Abstract
A Burkholderia pseudomallei-like organism has recently been identified among some soil isolates of B. pseudomallei in an area with endemic melioidosis. This organism is almost identical to B. pseudomallei in terms of morphological and biochemical profiles, except that it differs in ability to assimilate l-arabinose. These Ara+ isolates are also less virulent than the Ara− isolates in animal models. In addition, clinical isolates of B. pseudomallei available to date are almost exclusively Ara−. These features suggested that these two organisms may belong to distinctive species. In this study, the 16S rRNA-encoding genes from five clinical (four Ara− and one Ara+) and nine soil isolates (five Ara− and four Ara+) of B. pseudomallei were sequenced. The nucleotide sequences and phylogenetic analysis indicated that the 16S rRNA-encoding gene of the Ara+ biotype was similar to but distinctively different from that of the Ara− soil isolates, which were identical to the classical clinical isolates of B. pseudomallei. The nucleotide sequence differences in the 16S rRNA-encoding gene appeared to be specific for the Ara+ or Ara− biotypes. The differences were, however, not sufficient for classification into a new species within the genus Burkholderia. A simple and rapid multiplex PCR procedure was developed to discriminate between Ara− and Ara+ B. pseudomallei isolates. This new method could also be incorporated into our previously reported nested PCR system for detecting B. pseudomallei in clinical specimens.
Burkholderia pseudomallei is a causative agent of melioidosis, a severe and fatal infectious disease in humans which is known to be endemic in Southeast Asia and northern Australia (8). Sporadic cases are also reported throughout the world. Melioidosis is one of the most important causes of fatality in community-acquired septicemia in northeastern Thailand (6). The highest mortality occurs in patients with the septicemic form of melioidosis, which is characterized by dissemination of the bacteria in circulation and isolation of the bacteria from blood and from various organs. The clinical course of septicemic melioidosis often includes rapid deterioration and death that occurs within a few days after hospitalization. Rapid diagnosis and prompt treatment with appropriate antibiotics can significantly reduce the mortality (27).
B. pseudomallei is found in soil and water, especially in rice paddy fields (21). Most of the melioidosis patients in the areas of endemicity are rice farmers or others with direct contact with soil (14). Infection in humans is acquired by soil contamination through skin abrasions or wounds or by ingestion and inhalation. Recently, detailed comparative analysis of soil isolates and clinical isolates of B. pseudomallei revealed that there are two biotypes of the organism, which are almost identical in terms of phenotypes and biochemical profiles. The main distinctive characteristic of the two groups is the difference in their ability to assimilate l-arabinose (20). Almost all clinical isolates of B. pseudomallei are unable to assimilate arabinose as a single substrate (Ara−), while the soil isolates included both Ara+ and Ara− biotypes. Both biotypes were found in soil in the area of endemic melioidosis. In addition, evidence from our group and other groups demonstrated that the Ara+ isolates are much less virulent than the Ara− classical B. pseudomallei isolates. In animals experimentally infected with these organisms, the 50% lethal dose (LD50) for the Ara+ biotype was much higher than that for the Ara− biotype (3, 20). Their ribotyping patterns were also found to be distinctively different (24). These accumulated data suggest that the two organisms may belong to closely related but distinctive species. They also lead to a hypothesis that severe clinical melioidosis occurs following exposure to the virulent Ara− B. pseudomallei isolates but not to the nonvirulent Ara+ B. pseudomallei isolates. They may also provide an insight into the pathogenesis of clinical melioidosis and development of vaccines. In addition, a simple and rapid system to differentiate the two biotypes should be useful for epidemiological study and for confirmation of true infection or contamination of the clinical samples.
The present study describes the comparative analysis of nucleotide sequences of the 16S rRNA-encoding genes of the clinical and soil isolates of B. pseudomallei. Phylogenetic analysis was performed to demonstrate the relationship between the two biotypes, in relation to other Burkholderia species and other related bacteria. The signature biotype-specific nucleotides were identified. A simple and rapid multiplex PCR procedure for identification of and discrimination between the two biotypes of B. pseudomallei was also established. This system has been used for bacterial identification and also for detection of the virulent B. pseudomallei DNA in clinical specimens collected from acute melioidosis patients.
MATERIALS AND METHODS
Bacterial isolates.
Five clinical isolates and nine soil isolates of B. pseudomallei were used in this study. Four clinical isolates (S94004, S94017, K94050, and K96243) and five soil isolates (TRF684, TRF685, TRF688, TRF824, and TRF830) were of Ara− biotype. The other clinical isolate (S95019) was one of the extremely rare isolates collected from a human which can assimilate arabinose (Ara+). This isolate was obtained from the sputum of a melioidosis patient in Bangkok for whom the diagnosis of melioidosis was confirmed based on clinical characteristics and the response to treatment. The possibility of bacterial contamination was excluded. The other four soil isolates were arabinose assimilators (TRF681, TRF682, TRF683, and TRF686). The clinical isolates were obtained from the bacterial collection bank of T. Dharakul and S. Sirisinha, and the soil isolates were obtained from the recent survey of B. pseudomallei in the environment sponsored by the Thailand Research Fund. All isolates were identified as B. pseudomallei based on biochemical characteristics and antimicrobial susceptibility patterns. The analysis of arabinose assimilation by these isolates was performed as described (28). In addition, bacterial strains of Burkholderia cepacia, Pseudomonas putida, Pseudomonas aeruginosa, Haemophilus influenzae, Escherichia coli (ATCC 25922), Acinetobacter anitratus, Klebsiella pneumoniae, Enterobacter aerogenes, Proteus sp., Serratia marcescens, Aeromonas hydrophila, Staphylococcus aureus, group A and group B streptococci, and group D enterococci were provided by the Department of Microbiology, Faculty of Medicine, Siriraj Hospital, Bangkok, Thailand. The Pseudomonas fluorescens type strain DMS2589 and the Stenotrophomonas maltophilia type strain DMS904 were provided by the Department of Medical Sciences, Ministry of Public Health, Thailand.
Preparation of bacterial DNA for PCR.
Bacterial DNA was prepared by using a modified proteinase K digestion technique as previously described (11). Briefly, bacterial DNA (from 104 viable bacteria) was extracted in a total volume of 200 μl containing 10 mM Tris-HCl (pH 7.8), 5 mM EDTA, 0.5% sodium dodecyl sulfate, 0.5% Tween-20, and 0.2 mg of proteinase K/ml. The tube was incubated at 56°C for 2 h, and proteinase K was inactivated by heat denaturation at 95°C for 10 min. Twenty microliters of the reaction mixture was then taken for PCR amplification.
Nucleotide sequencing and phylogenetic analysis of the 16S rRNA gene.
The 16S rRNA-encoding gene was amplified from the bacterial DNA by using two sets of overlapping oligonucleotide primers. Primers Bps16S-U33 and Bps16S-OL731 were used for amplifying the 5′ end of the 16S rRNA, and primers Bps16S-683L and Bps16S-1460R were used for amplifying the 3′ end (Table 1). Fragments of 717 and 795 bp in size were purified and subjected to cycle sequencing in both directions. Nucleotide sequencing of the PCR-amplified products was performed in an automated nucleotide sequencer (model 310; Applied Biosystems, Foster City, Calif.), using a BigDye terminator sequencing system (Perkin-Elmer, Foster City, Calif.). Nucleotide sequences obtained were analyzed by using MacVector software (Oxford Biomolecular Group, Oxford, United Kingdom) and were compared to the sequences deposited in the GenBank database through the National Center for Biotechnology Information computer server (18a).
TABLE 1.
Nucleotide sequences of PCR primers for amplifying the 16S rRNA genes of Ara+ and Ara− B. pseudomallei isolates
| Primer | Positiona | Strand | Sequence (5′→3′) |
|---|---|---|---|
| Bps16S-U33b | 34–52 | Sense | AAGTCGAACGGCAGCACGG |
| Bps16S-OL731b | 751–733 | Antisense | TTTGCTCCCCACGCTTTCG |
| Bps16S-683L | 683–700 | Sense | GAATACCGATGGCGAAGG |
| Bps16S-1460R | 1477–1460 | Antisense | TACGGCTACCTTGTTACG |
| Bps16S-42L | 42–59 | Sense | CGGCAGCRCGGGCTTCGG |
| Bps16S-266R | 284–266 | Antisense | TGTGGCTGGTCGTCCTCTC |
| Bps16S-427R | 446–427 | Antisense | CACTCCGGGTATTAGCCAGA |
Phylogenetic analysis was performed by using the DNAdist, Neighbor, and Drawtree software in PHYLIP (phylogenetic inference package), version 3.572 (J. Felsenstein, University of Washington, Seattle, Wash.). The nucleotide sequences of the 16S rRNA-encoding genes of 44 other bacteria including 16 members of Burkholderia species, for which the complete or almost complete 16S rRNA gene sequences were available, were used for nucleotide sequence comparison and phylogenetic analysis. The species (and their GenBank accession numbers) were as follows: B. pseudomallei 1026b (U91839), Burkholderia thailandensis E264 (U91838), Burkholderia plantarii (U96933), Burkholderia glumae (U96931), Burkholderia vietnamensis (U96928), B. cepacia (U96927), Burkholderia cocovenenans (U96934), Burkholderia pyrrocinia (U96930), Burkholderia glathei (U96935), Burkholderia gladioli (X67038), Burkholderia graminis (U96939), Burkholderia phenazinium (U96936), Burkholderia vandii (U96932), Burkholderia caryophylli (X67039), Burkholderia andropogonis (X67037), Burkholderia caribiensis (Y17009), Herbaspirillum seropedicae (Y10146), Ralstonia eutropha (AF027407), Bordetella bronchiseptica (X57026), Ideonella dechloratans (X72724), Bordetella holmesii CDC F5101 (U04820), Zoogloea ramigera (X74914), Oxalobacter formigenes (U49754), Janthinobacterium lividum (Y08846), Pseudomonas syzygii R001 (U28237), denitrifying Fe-oxidizing bacteria (U51102), Duganella zoogloeoides (D14256), Delftia acidovorans (AF078774), the ultramicrobacterium ND5 (AB008506), Rubrivivax gelatinosus (D16214), Bordetella parapertussis ATCC 15311 (U04949), Leptothrix discophora SS-1 (L33975), Bordetella pertussis ATCC 9797 (U04950), Brachymonas denitrificans (D14320), Bordetella avium ATCC 35086 (U04947), Alcaligenes xylosoxidans (D88005), Azoarcus indigens (AF011345), Aquaspirillum sinuosum (AF078754), Alcaligenes faecalis (M22508), S. maltophilia (X95923), P. aeruginosa LMG1242T (Z76651), P. putida (Z76667), Salmonella typhi STRNA16 (Z47544), and E. coli EHEC ATCC 43895 (Z83205). The nucleotide sequences were aligned by using MacVector software, and the evolutionary distances were calculated by using the program DNAdist based on the maximum-likelihood algorithm. The phylogenetic trees were constructed by the Drawtree program from the distance matrix by using the neighbor-joining method.
Primer design.
A new set of multiplex PCR amplification primers was designed from the variable region of the 16S rRNA gene of the Ara+ and Ara− B. pseudomallei isolates obtained from this study. Primer Bps16S-42L is homologous to the sequences of both biotypes of B. pseudomallei but not to those of other organisms (Fig. 1). Primer Bps16S-427R was complementary to the sequence of only Ara− B. pseudomallei isolates and not to that of Ara+ B. pseudomallei isolates or other bacteria. Primer Bps16S-266R could bind to both organisms as well as to a few other organisms. PCR amplification of Ara− B. pseudomallei isolate DNA yields two amplified fragments, of 405 and 243 bp in length, but only the 243-bp fragment is obtained from the amplification of Ara+ B. pseudomallei isolate DNA. The primers do not amplify the 16S rRNA gene of other organisms.
FIG. 1.
Specificity of the PCR primers. Comparison of nucleotide sequences of the 16S rRNA genes corresponding to primers Bps16S-42L (A) and Bps16S-427R (B). The nucleotide sequence of Ara− B. pseudomallei isolates was compared with those of Ara+ B. pseudomallei isolates, other Burkholderia spp., H. seropedicae, R. eutropha, P. aeruginosa, P. putida, S. typhi, and E. coli. The sequences at the primer sites are underlined.
These multiplex oligonucleotide primers were also designed to be located internally to the outer primer set for amplifying the 16S rRNA gene and therefore were compatible with our previously described nested PCR procedure for detecting B. pseudomallei isolate DNA in clinical specimens (11). The new multiplex primers could be used as the nested primers by replacing the primers BS3L and BS4R with this new primer set.
Extraction of bacterial DNA from buffy-coat samples.
The bacterial DNA was extracted from buffy-coat samples collected from seven patients with suspected melioidosis, using the heat treatment and proteinase K digestion protocol as previously described (11). The diagnosis of melioidosis was confirmed by isolation of B. pseudomallei from five patients. For the other two patients culture was negative for B. pseudomallei, and these patients therefore served as negative controls.
PCR amplification.
PCR amplification was carried out in a total volume of 50 μl containing 10 μl of DNA extract, 5 μl of 10× amplification buffer (containing 500 mM KCl, 100 mM Tris-HCl [pH 8.3], 15 mM MgCl2, and 0.01% [wt/vol] gelatin), 4 μl of deoxynucleoside triphosphates (2.5 mM each), 20 pmol of each primer, and 2.5 U of Taq DNA polymerase (Perkin-Elmer). The tube was subjected to thermal cycling for 35 cycles, each comprised of 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min, and then subjected to a final extension step for 10 min at 72°C, in a GeneAmp DNA Thermal Cycler 2400 (Perkin-Elmer). Tris-EDTA buffer was used as the negative control to exclude amplicon contamination. The presence of amplified DNA product was analyzed on a 2% agarose gel, stained with ethidium bromide and visualized under a UV light. Strict laboratory precautions were carefully exercised to avoid contamination (13). For nested PCR amplification of DNA obtained from clinical specimens, the nested PCR procedure described previously (11) was performed. The three multiplex primers were used instead of the primers BS3L and BS4R.
Nucleotide sequence accession numbers.
The nucleotide sequences of 16S rRNA genes of the nine Ara− B. pseudomallei isolates and five Ara+ B. pseudomallei isolates obtained from this study have been deposited in GenBank under accession no. AF093047 to AF093060.
RESULTS
Nucleotide sequence analysis.
Nucleotide sequences obtained from PCR-amplified products of 14 B. pseudomallei isolates were compared (Fig. 2). The sequences of four Ara− clinical isolates and five Ara− soil isolates were almost identical, indicating that they were isolates of the same species, the classical B. pseudomallei. The only nucleotide difference among the classical isolates was located at position 157, where six B. pseudomallei isolates (three clinical and three soil isolates) had A but the other three (one clinical isolate and two soil isolates) had G. All of the Ara+ isolates in this study had G at position 157, and the published sequences of B. pseudomallei 1026b (Ara−) and B. thailandensis E264 (Ara+) also included G at position 157. In addition, the nucleotide sequences of the nine classical Ara− B. pseudomallei isolates obtained from this study included C at position 1292, which is different from the nucleotide (T) found at this position for B. pseudomallei 1026b.
FIG. 2.
Alignment of nucleotide sequences of 16S rRNA genes from four clinical isolates (K96243, K94050, S94004, and S94017) and five soil isolates (TRF684, TRF685, TRF688, TRF824, and TRF830) of Ara− B. pseudomallei, and one clinical isolate (S95019) and four soil isolates (TRF681, TRF682, TRF683, and TRF686) of Ara+ B. pseudomallei (GenBank accession no. AF093047 to AF093060). The published sequences of B. pseudomallei 1026b (U91839) and B. thailandensis E264 (U91838) are also included for comparison.
The 16S rRNA gene sequences were virtually identical among all five Ara+ B. pseudomallei isolates at all positions sequenced. These sequences were also identical to the published sequence of B. thailandensis E264 except at two positions (G replaced A at positions 1020 and 1141).
Among a total of 1,488 nucleotides analyzed, differences among isolates of B. pseudomallei were observed at 15 positions, 11 of which appeared to be biotype specific, as they were present in all Ara− biotype B. pseudomallei isolates but not in those with Ara+ biotype, and vice versa. The biotype-specific nucleotides identified are listed in Table 2. These sequences were different from those of the 16S rRNA genes of other Burkholderia spp., including B. vietnamensis, which was initially isolated from the rhizosphere of a rice plant from a region of Vietnam in close proximity to northeast Thailand (17).
TABLE 2.
Nucleotide sequence differences between Ara+ and Ara− isolates of B. pseudomallei
| Position | Nucleotide | No. of isolates with the nucleotide
|
|||
|---|---|---|---|---|---|
| Ara−
|
Ara+
|
||||
| Clinical (n = 4) | Soil (n = 5) | Clinical (n = 1) | Soil (n = 4) | ||
| 65 | T | 4 | 5 | 0 | 0 |
| C | 0 | 0 | 1 | 4 | |
| 157a | A | 3 | 3 | 0 | 0 |
| G | 1 | 2 | 1 | 4 | |
| 158 | A | 4 | 5 | 0 | 0 |
| T | 0 | 0 | 1 | 4 | |
| 427 | T | 4 | 5 | 0 | 0 |
| C | 0 | 0 | 1 | 4 | |
| 438 | C | 4 | 5 | 0 | 0 |
| A | 0 | 0 | 1 | 4 | |
| 443 | A | 4 | 5 | 0 | 0 |
| G | 0 | 0 | 1 | 4 | |
| 974 | G | 4 | 5 | 0 | 0 |
| T | 0 | 0 | 1 | 4 | |
| 977 | C | 4 | 5 | 0 | 0 |
| T | 0 | 0 | 1 | 4 | |
| 979 | A | 4 | 5 | 0 | 0 |
| C | 0 | 0 | 1 | 4 | |
| 986 | T | 4 | 5 | 0 | 0 |
| G | 0 | 0 | 1 | 4 | |
| 987 | T | 4 | 5 | 0 | 0 |
| C | 0 | 0 | 1 | 4 | |
| 991 | C | 4 | 5 | 0 | 0 |
| A | 0 | 0 | 1 | 4 | |
| 1020a | G | 4 | 5 | 1 | 4 |
| 1141a | G | 4 | 5 | 1 | 4 |
| 1292a | C | 4 | 5 | 1 | 4 |
Phylogenetic analysis.
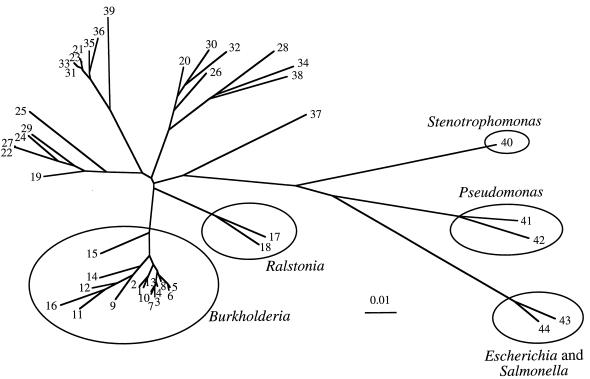
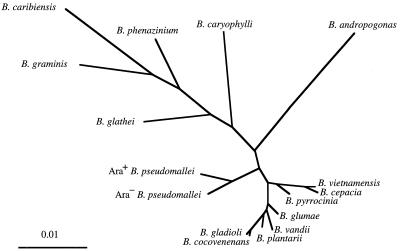
Phylogenetic analysis demonstrated that the two biotypes of B. pseudomallei were related but were clearly distinguishable (Fig. 3). The two biotypes were more closely related to each other than to other members of the genus Burkholderia (Fig. 4). It should be noted that B. vietnamensis was more closely related to B. cepacia than to B. pseudomallei.
FIG. 3.
Phylogenetic tree of the 16S rRNA genes shows the relationship of the Ara− and Ara+ B. pseudomallei isolates with other bacteria. The sequences used in this analysis are from the bacteria (1) Ara− B. pseudomallei, (2) Ara+ B. pseudomallei, (3) B. plantarii, (4) B. glumae, (5) B. vietnamensis, (6) B. cepacia, (7) B. cocovenenans, (8) B. pyrrocinia, (9) B. glathei, (10) B. gladioli, (11) B. graminis, (12) B. phenazinium, (13) B. vandii, (14) B. caryophylli, (15) B. andropogonis, (16) B. caribiensis, (17) R. eutropha, (18) H. seropedicae, (19) B. bronchiseptica, (20) I. dechloratans, (21) B. holmesii, (22) Z. ramigera, (23) O. formigenes, (24) J. lividum, (25) P. syzygii, (26) denitrifying Fe-oxidizing bacteria, (27) D. zoogloeoides, (28) D. acidovorans, (29) ultramicrobacterium, (30) R. gelatinosus, (31) B. parapertussis, (32) L. discophora, (33) B. pertussis, (34) B. denitrificans, (35) B. avium, (36) A. xylosoxidans, (37) A. indigens, (38) A. sinuosum, (39) A. faecalis, (40) S. maltophilia, (41) P. aerguinosa, (42) P. putida, (43) S. typhi, and (44) E. coli. The scale bar indicates a distance in substitutions per nucleotide.
FIG. 4.
Phylogenetic analysis of the 16S rRNA genes of 16 members in the genus Burkholderia for which the complete or almost-complete 16S rRNA sequences were available. The scale bar indicates a distance in substitutions per nucleotide.
Discrimination of Ara− and Ara+ B. pseudomallei isolates by multiplex PCR.
A new set of multiplex PCR primers was designed to differentiate the two biotypes of B. pseudomallei from other organisms. The forward primers Bps16S-42L bound only to the sequences of Ara− and Ara+ B. pseudomallei isolates. The reverse primer Bps16S-427L was complementary to the region covering nucleotides T 427 C (T substituted for C at position 427), C 438 A, and A 443 G, which were used as the targets for discrimination between Ara− and Ara+ B. pseudomallei. In the presence of Ara− B. pseudomallei isolate DNA, PCR-amplified products consisting of two bands of 405 and 243 bp could be visualized in an ethidium bromide-stained agarose gel (Fig. 5). However, only the 243-bp fragment was present when the Ara+ B. pseudomallei isolate DNA was amplified. These primers could amplify all clinical and soil isolates of B. pseudomallei used in this study. The DNA from other organisms, including B. cepacia, P. putida, P. aeruginosa, P. fluorescens, S. maltophilia, E. coli, A. anitratus, K. pneumoniae, E. aerogenes, Proteus sp., A. hydrophila, S. marcescens, H. influenzae, S. aureus, group A and group B streptococci, and group D enterococci, could not be amplified by these multiplex primers (data not shown).
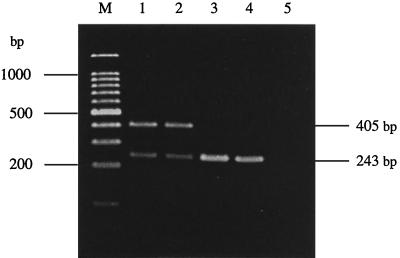
FIG. 5.
Ethidium bromide-stained agarose gel electrophoresis of the amplified 16S rRNA genes of Ara− and Ara+ B. pseudomallei isolates performed by using the multiplex primer set. M, 100-bp marker; lane 1, Ara− B. pseudomallei isolate K94050; lane 2, Ara− B. pseudomallei isolate K96243; lane 3, Ara+ B. pseudomallei isolate S95019; lane 4, Ara+ B. pseudomallei isolate TRF681; lane 5, negative control.
Detection of B. pseudomallei DNA in buffy-coat specimens.
The new set of multiplex primers was used as the inner primers in the nested PCR system, by replacing our previously described BS3L and BS4R primers (11). Buffy-coat specimens from seven clinically suspected cases of melioidosis were tested, five of which were confirmed to be melioidosis by bacterial cultures. All of the specimens from the five culture-proven melioidosis cases were PCR positive (resulting in two amplified bands), but the specimens from the other two patients without melioidosis were PCR negative.
DISCUSSION
The genus Burkholderia was proposed in 1992, and since then a number of former Pseudomonas species have been transferred to this genus (30). Several members of this genus are present in the environment and can be isolated from soil, water, and plants. In addition, two members of this genus are human pathogens, causing both acute diseases and opportunistic infection, such as in patients with cystic fibrosis. These pathogenic species include B. pseudomallei and B. cepacia. B. pseudomallei is the only member of this genus that causes severe disseminated disease, i.e., melioidosis. The emergence of a B. pseudomallei-like organism which was nonvirulent or less virulent than the classical B. pseudomallei (9, 16, 18) received much attention. A nonvirulent isolate was explored as a potential vaccine candidate for melioidosis (10).
The major biochemical difference between the virulent (classical) and nonvirulent isolates is the ability to assimilate l-arabinose. About 25 to 50% of the soil isolates of B. pseudomallei are Ara+ (20), but these Ara+ isolates are extremely rare in clinical specimens. Only 1 of the 400 clinical isolates in our collection and none of the 1,200 clinical isolates studied by Smith et al. (20) was an arabinose assimilator. This Ara+ clinical isolate was isolated from the sputum of a elderly diabetic Thai patient who had pneumonitis. The patient did not respond to the usual antibiotics and, after sputum cultured positive for B. pseudomallei, he was treated with ceftazidime (an antibiotic used for treatment of melioidosis). The patient responded to the treatment and fully recovered. B. pseudomallei was the only organism isolated from this patient. When subsequent analysis showed that this isolate was Ara+, in independent reviews of the case two infectious-disease specialists concluded that the pneumonitis in this patient was caused by this Ara+ B. pseudomallei. The isolation of Ara+ B. pseudomallei from a clinical case demonstrated that it can, although rarely, cause clinical illness, particularly in an immunocompromised host. This isolate demonstrated an LD50 of >109 CFU in mice. The nonvirulent or less-virulent status of the Ara+ isolates has been demonstrated in several animal models of experimental B. pseudomallei infection (3, 20). The arabinose assimilation characteristic of these two biotypes appeared to be stable, as demonstrated by the failure to convert the Ara phenotypes between the Ara− and Ara+ isolates by selective culture (20). The distinctive characteristics of these organisms led to the question of whether they were different variants or biotypes of the same species or belonged to a closely related but distinctive species. The 16S rRNA gene sequences are highly conserved within the same species and have been widely used for species confirmation (15, 22). Therefore, the nucleotide sequences of the 16S rRNA gene were compared and phylogenetic analysis was performed in the present study.
This study clearly showed that the 16S rRNA gene sequences of the two biotypes were distinctively different, but the difference was less than the differences among other species of the genus Burkholderia (Fig. 4). Eleven biotype-specific nucleotide positions were identified among the total of 1,488 nucleotides (99.26% homology), and these were totally conserved within the biotype. Differences in their phenotypes (biochemical assimilation of l-arabinose, adonitol, 5-ketogluconate, and d-xylose and inability to assimilate dulcitol, erythritol, and trehalose by the nonvirulent isolates) (28), colony morphology (3), virulence as demonstrated by the clinical severity in humans and experimental animals such as mice and hamsters (3, 20), genomic sequences such as their ribotyping patterns (24), and nucleotide sequences in the 16S rRNA gene have been demonstrated, and a distinctive species named B. thailandensis has been proposed for the Ara+ biotype (4). The data described herein provide evidence that, based on the current polyphasic taxonomy system encompassing phenotypic, genotypic, and phylogenetic information (7, 26), the degree of nucleotide differences in the 16S ribosomal RNA genes of the Ara− and Ara+ B. pseudomallei isolates may not be sufficient to warrant classification as distinctive species.
The nucleotide differences between the 16S rRNA genes of the Ara− and Ara+ B. pseudomallei isolates and those of other species clustered in three regions. These three hypervariable regions were also observed in other bacterial species and were therefore used for species identification (23, 29, 30). A new set of multiplex PCR primers was designed for differentiating the two biotypes from each other and from other bacteria. The primers were based on the sequences of the first two diverse regions, as used in our previously described PCR system (11). This rationale allowed the generation of PCR-amplified products of appropriate sizes (243 and 405 bp) which can be clearly detected. Detection of the rRNA gene by PCR has several advantages in terms of both sensitivity and specificity because of the multiple copies of targeted DNA sequences and the conservation of the sequences in the same species (1). The described system allowed one-step discrimination between the two biotypes. In addition, the multiplex primers could be used as the inner primers for nested PCR amplification of sequences from B. pseudomallei in clinical specimens. This PCR amplification system has been proven useful in clinical situations, in which the PCR results were obtained before bacterial cultures became positive. The PCR system appears to be almost as sensitive as standard bacterial culture for the detection of B. pseudomallei (5, 11). PCR amplification can be performed directly from the clinical samples, which may contain small numbers of the bacteria. This technique has proven valuable in identifying bacterial pathogens that are difficult to detect and identify by traditional microbiological methods (1). It should be noted that the 16S rRNA genes of B. pseudomallei and B. mallei are identical (2, 11, 25, 30), and therefore the two species cannot be differentiated using this system. As previously mentioned, this is not considered a problem in clinical situations since glanders, the disease caused by B. mallei, is a disease of the horse, mule, and donkey and is epidemiologically different from melioidosis (18). Only sporadic cases of glanders occur in animals in Asia, Africa, and South America (19). An extensive survey of soil and water samples collected in Thailand, an area with endemic melioidosis, failed to isolate B. mallei (unpublished data).
Our previously described sets of PCR primers for identifying B. pseudomallei have been used in the confirmation and in the diagnosis of melioidosis (11). Primers in the 16S rRNA gene region had sensitivity approaching 100% for clinical samples, and this level is higher than that for the primers for the 23S rRNA gene and for the intergenic region between the 16S and 23S rRNA genes (12). In addition, these primers were also useful for the isolation and identification of B. pseudomallei in soil (reference 5 and our unpublished data). PCR of soil samples has been proven to be robust and more sensitive than bacterial culture and is also a useful confirmatory test in determining the identity of the soil isolates where biochemical tests give inconsistent results (5). Since both Ara− and Ara+ B. pseudomallei organisms are present in soil samples from the same geographical region, a PCR system that can identify the species would be valuable in environmental surveys of B. pseudomallei.
ACKNOWLEDGMENTS
This work is a part of the Siriraj Burkholderia pseudomallei genome project, supported by the Faculty of Medicine, Siriraj Hospital, Mahidol University, Bangkok, Thailand. T. Dharakul and S. Songsivilai are supported by career development grants from the National Science and Technology Development Agency of Thailand and the Anandhamahidol Foundation.
We thank V. Thamlikitkul and the Thailand Research Fund for providing the soil isolates and D. Kanistanon for technical support of phylogenetic analysis.
REFERENCES
- 1.Anderson B. Broad range polymerase chain reaction for detection and identification of bacteria. J Fla Med Assoc. 1994;81:835–837. [PubMed] [Google Scholar]
- 2.Bauernfeind A, Roller C, Meyer D, Jungwirth R, Schneider I. Molecular procedure for rapid detection of Burkholderia mallei and Burkholderia pseudomallei. J Clin Microbiol. 1998;36:2737–2741. doi: 10.1128/jcm.36.9.2737-2741.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brett P J, DeShazer D, Woods D E. Characterization of Burkholderia pseudomallei and Burkholderia pseudomallei-like strains. Epidemiol Infect. 1997;118:137–148. doi: 10.1017/s095026889600739x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brett P J, DeShazer D, Woods D E. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int J Syst Bacteriol. 1998;48:317–320. doi: 10.1099/00207713-48-1-317. [DOI] [PubMed] [Google Scholar]
- 5.Brook M D, Currie B, Desmarchelier P M. Isolation and identification of Burkholderia pseudomallei from soil using selective culture techniques and the polymerase chain reaction. J Appl Microbiol. 1997;82:589–596. [PubMed] [Google Scholar]
- 6.Chaowagul W, White N J, Dance D A B, Wattanagoon Y, Naigowit P, Davis T M E, Looareesuwan S, Pitakwatchara N. Melioidosis: a major cause of community-acquired septicemia in northeastern Thailand. J Infect Dis. 1989;159:890–899. doi: 10.1093/infdis/159.5.890. [DOI] [PubMed] [Google Scholar]
- 7.Colwell R R. Polyphasic taxonomy of the genus Vibrio: numerical taxonomy of Vibrio cholerae, Vibrio parahaemolysis, and related Vibrio species. J Bacteriol. 1970;104:410–433. doi: 10.1128/jb.104.1.410-433.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dance D A B. Melioidosis: the tips of the iceberg? Clin Microbiol Rev. 1991;4:52–60. doi: 10.1128/cmr.4.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dannenberg A M, Jr, Scott E. Melioidosis: pathogenesis and immunity in mice and hamsters. II. Studies with avirulent strains of Malleomyces pseudomallei. Am J Pathol. 1958;34:1099–1121. [PMC free article] [PubMed] [Google Scholar]
- 10.Dannenberg A M, Jr, Scott E. Melioidosis: pathogenesis and immunity in mice and hamsters. III. Effects of vaccination with avirulent strains of Pseudomonas pseudomallei on the resistance to the establishment and the resistance to the progress of respiratory melioidosis caused by virulent strains: all-or-none aspects of this disease. J Immunol. 1959;84:233–246. [PubMed] [Google Scholar]
- 11.Dharakul T, Songsivilai S, Viriyachitra S, Luangwedchakarn V, Tassaneetritap B, Chaowagul W. Detection of Burkholderia pseudomallei DNA in patients with septicemic melioidosis. J Clin Microbiol. 1996;34:609–614. doi: 10.1128/jcm.34.3.609-614.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haase A, Brennan M, Barrett S, Wood Y, Huffam S, O’Brien D, Currie B. Evaluation of PCR for diagnosis of melioidosis. J Clin Microbiol. 1998;36:1039–1041. doi: 10.1128/jcm.36.4.1039-1041.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitchin P A, Szotyori Z, Fromholc C, Almond N. Avoidances of false positives. Nature (London) 1990;344:201. doi: 10.1038/344201a0. [DOI] [PubMed] [Google Scholar]
- 14.Leelarasamee A, Bovornkitti S. Melioidosis: review and update. Rev Infect Dis. 1989;11:413–425. doi: 10.1093/clinids/11.3.413. [DOI] [PubMed] [Google Scholar]
- 15.Ludwig W, Schleifer K-H. Bacterial phylogeny based on 16S and 23S rRNA sequence analysis. FEMS Microbiol Rev. 1994;15:155–173. doi: 10.1111/j.1574-6976.1994.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 16.McCormick J B, Weaver R E, Hayes P S, Boyce J M, Feldman R A. Wound infection by an indigenous Pseudomonas pseudomallei-like organism isolated from the soil: case report and epidemiologic study. J Infect Dis. 1977;135:103–107. doi: 10.1093/infdis/135.1.103. [DOI] [PubMed] [Google Scholar]
- 17.Meyer J M, Van V T, Stintzi A, Berge O, Winkelmann G. Ornibactin production and transport properties in strains of Burkholderia vietnamensis and Burkholderia cepacia (formerly Pseudomonas cepacia) Biometals. 1995;8:309–317. doi: 10.1007/BF00141604. [DOI] [PubMed] [Google Scholar]
- 18.Miller W R, Pannell L, Cravitz L, Tanner W A, Rosebury T. Studies on certain biological characteristics of Malleomyces mallei and Malleomyces pseudomallei. II. Virulence and infectivity for animals. J Bacteriol. 1948;55:127–135. [PMC free article] [PubMed] [Google Scholar]
- 18a.National Center for Biotechnology Information. 1 April 1999, revision date. Sequences. [Online.] National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, Md. www.ncbi.nlm.nih.gov. [6 April 1999, last date accessed.]
- 19.Sanford J P. Pseudomonas species (including melioidosis and glanders) In: Mandell G L, Douglas R G, Bennett J E, editors. Principles and practice of infectious diseases. 2nd ed. New York, N.Y: John Wiley & Sons, Inc.; 1985. pp. 1250–1254. [Google Scholar]
- 20.Smith M D, Angus B J, Wuthiekanun V, White N J. Arabinose assimilation defines a nonvirulent biotype of Burkholderia pseudomallei. Infect Immun. 1997;65:4319–4321. doi: 10.1128/iai.65.10.4319-4321.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith M D, Wuthiekanun V, Walsh A L, White N J. Quantitative recovery of Burkholderia pseudomallei from soil in Thailand. Trans R Soc Trop Med Hyg. 1995;89:488–490. doi: 10.1016/0035-9203(95)90078-0. [DOI] [PubMed] [Google Scholar]
- 22.Stackebrandt E, Goebel B M. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Microbiol. 1994;44:846–849. [Google Scholar]
- 23.Taghavi M, Hayward C, Sly L I, Fegan M. Analysis of the phylogenetic relationships of strains of Burkholderia solanacearum, Pseudomonas syzygii, and the blood disease bacterium of banana based on 16S rRNA gene sequences. Int J Syst Bacteriol. 1996;46:10–15. doi: 10.1099/00207713-46-1-10. [DOI] [PubMed] [Google Scholar]
- 24.Trakulsomboon S, Dance D A, Smith M D, White N J, Pitt T L. Ribotype differences between clinical and environmental isolates of Burkholderia pseudomallei. J Med Microbiol. 1997;46:565–570. doi: 10.1099/00222615-46-7-565. [DOI] [PubMed] [Google Scholar]
- 25.Tyler S D, Strathdee C A, Rozee K R, Johnson W M. Oligonucleotide primers designed to differentiate pathogenic pseudomonads on the basis of the sequencing of genes coding for 16S-23S rRNA internal transcribed spacers. Clin Diagn Lab Immunol. 1995;2:448–453. doi: 10.1128/cdli.2.4.448-453.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vandamme P, Pot B, Gillis M, De Vos P, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White N J, Dance D A B, Chaowagul W, Wattanagoon Y, Wuthiekanun V, Pitakwatchara N. Halving of mortality of severe melioidosis by ceftazidime. Lancet. 1989;ii:697–700. doi: 10.1016/s0140-6736(89)90768-x. [DOI] [PubMed] [Google Scholar]
- 28.Wuthiekanun V, Smith M D, Dance D A B, Walsh A L, Pitt T L, White N J. Biochemical characteristics of clinical and environmental isolates of Burkholderia pseudomallei. J Med Microbiol. 1996;45:408–412. doi: 10.1099/00222615-45-6-408. [DOI] [PubMed] [Google Scholar]
- 29.Xiang L, Dorsch M, Dot T D, Sly L I, Stackebrandt E, Hayward A C. Phylogenetic studies of the rRNA II pseudomonads based on 16S rRNA gene sequences. J Appl Bacteriol. 1993;74:324–329. [Google Scholar]
- 30.Yabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hashimoto Y, Ezaki T, Arakawa M. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol Immunol. 1992;36:1251–1275. doi: 10.1111/j.1348-0421.1992.tb02129.x. [DOI] [PubMed] [Google Scholar]