Abstract
Bacterial signaling histidine kinases (HKs) have long been postulated to function exclusively through linear signal transduction chains. However, several HKs have recently been shown to form complex multikinase networks (MKNs). The most prominent MKN, involving the enzymes RetS and GacS, controls the switch between the motile and biofilm lifestyles in the pathogenic bacterium Pseudomonas aeruginosa. While GacS promotes biofilm formation, RetS counteracts GacS using three distinct mechanisms. Two are dephosphorylating mechanisms. The third, a direct binding between the RetS and GacS HK regions, blocks GacS autophosphorylation. Focusing on the third mechanism, we determined the crystal structure of a cocomplex between the HK region of RetS and the dimerization and histidine phosphotransfer (DHp) domain of GacS. This is the first reported structure of a complex between two distinct bacterial signaling HKs. In the complex, the canonical HK homodimerization interface is replaced by a strikingly similar heterodimeric interface between RetS and GacS. We further demonstrate that GacS autophosphorylates in trans, thus explaining why the formation of a RetS-GacS complex inhibits GacS autophosphorylation. Using mutational analysis in conjunction with bacterial two-hybrid and biofilm assays, we not only corroborate the biological role of the observed RetS-GacS interactions, but also identify a residue critical for the equilibrium between the RetS-GacS complex and the respective RetS and GacS homodimers. Collectively, our findings suggest that RetS and GacS form a domain-swapped hetero-oligomer during the planktonic growth phase of P. aeruginosa before unknown signals cause its dissociation and a relief of GacS inhibition to promote biofilm formation.
Keywords: histidine kinase, bacterial signal transduction, protein-protein interaction, Pseudomonas aeruginosa, biofilm, multikinase network, RetS, GacS, two-component system, crosstalk
Abbreviations: BACTH, bacterial adenylate cyclase two-hybrid; DHp, dimerization and histidine phosphotransfer; HK, histidine kinase; HPt, histidine phosphotransfer; MKN, multikinase network; RR, response regulator
Sensor histidine kinase (HK)-linked signal transduction systems are the primary means whereby bacteria sense extracellular signals to shape an adaptive response (1, 2, 3). The classic two-component signaling system consists of autophosphorylation of the HK followed by phosphate transfer to a cognate response regulator (RR). In the closely related phosphorelay systems, there are two additional transfer steps. Here, the phosphate moves from the HK region to a receiver domain with no coupled output domain, then to a histidine phosphotransfer (HPt) protein, and from there finally to an RR (4, 5). The additional phosphotransfers allow for finer-tuned output regulation (2, 6). Hybrid signaling HKs contain a sensory domain, HK region, a receiver domain, and an HPt domain within a single polypeptide chain (5). Because the tethering of the HK region to the receiver domain confers specificity to the associated phosphotransfer step, the otherwise stringent evolutionary requirement for HK-RR complementarity is more relaxed in hybrid HKs (7). Cross talk between distinct phosphorelay chains was long thought to be undesirable and therefore forbidden (8, 9, 10). However, mounting evidence suggests the presence of intricately webbed multikinase networks (MKNs) (11). At this point, we have gained a reasonably clear understanding of how the linear phosphotransfer events are facilitated within a single relay, and we have only a cursory understanding of how such cross talk occurs and is regulated. To date, the best studied example of interactions within an MKN is perhaps the multilayered interplay between the HK family enzymes RetS and GacS in the opportunistic pathogen Pseudomonas aeruginosa. The hybrid HK GacS and its cognate RR GacA sit at the heart of the Gac/Rsm signal transduction pathway (12). This pathway allows P. aeruginosa to switch between a motile, invasive lifestyle—which causes an acute infection in a human host—and a sessile, biofilm-associated lifestyle—which often results in a chronic infection in a human host (13, 14). Once phosphorylated, GacA acts as a transcriptional activator, indirectly upregulating genes associated with the sessile biofilm lifestyle (15, 16, 17). Conversely, GacA indirectly downregulates genes associated with a motile, invasive lifestyle, such as the expression of flagella-mediated motility-related genes and Type Three Secretion System–related genes necessary for producing the observed cytotoxic effects in an acute infection (15, 16).
GacS is reciprocally regulated by two HK family proteins, LadS and RetS (11). LadS enhances the phosphotransfer activity of GacS via phosphorylation of the HPt domain of GacS (18, 19). RetS, on the other hand, inhibits GacS via three distinct mechanisms (summarized in Fig. S1) (11, 12, 20). RetS has an unusual architecture consisting of a periplasmic sensor domain, an HK region, and two receiver domains. RetS uses its HK region and C-terminal receiver domain to siphon phosphate groups from the receiver and DHp domains of GacS, respectively (11, 20). Mediated by direct interactions between the HK regions of the two enzymes, RetS also interferes with the initial autophosphorylation of GacS (11, 12, 21). This mode of inhibition is not well understood and is the focus of the present study. Initial models suggested that RetS might form a heterodimeric complex with GacS (12). However, our recent work demonstrated that the GacS dimer remains intact upon RetS binding (21), suggesting the formation of a larger heteromeric assembly, perhaps a tetramer (21). In the same study we also demonstrated that a structurally dynamic region of the RetS DHp domain is important for GacS binding and might be involved in regulating the interaction.
In the present study, we report the crystal structure of a complex between the RetS HK region and the GacS DHp domain. The RetS-GacS interface closely resembles the canonical interface in homodimeric enzymes. Consistent with the proposed role of helix cracking in the regulation of the interaction, the structurally dynamic helix of RetS DHp is fully formed and involved in GacS binding. We experimentally determined that GacS autophosphorylates in trans. Thus, the RetSHK-GacSDHp structure also answers the question how RetS prevents GacS autophosphorylation, because RetS binding disrupts the spatial arrangements needed for trans-autophosphorylation.
Results
RetS and GacS form a DHp-DHp interface that closely resembles the dimerization interface in canonical signaling histidine kinases
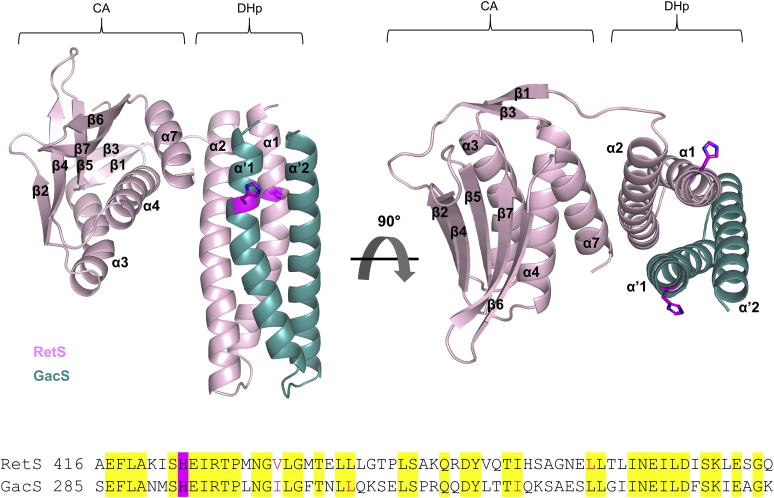
Cocrystallization of the HK region of RetS (RetSHK, amino acid residues 413–649) and the DHp domain of GacS (GacSDHp, amino acid residues 270–349) yielded crystals that gave X-ray diffraction data up to 2.3 Å resolution using a CC1/2 threshold of 0.3 as cutoff (data collection and structure refinement statistics are provided in Table 1) (22). The structure was solved via molecular replacement using a single molecule of the previously solved RetSHK dimer as search model (21). The complex consists of a 1:1 heterodimer wherein the DHp domain of RetS and GacS forms an extensive interface (Fig. 1). The final model contains residues 414–573, 604–639 of RetS and GacS residues 285–344. RetS residues 413, 574–603, and 640–649 appear to be structurally dynamic in the complex because no electron density was observed for these sections of the molecule. Similarly, there was no interpretable electron density for GacS residues 270–284 and 345–349. RetSHK consists of a CA and a DHp domain. The overall folds of the individual domains mirror those observed in the crystal structure of the RetSHK homodimer (21). The RetS-DHp domain assumes the canonical helix-loop-helix fold wherein the conserved histidine residue H424 is located on the α1 helix and solvent-exposed. The RetS-CA domain assumes the expected α/β sandwich fold comprising the α3–α4 helices, the α7 helix, and strands β1–β7. The residues that formed helices α5 and α6 helices in the RetSHK homodimer show no electron density in the RetS-GacS complex (residues 574–603) (Fig. S2). The DHp domain of GacS forms the anticipated helix-loop-helix structure. The conserved catalytic histidine residue H293 is also solvent-exposed.
Table 1.
X-ray diffraction data collection and refinement statistics
| X-ray diffraction data statistics | |
| Space Group | P 3(1) 2 1 |
| Unit Cell: a, b, c (Å) | 103.4, 103.4, 62.0 |
| Unit Cell: α, β, γ (º) | 90, 90, 120 |
| Resolution range (Å) | 44.76–2.29 (2.37–2.29) |
| Total reflections | 369,869 (36,066) |
| Unique reflections | 17,507 (1708) |
| Multiplicity | 21.1 (21.1) |
| Completeness (%) | 100 (100) |
| I/σ(I) | 16.9 (0.4) |
| Rmerge | 0.147 (13.7) |
| CC1/2 threshold | 0.3 |
| Refinement statistics | |
| Resolution (Å) | 44.76–2.30 |
| Rwork/Rfree | 0.2475/0.2662 |
| Root mean square bonds (Å) | 0.004 |
| Root mean square angles (º) | 0.815 |
| Average B factor (Å2) | 89.6 |
Outer shell statistics are provided in parentheses.
Figure 1.
Crystal structure of the 1:1 RetSHK-GacSDHpcomplex. The binding interface replaces the RetS and GacS homodimeric interfaces. RetS is shown in light pink. GacS is shown in turquoise. Catalytic histidine residues are shown in magenta. Shown underneath the structure is a sequence alignment of the DHp domains of RetS and GacS. The conserved catalytic histidine residues are highlighted in magenta. Residues conserved between RetS and GacS are highlighted in yellow. Residues selected for mutagenesis are colored red.
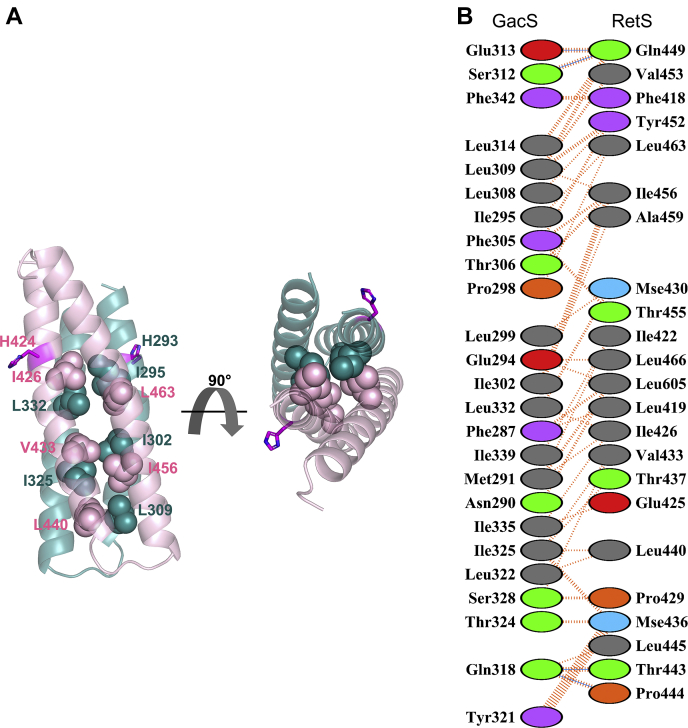
The DHp domains of RetS and GacS form a four-helix bundle, closely resembling the canonical interface observed in homodimeric bacterial HKs (1, 2, 23) (Fig. 2A). The same section of RetSHK also partakes in RetS homodimerization or, in the case of GacS, would also be predicted to form the binding surface in a GacS homodimer. Altogether 25 GacS residues corresponding to a surface area of 1351 Å2 and 23 RetS residues that cover 1348 Å2 of surface area form the extensive interface. The 89 nonbonded contacts are largely hydrophobic, containing only four hydrogen bonds (Fig. 2B).
Figure 2.
The RetS-GacS DHp-DHp interface.A, hydrophobic residues form the core of the RetS-GacS heterodimeric four helix bundle. RetS is shown in light pink. GacS is shown in turquoise. Catalytic histidine residues are shown in magenta. Hydrophobic interface residues shown as spheres. B, interacting interface residues. PDBsum was used to generate schematic cataloguing interactions between RetS and GacS. Dashed orange lines represent nonbonded contacts. Blue lines represent hydrogen bonds (62).
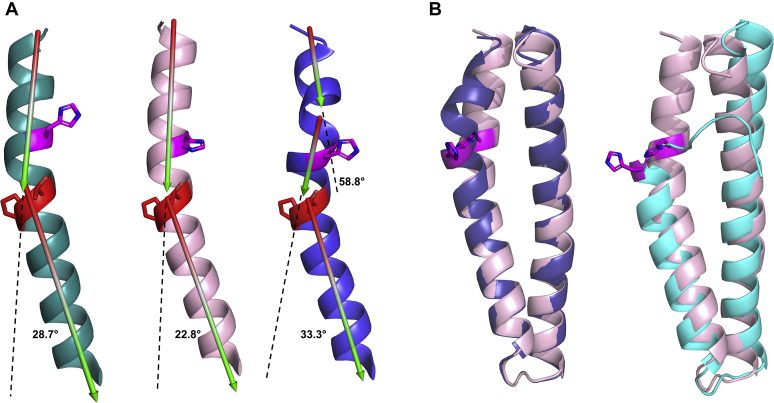
In HKs the α1 helix is kinked N-terminal at highly conserved threonine and proline residues. The kinking provides the plasticity needed for autophosphorylation, phosphotransfer, and phosphatase activity (24). This kink is also observable N-terminal to Thr428 and Pro429 of RetS and N-terminal to Thr297 and Pro298 in GacS (Fig. 3A). The kink angle for GacS is 28.7° and the angle for RetS is 22.8° (Fig. 3A). In the RetSHK homodimer, the corresponding section is unfolded in one molecule and displays a kink angle of 33.3° in the other molecule (Fig. 3). While RetS is not a functional kinase, the kink does play a role in regulating the equilibrium between a domain-swapped RetS-GacS oligomer and the individual homodimers (21). We previously demonstrated that this section is critical for GacS binding but not RetS homodimerization and predicted that it would be helical in the heteromeric complex (21). Consistent with these predictions, the dynamic N-terminal section of RetS α1 helix is now helical and forms part of the interface in the complex with GacS (Fig. 3B).
Figure 3.
Conserved DHp kink in RetS and GacS and fully folded RetS DHp.A, conserved kink in the N-terminal sections of the DHp domain α1 helix. Angles of conserved α1 helix kinks N-terminal to conserved threonine and proline residues (GacS Thr297 and Pro298, and RetS Thr428 and Pro429). GacS is shown in turquoise. RetS is shown in light pink. Shown in blue is the dually kinked structure of the corresponding section in molecule A of the homodimeric RetSHK structure (PDB 6dk7). The catalytic histidine residues are shown in magenta. The conserved threonine and proline residues are shown in red. B, RetS DHp is fully folded in the heterodimeric complex. Alignment of the DHp domain of RetS with the DHp domain of the RetS homodimer Chain A (PDB 6dk7) (RMSD = 0.539) visualizes the fully folded α1 helix of the DHp domain of RetS in the heterodimeric complex (left image). Alignment of the DHp domain of RetS with the DHp domain of the RetS homodimer Chain B (PDB 6dk7) (RMSD = 1.864) visualizes the fully folded α1 helix of the DHp domain of RetS in the heterodimeric complex (right image). RetS is shown in light pink. RetS homodimer Chain A is shown in blue. RetS homodimer Chain B is shown in cyan. The catalytic histidine residues are shown in magenta.
GacS binding forces conformational changes in RetSHK
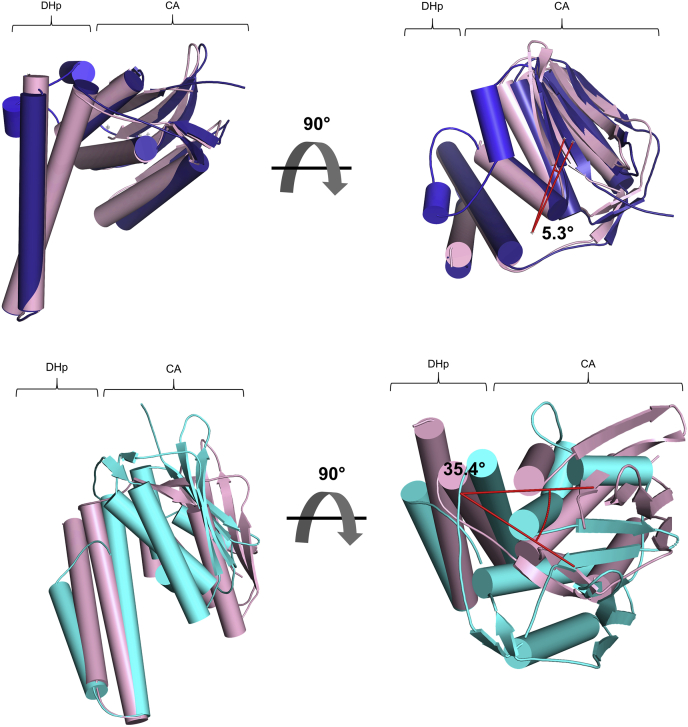
In the RetS-GacS complex, RetSHK assumes a distinct conformation compared to those observed in the RetSHK homodimer. Overall, it closely resembles Chain A (PDB code 6dk7) with an RMSD of 0.359 Å between the DHp domains. Here, the positions of the CA domains are very similar as demonstrated by the 5.3° angle of rotation between the two CA domains (Fig. 4). A larger angle of rotation between the two CA domains of 35.4° accounts for the larger overall RMSD of 1.53 Å between the RetSHK molecule in the RetS-GacS complex and Chain B of the RetSHK homodimer (Fig. 4).
Figure 4.
RetS CA domain movement. Alignment of the DHp domain of RetS with the DHp domain of the RetS homodimer Chain A (PDB 6dk7) (RMSD = 0.359) visualizes slight movement of the CA domain as demonstrated by the angle of rotation between the CA domains of 5.31° (top image). Alignment of the DHp domain of RetS with the DHp domain of the RetS homodimer Chain B (PDB 6dk7) (RMSD = 1.533) visualizes the greater movement of the CA domain as demonstrated by the angle of rotation between the CA domains of 35.41° (bottom image). RetS is shown in light pink. RetS homodimer Chain A is shown in blue. RetS homodimer Chain B is shown in cyan.
Beyond the relative movements of the CA and DHp domains, GacS binding significantly impacts regions that were previously implicated in the regulation of the RetS-GacS interaction. In the RetSHK-GacSDHp complex, residues 574–603 encompassing a section of the molecule containing the so-called ATP lid loop are not structured. In the asymmetric RetSHK homodimer, the ATP lid loop regions assume two distinct but well-defined conformations consisting of a short N-terminal helix and the lid loop (21) (Fig. S2). The ATP lid loop of other HKs also displays conformational plasticity, such as in the HK from Bacillus subtilis DesK (25). However, RetS has lost the ability to bind ATP (21). Instead, the lid region from one RetSHK molecule forms a short helix that displaces an unfolded section of α1 helix from the other molecule at the DHp-DHp interface (21). The biological significance of these interactions was corroborated in vitro and in vivo (21). In the RetS-GacS complex, the α1 helix of GacS DHp is fully folded, while the lid region is now dislodged from the interface and apparently unstructured.
GacS autophosphorylates in trans
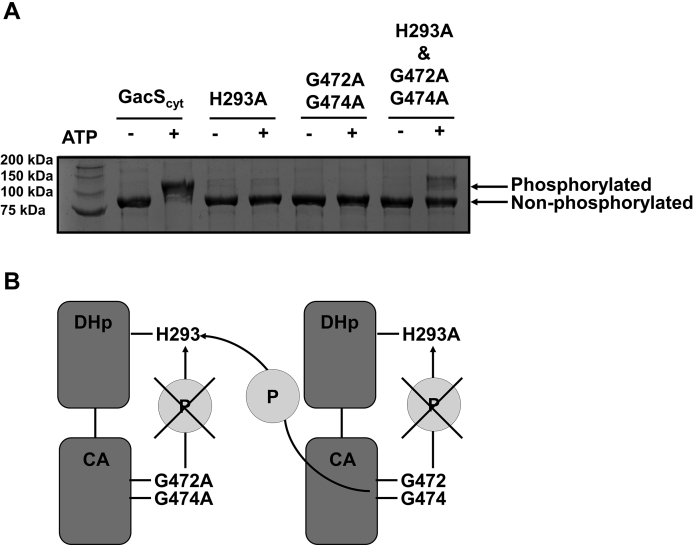
Because RetS binding disrupts the DHp–DHp interface of the GacS dimer, we reasoned, this interaction should interfere with GacS autophosphorylation if GacS actually autophosphorylates in trans. Many HKs autophosphorylate in trans, although some have been demonstrated to autophosphorylate in cis (26, 27). BarA, the GacS homolog in Escherichia coli autophosphorylates in trans (28). The handedness of the loop between the α1 and α2 helices of the DHp domain may also be used to predict whether an HK will autophosphorylate in trans or in cis (26). Given that the GacS homolog autophosphorylates in trans and, in the structure, GacS has a right-handed loop between the α1 and α2 helices of the DHp domain, GacS was predicted to autophosphorylate in trans as well. To test this prediction, an autophosphorylation assay followed by Zn2+-Phos-tag SDS-PAGE was performed (Fig. 5). GacScyt (GacS 219–925) and the GacScyt variants H293A and G472A G474A were examined in the assay. Variation of the conserved HK region histidine residue (H293) to an alanine inhibits GacS autophosphorylation (28). The variation of conserved G2 box residues G472 and G474 to alanines inhibits the ability of GacScyt to bind ATP. The G2 box is a conserved region in HKs located within the CA domain, which binds ATP (28). Individual variant constructs (GacScyt H293A and GacScyt G472A G474A) are unable to autophosphorylate in cis, but when both variant constructs are introduced into the autophosphorylation assay, they can autophosphorylate in trans. The observed mobility shift when both GacScyt H293A and GacScyt G272A G474A were present in the autophosphorylation assay demonstrated that GacS autophosphorylates in trans, thus providing evidence supporting the proposed mechanism by which RetS inhibits GacS autophosphorylation (Fig. 5).
Figure 5.
GacS autophosphorylates in trans.A, an autophosphorylation assay followed by Zn2+-Phos-tag SDS-PAGE was used to examine the autophosphorylation of GacScyt and GacScyt variants in the absence and presence of ATP. GacScyt wild-type, GacScyt H293A (a variant that cannot undergo autophosphorylation), GacScyt G472A G747A (a variant that cannot bind ATP), and an equimolar ratio of GacScyt H293A and GacScyt G472A G474A were used to assess the ability of GacS to autophosphorylate in trans or in cis. GacScyt and the GacScyt variants are 77 kDa. Each lane contains 7.74 μg protein. GacScyt has three potential phosphorylation sites (the catalytic histidine in the DHp domain, the conserved aspartate in the receiver domain, and the conserved histidine in the Hpt domain). B, autophosphorylation assay. Individual variant constructs (GacScyt H293A and GacScyt G472A G474A) are unable to undergo cis autophosphorylation, but when both variant constructs are introduced into the autophosphorylation assay, they can autophosphorylate in trans. The histidine kinase region is shown for clarity even though the assay was performed with the cytosolic region of GacS.
GacS L309 and I302 are critical for promoting complex formation with RetS
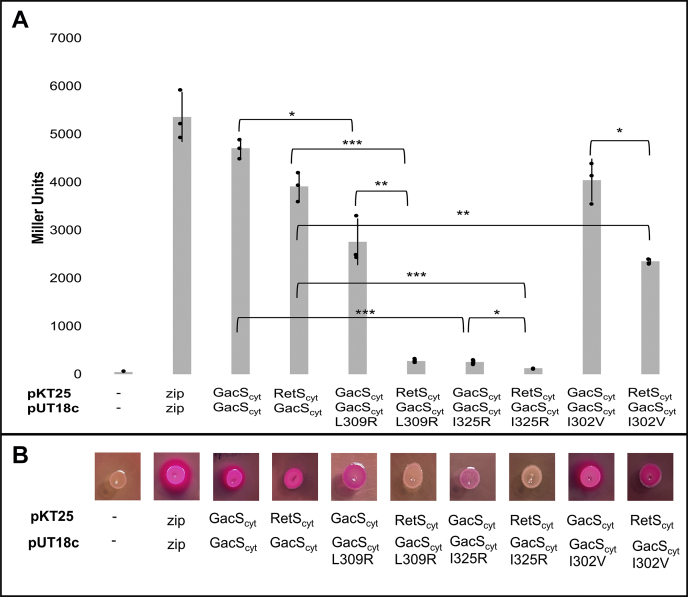
In order to experimentally corroborate the RetS-GacS interface found in the crystal structure, a number of interface residues were mutated and the variants examined via the bacterial adenylate cyclase two-hybrid (BACTH) assay. The DHp domains of RetS and GacS share a 56.9% amino acid sequence identity (Fig. 1), whereas the entire HK regions of RetS and GacS share 40.4% sequence identity. The high degree of conservation in the DHp domain might explain the overall complementarity of their molecular surfaces. Yet, we also sought to identify distinctive residues that promote the formation of a RetS-GacS complex at the DHp-DHp interface over the formation of the typical homodimers. The interactions of the cytoplasmic region of GacS (GacScyt) with itself and with the cytoplasmic region of RetS (RetScyt) were used as positive controls and reference points (Fig. 6). The GacS homodimer is expected to contain multiple dimerization interfaces including HAMP-HAMP and DHp-DHp domains (29). Therefore, the RetS-GacS interactions are expected to be more sensitive to mutations disrupting the interactions between the DHp domains than the more extensively paired GacS homodimer. A complicating factor in this analysis was the observation we made in prior experiments that monomeric RetS and GacS are not stable proteins. A variant that has completely lost the ability to dimerize could either be misfolded or simply be unstable because it can no longer dimerize. Therefore, we were particularly interested in identifying GacS residues that are critical for RetS binding but not for GacS dimerization. A number of residues were probed. The GacS L309R mutation attenuated homodimerization but completely disrupted the RetScyt-GacScyt interaction (Figs. 6 and S3) confirming that L309 is important in the RetS-GacS complex. The GacScyt I325R mutation abrogated both GacScyt homodimerization and the RetScyt-GacScyt interaction (Figs. 6 and S3). GacScyt I302V formed a stable homodimer, while binding to RetScyt was partially disrupted (Figs. 6 and S3). This suggests that the GacScytI302V construct is stable and, as predicted by the crystal structure, that I302 is involved in RetS binding. Two RetScyt variant constructs were also examined in the BACTH assay. RetScyt L463R could no longer form homodimers or bind to GacS (Fig. S4). The RetScyt V433I mutation was too subtle as the substitution had no significant impact on RetScyt homodimerization or the RetScyt-GacScyt interface (Fig. S4).
Figure 6.
Interface variants in the BACTH assay.A, examination of interface variants in the BACTH β-galactosidase assay. Interface variants were examined in the BACTH β-galactosidase assay after 24 h incubation at 30 °C. Assay was performed in triplicate. Statistical significance was determined by a two-tailed, nonpaired Student's t test. ∗p < 0.01, ∗∗p < 0.001, ∗∗∗p < 0.0001. B, examination of interface variants in the BACTH MacConkey agar assay. Interface variants were examined in the BACTH MacConkey agar assay after 24 h incubation at 32 °C. Assay was performed in triplicate.
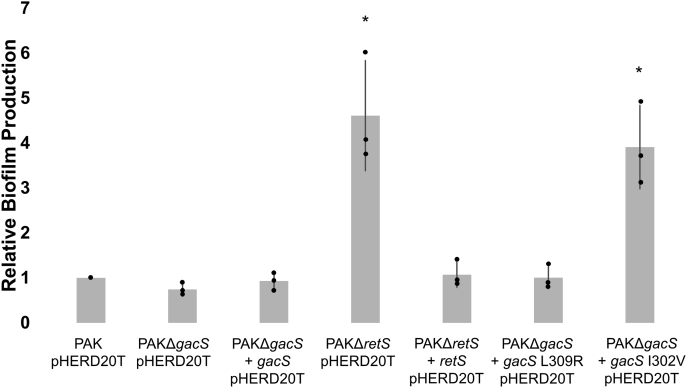
Because the GacS L309R and GacS I302V mutations showed differential binding profiles in the BACTH assays, we decided to examine their impact on P. aeruginosa biofilm formation. Both substitutions were introduced into the full-length gacS gene and the resulting proteins were expressed in trans in a PAKΔgacS strain. A crystal violet assay was used to monitor production of biofilm formation-associated carbohydrates. At the early growth stage, biofilm formation is closely held in check by RetS and the gacS deletion strain is essentially indistinguishable from the wild-type or a complemented strain (Fig. 7). However, we predicted that GacS mutations that selectively prevent RetS binding but not GacS dimerization should cause a hyperbiofilm phenotype akin to what is observed in a ΔretS strain because here RetS would no longer be able to block GacS autophosphorylation. Indeed, the I302V mutation, which had resulted in a moderate disruption to the heterodimeric interface in the BACTH assay, produced a gain-of-function phenotype comparable to the hyperbiofilm phenotype of the retS mutant (Fig. 7) (13). The L309R variant, on the other hand, produced no phenotypic change, suggesting that the local impact of this substitution on the DHp-DHp interface not only interferes with RetS binding but also prevents GacS autophosphorylation (Fig. 7).
Figure 7.
In vivo biofilm assay assessing GacS variants. Relative biofilm production assessed by the crystal violet biofilm assay. PAKΔgacS + pHERD20T-gacS I302V was demonstrated to have a phenotype comparable to that of PAKΔretS pHERD20T in the crystal violet biofilm assay after incubation at 37 °C for 6 h. The complemented strains and PAKΔgacS + pHERD20T-gacS L309R were demonstrated not to be significantly different from PAK pHERD20T. Assay was performed in triplicate. ∗ indicates that the strain demonstrated significantly more biofilm production than PAK pHERD20T. Statistical significance was determined by a two-tailed, nonpaired Student's t test. ∗p < 0.01.
Discussion
Initially, the Gac/Rsm pathway was discovered as a central signal transduction pathway in Pseudomonads that regulates the production of secondary metabolites (e.g., antimicrobials, hydrogen cyanide, siderophores) and also the switch between a motile, invasive lifestyle and a sessile biofilm-associated lifestyle (16, 30, 31, 32, 33). However, beyond the biological significance of this particular signaling pathway, the GacS-GacA system has become the model for studying crosswise interactions between multiple signaling kinases. HKs have been demonstrated to maintain a high degree of fidelity for their cognate RRs and vice versa, but we are beginning to recognize that MKNs are often necessary to control complex outputs (9, 34). Such MKNs were once postulated to be prohibited, but it now appears many bacterial species use them to integrate diverse extracellular signals to regulate adaptive responses (34). MKNs control transitions associated with virulence, response to switching from aerobic to anaerobic conditions, the integration of diverse quorum sensing signals, as well as sporulation and fruiting body formation (34). However, none is more complex than the MKN associated with the regulation of the GacS-GacA system. At least seven HKs coordinate their signaling to fine-tune P. aeruginosa gene expression. LadS and RetS do so through direct interactions with GacS (11, 12, 18, 20), while PA1611 appears to sequester RetS to promote GacS signaling (35, 36). However, RetS, PA1611, ErcS, and SagS all appear to also interact with HptB to modulate RsmY levels (37, 38, 39, 40). SagS interacts with BfiS to integrate the BfiS/BfiR system, which promotes biofilm formation into the MKN (39). The mechanism whereby these signaling pathways integrate are varied and, in some cases, multifaceted as the interactions can be both activating and suppressing. Often, the molecular mechanisms underlying the MKNs can be readily understood as they involve well-characterized protein–protein interactions mirroring canonical signaling pathways. The basis for the direct pairwise interactions of the HK regions observed for RetS-GacS but also RetS-PA1611 and SagS-BfiS in P. aeruginosa, as well as the DivL-CckA interactions in Caulobacter crescentus are less well understood (41). The present evidence of domain-swapping between DHp domains in the RetS-GacS complex suggests that once again MKN signaling evolved from known contact interfaces of regular linear HK systems. Although, earlier work suggests that the interface formed between PA1611 and RetS involves the DHp domain of PA1611 and the beta-sheet of the CA domain of RetS, suggesting an interface that does not have an equivalent in known HK contacts (36). The present structure may broadly represent the basis for how heteromeric HK-HK interactions inhibit autophosphorylation in MKNs.
The RetSHK-GacSDHp complex structure answers the question of how binding between the RetS and GacS DHp domains prevents GacS autophosphorylation because the formation of a heterodimeric DHp-DHp interface should inhibit GacS trans-autophosphorylation (11, 12). However, perhaps this inhibition is not complete, thus explaining why RetS uses not one but three distinct mechanisms to inhibit GacS signaling (Fig. S1). Potential trans-autophosphorylation of GacS in a heteromeric RetS-GacS complex is at this point only speculative; however, the siphoning of phosphates from the catalytic histidine in GacS-HK by the second receiver domain of RetS would otherwise appear to be redundant. Yet, Francis et al. (11) demonstrated that this phosphatase activity is critical for inhibiting GacS signaling in vivo. The RetS HK region also dephosphorylates the receiver domain of GacS in a manner similar to transmitter phosphatase activity (2, 11, 42). It is not known if RetS-GacS binding through the DHp-DHp increases the efficiency or is in fact a prerequisite for the efficient working of the two other inhibitory mechanisms. In recent years, some progress has been made toward elucidating the roles of periplasmic sensory domains of RetS and GacS in regulating their interplay. Remarkably, the sensory domain of RetS appears to promote the inhibition of GacS when exposed to host cell-derived mucins, while P. aeruginosa lysis releases a molecular signal, also recognized by the RetS sensory domain that causes GacS activation (43, 44). The sensory domain of GacS, on the other hand, is required for GacS activation, but the longstanding hunt for the elusive ligand is ongoing (32).
Overall, the present study has uncovered the novel heterodimeric DHp-DHp interface in the RetS-GacS complex, which readily explains how direct binding of RetS-HK to GacS-HK interferes with GacS trans-autophosphorylation. The observed RetSHK-GacSDHp structure is also consistent with the proposed model for regulation of RetS-GacS binding via RetS helix-cracking, which predicted that a structurally dynamic section of RetS would form the N-terminal end of the DHp α1 helix and interact with GacS (21). Another structurally dynamic feature of the RetS-HK dimer, the so-called ATP lid loop was shown to play an important role in stabilizing the RetS homodimer but not the RetS-GacS complex (21). Consistent with this prediction, the ATP lid loop region and a short α helix N-terminal to the ATP lid loop region of the RetS CA domain are unstructured in the RetSHK-GacSDHp complex. The mutational analysis of the DHp-DHp interface offered additional insight into which residues might be critical in providing specificity for the unusual heteromeric RetS-GacS interactions in favor of the RetS-RetS and GacS-GacS interfaces. We demonstrated upon variation of select residues (GacS I302, GacS L309) an inhibition to binding in the heterodimeric interface of the cytoplasmic regions, but not an equivalent inhibition to binding in the homodimeric interface of the cytoplasmic regions. We also demonstrated a phenotype comparable to the hyperbiofilm retS mutant for the GacS I302V strain in an in vivo assay, demonstrating the importance of I302 in RetS binding (13).
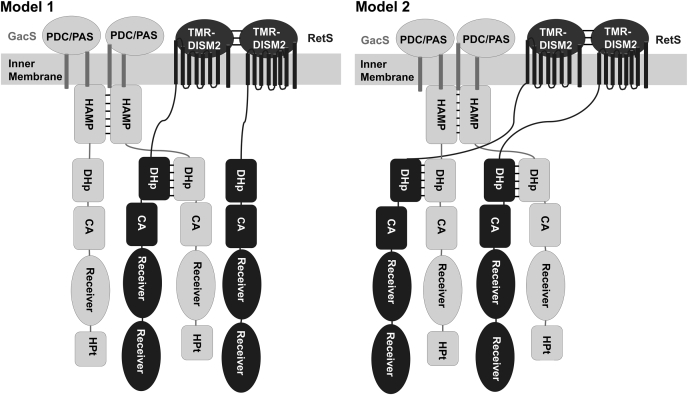
There is a disparity between the observation that RetS disrupts the GacS DHp-DHp dimerization interface and our previous finding that RetS overall does not disrupt the GacS homodimer (20). This apparent contradiction may be explained by the fact that the GacS protein construct used in the original FRET measurements included not only the histidine kinase region but also the HAMP domain of GacS (21). HAMP domains are ubiquitous signaling domains of signaling HKs and methyl accepting chemotaxis proteins and facilitate homodimerization and signal transduction by forming structurally dynamic intermolecular four-helix bundles (45). The GacS HAMP domain appears to maintain GacS-GacS association even in the presence of RetS (Fig. 8). This observation is also consistent with the finding that the HAMP domain is required for GacS homodimerization in Pseudomonas fluorescens (29). Similarly, the periplasmic domain of RetS has also been demonstrated to dimerize in vitro (46, 47). If and how this interaction is affected by GacS binding is unknown.
Figure 8.
Models for RetS-GacS tetramers. Binding between the two proteins could be asymmetric (Model 1), where only one DHp-DHp interface forms. This model allows for the possible formation of a polymer consisting of repeat units of asymmetric tetramers linked through DHp-DHp interactions. Alternatively, RetS and GacS could form a symmetric tetramer with two DHp-DHp interfaces (Model 2).
Collectively, the present work and previous results support a model in which RetS and GacS form a domain-swapped complex (Fig. 8). The exact stoichiometry and size of this complex remain to be determined. While the presence of additional GacS-GacS and RetS-RetS interfaces might make it tempting to propose the formation of a symmetric heterotetramer (Fig. 8, Model 2), steric factors may create an asymmetric complex, cause dissociation of the RetS dimer, or may even facilitate the formation of a larger polymeric structure consisting of alternating RetS and GacS dimers.
Experimental procedures
Cloning and site-directed mutagenesis
The plasmid construct for the expression of RetSHK (residues 413–649) from the pDEST-HisMBP plasmid was created previously (21). For the expression of the GacSDHp protein, the section of the gacS gene that encodes residues 270–349 was amplified via PCR from pDEST-HisMBP-GacSHK using GacS_350_stop_F and GacS_350_stop_R primers. The PCR product was cloned into pDONR221 and from there into pDEST-HisMBP using Gateway recombinational cloning (Thermo Fisher) to create pDEST-HisMBP-GacSDHp.
Constructs for the BACTH assay were generated using standard cloning protocols. pKT25-RetScyt-GFP, encoding RetS residues 387–942 in frame with the gfp gene, was generated previously (21). pKT25-RetScyt-GFP L463R and V433I variants were created using Agilent QuickChange XL site-directed mutagenesis kit (Agilent) following the manufacturer's protocol. A pUT18c-GacSHK plasmid was created by introducing a stop codon after gacS codon 509 into the previously generated pUT18c-GacScyt (GacScyt includes residues 219–925) plasmid using the Agilent QuickChange XL site-directed mutagenesis kit (Agilent) and GacS509STPx2_F and GacS509STPx2_R primers following the manufacturer's protocol (21). Agilent QuickChange XL site-directed mutagenesis kit (Agilent) was also used to create the pUT18c-GacSHK constructs expressing variants L309R, I325R, and I302V following the manufacturer's protocol. Ultimately, the pUT18c-GacSHK constructs expressing the original gacS sequence and the three mutated genes were only used in this study as templates to cut-and-paste the L309R, I325R, and I302V associated mutations into the pUT18c-GacScyt vector via internal restriction sites (XbaI and StuI) using standard restriction cloning protocols.
GacScyt variants GacScyt H293A and GacScyt G472A G474A used for the in vitro autophosphorylation assays were also generated with the Agilent QuickChange XL site-directed mutagenesis kit (Agilent) using the pQE60-GacScytvector as template, which was previously generated and generously shared with us by Dr Steven Porter's group (University of Exeter) (11).
The Agilent QuickChange XL site-directed mutagenesis kit (Agilent) was also used to create pHERD20T-gacS L309R and pHERD20T-gacS I302V variants using the previously generated pHERD20T-gacS vector as a template following the manufacturer's protocol (48, 49). Cloning and site-directed mutagenesis primers are listed in Table S1. Recombinant DNA used in this study is listed in Table S2.
Recombinant protein expression and purification
BL-21(DE3)(RIL) pDEST-HisMBP-RetSHK was grown in 6 L Lysogeny broth (LB) with 100 μg/ml ampicillin, 30 μg/ml chloramphenicol, and 10 g glucose/l at 37 °C, shaking at 250 rpm. The cultures were induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) after the OD600 reached 0.6, and incubated at 18 °C for 18 h, again shaking at 250 rpm. A 35.5 g cell pellet was resuspended in 200 ml NiNTA A buffer (buffer compositions are listed in Table S3) and 0.3 mM phenylmethanesulfonyl fluoride (PMSF) and lysed via sonication (21). The soluble fraction was collected via centrifugation for 1 h at 100,000g, 4 °C. HisMBP-RetSHK was purified via Ni-NTA affinity chromatography using a 30 ml Ni-NTA Superflow column (Qiagen) and a 150 ml linear gradient elution with Ni-NTA B buffer. SDS-PAGE was used to assess purification of HisMBP-RetSHK. To remove the HisMBP-tag, the collected protein was incubated with 1 mg His6TEV protease per 50 mg protein, as estimated by the UV280 absorption and extinction coefficient of the fusion protein, and dialyzed into Ni-NTA A buffer. A second 30 ml Ni-NTA Superflow column (Qiagen) was used to separate RetSHK from the HisMBP tag and the His6TEV protease. SDS-PAGE was used to assess purification of RetSHK. RetSHK was further purified using a HiTrap Q HP column (GE Life Sciences) with a 150 ml linear gradient of anion exchange buffer A and anion exchange buffer B. SDS-PAGE was used to assess purification of RetSHK. The eluted sample was polished on a 26/60 Superdex 200 column (GE Life Sciences) pre-equilibrated in RetSHK gel filtration buffer (21). SDS-PAGE was used to assess purification of RetSHK. Expression and purification protocols for GacSDHp from the pDEST-HisMBP-GacSDHp plasmid followed the same protocol as that for RetSHK (21).
Selenomethionine-substituted RetSHK was produced by altering the growth conditions prior to protein purification. BL-21(DE3)(RIL) pDEST-HisMBP-RetSHK was grown in 1 L Lysogeny broth (LB) with 100 μg/ml ampicillin, 30 μg/ml chloramphenicol and 10 g glucose at 37 °C, shaking at 250 rpm until the OD600 was 0.5. The culture was incubated on ice for 30 min, after which the cells were pelleted via centrifugation at 4000g, 4 °C for 15 min. The cell pellet was resuspended in 100 ml of M9 salts plus Medicilon noninhibitory amino acid cocktail (NIAAC), pelleted a second time at 4000g, 4 °C for 15 min, and then resuspended in 100 ml of M9 salts plus Medicilon NIAAC (Medicilon). Medicilon selenomethionine M9 medium (Medicilon) with 100 μg/ml ampicillin and 30 μg/ml chloramphenicol was inoculated with the resuspended pellet. The cultures were incubated at 37 °C for 30 min, after which time protein expression was induced with 1 mM IPTG, and the cultures were incubated at 18 °C for 18 h. Protein purification of selenomethionine-substituted RetSHK followed the same protocol as that for RetSHK (21).
To produce GacScyt the pQE60-GacScyt plasmid was transformed into the E. coli JM109 cell line. Cells were grown in LB medium with 100 μg/ml ampicillin at 37 °C for approximately 3 h until the OD600 was 0.6. The temperature was reduced to 18 °C and expression was induced with the addition of 1 mM IPTG. Induction continued for 18 h, after which cell pellets were harvested. Cell pellets were resuspended in 10 ml GacScyt NiNTA A buffer per gram cell pellet with 0.3 mM PMSF and then lysed via sonication. After centrifugation at 100,000g, 4 °C for 1 h, the supernatant was applied to a Ni-NTA Superflow column (Qiagen) for FPLC and eluted using a 150 ml linear gradient of GacScyt Ni-NTA A buffer and GacScyt Ni-NTA B buffer. SDS-PAGE was used to assess the purification of GacScyt. The sample was then applied to a 26/60 Superdex 200 gel filtration column (GE Life Sciences) for the final purification step using GacScyt gel filtration buffer. SDS-PAGE was used to assess GacScyt purification. (Buffer compositions are listed in Table S3.)
GacScyt pQE60 variants GacScyt H293A and GacScyt G472A G474A were expressed in E. coli JM109 cell line following the same protocol as for GacScyt. The purification followed the same protocol as for GacScyt.
RetSHK-GacSDHp crystallization and structure determination
Crystals containing the complex of seleno-methionine-substituted RetSHK with GacSDHp were grown by vapor diffusion in a 6 μl hanging drop containing a 5:1 volume ratio of protein:mother liquor, with 440 μM of each RetSHK and GacSDHp in gel filtration buffer. The mother liquor was composed of 30% PEG3350, 0.2 M Li2SO4, 0.1 M Tris-HCl, pH 8.5. The reservoir contained 0.4 ml of mother liquor. Crystals grew over a 2-week period at room temperature. The crystals were loop-mounted and flash frozen in liquid nitrogen. X-ray diffraction data were collected at Advanced Light Source Beamline 4.2.2 at Lawrence Berkeley National Laboratory. The diffraction images were processed and integrated with XDS and intensities converted to amplitudes using Aimless (50, 51, 52). The resolution was cut off at 2.3 Å using a CC1/2 threshold of 0.3 (22). The anomalous signal of the selenium atoms was not strong enough to facilitate structure solution. Therefore, the structure was solved with the PHASER molecular replacement tool within the Phenix suite using the structure of RetSHK (PDB 6DK8) as the search model (21, 53, 54). Iterative cycles of model building in Coot and automated refinements in Phenix and RefMac were used to build the RetSHK-GacSDHp structure (53, 55, 56, 57). The degree of rotation between the CA domain of RetS and the CA domains of the RetS homodimer (PDB 6dk7) was estimated via PyMOL(58). PyMOL was used to determine the kink angle in the α1 helix of GacS, RetS, and the RetS homodimer (PDB 6dk7) (58). PyMOL was also used to generate Figure 1, Figure 2, Figure 3, Figure 4 and S2 (58).
Bacterial adenylate cyclase two-hybrid (BACTH) assay
Interface variants were examined in the BACTH MacConkey agar assay and the BACTH β-galactosidase assay (59, 60). For the BACTH MacConkey agar assay, LB cultures containing 100 μg/ml ampicillin and 50 μg/ml kanamycin were incubated for 18 h at 37 °C. The OD600 was adjusted to 1.0. Two microliter of culture was dispensed onto MacConkey agar containing 0.5 mM IPTG, 1 (w/v) % maltose, 100 μg/ml ampicillin, and 50 μg/ml kanamycin. The MacConkey agar plates were incubated at 32 °C for 24 h. The BACTH MacConkey agar assay was performed in triplicate. For the BACTH β-galactosidase assay, LB cultures with 100 μg/ml ampicillin, 50 μg/ml kanamycin, and 0.5 mM IPTG were incubated for 24 h at 30 °C. OD600 was measured for each culture. Twenty microliter of culture was added to 80 μl permeabilization solution (100 mM Na2HPO4, 20 mM KCl, 2 mM MgSO4, 0.8 mg/ml cetrimonium bromide, 0.4 mg/ml sodium deoxycholate, 5.4 μl β-mercaptoethanol) (60). Samples were incubated for 30 min at 30 °C, after which, 0.6 ml of substrate solution (60 mM Na2PO4, 40 mM NaH2PO4, 1 mg/ml o-nitrophenyl β-D-galactoside, 2.7 μl β-mercaptoethanol) was added to the samples with the time noted (60). The reaction was stopped via the addition of 1 M Na2CO3 and the time was recorded (60). The samples were centrifuged at 16,000g for 10 min and absorbance at 420 nm was recorded (60). Miller Units were calculated using the following formula: 1000 × (Abs420/(OD600 × volume of sample in mL × reaction time in minutes)) (60). The BACTH β-galactosidase assay was performed in triplicate.
Autophosphorylation assay and Zn2+ Phos-tag SDS-PAGE
The Zn2+-Phos-tag assay closely followed the steps provided in the manufacturer's protocol, and all solutions were prepared according to the manufacturer's protocol (FUJIFILM Wako Chemicals). Autophosphorylation was performed via the incubation of 5 μM GacScyt or GacScyt variant proteins GacScyt H293A and GacScyt G472A G474A with 2 mM ATP for 30 min at 21 °C (11). The reaction was stopped via the addition of 3× loading buffer to a final concentration of 1× following manufacturer's protocol (FUJIFILM Wako Chemicals). The samples were analyzed via Zn2+-Phos-tag SDS-PAGE following manufacturer's protocol using 100 μM Phos-tag in a 10% acrylamide Zn2+-Phos-tag SDS-PAGE gel (FUJIFILM Wako Chemicals) (28). The autophosphorylation assay and Zn2+-Phos-tag SDS-PAGE were performed in triplicate.
Crystal violet biofilm assay
P. aeruginosa PAK strains were examined in the crystal violet biofilm assay. The strains were plated to LB agar containing 300 μg/ml carbenicillin. Individual colonies were used to inoculate LB containing 300 μg/ml carbenicillin, which was then incubated at 37 °C overnight while shaking at 250 rpm. The strains were subcultured into modified M63 media containing 0.5% arabinose and 300 μg/ml carbenicillin, which were incubated at 37 °C overnight while shaking at 250 rpm (61). The OD600 of the cultures was adjusted to 0.05 using modified M63 media containing 0.5% arabinose and 300 μg/ml carbenicillin. Hundred microliter of each culture was dispensed into 96-well plates (Corning # 2797) (61). Plates were covered with aluminum foil and incubated at 37 °C for 6 h. Following incubation, the media were removed via pipette and the wells were washed with water via pipette. The wells were stained with 0.1% crystal violet for 10 min and then washed with water via pipette (61). The remaining crystal violet-stained cells were solubilized in 125 μl of 30% acetic acid for 15 min (61). Hundred microliter of the solution was transferred to a 96-well plate (Corning # 3370) and absorbance at 600 nm was measured using a Tecan M200 plate reader.
Data availability
All data are contained within the manuscript.
Supporting information
This article contains supporting information (10, 11, 19).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We would like to thank Dr Stephen Porter (University of Exeter) for his gift of pQE60-GacScyt. We would also like to thank Dr Alain Filloux (Imperial College London) for his gift of P. aeruginosa PAK strains. Permission for the use of Lawrence Berkeley National Laboratory Advanced Light Source beamline 4.2.2 was provided by the US Department of Energy under contract DE-AC02-05CH11231.
Author contributions
K. M. R. K. and F. D. S. conceptualization; K. M. R. K., J. C. N., and F. D. S. data curation; K. M. R. K., J. C. N., and F. D. S. formal analysis; F. D. S. funding acquisition; K. M. R. K. investigation; K. M. R. K., J. C. N., and F. D. S. methodology; F. D. S. project administration; K. M. R. K., J. C. N., and F. D. S. resources; F. D. S. supervision; K. M. R. K. visualization; K. M. R. K. and F. D. S. writing–original draft; K. M. R. K. and F. D. S. writing–review and editing.
Funding and additional information
This study was supported by NIH grant R21AI128255-01A1 to F. D. S. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Chris Whitfield
Supporting information
References
- 1.Stock A.M., Robinson V.L., Goudreau P.N. Two-component signal transduction. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 2.Gao R., Stock A.M. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zschiedrich C.P., Keidel V., Szurmant H. Molecular mechanisms of two-component signal transduction. J. Mol. Biol. 2016;428:3752–3775. doi: 10.1016/j.jmb.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodrigue A., Quentin Y., Lazdunski A., Méjean V., Foglino M. Two-component systems in Pseudomonas aeruginosa: Why so many? Trends Microbiol. 2000;8:498–504. doi: 10.1016/s0966-842x(00)01833-3. [DOI] [PubMed] [Google Scholar]
- 5.Buschiazzo A., Trajtenberg F. Two-component sensing and regulation: How do histidine kinases talk with response regulators at the molecular level? Annu. Rev. Microbiol. 2019;73:507–528. doi: 10.1146/annurev-micro-091018-054627. [DOI] [PubMed] [Google Scholar]
- 6.West A.H., Stock A.M. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- 7.Capra E.J., Perchuk B.S., Ashenberg O., Seid C.A., Snow H.R., Skerker J.M., Laub M.T. Spatial tethering of kinases to their substrates relaxes evolutionary constraints on specificity. Mol. Microbiol. 2012;86:1393–1403. doi: 10.1111/mmi.12064. [DOI] [PubMed] [Google Scholar]
- 8.Capra E.J., Perchuk B.S., Skerker J.M., Laub M.T. Adaptive mutations that prevent crosstalk enable the expansion of paralogous signaling protein families. Cell. 2012;150:222–232. doi: 10.1016/j.cell.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laub M.T., Goulian M. Specificity in two-component signal transduction pathways. Annu. Rev. Genet. 2007;41:121–145. doi: 10.1146/annurev.genet.41.042007.170548. [DOI] [PubMed] [Google Scholar]
- 10.Podgornaia A.I., Laub M.T. Determinants of specificity in two-component signal transduction. Curr. Opin. Microbiol. 2013;16:156–162. doi: 10.1016/j.mib.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Francis V.I., Waters E.M., Finton-James S.E., Gori A., Kadioglu A., Brown A.R., Porter S.L. Multiple communication mechanisms between sensor kinases are crucial for virulence in Pseudomonas aeruginosa. Nat. Commun. 2018;9 doi: 10.1038/s41467-018-04640-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodman A.L., Merighi M., Hyodo M., Ventre I., Filloux A., Lory S. Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen. Genes Dev. 2009;23:249–259. doi: 10.1101/gad.1739009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodman A.L., Kulasekara B., Rietsch A., Boyd D., Smith R.S., Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell. 2004;7:745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 14.Brencic A., McFarland K.A., McManus H.R., Castang S., Mogno I., Dove S.L., Lory S. The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 2009;73:434–445. doi: 10.1111/j.1365-2958.2009.06782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bordi C., Lamy M.C., Ventre I., Termine E., Hachani A., Fillet S., Roche B., Bleves S., Méjean V., Lazdunski A., Filloux A. Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Mol. Microbiol. 2010;76:1427–1443. doi: 10.1111/j.1365-2958.2010.07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lapouge K., Schubert M., Allain F.H.T., Haas D. Gac/Rsm signal transduction pathway of γ-proteobacteria: From RNA recognition to regulation of social behaviour. Mol. Microbiol. 2008;67:241–253. doi: 10.1111/j.1365-2958.2007.06042.x. [DOI] [PubMed] [Google Scholar]
- 17.Moradali M.F., Ghods S., Rehm B.H.A. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 2017;7 doi: 10.3389/fcimb.2017.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chambonnier G., Roux L., Redelberger D., Fadel F., Filloux A., Sivaneson M., de Bentzmann S., Bordi C. The hybrid histidine kinase LadS forms a multicomponent signal transduction system with the GacS/GacA two-component system in Pseudomonas aeruginosa. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1006032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ventre I., Goodman A.L., Vallet-Gely I., Vasseur P., Soscia C., Molin S., Bleves S., Lazdunski A., Lory S., Filloux A. Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc. Natl. Acad. Sci. U. S. A. 2006;103:171–176. doi: 10.1073/pnas.0507407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laskowski M.A., Kazmierczak B.I. Mutational analysis of RetS, an unusual sensor kinase-response regulator hybrid required for Pseudomonas aeruginosa virulence. Infect. Immun. 2006;74:4462–4473. doi: 10.1128/IAI.00575-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mancl J.M., Ray W.K., Helm R.F., Schubot F.D. Helix cracking regulates the critical interaction between RetS and GacS in Pseudomonas aeruginosa. Structure. 2019;27:785–793.e5. doi: 10.1016/j.str.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 22.Assmann G., Brehm W., Diederichs K. Identification of rogue datasets in serial crystallography. J. Appl. Crystallogr. 2016;49:1021–1028. doi: 10.1107/S1600576716005471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulrich L.E., Zhulin I.B. Four-helix bundle: A ubiquitous sensory module in prokaryotic signal transduction. Bioinformatics. 2005;21 Suppl 3:iii45–iii48. doi: 10.1093/bioinformatics/bti1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhate M.A.P., Molnar K.A.S., Goulian M., Degrado W.F. Signal transduction in histidine kinases: Insights from new structures. Structure. 2015;23:981–994. doi: 10.1016/j.str.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Albanesi D., Martín M., Trajtenberg F., Mansilla M.C., Haouz A., Alzari P.M., De Mendoza D., Buschiazzo A. Structural plasticity and catalysis regulation of a thermosensor histidine kinase. Proc. Natl. Acad. Sci. U. S. A. 2009;106:16185–16190. doi: 10.1073/pnas.0906699106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashenberg O., Keating A.E., Laub M.T. Helix bundle loops determine whether histidine kinases autophosphorylate in cis or in trans. J. Mol. Biol. 2013;425:1198–1209. doi: 10.1016/j.jmb.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casino P., Miguel-Romero L., Marina A. Visualizing autophosphorylation in histidine kinases. Nat. Commun. 2014;5 doi: 10.1038/ncomms4258. [DOI] [PubMed] [Google Scholar]
- 28.Kinoshita-Kikuta E., Kinoshita E., Eguchi Y., Koike T. Validation of cis and trans modes in multistep phosphotransfer signaling of bacterial tripartite sensor kinases by using phos-tag SDS-pagE. PLoS One. 2016;11 doi: 10.1371/journal.pone.0148294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Workentine M.L., Chang L., Ceri H., Turner R.J. The GacS-GacA two-component regulatory system of Pseudomonas fluorescens: A bacterial two-hybrid analysis. FEMS Microbiol. Lett. 2009;292:50–56. doi: 10.1111/j.1574-6968.2008.01445.x. [DOI] [PubMed] [Google Scholar]
- 30.Yan Q., Lopes L.D., Shaffer B.T., Kidarsa T.A., Vining O., Philmus B., Song C., Stockwell V.O., Raaijmakers J.M., McPhail K.L., Andreote F.D., Chang J.H., Loper J.E. Secondary metabolism and interspecific competition affect accumulation of spontaneous mutants in the GacS-GacA regulatory system in Pseudomonas protegens. MBio. 2018;9 doi: 10.1128/mBio.01845-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei X., Huang X., Tang L., Wu D., Xu Y. Global control of GacA in secondary metabolism, primary metabolism, secretion systems, and motility in the rhizobacterium Pseudomonas aeruginosa M18. J. Bacteriol. 2013;195:3387–3400. doi: 10.1128/JB.00214-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Latour X. The evanescent gacs signal. Microorganisms. 2020;8:1–25. doi: 10.3390/microorganisms8111746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sonnleitner E., Haas D. Small RNAs as regulators of primary and secondary metabolism in Pseudomonas species. Appl. Microbiol. Biotechnol. 2011;91:63–79. doi: 10.1007/s00253-011-3332-1. [DOI] [PubMed] [Google Scholar]
- 34.Francis V.I., Porter S.L. Multikinase networks: Two-component signaling networks integrating multiple stimuli. Annu. Rev. Microbiol. 2019;73:199–223. doi: 10.1146/annurev-micro-020518-115846. [DOI] [PubMed] [Google Scholar]
- 35.Kong W., Chen L., Zhao J., Shen T., Surette M.G., Shen L., Duan K. Hybrid sensor kinase PA1611 in Pseudomonas aeruginosa regulates transitions between acute and chronic infection through direct interaction with RetS. Mol. Microbiol. 2013;88:784–797. doi: 10.1111/mmi.12223. [DOI] [PubMed] [Google Scholar]
- 36.Bhagirath A.Y., Pydi S.P., Li Y., Lin C., Kong W., Chelikani P., Duan K. Characterization of the direct interaction between hybrid sensor kinases PA1611 and RetS that controls biofilm formation and the type III secretion system in Pseudomonas aeruginosa. ACS Infect. Dis. 2017;3:162–175. doi: 10.1021/acsinfecdis.6b00153. [DOI] [PubMed] [Google Scholar]
- 37.Lin C.T., Huang Y.J., Chu P.H., Hsu J.L., Huang C.H., Peng H.L. Identification of an HptB-mediated multi-step phosphorelay in Pseudomonas aeruginosa PAO1. Res. Microbiol. 2006;157:169–175. doi: 10.1016/j.resmic.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 38.Hsu J.L., Chen H.C., Peng H.L., Chang H.Y. Characterization of the histidine-containing phosphotransfer protein B-mediated multistep phosphorelay system in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 2008;283:9933–9944. doi: 10.1074/jbc.M708836200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petrova O.E., Sauer K. SagS contributes to the motile-sessile switch and acts in concert with BfiSR to enable Pseudomonas aeruginosa biofilm formation. J. Bacteriol. 2011;193:6614–6628. doi: 10.1128/JB.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bouillet S., Ba M., Houot L., Iobbi-Nivol C., Bordi C. Connected partner-switches control the life style of Pseudomonas aeruginosa through RpoS regulation. Sci. Rep. 2019;9:1–11. doi: 10.1038/s41598-019-42653-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mann T.H., Shapiro L. Integration of cell cycle signals by multi-PAS domain kinases. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E7166–E7173. doi: 10.1073/pnas.1808543115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huynh T.A.N., Stewart V. Negative control in two-component signal transduction by transmitter phosphatase activity. Mol. Microbiol. 2011;82:275–286. doi: 10.1111/j.1365-2958.2011.07829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang B.X., Wheeler K.M., Cady K.C., Lehoux S., Cummings R.D., Laub M.T., Ribbeck K. Mucin glycans signal through the sensor kinase RetS to inhibit virulence-associated traits in Pseudomonas aeruginosa. Curr. Biol. 2021;31:90–102.e7. doi: 10.1016/j.cub.2020.09.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Le Roux M., Kirkpatrick R.L., Montauti E.I., Tran B.Q., Brook Peterson S., Harding B.N., Whitney J.C., Russell A.B., Traxler B., Goo Y.A., Goodlett D.R., Wiggins P.A., Mougous J.D. Kin cell lysis is a danger signal that activates antibacterial pathways of Pseudomonas aeruginosa. Elife. 2015;2015:1–65. doi: 10.7554/eLife.05701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parkinson J.S. Signaling Mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu. Rev. Microbiol. 2010;64:101–122. doi: 10.1146/annurev.micro.112408.134215. [DOI] [PubMed] [Google Scholar]
- 46.Vincent F., Round A., Reynaud A., Bordi C., Filloux A., Bourne Y. Distinct oligomeric forms of the Pseudomonas aeruginosa RetS sensor domain modulate accessibility to the ligand binding site. Environ. Microbiol. 2010;12:1775–1786. doi: 10.1111/j.1462-2920.2010.02264.x. [DOI] [PubMed] [Google Scholar]
- 47.Jing X., Jaw J., Robinson H.H., Schubot F.D. Crystal structure and oligomeric state of the Ret S signaling kinase sensory domain. Proteins Struct. Funct. Bioinforma. 2010;78:1631–1640. doi: 10.1002/prot.22679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiu D., Damron F.H., Mima T., Schweizer H.P., Yu H.D. PBAD-based shuttle vectors for functional analysis of toxic and highly regulated genes in Pseudomonas and Burkholderia spp. and other bacteria. Appl. Environ. Microbiol. 2008;74:7422–7426. doi: 10.1128/AEM.01369-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mancl J.M. Virginia Polytechnic Institute and State University; Blacksburg, VA: 2018. Molecular investigations of protein assemblies involved in prokaryotic virulence. PhD thesis. [Google Scholar]
- 50.Evans P.R., Murshudov G.N. How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Evans P.R. An introduction to data reduction: Space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Echols N., Headd J.J., Hung L.W., Jain S., Kapral G.J., Grosse Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R.D., Read R.J., Richardson D.C. The Phenix software for automated determination of macromolecular structures. Methods. 2011;55:94–106. doi: 10.1016/j.ymeth.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guss J.M. Biomolecular crystallography: Principles, practice, and application to structural biology, by Bernard Rupp. Crystallogr. Rev. 2011;17:65–67. [Google Scholar]
- 57.Vagin A.A., Steiner R.A., Lebedev A.A., Potterton L., McNicholas S., Long F., Murshudov G.N. REFMAC5 dictionary: Organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 58.DeLano W.L. Schrödinger LLC; San Carlos, CA: 2020. The PyMOL Molecular Graphics System, Version 2.3. [Google Scholar]
- 59.Karimova G., Dautin N., Ladant D. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 2005;187:2233–2243. doi: 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang X., Bremer H. Control of the Escherichia coli rrnB P1 promoter strength by ppGpp. J. Biol. Chem. 1995;270:11181–11189. doi: 10.1074/jbc.270.19.11181. [DOI] [PubMed] [Google Scholar]
- 61.O'Toole G.A. Microtiter dish Biofilm formation assay. J. Vis. Exp. 2010 doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laskowski R.A., Jabłońska J., Pravda L., Vařeková R.S., Thornton J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. 2018;27:129–134. doi: 10.1002/pro.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript.