Abstract
The Mycobacterium tuberculosis complex includes M. tuberculosis, M. bovis, M. africanum, and M. microti. Most clinical isolates are M. tuberculosis or M. bovis. These species can be distinguished by phenotypes and genotypes. However, there is no simple definition of M. africanum, and some authors question the validity of this species. We analyzed 17 human isolates from Sierra Leone, identified as M. africanum by biochemical and growth characteristics. We sequenced polymorphic genes and intergenic regions. We amplified DNA from six loci with variable numbers of tandem repeats (VNTRs) and determined the exact number of repeats at each locus in each strain. All M. africanum isolates had the ancestral CTG Leu at katG codon 463. Drug-resistant M. africanum isolates had katG and rpoB mutations similar to those found in drug-resistant M. bovis and M. tuberculosis. Fourteen Sierra Leone M. africanum isolates (designated group A) had katG codon 203 ACC Thr, also found in M. africanumT (the T indicates type strain) from Senegal. Group A isolates clustered with M. africanumT by VNTR analysis. Three M. africanum isolates (group B) had katG codon 203 ACT Thr, found in M. tuberculosisT, and clustered with M. tuberculosisT by VNTR analysis. Phenotypic identification of M. africanum yielded a heterogeneous collection of strains. Genotypic analyses identified a cluster (M. africanum group A) which included M. africanumT and was distinct from the rest of the M. tuberculosis complex. Future studies of M. africanum should include both phenotypic and genotypic analyses.
The Mycobacterium tuberculosis complex includes four recognized species (M. tuberculosis, M. bovis, M. africanum, and M. microti), which together cause human and animal tuberculosis (37). M. tuberculosis and M. bovis are the most common isolates in clinical laboratories and can be distinguished by biochemical tests, growth phenotypes, and several genetic markers. However, M. africanum isolates have substantial phenotypic heterogeneity with some strains resembling M. bovis and others resembling M. tuberculosis (4, 10, 12, 15, 36). This has led several authors to question the validity of the species designation M. africanum (33, 37, 39).
We conducted a detailed genotypic analysis of a panel of 17 recent M. africanum isolates. All were isolated from humans in Sierra Leone between 1991 and 1993 and were identified as M. africanum on the basis of biochemical tests and growth patterns. These strains are a subset of a collection described previously (12, 14, 29). We sequenced portions of several genes and intergenic regions which are known to have polymorphisms in M. tuberculosis and M. bovis. We also analyzed six loci with variable numbers of tandem repeats (VNTRs). Our goal was to determine whether M. africanum strains have distinct genotypes which set them apart from M. tuberculosis and M. bovis.
MATERIALS AND METHODS
Mycobacterial strains.
Mycobacterial strains are listed in Tables 1 and 2. M. africanum isolates were obtained from humans in Sierra Leone with pulmonary tuberculosis (12, 29). These strains were part of a large collection of 96 strains originally collected for the purpose of drug resistance surveillance organized by the National Tuberculosis Control Program in Sierra Leone and supported by the German Leprosy Relief Association. Isolates were characterized as M. africanum based on biochemical and biophysical properties as described previously (12). Susceptibility testing was performed by the proportion method using the critical concentrations of 40 μg/ml for rifampin, 0.2 μg/ml for isoniazid (INH), 4 μg/ml for streptomycin, and 2 μg/ml for ethambutol. Pyrazinamidase production was determined as described by Wayne (35). The 17 M. africanum isolates in this study were selected from the larger collection to include isolates with a maximal divergence in antibiotic susceptibility phenotypes. M. africanum ATCC 25420T (the T indicates type strain), M. bovis ATCC 19210T, M. microti ATCC 19422T, M. tuberculosis ATCC 27294T, and M. bovis ATCC 35734 (BCG Pasteur) were obtained from the American Type Culture Collection in Manassas, Va.
TABLE 1.
Phenotypes of 17 M. africanum clinical isolates from Sierra Leonea
| Groupb | Strain no. | Nitrate reduction | Niacin accumulation | Catalase activity at 22°C (mm) | Niveau depth (mm) | Pyrazinamidase activity | Susceptibility to:
|
|||
|---|---|---|---|---|---|---|---|---|---|---|
| Isoniazid | Rifampin | Streptomycin | Ethambutol | |||||||
| A | 014 | Weakly positive | Weakly positive | 3 | 7 | Negative | R | R | R | S |
| A | 031 | Weakly positive | Weakly positive | 0 | 8 | Weak | R | R | R | S |
| A | 034 | Weakly positive | Weakly positive | 2 | 8 | Positive | R | R | R | S |
| A | 037 | Weakly positive | Negative | 10 | 13 | ND | R | R | R | R |
| A | 073 | Weakly positive | Negative | 1 | 12 | Positive | R | R | R | R |
| A | 213 | Weakly positive | Weakly positive | 1 | 5 | Negative | R | R | R | R |
| A | 279 | Weakly positive | Weakly positive | 1 | 8 | Positive | S | S | ND | S |
| A | 354 | Weakly positive | Negative | 1 | 8 | Positive | S | S | ND | S |
| A | 370 | Weakly positive | Weakly positive | 1 | 4 | Positive | S | S | ND | S |
| A | 481 | Weakly positive | Weakly positive | 1 | 6 | Positive | R | S | S | S |
| A | 556 | Weakly positive | Weakly positive | 2 | 5 | Weak | R | S | ND | S |
| A | 566 | Weakly positive | Negative | 0 | 9 | Positive | R | S | ND | S |
| A | 736 | Weakly positive | ND | 1 | 6 | Positive | S | S | S | S |
| A | 754 | Weakly positive | Weakly positive | 1 | 10 | Positive | R | R | ND | B |
| B | 053 | Weakly positive | Weakly positive | 2 | 10 | Positive | R | R | ND | S |
| B | 203 | Positive | Weakly positive | 1 | ND | Positive | B | S | ND | S |
| B | 246 | Positive | Positive | 16 | 8 | Positive | S | S | ND | S |
All 17 strains had no catalase activity at 68°C, produced no color change on bromcresol medium, and displayed dysgonic growth on Löwenstein-Jensen medium. Abbreviations: ND, not done; R, resistant; B, borderline; S, susceptible.
M. africanum group based on the sequence of katG codon 203.
TABLE 2.
Genotypes of 17 M. africanum clinical isolates from Sierra Leone and five reference strains of the M. tuberculosis complex
| Groupa | Species and strain | Polymorphism not associated with drug resistance
|
Polymorphism associated with drug resistance
|
||||
|---|---|---|---|---|---|---|---|
| katG codon 203 sequence | katG codon 463 sequence | VNTR allele profileb | Mixed-linker clusterc | katG codon 315 sequence | rpoB genotype | ||
| A | M. africanum 014 | ACT Thr | CTG Leu | 663535 | 3 | AGC Ser | Wild type |
| A | M. africanum 031 | ACT Thr | CTG Leu | 564545 | 1 | AGC Ser | Codon 519 Asn deletion CAA |
| A | M. africanum 034 | ACT Thr | CTG Leu | 564544 | 2 | ACC Thr | Codon 526 His→Arg (CAC→CGC) |
| A | M. africanum 037 | ACT Thr | CTG Leu | 564544 | 2 | ACC Thr | Codon 526 His→Arg (CAC→CGC) |
| A | M. africanum 073 | ACT Thr | CTG Leu | 663535 | 3 | ACC Thr | Codon 526 His→Asp (CAC→GAC) |
| A | M. africanum 213 | ACT Thr | CTG Leu | 664535 | Unique | ATC Ile | Codon 526 His→Tyr (CAC→TAC) |
| A | M. africanum 279 | ACT Thr | CTG Leu | 564544 | 1 | AGC Ser | Wild type |
| A | M. africanum 354 | ACT Thr | CTG Leu | 565544 | 1 | AGC Ser | Wild type |
| A | M. africanum 370 | ACT Thr | CTG Leu | 664536 | Unique | AGC Ser | Wild type |
| A | M. africanum 481 | ACT Thr | CTG Leu | 564536 | 1 | AAC Asn | Wild type |
| A | M. africanum 556 | ACT Thr | CTG Leu | 663535 | 4 | ACC Thr | Codon 533 Leu→Pro (CTG→CCG) |
| A | M. africanum 566 | ACT Thr | CTG Leu | 564544 | Unique | AGC Ser | Wild type |
| A | M. africanum 736 | ACT Thr | CTG Leu | 564544 | 1 | AGC Ser | Wild type |
| A | M. africanum 754 | ACT Thr | CTG Leu | 663535 | 4 | ACC Thr | Codon 531 Ser→Leu (TCG→TTG) |
| B | M. africanum 053d | ACC Thr | CTG Leu | 642432 | Unique | AGC Ser | Codon 531 Ser→Leu (TCG→TTG) |
| B | M. africanum 203 | ACC Thr | CTG Leu | 642432 | NDe | AGC Ser | Wild type |
| B | M. africanum 246 | ACC Thr | CTG Leu | 642432 | Unique | AGC Ser | Wild type |
| M. africanumT | ACT Thr | CTG Leu | 564544 | ND | AGC Ser | Wild type | |
| M. bovis BCG Pasteur | ACT Thr | CTG Leu | 655623 | ND | AGC Ser | Wild type | |
| M. bovisT | ACT Thr | CTG Leu | 675542 | ND | AGC Ser | Wild type | |
| M. microtiT | ACT Thr | CTG Leu | 653571 | ND | AGC Ser | Wild type | |
| M. tuberculosisT | ACC Thr | CGG Arg | 633433 | ND | AGC Ser | Wild type | |
M. africanum group based on the sequence of katG codon 203.
The number of tandem DNA repeats at each of six genetic loci (MPTR-A, ETR-A, ETR-B, ETR-C, ETR-D, and ETR-E) was determined in each strain (9). The results were combined to yield a six-digit allele profile. Each digit represents the number of tandem repeats at a particular locus (except locus MPTR-A, for which the number of copies is reduced by 10). For example, the M. africanum type strain has the VNTR allele 564544. This strain has 15 tandem repeat copies at locus MPTR-A, 6 copies at locus ETR-A, 4 copies at locus ETR-B, 5 copies at locus ETR-C, 4 copies at locus ETR-D, and 4 copies at locus ETR-E.
Mixed-linker fingerprints were previously published (12). Eleven strains fell into one of four clusters (designated cluster 1, 2, 3, or 4), and five strains had unique fingerprints.
This strain also had a nucleotide substitution at katG codon 253 and a frameshift mutation at katG codon 254. This genotype is predicted to be associated with INH resistance.
ND, not done.
DNA preparation.
Mycobacterial isolates were killed by immersion in 70% (vol/vol) ethanol (40). DNA was released from bacterial cells by snap freeze-thawing (41).
DNA sequence analysis.
A portion of the rpoB gene was amplified by PCR and sequenced as described previously (41). Similarly, the first 500 codons of the katG gene, the upstream noncoding region of inhA, and the oxyR-ahpC intergenic region were amplified and sequenced as described previously (21, 26). Sequences were compared to wild-type sequences found in GenBank under accession no. L27989 (rpoB), Z97193 (katG), U02492 and Z79701 (inhA), and U16243 (oxyR-ahpC). We did not sequence the gyrA gene. Although polymorphisms at gyrA codon 95 are useful for separating isolates of M. tuberculosis, this codon is not variable in M. africanum (26).
VNTR analysis.
Six loci containing variable numbers of tandem repeats (VNTRs) were amplified by PCR as previously described (9). The exact number of tandem repeats at each locus in each strain was determined by the length of the PCR products on agarose gels. One of these loci (MPTR-A) contains the major polymorphic tandem repeat (MPTR), and five loci (ETR-A through ETR-E) contained exact tandem repeats (ETRs). Primers, PCR conditions, map locations, and other details related to these VNTR loci were published previously (9).
Phylogenetic analysis.
Phylogenetic trees were constructed based on the VNTR allele profiles by maximum-parsimony analysis using PAUP software (30). Maximum parsimony and other methods for phylogenetic analysis are described in recent reviews (22, 28). The data were resampled with 10,000 bootstrap replications (6), and a majority-rule consensus was computed from the most-parsimonious trees. The analysis was conducted twice, treating character states as either ordered or unordered. Treating the character states as ordered assumes single-step transitions in the number of tandem repeats at each locus. This assumption is consistent with the results of sequence analysis of the MPTR-A locus (7). Genetic distances between pairs of VNTR allele profiles were calculated based on the minimum number of single-step transitions.
Mixed-linker PCR.
Genomic DNA was digested, ligated with mixed linkers, and amplified in a nested PCR as previously described (13). The PCR products were separated by agarose electrophoresis to generate a DNA fingerprint based on the mobile IS6110 insertion element. Mixed-linker fingerprints for this collection were previously published (12).
Other genetic analyses.
RD1 is a 9.5-kb region deleted in substrains of M. bovis BCG. Two primers which flank RD1 and one primer within RD1 were combined in a multiplex-PCR amplification as previously described (31). Strains containing the RD1 region yield a 150-bp product, while strains with deletion of RD1 (all BCG substrains) yield a 200-bp product. Substrains of M. bovis BCG also have a sequence polymorphism at codon 252 of a 1,551-bp open reading frame (orf1) in GenBank under accession no. M15467 (gene homologous to Rv0422c). A portion of this gene was amplified by PCR, and the sequence at this codon was determined by digestion with the restriction enzyme BanI as previously described (7).
RESULTS
Tables 1 and 2 list phenotypic and genotypic results for 17 M. africanum isolates from Sierra Leone. These isolates were identified as M. africanum on the basis of biochemical and biophysical properties as listed in Table 1 (12, 36). M. africanum isolates grew as smooth, dysgonic, nonpigmented colonies. Growth was inhibited by thiophene-2-carboxylic hydrazide (TCH), with the exception of growth by strains resistant to INH. All strains had pyrazinamidase activity except two isolates (014 and 213) which were resistant to multiple drugs. Acquisition of pyrazinamide resistance is associated with the loss of pyrazinamidase activity. Most M. africanum isolates had catalase activity less than or equal to 10 mm (height of bubbles above agar) at 22°C, and all were negative for catalase activity at 68°C. All isolates except 370 grew at least 5 mm below the surface (niveau depth) on Lebek’s semisolid medium, indicating microaerophilic growth. All isolates produced no color change when grown on bromcresol medium. The nitrate reduction and niacin accumulation phenotypes were variable, as described for other collections of M. africanum strains (4, 36).
katG sequence.
katG codes for catalase peroxidase. Loss of catalase peroxidase activity can result from deletions or point mutations in katG and is associated with INH resistance. We sequenced the entire katG gene in the 17 M. africanum strains and the 5 reference strains and compared the sequences to the wild-type sequence (GenBank accession no. Z97193). Results are listed in Table 2. We found mutations at five katG codons (203, 253, 254, 315, and 463). Eleven strains were INH resistant; of these strains, seven had polymorphisms at katG codon 315. One INH-resistant strain had both a point mutation at codon 253 and a frameshift mutation at codon 254. Identical or similar katG mutations are associated with INH resistance in M. tuberculosis (21).
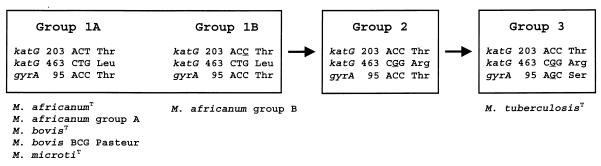
We found polymorphisms at katG codons 203 and 463 which were not associated with INH resistance. Sreevatsan et al. (26) used polymorphisms at katG codon 463 and gyrA codon 95 to define a broad evolutionary scenario for the M. tuberculosis complex, as shown in Fig. 1. katG codon 463 CTG Leu is the ancestral genotype; it was found in all strains of M. bovis (109 strains), M. microti (9 strains), and M. intracellulare (1 strain) (26). Some M. tuberculosis strains (group 1) also have CTG Leu at this locus, but the majority of M. tuberculosis strains (including M. tuberculosisT) have CGG Arg (groups 2 and 3) (26). All 17 M. africanum strains from Sierra Leone and M. africanumT from Senegal had the ancestral CTG Leu and thus belong to group 1 of the M. tuberculosis complex.
FIG. 1.
Broad evolutionary scenario for M. tuberculosis complex organisms. Sreevatsan et al. (26) defined groups 1, 2, and 3 based on the sequence of katG codon 463 and gyrA codon 95. We subdivided group 1 strains based on the sequence of katG codon 203. The distribution of the 22 strains described in this manuscript is shown.
The only site which varied among the M. africanum strains and which was not associated with drug resistance was katG codon 203. This synonymous polymorphism was used to divide the M. africanum strains into two groups (Fig. 1). Fourteen M. africanum strains from Sierra Leone (designated group A) had katG codon 203 ACT Thr. This genotype appears to be the ancestral genotype; it is found in M. africanumT, M. bovisT, M. microtiT, M. bovis BCG, and clinical M. bovis isolates. Three of the M. africanum strains (designated group B) had katG codon 203 ACC Thr, which is also found in M. tuberculosisT and in all of several hundred clinical M. tuberculosis isolates sequenced to date.
rpoB sequence.
rpoB codes for the β subunit of the DNA-dependent RNA polymerase which is the target of rifampin activity. Mutations in rpoB codons 509 to 533 are strongly associated with rifampin resistance in pathogenic mycobacteria, including M. tuberculosis, M. leprae, and M. avium (17, 41). Eight of the M. africanum isolates from Sierra Leone, seven of which had rpoB mutations, were resistant to rifampin. Nine of the M. africanum isolates were rifampin susceptible, and eight of these had the wild-type rpoB sequence in this segment of the gene. Interestingly, M. africanum 556 was rifampin susceptible but had a codon 533 Leu-to-Pro (Leu→Pro) mutation previously associated with rifampin resistance in clinical M. tuberculosis isolates (41). M. africanum 556 is no longer viable, so we were unable to repeat these analyses. Overall, we observed six distinct rpoB mutations in these M. africanum strains, all of which have been described in rifampin-resistant M. tuberculosis strains.
VNTR analysis.
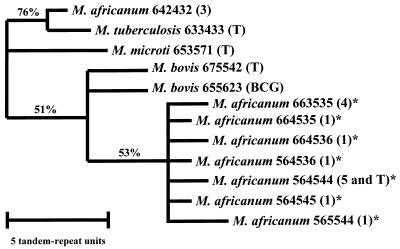
We amplified DNA at six loci with variable numbers of tandem repeats (9). The number of tandem DNA repeats at each locus in each strain was determined by the length of the PCR product on an agarose gel. The results of the six loci (MPTR-A, ETR-A, ETR-B, ETR-C, ETR-D, and ETR-E) were combined to yield a six-digit VNTR allele profile (9). Each digit represents the number of tandem repeats at a particular locus, with the exception of locus MPTR-A, where it represents the number of copies minus 10. For example, M. africanumT has the VNTR allele profile 564544. This strain has 15 tandem repeat copies at locus MPTR-A, 6 copies at locus ETR-A, 4 copies at locus ETR-B, 5 copies at locus ETR-C, 4 copies at locus ETR-D, and 4 copies at locus ETR-E. VNTR allele profiles are listed in Table 2. Figure 2 displays phylogenetic relationships among the VNTR allele profiles, and Table 3 shows genetic distances between pairs of VNTR allele profiles.
FIG. 2.
Parsimony phylogenetic tree based on VNTR allele profiles. The tree includes 12 VNTR allele profiles generated from 22 strains. The number of clinical isolates is indicated in parentheses, as are the profiles associated with the type strains (T) and BCG Pasteur (BCG). VNTR allele profiles found in M. africanum group A are marked by an asterisk. Horizontal lengths represent genetic distances; vertical lengths are not meaningful. All clusters shown on this tree were found in 100% of the most-parsimonious trees. The data were resampled with 10,000 bootstrap replications (6, 30). The percentage of bootstrap replications which yielded each grouping is indicated. The tree was constructed with the assumption that the number of tandem repeats in each strain changes by a single copy at a time (ordered character states). When we conducted the same analysis with unordered character states, the M. africanum group A isolates remained clustered.
TABLE 3.
Genetic distances between pairs of VNTR allele profilesa
| VNTR allele profile | M. africanum group B 642432 | M. tuber-culosis type 633433 | M. microti type 653571 | M. bovis type 675542 | M. bovis BCG 655623 |
M. africanum group A
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 663535 | 664535 | 664536 | 564536 | 564544 | 564545 | 565544 | ||||||
| M. africanum group B 642432 | 0 | 3 | 8 | 8 | 8 | 7 | 8 | 9 | 10 | 9 | 10 | 10 |
| M. tuberculosis type 633433 | 3 | 0 | 9 | 9 | 7 | 6 | 7 | 8 | 9 | 8 | 9 | 9 |
| M. microti type 653571 | 8 | 9 | 0 | 8 | 10 | 9 | 10 | 11 | 12 | 9 | 10 | 10 |
| M. bovis type 675542 | 8 | 9 | 8 | 0 | 6 | 7 | 6 | 7 | 8 | 5 | 6 | 4 |
| M. bovis BCG 655623 | 8 | 7 | 10 | 6 | 0 | 7 | 6 | 7 | 8 | 7 | 8 | 6 |
| M. africanum group A 663535 | 7 | 6 | 9 | 7 | 7 | 0 | 1 | 2 | 3 | 4 | 3 | 5 |
| M. africanum group A 664535 | 8 | 7 | 10 | 6 | 6 | 1 | 0 | 1 | 2 | 3 | 2 | 4 |
| M. africanum group A 664536 | 9 | 8 | 11 | 7 | 7 | 2 | 1 | 0 | 1 | 4 | 3 | 5 |
| M. africanum group A 564536 | 10 | 9 | 12 | 8 | 8 | 3 | 2 | 1 | 0 | 3 | 2 | 4 |
| M. africanum group A 564544 | 9 | 8 | 9 | 5 | 7 | 4 | 3 | 4 | 3 | 0 | 1 | 1 |
| M. africanum group A 564545 | 10 | 9 | 10 | 6 | 8 | 3 | 2 | 3 | 2 | 1 | 0 | 2 |
| M. africanum group A 565544 | 10 | 9 | 10 | 4 | 6 | 5 | 4 | 5 | 4 | 1 | 2 | 0 |
The box shows distances within the group A cluster of M. africanum strains from Sierra Leone. All three strains in M. africanum group B had an identical VNTR allele profile.
The 14 M. africanum group A isolates had seven VNTR allele profiles, indicated by asterisks in Fig. 2. One of the group A VNTR profiles was identical to that of M. africanumT (564544). The group A isolates, including M. africanumT, clustered by VNTR analysis. They were more closely related to M. bovisT than to the rest of the M. tuberculosis complex (Fig. 2 and Table 3). In contrast, the three M. africanum group B isolates had the VNTR allele profile 642432 and clustered with M. tuberculosisT.
Mixed-linker PCR fingerprints.
The mixed-linker PCR fingerprints for these strains were previously published (12). Eleven of the M. africanum isolates in group A were clustered by mixed-linker analysis, and three were unique. Two of the group B isolates were unique, and the third was not analyzed by mixed-linker PCR. Cluster 1 and cluster 2 have mixed-linker PCR fingerprints which differ by a single band and together accounted for 40% of all M. africanum isolates from Sierra Leone in a previous report (12).
The clustering of strains by mixed-linker analysis and VNTR analysis was consistent with the results of the other genotypic methods. Mixed-linker cluster 2 contained two strains with identical VNTR allele profiles (564544), identical katG codon 315 mutations, and identical rpoB mutations. The two strains in mixed-linker cluster 3 and the two strains in cluster 4 all shared the VNTR allele profile 663535, though their drug resistance mutations varied. The five strains in mixed-linker cluster 1 had four closely related VNTR allele profiles.
Other analyses.
All 17 M. africanum isolates had wild-type sequences in the upstream noncoding region of inhA and the oxyR-ahpC intergenic region (26). All 17 M. africanum strains had the wild-type sequence AGC Ser at orf1 codon 252 (7). All 17 M. africanum strains contained RD1, as previously reported (31).
DISCUSSION
The M. tuberculosis complex includes four species on the Approved List of Bacterial Names: M. tuberculosis, M. bovis, M. africanum, and M. microti (37). A fifth group, characterized by the Canetti strain, has distinct features, but has not been proposed as a separate species (34). Strains from all four species show >85% DNA-DNA relatedness and have identical 16S rDNA and 16S-23S rDNA internal transcribed spacer sequences (8, 16, 18). Comparative sequence analysis of multiple structural genes revealed minimal polymorphism among strains of all four species (26). Although several authors have suggested that the four species should be reclassified as subspecies of M. tuberculosis, no formal proposal has been made (34, 37).
M. tuberculosis is the major cause of human tuberculosis worldwide. M. bovis causes tuberculosis in multiple mammalian species, including humans. The distinction between M. tuberculosis and M. bovis has epidemiologic implications, since infected animals are often a source for human M. bovis infections, and therapeutic implications, since M. bovis strains are resistant to pyrazinamide. M. bovis may be less virulent than M. tuberculosis in humans, but it is clearly more virulent in some animal models. Strains of M. tuberculosis and M. bovis can be distinguished on the basis of epidemiology, biochemical tests, growth phenotypes, and several genetic markers. M. tuberculosis strains nearly always have a functional nicotinamidase, produce niacin, reduce nitrate, have enhanced growth in the presence of glycerol, and are resistant to TCH (36, 37). In contrast, M. bovis strains lack nicotinamidase function, do not produce niacin or reduce nitrate, are not stimulated by glycerol, and are sensitive to TCH (36, 37). M. tuberculosis and M. bovis strains can be distinguished by the sequence of oxyR nucleotide 285 (5, 25). Also, most, but not all, M. bovis strains have a guanine at pncA nucleotide 169 and lack the mtp40 gene (5, 20, 24, 27, 38).
M. africanum was described by Castets and colleagues in 1968 (2). The type strain (ATCC 25420) was isolated from the sputum of a tuberculosis patient in Senegal. Strains of M. africanum display substantial phenotypic heterogeneity, and there is no simple phenotypic definition of the species (10, 12, 15, 36, 37). Difficulties in precisely defining or identifying M. africanum have complicated recent studies of this species (15). Both genotypes and phenotypes of M. africanum vary by geographic area (4, 10, 12, 14), so reports from different regions are not directly comparable. Collins et al. (4) used the nitrate reduction assay to divide M. africanum strains into two groups, African I (negative for nitrate reduction) and African II (positive for nitrate reduction). African I isolates are generally associated with West Africa, and African II isolates are generally associated with East Africa (4, 11, 12). However, a recent survey identified similar numbers of African I and African II isolates in the West African nation of Guinea-Bissau (15).
Several researchers have questioned whether M. africanum is actually distinct from the other members of the M. tuberculosis complex (33, 37, 39). However, the existing M. africanum literature suggests that it differs from M. tuberculosis and M. bovis in host range, source of infection, geographic distribution, virulence, and pathogenesis. M. africanum has been reported in monkeys as well as humans (32). It is a common cause of human tuberculosis in both East and West Africa (12, 19, 23). Strains resembling M. africanum are isolated at low rates from humans on other continents (4, 11, 42). These strains cause a different pattern of disease than M. tuberculosis. For example, they are rarely isolated from humans with genitourinary tuberculosis (11). In experimental models, M. africanum has reduced virulence compared to M. tuberculosis (1, 3).
We identified isolates from Sierra Leone as M. africanum on the basis of biochemical and biophysical properties. In general, these strains differed from M. tuberculosis and M. microti in having negative or weak niacin production and differed from M. bovis in their pyrazinamidase production and their susceptibility to TCH. However, these phenotypes were variable (Table 1), as has been reported by other investigators. Phenotypic analysis is complicated by some changes associated with drug resistance. Acquisition of INH resistance is associated with TCH resistance (12), and acquisition of pyrazinamide resistance is associated with a loss of pyrazinamidase (24, 27).
These 17 M. africanum strains shared a number of genotypes with the rest of the M. tuberculosis complex, including the wild-type sequences of inhA, the oxyR-ahpC intergenic region, orf1 codon 252, and the presence of RD1. These M. africanum strains have mutations leading to INH and rifampin resistance similar to those seen in M. tuberculosis and M. bovis.
The 17 M. africanum strains fell into two distinct genotypic groups (Fig. 1). Fourteen group A strains had katG codon 203 ACT Thr and codon 463 CTG Leu and had closely related VNTR allele profiles. Their genotypes and phenotypes were similar to those of M. africanumT from Senegal. The geographic distribution of group A M. africanum strains is unknown. In addition to the isolates described here, we have identified M. africanum strains from New York and The Netherlands with VNTR allele profiles that cluster with group A (data not shown). In contrast, the three group B strains had katG codon 203 ACC Thr and VNTR allele profile 642432. In these respects, they cluster with M. tuberculosisT.
VNTR analysis and mixed-linker PCR fingerprinting are based on different genetic targets and were independently informative for strain differentiation. The 14 group A M. africanum strains formed 7 clusters by VNTR analysis, 7 clusters by mixed-linker analysis, and 10 clusters when the results of both analyses were combined. VNTR analysis and the sequence at katG codon 203 yielded identical separation of the M. africanum strains into genotypic groups A and B. Each cluster identified by mixed-linker analysis was also restricted to a single genotypic group. VNTR analysis may be useful for evolutionary study when combined with other techniques. VNTR analysis samples multiple independent genomic loci. VNTR results are digital and are amenable to phylogenetic and genetic distance analyses as shown in Fig. 2 and Table 3. However, the number of alleles at each VNTR locus is limited, so genetic convergence may occur frequently. Convergence would make strains appear more closely related than they actually are.
All 14 M. africanum group A strains had typical M. africanum phenotypes. One group B strain had two features more typical of M. tuberculosis, namely, it was positive for niacin production and showed 16 mm of catalase activity. Under the classification system proposed by Collins et al. (4), all 14 group A strains and one group B strain are African I (negative or weakly positive for nitrate reduction) and two group B strains are African II (positive for nitrate reduction). However, there was no phenotype which clearly separated the genotypic groups A and B.
Strains with M. africanum phenotypes have substantial genotypic variability. M. africanum group A isolates had M. africanum phenotypes and clustered with M. africanumT in genotypic analyses. M. africanum group B strains also had M. africanum phenotypes but clustered with M. tuberculosis strains in genotypic analyses. Haas et al. (14) identified strains from Uganda with M. africanum phenotypes and katG codon 463 CGG Arg, a sequence previously found only in M. tuberculosis (26). These data suggest that multiple genetically distinct strains have converged toward an M. africanum phenotype. Our results support the retention of the designation M. africanum for the group A strains, since they cluster with M. africanumT in multiple analyses and are distinct from the rest of the M. tuberculosis complex. Future studies of M. africanum should include both phenotypic and genotypic analyses.
ACKNOWLEDGMENTS
This work was supported by NIH grants AI35230 (R.F.) and AI37004 (J.M.M.), the Durham VA Medical Center’s Research Center on AIDS and HIV Infection (R.F.), and the Department of Veterans Affairs (R.F.).
REFERENCES
- 1.Castets M. Mycobacterium africanum. Med Trop. 1979;39:145–148. [PubMed] [Google Scholar]
- 2.Castets M, Boisvert H, Grumbach F, Brunel M, Rist N. Tuberculosis bacilli of the African type: preliminary note. Rev Tuberc Pneumol. 1968;32:179–184. [PubMed] [Google Scholar]
- 3.Castets M, Sarrat H. Experimental study of the virulence of Mycobacterium africanum: preliminary note. Bull Soc Med Afr Noire Lang Fr. 1969;14:693–696. [PubMed] [Google Scholar]
- 4.Collins C H, Yates M D, Grange J M. Subdivision of Mycobacterium tuberculosis into five variants for epidemiological purposes: methods and nomenclature. J Hyg Camb. 1982;89:235–242. doi: 10.1017/s0022172400070765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espinosa de los Monteros L E, Galán J C, Gutiérrez M, Samper S, García Marín J F, Martín C, Domínguez L, de Rafael L, Baquero F, Gómez-Mampaso E, Blázquez J. Allele-specific PCR method based on pncA and oxyR sequences for distinguishing Mycobacterium bovis from Mycobacterium tuberculosis: intraspecific M. bovis pncA sequence polymorphism. J Clin Microbiol. 1998;36:239–242. doi: 10.1128/jcm.36.1.239-242.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 7.Frothingham R. Differentiation of strains in Mycobacterium tuberculosis complex by DNA sequence polymorphisms, including rapid identification of M. bovis BCG. J Clin Microbiol. 1995;33:840–844. doi: 10.1128/jcm.33.4.840-844.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frothingham R, Hills H G, Wilson K H. Extensive DNA sequence conservation throughout the Mycobacterium tuberculosis complex. J Clin Microbiol. 1994;32:1639–1643. doi: 10.1128/jcm.32.7.1639-1643.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frothingham R, Meeker-O’Connell W A. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology. 1998;144:1189–1196. doi: 10.1099/00221287-144-5-1189. [DOI] [PubMed] [Google Scholar]
- 10.Frottier J, Eliaszewicz M, Arlet V, Gaudillat C. Infections caused by Mycobacterium africanum. Bull Acad Natl Med. 1990;174:29–33. [PubMed] [Google Scholar]
- 11.Grange J M, Yates M D. Incidence and nature of human tuberculosis due to Mycobacterium africanum in South-East England. Epidemiol Infect. 1989;103:127–132. doi: 10.1017/s0950268800030429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haas W H, Bretzel G, Amthor B, Schilke K, Krommes G, Rüsch-Gerdes S, Sticht-Groh V, Bremer H J. Comparison of DNA fingerprint patterns of isolates of Mycobacterium africanum from East and West Africa. J Clin Microbiol. 1997;35:663–666. doi: 10.1128/jcm.35.3.663-666.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haas W H, Butler W R, Woodley C L, Crawford J T. Mixed-linker polymerase chain reaction: a new method for rapid fingerprinting of isolates of the Mycobacterium tuberculosis complex. J Clin Microbiol. 1993;31:1293–1298. doi: 10.1128/jcm.31.5.1293-1298.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haas W H, Schilke K, Brand J, Amthor B, Weyer K, Fourie P B, Bretzel G, Sticht-Groh V, Bremer H J. Molecular analysis of katG gene mutations in strains of Mycobacterium tuberculosis complex from Africa. Antimicrob Agents Chemother. 1997;41:1601–1603. doi: 10.1128/aac.41.7.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffner S E, Svenson S B, Norberg R, Dias F, Ghebremichael S, Källenius G. Biochemical heterogeneity of Mycobacterium tuberculosis complex isolates in Guinea-Bissau. J Clin Microbiol. 1993;31:2215–2217. doi: 10.1128/jcm.31.8.2215-2217.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imaeda T. Deoxyribonucleic acid relatedness among selected strains of Mycobacterium tuberculosis, Mycobacterium bovis, Mycobacterium bovis BCG, Mycobacterium microti, and Mycobacterium africanum. Int J Syst Bacteriol. 1985;35:147–150. [Google Scholar]
- 17.Kapur V, Li L-L, Iordanescu S, Hamrick M R, Wanger A, Kreiswirth B N, Musser J M. Characterization by automated DNA sequencing of mutations in the gene (rpoB) encoding the RNA polymerase β subunit in rifampin-resistant Mycobacterium tuberculosis strains from New York City and Texas. J Clin Microbiol. 1994;32:1095–1098. doi: 10.1128/jcm.32.4.1095-1098.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirschner P, Springer B, Vogel U, Meier A, Wrede A, Kiekenbeck M, Bange F-C, Böttger E C. Genotypic identification of mycobacteria by nucleic acid sequence determination: report of a 2-year experience in a clinical laboratory. J Clin Microbiol. 1993;31:2882–2889. doi: 10.1128/jcm.31.11.2882-2889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ledru S, Cauchoix B, Yameogo M, Zoubga A, Lamande-Chiron J, Portaels F, Chiron J P. Impact of short-course therapy on tuberculosis drug resistance in South-West Burkina Faso. Tubercle Lung Dis. 1996;77:429–436. doi: 10.1016/s0962-8479(96)90116-1. [DOI] [PubMed] [Google Scholar]
- 20.Liébana E, Aranaz A, Francis B, Cousins D. Assessment of genetic markers for species differentiation within the Mycobacterium tuberculosis complex. J Clin Microbiol. 1996;34:933–938. doi: 10.1128/jcm.34.4.933-938.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Musser J M, Kapur V, Williams D L, Kreiswirth B N, van Soolingen D, van Embden J D A. Characterization of the catalase-peroxidase gene (katG) and inhA locus in isoniazid-resistant and -susceptible strains of Mycobacterium tuberculosis by automated DNA sequencing: restricted array of mutations associated with drug resistance. J Infect Dis. 1996;173:196–202. doi: 10.1093/infdis/173.1.196. [DOI] [PubMed] [Google Scholar]
- 22.Nei M. Phylogenetic analysis in molecular genetics. Annu Rev Genet. 1996;30:371–403. doi: 10.1146/annurev.genet.30.1.371. [DOI] [PubMed] [Google Scholar]
- 23.Schwander S, Rusch-Gerdes S, Mateega A, Lutalo T, Tugume S, Kityo C, Rubaramira R, Mugyenyi P, Okwera A, Mugerwa R. A pilot study of antituberculosis combinations comparing rifabutin with rifampicin in the treatment of HIV-1 associated tuberculosis. A single-blind randomized evaluation in Ugandan patients with HIV-1 infection and pulmonary tuberculosis. Tubercle Lung Dis. 1995;76:210–218. doi: 10.1016/s0962-8479(05)80007-3. [DOI] [PubMed] [Google Scholar]
- 24.Scorpio A, Collins D, Whipple D, Cave D, Bates J, Zhang Y. Rapid differentiation of bovine and human tubercle bacilli based on a characteristic mutation in the bovine pyrazinamidase gene. J Clin Microbiol. 1997;35:106–110. doi: 10.1128/jcm.35.1.106-110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sreevatsan S, Escalante P, Pan X, Gillies II D A, Siddiqui S, Khalaf C N, Kreiswirth B N, Bifani P, Adams L G, Ficht T, Perumaalla V S, Cave M D, van Embden J D A, Musser J M. Identification of a polymorphic nucleotide in oxyR specific for Mycobacterium bovis. J Clin Microbiol. 1996;34:2007–2010. doi: 10.1128/jcm.34.8.2007-2010.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sreevatsan S, Pan X, Stockbauer K E, Connell N D, Kreiswirth B N, Whittam T S, Musser J M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sreevatsan S, Pan X, Zhang Y, Kreiswirth B N, Musser J M. Mutations associated with pyrazinamide resistance in pncA of Mycobacterium tuberculosis complex organisms. Antimicrob Agents Chemother. 1997;41:636–640. doi: 10.1128/aac.41.3.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart C B. The powers and pitfalls of parsimony. Nature (London) 1993;361:603–607. doi: 10.1038/361603a0. [DOI] [PubMed] [Google Scholar]
- 29.Sticht-Groh V, Rüsch-Gerdes S, Remillieux M. Multidrug-resistant strains of tubercle bacilli isolated from pulmonary tuberculosis retreatment patients in Sierra Leone, West Africa. Tubercle Lung Dis. 1993;74:411–412. doi: 10.1016/0962-8479(93)90091-b. [DOI] [PubMed] [Google Scholar]
- 30.Swofford D L. PAUP: phylogenetic analysis using parsimony and other methods, version 4. Sunderland, Mass: Sinauer Associates; 1998. [Google Scholar]
- 31.Talbot E A, Williams D L, Frothingham R. PCR identification of Mycobacterium bovis BCG. J Clin Microbiol. 1997;35:566–569. doi: 10.1128/jcm.35.3.566-569.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thorel M F. Isolation of Mycobacterium africanum from monkeys. Tubercle. 1980;61:101–104. doi: 10.1016/0041-3879(80)90018-5. [DOI] [PubMed] [Google Scholar]
- 33.Tsukamura M, Mizuno S, Toyama H. Taxonomic studies on the Mycobacterium tuberculosis series. Microbiol Immunol. 1985;29:285–299. doi: 10.1111/j.1348-0421.1985.tb00827.x. [DOI] [PubMed] [Google Scholar]
- 34.van Soolingen D, Hoogenboezem T, de Haas P E, Hermans P W, Koedam M A, Teppema K S, Brennan P J, Besra G S, Portaels F, Top J, Schouls L M, van Embden J D A. A novel pathogenic taxon of the Mycobacterium tuberculosis complex, Canetti: characterization of an exceptional isolate from Africa. Int J Syst Bacteriol. 1997;47:1236–1245. doi: 10.1099/00207713-47-4-1236. [DOI] [PubMed] [Google Scholar]
- 35.Wayne L G. Simple pyrazinamidase and urease tests for routine identification of mycobacteria. Am Rev Respir Dis. 1974;109:147–151. doi: 10.1164/arrd.1974.109.1.147. [DOI] [PubMed] [Google Scholar]
- 36.Wayne L G. Microbiology of tubercle bacilli. Am Rev Respir Dis. 1982;125(3 part 2):31–41. doi: 10.1164/arrd.1982.125.3P2.31. [DOI] [PubMed] [Google Scholar]
- 37.Wayne L G, Kubica G P. The mycobacteria. In: Sneath P H A, Holt J G, editors. Bergey’s manual of systemic bacteriology. Vol. 2. Baltimore, Md: The Williams & Wilkins Co.; 1986. pp. 1435–1457. [Google Scholar]
- 38.Weil A, Plikaytis B B, Butler W R, Woodley C L, Shinnick T M. The mtp40 gene is not present in all strains of Mycobacterium tuberculosis. J Clin Microbiol. 1996;34:2309–2311. doi: 10.1128/jcm.34.9.2309-2311.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wieten G, Haverkamp J, Groothuis D G, Berwald L G, David H L. Classification and identification of Mycobacterium africanum by pyrolysis mass spectrometry. J Gen Microbiol. 1983;129:3679–3688. doi: 10.1099/00221287-129-12-3679. [DOI] [PubMed] [Google Scholar]
- 40.Williams D L, Gillis T P, Dupree W G. Ethanol fixation of sputum sediments for DNA-based detection of Mycobacterium tuberculosis. J Clin Microbiol. 1995;33:1558–1561. doi: 10.1128/jcm.33.6.1558-1561.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Williams D L, Waguespack C, Eisenach K, Crawford J T, Portaels F, Salfinger M, Nolan C M, Abe C, Sticht-Groh V, Gillis T P. Characterization of rifampin-resistance in pathogenic mycobacteria. Antimicrob Agents Chemother. 1994;38:2380–2386. doi: 10.1128/aac.38.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yates M D, Grange J M. Bacteriological survey of tuberculosis lymphadenitis in southeast England, 1981–1989. J Epidemiol Community Health. 1992;46:332–335. doi: 10.1136/jech.46.4.332. [DOI] [PMC free article] [PubMed] [Google Scholar]