Abstract
Mitochondria undergo continuous cycles of fission and fusion to promote inheritance, regulate quality control, and mitigate organelle stress. More recently, this process of mitochondrial dynamics has been demonstrated to be highly sensitive to nutrient supply, ultimately conferring bioenergetic plasticity to the organelle. However, whether regulators of mitochondrial dynamics play a causative role in nutrient regulation remains unclear. In this study, we generated a cellular loss-of-function model for dynamin-related protein 1 (DRP1), the primary regulator of outer membrane mitochondrial fission. Loss of DRP1 (shDRP1) resulted in extensive ultrastructural and functional remodeling of mitochondria, characterized by pleomorphic enlargement, increased electron density of the matrix, and defective NADH and succinate oxidation. Despite increased mitochondrial size and volume, shDRP1 cells exhibited reduced cellular glucose uptake and mitochondrial fatty acid oxidation. Untargeted transcriptomic profiling revealed severe downregulation of genes required for cellular and mitochondrial calcium homeostasis, which was coupled to loss of ATP-stimulated calcium flux and impaired substrate oxidation stimulated by exogenous calcium. The insights obtained herein suggest that DRP1 regulates substrate oxidation by altering whole-cell and mitochondrial calcium dynamics. These findings are relevant to the targetability of mitochondrial fission and have clinical relevance in the identification of treatments for fission-related pathologies such as hereditary neuropathies, inborn errors in metabolism, cancer, and chronic diseases.
Keywords: dynamin-related protein 1, mitochondrial dynamics, β-oxidation, calcium signaling, skeletal muscle
Abbreviations: BSA, bovine serum albumin; CG, Calcium Green-1 AM; CST, Cell Signaling Technology; DMEM, Dulbecco’s modified Eagle medium; DRP1, dynamin-related protein 1; ET, electron transfer; MTR, MitoTracker Red FM; OPA1, optic atrophy 1; OXPHOS, oxidative phosphorylation; TMPD, N,N,N',N'-Tetramethyl-p-phenylenediamine dihydrochloride
Mitochondria regulate energy production in response to cellular demand by producing adenosine triphosphate (ATP) through a series of complex biochemical reactions known as oxidative phosphorylation (OXPHOS) (1). Through glycolysis and β-oxidation, catabolism of glucose and fatty acid substrates promotes entry to the tricarboxylic acid cycle, leading to the generation of reducing equivalents for OXPHOS (1, 2). In addition to regulating OXPHOS, mitochondria regulate substrate entry and respond to cellular stress by means of fission and fusion (3, 4). This morphological plasticity, known as mitochondrial dynamics, allows the organelle to integrate and remove individual mitochondrion to maintain network integrity, regulate substrate supply, and preserve respiratory function (5, 6, 7).
Mitochondrial fission is regulated by the nuclear-encoded, cytosolic guanosine triphosphatase (GTPase) dynamin-related protein 1 (DRP1) (1, 8, 9). Although the mechanisms of DRP1 activation remain incomplete, it is understood that DRP1 is recruited to the mitochondrial outer membrane and bound by anchored receptors that include mitochondrial fission factor (Mff) along with mitochondrial dynamics proteins of 49 and 51 kDa (MiD49 and MiD51) (10). DRP1 then oligomerizes and forms a ring-like structure around the mitochondrion (7, 11, 12). A GTP hydrolysis-induced conformational change in DRP1 shortens the protein, causing it to wrap tightly into a closed ring and constrict the mitochondrial membrane (13). Ultimately, the formation of mitochondrial constriction machinery along with scission mediators results in physical division of the mitochondrion into two daughter mitochondria (7, 14). Mitochondria with intact membrane potential may be reintegrated into the network, whereas damaged or depolarized organelles are recycled by PTEN-induced kinase 1 (Pink1) and Parkin-mediated autophagy (mitophagy) (15). Mitochondrial fusion occurs when adjacent or neighboring mitochondria form elongated, tubular structures (5, 7). Fusion events are achieved by coordination of mitofusins 1 and 2 (MFN1 and MFN2, respectively) on the outer mitochondrial membrane as well as optic atrophy 1 (OPA1) on the inner mitochondrial membrane (16).
Increased mitochondrial fission has been observed in a number of metabolic diseases, including obesity, type 2 diabetes, Alzheimer’s disease, Parkinson’s disease, and pulmonary arterial hypertension (17). However, it remains unclear as to what extent fission is required in skeletal muscle for routine bioenergetic function, substrate metabolism, and quality control. Recently, it has been observed that humans with defects in fatty acid oxidation display more round, punctate mitochondria (18). This is supported by previous observations of mitochondria increasingly favoring a punctate morphology in conditions of nutrient oversupply (19, 20, 21). Conversely, in situations of high energy demand as observed following exercise training, fasting, or G1/S cell cycle phase, mitochondria form highly elongated reticular networks with increased respiratory capacity (21, 22, 23, 24). Furthermore, we have previously demonstrated that prolonged muscle contraction restricts mitochondrial fission in human skeletal muscle, which correlated with the degree of improvement in fatty acid oxidation and insulin sensitivity (25, 26).
The purpose of this study was to determine the effect of DRP1 on skeletal muscle mitochondrial nutrient handling. We observed that DRP1 is partially required for glucose and fatty acid oxidation mediated primarily by stabilization of whole-cell and mitochondrial Ca2+ dynamics. Our findings signify the delicacy of fission and fusion balance for maintaining optimal mitochondrial networks. Furthermore, restoration of defects in nutrient oxidation may alleviate inheritable mutations or late-onset diseases related to impaired mitochondrial fission.
Results
DRP1 contributes to NADH- and succinate-linked respiration and mitochondrial ultrastructure formation
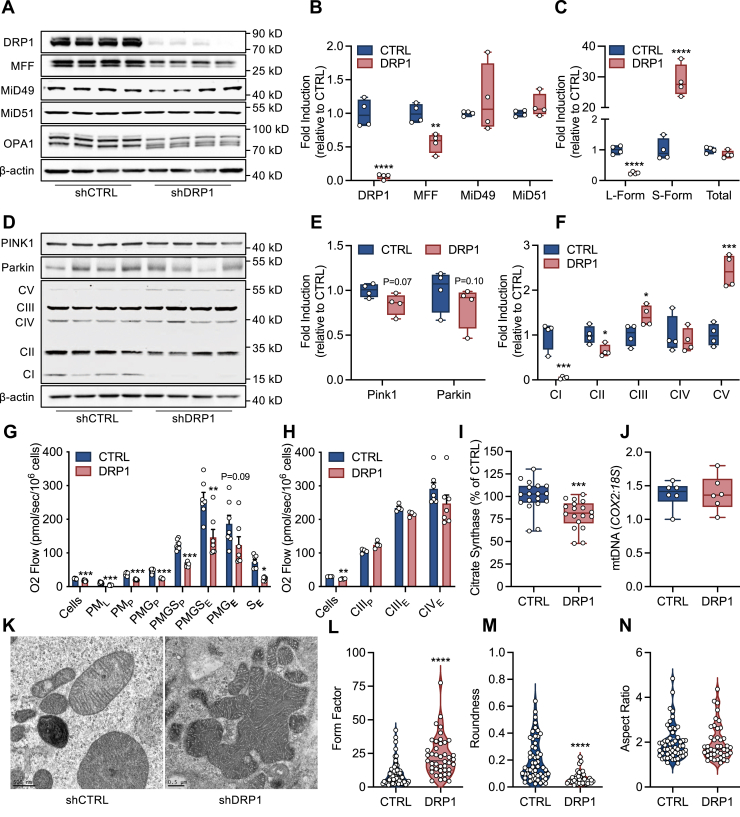
Plasmid transfection of wildtype C2C12 cells with shRNA targeting DRP1 resulted in ∼90% reduction in protein expression (Fig. 1, A and B). In addition, shDRP1 cells exhibited a ∼50% reduction in MFF with no change in MiD49 or MiD51 expression (Fig. 1, A and B). OPA1 is encoded by eight mRNA splice forms that result in at least five distinguishable protein variants that can undergo proteolytic processing to enhance mitochondrial fission (27). shDRP1 cells displayed degraded expression of the long isoform of OPA1 (L-OPA1) coupled to increased expression of the soluble short form (S-OPA1) which localizes in the intermembrane space (Fig. 1C). Pink and Parkin expression tended to decline in shDRP1 cells (Fig. 1, D and E). Alterations in fission and fusion protein expression were associated with reduced expression of mitochondrial complex I and II, as well as increased expression of CIII and CV (Fig. 1F). Since mitochondrial fission is linked to energy production (21), we then evaluated OXPHOS and electron transfer (ET) capacity. We found that routine (intact cells) and leak respiration, as well as NADH- and succinate-linked OXPHOS, were decreased (Fig. 1G). The respiratory defect in shDRP1 cells was not relieved by stimulation of maximal electron flow with FCCP, indicating that oxidation was rate limiting. OXPHOS and maximal electron flow supported by duroquinol, which feeds electrons to complex III, was unaltered by shDRP1 (Fig. 1H). Furthermore, maximal electron flow supported by ascorbate and N,N,N',N'-Tetramethyl-p-phenylenediamine dihydrochloride (TMPD) through complex IV was unaltered by shDRP1, indicating that the oxidative defect present in shDRP1 cells was likely upstream of complex III. We also observed that DRP1 lowered citrate synthase activity (Fig. 1I) with no change in mitochondrial DNA content (Fig. 1J). Transmission electron microscopy was then employed to evaluate mitochondrial ultrastructure (Fig. 1K). shCTRL mitochondria were pleomorphic and somewhat polygonal. Each cell had a relatively large, round, centrally placed nucleus, which had marginated heterochromatin that was regularly punctated by nuclear pores, as well as one or several nucleoli in its nucleoplasm. The cytoplasm encompassed an unusual number of myelin bodies. These membrane-bound structures consisted largely of very dense, tightly furled laminae that resembled myelin, hence the application. Irrespective of shape most of the mitochondria had lamelliform crista, although digiform (finger-like, tubular) crista sporadically appeared. In shDRP1 cells the plasmalemma lacked an array of microvilli, although a few of these structures were present, along with an occasional fimbria. The cytoplasmic matrix was suffused by a sea of free polysomes. Small numbers of individual elements of rough endoplasmic reticulum were scattered throughout the cytoplasm. Several mitochondria formed clusters that appeared to be the forerunner of a fusion process that eventually may lead to the formation of megamitochondria, although mature forms of the latter were infrequently observed (Fig. 1, L–N). Collectively, these data indicate that DRP1 contributes to bioenergetic efficiency as well as ultrastructure formation.
Figure 1.
DRP1 contributes to NADH- and succinate-linked respiration and mitochondrial ultrastructure formation.A–C, representative immunoblots and quantitation of DRP1, MFF, MiD49, MiD51, and OPA1 expression (N = 4 per condition). D–F, representative immunoblots and quantitation of PINK1, Parkin, and CI-V expression (N = 4 per condition). G, respiration supported by pyruvate and malate (PM), glutamate (G), and succinate (S) in the presence of ADP (P) or FCCP (E) in digitonin-permeabilized cells (N = 8 per condition). H, respiration supported by duroquinol (CIII) and ascorbate and TMPD (CIV) in the presence of ADP (P) or FCCP (E) in digitonin-permeabilized cells (N = 8 per condition). I, enzymatic activity of citrate synthase (N = 10 per condition). J, mtDNA content (COX2:18S) (N = 6 per condition). K, representative transmission electron microscopy (TEM) of phosphate-buffered aldehyde-fixed cells (500 nm). L–N, quantitation of mitochondrial branching and complexity (form factor), roundness, and length (aspect ratio) (shCTRL: 56 mitochondria, shDRP1: 43 mitochondria). B, C, E, F, I, and J are displayed as a box (mean ± 5–95% confidence interval) and whiskers (minimum to maximum) plot. G and H are displayed as the mean ± SEM with all points. L–N are displayed as a violin plot. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.01, ∗∗∗∗p < 0.001. B, C, E, F, I, and J were assessed by unpaired Student’s t test. G and H were assessed by one-way ANOVA with Tukey’s multiple comparisons. L–N were assessed by Mann–Whitney U test.
DRP1 contributes to cellular glucose uptake but not glycolysis in an insulin-independent manner
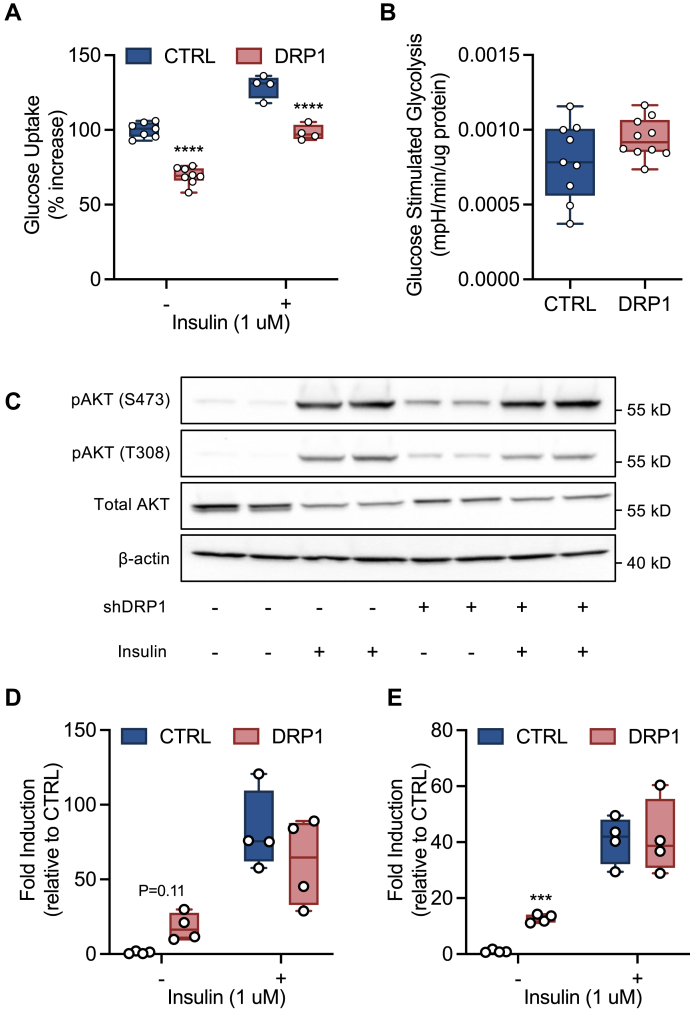
Based on our observation that DRP1 markedly alters mitochondrial structure and function, we sought to determine whether loss of function perturbed cellular glucose metabolism. shDRP1 cells displayed a ∼40% decrease in glucose uptake compared with shCTRL (Fig. 2A). shDRP1 and shCTRL cells were equally responsive to insulin stimulation (30% of non-stimulated rate). As a result, shDRP1 cells displayed reduced insulin-stimulated glucose uptake compared with shCTRL. Changes in cellular glucose uptake were not attributable to glycolysis, which did not differ between shDRP1 and shCTRL (Fig. 2B). Furthermore, insulin-stimulated AKT phosphorylation at serine 473 and threonine 308 was unchanged in shDRP1 cells, suggesting that insulin signaling was unaffected (Fig. 2, C–E). However, insulin-independent activation of AKT, as evidenced by an increase in phosphorylation in the absence of insulin, was elevated in shDRP1 relative to shCTRL (Fig. 2, C–E). Total AKT expression decreased with insulin stimulation but was unaltered in shDRP1 cells (Fig. 2C). Taken together, these data indicate that DRP1 is not required for insulin-stimulated glucose uptake but contributes to insulin-independent activation of AKT.
Figure 2.
DRP1 contributes to cellular glucose uptake but not glycolysis in an insulin-independent manner.A, [3-3H]glucose uptake ±30 min of insulin stimulation (N = 6 per condition). B, rate of glucose-stimulated extracellular acidification (N = 6 per condition). C–E, representative immunoblots and quantitation of pAKT (T308) and pAKT (S473) activation (N = 4 per condition). A, B, D, and E are displayed as a box (mean ± 5–95% confidence interval) and whiskers (minimum to maximum) plot. ∗∗∗p < 0.01, ∗∗∗∗p < 0.001. A, D, and E were assessed by a one-way ANOVA with Tukey’s multiple comparisons. B was assessed by unpaired Student’s t test.
DRP1 is required for fatty-acid stimulated glycolysis and contributes to β-oxidation
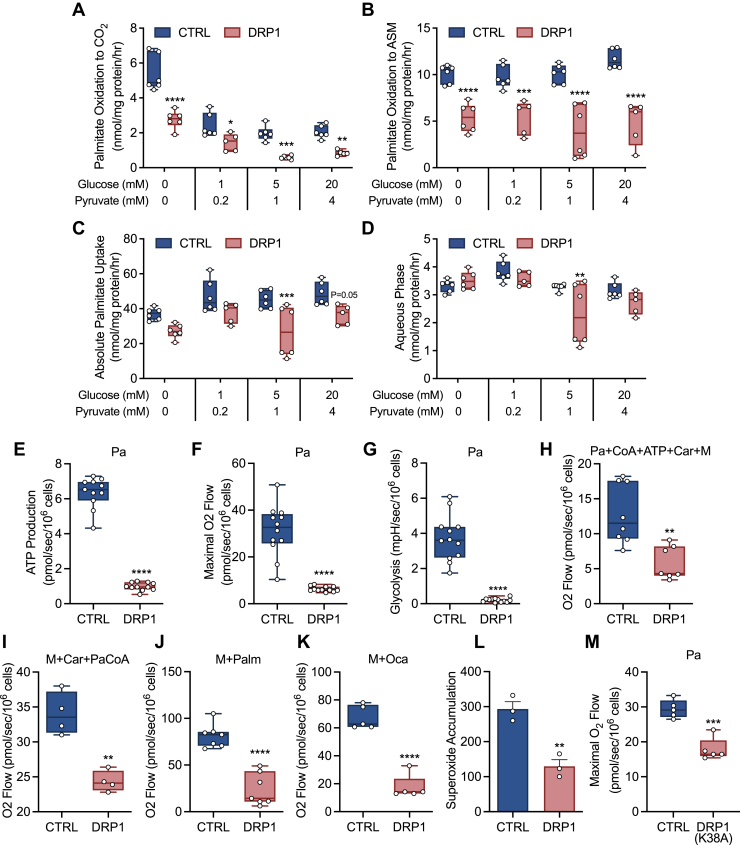
To determine if mitochondrial β-oxidation was impaired in shDRP1 cells, we first assessed utilization and uptake of [14C]palmitic acid. We observed that fatty acid oxidation to both CO2 and acid-soluble metabolites was lower in shDRP1 cells (Fig. 3, A and B). To further confirm specificity to fatty acid oxidation, we titrated glucose and pyruvate back into the medium and observed similar suppression of oxidation in both CTRL and shDRP1 cells (Fig. 3, A–D). We then measured ATP production and glycolysis in living cells acutely challenged with palmitate (Pa). shDRP1 cells exhibited lower ATP production rates and maximal respiration supported by Pa (Fig. 3, E and F). Furthermore, Pa-stimulated glycolysis was also impaired in shDRP1 cells (Fig. 3G). Based on our observations that Pa utilization was reduced in shDRP1 cells, we used a palmitate:bovine serum albumin (BSA) conjugate in the presence of CoA, ATP, and carnitine (Car) to evaluate palmitoyl-coenzyme A synthesis, acyl-CoA entry into the IMS, CPT1 synthesis of palmitoylcarnitine, and carnitine:acylcarnitine translocase entry into the matrix, which was reduced (Fig. 3H). We then evaluated formation of palmitoylcarnitine by CPT1 and carnitine:acylcarnitine translocase entry into the mitochondrial matrix (28) by addition of palmitoyl-coenzyme A, which was also reduced in shDRP1 cells (Fig. 3I). Palmitoylcarnitine, which requires the carnitine:acylcarnitine translocase and carnitine palmitoyltransferase II (CPTII) for transport into the mitochondrial matrix (28), was lower in shDRP1 cells (Fig. 3J). Similarly, octanoate, which passively diffuses into the mitochondria for oxidation, was lower in shDRP1 cells (Fig. 3K). Since oxidation of fatty acids is coupled to generation of free radicals, we evaluated superoxide formation and found that this was also diminished in shDRP1 cells (Fig. 3L). To further confirm the role of DRP1 in fatty acid oxidation, we generated C2C12s expressing a DRP1-K38A dominant negative mutation. Unlike shRNA silencing, the K38A mutation is translated but inhibits GTPase activity and oligomerization, ultimately limiting DRP1 activity. Similar to shDRP1 cells, the K38A mutant displayed impaired fatty acid oxidation compared with CTRL (Fig. 3M). Taken together, these data suggest that DRP1 contributes to β-oxidation by mediating transfer and uptake of fatty acids.
Figure 3.
DRP1 is required for fatty acid–stimulated glycolysis and contributes to β-oxidation.A, [1-14C]palmitate oxidation to CO2, (B) acid-soluble metabolite (ASM), (C) absolute uptake, and (D) aqueous phase ±0, 1, 5, and 20 mM glucose and 0, 0.2, 1, and 4 mM pyruvate (N = 6 per condition). E–G, rate of ATP production (N = 12 per condition), respiration in the presence of FCCP (N = 12 per condition), and glycolytic rate in the presence of oligomycin supported by palmitate (Pa) in living cells (N = 12 per condition). H, respiration of palmitate (Pa), coenzyme A (CoA), ATP, carnitine (Car), and malate (M) (N = 8 per condition), (I) palmitoyl-CoA (PaCoA), M, and Car (N = 8 per condition), and (J) M and palmitoylcarnitine (Palm) (N = 8 per condition), and (K) M and octanoate (Oca) in the presence of ADP in digitonin-permeabilized cells. L, accumulation of extracellular superoxide after 16-h incubation with 5,5-dimethyl-pyrroline N-oxide (DMPO). M, respiration supported by Pa and Car in CTRL or K38A DRP1 cells. A–K, and M are displayed as a box (mean ± 5–95% confidence interval) and whiskers (minimum to maximum) plot. L is displayed as the mean ± SEM with all points. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.01, ∗∗∗∗p < 0.001. A–D were assessed by two-way ANOVA with Tukey’s multiple comparisons. E–K, and M were assessed by unpaired Student’s t test.
DRP1 is required for expression of genes that regulate calcium signaling
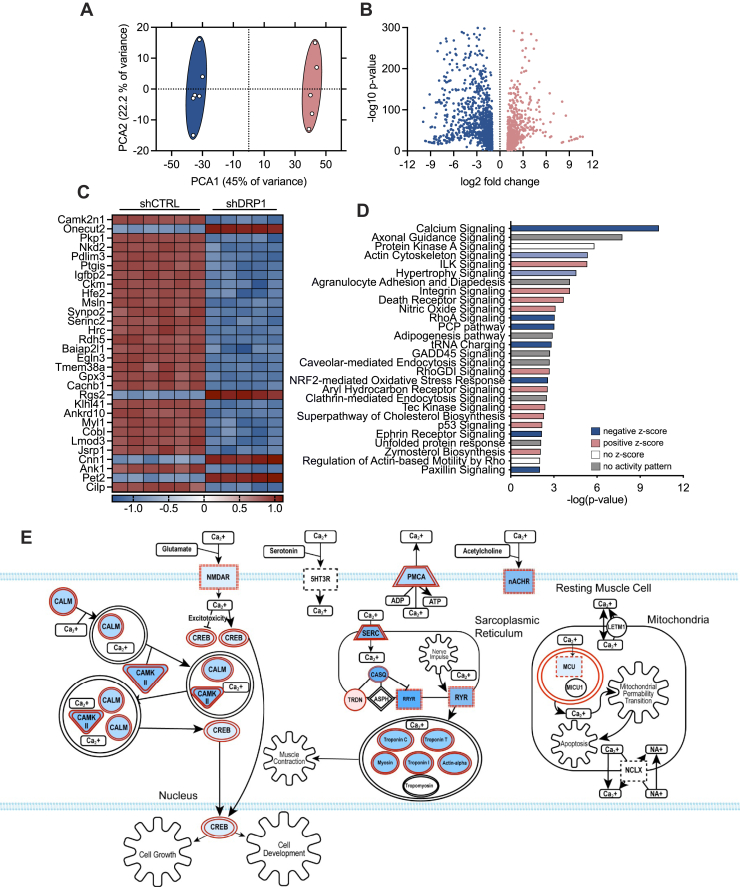
To elucidate the mechanisms whereby DRP1 regulates mitochondrial nutrient uptake and β-oxidation, untargeted transcriptomic profiling was employed to identify differentially expressed genes and signaling pathways. Principal component analysis revealed similar clustering of shCTRL and shDRP1 cells, which differed highly from one another (Fig. 4A). RNA sequencing revealed 2019 transcripts differentially regulated by DRP1, of which 917 were upregulated and 1102 were downregulated (Fig. 4B) (10.5281/zenodo.4641243). The top 30 differentially expressed genes were then visualized in a heat map (Fig. 4, C and Fig. S1). Several transcripts required for mitochondrial respiratory complex assembly and function such as NDUFB8, SDHB, and UQCRC2 were decreased in shDRP1 cells, whereas MT-CO1, the mitochondrially encoded component of cytochrome c oxidase, was increased (Fig. S1). Transcriptional regulators of mitochondrial dynamics and quality control such as MFF, MIEF2, OPA1, MFN1, MFN2, and PINK1 were uniformly downregulated in shDRP1 cells (Fig. S1). Transcriptional regulation of fatty acid oxidation was largely unaltered with exception to CPT1A and CPT1C, both of which were increased in shDRP1 cells (Fig. S1). Several transcripts required for glycolysis were decreased including PFKM, ALDOA, PGAM2, PCK2, GOT1, PCX, GOT2, MDH2, and PDHX, whereas LDHA and ENO1 expressions were increased in shDRP1 cells (Fig. S1). Based on our observations, we subjected differentially expressed genes to pathway analysis and identified 27 significantly enriched canonical signaling pathways in shDRP1 cells (Fig. 4D). Individual z-scores were then generated to identify directional expression patterns that revealed marked downregulation of Ca2+ signaling (Fig. 4D). Differentially regulated genes involved in Ca2+ signaling include 53 mRNA transcripts (p = 7.90 × 10−14, 25.7% overlap with canonical signaling) notably impacting N-methyl-d-aspartic acid receptor, plasma-membrane Ca2+ ATPase, nicotinic acetylcholine receptor, sarcoplasmic reticulum, and mitochondrial signaling pathways (Fig. 4E). Based on the observation that genes associated with Ca2+ transport were reduced by shDRP1, we then hypothesized that reductions in fatty acid oxidation were attributable to dysfunctional mitochondrial Ca2+ dynamics.
Figure 4.
DRP1 is required for expression of genes that regulate calcium signaling.A, principal component analysis of untargeted transcriptomic profiling of shCTRL (blue) and shDRP1 (pink) cells. B, volcano plot of genes differentially regulated by shDRP1. C, heat map visualization of top 30 differentially regulated genes by shDRP1. D, pathway enrichment analysis of differentially regulated genes. E, schematic illustration of differentially regulated genes involved in calcium signaling and homeostasis. Structures highlighted in shades of red were differentially regulated by shDRP1. Darker shades of red correlate to increased activation.
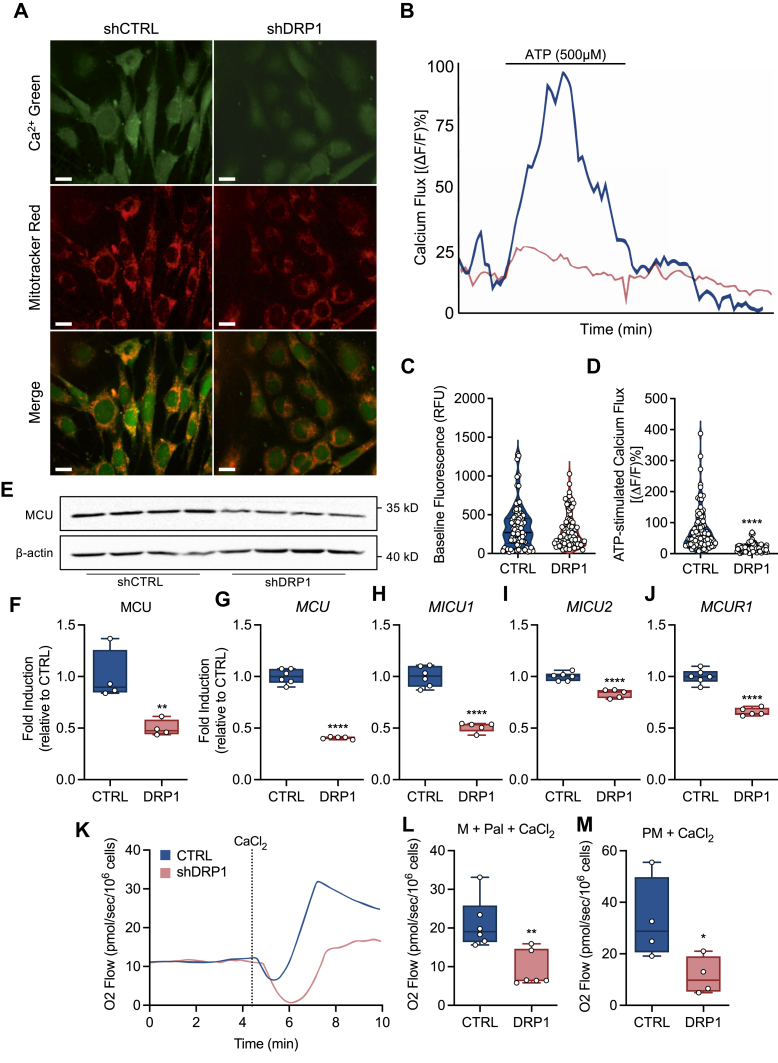
DRP1 is required for mitochondrial Ca2+ influx
To determine if the diminution of gene expression contributed to impaired mitochondrial function, we performed live cell imaging before, during, and after stimulation with ATP, a Ca2+ stimulating agonist (Fig. 5, A and B). Although there was no difference in baseline Ca2+ fluorescence (Fig. 5, A–C), stimulation with ATP revealed a severe defect in mitochondrial Ca2+ flux in shDRP1 cells (Fig. 5, A–E and Videos S1 and S2). Since the primary pathway for Ca2+ entry occurs via the mitochondrial calcium uniporter (29), we evaluated changes in protein and gene expression. Consistent with RNA sequencing, protein (Fig. 5F) and gene expression (Fig. 5G) of the mitochondrial calcium uniporter was decreased in shDRP1 cells. In addition to the MCU gene, other genes required for MCU activity such as MICU1, MICU2, and MICUR1 were decreased in shDRP1 cells (Fig. 5, H–J). To further determine the role of mitochondrial Ca2+ entry in substrate oxidation, we evaluated oxidative capacity driven by Ca2+ in permeabilized cells. We observed that calcium-mediated oxidation of palmitoylcarnitine and the combination of pyruvate and malate was also decreased in shDRP1 cells (Fig. 5, K–M). Collectively, these data indicate that DRP1 contributes to stabilization of the mitochondrial calcium uniporter and Ca2+-mediated substrate oxidation.
Figure 5.
DRP1 is required for mitochondrial calcium flux.A, representative confocal micrographs illustrating localization of Ca2+ (green), MitoTracker red (red), and merge (the scale bar represents 20 μm). B, representative tracing of mitochondrial Ca2+ flux before, during, and after perfusion with ATP (shCTRL = blue line, shDRP1 = pink line). C and D, quantification of baseline and ATP-stimulated calcium flux (shCTRL, 90 mitochondria; shDRP1, 92 mitochondria). E and F representative immunoblots and quantitation of MCU protein expression (N = 4 per condition). G and H, MCU, MICU1, MICU2, and MCUR1 gene expression (CTRL N = 6, shDRP1 N = 5). K, representative plot of O2 flow before and after stimulation with calcium chloride in permeabilized cells. L, calcium-stimulated oxidation of palmitoylcarnitine + malate and (M) pyruvate + malate in permeabilized cells. C and D are displayed as a violin plot. F–J, L, and M are displayed as a box (mean ± 5–95% confidence interval) and whiskers (minimum to maximum) plot. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.001. C, D, F–J, L, and M were assessed by an unpaired Student’s t test.
Discussion
DRP1 is the primary mediator of mitochondrial fission, and dysregulation of its activity has been attributed to a number of chronic metabolic and neurodegenerative diseases (30). Furthermore, homozygous deletion or dominant negative mutations of DNML1, the protein-coding gene for DRP1, result in embryonic lethality or neonatal mortality in both rodents and humans (31, 32). Here we demonstrate that, in addition to mitochondrial fission, DRP1 is required for β-oxidation in myoblasts, which was associated with severe destabilization of intracellular Ca2+ homeostasis. Taken together, these findings highlight a noncanonical mechanism whereby mitochondrial dynamics regulates substrate transport and utilization.
DRP1-mediated fission events are required for routine mitochondrial quality control by facilitating mechanical removal of damaged organelles and limiting re-entry into the intact network (5, 6, 7). We observed that loss of DRP1 resulted in the formation of large, multilobular mitochondria characterized by grossly unorganized and disrupted cristae and impaired LC3 and Pink1 expression, indicating that mitochondrial turnover was limited. This is supported by previous reports whereby inhibition of DRP1 resulted in the formation of highly dense mitochondrial aggregates resembling megamitochondria in multiple cell lineages (8, 33, 34). Furthermore, mitochondria favor hyperconnectivity during periods of nutrient starvation, which maximizes cristae volume and ATP synthase activity (22).
A central finding of our study was the impairment of both glucose uptake and fatty acid oxidation following loss of DRP1, ultimately diminishing ATP production. In conditions of nutrient excess such as lipid overload, DRP1 undergoes posttranslational modifications to induce fragmentation and limit glucose and fatty acid oxidation, a process that is reversed by pharmacologic or genetic inhibition (20, 26, 35, 36). Conversely, during periods where nutrient demand exceeds supply such as G1/S transition, mitochondrial network connectivity and fusion are increased to support maximal rates of oxidative phosphorylation (23). As such, the suppression of nutrient uptake by loss of DRP1 appears to be a function of alteration in membrane structures that limit substrate entry.
Untargeted transcriptomic profiling and subsequent functional validation revealed severe downregulation of both cellular and mitochondrial Ca2+ dynamics in DRP1-deficient cells. Intracellular concentrations of Ca2+ are tightly regulated between the endoplasmic reticulum, mitochondria, and to a lesser extent in the cytoplasm (37). When cellular Ca2+ signals are produced, ionic entry to the mitochondria can occur directly by the outer membrane voltage-dependent anion channels, the MCU, or Na+/Ca2+/Li+ exchanger, which can then drive metabolic processes such as ATP production (38). For example, Ca2+ flux and CAMKK’s can regulate glucose uptake in skeletal muscle independent of catabolic nutrient sensors such as Akt and 5′ AMP-activated protein kinase (39) and caffeine- or ionomycin-induced Ca2+ flux can stimulate fatty acid oxidation in skeletal muscle (40, 41). Furthermore, deletion of CaV1.1, the voltage-dependent calcium channel, has been shown to limit palmitate and carnitine oxidation (42). As such, it appears that when mitochondrial fission potential is restricted, altered membrane dynamics may destabilize Ca2+ efflux, which ultimately limits fatty acid uptake and entry to the mitochondria. This is supported by observations in mice bearing a muscle-specific deletion of DRP1, which exhibit depolarized mitochondrial membrane potential and respiratory complex assembly (43). However, the precise mechanisms whereby mitochondrial dynamics contribute to calcium-mediated substrate uptake and utilization remain unclear. For example, DRP1 may interact with other quality control proteins or cell surveillance programs to mediate metabolic effects (44). Furthermore, diminished respiratory function may be an indirect response to mitochondrial morphological and functional impairments that occur in the absence of DRP1 (43).
Taken together, inhibition of DRP1-mediated mitochondrial fission restricts substrate oxidation. Limitations in substrate oxidation were partially attributable to reductions in cellular and mitochondrial calcium homeostasis. These findings underscore the importance of maintaining healthy and dynamic mitochondrial networks within metabolically active cells to ensure nutrient uptake and supply and may have clinical relevance in the treatment of mitochondrial diseases.
Experimental procedures
Cell culture and transfection
C2C12 cells were purchased from American Type Culture Collection and expanded in Dulbecco’s modified Eagle medium (DMEM) supplemented with 4.5 g/l d-glucose (25 mM), 50 U/ml penicillin/streptomycin, 0.5 μg/ml amphotericin B, and 10% fetal bovine serum. Cells were incubated at 37 °C, 5% CO2 in a humidified incubator. After reaching 100% confluence, cells were induced to differentiate by replacing the growth medium with high-glucose DMEM supplemented with 2% horse serum and allowing cells to differentiate for 48 h unless otherwise indicated. In both growth and differentiation stages, the medium was replenished daily. A stable cell line expressing shRNA against DRP1 was created by transfecting MISSION TRC shRNA clone (Sigma) using TransIT-2020 (Mirus Bio) according to the manufacturer’s protocol. In brief, C2C12 cells were transfected with a MISSION DRP1 shRNA construct (Clone ID: NM_152816.2-1665s21c1, Sigma) or a MISSION control shRNA construct (shCTRL; pLKO.1-puro empty vector control plasmid, Sigma), and after 24 h the medium was replaced with DMEM and puromycin to select cells expressing shDRP1 or shCTRL. Stable cell lines were selected with puromycin (2 μg/ml) for 10 days, and transfection efficiency was confirmed by Western blot. A stable cell line expressing a dominant negative K38A DRP1 mutation was created by transfecting the dominant negative K38A DRP1 plasmid using X-tremeGENE HP (Sigma) according to the manufacturer’s protocol. In brief, C2C12 cells were transfected and after 48 h, the medium was replaced with growth medium and geneticin (1.25 mg/ml) to select cells expressing the dominant negative K38A DRP1 mutation. Mouse Drp1(K38A) was a gift from David Chan (Addgene plasmid # 26049; http://n2t.net/addgene:26049; RRID:Addgene_26049).
Transmission electron microscopy
Mitochondrial ultrastructure and content were evaluated by transmission electron microscopy as described (45, 46). Briefly, cells were seeded onto 0.1% gelatin-coated 0.4-μm polyethylene terephthalate membranes (Falcon) and cultured as described above. After 48 h of differentiation, cells were washed in serum-free medium, fixed at room temperature in 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.4), and washed with cold phosphate buffered saline (PBS). Membranes were then stored at 4 °C in PBS until processing. The cells were then rinsed in 0.1 M phosphate buffer (pH 7.4), postfixed for 2 h in an unbuffered 1:1 mixture of 2% osmium tetroxide and 3% potassium ferricyanide and soaked overnight in an acidified solution of 0.25% uranyl acetate. Cells were then dehydrated in ascending concentrations of ethanol, passed through propylene oxide, and embedded in Embed-812 resin mixture (Electron Microscopy Sciences). Thin sections were sequentially stained with acidified uranyl acetate followed by a modification of Sato’s triple lead stain and examined in an FEI Tecnai Spirit (T12) with a Gatan US4000 4k × 4k CCD. Descriptors were quantitated using the Shape Descriptors plugin for ImageJ v4.0.
Citrate synthase activity
Citrate synthase activity was determined by commercially available enzymatic assay (Sigma). Briefly, cells were washed twice in ice-cold PBS and lysed (CelLytic M Cell Lysis Reagent; Sigma) in the presence of a protease inhibitor cocktail. Lysed cells were centrifuged at 20,000g for 15 min, and the supernatant was transferred to a fresh tube for determination of total protein content. Protein lysate, 8 μg, was combined with 1× assay buffer, 30 mM acetyl CoA solution, and 10 mM DTNB and plated in triplicate in a 96-well plate. Basal absorbance was measured on a plate reader set on a kinetic program (1.5 min duration, 10 s interval) at 412 nm. 10 mM oxaloacetate was then added to each well, the plate was briefly shaken, and the absorbance was read again at 412 nm on the same kinetic program.
mtDNA content
Mitochondrial DNA content was determined by RT-qPCR using Power SYBR Green (Thermo Fisher Scientific) as described (47) with primers directed against the mitochondrial encoded cytochrome c oxidase subunit II (Cox2) and the nuclear encoded 18S (48). Primer sequences are listed in Table S1. All reactions were carried out in quadruplicate. Ct values were obtained and normalized to 18S expression. Relative fold expressions from RT-qPCR were determined using the comparative ΔΔCt method.
Glucose uptake
Glucose uptake was measured as described (49). Briefly, cells were incubated in serum-free low-glucose DMEM supplemented with 1% bovine serum albumin for 4 h prior to insulin stimulation. Cells were stimulated with 1 μM insulin (Novolin-R 100, Novo Nordisk) in Krebs-Ringers-Hepes (KRH) buffer (20 mM Hepes, 136 mM NaCl, 4.7 mM KCl, 1.25 mM MgSO4·7H2O, 1.25 mM CaCl2, pH 7.40) for 30 min. The insulin stimulation medium was then removed and replaced with KRH containing 1 μCi/ml [3H]-2-deoxy-d-glucose (PerkinElmer) and 2 mM 2-deoxy-d-glucose. After 10 min of radiolabeled glucose uptake, cells were washed with ice-cold PBS three times and then lysed with 0.1% sodium dodecyl sulfate (SDS) in ddH2O for 30 min with gentle shaking. Lysates were then used for protein quantification and scintillation counting in Ultima Gold scintillation fluid (PerkinElmer).
Live cell respirometry
Living cell respiration was determined on a Seahorse XFe24 bioanalyzer (Agilent Technologies, Inc) as described (47). For all assays, cells were seeded into a Seahorse XFe24 plate at a density of 1.0 × 104 cells per well and grown to confluence, after which they were induced to differentiate for 48 h as described above. Exogenous fatty acid oxidation was measured by incubation in a substrate-limited medium (0.5 mM glucose, 0.5 mM L-carnitine, 1 mM glutamine, and 1% FBS in Agilent modified DMEM, pH 7.40) overnight. On the day of the assay, the medium was changed to Krebs–Henseleit buffer (111 mM NaCl, 4.7 mM KCl, 1.25 mM CaCl2, 2 mM MgSO4, 1.2 mM NaH2PO4, pH 7.4) supplemented with 2.5 mM glucose, 0.5 mM L-carnitine, and 5 mM Hepes and incubated for 1 h prior to assay at 37 °C without CO2. Immediately prior to assay, 200 μM palmitate–BSA conjugate or BSA control was added to each well. Maximal glycolytic activity was determined by injection of glucose (10 mM) and oligomycin (1 μM) into glucose-free medium (2 mM glutamine in Agilent Base DMEM, pH 7.40). Oxygen consumption rates and extracellular acidification rates for each assay were normalized to total protein content.
Assessment of OXPHOS and ET capacity in permeabilized cells
OXPHOS and ET capacity were determined in permeabilized cells as described (50). Briefly, cells were collected in trypsin-EDTA, transferred into conical tubes of serum-supplemented medium, and centrifuged at 800g for 10 min at 25 °C. The cell pellet was then resuspended in Mir05 medium. A 2-ml suspension containing ∼500K cells/ml was added into each chamber of an O2K system (OROBOROS). Cells were permeabilized by titrating digitonin (4–10 μg/ml) in dimethyl sulfoxide. Respiratory function was determined using the following concentrations of substrates, uncouplers, and inhibitors: malate (2 mM), pyruvate (2.5 mM), ADP (2.5 mM), glutamate (10 mM), succinate (10 mM), palmitate (25 μM), coenzyme A (0.5 mM), ATP (2.5 mM), L-carnitine (1 mM), palmitoylcarnitine (20 μM), palmitoyl-CoA (10 μM), tetramethyl-p-phenylenediamine (TMPD, 0.5 μM), ascorbate (2 mM), carbonylcyanide-p-trifluoromethoxyphenylhydrazone (FCCP, 0.5 μM increment), rotenone (75 nM), antimycin A (125 nM), and sodium azide (200 mM) as indicated. For calcium-stimulated oxidation experiments, calcium chloride (CaCl2, 0.6 mM; 103 μM free calcium) was titrated into permeabilized cells and corrected by the rotenone sensitive rate. NADH- and succinate-linked OXPHOS and ET capacity were corrected for residual oxygen consumption by the antimycin A sensitive rate. Fatty acid oxidation was corrected by the rotenone sensitive rate. Oxidation of reduced TMPD was corrected by the sodium azide sensitive rate.
Lipid oxidation
Palmitate oxidation was assessed by [1-14C]-palmitate labeling as described with minor modifications (51, 52, 53, 54). Briefly, cells were incubated in DMEM supplemented with 12.5 mM Hepes, 1 mM L-carnitine, and 100 μM [1-14C]-palmitate along with increasing concentrations of both glucose (0, 1, 5, and 20 mM) and pyruvate (0, 0.2, 1, and 4 mM) for 2 h at 37 °C. After incubation, cells were placed on ice and 400 μl of incubation medium was transferred to borosilicate tubes. A secondary “trap” tube containing 1 N NaOH was then added, the borosilicate tubes were sealed with a rubber stopper, and 100 μl of 70% perchloric acid was added. Following a 1-h incubation at room temperature, liberated 14CO2 that had been captured in 1 N NaOH was detected through scintillation counting. The now acidified remaining incubation medium was collected and used to detect acid-soluble metabolites. After an overnight incubation at 4 °C, the collected media were centrifuged at 14,000g for 10 min, and an aliquot of the supernatant was removed for scintillation counting. Following removal of medium from the plates, cells were washed with PBS (×3), lysed in 0.1% SDS, and collected after 20 min of shaking at room temperature. A chloroform:methanol (1:2) extraction was performed to determine partitioning of radiolabeled substrate into the lipid pool (organic phase) or other metabolic pathways (aqueous phase). All data were normalized to protein content.
Western blotting
Immunoblotting was performed as described (49). Briefly, cells were harvested after treating with or without 1 μM insulin for 10 min, placed on ice and washed three times with ice-cold PBS, scraped into cell extraction buffer (5% protease inhibitor, 5% PhosStop, 1 μM sodium orthovanadate, and 5.1 μM phenylmethanesulfonyl fluoride), and subjected to one freeze–thaw cycle. A BCA assay was performed on the supernatant to determine protein content. Protein lysate, 25 μg, for each condition was prepared in Laemmli buffer with 2% β-mercaptoethanol and heated to 100 °C for 5 min. Samples were loaded in Tris-glycine gels, and proteins were separated by electrophoresis at 125 V. Proteins were then blotted onto methanol-activated PVDF membrane in ice-cold transfer buffer, cut to size, and blocked in 5% BSA for 1 h at room temperature. Primary antibodies (anti-DRP1, Cell Signaling Technology [CST] cat# 8570; anti-MFF, CST cat# 84580; anti-Total Rodent OXPHOS, Abcam cat# ab110413 [CV: ATP5A, CIII: UQCRC2, CIV: MTCO1, CII: SDHB, CI: NDUFB8]; anti-MiD51, Proteintech cat# 20164-1-AP; anti-MiD49, Proteintech cat# 16413-1-AP; anti-OPA1, CST cat# 80471; anti-PINK1, Santa Cruz Biotechnology, cat# sc-33796; anti-LC3B, Novus Biologicals, cat# NB100-2220, diluted 1:5000; anti-Parkin, Proteintech, cat# 14060-1-AP; anti-phospho-Akt Ser473, CST cat# 4060; anti-phospho-Akt Thr308, CST cat# 13038, anti-Akt, CST cat# 9272; anti-MCU, CST cat# 14997; anti-β-actin, Proteintech cat# 66009-1-Ig) were diluted 1:1000 unless otherwise noted above in 5% BSA and incubated overnight at 4 °C. Blots were then washed, incubated for 1 h at room temperature in 1:10,000 diluted secondary antibody conjugated to horseradish peroxidase, and imaged using enhanced chemiluminescence (iBright CL1000; ThermoFisher). Image analysis and quantification was performed using ImageJ. All bands present in the representative images were subject to quantification.
RNA sequencing
Differentiated cells were lysed and total RNA was extracted using RNeasy Mini Kit (Qiagen). RNA was quantified using NanoDrop and normalized to 200 ng/μl in nuclease-free water. RNA integrity was assessed with Agilent Bioanalyzer 2100. Libraries were constructed and sequenced using Lexogen QuantSeq. Briefly, library generation was performed using an oligodT primer, and double-stranded cDNA was purified with magnetic beads. Libraries were amplified using PCR, and transcripts were forward sequenced at 75 bp using NextSeq 500 (Illumina). BlueBee software was used to analyze alignment, and DESeq2 was used for differential expression analysis.
Superoxide production
Superoxide production was determined by addition of 100 mM of spin trap 5,5-dimethyl-1-pyrroline-N-oxide (Enzo Life Sciences) into the cell culture medium for the duration of treatment (55). The resulting conditioned media and crude cell extract were homogenized by sonication on ice and stored at −20 °C until time of assay. Superoxide radicals were detected by electron paramagnetic resonance (Bruker EMX Plus spectroscope) in a quartz flat cell at room temperature. Instrument parameters were 20 mW microwave power, 1.0 G modulation amplitude, 1 × 105 gain, 0.163 s time constant, and 80 G scan range. To improve signal-to-noise ratio, spectra were accumulated 4× for each sample. Quantitation was carried out by measuring and comparing the amplitudes of the first peaks on each spectrum.
Calcium flux
Live cell calcium flux was assessed as described with minor modifications (56). Briefly, cells were seeded onto 12-mm gelatin-coated glass coverslips and stained with 10 μM Calcium Green-1 AM (CG; Thermo Fisher Scientific) in 20% Pluronic F127 in dimethyl sulfoxide and counterstained with MitoTracker Red FM (MTR; Thermo Fisher Scientific) in carbogenated (95% O2-5% CO2) normal Krebs solution (124 mM NaCl, 25 mM NaHCO3, 3 mM KCl, 1 mM MgSO4·7H2O, 1.5 mM NaH2PO4, 2 mM glucose, and 2 mM CaCl2·2H2O) for 30 min at 29 °C. Coverslips were then transferred to a recording chamber, cells were illuminated with a 488-nm laser, and CG fluorescence was collected at 509 nm. MTR was excited/emitted at 581/644 nm. Cells were visualized with a 40× water immersion objective controlled by Improvision (PerkinElmer) software. Cells on coverslips were constantly perfused with normal Krebs solution at 29 °C at a rate of 2 ml/min. After approximately 10 min, the perfusion solution was switched to normal Krebs with 500 μM ATP for 5 min followed by a 5-min washout period. Images of CG- and MTR-labeled cells were continuously collected during the perfusion and quantified as change in fluorescence from baseline.
Statistical analysis
Statistical analyses were performed with GraphPad Prism (version 8). Statistical procedures from individual experiments are detailed in the respective figure legends. Normality of the models was assessed using a Kolmogorov–Smirnov and D'Agostino–Pearson test where appropriate. Significance was accepted as p < 0.05. Ingenuity (QIAGEN) was used to conduct untargeted pathway analysis using the following parameters: q < 0.01, fold induction >2 and <−2, and base mean >50. Differential analysis of RNA read count data was performed using DESeq2 software, which models read counts as a negative binomial distribution and uses an empirical Bayes shrinkage-based method to estimate signal dispersion and fold-changes as described (57). Gene expression signals were logarithmically transformed (to base 2) for all downstream analyses (the lowest expression value being set to 1 for this purpose). Statistical analysis of RNA sequencing was performed using R.
Data availability
The RNA sequencing data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (58) and are accessible through GEO Series accession number GSE162983.
Supporting information
This article contains supporting information.
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
The authors wish to thank Jacob R. Hall, Rachel Marino, and Cameron A. Matherne for technical support. The authors also wish to thank Richard Carmouche, Sara Webb, Jasleen Kaur, and Susan Newman of the Pennington Biomedical Research Center Genomics Core Facility for performing RNA sequencing. This research utilized core facilities funded in part by grants from the United States National Institutes of Health P20 GM135002, P20 GM103528, P30 DK072476, and U54 GM104940.
Author contributions
W. T. K., C. L. A., G. D., C. L. H., and J. P. K. conceptualization; W. T. K., C. L. A., E. R. M. Z., R. C. N., H. F., R. C. R., K. S., C. L. H., and J. P. K. methodology; W. T. K., C. L. A., E. R. M. Z., R. C. N., G. D., H. F., B. T., G. E. H., R. C. R., W. S. D., K. S., C. L. H., and J. P. K. formal analysis; W. T. K., E. R. M. Z., R. C. N., G. D., H. F., K. P., R. C. R., S. L.-D, W. S. D., K. S., and C. L. H. investigation; W. T. K. and C. L. A. data curation; W. T. K. and C. L. A. writing – original draft; W. T. K., C. L. A., E. R. M. Z., R. C. N., G. D., H. F., B. T., K. P., G. E. H., R. C. R., S. L.-D., W. S. D., K. S., C. L. H., and J. P. K. writing – review & editing; W. T. K., C. L. A., R. C. N., K. P., G. E. H., and R. C. R. visualization; C. L. A., C. L. H., and J. P. K. supervision; W. T. K., C. L. A., and J. P. K. project administration; R. C. N., R. C. R., K. S., and J. P. K. funding acquisition.
Funding and additional information
This research was funded, in part, by grants from the United States National Institutes of Health R01 DK108089 (J. P. K.), R01 DK103860 (R. C. N.), R01 DK108765 (R. C. R.), and R01 DK115749 (K. S.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Qi-Qun Tang
Supporting information
References
- 1.van der Bliek A.M., Sedensky M.M., Morgan P.G. Cell biology of the mitochondrion. Genetics. 2017;207:843–871. doi: 10.1534/genetics.117.300262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Houten S.M., Wanders R.J. A general introduction to the biochemistry of mitochondrial fatty acid beta-oxidation. J. Inherit. Metab. Dis. 2010;33:469–477. doi: 10.1007/s10545-010-9061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan D.C. Mitochondria: Dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Okamoto K., Shaw J.M. Mitochondrial morphology and dynamics in yeast and multicellular eukaryotes. Annu. Rev. Genet. 2005;39:503–536. doi: 10.1146/annurev.genet.38.072902.093019. [DOI] [PubMed] [Google Scholar]
- 5.Bereiter-Hahn J., Voth M. Dynamics of mitochondria in living cells: Shape changes, dislocations, fusion, and fission of mitochondria. Microsc. Res. Tech. 1994;27:198–219. doi: 10.1002/jemt.1070270303. [DOI] [PubMed] [Google Scholar]
- 6.Detmer S.A., Chan D.C. Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 2007;8:870–879. doi: 10.1038/nrm2275. [DOI] [PubMed] [Google Scholar]
- 7.Westermann B. Mitochondrial fusion and fission in cell life and death. Nat. Rev. Mol. Cell Biol. 2010;11:872–884. doi: 10.1038/nrm3013. [DOI] [PubMed] [Google Scholar]
- 8.Smirnova E., Shurland D.L., Ryazantsev S.N., van der Bliek A.M. A human dynamin-related protein controls the distribution of mitochondria. J. Cell Biol. 1998;143:351–358. doi: 10.1083/jcb.143.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smirnova E., Griparic L., Shurland D.L., van der Bliek A.M. Dynamin-related protein Drp1 is required for mitochondrial division in mammalian cells. Mol. Biol. Cell. 2001;12:2245–2256. doi: 10.1091/mbc.12.8.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Loson O.C., Song Z., Chen H., Chan D.C. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol. Biol. Cell. 2013;24:659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kashatus J.A., Nascimento A., Myers L.J., Sher A., Byrne F.L., Hoehn K.L., Counter C.M., Kashatus D.F. Erk2 phosphorylation of Drp1 promotes mitochondrial fission and MAPK-driven tumor growth. Mol. Cell. 2015;57:537–551. doi: 10.1016/j.molcel.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taguchi N., Ishihara N., Jofuku A., Oka T., Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- 13.Mears J.A., Lackner L.L., Fang S., Ingerman E., Nunnari J., Hinshaw J.E. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat. Struct. Mol. Biol. 2011;18:20–26. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kalia R., Wang R.Y., Yusuf A., Thomas P.V., Agard D.A., Shaw J.M., Frost A. Structural basis of mitochondrial receptor binding and constriction by DRP1. Nature. 2018;558:401–405. doi: 10.1038/s41586-018-0211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geisler S., Holmstrom K.M., Skujat D., Fiesel F.C., Rothfuss O.C., Kahle P.J., Springer W. PINK1/parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 16.Chen H., Detmer S.A., Ewald A.J., Griffin E.E., Fraser S.E., Chan D.C. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Archer S.L. Mitochondrial dynamics--mitochondrial fission and fusion in human diseases. N. Engl. J. Med. 2013;369:2236–2251. doi: 10.1056/NEJMra1215233. [DOI] [PubMed] [Google Scholar]
- 18.Hagenbuchner J., Scholl-Buergi S., Karall D., Ausserlechner M.J. Very long-/and long chain-3-hydroxy acyl CoA dehydrogenase deficiency correlates with deregulation of the mitochondrial fusion/fission machinery. Sci. Rep. 2018;8:3254. doi: 10.1038/s41598-018-21519-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molina A.J., Wikstrom J.D., Stiles L., Las G., Mohamed H., Elorza A., Walzer G., Twig G., Katz S., Corkey B.E., Shirihai O.S. Mitochondrial networking protects beta-cells from nutrient-induced apoptosis. Diabetes. 2009;58:2303–2315. doi: 10.2337/db07-1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jheng H.F., Tsai P.J., Guo S.M., Kuo L.H., Chang C.S., Su I.J., Chang C.R., Tsai Y.S. Mitochondrial fission contributes to mitochondrial dysfunction and insulin resistance in skeletal muscle. Mol. Cell. Biol. 2012;32:309–319. doi: 10.1128/MCB.05603-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liesa M., Shirihai O.S. Mitochondrial dynamics in the regulation of nutrient utilization and energy expenditure. Cell Metab. 2013;17:491–506. doi: 10.1016/j.cmet.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomes L.C., Di Benedetto G., Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 2011;13:589–598. doi: 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitra K., Wunder C., Roysam B., Lin G., Lippincott-Schwartz J. A hyperfused mitochondrial state achieved at G1-S regulates cyclin E buildup and entry into S phase. Proc. Natl. Acad. Sci. U. S. A. 2009;106:11960–11965. doi: 10.1073/pnas.0904875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rambold A.S., Kostelecky B., Elia N., Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. U. S. A. 2011;108:10190–10195. doi: 10.1073/pnas.1107402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Axelrod C.L., Fealy C.E., Mulya A., Kirwan J.P. Exercise training remodels human skeletal muscle mitochondrial fission and fusion machinery towards a pro-elongation phenotype. Acta Physiol. (Oxf.) 2019;225 doi: 10.1111/apha.13216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fealy C.E., Mulya A., Lai N., Kirwan J.P. Exercise training decreases activation of the mitochondrial fission protein dynamin-related protein-1 in insulin-resistant human skeletal muscle. J. Appl. Physiol. (1985) 2014;117:239–245. doi: 10.1152/japplphysiol.01064.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Akepati V.R., Muller E.C., Otto A., Strauss H.M., Portwich M., Alexander C. Characterization of OPA1 isoforms isolated from mouse tissues. J. Neurochem. 2008;106:372–383. doi: 10.1111/j.1471-4159.2008.05401.x. [DOI] [PubMed] [Google Scholar]
- 28.Schulz H. Beta oxidation of fatty acids. Biochim. Biophys. Acta. 1991;1081:109–120. doi: 10.1016/0005-2760(91)90015-a. [DOI] [PubMed] [Google Scholar]
- 29.Patron M., Raffaello A., Granatiero V., Tosatto A., Merli G., De Stefani D., Wright L., Pallafacchina G., Terrin A., Mammucari C., Rizzuto R. The mitochondrial calcium uniporter (MCU): Molecular identity and physiological roles. J. Biol. Chem. 2013;288:10750–10758. doi: 10.1074/jbc.R112.420752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serasinghe M.N., Chipuk J.E. Mitochondrial fission in human diseases. Handb. Exp. Pharmacol. 2017;240:159–188. doi: 10.1007/164_2016_38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishihara N., Nomura M., Jofuku A., Kato H., Suzuki S.O., Masuda K., Otera H., Nakanishi Y., Nonaka I., Goto Y., Taguchi N., Morinaga H., Maeda M., Takayanagi R., Yokota S. Mitochondrial fission factor Drp1 is essential for embryonic development and synapse formation in mice. Nat. Cell Biol. 2009;11:958–966. doi: 10.1038/ncb1907. [DOI] [PubMed] [Google Scholar]
- 32.Waterham H.R., Koster J., van Roermund C.W., Mooyer P.A., Wanders R.J., Leonard J.V. A lethal defect of mitochondrial and peroxisomal fission. N. Engl. J. Med. 2007;356:1736–1741. doi: 10.1056/NEJMoa064436. [DOI] [PubMed] [Google Scholar]
- 33.Palma E., Ma X., Riva A., Iansante V., Dhawan A., Wang S., Ni H.M., Sesaki H., Williams R., Ding W.X., Chokshi S. Dynamin-1-like protein inhibition drives megamitochondria formation as an adaptive response in alcohol-induced hepatotoxicity. Am. J. Pathol. 2019;189:580–589. doi: 10.1016/j.ajpath.2018.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santoro A., Campolo M., Liu C., Sesaki H., Meli R., Liu Z.W., Kim J.D., Diano S. DRP1 suppresses leptin and glucose sensing of POMC neurons. Cell Metab. 2017;25:647–660. doi: 10.1016/j.cmet.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu Q., Zhang H., Gutierrez Cortes N., Wu D., Wang P., Zhang J., Mattison J.A., Smith E., Bettcher L.F., Wang M., Lakatta E.G., Sheu S.S., Wang W. Increased Drp1 acetylation by lipid overload induces cardiomyocyte death and heart dysfunction. Circ. Res. 2020;126:456–470. doi: 10.1161/CIRCRESAHA.119.315252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsushima K., Bugger H., Wende A.R., Soto J., Jenson G.A., Tor A.R., McGlauflin R., Kenny H.C., Zhang Y., Souvenir R., Hu X.X., Sloan C.L., Pereira R.O., Lira V.A., Spitzer K.W. Mitochondrial reactive oxygen species in lipotoxic hearts induce post-translational modifications of AKAP121, DRP1, and OPA1 that promote mitochondrial fission. Circ. Res. 2018;122:58–73. doi: 10.1161/CIRCRESAHA.117.311307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rizzuto R., Marchi S., Bonora M., Aguiari P., Bononi A., De Stefani D., Giorgi C., Leo S., Rimessi A., Siviero R., Zecchini E., Pinton P. Ca(2+) transfer from the ER to mitochondria: When, how and why. Biochim. Biophys. Acta. 2009;1787:1342–1351. doi: 10.1016/j.bbabio.2009.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanwar J., Singh J.B., Motiani R.K. Molecular machinery regulating mitochondrial calcium levels: The nuts and bolts of mitochondrial calcium dynamics. Mitochondrion. 2020;57:9–22. doi: 10.1016/j.mito.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Witczak C.A., Fujii N., Hirshman M.F., Goodyear L.J. Ca2+/calmodulin-dependent protein kinase kinase-alpha regulates skeletal muscle glucose uptake independent of AMP-activated protein kinase and Akt activation. Diabetes. 2007;56:1403–1409. doi: 10.2337/db06-1230. [DOI] [PubMed] [Google Scholar]
- 40.Balu D., Ouyang J., Parakhia R.A., Pitake S., Ochs R.S. Ca(2+) effects on glucose transport and fatty acid oxidation in L6 skeletal muscle cell cultures. Biochem. Biophys. Rep. 2016;5:365–373. doi: 10.1016/j.bbrep.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright D.C., Hucker K.A., Holloszy J.O., Han D.H. Ca2+ and AMPK both mediate stimulation of glucose transport by muscle contractions. Diabetes. 2004;53:330–335. doi: 10.2337/diabetes.53.2.330. [DOI] [PubMed] [Google Scholar]
- 42.Georgiou D.K., Dagnino-Acosta A., Lee C.S., Griffin D.M., Wang H., Lagor W.R., Pautler R.G., Dirksen R.T., Hamilton S.L. Ca2+ binding/permeation via calcium channel, CaV1.1, regulates the intracellular distribution of the fatty acid transport protein, CD36, and fatty acid metabolism. J. Biol. Chem. 2015;290:23751–23765. doi: 10.1074/jbc.M115.643544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Favaro G., Romanello V., Varanita T., Andrea Desbats M., Morbidoni V., Tezze C., Albiero M., Canato M., Gherardi G., De Stefani D., Mammucari C., Blaauw B., Boncompagni S., Protasi F., Reggiani C. DRP1-mediated mitochondrial shape controls calcium homeostasis and muscle mass. Nat. Commun. 2019;10:2576. doi: 10.1038/s41467-019-10226-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cho H.M., Ryu J.R., Jo Y., Seo T.W., Choi Y.N., Kim J.H., Chung J.M., Cho B., Kang H.C., Yu S.W., Yoo S.J., Kim H., Sun W. Drp1-Zip1 interaction regulates mitochondrial quality surveillance system. Mol. Cell. 2019;73:364–376.e8. doi: 10.1016/j.molcel.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 45.Fujioka H., Tandler B., Cohen M., Koontz D., Hoppel C.L. Multiple mitochondrial alterations in a case of myopathy. Ultrastruct. Pathol. 2014;38:204–210. doi: 10.3109/01913123.2014.888114. [DOI] [PubMed] [Google Scholar]
- 46.Fujioka H., Tandler B., Rosca M., McCandless S.E., Katirji B., Cohen M.L., Rapisuwon S., Hoppel C.L. Multiple muscle cell alterations in a case of encephalomyopathy. Ultrastruct. Pathol. 2014;38:13–25. doi: 10.3109/01913123.2013.831158. [DOI] [PubMed] [Google Scholar]
- 47.Axelrod C.L., King W.T., Davuluri G., Noland R.C., Hall J., Hull M., Dantas W.S., Zunica E.R., Alexopoulos S.J., Hoehn K.L., Langohr I., Stadler K., Doyle H., Schmidt E., Nieuwoudt S. BAM15-mediated mitochondrial uncoupling protects against obesity and improves glycemic control. EMBO Mol. Med. 2020;12 doi: 10.15252/emmm.202012088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoon J.C., Ng A., Kim B.H., Bianco A., Xavier R.J., Elledge S.J. Wnt signaling regulates mitochondrial physiology and insulin sensitivity. Genes Dev. 2010;24:1507–1518. doi: 10.1101/gad.1924910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nieuwoudt S., Mulya A., Fealy C.E., Martelli E., Dasarathy S., Naga Prasad S.V., Kirwan J.P. In vitro contraction protects against palmitate-induced insulin resistance in C2C12 myotubes. Am. J. Physiol. Cell Physiol. 2017;313:C575–C583. doi: 10.1152/ajpcell.00123.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye F., Hoppel C.L. Measuring oxidative phosphorylation in human skin fibroblasts. Anal. Biochem. 2013;437:52–58. doi: 10.1016/j.ab.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 51.Galgani J.E., Johannsen N.M., Bajpeyi S., Costford S.R., Zhang Z., Gupta A.K., Ravussin E. Role of skeletal muscle mitochondrial density on exercise-stimulated lipid oxidation. Obesity (Silver Spring) 2012;20:1387–1393. doi: 10.1038/oby.2011.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muoio D.M., Noland R.C., Kovalik J.P., Seiler S.E., Davies M.N., DeBalsi K.L., Ilkayeva O.R., Stevens R.D., Kheterpal I., Zhang J., Covington J.D., Bajpeyi S., Ravussin E., Kraus W., Koves T.R. Muscle-specific deletion of carnitine acetyltransferase compromises glucose tolerance and metabolic flexibility. Cell Metab. 2012;15:764–777. doi: 10.1016/j.cmet.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Covington J.D., Noland R.C., Hebert R.C., Masinter B.S., Smith S.R., Rustan A.C., Ravussin E., Bajpeyi S. Perilipin 3 differentially regulates skeletal muscle lipid oxidation in active, sedentary, and type 2 diabetic Males. J. Clin. Endocrinol. Metab. 2015;100:3683–3692. doi: 10.1210/JC.2014-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koves T.R., Ussher J.R., Noland R.C., Slentz D., Mosedale M., Ilkayeva O., Bain J., Stevens R., Dyck J.R., Newgard C.B., Lopaschuk G.D., Muoio D.M. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab. 2008;7:45–56. doi: 10.1016/j.cmet.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 55.Shi H., Timmins G., Monske M., Burdick A., Kalyanaraman B., Liu Y., Clément J.-L., Burchiel S., Liu K.J. Evaluation of spin trapping agents and trapping conditions for detection of cell-generated reactive oxygen species. Arch. Biochem. Biophys. 2005;437:59–68. doi: 10.1016/j.abb.2005.02.028. [DOI] [PubMed] [Google Scholar]
- 56.Burke S.J., Stadler K., Lu D., Gleason E., Han A., Donohoe D.R., Rogers R.C., Hermann G.E., Karlstad M.D., Collier J.J. IL-1beta reciprocally regulates chemokine and insulin secretion in pancreatic beta-cells via NF-kappaB. Am. J. Physiol. Endocrinol. Metab. 2015;309:E715–E726. doi: 10.1152/ajpendo.00153.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghosh S., Wicks S.E., Vandanmagsar B., Mendoza T.M., Bayless D.S., Salbaum J.M., Dearth S.P., Campagna S.R., Mynatt R.L., Noland R.C. Extensive metabolic remodeling after limiting mitochondrial lipid burden is consistent with an improved metabolic health profile. J. Biol. Chem. 2019;294:12313–12327. doi: 10.1074/jbc.RA118.006074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgar R., Domrachev M., Lash A.E. Gene expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (58) and are accessible through GEO Series accession number GSE162983.