Abstract
Studies evaluating neuroimaging, genetically predicted gene expression, and pre-clinical genetic models of PTSD, have identified PTSD-related abnormalities in the prefrontal cortex (PFC) of the brain, particularly in dorsolateral and ventromedial PFC (dlPFC and vmPFC). In this study, RNA sequencing was used to examine gene expression in the dlPFC and vmPFC using tissue from the VA National PTSD Brain Bank in donors with histories of PTSD with or without depression (dlPFC n = 38, vmPFC n = 35), depression cases without PTSD (n = 32), and psychopathology-free controls (dlPFC n = 24, vmPFC n = 20). Analyses compared PTSD cases to controls. Follow-up analyses contrasted depression cases to controls. Twenty-one genes were differentially expressed in PTSD after strict multiple testing correction. PTSD-associated genes with roles in learning and memory (FOS, NR4A1), immune regulation (CFH, KPNA1) and myelination (MBP, MOBP, ERMN) were identified. PTSD-associated genes partially overlapped depression-associated genes. Co-expression network analyses identified PTSD-associated networks enriched for immune-related genes across the two brain regions. However, the immune-related genes and association patterns were distinct. The immune gene IL1B was significantly associated with PTSD in candidate-gene analysis and was an upstream regulator of PTSD-associated genes in both regions. There was evidence of replication of dlPFC associations in an independent cohort from a recent study, and a strong correlation between the dlPFC PTSD effect sizes for significant genes in the two studies (r = 0.66, p < 2.2 × 10−16). In conclusion, this study identified several novel PTSD-associated genes and brain region specific PTSD-associated immune-related networks.
Keywords: PTSD, RNAseq, vmPFC, dlPFC, Expression
1. Introduction
Genetic studies of posttraumatic stress disorder (PTSD) and related conditions have historically focused primarily on DNA variation, DNA methylation, and gene expression in peripheral cells. While these have yielded important insights into biological correlates of PTSD, most notably implicating alterations in inflammatory (Michopoulos et al., 2017; Miller et al., 2018), immunological (Bhatt et al., 2020; Michopoulos et al., 2020), and glucocorticoid-related processes (Dunlop and Wong, 2019), it remains unclear to what extent peripheral samples can inform understanding of mechanisms of PTSD in the brain. Recently, the first large-scale transcriptome-wide study of PTSD in human post-mortem brain tissue was published (Girgenti et al., 2021) using samples from the VA National PTSD Brain Bank (Friedman et al., 2017). Girgenti et al. measured gene expression through RNA sequencing of tissue from patients with histories of PTSD (n = 52) and depression (n = 45) versus controls (n = 46) in four regions of interest from the prefrontal cortex (PFC): dorsolateral PFC (dlPFC; Brodmann area 9/46), medial orbitofrontal cortex (Brodmann area 11), dorsal anterior cingulate (Brodmann area 24), and subgenual PFC (Brodmann area 25). Analyses revealed significant alterations in gene networks related to GABA interneurons and inflammatory processes that distinguished PTSD cases from depressed cases and controls, particularly in the dlPFC. Sex-specific analyses also showed marked sexual dimorphism in several of the PTSD-associated networks, and region-based analyses showed distinct patterns of association across several regions of the frontal cortex.
At the same time, we undertook an independent RNA sequencing study of cortical tissue from a group of donors that partially overlapped with those of Girgenti et al. (approximately 50%, details below). We examined a portion of vmPFC (BA 12/32) that was not examined by Girgenti et al., and a distinct but adjacent section of dlPFC (BA 9/46). We focused on dlPFC and vmPFC for several reasons (Selemon et al., 2019). The prefrontal cortex has well-established roles in emotion regulation and executive-functioning. Functional magnetic resonance imaging (fMRI) studies have found reduced connectivity in dlPFC and medial PFC of PTSD cases (Reuveni et al., 2016; Holmes et al., 2018). Structural neuroimaging studies of patients with PTSD have found reduced cortical thickness in these areas (Sadeh et al., 2016; Miller et al., 2015). Pre-clinical and clinical studies of various types have shown the vmPFC to be involved in fear extinction (Milad et al., 2007), and there are reciprocal connections between dlPFC and subcortical regions implicated in fear and PTSD such as the hippocampus and amygdala (Hartley and Phelps, 2010). Importantly, functional analyses of PTSD genome wide association studies using partitioning heritability and transcriptomic imputation revealed that PFC gene expression is involved in genetic risk of PTSD (Gelernter et al., 2019; Huckins et al., 2020; Stein et al., 2021). Together with the transcriptomic differences noted in Girgenti et al. (2021), these findings point towards the PFC as an important locus of PTSD pathogenesis and/or disease maintenance.
2. Materials and methods
RNA sequencing was used to measure gene expression in dlPFC and vmPFC using tissue from donors assessed for PTSD, depression, and other conditions. This included PTSD cases with and without depression (dlPFC n = 38, vmPFC n = 35), PTSD-free depressed cases (n = 32 for both regions), and controls without PTSD or depression (dlPFC n = 24, vmPFC n = 20). The sex-stratified totals and demographic information for the cohort are presented in Table 1, Table 2, and sex-stratified demographic tables are presented in Supplementary Table 1. The tissue quality scores are summarized in Supplementary Table 2, and the number of mapped reads is summarized in Supplementary Table 3.
Table 1.
Sample size for PTSD, depression, and control comparison groups.
| Region | Sample Size |
|||
|---|---|---|---|---|
| PTSD (M/F) | Depression (M/F) | Control (M/F) | Total (M/F) | |
| dlPFC | 38 (18/20) | 32 (21/11) | 24 (16/8) | 94 (55/39) |
| vmPFC | 35 (16/19) | 32 (21/11) | 20 (13/7) | 87 (50/37) |
Table 2.
Descriptive Statistics for the analyzed PTSD cases, depression cases, and controls.
| Region | Control | PTSD | p (PTSD vs Control) | MDD | p (MDD vs Control) | |
|---|---|---|---|---|---|---|
| dlPFC |
N | 24 | 38 | 32 | ||
| Sex = M (%) | 16 (66.67) | 18 (47.37) | 0.22 | 21 (65.63) | 1.00 | |
| AgeDeath mean (SD) | 46.53 (9.97) | 40.98 (11.64) | 0.059 | 41.24 (10.90) | 0.068 | |
| PMI mean (SD) | 29.56 (7.02) | 28.37 (8.24) | 0.56 | 27.31 (7.50) | 0.26 | |
| White non-Hispanic ancestrya n, (%) | 15 (62.50) | 31 (81.60) | 0.17 | 27 (84.40) | 0.12 | |
| Smoking (%) | 7 (29.17) | 28 (73.68) | 0.0015 | 22 (68.75) | 0.0077 | |
| Military Service (%) | 1 (4.17) | 15 (39.47) | 0.0052 | 4 (12.50) | 0.54 | |
| Suicide Death (%) | 0 (0.00) | 8 (21.05) | 0.043 | 5 (15.63) | 0.12 | |
| Alcohol or drug death (%) | 0 (0.00) | 25 (65.79) | 1.072 × 10−6 | 19 (59.36) | 1.31 × 10−5 | |
| vmPFC | N | 20 | 35 | 32 | ||
| Sex = M (%) | 13 (65.00) | 16 (45.70) | 0.27 | 21 (65.60) | >0.99 | |
| AgeDeath mean (SD) | 47.25 (10.38) | 41.11 (11.88) | 0.059 | 41.24 (10.90) | 0.054 | |
| PMI mean (SD) | 30.10 (7.22) | 28.10 (8.50) | 0.38 | 27.31 (7.50) | 0.19 | |
| White non-Hispanic ancestrya n (%) | 14 (70.00) | 28 (80.00) | 0.61 | 27 (84.40) | 0.38 | |
| Smoking (%) | 5 (25.00) | 26 (74.30) | 0.0011 | 22 (68.80) | 0.0053 | |
| Military service (%) | 1 (5.00) | 13 (37.10) | 0.021 | 4 (12.50) | 0.68 | |
| Suicide death (%) | 0 (0.00) | 7 (20.00) | 0.085 | 5 (15.60) | 0.17 | |
| Alcohol or drug death (%) | 0 (0.00) | 24 (68.60) | 3.32E-06 | 19 (59.40) | 0.000056 | |
This cohort subjects had primarily white non-Hispanic or African American ancestry. The exception was one control with mixed ancestry. P value for the proportion of ancestry tests the proportion of WNH vs non-WNH ancestry per group.
We performed transcriptome-wide association analyses comparing PTSD cases to the controls both overall and in men and women separately. Analyses included covariates for age, sex, post-mortem interval (PMI), estimates of cell proportion (Hagenauer et al., 2018), indicators of tissue quality (Jaffe et al., 2017), and sequencing-run ID (see Supplementary Methods for details). A false discovery rate (FDR) corrected p-value is reported (pcor), correcting over the analyses of the two different regions and analyses of the full cohort and stratified analyses (6 transcriptome-wide analyses). Follow-up analysis in PTSD-associated genes compared the depressed cases to controls, and overlap was examined between depression-associated and PTSD-associated genes. We performed cell-type enrichment analyses, gene-network analyses, confirmatory analyses of previously-implicated candidate genes, and upstream-regulator analysis. Finally, we compared our dlPFC results to those presented in Girgenti et al. Of the donors included in our dlPFC and vmPFC analyses, 50/94 (53.19%) and 46/87 (52.87%) respectively, were also included in the Girgenti cohort. We additionally compared results to the University of Pittsburgh Medical Center (UPMC) cohort, a subset of the Girgenti cohort which is independent of the cohort examined here. See the Supplementary Materials for a detailed description of the methods.
3. Results
3.1. Cell type proportions
We obtained cell type proportion estimates obtained using two different methods: BrainInABlender and CIBERSORTx. For cell types estimated using both methods, the correlation varied (Supplementary Table 4) and was very high for oligodendrocytes (r = 0.95) and astrocytes (r = 0.93), but lower for microglia (r = 0.45) where CIBERSORTx estimated the cell proportion for almost all samples to be 0. We observed two significant associations between cell-type proportion scores and PTSD using BrainInABlender (Supplementary Table 5): (a) lower microglia scores in the dlPFC of female PTSD cases (p = 0.0014), with a trend decrease for males (p = 0.052), and (b) lower endothelial cell scores in the vmPFC (p = 0.039), although we note that these associations would not survive a correction for multiple testing. A similar examination of cell type proportions estimated using CIBERSORTx did not reveal any PTSD-associated cell types (Supplementary Table 6). We used the cell type scores from BrainInABlender as covariates in subsequent analyses to control for cell population differences.
3.2. Transcriptome-wide differential expression
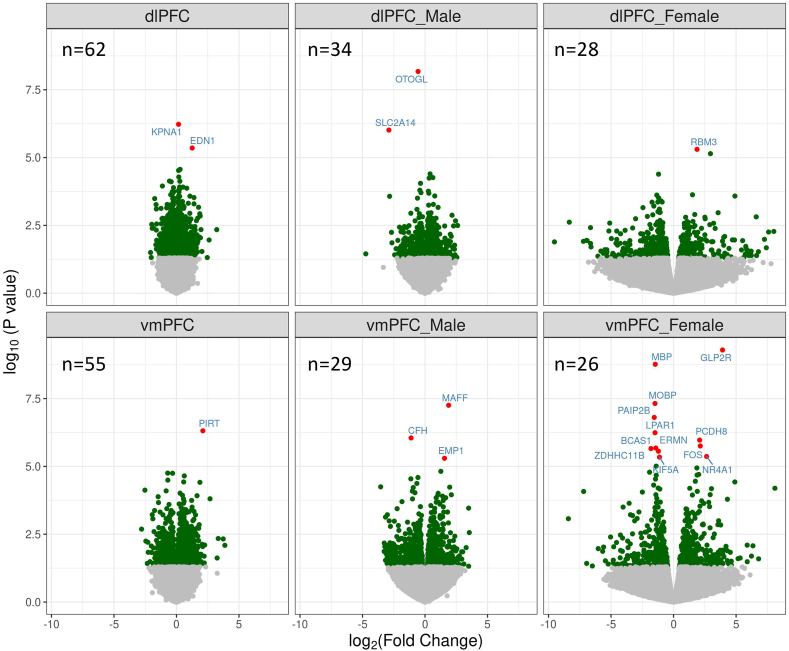
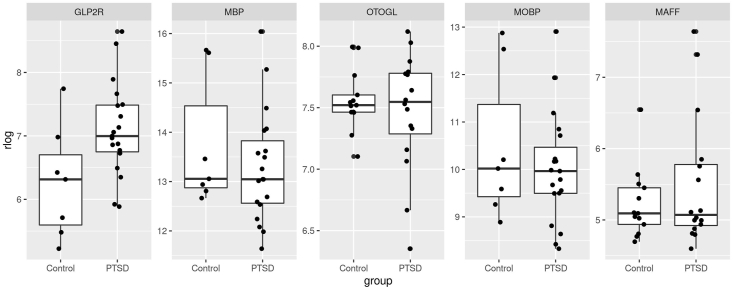
A total of 190,346 single-gene analyses were performed, including PTSD analyses in the dlPFC, vmPFC, and sex-stratified analyses within both regions. QQ-plots did not show evidence of inflation (Supplementary Fig. 1). We identified 21 genes associated with PTSD at the multiple-testing corrected significance level (FDR correction across all 190,346 analyses). The most significant associations were observed in female vmPFC (Table 3, Fig. 1, Supplementary Fig. 2). Boxplots for the top associated genes are presented in Fig. 2. The top result was an upregulation of glucose-related gene GLP2R in the vmPFC of female PTSD cases (logfc = 3.90, p = 5.10 × 10−10, pcor = 9.80 × 10−5). This association was nominally-significant in the full cohort, both in the dlPFC (logfc = 0.50, p = 0.042) and vmPFC (logfc = 0.67, p = 0.033), but not in the dlPFC in the women (p = 0.65; Fig. 3A; see Supplementary Tables 7 and 8 for subgroup analyses). We observed downregulation of several genes specifically in the vmPFC in female PTSD cases, for example (MBP, p = 1.7 × 10−9; MOBP, 4.80 × 10−8). There were several experiment-wide significant PTSD associations in men, including downregulation of OTOGL in the dlPFC (logfc = −0.55, p = 6.70 × 10−9, pcor = 0.00043) and downregulation of the immune gene CFH in the vmPFC (logfc = −1.1, p = 8.90 × 10−7, pcor = 0.017). The gene EDN1 was significantly upregulated in the dlPFC of PTSD cases (logfc = 1.3, p = 4.40 × 10−6, pcor = 0.046). Nominally significant upregulation of EDN1 was observed in both sexes, and in both the dlPFC and vmPFC. Follow-up analyses examined the effects of PTSD after controlling for potential confounders: suicide, opiates, SRI use, probable concussion/TBI, and smoking. These are summarized in Supplementary Table 9. All Table 3 associations remained significant (p < 0.05) after the inclusion of the potential confounder as a covariate, with one exception. The association between NR4A1 in the vmPFC of women PTSD cases was not significant in a model which included the effect of smoking (p = 0.077). In addition, we examined the potential confounding effect of ancestry. As PTSD status is confounded with ancestry in the female PTSD cases (see Supplementary Table 2), we could not include it as a covariate in the analyses. However, we examined the role of non-European ancestry in the Top genes (Table 3) for both the dlPFC and vmPFC by analyzing ancestry within each of the case groups (PTSD, depression, and controls) and then meta-analyzing the estimates across the three groups (see Supplementary Methods for details and Supplementary Table 10 for results). We found that two of the genes had corrected-significant associations with ancestry in the dlPFC (LPAR1 and BCAS1), and two had nominally significant associations with ancestry in the dlPFC (PAIP2B and ERMN). However, these four genes were associated with PTSD in the vmPFC, and these genes were not significantly associated with ancestry in the vmPFC, so it is unlikely the PTSD associations for these four genes could be ascribed to confounding by ancestry.
Table 3.
Significant gene expression difference in the analysis of PTSD cases and controls.
| Gene | Region | Sample | logfc | pvalue | pcor |
|---|---|---|---|---|---|
| GLP2R | vmPFC | Female | 3.92 | 5.12E-10 | 9.80E-05 |
| MBP | vmPFC | Female | −1.46 | 1.72E-09 | 0.00016 |
| OTOGLa | dlPFC | Male | −0.55 | 6.72E-09 | 0.00043 |
| MOBPa | vmPFC | Female | −1.49 | 4.80E-08 | 0.0021 |
| MAFF | vmPFC | Male | 1.89 | 5.58E-08 | 0.0021 |
| PAIP2B | vmPFC | Female | −1.55 | 1.56E-07 | 0.005 |
| PIRT | vmPFC | Complete | 2.11 | 4.87E-07 | 0.013 |
| LPAR1 | vmPFC | Female | −1.48 | 5.75E-07 | 0.013 |
| KPNA1 | dlPFC | Complete | 0.17 | 5.95E-07 | 0.013 |
| CFHa | vmPFC | Male | −1.11 | 8.93E-07 | 0.017 |
| SLC2A14 | dlPFC | Male | −2.88 | 9.71E-07 | 0.017 |
| PCDH8 | vmPFC | Female | 2.08 | 1.07E-06 | 0.017 |
| FOSa | vmPFC | Female | 2.14 | 1.76E-06 | 0.026 |
| BCAS1 | vmPFC | Female | −1.42 | 2.11E-06 | 0.028 |
| ZDHHC11B | vmPFC | Female | −1.81 | 2.20E-06 | 0.028 |
| ERMN | vmPFC | Female | −1.21 | 2.75E-06 | 0.033 |
| NR4A1 | vmPFC | Female | 2.64 | 4.31E-06 | 0.046 |
| EDN1a | dlPFC | Complete | 1.3 | 4.40E-06 | 0.046 |
| KIF5A | vmPFC | Female | −1.13 | 4.58E-06 | 0.046 |
| RBM3 | dlPFC | Female | 1.88 | 4.98E-06 | 0.046 |
| EMP1 | vmPFC | Male | 1.56 | 5.03E-06 | 0.046 |
Indicates that the association was supported in the data from the dlPFC in Girgenti et al. See Results: Comparison to Prior Results section for details.
Fig. 1.
Volcano plot of PTSD-associated gene expression. Nominally significant (p < 0.05) genes are in green, and corrected significant genes are in red. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 2.
Boxplots of the rlog normalized expression for PTSD cases and controls for the five most significant PTSD-associated genes.
Fig. 3.
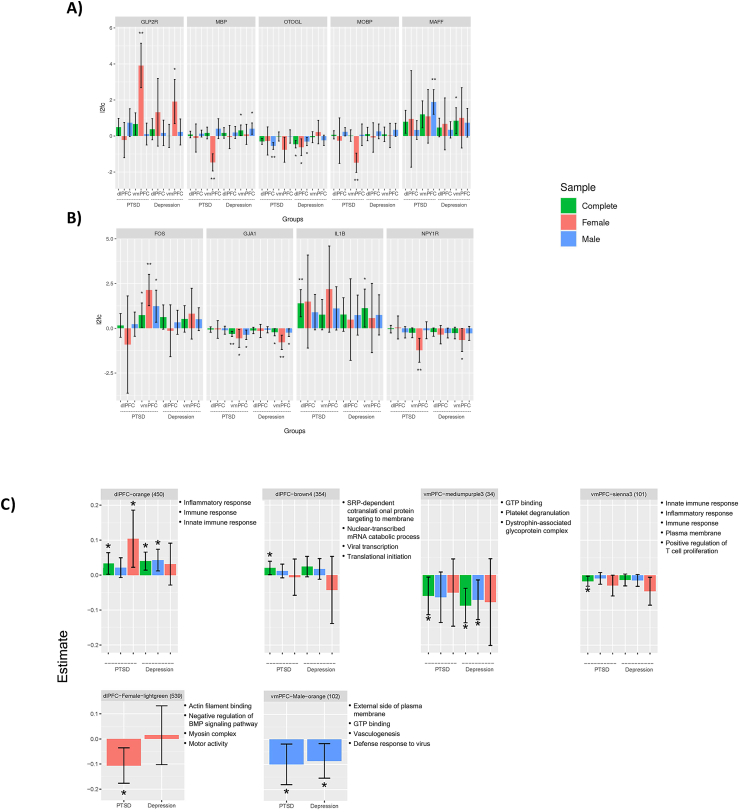
Effect sizes for PTSD and Depression for A) the five most significant PTSD-associated genes; B) candidate genes significant at the candidate-gene adjusted level (pcor-candidate<0.05); and C). Gene networks significantly associated with PTSD.
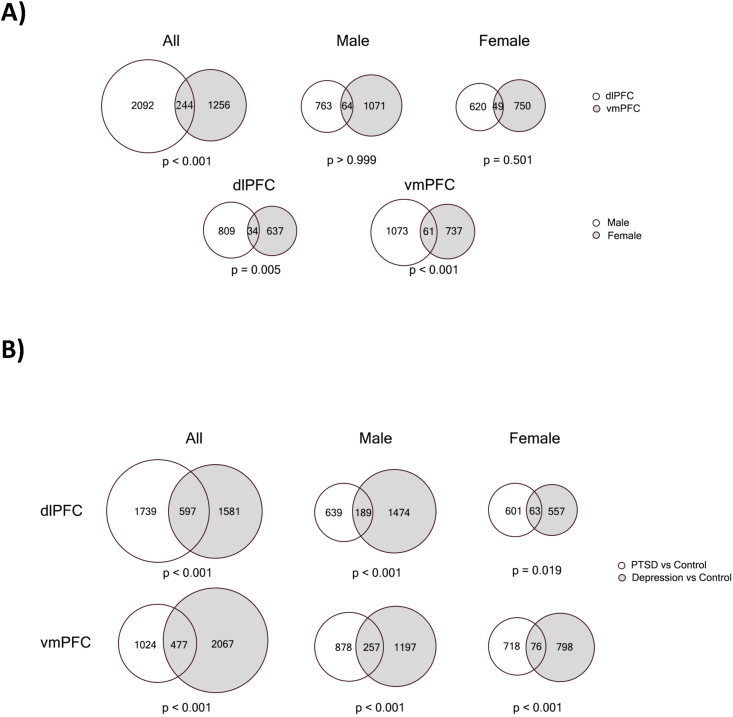
Fig. 4A depicts the overlap in PTSD-associated genes for the PFC regions and for men versus women. Of the genes that were nominally significant in the dlPFC, 10.45% were significant in the vmPFC. While this indicates that many associations are regionally distinct, simulations indicated that there was a greater overlap than would be expected by chance (p < 0.001).
Fig. 4.
Venn diagrams of the overlap of nominally significant (p < 0.05) PTSD-associated genes A) across brain regions and sex and B) Across PTSD and depression. Note: p-values are based on permutation test with 1000 replicates.
Next, we examined specificity of the PTSD-associated genes through a case-control analysis of depressed cases (n = 32 in both regions) vs controls (in dlPFC n = 24, in vmPFC n = 20). Several of the top PTSD-associated genes were nominally associated with depression (see Supplementary Tables 10 and 11 for dlPFC and vmPFC results, Fig. 3A, and Supplementary Fig. 2). For example, the upregulation of GLP2R in the vmPFC of women with PTSD was mirrored in the vmPFC of women with depression (logfc = 1.90, p = 0.0022). Upregulation of EDN1 observed in the dlPFC and vmPFC of PTSD cases was evident in the dlPFC of depression cases (logfc = 0.98, p = 0.0024), in both men and women (p = 0.03 and p = 0.01 respectively), and in the vmPFC of women with depression (logfc = 2.1, p = 0.01). Overlap between PTSD- and depression-associated genes is shown in Fig. 4B. In the dlPFC, 25.56% of the nominally significant PTSD associated genes were also associated with depression (p < 0.01). In the vmPFC, 31.78% of the nominally significant PTSD associated genes were also associated with depression (p < 0.001).
3.3. Overrepresentation analysis
Overrepresentation analysis of top PTSD-associated genes yielded enriched GO terms in the vmPFC in the overall and sex-stratified analyses and in the dlPFC of women (Supplementary Table 15). Several pathways were noteworthy, including GO:0006950 response to stress (pcor = 0.0043) and GO:0080,134 regulation of response to stress (pcor = 0.0026), which were significantly overrepresented in the vmPFC PTSD-associated genes. The most significantly enriched pathway in the female vmPFC results was the immune-related GO:0042,611 MHC protein complex (pcor = 4.90 × 10−8).
We next performed overrepresentation analysis of PTSD associated genes in cell type markers. A GSEA-based analysis yielded multiple enrichments (pcor<0.05) including enrichment for excitatory neuron, endothelial cell, oligodendrocyte, oligodendrocyte precursor cell, and pericyte markers (Supplementary Table 16). A GOseq-based analysis (Supplementary Table 17) identified nominally significant enrichment of endothelial cell markers and oligodendrocyte markers and corrected-significant associations for excitatory neuron and pericyte markers.
3.4. PTSD candidate genes
Candidate-gene analysis of previously implicated genes (a literature-review based list of 143 PTSD genes presented in the Supplement of (Huckins et al., 2020)) yielded four associations that exceeded a candidate gene significance threshold (pcor-candidate, corrected for 143 genes; see Fig. 3B, Supplementary Tables 13 and 14): FOS (also a top gene) was upregulated in the vmPFC of women with PTSD (logfc = 2.14, p = 1.76 × 10−6, pcor-candidate = 0.0011), GJA1 which was downregulated in the vmPFC (logfc = −0.31, p = 1.66 × 10−5, pcor-candidate = 0.0053), IL1B was upregulated in the dlPFC (logfc = 1.40, p = 0.00028, pcor-candidate = 0.031), and NPY1R was downregulated in the vmPFC of women with PTSD (logfc = −1.20, p = 0.00032, pcor-candidate = 0.017).
3.5. Gene networks
Weighted gene co-expression network analysis (WGCNA) was performed in the full cohort to generate dlPFC and vmPFC co-expression networks. In the sex-stratified cohorts, WGCNA generated dlPFC-Male, dlPFC-Female, vmPFC-Male, and vmPFC-Female networks. Identified networks are labeled with an arbitrary color name. No networks were associated with PTSD at a multiple testing-corrected significance level. Two dlPFC and two vmPFC networks were nominally significantly associated with PTSD (Supplementary Table 18). Analysis of the sex-stratified networks yielded one dlPFC-Female and one vmPFC-Male PTSD-associated network. Fig. 3C summarizes the associations of these networks with PTSD and depression. The GO terms and cell-type markers associated with each network are presented in Supplementary Tables 19 and 20 respectively. The dlPFC-orange network and vmPFC-sienna3 networks are discussed below, as they were the dlPFC and vmPFC networks which displayed overlapping GO terms, indicating an overlap in function. The remainder of the PTSD-associated networks are described in the Supplementary Results.
The dlPFC-orange network contains 122 genes associated (p < 0.05) with PTSD in the dlPFC—the most in any of the PTSD associated networks. Of these, 120 were upregulated in PTSD cases, including the experiment-wide significant gene EDN1. This network contains 8 genes from our PTSD candidate gene list including FKBP5, FOS, IL1B, OXTR, EPHB4, S100A10, SERPINA1, and VCL. Among these, two were associated with PTSD in the dlPFC: 1L1B (logfc = 1.41, p = 2.80 × 10−4) and VCL (logfc = 0.20, p = 0.0017). The dlPFC-orange network is enriched for several immune related GO terms (Supplementary Table 18), including GO:0006954 inflammatory response (pcor = 6.38 × 10−18), GO:0006955 immune response (pcor = 6.39 × 10−10), and GO:0045087 innate immune response (pcor = 5.86 × 10−07). Enrichment analysis (Supplementary Table 17) indicated that dlPFC-orange is enriched for endothelial (pcor = 1.60 × 10−12) and pericyte markers (pcor = 0.023). Follow-up analyses indicated that the dlPFC-orange network is also upregulated in depression cases (Fig. 3C).
The vmPFC-sienna3 network contains 6 genes that were nominally significant in the vmPFC analysis, all but one of which were downregulated in the PTSD cases. The most significant was the HLA-DOA gene (logfc = −0.61, p = 0.0094). In follow-up analyses, the vmPFC-sienna3 network was not significant in either of the sex-stratified analyses nor was it significantly associated with depression (Fig. 3C). Cell-marker enrichment analysis (Supplementary Table 20) indicated that vmPFC-sienna3 is strongly enriched for microglial markers (pcor = 7.13 × 10−29). Although they only have 5 genes in common, vmPFC-sienna3 is enriched for the same immune-related GO terms as dlPFC-orange: inflammatory response (pcor = 1.64 × 10−12), immune response (pcor = 9.89 × 10−10), and innate immune response (pcor = 2.74 × 10−13). Comparing the immune related genes in both networks (Supplementary Table 19), there are proportionally more interleukin and tumor necrosis factor genes in dlPFC-orange, and there are more compliment factor and human leukocyte antigen genes in vmPFC-sienna3. Hence the dlPFC-orange and vmPFC-sienna3 networks appear to characterize different aspects of the immune response.
3.6. Upstream regulator analysis
Upstream regulator analyses were performed on the set of dlPFC and vmPFC results to identify genes potentially underlying the observed associations (Supplementary Table 21). See Supplementary Methods for analysis details. In the dlPFC, 18 upstream regulators were identified. Interestingly, the immune-related PTSD candidate gene IL1B is predicted to be an upstream regulator in the dlPFC, where it is predicted to regulate the top gene EDN1. An examination of the targets of the remaining predicted upstream regulators indicates that IL1B may be central to the regulatory network, as it is a target of 11 of the remaining 17 upstream regulators. In the vmPFC, 35 upstream regulators were identified. The top upstream regulators are TNF, IL1B, and NR3C1. The majority of the upstream regulators in the vmPFC (25 of the 35 regulators) target FOS. EDN1 is also well represented in the target lists of the dlPFC and vmPFC upstream regulators, being the target of 7 dlPFC regulators and vmPFC 11 regulators. There was a greater overlap in the upstream regulators in the two regions than the individual gene expression results. Of the 18 dlPFC regulators, half are vmPFC regulators, compared to only 10% of nominally significant genes in the dlPFC being observed in the vmPFC. Finally, we also examined upstream regulators of the significant PTSD-associated networks in the dlPFC and vmPFC. There were four predicted upstream regulators of dlPFC-orange, including IL1B (Supplementary Table 21), which was embedded in a master of a regulatory network (Supplementary Fig. 3) including other immune (e.g., TNF and NFKB) and stress (e.g., NR3C1) regulators.
3.7. Comparison to Prior Results
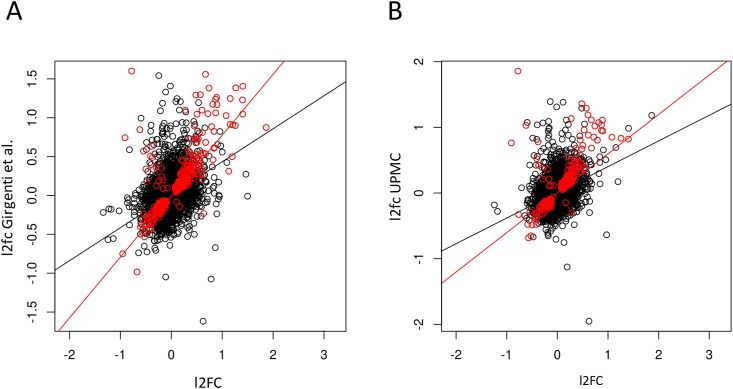
We compared our dlPFC findings to (a) results from the whole Girgenti et al. cohort, which partially overlaps the cohort studied here, and (b) the UPMC cohort, a subset of the Girgenti cohort which is entirely independent of the cohort examined here. We note that many of the dlPFC PTSD associations highlighted in Girgenti et al. were also observed in our dlPFC results including GADD45b (logfc = 0.36, p = 0.017), UBA7 (logfc = 0.15,p = 0.0076), and ELFN1 in women (logfc = −0.91, p = 0.030). Of genes that were nominally significant in our dlPFC analyses, 17% were nominally significant in the Girgenti et al. cohort and 17% were significant in the UPMC cohort—significantly greater than would be expected by chance (p < 0.001). However, this may underestimate the degree of correspondence between the cohorts. When we examined the logfc values in our analysis and the Girgenti cohort (Fig. 5), there was significant overall correlation (r = 0.40, p < 2.2 × 10−16) which was higher in the genes which were nominally significantly associated in both groups (r = 0.75, p < 2.2 × 10−16). When we compared our dlPFC logfc values to the UPMC cohort there was a significant correlation overall (r = 0.39, p < 2.2 × 10−16), and higher correlation in the genes that were significant in both cohorts (r = 0.66, p < 2.2 × 10−16). When we compared the direction of effects in our results with the UPMC cohort, we found that for the 272 genes that were nominally significant in both analyses, the direction of effect was the same in 257 of them (94.5%), much more than expected by chance (p < 2.2 × 10−16). We also examined our top associations (Table 3) in the Girgenti and UPMC cohort. The most significant dlPFC association that we found, with the gene OTOGL, was similarly downregulated in the dlPFC in the Girgenti (logfc = −0.20, p = 0.0059) and UPMC cohorts (logfc = −0.32, p = 0.00036). The observed female-specific increase in GLP2R in dlPFC was similarly up-regulated only in females in Girgenti et al. (logfc = 0.46, p = 0.036). The observed association with EDN1 was also significant in the Girgenti (logfc = 0.90, p = 8.79 × 10−7) and UPMC cohort (logfc = 0.83, p = 0.0037) with a consistent direction of effect. Similarly, IL1B was corrected significant in Girgenti et al. with the same direction of effect (logfc = 1.23, p = 0.0037), but was not reported in the UPMC cohort. While Girgenti et al. did not have data on gene expression in the vmPFC, several of the genes that were associated with PTSD in the our vmPFC analyses were nominally significant in the UPMC dlPFC analysis including FOS (p = 0.0036), MAFF (p = 0.0046), and CFH (p = 0.018). Finally, we examined the data from a qPCR candidate-gene study of immune genes performed in a subset (n = 50) of the dlPFC tissue examined in this study. These analyses indicate a consistent estimate of the effect of IL1B effect size and a correlation between IL1B expression measured by RNAseq and qPCR (see Supplementary Materials for details).
Fig. 5.
Comparing logfc from our dlPFC analyses to A) those presented in Girgenti et al. (2021). and B) the UPMC subset of that study which is independent of our cohort. Genes that are nominally significant (p < 0.05) in both studies are in red. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
In this study, we performed a transcriptome-wide and candidate-gene RNA seq analysis in PTSD versus control cases in the dlPFC and vmPFC. We also examined gene co-expression networks and neural cell types proportions for association with PTSD. Glucagon Like Peptide 2 Receptor (GLP2R) was the most robustly affected gene transcript in PTSD, selectively up-regulated in the vmPFC of women. GLP2R is highly expressed in the cerebral cortex where it is involved in the regulation of appetite and glucose homeostasis (https://www.proteinatlas.org/ENSG00000065325-GLP2R/tissue). (Guan, 2014). Recently, GLP2R expression was observed to be higher in the dlPFC and directly associated with body mass index in mood disorder and psychiatric cases (Mansur et al., 2019).
Several differentially expressed genes are activated in cortical regions in fear conditioning paradigms. FOS (also known as c-Fos), which was selectively elevated in the vmPFC of female PTSD cases vs controls, is often used as an indirect measure of neuronal activity (Bullitt, 1990). Zhang et al. (2002) showed that c-Fos regulates BDNF (Zhang et al., 2002). It is upregulated in regions of the hippocampus, amygdala, and cortex of mice exposed to a fear conditioning paradigm (Milanovic et al., 1998). We also observed upregulation of the gene NR4A1 (which encodes Nerve growth factor IB) in the vmPFC of female PTSD cases. NR4A1 is elevated in the hippocampus (von Hertzen and Giese, 2005), cortex and amygdala (Malkani and Rosen, 2000) during shock memory consolidation. Although it should be noted that the NR4A1 association was not significant after controlling for smoking status. Considering the suspected connections between failed fear extinction and the maintenance of PTSD symptoms, additional study of these genes is warranted.
Several oligodendrocyte-related genes were downregulated in the vmPFC of women with PTSD, including myelin basic protein (MBP), myelin associated oligodendrocyte basic protein (MOBP), and ermin (ERMN). Deficits in myelin structure and function have been previously implicated in PTSD (Chao et al., 2015). Restraint stress in mice has been associated with downregulation of oligodendrocyte genes, including MBP and ERMN (Chu et al., 2016). The downregulation of these genes is consistent with alterations in myelination and oligodendrocyte function in the mPFC observed in social-defeat stress models in mice (Bonnefil et al., 2019). However, our analyses did not indicate reduced ratios of oligodendrocyte cells in female vmPFC cases (p = 0.55), and the PTSD association is observed in models controlling for the estimated oligodendrocyte proportions, which suggests that oligodendrocytes are producing lower amounts of these proteins in PTSD cases. We also found evidence of downregulation of endothelial cells in the vmPFC and microglia in the dlPFC, which indicates that these cell types may be disrupted in PTSD cases, although it should be noted that these associations would not survive a multiple testing correction and further corroboration is necessary. Enrichment analysis of cell markers implicated excitatory neurons, pericytes, and endothelial cells. We note that our justifications for selecting these areas for study were prior associations between PTSD, cortical thickness, and connectivity in these cortical areas (Reuveni et al., 2016; Sadeh et al., 2016; Miller et al., 2015; Holmes et al., 2018). It is possible that these gene expression differences are related to or contribute to these structural associations, as myelination has been linked to connectivity (Hunt et al., 2016), and cortical thinning has been linked to reduced proportions of several cell types including microglial cells (Vidal-Pineiro et al., 2020). However, more work is needed to directly link the observed associations to specific neuropathological and imaging findings. Nevertheless, these findings provide strong evidence of oligodendrocyte and myelination gene involvement in PTSD, especially in the female vmPFC. They also provide evidence for the involvement of excitatory neurons and cell types involved in regulation of the blood-brain barrier and neuroprotection in PTSD.
Our study indicates a role for the immune system and cytokines based on RNA measured in brain tissue, produced by brain cells. Indeed, microglia, neurons, and astrocytes can express cytokine genes and produce cytokines proteins which can impact neurotransmitter expression and functioning (Miller et al., 2013). Neuroinflammation is a process that can be either adaptive, as when responding to injury and promoting recovery, or maladaptive, e.g. as neuroinflammation is also associated with processes such as Alzheimer's disease (DiSabato et al., 2016). It has been known that psychological stress can activate the immune system, and numerous studies have found evidence of peripheral inflammation in subjects with PTSD and anxiety (Michopoulos et al., 2017; Tursich et al., 2014). Peripheral immune system activation can cause a corresponding inflammatory response in microglia (the primary immune cell in the brain) which can have behavioral effects and impact neuronal function (Marin and Kipnis, 2017). Cytokine signaling in the brain and microglia also play a role in absence of a pathogen, and impact tissue health and maintenance (Marin and Kipnis, 2017) Microglia have also been implicated in brain development and learning, for example through synaptic pruning, which continues into adulthood (Marin and Kipnis, 2017). The relationship between PTSD and inflammation may be complex, and multiple lines of inquiry have pointed to ways that anxiety and stress may influence inflammation, both in the periphery and the brain (e.g through the NFKB pathway (Koo et al., 2010)), and also ways in which the immune system could itself play a role in behavior and mood leading to psychiatric disorder susceptibility (e.g. through IL6-mediated susceptibility to stress-induced depression (Hodes et al., 2014) or inherited major histocompatibility complex variation and its role in susceptibility to schizophrenia (Ripke et al., 2013)).
The present study also yielded important new information about the relationship between PTSD and immune system genes. While a vmPFC immune-related network enriched for microglia markers was downregulated in PTSD cases, we additionally identified a dlPFC immune-related WGCNA network that was largely up-regulated in PTSD cases. Other immune-related transcripts from the top 20 PTSD-associated genes included KPNA1 (upregulated in the dlPFC) and Complement Factor H (CFH; downregulated in the vmPFC). CFH is particularly intriguing in light of the recent awareness of roles for complement pathways and microglia in synaptic remodeling (Stephan et al., 2012). Altered complement expression and cortical dendritic retraction is observed in preclinical models of trauma-associated (shock) exposure with lasting contextual fear and exaggerated acoustic startle responses (Smith et al., 2019). CFH is selectively expressed in human endothelial cells, which further evidence of endothelial cell involvement in PTSD (Zhang et al., 2014). The NR4A1 gene, involved in memory formation and a top 20 up-regulated gene, is also induced during immune responses to lipopolysaccharide (Pei et al., 2005). The immune response to lipopolysaccharide pathway has recently been implicated in an epigenome-wide association study of PTSD (Logue et al., 2020) and is a popular animal model for stress. Overrepresentation analysis of PTSD associated genes in the female vmPFC implicated the MHC protein complex. We observed nominally significant evidence for reductions in microglial-specific transcripts in both male and female PTSD cases, with females being more robustly affected. Additionally, two of the PTSD-associated gene networks generated by WGCNA were enriched for several immune-related GO terms. Interestingly, although the same GO terms were implicated by these two networks, different genes were associated with PTSD in each of these networks. Moreover, the association went in opposite directions, with PTSD-associated genes in the dlPFC-orange network upregulated in the dlPFC of PTSD cases, while PTSD-associated genes in the vmPFC-sienna3 network were downregulated in the vmPFC of PTSD cases. The immune related genes in each network were distinct, with more TNF and IL genes in dlPFC-orange (including IL1B), and more complement factor and human leukocyte antigen genes in vmPFC-sienna3. This suggests important differences in immune system functioning in these regions and differences in the role immune genes play in PTSD across different regions of the brain. The involvement of immune system genes in both regions is reinforced by the upstream regulator analysis. IL1B is noted as an upstream regulator of PTSD-associated expression differences in the dlPFC, mirroring a prior finding of IL1B as an upstream regulator of PTSD in subjects with high BMI in the dlPFC (Stone et al., 2021). IL1B and TNF were the top upstream regulators of PTSD associated genes in the vmPFC. These data add to a robust line of evidence associating IL1B and TNF to PTSD in human patients and preclinical rodent studies of stress- and fear-related behaviors (Jones et al., 2015; Hovhannisyan et al., 2017; Hussein et al., 2017; Bruenig et al., 2017). Hence, while the associated genes in the dlPFC and vmPFC are distinct, they are both regulated by and include immune-related genes. The bidirectionality (both up and down regulation) of these responses in different regions suggest a complex, rather than a simple dysregulation of immune pathways in PTSD. As our investigation is cross sectional, we cannot infer whether the inflammatory marker associations with PTSD are a risk factor for PTSD, a consequence of PTSD, or both.
Finally, our results showed strong correspondence between the present dlPFC results and those of Girgenti et al. suggesting correspondence between the two studies, which used different labs, different sequencing depth, independent analysis pipelines, different covariates while using adjacent tissue sections from the same hemisphere. Results from the UPMC subset of the Girgenti et al. cohort provides independent replication for OTOGL and EDN1, two of the top associated dlPFC genes in our sample. Finally, the high correlation between effect size estimates (r = 0.66) and the same direction of effect observed in 95% of nominally significant genes across the two independent cohorts support the existence of a consistent and replicable PTSD gene association signature in the dlPFC that extends beyond the genes exceeding the multiple testing correction thresholds for both studies. Additionally, our examination of overlap of PTSD associated genes mirror findings of Girgenti et al. (2021), which indicate heterogeneity of associations by sex and tissue, and indicates that some PTSD and depression associated genes are distinct.
4.1. Limitations
Our study had several limitations. Several of the subjects that were sequenced had relatively lower RIN scores (as low as 3.3 for vmPFC). Although these numbers were less than ideal, these samples were only used when they passed the QC filtering procedure of the sequencing center. Additionally, all of our analysis models included quality SVAs as covariates to control for tissue degeneration. As is common in human postmortem studies, there was a high level of potentially significant comorbidity in the psychiatric groups. Common co-morbidity between PTSD and depression could account for overlapping findings. This study is cross sectional in nature, and therefore we cannot infer whether the observed associations are causes of PTSD, consequences of PTSD, or both. However, as we examined brain tissue, the associated genes may be more centrally linked to PTSD pathogenesis than those implicated in studies of peripheral tissue. It is likely that other regions such as the hippocampus and the amygdala also play critical roles in the development and maintenance of PTSD, and that different genes will be implicated in these regions. Additionally, we note that while the novelty of our results is enhanced by examining gene expression in the vmPFC, which wasn't examined in the Girgenti et al. study, this novelty implies that we do not have replication data for that region. Therefore, our vmPFC results must be considered provisional until additional corroborating vmPFC data is obtained. This is especially true for subgroup analyses (e.g. men/women) due to the smaller sample sizes for those analyses. In addition, while our cohort is primarily white non-Hispanic, there is a large proportion of non-European ancestry within the female controls. This complicated our efforts to test whether associations in the stratified analyses were due to ancestry. However, follow-up analyses indicated that only four of our top (Table 3) genes had evidence that gene expression was associated with ancestry (PAIP2B, LPAR1, BCAS1, and ERMN; see Supplementary Table 10), and these ancestry associations were observed in the dlPFC, whereas female-specific PTSD associations were observed primarily in the vmPFC (Table 3). Nevertheless, we think that this is another reason that female-specific results should currently be considered provisional.
5. Conclusion
The present data support association between immune system/inflammation in PTSD repeatedly seen in peripheral tissues (Bellavance and Rivest, 2014), but with a complex pattern of association with evidence for regional and gender interactions indicating complex immune/inflammatory dysregulation in PTSD, with IL1B emerging as a key modulator. The genes identified in this study have the potential to serve as biomarkers of PTSD, targets of treatment, and may be useful in guiding other gene expression and DNAm studies of PTSD. Additionally, they offer important new clues about immune system action in PTSD and heterogeneity of association across the different regions of the brain.
CRediT authorship contribution statement
Mark W. Logue: Conceptualization, Funding acquisition, Formal analysis, Writing – original draft, Writing – review & editing. Zhenwei Zhou: Formal analysis, Conceptualization, Data curation, Visualization, Writing – original draft. Filomene G. Morrison: Data curation, Validation, Writing – review & editing. Erika J. Wolf: Conceptualization, Writing – original draft, Writing – review & editing. Nikolaos P. Daskalakis: Conceptualization, Formal analysis, Methodology, Writing – original draft, Writing – review & editing. Christos Chatzinakos: Formal analysis. Foivos Georgiadis: Formal analysis. Adam T. Labadorf: Conceptualization, Writing – review & editing. Matthew J. Girgenti: Validation, Writing – review & editing. Keith A. Young: Writing – review & editing. Douglas E. Williamson: Writing – review & editing. Xiang Zhao: Formal analysis. Jaclyn Garza Grenier: Investigation, the Traumatic Stress Brain Research Group, Resources. Bertrand Russell Huber: Resources, Conceptualization, Investigation, Writing – review & editing. Mark W. Miller: Conceptualization, Resources, Funding acquisition, Writing – original draft, Writing – review & editing.
Acknowledgements
The Traumatic Stress Brain Research Group are Matthew Friedman, M.D., Ph.D. – PTSD BB, Neil Kowall, M.D, Christopher Brady, Ph.D., Ann McKee, M.D., Thor Stein, M.D., Ph.D., Bertrand Huber, M.D., Ph.D., Paul Holtzheimer, M.D., Victor Alvarez, M.DDavid Benedek, M.D., Robert J. Ursano, MD, Douglas Williamson, PhD, Brian Marx, PhD, Terence M. Keane, PhD, Dianne Cruz, M.S, Keith A. Young, PhD, John Krystal, MD, Deborah Mash, MD, Melanie Hardegree, RN, William Scott, Ph.D, David Davis Ph., Matthew Girgenti, and Gayle Serlin, PhD.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ynstr.2021.100398.
Funding
This work was funded by I01BX003477, a VA BLR&D grant to MWL, 1R03AG051877, 1R21AG061367-01, and 1I01CX001276-01A2 to EJW, 5T32MH019836-18 to FGM, R21MH102834 to MWM. Genotype data for the Brain Bank Cohort was generated with the support of resources and of facilities at the Pharmacogenomics Analysis Laboratory (Research Service, Central Arkansas Veterans Healthcare System, Little Rock, Arkansas), a core research laboratory funded by the Cooperative Studies Program, Research and Development, Department of Veterans Affairs. NPD and CC were supported by NIMH P50MH115874 and R01MH117292. NPD was also supported by an appointed KL2 award from Harvard Catalyst | The Harvard Clinical and Translational Science Center (NCATS KL2TR002542, UL1TR002541), Brain & Behavior Research FoundationBrain & Behavior Research Foundation NARSAD Young Investigator Grant, and McLean Hospital Jonathan Edward Brooking Mental Health Research Fellowship. VA salary funding and/or resources were obtained from NCPTSD/VABHS (MWL, FGM, EJW, XZ, BRH, MWM), Durham VAMC (DEW, DC), CTVHCS (KAY) and VACHCS (MJG).
Disclosures
FGM participated in this study while a postdoc at Boston University/VA Boston, but is currently an employee of BlackThorn Therapeutics. Over the past 3 years, NPD has held a part-time paid position at Cohen Veteran Biosciences, has been a consultant for Sunovion Pharmaceuticals and is on the scientific advisory board for Sentio Solutions for unrelated work. The remaining individually-named authors (MWL, ZZ, EJW, CC, FG, ATL, MJG, KAY, DEW, XZ, JGG, BRH, and MWM), have nothing to disclose. From the members of the Traumatic Stress Brain Research Group, PH consulting fees from Abbott (for DBS work) and royalties from UpToDate and Oxford University Press. JK has consulting agreements (less then $10,000 per year) with the following: AstraZeneca Pharmaceuticals, Biogen, Idec, MA, Biomedisyn Corporation, Bionomics, Limited (Australia), Boehringer Ingelheim International, COMPASS Pathways, Limited, United Kingdom, Concert Pharmaceuticals, Inc., Epiodyne, Inc. EpiVario, Inc., Heptares Therapeutics, Limited (UK), Janssen Research & Development, Otsuka America, Pharmaceutical, Inc., Perception Neuroscience Holdings, Inc., Spring Care, Inc., Sunovion Pharmaceuticals, Inc., Takeda Industries, Taisho Pharmaceutical Co., Ltd. JHK. serves on the scientific advisory boards of Bioasis Technologies, Inc., Biohaven Pharmaceuticals, BioXcel Therapeutics, Inc. (Clinical Advisory Board), BlackThorn Therapeutics, Inc., Cadent Therapeutics (Clinical Advisory Board), Cerevel Therapeutics, LLC., EpiVario, Inc., Lohocla Research Corporation, PsychoGenics, Inc., is on the board of directors of Inheris Biopharma, Inc. has stock options with Biohaven Pharmaceuticals Medical Sciences, BlackThorn Therapeutics, Inc., EpiVario, Inc., and Terran Life Sciences and is editor of Biological Psychiatry with income greater then $10,000. The remaining members (excluding those authors already named, MF, NK, CB, AM, TS, VA, DB, RJU, BM, TMK, DC, DM, MH, WS, DD, and GS) have nothing to disclose. The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of the Department of Veterans Affairs, the Department of Defense, NIMH, or the U.S. government.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Bellavance M.A., Rivest S. The HPA - immune Axis and the immunomodulatory actions of glucocorticoids in the brain. Front. Immunol. 2014;5:136. doi: 10.3389/fimmu.2014.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S. PTSD is associated with neuroimmune suppression: evidence from PET imaging and postmortem transcriptomic studies. Nat. Commun. 2020;11:2360. doi: 10.1038/s41467-020-15930-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnefil V. Region-specific myelin differences define behavioral consequences of chronic social defeat stress in mice. Elife. 2019;8 doi: 10.7554/eLife.40855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruenig D. Genetic and serum biomarker evidence for a relationship between TNFα and PTSD in Vietnam war combat veterans. Compr. Psychiatr. 2017;74:125–133. doi: 10.1016/j.comppsych.2017.01.015. [DOI] [PubMed] [Google Scholar]
- Bullitt E. Expression of c-fos-like protein as a marker for neuronal activity following noxious stimulation in the rat. J. Comp. Neurol. 1990;296:517–530. doi: 10.1002/cne.902960402. [DOI] [PubMed] [Google Scholar]
- Chao L.L., Tosun D., Woodward S.H., Kaufer D., Neylan T.C. Preliminary evidence of increased hippocampal myelin content in veterans with posttraumatic stress disorder. Front. Behav. Neurosci. 2015;9:333. doi: 10.3389/fnbeh.2015.00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu X. 24-hour-restraint stress induces long-term depressive-like phenotypes in mice. Sci. Rep. 2016;6 doi: 10.1038/srep32935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiSabato D.J., Quan N., Godbout J.P. Neuroinflammation: the devil is in the details. J. Neurochem. 2016;139(Suppl. 2):136–153. doi: 10.1111/jnc.13607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlop B.W., Wong A. The hypothalamic-pituitary-adrenal axis in PTSD: pathophysiology and treatment interventions. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2019;89:361–379. doi: 10.1016/j.pnpbp.2018.10.010. [DOI] [PubMed] [Google Scholar]
- Friedman M.J. VA's national PTSD Brain Bank: a national resource for research. Curr. Psychiatr. Rep. 2017;19:73. doi: 10.1007/s11920-017-0822-6. [DOI] [PubMed] [Google Scholar]
- Gelernter J.A.-O. Genome-wide association study of post-traumatic stress disorder reexperiencing symptoms in >165,000 US veterans. Nat. Neurosci. 2019;22:1394–1401. doi: 10.1038/s41593-019-0447-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girgenti M.J. Transcriptomic organization of the human brain in posttraumatic stress disorder. Nat. Neurosci. 2021;24:24–33. doi: 10.1038/s41593-020-00748-7. [DOI] [PubMed] [Google Scholar]
- Guan X. The CNS glucagon-like peptide-2 receptor in the control of energy balance and glucose homeostasis. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2014;307:R585–R596. doi: 10.1152/ajpregu.00096.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagenauer M.H. Inference of cell type content from human brain transcriptomic datasets illuminates the effects of age, manner of death, dissection, and psychiatric diagnosis. PloS One. 2018;13 doi: 10.1371/journal.pone.0200003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley C.A., Phelps E.A. Changing fear: the neurocircuitry of emotion regulation. Neuropsychopharmacology. 2010;35:136–146. doi: 10.1038/npp.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodes G.E. Individual differences in the peripheral immune system promote resilience versus susceptibility to social stress. Proc. Natl. Acad. Sci. U. S. A. 2014;111:16136–16141. doi: 10.1073/pnas.1415191111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes S.E. Cerebellar and prefrontal cortical alterations in PTSD: structural and functional evidence. Chronic Stress (Thousand Oaks) 2018:2. doi: 10.1177/2470547018786390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovhannisyan L., Stepanyan A., Arakelyan A. Genetic variability of interleukin-1 beta as prospective factor from developing post-traumatic stress disorder. Immunogenetics. 2017;69:703–708. doi: 10.1007/s00251-017-1016-4. [DOI] [PubMed] [Google Scholar]
- Huckins L.M. Analysis of genetically regulated gene expression identifies a prefrontal PTSD gene, SNRNP35, specific to military cohorts. Cell Rep. 2020;31 doi: 10.1016/j.celrep.2020.107716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt B.A.E. Relationships between cortical myeloarchitecture and electrophysiological networks. Proc. Natl. Acad. Sci. U. S. A. 2016;113:13510–13515. doi: 10.1073/pnas.1608587113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussein S., Dalton B Fau - Willmund G.D., Willmund Gd Fau - Ibrahim M.A.A., Ibrahim Maa Fau - Himmerich H., Himmerich H. A systematic review of tumor necrosis factor-α in post-traumatic stress disorder: evidence from human and animal studies. Psychiatr. Danub. 2017;29:407–420. doi: 10.24869/psyd.2017.407. [DOI] [PubMed] [Google Scholar]
- Jaffe A.E. qSVA framework for RNA quality correction in differential expression analysis. Proc. Natl. Acad. Sci. U. S. A. 2017;114:7130–7135. doi: 10.1073/pnas.1617384114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M.E., Lebonville C.L., Barrus D., Lysle D.T. The role of brain interleukin-1 in stress-enhanced fear learning. Neuropsychopharmacology. 2015;40:1289–1296. doi: 10.1038/npp.2014.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo J.W., Russo S.J., Ferguson D., Nestler E.J., Duman R.S. Nuclear factor-kappaB is a critical mediator of stress-impaired neurogenesis and depressive behavior. Proc. Natl. Acad. Sci. U. S. A. 2010;107:2669–2674. doi: 10.1073/pnas.0910658107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logue M.W. An epigenome-wide association study of posttraumatic stress disorder in US veterans implicates several new DNA methylation loci. Clin. Epigenet. 2020;12:46. doi: 10.1186/s13148-020-0820-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkani S., Rosen J.B. Induction of NGFI-B mRNA following contextual fear conditioning and its blockade by diazepam. Brain Res Mol Brain Res. 2000;80:153–165. doi: 10.1016/s0169-328x(00)00130-3. [DOI] [PubMed] [Google Scholar]
- Mansur R.B. The effect of body mass index on glucagon-like peptide receptor gene expression in the post mortem brain from individuals with mood and psychotic disorders. Eur. Neuropsychopharmacol. 2019;29:137–146. doi: 10.1016/j.euroneuro.2018.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin I.A., Kipnis J. Central nervous system: (immunological) ivory tower or not? Neuropsychopharmacology. 2017;42:28–35. doi: 10.1038/npp.2016.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michopoulos V., Powers A., Gillespie C.F., Ressler K.J., Jovanovic T. Inflammation in fear- and anxiety-based disorders: PTSD, GAD, and beyond. Neuropsychopharmacology. 2017;42:254–270. doi: 10.1038/npp.2016.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michopoulos V. Association of prospective risk for chronic PTSD symptoms with low TNFalpha and IFNgamma concentrations in the immediate aftermath of trauma exposure. Am. J. Psychiatr. 2020;177:58–65. doi: 10.1176/appi.ajp.2019.19010039. [DOI] [PubMed] [Google Scholar]
- Milad M.R. Recall of fear extinction in humans activates the ventromedial prefrontal cortex and hippocampus in concert. Biol. Psychiatr. 2007;62:446–454. doi: 10.1016/j.biopsych.2006.10.011. [DOI] [PubMed] [Google Scholar]
- Milanovic S. Production of the Fos protein after contextual fear conditioning of C57BL/6N mice. Brain Res. 1998;784:37–47. doi: 10.1016/s0006-8993(97)01266-3. [DOI] [PubMed] [Google Scholar]
- Miller A.H., Haroon E., Raison C.L., Felger J.C. Cytokine targets in the brain: impact on neurotransmitters and neurocircuits. Depress. Anxiety. 2013;30:297–306. doi: 10.1002/da.22084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.W., Lin A.P., Wolf E.J., Miller D.R. Oxidative stress, inflammation, and neuroprogression in chronic PTSD. Harv. Rev. Psychiatr. 2018;26:57–69. doi: 10.1097/HRP.0000000000000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller M.W. A novel locus in the oxidative stress-related gene ALOX12 moderates the association between PTSD and thickness of the prefrontal cortex. Psychoneuroendocrinology. 2015;62:359–365. doi: 10.1016/j.psyneuen.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei L., Castrillo A., Chen M., Hoffmann A., Tontonoz P. Induction of NR4A orphan nuclear receptor expression in macrophages in response to inflammatory stimuli. J. Biol. Chem. 2005;280:29256–29262. doi: 10.1074/jbc.M502606200. [DOI] [PubMed] [Google Scholar]
- Reuveni I. Anatomical and functional connectivity in the default mode network of post-traumatic stress disorder patients after civilian and military-related trauma. Hum. Brain Mapp. 2016;37:589–599. doi: 10.1002/hbm.23051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeh N. SKA2 methylation is associated with decreased prefrontal cortical thickness and greater PTSD severity among trauma-exposed veterans. Mol. Psychiatr. 2016;21:357–363. doi: 10.1038/mp.2015.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selemon L.D., Young K.A., Cruz D.A., Williamson D.E. Frontal lobe circuitry in posttraumatic stress disorder. Chronic stress (Thousand Oaks, Calif.) 2019;3 doi: 10.1177/2470547019850166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K.L. Microglial cell hyper-ramification and neuronal dendritic spine loss in the hippocampus and medial prefrontal cortex in a mouse model of PTSD. Brain Behav. Immun. 2019;80:889–899. doi: 10.1016/j.bbi.2019.05.042. [DOI] [PubMed] [Google Scholar]
- Stein M.B. Genome-wide association analyses of post-traumatic stress disorder and its symptom subdomains in the Million Veteran Program. Nat. Genet. 2021;53:174–184. doi: 10.1038/s41588-020-00767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan A.H., Barres Ba Fau - Stevens B., Stevens B. The complement system: an unexpected role in synaptic pruning during development and disease. Annu. Rev. Neurosci. 2012;35:369–389. doi: 10.1146/annurev-neuro-061010-113810. [DOI] [PubMed] [Google Scholar]
- Stone L.A. Cortical transcriptomic alterations in association with appetitive neuropeptides and body mass index in posttraumatic stress disorder. Int. J. Neuropsychopharmacol. 2021;24:118–129. doi: 10.1093/ijnp/pyaa072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tursich M. Association of trauma exposure with proinflammatory activity: a transdiagnostic meta-analysis. Transl. Psychiatry. 2014;4:e413. doi: 10.1038/tp.2014.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal-Pineiro D. Cellular correlates of cortical thinning throughout the lifespan. Sci. Rep. 2020;10:21803. doi: 10.1038/s41598-020-78471-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hertzen L.S., Giese K.P. Memory reconsolidation engages only a subset of immediate-early genes induced during consolidation. J. Neurosci. 2005;25:1935–1942. doi: 10.1523/JNEUROSCI.4707-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. c-fos regulates neuronal excitability and survival. Nat. Genet. 2002;30:416–420. doi: 10.1038/ng859. [DOI] [PubMed] [Google Scholar]
- Zhang Y. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;3:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.