Abstract
SARS-CoV-2 main protease is a possible target for protection against viral infection. This study examined the inhibitory effect of food phytochemicals on the main protease of SARS-CoV-2 by determining a cleaved product after chromatographic separation. First, 37 phytochemicals, including glycosides and metabolites, were screened at 20 µM; epigallocatechin gallate, myricetin, theaflavin, herbacetin, piceatannol, myricitrin, and isothiocyanates inhibited the enzyme in varying degrees. The IC50 values were estimated from 0.4 to 33.3 µM against the 0.5-µM enzyme. The dose-dependent adduction of epigallocatechin gallate and myricetin was confirmed by quinone staining of protein blotted onto a membrane. The enzyme activity was decreased by increasing the concentration of the two phytochemicals, accompanied by increasing the respective adducted molecule estimated by intact mass spectrometry. Reduced glutathione canceled the formation of conjugate and the inhibitory effect of epigallocatechin gallate or myricetin on the enzyme, suggesting that the formation of the quinone moiety in the phytochemicals is critical for the inhibition. The covalent binding of epigallocatechin gallate or myricetin to the cysteine residue at the active site was confirmed by analyzing peptides from the chymotrypsin-digested main protease.
Keywords: Coronavirus main protease, Food phytochemical, EGCG, Myricetin, Autoxidation, Covalent modification
Introduction
Coronavirus disease 2019 (COVID-19) caused by SARS-CoV-2 has rapidly spread globally and threatens human life. There are some promising target molecules to prevent the virus infection or proliferation. This study focuses on the coronavirus main protease (Mpro, alternatively 3C-like protease), which mediates the scission of viral protein to replicate. The Mpro protease inhibitor, GC376, reduced the viral titer in cats with feline infectious peritonitis virus from feline enteric coronavirus [1]. Also, a similar peptidergic Mpro inhibitor improved the survival of mice infected by MERS-CoV [2]. Judging from the similarity among the coronavirus family, the SARS-CoV-2 Mpro is a promising target for preventing COVID-19. After reports about the crystal structure of Mpro from SARS-CoV-2 with an inhibitor [3,4], many attempts to evaluate inhibitors, including natural chemicals, have proceeded in in silico study.
Food is a good source for health promotion; it can supply beneficial phytochemicals to humans. Thus, the daily consumption of phytochemicals could help strengthen our body against diseases, such as COVID-19. Formerly, experimental studies for the inhibitory effect of flavonoids on the recombinant protease from SARS-CoV-1 (classic) or MERS-CoV have been reported; some flavonoids, including herbacetin and quercetin 3-O-glucoside, inhibited MERS-CoV Mpro [5]. SARS-CoV-1 Mpro was inhibited by flavonoids and flavones, such as herbacetin, rhoifolin, pectolinarin, apigenin, luteolin, quercetin, and amentoflavone [6,7]. Concerning SARS-CoV-2 Mpro, epigallocatechin gallate (EGCG) [8,9], theaflavin [8], myricetin [10], quercetin [11], herbacetin, pectolinarin, baicalin [12], rutin, baicalein [13], and cyanidin-3-glucoside [14] shows inhibitory effects on the enzyme in vitro. Most research was performed by fluorescence resonance energy transfer (FRET) assay, which sometimes results in false negatives or positives due to the interference of fluorescence signals derived from test chemicals [15]; a competitive binding assay using the fluorescent probe 8-anilinonaphthalene-1-sulfonate is also reported [13].
The study examines the inhibitory effect of food phytochemicals by directly measuring the digested peptide substrate after high-performance liquid chromatography (HPLC) separation. Additionally, the mechanism of inhibition by covalent adduction of phytochemicals, such as EGCG and myricetin, was estimated by quinone staining and mass spectrometry (MS).
Materials and methods
Materials
Dabcyl-KTSAVLQSGFRKME-Edans was purchased from GL Biochem, Ltd. (Shanghai, China). GC376 and herbacetin were obtained from Cayman Chemical (MI, USA). Kaempferol, quercetin, theaflavin, isoquercetin (quercetin-3-O-glucoside), cyanidine chloride, and nitroblue tetrazolium (NBT) were from Fujifilm-Wako Pure Chemical Co. (Osaka, Japan). Pectolinarin was purchased from Merck Sigma-Aldrich (MA, USA). Cyanidine-3-glucoside chloride, cyanidine-3-rutinoside chloride, and luteolin-7-O-glucoside were obtained from Extrasynthese S.A. (Lyon, France). Cyanidine-3-sophoroside chloride and cyanidine-3-O-glucosyl-rutinoside (C-3-GR) chloride were from Tokiwa Phytochemical Co., Ltd. (Chiba, Japan). Allyl isothiocyanate (AITC), benzyl isothiocyanate (BITC), phenethyl isothiocyanate (PEITC), ebselen, resveratrol, luteolin, isorhamnetin (IR), baicalein, myricitrin, piceatannol, rhapontigenin, and isorhapontigenin were obtained from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). Luteolin-7-O-glucuronide was from MedChemExpress (NJ, USA). Rutin (quercetin-3-O-rutinoside) was purchased from Nacalai Tesque, Inc. (Kyoto, Japan). EGCG was purchased from LKT Labs, Inc. (MN, USA). Quercetin-7-O-glucuroinde, quercetin-3-O-glucuroinde, quercetin-4’-O-glucuroinde, quercetin-3’-O-glucuroinde, isorhamnetin-7-O-glucuronide, isorhamnetin-3-O-glucuronide, isorhamnetin-4’-O-glucuronide, quercetin-7-O-sulfate, quercetin-3’-O-sulfate, and quercetin-4’-O-sulfate were prepared as described previously [16,17]. Chymotrypsin (sequencing grade) and ProteaseMAX were purchased from Promega (WI, USA). Amylose resin and Factor Xa were obtained from New England Biolabs Japan Inc. (Tokyo, Japan). Other chemicals used were of high-quality grades, unless otherwise indicated.
Preparation of maltose-binding protein (MBP)-tagged main protease and its MBP-cleaved protease
The MBP-tagged main protease containing SARS-CoV-2 Mpro (GenBank Accession No. YP_009725301; Met1–Gln306) was constructed using pMAL-c2 expression vector (New England Biolabs Japan Inc.). The gene encoding SARS-CoV-2 Mpro with E. coli codon usage was synthesized by Eurofins Genomics (Tokyo, Japan). The fusion protein was purified by amylose-conjugated affinity chromatography. MBP-cleaved Mpro was prepared from MBP-tagged Mpro by Factor Xa protease digestion. The prepared Mpro showed a single band (MBP-tagged Mpro, 77.5 kDa; MBP-cleaved Mpro, 34.7 kDa) (Supplemental Fig. 1). The concentration of each protein was estimated by bicinchoninic acid protein assay.
Measurement of enzyme activity by HPLC fluorescence detection
The enzymatic reaction was performed in a 20-mM Tris-HCl buffer containing 1-mM ethylenediaminetetraacetic acid (EDTA), 150-mM NaCl, and 0.01% Triton X-100 (pH 7.5). All tested chemicals were dissolved in dimethyl sulfoxide (DMSO), and the concentration of DMSO was 2% in the reaction mixture, unless otherwise indicated. The enzyme protease (0.5 µM) was preincubated with an inhibitor (0–200 µM) or vehicle at 37°C for 30 min, unless otherwise indicated. After that, 10-µM Dabcyl-KTSAVLQSGFRKME-Edans was added to the solution and further incubated for 1 h. The reaction was terminated by adding an equal volume of CH3CN containing 0.1% formic acid, and 10 µL of the mixture was injected into the Shimadzu Prominence HPLC connected with a photodiode array detector (PDA) and an RF-10XL fluorescence detector (ex. 336 nm, em. 490 nm) in series (Shimadzu Co., Kyoto, Japan). PDA was used to check possible co-elution with the fluorescent peptide and an additive. The fluorescence detector was used to quantify cleaved fluoresce peptide. The separation was done using a Phenomenex Kinetex XB-C18 (4.6 × 50 mm, 2.6 µm) with gradient elution using 0.1% trifluoroacetic acid in water (solvent A) and CH3CN (solvent B) at a flow rate of 1.0 mL/min. The gradient program was as follows: initial B10%, 3 min B100%, 4 min B10%, and 10 min B10%. The analysis was performed in triplicate. The IC50 value was calculated using Prism v.9.2 (GraphPad Software, LLC., CA, USA).
The scission products were confirmed using a quadrupole-time-of-flight mass spectrometer [Q-TOF, X500R, Sciex (MA, USA)] connected with ultra-high-performance liquid chromatography (UHPLC, Exion, Sciex). After the reaction, the reaction mixture was then separated using a Thermo Scientific Hypersil Gold column (2.1 × 100 mm, 1.9 µm) by gradient elution using 0.1% formic acid in water (solvent A) and CH3CN (solvent B) at a flow rate of 0.4 mL/min. The gradient program was as follows: initial B10%, 2 min B100%, 2.1 min B10%, and 4 min B10%. Finally, the eluate was introduced into the Q-TOF mass spectrometer and analyzed using information-dependent analysis mode.
Intact MS and deconvolution of the MS spectrum
The MBP-cleaved enzyme (0.5 µM) was incubated with a test chemical (0–5 µM, typically 1 µM) in Tris-HCl buffer as shown above for 30 min at 37°C. The protein (10 µL) was separated using a Phenomenex bioZen Intact XB-C8 column (2.1 × 100 mm, 3.6 µm) for gradient elution using 0.1% formic acid in water (solvent A) and 0.1% formic acid in CH3CN (solvent B) at a flow rate of 0.3 mL/min connected to the MS. The gradient program was as follows: initial B5%, 0.5 min B5%, 4 min B90%, 5 min B5%, and 8 min B5%. The eluate was introduced into the MS using TOF-positive mode. This experiment was performed twice. The deconvolution of the mass spectrum was acquired by UniDec (v.4.4.1) [18].
Detection of quinones on the blotted membrane by redox-cycling staining
After incubating the enzyme with the test chemical, 10-mM dithiothreitol and × 4 Laemmli sample loading buffer (Bio-Rad, CA, USA) were added to the reaction mixture. The sample (60-ng protein/lane) and Precision Plus Protein Dual Color Standard (Bio-Rad) were then applied to an SDS-polyacrylamide gel (SDS-PAGE, 10% acrylamide) and then migrated by electrophoresis. The proteins in gels were blotted onto a PVDF membrane (Immobilon-P, Merck), and quinones on the membrane were stained using NBT (0.24 mM) in 2-M glycine-NaOH (pH 10) as described previously [19,20]. The same samples were separated using a gel, and the proteins in the gel were stained using Flamingo Stain (Bio-Rad) according to the manufactures’ protocol. Detection was performed using a LAS1000 plus (Fujifilm, Tokyo, Japan). This experiment was performed twice.
Estimation of free thiols by biotin incorporation into the protein
Detection of free thiols in the enzyme was performed as described previously with some modifications [20]. First, protein samples were incubated with 0.25-mM EZ-link iodoacetyl-LC-biotin (Thermo Scientific, IL, USA) for 30 min in the dark and then mixed with 10-mM of dithiothreitol and the × 4 loading buffer. Next, the sample (20-ng protein/lane) was separated by 10% SDS-PAGE and the gels were blotted onto a PVDF membrane; the membrane was then blocked by EzBlock Chemi (ATTO Co., Tokyo, Japan) for 30 min. Finally, the membrane was treated with streptavidin-peroxidase polymer (Merck, ultrasensitive, 1/100,000) for 30 min, and the binding was visualized by Chemi-Lumi One L (Nacalai Tesque, Inc.). This experiment was performed twice.
Identification of an adducted site in Mpro by digestion with chymotrypsin
The MBP-cleaved enzyme (5 µM) was incubated with EGCG/myricetin (10 µM) in the Tris-HCl buffer at 37°C for 30 min. The reaction mixture (50 µL) was then applied to a tube for centrifugal ultrafiltration (Amicon Ultra 10K, Merck). Then, 450-µL 0.1-M Tris-HCl containing 10-mM CaCl2 (pH 8.0) was added and centrifuged to exchange the buffer for digestion. To the protein fraction, 100-µL chymotrypsin (1.7 µg) in buffer containing 0.025% ProteaseMAX was added and then further incubated at 25°C for 6 h. The digested sample was purified using a peptide desalting and enrichment Tip, GL-Tip SDB (GL Sciences Inc., Tokyo, Japan) according to the manufacturers’ protocol. Peptide separation was done using a Phenomenex Jupiter PROTEO C18 column (2 × 150 mm, 5 µm) with a gradient elution system at 0.2 ml/min flow rate. Solvent A was 0.1% formic acid in H2O, and solvent B was 0.1% formic acid in CH3CN. The linear gradient program was as follows: initial B2%, 4 min B2%, 55 min B40%, 58 min B90%, 59 min B90%, 60 min B2%, and 75 min B2%. The eluate was introduced into the MS with positive mode. MS-Digest, ProteinSpector (https://prospector.ucsf.edu/prospector/mshome.htm) was used to calculate the theoretical mass numbers of modified peptides. This experiment was performed three times, and the typical result was shown.
Results
To examine the inhibitory effect of 37 phytochemicals on Mpro (MBP-tagged Mpro), Dabcyl-KTSAVLQSGFRKME-Edans was used as a substrate, and the formed cleaved peptide (SGFRKME-Edans) was then estimated using an HPLC fluorescence detector. Before the screening, the generation of cleaved peptides (Dabcyl-KTSAVLQ and SGFRKME-Edans), as products, was confirmed by TOF-MS. The scission fragment molecule of Dabcyl-KTSAVLQ has C47H72N12O12 ([M+H]+ 996.54), and the other fragment of SGFRKME-Edans has C48H71N13O13S2 ([M+H]+ 1101.47).
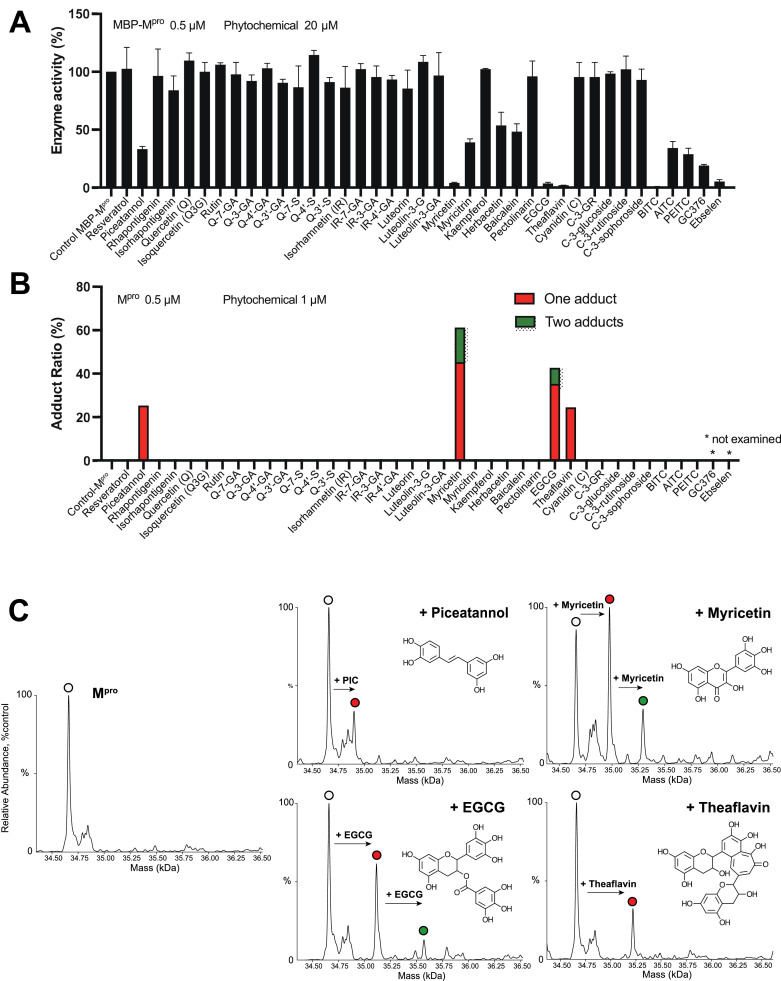
At 20 µM, several phytochemicals had a considerable suppressive effect on the protease activity of the enzyme (0.5 µM) (Fig. 1 A). The IC50s for the phytochemicals were estimated as EGCG (0.4 µM), myricetin (0.9 µM), theaflavin (6.1 µM), piceatannol (13.8 µM), herbacetin (19.2 µM), baicalein (20.3 µM), myricitrin (21.7 µM), BITC (4.5 µM), PEITC (14.2 µM), and AITC (33.3 µM) (Supplemental Fig. 2). Under the conditions, IC50s of the positive controls, ebselen and GC376, were 0.4 and 18.8 µM, respectively. However, quercetin, pectolinarin, rutin, and cyanidine-3-glucoside, in which their inhibitory activity to Mpro has already been reported [11], [12], [13], [14], did not show suppressive effect at the concentration examined (20 µM).
Fig. 1.
The effect of various phytochemicals on the enzyme activity and its mass. (A) MBP-tagged Mpro (0.5 µM) was incubated with phytochemicals (20 µM) at 37°C for 30 min. After the reaction, a peptide substrate was added. The enzyme activity was then analyzed by measuring the cleaved fluoresce peptide generated from the substrate as described in Materials and Methods. (B) Mpro (MBP-removed, 0.5 µM) was incubated with the phytochemicals (1 µM) at 37°C for 30 min. After the reaction, changes in the mass of Mpro were examined by intact MS. The deconvolution of the mass spectrum was performed using the UniDec software, and the mass shifts of the enzyme by the adduction of one or more phytochemicals were evaluated by measuring the respective peak area. The deconvoluted mass chromatograms of piceatannol, myricetin, EGCG, and theaflavin with the individual chemical structure are also shown. Abbreviations used: Q, quercetin; G, glucose; glucuronide, GA; S, sulfate; IR, isorhamnetin; EGCG, epigallocatechin gallate; C, cyanidine; GR, glucosyl-rutinoside; BICT, benzyl isothiocyanate; AITC, allyl isothiocyanate; PEITC, phenethyl isothiocyanate.
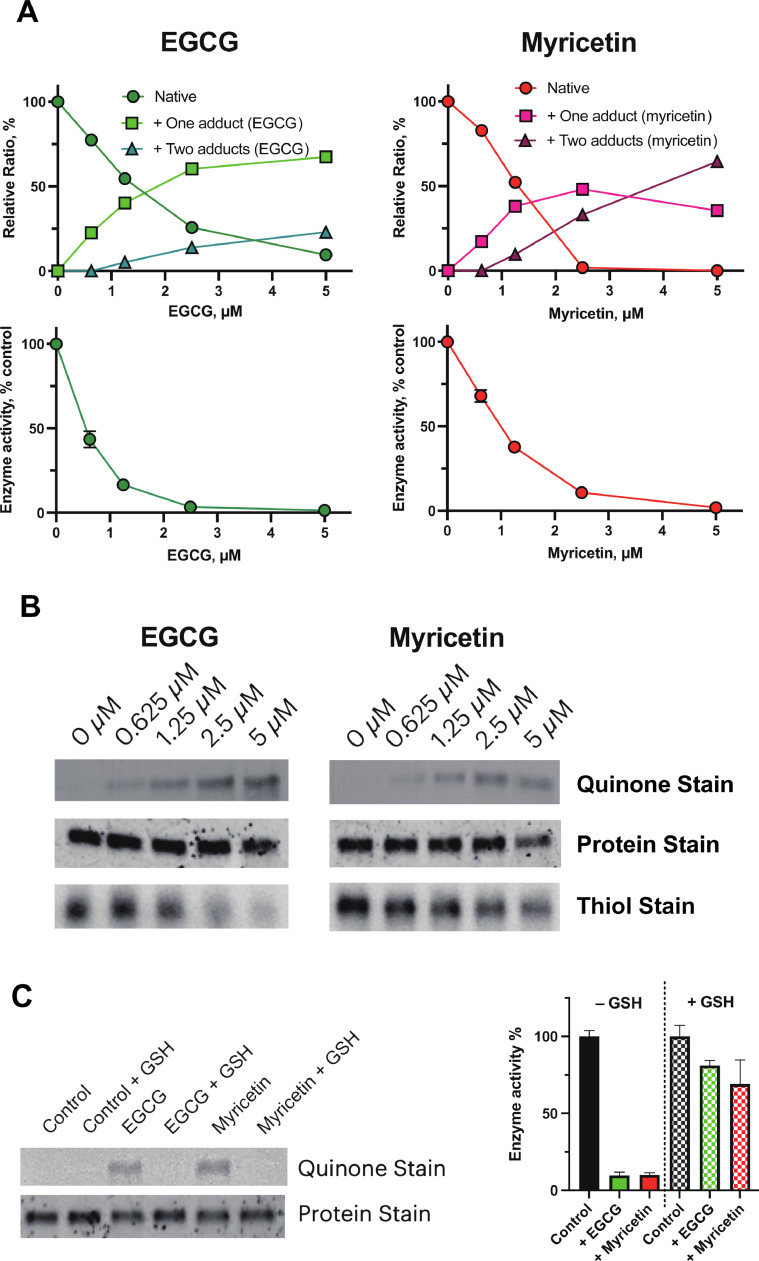
The changes in the mass of Mpro by incubation with the phytochemicals were examined. Because MBP-Mpro (77.5 kDa) signal by the intact MS was not clearly observed, MBP-cleaved Mpro (34.7 kDa) was prepared and used after that. The mass changes of the Mpro by phytochemical conjugation (1 µM) were shown in Fig. 1B. One or two molecule(s) of piceatannol, myricetin, EGCG, and theaflavin were adducted onto the Mpro. Herbacetin, baicalein, myricitrin, and ITCs showed significant inhibitory effects, but their covalent adducts were not observed at 1 µM (Fig. 1B). The adductions by myricetin and EGCG (0–5 µM) were dose-dependently observed, and the enzyme activities were decreased accordingly (Fig. 2 A).
Fig. 2.
Dose-dependent changes in the mass and activity of the enzyme incubated with EGCG or myricetin. (A) After incubating the Mpro enzyme (0.5 µM) with 1–5-µM myricetin/EGCG, adduct formation was estimated by intact MS. The dose-dependent effects of EGCG and myricetin on enzyme activity were also examined. The reaction of the enzyme with the substrate was set for 15 min. (B) After incubation, the proteins were separated by SDS-PAGE. Gel/blot images for the enzyme exposed to EGCG or myricetin were shown. For protein staining, the gel was stained using Flamingo gel stain. Additionally, the gel was blotted onto a PVDF membrane for quinone staining and then stained using nitroblue tetrazolium (NBT) in alkaline glycinate buffer. The enzyme was first derivatized by iodoacetamide-LC-biotin and then applied to a gel and blotted for free thiol staining. The blotted membrane was incubated with streptavidin-peroxidase followed by adding a chemiluminescence reagent for evaluation. (C) The enzyme (0.5 µM) was incubated with 2.5-µM EGCG or myricetin in the presence or absence of 1-mM reduced glutathione (GSH) for 30 min. For the control, DMSO (final 1%) was used. The enzyme activity (%) was expressed as %control or %control with GSH. The enzyme was also separated by SDS-PAGE and blotted onto the membrane. The membrane was then stained by NBT as described in (B).
Next, the adduction of EGCG and myricetin to the enzyme was confirmed by quinone staining. After the reaction of the Mpro enzyme with the two phytochemicals, the Mpro was subjected to gel electrophoresis, blotted onto a PVDF membrane, and then stained with NBT (Fig. 2B). From 0.625-µM phytochemicals, NBT-reactive quinone moieties at the location of Mpro were observed dose-dependently. Simultaneously the free thiols in the Mpro were decreased by increasing the concentration of phytochemicals. Most of the enzyme activity was retained when reduced glutathione (GSH, 1 mM) was added to the buffer in advance (Fig. 3 C). Additionally, the control enzyme activity was approximately increased 1.3-fold by GSH supplementation (data not shown). Quinone staining by EGCG/myricetin was also canceled when incubated in the presence of GSH.
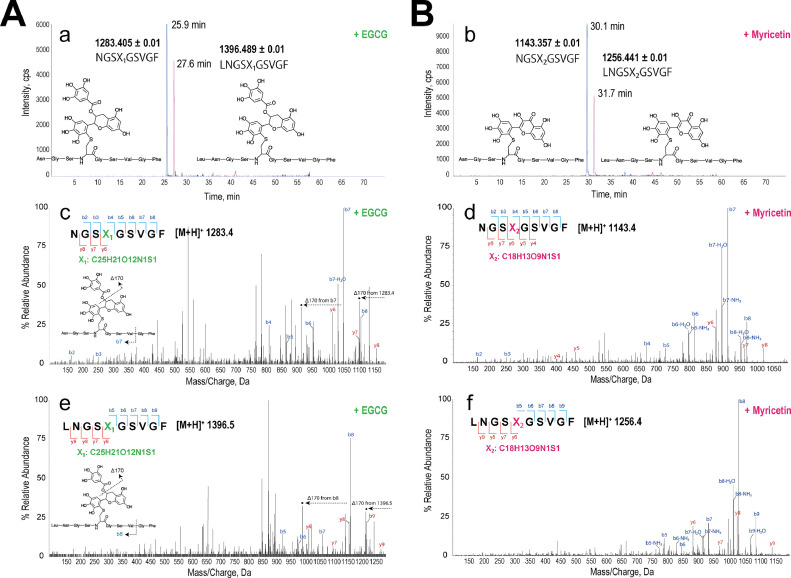
Fig. 3.
MS(/MS) analysis of covalently modified peptides at the active site. A; EGCG. B; Myricetin. After incubation, Mpro was digested with chymotrypsin. Extract ion chromatograms of the theoretical mass of EGCG- or myricetin-conjugated peptides with proposed chemical structures (a, b). X1 indicates EGCG-conjugated cysteine, and X2 indicates myricetin-conjugated cysteine. NGSX1/2GSVGF (1283.405 ± 0.01 (EGCG), 1143.357 ± 0.01 (myricetin)) and LNGSX1/2GSVGF (1396.489 ± 0.01 (EGCG), 1256.441 ± 0.01 (myricetin)) were the theoretical chymotrypsin-cleavable sequences (possible conjugated peptides) located at the active site. The MS/MS chromatograms generated from NGSX1/2GSVGF are shown in c (EGCG) and d (myricetin). Those of LNGSXGSVGF (1 missed cleavage) are shown in e (EGCG) and f (myricetin). Inset fragmentation schemes, c and e: EGCG-dependent characteristic fragments (Δ170) from the liberation of the galloyl moiety at the D ring in EGCG by collision-induced dissociation of EGCG-adducted peptides.
The enzyme (5 µM) was digested with chymotrypsin after incubation with EGCG or myricetin (10 µM). The peptide digests were separated by HPLC and then analyzed by MS(/MS). If adducted, the mass of the peptide conjugated with EGCG or myricetin should increase by 456 or 316 Da, corresponding to the conjugation of EGCG (MW 458) or myricetin (MW 318). The presumed signals of EGCG adducted to NGSCGSVGF (25.9 min) and LNGSCGSVGF (27.6 min) were detected on the extract ion chromatograms (Fig. 3A(a)). Additionally, the sequence, LNGSCGSVGF, is formed by one missed cleavage with chymotrypsin. Their sequences of the fragments induced by collision-induced dissociation were assigned as the peptides adducted by EGCG at the C (Cys) of the (L)NGSCGSVGF (Fig. 3A (c, e)) by comparing the prediction by the MS-Digest. The signals (–170 Da) from the EGCG-adducted sequences were also observed, and the mass difference was probably formed by liberating galloyl moiety at the D ring of EGCG [21]. This indicates that the thiol was covalently conjugated with the B ring of EGCG, but not the D ring. The myricetin-adducted peptides (+316 Da), corresponding to the NGSCGSVGF (30.1 min) and LNGSCGSVGF (31.7 min), were observed (Fig. 3B (b)), and the fragmentation patterns (Fig. 3B (d, f)) were matched with the predicted myricetin-adducted NGSCGSVGF and LNGSCGSVGF.
Discussion
The viral Mpro is a promising target for preventing the infection of coronavirus, i.e., the Mpro inhibitor GC376 could prevent viral infections [1,22,23]. Synthetic α-ketoamide compounds inhibited the SARS-CoV-2 Mpro, possibly by covalent adduction to Cys145 of the enzyme [3,4]. A clinical candidate, PF-07321332, targeted to the enzyme has recently proceeded to Phase 1 trial [24]. Apart from medicine, various components in food could be beneficial to life. However, the boundary between medicine and food is ambiguous because some foods, such as herbs, are used as folk medicine. Indeed, as introduced, many phytochemicals prevent the recombinant viral enzyme in vitro. However, the mechanism of inhibiting phytochemicals is unknown in detail. Hence, our preliminary study indicated that some phytochemicals could not be applied to the FRET assay with a fluorescent plate reader because of its fluorescence quenching ability or self-fluorescence as supposed [15]. We then evaluated the enzyme activity by analyzing the cleaved fluoresce peptide after separation by conventional HPLC. A drawback of this assay is the run time for analysis, 10 min/sample. In contrast, chromatography separates the fluoresce peptide with a test chemical, avoiding the possible interference between them.
Of the chemicals tested, the inhibitory activities of some phytochemicals have already been examined at a 200-µM concentration by FRET assay [25]. In contrast, current results using 37 compounds differ from the outcome in some points. It might be due to the difference in the evaluation method of enzyme activity. In this study, at 20 µM, myricetin, EGCG, theaflavin, and BITC considerably inhibited the viral protease, and to a lesser extent, piceatannol, PEITC, herbacetin, baicalein, myricitrin, and AITC. Furthermore, the patterns of inhibition and adduct formation were relatively matched (Fig. 1AB). In addition, as mentioned before, most of these phytochemicals have been reported as possible inhibitors of Mpro.
This study also focused on intact phytochemicals and their metabolites because ingested phytochemicals are often metabolized into conjugates, such as glucuronide or sulfate formed by intracellular phase II enzymes in cells. Possible inhibitory effects of these metabolites by a flavonoid have already been supposed by in silico study [26,27], and baicalin, a glucuronide of baicalein, inhibits SARS-CoV-2 recombinant Mpro in vitro [12]. However, most metabolites or the glucosides of flavonoids examined, except for myricitrin (a rhamnoside of myricetin), did not inhibit the enzyme at the concentration tested (20 µM). Among the circulating metabolites, the glucuronides could be cleaved at the inflammatory site by a secreted β-glucuronidase and then regenerate aglycone [28,29]. The sulfate form of phytochemicals could restore the original phytochemical in cells [30]. These probably suggest that glucuronides or sulfates of phytochemicals, which are often abundantly present in blood compared with phytochemical (aglycone), are possibly converted to the active hydrophobic aglycone form and then incorporated into an infected cell.
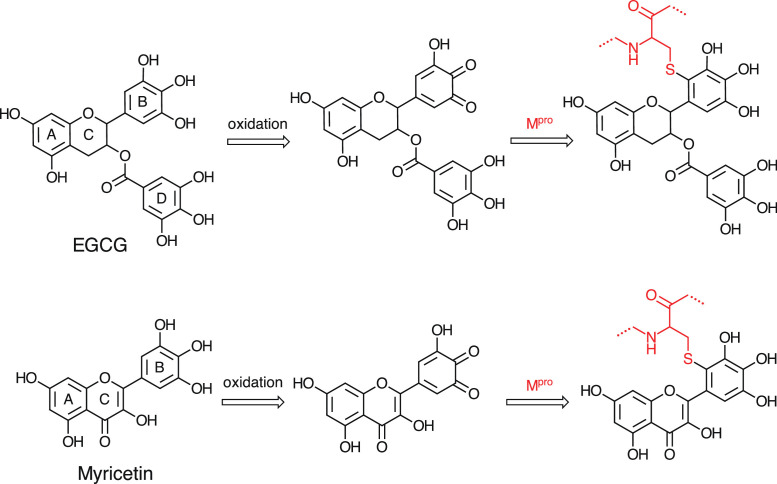
When EGCG, myricetin, theaflavin, and piceatannol (1 µM) were mixed with 0.5-µM enzyme for 30 min at neutral pH, the adduct was formed (Fig. 1B). A former study showed that EGCG might have possibly covalently modified a cysteinyl thiol residue of glyceraldehyde-3-phosphate dehydrogenase and inactivated the enzyme at 37°C for 30 min [21]. We have also found that myricetin and EGCG inhibit the Mpro enzyme accompanying the formation of one or two molecules adducted to the enzyme molecule dose-dependently (Fig. 2). The autoxidation of catechin or flavonoid generates a quinone moiety. The formed quinone could react with a thiol of the enzyme, resulting in the enzyme conjugation with the phytochemical (Fig. 4 ) [31]. Piceatannol is abundantly present in the fruit of Passiflora edulis [32] and is a metabolite of resveratrol. In this study, we have discovered that piceatannol inhibits the enzyme; the catechol structure of piceatannol is presumably the critical structure for conjugation.
Fig. 4.
Proposed reaction scheme for adduction of EGCG or myricetin with Mpro.
Pyrogallol (myricetin and its synthetic derivative) is considered an alternative warhead in the design of targeted covalent ligands [10]. EGCG has trihydroxy benzoyl moieties at the B and D rings. The liberation of the galloyl moiety (Δ170) at the D ring was observed in fragments of digested peptides conjugated with EGCG (Fig. 3A (c, e)), and similar liberation of the galloyl moiety (Δ170) from EGCG-adducted MHRQETVDC was already reported [21]. In contrast, a signal generated from the liberation of Δ290 from the opposite side (A, B, and C rings) was not observed (data not shown). Thus, this indicates that EGCG was mainly conjugated with a protein thiol via the galloyl moiety at the B ring. Additionally, by comparing myricetin with myricitrin, sugar moiety in the phytochemical weakens its inhibitory activity. Formerly, in a lipid oxidation system, myricetin and thiol conjugates were identified [33]. Recently, the conjugation (Δ316 Da) of one molecule of myricetin to Mpro evaluated by intact MS has been reported [10], and the mechanism of conjugation of myricetin with protein thiol via the formation of o-quinone is also proposed.
Quinone staining showed that a reduced type of glutathione (GSH) suppressed the conjugation of EGCG/myricetin (Fig. 2C). Furthermore, enzyme activity loss by EGCG/myricetin was canceled in the presence of GSH. These results suggested that GSH reacted with the o-quinone moiety of the phytochemical instead of a thiol in the enzyme. Thus, quinones were probably reactants to the enzyme.
We attempted to identify EGCG or myricetin-adducted site in the enzyme by chymotrypsin digestion (Fig. 3). A former study showed that ebselen, PX-12, and carmofur adduct to Cys145, which was assigned by analyzing TIKGSFLNGSCGSVGF or FTIKGSFLNGSCGSVGF digest by chymotrypsin [3]. We have found that the EGCG/myricetin-adducted peptides (0 and 1 missed cleavage) are located at the active site of Mpro, (L)NGSCGSVGF. Furthermore, this is the first report that phytochemicals covalently adduct the active site of Mpro.
The inhibitors, ebselen, disulfiram, carmofur, PX-12, tideglusib and shikonin, were found to be nonspecific promiscuous SARS-CoV-2 main protease inhibitors because they lose their activities in the presence of a reducing reagent like dithiothreitol [34]. In contrast, ebselen and its derivative possess Mpro inhibitory and antiviral activities [35]. The favored reaction between quinone and an enzyme thiol may lead to off-target effects; quinones of phytochemicals may be trapped by intravital GSH, cysteine, or protein thiols before approaching the viral enzyme in infected cells. Then, a thiol-sensitive oxidized phytochemical may be classified as a nonspecific inhibitor of the Mpro enzyme. However, it has also been reported that the reaction rate of myricetin with GSH is slow [10]. Additionally, in vivo environment is more hypoxic (reductive) than in vitro test tubes. Furthermore, there are many causes of oxidation of phytochemicals in intra- and extracellular environments [36]; therefore, quinone form of phytochemicals could react with the viral thiol enzyme under higher oxidative stress.
The inhibitory effect of ITC including PEITC on Mpro has been reported [37,38]. This study also shows that three ITCs prevent cleaved peptide generation. As already known, ITC is highly reactive to thiols, forming a covalent ITC-thiol adduct. Therefore, the modification of cysteine at the active site by ITC would inactivate the enzyme. However, the adducts were not observed at least at 1-µM ITC (Fig. 1B), probably because the thiocarbamoyl linkage between ITC and thiol is unstable. ITC also reacts with amines to a lesser extent [39,40]. Therefore, the study on the effect of ITC on the enzyme remains unelucidated.
It has been reported that the plasma concentration of total polyphenol metabolites ranged from 0 to 4 µM with an intake of 50-mg aglycone equivalents [41]. Therefore, it may not be enough to deliver these possible functional phytochemicals to infected cells. However, current results have shown that some phytochemicals at a physiological concentration, a few µMs, inhibit the viral enzyme in vitro. Furthermore, former studies have shown that cellular proteins were modified by 5–40-µM catechins, including EGCG, supplemented to the culture medium [21,42], suggesting that EGCG (and other phytochemicals) could penetrate the cell and covalently modify cellular proteins. Besides, nanodelivery systems with food phytochemicals may also overcome the issue of lower bioavailability [43].
A limitation of this study is that the inhibition was tested by incubating the recombinant enzyme with pure phytochemicals in vitro. Therefore, using SARS-CoV-2-infected cells with a test compound [44] is preferable; however, it requires a higher biosafety, such as biosafety level-3 environment. Alternatively, a cell-based Flip-GFP and cell-based luciferase-Glo assays for Mpro have been developed [45], [46], [47], [48], [49]. These novel assay systems could validate the inhibitory effects of the phytochemicals.
Conclusively, we have estimated the inhibitory effect of the phytochemicals on the virus recombinant main protease by measuring the generation of the intact fluorescent-cleaved peptide substrate. It was found that some phytochemicals, including myricetin and EGCG, significantly inhibited the enzyme even at a physiological concentration. Furthermore, the covalent phytochemical–Mpro adducts were observed along with the decrease in enzyme activity, and the adduction happened at the Cys of the active site of Mpro. Finally, verifying in vivo occurrences of the reaction between phytochemicals and the viral enzyme should be a subject for future studies.
CRediT authorship contribution statement
Yoji Kato: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – original draft, Writing – review & editing. Akari Higashiyama: Investigation, Writing – original draft. Emi Takaoka: Investigation, Writing – original draft. Miyu Nishikawa: Investigation, Writing – original draft. Shinichi Ikushiro: Investigation, Methodology, Supervision, Writing – review & editing.
Declaration of Competing Interest
There is no conflict of interest.
Acknowledgments
This work is partly supported by grants from the Ijima Memorial Foundation for the Promotion of Food Science and Technology. We are thankful to Motonori Tsuji (Institute of Molecular Function) for his preliminary in silico study and helpful advice. We are also grateful to Chisato Yakemoto and Professor Mikiko Ito for their contribution to a preliminary study.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.arres.2021.100021.
Appendix. Supplementary materials
References
- 1.Kim Y., Liu H., Galasiti Kankanamalage A.C., Weerasekara S., Hua D.H., Groutas W.C., Chang K.O., Pedersen N.C. Reversal of the progression of fatal coronavirus infection in cats by a broad-spectrum coronavirus protease Inhibitor. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rathnayake A.D., Zheng J., Kim Y., Perera K.D., Mackin S., Meyerholz D.K., Kashipathy M.M., Battaile K.P., Lovell S., Perlman S., Groutas W.C., Chang K.O. 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.abc5332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of M(pro) from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 4.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jo S., Kim H., Kim S., Shin D.H., Kim M.S. Characteristics of flavonoids as potent MERS-CoV 3C-like protease inhibitors. Chem. Biol. Drug Des. 2019;94:2023–2030. doi: 10.1111/cbdd.13604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jo S., Kim S., Shin D.H., Kim M.S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme Inhib. Med. Chem. 2020;35:145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryu Y.B., Jeong H.J., Kim J.H., Kim Y.M., Park J.Y., Kim D., Nguyen T.T., Park S.J., Chang J.S., Park K.H., Rho M.C., Lee W.S. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CL(pro) inhibition. Bioorg. Med. Chem. 2010;18:7940–7947. doi: 10.1016/j.bmc.2010.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jang M., Park Y.I., Cha Y.E., Park R., Namkoong S., Lee J.I., Park J. Tea polyphenols EGCG and theaflavin inhibit the activity of SARS-CoV-2 3CL-protease in vitro. Evid. Based Complement Alternat. Med. 2020;2020 doi: 10.1155/2020/5630838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiou W.C., Chen J.C., Chen Y.T., Yang J.M., Hwang L.H., Lyu Y.S., Yang H.Y., Huang C. The inhibitory effects of PGG and EGCG against the SARS-CoV-2 3C-like protease. Biochem. Biophys. Res. Commun. 2021 doi: 10.1016/j.bbrc.2020.12.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Su H., Yao S., Zhao W., Zhang Y., Liu J., Shao Q., Wang Q., Li M., Xie H., Shang W., Ke C., Feng L., Jiang X., Shen J., Xiao G., Jiang H., Zhang L., Ye Y., Xu Y. Identification of pyrogallol as a warhead in design of covalent inhibitors for the SARS-CoV-2 3CL protease. Nat. Commun. 2021;12:3623. doi: 10.1038/s41467-021-23751-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abian O., Ortega-Alarcon D., Jimenez-Alesanco A., Ceballos-Laita L., Vega S., Reyburn H.T., Rizzuti B., Velazquez-Campoy A. Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening. Int. J. Biol. Macromol. 2020;164:1693–1703. doi: 10.1016/j.ijbiomac.2020.07.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jo S., Kim S., Kim D.Y., Kim M.S., Shin D.H. Flavonoids with inhibitory activity against SARS-CoV-2 3CLpro. J. Enzyme Inhib. Med. Chem. 2020;35:1539–1544. doi: 10.1080/14756366.2020.1801672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deetanya P., Hengphasatporn K., Wilasluck P., Shigeta Y., Rungrotmongkol T., Wangkanont K. Interaction of 8-anilinonaphthalene-1-sulfonate with SARS-CoV-2 main protease and its application as a fluorescent probe for inhibitor identification. Comput. Struct. Biotechnol. J. 2021;19:3364–3371. doi: 10.1016/j.csbj.2021.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pitsillou E., Liang J., Karagiannis C., Ververis K., Darmawan K.K., Ng K., Hung A., Karagiannis T.C. Interaction of small molecules with the SARS-CoV-2 main protease in silico and in vitro validation of potential lead compounds using an enzyme-linked immunosorbent assay. Comput. Biol. Chem. 2020;89 doi: 10.1016/j.compbiolchem.2020.107408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gribbon P., Sewing A. Fluorescence readouts in HTS: no gain without pain? Drug Discov. Today. 2003;8:1035–1043. doi: 10.1016/s1359-6446(03)02895-2. [DOI] [PubMed] [Google Scholar]
- 16.Tanaka S., Trakooncharoenvit A., Nishikawa M., Ikushiro S., Hara H. Comprehensive analyses of quercetin conjugates by LC/MS/MS revealed that isorhamnetin-7- O-glucuronide-4′- O-sulfate Is a major metabolite in plasma of rats fed with quercetin glucosides. J. Agric. Food Chem. 2019;67:4240–4249. doi: 10.1021/acs.jafc.8b06929. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura T., Kinjo C., Nakamura Y., Kato Y., Nishikawa M., Hamada M., Nakajima N., Ikushiro S., Murota K. Lymphatic metabolites of quercetin after intestinal administration of quercetin-3-glucoside and its aglycone in rats. Arch. Biochem. Biophys. 2018;645:126–136. doi: 10.1016/j.abb.2018.03.024. [DOI] [PubMed] [Google Scholar]
- 18.Marty M.T., Baldwin A.J., Marklund E.G., Hochberg G.K., Benesch J.L., Robinson C.V. Bayesian deconvolution of mass and ion mobility spectra: from binary interactions to polydisperse ensembles. Anal. Chem. 2015;87:4370–4376. doi: 10.1021/acs.analchem.5b00140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paz M.A., Flückiger R., Boak A., Kagan H.M., Gallop P.M. Specific detection of quinoproteins by redox-cycling staining. J. Biol. Chem. 1991;266:689–692. [PubMed] [Google Scholar]
- 20.Kato Y., Peskin A.V., Dickerhof N., Harwood D.T., Kettle A.J. Myeloperoxidase catalyzes the conjugation of serotonin to thiols via free radicals and tryptamine-4,5-dione. Chem. Res. Toxicol. 2012;25:2322–2332. doi: 10.1021/tx300218f. [DOI] [PubMed] [Google Scholar]
- 21.Ishii T., Mori T., Tanaka T., Mizuno D., Yamaji R., Kumazawa S., Nakayama T., Akagawa M. Covalent modification of proteins by green tea polyphenol (-)-epigallocatechin-3-gallate through autoxidation. Free Radic. Biol. Med. 2008;45:1384–1394. doi: 10.1016/j.freeradbiomed.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 22.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vuong W., Khan M.B., Fischer C., Arutyunova E., Lamer T., Shields J., Saffran H.A., McKay R.T., van Belkum M.J., Joyce M.A., Young H.S., Tyrrell D.L., Vederas J.C., Lemieux M.J. Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2020;11:4282. doi: 10.1038/s41467-020-18096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Owen D.R., Allerton C.M.N., Anderson A.S., Aschenbrenner L., Avery M., Berritt S., Boras B., Cardin R.D., Carlo A., Coffman K.J., Dantonio A., Di L., Eng H., Ferre R., Gajiwala K.S., Gibson S.A., Greasley S.E., Hurst B.L., Kadar E.P., Kalgutkar A.S., Lee J.C., Lee J., Liu W., Mason S.W., Noell S., Novak J.J., Obach R.S., Ogilvie K., Patel N.C., Pettersson M., Rai D.K., Reese M.R., Sammons M.F., Sathish J.G., Singh R.S.P., Steppan C.M., Stewart A.E., Tuttle J.B., Updyke L., Verhoest P.R., Wei L., Yang Q., Zhu Y. An oral SARS-CoV-2 Mpro inhibitor clinical candidate for the treatment of COVID-19. medRxiv (2021) 2021 doi: 10.1101/2021.07.28.21261232. 2007.2028. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen T.T.H., Jung J.-H., Kim M.-K., Lim S., Choi J.-M., Chung B., Kim D.-W., Kim D. The Inhibitory effects of plant derivate polyphenols on the main protease of SARS coronavirus 2 and their structure–activity relationship. Molecules. 2021;26:1924. doi: 10.3390/molecules26071924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.da Silva F.M.A., da Silva K.P.A., de Oliveira L.P.M., Costa E.V., Koolen H.H., Pinheiro M.L.B., de Souza A.Q.L., de Souza A.D.L. Flavonoid glycosides and their putative human metabolites as potential inhibitors of the SARS-CoV-2 main protease (Mpro) and RNA-dependent RNA polymerase (RdRp) Mem. Inst. Oswaldo Cruz. 2020;115 doi: 10.1590/0074-02760200207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cherrak S.A., Merzouk H., Mokhtari-Soulimane N. Potential bioactive glycosylated flavonoids as SARS-CoV-2 main protease inhibitors: A molecular docking and simulation studies. PLoS One. 2020;15 doi: 10.1371/journal.pone.0240653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimoi K., Saka N., Nozawa R., Sato M., Amano I., Nakayama T., Kinae N. Deglucuronidation of a flavonoid, luteolin monoglucuronide, during inflammation. Drug Metab. Dispos. 2001;29:1521–1524. doi. [PubMed] [Google Scholar]
- 29.Kawai Y., Nishikawa T., Shiba Y., Saito S., Murota K., Shibata N., Kobayashi M., Kanayama M., Uchida K., Terao J. Macrophage as a target of quercetin glucuronides in human atherosclerotic arteries: implication in the anti-atherosclerotic mechanism of dietary flavonoids. J. Biol. Chem. 2008;283:9424–9434. doi: 10.1074/jbc.M706571200. [DOI] [PubMed] [Google Scholar]
- 30.Kato Y., Kawai M., Kawai S., Okano Y., Rokkaku N., Ishisaka A., Murota K., Nakamura T., Nakamura Y., Ikushiro S. Dynamics of the cellular metabolism of leptosperin found in manuka honey. J. Agric. Food Chem. 2019;67:10853–10862. doi: 10.1021/acs.jafc.9b03894. [DOI] [PubMed] [Google Scholar]
- 31.Kato Y., Suga N. Covalent adduction of endogenous and food-derived quinones to a protein: its biological significance. J. Clin. Biochem. Nutr. 2018;62:213–220. doi: 10.3164/jcbn.18-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsui Y., Sugiyama K., Kamei M., Takahashi T., Suzuki T., Katagata Y., Ito T. Extract of passion fruit (Passiflora edulis) seed containing high amounts of piceatannol inhibits melanogenesis and promotes collagen synthesis. J. Agric. Food Chem. 2010;58:11112–11118. doi: 10.1021/jf102650d. [DOI] [PubMed] [Google Scholar]
- 33.Masuda T., Miura Y., Inai M., Masuda A. Enhancing effect of a cysteinyl thiol on the antioxidant activity of flavonoids and identification of the antioxidative thiol adducts of myricetin. Biosci., Biotechnol. Biochem. 2013;77:1753–1758. doi: 10.1271/bbb.130315. [DOI] [PubMed] [Google Scholar]
- 34.Ma C., Hu Y., Townsend J.A., Lagarias P.I., Marty M.T., Kolocouris A., Wang J., Ebselen Disulfiram. Carmofur, PX-12, Tideglusib, and Shikonin are nonspecific promiscuous SARS-CoV-2 main protease inhibitors. ACS Pharmacol. Translat. Sci. 2020;3:1265–1277. doi: 10.1021/acsptsci.0c00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amporndanai K., Meng X., Shang W., Jin Z., Rogers M., Zhao Y., Rao Z., Liu Z.J., Yang H., Zhang L., O'Neill P.M., Samar Hasnain S. Inhibition mechanism of SARS-CoV-2 main protease by ebselen and its derivatives. Nat. Commun. 2021;12:3061. doi: 10.1038/s41467-021-23313-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bolton J.L., Dunlap T. Formation and biological targets of quinones: cytotoxic versus cytoprotective effects. Chem. Res. Toxicol. 2017;30:13–37. doi: 10.1021/acs.chemrestox.6b00256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peñalver L., Schmid P., Szamosvári D., Schildknecht S., Globisch C., Sawade K., Peter C., Böttcher T. A Ligand Selection Strategy Identifies Chemical Probes Targeting the Proteases of SARS-CoV-2. Angew. Chem. Int. Ed. 2021;60:6799–6806. doi: 10.1002/anie.202016113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuzikov M., Costanzi E., Reinshagen J., Esposito F., Vangeel L., Wolf M., Ellinger B., Claussen C., Geisslinger G., Corona A., Iaconis D., Talarico C., Manelfi C., Cannalire R., Rossetti G., Gossen J., Albani S., Musiani F., Herzog K., Ye Y., Giabbai B., Demitri N., Jochmans D., Jonghe S.D., Rymenants J., Summa V., Tramontano E., Beccari A.R., Leyssen P., Storici P., Neyts J., Gribbon P., Zaliani A. Identification of inhibitors of SARS-CoV-2 3CL-Pro enzymatic activity using a small molecule in vitro repurposing screen. ACS Pharmacol. Transl. Sci. 2021;4:1096–1110. doi: 10.1021/acsptsci.0c00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura T., Kawai Y., Kitamoto N., Osawa T., Kato Y. Covalent modification of lysine residues by allyl isothiocyanate in physiological conditions: plausible transformation of isothiocyanate from thiol to amine. Chem. Res. Toxicol. 2009;22:536–542. doi: 10.1021/tx8003906. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura T., Hirakawa M., Nakamura Y., Ishisaka A., Kitamoto N., Murakami A., Kato Y. Covalent modification of phosphatidylethanolamine by benzyl isothiocyanate and the resultant generation of ethanolamine adduct as its metabolite. Chem. Res. Toxicol. 2019;32:638–644. doi: 10.1021/acs.chemrestox.8b00331. [DOI] [PubMed] [Google Scholar]
- 41.Manach C., Williamson G., Morand C., Scalbert A., Remesy C. Bioavailability and bioefficacy of polyphenols in humans. I. Review of 97 bioavailability studies. Am. J. Clin. Nutr. 2005;81:230S–242S. doi: 10.1093/ajcn/81.1.230S. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka T., Ishii T., Mizuno D., Mori T., Yamaji R., Nakamura Y., Kumazawa S., Nakayama T., Akagawa M. (-)-Epigallocatechin-3-gallate suppresses growth of AZ521 human gastric cancer cells by targeting the DEAD-box RNA helicase p68. Free Radic. Biol. Med. 2011;50:1324–1335. doi: 10.1016/j.freeradbiomed.2011.01.024. [DOI] [PubMed] [Google Scholar]
- 43.Dai W., Ruan C., Zhang Y., Wang J., Han J., Shao Z., Sun Y., Liang J. Bioavailability enhancement of EGCG by structural modification and nano-delivery: A review. J. Funct. Foods. 2020;65 doi: 10.1016/j.jff.2019.103732. [DOI] [Google Scholar]
- 44.Hattori S.-i., Higashi-Kuwata N., Hayashi H., Allu S.R., Raghavaiah J., Bulut H., Das D., Anson B.J., Lendy E.K., Takamatsu Y., Takamune N., Kishimoto N., Murayama K., Hasegawa K., Li M., Davis D.A., Kodama E.N., Yarchoan R., Wlodawer A., Misumi S., Mesecar A.D., Ghosh A.K., Mitsuya H. A small molecule compound with an indole moiety inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nat. Commun. 2021;12:668. doi: 10.1038/s41467-021-20900-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xia Z., Sacco M., Hu Y., Ma C., Meng X., Zhang F., Szeto T., Xiang Y., Chen Y., Wang J. Rational Design of Hybrid SARS-CoV-2 main protease inhibitors guided by the superimposed cocrystal structures with the peptidomimetic inhibitors GC-376, Telaprevir, and Boceprevir. ACS Pharmacol. Translat. Sci. 2021;4:1408–1421. doi: 10.1021/acsptsci.1c00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rothan H.A., Teoh T.C. Cell-based high-throughput screening protocol for discovering antiviral inhibitors against SARS-COV-2 main protease (3CLpro) Mol. Biotechnol. 2021;63:240–248. doi: 10.1007/s12033-021-00299-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rawson J.M.O., Duchon A., Nikolaitchik O.A., Pathak V.K., Hu W.S. Development of a cell-based luciferase complementation assay for identification of SARS-CoV-2 3CL(pro) Inhibitors. Viruses. 2021;13 doi: 10.3390/v13020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma C., Tan H., Choza J., Wang Y., Wang J. Validation and invalidation of SARS-CoV-2 main protease inhibitors using the Flip-GFP and Protease-Glo luciferase assays. bioRxiv. 2021 doi: 10.1101/2021.08.28.458041. 2021.2008.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gerber P.P., Duncan L.M., Greenwood E.J., Marelli S., Naamati A., Teixeira-Silva A., Crozier T.W., Gabaev I., Zhan J.R., Mulroney T.E., Horner E.C., Doffinger R., Willis A.E., Thaventhiran J.E., Protasio A.V., Matheson N.J. A protease-activatable luminescent biosensor and reporter cell line for authentic SARS-CoV-2 infection. bioRxiv. 2021 doi: 10.1101/2021.03.22.435957. 2021.2003.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.