Abstract
Cyclin-dependent kinase 7 (CDK7) is a master regulatory kinase that drives cell cycle progression and stimulates expression of oncogenes in a myriad of cancers. Inhibitors of CDK7 (CDK7i) are currently in clinical trials; however, as with many cancer therapies, patients will most likely experience recurrent disease due to acquired resistance. Identifying targets underlying CDK7i resistance will facilitate prospective development of new therapies that can circumvent such resistance. Here we utilized triple-negative breast cancer as a model to discern mechanisms of resistance as it has been previously shown to be highly responsive to CDK7 inhibitors. After generating cell lines with acquired resistance, high-throughput RNA sequencing revealed significant upregulation of genes associated with efflux pumps and transforming growth factor-beta (TGF-β) signaling pathways. Genetic silencing or pharmacological inhibition of ABCG2, an efflux pump associated with multidrug resistance, resensitized resistant cells to CDK7i, indicating a reliance on these transporters. Expression of activin A (INHBA), a member of the TGF-β family of ligands, was also induced, whereas its intrinsic inhibitor, follistatin (FST), was repressed. In resistant cells, increased phosphorylation of SMAD3, a downstream mediator, confirmed an increase in activin signaling, and phosphorylated SMAD3 directly bound the ABCG2 promoter regulatory region. Finally, pharmacological inhibition of TGF-β/activin receptors or genetic silencing of SMAD4, a transcriptional partner of SMAD3, reversed the upregulation of ABCG2 in resistant cells and phenocopied ABCG2 inhibition. This study reveals that inhibiting the TGF-β/Activin-ABCG2 pathway is a potential avenue for preventing or overcoming resistance to CDK7 inhibitors.
Keywords: activin, ABCG2, multidrug transporters, cyclin-dependent kinase 7, CDK7, triple-negative breast cancer, TNBC, TGF-β, THZ1, SY-1365
Abbreviations: ABC, ATP-binding cassette; CDK, cyclin-dependent kinase; CDK7i, inhibitor of CDK7; EMT, epithelial to mesenchymal transition; FST, follistatin; GSEA, Gene Set Enrichment Analysis; TGF-β, transforming growth factor beta; TNBC, triple-negative breast cancer
Cyclin-dependent kinases (CDKs) belong to the serine/threonine phosphotransferase family, which are activated by cyclins (1). They control progression through the cell cycle and ensure genome integrity (2). Over the last 2 decades, inhibitors of the cyclin/cyclin-dependent kinase holoenzymes have been utilized in many different neoplasms including patients with breast cancer (3). Although these agents are efficacious in reducing cell viability and growth, they have dose-limiting toxicities and are frequently thwarted by therapeutic resistance, necessitating safer and more efficacious alternatives (3). Addressing this need, inhibitors of cyclin-dependent kinase 7 (CDK7) have recently entered clinical trials (https://clinicaltrials.gov/ct2/show/NCT03134638, https://clinicaltrials.gov/ct2/show/NCT03134638, https://clinicaltrials.gov/ct2/show/NCT04247126, https://clinicaltrials.gov/ct2/show/NCT03770494). CDK7 is a unique member of this family that controls the transcription of cell identity genes as well as regulates the activity of other CDKs (4). CDK7 interacts with its partner proteins ménage-à-trois 1 (MAT1) and cyclin H to form the cyclin-dependent kinase activating kinase complex (5). This complex promotes cell cycle progression via the phosphorylation of the T-loop of CDKs 1, 2, 4, 6, 9, and 12, leading to their activation and cell cycle progression (6). CDK7 also phosphorylates the C-terminal domain of RNA polymerase II, enabling transcription initiation (4). This stimulates the expression of genes, including oncogenes, via the formation of large DNA–protein complexes called super-enhancers (7, 8). Given its dual roles, CDK7 is a target of interest for the treatment of cancer. Compounds such as the prototypical CDK7 inhibitor (CDK7i), THZ1, as well as those in clinical trials (e.g., SY-1365 and CT7001), selectively inhibit CDK7 activity, leading to a reduction in tumor size and cancer burden in mouse models of glioma, lung, leukemia, and breast cancer (8, 9, 10, 11, 12, 13, 14). Despite this initial promise, it is likely that many cancers will ultimately develop resistance to CDK7i, making it imperative to prospectively identify signaling pathways that drive therapeutic resistance. These potential vulnerabilities could then be leveraged in the design of synergistic or sequential therapies to prevent progression to resistance.
In this study, we focused on identifying mechanisms of CDK7 inhibitor (CDK7i) resistance in triple-negative breast cancer (TNBC). TNBC is a highly aggressive subtype of breast cancer that represents approximately 15% to 20% of breast cancer diagnoses in the United States (15). Unlike the luminal and HER2-enriched subtypes, TNBC lacks expression of the receptors for estrogen and progesterone, as well as the human epidermal growth factor 2 receptor. Thus, therapies targeted to these oncogenic drivers are ineffective in TNBC, leaving patients with limited treatment options (16). The current standards of care for this disease involve cytotoxic chemotherapies, including antimitotics such as paclitaxel and cyclophosphamide (17). Although these therapies are initially very effective in reversing disease course, recurrent disease is common and in many cases tumors become refractory to the initial chemotherapies (18). Upon detection of metastatic disease, the median patient survival is only 13 months (19). This highlights the significant clinical unmet need to discern the molecular mechanisms that drive these highly aggressive cancers with the intent of identifying new therapeutic modalities.
Models of TNBC have been shown to be particularly sensitive to CDK7 inhibitors (9, 17). These drugs are proposed to disrupt super-enhancers that drive expression of genes necessary for survival, called Achilles’ genes. By dismantling these super-enhancers, CDK7is induce cell death in vivo (7, 9, 11, 20). Despite the effectiveness of CDK7i in preclinical models of TNBC and many other cancers, human tumors are likely to experience resistance owing to selective pressures leading to adaptive alternative pathway activation or evolutionary changes (21). Proactively identifying mechanisms of resistance should reveal novel biomarkers of resistance and accelerate development of combination approaches to prevent these processes (21, 22). Herein, we elucidate a new pathway by which TNBC cells acquire resistance to CDK7i through increasing signaling by the transforming growth factor beta (TGF-β) family of ligands, particularly activin. This, in turn, promotes the expression of efflux proteins, such as ABCG2. Supporting an essential role for this pathway in maintaining resistance, inhibiting ABCG2 or TGF-β/activin signaling resensitizes resistant cells to CDK7i. This study elucidates a novel mechanism of CDK7 inhibitor resistance that may inform future studies discovering biomarkers of resistance and approaches to prevent its acquisition in human disease.
Results
Acquisition of CDK7i resistance in TNBC cells
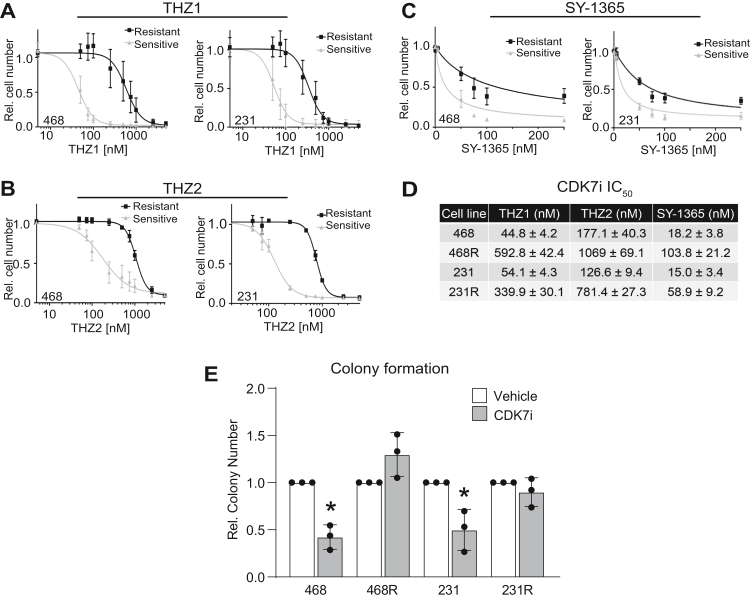
TNBC cell lines have previously been reported to be sensitive to CDK7 inhibition (9). To establish CDK7i resistant lines, we treated sensitive/parental MDA-MB-468 and MDA-MB-231 cells with the CDK7 inhibitor, THZ1, using stepwise dose escalation. This resulted in resistant cell lines (MDA-MB-468R and MDA-MB-231R) with IC50s that are 5 to 10 times higher than their sensitive counterparts (Fig. 1A). To test whether THZ1 resistance extends to other CDK7i, including those that are currently being evaluated in clinical trials, we treated MDA-MB-468 and MDA-MB-231-sensitive/parental and THZ1-resistant cells with increasing doses of THZ2, a THZ1 analog (Fig. 1B), and SY-1365 (Fig. 1C). For both cell lines, THZ1-resistant cells maintained resistance to THZ2, and SY-1365 with ∼4 to 10× shifts in IC50s (Fig. 1D). To assess the long-term consequence of THZ1 exposures, we used a colony formation assay of sensitive/parental and resistant cells. Cells were exposed to THZ1 for 72 h; equal numbers of viable cells were then replated and maintained in drug-free media for 8 days. THZ1 caused a sustained reduction in cell growth/colony formation in the sensitive cell lines even after removal of the drug (Fig. 1E). However, the MDA-MB-231R and MDA-MB-468R lines exhibited no response, affirming their sustained resistance to THZ1.
Figure 1.
Acquisition of CDK7i resistance in TNBC cells. CDK7 inhibitor–sensitive/parental TNBC cell lines MDA-MB-468 (468) and MDA-MB-231 (231) and resistant MDA-MB-468 (468R) and MDA-MB-231 (231R) cells were treated with either vehicle or the following CDK7 inhibitors at increasing doses: (A) THZ1, (B) THZ2, or (C) SY-1365 for 72 h and the relative cell number was assessed using crystal violet staining. For these experiments, the dose–response curves were significantly different between the sensitive/parental lines compared with the isogenic resistant lines (p < 0.05). D, table of IC50s from the CDK7i dose–response curves. E, sensitive/parental and resistant MDA-MB-468 cells were treated with dimethyl sulfoxide or 50 nM THZ1 for 72 h, whereas MDA-MB-231-sensitive/parental and resistant MDA-MB-231 cells were treated with dimethyl sulfoxide or 75 nM THZ1. All cell lines were then replated and analyzed for colony formation after 8 days of growth in drug-free, complete media. For all experiments, ∗p < 0.05. n = 3 to 4 experiments, each completed in triplicate. Values are means ± SD.
CDK7i-resistant cells upregulate the expression of extracellular transport genes
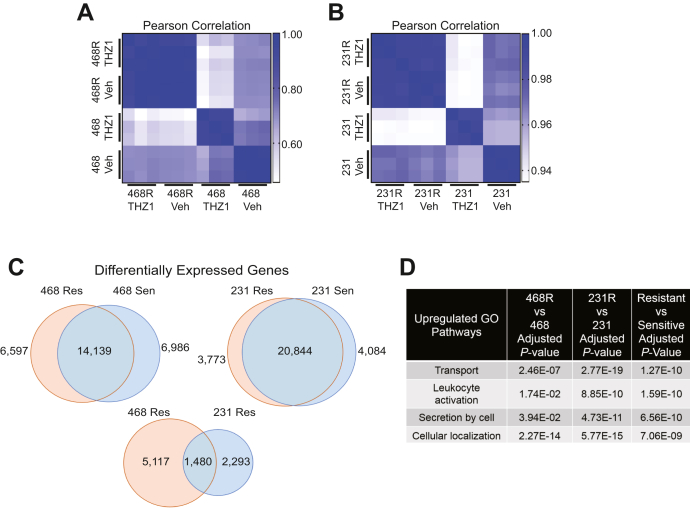
To investigate the mechanisms underlying CDK7i resistance, we used RNA-Seq to identify transcriptomic differences between vehicle (dimethyl sulfoxide [DMSO]) or THZ1-treated cell lines that were sensitive or resistant to CDK7i. As expected, Pearson correlation analysis of the gene expression profiles revealed that sensitive/parental cells exhibited substantial transcriptomic shifts following exposure to THZ1. In contrast, treatment with THZ1 had a minimal impact on gene expression in resistant cells compared with their vehicle-treated counterparts, again confirming resistance (Fig. 2, A and B). Comparisons of the transcriptomes of vehicle-treated cell lines revealed 3773 and 6597 upregulated transcripts in MDA-MB-231R and MDA-MB-468R cells, respectively, compared with their sensitive/parental counterparts (Fig. 2C and Table S1). In addition, the MDA-MB-231R and MDA-MB-468R cells shared 1480 upregulated transcripts (Fig. 2C). To elucidate the underlying biological pathways contributing to CDK7i, we performed Gene Ontology (GO) on the shared genes (Fig. 2D). Transport, leukocyte activation, and secretion were among the most enriched gene ontology pathways (Fig. 2D). These pathways were also enriched when comparing the upregulated genes individually in each resistant line. Examination of the individual genes within these networks uncovered several that are involved in intracellular/extracellular transport such as ABCG2, ABCC1, and TAP1 that are upregulated with the acquisition of resistance. These data suggested that drug transport may contribute to CDK7i resistance in TNBC cells.
Figure 2.
CDK7i-resistant cells upregulate the expression of extracellular transport genes.A and B, Pearson correlation analysis using RNA-Seq gene expression data from the sensitive/parental and resistant cells treated with vehicle or THZ1 (50–75 nM) for 48 h: MDA-MB-231 Vehicle (231 Veh), MDA-MB-231 THZ1 (231 THZ1), MDA-MB-231R Vehicle (231R Veh), MDA-MB-231R THZ1 (231R THZ1), MDA-MB-468 Vehicle (468 Veh), MDA-MB-468 THZ1 (468 THZ1), MDA-MB-468R Vehicle (468R Veh), and MDA-MB-468R THZ1 (468R THZ1). C, Venn diagrams indicating the number of differentially expressed transcripts when comparing the sensitive/parental expression profiles with those observed in the resistant MDA-MB-231R and MDA-MB-468R cells. D, list of significantly upregulated Gene Ontology pathways in resistant cells.
ABCG2 is upregulated in CDK7i-resistant cells and correlates with worse overall survival in TNBC
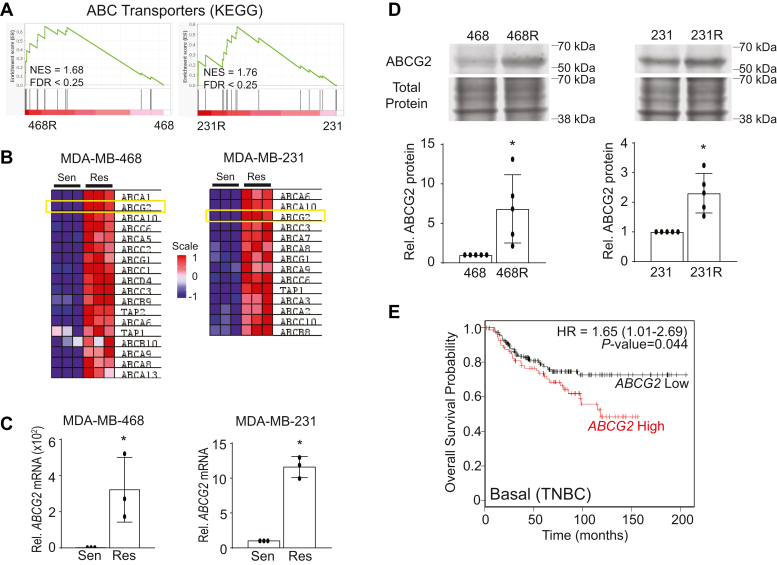
We used the Kyoto Encyclopedia of Genes and Genomes curated gene signature for ABC transporters (23) to conduct Gene Set Enrichment Analysis (GSEA) on the individual MDA-MB-468 and MDA-MB-231 gene lists from resistant compared with sensitive/parental cells. This showed that CDK7i-resistant cell lines were significantly enriched in the expression of ABC transporter genes (Fig. 3, A and B). ATP-binding cassette (ABC) transporters such as ABCG2 are well-established mediators of resistance to cytotoxic chemotherapies in breast cancer (24, 25, 26). Of the ABC transporter genes, ABCG2 was the most highly induced gene that was shared between the two CDK7i-resistant cell lines. Of note, ABCG2 was first identified as Breast Cancer Resistance Protein or BCRP (25); it is an efflux pump that conveys multidrug resistance in this and other cancers. Over 200 substrates have been identified for ABCG2 that have varied chemical structures. Hence, ABCG2 can confer resistance to diverse and commonly utilized chemotherapeutics such as etoposide, methotrexate, and doxorubicin (27). Moreover, a recent report has revealed that enforced overexpression of ABCG2 can induce resistance to THZ1 in HEK293 cells, and in silico docking studies suggest that THZ1 can directly bind to ABCG2 (28). We confirmed that ABCG2 mRNA (Fig. 3C) and protein (Fig. 3D) were upregulated in both MDA-MB-231R and MDA-MB-468R cells compared with the CDK7i-sensitive/parental cells. To determine if expression levels of ABCG2 are associated with TNBC patient survival, we used Kaplan–Meier Plotter to conduct a meta-analysis of several breast cancer gene expression datasets (29). TNBC (basal) patients with high ABCG2 expression had a reduced probability of overall survival compared with those with low ABCG2 (Fig. 3E), indicating that the ABCG2 efflux pump may negatively impact outcomes of patients with this disease.
Figure 3.
ABCG2 is upregulated in CDK7i-resistant cells and correlates with worse overall survival in TNBC.A, Gene Set Enrichment Analysis (GSEA) of upregulated genes using the ABC transporter gene signature from Kyoto Encyclopedia of Genes and Genomes (KEGG) (23) in the CDK7i-sensitive/parental and resistant MDA-MB-468 and MDA-MB-231 cells. B, heatmap of the upregulated ABC transporter genes identified using GSEA. C and D, verification of changes in ABCG2 mRNA (C) and protein (D) expression in sensitive/parental and resistant MDA-MB-468 and MDA-MB-231 cells. Quantitation of ABCG2 expression in MDA-MB-468 and MDA-MB-231 cells are from three independent experiments. Bars are means ± SD. Results are expressed relative to total protein. ∗p < 0.05. E, overall survival curves in patients with basal (predominant subtype of TNBC) breast cancer generated with KM Plotter that were stratified based upon the expression of ABCG2 (high = 83 patients, low = 158 patients).
ABCG2 inhibition restores CDK7i responsiveness in resistant TNBC cells
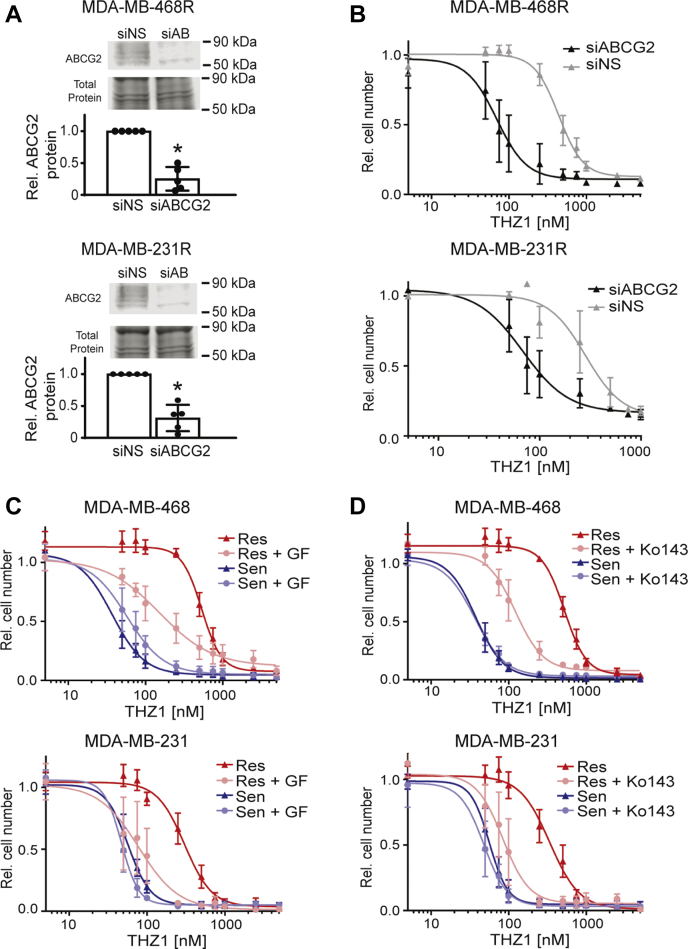
To determine if the increased expression of ABCG2 observed in CDK7i-resistant cells contributes to drug resistance, we silenced its expression by transiently transfecting siRNA to ABCG2 (siABCG2) or a nonsilencing (siNS) control in the resistant lines (MDA-MB-468R or MDA-MB-231R) (Fig. 4A). We then generated dose–response curves for THZ1 (Fig. 4B). Silencing ABCG2 expression had no impact on the response of sensitive/parental cells to THZ1. In contrast, reducing ABCG2 expression in the resistant lines shifted their THZ1 dose–response curve to the left, with the IC50 for THZ1 shifting from 440 nM in the 468R control (siNS) cells to 70 nM in the ABCG2 silenced cells and from 285 nM in the 231R control cells to 68 nM upon ABCG2 silencing. To determine if blocking AGCG2 activity could be used as a pharmacological approach to restore THZ1 sensitivity in resistant cells, we repeated these experiments using pharmacological inhibitors of ABCG2, GF120918 (Elacridar) (30), and Ko143 (31, 32, 33). Both ABCG2 inhibitors resensitized the resistant cells to THZ1. This was manifested as a shift in the THZ1 IC50 from 535 to 152 nM upon cotreatment with GF120918 in the MDA-MB-468-resistant cells (Fig. 4C); a similar leftward shift was observed in the MDA-MB-231-resistant cells with the IC50 for THZ1 changing from 304 to 78 nM with GF120918 cotreatment (Fig. 4C). Similar results were observed with a second, more selective ABCG2 inhibitor, Ko143. The THZ1 IC50 for MDA-MB-468-resistant cells shifted from 524 to 123 nM (Fig. 4D) and the THZ1 IC50 of the MDA-MB-231-resistant cells shifted from 339 to 85 nM (Fig. 4D). Hence, using genetic and pharmacological approaches, these data indicate that ABCG2 is necessary to sustain acquired resistance to CDK7i in these models and supports future assessment of clinically effective ABCG2 inhibitors in combination with CDK7i in patients.
Figure 4.
ABCG2 inhibition restores CDK7i responsiveness in resistant TNBC cells.A, protein verification of siRNA-mediated silencing of ABCG2 in MDA-MB-468R and MDA-MB-231R cells after 72 h. B, seventy-two–hour THZ1 dose–response curve in MDA-MB-468R and MDA-MB-231R cells initiated 48 h after transfection. The siABCG2 curves are significantly different than the siNS curves according to a sum-of-squares F test with p-value <0.5. C, THZ1 dose–response curves for MDA-MB-468 and MDA-MB-231-sensitive/parental and resistant cells that were cotreated with either vehicle or GF120918 (1.5 μM) for 72 h. D, THZ1 dose–response curves for MDA-MB-468 and MDA-MB-231-sensitive/parental and resistant cells that were cotreated with vehicle or Ko143 (100 nM) for 72 h. The GF and Ko143 cotreatment curves are significantly different from their resistant curve counterparts treated with vehicle, according to a sum-of-squares F test with a p-value <0.05.
EMT signature genes are upregulated in CDK7i-resistant TNBC cells
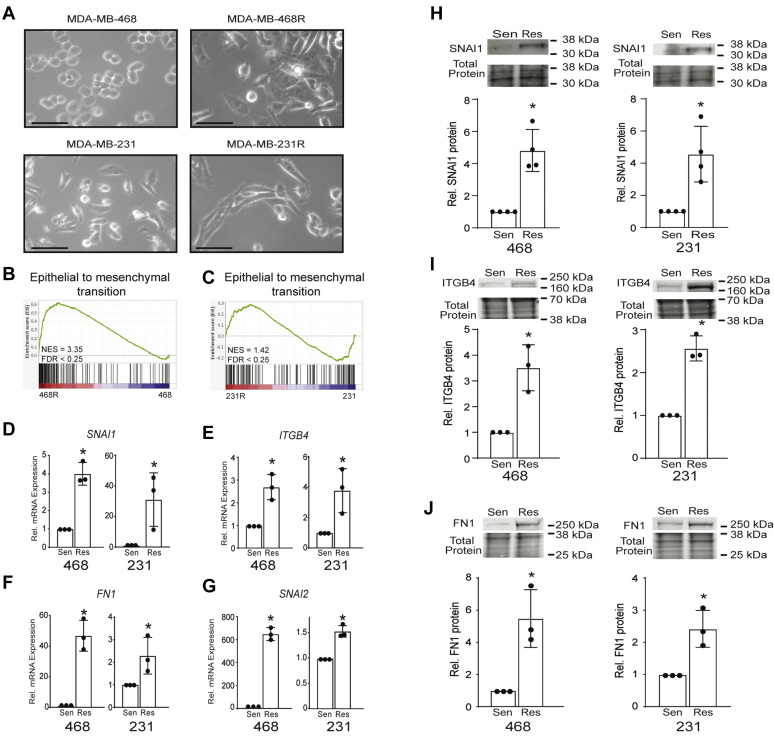
We next sought to identify the mechanism(s) underlying the induction of ABCG2 expression that occurs with CDK7i resistance. Identifying such mechanisms should facilitate developing treatment approaches that prevent acquisition of CDK7i resistance via ABCG2 upregulation. It is becoming increasingly apparent that cellular plasticity, including epithelial to mesenchymal transition (EMT), underlies chemotherapeutic resistance in several cancer models. Hybrid epithelial/mesenchymal states in human cancers has further demonstrated the clinical relevance of EMT in these diseases (34, 35, 36, 37, 38). In this regard, we observed gross morphological changes in the resistant cell lines. They appeared more elongated than sensitive/parental cells, suggesting an acquisition of mesenchymal features (Fig. 5A). Of note, the expression of ABCG2 can be induced by EMT but its regulation in the setting of CDK7i resistance is unknown (39, 40, 41). We hypothesized that cells undergo EMT in the process of becoming resistant to CDK7i and that this may lead to ABCG2 induction. Indeed, gene set enrichment analysis of the RNA-Seq data revealed significant enrichment of EMT-related gene signatures in both MDA-MB-468R and MDA-MB-231R cells (Fig. 5, B and C and Fig. S1). Changes in the expression of selected EMT genes including Snail (SNAI1), Slug (SNAI2), Fibronectin (FN1), and Integrin β4 (ITGB4) was confirmed (Fig. 5, D–G) in both the resistant MDA-MB-468 and MDA-MB-231-resistant cells. The increased gene expression was further translated into elevated expression of the corresponding proteins that were examined (SNAI1, ITGB4, and FN1, Fig. 5, H–J) (42, 43, 44, 45, 46, 47). These data indicate that the acquisition of CDK7i resistance and elevated expression of ABCG2 is associated with the activation of an EMT program in TNBC cells. Moreover, they align with prior studies demonstrating that EMT-associated transcription factors can induce the expression of the ABCG2 gene when they are overexpressed (48).
Figure 5.
Epithelial to mesenchymal transition (EMT) signature genes are upregulated in CDK7i-resistant TNBC cells.A, phase contrast images of sensitive/parental (MDA-MB-468 and MDA-MB-231) and resistant (MDA-MB-468R and MDA-MB-231R) cells. The scale bar represents 30 μm. B, Gene Set Enrichment Analysis of upregulated genes associated with EMT (Hallmark v7.4) is shown for the MDA-MB-468R and C, MDA-MB-231R cells. D–G, quantitative RT-PCR confirmation of the upregulation of SNAI1 (D), ITGB4 (E), FN1 (F), and SNAI2 (G) with the acquisition of CDK7i resistance. H–J, expression of EMT-associated proteins SNAI1 (H), ITGB4 (I), and FN1 (J) in MDA-MB-468 and MDA-MB-231-sensitive/parental and CDK7i-resistant cells. Cell line data are the results of three independent experiments. Bars are means ± SD. Protein results are expressed relative to the total protein. ∗p < 0.05.
TGF-β/activin signaling is upregulated and necessary for maintaining CDK7i resistance
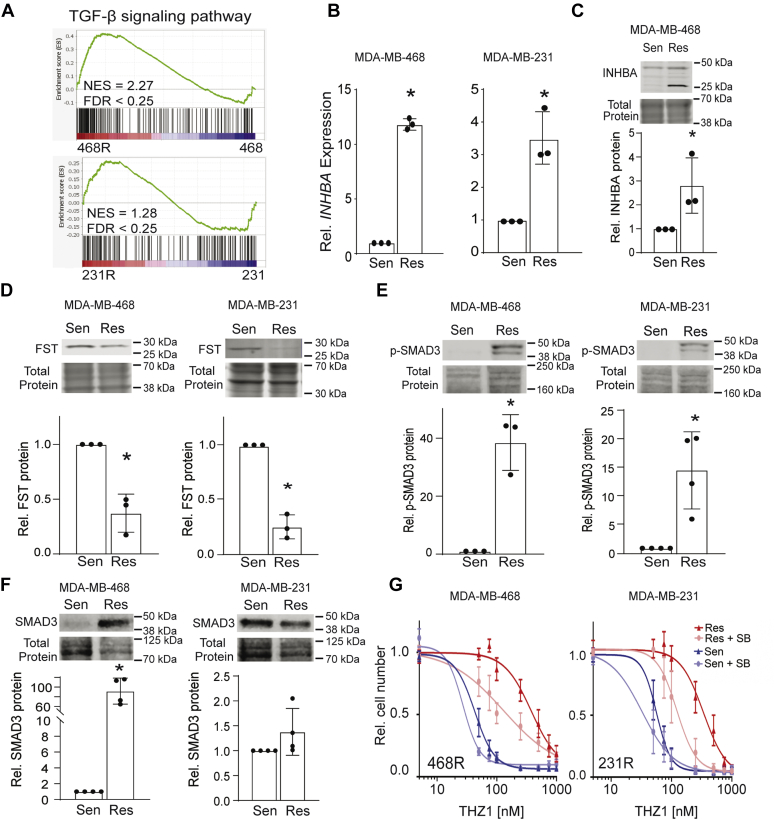
The transforming growth factor-beta (TGF-β) family of ligands, specifically TGF-β and activin, are widely known drivers of EMT in breast cancer (49, 50) and have been reported to promote resistance to cancer therapies (51, 52). Further interrogation of the overlapping resistance gene set from the RNA-Seq data by GSEA revealed that the TGF-β family signaling pathway is upregulated in CDK7i-resistant cells compared with their sensitive/parental counterparts (Fig. 6A and Fig. S1). Moreover, the EMT genes/proteins evaluated in Figure 5 (SNAI1, ITGB4, FN1, and SNAI2) are downstream targets of TGF-β/activin signaling (53, 54). Although TGF-β1 ligand gene expression (TGFB1) was only significantly induced in the RNA-Seq data for one of the resistant cell lines compared with sensitive/parental cells, we noted a substantial increase in the expression of INHBA (activin A) in both lines (data not shown). Increased expression of INHBA mRNA and protein in resistant compared with sensitive/parental was confirmed (Fig. 6, B and C). Moreover, expression of follistatin (FST), an intrinsic inhibitor of activin, was suppressed in resistant cells (Fig. 6D). Like TGF-β, activin induces the canonical SMAD signaling pathway involving SMAD2/3 phosphorylation and activation of SMAD4 transcriptional activity (55). We found that resistant cells have increased SMAD3 phosphorylation (p-SMAD3) compared with sensitive/parental cells (Fig. 6E), further supporting activation of activin signaling upon acquisition of resistance. In MDA-MB-468 cells, the increase in SMAD3 phosphorylation was associated with an increase in SMAD3 levels. This may be due to stabilization of SMAD3 protein or increased transcription of the SMAD3 gene in these cells with the acquisition of resistance but still demonstrates that resistance involves an increase in the total activity of SMAD3 (Fig. 6F). Together, these data indicate that TGF-β family signaling, particularly activin, is upregulated in CDK7i-resistant cells and suggests that this pathway may be an essential mechanism establishing resistance. There are currently no small molecule inhibitors that can discriminate between TGF-β and activin receptors (56). Thus, to determine if signaling from this family of receptors is essential for the phenotypes observed in CDK7i-resistant cells, we used SB431542, an inhibitor of both TGF-β and activin receptors. Treatment with a low dose of SB431542 (5.6 or 11 μM, respectively) had no significant effect on the THZ1 dose–response relationship in sensitive/parental MDA-MB-231 or MDA-MB-468 cells. In contrast, in resistant cells, blocking TGF-β/activin receptor activity with the SB compound increased THZ1 responsiveness by shifting the dose–response curve to the left compared with resistant cells treated with THZ1 plus vehicle (Fig. 6G). The MDA-MB-468-resistant cells shifted from an IC50 of 340 nM in the vehicle-treated cells to 123 nM in the SB431542-treated cells (Fig. 6G). The MDA-MB-231-resistant cells shifted from a THZ1 IC50 of 380 nM in the vehicle-treated cells to 130 nM in the SB431542-treated cells (Fig. 6G). These data indicate that blocking TGF-β/activin signaling resensitizes resistant TNBC cell lines to CDK7i.
Figure 6.
TGF-β/activin signaling is upregulated and necessary for maintaining CDK7i resistance.A, Gene Set Enrichment Analysis showing the enrichment of the TGF-β family signaling gene signature (79) in MDA-MB-468R and MDA-MB-231R cells compared with their respective sensitive/parental cells. B, changes in the expression of activin A mRNA (INHBA) was confirmed in MDA-MB-468 and MDA-MB-231 cells. C, western blot analysis of INHBA protein expression in resistant compared with sensitive/parental MDA-MB-468 cells. The larger-molecular-weight protein band for INHBA is the glycosylated isoform. D, protein expression of the intrinsic inhibitor of activin, follistatin (FST) was reduced in resistant compared with sensitive/parental MDA-MB-468 and MDA-MB-231 cells. E, phosphorylation of SMAD3 is induced in resistant compared with sensitive/parental cells. F, western blot analysis of total SMAD3 expression in MDA-MB-468 and MDA-MB-231-sensitive/parental and resistant cells. G, resistant and sensitive/parental cells were treated with 11.2 μM SB431542 for MDA-MB-468 or 5.6 μM SB431542 for MDA-MB-231 or vehicle for 48 h prior to generating 72-h THZ1 dose–response curves. The THZ1 dose–response curves are significantly different when cotreated with the SB compound compared with vehicle in the resistant cells according to a sum-of-squares F test with a p-value <0.05. These data are the results of three independent experiments. Bars are means ± SD. Protein expression is quantified relative to the total protein. ∗p < 0.05.
TGF-β/activin family signaling maintains expression of ABCG2 in CDK7i-resistant cells
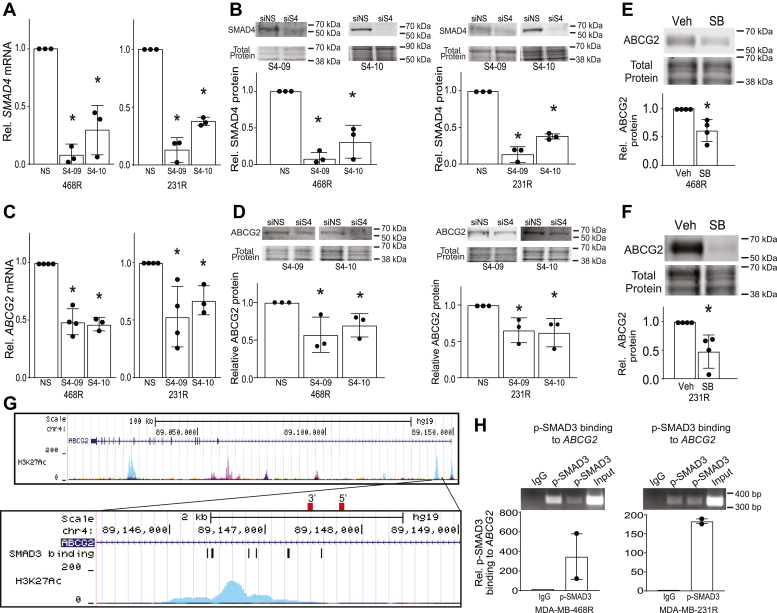
Signaling by TGF-β has been reported to induce ABCG2 expression (39, 40), suggesting that the TGF-β/activin pathway may underlie the upregulation of ABCG2 that occurs with acquisition of CDK7i resistance. To determine if resistance to CDK7i was due to increased canonical signaling by TGF-β/activin that in turn elevates ABCG2 expression, we used siRNA to suppress the expression of SMAD4 (Mothers Against Decapentaplegic Homolog 4). SMAD4 is the downstream transcription factor that integrates signaling from TGF-β/activin receptors (57, 58, 59). Transient transfection with siSMAD4 decreased SMAD4 mRNA and protein expression compared with the nonsilencing control (Fig. 7, A and B). This resulted in a significant reduction of ABCG2 mRNA and protein (Fig. 7, C and D) expression in both resistant cell lines, indicating that sustained SMAD4 was necessary to maintain ABCG2 gene expression in models of CDK7i acquired resistance. We then used a pharmacological approach to further assess the interplay between TGF-β/activin family signaling and ABCG2 expression in CDK7i resistance. Treatment of MDA-MB-468R or MDA-MB-231R cells with SB4321542 resulted in decreased ABCG2 protein expression (Fig. 7, E and F), affirming the importance of TGF-β/activin signaling in upregulating ABCG2 levels in resistant cells. To determine if TGF-β/activin signaling directly regulates ABCG2 gene expression, we used publicly available H3K27Ac chromatin immunoprecipitation (ChIP)-Seq data to identify regions of the gene that are within open chromatin and available to bind SMAD proteins (60). We identified a region of the ABCG2 gene locus that was associated with the largest H3K27Ac peak near the transcription start site (Fig. 7G). We then queried this region for predicted consensus binding motifs for SMAD2/SMAD3/SMAD4 using JASPER (Table S2) (61, 62) and identified several potential SMAD-binding sites. SMAD4 is the common DNA-binding partner of SMADs 2 and 3 (mediators of TGF-β/activin signaling) as well as SMADs 1, 5, and 8 (mediators of bone morphogenetic protein signaling). Thus, to specifically test the potential for TGF-β/activin to control ABCG2 expression, we assessed the binding of phosphorylated SMAD3 to the ABCG2 locus rather than SMAD4. Chromatin immunoprecipitation was conducted using an antibody for phosphorylated SMAD3 followed by PCR with primers to this region of the ABCG2 gene. This analysis revealed that phosphorylated SMAD3 binds to the ABCG2 gene in resistant MDA-MB-468 and MDA-MB-231 TNBC cell lines (Fig. 7H). Together these studies reveal that acquisition of CDK7i resistance involves activation of TGF-β/activin signaling that then leads to induction of the expression of the multidrug transporter ABCG2 (Fig. 8). Of note, elements of this pathway can be repressed with small molecule inhibitors to ABCG2 or TGF-β/activin receptors, suggesting a therapeutic avenue for preventing and/or reversing resistance to the novel class of inhibitors that target CDK7.
Figure 7.
TGF-β/activin signaling sustains expression of ABCG2 in CDK7i-resistant cells.A, quantitation of SMAD4 mRNA and B, protein expression in MDA-MB-468 and MDA-MB-231 in resistant cells after siRNA-mediated silencing of SMAD4 with two different siRNAs, SMAD4-09 (S4-09) and SMAD4-10 (S4-10), or transfection with a nonsilencing control (siNS). Quantitation of SMAD4 protein expression relative to total protein for the experiments in triplicate is shown in the lower panel. C, SMAD4 silencing reduces ABCG2 mRNA and D, protein expression in MDA-MB-468 and MDA-MB-231-resistant cells. Quantitation of ABCG2 protein expression relative to total protein for the experiments in triplicate is shown in the lower panel. E, SB431542 (11.2 μM) treatment of resistant MDA-MB-468 reduces ABCG2 protein expression. F, SB431542 (5.6 μM) treatment of resistant MDA-MB-231 cells reduces ABCG2 protein expression. Quantitation of ABCG2 protein expression relative to total protein for the experiments in triplicate is shown in the lower panel. G, genome region depicting open chromatin as indicated by publicly available H3K27Ac ChIP-Seq data of the ABCG2 gene locus. The red rectangles show the positions of the primers used in the ChIP-PCR. H, p-Smad3 ChIP-PCR of a predicted binding site in the ABCG2 promoter regulatory region in resistant MDA-MB-468 and MDA-MB-231 cells. Error bars show the range of the p-SMAD3 binding as quantified by PCR from experimental duplicates. For all other data, error bars are means ± SD. ∗p < 0.05
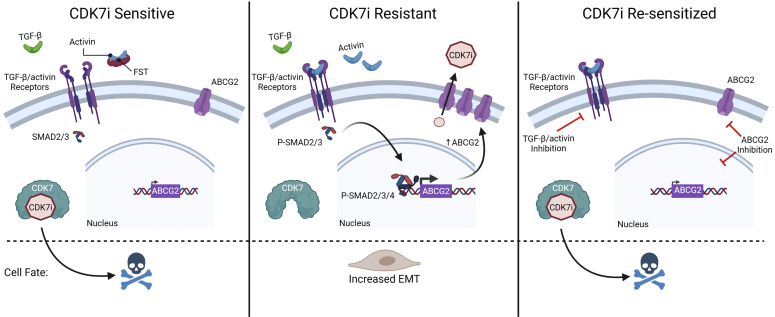
Figure 8.
A model of CDK7i resistance in TNBC cells. CDK7i resistance in TNBC can be driven by increased TGF-β/activin signaling compared with the parental/sensitive TNBC cells. This leads to increased EMT and ABCB2 expression. By inhibiting TGF-β/activin signaling or ABCG2 the CDK7i-resistant TNBC cells can be resensitized.
Discussion
CDK7 inhibitors are promising new therapeutics that are currently being evaluated in clinical trials, including those focused on TNBC. Like most targeted cancer therapies, resistance to this class of drugs is expected with long-term use. In the current study, we prospectively discovered that acquired resistance to CDK7 inhibitors can occur through adaptive activation of TGF-β family signaling, particularly induction of activin and repression of its intrinsic inhibitor, follistatin. Increased canonical signaling through SMADs then induces the expression of ABCG2 through direct binding of activated/phosphorylated SMAD3 to the gene. ABCG2 is a multidrug resistance protein that mediates resistance to several cytotoxic chemotherapies (24, 25, 26, 40). Blocking ABCG2 expression or activity in the current study demonstrates that it is necessary to sustain resistance to CDK7i. More recently, ABCG2 has been reported to induce resistance to CDK7i in MCF-7 cells (28), which represent a different subtype of breast cancer (21, 63, 64, 65). The mechanisms leading to ABCG2 upregulation remained unknown until the discovery reported herein revealing that induction of TGF-β/activin signaling in resistant cells is necessary to sustain ABCG2 expression through canonical SMAD signaling.
We used two cell lines in this study that represent two distinct subtypes of TNBC, mesenchymal stem–like (MSL, MDA-MB-231) and basal-like 1 (BL1, MDA-MB-468). Of note, acquired resistance in both subtypes involved the acquisition of an EMT gene expression signature and a more mesenchymal phenotype. This suggests that induction of EMT may be a common mechanism of resistance that could span cell lineages. Of note, TGF-β/activin signaling and EMT induce properties of stem cells (66). We observed an increase in the expression of Snail (Snai1) and Integrin β4 (ITGB4), two mediators of TGF-β/activin signaling that promote stem cell phenotypes (43, 53, 67, 68). Moreover, activin signaling has also been shown to increase stem cell properties in development and cancer (69, 70, 71). These data suggest that the development of acquired resistance may involve the activation of stem cell pathways. The CDK7 signaling pathway also plays a role in maintaining stem cells. Genetic disruption of CDK7 causes premature aging owing to adult stem cell exhaustion, whereas cyclin-dependent kinase activating kinase has been shown to be required for pluripotency of murine embryonic stem cells (5, 72). The studies presented herein did not directly assess whether CDK7i resistance involves an enrichment of cancer stem cells, which would require limiting dilution assays in mouse models. However, they do suggest that development of CDK7i resistance may overcome the requirement for CDK7 in maintaining stemness. Determining whether TGF-β and/or activin can substitute for CDK7 signaling in controlling stem cell properties will reveal how these pathways interact and their specific contributions to cancer stem cells and disease progression.
Although activin and TGF-β are distinct and have unique functions, they also have many overlapping activities including their ability to activate SMAD2/3 and induction of EMT, albeit at differing potencies (69). RNA-Seq data revealed a ∼3- to 7-fold induction of INHBA in the absence of similar changes in the expression of genes encoding TGF-β ligands. Homodimers of INHBA form activin A. Unlike TGF-β, the interaction of activin with its receptor can be prevented by follistatin binding. We found that FST expression was repressed with the acquisition of CDK7i resistance. Together, these data suggest that activin signaling may underlie the acquisition of resistance in these cell line models. However, we cannot rule out the possibility that signaling by TGF-β ligands may also be involved. We found that SMAD3 phosphorylation, a target of both activin and TGF-β signaling, is elevated in resistant cells and that its partner, SMAD4, is necessary to maintain expression of ABCG2 in resistant cells. Given the complexity of this family, we used a small molecule inhibitor of both activin and TGF-β receptors to block their activity and found that this partially restored sensitivity to THZ1. The lack of selective inhibitors for the activin and TGF-β receptors suggests that restoration of CDK7i response in resistant tumors would likely involve blockade of both receptors. Our studies also did not demonstrate whether activation of TGF-β and/or activin receptors is sufficient to induce resistance. Stimulation of the sensitive/parental cells with TGF-β1 alone was unable to induce resistance to THZ1 (data not shown); however this could be due to several factors. Some of these include a potential requirement for multiple TGF-β ligands, a need for an unknown but specific time frame of ligand activation to convey resistance, or the activation of unrelated pathways that remain to be discovered that collaborate with TGF-β ligands.
We focused on the role of ABCG2 in mediating resistance to CDK7i because it was commonly highly upregulated in both cell lines. However, many ABC transporter genes were induced with the acquisition of resistance and TGF-β signaling can increase the expression of multiple ABC transporters in other models (73). Although inhibition of ABCG2 restored sensitivity to THZ1, it is possible that the other transporters could also contribute to resistance to various CDK7 inhibitors. These transporters have differing degrees of target selectivity based on the molecular composition of the targets. This may underlie the distinct shifts in drug resistance that were observed with the different CDK7 inhibitors used in this study, where resistance to SY-1365 was less pronounced in the MDA-MB-231R cells than the other inhibitors. In this case, maximal resistance to SY-1365 may require upregulation of additional ABC transporters. To avoid such resistance, the use of broad-spectrum inhibitors of ABC transporters may be more effective than those that target individual transporters. Alternatively, the development of resistance to one CDK7i following the upregulation of a subset of transporters may still allow some degree of sensitivity to other, molecularly distinct CDK7is that are not substrates for those efflux pumps.
Although we observed an upregulation of activin (INHBA) gene expression with the acquisition of CDK7i resistance, the specific mechanisms that lead to this induction are not yet known. The CDK7i YKL-5-124 has recently been reported to induce DNA double-strand breaks and genome instability in small cell lung cancer (74). It is notable that double-strand breaks or telomere dysfunction has been shown to induce activin (INHBA) expression in variant human mammary epithelial cells (75). We anticipate that the DNA damage that has been reported for CDK7i in lung cancer cells may also occur in breast cancer cells. If so, this could lead to the rapid and sustained activation of INHBA expression observed with CDK7 inhibition. This would suggest that CDK7 inhibitors intrinsically induce their own resistance through a pathway that involves DNA damage followed by increased activin expression and signaling.
In summary, we have uncovered a mechanism of CDK7 inhibitor resistance involving activation of the TGF-β family signaling pathway, particularly activin, that then induces the expression of the ABCG2 multidrug resistance protein. We further found that resistance can be pharmacologically reversed using either inhibitors of ABCG2 or TGF-β/activin receptors. Understanding mechanisms of resistance will be essential for realizing the full utility of CDK7 inhibitors by blocking the development of resistance through sequential or combinatorial therapies. Our studies suggest two approaches for ensuring responsiveness to CDK7i in TNBC.
Experimental procedures
Cell culture and reagents
The MDA-MB-231 and MDA-MB-468 cell lines were purchased from ATCC between 2015 and 2018 and maintained as directed. Cell lines were expanded, aliquoted, cryogenically stored, and used within 12 passages after thaw. All cell lines were tested for Mycoplasma pulmonis and Mycoplasma spp using MycoAlertTM Plus Mycoplasma Detection Kit (Lonza, LT07-703). CDK7i-resistant cell lines were developed by treating the sensitive/parental cell lines with increasing doses of THZ1 over a 2-month period to a final concentration of 250 nM THZ1. Cells were imaged at 40× magnification using phase contrast on a Leica DMS200 microscope or Keyence BZ-X810. THZ1 (ApexBio, A8882), THZ2 (ApexBio, A8717), SY-1365 (MedChemExpress, HY-128587), Ko143 (Sigma, K2144-1), SB431542 (Sigma, S4317), and GF120918 (Cellagen Tech, C4312) were suspended in DMSO and then diluted in media.
RNA silencing
Cells were seeded in a 10-cm plate at 1 × 106 cells and reverse transfected with lipofectamine 2000 (30 μl) with Dharmacon siRNAs (30 μl of 20 μM) targeting ABCG2 (L-009924-00-0005), SMAD4 (J-003902-09-0005 or J-003902-10-0005), or a nontargeting control (D-001210-02-50). The next day, the cells were replated in a 24-well plate at 40,000 cells/well. Two days after replating, cells were treated with THZ1 (75 and 50 nM for MDA-MB-231 and MDA-MB-468 cells, respectively) and collected 48 h post treatment for gene silencing verification via quantitative real-time PCR and Western blots.
Reverse transcription–polymerase chain reaction
Total RNA was isolated with Trizol Reagent (Invitrogen). mRNA expression was measured as previously described (76). ThermoFisher probes used were: ABCG2 (Hs01053790_m1), SNAI1 (Hs00195591_m1), ITGB4 (Hs01103158_m1), SMAD4 (Hs00929647_m1), INHBA (Hs00170103_m1), FN1 (Hs00365052_m1), and SNAI2 (Hs00950344_m1). GAPDH (Hs02786624_g1) was used for normalization in the RT-PCR assays.
Dose–response curves
Cells were plated in a 24-well plate at 40,000 cells/well and treated the following day with vehicle (DMSO), CDK7i, or drug combinations of CDK7i with ABCG2 or TGF-β/activin receptor inhibitors. After 72 h, cells were stained with 0.05% crystal violet for quantitation. Absorbance of the crystal violet was assessed at 590 nM using a Promega GloMax Explorer plate reader.
RNA-Seq analysis
RNA was isolated using Trizol reagent followed by treatment with DNAse I (DNA-free kit, Ambion) per manufacturer’s instructions. For RNA-Seq, sensitive and resistant MDA-MB-231 and MDA-MB-468 cells were cultured in complete RPMI media for 48 h. Library preparation, sequencing, and analysis were completed by Novogene Corporation Inc using the Illumina platform with paired-end, 150-bp reads mapped to hg19. Differential expression was determined using the DESeq2 R package, where differentially expressed genes were deemed significantly changed if the Benjamini–Hochberg adjusted p-value was <0.05. The threshold for differential expression was a corrected p-value of 0.05 and absolute fold change of 2. To assess repeatability and cell line associations, the RNA-Seq gene lists were evaluated using the square of the Pearson correlation coefficient of the individual replicates.
GSEA (77) was used to assess the extent of enrichment of ABC transporters (Kyoto Encyclopedia of Genes and Genomes) (23), EMT (Hallmark v7.4) (78), and TGF-β family signaling pathways (79) with signatures obtained from the GSEA portal molecular signatures database (78). GO (http://www.geneontology.org/) was used to evaluate biological processes that were altered with CDK7i resistance. GO pathways with significantly overlapping genes were consolidated with NaviGO (80) based upon similarity scores. Resnik similarity scores >1.5 indicated high levels of similarity between pathways.
Public database analyses
Overall survival was evaluated using KM Plotter (29) to interrogate gene expression microarray data of 241 patients with basal breast cancer, which comprises 80% of all TNBC cases (81); patients were stratified based on the expression of ABCG2 (AffymetrixID, 209735_at). The overall survival of ABCG2 high- and low-expression groups was calculated with the optimal cutoff of 136 (range = 2–812) and a patient population of low (n = 158) and high (n = 83). KM Plotter uses patient data from a combination of data sets within The Cancer Genome Atlas, the Gene Expression Omnibus, and the European Genome-Phenome Archive. Patients were included regardless of treatment history.
Trypan blue viability assay and crystal violet staining
Viability was assessed with trypan blue exclusion as described (82). For colony formation assays, cells were treated with either vehicle (DMSO) or THZ1 (75 nM for MDA-MB-231 and 50 nM for MDA-MB-468) for 3 days. Eight hundred live cells were collected per well and then seeded in 24-well plates with complete media in the absence of drug, with the cell media replaced every 3 days. After 8 days, cells were stained with 0.05% crystal violet. Acetic acid, 10%, was added to the plates, and absorbance was read at 590 nm.
Protein quantitation
Abundance of specific proteins was quantified using Western blot analysis as described (76) with the following primary antibodies: ABCG2 (Cell Signaling, 4477S, 1:250), SMAD4 (Santa Cruz, sc-7966, 1:500), phosphorylated SMAD3 (Cell Signaling, 9520S, 1:500), SMAD3 (Cell Signaling, 9523S, 1:500), fibronectin (Cell Signaling, 26836S, 1:500), SNAI1 (Cell Signaling, C15D3, 1:500), ITGB4 (Cell Signaling, 14803S, 1:500), INHBA (Abcam, ab56057, 1:500), and FST (Santa Cruz, sc365003, 1:500). The following secondary antibodies were utilized to detect bound primary antibodies: IRDye 680RD or 800CW goat anti-rabbit or mouse IgM, 1:5000, LI-COR, 926-32211, 926-68070, 926-68071. Images of Western blots probed with the fluorescent secondary antibody were collected with a LI-COR Odyssey Fc or CLx, and target proteins were quantified and normalized to total protein present. Total protein was ascertained utilizing REVERT staining. Total protein staining was used for normalization of Western blots aligning with reports demonstrating that total protein staining is a more accurate loading control than the use of housekeeping proteins (83, 84, 85). Western blots were quantified using Image studio v5.2.
Chromatin immunoprecipitation–PCR
Using JASPAR, SMAD2/SMAD3/SMAD4-binding sites were predicted in the ABCG2 gene locus (hg:19, Chr4:89146000-89148000). Using UCSC Genome browser (http://genome.ucsc.edu), and publicly available H3K27Ac data, we additionally overlapped transcriptionally active and open DNA with the predicted binding sites (61, 62). An antibody against phosphorylated SMAD3 (Cell Signaling, 9520S, 40 μg) was used to immunoprecipitate this region of the ABCG2 gene. The ChIP-PCR protocol was completed as described (82). Primers were made to the region hg:19, Chr4:89147545-89147842 and were as follows: Left primer- 5′ taaacctggggcaacaccta 3’; right primer- 5′ catgccagaattccttggtt 3’.
Statistical analyses
Unless otherwise stated, all data are presented as means ± SD of three or more independent experiments that were each performed with either triplicate or duplicate technical replicates. Statistical significance was determined using a one or two-tailed Student’s t test or sum-of-squares F test with a p-value <0.05.
Data availability
All data are included in the article except the RNA-Seq data that was submitted to the Gene Expression Omnibus (GEO) with the accession number GSE175727.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgements
This work was also supported, in part, by shared resources of the Case Comprehensive Cancer Center (NIH P30CA043703).
Author contributions
B. M. W., B. L. B., E. W.-M., and R. A. K. conceptualization; B. M. W., B. L. B., E. W.-M., J. R. B., D. D. S., L. J. A., and R. A. K. methodology; B. M. W. and B. L. B. software; B. M. W., B. L. B., E. W.-M., and J. R. B. validation; B. M. W., B. L. B., E. W.-M., J. R. B., D. D. S., and L. J. A. formal analysis; B. M. W., B. L. B., and R. A. K. investigation; B. M. W., B. L. B., and R. A. K. resources; B. M. W., B. L. B., E. W.-M., J. R. B., D. D. S., L. J. A., and R. A. K. data curation; B. M. W. and B. L. B. writing – original draft; B. M. W., B. L. B., E. W.-M., J. R. B., D. D. S., L. J. A., and R. A. K. writing – review & editing; B. M. W., B. L. B., E. W.-M., J. R. B., D. D. S., L. J. A., and R. A. K. visualization; B. M. W., B. L. B., and R. A. K. supervision; B. M. W., B. L. B., and R. A. K. project administration; B. M. W., B. L. B., and R. A. K. funding acquisition.
Funding and additional information
This work was supported by the following grants from the NIH: R01CA206505 (R. A. K.), F31CA224809 (B. M. W.), T32GM008803 (B. M. W.), T32GM007250 (B. M. W.), T32GM135081 (J. R. B.); and from the DOD: W81XWH-18-1-0455 (L. J. A.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Edited by Alex Toker
Supporting information
References
- 1.Malumbres M. Cyclin-dependent kinases. Genome Biol. 2014;15:122. doi: 10.1186/gb4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ding L., Cao J., Lin W., Chen H., Xiong X., Ao H., Yu M., Lin J., Cui Q. The roles of cyclin-dependent kinases in cell-cycle progression and therapeutic strategies in human breast cancer. Int. J. Mol. Sci. 2020;21:1960. doi: 10.3390/ijms21061960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanchez-Martinez C., Lallena M.J., Sanfeliciano S.G., de Dios A. Cyclin dependent kinase (CDK) inhibitors as anticancer drugs: Recent advances (2015-2019) Bioorg. Med. Chem. Lett. 2019;29:126637. doi: 10.1016/j.bmcl.2019.126637. [DOI] [PubMed] [Google Scholar]
- 4.Kelso T.W., Baumgart K., Eickhoff J., Albert T., Antrecht C., Lemcke S., Klebl B., Meisterernst M. Cyclin-dependent kinase 7 controls mRNA synthesis by affecting stability of preinitiation complexes, leading to altered gene expression, cell cycle progression, and survival of tumor cells. Mol. Cell. Biol. 2014;34:3675–3688. doi: 10.1128/MCB.00595-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patel S.A., Simon M.C. Functional analysis of the Cdk7.cyclin H.Mat1 complex in mouse embryonic stem cells and embryos. J. Biol. Chem. 2010;285:15587–15598. doi: 10.1074/jbc.M109.081687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher R.P. Secrets of a double agent: CDK7 in cell-cycle control and transcription. J. Cell Sci. 2005;118:5171–5180. doi: 10.1242/jcs.02718. [DOI] [PubMed] [Google Scholar]
- 7.Chipumuro E., Marco E., Christensen C.L., Kwiatkowski N., Zhang T., Hatheway C.M., Abraham B.J., Sharma B., Yeung C., Altabef A., Perez-Atayde A., Wong K.K., Yuan G.C., Gray N.S., Young R.A. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell. 2014;159:1126–1139. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Augert A., MacPherson D. Treating transcriptional addiction in small cell lung cancer. Cancer Cell. 2014;26:783–784. doi: 10.1016/j.ccell.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 9.Wang Y., Zhang T., Kwiatkowski N., Abraham B.J., Lee T.I., Xie S., Yuzugullu H., Von T., Li H., Lin Z., Stover D.G., Lim E., Wang Z.C., Iglehart J.D., Young R.A. CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell. 2015;163:174–186. doi: 10.1016/j.cell.2015.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z., Peng H., Wang X., Yin X., Ma P., Jing Y., Cai M.C., Liu J., Zhang M., Zhang S., Shi K., Gao W.Q., Di W., Zhuang G. Preclinical efficacy and molecular mechanism of targeting CDK7-dependent transcriptional addiction in ovarian cancer. Mol. Cancer Ther. 2017;16:1739–1750. doi: 10.1158/1535-7163.MCT-17-0078. [DOI] [PubMed] [Google Scholar]
- 11.Greenall S.A., Lim Y.C., Mitchell C.B., Ensbey K.S., Stringer B.W., Wilding A.L., O'Neill G.M., McDonald K.L., Gough D.J., Day B.W., Johns T.G. Cyclin-dependent kinase 7 is a therapeutic target in high-grade glioma. Oncogenesis. 2017;6 doi: 10.1038/oncsis.2017.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cayrol F., Praditsuktavorn P., Fernando T.M., Kwiatkowski N., Marullo R., Calvo-Vidal M.N., Phillip J., Pera B., Yang S.N., Takpradit K., Roman L., Gaudiano M., Crescenzo R., Ruan J., Inghirami G. THZ1 targeting CDK7 suppresses STAT transcriptional activity and sensitizes T-cell lymphomas to BCL2 inhibitors. Nat. Commun. 2017;8:14290. doi: 10.1038/ncomms14290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y., Zhou L., Bandyopadhyay D., Sharma K., Allen A.J., Kmieciak M., Grant S. The covalent CDK7 inhibitor THZ1 potently induces apoptosis in multiple myeloma cells in vitro and in vivo. Clin. Cancer Res. 2019;25:6195–6205. doi: 10.1158/1078-0432.CCR-18-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao X., Dang L., Zheng X., Lu Y., Lu Y., Ji R., Zhang T., Ruan X., Zhi J., Hou X., Yi X., Li M.J., Gu T., Gao M., Zhang L. Targeting super-enhancer-driven oncogenic transcription by CDK7 inhibition in anaplastic thyroid carcinoma. Thyroid. 2019;29:809–823. doi: 10.1089/thy.2018.0550. [DOI] [PubMed] [Google Scholar]
- 15.Lehmann B.D., Jovanovic B., Chen X., Estrada M.V., Johnson K.N., Shyr Y., Moses H.L., Sanders M.E., Pietenpol J.A. Refinement of triple-negative breast cancer molecular subtypes: Implications for neoadjuvant chemotherapy selection. PLoS One. 2016;11 doi: 10.1371/journal.pone.0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bianchini G., Balko J.M., Mayer I.A., Sanders M.E., Gianni L. Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 2016;13:674–690. doi: 10.1038/nrclinonc.2016.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B., Ni Chonghaile T., Fan Y., Madden S.F., Klinger R., O'Connor A.E., Walsh L., O'Hurley G., Mallya Udupi G., Joseph J., Tarrant F., Conroy E., Gaber A., Chin S.F., Bardwell H.A. Therapeutic rationale to target highly expressed CDK7 conferring poor outcomes in triple-negative breast cancer. Cancer Res. 2017;77:3834–3845. doi: 10.1158/0008-5472.CAN-16-2546. [DOI] [PubMed] [Google Scholar]
- 18.Zeichner S.B., Terawaki H., Gogineni K. A review of systemic treatment in metastatic triple-negative breast cancer. Breast Cancer (Auckl.) 2016;10:25–36. doi: 10.4137/BCBCR.S32783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kassam F., Enright K., Dent R., Dranitsaris G., Myers J., Flynn C., Fralick M., Kumar R., Clemons M. Survival outcomes for patients with metastatic triple-negative breast cancer: Implications for clinical practice and trial design. Clin. Breast Cancer. 2009;9:29–33. doi: 10.3816/CBC.2009.n.005. [DOI] [PubMed] [Google Scholar]
- 20.Wang H., Jo Y.J., Sun T.Y., Namgoong S., Cui X.S., Oh J.S., Kim N.H. Inhibition of CDK7 bypasses spindle assembly checkpoint via premature cyclin B degradation during oocyte meiosis. Biochim. Biophys. Acta. 2016;1863:2993–3000. doi: 10.1016/j.bbamcr.2016.09.020. [DOI] [PubMed] [Google Scholar]
- 21.Heiser L.M., Sadanandam A., Kuo W.L., Benz S.C., Goldstein T.C., Ng S., Gibb W.J., Wang N.J., Ziyad S., Tong F., Bayani N., Hu Z., Billig J.I., Dueregger A., Lewis S. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc. Natl. Acad. Sci. U. S. A. 2012;109:2724–2729. doi: 10.1073/pnas.1018854108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chun K.H., Park J.H., Fan S. Predicting and overcoming chemotherapeutic resistance in breast cancer. Adv. Exp. Med. Biol. 2017;1026:59–104. doi: 10.1007/978-981-10-6020-5_4. [DOI] [PubMed] [Google Scholar]
- 23.Kanehisa M., Sato Y., Kawashima M., Furumichi M., Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44:D457–462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi Y.H., Yu A.M. ABC transporters in multidrug resistance and pharmacokinetics, and strategies for drug development. Curr. Pharm. Des. 2014;20:793–807. doi: 10.2174/138161282005140214165212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amawi H., Sim H.M., Tiwari A.K., Ambudkar S.V., Shukla S. ABC transporter-mediated multidrug-resistant cancer. Adv. Exp. Med. Biol. 2019;1141:549–580. doi: 10.1007/978-981-13-7647-4_12. [DOI] [PubMed] [Google Scholar]
- 26.Liu X. ABC family transporters. Adv. Exp. Med. Biol. 2019;1141:13–100. doi: 10.1007/978-981-13-7647-4_2. [DOI] [PubMed] [Google Scholar]
- 27.Orlando B.J., Liao M. ABCG2 transports anticancer drugs via a closed-to-open switch. Nat. Commun. 2020;11:2264. doi: 10.1038/s41467-020-16155-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sava G.P., Fan H., Fisher R.A., Lusvarghi S., Pancholi S., Ambudkar S.V., Martin L.A., Charles Coombes R., Buluwela L., Ali S. ABC-transporter upregulation mediates resistance to the CDK7 inhibitors THZ1 and ICEC0942. Oncogene. 2020;39:651–663. doi: 10.1038/s41388-019-1008-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gyorffy B., Lanczky A., Eklund A.C., Denkert C., Budczies J., Li Q., Szallasi Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2010;123:725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 30.de Bruin M., Miyake K., Litman T., Robey R., Bates S.E. Reversal of resistance by GF120918 in cell lines expressing the ABC half-transporter, MXR. Cancer Lett. 1999;146:117–126. doi: 10.1016/s0304-3835(99)00182-2. [DOI] [PubMed] [Google Scholar]
- 31.Palasuberniam P., Yang X., Kraus D., Jones P., Myers K.A., Chen B. ABCG2 transporter inhibitor restores the sensitivity of triple negative breast cancer cells to aminolevulinic acid-mediated photodynamic therapy. Sci. Rep. 2015;5:13298. doi: 10.1038/srep13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu K., Zhu J., Huang Y., Li C., Lu J., Sachar M., Li S., Ma X. Metabolism of KO143, an ABCG2 inhibitor. Drug Metab. Pharmacokinet. 2017;32:193–200. doi: 10.1016/j.dmpk.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 33.Wu C.P., Hung C.Y., Lusvarghi S., Huang Y.H., Tseng P.J., Hung T.H., Yu J.S., Ambudkar S.V. Overexpression of ABCB1 and ABCG2 contributes to reduced efficacy of the PI3K/mTOR inhibitor samotolisib (LY3023414) in cancer cell lines. Biochem. Pharmacol. 2020;180:114137. doi: 10.1016/j.bcp.2020.114137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng X., Carstens J.L., Kim J., Scheible M., Kaye J., Sugimoto H., Wu C.C., LeBleu V.S., Kalluri R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature. 2015;527:525–530. doi: 10.1038/nature16064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischer K.R., Durrans A., Lee S., Sheng J., Li F., Wong S.T., Choi H., El Rayes T., Ryu S., Troeger J., Schwabe R.F., Vahdat L.T., Altorki N.K., Mittal V., Gao D. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472–476. doi: 10.1038/nature15748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.George J.T., Jolly M.K., Xu S., Somarelli J.A., Levine H. Survival outcomes in cancer patients predicted by a partial EMT gene expression scoring metric. Cancer Res. 2017;77:6415–6428. doi: 10.1158/0008-5472.CAN-16-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neel D.S., Bivona T.G. Secrets of drug resistance in NSCLC exposed by new molecular definition of EMT. Clin. Cancer Res. 2013;19:3–5. doi: 10.1158/1078-0432.CCR-12-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jolly M.K., Somarelli J.A., Sheth M., Biddle A., Tripathi S.C., Armstrong A.J., Hanash S.M., Bapat S.A., Rangarajan A., Levine H. Hybrid epithelial/mesenchymal phenotypes promote metastasis and therapy resistance across carcinomas. Pharmacol. Ther. 2019;194:161–184. doi: 10.1016/j.pharmthera.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 39.Yin L., Castagnino P., Assoian R.K. ABCG2 expression and side population abundance regulated by a transforming growth factor beta-directed epithelial-mesenchymal transition. Cancer Res. 2008;68:800–807. doi: 10.1158/0008-5472.CAN-07-2545. [DOI] [PubMed] [Google Scholar]
- 40.Ehata S., Johansson E., Katayama R., Koike S., Watanabe A., Hoshino Y., Katsuno Y., Komuro A., Koinuma D., Kano M.R., Yashiro M., Hirakawa K., Aburatani H., Fujita N., Miyazono K. Transforming growth factor-beta decreases the cancer-initiating cell population within diffuse-type gastric carcinoma cells. Oncogene. 2011;30:1693–1705. doi: 10.1038/onc.2010.546. [DOI] [PubMed] [Google Scholar]
- 41.Mato E., Gonzalez C., Moral A., Perez J.I., Bell O., Lerma E., de Leiva A. ABCG2/BCRP gene expression is related to epithelial-mesenchymal transition inducer genes in a papillary thyroid carcinoma cell line (TPC-1) J. Mol. Endocrinol. 2014;52:289–300. doi: 10.1530/JME-14-0051. [DOI] [PubMed] [Google Scholar]
- 42.Larue L., Bellacosa A. Epithelial-mesenchymal transition in development and cancer: Role of phosphatidylinositol 3' kinase/AKT pathways. Oncogene. 2005;24:7443–7454. doi: 10.1038/sj.onc.1209091. [DOI] [PubMed] [Google Scholar]
- 43.Bierie B., Pierce S.E., Kroeger C., Stover D.G., Pattabiraman D.R., Thiru P., Liu Donaher J., Reinhardt F., Chaffer C.L., Keckesova Z., Weinberg R.A. Integrin-beta4 identifies cancer stem cell-enriched populations of partially mesenchymal carcinoma cells. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E2337–E2346. doi: 10.1073/pnas.1618298114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma B., Zhang L., Zou Y., He R., Wu Q., Han C., Zhang B. Reciprocal regulation of integrin beta4 and KLF4 promotes gliomagenesis through maintaining cancer stem cell traits. J. Exp. Clin. Cancer Res. 2019;38:23. doi: 10.1186/s13046-019-1034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ruan S., Lin M., Zhu Y., Lum L., Thakur A., Jin R., Shao W., Zhang Y., Hu Y., Huang S., Hurt E.M., Chang A.E., Wicha M.S., Li Q. Integrin beta4-targeted cancer immunotherapies inhibit tumor growth and decrease metastasis. Cancer Res. 2020;80:771–783. doi: 10.1158/0008-5472.CAN-19-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park J., Schwarzbauer J.E. Mammary epithelial cell interactions with fibronectin stimulate epithelial-mesenchymal transition. Oncogene. 2014;33:1649–1657. doi: 10.1038/onc.2013.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li C.L., Yang D., Cao X., Wang F., Hong D.Y., Wang J., Shen X.C., Chen Y. Fibronectin induces epithelial-mesenchymal transition in human breast cancer MCF-7 cells via activation of calpain. Oncol. Lett. 2017;13:3889–3895. doi: 10.3892/ol.2017.5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saxena M., Stephens M.A., Pathak H., Rangarajan A. Transcription factors that mediate epithelial-mesenchymal transition lead to multidrug resistance by upregulating ABC transporters. Cell Death Dis. 2011;2 doi: 10.1038/cddis.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu X., Zhang L., He X., Zhang P., Sun C., Xu X., Lu Y., Li F. TGF-beta plays a vital role in triple-negative breast cancer (TNBC) drug-resistance through regulating stemness, EMT and apoptosis. Biochem. Biophys. Res. Commun. 2018;502:160–165. doi: 10.1016/j.bbrc.2018.05.139. [DOI] [PubMed] [Google Scholar]
- 50.Bashir M., Damineni S., Mukherjee G., Kondaiah P. Activin-A signaling promotes epithelial-mesenchymal transition, invasion, and metastatic growth of breast cancer. NPJ Breast Cancer. 2015;1:15007. doi: 10.1038/npjbcancer.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jakowlew S.B. Transforming growth factor-beta in cancer and metastasis. Cancer Metastasis Rev. 2006;25:435–457. doi: 10.1007/s10555-006-9006-2. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y., Zeng Y., Liu T., Du W., Zhu J., Liu Z., Huang J.A. The canonical TGF-beta/Smad signalling pathway is involved in PD-L1-induced primary resistance to EGFR-TKIs in EGFR-mutant non-small-cell lung cancer. Respir. Res. 2019;20:164. doi: 10.1186/s12931-019-1137-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X.L., Liu L., Li D.D., He Y.P., Guo L.H., Sun L.P., Liu L.N., Xu H.X., Zhang X.P. Integrin beta4 promotes cell invasion and epithelial-mesenchymal transition through the modulation of Slug expression in hepatocellular carcinoma. Sci. Rep. 2017;7:40464. doi: 10.1038/srep40464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Steinbichler T.B., Dudas J., Ingruber J., Glueckert R., Sprung S., Fleischer F., Cidlinsky N., Dejaco D., Kofler B., Giotakis A.I., Skvortsova I.I., Riechelmann H. Slug is a surrogate marker of epithelial to mesenchymal transition (EMT) in head and neck cancer. J. Clin. Med. 2020;9:2061. doi: 10.3390/jcm9072061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tarasewicz E., Jeruss J.S. Phospho-specific Smad3 signaling: Impact on breast oncogenesis. Cell Cycle. 2012;11:2443–2451. doi: 10.4161/cc.20546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cui X., Shang S., Lv X., Zhao J., Qi Y., Liu Z. Perspectives of small molecule inhibitors of activin receptorlike kinase in antitumor treatment and stem cell differentiation (review) Mol. Med. Rep. 2019;19:5053–5062. doi: 10.3892/mmr.2019.10209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xia X., Wu W., Huang C., Cen G., Jiang T., Cao J., Huang K., Qiu Z. SMAD4 and its role in pancreatic cancer. Tumour Biol. 2015;36:111–119. doi: 10.1007/s13277-014-2883-z. [DOI] [PubMed] [Google Scholar]
- 58.Zhao M., Mishra L., Deng C.X. The role of TGF-beta/SMAD4 signaling in cancer. Int. J. Biol. Sci. 2018;14:111–123. doi: 10.7150/ijbs.23230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCarthy A.J., Chetty R. Smad4/DPC4. J. Clin. Pathol. 2018;71:661–664. doi: 10.1136/jclinpath-2018-205095. [DOI] [PubMed] [Google Scholar]
- 60.Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Raney B.J., Dreszer T.R., Barber G.P., Clawson H., Fujita P.A., Wang T., Nguyen N., Paten B., Zweig A.S., Karolchik D., Kent W.J. Track data hubs enable visualization of user-defined genome-wide annotations on the UCSC Genome Browser. Bioinformatics. 2014;30:1003–1005. doi: 10.1093/bioinformatics/btt637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sorlie T., Perou C.M., Tibshirani R., Aas T., Geisler S., Johnsen H., Hastie T., Eisen M.B., van de Rijn M., Jeffrey S.S., Thorsen T., Quist H., Matese J.C., Brown P.O., Botstein D. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neve R.M., Chin K., Fridlyand J., Yeh J., Baehner F.L., Fevr T., Clark L., Bayani N., Coppe J.P., Tong F., Speed T., Spellman P.T., DeVries S., Lapuk A., Wang N.J. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., Campbell L.L., Polyak K., Brisken C., Yang J., Weinberg R.A. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dhasarathy A., Phadke D., Mav D., Shah R.R., Wade P.A. The transcription factors Snail and Slug activate the transforming growth factor-beta signaling pathway in breast cancer. PLoS One. 2011;6 doi: 10.1371/journal.pone.0026514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu X.L., Kapoun A.M. Heterogeneous activation of the TGFbeta pathway in glioblastomas identified by gene expression-based classification using TGFbeta-responsive genes. J. Transl. Med. 2009;7:12. doi: 10.1186/1479-5876-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Katsuno Y., Derynck R. Epithelial plasticity, epithelial-mesenchymal transition, and the TGF-beta family. Dev. Cell. 2021;56:726–746. doi: 10.1016/j.devcel.2021.02.028. [DOI] [PubMed] [Google Scholar]
- 70.Lonardo E., Hermann P.C., Mueller M.T., Huber S., Balic A., Miranda-Lorenzo I., Zagorac S., Alcala S., Rodriguez-Arabaolaza I., Ramirez J.C., Torres-Ruiz R., Garcia E., Hidalgo M., Cebrian D.A., Heuchel R. Nodal/activin signaling drives self-renewal and tumorigenicity of pancreatic cancer stem cells and provides a target for combined drug therapy. Cell Stem Cell. 2011;9:433–446. doi: 10.1016/j.stem.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 71.Togashi Y., Kogita A., Sakamoto H., Hayashi H., Terashima M., de Velasco M.A., Sakai K., Fujita Y., Tomida S., Kitano M., Okuno K., Kudo M., Nishio K. Activin signal promotes cancer progression and is involved in cachexia in a subset of pancreatic cancer. Cancer Lett. 2015;356:819–827. doi: 10.1016/j.canlet.2014.10.037. [DOI] [PubMed] [Google Scholar]
- 72.Ganuza M., Saiz-Ladera C., Canamero M., Gomez G., Schneider R., Blasco M.A., Pisano D., Paramio J.M., Santamaria D., Barbacid M. Genetic inactivation of Cdk7 leads to cell cycle arrest and induces premature aging due to adult stem cell exhaustion. EMBO J. 2012;31:2498–2510. doi: 10.1038/emboj.2012.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hu Y.W., Wang Q., Ma X., Li X.X., Liu X.H., Xiao J., Liao D.F., Xiang J., Tang C.K. TGF-beta1 up-regulates expression of ABCA1, ABCG1 and SR-BI through liver X receptor alpha signaling pathway in THP-1 macrophage-derived foam cells. J. Atheroscler. Thromb. 2010;17:493–502. doi: 10.5551/jat.3152. [DOI] [PubMed] [Google Scholar]
- 74.Zhang H., Christensen C.L., Dries R., Oser M.G., Deng J., Diskin B., Li F., Pan Y., Zhang X., Yin Y., Papadopoulos E., Pyon V., Thakurdin C., Kwiatkowski N., Jani K. CDK7 inhibition potentiates genome instability triggering anti-tumor immunity in small cell lung cancer. Cancer Cell. 2020;37:37–54.e39. doi: 10.1016/j.ccell.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fordyce C., Fessenden T., Pickering C., Jung J., Singla V., Berman H., Tlsty T. DNA damage drives an activin a-dependent induction of cyclooxygenase-2 in premalignant cells and lesions. Cancer Prev. Res. (Phila) 2010;3:190–201. doi: 10.1158/1940-6207.CAPR-09-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seachrist D.D., Hannigan M.M., Ingles N.N., Webb B.M., Weber-Bonk K.L., Yu P., Bebek G., Singh S., Sizemore S.T., Varadan V., Licatalosi D.D., Keri R.A. The transcriptional repressor BCL11A promotes breast cancer metastasis. J. Biol. Chem. 2020;295:11707–11719. doi: 10.1074/jbc.RA120.014018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Plasari G., Calabrese A., Dusserre Y., Gronostajski R.M., McNair A., Michalik L., Mermod N. Nuclear factor I-C links platelet-derived growth factor and transforming growth factor beta1 signaling to skin wound healing progression. Mol. Cell. Biol. 2009;29:6006–6017. doi: 10.1128/MCB.01921-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wei Q., Khan I.K., Ding Z., Yerneni S., Kihara D. NaviGO: Interactive tool for visualization and functional similarity and coherence analysis with gene ontology. BMC Bioinformatics. 2017;18:177. doi: 10.1186/s12859-017-1600-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Prat A., Karginova O., Parker J.S., Fan C., He X., Bixby L., Harrell J.C., Roman E., Adamo B., Troester M., Perou C.M. Characterization of cell lines derived from breast cancers and normal mammary tissues for the study of the intrinsic molecular subtypes. Breast Cancer Res. Treat. 2013;142:237–255. doi: 10.1007/s10549-013-2743-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sahni J.M., Gayle S.S., Bonk K.L., Vite L.C., Yori J.L., Webb B., Ramos E.K., Seachrist D.D., Landis M.D., Chang J.C., Bradner J.E., Keri R.A. Bromodomain and extraterminal protein inhibition blocks growth of triple-negative breast cancers through the suppression of aurora kinases. J. Biol. Chem. 2016;291:23756–23768. doi: 10.1074/jbc.M116.738666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eaton S.L., Roche S.L., Llavero Hurtado M., Oldknow K.J., Farquharson C., Gillingwater T.H., Wishart T.M. Total protein analysis as a reliable loading control for quantitative fluorescent western blotting. PLoS One. 2013;8 doi: 10.1371/journal.pone.0072457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Collins M.A., An J., Peller D., Bowser R. Total protein is an effective loading control for cerebrospinal fluid western blots. J. Neurosci. Methods. 2015;251:72–82. doi: 10.1016/j.jneumeth.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aldridge G.M., Podrebarac D.M., Greenough W.T., Weiler I.J. The use of total protein stains as loading controls: An alternative to high-abundance single-protein controls in semi-quantitative immunoblotting. J. Neurosci. Methods. 2008;172:250–254. doi: 10.1016/j.jneumeth.2008.05.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the article except the RNA-Seq data that was submitted to the Gene Expression Omnibus (GEO) with the accession number GSE175727.