TR3 transgenic mice. Circulation. 2002;106:1530–1535. doi: 10.1161/01.CIR.0000028811.03056.BF. [DOI] [PubMed] [Google Scholar]
- 336.Pires NM, et al. Activation of nuclear receptor Nur77 by 6-mercaptopurine protects against neointima formation. Circulation. 2007;115:493–500. doi: 10.1161/CIRCULATIONAHA.106.626838. [DOI] [PubMed] [Google Scholar]
- 337.Fassett MS, Jiang W, D’Alise AM, Mathis D, Benoist C. Nuclear receptor Nr4a1 modulates both regulatory T-cell (Treg) differentiation and clonal deletion. Proc. Natl Acad. Sci. USA. 2012;109:3891–3896. doi: 10.1073/pnas.1200090109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Hanna RN, et al. The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C- monocytes. Nat. Immunol. 2011;12:778–785. doi: 10.1038/ni.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Maxwell MA, et al. Nur77 regulates lipolysis in skeletal muscle cells. Evidence for cross-talk between the beta-adrenergic and an orphan nuclear hormone receptor pathway. J. Biol. Chem. 2005;280:12573–12584. doi: 10.1074/jbc.M409580200. [DOI] [PubMed] [Google Scholar]
- 340.Myers SA, Eriksson N, Burow R, Wang SC, Muscat GE. Beta-adrenergic signaling regulates NR4A nuclear receptor and metabolic gene expression in multiple tissues. Mol. Cell. Endocrinol. 2009;309:101–108. doi: 10.1016/j.mce.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 341.Pei L, et al. NR4A orphan nuclear receptors are transcriptional regulators of hepatic glucose metabolism. Nat. Med. 2006;12:1048–1055. doi: 10.1038/nm1471. [DOI] [PubMed] [Google Scholar]
- 342.Taniguchi J, et al. A synthetic DNA-binding inhibitor of SOX2 guides human induced pluripotent stem cells to differentiate into mesoderm. Nucleic Acids Res. 2017;45:9219–9228. doi: 10.1093/nar/gkx693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Wang L, et al. Imaging of Neurite Network with an Anti-L1CAM Aptamer Generated by Neurite-SELEX. J. Am. Chem. Soc. 2018;140:18066–18073. doi: 10.1021/jacs.8b10783. [DOI] [PubMed] [Google Scholar]
- 344.Shah B, Yin PT, Ghoshal S, Lee KB. Multimodal magnetic core-shell nanoparticles for effective stem-cell differentiation and imaging. Angew. Chem. Int. Ed. Engl. 2013;52:6190–6195. doi: 10.1002/anie.201302245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Shan J, Tenhu H. Recent advances in polymer protected gold nanoparticles: synthesis, properties and applications. Chem. Commun. (Camb.) 2007;28:4580–4598. doi: 10.1039/b707740h. [DOI] [PubMed] [Google Scholar]
- 346.Ghosh Chaudhuri R, Paria S. Core/shell nanoparticles: classes, properties, synthesis mechanisms, characterization, and applications. Chem. Rev. 2012;112:2373–2433. doi: 10.1021/cr100449n. [DOI] [PubMed] [Google Scholar]
- 347.Balasubramanian B, et al. Synthesis of monodisperse TiO2-paraffin core-shell nanoparticles for improved dielectric properties. ACS Nano. 2010;4:1893–1900. doi: 10.1021/nn9016422. [DOI] [PubMed] [Google Scholar]
- 348.Zhang H, Lee MY, Hogg MG, Dordick JS, Sharfstein ST. Gene delivery in three-dimensional cell cultures by superparamagnetic nanoparticles. ACS Nano. 2010;4:4733–4743. doi: 10.1021/nn9018812. [DOI] [PubMed] [Google Scholar]
- 349.Shao X, et al. Treatment of Alzheimer’s disease with framework nucleic acids. Cell Prolif. 2020;53:e12787. doi: 10.1111/cpr.12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Wackerhage H, Del Re,DP, Judson RN, Sudol M, Sadoshima J. The Hippo signal transduction network in skeletal and cardiac muscle. Sci. Signal. 2014;7:re4. doi: 10.1126/scisignal.2005096. [DOI] [PubMed] [Google Scholar]
- 351.Lin KC, et al. Regulation of Hippo pathway transcription factor TEAD by p38 MAPK-induced cytoplasmic translocation. Nat. Cell Biol. 2017;19:996–1002. doi: 10.1038/ncb3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Xin M, Olson EN, Bassel-Duby R. Mending broken hearts: cardiac development as a basis for adult heart regeneration and repair. Nat. Rev. Mol. Cell Biol. 2013;14:529–541. doi: 10.1038/nrm3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Addis RC, Epstein JA. Induced regeneration–the progress and promise of direct reprogramming for heart repair. Nat. Med. 2013;19:829–836. doi: 10.1038/nm.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Qian L, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Collins CA, et al. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell. 2005;122:289–301. doi: 10.1016/j.cell.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 356.Dumont NA, Wang YX, Rudnicki MA. Intrinsic and extrinsic mechanisms regulating satellite cell function. Development. 2015;142:1572–1581. doi: 10.1242/dev.114223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Day K, Shefer G, Richardson JB, Enikolopov G, Yablonka-Reuveni Z. Nestin-GFP reporter expression defines the quiescent state of skeletal muscle satellite cells. Dev. Biol. 2007;304:246–259. doi: 10.1016/j.ydbio.2006.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Kim JH, et al. Neural cell integration into 3D bioprinted skeletal muscle constructs accelerates restoration of muscle function. Nat. Commun. 2020;11:1025. doi: 10.1038/s41467-020-14930-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Qazi TH, Mooney DJ, Pumberger M, Geissler S, Duda GN. Biomaterials based strategies for skeletal muscle tissue engineering: existing technologies and future trends. Biomaterials. 2015;53:502–521. doi: 10.1016/j.biomaterials.2015.02.110. [DOI] [PubMed] [Google Scholar]
- 360.Del Carmen Ortuno-Costela M, Garcia-Lopez M, Cerrada V, Gallardo ME. iPSCs: a powerful tool for skeletal muscle tissue engineering. J. Cell. Mol. Med. 2019;23:3784–3794. doi: 10.1111/jcmm.14292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Singh VK, Kumar N, Kalsan M, Saini A, Chandra R. Mechanism of induction: induced pluripotent stem cells (iPSCs) J. Stem Cells. 2015;10:43–62. [PubMed] [Google Scholar]
- 362.Gorecka A, et al. Autologous transplantation of adipose-derived stem cells improves functional recovery of skeletal muscle without direct participation in new myofiber formation. Stem Cell Res. Ther. 2018;9:195. doi: 10.1186/s13287-018-0922-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Beldjilali-Labro M, et al. Biomaterials in tendon and skeletal muscle tissue engineering: current trends and challenges. Materials. 2018;11:4580–98. doi: 10.3390/ma11071116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Blumenfeld CM, et al. Drug capture materials based on genomic DNA-functionalized magnetic nanoparticles. Nat. Commun. 2018;9:2870. doi: 10.1038/s41467-018-05305-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Ueki R, et al. A chemically unmodified agonistic DNA with growth factor functionality for in vivo therapeutic application. Sci. Adv. 2020;6:eaay2801. doi: 10.1126/sciadv.aay2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Zhang M, et al. Cardioprotection of tetrahedral DNA nanostructures in myocardial ischemia-reperfusion injury. ACS Appl. Mater. interfaces. 2019;11:30631–30639. doi: 10.1021/acsami.9b10645. [DOI] [PubMed] [Google Scholar]
- 367.Zhang X, et al. The Nrf-2/HO-1 signaling axis: a ray of hope in cardiovascular diseases. Cardiol. Res. Pract. 2020;2020:5695723. doi: 10.1155/2020/5695723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Shen Y, Liu X, Shi J, Wu X. Involvement of Nrf2 in myocardial ischemia and reperfusion injury. Int. J. Biol. Macromol. 2019;125:496–502. doi: 10.1016/j.ijbiomac.2018.11.190. [DOI] [PubMed] [Google Scholar]
- 369.Seifert AW, et al. Skin shedding and tissue regeneration in African spiny mice (Acomys) Nature. 2012;489:561–565. doi: 10.1038/nature11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Metcalfe AD, Ferguson MW. Tissue engineering of replacement skin: the crossroads of biomaterials, wound healing, embryonic development, stem cells and regeneration. J. R. Soc. Interface. 2007;4:413–437. doi: 10.1098/rsif.2006.0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Fuchs E. Epithelial skin biology: three decades of developmental biology, a hundred questions answered and a thousand new ones to address. Curr. Top. Dev. Biol. 2016;116:357–374. doi: 10.1016/bs.ctdb.2015.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Roger M, et al. Bioengineering the microanatomy of human skin. J. Anat. 2019;234:438–455. doi: 10.1111/joa.12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Driskell RR, et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. 2013;504:277–281. doi: 10.1038/nature12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Kim BS, Gao G, Kim JY, Cho DW. 3D cell printing of perfusable vascularized human skin equivalent composed of epidermis, dermis, and hypodermis for better structural recapitulation of native skin. Adv. Healthc. Mater. 2019;8:e1801019. doi: 10.1002/adhm.201801019. [DOI] [PubMed] [Google Scholar]
- 375.Jiang Y, Lu S. Three-dimensional insights into dermal tissue as a cue for cellular behavior. Burns. 2014;40:191–199. doi: 10.1016/j.burns.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 376.Hurtley S, Hines PJ, Mueller KL, Culotta E. Skin. From bench to bedside. Introduction. Science. 2014;346:932–933. doi: 10.1126/science.346.6212.932. [DOI] [PubMed] [Google Scholar]
- 377.Khavkin J, Ellis DA. Aging skin: histology, physiology, and pathology. Facial Plast. Surg. Clin. North Am. 2011;19:229–234. doi: 10.1016/j.fsc.2011.04.003. [DOI] [PubMed] [Google Scholar]
- 378.Parna E, Aluoja A, Kingo K. Quality of life and emotional state in chronic skin disease. Acta Derm. Venereol. 2015;95:312–316. doi: 10.2340/00015555-1920. [DOI] [PubMed] [Google Scholar]
- 379.Mendes AL, Miot HA, Haddad VJ. Diabetes mellitus and the skin . Bras. Dermatol. 2017;92:8–20. doi: 10.1590/abd1806-4841.20175514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Robinson KP, Chan JJ. Colchicine in dermatology: a review. Australas. J. Dermatol. 2018;59:278–285. doi: 10.1111/ajd.12795. [DOI] [PubMed] [Google Scholar]
- 381.Morton LM, Phillips TJ. Wound healing and treating wounds: differential diagnosis and evaluation of chronic wounds. J. Am. Acad. Dermatol. 2016;74:589–605. doi: 10.1016/j.jaad.2015.08.068. [DOI] [PubMed] [Google Scholar]
- 382.Tottoli EM, et al. Skin wound healing process and new emerging technologies for skin wound care and regeneration. Pharmaceutics. 2020;12:735. doi: 10.3390/pharmaceutics12080735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Chouhan D, Dey N, Bhardwaj N, Mandal BB. Emerging and innovative approaches for wound healing and skin regeneration: current status and advances. Biomaterials. 2019;216:119267. doi: 10.1016/j.biomaterials.2019.119267. [DOI] [PubMed] [Google Scholar]
- 384.Wang C, et al. Engineering bioactive self-healing antibacterial exosomes hydrogel for promoting chronic diabetic wound healing and complete skin regeneration. Theranostics. 2019;9:65–76. doi: 10.7150/thno.29766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Chouhan D, Mandal BB. Silk biomaterials in wound healing and skin regeneration therapeutics: from bench to bedside. Acta Biomaterialia. 2020;103:24–51. doi: 10.1016/j.actbio.2019.11.050. [DOI] [PubMed] [Google Scholar]
- 386.Sapru S, Das S, Mandal M, Ghosh AK, Kundu SC. Prospects of nonmulberry silk protein sericin-based nanofibrous matrices for wound healing-In vitro and in vivo investigations. Acta Biomaterialia. 2018;78:137–150. doi: 10.1016/j.actbio.2018.07.047. [DOI] [PubMed] [Google Scholar]
- 387.Chouhan D, et al. Recombinant spider silk functionalized silkworm silk matrices as potential bioactive wound dressings and skin grafts. ACS Appl. Mater. Interfaces. 2018;10:23560–23572. doi: 10.1021/acsami.8b05853. [DOI] [PubMed] [Google Scholar]
- 388.Guo R, Xu S, Ma L, Huang A, Gao C. The healing of full-thickness burns treated by using plasmid DNA encoding VEGF-165 activated collagen-chitosan dermal equivalents. Biomaterials. 2011;32:1019–1031. doi: 10.1016/j.biomaterials.2010.08.087. [DOI] [PubMed] [Google Scholar]
- 389.Guo R, Xu S, Ma L, Huang A, Gao C. Enhanced angiogenesis of gene-activated dermal equivalent for treatment of full thickness incisional wounds in a porcine model. Biomaterials. 2010;31:7308–7320. doi: 10.1016/j.biomaterials.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 390.Yang Y, et al. Electrospun fibers with plasmid bFGF polyplex loadings promote skin wound healing in diabetic rats. Mol. Pharm. 2012;9:48–58. doi: 10.1021/mp200246b. [DOI] [PubMed] [Google Scholar]
- 391.Lou D, Luo Y, Pang Q, Tan WQ, Ma L. Gene-activated dermal equivalents to accelerate healing of diabetic chronic wounds by regulating inflammation and promoting angiogenesis. Bioact. Mater. 2020;5:667–679. doi: 10.1016/j.bioactmat.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Bao P, et al. The role of vascular endothelial growth factor in wound healing. J. Surg. Res. 2009;153:347–358. doi: 10.1016/j.jss.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Wang P, et al. In situ formed anti-inflammatory hydrogel loading plasmid DNA encoding VEGF for burn wound healing. Acta Biomaterialia. 2019;100:191–201. doi: 10.1016/j.actbio.2019.10.004. [DOI] [PubMed] [Google Scholar]
- 394.Yang Y, et al. Promotion of skin regeneration in diabetic rats by electrospun core-sheath fibers loaded with basic fibroblast growth factor. Biomaterials. 2011;32:4243–4254. doi: 10.1016/j.biomaterials.2011.02.042. [DOI] [PubMed] [Google Scholar]
- 395.He S, et al. Multiple release of polyplexes of plasmids VEGF and bFGF from electrospun fibrous scaffolds towards regeneration of mature blood vessels. Acta Biomaterialia. 2012;8:2659–2669. doi: 10.1016/j.actbio.2012.03.044. [DOI] [PubMed] [Google Scholar]
- 396.Losi P, et al. Fibrin-based scaffold incorporating VEGF- and bFGF-loaded nanoparticles stimulates wound healing in diabetic mice. Acta Biomaterialia. 2013;9:7814–7821. doi: 10.1016/j.actbio.2013.04.019. [DOI] [PubMed] [Google Scholar]
- 397.Mo Y, et al. Controlled dual delivery of angiogenin and curcumin by electrospun nanofibers for skin regeneration. Tissue Eng. Part A. 2017;23:597–608. doi: 10.1089/ten.tea.2016.0268. [DOI] [PubMed] [Google Scholar]
- 398.Szczesny G, et al. Bacteriology of callus of closed fractures of tibia and femur. J. Trauma. 2008;65:837–842. doi: 10.1097/TA.0b013e3181469d44. [DOI] [PubMed] [Google Scholar]
- 399.Wiraja C, et al. Framework nucleic acids as programmable carrier for transdermal drug delivery. Nat. Commun. 2019;10:1147. doi: 10.1038/s41467-019-09029-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Son J, et al. DNA aptamer immobilized hydroxyapatite for enhancing angiogenesis and bone regeneration. Acta Biomaterialia. 2019;99:469–478. doi: 10.1016/j.actbio.2019.08.047. [DOI] [PubMed] [Google Scholar]
- 401.Mao Z, et al. Enhanced angiogenesis of porous collagen scaffolds by incorporation of TMC/DNA complexes encoding vascular endothelial growth factor. Acta Biomaterialia. 2009;5:2983–2994. doi: 10.1016/j.actbio.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 402.Nillesen ST, et al. Increased angiogenesis and blood vessel maturation in acellular collagen-heparin scaffolds containing both FGF2 and VEGF. Biomaterials. 2007;28:1123–1131. doi: 10.1016/j.biomaterials.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 403.Nillesen ST, et al. Increased angiogenesis in acellular scaffolds by combined release of FGF2 and VEGF. J. Control. Release. 2006;116:e88–e90. doi: 10.1016/j.jconrel.2006.09.065. [DOI] [PubMed] [Google Scholar]
- 404.Qin X, et al. Tetrahedral framework nucleic acids prevent retina ischemia-reperfusion injury from oxidative stress via activating the Akt/Nrf2 pathway. Nanoscale. 2019;11:20667–20675. doi: 10.1039/C9NR07171G. [DOI] [PubMed] [Google Scholar]
- 405.Zhao D, et al. Tetrahedral DNA nanostructure promotes endothelial cell proliferation, migration, and angiogenesis via Notch signaling pathway. ACS Appl. Mater. Interfaces. 2018;10:37911–37918. doi: 10.1021/acsami.8b16518. [DOI] [PubMed] [Google Scholar]
- 406.Zhao D, et al. Tetrahedral framework nucleic acid promotes the treatment of bisphosphonate-related osteonecrosis of the jaws by promoting angiogenesis and M2 polarization. ACS Appl. Mater. interfaces. 2020;12:44508–44522. doi: 10.1021/acsami.0c13839. [DOI] [PubMed] [Google Scholar]
- 407.Cui W, et al. Preventive effect of tetrahedral framework nucleic acids on bisphosphonate-related osteonecrosis of the jaw. Nanoscale. 2020;12:17196–17202. doi: 10.1039/D0NR03731A. [DOI] [PubMed] [Google Scholar]
- 408.Willem de Vries J, et al. DNA nanoparticles for ophthalmic drug delivery. Biomaterials. 2018;157:98–106. doi: 10.1016/j.biomaterials.2017.11.046. [DOI] [PubMed] [Google Scholar]
- 409.Rodier JT, et al. Linear polyethylenimine-DNA nanoconstruct for corneal gene delivery. J. Ocul. Pharmacol. Ther. 2019;35:23–31. doi: 10.1089/jop.2018.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Trigueros S, Domenech EB, Toulis V, Marfany G. In vitro gene delivery in retinal pigment epithelium cells by plasmid dna-wrapped gold nanoparticles. Genes. 2019;10:289. doi: 10.3390/genes10040289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Gao S, et al. Tetrahedral framework nucleic acids induce immune tolerance and prevent the onset of type 1 diabetes. Nano Lett. 2021;21:4437–4446. doi: 10.1021/acs.nanolett.1c01131. [DOI] [PubMed] [Google Scholar]
- 412.Xia K, et al. Systematic study in mammalian cells showing no adverse response to tetrahedral DNA nanostructure. ACS Appl. Mater. Interfaces. 2018;10:15442–15448. doi: 10.1021/acsami.8b02626. [DOI] [PubMed] [Google Scholar]
- 413.Pei H, Zuo X, Zhu D, Huang Q, Fan C. Functional DNA nanostructures for theranostic applications. Acc. Chem. Res. 2014;47:550–559. doi: 10.1021/ar400195t. [DOI] [PubMed] [Google Scholar]
- 414.Liang L, et al. Single-particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells. Angew. Chem. Int. Ed. Engl. 2014;53:7745–7750. doi: 10.1002/anie.201403236. [DOI] [PubMed] [Google Scholar]
- 415.Yang D, Tan Z, Mi Y, Wei B. DNA nanostructures constructed with multi-stranded motifs. Nucleic Acids Res. 2017;45:3606–3611. doi: 10.1093/nar/gkx121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Bagalkot V, Farokhzad OC, Langer R, Jon S. An aptamer-doxorubicin physical conjugate as a novel targeted drug-delivery platform. Angew. Chem. Int. Ed. Engl. 2006;45:8149–8152. doi: 10.1002/anie.200602251. [DOI] [PubMed] [Google Scholar]
- 417.Erben CM, Goodman RP, Turberfield AJ. Single-molecule protein encapsulation in a rigid DNA cage. Angew. Chem. Int. Ed. 2006;45:7414–7417. doi: 10.1002/anie.200603392. [DOI] [PubMed] [Google Scholar]
- 418.Chou LY, Zagorovsky K, Chan WC. DNA assembly of nanoparticle superstructures for controlled biological delivery and elimination. Nat. Nanotechnol. 2014;9:148–155. doi: 10.1038/nnano.2013.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.He Y, et al. Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra. Nature. 2008;452:198–201. doi: 10.1038/nature06597. [DOI] [PubMed] [Google Scholar]
- 420.Iinuma R, et al. Polyhedra self-assembled from DNA tripods and characterized with 3D DNA-paint. Science. 2014;344:65–69. doi: 10.1126/science.1250944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Peng R, et al. Facile assembly/disassembly of DNA nanostructures anchored on cell-mimicking giant vesicles. J. Am. Chem. Soc. 2017;139:12410–12413. doi: 10.1021/jacs.7b07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Liu X, Yan H, Liu Y, Chang Y. Targeted cell-cell interactions by DNA nanoscaffold-templated multivalent bispecific aptamers. Small. 2011;7:1673–1682. doi: 10.1002/smll.201002292. [DOI] [PubMed] [Google Scholar]
- 423.Shi P, et al. In situ synthesis of an aptamer-based polyvalent antibody mimic on the cell surface for enhanced interactions between immune and cancer cells. Angew. Chem. Int. Ed. Engl. 2020;59:11892–11897. doi: 10.1002/anie.202004206. [DOI] [PubMed] [Google Scholar]
- 424.Yong-Xing Z, et al. DNA origami delivery system for cancer therapy with tunable release properties. ACS Nano. 2012;6:8684–8691. doi: 10.1021/nn3022662. [DOI] [PubMed] [Google Scholar]
- 425.Qian Z, et al. DNA origami as an in vivo drug delivery vehicle for cancer therapy. ACS Nano. 2014;8:6633–6643. doi: 10.1021/nn502058j. [DOI] [PubMed] [Google Scholar]
- 426.Zhang G, Zhang Z, Yang. J. DNA tetrahedron delivery enhances doxorubicin-induced apoptosis of HT-29 colon cancer cells. Nanoscale Res. Lett. 2017;12:495. doi: 10.1186/s11671-017-2272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Li S, et al. A DNA nanorobot functions as a cancer therapeutic in response to a molecular trigger in vivo. Nat. Biotechnol. 2018;36:258–264. doi: 10.1038/nbt.4071. [DOI] [PubMed] [Google Scholar]
- 428.Vindigni G, et al. Receptor-mediated entry of pristine octahedral DNA nanocages in mammalian cells. ACS Nano. 2016;10:5971–5979. doi: 10.1021/acsnano.6b01402. [DOI] [PubMed] [Google Scholar]
- 429.Chang M, Yang C-S, Huang. D-M. Aptamer-conjugated DNA icosahedral nanoparticles as a carrier of doxorubicin for cancer therapy. ACS Nano. 2011;5:6156–6163. doi: 10.1021/nn200693a. [DOI] [PubMed] [Google Scholar]
- 430.Taghdisi SM, et al. Double targeting and aptamer-assisted controlled release delivery of epirubicin to cancer cells by aptamers-based dendrimer in vitro and in vivo. Eur. J. Pharm. Biopharm. 2016;102:152–158. doi: 10.1016/j.ejpb.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 431.Lee H, et al. Molecularly self-assembled nucleic acid nanoparticles for targeted in vivo siRNA delivery. Nat. Nanotechnol. 2012;7:389–393. doi: 10.1038/nnano.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Liu X, et al. A DNA nanostructure platform for directed assembly of synthetic vaccines. Nano Lett. 2012;12:4254–4259. doi: 10.1021/nl301877k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Yang J, et al. Self-assembled double-bundle DNA tetrahedron for efficient antisense delivery. ACS Appl Mater. Interfaces. 2018;10:23693–23699. doi: 10.1021/acsami.8b07889. [DOI] [PubMed] [Google Scholar]
- 434.Kim K-R, et al. Sentinel lymph node imaging by a fluorescently labeled DNA tetrahedron. Biomaterials. 2013;34:5226–5235. doi: 10.1016/j.biomaterials.2013.03.074. [DOI] [PubMed] [Google Scholar]
- 435.Goodman RP, Berry RM, Turberfield AJ. The single-step synthesis of a DNA tetrahedron. Chem. Commun. (Camb) 2004;21:1372–1373. doi: 10.1039/b402293a. [DOI] [PubMed] [Google Scholar]
- 436.Li Z, et al. A replicable tetrahedral nanostructure self-assembled from a single DNA strand. J. Am. Chem. Soc. 2009;131:13093–13098. doi: 10.1021/ja903768f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Zhang Y, et al. DNA-affibody nanoparticles for inhibiting breast cancer cells overexpressing HER2. Chem. Commun. (Camb.) 2017;53:573–576. doi: 10.1039/C6CC08495H. [DOI] [PubMed] [Google Scholar]
- 438.Sun P, et al. SL2B aptamer and folic acid dual-targeting DNA nanostructures for synergic biological effect with chemotherapy to combat colorectal cancer. Int. J. Nanomed. 2017;12:2657–2672. doi: 10.2147/IJN.S132929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 439.Ma W, et al. An intelligent DNA nanorobot with in vitro enhanced protein lysosomal degradation of HER2. Nano Lett. 2019;19:4505–4517. doi: 10.1021/acs.nanolett.9b01320. [DOI] [PubMed] [Google Scholar]
- 440.Loibl S, Gianni L. HER2-positive breast cancer. Lancet. 2017;389:2415–2429. doi: 10.1016/S0140-6736(16)32417-5. [DOI] [PubMed] [Google Scholar]
- 441.Hyojin Lee D, et al. Enhanced human epidermal growth factor receptor 2 degradation in breast cancer cells by lysosome-targeting gold nanoconstructs. ACS Nano. 2015;9:9859–9867. doi: 10.1021/acsnano.5b05138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Mahlknecht G, et al. Aptamer to ErbB-2/HER2 enhances degradation of the target and inhibits tumorigenic growth. Proc. Natl Acad. Sci. USA. 2013;110:8170–8175. doi: 10.1073/pnas.1302594110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009;86:151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Reyes-Reyes EM, Salipur FR, Shams M, Forsthoefel MK, Bates PJ. Mechanistic studies of anticancer aptamer AS1411 reveal a novel role for nucleolin in regulating Rac1 activation. Mol. Oncol. 2015;9:1392–1405. doi: 10.1016/j.molonc.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Charoenphol P, Bermudez H. Aptamer-targeted DNA nanostructures for therapeutic delivery. Mol. Pharm. 2014;11:1721–1725. doi: 10.1021/mp500047b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Xia Z, et al. Tumor-penetrating peptide-modified DNA tetrahedron for targeting drug delivery. Biochemistry. 2016;55:1326–1331. doi: 10.1021/acs.biochem.5b01181. [DOI] [PubMed] [Google Scholar]
- 447.Setyawati MI, Kutty RV, Leong DT. DNA nanostructures carrying stoichiometrically definable antibodies. Small. 2016;12:5601–5611. doi: 10.1002/smll.201601669. [DOI] [PubMed] [Google Scholar]
- 448.Miao P, Wang B, Chen X, Li X, Tang Y. Tetrahedral DNA nanostructure-based microRNA biosensor coupled with catalytic recycling of the analyte. ACS Appl Mater. Interfaces. 2015;7:6238–6243. doi: 10.1021/acsami.5b01508. [DOI] [PubMed] [Google Scholar]
- 449.Liu X, Wu L, Wang L, Jiang W. A dual-targeting DNA tetrahedron nanocarrier for breast cancer cell imaging and drug delivery. Talanta. 2018;179:356–363. doi: 10.1016/j.talanta.2017.11.034. [DOI] [PubMed] [Google Scholar]
- 450.Fu W, et al. Enhanced efficacy of temozolomide loaded by a tetrahedral framework DNA nanoparticle in the therapy for glioblastoma. ACS Appl Mater. Interfaces. 2019;11:39525–39533. doi: 10.1021/acsami.9b13829. [DOI] [PubMed] [Google Scholar]
- 451.Chen CH, et al. Aptamer-based endocytosis of a lysosomal enzyme. Proc. Natl Acad. Sci. USA. 2008;105:15908–15913. doi: 10.1073/pnas.0808360105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Tian Y, Huang Y, Gao P, Chen T. Nucleus-targeted DNA tetrahedron as a nanocarrier of metal complexes for enhanced glioma therapy. Chem. Commun. (Camb.) 2018;54:9394–9397. doi: 10.1039/C8CC04021D. [DOI] [PubMed] [Google Scholar]
- 453.Bugaj AM. Targeted photodynamic therapy–a promising strategy of tumor treatment. Photochem Photobio. Sci. 2011;10:1097–1109. doi: 10.1039/c0pp00147c. [DOI] [PubMed] [Google Scholar]
- 454.Kim K-R, Bangb D, Ahn. D-R. Nano-formulation of a photosensitizer using a DNA tetrahedron and its potential for in vivo photodynamic therapy. Biomater. Sci. 2016;4:605–609. doi: 10.1039/C5BM00467E. [DOI] [PubMed] [Google Scholar]
- 455.Zhuang X, et al. A photosensitizer-loaded DNA origami nanosystem for photodynamic therapy. ACS Nano. 2016;10:3486–3495. doi: 10.1021/acsnano.5b07671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Bujold KE, Hsu JCC, Sleiman HF. Optimized DNA “nanosuitcases” for encapsulation and conditional release of siRNA. J. Am. Chem. Soc. 2016;138:14030–14038. doi: 10.1021/jacs.6b08369. [DOI] [PubMed] [Google Scholar]
- 457.Ren K, et al. A DNA dual lock-and-key strategy for cell-subtype-specific siRNA delivery. Nat. Commun. 2016;7:13580. doi: 10.1038/ncomms13580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Zhang Q, et al. Anti-inflammatory and antioxidative effects of tetrahedral dna nanostructures via the modulation of macrophage responses. ACS Appl. Mater. interfaces. 2018;10:3421–3430. doi: 10.1021/acsami.7b17928. [DOI] [PubMed] [Google Scholar]
- 459.Zhang Y, et al. Inhibiting methicillin-resistant Staphylococcus aureus by tetrahedral DNA nanostructure-enabled antisense peptide nucleic acid delivery. Nano Lett. 2018;18:5652–5659. doi: 10.1021/acs.nanolett.8b02166. [DOI] [PubMed] [Google Scholar]
- 460.Zhang Y, et al. Multi-targeted antisense oligonucleotide delivery by a framework nucleic acid for inhibiting biofilm formation and virulence. Nano-Micro Lett. 2020;12:74. doi: 10.1007/s40820-020-0409-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Gerling T, Wagenbauer KF, Neuner AM, Dietz H. Dynamic DNA devices and assemblies formed by shape-complementary, non-base pairing 3D components. Science. 2015;347:1446–1452. doi: 10.1126/science.aaa5372. [DOI] [PubMed] [Google Scholar]
- 462.Wu C, et al. Building a multifunctional aptamer-based DNA nanoassembly for targeted cancer therapy. J. Am. Chem. Soc. 2013;135:18644–18650. doi: 10.1021/ja4094617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Zhu G, et al. Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics. Proc. Natl Acad. Sci. USA. 2013;110:7998–8003. doi: 10.1073/pnas.1220817110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Lv Y, et al. Preparation and biomedical applications of programmable and multifunctional DNA nanoflowers. Nat. Protoc. 2015;10:1508–1524. doi: 10.1038/nprot.2015.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Da H, et al. Engineering a cell-surface aptamer circuit for targeted and amplified photodynamic cancer therapy. ACS Nano. 2013;7:2312–2319. doi: 10.1021/nn305484p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Li W, et al. Self-assembled DNA nanocentipede as multivalent drug carrier for targeted delivery. ACS Appl. Mater. Interfaces. 2016;8:25733–25740. doi: 10.1021/acsami.6b08210. [DOI] [PubMed] [Google Scholar]
- 467.Zadegan RM, et al. Construction of a 4 zeptoliters switchable 3D DNA box origami. ACS Nano. 2012;6:10050–10053. doi: 10.1021/nn303767b. [DOI] [PubMed] [Google Scholar]
- 468.Douglas SM, Bachelet I, Church. GM. A logic-gated nanorobot for targeted transport of molecular payloads. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 469.Wang Y, Jiang LP, Zhou S, Bi S, Zhu JJ. DNA polymerase-directed hairpin assembly for targeted drug delivery and amplified biosensing. ACS Appl Mater. Interfaces. 2016;8:26532–26540. doi: 10.1021/acsami.6b08597. [DOI] [PubMed] [Google Scholar]
- 470.Bi S, Xiu B, Ye J, Dong Y. Target-catalyzed DNA four-way junctions for cret imaging of microrna, concatenated logic operations, and self-assembly of DNA nanohydrogels for targeted drug delivery. ACS Appl Mater. Interfaces. 2015;7:23310–23319. doi: 10.1021/acsami.5b07827. [DOI] [PubMed] [Google Scholar]
- 471.Li J, et al. Self-assembly of DNA nanohydrogels with controllable size and stimuli-responsive property for targeted gene regulation therapy. J. Am. Chem. Soc. 2015;137:1412–1415. doi: 10.1021/ja512293f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Daljit Singh JK, Luu MT, Abbas A, Wickham SFJ. Switchable DNA-origami nanostructures that respond to their environment and their applications. Biophys. Rev. 2018;10:1283–1293. doi: 10.1007/s12551-018-0462-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 473.Endo M, Sugiyama H. DNA origami nanomachines. Molecules. 2018;23:1766. doi: 10.3390/molecules23071766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Kuzuya A, Sakai Y, Yamazaki T, Xu Y, Komiyama M. Nanomechanical DNA origami ‘single-molecule beacons’ directly imaged by atomic force microscopy. Nat. Commun. 2011;2:449. doi: 10.1038/ncomms1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Bernard Y, et al. A DNA-fuelled molecular machine made of DNA. nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 476.Kuzuya A, et al. Nanomechanical DNA Origami pH. Sens. Sens. 2014;14:19329–19335. doi: 10.3390/s141019329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Zadegan RM, Jepsen MD, Hildebrandt LL, Birkedal V, Kjems J. Construction of a fuzzy and Boolean logic gates based on DNA. Small. 2015;11:1811–1817. doi: 10.1002/smll.201402755. [DOI] [PubMed] [Google Scholar]
- 478.Amir Y, et al. Universal computing by DNA origami robots in a living animal. Nat. Nanotechnol. 2014;9:353–357. doi: 10.1038/nnano.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Bao-an C, et al. Reversal of multidrug resistance by magnetic Fe3O4 nanoparticle copolymerizating daunorubicin and MDr1 shrNA expression vector in leukemia cells. Int. J. Nanomed. 2010;5:437–444. doi: 10.2147/ijn.s10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Mariagrazia M, et al. P-glycoprotein (PGP), lung resistance-related protein (LRP) and multidrug resistance-associated protein (MRP) expression in acute promyelocytic leukaemia. Br. J. Haematol. 2000;108:703–709. doi: 10.1046/j.1365-2141.2000.01957.x. [DOI] [PubMed] [Google Scholar]
- 481.Morris SA, Farrell D, Grodzinski P. Nanotechnologies in cancer treatment and diagnosis. J. Natl Compr. Cancer Netw. 2014;12:1727–1733. doi: 10.6004/jnccn.2014.0175. [DOI] [PubMed] [Google Scholar]
- 482.Qiao J, et al. DNA origami as a carrier for circumvention of drug resistance. J. Am. Chem. Soc. 2012;134:13396–13403. doi: 10.1021/ja3003183. [DOI] [PubMed] [Google Scholar]
- 483.Liu J, et al. Multifunctional aptamer-based nanoparticles for targeted drug delivery to circumvent cancer resistance. Biomaterials. 2016;91:44–56. doi: 10.1016/j.biomaterials.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 484.Mei L, et al. Self-assembled multifunctional DNA nanoflowers for the circumvention of multidrug resistance in targeted anticancer drug delivery. Nano Res. 2015;8:3447–3460. doi: 10.1007/s12274-015-0841-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 485.Fu J, et al. DNA-scaffolded proximity assembly and confinement of multienzyme reactions. Top. Curr. Chem. (Cham) 2020;378:38. doi: 10.1007/s41061-020-0299-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Hiroaki H, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 487.Arthur MK, et al. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- 488.Gupta GK, Agrawal DK. CpG oligodeoxynucleotides as TLR9 agonists therapeutic application in allergy and asthma. Biodrugs. 2010;24:225–235. doi: 10.2165/11536140-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 489.Vollmer J, Krieg AM. Immunotherapeutic applications of CpG oligodeoxynucleotide TLR9 agonists. Adv. Drug Deliv. Rev. 2009;61:195–204. doi: 10.1016/j.addr.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 490.Schu ¨ ller VJ, et al. Cellular immunostimulation by CpG-sequence-coated DNA origami structures. ACS Nano. 2011;5:9696–9702. doi: 10.1021/nn203161y. [DOI] [PubMed] [Google Scholar]
- 491.Sellner S, et al. DNA nanotubes as intracellular delivery vehicles in vivo. Biomaterials. 2015;53:453–463. doi: 10.1016/j.biomaterials.2015.02.099. [DOI] [PubMed] [Google Scholar]
- 492.Rattanakiat S, Nishikawa M, Funabashi H, Luo D, Takakura Y. The assembly of a short linear natural cytosine-phosphate-guanine DNA into dendritic structures and its effect on immunostimulatory activity. Biomaterials. 2009;30:5701–5706. doi: 10.1016/j.biomaterials.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 493.Mohri K, et al. Self-assembling DNA dendrimer for effective delivery of immunostimulatory CpG DNA to immune cells. Biomacromolecules. 2015;16:1095–1101. doi: 10.1021/bm501731f. [DOI] [PubMed] [Google Scholar]
- 494.Jiang LI, et al. Self-assembled multivalent DNA nanostructures for noninvasive intracellular delivery of immunostimulatory CpG oligonucleotides. ACS Nano. 2011;5:8783–8789. doi: 10.1021/nn202774x. [DOI] [PubMed] [Google Scholar]
- 495.Ouyang X, et al. Rolling circle amplification-based DNA origami nanostructrures for intracellular delivery of immunostimulatory drugs. Small. 2013;9:3082–3087. doi: 10.1002/smll.201300458. [DOI] [PubMed] [Google Scholar]
- 496.de Vries JW, Zhang F, Herrmann A. Drug delivery systems based on nucleic acid nanostructures. J. Control Release. 2013;172:467–483. doi: 10.1016/j.jconrel.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 497.Nishikawa M, Matono M, Rattanakiat S, Matsuoka N, Takakura Y. Enhanced immunostimulatory activity of oligodeoxynucleotides by Y-shape formation. Immunology. 2008;124:247–255. doi: 10.1111/j.1365-2567.2007.02762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.Kohta M, et al. Design and development of nanosized DNA assemblies in polypod-like structures as efficient vehicles for immunostimulatory CpG motifs to immune cells. ACS Nano. 2012;6:5931–5940. doi: 10.1021/nn300727j. [DOI] [PubMed] [Google Scholar]
- 499.Hu J, Liu MH, Zhang CY. Construction of tetrahedral DNA-quantum dot nanostructure with the integration of multistep forster resonance energy transfer for multiplex enzymes assay. ACS Nano. 2019;13:7191–7201. doi: 10.1021/acsnano.9b02679. [DOI] [PubMed] [Google Scholar]
- 500.Ma F, Zhang Q, Zhang CY. Catalytic self-assembly of quantum-dot-based MicroRNA nanosensor directed by toehold-mediated strand displacement cascade. Nano Lett. 2019;19:6370–6376. doi: 10.1021/acs.nanolett.9b02544. [DOI] [PubMed] [Google Scholar]
- 501.Shiu SC, Fraser LA, Ding Y, Tanner JA. Aptamer display on diverse DNA polyhedron supports. Molecules. 2018;23:1695. doi: 10.3390/molecules23071695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 502.Tian T, et al. Targeted imaging of brain tumors with a framework nucleic acid probe. ACS Appl. Mater. Interfaces. 2018;10:3414–3420. doi: 10.1021/acsami.7b17927. [DOI] [PubMed] [Google Scholar]
- 503.Zhang K, et al. DNA tetrahedron based biosensor for argonaute2 assay in single cells and human immunodeficiency virus type-1 related ribonuclease H detection in vitro. Anal. Chem. 2019;91:7086–7096. doi: 10.1021/acs.analchem.9b00011. [DOI] [PubMed] [Google Scholar]
- 504.Li N, et al. A DNA tetrahedron nanoprobe with controlled distance of dyes for multiple detection in living cells and in vivo. Anal. Chem. 2017;89:6670–6677. doi: 10.1021/acs.analchem.7b00889. [DOI] [PubMed] [Google Scholar]
- 505.Li C, et al. Design of DNA nanostructure-based interfacial probes for the electrochemical detection of nucleic acids directly in whole blood. Chem. Sci. 2018;9:979–984. doi: 10.1039/C7SC04663D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 506.Jiang D, et al. Multiple-armed tetrahedral DNA nanostructures for tumor-targeting, dual-modality in vivo imaging. ACS Appl Mater. Interfaces. 2016;8:4378–4384. doi: 10.1021/acsami.5b10792. [DOI] [PubMed] [Google Scholar]
- 507.Feng QM, Zhu MJ, Zhang TT, Xu JJ, Chen HY. A novel DNA tetrahedron-hairpin probe for in situ“off-on” fluorescence imaging of intracellular telomerase activity. Analyst. 2016;141:2474–2480. doi: 10.1039/C6AN00241B. [DOI] [PubMed] [Google Scholar]
- 508.Bi X, Yin J, Chen Guanbang A, Liu CF. Chemical and enzymatic strategies for bacterial and mammalian cell surface engineering. Chemistry (Easton) 2018;24:8042–8050. doi: 10.1002/chem.201705049. [DOI] [PubMed] [Google Scholar]
- 509.Aghebat Rafat A, Sagredo S, Thalhammer M, Simmel FC. Barcoded DNA origami structures for multiplexed optimization and enrichment of DNA-based protein-binding cavities. Nat. Chem. 2020;12:852–859. doi: 10.1038/s41557-020-0504-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Gartner ZJ, Bertozzi CR. Programmed assembly of 3-dimensional microtissues with defined cellular connectivity. Proc. Natl Acad. Sci. USA. 2009;106:4606–4610. doi: 10.1073/pnas.0900717106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 511.Akbari, E. et al. Engineering cell surface function with DNA origami. Adv. Mater.29, 10.1002/adma.201703632(2017). [DOI] [PMC free article] [PubMed]
- 512.Sacca B, et al. Orthogonal protein decoration of DNA origami. Angew. Chem. Int. Ed. Engl. 2010;49:9378–9383. doi: 10.1002/anie.201005931. [DOI] [PubMed] [Google Scholar]
- 513.Li H, Han M, Weng X, Zhang Y, Li J. DNA-tetrahedral-nanostructure-based entropy-driven amplifier for high-performance photoelectrochemical biosensing. ACS Nano. 2021;15:1710–1717. doi: 10.1021/acsnano.0c09374. [DOI] [PubMed] [Google Scholar]
- 514.Kizer ME, Linhardt RJ, Chandrasekaran AR, Wang X. A Molecular hero suit for in vitro and in vivo dna nanostructures. Small. 2019;15:e1805386. doi: 10.1002/smll.201805386. [DOI] [PubMed] [Google Scholar]
- 515.Tian T, et al. Proteomic exploration of endocytosis of framework nucleic acids. Small. 2021;17:e2100837. doi: 10.1002/smll.202100837. [DOI] [PubMed] [Google Scholar]
- 516.Li J, Fan C, Pei H, Shi J, Huang Q. Smart drug delivery nanocarriers with self-assembled DNA nanostructures. Adv. Mater. 2013;25:4386–4396. doi: 10.1002/adma.201300875. [DOI] [PubMed] [Google Scholar]
- 517.Walsh AS, Yin H, Erben CM, Wood MJ, Turberfield AJ. DNA cage delivery to mammalian cells. ACS Nano. 2011;5:5427–5432. doi: 10.1021/nn2005574. [DOI] [PubMed] [Google Scholar]
- 518.Yuan Y, Gu Z, Yao C, Luo D, Yang D. Nucleic acid-based functional nanomaterials as advanced cancer therapeutics. Small. 2019;15:e1900172. doi: 10.1002/smll.201900172. [DOI] [PubMed] [Google Scholar]
- 519.Sun Y, et al. Erythromycin loaded by tetrahedral framework nucleic acids are more antimicrobial sensitive against Escherichia coli (E. coli) Bioact. Mater. 2021;6:2281–2290. doi: 10.1016/j.bioactmat.2020.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Birkholz O, et al. Multi-functional DNA nanostructures that puncture and remodel lipid membranes into hybrid materials. Nat. Commun. 2018;9:1521. doi: 10.1038/s41467-018-02905-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.De Leo V, Milano F, Agostiano A, Catucci L. Recent advancements in polymer/liposome assembly for drug delivery: from surface modifications to hybrid vesicles. Polymers. 2021;13:1027. doi: 10.3390/polym13071027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Bhalla N, Pan Y, Yang Z, Payam AF. Opportunities and challenges for biosensors and nanoscale analytical tools for pandemics: COVID-19. ACS Nano. 2020;14:7783–7807. doi: 10.1021/acsnano.0c04421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 523.Shen Q, Grome MW, Yang Y, Lin C. Engineering lipid membranes with programmable DNA nanostructures. Adv. Biosyst. 2020;4:1900215. doi: 10.1002/adbi.201900215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Ji X, et al. Epigenetic remodeling hydrogel patches for multidrug-resistant triple-negative breast cancer. Adv. Mater. 2021;33:e2100949. doi: 10.1002/adma.202100949. [DOI] [PubMed] [Google Scholar]
- 525.Zhou Y, et al. An organelle-specific nanozyme for diabetes care in genetically or diet-induced models. Adv. Mater. 2020;32:e2003708. doi: 10.1002/adma.202003708. [DOI] [PubMed] [Google Scholar]
- 526.Han J, Cui Y, Li F, Gu Z, Yang D. Responsive disassembly of nucleic acid nanocomplex in cells for precision medicine. Nano Today. 2021;39:101160. doi: 10.1016/j.nantod.2021.101160. [DOI] [Google Scholar]