Abstract
Close to half of the 878 methicillin-resistant Staphylococcus aureus (MRSA) strains recovered between 1992 and 1997 from the pediatric hospital in Lisbon were bacteria in which antibiotic resistance was limited to β-lactam antibiotics. The other half were multidrug resistant. The coexistence of MRSA with such unequal antibiotic resistance profiles prompted us to use molecular typing techniques for the characterization of the MRSA strains. Fifty-three strains chosen randomly were typed by a combination of genotypic methods. Over 90% of the MRSA strains belonged to two clones: the most frequent one, designated the “pediatric clone,” was reminiscent of historically “early” MRSA: most isolates of this clone were only resistant to β-lactam antimicrobials and remained susceptible to macrolides, quinolones, clindamycin, spectinomycin, and tetracycline. They showed heterogeneous and low-level resistance to methicillin (MIC, 1.5 to 6 μg/ml), carried the ClaI-mecA polymorph II, were free of the transposon Tn554, and showed macrorestriction pattern D (clonal type II::NH::D). The second major clone was the internationally spread and multiresistant “Iberian” MRSA with homogeneous and high-level resistance to methicillin (MIC, >200 μg/ml) and clonal type I::E::A. Surprisingly, the multidrug-resistant and highly epidemic Iberian MRSA did not replace the much less resistant pediatric clone during the 6 years of surveillance. The pediatric clone was also identified among contemporary MRSA isolates from Poland, Argentina, The United States, and Colombia, and the overwhelming majority of these were also associated with pediatric settings. We propose that the pediatric MRSA strain represents a formerly widely spread archaic clone which survived in some epidemiological settings with relatively limited antimicrobial pressure.
The prevalence of antimicrobial agent-resistant bacteria has been increasing rapidly in Portuguese hospitals during the last few years. According to a multicenter study of 10 Portuguese hospitals performed in 1996 to 1997, the prevalence of methicillin-resistant Staphylococcus aureus (MRSA) was estimated as 48%, one of the highest in Europe (26, 46). This is of great concern, because it is a common experience that once MRSA is introduced in a hospital, it is difficult to eradicate it (4, 15). In order to establish adequate infection control programs in hospitals, it is of major importance to find the sources of MRSA isolates and their origin and transmission routes, to identify their clonal identities, and to trace their geographic spread (5).
In an effort to keep track of MRSA clones, the Center for Molecular Epidemiology (Centro de Epidemiologia Molecular [CEM]) has organized a collaborative network (CEM/NET) with 14 hospitals in Portugal and with medical centers located in several Mediterranean countries, Eastern and Central Europe, Iceland, South America, and Asia (45). In this project, CEM has been functioning as a reference laboratory in which MRSA clinical isolates collected in collaborating institutions are characterized by a combination of phenotypic and genotypic typing methods (2, 30). The information obtained is then forwarded to the hospital in order to use the data for the design of improved infection control policies (39). The large database generated in these studies is of interest for global surveillance (3, 23). It was through such studies that highly epidemic multiresistant clones of MRSA capable of massive geographic expansion such as the Iberian and the Brazilian clones were identified (1, 6, 13, 24, 37, 42).
The aim of the present study was to test the nature of MRSA isolates collected between 1992 and 1997 from the pediatric hospital in Lisbon (Hospital Dona Estefânia) and compare the findings with the information about S. aureus stored in the CEM/NET database.
MATERIALS AND METHODS
Hospital.
Hospital Dona Estefânia, the pediatric hospital of the capital of Portugal, opened in Lisbon in 1877. It is of medium size (407 beds), with six services in a total of 15 wards, including five surgical units and two intensive care units (ICUs). The hospital receives patients from a tertiary care hospital in Lisbon and from all district hospitals located in the southern part of the country (covering approximately 50% of the territory). The first MRSA isolate was detected in this hospital in 1983 (27), and the prevalence of MRSA was high in 1992 (46.9%), 1993 (44.9%), and 1994 (41.0%). In 1994, drastic infection control measures were introduced into the hospital, which included routine cleaning of all areas, new rules on the use of disinfectants and antiseptics, and control of prescription of antimicrobial agents; frequent hand washing and the use of gloves and gowns were enforced. Patients with MRSA were assigned to cohorts, or, if that was impractical, strict barrier precautions were introduced. Periodic surveillance of hand carriage of MRSA was done, and a group of nurses were dedicated exclusively to the care of MRSA-infected patients. Following implementation of this policy, the prevalence of MRSA began to decline from 40.0% in 1995 to 29.7% in 1996 and 17.0% in 1997.
Bacterial strains.
The total numbers of MRSA strains isolated in each year between 1992 and 1997 were 127, 267, 158, 194, 86, and 46 strains, respectively. Characterization of these isolates by antibiograms was performed in the hospital microbiology laboratory. Fifty-three randomly selected strains from this collection were used in the molecular characterization of MRSA at the research laboratory (CEM). Isolates were recovered from a variety of body sites, such as sputum (42%), as well as a variety of exudates (30%), secretions (19%), blood (2%), urine (2%), cerebrospinal fluid (2%), and catheters (2%). Primary characterization of the isolates was done with the Slidex Staph kit test (bioMérieux, Marcy l’Etoile, France), coagulase tube test (Difco, Detroit, Mich.), and by observing mannitol fermentation (Difco). Only one isolate per patient was considered. Other strains belonging to the Instituto de Technologia Química e Biológica and the Rockefeller University culture collections were used as controls and for comparison of results.
Antimicrobial susceptibility testing.
Antibiograms were performed by the hospital clinical laboratory using the Vitek system (bioMérieux). The antimicrobial agents tested were ampicillin-sulbactam, cephalothin, ciprofloxacin, clindamycin, erythromycin, oxacillin, penicillin G, tetracycline, trimethoprim-sulfamethoxazole, and vancomycin. For MRSA isolates the Kirby-Bauer disc-diffusion method was also applied according to the National Committee for Clinical Laboratory Standards guidelines (28). MRSA medium (Becton and Dickinson Microbiology Europe, Meylan, France) was used as an additional method for detection of methicillin resistance. High-level resistance to spectinomycin (500 μg/ml), which is associated in the majority of the MRSA strains with carriage of one or more copies of the transposon Tn554 (32), was tested by spotting 5 μl of late-log-phase cultures onto Trypticase soy agar plates containing 500 μg of spectinomycin per ml. Results were read after 24 h of incubation at 37°C. COL and BM79, which are known to be susceptible and resistant to spectinomycin, respectively, were used as controls.
Assays for methicillin resistance and (PAPs).
When receiving the strains in the research laboratory, the methicillin resistance phenotype of all strains was further evaluated by the 1-mg methicillin disc diffusion method (9). In short, overnight cultures were spread with a sterile swab on Trypticase soy agar (Difco) plates, and a 1-mg methicillin disc was placed in the center of each plate, which was incubated at 37°C for 24 h. On the basis of the diameters of inhibition halos, the strains were assigned to tentative phenotypic expression classes. Five reference strains, including COL, BM79, NYHB3, and CDC1 (expressing different levels of resistance to methicillin) and the methicillin-susceptible S. aureus strain RN2677, were used as controls (7). The preliminary classification was confirmed by population analysis profiles (PAPs) of representative strains as previously described (7, 44). Aerobically grown overnight (18 h) cultures (109 to 1010 CFU/ml) were plated at four dilutions (100, 10−2, 10−4, and 10−6) on agar plates containing serial (twofold) dilutions of methicillin at concentrations of 0 and 0.75 to 800 μg/ml. Colonies were counted after incubation for 48 h at 37°C. A graphic representation (PAP) was constructed by plotting colony counts against the concentration of methicillin. The profiles obtained provided information about the homogeneity or heterogeneity of the methicillin-resistant phenotype. The MIC was defined as the lowest concentration of the antibiotic that inhibited 99.9% of the cells.
PFGE.
Chromosomal DNAs were prepared as described previously (10), digested with SmaI nuclease (New England Biolabs, Beverly, Mass.), and separated in a CHEF-DR II (contour-clamped homogeneous electric field) apparatus (Bio-Rad, Birmingham, United Kingdom) for 23 h. The running parameters were as follows: initial pulse, 5 s; final pulse, 35 s; voltage, 6 V/cm; and temperature, 13°C. Standard methodologies were used for staining and photographing the gels (35). MRSA isolates CPS23 (10), HPV107 (37), and HU25 (42), representatives of the major clones described in Portuguese hospitals, were also run for comparison. Strain NCTC8325 (31) and pulsed-field gel electrophoresis (PFGE) lambda marker (New England Biolabs) were used as molecular size standards. The analysis of the SmaI macrorestriction profiles was done by visual inspection of the patterns by using the criteria of Tenover et al. (43): isolates showing six or less fragment differences were considered to be subtypes of a major pattern. PFGE patterns were also evaluated by computer-assisted comparison with the Whole Band Analyzer version 3.3 (BioImage, Ann Harbor, Mich.) software for Unix SparcStation 4 running under a SunOS version 5.5.1 operating system. Similarities between patterns were determined by generation of dendrograms. The Dice similarity coefficient was used, and the patterns were clustered by the minimum linkage method.
Conventional gels.
Chromosomal DNAs were digested with the restriction endonuclease Bsp 106 (isoschizomer of ClaI) (Stratagene, La Jolla, Calif.) and run in a conventional gel electrophoresis apparatus (10, 35). ClaI-restricted DNAs from strains representing ClaI patterns previously described were also run along as standards (1, 13, 19, 30).
Southern hybridization.
ClaI and SmaI DNA fragments in conventional and PFGE gels were transferred as previously described (10). Membranes of conventional gels were hybridized with mecA (10, 25) and Tn554-specific probes (14, 19). Membranes of the SmaI-PFGE gels were hybridized with the mecA probe in order to identify the SmaI fragment containing the mecA gene. For probe labeling and hybridization, an enhanced chemiluminescence nonradioactive labeling kit, RPN3040 (Amersham), was used according to the manufacturer’s instructions.
RESULTS
Antibiotic resistance profiles of MRSA isolates recovered between 1992 and 1997 at the pediatric hospital.
The distribution of S. aureus isolates during the 6-year surveillance period in Hospital Dona Estefânia is shown in Table 1. A surprisingly large proportion of the MRSA isolates—381 of 878—were only resistant to β-lactam antibiotics (ampicillin-sulbactam, cephalothin, oxacillin, and penicillin) but were susceptible to all other antimicrobial agents tested, including spectinomycin. The rest of the MRSA isolates were multiresistant, which included—in addition to β-lactam antibiotics—resistance to ciprofloxacin, clindamycin, erythromycin, tetracycline, sulfamethoxazole-trimethoprim, and spectinomycin in a variety of different combinations.
TABLE 1.
Prevalence of MRSA in the pediatric Hospital Dona Estefânia, Lisbon, Portugal, between 1992 and 1997
| Yr | No. of S. aureus isolates | No. (%) of MRSA isolates | No. of MRSA isolates resistant to β-lactam antibiotics only |
|---|---|---|---|
| 1992 | 271 | 127 (46.9) | 76 |
| 1993 | 594 | 267 (44.9) | 142 |
| 1994 | 385 | 158 (41.0) | 76 |
| 1995 | 485 | 194 (40.0) | 44 |
| 1996 | 290 | 86 (29.7) | 40 |
| 1997 | 270 | 46 (17.0) | 3 |
| Total | 2,295 | 878 (38.0) | 381 |
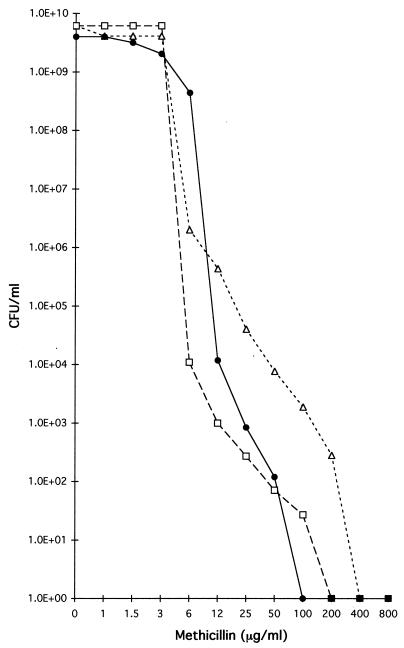
A group of 53 MRSA isolates were selected randomly from among the MRSA isolates in order to clarify the nature of MRSA strains by the use of molecular fingerprinting techniques. The majority (41 of 53) MRSA isolates could be classified into two sharply defined groups, on the basis of antimicrobial susceptibility pattern and methicillin resistance phenotype. Isolates belonging to the first group (25 strains) were resistant to ampicillin-sulbactam, cephalothin, penicillin, and oxacillin, but were susceptible to all other antimicrobial agents tested, including spectinomycin, suggesting the absence of Tn554, which was confirmed by the negative results in hybridization with the Tn554 DNA probe. The 1-mg methicillin disc assay showed that this group belonged to heterogeneous methicillin-resistant phenotypic class 1 or 2, since large inhibition halos typical of these classes were obtained (Table 2). This was confirmed by PAPs of three strains of this group (HDE6, HDE65, and HDE281), whose MICs were found to be between 1.5 and 6 μg/ml (Fig. 1).
TABLE 2.
Epidemiological data of MRSA from the pediatric Hospital Dona Estefânia, Lisbon, Portugal
| Yr(s) and isolate no. | Ward | Antibiogram result (resistant to)a | Methicillin disc assay resultb | Spectinomycin assay resultc | Clonal type (mecA::Tn554::PFGE)d |
|---|---|---|---|---|---|
| 1992 | |||||
| HDE1 | NAe | AMS, Cft, Oxa, Pen | 29 | S | II::NH::D1 |
| HDE2 | NA | AMS, Cft, Oxa, Pen | 30 | S | II::NH::D1 |
| HDE3 | NA | AMS, Cft, Oxa, Pen | 31 | S | II::NH::D1 |
| HDE4 | NA | AMS, Cft, Oxa, Pen | 25 | R | I::E::E |
| HDE5 | NA | AMS, Cft, Oxa, Pen | 27 | S | II::NH::D3 |
| HDE6 | NA | AMS, Cft, Oxa, Pen | 31 | S | II::NH::D1 |
| HDE7 | NA | AMS, Cft, Oxa, Pen, Ery, Cip, Cld, Te | 14 | R | I::E::E |
| HDE9 | NA | AMS, Cft, Oxa, Pen | 33 | S | II::NH::D1 |
| HDE11 | NA | AMS, Cft, Oxa, Pen | 23 | S | II::NH::D1 |
| HDE12 | NA | AMS, Cft, Oxa, Pen, Ery, Cip, Cld, Te | 15 | R | I::E::A5 |
| HDE14 | NA | AMS, Cft, Oxa, Pen, Ery, Cip, Cld, Te | 19 | R | I::E::A2 |
| HDE15 | NA | AMS, Cft, Oxa, Pen | 30 | S | II::NH::D3 |
| 1993–1994 | |||||
| HDE63 | ICU neonatology | AMS, Cft, Oxa, Pen, Ery, Cip, Cld, Te | 18 | R | I::E::A3 |
| HDE65 | Surgery 1 | AMS, Cft, Oxa, Pen | 31 | S | II::NH::D2 |
| HDE66 | Medicine urgency | AMS, Cft, Oxa, Pen | 32 | S | II::NH::D3 |
| HDE67 | Nursery | AMS, Cft, Oxa, Pen | 30 | S | II::NH::D3 |
| HDE68 | ICU neonatology | AMS, Cft, Oxa, Pen, Ery, Cip, Cld, Te | 16 | R | I::E::A4 |
| HDE71 | Delivery room | AMS, Cft, Oxa, Pen | 31 | S | II::NH::D3 |
| HDE72 | ICU neonatology | AMS, Cft, Oxa, Pen, Ery, Cip, Cld, Te | 13 | R | I::E::A4 |
| HDE73 | ICU neonatology | AMS, Cft, Oxa, Pen, Ery, Cip, Cld, Te | 16 | R | I::E::A4 |
| HDE75 | Surgery 1 | AMS, Cft, Oxa, Pen, Ery, Cip, Cld, Te | 14 | R | I::J::A1 |
| HDE77 | Surgery 4 | AMS, Cft, Oxa, Pen | 29 | S | II::NH::D3 |
| HDE83 | ICU pediatry | AMS, Cft, Oxa, Pen, Ery, Cip, Cld, Te | 13 | R | I::E::A4 |
| HDE84 | ICU neonatology | AMS, Cft, Oxa, Pen, Ery | 30 | S | II::NH::D3 |
| HDE91 | Surgery 2 | AMS, Cft, Oxa, Pen | 29 | S | II::NH::D1 |
| HDE281 | Pediatric surgery | AMS, Cft, Oxa, Pen | 29 | S | II::NH::D6 |
| HDE333 | Nursery | AMS, Cft, Oxa, Pen | 26 | S | II::NH::D3 |
| 1995 | |||||
| HDE163 | Delivery room | AMS, Cft, Oxa, Pen | 23 | S | II::NH::D4 |
| HDE174 | Pneumology | AMS, Cft, Oxa, Pen | 25 | S | II::NH::D3 |
| HDE203 | Surgery 1 | AMS, Cft, Oxa, Pen, Ery, Cip, Cld, Te | 12 | R | I::E::A6 |
| HDE218 | ICU neonatology | AMS, Cft, Oxa, Pen, Ery, Cip, Cld | 13 | R | I::E::A7 |
| HDE220 | ICU pediatry | AMS, Cft, Oxa, Pen, Ery, Cip, Cld, Te | 10 | R | I::E::A8 |
| HDE221 | Pediatry | AMS, Cft, Oxa, Pen | 27 | S | II::NH::D3 |
| HDE231 | Medicine urgency | AMS, Cft, Oxa, Pen | 25 | S | II::NH::D3 |
| HDE232 | Surgery 2 | AMS, Cft, Oxa, Pen | 24 | S | II::NH::D1 |
| HDE233 | Surgery 1 | AMS, Cft, Oxa, Pen, Ery, Cip, Cld, Te, SXT | 31 | S | XII::χ::F |
| HDE255 | Surgery 1 | AMS, Cft, Oxa, Pen | 24 | S | II::NH::D5 |
| HDE266 | Otorrhinolaringology | AMS, Cft, Oxa, Pen, Ery, Cld | 27 | S | II::NH::D6 |
| HDE318 | Nursery | AMS, Cft, Oxa, Pen, Ery, Cld (i) | 24 | S | II::NH::D3 |
| 1996 | |||||
| HDE287 | Pediatry | AMS, Cft, Oxa, Pen, Cld (i) | 26 | S | II::NH::D3 |
| HDE288 | Pneumology | AMS, Cft, Oxa, Pen | 26 | S | II::NH::D3 |
| HDE301 | Surgery 4 | AMS, Cft, Oxa, Pen, Ery, Cip, Cld | 16 | S | I::NH::A3 |
| HDE309 | Surgery 1 | AMS, Cft, Oxa, Pen, Ery | 27 | S | II::NH::D7 |
| HDE325 | Surgery 3 | AMS, Cft, Oxa, Pen | 27 | S | XVII::NH::G |
| HDE340 | Infectious diseases | AMS, Cft, Oxa, Pen, Ery, Cip, Cld, Te | 11 | R | I::E::A9 |
| HDE362 | Surgery 2 | AMS, Cft, Oxa, Pen, Ery | 31 | S | II::NH::D1 |
| HDE364 | ICU neonatology | AMS, Cft, Oxa, Pen, Ery, Cip, Cld, Te | 12 | R | I::E::A7 |
| HDE386 | ICU neonatology | AMS, Cft, Oxa, Pen, Ery, Cip, Cld, Te | 9 | R | I::E::A7 |
| 1997 | |||||
| HDE366 | ICU neonatology 3 | AMS, Cft, Oxa, Pen, Ery, Cip, Cld, Te | 9 | R | I::E::A7 |
| HDE371 | ICU pediatry | AMS, Cft, Oxa, Pen, SXT | 26 | S | XI::NH::H |
| HDE372 | ICU pediatry | AMS, Cft, Oxa, Pen, Ery, Cip, Te | 9 | R | I::E::A7 |
| HDE373 | ICU neonatology | AMS, Cft, Oxa, Pen, Ery, Te(i) | 23 | S | II::NH::D1 |
| HDE383 | Otorrhinolaringology | AMS, Cft, Oxa, Pen, Ery(i), Cld | 24 | S | II::NH::D5 |
Antibiogram results represent resistance to the antibiotics ampicillin-sulbactam (AMS), cephalothin (Cft), oxacillin (Oxa), penicillin G (Pen), erythromycin (Ery), ciprofloxacin (Cip), clindamycin (Cld), tetracycline (Te), trimethoprim-sulfamethoxazole (SXT), and vancomycin (V). (i), intermediate resistance.
Results of the 1-mg methicillin disc assay. Numbers indicate inhibition halos in millimeters.
Isolates were tested for their ability to grow in the presence of 500 μg of spectinomycin per ml. R, resistant (confluent growth); S, susceptible (no growth).
Clonal types were defined on the basis of ClaI-mecA polymorphs, ClaI-Tn554 insertion patterns, and SmaI-PFGE profiles. NH, no homology with the Tn554 probe.
NA, data not available.
FIG. 1.
Population analysis profiles of some MRSA isolates. Cultures of strains HDE6 (□), HDE65 (●), and HDE281 (▵) were plated at various dilutions on agar containing increasing concentrations of methicillin to determine the mode of phenotypic expression of resistance by population analysis.
The second group of MRSA isolates (16 strains) was resistant not only to β-lactam antimicrobials (ampicillin-sulbactam, cephalothin, oxacillin, and penicillin), but also to erythromycin, ciprofloxacin, clindamycin, tetracycline, and spectinomycin. Testing with the 1-mg methicillin disc showed small inhibition halos, indicating high-level and homogeneous methicillin resistance.
The 12 remaining MRSA isolates had small variations of the two main antibiogram patterns, including one isolate which was resistant to trimethoprim-sulfamethoxazole (Table 2). All MRSA isolates were susceptible to vancomycin.
Molecular typing.
Analysis of the vicinity of the mecA gene identified four ClaI::mecA polymorphs previously described (I, II, XI, and XII) and a new one named XVII (molecular sizes of hybridization bands are approximately 1.6 and 4.1 kb [data not shown]). ClaI::mecA polymorphs I and II accounted for 36% (19 isolates) and 59% (31 isolates) of the strains, respectively. Patterns XI, XII, and XVII were found in single isolates only.
Hybridization of the ClaI restriction digests with a Tn554-specific probe showed that 34 strains (64%) lacked this transposon, since no hybridization signal was obtained (NH). Tn554 pattern E (19) was found in 17 strains (32%). Patterns χ and J, corresponding to single and double insertions of the transposon, respectively, were found in single isolates only.
Separation of the SmaI DNA fragments of the 53 MRSA strains by PFGE generated six PFGE types. The majority of the isolates (31 of 53) shared a common PFGE type, D, in one of its subtypes, D1 through D7. PFGE type A was represented by 17 isolates (subtypes A1 through A9). PFGE type E was assigned to two isolates, and PFGE profiles F, G, and H were found in single isolates only (Table 2).
Hybridization of the PFGE membranes with the mecA probe indicated that, for strains with PFGE type D, the SmaI-mecA hybridization fragment had a molecular size of approximately 180 kb, whereas for PFGE type A, it was ca. 210 kb (data not shown).
Clonal analysis.
Clonal types were assigned to the isolates by combining the results obtained by the three molecular typing techniques: ClaI::mecA–ClaI::Tn554–SmaI::PFGE types.
The largest group of isolates (31 of 53 [59%]) carried the ClaI-mecA polymorph II, did not hybridize with Tn554 (NH), and shared a common PFGE type, D. The molecular fingerprinting properties of these strains (II::NH::D) overlapped with the distinct, limited antimicrobial resistance pattern already referred to above: this group of MRSA strains was only resistant to β-lactam antimicrobials (strains resistant to ampicillin-sulbactam, cephalothin, oxacillin, and penicillin), the MIC of methicillin for them was low and they showed heterogeneous methicillin resistance. Because of its preponderance in the pediatric hospital in Lisbon, we designated this MRSA the “pediatric” clone.
The second group of isolates (15 of 53 [35%]) showed typical features of the multiresistant and widely spread Iberian clone: it had clonal type I::E::A, was resistant to most antimicrobial agents tested, and showed high and homogeneous resistance to methicillin. Two additional MRSA isolates (HDE75 and HDE301) differed only in the Tn554 pattern, J and NH, indicating an additional insertion and/or absence of this transposon, respectively.
Four additional clonal types, I::E::E, XII::χ::F, XVII::NH::G, and XI::NH::H, were represented in this collection by five sporadic strains (Table 2).
Prevalence and in-hospital distribution of the pediatric MRSA clone.
Among the 53 MRSA isolates tested by the molecular fingerprinting methods during the 6-year survey, 31 isolates showed features of the pediatric MRSA clone. The majority of these showed a common pattern of antibiotic resistance limited to β-lactam antibiotics. The pediatric clone represented 8 of 12, 9 of 15, 8 of 12, 4 of 9, and 2 of 5 of the MRSA isolates in the samples analyzed in 1992, 1993 to 1994, 1995, 1996, and 1997, respectively. Based on antibiotyping alone, which was performed for the total number of MRSA isolates collected in the hospital in this period, MRSA isolates with the pediatric antibiotype were present in substantial numbers throughout the surveillance period (Table 1). MRSA isolates with antibiotic resistance restricted to β-lactam antibiotics (pediatric clone) were particularly frequent in 1993, when 142 of 267 MRSA isolates belonged to this group. This was related to an outbreak which began in August 1993 at the hospital nursery that lasted 2 months, and the strain responsible was isolated from 120 patients. The outbreak was controlled after the unit had been cleaned, specific recommendations for the care of newborns had been created, and the need for frequent hand washing had been reinforced. Since then, severe outbreaks have not been registered, although there is occasional evidence for cross-transmission of MRSA, mainly by hand carriage. The proportion of isolates of the pediatric clone declined sharply in 1997, when only 3 of the 40 MRSA isolates had the pediatric antibiotype. The 31 pediatric MRSA isolates identified by molecular typing techniques were recovered from as many as nine different hospital wards, including the medicine, delivery, neonatology, pediatrics, surgery, and otorhinolaryngology wards. In contrast, 14 of the 15 MRSA isolates fingerprinted as the Iberian clone between 1993 and 1997 were all from ICUs or surgery wards.
Detection of the pediatric clone of MRSA in international samples.
In contrast to the Iberian MRSA strains with their I::E::A clonal type, which were found to be widespread in Portuguese hospitals (1, 2, 30, 36–40), MRSA with the clonal type II::NH::D characteristic of the pediatric clone was extremely rare, and its detection among MRSA from Portuguese hospitals required a systematic search of the CEM/NET database.
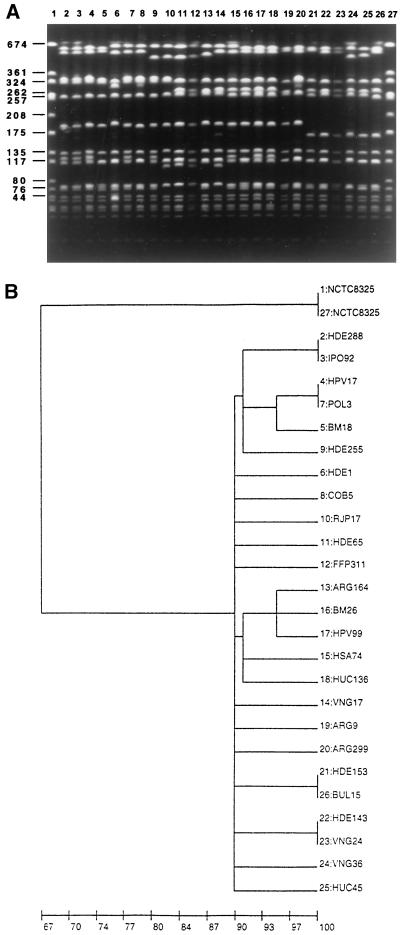
Through such a search, 8 isolates from Portugal were identified among the more than 1,000 genetically characterized MRSA isolates deposited in the database. These eight strains came from hospitals located in the north, center, and south of Portugal and shared a closely related clonal type (Fig. 2). An additional search of the CEM/NET database extended to international samples identified a substantial number of isolates with the pediatric clonal type among MRSA isolates from Poland (22), Argentina (6), New York (8, 11, 29), and Colombia (41) (Fig. 2). All of these strains shared (i) the same ClaI::mecA::Tn554 clonal profile, II::NH (with the exception of HPV17 from Portugal and strains from Poland, which were I::NH). (ii) All had a similar PFGE pattern, as observed by visual comparison of the isolates and by computer-assisted analysis by generating a dendrogram (Fig. 2A and 2B). (iii) The majority were resistant only to β-lactam antimicrobials (ampicillin-sulbactam, cephalothin, oxacillin, and penicillin). (iv) The strains showed low-level and heterogeneous resistance to methicillin (produced overlapping PAPs), with the exception of COB5, for which the MIC was higher (25 μg/ml). Relevant data about these strains are presented in Table 3.
FIG. 2.
(A) PFGE illustrative of the international dissemination of the pediatric clone. Representatives from different hospitals and countries are shown. Given below in parentheses is the origin of the isolate. PFGE patterns of the infrequent (eight isolates) pediatric MRSA strains identified in Portuguese hospitals other than Hospital Dona Estefânia (HDE) are also shown. MRSA strains from Argentina, Poland and Colombia are from the collections listed in Table 3. MRSA strains from New York are from the hospital designated in Table 3 as BM. Methicillin-susceptible S. aureus strains are described in the text. MRSA strains (by lane): 2, HDE288 (Lisbon, Portugal); 3, IPO92 (Lisbon, Portugal); 4, HPV17 (Lisbon, Portugal); lane 5, BM18 (New York, N.Y.); 6, POL3 (Warsaw, Poland); lane 7, HDE1 (Lisbon, Portugal); 8, COB5 (Colombia); 9, HDE255 (Lisbon, Portugal); 10, RJP17 (Porto, Portugal); 11, HDE65 (Lisbon, Portugal); 12, FFP311 (Porto, Portugal); 13, ARG164 (Buenos Aires, Argentina); 14, VNG17 (Vila Nova de Gaia, Portugal); 15, HSA74 (Porto, Portugal); 16, BM26 (New York, N.Y.); 17, HPV99 (Lisbon, Portugal); 18, HUC136 (Coimbra, Portugal); 19, ARG9 (Buenos Aires, Argentina); 20, ARG299 (La Plata, Argentina). Methicillin-susceptible S. aureus strains (by lane): 21, HDE153 (Lisbon, Portugal); 22, HDE143 (Lisbon, Portugal); 23, VNG24 (Vila Nova de Gaia, Portugal); 24, VNG36 (Vila Nova de Gaia, Portugal); 25, HUC45 (Coimbra, Portugal); 26, BUL15 (Bulgaria). Lanes 1 and 27 contain reference strain NCTC8325, used as a molecular size marker. Numbers to the left show molecular size in kilobases. (B) Computer-generated dendrogram of the PFGE of Fig. 2A. Results are from analysis of similarity by Dice’s coefficient; clustering was done by the minimum linkage method.
TABLE 3.
Detection of the pediatric clone of MRSA in international samples
| Origin | Isolation period (yr) | No. of hospitals | Hospital code | Type of hospital | No. of MRSA isolates
|
Ward(s) from which pediatric clone was recovered | Primary source of MRSA | Source or reference | |
|---|---|---|---|---|---|---|---|---|---|
| Total | Pediatric clone | ||||||||
| Lisbon, Portugal | 1992–1997 | 1 | HDE | Pediatric | 53 | 32 | Several | Children | This study |
| Colombiaa | 1996 | NAb | NA | NA | 10 | 10 | NA | NA | 41 |
| Poland | 1990–1996 | 18 | 158 | 75 | 22 | ||||
| 1993–1994 | WAW1 | Teaching | 39 | 33 | Pediatric | Children | |||
| 1991–1992, 1996 | WAW2 | University children’s | 30 | 11 | NA | Children | |||
| 1993, 1995 | WAW3 | Gynecological-obstetric university | 22 | 20 | Neonatalogy | Children | |||
| 1994 | WAW4 | Pediatric | 9 | 2 | NA | Children | |||
| 1994 | WAW5 | NA | 5 | 3 | NA | NA | |||
| 1992 | WAW6 | Pediatric | 2 | 1 | NA | Children | |||
| 1994 | GDK1 | Municipal | 4 | 2 | NA | NA | |||
| 1991 | LOD1 | University | 1 | 1 | NA | NA | |||
| Argentina | 1994–1996 | 13 | 148 | 23 | 6 | ||||
| 1996 | SOR | Pediatric | 14 | 7 | Several | Children | |||
| 1996 | GAR | Pediatric | 15 | 5 | Several | Children | |||
| 1995–1996 | ICBA | Cardiology | 10 | 2 | Surgery | NA | |||
| 1996 | BAZ | Community | 17 | 2 | Neonatology, medicine | Child, NA | |||
| 1994–1996 | CEM | Community | 14 | 3 | Ambulatory, medicine | NA | |||
| 1996 | CLI | Community | 10 | 2 | ICU, ambulatory | NA | |||
| 1994–1996 | FER | Community | 12 | 1 | Neonatology | Child | |||
| 1994–1996 | FLE | Neurology | 8 | 1 | ICU | NA | |||
| New York, N.Y. | 1989 | 1 | BM | Community | 55 | 38 | NA | NA | 11 |
| 1989 | 1 | MMC | Community | 79 | 15 (12)c | Labor and delivery | Children | 20 | |
| 1988–1991 | 1 | CMH | University | 37 | 12d | Pediatric | Children | 8, 14 | |
Study not complete.
NA, data not available.
Twelve of the 15 II:NH isolates were recovered from the labor and delivery wards. PFGE patterns were not available for comparison.
This study was an investigation of an outbreak in the neonatal ICU of the hospital. The outbreak was caused by the pediatric clone that was also recovered from the well-baby nurseries. All isolates recovered from the adult wards belonged to a different clone.
Deletion of mecA.
Six S. aureus strains collected in Hospital Dona Estefânia (HDE112, HDE143, and HDE153) and in two other Portuguese hospitals (HUC45, VNG24, and VNG36) (30, 38) and one MSSA strain from Bulgaria (BUL15) were susceptible to methicillin by the microbiological assay and did not react with the mecA DNA probe. On the other hand, these isolates showed a PFGE pattern that was very similar to the PFGE pattern D characteristic of the pediatric clone of MRSA (Fig. 2). The major difference was found in the SmaI hybridization fragment, which contains the mecA gene: in the six MSSA isolates, this fragment had a smaller molecular size (135 instead of 180 kb) and did not hybridize with the mecA probe, indicating a deletion of approximately 45 kb, which must have included both the mecA gene and flanking DNA. Interestingly, two of these MSSA strains (HUC45 and BUL15) had been initially classified as MRSA by the clinical laboratories of the collaborating hospitals, and only later, at the time of the molecular typing, were they found to be mecA negative.
DISCUSSION
Long-range coexistence of two MRSA clones in Hospital Dona Estefânia.
The findings described in this study document the long-term coexistence of MRSA strains of very different degrees of antimicrobial resistance profiles in a single hospital: Hospital Dona Estefânia, the pediatric hospital in Lisbon. Molecular typing indicates that strains with limited antimicrobial resistance belonged to the clonal type II::NH::D, which we designated pediatric MRSA. The MIC of methicillin for MRSA of this clonal type was low, and the strains were resistant only to β-lactam antimicrobials. A large proportion of the multiresistant strains were shown to be representatives of the highly-methicillin-resistant internationally spread Iberian clone. Instead of the expected “takeover” by the multiresistant epidemic clone (which was demonstrated in several previous surveillance studies), the pediatric MRSA clone retained a substantial prevalence throughout the 6 years of surveillance in Hospital Dona Estefânia.
The multidrug-resistant Iberian clone was introduced in the early 1990s in Portugal, where, due to its large capacity for dissemination, it became dominant in several hospitals soon after its appearance (1, 30, 35–39). More recently, MRSA isolates with molecular features similar to those of the Iberian clone were recovered in hospitals of several European countries (24) and in two New York City hospitals (33, 34), further documenting the superior capacity of this clone to spread and replace other MRSA lineages. Clearly no displacement of the pediatric clone could be detected in Hospital Dona Estefânia during 1992 to 1996.
One can only speculate about the reasons which allowed the continued high prevalence of the pediatric clone in Hospital Dona Estefânia despite its relatively limited drug resistance profile. In hospital wards (Table 2) in which MRSA isolates with the pediatric molecular or antibiotype properties were recovered most frequently, antimicrobial use is controlled through a strict policy of prescription which is limited to amoxicillin, amoxicillin-clavulanic acid, and expanded-spectrum cephalosporins. Such antibacterial agents as wide-spectrum cephalosporins are avoided. In contrast to the pediatric clone, 14 of the 15 MRSA isolates identified as the Iberian clone by molecular fingerprinting techniques were recovered in the ICUs and surgery units, which are the only units of the hospital where the hospital’s prescription policy does not put restrictions on antimicrobial use and “last resort” antibiotics are frequently prescribed. The two ICUs and surgery units of Hospital Dona Estefânia also admit patients from several regions of the country which may be the source for the introduction into the hospital of new MRSA clones (such as the Iberian clone) which are most frequent nationwide. The source of sporadic isolates of MRSA with other clonal types, such as I::E::E, XII::χ::F, XVII::NH::G, and XI::NH::H, found in the surgery wards and in the ICU of Hospital Dona Estefânia may also be related to the high patient turnover in these units.
Pediatric MRSA clone in international samples.
It is possible that the factors proposed to contribute to the persistence of the pediatric MRSA in Hospital Dona Estefânia are similar to factors that also allowed survival of this clone in international samples. Analysis of the patient population from which the international strains were recovered showed that they were frequently isolated from children and newborns. This was particularly evident in the case of MRSA collections from Poland (22), Argentina (6), and the Cornell Medical University Hospital in New York (8, 29), where the dominance of this clone in pediatric settings (pediatric hospitals or pediatric wards) was reported, whereas it was nonexistent or only sporadically found in other settings (Table 3). The New York study referred to above demonstrated that the pediatric clone of MRSA has the potential to cause severe disease and was responsible for the death of at least one child. Another study involving a teaching hospital in Brooklyn documented the existence of a single MRSA clone which carried the mecA polymorph II and lacked the transposon Tn554 (II::NH) and was recovered in the labor and delivery wards (20). PFGE patterns were not determined in this particular study, thus preventing a more accurate identification.
In a recent study in 12 New York City Hospitals, an MRSA strain with the clonal type V::NH::D* was frequently isolated from AIDS patients (34). The ClaI-mecA polymorph V is a close relative of polymorph II, and the PFGE pattern of these strains was similar enough to pattern D to suggest that V::NH::D* evolved from the same lineage as the pediatric clone.
The pediatric MRSA—an “archaic” clone?
Detection of the pediatric MRSA clone in the international MRSA samples requires an explanation. We propose that this clone represents the relics of a historically early MRSA lineage which, at that time, was widely distributed both in Europe and on the American continent (12). The low-level and heterogeneous methicillin resistance and resistance profile limited to β-lactams and the presence of a clonal type that is infrequent in most contemporary samples are consistent with this proposal. Similarities in the epidemiological setting in which this clone was detected (i.e., low antimicrobial pressure, low patient turnover) also support this model. Pediatric settings with relatively low antimicrobial pressure may be reservoirs of other historically early MRSA clones as well.
Instability of the mecA region.
Identification of six methicillin-susceptible S. aureus strains with widely different geographic origins but with a PFGE type similar to PFGE profile D suggests that they were derived from the pediatric clone by deletion of mecA and also suggests that this MRSA clone may have an inherent genetic instability. Spontaneous loss of the mecA region has already been described in vitro after long storage periods of MRSA strains (16) and was also observed in vivo (13, 17, 21). Deletion of mecA was recently shown to be inducible due to the action of the ccr (cassette chromosome recombinase) genes (18).
Prevalence of MRSA and infection control.
The surveillance study in Hospital Dona Estefânia described here demonstrates the coexistence of MRSA strains with very unequal antimicrobial resistant profiles in the same hospital over a prolonged time period. Reduction in the frequency of the two types of MRSA (the highly resistant MRSA associated with areas of high antimicrobial use and patient turnover and the endemic MRSA resident in areas of less aggressive antimicrobial use) may require different types of interventions. Keeping the frequency of highly resistant strains introduced through high patient turnover to ICUs and surgery wards low may require early screening and, if possible, assignment to cohorts at the time of admission. Decreasing the frequency of endemic strains may depend more on strict in-hospital infection control and low antimicrobial usage.
ACKNOWLEDGMENTS
Partial support for this work was provided by the CEM/NET initiative CEM/NET Project 31 from IBET (Portugal), contract PRAXIS XXI-2/2.1/BIO/1154/95 (Portugal), contract PECS/C/SAU/145/95 from Junta Nacional de Investigação Científica (JNICT) (Portugal), and a grant from Fundação Calouste Gulbenkian (Portugal) awarded to H. de Lencastre. The study was also supported by contract STRDA/C/BIO/360/92 (JNICT) awarded to H. de Lencastre and by grant PSAU/SAU/1591/92 from Ministério da Saúde (Portugal) awarded to I. Santos Sanches. R. Sá-Leão was supported by grants PRODEP from Faculdade de Ciências e Tecnologia da Universidade Nova de Lisboa and FMRH/BIC 1695/95 from JNICT. D. Dias was supported by grant BJI 1091/95 from JNICT.
We thank Inger Adamsson and Idalina Bonfim for technical assistance with isolates recovered in 1997 and Alexander Tomasz for critical reading of the manuscript and suggestions concerning the interpretation of findings.
REFERENCES
- 1.Aires de Sousa M, Santos Sanches I, Ferro M L, Vaz M J, Saraiva Z, Tendeiro T, Serra J, de Lencastre H. Intercontinental spread of a multidrug-resistant methicillin-resistant Staphylococcus aureus clone. J Clin Microbiol. 1998;36:2590–2596. doi: 10.1128/jcm.36.9.2590-2596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aires de Sousa M, Santos Sanches I, van Belkum A, van Leeuwen W, Verbrugh H, de Lencastre H. Characterization of methicillin-resistant Staphylococcus aureus isolates from Portuguese hospitals by multiple genotyping methods. Microb Drug Resist. 1996;2:331–341. doi: 10.1089/mdr.1996.2.331. [DOI] [PubMed] [Google Scholar]
- 3.Ayliffe G A J. The progressive intercontinental spread of methicillin-resistant Staphylococcus aureus. Clin Infect Dis. 1997;24:S74–S79. doi: 10.1093/clinids/24.supplement_1.s74. [DOI] [PubMed] [Google Scholar]
- 4.Barrett S P, Mummery R V, Chattopadhyay B. Trying to control MRSA causes more problems than it solves. J Hosp Infect. 1998;39:85–93. doi: 10.1016/s0195-6701(98)90322-x. [DOI] [PubMed] [Google Scholar]
- 5.Boyce J M. Methicillin-resistant Staphylococcus aureus: a continuing infection control challenge. Eur J Clin Microbiol Infect Dis. 1994;13:45–49. doi: 10.1007/BF02026126. [DOI] [PubMed] [Google Scholar]
- 6.Corso A, Santos Sanches I, Aires de Sousa M, Rossi A, de Lencastre H. Spread of a methicillin-resistant and multiresistant epidemic clone of Staphylococcus aureus in Argentina. Microb Drug Resist. 1998;4:277–288. doi: 10.1089/mdr.1998.4.277. [DOI] [PubMed] [Google Scholar]
- 7.de Lencastre H, Figueiredo A M S, Urban C, Rahal J, Tomasz A. Multiple mechanisms of methicillin resistance and improved methods for detection in clinical isolates of Staphylococcus aureus. Antimicrob Agents Chemother. 1991;35:632–639. doi: 10.1128/aac.35.4.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Lencastre H, Figueiredo A M S, Tomasz A. Genetic control of population structure in heterogeneous strains of methicillin-resistant Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 1993;12(Suppl. 1):S13–S18. doi: 10.1007/BF02389872. [DOI] [PubMed] [Google Scholar]
- 9.de Lencastre H, Tomasz A. Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1994;38:2590–2598. doi: 10.1128/aac.38.11.2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lencastre H, Couto I, Santos I, Melo-Cristino J, Torres-Pereira A, Tomasz A. Methicillin-resistant Staphylococcus aureus disease in a Portuguese hospital: characterization of clonal types by a combination of DNA typing methods. Eur J Clin Microbiol Infect Dis. 1994;13:64–73. doi: 10.1007/BF02026129. [DOI] [PubMed] [Google Scholar]
- 11.de Lencastre H, de Lencastre A, Tomasz A. Methicillin-resistant Staphylococcus aureus isolates recovered from a New York City hospital: analysis by molecular fingerprinting techniques. J Clin Microbiol. 1996;34:2121–2124. doi: 10.1128/jcm.34.9.2121-2124.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Lencastre, H., and H. Westh. Unpublished data.
- 13.Dominguez M A, de Lencastre H, Linares J, Tomasz A. Spread and maintenance of a dominant methicillin-resistant Staphylococcus aureus (MRSA) clone during an outbreak of MRSA disease in a Spanish hospital. J Clin Microbiol. 1994;32:2081–2087. doi: 10.1128/jcm.32.9.2081-2087.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Figueiredo A M S, Ha E, Kreiswirth B N, de Lencastre H, Noel G J, Senterfit L, Tomasz A. In vivo stability of heterogeneous expression classes in clinical isolates of methicillin-resistant staphylococci. J Infect Dis. 1991;164:883–887. doi: 10.1093/infdis/164.5.883. [DOI] [PubMed] [Google Scholar]
- 15.Humphreys H, Duckworth G. Methicillin-resistant Staphylococcus aureus (MRSA)—a re-appraisal of control measures in the light of changing circumstances. J Hosp Infect. 1997;36:167–170. doi: 10.1016/s0195-6701(97)90191-2. [DOI] [PubMed] [Google Scholar]
- 16.Hürlimann-Dalel R L, Ryffel C, Kayser F H, Berger-Bächi B. Survey of the methicillin resistance-associated genes mecA, mecR1-mecI, and femA-femB in clinical isolates of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1992;36:2617–2621. doi: 10.1128/aac.36.12.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inglis B, el-Adhami W, Stewart P R. Methicillin-sensitive and -resistant homologues of Staphylococcus aureus occur together among clinical isolates. J Infect Dis. 1993;167:323–328. doi: 10.1093/infdis/167.2.323. [DOI] [PubMed] [Google Scholar]
- 18.Ito T, Katayma Y, Hiramatsu K. Program and abstracts of the 38th Interscience Congress on Antimicrobial Agents and Chemotherapy. Washington, D.C: American Society for Microbiology; 1998. Overnight conversion of MRSA and MSSA with ccr (cassette chromosome recombinase) genes, abstr. C-62; p. 86. [Google Scholar]
- 19.Kreiswirth B, Kornblum J, Arbeit R D, Eisner W, Maslow J N, McGeer A, Low D E, Novick R P. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science. 1993;259:227–230. doi: 10.1126/science.8093647. [DOI] [PubMed] [Google Scholar]
- 20.Kreiswirth B J, Lutwick S M, Chapnick E K, Gradon J D, Lutwick L I, Sepkowitz D V, Eisner W, Levi M H. Tracing the spread of methicillin-resistant Staphylococcus aureus by southern blot hybridization using gene-specific probes of mec and Tn554. Microb Drug Resist. 1995;1:307–313. doi: 10.1089/mdr.1995.1.307. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence C, Cosseron M, Durand P, Costa Y, Leclercq R. Consecutive isolation of homologous strains of methicillin-resistant and methicillin-susceptible Staphylococcus aureus from a hospitalized child. J Hosp Infect. 1996;33:49–53. doi: 10.1016/s0195-6701(96)90028-6. [DOI] [PubMed] [Google Scholar]
- 22.Leski T, Oliveira D, Trzcinski K, Santos Sanches I, Aires de Sousa M, Hryniewicz W, de Lencastre H. Clonal distribution of methicillin-resistant Staphylococcus aureus in Poland. J Clin Microbiol. 1998;36:3532–3539. doi: 10.1128/jcm.36.12.3532-3539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lowy F D. Staphylococcus aureus infection. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 24.Mato R, Santos Sanches I, Venditti M, Platt D J, Brown A, Chung M, de Lencastre H. Spread of the multiresistant Iberian clone of methicillin-resistant Staphylococcus aureus (MRSA) to Italy and Scotland. Microb Drug Resist. 1998;4:107–112. doi: 10.1089/mdr.1998.4.107. [DOI] [PubMed] [Google Scholar]
- 25.Matthews P, Tomasz A. Insertional inactivation of the mec gene in a transposon mutant of a methicillin-resistant clinical isolate of Staphylococcus aureus. Antimicrob Agents Chemother. 1990;34:1777–1779. doi: 10.1128/aac.34.9.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melo-Cristino J the POSGAR. Antimicrobial resistance in staphylococci and enterococci in 10 Portuguese hospitals in 1996 and 1997. Microb Drug Resist. 1998;4:319–324. doi: 10.1089/mdr.1998.4.319. [DOI] [PubMed] [Google Scholar]
- 27.Melo Cristino J A G, Torres Pereira A, Afonso F. Infection with methicillin-gentamicin-resistant Staphylococcus aureus strains in a paediatric surgical unit in Lisbon. J Hosp Infect. 1985;6:426–428. doi: 10.1016/0195-6701(85)90060-x. [DOI] [PubMed] [Google Scholar]
- 28.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disk susceptibility test. Villanova, Pa: National Committee for Clinical Laboratory Standards; 1995. [Google Scholar]
- 29.Noel G J, Kreiswirth B N, Edelson P J, Nesin M, Projan S, Eisner W, Bauer D J, de Lencastre H, Figueiredo A M S, Tomasz A. Multiple methicillin-resistant Staphylococcus aureus strains as a cause of a single outbreak of severe disease in hospitalized neonates. Pediatr Infect Dis J. 1992;11:184–188. doi: 10.1097/00006454-199203000-00002. [DOI] [PubMed] [Google Scholar]
- 30.Oliveira D, Santos-Sanches I, Mato R, Tamayo M, Ribeiro G, Costa D, de Lencastre H. Virtually all methicillin-resistant Staphylococcus aureus (MRSA) infections in the largest teaching Portuguese hospital are caused by two internationally spread multiresistant strains: the “Iberian” and the “Brazilian” clones of MRSA. Clin Microbiol Infect. 1998;4:373–384. doi: 10.1111/j.1469-0691.1998.tb00081.x. [DOI] [PubMed] [Google Scholar]
- 31.Pattee P A, Lee H-C, Bannantine J P. Genetic and physical mapping of the chromosome of Staphylococcus aureus. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCH Publishers; 1990. pp. 41–58. [Google Scholar]
- 32.Phillips S, Novick R P. Tn554-a site-specific repressor-controlled transposon in Staphylococcus aureus. Nature. 1979;278:476–478. doi: 10.1038/278476a0. [DOI] [PubMed] [Google Scholar]
- 33.Roberts R B, Tennenberg A M, Eisner W, Hargrave J, Drusin L M, Yurt R, Kreiswirth B N. Outbreak in a New York city teaching hospital caused by the Iberian epidemic clone of MRSA. Microb Drug Resist. 1998;4:175–183. doi: 10.1089/mdr.1998.4.175. [DOI] [PubMed] [Google Scholar]
- 34.Roberts R B, de Lencastre A, Eisner W, Severina E P, Shopsin B, Kreiswirth B N, Tomasz A the MRSA Collaborative Study Group. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in 12 New York Hospitals. J Infect Dis. 1998;178:164–171. doi: 10.1086/515610. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Santos Sanches I, Leão R S, Oliveira D, Dias D, Mato R, de Lencastre H. Program and abstract of the 12th European Meeting on Bacterial Gene Transfer and Expression, Siena, Italy. 1996. Epidemic Iberian multidrug-resistant clone of Staphylococcus aureus in three hospitals in Portugal, abstr. 101. [Google Scholar]
- 37.Santos Sanches I, Ramirez M, Troni H, Abecassis M, Padua M, Tomasz A, de Lencastre H. Evidence for the geographic spread of a methicillin-resistant Staphylococcus aureus (MRSA) clone between Portugal and Spain. J Clin Microbiol. 1995;33:1243–1246. doi: 10.1128/jcm.33.5.1243-1246.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santos Sanches I, Aires de Sousa M, Sobral L, Calheiros I, Felicio L, Pedra I, de Lencastre H. Multidrug-resistant Iberian epidemic clone of methicillin-resistant Staphylococcus aureus endemic in a hospital in Northern Portugal. Microb Drug Resist. 1996;1:299–306. doi: 10.1089/mdr.1995.1.299. [DOI] [PubMed] [Google Scholar]
- 39.Santos Sanches I, Aires de Sousa M, Cleto L, Baeta de Campos M, de Lencastre H. Tracing the origin of an outbreak of methicillin-resistant Staphylococcus aureus infections in a Portuguese hospital by molecular fingerprinting methods. Microb Drug Resist. 1996;2:319–329. doi: 10.1089/mdr.1996.2.319. [DOI] [PubMed] [Google Scholar]
- 40.Santos Sanches I, Saraiva Z, Tendeiro T, Serra J, Velazquez Meza M E, Dias D, de Lencastre H. Extensive intra-hospital spread of a methicillin-resistant staphylococcal clone. Int J Infect Dis. 1998;3:26–31. doi: 10.1016/s1201-9712(98)90091-1. [DOI] [PubMed] [Google Scholar]
- 41.Tamayo, M., et al. Unpublished data.
- 42.Teixeira L A, Resende C A, Ormonde L R, Rosenbaum R, Figueiredo A M S, de Lencastre H, Tomasz A. Geographic spread of epidemic multiresistant Staphylococcus aureus clone in Brazil. J Clin Microbiol. 1995;33:2400–2404. doi: 10.1128/jcm.33.9.2400-2404.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomasz A, Nachman S, Leaf H. Stable classes of phenotypic expression in methicillin-resistant clinical isolates of staphylococci. Antimicrob Agents Chemother. 1991;35:124–129. doi: 10.1128/aac.35.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomasz A, de Lencastre H. Molecular microbiology and epidemiology: coexistence or alliance? In: Wenzel R P, editor. Prevention and control of nosocomial infections. Baltimore, Md: Williams & Wilkins; 1997. pp. 309–321. [Google Scholar]
- 46.Voss A, Milatovic D, Wallrauch-Schwarz C, Rosdahl V T, Braveny I. Methicillin-resistant Staphylococcus aureus in Europe. Eur J Clin Microbiol Infect Dis. 1994;13:50–55. doi: 10.1007/BF02026127. [DOI] [PubMed] [Google Scholar]