Abstract
Pulsed-field gel electrophoresis (PFGE) is a powerful molecular biology technique which has provided important insights into the epidemiology and population biology of many pathogens. However, few studies have used PFGE for the molecular epidemiology of Mycobacterium tuberculosis. A laboratory protocol was developed to determine the typeability, stability, and reproducibility of PFGE typing of M. tuberculosis. Formal data-analytical techniques were used to assess the genetic diversity elucidated by PFGE analyses using four separate restriction enzymes and by IS6110 RFLP analyses, as well as to assess the concordance among these typing methods. One hundred epidemiologically characterized clinical isolates of M. tuberculosis were genotyped with four different PFGE enzymes (AseI, DraI, SpeI, and XbaI), as well as by RFLP analysis with IS6110. Identical patterns were found among 34 isolates known to be genetically related, suggesting that the PFGE protocol is robust and reproducible. Among 66 isolates representing population-sampled cases, heterozygosity and information content dependency estimates indicate that all five genotyping systems capture quantitatively similar levels of genetic diversity. Nevertheless, comparisons between PFGE analyses and IS6110 typing reveals that PFGE provided more discrimination among isolates with fewer than five copies of IS6110 and less clustering in isolates with five or more copies. The comparisons confirm the hypothesis that the resolution of IS6110 RFLP genotyping is dependent upon the number of IS6110 elements in the genome of isolates. The general concordance among the results obtained with four independent enzymes suggests that M. tuberculosis is a clonal organism. The availability of a robust genotyping technique largely independent of repetitive elements has implications for the molecular epidemiology of M. tuberculosis.
Variability in the genomic copy number of the repetitive sequence IS6110 and polymorphism of the flanking PvuII restriction site has been exploited to generate strain-specific genotypes for Mycobacterium tuberculosis. The IS6110 genotyping system has been widely used in epidemiologic studies of tuberculosis (7). However, the inadequacies and complexity of restriction fragment length polymorphism (RFLP) analysis with IS6110 have created the need for secondary typing techniques. Isolates of M. tuberculosis containing few copies of IS6110 present a problem, as limited information is available from which to infer genetic relatedness (3, 6, 13, 19). There are also reports of M. tuberculosis isolates that completely lack IS6110 elements (1, 13, 19). In addition, the technical complexity of the IS6110 method has restricted its use largely to research and reference laboratories. Finally, the ability to infer genetic relatedness between isolates by using IS6110 has been limited by the lack of information on the variability in the rates of biologic events that change IS6110 RFLP patterns (15).
Pulsed-field gel electrophoresis (PFGE) has been widely used to type various microorganisms in both outbreak and population-based studies and is available in many clinical laboratories (16). However, to date, PFGE has not been commonly employed in epidemiological investigations of M. tuberculosis. Most published PFGE protocols for M. tuberculosis are technically challenging. Biosafety considerations and the unique cell wall composition of the organism have led to the development of protocols that are highly complex and difficult to reproduce. Little has been done to develop standardized methods for analyses of M. tuberculosis PFGE patterns. The lack of standardized methods to generate, store, and compare PFGE patterns has also limited the use of PFGE for population-based molecular epidemiologic studies of M. tuberculosis. Finally, previous investigators using PFGE to type M. tuberculosis have presented contradictory reports on the genetic diversity captured by PFGE and on its utility for molecular epidemiology (5, 11, 20).
In this study, we applied the protocol for PFGE described in this report to a rationally selected panel of well-characterized M. tuberculosis isolates. In addition, we compared the typeability, reproducibility, discriminatory power, and ease of interpretation of PFGE with those of IS6110 RFLP. Our results indicate that PFGE is a valid approach to the genotyping of M. tuberculosis that also provides insight into the biology of M. tuberculosis.
MATERIALS AND METHODS
Organisms.
All 112 isolates of M. tuberculosis in the study were selected from an ongoing molecular epidemiology project in San Francisco (9). Of the 112 isolates, 34 were known to be genetically related. They consist of 10 isolates from a laboratory cross-contamination episode, 10 isolates from five patients who persistently secreted M. tuberculosis, and 14 isolates subcultured to represent growth from a single cell of one clinical isolate. The remaining 78 isolates represent available viable cultures from all tuberculosis patients reported to the San Francisco Department of Public Health between 25 December 1995 and 10 June 1996. Routine demographic information such as age, sex, race or ethnicity, country of birth, and number of years in the United States were compared among these 78 patients and all of the microbiologically confirmed cases during this time period.
Preparation of organisms for PFGE.
Each isolate was grown in 15 ml of 7H9 liquid medium supplemented with 10% ADC (Difco Laboratories, Detroit, Mich.) for 2 to 3 weeks at 37°C in 5% CO2. The cells were harvested by centrifugation and inactivated in 70% ethanol for 1 h at 25°C (18). Inactivated cells were washed with sterile Tris-EDTA (TE) buffer (Bio-Rad Laboratories, Hercules, Calif.) and centrifuged, and pellets were frozen at −20°C until used for sample preparation. The 70% ethanol and the TE buffer were poured out without the use of a vacuum. Each inactivated cell pellet was air dried at 25°C, weighed, and resuspended in 150 μl of GenePath Cell Suspension Buffer (Bio-Rad Laboratories) per 10 mg of cells. The mean weight of the cell pellet from a 15-ml liquid culture was 43.6 mg with a standard deviation of 14.6 (n = 102). The resuspended cells were vortexed with 3-mm sterile glass beads for 2 min and then allowed to settle for 2 min. A 150-μl aliquot of the suspension was freeze-thawed three times and then added to an equal volume of 1.2% agarose (Bio-Rad Laboratories) equilibrated at 60°C. The agarose-cell suspension mixture was poured into disposable plug molds (Bio-Rad Laboratories). The plugs were incubated in 1.5 ml of 10-mg/ml lysozyme (Sigma Chemical Co., St. Louis, Mo.) in TE buffer for at least 12 h at 37°C in 2-ml microcentrifuge tubes. They were washed in 1.5 ml of 0.1× GenePath Wash Buffer (Bio-Rad Laboratories) twice for 30 min each time. The plugs were further incubated in 1.5 ml of GenePath Proteinase K Buffer (Bio-Rad Laboratories) containing 1-mg/ml proteinase K for 12 to 16 h at 55°C. The proteinase K was removed from the plugs with incubations in 1.5 ml of 1× GenePath Wash Buffer without agitation at least five times for each of the following durations: 10 min, 30 min, and 1 h. The plugs were then incubated in 1.5 ml of 0.1× GenePath Wash Buffer for at least 30 min. The washed plugs were stored at 4°C until used for PFGE analysis.
Restriction endonuclease digestion.
The restriction enzymes (REs) used for PFGE sample preparation include AseI and XbaI (New England Biolabs, Beverly, Mass.), DraI (Boehringer Mannheim, Indianapolis, Ind.), and SpeI (Bio-Rad Laboratories). For digests using AseI, XbaI, and SpeI, a 1.5-mm section of each plug was incubated in 150 μl of the appropriate RE buffer for at least 30 min. Each plug section was then resuspended in 100 μl of RE buffer containing 25 U of SpeI or XbaI or 40 U of AseI and incubated for 12 to 16 h at 37°C. For DraI digests, the 1.5-mm plug section was incubated twice in 500 μl of DraI buffer for 1 h and then resuspended in 100 μl of DraI buffer containing 40 U of DraI and incubated for 12 h at 37°C. Another 100 μl of buffer with 40 U of DraI was added, and the mixture was incubated for another 6 h at 37°C.
PFGE.
Plug sections were loaded in gels made from GenePath Gel Kits (Bio-Rad Laboratories). PFGE was conducted by using a CHEF-DRIII (Bio-Rad Laboratories) for samples digested with DraI and SpeI, a GenePath System with Open Channel version 2.0 software (Bio-Rad Laboratories) for samples digested with AseI, or a CHEF-DRII (Bio-Rad Laboratories) for samples digested with XbaI in 1× GenePath Running Buffer (Bio-Rad Laboratories) cooled to 14°C. For all separations, instruments were set at 6 V/cm (200 V) and a 120° angle. For the separation of DraI digests, pulse times of 3 to 15.8 s for 11 h, 15.8 to 33.4 s for 6.4 h, and 33.4 to 63.8 s for 4.6 h were used. For the separation of AseI digests, pulse times of 1 to 5.6 s for 10 h, 5.6 to 16.6 s for 6 h, and 16.6 to 23 s for 4 h were used. For the separation of SpeI digests, pulse times of 0.5 to 4 s for 10 h, 4 to 12.2 s for 6 h, and 12.2 to 17 s for 4 h were used. For the separation of XbaI, pulse times of 0.5 to 10 s for 16 h and 10 to 17 s for 4 h were used.
Following electrophoresis, the gels were stained with ethidium bromide and imaged with a Gel Doc 1000 Documentation System (Bio-Rad Laboratories).
PFGE pattern analysis.
A TIFF image of each gel was exported to the Bio Image Whole Band Analyzer (Bio-Image, Ann Arbor, Mich.) for analysis. Molecular weight standards and reference strains were included on each gel to facilitate intergel comparisons. For molecular weight assignments, the two outermost and center lanes of each gel included a SmaI digest of Staphylococcus aureus NCTC 8325 (fragment sizes: 674, 361, 324, 262, 257, 208, 175, 135, 117, 80, 76, 44, 36, 19, 16, and 9 kbp; Bio-Rad Laboratories). The reference strain, M. tuberculosis H37Rv (San Francisco Public Health Department), was run in three random lanes on each gel.
M. tuberculosis H37Rv was run on multiple occasions, and the patterns were compared to determine the amount of variation in fragment length. Using BioImage Whole Band Analyzer software (version 3.3), a molecular weight was assigned to each band of RE-digested M. tuberculosis H37Rv based on comparison with the S. aureus 8325 standard. For each RE, 8 H37Rv patterns from four different gels were used to make 28 pairwise comparisons to compute the mean and variance in proportional error for each fragment of H37Rv and as performed by SCTR-MFA software (13a, 14). Fragment sizes with a pairwise error of greater than 5% were excluded from the PFGE pattern analysis.
Analyses of M. tuberculosis isolates were made in a blinded fashion, with the exception of reference strain H37Rv. In order to facilitate the comparison of over 400 PFGE patterns, an automated approach was used to identify similar PFGE patterns. The similar PFGE patterns were then visually analyzed. In the automated analysis, the PFGE patterns were compared by using the Dice coefficient with 5% band tolerance. A similarity threshold of 80% was used to identify similar PFGE patterns. Next, the lanes with similar PFGE patterns were analyzed visually and assigned to clusters if the numbers and molecular weights of the fragments were identical.
Genetic diversity.
In order to assess the discriminatory power of each molecular typing system, we measured the genetic diversity defined by that typing system. To compare typing systems, we restricted analysis to samples typed with all five systems. Genetic diversity was calculated for each typing system by using the estimated heterozygosity, h, where h = [1 − sum (ni/N)2] (N/N − 1). For each pattern, i = 1…k, occurring ni = n1…nk times in the data, respectively. That is, ni is the number of times a pattern of type i is observed and N is the sample size.
The diversity measurement estimates the probability that two fingerprints selected at random will be different. It is minimized if all samples have one pattern and maximized if all samples have different patterns. For a given number of patterns observed among the N samples, h will be greater when patterns occur at equal frequency and will be lower when one or a few patterns are frequently observed, while others are rare patterns. Measuring higher values of h for molecular fingerprint data indicates more discriminatory power for the typing system. The variances in these estimates were calculated as described by Nei (10).
Information content dependency.
Often it is useful to know whether secondary genotyping provides more information than a primary genotyping system. In order to assess the increase in information provided by a second genotyping system, we introduce an entropy-based measure. Entropy is a measure of the average uncertainty of an outcome (4, 12). Entropy increases as more outcomes are possible and as these occur at more even frequency.
The information content uncertainty coefficients were calculated to measure the dependency of each typing system on the others. The dependency of typing system y on typing system x was calculated as follows: U(y|x) = [H(y) − H(y|x)]/H(y), where H(y) is the entropy of the set of pattern measurements with typing system y and H(y|x) is the entropy in pattern measurements with typing system y given x. An introduction to measures of association based on entropy in reference 12 guides one through these standard calculations. The quantitative assessment of the new information gained from a second typing method (Z) given the results of the first typing method was calculated as a percentage as follows: Z = [1 − U(y|x)] · 100. These values are presented in Tables 5 and 6.
TABLE 5.
Information gained by using a second typing method for the isolates included in this study
| First method | % of new information gained by using the following second method:
|
|||||
|---|---|---|---|---|---|---|
| IS6110 | DraI | AseI | SpeI | XbaI | Four-RE PFGE | |
| IS6110 | 2.9 | 2.1 | 2.9 | 4.6 | 4.5 | |
| DraI | 7.5 | 8.0 | 2.0 | 7.7 | 8.5 | |
| AseI | 6.6 | 7.7 | 7.6 | 8.5 | 8.4 | |
| SpeI | 6.2 | 0.6 | 6.5 | 6.4 | 7.2 | |
| XbaI | 2.1 | 0.6 | 1.6 | 0.6 | 1.5 | |
| Four-RE PFGE | 0.6 | 0 | 0 | 0 | 0 | |
TABLE 6.
Information gained by using a second typing method for the isolates with few copies of IS6110
| First method | % of new information gained by using the following second method:
|
|||||
|---|---|---|---|---|---|---|
| IS6110 | DraI | AseI | SpeI | XbaI | Four-RE PFGE | |
| IS6110 | 24.1 | 21.1 | 24.1 | 35.0 | 35.0 | |
| DraI | 0 | 6.2 | 0 | 14.4 | 14.4 | |
| AseI | 16.4 | 24.6 | 24.6 | 31.1 | 31.1 | |
| SpeI | 0 | 0 | 6.2 | 14.1 | 14.4 | |
| XbaI | 0 | 0 | 0 | 0 | 0 | |
| Four-RE PFGE | 0 | 0 | 0 | 0 | 0 | |
RFLP analysis with IS6110.
RFLP analysis was performed on the study isolates by using an internationally standardized procedure (17). In brief, bacteria were lysed and the isolated genomic DNA was digested with PvuII. The resulting DNA fragments were separated by gel electrophoresis, transferred to nylon membranes, probed with a horseradish peroxidase-labeled 245-bp fragment of insertion sequence IS6110, and exposed to X-ray film. The resulting images were compared by using the BioImage Whole Band Analyzer. Isolates were analyzed by using the Dice coefficient with a 10% band tolerance. Those isolates that were 80% similar were analyzed visually and assigned to clusters if the numbers and molecular weights of the fragments were identical.
RESULTS
In order to compare PFGE patterns on different gels and to determine the range of interpretable fragment sizes, the error in fragment length assignments from multiple runs of M. tuberculosis H37Rv was measured. Predictably, the fragment length variation was largest at the extremes of the gel for the four REs used. Based on the error analysis of fragments generated with each RE, fragment sizes with a mean proportional error of greater than 5% were excluded from PFGE pattern analysis. The number of bands and a qualitative description of the bands excluded from PFGE pattern analysis are presented in Table 1. With the exception of DraI PFGE patterns, little variation was observed in the number and position of the bands excluded from further analysis.
TABLE 1.
Bands excluded from PFGE pattern analysis
| RE | Size (kbp) excluded
|
Mean no. (range) of bands excluded | Variation in bands excludeda | |
|---|---|---|---|---|
| Above | Below | |||
| DraI | 76 | 5 (2–8) | Yb | |
| AseI | 361 | 36 | 8 (7–8) | Nc |
| SpeI | 257 | 36 | 4 (3–6) | Ld |
| XbaI | 257 | 36 | 5 (4–6) | L |
Variation in the number and molecular size position of bands excluded from analysis.
Y, variation present.
N, no variation.
L, limited variation.
Typeability.
It is essential that a reliable typing system yield interpretable information for a large proportion of isolates. Of the 78 isolates typed by using PFGE, 68 were typed with DraI (87%), 73 were typed with AseI (94%), 71 were typed with SpeI (91%), and 66 were typed with XbaI (85%). All 78 isolates (100%) were typed with IS6110 RFLP. Of the 78 isolates, 66 gave good-quality PFGE patterns with all four restriction endonucleases, and these were used in the study comparisons. Of the 34 genetically related isolates, all 34 (100%) were typed with all four PFGE enzymes and IS6110 analysis.
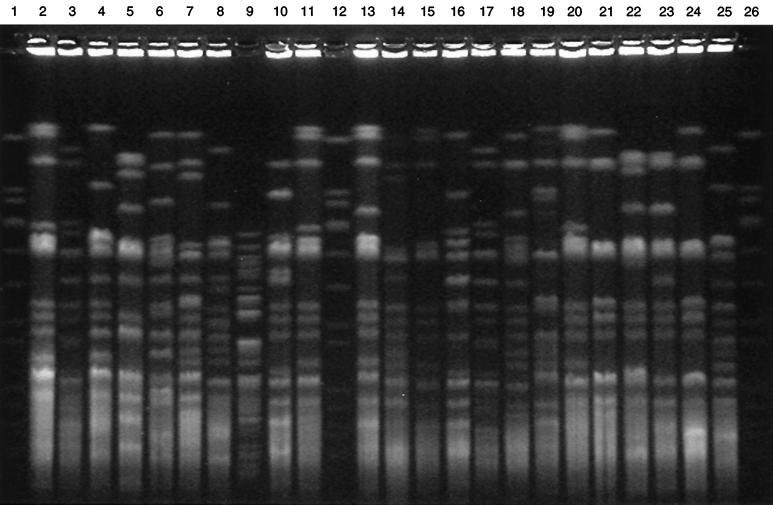
From an analytical perspective, PFGE patterns are easiest to compare when there are a manageable number of bands spread over a maximum size range. Of the four enzymes used in this study, DraI PFGE patterns were the easiest to analyze, as seen in Fig. 1. These patterns exhibited the fewest fragments (mean = 13 fragments) separated over the widest molecular size range (76 to 850 kb). AseI generated PFGE patterns with a larger number of fragments (mean = 16 fragments) spread over a molecular size range of 36 to 361 kb. XbaI produced PFGE patterns with a number of fragments similar to that of AseI (mean = 16 fragments), and SpeI generated PFGE patterns with slightly fewer fragments (mean = 15 fragments) separated over a molecular size range of 36 to 257 kb.
FIG. 1.
DraI PFGE of M. tuberculosis. Lanes: 1, 12, and 26, SmaI digest of S. aureus 8325; 9, DraI digest of Pseudomonas aeruginosa ATCC 27853; 3 and 17, M. tuberculosis H37Rv; 2, 4–8, 10, 11, 13–16, and 18–25, clinical isolates from the 6-month study period.
Stability and reproducibility of PFGE patterns.
The stability and reproducibility of PFGE patterns were demonstrated by testing a collection of epidemiologically well-characterized isolates known to be genetically related. Ten isolates had initially been documented to be part of a laboratory cross-contamination outbreak with IS6110 RFLP analysis. These 10 isolates from the laboratory cross-contamination gave identical PFGE patterns with all four REs. Sequential isolates collected over the course of infection from the five San Francisco tuberculosis patients were examined. The initial isolate from each of the patients had a unique PFGE pattern with each RE, and the subsequent isolate from each patient had a pattern identical to that of the initial isolate. Finally, the 14 single cell colonies picked from one clinical isolate gave an identical PFGE pattern with each of the REs. The reproducibility of the PFGE protocol was characterized by processing multiple batches of M. tuberculosis H37Rv. The PFGE patterns obtained from the multiple runs of H37Rv were identical.
Discriminatory ability.
The diversity of PFGE patterns and IS6110 RFLP was determined by analyzing 66 isolates from San Francisco tuberculosis patients during a 6-month period. The number of genotypes, as well as the percentage of isolates placed in clusters, was similar with all of the methods. The diversity of patterns generated by each typing system was measured by estimating the probability that two fingerprints selected at random would be different; this is known as its heterozygosity. The values generated are similar to those obtained by using Simpson’s index of diversity, which is based on the probability that two unrelated strains will be placed into different clusters (8). As summarized in Table 2, all of the typing systems provide similar levels of diversity, indicating that all of the methods provide comparable strain typing data.
TABLE 2.
Discriminatory abilities of PFGE and IS6110
| Typing system | % of isolates in clusters | No. of genotypes | Heterozygosity (95% confidence interval) |
|---|---|---|---|
| AseI | 36 | 47 | 0.95 (0.90–0.99) |
| DraI | 42 | 46 | 0.96 (0.93–0.99) |
| SpeI | 39 | 46 | 0.96 (0.94–0.99) |
| XbaI | 32 | 50 | 0.98 (0.96–1.00) |
| IS6110 | 35 | 49 | 0.97 (0.94–0.99) |
Comparison of clustering by PFGE and IS6110 RFLP.
The clusters generated with each PFGE enzyme were compared to clusters defined by RFLP analysis with IS6110 for all isolates. Approximately 80 to 85% of the isolates were placed in identical clusters by PFGE and RFLP with IS6110. The isolates with different cluster designations by PFGE are presented in Table 3. The four isolates placed in clusters by IS6110 but categorized as unique by three or more PFGE enzymes had fewer than three copies of IS6110. The three isolates categorized as unique by IS6110 but clustered by three or more PFGE enzymes had 9 to 12 copies of IS6110. Of these three isolates with 9 to 12 IS6110 copies, two were from patients of Filipino background, pointing to a possible epidemiologic relationship.
TABLE 3.
Isolates placed in different clusters by PFGE and IS6110
| IS6110 result | No. of isolates unique (clustered) by:
|
|||
|---|---|---|---|---|
| Four PFGE REs | Three PFGE REs | Two PFGE REs | One PFGE RE | |
| Clustered (unique) | 3 (2) | 1 (1) | 1 (2) | 1 (9) |
The clustering of isolates with PFGE was investigated among isolates with few copies of IS6110. The 66 clinical isolates were stratified by their IS6110 copy numbers into two groups—isolates with fewer than five copies of IS6110 and isolates with five or more copies of IS6110. The percentage of isolates clustered by PFGE and IS6110 in each group is presented in Table 4. PFGE identifies more unique strains among those isolates with fewer than five copies of IS6110, as 56 to 67% cluster with PFGE and 89% cluster with IS6110. This is not surprising, as IS6110 RFLP tends to misclassify isolates with few copies of IS6110 as genetically related. In isolates with five or more copies of IS6110, PFGE tends to cluster more isolates than IS6110 RFLP analysis, as 23 to 33% cluster with PFGE and 15% cluster with IS6110.
TABLE 4.
Effect of IS6110 copy number on clustering
| Typing system | % of isolates with:
|
|
|---|---|---|
| <5 copies of IS6110 clustered | ≥5 copies of IS6110 clustered | |
| DraI | 67 | 33 |
| AseI | 67 | 25 |
| SpeI | 67 | 29 |
| XbaI | 56 | 23 |
| IS6110 | 89 | 15 |
Content dependencies of typing methods.
Often it is important to know if typing of isolates with more than one method will provide additional useful information that further divides clusters. By examining the information content dependencies of the typing methods tested here, the relationship between these methods was determined. The values presented in Table 5 represent the percentage of new information provided by a second typing method when the results of the first typing method are known. For the 66 study isolates typed first by IS6110 RFLP, subsequent PFGE typing using DraI provides 97% identical information and 3% novel information. Given the typing results obtained by using DraI, IS6110 RFLP genotyping provides roughly 93% of the same information. As demonstrated here, regardless of the first method of typing selected, little genetic information is gained from additional typing.
However, there is great value in relying on PFGE as a typing method for isolates with few copies of IS6110, as seen in Table 6. If PFGE is the first typing method performed, no additional information is gained from subsequent typing by IS6110 RFLP. Furthermore, PFGE typing provides 21 to 35% novel information when performed as a second typing method after IS6110 RFLP.
DISCUSSION
The PFGE protocol developed in this study yields interpretable results with four restriction endonucleases for over 90% of the isolates. The analysis of replicate runs of the reference strain M. tuberculosis H37Rv, as well as genetically related isolates, shows the PFGE results to be reproducible and stable over time. In addition, the systematic approach to PFGE pattern analysis permits comparison of isolates in a more rigorous manner than has been previously attempted.
The PFGE approach utilized in this study and analysis of the selected clinical isolates have clarified issues raised in previous reports regarding the use of PFGE for M. tuberculosis. Previously published reports have presented conflicting views on the ability of PFGE to demonstrate genetic diversity among clinical isolates of M. tuberculosis (5, 11, 20). Olson et al. found little polymorphism among 99 clinical isolates from Scotland (11). Although Zhang et al. and Feizabadi et al. demonstrated considerable strain diversity, their results differ regarding the discriminatory powers of various REs (5, 20). Zhang et al. found XbaI and SpeI to be more discriminatory than AseI and DraI. In contrast, Feizabadi et al. reported that DraI identifies more polymorphism in the genome than does XbaI.
By using improved technical and analytical approaches, we have found that considerable diversity is revealed by PFGE typing using each of four REs. In contrast to previous investigators’ results, the degrees of genetic diversity revealed by the four independent REs were similar. The use of electrophoresis conditions optimized separately for each enzyme in our methods allowed improved resolution of patterns. A formal error analysis precisely identified the portion of the patterns that could be reliably compared between different experiments. The semiautomated procedure used in this study for the comparison of PFGE patterns on different images enabled us to cluster identical patterns. In addition, the analytic approach using the heterozygosity and information content dependency formally quantified the differences among the five different genotyping methods. The general concordance among the genotyping results obtained with four independent PFGE enzymes and RFLP analysis with IS6110 suggests that M. tuberculosis is a clonal organism.
Comparison of the classification results from PFGE and the more widely used IS6110-based RFLP approach raises intriguing questions about the use of IS6110 as a marker in molecular epidemiologic studies. In general, our results demonstrate no significant difference in discriminatory power between PFGE typing with these enzymes or with RFLP analysis with IS6110. Thus, in many settings, logistical issues can be used to select a genotyping technique. However, the isolates placed in different categories by using PFGE in contrast to IS6110 were not only isolates with few copies of IS6110 but also included isolates with many copies of IS6110. Previous studies have demonstrated that RFLP analysis with IS6110 tends to misclassify unrelated strains with few copies of IS6110 as genetically related (3, 6, 13, 19). In contrast, typing with PFGE is independent of IS6110 copy number. Consequently, unrelated strains with few copies of IS6110 can be appropriately classified by PFGE.
The possibility that IS6110-based typing may misclassify isolates with many copies of IS6110 has received less attention. PFGE analysis clustered isolates with many copies of IS6110 which were classified as unique by IS6110 RFLP analysis. The PFGE results suggest that RFLP analysis with IS6110 may blur genetic relationships among isolates with many copies of IS6110. In addition, other studies have also pointed to the considerable instability of IS6110-based RFLP patterns (15). In sum, these observations raise the hypothesis that IS6110 genotypic stability may be a continuous function of IS6110 copy number and needs to be taken into consideration in its use as a marker for molecular epidemiology.
Controversy exists regarding the amount of bacterial genetic data required to convincingly demonstrate that two isolates are likely to be progeny of the same progenitor. Previously, we have suggested that the burden of proof depends on the question asked; less is needed for studies of transmission dynamics than for tracking of specific outbreaks (2). The data from this study suggest that the absolute number of isolates misclassified by PFGE is low, and thus, any one of these four enzymes would be adequate for molecular epidemiologic assessment of transmission dynamics and would also be appropriate for typing of M. tuberculosis in outbreak investigations. The availability of a robust genotyping technique such as PFGE largely independent of repetitive elements has important implications for the molecular epidemiology of M. tuberculosis.
ACKNOWLEDGMENTS
We are grateful for the assistance of Cristina Agasino, Melvin Javonillo, and Pawel Moldenhawer.
This research effort was supported by NIH grants AI34238 and AI40906 and by Bio-Rad Laboratories.
Footnotes
This paper is dedicated to the memory of K. V. Seth.
REFERENCES
- 1.Agasino C B, Ponce de Leon A, Jasmer B, Small P M. Epidemiology of M. tuberculosis which do not contain IS6110 in San Francisco. Int J Tuberc Lung Dis. 1998;2:518–520. [PubMed] [Google Scholar]
- 2.Behr M A, Small P M. Molecular fingerprinting of Mycobacterium tuberculosis: how can it help the clinician? Clin Infect Dis. 1997;25:806–810. doi: 10.1086/515550. [DOI] [PubMed] [Google Scholar]
- 3.Braden C R, Templeton G L, Cave M D, Valway S, Onorato I M, Castro K G, Moers D, Yang Z, Stead W W, Bates J H. Interpretation of restriction fragment length polymorphism analysis of Mycobacterium tuberculosis isolates from a state with a large rural population. J Infect Dis. 1997;175:1446–1452. doi: 10.1086/516478. [DOI] [PubMed] [Google Scholar]
- 4.Durbin R, Eddy S, Krogh A, Mitchison G. Biological sequence analysis. New York, N.Y: Cambridge University Press; 1998. [Google Scholar]
- 5.Feizabadi M M, Robertson I D, Edwards R, Cousins D V, Hampson D J. Genetic differentiation of Australian isolates of Mycobacterium tuberculosis by pulsed-field gel electrophoresis. J Med Microbiol. 1997;46:501–505. doi: 10.1099/00222615-46-6-501. [DOI] [PubMed] [Google Scholar]
- 6.Fomukong N G, Tang T H, Al-Maamary S, Ibrahim W A, Ramayah S, Yates M, Zainuddin Z F, Dale J W. Insertion sequence typing of Mycobacterium tuberculosis: characterization of a widespread subtype with a single copy of IS6110. Tuber Lung Dis. 1994;75:435–440. doi: 10.1016/0962-8479(94)90117-1. [DOI] [PubMed] [Google Scholar]
- 7.Hayward A C. Restriction fragment length polymorphism typing of Mycobacterium tuberculosis. Thorax. 1995;50:1211–1218. doi: 10.1136/thx.50.11.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hunter P R, Gaston M A. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jasmer R M, Ponce de Leon A, Hopwell P C, Alarcon R G, Moss A R, Paz E A, Schecter G F, Small P M. Tuberculosis in Mexican-born persons in San Francisco: reactivation, acquired infection and transmission. Int J Tuberc Lung Dis. 1997;1:536–541. [PubMed] [Google Scholar]
- 10.Nei M. Molecular evolutionary genetics. New York, N.Y: Columbia University Press; 1987. p. 180. [Google Scholar]
- 11.Olson E S, Forbes K J, Watt B, Pennington T H. Population genetics of Mycobacterium tuberculosis complex in Scotland analysed by pulsed-field gel electrophoresis. Epidemiol Infect. 1995;114:153–160. doi: 10.1017/s0950268800052006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Press W H, Teukolsky S A, Vatterling W T, Flannery B P. Numerical recipes in C: the art of scientific computing. 2nd ed. New York, N.Y: Cambridge University Press; 1994. pp. 632–636. [Google Scholar]
- 13.Sahadevan R, Narayanan S, Paramasivan C N, Prabhakar R, Narayanan P R. Restriction fragment length polymorphism typing of clinical isolates of Mycobacterium tuberculosis from patients with pulmonary tuberculosis in Madras, India, by use of direct-repeat probe. J Clin Microbiol. 1995;33:3037–3039. doi: 10.1128/jcm.33.11.3037-3039.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Salamon, H. October 1998, posting date. [Online.] SCTR-MFA software. Stanford Center for TB Research, Stanford, Calif. http://molepi.stanford.edu/free_software.html. [12 April 1999, last date accessed.]
- 14.Salamon H, Segal M R, Ponce de Leon A, Small P M. Accommodating error analysis in comparison and clustering of molecular fingerprints. Emerg Infect Dis. 1998;4:159–168. doi: 10.3201/eid0402.980203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salamon, H., M. A. Behr, J. T. Rhee, and P. M. Small. Genetic distances for the study of infectious disease epidemiology. Submitted for publication. [DOI] [PubMed]
- 16.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Embden J D A, Cave M D, Crawford J T, Dale J W, Eisenach K D, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick T, Small P M. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams D L, Gillis T P, Dupree W G. Ethanol fixation of sputum sediments for DNA-based detection of Mycobacterium tuberculosis. J Clin Microbiol. 1995;33:1558–1561. doi: 10.1128/jcm.33.6.1558-1561.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuen L K, Ross B C, Jackson K M, Dwyer B. Characterization of Mycobacterium tuberculosis strains from Vietnamese patients by Southern blot hybridization. J Clin Microbiol. 1993;31:1615–1618. doi: 10.1128/jcm.31.6.1615-1618.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Mazurek G H, Cave M D, Eisenach K D, Pang Y, Murphy D T, Wallace R J., Jr DNA polymorphisms in strains of Mycobacterium tuberculosis analyzed by pulsed-field gel electrophoresis: a tool for epidemiology. J Clin Microbiol. 1992;30:1551–1556. doi: 10.1128/jcm.30.6.1551-1556.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]