Abstract
The WNT signaling pathway plays a crucial role in oviduct/fallopian development. However, the specific physiological processes regulated by the WNT pathway in the fallopian/oviduct function remain obscure. Benefiting from the Lgr4 knockout mouse model, we report the regulation of oviduct epithelial secretion by LGR4. Specifically, the loss of Lgr4 altered the mouse oviduct size and weight, severely reduced the number of oviductal epithelial cells, and ultimately impaired the epithelial secretion. These alterations were mediated by a failure of CTNNB1 protein accumulation in the oviductal epithelial cytoplasm, by the modulation of WNT pathways, and subsequently by a profound change of the gene expression profile of epithelial cells. In addition, selective activation of the WNT pathway triggered the expression of steroidogenic genes, like Cyp11a1 and 3β-Hsd1, through the activation of the transcriptional factor NR5A2 in an oviduct primary cell culture system. As demonstrated, the LGR4 protein modulates a WNT-NR5A2 signaling cascade facilitating epithelial secretory cell maturation and steroidogenesis to safeguard oviduct development and function in mice.
Keywords: secretory cell, oviduct, LGR4, WNT/CTNNB1, NR5A2
Introduction
The oviduct, also known as the fallopian tube in humans, is a part of the female reproductive tract. For decades, a growing body of evidence indicates the oviduct serves not merely as a pipeline allowing the transit of gametes and embryo but also provides structural, environmental, and nutritional support for the conception (Menezo et al., 2015; Li and Winuthayanon, 2017; Lee et al., 2018; No et al., 2018; Wang and Larina, 2018). The physiological oviduct functions include the pickup of oocytes by the leaf-like folds in the infundibulum, the maturation of the ovum, and favoring the oocyte fertilization in the ampulla and early embryo development in the ampulla and isthmus (Li et al., 2019). Impairments of fallopian tube functions lead to frequent failures to conceive (Shaw and Horne, 2012; Hendriks et al., 2020). However, altogether the oviduct is less well understood for its contribution to reproduction compared with other female reproductive organs, like the ovary and uterus. This is because the assisted reproductive technologies (ARTs), such as in vitro fertilization (IVF) and intracytoplasmic sperm injection (ICSI), bypass the human fallopian tubes entirely.
A member of the WNT/CTNNB1 signaling pathway, the Wnt1, has been described for the first time about 35 years ago (Nusse and Varmus, 1982). Over the years the scientists recognized that this signaling pathway is one of the most fundamental pathways throughout the animal kingdom (Nusse and Clevers, 2017). The canonical WNT signaling pathway is initiated by the specific binding of WNTs to frizzled class receptors (FZDs), followed by the degradation of glycogen synthase kinase 3 beta (GSK3β) and the activation of CTNNB1 (β-catenin) in the cytoplasm. Consequently, accumulated CTNNB1 becomes able to translocate into the nucleus, where it interacts appropriately with a variety of transcription factors to modulate gene expression. Various studies revealed precisely that the canonical WNT plays a crucial role in ontogenesis. WNT signaling regulates the mouse Müllerian duct development, which is a necessary precursor of the female reproductive tract (Deutscher and Hung-Chang Yao, 2007; Prunskaite-Hyyrylainen et al., 2016). Moreover, the elongation and coiling of the oviduct are controlled by Wnt7a (Parr and McMahon, 1998). It also has been shown that an enhanced CTNNB1 is positively associated with a higher frequency of tubal pregnancies which accounts for 98% of ectopic pregnancies in humans (Li et al., 2013; Wang et al., 2019). Ciliary and secretory cells are the two most abundant cell types in the oviduct/fallopian epithelium, which are structurally pseudostratified on the stroma. However, selective ablation of CTNNB1 from the oviduct ciliated cells does not affect embryo transport, which might be because of compensatory mechanisms via intact CTNNB1 expression in adjacent epithelial secretory cells (Li et al., 2017). Further, an in vivo cell lineage tracing work reveals that the canonical WNT pathway is required for the homeostasis of the ratio of ciliated and secretory epithelial cells (Ghosh et al., 2017). These studies imply the secretory epithelial cells have vital physiological roles; however, the specific action of CTNNB1 in secretory cells remains to be deciphered.
A key regulator of WNT/CTNNB1 signaling is the leucine-rich repeat containing G protein-coupled receptors (LGRs). LGR4, LGR5, and LGR6, three closely related LGRs, function as the receptors for R-spondins, which modify precisely the WNT effect. LGR5 is identified as the mark of stem/progenitor cells in oviductal epithelia in mice (Ng et al., 2014). In addition, it has been shown that LGR6 is required for human fallopian organoid development (Kessler et al., 2015). We and others demonstrated that Lgr4 expresses in a variety of tissues and plays a significant role in amplifying the WNT/CTNNB1 signal in multiple tissues and organs, like hair follicles (Zak et al., 2016), ovaries (Pan et al., 2014), kidneys (Vidal et al., 2020), intestine (Liu et al., 2013), male reproductive tract (Qian et al., 2013), uterus (Sone et al., 2013), and many others (Wang et al., 2013; Han et al., 2014; Planas-Paz et al., 2016). However, to date, the role of LGR4 in the oviduct/fallopian tube is still obscure.
Here, we hypothesize that besides homeostasis, the canonical WNT pathway regulates the physiological function of secretory cells in mouse oviducts and that this regulation requires the precise action of LGR4.
Materials and Methods
Ethics Statement
All mice were kept under controlled photoperiod conditions (lights on 07:00–19:00 h) and were supplied with food and sterilized H2O ad libitum. Experiments were conformed to the regulations drafted by the Association for Assessment and Accreditation of Laboratory Animal Care in Shanghai. The ethics committee approval was obtained from the Institutional Ethics Committee of Shanghai Institute of Planned Parenthood Research Center (now Shanghai Institute for Biomedical and Pharmaceutical Technologies) for the commencement of the study.
Mice
The C57BL/6 Lgr4 gene-trap mouse strain was generated as previously described (Weng et al., 2008). Genomic DNA was extracted from tail biopsies and analyzed using the TaKaRa TaqTM Hot version (Cat# R007A). Primers for genotyping are listed in Supplementary Table 1. The 23- to 25-day-old females were sacrificed to perform a series of in vivo analyses.
Hematoxylin and Eosin Staining and Immunohistochemistry
Oviducts were collected and fixed in Bouin’s solution for hematoxylin and eosin (H&E) staining and in 4% paraformaldehyde (PFA) for immunohistochemistry (IHC). For IHC assay, sections (4–5 μm) were deparaffinized in xylene and rehydrated in gradient alcohol concentrations. After antigen retrieval, the sections were blocked by 5% bovine serum albumin (BSA). The sections were then incubated with primary antibodies overnight at 4°C. The next day, the sections were incubated with secondary antibodies for 20 min and then developed with 3,3′-diaminobenzidine and counterstained with hematoxylin. Antibodies were diluted as follows: OVGP1 at 1:100 (Santa Cruz, sc-377267), PAX8 at 1:100 (Proteintech Cat# 60145-4-lg), CTNNB1 at 1:100 (BD Transduction LaboratoriesTM, Cat# 610153), NR5A1 at 1:200 (Abcam Cat# ab217317), and NR5A2 at 1:500 (Abcam Cat# ab189876). Samples from at least three animals were evaluated to ensure reproducibility of the results.
Alcian Blue Staining
Deparaffinized and hydrated slides were incubated in 3% acetic acid for 3 min, followed by staining in 1% Alcian blue solution (pH 2.5) for 30–60 min. The sections were washed in running tap water for 2 min and rinsed in diH2O. After dehydration, tissue sections were mounted and observed under a microscope.
Periodic Acid–Schiff Staining
Deparaffinized and hydrated slides were incubated in 0.5% periodic acid solution for 5 min. After rinsing in three changes of distilled water, sections were placed in Schiff’s reagent for 15 min, followed by washing in tap water for 5 min. After dehydration, the sections were mounted and observed under a microscope.
β-Gal Staining
The frozen sections were fixed in 0.2% glutaraldehyde in phosphate-buffered saline (PBS) for 15 min on ice then washed in PBS for 10 min. Then the sections were immersed in 1 mg/ml X-gal staining solution overnight at 37°C in the dark. A post-fixation in 4% PFA for 10 min was required. After rinsing in PBS, the sections were counterstained with nuclear fast red then rinsed in distilled water for 2 min. After dehydration, sections were mounted and observed under a microscope.
Mouse Oviduct Epithelial Cell Isolation, Cell Culture, and ELISA Assay
Briefly, the mouse oviduct was surgically isolated and placed into an enzymatic media [Dulbecco’s minimum essential medium DMEM/F12 supplemented with 0.25% collagenase type IV (Invitrogen, 17104019), and 10 U/ml DNase I (Sigma, 10104159001)] for 15 min at 37°C. The softened tissue was then washed two times with PBS and transferred into a dissection medium [PBS with 0.5% penicillin/streptomycin and 1% fetal bovine serum (FBS)]. The oviduct tissue was then mechanically dissected using micro-scissors. The oviductal cell suspension was diluted with an equal volume of medium DMEM/F12 supplemented with 5% fetal bovine and centrifuged twice at 300 × g for 5 min at 4°C. The supernatant was removed, and the pellet of oviductal cells was re-suspended in culture media (DMEM/F12 supplemented with 10% FBS, 1% insulin, transferrin, and selenium solution) and seeded in a 24-well plate at 37°C and 5% CO2 in a humidified incubator. The culture medium was changed every 2 days. Once the adhered cells reached 70% confluence, we conducted the co-culture assay or E2 stimulated ELSA assays (OVGP1 kit: MyBioSource, Cat# abx576628, San Diego, CA, United States; CXCL8/IL-8 kit: R&D, Cat# D8000C). Before the E2 stimulation, cells were cultured in a medium without phenol red and FBS for at least 24 h.
Collection of Mouse Germinal Vesicle Oocytes and Co-culture
On the day of oocyte collection, the medium of the epithelial cell culture was changed from the DMEM/F12 to M2 medium (Sigma, M7167). Germinal vesicle stage oocytes (GV oocytes) were collected from 6-week-old C57BL/6 female mice injected with 5 IU of pregnant mare’s serum gonadotropin (PMSG; Sigma) 40 h before. Then the cumulus–oocyte complexes were obtained by puncturing the antral follicles, denuded by repeat pipetting using a stripper in 0.3 mg/ml hyaluronidase (Sigma H4272), and classified by GV stage. GV oocytes were co-cultured with epithelial cells in M2 media. Oocytes were observed at 12 and 24 h for oocyte maturation. Grading of oocytes was performed by microscopic examination. Progression to the metaphase II (MII) stage was identified by extrusion of the first polar body into the perivitelline space, which was observed under a microscope.
Chromatin Immunoprecipitation Assay
Mouse oviductal epithelial cells (MOECs) were treated with or without proteins WNT4, RSPONDIN2, or compound CT99021 overnight, and chromatin immunoprecipitation (ChIP) analysis was performed as previously described (Wu et al., 2007). DNA was precipitated with a polyclonal NR5A2 (Abcam Cat# ab189876) antibody or mouse IgG as a negative control. Precipitated DNA was amplified with the primers designed to the Cyp11a1 and Hsd3b1 promoter regions. See the detailed information of the primers in Supplementary Table 1. PCR products were confirmed by sequencing.
RNA-Seq Data Analysis
RNA-Seq Library Preparation
For RNA sequencing, four Lgr4 knockout (KO) and control mice were sacrificed, and the oviduct tissues were homogenized in TRIzol reagent and followed by RNA isolation. Isolation of mRNA was performed using the NEBNext PolyA mRNA Magnetic Isolation Module (New England Biolabs, Ipswich, MA, United States). The mRNA was then used for RNA-seq library preparation with the NEB Next Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, United States). The library was subjected to Illumina sequencing with paired-end 2 × 150 as the sequencing mode.
Quality Control and Alignment of Sequencing Data
Raw reads were filtered to obtain high-quality clean reads by removing sequencing adapters, short reads (length <35 bp), and low-quality reads using Cutadapt v1.18 and Trimmomatic v0.35. Then FastQC (with default parameters) is used to ensure high reads quality. The clean reads were mapped to the mm10 genome by HISAT2 v2.1.0.
Analysis of Differential Expression
Gene expression levels were estimated using fragments per kilobase of exon per million fragments mapped (FPKM) by StringTie v1.3.4d (Pertea et al., 2015). EdgeR v3.24.2, a R package, was used to measure differential gene expression (Robinson et al., 2010). The false discovery rate (FDR) control method was used to calculate the adjusted p-values in multiple testing to evaluate the significance of the differences. Genes with an adjusted p-value < 0.001, |log2FC| ≥ 2, and FPKM > 1 were used for subsequent analysis.
Functional Enrichment Analysis
A gene annotation file was retrieved from ensembl genome browser 96 database.1 To annotate genes with Gene Ontology (GO), clusterProfiler v3.4.4 (R package) was used.
The raw next-generation sequencing (NGS) data were deposited to the NCBI SRA database under project number (PRJNA691984). Reverse transcription polymerase chain reaction (RT-PCR) was performed to verify the mRNA-seq results using SYBR Premier EX Taq (TaKaRa). Genes were amplified with the indicated primers (Supplementary Table 1). Relative levels of mRNAs were calculated using MX3500pro software and normalized to the levels of endogenous actin in the same samples.
Statistical Analysis
Statistical data were analyzed with GraphPad Prism version 5. The statistical data of oviduct weight, numbers of specific stained secretory cells in the oviduct epithelium, and percentages of different stages of oocyte and RT-PCRs were presented as means ± SEM. Analysis of variance or Student’s t-test was used for statistical comparison to determine significance. Statistical significance was set as NS, p > 0.05; ∗p ≤ 0.05; ∗∗p ≤ 0.01; and ∗∗∗p ≤ 0.001. All presented results were from at least three independent experiments. The investigators were blinded to the group allocation during the experiment and when assessing the outcome.
Results
The Expression of Lgr4 in Mouse Oviduct
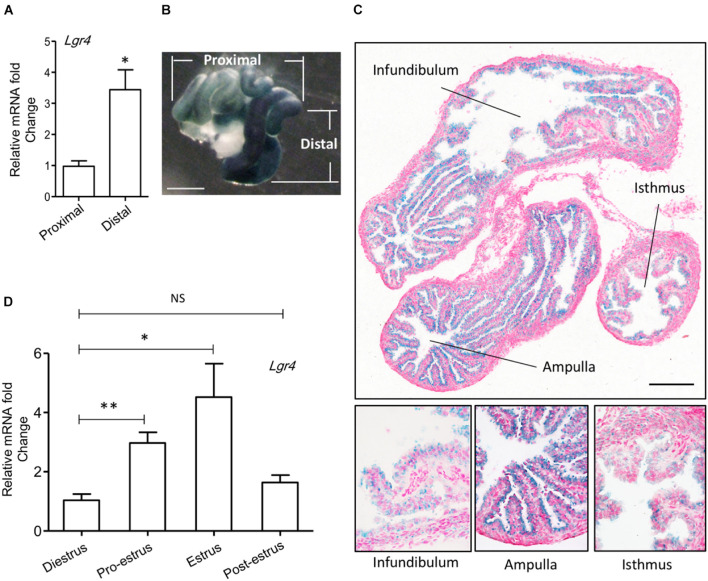
To study the role of Lgr4 in the oviduct, we analyzed its gene expression pattern. The mRNA level of Lgr4 was significantly enhanced by 3.52-fold in the distal region compared with the proximal region of the mouse oviduct (Figure 1A). As LGR4, LGR5, and LGR6 are the three most closely related leucine-rich repeat family of G-protein-coupled receptors (de Lau et al., 2012), we also determined the mRNA levels of Lgr5 and Lgr6. Unlike Lgr4, the expressions of Lgr5 and Lgr6 showed no differences between the distal and proximal regions in the mouse oviduct (Supplementary Figures 1A,B). To further identify the distribution of LGR4 in the oviduct, we took advantage of the β-gal reporter gene, which was inserted into the first intron of the Lgr4 genomic locus to track endogenous Lgr4 mRNA expression (Weng et al., 2008). The β-gal signal was enhanced strongly in the distal region (dark blue) compared with the slightly stained proximal region (Figure 1B). From the tissue morphology, the β-gal positive cells were oviductal epithelial cells. Although a positive signal could be detected in the epithelial layer of all three regions (infundibulum, ampulla, and isthmus), it was clear that the signal in the ampulla was the strongest compared with that in the other two regions (infundibulum and isthmus) (Figure 1C). Under specific hormonal controls during different stages of the estrus cycle, the oviduct produces different levels of fluid and macromolecules that optimize the microenvironment for gamete maturation. Therefore, we examined mRNA levels of Lgr4 expression. RT-PCR assay revealed that the Lgr4 expression was enhanced by more than 2.85-fold at the pro-estrus stage and then reached its peak level at 4.35-fold at the estrus stage compared with the di-estrus stage. This result suggested the expression of Lgr4 fluctuated dynamically during the estrus cycle (Figure 1D). The data showed that Lgr4 was present in the oviductal epithelium and with a significantly enhanced expression level at the distal region, especially in the ampulla, suggesting a physiological function of Lgr4 in this specific region within the estrus cycle.
FIGURE 1.
Expression of Lgr4 in oviducts of mice. (A) The relative expression of Lgr4 mRNA in the specific regions of the oviduct of the WT mice. (B) A whole mount LacZ staining represented the spatial expression pattern of Lgr4 in a heterozygous mouse oviduct. (C) A LacZ staining of a cryo-section of an Lgr4 heterozygous mouse oviduct. (D) The relative expression of Lgr4 mRNA in the mouse oviduct at different stages of the estrus cycle. At least three independent experiments were carried out. Data are presented as mean ± SEM of three mice. ∗p ≤ 0.05; ∗∗p ≤ 0.01. Scale bar, 1,000 μm in (B); 200 μm in (C).
Knockout of Lgr4 Led to Histological Deficiencies in the Oviduct
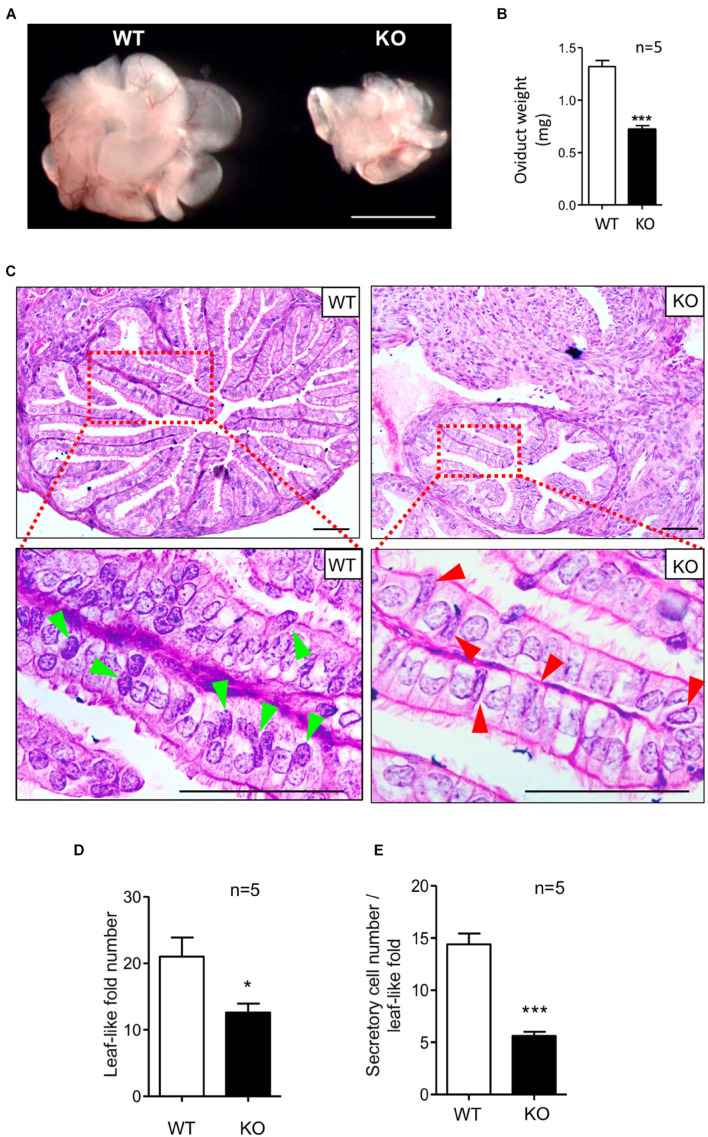
To determine the physiological role of the LGR4 protein in the murine oviduct, we compared the mutant and WT oviduct. The oviducts of KO female mice were significantly reduced in size (Figure 2A). In addition, the mean weight of isolated oviducts was 0.72 mg in the mutant mice, which was significantly lower than that in the WT group (1.32 mg) (Figure 2B). H&E staining was performed to determine the histological changes between the Lgr4 KO and WT oviducts. The cross-sectional area of the Lgr4 KO ampulla region was reduced in comparison with the WT control (Figure 2C, upper panel). The average number of the leaf-like folds in the WT ampulla was 21.00, while it was 12.60 in the mutant ampulla (Figure 2D). This result suggested that the loss of Lgr4 led to a reduced oviductal leaf-like folds. Furthermore, the columnar secretory cells with spot-like granules but no cilia were stained in dark blue in the WT (arrowed in bright green), while such type cells with less spot-like granules stained were reduced in the Lgr4-deficient oviduct (arrowed in red) (Figure 2C, lower panel). In the WT leaf-like fold, the average number of these secretory cells was 14.40, while in the mutant leaf-like fold, the number was reduced significantly to 5.60 (p < 0.001) (Figure 2E). As an ovarian dysfunction resulted from Lgr4 ablation (Pan et al., 2014), we removed the ovaries from both the WT and KO mice, and the ovariectomized female mice were then treated with daily injections of 100 ng per mouse of 17-β-estradiol or 1 mg per mouse of progesterone dissolved in 100 μl of sesame oil according to a previous report (Xiao et al., 2002). Histological defects with reduced numbers of leaf-like folds and secretory cells were still observed in the mutant ampulla (Supplementary Figures 2A–C). Together, these results suggested that the Lgr4 gene has an important role in the secretory cells in mouse oviduct.
FIGURE 2.
Defects of oviducts in response to Lgr4 knockout. (A) Morphology of oviducts. (B) The mean weights of the Lgr4 knockout and WT mouse adult oviducts. (C) Histology of the WT and Lgr4 KO oviducts was determined by hematoxylin and eosin staining. The red boxed areas are shown at higher magnification beneath. (D) The number of the leaf-like folds. (E) The number of the secretory cells per leaf-like folds. *p ≤ 0.05; ***p ≤ 0.001. Scale bar, 1,000 μm in (A); 50 μm in (C).
Ablation of Lgr4 Resulted in Secretory Deficiency of the Oviduct
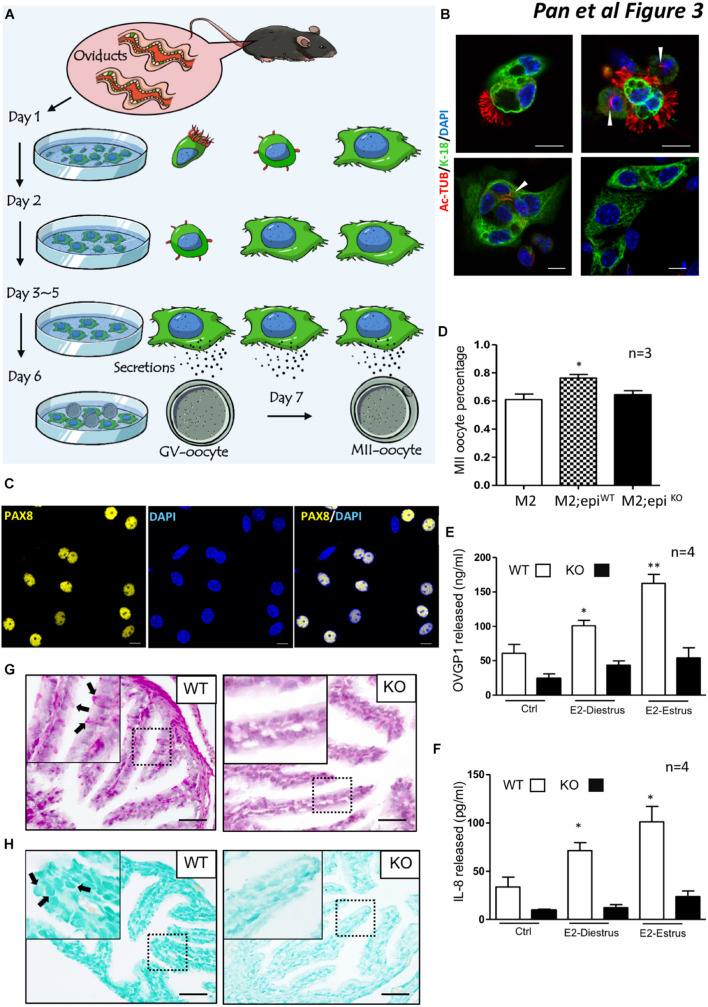
One fundamental role of the oviduct is to secrete factors to support and promote oocyte maturation. We used a two-type cell co-culture assay as outlined in the methods to detect whether ablation of Lgr4 led to a deficiency in oviduct secretion (Figure 3A). Before the examination, we measured the cytokeratin (cytokeratin-18) and alpha-tubulin (Ac-Tub) levels in MOEC cultures. One day after seeding, free-floating columnar MOECs with apparent cilia on the external surface (left) and adhered MOECs with reduced cilia (right) were observed (Figure 3B, upper panel). Two days later, the main cell type in the well was the adhered MOECs with (left) or without reduced cilia (right) (Figure 3B, lower panel). Three days later, almost all the adherent MOECs lost their cilia. PAX8 is reported to be a typical secretory cell marker in the oviduct (Ghosh et al., 2017; Ito et al., 2020). Most of the adhered cells at this time expressed PAX8 (Figure 3C). The data suggested that the cultured oviductal epithelial cells retained their secretory cell characteristic without cilial cells. As the co-culture system is widely used to promote the maturation of oocytes (Lee et al., 2011, 2017; Dadashpour Davachi et al., 2016), it can also show the secretory function of the oviduct epithelial cell in promoting oocyte maturation. When both the WT and KO adhered MOECs reached 70% confluence, we co-cultured the MOECs with the mouse GV oocytes for 12–24 h. The percentage of the GV oocytes that developed to the MII stage (MII oocyte) in the MII medium only was 61.00%. When the GV oocytes were co-cultured with WT mouse oviduct epithelial cells, the percentage enhanced significantly to 76.26%. However, when the GV oocytes were co-cultured with the Lgr4 mutant MOECs, the percentage upregulated slightly but not significantly to 64.57% compared with the MII medium only group (Figure 3D). The oviduct epithelium is covered with a mucosal surface containing mucins (MUCs) at its apical site, which provides a physical barrier against invading pathogens. After pathogen recognition through Toll-like receptors, epithelial cells release several chemokines, e.g., interleukin 8 (IL-8), to attract the immune system (Lagow et al., 1999; Zerbe et al., 2003; Chaturvedi et al., 2008; Kowsar et al., 2013). Oviduct-specific glycoprotein 1 (OVGP1), also named MUC9, is one of the most important glycoproteins secreted directly by the oviduct epithelial cells (Aviles et al., 2010; Saint-Dizier et al., 2014; Algarra et al., 2016; Lamy et al., 2018; Zhao and Kan, 2019). Therefore, we examined the protein levels of released OVGP1 and IL-8 in the WT and KO cultured MOEC supernatants. The oviduct is highly sensitive to the fluctuating ovarian hormone levels, so we supplemented the cultured MOECs with 17-β-estradiol (E2) mimicking the estrus effect, as the E2 influences the secretion of MOECs positively (Chen et al., 2013, 2018). The average OVGP1 level of the WT MOEC supernatant was 60.88 ng/ml, while this protein level was induced significantly to 100.89 ng/ml at the E2-diestrus stage (E2 = 10.00 pg/ml). The level was further enhanced to 162.45 ng/ml in the WT MOEC supernatant at the E2-estrus stage (E2 = 50.00 pg/ml). However, the average OVGP1 levels of the KO MOEC supernatant were 24.27, 43.51, and 54.30 pg/ml at each specific stage without any significant changes (Figure 3E). Similarly, the average IL-8 levels were 33.70, 71.45, and 101.22 ng/ml specifically, which were enhanced significantly under the stimulation of different concentrations of E2 in the WT MOEC supernatant. The average IL-8 levels of the KO MOEC supernatant were 9.65, 12.32, and 23.82 pg/ml at each specific stage without significant changes (Figure 3F).
FIGURE 3.
Secretion deficiencies in the oviducts of Lgr4 KO mice. (A) A schematic showing the procedure for mouse oviduct epithelial cell–oocyte co-culture system. (B) Immunofluorescence analysis of cytokeratin 18 (KRT-18) and alpha-tubulin (Ac-Tub) to the mouse oviduct epithelial cells. (C) Immunofluorescence analysis of PAX8. (D) The average percentage of the MII oocytes in response to different treatments. (E) The released protein level of OVGP1. (F) The released protein level of IL-8. (G) The periodic acid–Schiff (PAS) staining between the WT and Lgr4 knockout oviduct. (H) The Alcian blue staining between the WT and Lgr4 knockout oviduct. ∗p ≤ 0.05; ∗∗p ≤ 0.01. Scale bar, 10 μm in (B,C), 25 μm in (G,H). At least three independent experiments were carried out.
To further examine the role of Lgr4 in the secretion function of the mouse oviduct in vivo, we performed a set of histochemistry assays on the WT and Lgr4 KO oviduct in the estrus stage. The periodic acid–Schiff (PAS) staining was utilized to detect the presence of carbohydrates and carbohydrate compounds such as glycogen in the oviduct. The specific staining was observed in the WT group (arrowed in black), while in the Lgr4-deficient group only a low number of the specific stainings were detected (Figure 3G). In the Lgr4 KO group, the average number of the positive PAS staining signal was 2.00 per leaf-like fold, which was significantly lower than the wild type counterpart, 10.16 (Supplementary Figure 3A). A similar staining pattern was also found in the Alcian blue assay which was used to detect the acid mucosubstances and MUCs. Specifically, apparent signals were observed in the WT group (arrowed in black), while in the deficient group, the signals were reduced severely (Figure 3H). In the Lgr4 KO group, the average number of the positive Alcian blue staining signal was 2.33 per leaf-like fold, which was significantly lower than that in the WT, 23.00 (Supplementary Figure 3B). Together, all the in vitro and in vivo assays suggested that the secretion function of the MOECs was impaired by a deletion of the Lgr4 gene.
Deletion of Lgr4 Altered the Expression of Genes Involved in Oviductal Epithelial Secretion
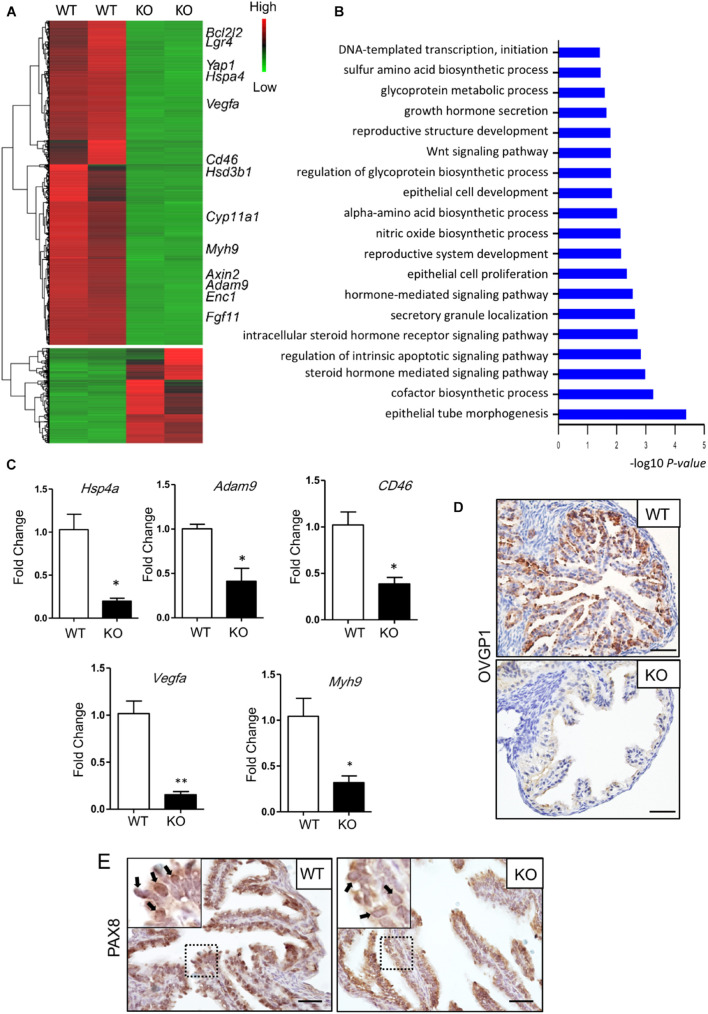
To reduce the influence of the estrus cycle on the transcriptom and to study the effect of Lgr4 deficiency on the transcriptom, we analyzed the wild type and mutant oviducts in the estrus stage. The RNA-seq revealed a downregulation of 2,597 genes and an upregulation of 762 genes with padj value < 0.001, |log2fold change| > 2, and FPKM > 1 (Figure 4A and Supplementary Table 2). GO highlighted that differentially expressed genes (DEGs) were mainly involved in the regulation of epithelial tube morphogenesis; biosynthetic and secretory process, like sulfur amino acid, glycoprotein, and hormones; WNT signaling pathway; DNA-templated transcription, and others (Figure 4B). A set of secreted factor genes was checked between the WT and KO oviducts also at the estrus stage (Figures 4C,D). The mRNA level of Hspa4, the heat-shock protein family A (HSP70) member 4 (HSPA4), which is important for the sperm fertilizing ability, was downregulated significantly by 5.20-fold in the Lgr4 mutant oviduct compared with the WT oviduct. The mRNA levels of disintegrin and metalloproteinase domain 9 (Adam9), and CD46 molecule (CD46), which were reported to have functions in the fertilization process, respectively (Alminana et al., 2017), were both significantly reduced by 2.42- and 2.64-fold. Another downregulated gene was Vegfa. VEGFA is a critical factor in the oviduct throughout the animal estrous cycle regulating the dynamics of oviduct fluid secretion and tubal contractibility. The mRNA of Vegfa was also detected to be significantly attenuated by 6.60-fold in the Lgr4 mutant oviduct. Myosin-9, also known as MYH9, has been identified on human sperm as a binding partner of OVGP1 (Kadam et al., 2006). The mRNA level of Myh9 was decreased significantly by 3.27-fold in the Lgr4 mutant oviduct compared with the WT oviduct. Accordantly, we checked the protein level of OVGP1, observing that this protein was reduced significantly in the oviducts of Lgr4 KO mice compared with the WT (Figure 4D). The IHC assay on the PAX8 protein showed specific 3,3′-diaminobenzidine (DAB) signals in the WT oviductal epithelial nuclei, while no apparent positive signals were observed in the KO mouse epithelial nuclei (Figure 4E, arrowed in black) at the stage of estrus. Collectively, the data demonstrated that Lgr4 deficiency led to a hypofunction of oviducts via downregulating of genes linked to secretory processes.
FIGURE 4.
Molecular defects in the oviduct of Lgr4-deficient mice. (A) RNA-seq for mouse oviducts (the estrus stage). Clustered heat map of log2-transformed FPKMs showing the differentially expressed genes after LGR4 deletion. Indicated genes are marked on the right side of the heat map. (B) Gene Ontology analysis. (C) RT-PCR assay showing the relative changes of some representative small molecular factor genes. (D) IHC assays to detect the protein level of OVGP1 in the WT or Lgr4-deficient oviducts. (E) IHC assays to detect the location of PAX8 in the WT and Lgr4-deficient oviductal epithelial cells. ∗p ≤ 0.05; ∗∗p ≤ 0.01. Scale bar, 25 μm in (D,E). RT-PCR, reverse transcription polymerase chain reaction; IHC, immunohistochemistry.
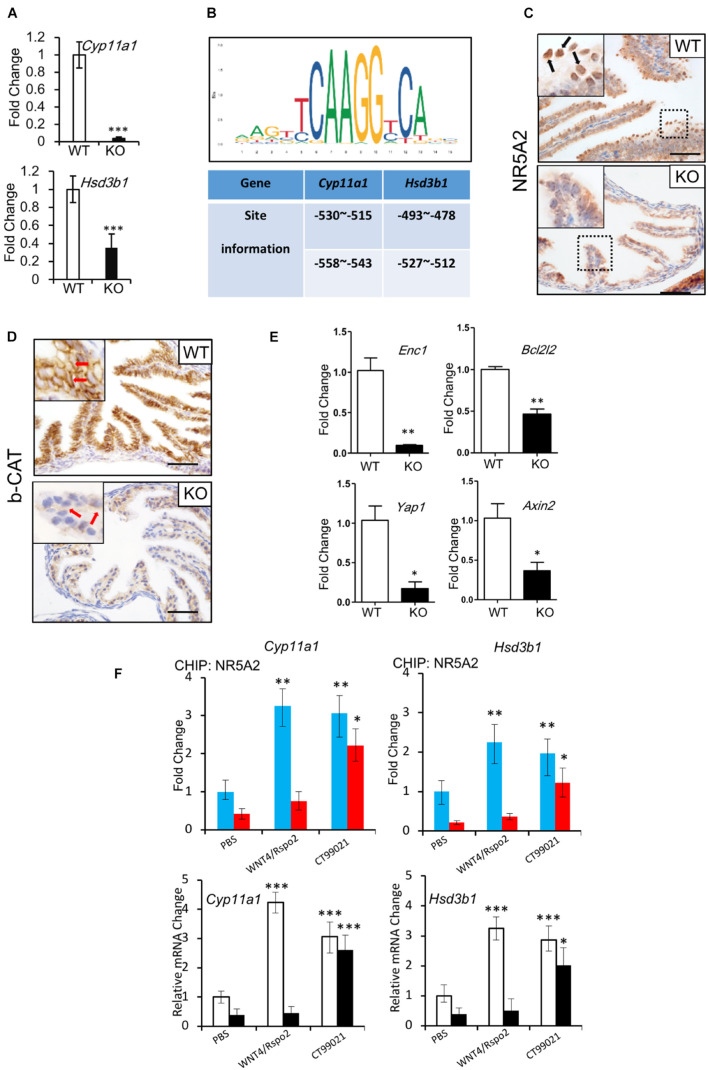
Lgr4 Deficiency Attenuated the CTNNB1/NR5A2 Signaling in Oviductal Secretory Cells
Hormone synthesis is predominantly controlled by members of the cytochrome P-450 (CYP) enzyme family and hydroxysteroid dehydrogenases (HSDs) (Chatuphonprasert et al., 2018). CYP11A1 and 3β-HSD are responsible for the formation of almost all the steroid hormones (Luu-The, 2013). Since we observed deficits in the secretory function of the oviducts of the Lgr4 KO mice, we evaluated mRNA levels of the Cyp11a1 and 3β-Hsd1 genes. The relative mRNA levels of the two genes were downregulated significantly (Figure 5A and Supplementary Table 2) at the stage of estrus. To determine how LGR4 regulated these steroidogenesis genes, we identified the WNT response elements in the Cyp11a1 and 3β-Hsd1 gene promoters. By the method of in silico analysis,2 we found that the two genes had two putative highly conserved NR5A1/2 binding sites in the proximal promoter regions (Figure 5B). Although the NR5A1 and NR5A2 share the same DNA binding sequences, these two transcription factors work in a non-overlapping way. These might suggest that NR5A1 and NR5A2 are presented in different systems. To demonstrate, we checked the presence of NR5A1 and NR5A2 in oviducts, respectively. By the IHC assay, we detected the apparent presence of NR5A2 but not NR5A1 in the WT mouse oviductal epithelium. Note that the corresponding signal was reduced severely in the mutant oviductal epithelial nuclei (Figure 5C, arrowed in black and Supplementary Figures 4A,B). The mis-location of NR5A2 protein observed in nuclei implied an impairment of the DNA binding ability with the transcriptional factor NR5A2.
FIGURE 5.
Regulation of NR5A2 activity by LGR4 mediated canonical WNT/CTNNB1 signaling pathway. (A) The mRNA levels of Cyp11a1 and Hsd3b1 in the WT and LGR4 mutant oviducts at the estrus stage. (B) Diagram of conserved NR5A1/2 binding sites in the Cyp11a1 and Hsd3b1 promoters predicted in silico. (C) IHC assays to detect the NR5A2 protein location in the WT or Lgr4-deficient oviducts. (D) IHC assays to detect the CTNNB1 protein level in the WT or Lgr4-deficient oviducts. (E) The mRNA levels of WNT target genes in WT and LGR4 mutant oviducts at estrus stage. (F) The chromatin of primary oviductal epithelial cells from WT or Lgr4 mutant mice was immunoprecipitated with a NR5A2 antibody. Immunoprecipitated DNA was amplified with the sets of specific primers for the indicated NR5A2 binding sites in Cyp11a1 and Hsd3b1 promoters (upper panel). The relative fold change of Cyp11a1 and Hsd3b1 genes of the primary oviductal epithelial cells after the treatment of WNT4/RSPO2 or CT99201 (lower panel) are presented. ∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001. Scale bar, 50 μm in (C,D). IHC, immunohistochemistry.
LGR4 has been widely considered as a WNT effector amplifier. To study if LGR4 has a similar function in our model, we compared the protein level of CTNNB1 between the WT and KO mice. The accumulation of CTNNB1 in the cytoplasm of cells was detectable in oviduct epithelial cells of WT mice at the stage of estrus. In contrast, the accumulation of CTNNB1 in Lgr4 KO oviductal epithelial cells was significantly downregulated (Figure 5D, arrowed in red). In addition, a set of WNT/CTNNB1 signaling target genes like Enc1 (Fujita et al., 2001), Bcl2l2 (Lapham et al., 2009), Yap1 (Konsavage et al., 2012), and Axin2 (Yan et al., 2001; Jho et al., 2002; Lustig et al., 2002) were also downregulated significantly in the Lgr4 KO oviducts compared with the WT control at the estrus stage (Figure 5E). These data further confirmed the regulatory function of LGR4 on the canonical WNT signaling pathway in the oviduct epithelium.
We hypothesized that the activated CTNNB1 might increase NR5A2 binding to the Cyp11a1 and 3β-Hsd1 gene promoters in the mouse oviduct. To this end, we performed the ChIP-qPCR assay utilizing primary culture cells of the WT and Lgr4 mutant mouse oviducts. In the WT, the capacity of the transcription factor NR5A2 to bind with Cyp11a1 and 3β-Hsd1 promoter regions was improved by ∼3.26- and ∼2.25-fold, respectively, in response to the stimulation of factors WNT4/RSPO2. Such improved binding capacity of NR5A2 was not detectable in cultured cells of Lgr4 mutant mice. However, CT99021, a commercial GSK-3α/β inhibitor that activates the canonical WNT pathway, enhanced the DNA binding abilities in WT and mutant primary oviductal epithelial cells. Specifically, in the WT group, the capacity of DNA binding to the Cyp11a1 and 3β-Hsd1 promoters of the transcription factor NR5A2 was increased by ∼3.02- and ∼1.97-fold, respectively, while in the mutant group, such ability of NR5A2 to the Cyp11a1 and 3β-Hsd1 promoters was rescued by ∼5.11- and ∼5.54-fold, respectively (Figure 5F, upper panel). The relative mRNA fold change of the Cyp11a1 and 3β-Hsd1 genes after WNT4/RSPO2 or CT99021 application in the WT or Lgr4 mutant oviductal epithelial cells further suggested the activity of the transcription factor NR5A2 was regulated by the canonical WNT cascade (Figure 5F, lower panel).
Discussion
A previous study showed that the epithelial cell-specific [keratin-5 (K5) Cre] knockout of Lgr4 in female mice resulted in subfertility. The researchers pointed out that this reproductive defect was due to alterations of epithelial differentiation characterized by a reduced uterine gland number (Sone et al., 2013). Although keratin-5 is expressed in the oviduct (Paik et al., 2012; Stadnicka et al., 2018), they did not mention the oviduct. The oviduct is derived from the Müllerian duct in mice (Stewart and Behringer, 2012). Lgr4 K5-Cre knockout reduced the numbers of fourfold and 8-cell–stage embryos and impaired embryo morphology at the blastocyst stage (Mohri et al., 2010). These observations indicated a potential involvement of Lgr4 in the functionality of oviducts. In this present study, we elucidated the physiological role of LGR4 in oviducts of mice in greater detail. We showed that Lgr4 was expressed in the epithelium of mouse oviducts and inducible at the estrus stage in ampulla region. Knockout of the Lgr4 gene resulted in a deficiency of the mouse oviduct structure and led to oviductal hypo-secretion. This was mediated by the impairment of the CTNNB1 protein accumulation in the cytoplasm of the oviductal secretory cells. In addition, the Lgr4 knockout caused a decrease in the WNT signaling that was required for the activation of the downstream transcriptional factor NR5A2.
The NR5A receptors are the orphan nuclear receptor proteins that act as transcription factors. They are evolutionarily conserved, as orthologs and paralogs are found in metazoans, from roundworms to mammals (Meinsohn et al., 2019). Despite the structural similarities between NR5A1 (steroidogenic factor-1 or SF-1) and NR5A2 (liver receptor homolog-1 or LRH-1), as well as their demonstrated ability to bind to the same DNA sequences, these two transcriptional factors are involved in the regulation of distinct physiological functions and often cannot compensate for each other (Duggavathi et al., 2008; Meinsohn et al., 2019). The two members are differentially expressed across a wide range of organs. NR5A1 exists in steroidogenic organs like the hypothalamus, pituitary gland, and testis. Both NR5A2 and NR5A1 regulated steroid synthesis, but the NR5A2 distribution was more widespread including in the liver (Xu et al., 2016; Stein et al., 2017; Miranda et al., 2018), pancreas (Benod et al., 2011; Holmstrom et al., 2011), intestine (Bayrer et al., 2015, 2018), ovary (Kawabe et al., 2013; Zhang et al., 2013; Bertolin et al., 2014), and uterus (Brosens et al., 2013; Mouzat et al., 2013). Here we showed the NR5A2, but not NR5A1, is localized in the mouse oviduct epithelium. According to the reported studies, both of the two transcription factors are co-activated by the CTNNB1. For example, NR5A1 and CTNNB1 interact in the signaling pathway to produce testosterone in Leydig cells (Jordan et al., 2003). NR5A2 interacts with CTNNB1 to promote cell cycle-dependent gene expression and regulates cell proliferation in the intestinal crypt and granulosa cells (Botrugno et al., 2004; Meinsohn et al., 2018). In addition, we show that the transcriptional control by NR5A2 of steroidogenesis required the activation of the canonical WNT pathway in oviductal epithelial cells.
The canonical WNT pathway occupies a multitude of essential roles in cell fate and tissue homeostasis. Misregulation of WNT signaling has been associated with a variety of human diseases (Clevers, 2006; Clevers and Nusse, 2012; Nusse and Clevers, 2017). Research using patient tissues detected that the CTNNB1 expression was enhanced in the oviductal epithelium in ectopic tubal pregnancies. The ectopic implantation of embryos escalated the canonical WNT signaling, which changed the environment in the fallopian tube, such as reduced E-cadherin, induced angiogenesis, and glycogen accumulation (Li et al., 2013). In this present study, we demonstrated that insufficient WNT caused secretory deficiencies in the oviduct. Hence, our study, together with previous reports, suggested that the WNT cascade required a precise regulation in the fallopian tube. Unlike Lgr5 and Lgr6, we showed the Lgr4 gene was expressed broadly in the mouse oviduct epithelium. And the RNA-seq data also suggested the level of Lgr4 was much higher than those of Lgr5 and Lgr6. Knockout of Lgr4 severely reduced the number of secretory cells. Interestingly, the mRNA-seq revealed insignificant changes in the levels of Lgr5 and Lgr6. Taking into the account that the WNT/CTNNB1 signaling is essential for the self-renewal of secretory cells in the oviduct and our data, we conclude that LGR4 but not LGR5 or LGR6 controls the canonical WNT signaling pathway in the secretory cells.
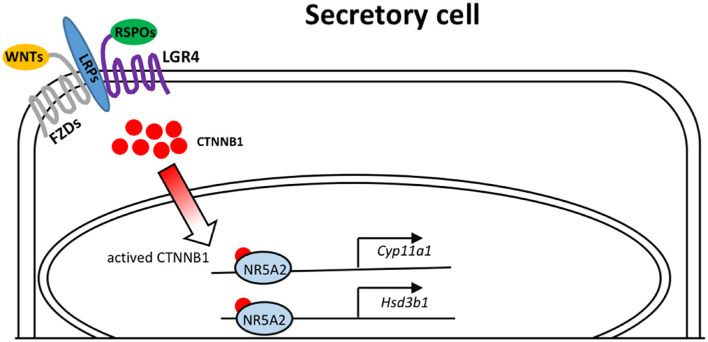
In summary, we presented data indicating a dominant role of Lgr4 in the regulation of oviductal epithelial secretion (Figure 6). As the fallopian tube pathologies underlie a total number of 30% of infertile women worldwide (Briceag et al., 2015), identifying the essential roles of LGR4 in these patients represents an advancement that has implications for the development of diagnostic tools and as a potential drug target to treat fallopian tube secretion insufficiency.
FIGURE 6.
Model depicting the molecular mechanisms of LGR4 mediated WNT signaling that activates the transcription factor NR5A2 to bind on the promoters of the steroidogenic genes, Cyp11a1 and Hsd3b1.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: BioProject (PRJNA691984) in NCBI, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA691984.
Ethics Statement
The animal study was reviewed and approved by the Institutional Ethics Committee of Shanghai Institute of Planned Parenthood Research Center (Shanghai Institute of Biomedical and Pharmaceutical Technologies now).
Author Contributions
XT carried out all the experiments and drafted the manuscript. LZ carried out all the analysis of bio-informatics data. TL, JZ, and KQ maintained and genotyped the mice. HW and SS prepared the slides and carried out the PAS, Alcian blue, HE, and IHC staining. MH and FZ performed the isolated and maintained the mouse tubal epithelial cells. MZ and CL carried out the oocyte–epithelial co-culture assay. RL and HP provided critical suggestions, designed the research, and supervised the project. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank Dali Li and Mingyao Liu for their Lgr4 knockout mouse as a gift. We also thank Thomas Behnisch and Yinghan Zhuang for their comments and advice.
Footnotes
Funding
This work was partially supported by grants from the National Natural Science Foundation of China [grant nos. 31701303 (to HP), 81971443 (to RL), and 31900591 (to MH)]. An Open Project Fund from Key Laboratory of Reproduction Regulation of NHC [KF2018-02 (to CL)]. Shanghai Municipal Committee of Science and Technology [21140903600 (to RL)]. Shanghai Municipal Commission of Health and Family Planning [20194Y0237 (to MZ)].
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcell.2021.666303/full#supplementary-material
The mRNA level of Lgr5 and Lgr6 in mouse oviducts. (A) The mRNA levels of Lgr5 in the specific regions of the oviduct. (B) The mRNA levels of Lgr6 in the specific regions of the oviduct. At least three independent experiments were carried out. Data were presented as mean ± SEM. NS > 0.05.
Loss of LGR4 led to histological abnormalities of oviducts. (A) Histological analysis of the WT and Lgr4 KO oviducts were performed by hematoxylin and eosin staining after the supplement of E2. (B) The average number of leaf-like folds per section after the supplement of E2. (C) The average number of the secretory cells per leaf-like fold after the supplement of E2. ∗p ≤ 0.05; ∗∗∗p ≤ 0.001. Scale bar, 50 μm.
Loss of LGR4 led to a deficiency in oviductal secretion. (A) The bar graphs summarized the number of positively labeled cells of PAS stained oviduct slides. (B) The number of the positively Alcian blue stained cells in oviducts were presented. At least three independent experiments were carried out. ∗∗∗p ≤ 0.001.
Detection of NR5A1 protein in mouse oviduct. (A) IHC assay of NR5A1 in the WT and Lgr4 knockout mouse oviducts. (B) IHC assay of NR5A1 in the WT testis as control. Note, the positive signals in the enclosed area by red dashed lines. Scale bar, 50 μm.
Primer list.
Differential expressed genes between the Lgr4 deletion and control oviducts. Differential gene expression analysis between Lgr4 deletion and control oviducts (the estrus stage) revealed a number of significantly different expressed genes.
References
- Algarra B., Han L., Soriano-Ubeda C., Aviles M., Coy P., Jovine L., et al. (2016). The C-terminal region of OVGP1 remodels the zona pellucida and modifies fertility parameters. Sci. Rep. 6:32556. 10.1038/srep32556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alminana C., Corbin E., Tsikis G., Alcantara-Neto A. S., Labas V., Reynaud K., et al. (2017). Oviduct extracellular vesicles protein content and their role during oviduct-embryo cross-talk. Reproduction 154 153–168. 10.1530/REP-17-0054 [DOI] [PubMed] [Google Scholar]
- Aviles M., Gutierrez-Adan A., Coy P. (2010). Oviductal secretions: will they be key factors for the future ARTs? Mol. Hum. Reprod. 16 896–906. 10.1093/molehr/gaq056 [DOI] [PubMed] [Google Scholar]
- Bayrer J. R., Mukkamala S., Sablin E. P., Webb P., Fletterick R. J. (2015). Silencing LRH-1 in colon cancer cell lines impairs proliferation and alters gene expression programs. Proc. Natl. Acad. Sci. U. S. A. 112 2467–2472. 10.1073/pnas.1500978112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayrer J. R., Wang H., Nattiv R., Suzawa M., Escusa H. S., Fletterick R. J., et al. (2018). LRH-1 mitigates intestinal inflammatory disease by maintaining epithelial homeostasis and cell survival. Nat. Commun. 9:4055. 10.1038/s41467-018-06137-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benod C., Vinogradova M. V., Jouravel N., Kim G. E., Fletterick R. J., Sablin E. P. (2011). Nuclear receptor liver receptor homologue 1 (LRH-1) regulates pancreatic cancer cell growth and proliferation. Proc. Natl. Acad. Sci. U. S. A. 108 16927–16931. 10.1073/pnas.1112047108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolin K., Gossen J., Schoonjans K., Murphy B. D. (2014). The orphan nuclear receptor Nr5a2 is essential for luteinization in the female mouse ovary. Endocrinology 155 1931–1943. 10.1210/en.2013-1765 [DOI] [PubMed] [Google Scholar]
- Botrugno O. A., Fayard E., Annicotte J. S., Haby C., Brennan T., Wendling O., et al. (2004). Synergy between LRH-1 and beta-catenin induces G1 cyclin-mediated cell proliferation. Mol. Cell 15 499–509. 10.1016/j.molcel.2004.07.009 [DOI] [PubMed] [Google Scholar]
- Briceag I., Costache A., Purcarea V. L., Cergan R., Dumitru M., Briceag I., et al. (2015). Fallopian tubes–literature review of anatomy and etiology in female infertility. J. Med. Life 8 129–131. [PMC free article] [PubMed] [Google Scholar]
- Brosens J. J., Blanks A. M., Lucas E. S. (2013). LRH-1: orphaned, adopted and needed for pregnancy. Nat. Med. 19 968–969. 10.1038/nm.3271 [DOI] [PubMed] [Google Scholar]
- Chatuphonprasert W., Jarukamjorn K., Ellinger I. (2018). Physiology and Pathophysiology of Steroid Biosynthesis, Transport and Metabolism in the Human Placenta. Front. Pharmacol. 9:1027. 10.3389/fphar.2018.01027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi P., Singh A. P., Batra S. K. (2008). Structure, evolution, and biology of the MUC4 mucin. FASEB J. 22 966–981. 10.1096/fj.07-9673rev [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Einspanier R., Schoen J. (2013). In vitro mimicking of estrous cycle stages in porcine oviduct epithelium cells: estradiol and progesterone regulate differentiation, gene expression, and cellular function. Biol. Reprod. 89:54. 10.1095/biolreprod.113.108829 [DOI] [PubMed] [Google Scholar]
- Chen S., Palma-Vera S. E., Kempisty B., Rucinski M., Vernunft A., Schoen J. (2018). In Vitro Mimicking of Estrous Cycle Stages: dissecting the Impact of Estradiol and Progesterone on Oviduct Epithelium. Endocrinology 159 3421–3432. 10.1210/en.2018-00567 [DOI] [PubMed] [Google Scholar]
- Clevers H. (2006). Wnt/beta-catenin signaling in development and disease. Cell 127 469–480. 10.1016/j.cell.2006.10.018 [DOI] [PubMed] [Google Scholar]
- Clevers H., Nusse R. (2012). Wnt/beta-catenin signaling and disease. Cell 149 1192–1205. 10.1016/j.cell.2012.05.012 [DOI] [PubMed] [Google Scholar]
- Dadashpour Davachi N., Zare Shahneh A., Kohram H., Zhandi M., Shamsi H., Hajiyavand A. M., et al. (2016). Differential influence of ampullary and isthmic derived epithelial cells on zona pellucida hardening and in vitro fertilization in ovine. Reprod. Biol. 16 61–69. 10.1016/j.repbio.2015.11.002 [DOI] [PubMed] [Google Scholar]
- de Lau W. B., Snel B., Clevers H. C. (2012). The R-spondin protein family. Genome Biol. 13:242. 10.1186/gb-2012-13-3-242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher E., Hung-Chang Yao H. (2007). Essential roles of mesenchyme-derived beta-catenin in mouse Mullerian duct morphogenesis. Dev. Biol. 307 227–236. 10.1016/j.ydbio.2007.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggavathi R., Volle D. H., Mataki C., Antal M. C., Messaddeq N., Auwerx J., et al. (2008). Liver receptor homolog 1 is essential for ovulation. Genes Dev. 22 1871–1876. 10.1101/gad.472008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Furukawa Y., Tsunoda T., Tanaka T., Ogawa M., Nakamura Y. (2001). Up-regulation of the ectodermal-neural cortex 1 (ENC1) gene, a downstream target of the beta-catenin/T-cell factor complex, in colorectal carcinomas. Cancer Res. 61 7722–7726. [PubMed] [Google Scholar]
- Ghosh A., Syed S. M., Tanwar P. S. (2017). In vivo genetic cell lineage tracing reveals that oviductal secretory cells self-renew and give rise to ciliated cells. Development 144 3031–3041. 10.1242/dev.149989 [DOI] [PubMed] [Google Scholar]
- Han X. H., Jin Y. R., Tan L., Kosciuk T., Lee J. S., Yoon J. K. (2014). Regulation of the follistatin gene by RSPO-LGR4 signaling via activation of the WNT/beta-catenin pathway in skeletal myogenesis. Mol. Cell. Biol. 34 752–764. 10.1128/mcb.01285-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks E., Rosenberg R., Prine L. (2020). Ectopic Pregnancy: diagnosis and Management. Am. Fam. Physician 101 599–606. [PubMed] [Google Scholar]
- Holmstrom S. R., Deering T., Swift G. H., Poelwijk F. J., Mangelsdorf D. J., Kliewer S. A., et al. (2011). LRH-1 and PTF1-L coregulate an exocrine pancreas-specific transcriptional network for digestive function. Genes Dev. 25 1674–1679. 10.1101/gad.16860911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Yamamoto Y., Kimura K. (2020). Analysis of ciliogenesis process in the bovine oviduct based on immunohistochemical classification. Mol. Biol. Rep. 47 1003–1012. 10.1007/s11033-019-05192-w [DOI] [PubMed] [Google Scholar]
- Jho E. H., Zhang T., Domon C., Joo C. K., Freund J. N., Costantini F. (2002). Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 22 1172–1183. 10.1128/MCB.22.4.1172-1183.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan B. K., Shen J. H., Olaso R., Ingraham H. A., Vilain E. (2003). Wnt4 overexpression disrupts normal testicular vasculature and inhibits testosterone synthesis by repressing steroidogenic factor 1/beta-catenin synergy. Proc. Natl. Acad. Sci. U. S. A. 100 10866–10871. 10.1073/pnas.1834480100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam K. M., D’souza S. J., Bandivdekar A. H., Natraj U. (2006). Identification and characterization of oviductal glycoprotein-binding protein partner on gametes: epitopic similarity to non-muscle myosin IIA. MYH 9. Mol. Hum. Reprod. 12 275–282. 10.1093/molehr/gal028 [DOI] [PubMed] [Google Scholar]
- Kawabe S., Yazawa T., Kanno M., Usami Y., Mizutani T., Imamichi Y., et al. (2013). A novel isoform of liver receptor homolog-1 is regulated by steroidogenic factor-1 and the specificity protein family in ovarian granulosa cells. Endocrinology 154 1648–1660. 10.1210/en.2012-2008 [DOI] [PubMed] [Google Scholar]
- Kessler M., Hoffmann K., Brinkmann V., Thieck O., Jackisch S., Toelle B., et al. (2015). The Notch and Wnt pathways regulate stemness and differentiation in human fallopian tube organoids. Nat. Commun. 6:8989. 10.1038/ncomms9989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konsavage W. M., Jr., Kyler S. L., Rennoll S. A., Jin G., Yochum G. S. (2012). Wnt/beta-catenin signaling regulates Yes-associated protein (YAP) gene expression in colorectal carcinoma cells. J. Biol. Chem. 287 11730–11739. 10.1074/jbc.M111.327767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowsar R., Hambruch N., Liu J., Shimizu T., Pfarrer C., Miyamoto A. (2013). Regulation of innate immune function in bovine oviduct epithelial cells in culture: the homeostatic role of epithelial cells in balancing Th1/Th2 response. J. Reprod. Dev. 59 470–478. 10.1262/jrd.2013-036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagow E., Desouza M. M., Carson D. D. (1999). Mammalian reproductive tract mucins. Hum. Reprod. Update 5 280–292. 10.1093/humupd/5.4.280 [DOI] [PubMed] [Google Scholar]
- Lamy J., Nogues P., Combes-Soia L., Tsikis G., Labas V., Mermillod P., et al. (2018). Identification by proteomics of oviductal sperm-interacting proteins. Reproduction 155 457–466. 10.1530/REP-17-0712 [DOI] [PubMed] [Google Scholar]
- Lapham A., Adams J. E., Paterson A., Lee M., Brimmell M., Packham G. (2009). The Bcl-w promoter is activated by beta-catenin/TCF4 in human colorectal carcinoma cells. Gene 432 112–117. 10.1016/j.gene.2008.12.002 [DOI] [PubMed] [Google Scholar]
- Lee H. J., Quaas A. M., Wright D. L., Toth T. L., Teixeira J. M. (2011). In vitro maturation (IVM) of murine and human germinal vesicle (GV)-stage oocytes by coculture with immortalized human fallopian tube epithelial cells. Fertil. Steril. 95 1344–1348. 10.1016/j.fertnstert.2010.08.034 [DOI] [PubMed] [Google Scholar]
- Lee S. H., Oh H. J., Kim M. J., Kim G. A., Choi Y. B., Jo Y. K., et al. (2017). Oocyte maturation-related gene expression in the canine oviduct, cumulus cells, and oocytes and effect of co-culture with oviduct cells on in vitro maturation of oocytes. J. Assist. Reprod. Genet. 34 929–938. 10.1007/s10815-017-0910-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., Oh H. J., Kim M. J., Kim G. A., Choi Y. B., Jo Y. K., et al. (2018). Effect of co-culture canine cumulus and oviduct cells with porcine oocytes during maturation and subsequent embryo development of parthenotes in vitro. Theriogenology 106 108–116. 10.1016/j.theriogenology.2017.09.015 [DOI] [PubMed] [Google Scholar]
- Li C., Wu Y. T., Zhu Q., Zhang H. Y., Huang Z., Zhang D., et al. (2019). TRPV4 is involved in levonorgestrel-induced reduction in oviduct ciliary beating. J. Pathol. 248 77–87. 10.1002/path.5233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Zhu W. J., Ma Z. L., Wang G., Peng H., Chen Y., et al. (2013). Enhanced beta-catenin expression and inflammation are associated with human ectopic tubal pregnancy. Hum. Reprod. 28 2363–2371. 10.1093/humrep/det246 [DOI] [PubMed] [Google Scholar]
- Li S., O’neill S. R., Zhang Y., Holtzman M. J., Takemaru K. I., Korach K. S., et al. (2017). Estrogen receptor alpha is required for oviductal transport of embryos. FASEB J. 31 1595–1607. 10.1096/fj.201601128R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Winuthayanon W. (2017). Oviduct: roles in fertilization and early embryo development. J. Endocrinol. 232 R1–R26. 10.1530/JOE-16-0302 [DOI] [PubMed] [Google Scholar]
- Liu S., Qian Y., Li L., Wei G., Guan Y., Pan H., et al. (2013). Lgr4 gene deficiency increases susceptibility and severity of dextran sodium sulfate-induced inflammatory bowel disease in mice. J. Biol. Chem. 288 8794–8803. 10.1074/jbc.M112.436204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig B., Jerchow B., Sachs M., Weiler S., Pietsch T., Karsten U., et al. (2002). Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol. Cell. Biol. 22 1184–1193. 10.1128/MCB.22.4.1184-1193.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu-The V. (2013). Assessment of steroidogenesis and steroidogenic enzyme functions. J. Steroid Biochem. Mol. Biol. 137 176–182. 10.1016/j.jsbmb.2013.05.017 [DOI] [PubMed] [Google Scholar]
- Meinsohn M. C., Morin F., Bertolin K., Duggavathi R., Schoonjans K., Murphy B. D. (2018). The Orphan Nuclear Receptor Liver Homolog Receptor-1 (Nr5a2) Regulates Ovarian Granulosa Cell Proliferation. J. Endocr. Soc. 2 24–41. 10.1210/js.2017-00329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinsohn M. C., Smith O. E., Bertolin K., Murphy B. D. (2019). The Orphan Nuclear Receptors Steroidogenic Factor-1 and Liver Receptor Homolog-1: structure, Regulation, and Essential Roles in Mammalian Reproduction. Physiol. Rev. 99 1249–1279. 10.1152/physrev.00019.2018 [DOI] [PubMed] [Google Scholar]
- Menezo Y., Guerin P., Elder K. (2015). The oviduct: a neglected organ due for re-assessment in IVF. Reprod. Biomed. Online 30 233–240. 10.1016/j.rbmo.2014.11.011 [DOI] [PubMed] [Google Scholar]
- Miranda D. A., Krause W. C., Cazenave-Gassiot A., Suzawa M., Escusa H., Foo J. C., et al. (2018). LRH-1 regulates hepatic lipid homeostasis and maintains arachidonoyl phospholipid pools critical for phospholipid diversity. JCI Insight 3:e96151. 10.1172/jci.insight.96151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohri Y., Umezu T., Hidema S., Tomisawa H., Akamatsu A., Kato S., et al. (2010). Reduced fertility with impairment of early-stage embryos observed in mice lacking Lgr4 in epithelial tissues. Fertil. Steril. 94 2878–2881. 10.1016/j.fertnstert.2010.05.050 [DOI] [PubMed] [Google Scholar]
- Mouzat K., Baron S., Marceau G., Caira F., Sapin V., Volle D. H., et al. (2013). Emerging roles for LXRs and LRH-1 in female reproduction. Mol. Cell. Endocrinol. 368 47–58. 10.1016/j.mce.2012.06.009 [DOI] [PubMed] [Google Scholar]
- Ng A., Tan S., Singh G., Rizk P., Swathi Y., Tan T. Z., et al. (2014). Lgr5 marks stem/progenitor cells in ovary and tubal epithelia. Nat. Cell Biol. 16 745–757. 10.1038/ncb3000 [DOI] [PubMed] [Google Scholar]
- No J., Zhao M., Lee S., Ock S. A., Nam Y., Hur T. Y. (2018). Enhanced in vitro maturation of canine oocytes by oviduct epithelial cell co-culture. Theriogenology 105 66–74. 10.1016/j.theriogenology.2017.09.002 [DOI] [PubMed] [Google Scholar]
- Nusse R., Clevers H. (2017). Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 169 985–999. 10.1016/j.cell.2017.05.016 [DOI] [PubMed] [Google Scholar]
- Nusse R., Varmus H. E. (1982). Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell 31 99–109. 10.1016/0092-8674(82)90409-3 [DOI] [PubMed] [Google Scholar]
- Paik D. Y., Janzen D. M., Schafenacker A. M., Velasco V. S., Shung M. S., Cheng D., et al. (2012). Stem-like epithelial cells are concentrated in the distal end of the fallopian tube: a site for injury and serous cancer initiation. Stem Cells 30 2487–2497. 10.1002/stem.1207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H., Cui H., Liu S., Qian Y., Wu H., Li L., et al. (2014). Lgr4 gene regulates corpus luteum maturation through modulation of the WNT-mediated EGFR-ERK signaling pathway. Endocrinology 155 3624–3637. [DOI] [PubMed] [Google Scholar]
- Parr B. A., McMahon A. P. (1998). Sexually dimorphic development of the mammalian reproductive tract requires Wnt-7a. Nature 395 707–710. [DOI] [PubMed] [Google Scholar]
- Pertea M., Pertea G. M., Antonescu C. M., Chang T. C., Mendell J. T., Salzberg S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33 290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas-Paz L., Orsini V., Boulter L., Calabrese D., Pikiolek M., Nigsch F. (2016). The RSPO-LGR4/5-ZNRF3/RNF43 module controls liver zonation and size. Nat. Cell Biol. 18 467–479. 10.1038/ncb3337 [DOI] [PubMed] [Google Scholar]
- Prunskaite-Hyyrylainen R., Skovorodkin I., Xu Q., Miinalainen I., Shan J., Vainio S. J. (2016). Wnt4 coordinates directional cell migration and extension of the Mullerian duct essential for ontogenesis of the female reproductive tract. Hum. Mol. Genet. 25 1059–1073. 10.1093/hmg/ddv621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y., Liu S., Guan Y., Pan H., Guan X., Qiu Z., et al. (2013). Lgr4-mediated Wnt/beta-catenin signaling in peritubular myoid cells is essential for spermatogenesis. Development 140 1751–1761. 10.1242/dev.093641 [DOI] [PubMed] [Google Scholar]
- Robinson M. D., Mccarthy D. J., Smyth G. K. (2010). edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Dizier M., Marnier C., Tahir M. Z., Grimard B., Thoumire S., Chastant-Maillard S., et al. (2014). OVGP1 is expressed in the canine oviduct at the time and place of oocyte maturation and fertilization. Mol. Reprod. Dev. 81 972–982. [DOI] [PubMed] [Google Scholar]
- Shaw J. L., Horne A. W. (2012). The paracrinology of tubal ectopic pregnancy. Mol. Cell. Endocrinol. 358 216–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone M., Oyama K., Mohri Y., Hayashi R., Clevers H., Nishimori K. (2013). LGR4 expressed in uterine epithelium is necessary for uterine gland development and contributes to decidualization in mice. FASEB J. 27 4917–4928. [DOI] [PubMed] [Google Scholar]
- Stadnicka K., Slawinska A., Dunislawska A., Pain B., Bednarczyk M. (2018). Molecular signatures of epithelial oviduct cells of a laying hen (Gallus gallus domesticus) and quail (Coturnix japonica). BMC Dev. Biol. 18:9. 10.1186/s12861-018-0168-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein S., Lemos V., Xu P., Demagny H., Wang X., Ryu D., et al. (2017). Impaired SUMOylation of nuclear receptor LRH-1 promotes nonalcoholic fatty liver disease. J. Clin. Invest. 127 583–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C. A., Behringer R. R. (2012). Mouse oviduct development. Results Probl. Cell Differ. 55 247–262. [DOI] [PubMed] [Google Scholar]
- Vidal V. P., Jian-Motamedi F., Rekima S., Gregoire E. P., Szenker-Ravi E., Leushacke M., et al. (2020). R-spondin signalling is essential for the maintenance and differentiation of mouse nephron progenitors. Elife 9:e53895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Larina I. V. (2018). In vivo three-dimensional tracking of sperm behaviors in the mouse oviduct. Development 145:dev157685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Lee C. L., Li R. H. W., Vijayan M., Duan Y. G., Yeung W. S. B., et al. (2019). Alteration of the immune cell profiles in the pathophysiology of tubal ectopic pregnancy. Am. J. Reprod. Immunol. 81:e13093. [DOI] [PubMed] [Google Scholar]
- Wang Y., Dong J., Li D., Lai L., Siwko S., Li Y., et al. (2013). Lgr4 regulates mammary gland development and stem cell activity through the pluripotency transcription factor Sox2. Stem Cells 31 1921–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng J., Luo J., Cheng X., Jin C., Zhou X., Qu J., et al. (2008). Deletion of G protein-coupled receptor 48 leads to ocular anterior segment dysgenesis (ASD) through down-regulation of Pitx2. Proc. Natl. Acad. Sci. U. S. A. 105 6081–6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Crampton S. P., Hughes C. C. (2007). Wnt signaling induces matrix metalloproteinase expression and regulates T cell transmigration. Immunity 26 227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L. J., Chang H., Ding N. Z., Ni H., Kadomatsu K., Yang Z. M. (2002). Basigin expression and hormonal regulation in mouse uterus during the peri-implantation period. Mol. Reprod. Dev. 63 47–54. 10.1002/mrd.10128 [DOI] [PubMed] [Google Scholar]
- Xu P., Oosterveer M. H., Stein S., Demagny H., Ryu D., Moullan N., et al. (2016). LRH-1-dependent programming of mitochondrial glutamine processing drives liver cancer. Genes Dev. 30 1255–1260. 10.1101/gad.277483.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Wiesmann M., Rohan M., Chan V., Jefferson A. B., Guo L., et al. (2001). Elevated expression of axin2 and hnkd mRNA provides evidence that Wnt/beta -catenin signaling is activated in human colon tumors. Proc. Natl. Acad. Sci. U. S. A. 98 14973–14978. 10.1073/pnas.261574498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zak M., Van Oort T., Hendriksen F. G., Garcia M. I., Vassart G., Grolman W. (2016). LGR4 and LGR5 Regulate Hair Cell Differentiation in the Sensory Epithelium of the Developing Mouse Cochlea. Front. Cell. Neurosci. 10:186. 10.3389/fncel.2016.00186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbe H., Schuberth H. J., Engelke F., Frank J., Klug E., Leibold W. (2003). Development and comparison of in vivo and in vitro models for endometritis in cows and mares. Theriogenology 60 209–223. 10.1016/S0093-691X(02)01376-6 [DOI] [PubMed] [Google Scholar]
- Zhang C., Large M. J., Duggavathi R., Demayo F. J., Lydon J. P., Schoonjans K., et al. (2013). Liver receptor homolog-1 is essential for pregnancy. Nat. Med. 19 1061–1066. 10.1038/nm.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Kan F. W. K. (2019). Human OVGP1 enhances tyrosine phosphorylation of proteins in the fibrous sheath involving AKAP3 and increases sperm-zona binding. J. Assist. Reprod. Genet. 36 1363–1377. 10.1007/s10815-019-01502-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The mRNA level of Lgr5 and Lgr6 in mouse oviducts. (A) The mRNA levels of Lgr5 in the specific regions of the oviduct. (B) The mRNA levels of Lgr6 in the specific regions of the oviduct. At least three independent experiments were carried out. Data were presented as mean ± SEM. NS > 0.05.
Loss of LGR4 led to histological abnormalities of oviducts. (A) Histological analysis of the WT and Lgr4 KO oviducts were performed by hematoxylin and eosin staining after the supplement of E2. (B) The average number of leaf-like folds per section after the supplement of E2. (C) The average number of the secretory cells per leaf-like fold after the supplement of E2. ∗p ≤ 0.05; ∗∗∗p ≤ 0.001. Scale bar, 50 μm.
Loss of LGR4 led to a deficiency in oviductal secretion. (A) The bar graphs summarized the number of positively labeled cells of PAS stained oviduct slides. (B) The number of the positively Alcian blue stained cells in oviducts were presented. At least three independent experiments were carried out. ∗∗∗p ≤ 0.001.
Detection of NR5A1 protein in mouse oviduct. (A) IHC assay of NR5A1 in the WT and Lgr4 knockout mouse oviducts. (B) IHC assay of NR5A1 in the WT testis as control. Note, the positive signals in the enclosed area by red dashed lines. Scale bar, 50 μm.
Primer list.
Differential expressed genes between the Lgr4 deletion and control oviducts. Differential gene expression analysis between Lgr4 deletion and control oviducts (the estrus stage) revealed a number of significantly different expressed genes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: BioProject (PRJNA691984) in NCBI, https://www.ncbi.nlm.nih.gov/bioproject/PRJNA691984.