Abstract
The progression of colorectal cancer (CRC) has been well studied and understood with the development of molecular and genetic techniques. However, specific marker(s) that could be used to predict lymph node (LN) involvement, which is the most important prognostic factor for CRC, have not been identified so far. Our previous study, in which network analysis of LN(+) and LN(−) CRC gene expression was carried out with data obtained from the Cancer Genome Atlas, led to the identification of AHA1. AHA1 is a co-chaperone activator of the Hsp90 ATPase activity. However, the role of AHA1 expression in cancer cells is still unclear. To investigate how AHA1 expression regulates the cancer cell progression and/or metastasis of human CRC, the expression levels of AHA1 and Hsp90 were examined in 105 CRC tissue samples and compared with those in paired normal tissue. The RNA expression levels of AHA1 and Hsp90aa1, but not Hsp90ab, were significantly higher in cancer tissues than in adjacent paired normal tissues (p = 0.032 and p = 0.0002, respectively). In particular, AHA1, but not Hsp90aa1 and Hsp90ab, was closely associated with the TNM stage, LN stage, and tumor metastasis (p = 0.035, p = 0.012, and p = 0.0003, respectively). Moreover, the expression of AHA1 was not only higher in the CRC cell lines than in the normal colon fibroblast cell line but was also associated with the progression of these CRC cell lines. Overexpression of AHA1 in SW480 cells increased, whereas suppression of AHA1 expression in HCT116 cells reduced cell migration and invasion through the regulation of Snail, E-cadherin, pSRC, and pAKT, which are associated with EMT signaling. Taken together, our study suggests that AHA1 contributes to the metastatic advantage of human CRC.
Subject terms: Cell biology, Biomarkers, Oncology, Cancer, Gastrointestinal cancer, Metastasis, Oncogenes, Tumour biomarkers
Introduction
Colorectal cancer (CRC) is a high incidence and high prevalence disease worldwide, which influence on human health1. The development and progression of CRC from adenoma into cancer are systemic process, which has been well understood2. With advances in molecular and genetic technologies, our understanding of the molecular mechanisms through which genetic changes, such as genomic instability and alterations, in DNA lead to a normal mucosa turning into CRC, has expanded3. Molecular genetic studies in CRC, in respect of tumorigenesis, have discovered several meaningful genes and pathways that can be used in the early diagnosis of CRC. Nevertheless, the tumor node metastasis (TNM) stage is still major prognostic factor in CRC4,5. Stage II and III cancers are primarily differentiated based on nodal (N) stage, pointing out the importance of lymph node (LN) involvement in tumor prognosis. Currently, the N stage is determined by the pathological examination of LNs obtained during surgery. However, sometimes pathological examination, even though its usefulness, can make mistake by under-staging, resulting in lost opportunity for adjuvant chemotherapy, and a higher risk of tumor recurrence in patient6,7. This makes searching other factor or/and method for prediction or diagnosis of lymph node involvement extremely important for patient care.
In our previous study, we explored the possible diagnostic and/or prognostic marker(s) that might help investigate LN involvement in CRC, by comparing the networks of gene expression in LN(+) and LN(−) CRC using datasets from the Cancer Genome Atlas (TCGA) database (https://cancergenome.nih.gov/), and identified significantly different genes in the gene networks8. One of the genes, namely AHSA2, which belongs to the activator of the Hsp90 ATPase (AHA) family and is also known as a pseudogene, did not show any difference in expression between LN(−) and LN(+) in the subsequent investigation of the samples. However, another AHA family gene, namely AHA1 (Ahsa1), which encodes protein that can activate the ATPase activity of Hsp90 as a co-chaperone9,10 was found to be upregulated in the LN(+) group compared to the LN(−) group. However, the role of AHA1 in cancer, including CRC is very limited and still unclear. In this study, we investigated how AHA1 affects cell migration, invasion, and the epithelial mesenchymal transition (EMT) signaling pathway in CRC cells, as well as whether AHA1 expression correlates with clinical pathological characteristics such as TNM stages and MIS, to investigate its potential as a prognostic marker of CRC.
Results
AHA1 is upregulated in progressive and metastatic CRC patients
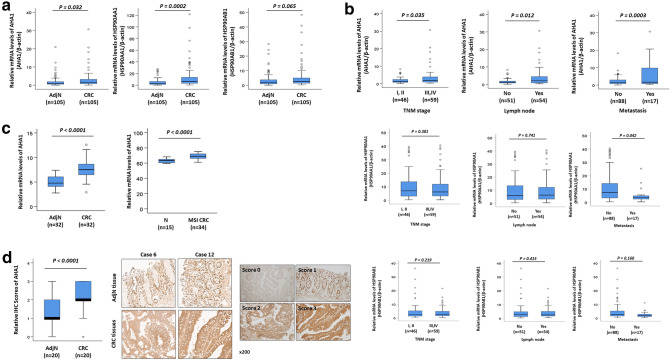
To confirm the mRNA expression levels of AHA1, Hsp90aa1, and Hsp90ab1 in CRC patients, qRT-PCR of these genes was performed for 105 paired CRC and adjacent normal tissues. The results showed that the expression of AHA1 and Hsp90aa1 was significantly increased in CRC tissues compared to that in adjacent normal tissues (p = 0.032, p = 0.002, respectively. Figure 1a). Further statistical analysis revealed that the mRNA expression of AHA1, but neither Hsp90aa1 nor Hsp90ab1, is significantly correlated with the clinicopathological characteristics such as TNM stage (p = 0.035), lymph node metastasis (p = 0.012), and metastasis (p = 0.0003) (Fig. 1b, Table 1). In addition, mRNA expression of AHA1 was analyzed using the GSE8671 and GSE24514 datasets11,12. The mRNA expression of AHA1 was significantly increased in 32 CRC tissues compared to that in 32 paired normal colonic mucosa tissues from the GSE8671 datasets, as well as in 32 MSI CRC tissues, compared to that in 15 normal colonic mucosa tissues from the GSE24514 datasets (Fig. 1c). Results obtained from IHC staining of AHA1 in CRC and adjacent normal tissues from 20 CRC patients were consistent with the mRNA expression data, in which AHA1 expression was significantly increased in tumors (average score = 2.15) compared to that in adjacent normal tissues (average score = 1.35, p < 0.0001) (Fig. 1d). However, survival analysis using public dataset of colorectal adenocarcinoma did not showed significant association of AHA1, HSP90AA1, and HSP90AB1 RNA expression level with survival (Supplementary Fig. S1)13. These results suggest that AHA1 may plays an important role in CRC progression and metastasis, however further investigation is necessary.
Figure 1.
AHA1 is up-regulated in progressive and metastatic CRC patients. (a) The mRNA expression of AHA1 and HSP90AA1, but not of HSP90AB1, was significantly higher in CRC tissues compared to that in paired adjacent normal tissues. (b) The higher mRNA expression of AHA1, but not HSP90AA1 and HSP90AB1, was associated with the progressive clinical characteristics, such as the TNM stage, LN involvement, and metastasis, in CRC patients. (c) The mRNA expression of AHA1 was significantly higher in 32 CRC tissues compared to that in 32 paired normal colonic mucosa tissues in GSE8671 datasets, as well as in 32 MSI CRC tissues, compared to that in normal colonic mucosa tissues in GSE24514 datasets. (d) AHA1 expression was higher in CRC tissues, compared to that in adjacent normal tissues, which was scored by the scoring system after IHC stating and quantified by assessing staining intensity using Leica’s Aperio ImagScope program (V12.4.0.5043, IL, USA). Representative IHC-stained photomicrograph images with AHA1 shown in CRC tissues, compared to adjacent normal tissues. AdjN adjacent normal specimen, CRC colorectal cancer specimen. Original magnification is ×200.
Table 1.
Clinicopathological characteristics of CRC patients, and the mRNA expression levels of genes.
| Characteristics | No. of cases | mRNA expression of Aha1 (%) | mRNA expression of HSP90AA1 (%) | mRNA expression of HSP90AB1 (%) | |||
|---|---|---|---|---|---|---|---|
| Median (range) | p-value | Median (range) | p-value | Median (range) | p-value | ||
| Adjacent normal | 105 | 1.80 (± 0.27) | 0.032 | 5.17 (± 0.61) | < 0.001 | 3.44 (± 0.45) | 0.065 |
| Colorectal cancer | 105 | 2.89 (± 0.42) | 12.50 (± 1.83) | 5.09 (± 0.76) | |||
| Age (years) | 0.278 | 0.641 | 0.669 | ||||
| ≤ 60 | 28 | 3.65 (± 1.11) | 11.07 (± 2.66) | 4.54 (± 1.35) | |||
| > 60 | 77 | 2.61 (± 0.42) | 13.02 (± 2.31) | 5.28 (± 0.92) | |||
| Gender | 0.172 | 0.203 | 0.455 | ||||
| Female | 39 | 3.64 (± 0.92) | 15.54 (± 3.34) | 5.83 (± 1.24) | |||
| Male | 66 | 2.44 (± 0.39) | 10.70 (± 2.13) | 4.64 (± 0.98) | |||
| Differentiation | 0.554 | 0.846 | 0.814 | ||||
| Well | 29 | 2.20 (± 0.41) | 14.07 (± 4.23) | 5.86 (± 1.84) | |||
| Moderate | 73 | 3.19 (± 0.58) | 11.79 (± 2.02) | 4.76 (± 0.83) | |||
| Poorly | 3 | 1.99 (± 1.27) | 14.47 (± 11.75) | 5.47 (± 3.73) | |||
| Location | 0.739 | 0.621 | 0.983 | ||||
| Colon | 72 | 2.79 (± 0.44) | 11.88 (± 2.08) | 5.10 (± 1.01) | |||
| Rectum | 33 | 3.09 (± 0.95) | 13.84 (± 3.68) | 5.06 (± 1.07) | |||
| Size (cm) | 0.072 | 0.48 | 0.242 | ||||
| ≤ 6 | 77 | 2.42 (± 0.42) | 15.62 (± 2.26) | 7.20 (± 1.01) | |||
| > 6 | 28 | 4.14 (± 1.08) | 12.15 (± 2.96) | 3.60 (± 0.66) | |||
| TNM stage | 0.035 | 0.34 | 0.487 | ||||
| I, II | 46 | 1.88 (± 0.28) | 14.49 (± 3.02) | 5.69 (± 1.27) | |||
| III, IV | 59 | 3.67 (± 0.71) | 10.95 (± 2.25) | 4.61 (± 0.94) | |||
| Lymp node | 0.012 | 0.6 | 0.745 | ||||
| 0 | 51 | 1.80 (± 0.26) | 13.49 (± 2.76) | 5.34 (± 1.15) | |||
| 1, 2 | 54 | 3.91 (± 0.77) | 11.56 (± 2.44) | 4.84 (± 1.02) | |||
| Invasion | 0.199 | 0.253 | 0.252 | ||||
| No | 85 | 2.62 (± 0.39) | 13.52 (± 2.21) | 5.51 (± 0.93) | |||
| Yes | 20 | 4.01 (± 1.50) | 8.17 (± 1.82) | 3.27 (± 0.62) | |||
| Metastasis | < 0.001 | 0.092 | 0.179 | ||||
| No | 88 | 2.23 (± 0.26) | 13.85 (± 2.14) | 5.54 (± 0.90) | |||
| Yes | 17 | 6.28 (± 2.10) | 5.49 (± 1.51) | 2.74 (± 0.65) | |||
TNM tumor, lymph node, metastasis.
High expression of AHA1 correlates with progressiveness of colon cancer cells
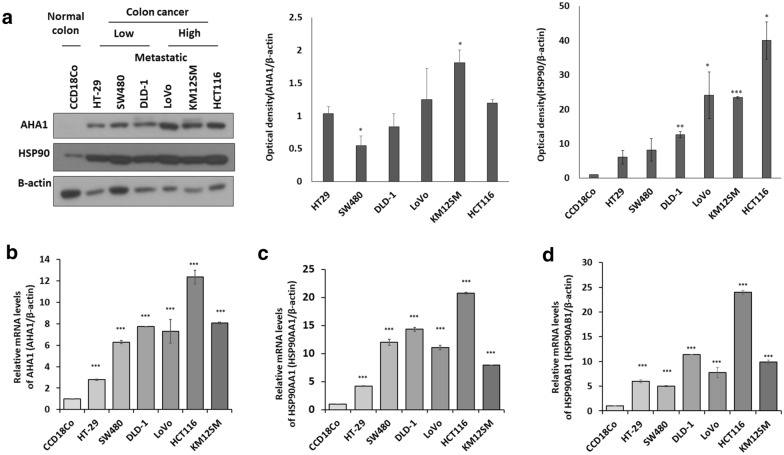
The endogenous expression levels of AHA1, Hsp90aa1, and Hsp90ab1 were examined in normal colon fibroblast cells (CCD18Co), early-stage colon cancer cells (HT-29), middle grade colon cancer cells (SW480, DLD-1), and highly metastatic colon cancer cells (Lovo, KM12SA, and HCT-116), using western blot and qRT-PCR analyses. The results showed that not only did all colon cancer cells have increased AHA1 expression compared to normal colon fibroblast cells (CCD18Co) but also the tendency of more progressive colon cancer cells to have a higher expression of AHA1 (Fig. 2a,b). On the other hand, the expression of Hsp90aa1 and Hsp90ab1 increased in colon cancer cells, regardless of the cancer grade, compared to that in normal colon fibroblast cells (CCD18Co) (Fig. 2c,d).
Figure 2.
Expression of AHA1 is associated with progressiveness of colon cancer cells. (a) The protein expression of AHA1 and HSP90 was higher in the CRC cells than in normal colon fibroblast cells (CCD18Co), as well as in high metastatic CRC cells (KM125M, and HCC116) than in low metastatic CRC cells (SW480, and DLD-1), which was examined with western blotting. The densitometry quantification of the western blot determined using Image J software (Ver. 1/52n, NIH). (b–d) The mRNA expression of AHA1, HSP90AA1, and HSP90AB1, was higher in the CRC cells than in normal colon fibroblast cells (CCD18Co), which was detected with qRT-PCR. *p < 0.05, **p < 0.01, ***p < 0.001 compared with control.
AHA1 controls the migration and invasion of colon cancer cells
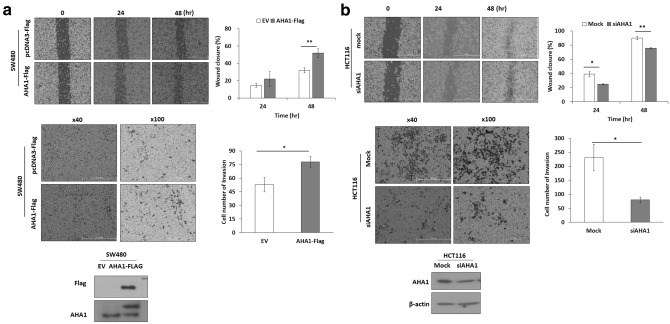
To investigate whether AHA1 affects the proliferation, migration, and invasion of colon cancer cells, cell counting, wound healing, and transwell invasion assays were performed in SW480 and HCT116 cells with different expression levels of AHA1. To verify that AHA1 enhances cell migration and invasion abilities, overexpression of AHA1 by exogenously introduced AHA1-flag in SW480 cells, and knockdown of AHA1 by siAHA1 in HCT116 cells, were performed. SW480 cells with overexpression of AHA1 showed earlier wound closure as well as significantly increased cell migration across the membrane coated with Matrigel, and HCT116 cell with downregulation of AHA1 showed opposite results, compared to that in the control after 24 h and 48 h (Fig. 3a,b). These results suggest that AHA1 enhances the migration and invasion of colon cancer cells.
Figure 3.
AHA1 expression is associated with cell migration and invasion in CRC cells. (a) Overexpression of AHA1 by transfection of AHA1-Flag for 24 h in SW480 enhanced cell migration and invasion. (b) Downregulation of AHA1 by transfection of siAHA1 for 48 h in HCT116 decreased cell migration and invasion. Cell migration was determined with wound healing assay for the indicated time point, and the degree of migration was quantified by calculating the area of migrated cells using the image processing software, ImageJ (Ver. 1/52n, NIH, Bethesda, MD, USA). Transwell assay was used to examine cell invasion. The representative images show membrane-associated cells stained using the Hemacolor rapid staining solution. *p < 0.05, **p < 0.01, ***p < 0.001 compared with control.
AHA1 regulates EMT signaling in colon cancer cells
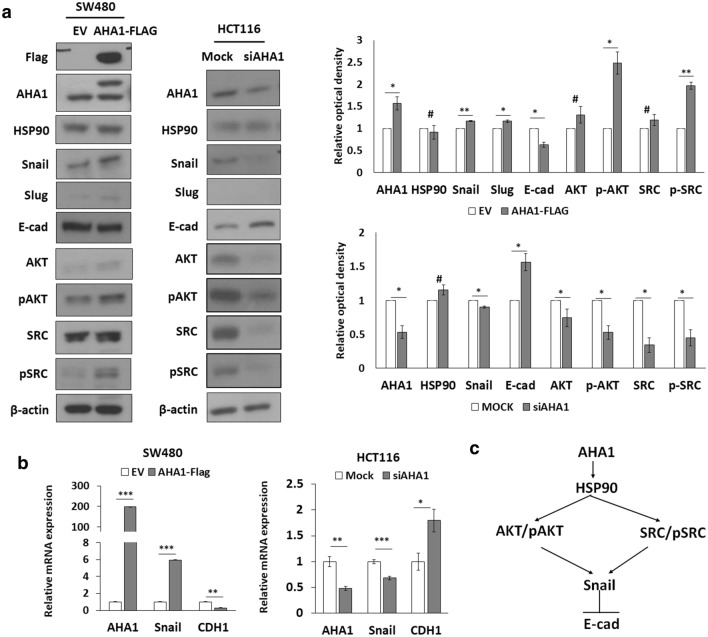
To identify the molecular pathways associated with AHA1 in colon cancer cell migration and invasion, various molecules were examined with western blotting and qRT-PCR. Overexpression of AHA1 by exogenously introduced AHA1-flag in SW480 decreased E-cadherin expression and increased Snail, pAkt, and pSrc expression. On the other hand, downregulation of AHA1 by siAHA1 in HCT116 increased E-cadherin expression and decreased Snail, pAkt, and pSrc expression (Fig. 4a,b). These results indicate that AHA1 regulates cell migration and invasion via the EMT signaling pathway through Snail and E-cadherin, as well as phosphorylation of Akt and Src in colon cancer cells (Fig. 4c).
Figure 4.
AHA1 regulates EMT signaling through Snail, E-cadherin, pAKT, and pSRC. (a,b) Overexpression of AHA1 by transfection of AHA1-Flag for 24 h in SW480 induced a decrease in the levels of E-cadherin, and an increase in the levels of Snail, pAkt, and pSrc expression. Downregulation of AHA1 by transfection of siAHA1 for 48 h in HCT116 induced an increase in the levels of E-cadherin, and a decrease in the levels of Snail, pAkt, and pSrc expression. Expression of proteins was determined with western blotting (a), and that of mRNA was determined using qRT-PCR (b). The densitometry quantification of the western blot determined using Image J software (Ver. 1/52n, NIH). *p < 0.05, **p < 0.01, ***p < 0.001, #p ≥ 0.05 compared with control. (c) Schematic diagram of AHA1 regulated signaling.
Discussion
AHA1 encodes a protein that activates the ATPase activity of Hsp90, leading to an increase in its chaperone activity9,10, and competes with inhibitory co-chaperones, such as Tsc1 and Fnip1, for binding to Hsp90aa114,15. Numerous AHA1-interacting proteins have been identified, which are involved in a variety of intracellular bioprocesses, including DNA maintenance, chromatin structure, translation, nucleocytoplasmic transport, and vesicle transport, among others, pointing to the participation of AHA1 in diverse Hsp90-regulated biological pathways to facilitate Hsp90 chaperone functions16,17. Most of the previous studies have focused on interactions of AHA1 with other chaperones of the Hsp90, and the consequent effects of the Hsp90 chaperone complex on various biological functions. Only limited research has so far been carried out on the association of AHA1 expression with, and its role in various diseases, especially cancer18–22. In this study, we focused on how AHA1 expression is associated with CRC cancer cell migration and invasion, which is one of the hallmarks of cancer cell progression. Our study showed that AHA1 was significantly overexpressed in tumor tissues, compared to that in adjacent normal tissues, in CRC patients, as well as in the analysis of GEO datasets. In addition, overexpression of AHA1, but not Hsp90, was significantly associated with higher TNM stage, LN involvement, and metastasis, implying its association with the progressiveness of CRC. To verify this finding, in vitro cell migration and invasion analysis was performed in context of AHA1 expression, using colon cancer cells. A comparison of AHA1 expression in colon cancer cells showed that more aggressive cells, such as HCT116 and KM125M, expressed more AHA1, along with Hsp90, than less aggressive cells, such as SW480, HT29, and normal colon fibroblast cells (CCD18Co). Also, HT-29 and SW480 cells were MSS, whereas DLD-1 was MSI23,24. Since AHA1 (RNA) expression is higher in DLD-1 cells than SW480, it may be interesting to further investigate association of AHA1 and MSI/MSS status. The relationship between AHA1 expression and migration and invasion was evaluated by overexpression and knockdown of AHA1, which showed that overexpression of AHA1 in SW480 increased, and knock-down of AHA1 in HCT116 decreased cell migration and invasion, a finding consistent with previous reports on osteosarcoma20–22. To determine which molecular signaling pathways are associated with AHA1 in colon cancer cell migration and invasion, various molecules were examined with western blotting after overexpression or knockdown of AHA1 in the colon cancer cells. Snails and E-cadherin, which are well-known EMT markers, as well as pSrc, and pAkt, showed changes in their levels in response to alteration of AHA1 expression. These results showed that EMT signaling, which is the most well-known mechanism of cancer metastasis in context of colon cancer cell migration and invasion, is regulated by AHA1. Src is a proto-oncogene non-receptor tyrosine kinase protein, whose activation is known to promote the survival, angiogenesis, proliferation, and invasion pathways in cancer, including CRC25. Elevated c-Src levels have also been reported to be correlated to advanced stages of the tumor, tumor size, metastatic potential of tumors in CRC, and control of the PI3K/AKT signaling pathway26,27, which indicates that both Src and Akt are directly or indirectly associated with EMT signaling28–30. Both Akt and c-Src are also known to be a client protein of Hsp9031,32. Studies have revealed that Hsp90 also binds to and stabilizes Snail, which is a driver of EMT signaling during tumor progression through E-cadherin, in response to DNA damage33. It is known that Hsp90 and its client proteins play important roles in cancer development through the regulation of various signaling proteins, including EMT signaling, and not only Hsp90, but also co-chaperons considered as an attractive target for anticancer therapy34,35. In addition, it has been reported that the increased expression of AHA1 alters the activity of Hsp90 client proteins and the phosphorylation status of key signaling proteins, including Akt, resulting in enhanced kinase activity36–38, and affects the efficacy of Hsp90 inhibitors, such as Tanespimycin (17-N-allylamino-17-demethoxygeldanamycin, 17-AAG), in cancer cells. This points to a potential therapeutic strategy to increase the sensitivity of cancer cells to Hsp90 inhibitors, by disrupting the Hsp90-AHA1 complex and targeting AHA138.
Taken together, our results show that the expression of AHA1 regulates CRC cell migration and invasion via EMT signaling, through pAkt, pSrc, Snail, and E-cadherin, which possibly occurs via regulation of Hsp90 activity in the HSP90/AHA1 complex. Activation of Hsp90 ATPase activity is the only known biochemical function of AHA1; although we have not confirmed the ATPase activity in this study, we cannot rule out alternative or additional effects of AHA1. In addition, AHA1 may serve as a potential prognostic marker associated with LN involvement and metastasis, as shown in our results, even though further verification is needed with a greater number of patient samples.
Methods
Tumor samples
One hundred and five cases of CRC and paired adjacent normal tissues were obtained from the Department of Colorectal Surgery, Korea University Medical Center. The diagnosis of CRC tissues was made based on pathology reports and histological evaluations. The tissues for this study were collected after obtaining approval from the Institutional Review Board of Korea University College of Medicine (IRB No. KU-IRB-13-84-A-1) and informed consent was obtained from all subjects or, if subjects are under 18, from a parent or legal guardian. Fresh tissue samples were frozen in liquid nitrogen after resection, and stored in a deep refrigerator at − 80 °C until use. Among these cases, patients who received preoperative treatment and those with no available paraffin block were excluded, and a total of 20 primary CRC and 20 metastatic CRC cases in patients, with or without metastasis, were reviewed. All hematoxylin-and-eosin (H&E)-stained slides of the cases were reviewed by pathologists and used for immunohistochemistry (IHC) staining. Clinicopathologic data, including age, sex, tumor location, tumor size, grade, pathologic T (T) category, pathologic N (N) stage, lymphovascular invasion, perineural invasion, LN status, distant metastasis, and resection marginal status, were reviewed. The TNM stages were adjusted to the specifications of the 8th American Joint Committee on Cancer Staging Manual. All experimental protocols were approved by the Institutional Review Boards of Korea University Anam Hospital (K2018-2161-001). All methods were carried out in accordance with the relevant guidelines and regulations. The clinicopathologic features of CRC patients are summarized in Table 1.
Analysis of Gene Expression Omnibus (GEO) datasets and clinical information
We used two datasets (GSE8671 and GSE24514), downloaded from the National Center of Biotechnology Information Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/gds/), to compare the expression of target genes between CRC tissues and normal tissues. GSE8671 included the transcriptome data of 32 colorectal adenomas with paired adjacent normal mucosa11. GSE24514 included the expression profiles of 34 MSI colorectal cancers and 15 normal colonic mucosae12. We performed survival analysis in colorectal cancer for AHA1, HSP90AA1, and HSP90AB1 using OncoLnc (http://www.oncolnc.org/). OncoLnc is a web-based analysis tool that can analyze survival correlation using The Cancer Genome Atlas (TCGA) data13.
Cell cultures and reagents
One normal colon fibroblast cell line (CCD18Co) and six colon cancer cell lines, HT-29, SW480, DLD-1, LoVo, HCT116, and KM12SM, were obtained from the American Type Culture Collection (Manassas, VA, USA). CCD18Co cells were cultured in Eagle’s minimum essential medium, and eight CRC cells were cultured in RPMI 1640 or Dulbecco’s modified Eagle’s medium, supplemented with 10% fetal bovine serum (FBS, WELGENE) and 1% penicillin/streptomycin (WELGENE) at 37 °C in a humidified atmosphere containing 5% CO2. The pcDNA3-Flag-AHA1 plasmid was constructed using the one-step SLIC method39. The oligo ribonucleotide sequences of human AHA1 siRNA (siAHA1) were as follows: 5′-AUU GGU CCA CGG AUA AGC U-3′ (sense) and 5′-GUG AGU AAG CUU GAU GGA G-3′ (antisense) (Shanghai GenePharma Co. Ltd., China).
Quantitative real time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from cells using TRIZOL (Invitrogen, Carlsbad, CA, USA). cDNA was synthesized, using a reverse transcription kit (Labopass, Cosmo Genetech, Seoul, South Korea) according to the manufacturer’s instructions, from the total RNA extracted from frozen tissue and cell lines. qRT-PCR was conducted using gene-specific primers with SYBR Green Q Master (Labopass) on an ABI 7500 Real Time PCR System (Applied Biosystems, Warrington, UK). The following PCR primers were used: HSP90AA1 sense: 5′-ACC CAG ACC CAA GAC CAA CCG-3′, antisense: 5′-ATT TGA AAT GAG CTC TCT CAG-3′; HSP90AB1 sense: 5′-GTG CAC CAT GGA GAG GAG-3′, antisense: 5′-ATT AGA GAT CAA CTC CCG AAG-3′; AHA1 sense: 5′-CAG AGG GAC ACT TTG CCA CCA-3′, antisense: 5′-CTC GAC CTT CCA TGC ACA GCT-3′; Snail sense: 5′-GAC CCC AAT CGG AAG CCT AAC TA-3′, antisense: 5′-AGC CTT TCC CAC TGT CCT CAT CT-3′; CDH1 sense: 5′-CGG GAA TGC AGT TGA GGA TC-3′, antisense: 5′-AGG ATG GTG TAA GCG ATG GC-3′; β-actin sense: 5′-AGA GCT ACG AGC TGC CTG AC-3′, antisense: 5′-AGC ACT GTG TTG GCG TAC AG-3; glyceraldehyde 3-phosphate dehydrogenase (GAPDH) sense: 5′-ACC CAC TCC TCC ACC TTT GA-3′, antisense: 5′-CTG TTG CTG TAG CCA AAT TCG T-3. The Ct values of the target genes were normalized to those of an endogenous reference genes (β-actin and GAPDH). Each gene was analyzed in triplicate in three independent experiments.
Immunohistochemistry (IHC) staining for AHA1
IHC studies were performed on formalin-fixed, paraffin-embedded slides of 20 CRC and adjacent normal tissues, to determine the expression of AHA1 in accordance with the manufacturer’s protocol, using rabbit polyclonal antibodies against AHA1 and HRP-labeled goat anti-rabbit polyclonal secondary antibody (Abcam plc, Cambridge, MA). Counterstaining was carried out using hematoxylin. IHC results were scored, with an average cytoplasmic staining intensity of 0 (no expression), 1 (mild intensity), 2 (moderate intensity), and 3 (strong intensity), by a pathologist, and staining intensity was quantified using Leica Aperio ImagScope (V12.4.0.5043, IL, USA).
Cell migration assay
Cell migration was studied using a wound-healing assay. Cells were seeded into 6-well plates and cultured to a confluent monolayer. The cell monolayer was scratched with a 200 μl sterile micropipette tip, and the wells were washed twice with phosphate-buffered saline (PBS) to remove detached cells. The cells were then cultured for indicated hours in RMPI-1640 supplemented with 2% FBS, to minimize cell proliferation during the period of assay40. The image of each scratch at the same location was captured after the indicated incubation time. The healed area was measured from the captured images using ImageJ (Ver. 1/52n, NIH, Bethesda, MD, USA).
Transwell invasion assays
The invasiveness of cells was evaluated with an invasion assay using a Transwell device (CT-3422, 8 μm pore size, 6.5 mm diameter, Corning Life Science, USA) coated with Matrigel (BD, 356230, 100 μg/ml, 15 µl/well). Cells (1 × 105) were seeded in the upper chamber of the Transwell device in serum-free media. The lower chamber of the transwell device was filled with cell growth medium containing 10% FBS as a chemoattractant. The invaded cells were fixed and stained with the Hemacolor rapid staining solutions (Merck, USA) for 5 min after removing the non-invading cells. The number of invaded cells was counted in five representative fields of the membrane under a light microscope (DP71, Olympus, Japan).
Western blotting analysis
The proteins from cell lysates were separated using SDS-PAGE and transferred to Immobilon-P PVDF Membrane (IPVH00010, Millipore, USA). These membranes were cut based on the size of the target protein (Supplementary Fig. S1) including loading control, and subsequently probed with the indicated primary antibodies and incubated with the appropriate goat anti-rabbit IgG or goat anti-mouse IgG (Cell Signaling Technology) secondary antibody conjugated with horseradish peroxidase (HRP), before signal detection using the enhanced chemiluminescence (ECL) system (Translab, Daejeon, South Korea) according to the manufacturer’s instructions. The blots were cut prior to hybridisation with antibodies (Supplementary Fig. S2). The primary antibody against AHA1 (GTX102312) was purchased from GeneTex (CA, USA). The antibody against Flag (F1804) was purchased from Merck. The antibody against HSP90 (ab13492) was purchased from Abcam. The antibodies against E-cadherin (14472) and Slug (9585) were purchased from Cell Signaling Technology (Danvers, MA, USA). The antibodies against Snail (sc-271977) and β-actin (sc-47778) were purchased from Santa Cruz Biotechnology. The densitometry quantification of the western blot determined using Image J software (Ver. 1/52n, NIH). The result of gels images was cropped and the unprocessed original blots are included in the Supplementary Figs. S2, S3, S4.
Statistical analysis
Results are indicated as mean ± standard deviation (SD) from at least three independent experiments. The significance of different mRNA expression values among CRC and adjacent normal tissues was determined by one-way analysis of variance (ANOVA) and the paired t-test using IBM SPSS Statistics version 25.0 (IBM Inc., Chicago, IL, USA). Comparisons between groups were made using a two-tailed Student’s t-test or ANOVA test. If the p-value obtained with ANOVA was < 0.05, the p-values between the groups were compared with post-test, Bonferroni, and Tukey HSD. p-values ≤ 0.05 were considered statistically significant.
Supplementary Information
Acknowledgements
This work was supported by the grant from korea University College of Medicine (K2110221), and the Basic Science Research Program Grants (Grant No. NRF-2018R1C1B6008346, NRF-2017R1A2B4003233, and NRF-2019R1A2C1083909) from the National Research Foundation of Korea, Republic of Korea.
Author contributions
J.L. and J.W.M. conceived and designed the experiments; D.K., J.W.M., D.H.M., E.S.K., and B.A. conducted the experiments; D.K., J.W.M., D.H.M., E.S.K., B.A., E.S.K., and J.L. analyzed and interpreted the results. D.K., J.W.M., and J.L. wrote the manuscript, and all authors reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Dasom Kim and Ji Wook Moon.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-99375-w.
References
- 1.Rawla P, Sunkara T, Barsouk A. Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Prz. Gastroenterol. 2019;14:89–103. doi: 10.5114/pg.2018.81072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, et al. Genetic alterations during colorectal-tumor development. N. Engl. J. Med. 1988;319:525–532. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 3.Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, et al. Colorectal cancer. Nat. Rev. Dis. Primers. 2015;1:15065. doi: 10.1038/nrdp.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N. Engl. J. Med. 2009;361:2449–2460. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vlad C, Kubelac P, Vlad D, Irimie A, Achimas CP. Evaluation of clinical, morphopathological and therapeutic prognostic factors in rectal cancer. Experience of a tertiary oncology center. J. BUON. 2015;20:92–99. [PubMed] [Google Scholar]
- 6.Edler D, Ohrling K, Hallström M, Karlberg M, Ragnhammar P. The number of analyzed lymph nodes—A prognostic factor in colorectal cancer. Acta Oncol. 2007;46:975–981. doi: 10.1080/02841860701203537. [DOI] [PubMed] [Google Scholar]
- 7.Sarli L, Bader G, Iusco D, Salvemini C, Mauro DD, Mazzeo A, et al. Number of lymph nodes examined and prognosis of TNM stage II colorectal cancer. Eur. J. Cancer. 2005;41:272–279. doi: 10.1016/j.ejca.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 8.Han SW, Ahn JY, Lee S, Noh YS, Jung HC, Lee MH, et al. Gene expression network analysis of lymph node involvement in colon cancer identifies AHSA2, CDK10, and CWC22 as possible prognostic markers. Sci. Rep. 2020;10:7170. doi: 10.1038/s41598-020-63806-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Panaretou B, Siligardi G, Meyer P, Maloney A, Sullivan JK, Singh S, et al. Activation of the ATPase activity of hsp90 by the stress-regulated cochaperone aha1. Mol. Cell. 2002;10:1307–1318. doi: 10.1016/S1097-2765(02)00785-2. [DOI] [PubMed] [Google Scholar]
- 10.Wolmarans A, Lee B, Spyracopoulos L, LaPointe P. The mechanism of Hsp90 ATPase stimulation by Aha1. Sci. Rep. 2016;6:33179. doi: 10.1038/srep33179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabates-Bellver J, Van der Flier LG, de Palo M, Cattaneo E, Maake C, Rehrauer H, et al. Transcriptome profile of human colorectal adenomas. Mol. Cancer Res. 2007;5:1263–1275. doi: 10.1158/1541-7786.MCR-07-0267. [DOI] [PubMed] [Google Scholar]
- 12.Alhopuro P, Sammalkorpi H, Niittymäki I, Biström M, Raitila A, Saharinen J, et al. Candidate driver genes in microsatellite-unstable colorectal cancer. Int. J. Cancer. 2012;130:1558–1566. doi: 10.1002/ijc.26167. [DOI] [PubMed] [Google Scholar]
- 13.Anaya J. OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. Peer J. Comput. Sci. 2016;2:e67. doi: 10.7717/peerj-cs.67. [DOI] [Google Scholar]
- 14.Woodford MR, Sager RA, Marris E, Dunn DM, Blanden AR, Murphy RL, et al. Tumor suppressor Tsc1 is a new Hsp90 co-chaperone that facilitates folding of kinase and non-kinase clients. EMBO J. 2017;36:3650–3665. doi: 10.15252/embj.201796700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woodford MR, Dunn DM, Blanden AR, Capriotti D, Loiselle D, Prodromou C, et al. The FNIP co-chaperones decelerate the Hsp90 chaperone cycle and enhance drug binding. Nat. Commun. 2016;7:12037. doi: 10.1038/ncomms12037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun L, Prince T, Manjarrez JR, Scroggins BT, Matts RL. Characterization of the interaction of Aha1 with components of the Hsp90 chaperone machine and client proteins. Biochim. Biophys. Acta. 2012;1823:1092–1101. doi: 10.1016/j.bbamcr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 17.Sun L, Hartson SD, Matts RL. Identification of proteins associated with Aha1 in HeLa cells by quantitative proteomics. Biochim. Biophys. Acta. 2015;1854:365–380. doi: 10.1016/j.bbapap.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 18.Closa A, Cordero D, Sanz-Pamplona R, Solé X, Crous-Bou M, Paré-Brunet L, et al. Identification of candidate susceptibility genes for colorectal cancer through eQTL analysis. Carcinogenesis. 2014;35:2039–2046. doi: 10.1093/carcin/bgu092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mangangcha IR, Malik MZ, Küçük Ö, Ali S, Singh RKB. Identification of key regulators in prostate cancer from gene expression datasets of patients. Sci. Rep. 2019;9:16420. doi: 10.1038/s41598-019-52896-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shao J, Wang L, Zhong C, Qi R, Li Y. AHSA1 regulates proliferation, apoptosis, migration, and invasion of osteosarcoma. Biomed. Pharmacother. 2016;77:45–51. doi: 10.1016/j.biopha.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 21.Guo QC, Shen JN, Jin S, Wang J, Huang G, Zhang LJ, et al. Comparative proteomic analysis of human osteosarcoma and SV40-immortalized normal osteoblastic cell lines. Acta Pharmacol. Sin. 2007;28:850–858. doi: 10.1111/j.1745-7254.2007.00603.x. [DOI] [PubMed] [Google Scholar]
- 22.Cao R, Shao J, Hu Y, Wang L, Li Z, Sun G, et al. microRNA-338-3p inhibits proliferation, migration, invasion, and EMT in osteosarcoma cells by targeting activator of 90 kDa heat shock protein ATPase homolog 1. Cancer Cell Int. 2018;18:49. doi: 10.1186/s12935-018-0551-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mouradov D, Sloggett C, Jorissen RN, Love CG, Li S, Burgess AW, et al. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res. 2014;74:3238–3247. doi: 10.1158/0008-5472.CAN-14-0013. [DOI] [PubMed] [Google Scholar]
- 24.Berg KCG, Eide PW, Eilertsen IA, Johannessen B, Bruun J, Danielsen SA, et al. Multi-omics of 34 colorectal cancer cell lines—A resource for biomedical studies. Mol. Cancer. 2017;6:116. doi: 10.1186/s12943-017-0691-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeatman TJ. A renaissance for SRC. Nat. Rev. Cancer. 2004;4:470–480. doi: 10.1038/nrc1366. [DOI] [PubMed] [Google Scholar]
- 26.Chen J, Elfiky A, Han M, Chen C, Saif MW. The role of Src in colon cancer and its therapeutic implications. Clin. Colorectal Cancer. 2014;13:5–13. doi: 10.1016/j.clcc.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Aligayer H, Boyd DD, Heiss MM, Abdalla EK, Curley SA, Gallick GE. Activation of Src kinase in primary colorectal carcinoma: An indicator of poor clinical prognosis. Cancer. 2002;94:344–351. doi: 10.1002/cncr.10221. [DOI] [PubMed] [Google Scholar]
- 28.Patel A, Sabbineni H, Clarke A, Somanath PR. Novel roles of Src in cancer cell epithelial-to-mesenchymal transition, vascular permeability, microinvasion and metastasis. Life Sci. 2016;157:52–61. doi: 10.1016/j.lfs.2016.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guarino M. Src signaling in cancer invasion. J. Cell Physiol. 2010;223:14–26. doi: 10.1002/jcp.22011. [DOI] [PubMed] [Google Scholar]
- 30.Xu W, Yang Z, Lu N. A new role for the PI3K/Akt signaling pathway in the epithelial–mesenchymal transition. Cell Adhes. Migr. 2015;9:317–324. doi: 10.1080/19336918.2015.1016686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Y, Singer MA, Lindquist S. Maturation of the tyrosine kinase c-src as a kinase and as a substrate depends on the molecular chaperone Hsp90. Proc. Natl. Acad. Sci. U. S. A. 1999;96:109–114. doi: 10.1073/pnas.96.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dunn DM, Woodford MR, Truman AW, Jensen SM, Schulman J, Caza T, et al. c-Abl mediated tyrosine phosphorylation of Aha1 activates its co-chaperone function in cancer cells. Cell Rep. 2015;12:1006–1018. doi: 10.1016/j.celrep.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun M, Guo X, Qian X, Wang H, Yang C, Brinkman KL, et al. Activation of the ATM-Snail pathway promotes breast cancer metastasis. J. Mol. Cell Biol. 2012;4:304–315. doi: 10.1093/jmcb/mjs048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jafari A, Rezaei-Tavirani M, Farhadihosseinabadi B, Taranejoo S, Zali H. HSP90 and co-chaperones: Impact on tumor progression and prospects for molecular-targeted cancer therapy. Cancer Investig. 2020;38:310–328. doi: 10.1080/07357907.2020.1752227. [DOI] [PubMed] [Google Scholar]
- 35.Edkins AL. Hsp90 co-chaperones as drug targets in cancer: Current perspectives. In: McAlpine SR, Edkins AL, editors. Heat shock protein inhibitors. Springer; 2016. pp. 21–54. [Google Scholar]
- 36.Baker-Williams AJ, Hashmi F, Budzyński MA, Woodford MR, Gleicher S, Himanen SV, et al. Co-chaperones TIMP2 and AHA1 competitively regulate extracellular HSP90: Client MMP2 activity and matrix proteolysis. Cell Rep. 2019;28:1894–906.e6. doi: 10.1016/j.celrep.2019.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lotz GP, Lin H, Harst A, Obermann WM. Aha1 binds to the middle domain of Hsp90, contributes to client protein activation, and stimulates the ATPase activity of the molecular chaperone. J. Biol. Chem. 2003;278:17228–17235. doi: 10.1074/jbc.M212761200. [DOI] [PubMed] [Google Scholar]
- 38.Holmes JL, Sharp SY, Hobbs S, Workman P. Silencing of HSP90 cochaperone AHA1 expression decreases client protein activation and increases cellular sensitivity to the HSP90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2008;68:1188–1197. doi: 10.1158/0008-5472.CAN-07-3268. [DOI] [PubMed] [Google Scholar]
- 39.Jeong JY, Yim HS, Ryu JY, Lee HS, Lee JH, Seen DS, et al. One-step sequence- and ligation-independent cloning as a rapid and versatile cloning method for functional genomics studies. Appl. Environ. Microbiol. 2012;78:5440–5443. doi: 10.1128/AEM.00844-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez LG, Wu X, Guan JL. Wound-healing assay. Methods Mol. Biol. 2005;294:23–29. doi: 10.1385/1-59259-860-9:023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.