ABSTRACT
Methanotrophs are an important group of microorganisms that counteract methane emissions to the atmosphere. Methane-oxidising bacteria of the Alpha- and Gammaproteobacteria have been studied for over a century, while methanotrophs of the phylum Verrucomicrobia are a more recent discovery. Verrucomicrobial methanotrophs are extremophiles that live in very acidic geothermal ecosystems. Currently, more than a dozen strains have been isolated, belonging to the genera Methylacidiphilum and Methylacidimicrobium. Initially, these methanotrophs were thought to be metabolically confined. However, genomic analyses and physiological and biochemical experiments over the past years revealed that verrucomicrobial methanotrophs, as well as proteobacterial methanotrophs, are much more metabolically versatile than previously assumed. Several inorganic gases and other molecules present in acidic geothermal ecosystems can be utilised, such as methane, hydrogen gas, carbon dioxide, ammonium, nitrogen gas and perhaps also hydrogen sulfide. Verrucomicrobial methanotrophs could therefore represent key players in multiple volcanic nutrient cycles and in the mitigation of greenhouse gas emissions from geothermal ecosystems. Here, we summarise the current knowledge on verrucomicrobial methanotrophs with respect to their metabolic versatility and discuss the factors that determine their diversity in their natural environment. In addition, key metabolic, morphological and ecological characteristics of verrucomicrobial and proteobacterial methanotrophs are reviewed.
Keywords: verrucomicrobial methanotrophs, geothermal ecosystems, methane, proteobacterial methanotrophs, hydrogen gas, metabolism, comparative genomic analysis
This review discusses the metabolic versatility of verrucomicrobial methanotrophs regarding the acidic volcanic ecosystems they thrive in and a comparison is made with the canonical proteobacterial methanotrophs.
INTRODUCTION
The atmospheric concentration of methane (CH4) has been increasing rapidly over the past quarter millennium due to anthropogenic activities (Etheridge et al. 1998; Turner, Frankenberg and Kort 2019). Currently, the amount of methane emitted from sources exceeds the amount of methane taken up by sinks, resulting in an imbalanced carbon cycle (Saunois et al. 2020). Consequently, the present atmospheric methane concentration (1.86 ppmv) is 2.6 times higher than the preindustrial concentration (Etheridge et al. 1998; Cai et al. 2016; Etminan et al. 2016; Saunois et al. 2020). Since methane is a powerful greenhouse gas, atmospheric methane is now estimated to contribute approximately 16% to global warming (Saunois et al. 2016). Moreover, an increase in global temperature can induce additional release of methane to the atmosphere, e.g. through permafrost thawing, causing positive climate feedback that results in an acceleration of climate change (Dean et al. 2018). Hence, it is important to understand the microbial sources and sinks of methane (Stein 2020).
Each year, 548 to 678 Tg (1012 g) CH4 is emitted from various sources into the atmosphere, of which 33%–54% and 46%–67% are from natural and anthropogenic origin, respectively (Kirschke et al. 2013; Dean et al. 2018). The majority of these sources contain methanogenic archaea that catalyse the final step of the biological degradation of organic matter by producing methane (Balch et al. 1979; Conrad 2020). The largest sources of methane are wetlands, which are anoxic, water-logged soils in which methanogenic archaea are present and active (Angel, Claus and Conrad 2012; Bridgham et al. 2013; Evans et al. 2019). Other natural sources of biogenic methane are aquatic systems (e.g. lakes), marine systems (especially coastal sediments), termites and wild animals (Jensen 1996; Reeburgh 2007; Bastviken et al. 2008; Brune 2010; Dean et al. 2018; Evans et al. 2019). Additional microbial processes that are implicated in methane production include conversion of methylphosphonates by Thaumarchaeota in the ocean and by cyanobacteria in freshwater and terrestrial ecosystems (Metcalf et al. 2012; Bižić et al. 2020). Natural sources in which methane is not produced microbially but through abiotic processes include wildfires and methane hydrates (located inside ocean floors and permafrost) (Buffett 2000; Vasileva and Moiseenko 2013; Dean et al. 2018). In addition, various geothermal systems such as fumaroles, mud volcanoes and hydrothermal vents emit 40 to 60 Tg CH4 annually (Etiope 2009). Methane released from these geothermal systems either has a thermogenic origin (e.g. formed through the thermogenic breakdown of organic matter over millions of years), an abiogenic origin (e.g. formed through the reduction of CO2 at high temperature in the Earth's crust) or even a biogenic origin, produced by methanogens in the deep subsurface (Sherwood Lollar, Lacrampe-Couloume and Slater 2006; Tassi et al. 2012; Stolper et al. 2015). As is the case for several natural sources, various man-made systems emit significant amounts of methane produced by methanogenic archaea. These sources include livestock (e.g. ruminants), landfills and rice paddies (Themelis and Ulloa 2007; Carlson et al. 2017; Dean et al. 2018; Chang et al. 2019). Other anthropogenic, non-biological sources are comprised of fossil fuels (through mining, combustion and industry) and biofuel/biomass (through combustion) (Hao and Ward 1993; Heede 2014; Hanaki and Portugal-Pereira 2018). The predominant sink conversion of methane occurs chemically in the troposphere by hydroxyl radicals (•OH) and to a lesser extent by chlorine and oxygen radicals (Allan, Struthers and Lowe 2007; Rigby et al. 2017). In various ecosystems, methane is produced in deeper anoxic layers that are covered by more oxidised sediments or water columns. In oxidised zones such as those present in wetlands, and notably at the oxic-anoxic interface, methanotrophic prokaryotes consume a major part of methane as energy and carbon source, prior to emission to the atmosphere (Brune, Frenzel and Cypionka 2000; Conrad 2009). These microorganisms therefore act as a biofilter for emissions of this potent greenhouse gas (La et al. 2018). In addition, specialised high-affinity methanotrophs present in soil seem to oxidise methane from the atmosphere, although only present at a very low concentration (Holmes et al. 1999; Cai et al. 2016; Tveit et al. 2019). Considering the role methanotrophs play in mitigating methane emissions, they are an important topic of study in understanding and counteracting global warming.
METHANOTROPHIC MICROORGANISMS
Aerobic methanotrophs conserve energy by oxidising CH4 with O2 to CO2. The first aerobic methanotrophs were discovered and isolated more than a century ago (Kaserer 1905; Söhngen 1906). In the remainder of the twentieth century, numerous novel isolates were described, all belonging to the Alpha- and Gammaproteobacteria (Whittenbury, Phillips and Wilkinson 1970; Hanson and Hanson 1996). Methanotrophy is not restricted to oxic environments, but can take place anaerobically as well. Already decades ago, methane consumption in anoxic, sulfate-rich marine sediments was observed, but the microorganisms responsible for this process remained elusive (Barnes and Goldberg 1976; Reeburgh 1976; Iversen and Jorgensen 1985). At the beginning of the current century, a marine consortium of methane-oxidising archaea and sulfate-reducing bacteria was discovered, mediating sulfate-dependent methane oxidation (Boetius et al. 2000). This finding was followed by the discovery of a consortium of Methylomirabilis bacteria and Methanoperedens archaea, capable of coupling anaerobic methane oxidation to denitrification of nitrite and nitrate (Raghoebarsing et al. 2006). Hereafter it was shown that both the bacterium and the archaeon were capable of methane oxidation independently. Methanoperedens archaea are able to couple methane oxidation to the reduction of nitrate (Haroon et al. 2013), iron (Beal, House and Orphan 2009; Ettwig et al. 2016; Cai et al. 2018) and manganese (Beal, House and Orphan 2009; Ettwig et al. 2016; Leu et al. 2020), whereas Methylomirabilis bacteria (of the candidate phylum NC10) couple methane oxidation to the reduction of nitrite (Ettwig et al. 2008, 2010; He et al. 2016; Versantvoort et al. 2018). Remarkably, Methylomirabilis bacteria possess the complete aerobic methane oxidation pathway and are postulated to produce oxygen internally by reducing nitrite to nitric oxide, which is subsequently dismutated into O2 and N2 (Ettwig et al. 2010, 2012). Most aerobic and anaerobic methanotrophs live in environments with moderate temperature and circumneutral pH (Bowman et al. 1993). Exceptions are several aerobic proteobacterial methanotrophs of the genera Methylocapsa, Methylocella, Methylocystis and Methylosinus (Dedysh et al. 1998; Dunfield and Dedysh 2010; Kip et al. 2011). These bacteria are moderate acidophiles growing in acidic peat environments with a pH as low as 4.2 and are frequently found as intracellular symbionts of Sphagnum mosses (Raghoebarsing et al. 2005; Kostka, Weston and Glass 2016; Kox et al. 2018). In addition, methanotrophs of the gammaproteobacterial genus Methylothermus are thermophiles, growing in hot springs at temperatures up to 72°C (Bodrossy et al. 1999; Hirayama et al. 2011; Houghton et al. 2019). In 2005, methane oxidation was observed in volcanic soils of the Solfatara volcano (Campi Flegrei, Pozzuoli, Italy) that are characterized by a high temperature (50 to 95°C) and an extreme acidity (pH 1.0) (Castaldi and Tedesco 2005). Two years later, thermoacidophilic methanotrophs were isolated from hot and acidic volcanic ecosystems at the Solfatara volcano (near Naples, Italy), in the Uzon Caldera (Kamchatka, Russia) and in Hell's gate (Tikitere, New Zealand) (Dunfield et al. 2007; Pol et al. 2007; Islam et al. 2008). Surprisingly, phylogenetic analyses revealed that these microbes belong to the phylum Verrucomicrobia, refuting the dogma that all aerobic methanotrophs are part of the phylum Proteobacteria.
METHANOTROPHS OF THE PHYLUM VERRUCOMICROBIA
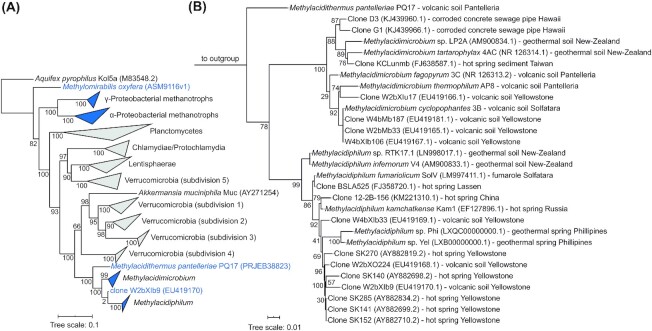
The first isolated verrucomicrobial methanotrophs were classified in the novel genus Methylacidiphilum (Op den Camp et al. 2009). These methanotrophs were observed to have temperature optima of 50 to 60°C (Dunfield et al. 2007; Pol et al. 2007; Islam et al. 2008). Currently, five Methylacidiphilum strains have been isolated that differ enough, based on DNA-DNA hybridisation and on the analysis of 16S rRNA and housekeeping genes, to be classified as separate species (Erikstad et al. 2019): Methylacidiphilum fumariolicum SolV, Methylacidiphilum infernorum V4, Methylacidiphilum kamchatkense Kam1, Methylacidiphilum sp. Yel and Methylacidiphilum sp. Phi (Table 1). The geothermal ecosystems they were isolated from have an extremely low pH, primarily as a result of the biogenic formation of sulfuric acid (H2SO4) from the oxidation of hydrogen sulfide (H2S) (Schoen and Rye 1970; Quatrini and Johnson 2018). Accordingly, the pH optima of all isolated Methylacidiphilum strains range from pH 2.0 to 3.0. Still, the pH range in which these strains can grow is much broader (Table 1). Methylacidiphilum fumariolicum SolV was shown to grow up to pH 6 when it was slowly adapted to higher pH (Mohammadi et al. 2017b). Moreover, this strain was shown to grow even below pH 1 (Pol et al. 2007) (Table 1). Through current culture-independent molecular methods, clones related to the isolated strains were found at geothermal sites across the globe, of which several could be novel species (Fig. 1).
Table 1.
Summary of the pH and temperature optima, isolation location and verified growth substrates of the isolated verrucomicrobial methanotrophs discussed in this review.
| Strain | pHoptimum (and range) | Toptimum (°C) (and range) | Verified growth substrates | Isolation location | References |
|---|---|---|---|---|---|
| Methylacidiphilum fumariolicum SolV | 2 (0.8–6.0) | 55 (40–65) | CH4, H2, methanol, propane, ethane | Acidic thermal mudpot, Solfatara, Italy | Pol et al. 2007; Mohammadi et al. 2017a; Picone et al. 2020b |
| Methylacidiphilum infernorum V4 | 2–2.5 (1.0–6.0) | 60 (40–60) | CH4, methanol | Acidic thermal soil, Tikitere, New Zealand | Dunfield et al. 2007; Hou et al. 2008 |
| Methylacidiphilum kamchatkense Kam1 | 2–2.5 (2.0–5.0) | 55 (37–60) | CH4, methanol | Acidic thermal spring, Kamchatka, Russia | Islam et al. 2008 |
| Methylacidiphilum sp. Phi | 3 | 55–65 | CH4 | Acidic hot spring, Makiling, The Philippines | Erikstad et al. 2019 |
| Methylacidiphilum sp. Yel | 2.8 | 50 | CH4 | Acidic hot spring, Yellowstone, USA | Erikstad et al. 2019 |
| Methylacidimicrobium tartarophylax 4AC | 1–3 (0.5–5.5) | 38 (?–43) | CH4, H2, methanol | Acidic soil, Solfatara, Italy | Van Teeseling et al. 2014; Mohammadi et al. 2019 |
| Methylacidimicrobium cyclopophantes 3B | 1.5–3 (0.6–5.5) | 44 (?–49) | CH4, methanol | Acidic soil, Solfatara, Italy | Van Teeseling et al. 2014 |
| Methylacidimicrobium fagopyrum 3C | 1.5–3 (0.6–5.5) | 35 (?–39) | CH4, methanol | Acidic soil, Solfatara, Italy | Van Teeseling et al. 2014 |
| Methylacidimicrobium sp. LP2A | 3.1 (1.0–5.2) | 30 (17–37) | CH4 | Acidic mud pool, Reporoa, New Zealand | Sharp et al. 2014 |
| Methylacidimicrobium thermophilum AP8 | 3–5 (1.5–5.5) | 50 (30–55) | CH4, H2 | Acidic geothermal soil, Pantelleria island, Italy | Picone 2020 |
Figure 1.
(A) Overview 16S rRNA gene phylogenetic tree of Verrucomicrobia, Lentisphaerae, Chlamydiae, Planctomycetes and methanotrophic Alpha- and Gammaproteobacteria. Blue shading indicates species that are methanotrophic. Accession numbers are indicated between brackets. 16S rRNA gene alignment was constructed using SINA v1.2.11 (Pruesse, Peplies and Glöckner 2012) and a tree that was constructed using FastTree v2.1 (Price, Dehal and Arkin 2010) with 1000 bootstraps and substitution model GTR-gamma. The tree was visualised using ITOL v5.5.1 (Letunic and Bork 2007). Bootstrap values are indicated as a proportion of 1000 re-samplings ranging from 1 to 100. (B) Detailed 16S rRNA gene tree of the verrucomicrobial methanotrophs. 16S rRNA gene alignment of verrucomicrobial methanotrophs together with an outgroup of Planctomycetes constructed using SINA v1.2.11 (Pruesse, Peplies and Glöckner 2012) and a tree that was constructed using FastTree v2.1 (Price, Dehal and Arkin 2010) with 1000 bootstraps and substitution model GTR-gamma. The tree was visualised using ITOL v5.5.1 (Letunic and Bork 2007). Bootstrap values are indicated as a proportion of 1000 re-samplings ranging from 1 to 100. Isolation location and accession number are indicated after each strain.
Since all isolated Methylacidiphilum strains were found in hot and acidic geothermal habitats, the question arose whether verrucomicrobial methanotrophs could have a more widespread distribution in habitats with different physicochemical parameters (Dunfield et al. 2007). Therefore, Sharp et al. (2014) conducted pyrosequencing of 16S rRNA genes on samples derived from geothermal habitats, acidic peat bogs and fens with a temperature range of 6.3 to 81.6°C and a pH range of 1.8 to 8.6. Surprisingly, 16S rRNA gene sequences of verrucomicrobial methanotrophs were detected in the full range of 22.5 to 81.6°C, but only below pH 5.0 (Sharp et al. 2014). From a sediment sample of 22°C and pH 2.6, the novel verrucomicrobial strain LP2A was enriched and isolated when incubated with methane as the sole energy source (Sharp et al. 2014). Interestingly, the 16S rRNA sequence of this newly isolated strain was only 90.6% identical to that of Methylacidiphilum infernorum V4, isolated from hot and acidic soil sediment (Dunfield et al. 2007). The novel mesophilic strain LP2A was subsequently placed in the novel genus Methylacidimicrobium within the phylum Verrucomicrobia (Van Teeseling et al. 2014). That same year, three additional mesophilic strains within this genus were isolated from geothermal soil at the Solfatara in Italy: Methylacidimicrobium tartarophylax 4AC, Methylacidimicrobium fagopyrum 3C and Methylacidimicrobium cyclopophantes 3B (Van Teeseling et al. 2014) (Fig. 1). These acidophiles have pH growth optima of 1.0 to 3.0, while strain 4AC can even grow at pH 0.5 (Van Teeseling et al. 2014) (Table 1). The major difference between Methylacidiphilum and Methylacidimicrobium strains was thought to be the growth temperature. Methylacidimicrobium strains were reported to be mesophiles, with temperature growth optima of 30 to 44°C and the apparent inability to grow at temperatures above 49°C (Sharp et al. 2014; Van Teeseling et al. 2014). However, recently, Methylacidimicrobium thermophilum AP8 was isolated from Pantelleria island (Italy) with a temperature optimum of 50°C (Picone 2020), questioning the clear temperature-based division between Methylacidiphilum and Methylacidimicrobium strains.
Apart from the strains isolated from geothermal soils in Italy, Russia and New-Zealand, 16S rRNA gene sequences belonging to strains of the Methylacidimicrobium genus were detected in hot springs and geothermal soil in other parts of the world (Fig. 1). In addition, a metagenome-assembled genome (MAG) representing a novel verrucomicrobial methanotroph genus and species, Ca. ‘Methylacidithermus pantelleriae’ PQ17, was obtained through sampling of geothermal soil on Pantelleria island (Picone et al. 2020a). Interestingly, verrucomicrobial methanotrophs do not seem to be restricted to geothermal habitats (Pagaling, Yang and Yan 2014). Clones related to Methylacidimicrobium strains were abundantly present in biofilms in the crown of corroded concrete sewage pipes on Hawaii, which suggests that verrucomicrobial methanotrophs have adapted to man-made systems (Pagaling, Yang and Yan 2014) (Fig. 1). These sewage pipes are characterised by a low pH and the presence of methane and sulfur compounds (e.g. H2S), which could explain the abundance of verrucomicrobial methanotrophs in this environment. Sequencing of samples from other acidic, methane-rich environments could reveal whether verrucomicrobial methanotrophs are present in other non-geothermal habitats.
Besides the importance of verrucomicrobial methanotrophs in the environment, they could also be relevant in biotechnology and industry, as they are capable of producing valuable molecules such as methanol, glycogen, polyhydroxybutyric acid, vitamins, compatible solutes and (thermostable) enzymes (Kalyuzhnaya, Puri and Lidstrom 2015; Strong, Xie and Clarke 2015; Bodelier et al. 2019). Although still in its infancy, notable examples of the biotechnological potential of verrucomicrobial methanotrophs are the recently shown improved methanol production (63% mole methanol produced per mole methane) by Methylacidiphilum fumariolicum SolV (Hogendoorn 2020; Hogendoorn et al. 2020) and the purification of a thermostable, high-affinity [NiFe] hydrogenase from the same bacterium (Schmitz et al. 2020a).
KEY SIMILARITIES AND DIFFERENCES BETWEEN VERRUCOMICROBIAL AND PROTEOBACTERIAL METHANOTROPHS
Proteobacterial methanotrophs can be found in diverse oxic environments in which methane is present, such as wetlands, soil, peat lands, marine sediments, landfills and rice paddies (Bodelier et al. 2019). On the contrary, verrucomicrobial methanotrophs are almost exclusively found in acidic (pH < 3.5) geothermal systems. Whether proteobacterial methanotrophs are present in such acidic habitats is unknown. However, phylogenetic analyses of pmoA genes present in acidic geothermal soil and mud pots show clustering with proteobacterial pmoA gene sequences (Pol et al. 2007; Gagliano et al. 2014). This finding suggests that verrucomicrobial and proteobacterial methanotrophs could share habitats, which could ultimately be proven by the isolation of a proteobacterial methanotroph from an acidic geothermal ecosystem where Methylacidiphilum or Methylacidimicrobium were isolated from. Although mostly living in vastly different habitats, they share their preference for the greenhouse gas methane. Besides this metabolic trait, several noteworthy differences exist between the two groups. Classically, an important property for the classification of proteobacterial methanotrophs is the type of intracytoplasmic membrane (ICM) structures they have in electron micrographs, either visible as membrane pairs or vesicular discs (Davies and Whittenbury 1970). In verrucomicrobial methanotrophs, ICM structures are mostly absent, although membrane stacks have been found in Methylacidimicrobium fagopyrum 3C and also in some cells of Methylacidiphilum infernorum V4 tubular membrane structures were observed (Dunfield et al. 2007; Van Teeseling et al. 2014). Another interesting difference between verrucomicrobial and proteobacterial methanotrophs are the phospholipid fatty acids (PLFAs) of their membranes (Bodelier et al. 2009; Op den Camp et al. 2009). Membranes of verrucomicrobial methanotrophs are almost exclusively made up of saturated fatty acids, whereas membranes of proteobacterial methanotrophs are mainly composed of unsaturated fatty acids (Bodelier et al. 2009; Op den Camp et al. 2009; Erikstad et al. 2019). Verrucomicrobial methanotrophs probably require a saturated membrane to minimise proton permeability in an extremely acidic environment (Siliakus, van der Oost and Kengen 2017).
Remarkably, the genome sizes of verrucomicrobial and proteobacterial methanotrophs differ significantly. The former typically have a genome size of 2.2 to 2.5 Mbp (Hou et al. 2008; Anvar et al. 2014; Kruse et al. 2019; Cremers et al. 2020) while the latter have larger genomes of 3.3 to 5.1 Mbp (Ward et al. 2004; Chen et al. 2010; Stein et al. 2010; Boden et al. 2011; Svenning et al. 2011; Vuilleumier et al. 2012). The larger genomes of proteobacterial methanotrophs could render them better adapted to more diverse environments in comparison with verrucomicrobial methanotrophs (Cobo-Simón and Tamames 2017). Interestingly, genome comparison of eleven verrucomicrobial methanotrophs reveals 317 core gene clusters that are shared by all these verrucomicrobial strains, whereas the same analysis of eleven proteobacterial methanotrophs reveals 74 core gene clusters shared by all these proteobacterial strains (Fig. 2). The smaller amount of core gene clusters shared by proteobacterial methanotrophs could be explained by the fact that these microbes are found in very distinct habitats under various circumstances. The verrucomicrobial methanotrophs, on the other hand, are adapted to a specific niche. The composition of Clusters of Orthologous Groups of proteins (COGS) of the core genomes of verrucomicrobial and proteobacterial methanotrophs show a similar distribution (Fig. 2). A notable example is the relatively large percentage unique gene clusters involved in cell wall/membrane/envelope biogenesis (COG group M) found in verrucomicrobial methanotrophs. This might be explained by the harsh environment in which these microbes thrive, which could require additional machinery to cope with acid stress.
Figure 2.
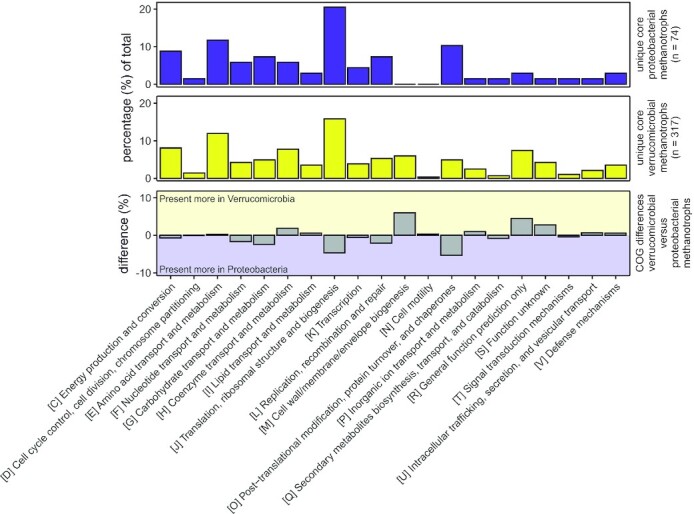
Bar graphs comparing the composition of Clusters of Orthologous Groups of proteins (COGS) of the unique core genomes of the verrucomicrobial methanotrophs and the proteobacterial methanotrophs. Core genomes were calculated by using usearch (Edgar 2010) with a cut-off value of 0.5 to group all genes into gene clusters. Clusters present in all genomes of the verrucomicrobial methanotrophs, as well as absent in all genomes of the proteobacterial methanotrophs, are defined as the unique core genome of verrucomicrobial methanotrophs. Clusters present in all genomes of the proteobacterial methanotrophs, as well as absent in all genomes of the verrucomicrobial methanotrophs, are defined as the unique core genome of proteobacterial methanotrophs. Analysis performed with 22 genomes in total, of which 11 verrucomicrobial methanotrophs: Methylacidiphilum fumariolicum SolV, Methylacidiphilum infernorum V4, Methylacidiphilum kamchatkense Kam1, Methylacidiphilum sp. Phi, Methylacidiphilum sp. Yel, Methylacidimicrobium cyclopophantes 3B, Methylacidimicrobium fagopyrum 3C, Methylacidimicrobium tartarophylax 4AC, Methylacidimicrobium thermophilum AP8, Methylacidimicrobium sp. LP2A and Methylacidithermus pantelleriae PQ17, and 11 proteobacterial methanotrophs: Methylobacterium extorquens AM1, Methylocaldum szegediense O-12, Methylocapsa acidiphila B2, Methylocella silvestris BL2, Methylococcus capsulatus Bath, Methylocystis rosea SV97T, Methyloferula stellata AR4, Methylomarinum vadi IT4, Methylomicrobium alcaliphilum 20Z, Methylomonas denitrificans FJG1 and Methylosinus trichosporium OB3b.
Although verrucomicrobial methanotrophs have smaller genomes, genomic analyses suggest verrucomicrobial methanotrophs are metabolically quite versatile microorganisms. In this review, we discuss the physiological and biochemical knowledge that has been obtained on the key metabolic pathways of verrucomicrobial methanotrophs since their discovery 13 years ago. As such, a connection between the metabolic potential in the genome and experimental observations is made. Metabolic genes investigated in this review involved in the oxidation of methane, ammonia, hydrogen and sulfur compounds, nitrogen assimilation, the respiratory chain and the synthesis of tetrahydrofolate and menaquinone can be found in Table S1 (Supporting Information).
METABOLIC VERSATILITY OF VERRUCOMICROBIAL METHANOTROPHS
Initially, the first isolated strains appeared to be obligate methylotrophs, growing only on methane and methanol (CH3OH) (Op den Camp et al. 2009). In fact, all described Methylacidiphilum and Methylacidimicrobium strains were isolated on methane (Dunfield et al. 2007; Pol et al. 2007; Islam et al. 2008; Sharp et al. 2014; Erikstad et al. 2019). However, after more than a decade of additional experimental research, we now know that verrucomicrobial methanotrophs are actually metabolically quite versatile microorganisms, able to metabolise a variety of compounds present in volcanic ecosystems, which together could influence methanotrophy.
Carbon and nitrogen assimilation
Similar to any other microorganism, methanotrophs need to assimilate carbon and nitrogen to generate biomass (Levicán et al. 2008). Methanotrophs use either formaldehyde (CH2O) or CO2 as a carbon source to generate biomass (Hanson and Hanson 1996). In general, gammaproteobacterial methanotrophs assimilate formaldehyde via the ribulose monophosphate pathway, whereas alphaproteobacterial methanotrophs employ the serine pathway, in which both formaldehyde and CO2 are used as a carbon source (Chistoserdova, Kalyuzhnaya and Lidstrom 2009). Verrucomicrobial methanotrophs lack a few essential genes for the complete serine cycle (i.e. hydroxypyruvate reductase and malyl-CoA lyase) or ribulose monophosphate pathway (i.e. 3-hexulose-6-phosphate synthase and 6-phospho-3-hexuloisomerase), but possess genes encoding the complete set of enzymes of the Calvin-Benson-Bassham (CBB) cycle (Khadem et al. 2011; Van Teeseling et al. 2014). In this cycle, the enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) is involved and CO2 is used as the sole carbon source. Several proteobacterial methanotrophs also harbour genes involved in the CBB cycle, although activity of this cycle has not yet been experimentally validated in these bacteria (Stein, Roy and Dunfield 2012; Khmelenina et al. 2018). Through 13C stable isotope measurements, Methylacidiphilum fumariolicum SolV and three Methylacidimicrobium strains were shown to indeed exclusively use CO2 as carbon source, not methane-derived formaldehyde (Khadem et al. 2011; Van Teeseling et al. 2014). The autotrophic CO2 fixation via the CBB cycle was also experimentally shown for the denitrifying methanotroph Ca. ‘Methylomirabilis oxyfera’ (Rasigraf et al. 2014). In geothermal habitats, high concentrations of ammonium can be present (Khadem et al. 2010; Holloway et al. 2011; Mohammadi et al. 2017b). Although methanotrophs cannot use ammonium as an energy source, this reduced form of fixed nitrogen is the preferred inorganic nitrogen source as it can be directly assimilated (Wang et al. 2016). Indeed, all known verrucomicrobial methanotrophs carry a gene encoding a highly-conserved ammonium transporter. Central molecules in nitrogen assimilation are glutamate and glutamine, from which other amino acids and purines and pyrimidines can be synthesised (Prusiner and Stadtman 1973; Zalkin and Smith 1998). Genes encoding glutamate dehydrogenase, glutamine synthetase and glutamate synthase are found in all isolated verrucomicrobial methanotrophs. The utilisation of these three enzymes for nitrogen assimilation has been validated experimentally in several proteobacterial methanotrophs (Murrell and Dalton 1983a; Lees, Owens and Murrell 1991).
Although ammonium is often present in the geothermal environment, exponentially growing microorganisms can rapidly deplete this source of nitrogen from their direct surroundings (Van Heeswijk, Westerhoff and Boogerd 2013). If insufficient ammonium is present in the environment, many microorganisms have the possibility to take up nitrate or nitrite from the environment and invest energy to produce ammonium (Moreno-Vivián et al. 1999). All isolated Methylacidiphilum strains possess a gene encoding a transporter that could transport nitrate and/or nitrite across the membrane. In addition, they harbour a gene encoding a cytoplasmic NAD(P)H-dependent nitrate reductase that reduces nitrate to nitrite and a cytoplasmic NAD(P)H-dependent nitrite reductase that catalyses the 6-electron reduction to ammonium. The assimilatory reduction of nitrate to ammonium is catalysed at the expense of four NAD(P)H molecules and is therefore only used when ammonium in the environment is scarce. In several Methylacidimicrobium strains, the pathway for nitrate assimilation is unclear, or could be absent.
All isolated verrucomicrobial methanotrophs possess the nifHDK operon, indicating their ability to fix N2 from the atmosphere. Methylacidiphilum fumariolicum SolV and Methylacidiphilum kamchatkense Kam1 were experimentally shown to use N2 as nitrogen source (Islam et al. 2008; Khadem et al. 2010). Diazotrophy has also been shown for several proteobacterial methanotrophs (Murrell and Dalton 1983b; Khmelenina et al. 2018). Although the ability to obtain nitrogen from the atmosphere provides a major advantage in an environment devoid of fixed nitrogen molecules, the fixation of nitrogen gas is very energy-demanding, requiring 16 ATP molecules per N2 molecule fixed (Dixon and Kahn 2004). As a result, the maximum specific growth rate of Methylacidiphilum fumariolicum SolV with N2 as nitrogen source is 2.8 times slower than when supplied with ammonium (Khadem et al. 2010). The absence of fixed nitrogen in the environment might therefore lead to a reduction in growth of verrucomicrobial methanotrophs and therefore to increased methane emissions.
Oxidation of methane and other hydrocarbons
Significant amounts of methane are emitted from mud pools, hot springs and fumaroles in terrestrial geothermal systems (Etiope and Klusman 2002; Castaldi and Tedesco 2005; Kvenvolden and Rogers 2005). Typically, geothermal gas consists of 0.01 to 1% (v/v) methane, although much higher concentrations have been detected (Giggenbach 1995; Etiope and Klusman 2002; D'Alessandro et al. 2009; Dunfield and Dedysh 2010). Under suitable conditions, verrucomicrobial methanotrophs could represent a significant filter against emissions of methane to the atmosphere in these environments (Op den Camp et al. 2009; Venturi et al. 2019). Studying in which circumstances verrucomicrobial methanotrophs consume more methane than in others is worth investigating to elucidate which factors enhance or inhibit methane oxidation.
The first step in methane oxidation is the conversion of methane to methanol (CH3OH), which can be catalysed by two genetically unrelated enzymes: the membrane-bound copper-containing particulate methane monooxygenase (pMMO) and the cytosolic iron-containing methane monooxygenase (sMMO) (Ross and Rosenzweig 2017). Several proteobacterial methanotrophs harbour genes encoding both the soluble and the particulate methane monooxygenase (Semrau, DiSpirito and Yoon 2010). However, none of the verrucomicrobial methanotrophs carry a gene encoding sMMO. Methylacidiphilum strains possess three complete but distinct pmoCAB operons and therefore in all probability oxidise methane to methanol via pMMO (Hou et al. 2008; Anvar et al. 2014; Van Teeseling et al. 2014; Kruse et al. 2019) (Figs 3 and 4). As exception, Methylacidiphilum sp. Yel only carries a single complete pmoCAB operon most closely related to the pmoCAB3 of the other Methylacidiphilum strains. All pMMOs characterised thus far show an (αβγ)3 oligomeric state, consisting of the two membrane-spanning subunits PmoC and PmoA and a predominantly periplasmic subunit PmoB, which is tethered to the membrane by two transmembrane helices (Lieberman and Rosenzweig 2005; Ross and Rosenzweig 2017). The enzyme activates O2 to break the C-H bond of methane, producing methanol and water, via a yet unresolved mechanism (Ross and Rosenzweig 2017; Ross et al. 2019) (Fig. 4). Interestingly, similarities between two pmo operon structures of proteobacterial and verrucomicrobial methanotrophs indicate evolution from a common ancestor (Op den Camp et al. 2009; Van Teeseling et al. 2014). Besides, the most distant pmoCAB3 operon seems to be obtained through horizontal gene transfer from an unknown microorganism (Dunfield et al. 2007). In fact, a large variety of key metabolic genes of verrucomicrobial methanotrophs are thought to be derived via horizontal gene transfer, especially from Proteobacteria (Sharp et al. 2013).
Figure 3.
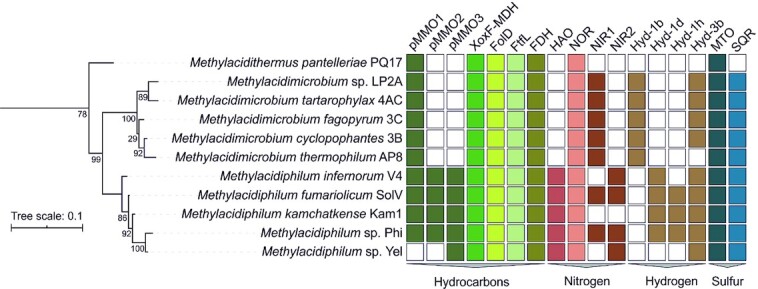
16S rRNA gene phylogenetic tree of verrucomicrobial methanotrophs and the presence or absence of genes involved in the oxidation of hydrocarbons, ammonia, hydrogen and sulfur compounds. Planctomycetes were used as an outgroup. Sequences were aligned using SINA v1.2.11 (Pruesse, Peplies and Glöckner 2012) and a tree was constructed using FastTree v2.1 (Price, Dehal and Arkin 2010) with 1000 bootstraps and substitution model GRT-GAMMA. The tree was visualised using ITOL v5.5.1 (Letunic and Bork 2007). Bootstrap values are indicated as a proportion of 1000 re-samplings ranging from 1 to 100. Presence of genes of interest was examined using BLASTp and of each hit the amino acid sequence was manually inspected for domains and identity. The result table was transformed to a binary ITOL dataset and both the 16S tree and the table were visualised using ITOL v5.5.1. pMMO: particulate methane monooxygenase; XoxF-MDH: lanthanide-dependent methanol dehydrogenase; FolD: methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase; FtfL: formate-tetrahydrofolate ligase; FDH: formate dehydrogenase; HAO: hydroxylamine oxidoreductase; NOR: nitric oxide reductase; NIR: nitrite reductase; Hyd: type of [NiFe] hydrogenase (based on Søndergaard, Pedersen and Greening 2016); MTO: methanethiol oxidase; SQR: sulfide:quinone oxidoreductase.
Figure 4.
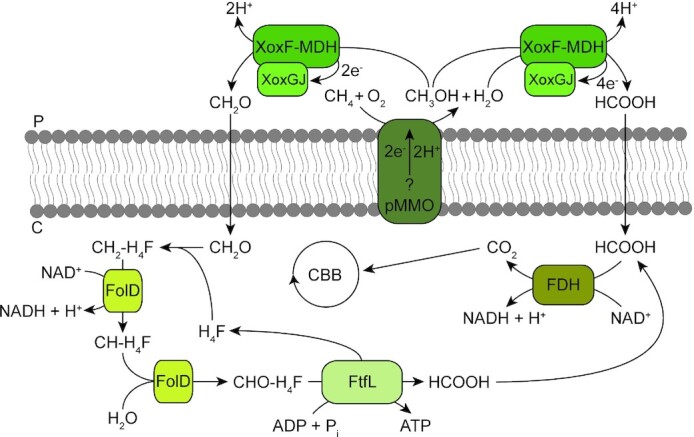
Possible pathways for methane oxidation in verrucomicrobial methanotrophs. pMMO oxidises methane to methanol (CH3OH), while an unknown electron donor is oxidised. The lanthanide-dependent XoxF methanol dehydrogenase (MDH) could subsequently oxidise methanol to either formate (HCOOH) or formaldehyde (CH2O), while donating electrons to its redox partner XoxGJ. If formate is produced it could diffuse into the cytoplasm and be converted to CO2 by the NAD+-dependent formate dehydrogenase (FDH). CO2 is fixed into biomass via the Calvin-Benson-Bassham (CBB) cycle. Alternatively, if formaldehyde is produced, it could bind to tetrahydrofolate (H4F) spontaneously or enzymatically by an unidentified enzyme, to form methylene-tetrahydrofolate (CH2-H4F). The enzyme FolD converts CH2-H4F to methenyl-tetrahydrofolate (CH-H4F), which is subsequently converted to formyl-tetrahydrofolate (CHO-H4F). This product is then converted to H4F and formate, while producing ATP. P: periplasm; C: cytoplasm.
In Methylacidiphilum fumariolicum SolV, the oxygen concentration seems to regulate the expression of the pmoCAB1 and pmoCAB2 operons (Khadem et al. 2012). Notably, the pmoCAB3 operon is barely expressed in Methylacidiphilum fumariolicum SolV when grown on methane (Khadem et al. 2012). Recently, Methylacidiphilum fumariolicum SolV was shown to grow on ethane and propane but not on butane (Picone et al. 2020b). pmoCAB3 could be specifically tuned towards the oxidation of alkanes, enabling Methylacidiphilum fumariolicum SolV to utilise gaseous hydrocarbons other than methane (Picone et al. 2020b). However, Methylacidiphilum sp. Yel only carries pmoCAB3 and is able to grow on methane, suggesting the enzyme encoded by this operon is at least also involved in the oxidation of methane (Erikstad et al. 2019). In contrast to Methylacidiphilum strains, Methylacidimicrobium strains and Ca. ‘Methylacidithermus pantelleriae’ PQ17 possess a single pmoCAB operon, related to the closely related pmoCAB1 and pmoCAB2 types of Methylacidiphilum species. As exception, Methylacidimicrobium thermophilum AP8 and Methylacidimicrobium sp. LP2A possess two almost identical operons (Sharp et al. 2014).
For a long time, the well-studied calcium-dependent methanol dehydrogenase (MDH) MxaFI was thought to be the key enzyme for methanol oxidation in both methanotrophs and methylotrophs (Anthony 2004). This periplasmic enzyme contains a pyrroloquinoline quinone (PQQ) prosthetic group at the catalytic centre and converts methanol to formaldehyde (CH2O) (Anthony 2004; Keltjens et al. 2014). The alphaproteobacterial methylotroph Methylobacterium extorquens AM1 carries genes encoding MxaFI and an MDH homolog named XoxF (Nakagawa et al. 2012). A ΔmxaF mutant strain was unable to grow on methanol when supplied with calcium, but growth on methanol was restored when cells were supplied with the lanthanide lanthanum (La3+) in the medium. Similarly, Methylacidiphilum fumariolicum SolV is only able to grow on methane when lanthanide-containing mud pot water (from the environment where the strain was isolated from) is added to the growth medium, or when the mineral medium contains micromolar concentrations of one or more lanthanides (Pol et al. 2014). These lanthanides are essential for verrucomicrobial methanotrophs when using methanol as an energy source, because they do not possess genes encoding the canonical calcium-dependent MxaFI, but a gene encoding the novel lanthanide-dependent MDH (XoxF) (Pol et al. 2014) (Fig. 3). Remarkably, the biological relevance of these rare earth elements was previously deemed unthinkable (Lim and Franklin 2004). Genomic analyses surprisingly revealed xoxF to be widespread in methylotrophs and methanotrophs, and is classified into five different clades (Chistoserdova 2011; Keltjens et al. 2014; Pol et al. 2014; Wu et al. 2015; Versantvoort et al. 2018; Kato et al. 2020). In fact, XoxF-type MDHs are actually much more abundant in nature than the MxaFI-type (Keltjens et al. 2014, Chistoserdova and Kalyuzhnaya 2018). The MxaF protein (large subunit) seems to have descended from a XoxF prototype (Keltjens et al. 2014) and all methylotrophs that harbour genes encoding MxaFI also possess a gene encoding the XoxF-type MDH (Chistoserdova and Kalyuzhnaya 2018). Dissolved lanthanide concentrations in most environments are very low (nM range) and its limitation may have driven the evolutionary adaptation of the proteobacterial methylotrophs towards an MDH variant that dependents on calcium, present in excess. The discovery of lanthanide-dependent XoxF-type MDHs has led to a completely new field of research, including the differential regulation in bacteria containing both types of MDHs (Zheng et al. 2018; Daumann 2019; Good et al. 2019; Picone and Op den Camp 2019; Good et al. 2020; Yanpirat et al. 2020). As example, the alphaproteobacterium Methylobacterium extorquens AM1 carries genes encoding both MDH types and was shown to upregulate xoxF expression already at a lanthanide concentration of 2.5 nM and downregulate mxaF expression at a lanthanide concentration of 25 nM or above (Vu et al. 2016; Daumann 2019).
Electrons yielded from methanol oxidation by XoxF are first transferred to a dedicated cytochrome partner (Zheng et al. 2018; Versantvoort et al. 2019) homologous to cytochrome cL, the electron acceptor of MxaFI-type MDH (Anthony 1992) (Fig. 4). In Methylacidiphilum strains, this cytochrome is a fusion protein of the cytochrome c protein XoxG and the periplasmic substrate binding protein XoxJ, but they also harbour a separate xoxJ gene. In Methylacidimicrobium strains, XoxG and XoxJ are encoded by two separate genes. Methylacidiphilum strains possess only one XoxF enzyme, whereas Methylacidimicrobium strains carry genes encoding for a copy of both the XoxF1 and XoxF2 clade that are less than 50% identical, which suggests that they could be expressed differentially under different conditions (Chistoserdova 2011; Keltjens et al. 2014; Picone and Op den Camp 2019). In addition, Methylacidimicrobium sp. LP2A harbours two genes encoding XoxF1 that are 96% identical to each other, presumably as a result of a relatively recent gene duplication (Van Teeseling et al. 2014; Op den Camp et al. 2018).
Similar to MxaFI-type MDH, XoxF-type MDH also contains a PQQ cofactor, and its overall structure is conserved in comparison with the MxaFI-type MDH (Keltjens et al. 2014). From Methylacidiphilum fumariolicum SolV, cultivated on lanthanide-containing mud pot water, XoxF was crystallised as a homodimer with a cerium ion coordinating with the PQQ cofactor (PDB: 4MAE; Pol et al. 2014). In addition, XoxF-type MDHs with a europium or lanthanum ion in the active site were crystallised (Deng, Ro and Rosenzweig 2018; Jahn et al. 2018; Good et al. 2020). The lanthanide coordinating with the PQQ cofactor of XoxF seems to render this enzyme catalytically more efficient compared to MxaF, since lanthanides are stronger Lewis acids than calcium, facilitating the hydride transfer (Keltjens et al. 2014; Daumann 2019). MxaFI-type MDHs produce formaldehyde from methanol oxidation, but XoxF was shown to produce formate (HCOOH) in vitro (Pol et al. 2014). Moreover, XoxF from Methylacidiphilum fumariolicum SolV can oxidise formaldehyde with high affinity and MxaFI-type MDHs in general are also able to oxidise formaldehyde (Keltjens et al. 2014). However, recently the XoxF5 of the alphaproteobacterium Methylobacterium extorquens AM1 was shown to oxidise methanol to formaldehyde in vivo, not to formate (Good et al. 2019). Currently, it is unresolved how formaldehyde would be oxidised further to formate in verrucomicrobial methanotrophs (Chistoserdova 2011). Formaldehyde could be oxidised directly to formate by formaldehyde dehydrogenase (Chistoserdova, Kalyuzhnaya and Lidstrom 2009), but verrucomicrobial methanotrophs do not possess a gene encoding this enzyme. Alternatively, methylotrophs can make use of the tetrahydromethanopterin (H4MPT) or the tetrahydrofolate (H4F) pathway for formaldehyde oxidation, as found in proteobacterial methanotrophs (Vorholt 2002; Marx, van Dien and Lidstrom 2005; Chistoserdova 2011). H4F is synthesised by bacteria de novo (Bermingham and Derrick 2002). Indeed, genes encoding all enzymes involved in tetrahydrofolate biosynthesis are present in the genomes of the verrucomicrobial methanotrophs. Formaldehyde produced by periplasmic XoxF could diffuse into the cytoplasm and bind spontaneously to H4F to form methylene-H4F (Kallen and Jencks 1966). Whether this condensation is of sufficient high rate for bacterial growth in vivo is under debate (Crowther, Kosály and Lidstrom 2008; He et al. 2020). In the proteobacterium Methylobacterium extorquens AM1, the formaldehyde-activating enzyme actually catalyses this reaction at high rate (Vorholt et al. 2000). Whereas this enzyme is present in proteobacterial methanotrophs, it is absent in verrucomicrobial methanotrophs, suggesting another enzyme should be involved in the condensation of formaldehyde and H4F. All verrucomicrobial methanotrophs carry a gene encoding the bifunctional enzyme FolD (Fig. 3), which is known to convert methylene-H4F to methenyl-H4F, and produce NAD(P)H, similar to the methylene-H4F dehydrogenase (Pawelek and MacKenzie 1998; Hou et al. 2008; Chistoserdova 2011; Eadsforth, Maluf and Hunter 2012). Subsequently, FolD acts as a methenyl-H4F cyclohydrolase, converting methenyl-H4F to formyl-H4F (Fig. 4). Ultimately, the enzyme formate-tetrahydroformate ligase FtfL, found in all verrucomicrobial methanotrophs, could convert formyl-H4F to tetrahydrofolate and formate, while producing one ATP molecule (Marx, Laukel and Vorholt 2003). If this pathway oxidises formaldehyde instead of XoxF, fewer electrons are donated to the cytochrome c protein XoxGJ, which is postulated to donate its electrons to the terminal oxidase to fuel respiration (Versantvoort et al. 2019). Nevertheless, the production of formaldehyde instead of formate by XoxF seems logical since formaldehyde oxidation via the H4F pathway provides valuable reducing equivalents in the form of NAD(P)H and ATP. Finally, in the last step of aerobic methane oxidation in verrucomicrobial methanotrophs, formate is oxidised to CO2 (Pol et al. 2014). All verrucomicrobial methanotrophs possess one complete fdsDABG operon, encoding a cytosolic formate dehydrogenase that is thought to oxidise formate, while producing NADH needed for the generation of biomass (Op den Camp et al. 2018). CO2 can ultimately be assimilated via the CBB cycle. The key enzyme RuBisCO found in verrucomicrobial methanotrophs forms a novel cluster within the form I RuBisCO phylogenetic tree and is most closely related to form IC RuBisCOs found in various Proteobacteria (Khadem et al. 2011).
Coping with toxic nitrogen compounds from ammonia oxidation
In the geothermal habitat where Methylacidiphilum fumariolicum SolV was isolated from, high concentrations (1–28 mM) of ammonium (NH4+) are present (Khadem et al. 2010; Mohammadi et al. 2017b). Methane and ammonia (NH3) are highly-reduced molecules with structural similarities. Nitrifying microorganisms make a living from the oxidation of ammonia, initiated via the oxidation of ammonia to hydroxylamine (NH2OH), catalysed by ammonia monooxygenase (AMO) (Klotz and Stein 2008). AMO is a membrane-bound copper-dependent enzyme homologous to pMMO of methanotrophs (Tavormina et al. 2011). Consequently, many nitrifiers can oxidise methane and many methanotrophs can oxidise ammonia (Bédard and Knowles 1989). Methanotrophs use ammonium as nitrogen source; however, ammonia in the periplasm can also fortuitously be oxidised by pMMO to hydroxylamine, which is a toxic compound (Nyerges and Stein 2009). The affinity of pMMO for ammonia decreases with increasing CH4 concentrations, which indicates competitive substrate inhibition (Nyerges, Han and Stein 2010. Hydroxylamine could impede various cellular processes by damaging proteins or DNA and strongly inhibit MDHs (Kaplan and Ciotti 1953; Duine and Frank 1980; Versantvoort et al. 2020). Therefore, verrucomicrobial and proteobacterial methanotrophs have developed mechanisms to balance the assimilation of ammonium and the detoxification of deleterious nitrogen compounds.
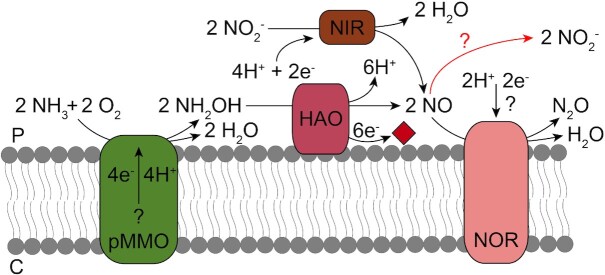
In order to detoxify hydroxylamine, many methanotrophs possess a gene encoding a hydroxylamine oxidoreductase (HAO), oxidising hydroxylamine to nitric oxide (NO) (Fig. 5) (Caranto and Lancaster 2017; Versantvoort et al. 2020). All known Methylacidiphilum strains carry a gene encoding an HAO (Fig. 3), as do many proteobacterial methanotrophs (Campbell et al. 2011; Stein and Klotz 2011; Versantvoort et al. 2020). In contrast, HAO is not found in any of the Methylacidimicrobium strains and as such, these strains might be more sensitive to ammonium (Stein 2018). Alternatively, Methylacidimicrobium strains could have developed a different strategy to deal with ammonia that does not require an HAO, analogous to the gammaproteobacterial methanotroph Methylomonas methanica. This methanotroph also lacks an HAO-like protein (Campbell et al. 2011) and was unable to oxidise ammonia to nitrite, although its methane oxidation rate was inhibited by ammonia (Nyerges and Stein 2009). This could indicate that its pMMO is unable to oxidise ammonia to hydroxylamine or the formed hydroxylamine is converted to a product by an unknown enzyme other HAO. In either case, an HAO-like protein is not necessary to deal with ammonia toxicity. In nitrifiers, electrons yielded from the oxidation of hydroxylamine are transferred to the quinone pool for energy conservation via the hydroxylamine:ubiquinone reductase module (Klotz and Stein 2008). However, methanotrophs are thought to lack this module. Still, per ammonia molecule oxidised, four electrons are yielded when ammonia is oxidised to nitrite (Fig. 5). Since two electrons are needed to activate O2 for the oxidation of ammonia to hydroxylamine by pMMO, the remaining two electrons might be used at the terminal oxidase for energy conservation (Stein and Klotz 2011).
Figure 5.
Ammonia (NH3) oxidation by pMMO and fate of the reaction products. pMMO fortuitously oxidises ammonia to hydroxylamine. Subsequently, hydroxylamine oxidoreductase (HAO) oxidises hydroxylamine to nitric oxide, donating electrons to an unknown cytochrome c protein (indicated by the red diamond). Nitric oxide can be reduced to inert N2O by the nitric oxide reductase (NOR), using an unknown electron donor. Alternatively, nitric oxide is converted chemically to nitrite in the presence of oxygen, or by an unknown enzyme (red arrow). Under anaerobic conditions, nitrite could be utilised as alternative electron acceptor. The nitrite reductase (NIR) could then reduce nitrite to nitric oxide, which is subsequently reduced to N2O. P: periplasm; C: cytoplasm.
Nitric oxide produced by HAO is a toxic compound. However, when ammonia is oxidised by Methylacidiphilum fumariolicum SolV under aerobic conditions, nitrite is observed as the main end product (Mohammadi et al. 2017b), similar to aerobic ammonia oxidisers and various proteobacterial methanotrophs (Nyerges, Han and Stein 2010; Campbell et al. 2011; Lehtovirta-Morley 2018). These cases necessitate an additional enzyme that oxidises the NO produced by HAO to nitrite. The nature of this nitric oxide-oxidising enzyme in methanotrophs and ammonia-oxidisers is unknown, although it is postulated that the copper-dependent nitrite reductase NirK could function in the opposite direction to oxidise NO to nitrite (Caranto and Lancaster 2017). In addition, NO can rapidly react with oxygen resulting in the formation of nitrite and nitrate in aqueous solutions (Udert, Larsen and Gujer 2005; Hughes 2008) and therefore the production of nitrite observed for Methylacidiphilum fumariolicum SolV could also be non-enzymatic (Fig. 5). Under anoxic conditions, nitrite reduction rates in Methylacidiphilum fumariolicum SolV are higher compared with rates under oxic conditions (Mohammadi et al. 2017b). As such, the alternative electron acceptor nitrite could be converted to NO by a nitrite reductase, of which one or two orthologues are present in most verrucomicrobial methanotrophs (Fig. 3). Nitric oxide can subsequently be reduced to N2O, as was shown for the gammaproteobacterial methanotrophs Methylomicrobium album and Methylomonas denitrificans and for Methylacidiphilum fumariolicum SolV (Nyerges, Han and Stein 2010; Kits, Klotz and Stein 2015; Mohammadi et al. 2017b). The membrane-bound nitric oxide reductase NorCB reduces NO to N2O and is found in all known verrucomicrobial methanotrophs (Figs 3 and 5). It is therefore conceivable that verrucomicrobial methanotrophs detoxify nitric oxide to inert N2O under anoxic or oxygen-limited conditions (Acton and Baggs 2011; Bodelier and Steenbergh 2014). In addition, several proteobacterial methanotrophs were shown to produce N2O without the initial formation of nitrite (Hoefman, van der Ha and Boon 2014). Since N2O is strong greenhouse gas with a significant role in global warming, methanotrophs might not only have a mitigating effect on climate change by oxidising methane, but also aggravate this change by producing N2O (Acton and Baggs 2011; Versantvoort et al. 2020). Altogether, the accidental oxidation of ammonia by pMMO results in the formation of various toxic compounds, for which Methylacidiphilum strains typically appear to have multiple detoxification pathways.
Oxidation of hydrogen gas
Multiple proteobacterial methanotrophs carry hydrogenase genes and were shown to consume H2 (De Bont 1976; Csáki et al. 2001; Kelly, Anthony and Murrell 2005). These methanotrophs were proposed to use these hydrogenases to produce reducing equivalents to drive the energy-demanding conversion of methane to methanol by pMMO (Hanczár et al. 2002). Besides, various proteobacterial methanotrophs possess genes encoding both a hydrogenase and ribulose-1,5-biphosphate carboxylase/oxygenase (RuBisCO), suggesting autotrophic growth on H2 is possible (Mohammadi et al. 2019). However, autotrophic growth on H2 in liquid media without methane has never been observed for proteobacterial methanotrophs (Taylor, Dalton and Dow 1981; Baxter et al. 2002).
Hydrogen gas (H2) is typically emitted in geothermal habitats in high concentrations (often > 1% v/v) and is a potential alternative energy source for various microorganisms (Aragno 1992; Chiodini et al. 2001; Carere et al. 2017; Mohammadi et al. 2019). Indeed, Methylacidiphilum fumariolicum SolV was experimentally shown to make a living as autotrophic hydrogenotroph in the absence of methane (Mohammadi et al. 2017a). Since methane emissions can heavily fluctuate in geothermal habitats, the ability to grow on another emitted, energy-rich gas is highly advantageous. Methylacidiphilum sp. RTK17.1 (an isolate almost identical to Methylacidiphilum infernorum V4) was shown to grow as a mixotroph on H2 and CH4, a trait hypothesised to have driven niche expansion, which could explain the dominance of verrucomicrobial methanotrophs in geothermal habitats (Carere et al. 2017; Mohammadi et al. 2019). Recently, this mixotrophic lifestyle on H2 and CH4 was also demonstrated by the alphaproteobacterium Methylocystis sp. strain SC2 (Hakobyan, Zhu and Glatter 2020).
Hydrogenases are a very diverse group of enzymes that convert H2 into two protons and two electrons, or vice versa (Lubitz et al. 2014). Three different metal compositions are known in the active site: [NiFe], [FeFe] and [Fe]. Genome comparisons of several verrucomicrobial methanotrophs revealed that all these microbes carry genes encoding one or more different [NiFe] hydrogenases, except for Ca. ‘Methylacidithermus pantelleriae’ PQ17 (Fig. 3) (Mohammadi et al. 2019). It has to be noted that the genome of strain PQ17 was retrieved from a metagenome, not from an isolate. It might therefore still possess several genes that are not directly found through genetic analyses, although the genome is more than 98% complete and shows little contamination (Picone 2020). All isolated Methylacidiphilum strains carry genes encoding the group 1d [NiFe] hydrogenase, except for Methylacidiphilum sp. Yel. This membrane-bound enzyme has a relatively high O2 tolerance compared to other hydrogenases and is involved in the aerobic respiration of H2 (Greening et al. 2015). Both Methylacidiphilum sp. RTK17.1 and Methylacidiphilum fumariolicum SolV were experimentally shown to grow on H2 using this hydrogenase under various oxygen concentrations (Carere et al. 2017; Mohammadi et al. 2017a).
All of the Methylacidimicrobium strains and none of the Methylacidiphilum strains possess genes encoding the oxygen-sensitive group 1b [NiFe] hydrogenase (Fig. 3) (Mohammadi et al. 2019). Methylacidimicrobium tartarophylax 4AC was shown to grow as an autotroph on H2 using this enzyme under microoxic conditions (Mohammadi et al. 2019). The group 1b [NiFe] hydrogenase, typically found in Proteobacteria that perform anaerobic respiration (Greening et al. 2015), therefore enables Methylacidimicrobium strains to respire H2 aerobically. In addition, all Methylacidimicrobium and Methylacidiphilum strains, except for Methylacidimicrobium thermophilum AP8, carry genes encoding a group 3b [NiFe] hydrogenase. This heterotetrameric cytosolic enzyme is known to couple the oxidation of NADPH to the production of H2 during fermentation (Berney et al. 2014). However, in the verrucomicrobial methanotrophs it is hypothesised to oxidise H2 and produce NADH for CO2 fixation (Carere et al. 2017). This proposed catalysis was demonstrated by the group 3b [NiFe] hydrogenase of Hydrogenobacter thermophilus (Yoon et al. 1996).
Most hydrogen-oxidising microorganisms live in habitats with relatively high H2 concentrations, such as animal guts and leguminous soils (Conrad and Seiler 1979; Pester and Brune 2007). In addition, several soil-inhabiting Actinobacteria were shown to consume H2 present in the atmosphere (Constant, Poissant and Villemur 2008; Constant et al. 2010; Constant et al. 2011). Although the atmospheric H2 concentration is very low (0.53 ppmv H2), soil systems are the largest sink of atmospheric H2, consuming 75 Tg H2 annually (Novelli et al. 1999). These H2 scavengers possess genes encoding a putative high-affinity [NiFe] hydrogenase, of which the oxygen-tolerant group 1h [NiFe] hydrogenase seems to be dominant. Remarkably, this group 1h [NiFe] hydrogenase is also found in the genomes of Methylacidiphilum fumariolicum SolV, Methylacidiphilum kamchatkense Kam1 and Methylacidiphilum sp. Phi (Fig. 3) (Mohammadi et al. 2017a; Kruse et al. 2019). In the related Methylacidiphilum infernorum V4, this gene may have been lost over time due to the movement of transposable elements in the genome (Kruse et al. 2019). Methylacidiphilum fumariolicum SolV was shown to express both the group 1d and group 1h [NiFe] hydrogenase to grow on H2 (Mohammadi et al. 2017a). At dissolved oxygen concentrations between 0.2 and 1.5%, only the group 1h [NiFe] hydrogenase supported growth, up to a growth rate of 0.03 h−1, which is almost 40% of the growth rate on methane (Mohammadi et al. 2017a). The enzyme has a remarkable tolerance towards O2, suggested to be the result of the unique coordination of the proximal [4Fe4S] clusters by three cysteines and an aspartate residue, instead of the usual coordination by four cysteine residues (Schäfer, Friedrich and Lenz 2013; Schäfer et al. 2016). The group 1h [NiFe] hydrogenase was purified from Methylacidiphilum fumariolicum SolV and kinetic experiments revealed an unusually high affinity for H2, enabling this methanotroph to oxidise atmospheric H2 (Schmitz et al. 2020a). It is hypothesised that this hydrogenase in particular could aid verrucomicrobial methanotrophs to thrive in geothermal systems (Carere et al. 2017; Schmitz et al. 2020a).
Oxidation of sulfur compounds
A variety of inorganic sulfur compounds such as hydrogen sulfide (H2S) and sulfur dioxide (SO2) are present in or released from geothermal systems (Spiro, Jacob and Logan 1992; Vasilakos et al. 2005). Additionally, organic sulfur compounds such as methanethiol (CH3SH) could be present, but data on its presence in terrestrial volcanic ecosystems are lacking. In seafloor hydrothermal systems, methanethiol is formed abiotically from CO2, H2S and H2, derived from organic matter below the ocean floor (Rogers and Schulte 2012; Reeves et al. 2014). It is conceivable that in terrestrial mud volcanoes with comparable conditions, thermogenic production of methanethiol could occur as well. This compound could then be used by methylotrophs such as the verrucomicrobial methanotrophs as carbon and sulfur source, or even as energy source, as observed in the methylotrophic alphaproteobacterium Hyphomicrobium sp. VS (Pol et al. 1994). Studying (organic) sulfur compounds and how microorganisms cope with these compounds is important, as several of these compounds have profound effects on the environment, causing acid precipitation and cloud formation (Lomans, Pol and Op den Camp 2002).
Little is known about the utilisation of sulfur compounds as energy sources in methanotrophs. Genomic comparison of verrucomicrobial methanotrophs revealed that all known strains carry a gene encoding methanethiol oxidase (annotated as selenium-binding protein 56) (Eyice et al. 2018) (Fig. 3). This enzyme oxidises methanethiol to form formaldehyde, hydrogen sulfide and hydrogen peroxide (H2O2) (Suylen et al. 1987). The oxidation of methanethiol could render the verrucomicrobial methanotrophs with useful products. Formaldehyde could be converted to formate via FolD and FtfL and to CO2 by the formate dehydrogenase (Fig. 4). H2S can be used as a sulfur source and could even be used as an energy source. In addition, a gene encoding sulfide:quinone oxidoreductase (SQR) is found in the genomes of all verrucomicrobial methanotrophs, and highly expressed in Methylacidiphilum fumariolicum SolV (Mohammadi et al. 2017a). More specifically, it contains a type III SQR of which little is known, that could be involved in aerobic H2S respiration (Marcia, Ermler and Peng 2010). The third product of methanethiol oxidation, hydrogen peroxide, is toxic and could be converted to water and oxygen by the enzyme catalase (Zamocky, Furtmüller and Obinger 2008). A gene encoding catalase is only found in Methylacidiphilum infernorum V4 and all Methylacidimicrobium strains. However, several peroxidases have been found in the genomes that might substitute the catalase activity.
The effect of intracellular H2S and methanethiol in methanotrophs is currently not well understood. In some microorganisms, methane oxidation seems to be enhanced or unaffected by the presence of methanethiol or hydrogen sulfide (Lee, Kim and Cho 2012), whereas in other strains methane oxidation is inhibited by these compounds (Börjesson 2001; Lee et al. 2015). Since all verrucomicrobial methanotrophs possess a gene encoding an SQR and because H2S is generally emitted at high concentrations from geothermal systems (Chiodini et al. 2001), it is conceivable that also H2S could be an important energy source for these microorganisms. Alternatively, sulfide-oxidising enzymes could be present as a means of detoxification. Future experiments have to resolve whether energy conservation from the oxidation of H2S is indeed possible.
The electron transport chain and energy conservation
The main location for energy conservation in prokaryotic cells is the cytoplasmic membrane. Quinones inside the cytoplasmic membrane are essential parts of the electron transport chain and therefore essential to energy conservation (Kurosu and Begari 2010). Verrucomicrobial methanotrophs seem to synthesise menaquinones (MK) via an alternative pathway that involves the intermediate futalosine (Hiratsuka et al. 2008). However, the gene mqnB involved in menaquinone biosynthesis was not detected in any of the genomes of verrucomicrobial methanotrophs (Kruse et al. 2019), but pathway variations are known to exist (Arakawa et al. 2010).
All verrucomicrobial methanotrophs described in this review possess five different complexes involved in energy conservation (Fig. 6). The NADH:quinone oxidoreductase (Complex I) is a membrane-bound protein complex that oxidises NADH, transfers the freed electrons to the quinone pool, and couples this reaction to the translocation of protons across the membrane, contributing to a proton motive force (Friedrich and Scheide 2000). The bacterial enzyme typically consists of 14 subunits, which are all encoded by the verrucomicrobial methanotrophs (Berrisford, Baradaran and Sazanov 2016). Complex II, or succinate dehydrogenase, is involved in both the electron transport chain and the tricarboxylic acid (TCA) cycle (Cecchini et al. 2002). In this cycle, Complex II couples the oxidation of succinate to the reduction of quinone (Fig. 6). Whereas Complex I is also involved in the translocation of protons across the membrane, Complex II is only involved in electron transfer. The quinols produced from the reduction of quinones by both complexes are typically oxidised by the bc1 complex (Complex III) (Trumpower 1990). In that case, electrons yielded from the oxidation of quinols are subsequently transferred via an additional cytochrome c protein to a cytochrome c oxidase (complex IV or terminal oxidase). However, the verrucomicrobial methanotrophs do not carry genes encoding Complex III. Instead, these microorganisms possess genes encoding a structurally different protein complex with similar function: the Alternative Complex III (ACIII; Fig. 6) (Pereira et al. 2007; Refojo et al. 2010). The classical Complex III contributes to the proton motive force by translocating protons over the membrane via the quinone cycle. Whether ACIII also translocates protons across the membrane for energy conservation is under debate (Refojo, Teixeira and Pereira 2012; Sun et al. 2018). Ultimately, Complex IV reduces the terminal electron acceptor O2 to water, with electrons retrieved from a c-type haem (Sun et al. 2018). All isolated verrucomicrobial methanotrophs, except for Ca. ‘Methylacidithermus pantelleriae’ PQ17, possess genes encoding three distinct Complexes IV: a cbb3-type, an aa3-type and a ba3-type. These cytochrome c oxidases are classified based on the type of haems they contain and are known to have different affinities for O2 and can be differentially used for various substrates (García-Horsman et al. 1994). In addition, Methylacidiphilum fumariolicum SolV and Methylacidiphilum sp. Phi possess a second ba3-type cytochrome c oxidase. Interestingly, ACIII of Flavobacterium johnsoniae can form a supercomplex with Complex IV, in which an additional cytochrome c protein for electron transfer is not needed (Sun et al. 2018). With this in mind, it is interesting to note that in verrucomicrobial methanotrophs the genes encoding the different subunits composing ACIII are found directly adjacent to the genes encoding the cbb3-type cytochrome c oxidase. Likewise, in other microorganisms the genes encoding ACIII are often found directly adjacent to genes encoding a cytochrome c oxidase (Refojo et al. 2010). However, only in verrucomicrobial methanotrophs and Opitutaceae a C-type cytochrome c oxidase is found together with ACIII, whereas typically these are A-type or B-type cytochrome c oxidases (Refojo et al. 2010). The different cytochrome c oxidases encoded by Methylacidiphilum fumariolicum SolV are differentially expressed, underscoring the metabolic versatility of verrucomicrobial methanotrophs (Khadem et al. 2012; Mohammadi et al. 2017a). Interestingly, the ba3-type cytochrome c oxidase of Aquifex aeolicus is able to use ubiquinol as electron donor, reduced by electrons yielded from sulfide oxidation by the sulfide:quinone oxidoreductase (SQR) (Gao et al. 2012). Since the verrucomicrobial methanotrophs encode for both a ba3-type cytochrome c oxidase and SQR, these bacteria might utilise a specific system to conserve energy from H2S oxidation, but this needs experimental validation. The proton motive force created by the transfer of electrons through the different membrane complexes is used for the synthesis of ATP via the ATP synthase (Complex V). The verrucomicrobial methanotrophs, except for Ca. ‘Methylacidithermus pantelleriae’ PQ17, carry genes encoding two ATP synthases (ATPases). One ATPase is related to ATPases of other Verrucomicrobia, while the other is related to the gammaproteobacterial ATPase (Hou et al. 2008; Kruse et al. 2019).
Figure 6.
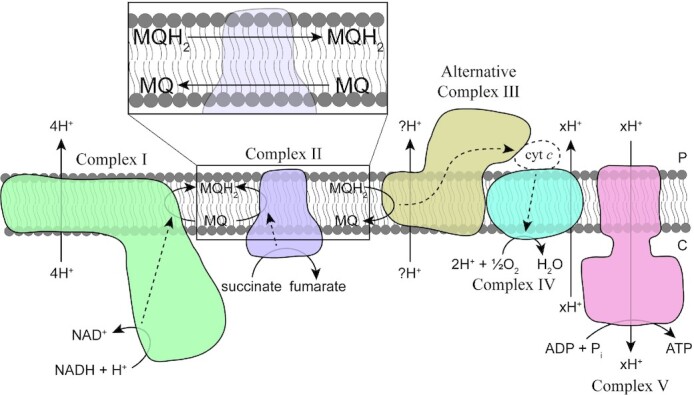
Schematic overview of the typical electron transport chain of verrucomicrobial methanotrophs. NADH generated through catabolic processes is oxidised by the NADH:quinone oxidoreductase (Complex I), transferring electrons to menaquinone (MQ) to form menaquinol (MQH2) while translocating protons across the membrane to generate a proton motive force. The succinate dehydrogenase (Complex II) oxidises succinate to fumarate as a part of the tricarboxylic acid cycle and transfers electrons to MQ to form MQH2. Alternative complex III oxidises MQH2 back to MQ, transferring electrons to a cytochrome c oxidase (Complex IV), either via an external periplasmic cytochrome c protein (cyt c) or directly. Whether protons are translocated during this electron transfer is unknown. Ultimately, electrons are used to reduce O2 to water, while protons are translocated over the membrane, contributing to the proton motive force that is used by an ATP synthase (Complex V) to synthesise ATP. Dashed lines indicate electron flow. Inset: transfer of oxidized and reduced quinones inside the bilayer (the quinone pool). C: cytoplasmic side of the membrane; P: periplasmic side of the membrane.
VERRUCOMICROBIAL METHANOTROPHS IN THE ENVIRONMENT
Verrucomicrobial methanotrophs are generally found in acidic volcanic ecosystems. Interestingly, global biogeographic distributions of closely related strains of verrucomicrobial methanotrophs suggest allopatric evolution over time (Erikstad et al. 2019). Additionally, evolutionary genomic analyses of verrucomicrobial methanotrophs revealed multiple horizontal gene transfer events (Sharp et al. 2013). Many genes have a relatively high similarity to those of the phylum Proteobacteria, but also to genes of Aquificae, Thermus/Deinococcus and archaea (Sharp et al. 2013; Schmitz et al. 2020b). Over the past 13 years, physiological and biochemical experiments have rendered valuable information that allows us to predict in which habitats we could encounter verrucomicrobial methanotrophs. Ecophysiological experiments could shed light on how verrucomicrobial methanotrophs interact with other microorganisms in the natural environment.
Acidic volcanic ecosystems are characterised by various morphological features. In general, these features are formed in places where mud, gas and water are expelled due to high fluid pressure from the subsurface, creating hot springs, mud pools and fumaroles (Cioni, Corazza and Marini 1984; Feyzullayev and Movsumova 2010; Benson et al. 2011). Where reduced sulfur compounds are present, often sulfuric acid (H2SO4) is produced by (thermo)acidophilic microorganisms. The microbial production of sulfuric acid is environmentally important because it creates a low pH, which is characteristic for the Solfatara, several parts of Yellowstone National Park and various other geothermal ecosystems (Schoen and Rye 1970; Quatrini and Johnson 2018). A low pH seems to be a key determinant of verrucomicrobial methanotrophs to be present in geothermal habitats (Sharp et al. 2014). 16S rRNA and pmoA gene sequences of verrucomicrobial methanotrophs have so far not been detected in habitats with a pH > 5.0 and all isolates have a pH optimum around 3. Physicochemical parameters within geothermal habitats such as temperature and the oxygen concentration can differ locally within a few meters. Many of the thermophilic Methylacidiphilum strains were isolated from hot geothermal areas in close proximity to the moderate temperature geothermal areas where the Methylacidimicrobium strains were isolated from (Sharp et al. 2014). Although bogs and fens are also characterised by a high acidity (pH 3.3 to 4.9), no 16S rRNA or pmoA gene sequences of verrucomicrobial methanotrophs have been detected in these habitats, which may be explained by the relatively low concentration of lanthanides in peat bogs (Vodyanitskii et al. 2012; Sharp et al. 2014). Based on environmental sequencing and activity studies, the methanotrophic activity can be attributed to Proteobacteria, at a pH as low as 3.5 (Dunfield and Dedysh 2010). Proteobacteria such as strains of the genus Acidithiobacillus have been found at lower pH, but this does not include proteobacterial methanotrophs (Crognale et al. 2018). With current knowledge, methane-rich habitats below pH 3.5, including the aforementioned crown of rusted sewer pipes, seem to be better suited for verrucomicrobial methanotrophs than proteobacterial methanotrophs.
Methane and hydrogen gas are often emitted from geothermal habitats and these two compounds seem to be the major energy sources for verrucomicrobial methanotrophs. All verrucomicrobial methanotrophs were isolated on methane and several strains were shown to grow as autotrophs on H2 (Carere et al. 2017; Mohammadi et al. 2017a; Mohammadi et al. 2019). Multiple verrucomicrobial methanotrophs possess more than one particulate methane monooxygenase. The three pmo operons within the Methylacidiphilum strains differ significantly in amino acid sequences, suggesting selection pressure for different functions and adaptation to changing environmental conditions (Op den Camp et al. 2009). In addition, short chain alkenes could be oxidised by one or more of the pMMOs found in Methylacidiphilum strains (Picone et al. 2020b). For methane oxidation to occur, a sufficient concentration of lanthanides must be present in the environment, because the verrucomicrobial methanotrophs only carry genes encoding the lanthanide-dependent XoxF-type methanol dehydrogenase. This does not necessarily mean that verrucomicrobial methanotrophs are only present in lanthanide-rich environments. If H2 and oxygen are present at suitable concentrations, they could grow as Knallgas bacteria without the need for lanthanides (Carere et al. 2017; Mohammadi et al. 2017a; Mohammadi et al. 2019). The concentration of H2 and O2 in the environment could partially determine which strains of verrucomicrobial methanotrophs are present in the environment, because the different kinds of [NiFe] hydrogenases found in their genomes may have very different tolerances towards oxygen and possess different affinities for H2. As example, Methylacidimicrobium tartarophylax 4AC only respires H2 at microoxic conditions with the group 1b [NiFe] hydrogenase (Mohammadi et al. 2019), whereas Methylacidiphilum fumariolicum SolV can respire H2 at wide range of oxygen concentrations catalysed by the group 1d and group 1h [NiFe] hydrogenases (Mohammadi et al. 2017a). In addition, all isolates carry a gene encoding a sulfide:quinone oxidoreductase (SQR), indicating that verrucomicrobial methanotrophs might even grow in acidic ecosystems where H2S is present and CH4 and H2 are absent.
The main difference between Methylacidiphilum and Methylacidimicrobium strains seems to be their growth temperature, with the former being thermophiles and the being latter mesophiles. However, the isolation of Methylacidimicrobium thermophilum AP8 with an optimum growth temperature of 50°C muddles this clear division (Picone 2020). Still, Methylacidiphilum and Methylacidimicrobium strains are often found within a few meters of each other, but at different temperatures (Sharp et al. 2014). In the environment, the concentration of ammonium could be a key factor in determining which verrucomicrobial methanotrophs are present. In general, ammonia is a competitive inhibitor of pMMO, potentially reducing methane oxidation rates. Additionally, high ammonium concentrations can reduce the methane oxidation rate of microorganisms that are unable to detoxify the oxidised products of ammonia oxidation (D'Alessandro et al. 2009; Nyerges, Han and Stein 2010). Since only Methylacidiphilum strains possess a gene encoding a hydroxylamine oxidoreductase, they are well-equipped to detoxify hydroxylamine, intracellularly produced through ammonia oxidation by pMMO. Consequently, Methylacidimicrobium strains are expected to be primarily found at moderate temperatures and habitats with relatively low ammonium concentrations. However, different pMMOs could have different affinities for ammonia, or could not be able to oxidise ammonia at all (Nyerges and Stein 2009).
Little is known about the interaction of verrucomicrobial methanotrophs with other microorganisms in their natural geothermal habitat. Trough δ13C analysis of CH4, the origin of this gas in geothermal systems can be deduced, since CH4 produced by archaea is relatively light compared to thermogenic or abiotic methane (Op den Camp et al. 2009). Typically, methane emitted from geothermal ecosystems is from a non-microbial source, produced through a chemical reaction of H2 and CO or by the non-microbial decomposition of buried organic matter (Giggenbach 1995; Etiope and Klusman 2002; Fiebig et al. 2007). However, methanogenic archaea have been found in anoxic parts of several geothermal ecosystems, but mostly at a pH of 5 or higher (Zeikus, Ben-Bassat and Hegge 1980; Berghuis et al. 2019). Remarkably, a metagenome-assembled genome (MAG) closely related to the methanogen Methanocella conradii was abundantly present in the metagenome of the hot, acidic, and methane-rich geothermal soil on the island Pantelleria, Italy (Picone et al. 2020a). This finding suggests that a larger part of methane emitted from hot and acidic geothermal ecosystems could be of microbial origin than initially thought (D'Alessandro et al. 2009). In contrast, H2 utilised by verrucomicrobial methanotrophs does not seem to be produced by other microorganisms in their habitat, but rather through abiotic or thermogenic processes in the Earth's crust (Aragno 1992; Lindsay et al. 2019).
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
Since the isolation of verrucomicrobial methanotrophs from hot and acidic geothermal ecosystems 13 years ago, significant progress had been made in our understanding of these microorganisms living in extreme environments. Verrucomicrobial methanotrophs are much more than their name suggests: these extremophiles are actually metabolically versatile microorganisms. In fact, the same could be true for their proteobacterial counterparts. Whereas several verrucomicrobial methanotrophs were shown to grow as Knallgas bacteria, hydrogen consumption by proteobacterial methanotrophs has only been demonstrated as part of a mixotrophic lifestyle. However, proteobacterial methanotrophs that possess the CBB cycle and an uptake hydrogenase could be Knallgas bacteria as well. In recent years, scientists have mainly focused on the metabolism of inorganic compounds by verrucomicrobial methanotrophs, not of organic compounds, as was done for several proteobacterial methanotrophs. Originally thought to be obligate methanotrophs, a large variety of multi-carbon compounds such as acetate, succinate and ethanol were shown to be consumed by alphaproteobacterial methanotrophs (Dunfield and Dedysh 2014). The first attempts to grow verrucomicrobial methanotrophs on several organic compounds such as glucose, citrate and malate were unsuccessful, but later Methylacidiphilum fumariolicum SolV was shown to grown on ethane and butane (Op den Camp et al. 2009; Picone et al. 2020b). The major questions for future research regarding metabolism are whether proteobacterial methanotrophs can grow as Knallgas bacteria and whether verrucomicrobial methanotrophs can incorporate organic compounds into their diet.
Since multiple verrucomicrobial methanotrophs were shown to be excellent hydrogen-oxidisers, it would be worthwhile to see if new enrichment strategies using hydrogen gas, carbon dioxide and low oxygen would result in novel verrucomicrobial isolates. The exciting discovery of 16S rRNA genes related to verrucomicrobial methanotrophs outside of terrestrial volcanic ecosystems suggests a distribution outside of geothermal habitats. Several factors such as pH and temperature, but also the oxygen and ammonium concentration and the presence of methane, hydrogen gas, hydrogen sulfide and lanthanides could be used as parameters to predict the presence of verrucomicrobial methanotrophs in nature. Methane and hydrogen gas seem to be the main energy sources for verrucomicrobial methanotrophs. In addition, hydrogen sulfide could be a potent energy source, but additional physiological and biochemical experiments are needed to clarify this. From soil it is known that up to 90% of methane produced in these systems is consumed before it reaches the atmosphere, thereby contributing significantly to the mitigation of greenhouse gas emissions from the biosphere (Oremland and Culbertson 1992; Singh et al. 2010). To what extent verrucomicrobial methanotrophs alleviate methane emissions from geothermal environments is unknown. With the annual seepage of 40 to 60 Tg methane from mud volcanoes, fumaroles and hydrothermal vents (Etiope 2009), it would be interesting to investigate verrucomicrobial methanotrophy in the environment, especially if the use alternative energy sources such as hydrogen gas and hydrogen sulfide could enhance this process.
Supplementary Material
ACKNOWLEDGEMENTS
RS, NP and HOdC were supported by the European Research Council (ERC Advanced Grant project VOLCANO 669371). MSMJ was supported by the European Research Council (ERC Advanced Grant project EcoMoM 339880). WV was supported by the Netherlands Organisation for Scientific Research (NWO) grant VI.Vidi.192.001. SP was supported by the Netherlands Organisation for Scientific Research (NWO) grant ALWOP.308.
Contributor Information
Rob A Schmitz, Department of Microbiology, Institute for Water and Wetland Research, Radboud University, Heyendaalseweg 135, 6525 AJ, Nijmegen, The Netherlands.
Stijn H Peeters, Department of Microbiology, Institute for Water and Wetland Research, Radboud University, Heyendaalseweg 135, 6525 AJ, Nijmegen, The Netherlands.
Wouter Versantvoort, Department of Microbiology, Institute for Water and Wetland Research, Radboud University, Heyendaalseweg 135, 6525 AJ, Nijmegen, The Netherlands.
Nunzia Picone, Department of Microbiology, Institute for Water and Wetland Research, Radboud University, Heyendaalseweg 135, 6525 AJ, Nijmegen, The Netherlands.
Arjan Pol, Department of Microbiology, Institute for Water and Wetland Research, Radboud University, Heyendaalseweg 135, 6525 AJ, Nijmegen, The Netherlands.
Mike S M Jetten, Department of Microbiology, Institute for Water and Wetland Research, Radboud University, Heyendaalseweg 135, 6525 AJ, Nijmegen, The Netherlands.
Huub J M Op den Camp, Department of Microbiology, Institute for Water and Wetland Research, Radboud University, Heyendaalseweg 135, 6525 AJ, Nijmegen, The Netherlands.
Conflicts of interest
None declared.
REFERENCES
- Acton SD, Baggs EM. Interactions between N application rate, CH4 oxidation and N2O production in soil. Biogeochemistry. 2011;103:15–26. [Google Scholar]
- Allan W, Struthers H, Lowe DC. Methane carbon isotope effects caused by atomic chlorine in the marine boundary layer: Global model results compared with Southern Hemisphere measurements. J Geophys Res. 2007;112:D04306. [Google Scholar]
- Angel R, Claus P, Conrad R. Methanogenic archaea are globally ubiquitous in aerated soils and become active under wet anoxic conditions. ISME J. 2012;6:847–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony C. The c-type cytochromes of methylotrophic bacteria. Biochim Biophys Acta. 1992;1099:1–15. [PubMed] [Google Scholar]
- Anthony C. The quinoprotein dehydrogenases for methanol and glucose. Arch Biochem Biophys. 2004;428:2–9. [DOI] [PubMed] [Google Scholar]
- Anvar SY, Frank J, Pol Aet al. The genomic landscape of the verrucomicrobial methanotroph Methylacidiphilum fumariolicum SolV. BMC Genomics. 2014;15:914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragno M. Thermophilic, aerobic, hydrogen-oxidizing (Knallgas) bacteria. In: Balows A, Trüper HG, Dworkin M, Harder W, Schleifer KH (eds). The Prokaryotes. New York: Springer, 1992, 3917–33. [Google Scholar]
- Arakawa C, Kuratsu M, Furihata Ket al. Diversity of the early step of the futasoline pathway. Antimicrob Agents Chemother. 2010;55:913–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Fox GE, Magrum LJet al. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes RO, Goldberg ED. Methane production and consumption in anoxic marine sediments. Geology. 1976;4:297–300. [Google Scholar]
- Bastviken D, Cole JJ, Pace MLet al. Fates of methane from different lake habitats: Connecting whole-lake budgets and CH4 emissions. J Geophys Res Biogeo. 2008;113:G02024. [Google Scholar]
- Baxter NJ, Hirt RP, Bodrossy Let al. The ribulose-1,5-bisphosphate carboxylase/oxygenase gene cluster of Methylococcus capsulatus (Bath). Arch Microbiol. 2002;177:279–89. [DOI] [PubMed] [Google Scholar]
- Beal EJ, House CH, Orphan VJ. Manganese- and iron-dependent marine methane oxidation. Science. 2009;325:184–7. [DOI] [PubMed] [Google Scholar]
- Benson CA, Bizzoco RW, Lipson DAet al. Microbial diversity in nonsulfur, sulfur and iron geothermal steam vents. FEMS Microbiol Ecol. 2011;76:74–88. [DOI] [PubMed] [Google Scholar]
- Berghuis BA, Yu FB, Schulz Fet al. Hydrogenotrophic methanogenesis in archaeal phylum Verstraetearchaeota reveals the shared ancestry of all methanogens. Proc Natl Acad Sci USA. 2019;116:5037–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermingham A, Derrick JP. The folic acid biosynthesis pathway in bacteria: evaluation of potential for antibacterial drug discovery. Bioessays. 2002;24:637–48. [DOI] [PubMed] [Google Scholar]
- Berney M, Greening C, Hards Ket al. Three different [NiFe] hydrogenases confer metabolic flexibility in the obligate aerobe Mycobacterium smegmatis. Environ Microbiol. 2014;16:318–30. [DOI] [PubMed] [Google Scholar]
- Berrisford JM, Baradaran R, Sazanov LA. Structure of bacterial respiratory complex I. Biochim Biophys Acta. 2016;1857:892–901. [DOI] [PubMed] [Google Scholar]
- Bižić M, Klintzsch T, Ionescu Det al. Aquatic and terrestrial cyanobacteria produce methane. Sci Adv. 2020;6:eaax5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodelier PLE, Gillisen MJ, Hordijk Ket al. A reanalysis of phospholipid fatty acids as ecological biomarkers for methanotrophic bacteria. ISME J. 2009;3:606–17. [DOI] [PubMed] [Google Scholar]
- Bodelier PLE, Pérez G, Veraart AJet al. Methanotroph ecology, environmental distribution and functioning. In: Lee E (ed). Methanotrophs: Microbiology Fundamentals and Biotechnological Applications. Cham: Springer, 2019, 1–38. [Google Scholar]
- Bodelier PLE, Steenbergh AK. Interactions between methane and the nitrogen cycle in light of climate change. Curr Opin Environ Sustain. 2014;10:26–36. [Google Scholar]
- Boden R, Cunliffe M, Scanlan Jet al. Complete genome sequence of the aerobic marine methanotroph Methylomonas methanica MC09. J Bacteriol. 2011;193:7001–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodrossy L, Kovács KL, McDonald IRet al. A novel thermophilic methane-oxidising γ-Proteobacterium. FEMS Microbiol Lett. 1999;170:335–41. [Google Scholar]
- Boetius A, Ravenschlag K, Schubert CJet al. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature. 2000;407:623–6. [DOI] [PubMed] [Google Scholar]
- Bowman JP, Sly LI, Nichols PDet al. Revised taxonomy of the methanotrophs: description of Methylobacter gen. nov., emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the group I methanotrophs. Int J Syst Bacteriol. 1993;43:735–3. [Google Scholar]
- Bridgham SD, Cadillo-Quiroz H, Keller JKet al. Methane emissions from wetlands: biogeochemical, microbial, and modeling perspectives from local to global scales. Glob Chang Biol. 2013;19:1325–46. [DOI] [PubMed] [Google Scholar]
- Brune A, Frenzel P, Cypionka H. Life at the oxic-anoxic interface: microbial activities and adaptations. FEMS Microbiol Rev. 2000;24:691–710. [DOI] [PubMed] [Google Scholar]
- Brune A. Methanogens in the digestive tract of termites. In: Hackstein J (ed). (Endo)symbiotic Methanogenic Archaea. Heidelberg: Springer, 2010, 81–100. [Google Scholar]
- Buffett BA. Clathrate Hydrates. Annu Rev Earth Planet Sci. 2000;28:477–507. [Google Scholar]
- Bédard C, Knowles R. Physiology, biochemistry, and specific inhibitors of CH4, NH4+, and CO oxidation by methanotrophs and nitrifiers. Microbiol Rev. 1989;53:68–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börjesson G. Inhibition of methane oxidation by volatile sulfur compounds (CH3SH and CS2) in landfill cover soils. Waste Manag Res. 2001;19:314–9. [DOI] [PubMed] [Google Scholar]
- Cai C, Leu AO, Xie G-Jet al. A methanotrophic archaeon couples anaerobic oxidation of methane to Fe(III) reduction. ISME J. 2018;12:1929–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Zheng Y, Bodelier PLEet al. Conventional methanotrophs are responsible for atmospheric methane oxidation in paddy soils. Nat Comm. 2016;7:11728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell MA, Nyerges G, Kozlowski JAet al. Model of the molecular basis for hydroxylamine oxidation and nitrous oxide production in methanotrophic bacteria. FEMS Microbiol Let. 2011;322:82–9. [DOI] [PubMed] [Google Scholar]
- Caranto JD, Lancaster KM. Nitric oxide is an obligate bacterial nitrification intermediate produced by hydroxylamine oxidoreductase. Proc Natl Acad Sci USA. 2017;114:8217–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carere CR, Hards K, Houghton KMet al. Mixotrophy drives niche expansion of verrucomicrobial methanotrophs. ISME J. 2017;11:2599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson KM, Gerber JS, Mueller NDet al. Greenhouse gas emissions intensity of global croplands. Nat Clim Change. 2017;7:63–8. [Google Scholar]
- Castaldi S, Tedesco D. Methane production and consumption in an active volcanic environment of Southern Italy. Chemosphere. 2005;58:131–9. [DOI] [PubMed] [Google Scholar]
- Cecchini G, Schröder I, Gunsalus RPet al. Succinate dehydrogenase and fumarate reductase from Escherichia coli. Biochim Biophys Acta. 2002;1553:140–57. [DOI] [PubMed] [Google Scholar]
- Chang J, Peng S, Ciais Pet al. Revisiting enteric methane emissions from domestic ruminants and their δ13CCH4 source signature. Nat Comm. 2019;10:4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Crombie A, Rahman MTet al. Complete genome sequence of the aerobic facultative methanotroph Methylocella silvestris BL2. J Bacteriol. 2010;192:3840–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiodini G, Frondini F, Cardellini Cet al. CO2 degassing and energy release at Solfatara volcano, Campi Flegrei, Italy. J Geophys Res Solid Earth. 2001;106:16213–21. [Google Scholar]
- Chistoserdova L, Kalyuzhnaya MG, Lidstrom ME. The expanding world of methylotrophic metabolism. Annu Rev Microbiol. 2009;63:477–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L, Kalyuzhnaya MG. Current trends in methylotrophy. Trends Microbiol. 2018;26:703–14. [DOI] [PubMed] [Google Scholar]
- Chistoserdova L. Modularity of methylotrophy, revisited. Environ Microbiol. 2011;13:2603–22. [DOI] [PubMed] [Google Scholar]
- Cioni R, Corazza E, Marini L. The gas/steam ratio as indicator of heat transfer at the Solfatara fumaroles, Phlegraean Fields (Italy). Bull Vulcanol. 1984;47:295–302. [Google Scholar]
- Cobo-Simón M, Tamames J. Relating genomic characteristics to environmental preferences and ubiquity in different microbial taxa. BMC Genomics. 2017;18:499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R, Seiler W. Field measurements of hydrogen evolution by nitrogen-fixing legumes. Soil Biol Biochem. 1979;11:689–90. [Google Scholar]
- Conrad R. Methane production in soil environments – anaerobic biogeochemistry and microbial life between flooding and desiccation. Microorganisms. 2020;8:881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad R. The global methane cycle: recent advances in understanding the microbial processes involved. Environ Microbiol Rep. 2009;1:285–92. [DOI] [PubMed] [Google Scholar]
- Constant P, Chowdhury SP, Hesse Let al. Genome data mining and soil survey for the novel group 5 [NiFe]-hydrogenase to explore the diversity and ecological importance of presumptive high-affinity H2-oxidizing bacteria. Appl Environ Microbiol. 2011;77:6027–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constant P, Chowdhury SP, Pratscher Jet al. Streptomycetes contributing to atmospheric molecular hydrogen soil uptake are widespread and encode a putative high-affinity [NiFe]-hydrogenase. Environ Microbiol. 2010;12:821–9. [DOI] [PubMed] [Google Scholar]
- Constant P, Poissant L, Villemur R. Isolation of Streptomyces sp. PCB7, the first microorganism demonstrating high-affinity uptake of tropospheric H2. ISME J. 2008;2:1066–76. [DOI] [PubMed] [Google Scholar]
- Cremers G, Pol A, Jetten MSMet al. Draft genome sequences of two acidophilic, mesophilic verrucomicrobial methanotrophs contain only one pmoCAB operon. Microbiol Res Announc. 2020;9:e00315–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crognale S, Venturi S, Tassi Fet al. Microbiome profiling in extremely acidic soils affected by hydrothermal fluids: the case of the Solfatara Crater (Campi Flegrei, southern Italy). FEMS Microbiol Ecol. 2018;94:fiy90. [DOI] [PubMed] [Google Scholar]
- Crowther GJ, Kosály G, Lidstrom ME. Formate as the main branch point for methylotrophic metabolism in Methylobacterium extorquens AM1. J Bacteriol. 2008;190:5057–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csáki R, Hanczár T, Bodrossy Let al. Molecular characterization of structural genes coding for a membrane bound hydrogenase in Methylococcus capsulatus (Bath). FEMS Microbiol Lett. 2001;205:203–7. [DOI] [PubMed] [Google Scholar]
- D'Alessandro W, Bellomo S, Brusca Let al. Hydrothermal methane fluxes from the soil at Pantelleria island (Italy). J Volcanol Geotherm Res. 2009;187:147–57. [Google Scholar]
- Daumann LJ. Essential and ubiquitous: The emergence of lanthanide metallobiochemistry. Angew Chem Int Ed. 2019;58:12795–802. [DOI] [PubMed] [Google Scholar]
- Davies SL, Whittenbury R. Fine structure of methane and other hydrocarbon-utilizing bacteria. J Gen Microbiol. 1970;61:227–32. [DOI] [PubMed] [Google Scholar]
- Dean JF, Middelburg JJ, Röckmann Tet al. Methane feedbacks to the global climate system in a warmer world. Rev Geophys. 2018;56:207–50. [Google Scholar]
- De Bont JAM. Nitrogen fixation by methane-utilizing bacteria. Antonie Van Leeuwenhoek. 1976;42:245–53. [DOI] [PubMed] [Google Scholar]
- Dedysh SN, Panikov NS, Liesack Wet al. Isolation of acidophilic methane-oxidizing bacteria from northern peat wetlands. Science. 1998;282:281–4. [DOI] [PubMed] [Google Scholar]
- Deng YW, Ro SY, Rosenzweig AC. Structure and function of the lanthanide-dependent methanol dehydrogenase XoxF from the methanotroph Methylomicrobium buryatense 5GB1C. J Biol Inorg Chem. 2018;23:1037–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R, Kahn D. Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol. 2004;2:621–31. [DOI] [PubMed] [Google Scholar]
- Duine JA, Frank J Jr.. Studies on methanol dehydrogenase from Hyphomicrobium X. Isolation of an oxidized form of the enzyme. Biochem J. 1980;187:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunfield PF, Dedysh SN. Acidic environments. In: Timmis KN (ed). Handbook of Hydrocarbon and Lipid Microbiology, Berlin, Heidelberg: Springer, 2010, 2181–92. [Google Scholar]
- Dunfield PF, Dedysh SN. Methylocella: a gourmand among methanotrophs. Trends Microbiol. 2014;22:368–9. [DOI] [PubMed] [Google Scholar]
- Dunfield PF, Yuryev A, Senin Pet al. Methane oxidation by an extremely acidophilic bacterium of the phylum Verrucomicrobia. Nature. 2007;450:879–82. [DOI] [PubMed] [Google Scholar]
- Eadsforth TC, Maluf FV, Hunter WN. Acinetobacter baumannii FolD ligand complexes - potent inhibitors of folate metabolism and a re-evaluation of the structure of LY374571. FEBS J. 2012;279:4350–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. [DOI] [PubMed] [Google Scholar]
- Erikstad H-A, Ceballos RM, Smestad NBet al. Global biogeographic distribution patterns of thermoacidophilic Verrucomicrobia methanotrophs suggest allopatric evolution. Front Microbiol. 2019;10:1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge DM, Steele LP, Francey RJet al. Atmospheric methane between 1000 A.D. and present: Evidence of anthropogenic emissions and climatic variability. J Geophys Res. 1998;103:15979–93. [Google Scholar]
- Etiope G, Klusman RW. Geologic emissions of methane to the atmosphere. Chemosphere. 2002;49:777–89. [DOI] [PubMed] [Google Scholar]
- Etiope G. Natural emissions of methane from geological seepage in Europe. Atmos Environ. 2009;43:1430–43. [Google Scholar]
- Etminan M, Myhre G, Highwood EJet al. Radiative forcing of carbon dioxide, methane, and nitrous oxide: A significant revision of the methane radiative forcing. Geophys Res Lett. 2016;43:12614–23. [Google Scholar]
- Ettwig KF, Butler MK, Le Paslier Det al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–8. [DOI] [PubMed] [Google Scholar]
- Ettwig KF, Shima S, van de Pas-Schoonen KTet al. Denitrifying bacteria anaerobically oxidize methane in the absence of Archaea. Environ Microbiol. 2008;10:3164–73. [DOI] [PubMed] [Google Scholar]
- Ettwig KF, Speth DR, Reimann Jet al. Bacterial oxygen production in the dark. Front Microbiol. 2012;3:273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettwig KF, Zhu B, Speth Det al. Archaea catalyze iron-dependent anaerobic oxidation of methane. Proc Natl Acad Sci USA. 2016;113:12792–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PN, Boyd JA, Leu AOet al. An evolving view of methane metabolism in the Archaea. Nat Rev Microbiol. 2019;17:219–32. [DOI] [PubMed] [Google Scholar]
- Eyice Ö, Myronova N, Pol Aet al. Bacterial SBP56 identified as a Cu-dependent methanethiol oxidase widely distributed in the biosphere. ISME J. 2018;12:145–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyzullayev AA, Movsumova UA. The nature of the isotopically heavy carbon of carbon dioxide and bicarbonates in the waters of mud volcanoes in Azerbaijan. Geochem Int. 2010;48:517–22. [Google Scholar]
- Fiebig J, Woodland AB, Spangenberg Jet al. Natural evidence for rapid abiogenic hydrothermal generation of CH4. Geochim Cosmochim Acta. 2007;71:3028–39. [Google Scholar]
- Friedrich T, Scheide D. The respiratory complex I of bacteria, archaea and eukarya and its module common with membrane-bound multisubunit hydrogenases. FEBS Lett. 2000;479:1–5. [DOI] [PubMed] [Google Scholar]
- Gagliano AL, D'Alessandro W, Tagliavia Met al. Methanotrophic activity and diversity of methanotrophs in volcanic geothermal soils at Pantelleria (Italy). Biogeosciences. 2014;11:5865–75. [Google Scholar]
- Gao Y, Meyer B, Sokolova Let al. Heme-copper terminal oxidase using both cytochrome c and ubiquinol as electron donors. Proc Natl Acad Sci USA. 2012;109:3275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Horsman JA, Barquera B, Rumbley Jet al. The superfamily of heme-copper repiratory oxidases. J Bacteriol. 1994;176:5587–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giggenbach WF. Variations in the chemical and isotopic composition of fluids discharged from the Taupo Volcanic Zone, New Zealand. J Volcanol Geotherm Res. 1995;68:89–116. [Google Scholar]
- Good NM, Fellner M, Demirer Ket al. Lanthanide-dependent alcohol dehydrogenases require an essential aspartate residue for metal coordination and enzymatic function. J Biol Chem. 2020;295:8272–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good NM, Moore RS, Suriano CJet al. Contrasting in vitro and in vivo methanol oxidation activities of lanthanide-dependent alcohol dehydrogenases XoxF1 and ExaF from Methylobacterium extorquens AM1. Sci Rep. 2019;9:4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greening C, Biswas A, Carere CRet al. Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival. ISME J. 2015;10:761–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakobyan A, Zhu J, Glatter T. Hydrogen utilization by Methylocystis sp. strain SC2 expands the known metabolic versatility of type IIa methanotrophs. Metab Eng. 2020;61:181–96. [DOI] [PubMed] [Google Scholar]
- Hanaki K, Portugal-Pereira J. The effect of biofuel production on greenhouse gas emission reductions. In: Takeuchi K, Shiroyama H, Saito O, Matsuura M. (eds). Biofuels and Sustainability. Tokyo: Springer, 2018, 53–71. [Google Scholar]
- Hanczár T, Csáki R, Bodrossy Let al. Detection and localization of two hydrogenases in Methylococcus capsulatus (Bath) and their potential role in methane metabolism. Arch Microbiol. 2002;177:167–72. [DOI] [PubMed] [Google Scholar]
- Hanson RS, Hanson TE. Methanotrophic bacteria. Microbiol Rev. 1996;60:439–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao WM, Ward DE. Methane production from global biomass burning. J Geophys Res. 1993;98:20657–61. [Google Scholar]
- Haroon MF, Hu S, Shi Yet al. Anaerobic oxidation of methane coupled to nitrate reduction in a novel archaeal lineage. Nature. 2013;500:567–70. [DOI] [PubMed] [Google Scholar]
- Heede R. Tracing anthropogenic carbon dioxide and methane emissions to fossil fuel and cement producers, 1854–2010. Clim Change. 2014;122:229–41. [Google Scholar]
- He H, Noor E, Ramos-Parra PAet al. In vivo rate of formaldehyde condensation with tetrahydrofolate. Metabolites. 2020;10:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z, Cai C, Wang Jet al. A novel denitrifying methanotroph of the NC10 phylum and its microcolony. Sci Rep. 2016;6:32241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsuka T, Furihata K, Ishikawa Jet al. An alternative menaquinone biosynthetic pathway operating in microorganisms. Science. 2008;321:1670–3. [DOI] [PubMed] [Google Scholar]
- Hirayama H, Suzuki Y, Abe Met al. Methylothermus subterraneus sp. nov., a moderately thermophilic methanotroph isolated from a terrestrial subsurface hot aquifer. Int J Syst Evol Microbiol. 2011;61:2646–53. [DOI] [PubMed] [Google Scholar]
- Hoefman S, van der Ha D, Boon N. Niche differentiation in nitrogen metabolism among methanotrophs within an operational taxonomic unit. BMC Microbiol. 2014;14:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogendoorn C, Pol A, Nuijten GHLet al. Methanol production by “Methylacidiphilum fumariolicum” SolV under different growth conditions. Appl Environ Microbiol. 2020;86:e01188–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogendoorn C. Autotrophic microorganisms in acidic volcanic ecosystems. [dissertation]. Nijmegen, The Netherlands: Radboud University Nijmegen; 2020. [Google Scholar]
- Holloway JM, Nordstrom DK, Böhlke JKet al. Ammonium in thermal waters of Yellowstone National Park: Processes affecting speciation and isotope fractionation. Geochim Cosmochim Acta. 2011;75:4611–36. [Google Scholar]
- Holmes AJ, Roslev P, McDonald IRet al. Characterization of methanotrophic bacterial populations in soils showing atmospheric methane uptake. Appl Environ Microbiol. 1999;65:3312–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houghton KM, Carere CR, Stott MBet al. Thermophilic methanotrophs: In hot pursuit. FEMS Microbiol Ecol. 2019;95:fiz125. [DOI] [PubMed] [Google Scholar]
- Hou S, Makarova KS, Saw JHWet al. Complete genome sequence of the extremely acidophilic methanotroph isolate V4, Methylacidiphilum infernorum, a representative of the bacterial phylum Verrucomicrobia. Biol Direct. 2008;3:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MN. Chemistry of nitric oxide and related species. Methods Enzymol. 2008;436:3–19. [DOI] [PubMed] [Google Scholar]
- Islam T, Jensen S, Reigstad LJ. et al. Methane oxidation at 55°C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc Natl Acad Sci USA. 2008;105:300–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen N, Jorgensen BB. Anaerobic methane oxidation rates at the sulfate-methane transition in marine sediments from Kattegat and Skagerrak (Denmark). Limnol Oceanogr. 1985;30:944–55. [Google Scholar]
- Jahn B, Pol A, Lumpe Het al. Similar but not the same: First kinetic and structural analyses of a methanol dehydrogenase containing a europium ion in the active site. ChemBioChem. 2018;19:1147–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen BB. Methanogenesis in monogastric animals. Environ Monit Assess. 1996;42:99–112. [DOI] [PubMed] [Google Scholar]
- Kallen RG, Jencks WP. The mechanism of the condensation of formaldehyde with tetrahydrofolic acid. J Biol Chem. 1966;241:5851–63. [PubMed] [Google Scholar]
- Kalyuzhnaya MG, Puri AW, Lidstrom ME. Metabolic engineering in methanotrophic bacteria. Metab Eng. 2015;29:142–52. [DOI] [PubMed] [Google Scholar]
- Kaplan NO, Ciotti MM. Competitive inhibition of hydroxylamine on alcohol dehydrogenase. J Biol Chem. 1953;201:785–94. [PubMed] [Google Scholar]
- Kaserer H. Über die Oxidation des Wasserstoffes und des Methans durch Mikroorganismen. Z landw Versuchsw in Österreich. 1905;8:789–92. [Google Scholar]
- Kato S, Takashino M, Igarashi Ket al. Isolation and genomic characterization of a proteobacterial methanotroph requiring lanthanides. Microbes Environ. 2020;35:ME19128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly DP, Anthony C, Murrell JC. Insights into the obligate methanotroph Methylococcus capsulatus. Trends Microbiol. 2005;13:195–8. [DOI] [PubMed] [Google Scholar]
- Keltjens JT, Pol A, Reimann Jet al. PQQ-dependent methanol dehydrogenases: rare-earth elements make a difference. Appl Microbiol Biotechnol. 2014;98:6163–83. [DOI] [PubMed] [Google Scholar]
- Khadem AF, Pol A, Jetten MSMet al. Nitrogen fixation by the verrucomicrobial methanotroph “Methylacidiphilum fumariolicum” SolV. Microbiology. 2010;156:1052–9. [DOI] [PubMed] [Google Scholar]
- Khadem AF, Pol A, Wieczorek Aet al. Autotrophic methanotrophy in Verrucomicrobia: Methylacidiphilum fumariolicum SolV uses the Calvin-Benson-Bassham cycle for carbon dioxide fixation. J Bacteriol. 2011;193:4438–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khadem AF, Pol A, Wieczorek ASet al. Metabolic regulation of “Ca. Methylacidiphilum fumariolicum” SolV cells grown under different nitrogen and oxygen limitations. Front Microbiol. 2012;3:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelenina VN, Murrell CJ, Smith TJet al. Physiology and Biochemistry of the Aerobic Methanotrophs. In: Rojo F (ed). Aerobic Utilization of Hydrocarbons, Oils and Lipids. Handbook of Hydrocarbon and Lipid Microbiology. Cham: Springer, 2018, 1–25. [Google Scholar]
- Kip N, Ouyang W, van Winden Jet al. Detection, isolation, and characterization of acidophilic methanotrophs from Sphagnum mosses. Appl Environ Microbiol. 2011;77:5643–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschke S, Bousquet P, Ciais Pet al. Three decades of global methane sources and sinks. Nature Geosci. 2013;6:813–23. [Google Scholar]
- Kits KD, Klotz MG, Stein LY. Methane oxidation coupled to nitrate reduction under hypoxia by the Gammaproteobacterium Methylomonas denitrificans, sp. nov. type strain FJG1. Environ Microbiol. 2015;17:3219–32. [DOI] [PubMed] [Google Scholar]
- Klotz MG, Stein LY. Nitrifier genomics and evolution of the nitrogen cycle. FEMS Microbiol Lett. 2008;278:146–56. [DOI] [PubMed] [Google Scholar]
- Kostka JE, Weston DJ, Glass JB. The Sphagnum microbiome: new insights from an ancient plant lineage. New Phytol. 2016;211:57–64. [DOI] [PubMed] [Google Scholar]
- Kox MAR, Aalto SL, Penttilä Tet al. The influence of oxygen and methane on nitrogen fixation in subarctic Sphagnum mosses. AMB Express. 2018;8:76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T, Ratnadevi CM, Erikstad H-Aet al. Complete genome sequence analysis of the thermoacidophilic verrucomicrobial methanotroph “Candidatus Methylacidiphilum kamchatkense” strain Kam1 and comparison with its closest relatives. BMC Genomics. 2019;20:642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosu M, Begari E. Vitamine K2 in electron transport system: Are enzymes involved in vitamin K2 biosynthesis promising drug targets?. Molecules. 2010;15:1531–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvenvolden KA, Rogers BW. Gaia's breath—global methane exhalations. Mar Pet Geol. 2005;22:579–90. [Google Scholar]
- La H, Hettiaratchi JPA, Achari Get al. Biofiltration of methane. Bioresour Technol. 2018;268:759–72. [DOI] [PubMed] [Google Scholar]
- Lee E-H, Moon K-E, Kim TGet al. Inhibitory effects of sulfur compounds on methane oxidation by a methane-oxidizing consortium. J Biosci Bioeng. 2015;120:670–6. [DOI] [PubMed] [Google Scholar]
- Lee J-H, Kim TG, Cho K-S. Isolation and characterization of a facultative methanotroph degrading malodor-causing volatile sulfur compounds. J Hazard Mater. 2012;235-236:224–9. [DOI] [PubMed] [Google Scholar]
- Lees V, Owens NP, Murrell JC. Nitrogen metabolism in marine methanotrophs. Arch Microbiol. 1991;157:60–5. [Google Scholar]
- Lehtovirta-Morley LE. Ammonia oxidation: Ecology, physiology, biochemistry and why they must all come together. FEMS Microbiol Lett. 2018;365:fny058. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–8. [DOI] [PubMed] [Google Scholar]
- Leu AO, Cai C, McIlroy SJet al. Anaerobic methane oxidation coupled to manganese reduction by members of the Methanoperedenaceae. ISME J. 2020;14:1030–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levicán G, Ugalde JA, Ehrenfeld Net al. Comparative genomic analysis of carbon and nitrogen assimilation mechanisms in three indigenous bioleaching bacteria: predictions and validations. BMC Genomics. 2008;9:581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman RL, Rosenzweig AC. Crystal structure of a membrane-bound metalloenzyme that catalyses the biological oxidation of methane. Nature. 2005;434:177–82. [DOI] [PubMed] [Google Scholar]
- Lim S, Franklin SJ. Lanthanide-binding peptides and the enzymes that Might Have Been. Cell Mol Life Sci. 2004;61:2184–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay MR, Colman DR, Amenabar MJet al. Probing the geological source and biological fate of hydrogen in Yellowstone hot springs. Environ Microbiol. 2019;21:3816–30. [DOI] [PubMed] [Google Scholar]
- Lomans BP, Pol A, Op den Camp HJM. Microbial cycling of volatile organic sulfur compounds. Cell Mol Life Sci. 2002;59:575–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubitz W, Ogata H, Rüdiger Oet al. Hydrogenases. Chem Rev. 2014;114:4081–148. [DOI] [PubMed] [Google Scholar]
- Marcia M, Ermler U, Peng G. A new structure-based classification of sulfide:quinone oxidoreductases. Proteins. 2010;78:1073–83. [DOI] [PubMed] [Google Scholar]
- Marx CJ, Laukel M, Vorholt JA. Purification of the formate-tetrahydrofolate ligase from Methylobacterium extorquens AM1 and demonstration of its requirement for methylotrophic growth. J Bacteriol. 2003;185:7169–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx CJ, van Dien SJ, Lidstrom ME. Flux analysis uncovers key role of functional redundancy in formaldehyde metabolism. PLoS Biol. 2005;3:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf WW, Griffin BM, Cicchillo RMet al. Synthesis of methylphosphonic acid by marine microbes: a source for methane in the aerobic ocean. Science. 2012;337:1104–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi SS, Pol A, van Alen TAet al. Ammonia oxidation and nitrite reduction in the verrucomicrobial methanotroph Methylacidiphilum fumariolicum SolV. Front Microbiol. 2017b;8:1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi SS, Pol A, van Alen TAet al. Methylacidiphilum fumariolicum SolV, a thermoacidophilic ‘Knallgas’ methanotroph with both an oxygen-sensitive and -insensitive hydrogenase. ISME J. 2017a;11:945–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi SS, Schmitz RA, Pol Aet al. The acidophilic methanotroph Methylacidimicrobium tartarophylax 4AC grows as autotroph on H2 under microoxic conditions. Front Microbiol. 2019;10:2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Vivián C, Cabello P, Martínez-Luque Met al. Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J Bacteriol. 1999;181:6573–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murrell JC, Dalton H. Ammonia assimilation in Methylococcus capsulatus (Bath) and other obligate methanotrophs. Microbiology. 1983b;129: 1197–206. [Google Scholar]
- Murrell JC, Dalton H. Nitrogen fixation in obligate methanotrophs. Microbiology. 1983a;129:3481–86. [Google Scholar]
- Nakagawa T, Mitsui R, Tani Aet al. A catalytic role of XoxF1 as La3+-dependent methanol dehydrogenase in Methylobacterium extorquens strain AM1. PLoS One. 2012;7:e50480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novelli PC, Lang PM, Masarie KAet al. Molecular hydrogen in the troposphere: Global distribution and budget. J Geophys Res Atmos. 1999;104:30427–44. [Google Scholar]
- Nyerges G, Han SK, Stein LY. Effects of ammonium and nitrite on growth and competitive fitness of cultivated methanotrophic bacteria. Appl Environ Microbiol. 2010;76:5648–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyerges G, Stein LY. Ammonia cometabolism and product inhibition vary considerably among species of methanotrophic bacteria. FEMS Microbiol Lett. 2009;297:131–6. [DOI] [PubMed] [Google Scholar]
- Op den Camp HJM, Islam T, Stott MBet al. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ Microbiol Rep. 2009;1:293–306. [DOI] [PubMed] [Google Scholar]
- Op den Camp HJM, Mohammadi SS, Pol Aet al. Verrucomicrobial methanotrophs. In: Kalyuzhnaya MG, Xing X-H (eds). Methane biocatalysis: Paving the way to sustainability. Cham: Springer, 2018, 43–55. [Google Scholar]
- Oremland RS, Culbertson CW. Importance of methane-oxidizing bacteria in the methane budget as revealed by the use of a specific inhibitor. Nature. 1992;356:421–3. [Google Scholar]
- Pagaling E, Yang K, Yan T. Pyrosequencing reveals correlations between extremely acidophilic bacterial communities with hydrogen sulphide concentrations, pH and inert polymer coatings at concrete sewer crown surfaces. J Appl Microbiol. 2014;117:50–64. [DOI] [PubMed] [Google Scholar]
- Pawelek PD, MacKenzie RE. Methenyltetrahydrofolate cyclohydrolase is rate limiting for the enzymatic conversion of 10-formyltetrahydrofolate to 5,10-methylenetetrahydrofolate in bifunctional dehydrogenase-cyclohydrolase enzymes. Biochemistry. 1998;37:1109–15. [DOI] [PubMed] [Google Scholar]
- Pereira MM, Refojo PN, Hreggvidsson GOet al. The alternative complex III from Rhodothermus marinus – A prototype of a new family of quinol:electron acceptor oxidoreductases. FEBS Lett. 2007;581:4831–5. [DOI] [PubMed] [Google Scholar]
- Pester M, Brune A. Hydrogen is the central free intermediate during lignocellulose degradation by termite gut symbionts. ISME J. 2007;1:551–65. [DOI] [PubMed] [Google Scholar]
- Picone N, Hogendoorn C, Cremers Get al. Geothermal gases shape the microbial community of the volcanic soil of Pantelleria, Italy mSystems. 2020a;5:e00517–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picone N, Mohammadi SS, Waajen ACet al. More than a methanotroph: a broader substrate spectrum for Methylacidiphilum fumariolicum SolV. Front Microbiol. 2020b;11:604485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picone N, Op den Camp HJM. Role of rare earth elements in methanol oxidation. Curr Opin Chem Biol. 2019;49:39–44. [DOI] [PubMed] [Google Scholar]
- Picone N. Novel methane- and ammonia-oxidizing microorganisms from acidic environments. [dissertation]. Nijmegen, The Netherlands: Radboud University Nijmegen; 2020. [Google Scholar]
- Pol A, Barends TRM, Dietl Aet al. Rare earth metals are essential for methanotrophic life in volcanic mudpots. Environ Microbiol. 2014;16:255–64. [DOI] [PubMed] [Google Scholar]
- Pol A, Heijmans K, Harhangi HRet al. Methanotrophy below pH 1 by a new Verrucomicrobia species. Nature. 2007;450:874–8. [DOI] [PubMed] [Google Scholar]
- Pol A, Op den Camp HJM, Mees SGet al. Isolation of a dimethylsulfide-utilizing Hyphomicrobium species and its application in biofiltration of polluted air. Biodegradation. 1994;5:105–12. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glöckner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner S, Stadtman ER. The enzymes of glutamine metabolism. New York: Academic Press, Inc, 1973. [Google Scholar]
- Quatrini R, Johnson DB. Microbiomes in extremely acidic environments: functionalities and interactions that allow survival and growth of prokaryotes at low pH. Curr Opin Microbiol. 2018;43:139–47. [DOI] [PubMed] [Google Scholar]
- Raghoebarsing AA, Pol A, van de Pas-Schoonen KTet al. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature. 2006;440:918–21. [DOI] [PubMed] [Google Scholar]
- Raghoebarsing AA, Smolders AJ, Schmid MCet al. Methanotrophic symbionts provide carbon for photosynthesis in peat bogs. Nature. 2005;436:1153–6. [DOI] [PubMed] [Google Scholar]
- Rasigraf O, Kool DM, Jetten MSet al. Autotrophic carbon dioxide fixation via the Calvin-Benson-Bassham cycle by the denitrifying methanotroph “Candidatus Methylomirabilis oxyfera”. Appl Environ Microbiol. 2014;80:2451–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeburgh WS. Methane consumption in Cariaco Trench waters and sediments. Earth Planet Sci Lett. 1976;28:337–44. [Google Scholar]
- Reeburgh WS. Oceanic methane biogeochemistry. Chem Rev. 2007;107:486–513. [DOI] [PubMed] [Google Scholar]
- Reeves EP, McDermott JM, Seewald JSet al. The origin of methanethiol in midocean ridge hydrothermal fluids. Proc Natl Acad Sci USA. 2014;111:5474–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refojo PN, Sousa FL, Teixeira Met al. The alternative complex III: A different architecture using known building modules. Biochim Biophys Acta. 2010;1797:1869–76. [DOI] [PubMed] [Google Scholar]
- Refojo PN, Teixeira M, Pereira MM. The Alternative complex III: properties and possible mechanisms for electron transfer and energy conservation. Biochim Biophys Acta. 2012;1817:1852–9. [DOI] [PubMed] [Google Scholar]
- Rigby M, Montzka SA, Prinn RGet al. Role of atmospheric oxidation in recent methane growth. Proc Natl Acad Sci USA. 2017;114:5373–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers KL, Schulte MD. Organic sulfur metabolisms in hydrothermal environments. Geobiology. 2012;10:320–32. [DOI] [PubMed] [Google Scholar]
- Ross MO, MacMillan F, Wang Jet al. Particulate methane monooxygenase contains only mononuclear copper centers. Science. 2019;364:566–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MO, Rosenzweig AC. A tale of two methane monooxygenases. J Biol Inorg Chem. 2017;22:307–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunois M, Jackson RB, Bousquet Pet al. The growing role of methane in anthropogenic climate change. Environ Res Lett. 2016;11:120207. [Google Scholar]
- Saunois M, Stavert AR, Poulter Bet al. The global methane budget 2000–2017. Earth Syst Sci Data. 2020;12:1561–623. [Google Scholar]
- Schmitz RA, Dietl A, Müller Met al. Structure of the 4-hydroxy-tetrahydrodipicolinate synthase from the thermoacidophilic methanotroph Methylacidiphilum fumariolicum SolV and the phylogeny of the aminotransferase pathway. Acta Cryst F. 2020b;76:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz RA, Pol A, Mohammadi SSet al. The thermoacidophilic methanotroph Methylacidiphilum fumariolicum SolV oxidizes subatmospheric H2 with a high-affinity, membrane-associated [NiFe] hydrogenase. ISME J. 2020a;14:1223–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoen R, Rye RO. Sulfur isotope distribution in Solfataras, Yellowstone National Park. Science. 1970;170:1082–4. [DOI] [PubMed] [Google Scholar]
- Schäfer C, Bommer M, Hennig SEet al. Structure of an actinobacterial-type [NiFe]-hydrogenase reveals insight into O2-tolerant H2 oxidation. Structure. 2016;24:285–92. [DOI] [PubMed] [Google Scholar]
- Schäfer C, Friedrich B, Lenz O. Novel, oxygen-insensitive group 5 [NiFe]-hydrogenase in Ralstonia eutropha. Appl Environ Microbiol. 2013;79:5137–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semrau JD, DiSpirito AA, Yoon S. Methanotrophs and copper. FEMS Microbiol Rev. 2010;34:496–531. [DOI] [PubMed] [Google Scholar]
- Sharp CE, Op den Camp HJM, Tamas Iet al. Unusual members of the PVC superphylum: the methanotrophic Verrucomicrobia genus “Methylacidiphilum”. In: Fuerst J (ed). Planctomycetes: cell structure, origins and biology. Berlin: Springer-Verlag, 2013, 211–27. [Google Scholar]
- Sharp CE, Smirnova AV, Graham JMet al. Distribution and diversity of Verrucomicrobia methanotrophs in geothermal and acidic environments. Environ Microbiol. 2014;16:1867–78. [DOI] [PubMed] [Google Scholar]
- Sherwood Lollar B, Lacrampe-Couloume G, Slater GF. Unravelling abiogenic and biogenic sources of methane in the Earth's deep subsurface. Chem Geol. 2006;226:328–39. [Google Scholar]
- Siliakus MF, van der Oost J, Kengen SWM. Adaptations of archaeal and bacterial membranes to variations in temperature, pH and pressure. Extremophiles. 2017;21:651–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh BK, Bardgett RD, Smith Pet al. Microorganisms and climate change: terrestrial feedbacks and mitigation options. Nat Rev Microbiol. 2010;8:779–90. [DOI] [PubMed] [Google Scholar]
- Spiro PA, Jacob DJ, Logan JA. Global inventory of sulfur emissions with 1° × 1° resolution. J Geophys Res. 1992;97:6023–36. [Google Scholar]
- Stein LY, Klotz MG. Nitrifying and denitrifying pathways of methanotrophic bacteria. Biochem Soc Trans. 2011;39:1826–31. [DOI] [PubMed] [Google Scholar]
- Stein LY, Roy R, Dunfield PF. Aerobic methanotrophy and nitrification: processes and connections. eLS. 2012; DOI: 10.1002/9780470015902.a0022213. [DOI] [Google Scholar]
- Stein LY, Yoon S, Semrau JDet al. Genome sequence of the obligate methanotroph Methylosinus trichosporium strain OB3b. J Bacteriol. 2010;192:6497–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein LY. Proteobacterial methanotrophs, methylotrophs, and nitrogen. In: Kalyuzhnaya MG, Xing X-H (eds). Methane biocatalysis: Paving the way to sustainability. Cham: Springer, 2018, 57–66. [Google Scholar]
- Stein LY. The long-term relationship between microbial metabolism and greenhouse gases. Trends in Microbiol. 2020;28:500–11. [DOI] [PubMed] [Google Scholar]
- Stolper DA, Martini AM, Clog Met al. Distinguishing and understanding thermogenic and biogenic sources of methane using multiply substituted isotopologues. Geochim Cosmochim Acta. 2015;161:219–47. [Google Scholar]
- Strong PJ, Xie S, Clarke WP. Methane as a resource: Can the methanotrophs add value?. Environ Sci Technol. 2015;49:4001–18. [DOI] [PubMed] [Google Scholar]
- Sun C, Benlekbir S, Venkatakrishnan Pet al. Structure of the alternative complex III in a supercomplex with cytochrome oxidase. Nature. 2018;557:123–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suylen GMH, Large PJ, van Dijken JPet al. Methyl mercaptan oxidase, a key enzyme in the metabolism of methylated sulphur compounds by Hyphomicrobium EG. Microbiology. 1987;133:2989–97. [Google Scholar]
- Svenning MM, Hestnes AG, Wartiainen Iet al. Genome sequence of the Arctic methanotroph Methylobacter tundripaludum SV96. J Bacteriol. 2011;193:6418–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söhngen NL. Ueber Bakterien, welche Methan als Kohlenstoffnahrung und Energiequelle gebrauchen. Zentralbl Bakteriol B. 1906;15:513–7. [Google Scholar]
- Søndergaard D, Pedersen C, Greening C. HydDB: A web tool for hydrogenase classification and analysis. Sci Rep. 2016;6:34212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassi F, Fiebig J, Vaselli Oet al. Origins of methane discharging from volcanic-hydrothermal, geothermal and cold emissions in Italy. Chem Geol. 2012;310-311:36–48. [Google Scholar]
- Tavormina PL, Orphan VJ, Kalyuzhnaya MGet al. A novel family of functional operons encoding methane/ammonia monooxygenase-related proteins in gammaproteobacterial methanotrophs. Environ Microbiol Rep. 2011;3:91–100. [DOI] [PubMed] [Google Scholar]
- Taylor SC, Dalton H, Dow CS. Ribulose-1,5-bisphosphate carboxylase/oxygenase and carbon assimilation in Methylococcus capsulatus (Bath). Microbiology, 1981;122:89–94. [Google Scholar]
- Themelis NJ, Ulloa PA. Methane generation in landfills. Renew Energy. 2007;32:1243–57. [Google Scholar]
- Trumpower BL. Cytochrome bc1 complexes of microorganisms. Microbiol Rev. 1990;54:101–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner AJ, Frankenberg C, Kort EA. Interpreting contemporary trends in atmospheric methane. Proc Natl Acad Sci USA. 2019;116:2805–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tveit AT, Hestnes AG, Robinson SLet al. Widespread soil bacterium that oxidizes atmospheric methane. Proc Natl Acad Sci USA. 2019;116:8515–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udert KM, Larsen TA, Gujer W. Chemical nitrite oxidation in acid solutions as a consequence of microbial ammonium oxidation. Environ Sci Technol. 2005;39:4066–75. [DOI] [PubMed] [Google Scholar]
- Van Heeswijk WC, Westerhoff HV, Boogerd FC. Nitrogen assimilation in Escherichia coli: Putting molecular data into a systems perspective. Microbiol Mol Biol Rev. 2013;77:628–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Teeseling MCF, Pol A, Harhangi HRet al. Expanding the verrucomicrobial methanotrophic world: Description of three novel species of Methylacidimicrobium gen. nov. Appl Environ Microbiol. 2014;80:6782–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilakos C, Maggos T, Bartzis JGet al. Determination of atmospheric sulfur compounds near a volcanic area in Greece. J Atmos Chem. 2005;52:101–16. [Google Scholar]
- Vasileva A, Moiseenko K. Methane emissions from 2000 to 2011 wildfires in Northeast Eurasia estimated with MODIS burned area data. Atmos Environ. 2013;71:115–21. [Google Scholar]
- Venturi S, Tassi F, Magi Fet al. Carbon isotopic signature of interstitial soil gases reveals the potential role of ecosystems in mitigating geogenic greenhouse gas emissions: Case studies from hydrothermal systems in Italy. Sci Total Environ. 2019;655:887–98. [DOI] [PubMed] [Google Scholar]
- Versantvoort W, Guerrero-Cruz S, Speth DRet al. Comparative genomics of Candidatus Methylomirabilis species and description of Ca. Methylomirabilis lanthanidiphila. Front Microbiol. 2018;9:1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versantvoort W, Pol A, Daumann LJet al. Characterization of a novel cytochrome c as the electron acceptor of XoxF-MDH in the thermoacidophilic methanotroph Methylacidiphilum fumariolicum SolV. Biochim Biophys Acta Protein Proteom. 2019;1867:595–603. [DOI] [PubMed] [Google Scholar]
- Versantvoort W, Pol A, Jetten MSMet al. Multiheme hydroxylamine oxidoreductases produce NO during ammonia oxidation in methanotrophs. Proc Natl Acad Sci USA. 2020;117:24459–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodyanitskii YN, Savichev AT, Trofimov SYet al. Accumulation of heavy metals in oil-contaminated peat soils. Eurasian Soil Sci. 2012;45:977–82. [Google Scholar]
- Vorholt JA, Marx CJ, Lidstrom MEet al. Novel formaldehyde-activating enzyme in Methylobacterium extorquens AM1 required for growth on methanol. J Bacteriol. 2000;182:6645–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorholt JA. Cofactor-dependent pathways of formaldehyde oxidation in methylotrophic bacteria. Arch Microbiol. 2002;178:239–49. [DOI] [PubMed] [Google Scholar]
- Vu HN, Subuyuj GA, Vijayakumar Set al. Lanthanide-dependent regulation of methanol oxidation systems in Methylobacterium extorquens AM1 and their contribution to methanol growth. J Bacteriol. 2016;198:1250–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuilleumier S, Khmelenina VN, Bringel Fet al. Genome sequence of the haloalkaliphilic methanotrophic bacterium Methylomicrobium alcaliphilum 20Z. J Bacteriol. 2012;194:551–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Yan D, Dixon Ret al. Deciphering the principles of bacterial nitrogen dietary preferences: a strategy for nutrient containment. mBio. 2016;7:e00792–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward N, Larsen Ø, Sakwa Jet al. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2004;2:e303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittenbury R, Phillips KC, Wilkinson JF. Enrichment, isolation and some properties of methane-utilizing bacteria. Microbiology. 1970;61:205–18. [DOI] [PubMed] [Google Scholar]
- Wu ML, Wessels H, Pol Aet al. XoxF-type methanol dehydrogenase from the anaerobic methanotroph “Candidatus Methylomirabilis oxyfera”. Appl Environ Microbiol. 2015;81:1442–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanpirat P, Nakatsuji Y, Hiraga Set al. Lanthanide-dependent methanol and formaldehyde oxidation in Methylobacterium aquaticum strain 22A. Microorganisms. 2020;8:822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon K-S, Ueda Y, Ishii Met al. NADH:ferredoxin reductase and NAD-reducing hydrogenase activities in Hydrogenobacter thermophilus strain TK-6. FEMS Microbiol Lett. 1996;139:139–42. [Google Scholar]
- Zalkin H, Smith JL. Enzymes utilizing glutamine as an amide donor. Adv Enzymol Relat Areas Mol Biol. 1998;72:87–144. [DOI] [PubMed] [Google Scholar]
- Zamocky M, Furtmüller PG, Obinger C. Evolution of catalases from bacteria to humans. Antioxid Redox Signal. 2008;10:1527–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeikus JG, Ben-Bassat A, Hegge PW. Microbiology of methanogenesis in thermal, volcanic environments. J Bacteriol. 1980;143:432–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Huang J, Zhao Fet al. Physiological effect of XoxG(4) on lanthanide-dependent methanotrophy. mBio. 2018;9:e02430–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.