Abstract
A collection of 114 independent Streptococcus agalactiae strains, including 54 strains isolated from the cerebrospinal fluid (CSF) samples of neonates and 60 strains from asymptomatic patients, was characterized by pulsed-field gel electrophoresis (PFGE) of DNA restricted with SmaI and by PCR analysis of the hylB gene. All strains were previously studied by multilocus enzyme electrophoresis (MLEE) (R. Quentin, H. Huet, F.-S. Wang, P. Geslin, A. Goudeau, and R. K. Selander, J. Clin. Microbiol. 33:2576–2581, 1995). Among these 114 strains, there were 92 PFGE patterns. Eleven genetic groups (A to K) were identified with 38% divergence. A more homogeneous group (PFGE group A) was defined, consisting of 73% of the strains previously identified as belonging to a particular MLEE phylogenetic group. A 162-kb fragment was identified as a marker of strains that invaded the central nervous system of neonates. It was detected in 69% of the PFGE patterns obtained with CSF isolates and in only 1.8% of the PFGE patterns obtained with carrier strains. The hylB gene encoding hyaluronate lyase was amplified for all strains in our collection. Ten of 15 isolates belonging to an MLEE subgroup, previously described as being likely to cause invasive infection, had an insertion in the hylB gene (IS1548).
In France, Streptococcus agalactiae is responsible for 50% of severe maternal and neonatal infections (3, 23). In the United States, a rate of 1.8 cases of S. agalactiae sepsis per 1,000 live births has been reported (25). Severe infections during pregnancy and septicemia and meningitis in neonates caused by S. agalactiae are also of major concern in the United States (11). These infections are caused by bacteria colonizing the urogenital tracts of 15 to 25% of pregnant women (1). S. agalactiae infections have major consequences for public health, because they may cause neurological problems in newborns and endometritis and sterility in the mother. S. agalactiae may cause meningitis, septicemia, and prenatal inflammatory events associated with a high risk of periventricular leukomalacia (2). Strategies to prevent neonatal colonization and infection involve intrapartum antibiotic prophylaxis for all colonized mothers during labor and treatment of all colonized neonates (6). These strategies are extreme, involving the treatment of a large number of people, given the relatively small risk of infection. More accurate information about the basic pathophysiology of infection would make it possible to target prophylaxis effectively. There are three possible reasons for the difference between the high colonization rate and the lower rate of severe infection: (i) the host immune system is the determining factor in the development of invasive disease, (ii) the bacteria determine the nature of infection because they belong to more virulent groups of S. agalactiae, which have emerged within the species during evolution, and (iii) a combination of the above.
Ecological and epidemiological studies have shown that premature newborns are more susceptible to S. agalactiae infection than are full-term infants (1). Nevertheless, infections do occur in mature neonates. Phenotyping and genotyping are consistent with the involvement of particular virulent clones of S. agalactiae. Serotyping has shown that most strains responsible for meningitis are serotype III (1); nevertheless, capsular types Ia (16), Ib (10), and V (4, 27) are also regularly isolated in cases of neonatal sepsis.
Genetic studies have identified two phylogenetic groups within S. agalactiae species (17, 21, 24) and some specificity of the strains implicated in severe neonatal infections (7, 8, 21, 24). However, such results were obtained by studying limited parts of the bacterial genome, such as metabolic enzyme loci and rRNA genes (7, 24). In this work, the genome of S. agalactiae was studied as a whole, by using pulsed-field gel electrophoresis (PFGE). We assessed the genetic relationships between isolates to investigate genetic clustering and identify features typical of invasive strains of S. agalactiae. It has long been suspected that there are genetic subgroups, but no virulence factor accounting for the invasive properties of these strains has been described. The study of the hyaluronate lyase gene has led to the suggestion that alleles of this gene may be associated with virulence and may be characteristic of some strains of S. agalactiae (15). Indeed, an insertion of 1,317 nucleotides (IS1548) has been identified in this gene, which was associated with strains isolated from patients with endocarditis (15). We identified the hylB gene encoding the hyaluronate lyase and IS1548 in French genital and neonatal S. agalactiae populations and looked at the relationships between PFGE profiles, the phylogenetic distribution of strains, and the sites from which they were isolated.
MATERIALS AND METHODS
Bacterial strains.
The 114 strains of S. agalactiae studied were collected in France. A national collection of 54 strains isolated from the cerebrospinal fluid (CSF) of neonates suffering from meningitis was collected from 25 general hospitals. Forty-five of the 54 strains isolated were from newborns with early-onset disease, and nine were from babies with late-onset disease. Fifty-nine epidemiologically unrelated strains isolated from asymptomatic patients were analyzed and compared with invasive isolates: 37 strains were isolated from the vaginas of pregnant women, and 22 strains were isolated from the gastric fluids of neonates. The type strain (NCTC 8181T) was used as a reference.
Previous serotyping of these strains, on the basis of capsule polysaccharide and protein antigens, has identified 14 serotypes (13, 24) (Fig. 1): serotypes Ia (6 strains), Ia/c (10 strains), Ib (4 strains), Ib/c (8 strains), II (9 strains), II/c (7 strains), II/R (3 strains), III (18 strains), III/R (37 strains), IV/c (2 strains), IV/R (1 strain), V (1 strain), V/c (1 strain), and V/R (1 strain). Five strains were not typeable for capsule polysaccharide antigens (NT) but could be typed for protein antigen (c or R), two strains were NT/c, and three strains were NT/R. One strain was untypeable. These strains were also analyzed by multilocus enzyme electrophoresis (MLEE) (24), which identified two major phylogenetic groups (MLEE groups I and II). MLEE group I consisted mostly of CSF isolates, which identified this group as posing a high risk of infection in neonates. MLEE group II was more heterogeneous, but the CSF isolates of this phylogenetic group were mainly clustered into two electrophoretic types (ET): ET 11 and ET 12. Therefore, MLEE group I, ET 11, and ET 12 are three groups of strains that cause severe neonatal disease.
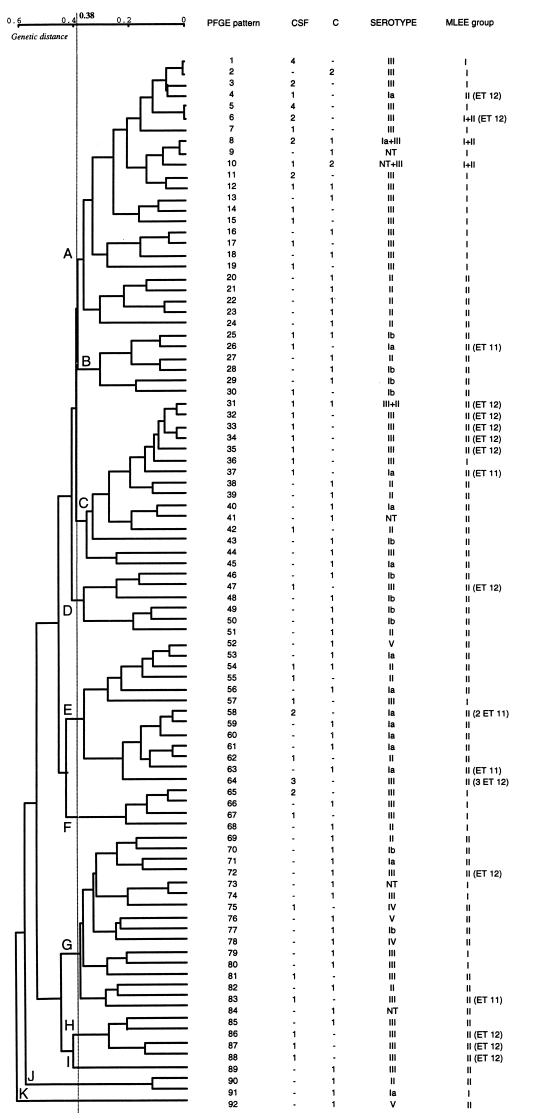
FIG. 1.
Schematic representation of PFGE patterns obtained after restriction with SmaI. For each pattern, the PFGE pattern, the number of strains isolated from the CSF of neonates (CSF) and from asymptomatic patients (C), and the corresponding MLEE phylogenetic group are indicated, and strains of ET 11 and 12 defined by MLEE as being virulent (24) are also indicated. The PFGE groups indicated are those identified by the unweighted pair group method with averages (see Fig. 2). Fragments that occurred in multiple patterns are also noted: a 183-kb fragment, a 162-kb fragment, and a quadruplet of 39, 51.5, 60, and 73-kb fragments.
Chromosome analysis by PFGE.
Plugs containing genomic DNA were prepared as described by Fasola et al. (12). Solidified plugs were incubated in 5 ml of Triton-EDTA (1% Triton, 0.05 M EDTA [pH 7.6]) for 2 h at 38°C and then in 5 ml of 0.5 M EDTA [pH 7.6] at 38°C for 3 h. The bacterial cells were lysed overnight at 37°C with 5 ml of a lysis solution (0.01 M Tris-HCl [pH 7.6], 1 M NaCl, 0.5% Sarkosyl, 1 mg of lysozyme per ml, 50 U of mutanolysin). Plugs were placed in 2.5 ml of 0.5 M EDTA [pH 7.6] containing 1% Sarkosyl for 1 h at 4°C, and 250 μl of proteinase K (20 mg/ml) was added. Plugs were incubated overnight at 37°C and then for 23 h at 48°C. Plugs were washed three times with TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 7.6]) for 30 min each at 4°C and were incubated for 1 h at 55°C with 5 ml of TE buffer [pH 7.6] containing 0.12 mg of phenylmethylsulfonyl fluoride per ml. They were then washed three times with TE buffer [pH 7.6] at 4°C for 30 min each. Plugs were stored in 0.5 M EDTA at 4°C. They were washed twice in 15 ml of TE buffer [pH 7.6] for 30 min each at 4°C and incubated for 2 h in 5 ml of 0.1% Triton at 4°C. The DNA was digested with 3 μl of SmaI (10 U/ml) (Boehringer, Mannheim, Germany) in 10 μl of a 10× enzyme buffer and 87 μl of sterile water, with incubation for 3 h at 4°C followed by 24 h at 25°C. The plugs were washed with 3 ml of 0.1 M EDTA, incubated in 10 ml of 0.1 M EDTA for 1 h at 4°C, and were subjected to electrophoresis in a 1% agarose gel (FMC BioProducts) in TBE (4.5 mM Tris-HCl, 4.5 mM borate, 0.125 mM EDTA [pH 7.6]) with a contour-clamped homogeneous electric field (9) (CHEF DRIII; Bio-Rad Laboratories). Pulse times were ramped from 3 to 55 s over 24 h at 200 V. PFGE patterns were detected by UV transillumination after ethidium bromide staining. Lambda phage concatemers were used as DNA size standards (Bio-Rad Laboratories). The patterns were digitized and analyzed by using the Taxotron package (Taxolab; Institut Pasteur, Paris, France) as described by Chatellier et al. (8).
Detection of hyaluronate lyase gene (hylB) and IS1548 by PCR.
The hylB gene was amplified with primers Hyal1 and Hyal2 (Table 1) as described by Lin et al. (18). IS1548 was amplified in a PCR mixture containing PCR buffer (10 mM Tris-HCl, 50 mM KCl, 1.5 mM MgCl2 [pH 8.3]), a 200 μM concentration of each deoxyribonucleoside triphosphate (Boehringer), 10 pmol of primers Hylis3 and Hylis4r (Table 1), 0.1 U of Taq DNA polymerase (Perkin-Elmer, Norwalk, Conn.), and 25 ng of the DNA template from tested strains prepared as previously described (5). The reaction procedure consisted of an initial denaturation step at 94°C for 30 s, followed by 25 cycles of denaturation at 94°C for 15 s, primer annealing at 55°C for 15 s, and extension at 72°C for 15 s (30 s for the last extension). The resulting amplified products were separated in a 1% agarose gel in TBE buffer (8.9 mM Tris, 8.9 mM borate, 0.25 mM EDTA [pH 8.0]) for 1 h 30 min at a constant voltage of 100 V. Amplified products were detected by UV transillumination with ethidium bromide staining. A 1-kb ladder (Life Technologies) was used as a molecular size standard. For the negative control, the DNA template was replaced with double-distilled water.
TABLE 1.
Primers used to amplify the hylB gene and IS1548
Sequencing.
Nucleotide sequences were determined by cycle sequencing based on the dideoxynucleoside termination chain method of Sanger (26). PCR products were sequenced by using the ThermoSequenase dye terminator cycle sequencing premix kit and following the manufacturer’s recommendations (Amersham Life Sciences, Cleveland, Ohio). Sequenced products were purified with Microcon concentrators (Amicon, Beverly, Mass.) to eliminate unused reagents. Products were subjected to electrophoresis in a 4.2% acrylamide/bis-acrylamide (19/1) gel (Appligène, Illkirch, France) containing 6 M urea, 0.7% N,N,N′,N′-tetramethylethylenediamine, and 0.05% ammonium persulfate in TBE buffer by using the Abi Prism 377 DNA sequencer according to the manufacturer’s instructions (Perkin-Elmer).
RESULTS
Genetic diversity of S. agalactiae strains as defined by PFGE.
PFGE patterns after restriction with SmaI were analyzed and showed the S. agalactiae population to be genetically diverse. Among the 114 strains, 92 PFGE patterns were identified. A schematic representation of all patterns in relation to the source of the strain, serotype, and MLEE group, including ET 11 and ET 12, which are thought to pose a high risk of infection (24), is presented in Fig. 1. The deduced genetic relationships between the 114 strains of S. agalactiae are shown in the dendrogram in Fig. 2. The 114 strains diverged by up to 60%. At a level of 38% dissimilarity, 11 PFGE groups, A to K (Fig. 2), were identified. Some PFGE fragments were present in multiple patterns (Fig. 1): a 183-kb fragment, a 162-kb fragment, and four fragments of 39, 51.5, 60, and 73 kb.
FIG. 2.
Genetic relationships between 114 S. agalactiae strains: 54 strains isolated from the CSF of neonates suffering from meningitis (CSF) and 60 strains from asymptomatic patients (C). The classification and divergence of isolates were calculated by the unweighted pair group method with averages from the PFGE results (Fig. 1). At 38% divergence, 11 PFGE groups, A to K, were identified. Strains of MLEE phylogenetic group I (24) mostly clustered in PFGE group A (73%), which consisted of 24 of the 54 CSF isolates, mostly of serotype III (53%).
Distribution of PFGE patterns, groups, and markers in relation to the sources of S. agalactiae strains.
The genetic diversity of strains isolated from the vaginas of pregnant women (36 PFGE patterns for 37 strains) and from the gastric fluid of neonates (22 PFGE patterns for 22 strains) was greater than that of strains isolated from the CSF of neonates (40 PFGE patterns for 54 strains) (Table 2). Forty-one of the 54 CSF isolates (76%) clustered into three groups: PFGE group A (24 strains), PFGE group E (9 strains), and PFGE group C (8 strains). A total of 61.5% of the 39 PFGE group A strains were isolated from CSF. The 183-kb fragment, one of the three potential PFGE markers (Fig. 1), was not significantly associated with the source of the strains. The 162-kb fragment and the quadruplet were mostly present in strains isolated from CSF. The 162-kb fragment was detected in 37 of the 54 strains isolated from CSF and in 15 of the 60 strains isolated from asymptomatic patients (χ2 test; P = 3 × 10−6). The quadruplet was detected in 24 of the 54 CSF isolates and in 13 of the 60 strains isolated from asymptomatic patients (χ2 test; P = 9 × 10−3).
TABLE 2.
Distribution of PFGE patterns, groups, and markers in relation to the sources of S. agalactiae strainsa
| Distribution of: | n | No. of isolates from:
|
||
|---|---|---|---|---|
| Vagina | Gastric fluid | CSF | ||
| No. of strains | 113 | 37 | 22 | 54 |
| No. of PFGE patterns | 92 | 36 | 22 | 40 |
| PFGE group | ||||
| A | 39 | 11 | 4 | 24 |
| B | 7 | 3 | 1 | 3 |
| C | 16 | 4 | 4 | 8 |
| D | 6 | 2 | 3 | 1 |
| E | 17 | 7 | 1 | 9 |
| Others (F to K) | 28 | 10 | 9 | 9 |
| PFGE marker | ||||
| 183 kb | 35 | 17 | 7 | 11 |
| 162 kb | 52 | 12 | 3 | 37 |
| Quadruplet | 37 | 10 | 3 | 24 |
The type strain, NCTC 8181T, isolated from milk was not included in this analysis.
Distribution of PFGE patterns, groups, and markers in relation to serotype.
PFGE patterns within serotypes differed greatly, with almost one PFGE pattern obtained per strain. The strains of serotype III were more homogeneous, with 41 PFGE patterns for 55 strains (Table 3). Serotypes Ia and III clustered in particular PFGE groups. Eleven of the 16 strains (69%) of serotype Ia belonged to PFGE groups E and C, and 36 of the 55 strains (65%) of serotype III belonged to PFGE groups A and C. The 183-kb PFGE marker was present in 14 of the 19 serotype II strains (χ2 test; P = 4 × 10−6) and 10 of the 16 serotype Ia strains (χ2 test; P = 2 × 10−3), and the 162-kb fragment was detected in 46 of the 55 serotype III strains (χ2 test; P = 3 × 10−15).
TABLE 3.
Distribution of PFGE patterns, groups, and markers in relation to the serotypes of S. agalactiae strains
| Distribution of: | n | No. of isolates of serotype
|
||||
|---|---|---|---|---|---|---|
| Ia | Ib | II | III | Others | ||
| No. of strains | 114 | 16 | 12 | 19 | 55 | 12 |
| No. of PFGE patterns | 92 | 15 | 11 | 18 | 41 | 10 |
| PFGE group | ||||||
| A | 39 | 2 | 0 | 5 | 29 | 3 |
| B | 7 | 1 | 5 | 1 | 0 | 0 |
| C | 16 | 3 | 1 | 4 | 7 | 1 |
| D | 6 | 0 | 4 | 1 | 1 | 0 |
| E | 17 | 8 | 0 | 4 | 4 | 1 |
| Others (F to K) | 29 | 2 | 2 | 4 | 14 | 7 |
| PFGE marker | ||||||
| 183 kb | 35 | 10 | 2 | 14 | 7 | 2 |
| 162 kb | 52 | 2 | 0 | 1 | 46 | 3 |
| Quadruplet | 37 | 6 | 0 | 3 | 25 | 3 |
Distribution of PFGE patterns, groups, and markers in relation to MLEE phylogenetic groups of S. agalactiae species.
MLEE group I was less genetically diverse than MLEE group II (Table 4). Twenty-nine PFGE patterns were identified among the 41 strains of MLEE group I, whereas 66 PFGE patterns were obtained from the 73 strains of MLEE group II. Thirty of the 41 (73%) MLEE group I strains clustered in PFGE group A, whereas 64 of the 73 (74%) MLEE group II strains were distributed among the other 10 PFGE groups (groups B to K). The 162-kb putative PFGE marker was significantly associated with MLEE group I, because 38 of the 41 strains of this MLEE group had this fragment in their PFGE patterns, versus 14 of the 73 strains of MLEE group II (χ2 test; P = 3 × 10−12). The 162-kb fragment was also detected in 67% of ET 12 strains but was not detected in the six strains of ET 11.
TABLE 4.
Distribution of PFGE patterns, groups, and markers in relation to MLEE phylogenetic groups of S. agalactiae strains (24)
| Distribution of: | n | No. of isolates in:
|
|||
|---|---|---|---|---|---|
| MLEE group I | MLEE group II | ET 11 | ET 12 | ||
| No. of strains | 114 | 41 | 73 | 6 | 15 |
| No. of PFGE patterns | 92 | 29 | 66 | 5 | 15 |
| PFGE group | |||||
| A | 39 | 30 | 9 | 0 | 2 |
| B | 7 | 0 | 7 | 1 | 0 |
| C | 16 | 1 | 15 | 1 | 5 |
| D | 6 | 0 | 6 | 0 | 1 |
| E | 17 | 1 | 16 | 3 | 3 |
| Others (F to K) | 29 | 9 | 20 | 1 | 4 |
| PFGE marker | |||||
| 183 kb | 35 | 3 | 32 | 3 | 5 |
| 162 kb | 52 | 38 | 14 | 0 | 10 |
| Quadruplet | 37 | 26 | 11 | 0 | 2 |
Detection of the hylB gene encoding hyaluronate lyase.
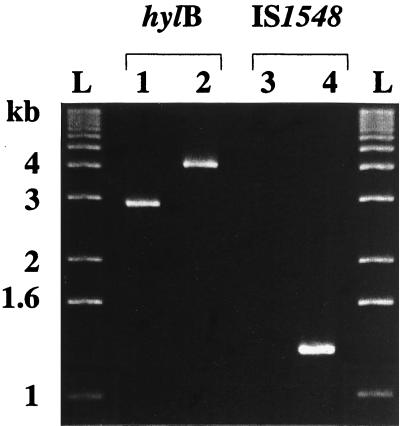
PCR with primers Hyal1 and Hyal2 flanking the hylB gene and DNA from each of the 114 strains of S. agalactiae amplified either a 2.9- or a 4.2-kb fragment (Fig. 3). The distribution of these amplified products in relation to the source, serotype, and MLEE group of the strain is reported in Table 5. The 4.2-kb fragment was significantly more frequent in strains isolated from the CSF of neonates suffering from meningitis (18.5%) (10 of 54 strains) than in strains isolated from asymptomatic patients (3%) (2 of 60 strains) (χ2 test; P = 8 × 10−3). This was due to the 2.9- and 4.2-kb fragments being associated with a high risk of infection (MLEE group I, ET 11, and ET 12). The 4.2-kb fragment was amplified from 10 of the 15 ET 12 strains (67%) but was not amplified from any of the 41 MLEE group I strains or the 6 strains of ET 11 (χ2 test; P = 3 × 10−14). All strains in which the 4.2-kb fragment was detected were of serotype III. The 1,100 bases at the 5′ and 3′ ends of the 2.9- and 4.2-kb fragments were sequenced and were found to be 99% identical to the hylB gene sequence reported by Lin et al. (18).
FIG. 3.
PCR products obtained with the Hyal1 and Hyal2 primers specific for the hylB gene and with the Hylis3 and Hylis4r primers specific for IS1548 (15). With Hyal1 and Hyal2 primers, a 2.9-kb (lane 1) or a 4.2-kb (lane 2) amplified product was obtained. With Hylis3 and Hylis4r primers, no (lane 3) or a 1.3-kb (lane 4) amplified product was obtained. Lanes L, 1-kb DNA molecular size ladder.
TABLE 5.
Distribution of the hylB gene and IS1548 in relation to the sources, serotypes, and MLEE phylogenetic groups of S. agalactiae strains (24)
| Distribution of: | n | No. of isolates with:
|
|||
|---|---|---|---|---|---|
|
hylB gene amplification
|
IS1548 amplification
|
||||
| 2.9-kb fragment | 4.2-kb fragment | No ampli-fication | 1.3-kb fragment | ||
| Sourcea | |||||
| Vagina | 37 | 36 | 1 | 36 | 1 |
| Gastric fluid | 22 | 21 | 1 | 21 | 1 |
| CSF | 54 | 44 | 10 | 44 | 10 |
| Total | 113 | 101 | 12 | 101 | 12 |
| Serotype | |||||
| Ia | 16 | 16 | 0 | 16 | 0 |
| Ib | 12 | 12 | 0 | 12 | 0 |
| II | 19 | 19 | 0 | 19 | 0 |
| III | 55 | 43 | 12 | 43 | 12 |
| Others | 12 | 12 | 0 | 12 | 0 |
| Total | 114 | 102 | 12 | 102 | 12 |
| MLEE phylogenetic group | |||||
| MLEE group I | 41 | 41 | 0 | 41 | 0 |
| MLEE group II | 73 | 61 | 12 | 61 | 12 |
| ET 11 | 6 | 6 | 0 | 6 | 0 |
| ET 12 | 15 | 5 | 10 | 5 | 10 |
| Total | 114 | 102 | 12 | 102 | 12 |
The type strain, NCTC 8181T, isolated from milk was not included in this analysis.
Detection of the IS1548 insertion sequence in the hylB gene.
A 1.3-kb fragment was amplified from 12 strains with primers Hylis3 and Hylis4r, which flank the IS1548 sequence (15) (Fig. 3). These strains were the same 12 strains from which we had previously amplified a 4.2-kb fragment with primers specific for the hylB gene. The 1,081 bases at the 5′ and 3′ ends of this 1.3-kb fragment were sequenced and were found to be 99.3% identical to the DNA sequence of IS1548 reported by Granlund et al. (15).
DISCUSSION
S. agalactiae is responsible for severe neonatal and maternal infections, our understanding of which may be improved by analyzing the genetic structure of the S. agalactiae species. The collection of 114 strains investigated by PFGE was previously analyzed by MLEE (24), ribotyping (7), and randomly amplified polymorphic DNA (RAPD) analysis (8). MLEE analysis defined two major phylogenetic groups (I and II) (24). Only analysis of genes encoding rRNA identified two separate genetic groups within S. agalactiae. RAPD analysis (8) and PFGE did not identify two different populations. However, quantitative analysis of PFGE data did demonstrate greater homogeneity among strains of PFGE group A, which corresponds to MLEE group I (24). This group contains serotype III strains and a large number of strains that cause meningitis (24). The nature of the genes studied may account for this difference in phylogenetic results. Metabolic genes and genes encoding rRNA, which are vital for bacterial survival, are more likely to be conserved and are therefore good markers of phylogenetic lineage within bacteria and species (20). RAPD analysis and PFGE randomly explore most of the bacterial genome and probably identify more minor parts of the genome not essential for bacterial survival.
Phylogenetic markers may be used to identify strains within the S. agalactiae population able to invade the central nervous systems of neonates. Several genetic peculiarities of invasive isolates were observed by MLEE, ribotyping, and RAPD analysis (7, 8, 24) and in this study by PFGE analysis. MLEE and ribotyping are reproducible. However, MLEE typing requires the study of at least 12 metabolic enzymes, and therefore this method can be done only by specialist laboratories (24). Similarly, ribotyping is a painstaking and laborious method that involves the use of three restriction enzymes to identify invasive strains of MLEE group I (7). RAPD analysis, which requires at least three primers to be discriminant for S. agalactiae species (8), is not very reproducible (22). PFGE typing requires only one restriction enzyme digestion (SmaI), and recently proposed modifications to the PFGE method may reduce the time required to less than 3 days (19). In addition, the reproducibility of the PFGE method is good (14). Thus, PFGE appears to be an easier method than the others for typing isolates of S. agalactiae. With this tool, we identified a 162-kb fragment which was a marker of the strains belonging to MLEE group I, which poses a high risk of meningitis. This marker is also able to detect 10 of the 27 CSF isolates belonging to MLEE group II. Another characteristic of the CSF isolates was also identified. It consists of a quadruplet of fragments (39, 51.5, 60, and 73 kb). However, a combination of these two markers did not increase the sensitivity and specificity of identification of invasive isolates, because the strains which exhibited the quadruplet also have the 162-kb fragment. The usefulness of the 162-kb marker to detect strains which pose a high risk of invasive infection in the vagina and gastric fluid needs to be confirmed in prospective studies.
Among the 52 isolates that possess the 162-kb fragment, 46 were serotype III, which has long been associated with a higher risk of meningitis. It is thus possible that this fragment may contain the type III capsule gene locus. This point could be clarified by PCR and Southern analysis.
An insertion sequence, IS1548, has been identified in the hylB gene of strains isolated from patients suffering from endocarditis, but this sequence was not present in the hylB gene of strains isolated from neonates with severe infections (15). Our results are partially consistent with this. IS1548 was specifically detected in ET 12, one of the three virulent clones we have previously identified by MLEE (24). This ET consisted of 15 strains, including 14 CSF isolates. Ten of these CSF isolates contained IS1548, whereas only two strains not belonging to ET 12 had this hylB gene insertion. IS1548 may be typical of S. agalactiae isolates with virulence traits specific for endocarditis, as suggested by Granlund et al. (15) and may also be a marker for a subgroup of S. agalactiae strains causing severe infections in neonates. Furthermore, insertion elements generally inactivate genes, suggesting that hyaluronate lyase is not required for neonatal invasive infection by strains of this virulent ET 12 clone.
PCR is the most appropriate technique for identifying isolates with no phenotypic peculiarities in medical laboratories. A PCR protocol is available to identify strains of one of the three virulent MLEE groups (ET 12) by amplifying IS1548. It should be possible to define primers for identifying the other two clones, for which the risk of invasive infection is high, by PCR. The 162-kb fragment must therefore be further characterized. This fragment may contain a gene or group of genes encoding S. agalactiae virulence factors.
ACKNOWLEDGMENTS
This work was supported by grants from the Région Centre to K.R. and from the Centre Hospitalier Universitaire de Tours to R.Q.
We thank the Collège de Bactériologie Virologie et Hygiène des Hôpitaux de France for supplying the CSF isolates via Pierre Geslin.
REFERENCES
- 1.Baker C J. Group B streptococcal infections. Clin Perinatol. 1997;24:59–70. [PubMed] [Google Scholar]
- 2.Baud O, d’Allest A M, Lacaze-Masmonteil T, Zupan V, Nedelcoux H, Boithias C, Delaveaucoupet J, Dehan M. The early diagnosis of periventricular leukomalacia in premature infants with positive rolandic sharp waves on serial electroencephalography. J Pediatr. 1998;132:813–817. doi: 10.1016/s0022-3476(98)70309-9. [DOI] [PubMed] [Google Scholar]
- 3.Blanc B, Blond M-H, Chaix C, Goffinet F, Guillaume S, Judlin P, Lenclen R, Philippe H-J, Pierre F, Poulain P, Quentin R, Terville J-P the Collège National des Gynécologues et Obstétriciens Français. Les infections cervico-vaginales au cours de la grossesse—recommandations pour la pratique clinique. Bull Soc Fr Microbiol. 1998;13:55–62. [Google Scholar]
- 4.Blumberg H M, Stephens D S, Modansky M, Erwin M, Elliot J, Facklam R R, Schuchat A, Baughman W, Farley M M. Invasive group B streptococcal disease: the emergence of serotype V. J Infect Dis. 1996;173:365–373. doi: 10.1093/infdis/173.2.365. [DOI] [PubMed] [Google Scholar]
- 5.Brenner D J, McWhorter A C, Leete Knutson J K, Steigerwalt A G. Escherichia vulneris: a new species of Enterobacteriaceae associated with human wounds. J Clin Microbiol. 1982;15:1133–1140. doi: 10.1128/jcm.15.6.1133-1140.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Prevention of perinatal group B streptococcal disease: a public health perspective. Morbid Mortal Weekly Rep. 1996;45:1–24. [PubMed] [Google Scholar]
- 7.Chatellier S, Huet H, Kenzi S, Rosenau A, Geslin P, Quentin R. Genetic diversity of rRNA operons of unrelated Streptococcus agalactiae strains isolated from cerebrospinal fluid of neonates suffering from meningitis. J Clin Microbiol. 1996;34:2741–2747. doi: 10.1128/jcm.34.11.2741-2747.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatellier S, Ramanantsoa C, Harriau P, Rolland K, Rosenau A, Quentin R. Characterization of Streptococcus agalactiae strains by randomly amplified polymorphic DNA analysis. J Clin Microbiol. 1997;35:2573–2579. doi: 10.1128/jcm.35.10.2573-2579.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chu G, Vollrath D, Davis R W. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science. 1986;234:1582–1585. doi: 10.1126/science.3538420. [DOI] [PubMed] [Google Scholar]
- 10.Chun C S Y, Brady L J, Boyle M D P, Dillon H C, Ayoub E M. Group B streptococcal C protein-associated antigens: association with neonatal sepsis. J Infect Dis. 1991;163:786–791. doi: 10.1093/infdis/163.4.786. [DOI] [PubMed] [Google Scholar]
- 11.Dillon H C, Khare S, Gray B M. Group B streptococcal carriage and disease: a 6-year prospective study. J Pediatr. 1987;110:31–36. doi: 10.1016/s0022-3476(87)80283-4. [DOI] [PubMed] [Google Scholar]
- 12.Fasola E, Livdahl C, Ferrieri P. Molecular analysis of multiple isolates of the major serotypes of group B streptococci. J Clin Microbiol. 1993;31:2616–2620. doi: 10.1128/jcm.31.10.2616-2620.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geslin P, Sissia G, Jelinkova J, Fremaux A, Motlova J. Serotype distribution of group B streptococci isolated from human sources in France over a 10-year period (1980–1989) Zentbl Bakteriol Suppl. 1992;22:484–485. [Google Scholar]
- 14.Gordillo M E, Singh K V, Baker C J, Murray B E. Typing of group B streptococci: comparison of pulsed-field gel electrophoresis and conventional electrophoresis. J Clin Microbiol. 1993;31:1430–1434. doi: 10.1128/jcm.31.6.1430-1434.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Granlund M, Öberg L, Sellin M, Norgren M. Identification of a novel insertion element, IS1548, in group B streptococci, predominantly in strains causing endocarditis. J Infect Dis. 1998;177:967–976. doi: 10.1086/515233. [DOI] [PubMed] [Google Scholar]
- 16.Harrison L H, Elliott J A, Dwyer D M, Libonati J P, Ferrieri P, Billmann L, Schuchat A. Serotype distribution of invasive group B streptococcal isolates in Maryland: implications for vaccine formulation. Maryland Emerging Infection Program. Infect Dis. 1998;177:998–1002. doi: 10.1086/515260. [DOI] [PubMed] [Google Scholar]
- 17.Helmig R, Uldbjerg N, Boris J, Kilian M. Clonal analysis of Streptococcus agalactiae isolated from infants with neonatal sepsis or meningitis and their mothers and from healthy pregnant women. J Infect Dis. 1993;168:904–909. doi: 10.1093/infdis/168.4.904. [DOI] [PubMed] [Google Scholar]
- 18.Lin B, Hollingshead S K, Coligan J E, Egan M L, Baker J R, Pritchard D G. Cloning and expression of the gene for group B streptococcal hyaluronate lyase. J Biol Chem. 1994;269:30113–30116. [PubMed] [Google Scholar]
- 19.Matushek M G, Bonten M J M, Hayden M K. Rapid preparation of bacterial DNA for pulsed-field gel electrophoresis. J Clin Microbiol. 1996;34:2598–2600. doi: 10.1128/jcm.34.10.2598-2600.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maynard Smith J, Smith N H, O’Rourke M, Spratt B G. How clonal are bacteria? Proc Natl Acad Sci USA. 1993;90:4384–4388. doi: 10.1073/pnas.90.10.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Musser J M, Mattingly S J, Quentin R, Goudeau A, Selander R K. Identification of a high-virulence clone of type III Streptococcus agalactiae (group B Streptococcus) causing invasive neonatal disease. Proc Natl Acad Sci USA. 1989;86:4731–4735. doi: 10.1073/pnas.86.12.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penner G A, Bush A, Wise R, Kim W, Domier L, Kasha K, Laroche A, Scoles G, Molnar S J, Fedak G. Reproducibility of random amplified polymorphic DNA (RAPD) analysis among laboratories. PCR Methods Appl. 1993;2:341–345. doi: 10.1101/gr.2.4.341. [DOI] [PubMed] [Google Scholar]
- 23.Pierre F, Quentin R, Gold F, Berger C. Encyclopédie Médico-Chirurgicale, Obstétrique. 5040C10. Paris, France: Editions Techniques; 1992. Infection bactérienne maternofoetale. [Google Scholar]
- 24.Quentin R, Huet H, Wang F-S, Geslin P, Goudeau A, Selander R K. Characterization of Streptococcus agalactiae strains by multilocus enzyme genotype and serotype: identification of multiple virulent clone families that cause invasive neonatal disease. J Clin Microbiol. 1995;33:2576–2581. doi: 10.1128/jcm.33.10.2576-2581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Regan J A, Klebanoff M A, Nugent R P, Eschenbach D A, Blackwelder W C, Lou Y, Gibbs R S, Rettig P J, Martin D H, Edelman R. Colonization with group B streptococci in pregnancy adverse outcome. VIP Study Group. Am J Obstet Gynecol. 1996;174:1354–1360. doi: 10.1016/s0002-9378(96)70684-1. [DOI] [PubMed] [Google Scholar]
- 26.Sanger F. Determination of nucleotide sequences in DNA. Science. 1981;214:1205–1210. doi: 10.1126/science.7302589. [DOI] [PubMed] [Google Scholar]
- 27.Suara R O, Adegbola R A, Baker C J, Secka O, Mulholland E K, Greenwood B M. Carriage of group B streptococci in pregnant Gambian mothers and their infants. Infect Dis. 1994;170:1316–1319. doi: 10.1093/infdis/170.5.1316. [DOI] [PubMed] [Google Scholar]