Abstract
Yanfeng 47 (YF47) is an elite japonica rice variety cultivated in China on nearly 2 million hectares over the past 20 years. However, YF47 is highly susceptible to rice blast (Magnaporthe oryzae), one of the most destructive rice diseases. In this study, we developed novel TPAP (tetra-primer ARMS-PCR) functional markers for the genes Pita, Pib, and Pid2, all of which afford broad-spectrum resistance to blast. A collection of 91 japonica rice germplasms with similar ecological characteristics to YF47 were screened, and Wuyunjing 27 (WYJ27) with Pita and Pib alleles and P135 with the Pid2 allele were identified. Furthermore, the corresponding positive Pita, Pib, and Pid2 alleles were transferred into YF47 using single, mutual, and backcrosses, together with molecular marker-assisted selection (MAS) and anther culture technology. These genetic materials, carrying one, two, or three functional alleles, were generated within 3 years, and compared to YF47, they all showed improved resistance to naturally inoculated rice blast. Further improved lines (IL) 1 to 5 (all containing Pita, Pib, and Pid2 alleles) were evaluated for yield performance, and when no fungicide was applied, all lines except IL-4 showed increased traits compared with those of YF47. IL-5, renamed Yanjing 144 (YJ144), showed yield increases in the Liaoning province regional variety comparison test and superior appearance quality compared to YF47. Our work provides a molecular design strategy for pyramiding multiple beneficial genes to rapidly improve rice blast resistance, yield, and quality using multiple breeding strategies.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11032-021-01259-4.
Keywords: Rice blast resistance, Molecular design breeding, TPAP functional marker, Multiple genes pyramiding, Anther culture technology
Introduction
Food security has become one of the world’s most important problems (Seck et al. 2012), and crop production faces unprecedented challenges, including climate change and the outbreak of COVID-19. Rice is the primary food crop for over half the world’s population (Pennisi, 2010), with close to 1.5 billion tons produced each year. However, extreme environmental conditions create biotic and abiotic stress that may severely affect production. Rice blast, caused by Magnaporthe oryzae, is one of the most destructive diseases affecting rice (Dean et al. 2012). Numerous studies have shown that it affects both yield and quality and yield loss estimates could reach 30–50% under temperature and humidity conditions favorable to outbreaks of M. oryzae. This is equivalent to destroying enough rice to feed more than 60 million people each year (Skamnioti and Gurr, 2009). From 2013 to 2017, the average annual area affected by rice blast in China was 75 million ha (https://www.natesc.org.cn, accessed 17 July 2020). The disease is broadly distributed and survives in a wide range of environmental conditions. Its predominant groups are complex and dynamic, which greatly increases the difficulty of prevention (Wang et al. 2017).
Over long-term co-evolution, rice has developed multiple mechanisms to resist pathogens, including pathogen-associated molecular pattern (PAMP)-induced immune responses (PAMP-triggered immunity, PTI) and pathogenic effector proteins (effector-induced immune response mechanism immunity, ETI) (Akerley et al. 1995; Nakahara and Masuta, 2014). The widespread incorporation in modern rice varieties of multiple resistance genes with durable resistance to rice blast is recognized as the most effective and environment-friendly approach (Miah et al. 2013). Multiple rice blast resistance genes have been cloned successfully: Pish, Pit, Pi37, Pib, Pi21, Pigm, Pi9, Piz-t, Pi2, Pid2, Pid3, Pi25, Pi36, Pi5/Pi3/Pi-i, Pi56, Pia, PiCO39, Pi1, Pik, Ptr, Pik-h/Pi54, Pik-p, Pik-m, pb1, and Pita (Li et al. 2015; Zhao et al. 2018; Zhou et al. 2018). Based on their encoded protein structures and molecular functions, these cloned genes can be divided into four categories: (1) nucleotide binding site–leucine-rich repeat (NBS-LRR) proteins, including the above genes with the exception of Pid2, Pi21, and Ptr; (2) receptor-like protein kinases, such as Pid2; (3) proline-rich domain proteins, including Pi21; (4) armadillo-rich repeat proteins, such as Ptr. Furthermore, most rice blast resistance genes have been identified and cloned in a dominant inheritance pattern, whereas only a small number function in a pattern of recessive inheritance, such as Pi21 (Chen et al. 2006; Fukuoka et al. 2009). The identification, cloning, and deep understanding of the mechanisms of these blast resistance alleles have already provided an enriched gene bank that has been widely used in traditional and hybrid rice breeding.
Marker-assisted selection (MAS) is a rapid, accurate, and effective tool for identifying target genes (Feng et al. 2019). In addition, it is little affected by environmental conditions (Ye et al. 2001; Chen et al. 2013). Currently, MAS technology is widely used to improve rice yield, optimize quality and enhance resistance and permit multiple breakthrough achievements (Wang et al. 2005; Chen et al. 2018). To date, MAS has also been widely used to improve rice blast resistance. Single resistance genes, though, cannot effectively deal with multiple pathotype infections simultaneously; thus, multiple studies used polymerized rice blast genes to improve the blast resistance of susceptible varieties (Jiang et al. 2012; Zhao et al. 2018), including enhanced Jin 23B and Kongyu 131 through Pi1 and Pi2 pyramiding, pyramiding blast resistance genes Piz5 and Pi54 into the elite Basmati rice restorer line PRR 78, and breeding the high-yielding and durable resistance rice variety Longliangyou 3189 by using Pigm (Koide et al. 2010; Singh et al. 2013; Deng et al. 2017; Yang et al. 2019; Yin et al. 2021). However, it is challenging to pyramid multiple favorable genes because of the laborious molecular detection techniques and long breeding periods involved (Yang et al. 2019). Anther culture technology produces haploid plants from pollen microspores that can become diploid either naturally or via treatment with active mitotic substances. This technique could homogenize breeding materials within two generations of rice breeding (Lee et al. 2003; Zhao et al. 2015). It was estimated that more than 40 rice varieties in China between 1980 and 2020 were successfully generated using this approach. However, few studies have reported where the combination of MAS with anther culture has generated superior rice varieties.
Accurate and convenient molecular markers are the basis of MAS technology (Jeppe and Thomas, 2003). At present, alleles of the multiple resistance genes in rice are demonstrated by the diversity of single-nucleotide polymorphism (SNP); therefore, cost-effective identification of functional molecular markers for SNP diversity is crucial (Abdiel et al. 2021). For this, cleaved amplification polymorphism sequence-tagged sites (CAPS), sequence-related amplified polymorphism (SPAP), and kompetitive allele-specific PCR (KASP) have been developed (Abdiel et al. 2021; Zhang et al. 2021). Among these, KASP is new, low-cost, and convenient, but it needs more expensive instrument support (Zhang et al. 2021). The others depend on DNA sequencing or restriction enzyme digestion, which is time-consuming (Jeppe and Thomas, 2003; Zhang et al. 2021). Tetra-primer ARMS-PCR (TPAP) is a rapid genotyping method for SNP identification and has been used in SNP analyses from plant science to medical science (Ye et al. 2001; Chen et al. 2013). In its design, two outer primers are designed to amplify the positive control DNA fragment, and two inner primers each containing a mismatched nucleotide are developed corresponding to the SNP diversity (Ye et al. 2001); ultimately, only one PCR amplification could identify the SNP diversity.
YF47, an elite japonica rice variety approved for the Chinese provinces of Liaoning and Shandong in 2001 and 2009, respectively, has high yield and wide adaptability characteristics (https://www.ricedata.cn/variety/varis/600002.htm, accessed 21 July 2020). This variety also received national certification in 2006 and is cultivated on nearly 2 million hectares in the Chinese provinces of Liaoning, Hebei, Shandong, and Jiangsu (Lu et al. 2017). However, because YF47 is highly susceptible to rice blast disease (Liu et al. 2014; Li et al. 2017), improving resistance is important for ensuring food security in northern China. Pyramiding multiple blast resistance genes is an effective way to improve resistance (Jiang et al. 2012; Zhao et al. 2018). It was reported that Pi2/Pi9/Pigm allele clusters mainly occur in indica rice with increased rice blast resistance (Deng et al. 2017; Xie et al. 2019), however, in the northeast, Pita presents a great opportunity to improve rice blast resistance due to the corresponding avirulence gene Avr-Pita, which is widespread in the area (Wang et al. 2014, 2016; Xie et al. 2019). Furthermore, owing to the prevalence of blast strain ZB15 in the area in recent years (Liu et al. 2014; Li et al. 2017), its corresponding high resistance gene Pid2 should be considered a good application prospect. Pib is resistant to Chinese strains ZB13 and ZC15, and regarding the progress of rice blast resistance breeding in Jiangsu province (the japonica rice area in eastern China) (Wang et al. 2012), pyramiding Pita and Pib achieved a good application value. Therefore, based on a comprehensive consideration of the defense mechanisms of resistance genes to improve the rice blast resistance of YF47, Pita, Pib, and Pid2 were selected as candidate genes. In this study, TPAP functional markers for Pita, Pib, and Pid2 alleles were developed and used to evaluate the alleles in a collection of 91 japonica rice germplasms. Two donor parents, WYJ27 with Pita and Pib and P135 with Pid2, were crossed with the receptor parent YF47, following a mutual intercross and backcross to YF47 to produce highly blast-resistant lines. A range of breeding strategies were employed, including multiple-gene pyramiding and anther culture. The derived materials with one, two, or three of them introduced with the Pita, Pib, and Pid2 genes showed improved resistance to rice blast on natural inoculation. One line IL-5 (also named YJ144) carrying three resistance genes was further selected and shown to exhibit improved resistance, yield, and quality. Taken together, this work provides a novel and rapid strategy in modern rice breeding to generate superior varieties with comprehensively improved traits.
Materials and methods
Plant materials
Plant materials comprised two groups. In group A were control rice varieties that were used to develop functional molecular markers: LJHG—susceptibility check, K013—resistance check for Pita, K014—resistance check for Pib, and Digu—resistance check for Pid2 (Wang et al. 2012, 2015). Group B varieties were used to improve the rice blast resistance of YF47: WYJ27 with Pita and Pib, P135 with Pid2 and the recurrent parent YF47. P135 is a japonica accession carrying Pid2 and produced by crossing YF47 with Digu. Detailed information on these rice varieties is shown in Table 1.
Table 1.
Plant materials used in this study
| Variety (or line) name | Subspecies | Origin | Genotype |
|---|---|---|---|
| LJHG | japonica | Yunnan, China | pita, pib, pid2 |
| K013 | indica | Jiangsu, China | Pita |
| K014 | indica | Jiangsu, China | Pib |
| Digu | indica | Sichuan, China | Pid2 |
| Yanfeng 47 (YF47) | japonica | Liaoning, China | pita, pib, pid2 |
| Wuyunjing 27 (WYJ27) | japonica | Jiangsu, China | Pita, Pib |
| P135 | japonica | Liaoning, China | Pid2 |
Design strategy for Pita, Pib, and Pid2 markers
Following the procedure in Ye et al. (2001), the online primer design software Primer 1 was used to design four primers for each gene: one pair of outer primers to amplify a short stretch of genomic DNA as a positive control; one forward inner primer and one reverse inner primer designed according to the functional SNP. One deliberate mismatched base at the -3 site of the 3′-end of the extension was introduced into each inner primer (Table 2).
Table 2.
Molecular markers used in this study
The deliberate introduction of a mismatched base is shown in red
Crossing and selection
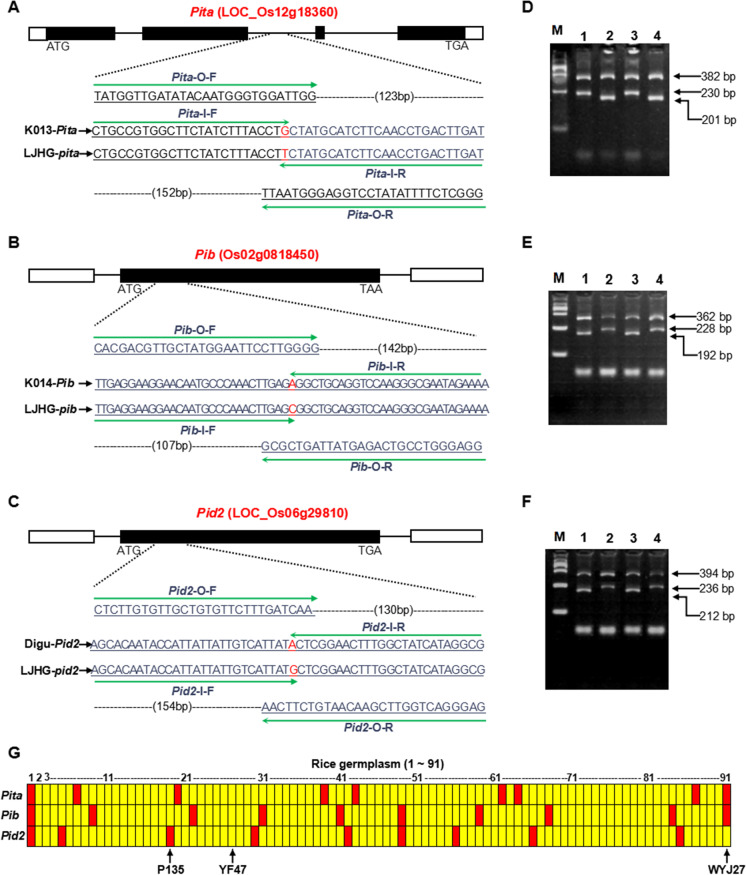
In 2013, WYJ27 and P135 were separately crossed with YF47 in Panjin (41.07° N, 122.03° E), and the seeds of F1-1 and F1-2 were planted in winter in Hainan (18.15° N, 109.30° E), followed by F1-1 × F1-2 to get the seeds of F1-3. In 2014, 25 strains of F1-3 plants carrying Pita/pita, Pib/pib, and Pid2/pid2 were obtained through molecular detection in Panjin; furthermore, YF47 was hybridized with F1-3, and BC1F1 lines (n = 102) were planted in Hainan. Based on molecular detection, BC1F1-7 (Pita/pita, Pib/pib, Pid2/pid2, n = 13) plants were identified and anther culture was carried out at the late uninucleate stage when 7000 anthers were collected. From these, about 630 embryoids were induced; callus induction was 9%. Finally, 151 green seedlings were successfully generated, giving a plant differentiation rate of 24%. In 2015, 115 H1 plants were obtained to carry out genotypic identification in Panjin. Ultimately, improved lines of no resistance gene (n = 6), Pita (n = 26), Pib (n = 15), Pid2 (n = 22), Pita + Pib (n = 22), Pita + Pid2 (n = 15), Pib + Pid2 (n = 10), and Pita + Pib + Pid2 (n = 5) were obtained.
Anther culture operation scheme
The culture medium composition followed the methods described by Lee et al. (2003): At the late uninucleate stage, the spike neck wrapped with young spikes were dropped out and pretreated at 10 °C for 5 h, N6 was used as basal media for the first step of callus induction, adding 2,4-D (2 mg/L), NAA (1 mg/L), KT (1 mg/L), LH (0.5 g/L), sucrose (50 g/L), and agar (4.2 g/L). Further, MS was used as the callus differentiation medium, adding 6-BA (2 mg/L), NAA (0.5 mg/L), IAA (0.5 mg/L), IBA (0.1 mg/L), inositol (0.1 g/L), MET (2 mg/L), LH (0.5 g/L), sucrose (30 g/L), and agar (4.2 g/L). The pH value of culture medium was 5.8.
Identification of rice blast resistance
Rice blast resistance was monitored in 2019 and 2020 in the city of Dandong (40.07° N, 124.23° E). This is a mountainous area with high humidity and heavy fog, with the most serious susceptibility to rice blast disease in Liaoning province. According to previous studies, 44 races of 6 groups (ZA, ZB, ZC, ZD, ZE, and ZF) of the fungus were prevalent in this area in recent years (Liu et al. 2014; Li et al. 2017). A total of 124 rice accessions (including 121 filial generation accessions, YF47, WYJ27, and P135) were tested in a randomized complete block design with three replications. Resistance identification was conducted by natural inoculation according to Jiang et al. (2012), with slight changes: every replication was planted at a high density (50 plants/m2) to create a hot and humid environment. Menggudao (susceptible variety) was planted at both sides of each row and around the entire population. Diseased straw collected from the previous year was used around each accession and also around Menggudao. The leaf blast disease index at the tillering and heading stage was recorded using the 0–5 scale rating system, and neck blast severity was recorded as a percentage of infection in the neck of rice panicle at the maturity stage (Bonman et al. 1986; Jiang et al. 2012).
Investigation of yield-related characters and rice quality
Yield-related characters were investigated in Panjin during 2019 and 2020. In 2019, the five improved lines carrying the three blast resistance genes were investigated with and without the application of fungicides, and with YF47 as the control. In 2020, yield-related characters of YJ144 and YF47 were investigated with and without the application of fungicides. Test materials were grown under irrigated conditions in a randomized complete block design with three replications, in six-row plots, with each row 10 m long, and a plant spacing of 30 × 13.3 cm. Ten plants were harvested to investigate yield traits: number of effective panicles, number of filled grains per panicle, and 1000 grain weight. Rice grain from a 1-m2 area was harvested and used to calculate yield. In 2020, YJ144 and YF47 were harvested and stored at room temperature for 3 months, according to Ren et al. (2016). A 600-g sample of grain was processed to analyze the milling quality, and 30 plump grains were selected to measure grain length and width. After processing into polished rice, the chalkiness degree was assessed using an SC-E rice appearance quality analyzer (Wanshen, China). The amylose content, alkali spread value, and gel consistency were determined as described previously (Butardo et al. 2011), and the protein content was calculated by using the N-protein conversion coefficient 5.95 described by Hu et al. (2014). The eating quality score was determined by sensory tasting, followed the methods described by Lestari et al. (2009).
The regional variety comparison test in Liaoning province was conducted both in 2019 and in 2020 with three replications in eight-row plots. Each row was 10 m long, with a plant spacing of 30 × 13.3 cm. This was replicated in six cities: Tieling (TL), Shenyang (SY), Liaozhong (LZ), Linghai (LH), Dandong (DD), and Panjin (PJ).
DNA isolation, PCR amplification, and electrophoresis detection
A plant genomic isolation kit (Tiangen, Wuhan, China) was used to extract rice genomic DNA. PCR was prepared in a total volume of 20 μL containing 10 μL of 2 × Tap PCR Master Mix (Tiangen, Wuhan, China), 0.3 μL of each outer primer (4 pmol/μL), 0.2 μL of each inner primer (4 pmol/μL), and 1 ng of genomic DNA diluted with ddH2O to 20 μL. The PCR procedure was as follows: 95 °C for 5 min; 32 cycles of 95 °C for 30 s, 55–58 °C for 30 s, 72 °C for 50 s; and a final extension at 72 °C for 10 min. Agarose gels (5%) were used to separate PCR products, which were then stained with ethidium bromide, and photographed with a GEL DOC 1000 system (BioRad, Hercules, CA, USA).
Statistical analyses
The data and chart analyses were performed using Graph Pad Prism v. 5.0 and Microsoft Office PowerPoint 2007.
Results
Developing novel TPAP molecular markers for Pita, Pib, and Pid2
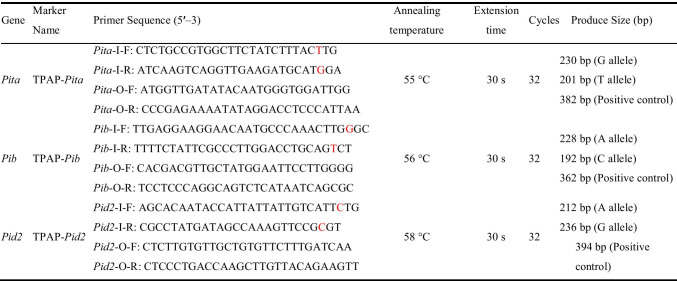
Pita encodes a cytoplasmic membrane receptor protein containing an NBS domain and a leucine-rich domain (LRD), and its disease-resisting mechanism is due to its detection of the expression product of avirulent gene Avr-Pita in rice blast, thus triggering rapid local defenses (Bryan et al. 2000). Two other useful alleles function in rice blast resistance. One is Pib, which belongs to the NBS-LRR disease-resistant gene group and encodes a protein product comprising one NBS and 17 LRR. The other is Pid2, which encodes a transmembrane receptor protein kinase (RLK) and confers high resistance to the Chinese rice blast strain ZB15 (Wang et al. 1999, 2015). One major task of rice breeders is to use these genes effectively to generate novel highly resistant varieties. Functional molecular markers have been developed for Pita and Pib detection, with two pairs of primers and double PCR amplification needed (Fjellstrom et al. 2004; Wang et al. 2012), while Pid2 allele detection still requires sequencing (Wang et al. 2015). However, the detection primers for Pita, Pib, and Pid2 are inefficient for pyramiding multiple genes because the process is time-consuming (Fjellstrom et al. 2004; Kim et al. 2016). Therefore, rapid, simple, and low-cost functional molecular markers for Pita, Pib, and Pid2 detection are urgently needed.
First, based on the functional nucleotide polymorphism sites of Pita/pita, Pib/pib, and Pid2/pid2 (LOC_Os12g18360 and LOC_Os06g29810 collected from the MSU database, and Os02g0818450 from RAP-DB), molecular markers were designed for TPAP–Pita, TPAP–Pib, and TPAP–Pid2 (Fig. 1A–C) (Wang et al. 1999, 2015; Bryan et al. 2000). Referring to the previous studies, K013, K014 and Digu were chosen as the resistance checks for Pita, Pib, and Pid2, respectively, and LJHG was used as the susceptibility check (Wang et al. 2012, 2015). For Pita, the marker TPAP–Pita amplified 230-bp and 201-bp DNA fragments corresponding to the resistance and susceptibility alleles, and a 382-bp fragment was the positive control (Fig. 1D). For Pib, the marker TPAP–Pib amplified 228-bp and 192-bp DNA fragments corresponding to the resistance and susceptibility alleles, and a 362-bp DNA fragment was the positive control (Fig. 1E). For Pid2, the marker TPAP–Pid2 amplified 212-bp and 236-bp DNA fragments corresponding to the resistance and susceptibility alleles, and a 394-bp DNA fragment was the positive control (Fig. 1F). To confirm the precise application of molecular markers, we sequenced the PCR products using outer primers of TPAP–Pita, TPAP–Pib, and TPAP–Pid2, and confirmed the accuracy of the above constructed molecular markers (Fig. S1–S3).
Fig. 1.
TPAP primer design and detection of Pita, Pib, and Pid2 alleles. Design strategy for TPAP–Pita (A), TPAP–Pib (B), and TPAP–Pid2 (C) markers. Functional SNPs are shown in red; underlines show primer binding sites. PCR assay of Pita (D) between resistant (K103, lanes 1 and 3) and susceptible (LJHG, lanes 2 and 4) genotypes in two repeats; Pib (E) between resistant (K104, lanes 2 and 4) and susceptible (LJHG, lanes 1 and 3) genotypes in two repeats; Pid2 (F) between resistant (Digu, lanes 1 and 3) and susceptible (LJHG, lanes 2 and 4) genotypes in two repeats. M, marker 2000. G Genotype analysis of Pita, Pib, and Pid2 on 91 rice germplasm using TPAP–Pita, TPAP–Pib, and TPAP–Pid2. Nos. 1–2 are the resistance check and susceptibility check of Pita, Pib, and Pid2, corresponding to K013 and LJHG, K014 and LJHG, and Digu and LJHG, respectively; Nos. 3–48, 49–63, 64–67, 68–82, 83–87, and 88–91 are japonica germplasms collected from Liaoning, Ningxia, Tianjin, Xinjiang, Jilin, and Jiangsu provinces, respectively. The yellow regions represent susceptible genotypes, and the red regions represent the resistance genotypes YF47, Yanfeng47, and WYJ27, Wuyunjing27
Generation and resistance analysis of blast resistance materials using the crosses of YF47 as a receptor parent, and WYJ27 and P135 as donor parents
To investigate the benefit that blast-resistant alleles confer, we studied 91 japonica varieties (or lines) from different provinces: Liaoning, Ningxia, Tianjin, Xinjiang, Jilin, and Jiangsu (Table S1). Most of them show similar ecological characteristics and were further used for genotype analysis of Pita, Pib, and Pid2. It was found that some cultivars contained one or two functional alleles; however, no one rice variety contained all three (Fig. 1G). Next, considering genetic relationship, plant architecture, rice yield, and quality, WYJ27 (with Pita and Pib alleles) and P135 (with the Pid2 allele) were selected as donor parents to improve rice blast resistance of YF47. As a result, a series of rice lines carrying one, two, or three resistant genes were identified using multiple strategies of rice breeding, including single, mutual, and backcrosses together with MAS (Fig. 2). To accelerate the breeding process further, the anther culture technique was used to generate these genetic materials. A total of 7000 anthers were collected from BC1F1–7 plants at the late uninucleate stage. From these, about 630 embryoids were induced; callus induction was 9%. Finally, 151 green seedlings were successfully generated, giving a plant differentiation rate of 24% (Fig. 2).
Fig. 2.
Construction of genetic materials carrying various rice blast resistance genes
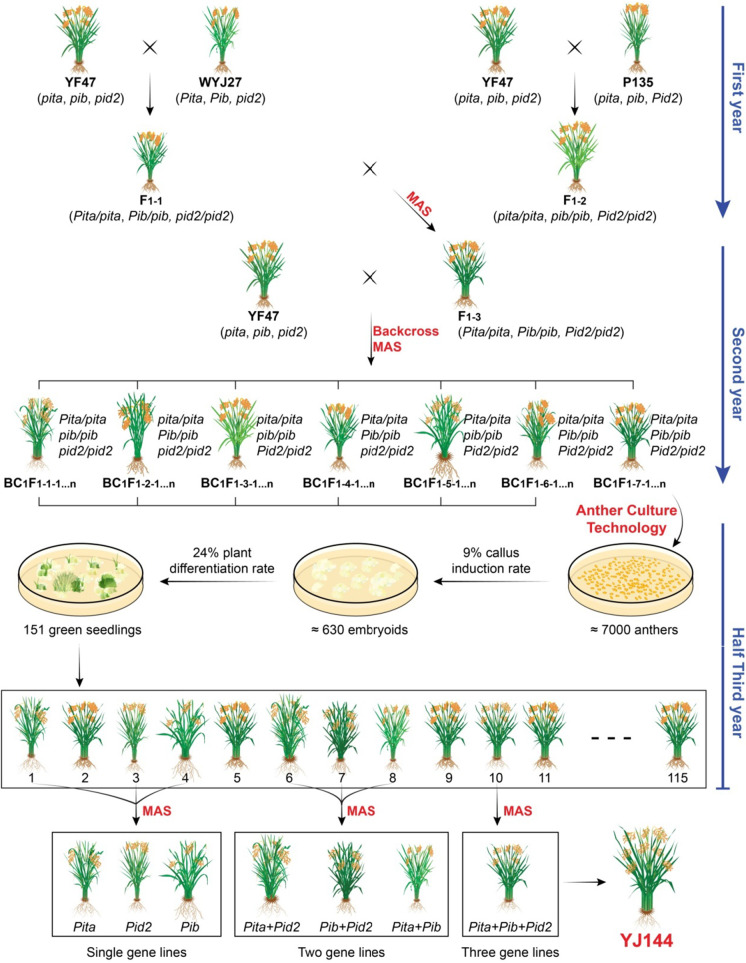
To further evaluate the resistance levels of those generated materials, we recorded a leaf and neck blast disease index in both 2019 and 2020 using natural inoculation (Fig. 3). In 2019, at the tillering stage, the leaf blast disease index for no resistance gene lines (NL, n = 6), Pita (n = 26), Pib (n = 15), Pid2 (n = 22), Pita + Pib (n = 22), Pita + Pid2 (n = 15), Pib + Pid2 (n = 10), and Pita + Pib + Pid2 (n = 5) was recorded as means of 4.1, 2.6, 3.7, 3.8, 1.8, 1.9, 2.8, and 0.4, respectively (Fig. 3A). At the heading stage, the corresponding means were 4.8, 3.4, 4.1, 4.1, 2.5, 2.8, 3.6, and 0.8, respectively (Fig. 3B). The receptor parent YF47 had a leaf blast disease index of 4 and 5 at the tillering and heading stages, respectively. The neck blast severity was recorded as infection percentages of 68.6, 35.7, 42.9, 44.7, 24.8, 28.7, 35.6, and 7.0%, respectively, whereas 68.0% was observed for YF47 (Fig. 3C). In 2020, the disease index increased slightly but showed a similar pattern that of 2019 (Fig. 3D–F). The other two donor parents, WYJ27 and P135, were also used to investigate the disease index. They both showed improved leaf blast disease index and reduced percentage of infection over the control parent YF47 (Fig. 3). Taken together, Pita showed the best improvement against rice blast, while Pib exhibited slightly stronger resistance than Pid2. It is noteworthy that blast resistance increased with the number of functional alleles in the line.
Fig. 3.
Determination of rice leaf blast resistance and neck blast resistance. Rice leaf disease index at the tillering (A) and heading stage (B) in 2019. C Rice neck blast scores at the maturity stage in 2019. Rice leaf disease index at the tillering (D) and heading (E) stages in 2020. F Rice neck blast scores at the maturity stage in 2020. The horizontal/vertical lines show mean ± SD. NL, no resistance gene lines
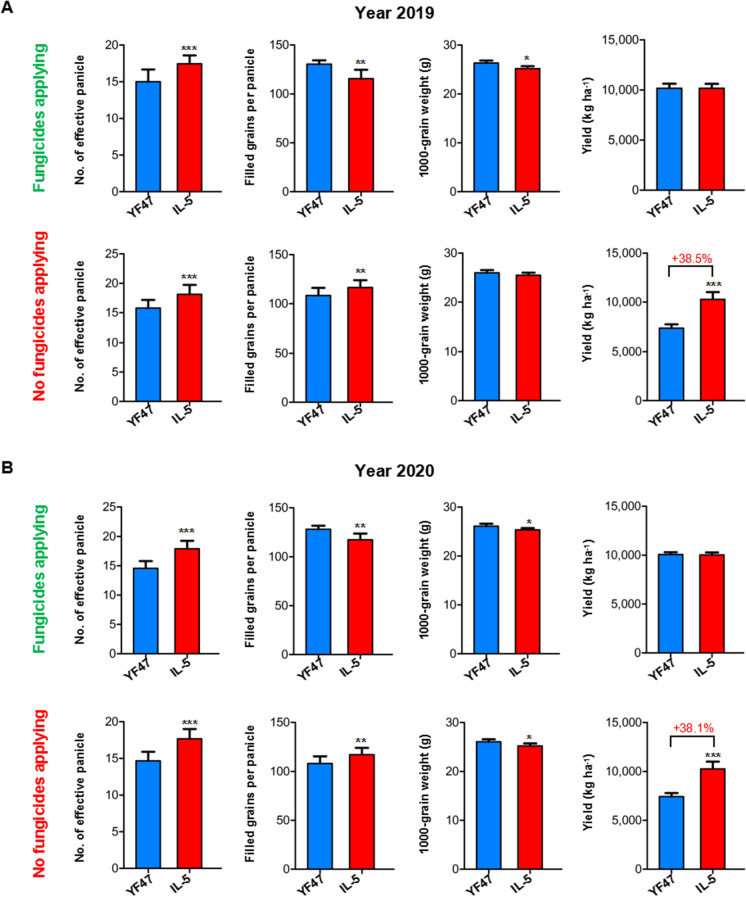
Agronomic traits of YF47 and the improved lines carrying three resistance genes with and without application of fungicides
The field performances in Liaoning province were tested for five improved lines IL-1 to IL-5, all with introduced Pita, Pib, and Pid2 alleles with YF47 as a control. Their yield-related traits, including the number of effective panicles, filled grains per panicle, 1000 grain weight, and total yield per hectare, were investigated with and without the application of fungicides. In 2019, under the application of fungicides, IL-5 showed an increased number of effective panicles, whereas IL-2 and 4 exhibited the opposite outcome. IL-3, 4, and 5 all showed reduced numbers of filled grains per panicle, while the five lines all showed decreased 1000 grain weight. As a result, each of the three lines (IL-1, 2, and 4) exhibited a reduction in yield compared to YF47. The other two lines, IL-3 and 5, had yields equal to that of YF47 (Fig. 4A and Fig. S4A). In 2019, without application of fungicides, IL-5 showed an increased number of effective panicles, whereas IL-1, 2, and 4 exhibited the opposite effect. IL-1, 2, 3, and 5 all had an increased number of filled grains per panicle. Also, lines IL-1 to 4 all showed decreased 1000 grain weight. As a result, after rice blast infection, all lines except IL-4 exhibited better yield performance compared to that of YF47 (Fig. 4A and Fig. S4B). Thus, IL-3 and 5 are two candidate lines with huge for rice breeding.
Fig. 4.
Yield-related trait comparison of YF47 and IL-5. Yield-related trait comparison of YF47 and IL-5 in 2019 (A) and 2020 (B), with and without application of fungicides. IL-5, improved line 5. The data shows mean ± SD; *p < 0.05, **p < 0.01, and ***p < 0.001
To evaluate further the performance of IL-5, with and without application of fungicides, similar experiments were conducted in 2020. We compared the yield-related parameters of YF47 and IL-5. As shown in Fig. 4B, under the application of fungicides, the effective panicle number of IL-5 was significantly higher than that of YF47 (p < 0.01), while the filled grains per panicle and 1000 grain weight were significantly lower than those of YF47 (p < 0.05), resulting in nearly the same yield. However, where fungicides were not applied, the effective panicle number and the filled grains per panicle of IL-5 were both significantly higher than those of YF47 (p < 0.01), while its 1000 grain weight was significantly lower (p < 0.05), resulting in higher yield for IL-5 (p < 0.001).
YJ144, a high blast resistance rice line, exhibited increased yield traits in the Liaoning province regional variety comparison tests
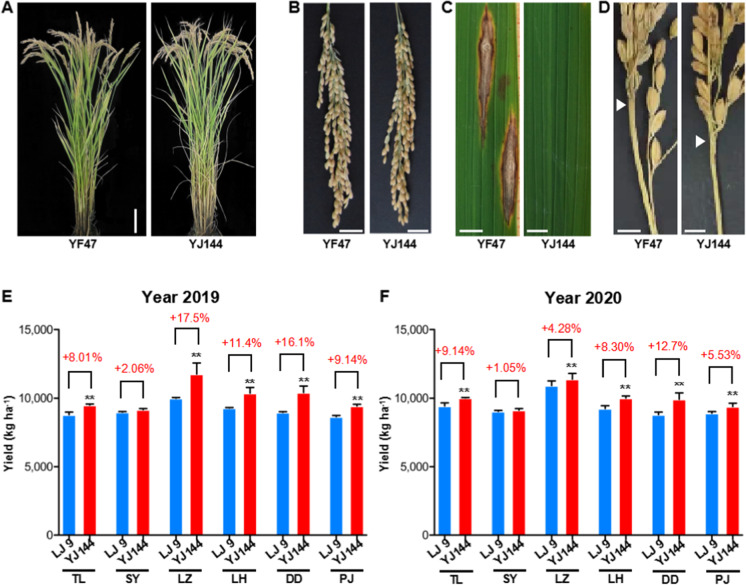
Of five improved lines, each containing three rice blast resistance genes, IL-5 had the minimum disease index for leaf blast and neck blast, and showed increased yield both in 2019 and 2020 (Figs. 3 and 4 and Fig. S4). We named this line YJ144. Then, we conducted phenotypic and morphological analyses of YF47 and YJ144. As shown in Fig. 5A–B, YJ144 showed similar plant architecture and panicle morphological architecture compared to YF47. Moreover, YJ144 had higher rice blast resistance (Fig. 5C–D), consistent with their relative leaf and neck blast disease index, as indicated in Fig. 3. To confirm that the three functional alleles (Pita, Pib, and Pid2) had been successfully introduced, we sequenced the appropriate functional sites of these genes within YF47 and YJ144. The results confirmed that YJ144 contained these novel functional sites, as shown in Fig. S1–S3.
Fig. 5.
Plant architecture, rice blast resistance, and yield performance of YJ144. A Plant architecture comparison of YJ144 and YF47. Scale bar, 10 cm. B Panicle architecture comparison of YJ144 and YF47. Scale bar, 2 cm. Rice leaf (C) and neck (D) blast disease index of YJ144 and YF47. Yield comparison of LJ9 and YJ144 in the regional variety comparison test in six cities of Liaoning province in 2019 (E) and 2020 (F). LJ9 (Liaojing9) was a control in the regional variety comparison test. TL, SY, LZ, LH, DD, and PJ represent the Liaoning cities of Tieling, Shenyang, Liaozhong, Linghai, Dandong, and Panjin, respectively. The data shows mean ± SD; **p < 0.01
To examine the performance of YJ144 in field trials, in 2019 and 2020, it was entered in the regional variety comparison tests in Liaoning province. Liaojing 9 (LJ9) is an officially designated control variety with medium yield used in regional comparison tests in Liaoning province. Its planting has covered nearly 0.6 million hectares over the past 30 years (https://www.ricedata.cn/variety/varis/600590.htm, accessed 25 July 2020). This trial encompassed tests in six Liaoning cities: Tieling (TL), Shenyang (SY), Liaozhong (LZ), Linghai (LH), Dandong (DD), and Panjin (PJ). In both 2019 and 2020, YJ144 exhibited significant yield increases of 4.3 to 17.5% at all six sites apart from SY. Yields at SY also increased in 2019 and 2020 but not significantly (Fig. 5E–F). These findings suggest that YJ144 exhibited superior yield capacity and could be used as an important japonica germplasm with both high blast resistance and yield.
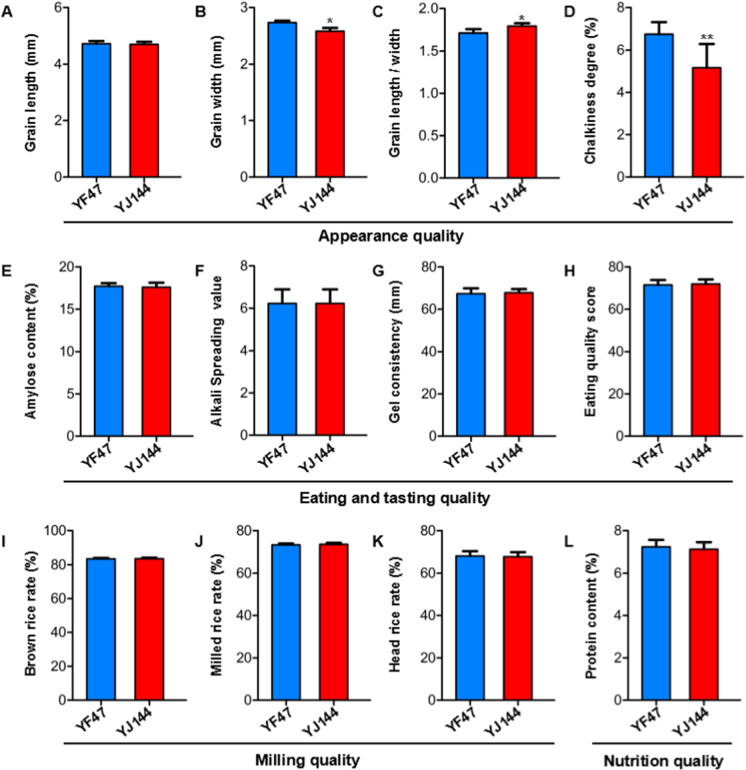
Appearance quality of YJ144 is improved compared to that of YF47
To ascertain whether the introduction of blast-resistant genes could affect rice quality, we determined some rice quality indicators: appearance, milling, nutrition, eating, and tasting. YJ144 and the control YF47, both harvested in 2020, were tested. As shown in Fig. 6A–C, the grain lengths of YJ47 and YJ144 were very similar, while the grain width of YF47 was greater than that of YJ144, which resulted in a significantly higher length-to-width ratio for YJ144. Additionally, the chalkiness of YJ144 was significantly lower than that of YF47 (Fig. 6D), probably resulting from its greater length-to-width ratio. However, grain shape and chalk differences between YF47 and YJ144 did not give rise to significant changes in milling quality, including the brown, milled, and head rice rates (Fig. 6E–H). To assess nutritional, eating and cooking quality, we compared protein and amylose contents, the alkali spreading value, and gel consistency, which are recognized as the most critical quality factors for eating and cooking. Ultimately, none of the characteristics showed any significant change between YF47 and YJ144 (Fig. 6H and I–L). Taken together, our findings demonstrated that YJ144 showed improved appearance quality compared to YF47.
Fig. 6.
Comparison of rice quality between YF47 and YJ144. Appearance quality comparison of YF47 and YJ144, including grain length (A), grain width (B), the ratio of grain length to width (C), and degree of chalkiness (D). Comparison of milling quality of YF47 and YJ144, including the brown rice (E), milled rice (F), and head rice rates (G). H Nutrition quality comparison of YF47 and YJ144. Eating and tasting quality comparison of YF47 and YJ144, including amylose content (I), alkali spread value (J), gel consistency (K), and eating quality score (L). The data show mean ± SD; *p < 0.05 and **p < 0.01
Discussion
Accurate and rapid detection of rice blast resistance is fundamental for resistance breeding. In previous studies, plants were artificially inoculated with pathogen races (Deng et al. 2006; Wang et al. 2014), but the type of pathogen inoculated was limited and the operation was complicated and easily affected by artificial factors. One problem was the nature of the fungus found in Liaoning province, which consists of at least 44 races in 6 groups (ZA, ZB, ZC, ZD, ZE, and ZF), the predominant ones of which are complex and dynamic (Liu et al. 2014; Li et al. 2017). In this study, we used natural infection through dense planting, application of diseased straw, and cultivation of a susceptible variety in the surrounding plot. Under these natural conditions, plants were infected by numerous M. oryzae pathotypes simultaneously; therefore, this is likely to reflect environmental factors and test the effects of resistance genes more accurately. Under these conditions, YF47 is readily susceptible to M. oryzae, whereas WYJ27 with Pita and Pib alleles, and P135 with the Pid2 allele, exhibited increased resistance (Fig. 3). In addition, resistance to rice blast improved with an increased number of resistance genes, including relevant Pita, Pib, and Pid2 alleles (Fig. 3). Natural infection showed good and stable results in rice blast resistance detection, similar to the conclusions reported by Jiang et al. (2012).
Numerous studies showed that the aggregation of multiple blast genes is prone to durable disease resistance, caused by the adaptation to different physiological races. The additive effect among genes could also lead to a high pyramiding effect (Xiao et al. 2020; Xu et al. 2021). In this study, improved lines with single a resistance gene represented susceptible or moderate resistance, but two or three pyramiding genes presented improved or superior resistance to rice blast, respectively. According to the previous study (Wang et al. 1999, 2015; Bryan et al. 2000), Pita and Pib belong to the NBS-LRR type, and Pid2 belongs to receptor-like protein kinases, resulting in different molecular functions. The encoded product of the Pita gene can interact with the AVR-Pita expression product of Magnaporthe grisea to induce disease resistance; the Pib is resistant to the Chinese strain ZB15, and Pid2 is resistant to Chinese strains ZB13 and ZC15. More resistance genes’ pyramiding could respond to broader physiological race changes, and, according to Wang et al. (2012), Pita has the better improved application value on rice blast resistance than Pib in Jiangshu province in China; this is similar to our findings; also, Pita + Pib pyramiding has a superior joint effect (Wang et al. 2012); thus, we speculated that the above factors are the main reasons for the high pyramiding effect as shown in Figs. 3, 4, and 5.
In this study, using TPAP technology, we developed a set of functional markers to detect Pita, Pib, and Pid2, where only one time PCR amplification was required for accurate identification at each locus. Compared with the previous study, Pib and Pid2 detection needed two pairs of primer amplification or sequencing (Fjellstrom et al. 2004; Wang et al. 2015), respectively. For Pita, Ramkumar et al. and Hayashi et al. developed allele-specific and linkage molecular markers (Hayashi et al. 2006; Ramkumar et al. 2015). Our markers had a mismatch deliberately inserted, which enhanced amplification accuracy by providing a lower annealing temperature, an advantage of the better amplification conditions. Subsequently, a total of 91 japonica rice germplasms were collected from northern districts that presented similar ecological environments, and were screened within the loci of Pita, Pib, and Pid2. WYJ27 was found to contain Pita and Pib alleles, while P135 had Pid2 alleles. Both were selected as donor parents to improve the rice blast resistance of YF47. Overall, the above work could provide both cost-effective TPAP molecular markers and japonica rice germplasms with Pita, Pib, and Pid2 alleles, which could be used as the basic elements for MAS in rice breeding.
Compared with traditional breeding strategies, MAS can significantly improve selection efficiency; however, pyramiding multiple genes still requires many generations (Jiang et al. 2012; Baliyan et al. 2018). In this study, using multiple breeding strategies (single cross, mutual intercross, and backcross) combined with MAS, we successfully obtained BC1F1–7 lines carrying Pita/pita, Pib/pib, and Pid2/pid2. Then, through anther culture, which is more commonly used in japonica rice than in indica rice due to its high rates of induction and differentiation (Lee et al. 2003; Wang et al. 2020), a series of derivative lines with different numbers of resistance genes were also created by using MAS (Fig. 2). As a result, the generation of stable materials was achieved within 3 years and showed much greater efficiency than conventional or marker-assisted breeding. In contrast to conventional breeding, CRISPR/Cas technology is widely used to modify gene components, in both the animal and plant kingdoms, by deleting negative genetic elements responsible for undesired traits or introducing gain-of-function mutations through precise genome editing. For example, the application of CRISPR/Cas has to date improved several rice crop characteristics, including yield, quality, and disease resistance (Wang et al. 2013; Zhou et al. 2015). However, it is difficult to edit three genes in their functional sites simultaneously. It also takes more than 1 year to eliminate the hygromycin B resistance gene and the noise background through one or two consecutive crosses between the transgenic line and the receptor parent. Further, the generated lines could be used only as intermediates in rice breeding. Our research provides a strategy for rapidly breeding rice cultivars with durable resistance to rice blast.
To date, large numbers of genes controlling vital agronomic traits have been characterized (www.ricedata.cn, accessed 29 July 2020). High yield, multiple resistance, and superior quality are primary objectives of genetic improvement; however, these goals are difficult to coordinate (Yu et al. 2018), such as Pi21 being separable from a closely linked gene conferring poor flavor (Fukuoka et al. 2009), and Pigm R reducing 1000 grain weight and rice yield. Recently, a rice genome navigation system, RiceNavi, was developed for quantitative trait nucleotides (QTN) pyramiding and breeding route optimization, and it is easy to employ multiple useful alleles effectively to generate novel rice varieties for rice breeders (Wei et al. 2021). In this study, we first developed a genetic improvement strategy by combining primer construction, applying various breeding methods, and conducting a comprehensive investigation of resistance, yield, and quality. Subsequently, for better coordinate yield, quality, and blast resistance, we identified Pita-, Pib-, and Pid2-resistant genes in 91 rice germplasm; next, a high-yield parent carrying Pita and Pib (WYJ27) and a superior quality parent possessing Pid2 (P135, with low chalkiness) were selected for genetic improvement, with a widely adaptable variety YF47 as the recurrent parent. Moreover, BC1F1 plants with Pita, Pib, and Pid2 were used for anther culture. Further, the blast resistance, yield components and rice quality of improved lines were investigated in depth. Ultimately, an ideal line, YJ144, was generated. Our molecular design strategy for pyramiding multiple beneficial genes generated superior varieties with comprehensively improved traits.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contribution
T.M. and G.S. designed and performed the experiments, and analyzed data; M.Z., S.A., Z.S., S.H., G.J., L.X., and S.T. participated in improved lines’ construction and data analysis; S.A., G.Y., X.W., G.S., and T.M. wrote the manuscript and prepared the illustrations. G.S. and P.H. conceived the idea and supervised the project. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by grants from the China National Key Research and Development Program (Grant No. 2020YFE0202300); the Central Public-Interest Scientific Institution Basal Research Fund of China (Grant No. Y2020YJ09); the International Science & Technology Innovation Program of Chinese Academy of Agricultural Sciences, China (Grant No. CAAS-ZDRW202109); and the PhD Research Startup Foundation of Liaoning Province (2020-BS-300).
Declarations
Data and materials availability
All data are given in the manuscript.
Not applicable.
Consent for publication
The authors give consent for the publication.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Peisong Hu, Email: peisonghu@126.com.
Gaoneng Shao, Email: shaogaoneng@caas.cn.
References
- Abdiel PG, Pumipat T, Julapark C, Hans JD, Anucha W, Waraporn S, Paweena C. Genetic diversity and population structure of ridge gourd (Luffa acutangula) accessions in a Thailand collection using SNP markers. Sci Rep. 2021;11:15311. doi: 10.1038/s41598-021-94802-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerley BJ, Cotter PA, Miller JF. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell. 1995;80:611–620. doi: 10.1016/0092-8674(95)90515-4. [DOI] [PubMed] [Google Scholar]
- Baliyan N, Malik R, Rani R, Mehta K, Vashisth U, Dhillon S, Boora KS. Integrating marker-assisted background analysis with foreground selection for pyramiding bacterial blight resistance genes into Basmati rice. CR Biol. 2018;341:1–8. doi: 10.1016/j.crvi.2017.11.003. [DOI] [PubMed] [Google Scholar]
- Bonman JM, Vergel de Dios TI, Khin MM. Physiologic specialization of Pyricularia oryzae in the Philippines. Plant Dis. 1986;70:767–769. doi: 10.1094/PD-70-767. [DOI] [Google Scholar]
- Bryan GT, Wu KS, Farrall L, Jia Y, Hershey HP, McAdams SA, Faulk KN, Donaldson GK, Tarchini R, Valent B. A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta. Plant Cell. 2000;12:2033–2046. doi: 10.1105/tpc.12.11.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butardo VM, Fitzgerald MA, Bird AR, Gidley MJ, Flanagan BM, Larroque O, Resurreccion AP, Laidlaw HK, Jobling SA, Morell MK, Rahman S. Impact of down-regulation of starch branching enzyme IIb in rice by artificial microRNA-and hairpin RNA-mediated RNA silencing. J Exp Bot. 2011;62:4927–4941. doi: 10.1093/jxb/err188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MJ, Liu GF, Yu H, Wang B, Li JY. Towards molecular design of rice plant architecture and grain quality. Chin Sci Bull. 2018;63:1276–1289. doi: 10.1360/N972018-00429. [DOI] [Google Scholar]
- Chen T, Luo MR, Zhang YD, Zhu Z, Zhao L, Zhao QY, Zhou LH, Yao S, Wang CL. Detection of wx-mq gene for low-amylose content by tetra-primer amplification refractory mutation system PCR in rice. Chin J Rice Sci. 2013;27:529–534. [Google Scholar]
- Chen XW, Shang JJ, Chen DX, Lei CL, Zou Y, Zhai WX, Liu GZ, Xu JC, Ling ZZ, Cao G, Ma BT, Wang WP, Zhao XF, Li SG, Zhu LH. A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 2006;46:794–804. doi: 10.1111/j.1365-313X.2006.02739.x. [DOI] [PubMed] [Google Scholar]
- Dean R, Kan JA, Pretorius ZA, Hammond-Kosack KE, Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R, Ellis J, Foster GD. The top 10 fungal pathogens in molecular plant pathology. Mol Plant Pathol. 2012;13:414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Zhu X, Shen Y, He Z. Genetic characterization and fine mapping of the blast resistance locus Pigm (t) tightly linked to Pi2 and Pi9 in a broad-spectrum resistant Chinese variety. Theor Appl Genet. 2006;113:705–713. doi: 10.1007/s00122-006-0338-7. [DOI] [PubMed] [Google Scholar]
- Deng YW, Zhai KR, Xie Z, Yang DY, Zhu XD, Lin JZ, Wang X, Qin P, Yang YZ, Zhang GM, Li Q, Zhang JF, Wu SQ, Milazzo LJ, Mao BZ, Wang ET, Xie HA, Tharreau D, He ZH. Epigenetic regulation of antagonistic receptors confers rice blast resistance with yield balance. Science. 2017;355:962–965. doi: 10.1126/science.aai8898. [DOI] [PubMed] [Google Scholar]
- Feng XM, Lin KX, Zhang WQ, Nan JZ, Zhang XH, Wang C, Wang RS, Jiang GQ, Yuan QB, Lin SY. Improving the blast resistance of the elite rice variety Kongyu-131 by updating the pi21 locus. BMC Plant Bio. 2019;19:249. doi: 10.1186/s12870-019-1868-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fjellstrom R, Concetta A, Bormans C, McClung AM, Marco A, Marchetti A, Shank R, Park WD. Development of DNA markers suitable for marker assisted selection of three genes conferring resistance to multiple Pathotypes. Crop Sci. 2004;44:1790–1798. doi: 10.2135/cropsci2004.1790. [DOI] [Google Scholar]
- Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebana K, Hayashi N, Takahashi A, Hirochika H, Okuno K, Yano M. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science. 2009;325:998–1001. doi: 10.1126/science.1175550. [DOI] [PubMed] [Google Scholar]
- Hayashi K, Yoshida H, Ashikawa I. Development of PCR-based allele-specific and InDel marker sets for nine rice blast resistance genes. Theor Appl Genet. 2006;113:251–260. doi: 10.1007/s00122-006-0290-6. [DOI] [PubMed] [Google Scholar]
- Hu B, Wang W, Ou SJ, Tang JY, Li H, Che RH, Zhang ZH, Chai XY, Wang HR, Wang YQ, Liang CZ, Liu LC, Piao ZZ, Deng QY, Deng K, Xu C, Liang Y, Zhang LH, Li LG, Chu CC. Variation in NRT1.1B contributes to nitrate-use divergence between rice subspecies. Nat Genet. 2014;47:834–838. doi: 10.1038/ng.3337. [DOI] [PubMed] [Google Scholar]
- Jeppe A, Thomas L. Functional markers in plants. Trends Plant Sci. 2003;11:554–560. doi: 10.1016/j.tplants.2003.09.010. [DOI] [PubMed] [Google Scholar]
- Jiang HC, Feng YT, Bao L, Li X, Gao GJ, Zhang QL, Xiao JH, Xu CG, He YP. Improving blast resistance of Jin 23B and its hybrid rice by marker-assisted gene pyramiding. Mol Breed. 2012;30:1679–1688. doi: 10.1007/s11032-012-9751-6. [DOI] [Google Scholar]
- Kim SR, Ramos J, Ashikari M, Parminder SV, Edgar AT, Eero N, Hechanova SL, Ramial M, Kshirod KJ. Development and validation of allele-specific SNP/indel markers for eight yield-enhancing genes using whole-genome sequencing strategy to increase yield potential of rice Oryza Sativa L. Rice. 2016;9:12. doi: 10.1186/s12284-016-0084-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide Y, Kawasaki A, Yanoria MJ, Hairmansis A, Nguyet NTM, Bigirimana J, Fujita D, Kobayashi N, Fukuta Y. Development of pyramided lines with two resistance genes, Pish and Pib, for blast disease (Magnaporthe oryzae B. Couch) in rice (Oryza sativa L.) Plant Breed. 2010;129:670–675. doi: 10.1111/j.1439-0523.2010.01781.x. [DOI] [Google Scholar]
- Lee SY, Lee JH, Kwon TO. Selection of salt-tolerant doubled haploids in rice anther culture. Plant Cell, Tissue Organ Cult. 2003;74:143–149. doi: 10.1023/A:1023956526669. [DOI] [Google Scholar]
- Lestari P, Ham TH, Lee HH, Woo MO, Jiang W, Chu SH, Kwon SW, Ma K, Lee JH, Cho YC, Koh HJ. PCR marker-based evaluation of the eating quality of japonica rice (Oryza sativa L.) J Agric Food Chem. 2009;57:2754–2762. doi: 10.1021/jf803804k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Sun YD, Liu H, Wang YY, Jia YL, Xu MH. Natural variation of rice blast resistance gene Pi-d2. Genet Mol Res. 2015;14:1235–1249. doi: 10.4238/2015.February.13.2. [DOI] [PubMed] [Google Scholar]
- Li SB, Wei SH, Luo WF, Zhang Y, Liu ZH. Dynamics of rice blast fungus population in Liaoning province in 2015–2016. J Shenyang Agric Univ. 2017;48:284–289. [Google Scholar]
- Liu ZH, Wang WS, Wei SH, Wang HN, Zheng WJ, Yang TZ. Dynamics of rice blast fungus population in Liaoning province during 2011–2012. J Shenyang Agric Univ. 2014;45:393–397. [Google Scholar]
- Lu NN, Yan LH, Zheng CK, Yin HB, Guo SL, Xie XZ. Effects of salt stress on growth and agronomic traits of Yanfeng 47 and Yanjing 456. Crops. 2017;5:106–111. [Google Scholar]
- Miah G, Rafii MY, Ismail MR, Puteh AB, Rahim HA, Asfaliza R, Latif MA. Blast resistance in rice: a review of conventional breeding to molecular approaches. Mol Biol Rep. 2013;40:2369–2388. doi: 10.1007/s11033-012-2318-0. [DOI] [PubMed] [Google Scholar]
- Nakahara K, Masuta C. Interaction between viral RNA silencing suppressors and host factors in plant immunity. Curr Opin Plant Biol. 2014;20:88–95. doi: 10.1016/j.pbi.2014.05.004. [DOI] [PubMed] [Google Scholar]
- Pennisi E. Armed and dangerous. Science. 2010;327:804–805. doi: 10.1126/science.327.5967.804. [DOI] [PubMed] [Google Scholar]
- Ramkumar G, Prahalada GD, Hechanova SL, Vinarao R, Jena KK. Development and validation of SNP-based functional codominant markers for two major disease resistance genes in rice ( O. sativa L.) Mol. Breed. 2015;35:129. doi: 10.1007/s11032-015-0323-4. [DOI] [Google Scholar]
- Ren DY, Rao YC, Huang LC, Leng YJ, Hu J, Lu M, Zhang GH, Zhu L, Gao ZY, Dong GJ, Guo LB, Qian Q, Zeng DL. Fine mapping identifies a new QTL for brown rice rate in rice (Oryza Sativa, L.) Rice. 2016;9:4. doi: 10.1186/s12284-016-0076-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seck PA, Diagne A, Mohanty S, Wopereis MCS. Crops that feed the world 7: Rice. Food Sec. 2012;4:7–24. doi: 10.1007/s12571-012-0168-1. [DOI] [Google Scholar]
- Singh VK, Singh A, Singh SP, Ellur RK, Choudhary V, Sarkel S, Singh D, Krishnan SG, Nagarajan M, Vinod KK, Singh UD, Rathore R, Prashanthi SK, Agrawal PK, Bhatt JC, Mohapatra T, Prabhu KV, Singh AK. Incorporation of blast resistance into “PRR78”, an elite Basmati rice restorer line, through marker assisted backcross breeding. Field Crop Res. 2012;128:8–16. doi: 10.1016/j.fcr.2011.12.003. [DOI] [Google Scholar]
- Singh VK, Singh A, Singh SP, Ellur RK, Singh D, Gopala Krishnan S, Bhowmick PK, Nagarajan M, Vinod KK, Singh UD, Mohapatra T, Prabhu KV, Singh AK. Marker-assisted simultaneous but stepwise backcross breeding for pyramiding blast resistance genes Piz5 and Pi54 into an elite Basmati rice restorer line ‘PRR 78’. Plant Breed. 2013;132:486–495. [Google Scholar]
- Skamnioti P, Gurr SJ. Against the grain: safe guarding rice from rice blast disease. Trends Biotechnol. 2009;27:141–150. doi: 10.1016/j.tibtech.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Wang BH, Ebbole DJ, Wang ZH. The arms race between Magnaporthe oryzae and rice: diversity and interaction of Avr and R genes. J Integr Agric. 2017;16:2746–2760. doi: 10.1016/S2095-3119(17)61746-5. [DOI] [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Casmediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Qu BY, Dou SJ, Li LY, Yin D, Pang ZQ, Zhou ZZ, Tian MM, Liu GZ, Xie Q, Tang DZ, Chen XW, Zhu LH. The E3 ligase OsPUB15 interacts with the receptor-like kinase PID2 and regulates plant cell death and innate immunity. BMC Plant Biol. 2015;15:49. doi: 10.1186/s12870-015-0442-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yang J, Yang JH, Fan FJ, Zhu JY, Chen ZD, Zhong WG. Analysis on breeding value of Pi-ta, Pi-b genes in japonica rice breeding with neck resistance in Jiangsu. Acta Agron Sin. 2012;27:141–145. [Google Scholar]
- Wang P, Bai YL, Wang MX, Hu BH, Pu ZG, Zhang ZY, Zhang Q, Xu DW, Luo WL, Chen ZQ. Breeding of CMS maintainer lines through anther culture assisted by high-resolution melting-based markers. J Integr Agric. 2020;19:2965–2973. doi: 10.1016/S2095-3119(20)63179-3. [DOI] [Google Scholar]
- Wang SW, Zheng WJ, Zhao JM, Wei SH, Wang Y, Zhao BH, Liu ZH. Identification and analysis of Magnaporthe oryzae Avirulence genes in Liaoning Province. J Integr Agric. 2014;47:462–472. [Google Scholar]
- Wang Y, Zhao JM, Zhang LX, Wang P, Wang SW, Wang H, Wang XX, Liu ZH, Zheng WJ. Analysis of the diversity and function of the alleles of the rice blast resistance genes Piz-t, Pita and Pik in 24 rice cultivars. J Integr Agric. 2016;15:1423–1431. doi: 10.1016/S2095-3119(15)61207-2. [DOI] [Google Scholar]
- Wang YH, Xue YB, Li JY. Towards molecular breeding and improvement of rice in China. Trends Plant Sci. 2005;10:610–614. doi: 10.1016/j.tplants.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Wang ZX, Yano M, Yamanouchi U, Iwamoto M, Monna L, Hayasaka H, Katayose Y, Sasaki T. The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J. 1999;19:55–64. doi: 10.1046/j.1365-313X.1999.00498.x. [DOI] [PubMed] [Google Scholar]
- Wei X, Qiu J, Yong KC, Fan JJ, Zhang Q, Hua H, Liu J, Wang Q, Olsen KM, Han B, Huang XH. A quantitative genomics map of rice provides genetic insights and guides breeding. Nat Genet. 2021;53:243–253. doi: 10.1038/s41588-020-00769-9. [DOI] [PubMed] [Google Scholar]
- Xiao N, Wu Y, Li A. Strategy for use of rice blast resistance genes in rice molecular breeding. Rice Sci. 2020;27:263–277. doi: 10.1016/j.rsci.2020.05.003. [DOI] [Google Scholar]
- Xie Z, Yan BX, Shou JY, Tang J, Wang X, Zhai KR, Liu JY, Li Q, Luo MZ, Deng YW, He ZH. A nucleotide-binding site-leucine-rich repeat receptor pair confers broad-spectrum disease resistance through physical association in rice. Philos Trans r Soc b-Biol Sci. 2019;374:20180308. doi: 10.1098/rstb.2018.0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu LH, Zhang PB, Li MQ, Zhang F, Liu F, Yao FY. Development of “multi-resistance rice” by pyramiding of insect (Cry1C) and blast resistance (Pi1 and Pi2) genes. Plant Diseases and Pests. 2021;12:25–28. [Google Scholar]
- Yang DB, Tang JH, Yang D, Chen Y, Ali JH, Mou TM. Improving rice blast resistance of Feng39S through molecular marker-assisted backcrossing. Rice. 2019;12:70. doi: 10.1186/s12284-019-0329-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye S, Dhillon S, Ke X, Collins AR, Day IN. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 2001;17:e88. doi: 10.1093/nar/29.17.e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin JJ, Zou LJ, Zhu XB, Cao YY, He M, Chen XW. Fighting the enemy: how rice survives the blast pathogen’s attack. Crop J. 2021;9:543–552. doi: 10.1016/j.cj.2021.03.009. [DOI] [Google Scholar]
- Yu H, Wang B, Chen MJ, Liu GF, Li JY. Research advance and perspective of rice breeding by molecular design. Chin Bull Life Sci. 2018;30:1032–1037. [Google Scholar]
- Zhang YF, Li T, Geng YK, Wang YM, Liu YC, Li HF, Hao CY, Wang HJ, Shang XW, Zhang XY. Identification and development of a KASP functional marker of TaTAP46-5A associated with kernel weight in wheat (Triticum aestivum) Plant Breed. 2021;140:585–594. doi: 10.1111/pbr.12922. [DOI] [Google Scholar]
- Zhao HJ, Wang XY, Jia YL, Minkenberg B, Wheatley M, Fan JB, Jia MH, Famoso A, Edwards JD, Wamishe Y, Valent B, Wang GL, Yang YO. The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat Commun. 2018;9:2039. doi: 10.1038/s41467-018-04369-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Liu L, Wang J, Guo H, Gu J, Zhao S, Li J, Xie Y. Development of a new wheat germplasm with high anther culture ability by using a combination of gamma-ray irradiation and anther culture. J Sci Food Agric. 2015;95:120–125. doi: 10.1002/jsfa.6691. [DOI] [PubMed] [Google Scholar]
- Zhou X, Liao H, Chern M, Yin J, Chen Y, Wang J, Zhu X, Chen Z, Chen Z, Yuan C, Zhao W, Wang J, Li W, He M, Ma B, Wang J, Qin P, Chen W, Wang Y, Liu J, Qian Y, Wang W, Wu X, Li P, Zhu L, Li S, Ronald PC, Chen X. Loss of function of a rice TPR-domain RNA-binding protein confers broad-spectrum disease resistance. PNAS. 2018;115:3174–3179. doi: 10.1073/pnas.1705927115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Xin J, Fan N, Zou Q, Huang J, Ouyang Z, Zhao Y, Zhao B, Liu Z, Lai S, Yi X, Guo L, Esteban MA, Zeng Y, Yang H, Lai L. Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cell Mol Life Sci. 2015;72:1175–1184. doi: 10.1007/s00018-014-1744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.