Abstract
Background: Accumulating evidence from observational studies suggested that circulating adiponectin levels are associated with the risk of rheumatoid arthritis (RA), but the causality remains unknown. We aimed to assess the causal relationship of adiponectin with RA risk.
Methods: Based on summary statistics from large-scale genome-wide association studies (GWAS), we quantified the genetic correlation between adiponectin and RA. Then bidirectional Mendelian randomization (MR) analysis was performed to assess the causal relationship. Twenty single-nucleotide polymorphisms (SNPs) associated with adiponectin were selected as instrumental variables from a recent GWAS (n = 67,739). We applied theses SNPs to a large-scale GWAS for RA (14,361 cases and 43,923 controls) with replication using RA data from the FinnGen consortium (6,236 cases and 147,221 controls) and the UK Biobank (5,201 cases and 457,732 controls). The inverse-variance weighted (IVW) and multiple pleiotropy-robust methods were used for two-sample MR analyses.
Results: Our analyses showed no significant genetic correlation between circulating adiponectin levels and RA [rG = 0.127, 95% confidence interval (CI): –0.012 to 0.266, P = 0.074]. In MR analyses, genetically predicted adiponectin levels were not significantly associated with the RA risk (odds ratio: 0.98, 95% CI: 0.88–1.09, P = 0.669). In the reverse direction analysis, there is little evidence supporting an association of genetic susceptibility to RA with adiponectin (β: 0.007, 95% CI: –0.003 to 0.018, P = 0.177). Replication analyses and sensitivity analyses using different models yielded consistent results.
Conclusions: Our findings provided no evidence to support the causal effect of adiponectin levels on RA risk and of RA on circulating adiponectin levels.
Keywords: rheumatoid arthritis, adiponectin, causality, Mendelian randomization, bidirectional
Introduction
Rheumatoid arthritis (RA) is an autoimmune disease with unclear etiology, mainly involving synovial joints, leading to joint malformation and a reduced quality of life (Lee and Bae, 2018). RA can be distributed at any age, and the high incidence age is 40–60 years old. The etiology of RA remains vague, but both genetic and environmental triggers likely result in the onset and progression of the disease (Bae and Lee, 2019a).
The most abundant adipokines in plasma is adiponectin, an endogenous bioactive polypeptide or protein secreted only by adipose tissue (Li and Wu, 2012; Dan et al., 2020). It stimulates endothelial cells, monocytic cells, and synovial fibroblasts to product interleukin-6 (IL-6) and metalloproteinase (MMP) (Lee and Bae, 2018). Moreover, adiponectin is presumed to be closely associated with the adjustment of inflammatory responses (Otero et al., 2006). Lots of studies have been conducted to explore the relationship between adiponectin and RA risk, but the results are inconsistent. Some studies had showed that circulating adiponectin levels in RA patients were higher than those in healthy controls (Lee and Bae, 2018), while others found an opposite significant or either null association (El-Hini et al., 2013; Li et al., 2015). Besides, the previous studies were all observational studies with a limited sample size, which is susceptible to confounding and reverse causality. Thus, whether circulating adiponectin levels have causal effect on risk of developing RA remains unclear.
Mendelian randomization (MR) utilizes genetic variants as instrumental variables (IVs), which is more dependable to test the latent causal association between exposures and diseases (Smith and Ebrahim, 2003; Lawlor et al., 2008; Burgess et al., 2015; Bennett and Holmes, 2017). MR analysis can overcome the reverse causality as the genotypes are essentially fixed since conception, and they are prior to the disease occurrence process (Lawlor et al., 2008; Pierce and Burgess, 2013). Moreover, MR analyses can eliminate the confounder on account of random assignment of alleles. In the present study, we selected available IVs based on the currently published genome-wide association studies (GWASs) to explore whether circulating adiponectin levels were causally associated with RA using bidirectional two-sample MR analysis.
Materials and Methods
Study Design
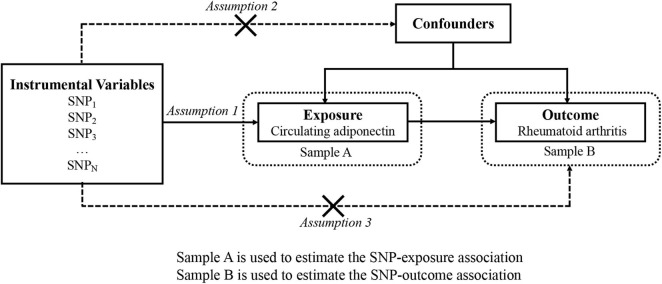
The overview of this research design is illustrated in Figure 1. Implementation of the MR analysis depends on three stringent assumptions:
FIGURE 1.
An overview of the study design with three fundamental assumptions about MR study.
-
•
Each genetic variant should be closely related to circulating adiponectin levels. We employed the F statistic and R2 to evaluate the strength of instruments and to evaluate the changes in circulating adiponectin expounded by the IV. If F statistic > 10 (Burgess and Thompson, 2011), it is considered that the association is strong enough to avoid weak instrument bias.
-
•
Each genetic variant should not be associated with confounders of the exposure–outcome association.
-
•
Each genetic variant should influence the risk for RA only by adiponectin instead of other pathway (no “pleiotropy”) (Bowden et al., 2016).
Data Sources
Our investigation did not require further ethical approval because this MR research was implemented on the basis of publicly available data. We selected 20 single nucleotide polymorphisms (SNPs) associated with adiponectin as IVs from the latest GWAS meta-analysis, including 67,739 individuals of participants (89% European ancestry). These SNPs for adiponectin are genome-wide significant (p < 5 × 10–8) and not in linkage disequilibrium (LD) (Dastani et al., 2012). Relevant genetic information was gathered for the 20 selected SNPs, such as effect alleles, non-effect alleles, effect allele frequency, effect size (β), standard error, and p-value. Of note, the standard errors that are not directly provided in the original paper were estimated based on the β and p-values.
The RA data were extracted from a large independent meta-analysis of interracial GWAS with more than 100,000 participants with European and Asian ancestry (Okada et al., 2014). To decrease the risk of population stratification bias, merely summary-level data from European individuals (14,361 cases and 43,923 controls) were obtained for analysis (Okada et al., 2014). RA patients had been certified by rheumatoid arthritis specialists or met the diagnostic criteria for rheumatism (Arnett et al., 1988). Detailed information of above-mentioned GWASs in this study was accessible from previous publications (Okada et al., 2014).
To validate the results, we retrieved two additional data sources for RA, the FinnGen consortium (R5 release, 2021) and the UK Biobank (Sudlow et al., 2015). In the FinnGen consortium, we used the fifth wave of the GWAS results on RA, including 6,236 cases and 147,221 controls. All cases were defined by the code M13 in International Classification of Diseases---Tenth Revision. In the UK Biobank study, we leveraged GWAS data on 5,201 RA cases and 457,732 controls, which were deposited in the MRC-IEU.1 Due to the limited number of diagnosed RA cases available in the UK Biobank, we used self-reported RA data to increase the sample size.
Linkage Disequilibrium Score Regression
To research the extent to which heritability of circulating adiponectin trait and RA is shared, we conducted LD score regression analysis via the LD Hub (Zheng et al., 2017). Compared with MR, LD score regression generally focuses on the number of genetic correlations, which could distinguish population stratification and polygenicity in GWAS.
Bidirectional Mendelian Randomization
To assess whether reverse causality exists, MR was employed in taking RA as exposure and the circulating adiponectin level as the outcome. Since summary-level data used to extract the SNP for adiponectin were not available, another GWAS summary data on adiponectin from ADIPOGen consortium were used, which included 39,883 individuals (Dastani et al., 2012).
Statistical Analyses
To sustain effect alleles always in connection with the same allele, we further adjusted the adiponectin and RA datasets. In the main analysis, the fixed-effects inverse-variance weighted (IVW) was performed to calculate the estimation of causal effect for different SNPs (Hartwig et al., 2016). Although numerous variants in an MR analysis can increase statistical power, pleiotropic genetic variation may lead to the presence of ineffective IVs (Hartwig et al., 2016). To adjust the potential pleiotropy, sensitivity analyses were utilized. More specifically, the WM method is effective in generating a consistent causal estimate when nearly one-half of the genetic variants were invalid IVs (Bowden et al., 2016). In the MR-Egger regression analysis (Bowden et al., 2015), we tested the existence of directional pluripotent effects on the basis of its intercept term (p < 0.05). The slope coefficient from Egger regression can be interpreted as the consistency of a causal estimate (Burgess and Thompson, 2017). Of note, such method may produce wider CIs with relatively lower precision (Bae and Lee, 2019b). To further test the existence of potential pleiotropy effects, we employed the MR-Pleiotropy residual sum and Outlier (MR-PRESSO) for outlier correction (Verbanck et al., 2018). In addition, hundreds of SNPs as IVs can be evaluated in the analysis by using the contamination mix method, which is not affected by the presence of invalid SNPs (Burgess et al., 2020).
Moreover, we manually searched secondary phenotypes of each selected SNP and its proxy in the PhenoScanner2 to further rule out potential pleiotropic effects (Supplementary Tables 1, 2). All the above MR analyses were reiterated after exclusion of SNPs associated with potential confounders at the genome-wide significance level.
To verify the statistical heterogeneity among SNPs used in IVW, Cochran’s Q test was employed (Egger et al., 1997). To probe the potential effect of an SNP on the causal estimates, we performed a ‘‘leave-one-out’’ sensitivity analysis with each SNP removed. The a priori statistical power was estimated using an online tool.3 Finally, the 20 SNPs for the adiponectin explained 1.17% of the variance. For a genetically predicted 1-SD increase in the circulating adiponectin level, our analyses had adequate power (>80%) to detect an OR of 1.29 for RA.
All statistical analyses were conducted in R (version 3.6.3, using the “TwoSampleMR,” “MendelianRandomization,” and “MRPRESSO” R packages). Statistical significance was set as bilateral p-values < 0.05.
Results
Linkage Disequilibrium Score Regression
Our analyses showed no significant genetic correlation Between circulating adiponectin levels and RA (rG = 0.127, 95% CI: –0.012 to 0.266, P = 0.074).
Adiponectin and Rheumatoid Arthritis
A total of 20 independent SNPs were used as IVs in MR analysis. Summary information about these 20 SNPs is shown in Supplementary Tables 3–5. The F statistics for these SNPs are >10, demonstrating that the weak IV bias did not exist.
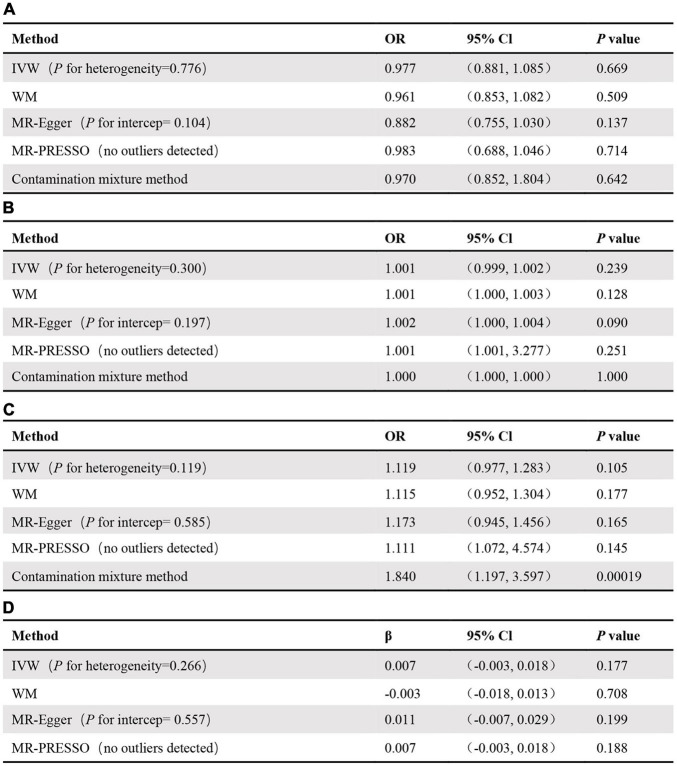
No significant relationship was observed between circulating adiponectin and the risk of RA [odds ratio (OR): 0.98, 95% confidence interval (CI): 0.88–1.09, p = 0.669] using the IVW method. Moreover, MR-Egger (OR: 0.88, 95% CI: 0.75–1.03, p = 0.137), weighted median (OR: 0.96, 95% CI: 0.85–1.08, p = 0.509), MR-PRESSO (OR: 0.98, 95% CI: 0.69–1.05, p = 0.713), and the contamination mix method (OR: 0.97, 95% CI: 0.85–1.80, p = 0.642) demonstrated consistent results (Figure 2A). The “leave-one-out analysis” indicated that no SNP can significantly promote the estimates of adiponectin on risk of RA, which showed that the results were reliable. We additionally discovered that several SNPs were related to obesity-related traits, such as high-density lipoprotein, low-density lipoprotein, and body mass index. Additionally, there was no evidence supporting a significant intercept about horizontal pleiotropy based on MR-Egger regression (intercept = 0.014, p = 0.104). No significant heterogeneity was found in the included independent SNP estimates using Cochran’s Q test. We applied the same analysis to the other two databases, which showed consistent results. The results of the analysis of the UK Biobank data and the FinnGen consortium are shown in Figures 2B,C, respectively.
FIGURE 2.
MR results for association of adiponectin and original-RA (A), the UK Biobank-RA (B), the FinnGen consortium-RA (C), as well as association of RA and adiponectin (D).
Rheumatoid Arthritis and Adiponectin
Similarly, we took RA as the exposure and circulating adiponectin levels as the outcome to verify whether RA was causally associated with circulating adiponectin levels. In the end, 29 SNPs with significant association (p < 5 × 10–8) for RA were included in this MR analysis (Supplementary Table 6). Based on the results, we observed no significant causal link between risk of RA and circulating adiponectin levels (β: 0.007, 95% CI: –0.003 to 0.018, p = 0.177) using the IVW method (Supplementary Figure 1), MR-Egger regression (β: 0.011, 95% CI: –0.007 to 0.029, p = 0.199), weighted-median method (β:–0.003, 95% CI: –0.018 to 0.013, p = 0.708), and MR PRESSO (β: 0.007, 95% CI: –0.003 to 0.018, p = 0.188) (Figure 2D). The leave-one-out analysis also confirmed that no SNP can drive the estimates of risk of RA on adiponectin (Supplementary Figure 2). In addition, there was no indication of a significant intercept about horizontal pleiotropy (intercept = –0.001, p = 0.557) (Supplementary Figure 3) and significant heterogeneity (Supplementary Figure 4).
Discussion
The result of the current research shows no indication supporting the fact that circulating adiponectin levels were causally associated with RA risk. Besides, the genetic susceptibility of RA was not causally associated with circulating adiponectin levels.
Adiponectin is a secretion of adipose tissue and plays an important role in regulating the inflammatory response in inflammation (Tsatsanis et al., 2005; Fantuzzi, 2008). Some observational studies showed that serum adiponectin levels were significantly higher in RA patients compared with healthy controls (Alkady et al., 2011; Khajoei et al., 2019). In contrary, a case-control study documented that plasma adiponectin levels were significantly lower in RA patients compared with healthy controls in Chinese population (Zhao et al., 2020). A previous meta-analysis has revealed that adiponectin levels in the RA group were significantly higher than those in the control group, and adiponectin may have an antiphlogistic effect in the pathogenesis of RA by merging 11 studies (Lee and Bae, 2018). Although accumulated evidence suggested that elevated serum adiponectin levels are associated with the risk of RA, the causal influence between the two remains not known. There was no evidence supporting the fact that circulating adiponectin levels were causally associated with RA by using MR analysis in our study. The discrepancy between the results of our study and those of previous studies may be due to the bias or confounders intrinsic to observational epidemiological studies, such as a small sample size, heterogeneity in demographic characteristics, reverse causation, and selection bias.
As far as we know, it is considered to be the first MR study to explore the bidirectional relationship between circulating adiponectin levels and the risk of RA. There are some limitations to be aware of. First, satisfying three assumptions is a prerequisite for obtaining effective results in MR studies. Nevertheless, due to the flaws of MR analysis, the second hypothesis and the third hypothesis cannot be accurately accessed (Arnett et al., 1988), which may lead to potential deviations. The second hypothesis states that genetic variants that should influence the risk of RA only depend on risk factors instead of any other pathway. Thus, we used ME-Egger to identify horizontal pleiotropy. In addition, the third hypothesis states that each genetic variant should not be associated with confounders of the risk factor–outcome association, and we manually excluded SNPs associated with confounders to ensure that we obtained robust main results. Second, our research data were based upon four massive GWASs. Because the detailed demographic data and clinical symptoms of the subjects were not available, subgroup analysis cannot be performed. In addition, this study selected 20 limited SNPs related to circulating adiponectin levels as IVs, which only explain limit proportion of variation and may not have enough ability to find an association, so we sought two additional data sources, the FinnGen consortium and the UK Biobank, to validate the results. Finally, we could not explain the complex feedback loop because of the body’s adaptability to early physiological changes, which may add more potential interference to the IV analysis.
Although LD score regression analysis showed no statistically significant association between adiponectin and RA, it suggested a potential association as the p-value reached borderline significance. In previous studies, we found that adiponectin could be inhibited by inflammatory cytokines such as tumor necrosis factor-a and IL-6 in CD4+ T cells as well as 3T3-L1 adipocytes (Fasshauer et al., 2003; Tilg and Moschen, 2006). In addition, the inhibition of adiponectin may be mediated in part by the P44/42 MAP kinase (Fasshauer et al., 2003), which may result in lower adiponectin levels in RA patients than in healthy individuals. Some studies have found that adiponectin also stimulates the production of inflammatory cell factors such as IL-6, IL-8, and IL-11 in RA synovium fibroblasts (Liu et al., 2015), suggesting that adiponectin exacerbates inflammation. To verify the causal association between adiponectin and RA, MR studies with more powerful instruments and larger sample sizes are needed.
In summary, the above bidirectional MR study shows that genetically predicted circulating adiponectin levels are not causally associated with RA and do not support the causal relationship between RA and circulating adiponectin. Further studies are still demanded to authenticate our results.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
CF has came up with the idea and contributed to the design of this study. HC has conducted the main analysis and drafted the manuscript. All authors have revised the manuscript and approved the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank all the researchers involved in this MR study. We also thank all the investigators for sharing these data.
Footnotes
Funding
This work was funded by grants from the National Natural Science Foundation of China (Grant No. 81703289).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2021.670282/full#supplementary-material
References
- Alkady E. A., Ahmed H. M., Tag L., Abdou M. A. (2011). [Serum and synovial adiponectin, resistin, and visfatin levels in rheumatoid arthritis patients. Relation to disease activity]. Z. Rheumatol. 70 602–608. 10.1007/s00393-011-0834-2 [DOI] [PubMed] [Google Scholar]
- Arnett F. C., Edworthy S. M., Bloch D. A., McShane D. J., Fries J. F., Cooper N. S., et al. (1988). The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 31 315–324. [DOI] [PubMed] [Google Scholar]
- Bae S. C., Lee Y. H. (2019a). Alcohol intake and risk of rheumatoid arthritis: a Mendelian randomization study. Z. Rheumatol. 78 791–796. 10.1007/s00393-018-0537-z [DOI] [PubMed] [Google Scholar]
- Bae S. C., Lee Y. H. (2019b). Alcohol intake and risk of systemic lupus erythematosus: a mendelian randomization study. Lupus 28 174–180. 10.1177/0961203318817832 [DOI] [PubMed] [Google Scholar]
- Bennett D. A., Holmes M. V. (2017). Mendelian randomisation in cardiovascular research: an introduction for clinicians. Heart 103 1400–1407. 10.1136/heartjnl-2016-310605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden J., Davey Smith G., Burgess S. (2015). Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44 512–525. 10.1093/ije/dyv080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden J., Davey Smith G., Haycock P. C., Burgess S. (2016). Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol. 40 304–314. 10.1002/gepi.21965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S., Daniel R. M., Butterworth A. S., Thompson S. G., Consortium E. P.-I. (2015). Network mendelian randomization: using genetic variants as instrumental variables to investigate mediation in causal pathways. Int. J. Epidemiol. 44 484–495. 10.1093/ije/dyu176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S., Foley C. N., Allara E., Staley J. R., Howson J. M. M. (2020). A robust and efficient method for mendelian randomization with hundreds of genetic variants. Nat. Commun. 11:376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S., Thompson S. G. (2011). Bias in causal estimates from mendelian randomization studies with weak instruments. Stat. Med. 30 1312–1323. 10.1002/sim.4197 [DOI] [PubMed] [Google Scholar]
- Burgess S., Thompson S. G. (2017). Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 32 377–389. 10.1007/s10654-017-0255-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dan Y. L., Wang P., Cheng Z., Wu Q., Wang X. R., Wang D. G., et al. (2020). Circulating adiponectin levels and systemic lupus erythematosus: a two-sample mendelian randomization study. Rheumatology (Oxford) 60 940–946. [DOI] [PubMed] [Google Scholar]
- Dastani Z., Hivert M. F., Timpson N., Perry J. R., Yuan X., Scott R. A., et al. (2012). Novel loci for adiponectin levels and their influence on type 2 diabetes and metabolic traits: a multi-ethnic meta-analysis of 45,891 individuals. PLoS Genet. 8:e1002607. 10.1371/journal.pgen.1002607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egger M., Smith G. D., Phillips A. N. (1997). Meta-analysis: principles and procedures. BMJ 315 1533–1537. 10.1136/bmj.315.7121.1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hini S. H., Mohamed F. I., Hassan A. A., Ali F., Mahmoud A., Ibraheem H. M. (2013). Visfatin and adiponectin as novel markers for evaluation of metabolic disturbance in recently diagnosed rheumatoid arthritis patients. Rheumatol. Int. 33 2283–2289. 10.1007/s00296-013-2714-3 [DOI] [PubMed] [Google Scholar]
- Fantuzzi G. (2008). Adiponectin and inflammation: consensus and controversy. J. Allergy Clin. Immunol. 121 326–330. 10.1016/j.jaci.2007.10.018 [DOI] [PubMed] [Google Scholar]
- Fasshauer M., Kralisch S., Klier M., Lossner U., Bluher M., Klein J., et al. (2003). Adiponectin gene expression and secretion is inhibited by interleukin-6 in 3T3-L1 adipocytes. Biochem. Biophys. Res. Commun. 301 1045–1050. 10.1016/s0006-291x(03)00090-1 [DOI] [PubMed] [Google Scholar]
- Hartwig F. P., Davies N. M., Hemani G., Davey Smith G. (2016). Two-sample Mendelian randomization: avoiding the downsides of a powerful, widely applicable but potentially fallible technique. Int. J. Epidemiol. 45 1717–1726. 10.1093/ije/dyx028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khajoei S., Hassaninevisi M., Kianmehr N., Seif F., Khoshmirsafa M., Shekarabi M., et al. (2019). Serum levels of adiponectin and vitamin D correlate with activity of rheumatoid arthritis. Mol. Biol. Rep. 46 2505–2512. 10.1007/s11033-019-04682-1 [DOI] [PubMed] [Google Scholar]
- Lawlor D. A., Harbord R. M., Sterne J. A., Timpson N., Davey Smith G. (2008). Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat. Med. 27 1133–1163. 10.1002/sim.3034 [DOI] [PubMed] [Google Scholar]
- Lee Y. H., Bae S. C. (2018). Circulating adiponectin and visfatin levels in rheumatoid arthritis and their correlation with disease activity: a meta-analysis. Int. J. Rheum Dis. 21 664–672. 10.1111/1756-185x.13038 [DOI] [PubMed] [Google Scholar]
- Li L., Wu L. L. (2012). Adiponectin and interleukin-6 in inflammation-associated disease. Vitam. Horm. 90 375–395. 10.1016/b978-0-12-398313-8.00014-2 [DOI] [PubMed] [Google Scholar]
- Li P., Yang L., Ma C. L., Liu B., Zhang X., Ding R., et al. (2015). Low-molecular-weight adiponectin is more closely associated with disease activity of rheumatoid arthritis than other adiponectin multimeric forms. Clin. Rheumatol. 34 1025–1030. 10.1007/s10067-015-2899-9 [DOI] [PubMed] [Google Scholar]
- Liu D., Luo S., Li Z. (2015). Multifaceted roles of adiponectin in rheumatoid arthritis. Int. Immunopharmacol. 28 1084–1090. 10.1016/j.intimp.2015.08.013 [DOI] [PubMed] [Google Scholar]
- Okada Y., Wu D., Trynka G., Raj T., Terao C., Ikari K., et al. (2014). Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature 506 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero M., Lago R., Gomez R., Lago F., Dieguez C., Gomez-Reino J. J., et al. (2006). Changes in plasma levels of fat-derived hormones adiponectin, leptin, resistin and visfatin in patients with rheumatoid arthritis. Ann. Rheum Dis. 65 1198–1201. 10.1136/ard.2005.046540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce B. L., Burgess S. (2013). Efficient design for mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am. J. Epidemiol. 178 1177–1184. 10.1093/aje/kwt084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R5 release (2021). Data Download. Available online at: https://finngen.gitbook.io (accessed July 30, 2021). [Google Scholar]
- Smith G. D., Ebrahim S. (2003). ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 32 1–22. 10.1093/ije/dyg070 [DOI] [PubMed] [Google Scholar]
- Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., et al. (2015). UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12:e1001779. 10.1371/journal.pmed.1001779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilg H., Moschen A. R. (2006). Adipocytokines: mediators linking adipose tissue, inflammation and immunity. Nat. Rev. Immunol. 6 772–783. 10.1038/nri1937 [DOI] [PubMed] [Google Scholar]
- Tsatsanis C., Zacharioudaki V., Androulidaki A., Dermitzaki E., Charalampopoulos I., Minas V., et al. (2005). Adiponectin induces TNF-alpha and IL-6 in macrophages and promotes tolerance to itself and other pro-inflammatory stimuli. Biochem. Biophys. Res. Commun. 335 1254–1263. 10.1016/j.bbrc.2005.07.197 [DOI] [PubMed] [Google Scholar]
- Verbanck M., Chen C. Y., Neale B., Do R. (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from mendelian randomization between complex traits and diseases. Nat. Genet. 50 693–698. 10.1038/s41588-018-0099-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. L., Zhang T. P., Wu J., Li B. Z., Li X. M., Pan H. F., et al. (2020). Association of adiponectin and adiponectin receptor gene polymorphisms with rheumatoid arthritis in a Chinese population. Postgrad. Med. J. 96 149–155. 10.1136/postgradmedj-2018-136372 [DOI] [PubMed] [Google Scholar]
- Zheng J., Erzurumluoglu A. M., Elsworth B. L., Kemp J. P., Howe L., Haycock P. C., et al. (2017). LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics (Oxford, England) 33 272–279. 10.1093/bioinformatics/btw613 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.