Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has necessitated the rapid development of antibody-based therapies and vaccines as countermeasures. Here, we use cryoelectron microscopy (cryo-EM) to characterize two protective anti-SARS-CoV-2 murine monoclonal antibodies (mAbs) in complex with the spike protein, revealing similarities between epitopes targeted by human and murine B cells. The more neutralizing mAb, 2B04, binds the receptor-binding motif (RBM) of the receptor-binding domain (RBD) and competes with angiotensin-converting enzyme 2 (ACE2). By contrast, 2H04 binds adjacent to the RBM and does not compete for ACE2 binding. Naturally occurring sequence variants of SARS-CoV-2 and corresponding neutralization escape variants selected in vitro map to our structurally defined epitopes, suggesting that SARS-CoV-2 might evade therapeutic antibodies with a limited set of mutations, underscoring the importance of combination mAb therapeutics. Finally, we show that 2B04 neutralizes SARS-CoV-2 infection by preventing ACE2 engagement, whereas 2H04 reduces host cell attachment without directly disrupting ACE2-RBM interactions, providing distinct inhibitory mechanisms used by RBD-specific mAbs.
Keywords: SARS-CoV-2, cryo-EM, biolayer interferometry, RBD, spike, variants of concern, antibody neutralization, ACE2, COVID-19
Graphical abstract

Errico et al. use cryoelectron microscopy to solve the structure of two murine-derived neutralizing antibodies against the SARS-CoV-2 receptor-binding domain, showing that they target discrete epitopes and utilize distinct mechanisms of neutralization.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiologic agent of coronavirus disease 2019 (COVID-19), is a positive-sense RNA virus in the Betacoronavirus genus. It is closely related to other highly pathogenic coronaviruses, most notably severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle Eastern respiratory syndrome virus (MERS) (Zhou et al., 2020b). SARS-CoV-2 emerged in Wuhan, China, in late 2019 (Guan et al., 2020; Zhou et al., 2020b). Since then, it has spread to nearly every country, causing more than 39 million infections and over 1 million deaths at the time of writing (Dong et al., 2020). Efforts to contain the spread of the virus have been limited to social distancing, mask wearing, hand hygiene practices, and avoidance of large gatherings, which has resulted in widespread societal disruption and economic damage. Numerous vaccines and therapeutics are in development against SARS-CoV-2 infection, although currently, no prophylactic countermeasures have been approved for use.
SARS-CoV-2 is a spherical, enveloped virion with a diameter of approximately 90 nm (Ke et al., 2020). The virion surface is decorated by the spike protein, which binds to human angiotensin-converting enzyme 2 (ACE2) on host cell surfaces, and mediates viral entry (Hoffmann et al., 2020; Walls et al., 2020). Spike monomers consist of S1 and S2 subunits, which form homotrimers (Wrapp et al., 2020). The S1 subunit is made up of 4 sub-domains, S1A through S1D (Barnes et al., 2020a). S1B encodes the receptor-binding domain (RBD), which directly interacts with ACE2. The S2 subunit contains a fusion peptide, which mediates fusion with host cell membranes after receptor binding by S1. The RBD is observed to have two distinct conformations, termed “up” and “down,” with the “up” position thought to be required for ACE2 binding. With three S1 subunits per trimer, this leads to 4 possible configurations for each trimeric spike protein: all down (D/D/D), one up with two down (U/D/D), two up with one down (U/U/D), and three up (U/U/U). In practice, the majority of spike trimers assume the D/D/D and U/D/D configurations (Walls et al., 2020; Wrapp et al., 2020), although U/U/D and U/U/U have been observed (Barnes et al., 2020a; Ke et al., 2020).
Many neutralizing antibodies targeting SARS-CoV-2 spike have been identified (Alsoussi et al., 2020; Barnes et al., 2020a; Brouwer et al., 2020; Chi et al., 2020; Liu et al., 2020b; Lv et al., 2020; Shi et al., 2020; Wang et al., 2020; Wu et al., 2020b; Zhou et al., 2020a; Zost et al., 2020). These antibodies typically recognize the RBD and function by competitively inhibiting ACE2 binding, although a subset of antibodies that display neutralizing activity do not inhibit receptor engagement (Pinto et al., 2020; Zhou et al., 2020a). Many antibodies targeting the receptor-binding motif (RBM) portion of the RBD display potent neutralizing efficacy in vitro, with low ng/mL IC50 (half maximal inhibitory concentration) values (Alsoussi et al., 2020; Liu et al., 2020b; Zost et al., 2020). Antibodies targeting alternative epitopes on spike, such as non-RBM portions of the RBD and the N-terminal domain, generally neutralize with lower potency via unknown mechanisms. Many anti-RBD antibodies confer protection in animal models of SARS-CoV-2 infection, highlighting their potential use as therapeutic agents (Alsoussi et al., 2020; Hassan et al., 2020).
As an RNA virus, SARS-CoV-2 can rapidly generate mutations in critical epitopes on spike, which could render clinical interventions ineffective. Indeed, a database of over 106,000 SARS-CoV-2 sequences isolated from humans reports over 400 amino acid substitutions in the RBD of spike alone (Singer et al., 2020). Understanding antibody responses to SARS-CoV-2 at the molecular level is critical to predicting vaccine efficacy and designing potent, durable antibody-based therapeutics. Recently, a panel of anti-SARS-CoV-2 antibodies was generated by immunizing mice with recombinant RBD and boosting with SARS-CoV-2 spike trimers (Alsoussi et al., 2020). Two monoclonal antibodies (mAbs) from this panel, 2B04 and 2H04, displayed potent neutralization in vitro (IC50 of 1.46 and 154 ng/mL, respectively) and conferred protection in vivo in a mouse model of SARS-CoV-2 infection (Alsoussi et al., 2020). Here, we use single-particle cryoelectron microscopy (cryo-EM) to characterize the epitopes targeted by these antibodies. Additionally, we identify viral mutations that have occurred naturally in human SARS-CoV-2 infections, which could lead to viral escape from inhibition by these mAbs, and we probe the sensitivity of neutralization of SARS-CoV-2 variants of concern by 2B04 and 2H04. Lastly, we uncover a novel basis for SARS-CoV-2 neutralization by 2H04 without direct RBM blockade.
Results
Anti-SARS-CoV-2 mAbs 2B04 and 2H04 bind to the RBD of the trimeric spike protein
To understand the structural basis for binding by 2B04 and 2H04, we solved the structures of antigen-binding fragments (Fab) of the mAbs in complex with the SARS-CoV-2 trimeric spike protein using single-particle cryo-EM. Initial datasets collected on standard lacey carbon grids showed a strong top-down preferred orientation of the particles, resulting in anisotropic maps, which prevented reliable model building. To overcome this limitation, we collected more data with complexes frozen on lacey carbon grids covered by an ultra-thin carbon film, which showed a preferred orientation for side views, and combined the two datasets (Figures S1A–S1D and S2). From these data, we solved the structure of 2B04 bound to the U/D/D conformational state of the spike protein to a resolution of 3.2 Å (Figures 1 A and S1E). The local resolution of the complex varied from 3 Å in the well-ordered S2 core of the spike to 8 Å at the outlying constant regions of the Fab molecule (Figure S1G). For 2H04, the majority of the particles were in the D/D/D conformation, allowing reconstruction with C3 symmetry to a resolution of 3.0 Å (Figures 1C and S1F), with local resolution varying from 3 Å in S2 core to approximately 8 Å at the tips of the Fabs (Figure S1H).
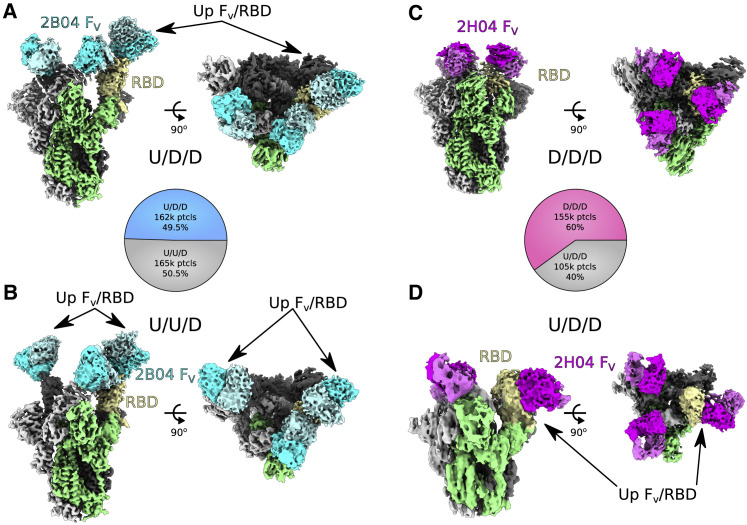
Figure 1.
Anti-SARS-CoV-2 mAbs 2B04 and 2H04 bind to the RBD of the trimeric spike (S) protein
(A) 2B04 up/down/down (U/D/D) density map. The S1/S2 portion of the spike excluding the RBD is colored green, with the “up” RBD subunit shown in yellow. 2B04 heavy chain is shown in cyan, with the light chain shown as pale turquoise.
(B) Map of 2B04 bound to SARS-CoV-2 trimeric spike in the up/up/down (U/U/D) configuration, with sections colored as in (A).
(C) 2H04 down/down/down (D/D/D) density map. The S1/S2 region of one spike monomer excluding the RBD is shown in green, with the RBD shown in yellow. 2H04 heavy chain is shown in magenta, with the light chain shown in violet.
(D) Map of 2H04 bound to SARS-CoV-2 trimeric spike in the U/D/D configuration, colored as in (C). Pie charts in between (A) and (B) and (C) and (D) represent distribution of particles belonging to the two 2B04 or two 2H04 maps, respectively. The Fab and RBD portions of the maps are contoured at a higher level than the core S1/S2 portions of the map for (A)–(C).
Both 2B04 and 2H04 were able to bind with full occupancy to the spike protein, regardless of RBD conformation (Figures 1 and S3A–S3D). The two major RBD configurations observed for 2B04 were U/D/D and U/U/D in roughly equal proportion, although a minority of D/D/D particles were observed in 3D classification (Figures 1A and 1B and S2A). For 2H04, the majority of particles were in the D/D/D conformational state (Figure 1C), with a smaller proportion found in a U/D/D conformation (Figure 1D).
While the S1/S2 regions of the maps were well resolved, the density at the interface between the RBD and Fab, and for the Fab itself, was weak, presumably due to conformational flexibility. To overcome this limitation, we performed a focused classification using a mask enclosing one “down” RBD and variable fragment (FV) of the Fab in the case of 2B04, or all three subunits in the case of 2H04, to identify populations of particles that had well-ordered RBD/FV regions relative to the rest of the spike protein (Figure S2). Local non-uniform refinement of the particles from the focused classification in cryoSPARC led to a 3.3 Å reconstruction of the RBD/FV complex of 2B04 and a 3.14 Å map for the RBD/FV complex of 2H04 (Figures S3E and S3F). Importantly, the maps for the RBD and FV of both reconstructions were significantly improved, depicting continuous backbone traces with clear secondary structure and well-defined density for most side chains (Figures S3G and S3H).
2B04 and 2H04 target distinct epitopes on the RBD
Using the maps generated by local refinement, we could identify specific residues and strands at the interface of the complexes for both mAbs. 2B04 binds to the RBM of the RBD (Figure 2 A). CDR1 and CDR2 of the heavy chain spread across the ridge formed by the β5 and β6 strands (residues 445–454 and 491–498, respectively) of the RBD and press up against the flexible RBM loop (residues 472–490), while CDR-H3 mainly contacts the RBM loop (Figure 2A). CDR1 and CDR3 of the light chain also interact with the flexible RBM loop (Figure 2A), although the majority of contacts derive from the heavy chain of 2B04 (Figure 2B). The total buried surface area of the complex is 656 Å2, with 92% of the area covered by the heavy chain. For 2H04, the epitope is formed by a discontinuous set of strands adjacent to the RBM. 2H04 CDR-H1, CDR-H2, and CDR-H3 contact a loop adjacent to the RBM encoded by residues 439–450 (Figure 2C). CDR-L1, CDR-L2, and CDR-L3 target the region encoded by residues 339–353 (Figure 2C). CDR-L3 and residues 60–62 of the heavy chain also make extensive contacts with the core fucose of the glycan on N343 (Figure S4), although it is unclear whether this interaction contributes to binding. Compared with 2B04, 2H04 shows more extensive utilization of the light chain (Figure 2D). The total buried surface area at the interface of 2H04 and the RBD is 914 Å2, with 60% coming from the interaction of the heavy chain.
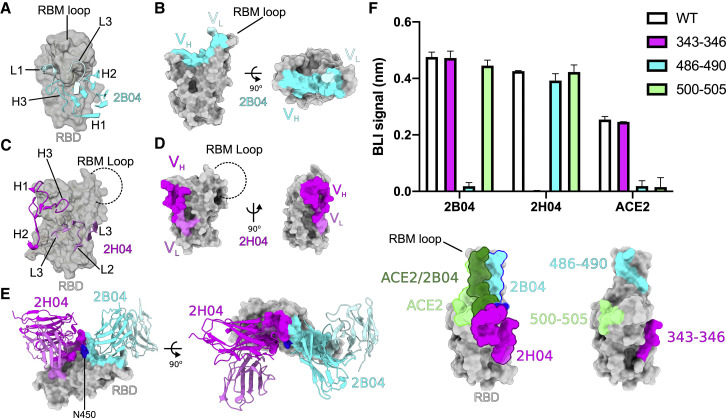
Figure 2.
2B04 and 2H04 target distinct epitopes on the RBD
(A) 2B04 targets an epitope centered on the RBM of the SARS-CoV-2 RBD. Complementarity-determining regions (CDRs) of 2B04 are shown as cartoon ribbons, with the heavy chain colored cyan and the light chain colored pale turquoise. The RBM is shown as a transparent surface with underlying ribbon diagram.
(B) Distribution of 2B04 heavy- versus light-chain contacts on the surface of the RBD. Heavy-chain contacts colored as in (A).
(C) 2H04 targets an epitope adjacent to the RBM on the SARS-CoV-2 RBD, distal from the RBM loop. CDR regions of 2H04 are shown as cartoon ribbons, with the heavy chain colored magenta and the light chain colored violet.
(D) Distribution of 2H04 heavy- versus light-chain contacts on the surface of the RBD. Contacts colored as in (C).
(E) Alignment of 2B04 and 2H04 models to the 2B04 RBD. 2B04 contacts are colored as in (A), and 2H04 contacts are colored as in (C). N450, the only shared contact between 2B04 and 2H04, is colored in blue on the RBD.
(F) BLI analysis of RBD mutants to 2B04, 2H04, and ACE2. Results represent mean (±standard deviation [SD]) from two independent experiments.
Alignment of the 2H04 bound RBD with the 2B04 bound RBD showed that the Fabs have largely distinct epitope footprints, with only one overlapping residue, N450 (Figure 2E). Binding of 2B04, but not 2H04, was significantly reduced when residues 486FNCYF490 of the RBD were mutated to 486AACAA490 (Figure 2F). By contrast, binding of 2H04, but not 2B04, to the RBD was eliminated by mutating residues 343–346 to alanines (Figure 2F). Alanine substitution of residues 486–490 or 500–505 was sufficient to abrogate ACE2 binding, showing that the 2B04 epitope overlaps significantly, but not completely, with the ACE2-binding footprint (Figure 2F). Substitution of residues 343–346 had no effect on ACE2 binding, suggesting that 2H04 might not compete with ACE2 for binding to the RBD.
2B04 and 2H04 share epitopes with human antibodies
Several groups have identified epitopes on the RBD of the SARS-CoV-2 spike protein (Barnes et al., 2020a, 2020b; Cao et al., 2020; Hansen et al., 2020; Hurlburt et al., 2020; Ju et al., 2020; Liu et al., 2020a, 2020b; Lv et al., 2020; Pinto et al., 2020; Shi et al., 2020; Wu et al., 2020a, 2020b; Yuan et al., 2020a, 2020b; Zhou et al., 2020a). We compared the epitopes of 2B04 and 2H04 with those of other antibodies to identify possible conserved modes of engagement. To do this, we aligned the structures of all available human antibody-bound SARS-CoV-2 spike or RBD structures published in the protein databank with our 2B04/RBD and 2H04/RBD complexes. For the RBM antibodies, there appeared to be five distinct clusters of epitopes on RBD (Figure 3 A), and 2B04 greatly overlapped with only one other antibody, 2-4 (Figure 3A, cyan group) (Liu et al., 2020b). The largest group targeted the opposite side of the RBM ridge from 2B04 and was comprised of mAbs CB6, B38, CC12.1, CC12.3, C105, COVA2-04, and CV30 (Figure 3A, red group) (Barnes et al., 2020a; Hurlburt et al., 2020; Shi et al., 2020; Wu et al., 2020a, 2020b; Yuan et al., 2020b). A smaller group adjacent to this was formed by COVA2-39 and REGN-10933 (Figure 3A, orange) (Hansen et al., 2020; Wu et al., 2020a). Two additional epitopes had one representative antibody each, made up by BD23 and P2B-2F6 (Cao et al., 2020; Ju et al., 2020). While all of these epitopes overlap significantly in the RBM ridge and directly block ACE2 binding, each group also has unique interactions with the RBM (Figure 3C). All five groups have footprints that overlap extensively with contact residues between the RBD and ACE2, consistent with their ability to neutralize SARS-CoV-2 by blocking interaction with ACE2 (Figure 3C).
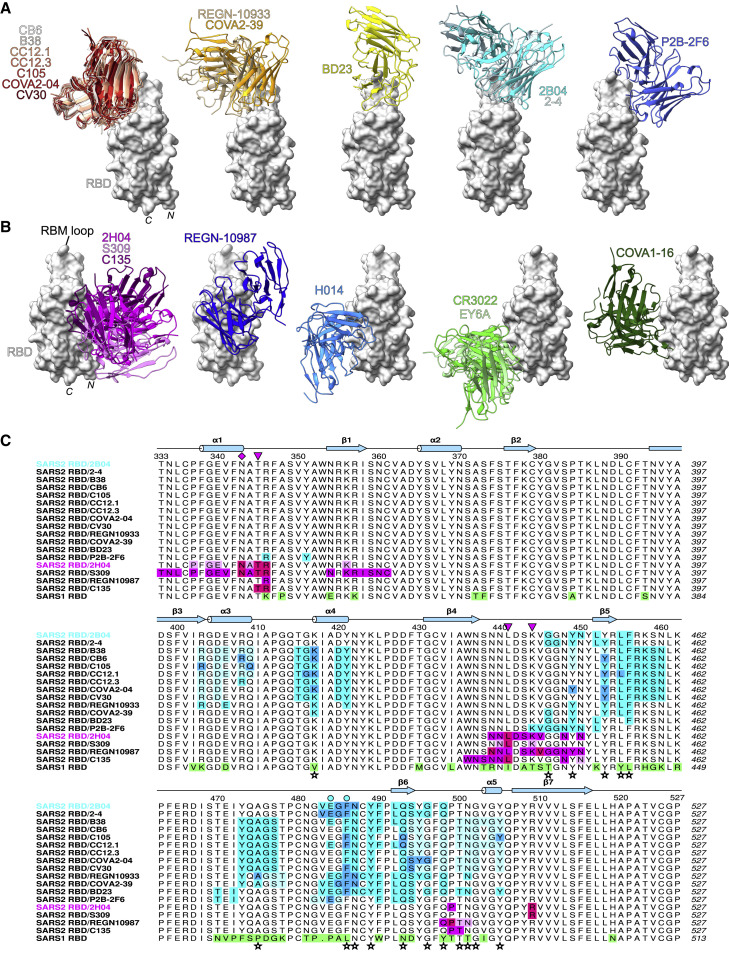
Figure 3.
2B04 and 2H04 share epitopes with human antibodies
(A) Comparison of human antibodies that target the RBM of the SARS-CoV-2 RBD and 2B04. mAb groups were formed based on overlap between their interfacial residues as determined by Proteins, Interfaces, Structures, and Assemblies (PISA) analysis.
(B) Comparison of human antibodies that target non-RBM epitopes of the SARS-CoV-2 RBD with 2H04. Groups are based on overlap between interfacial residues as determined by PISA analysis.
(C) Multiple sequence alignment of the SARS-CoV-2 RBD (residues 333–527) with binding footprints highlighted for human antibodies that are predicted to compete with 2B04 or 2H04 based on structural analysis. For RBM antibodies, heavy chain, light chain, and shared contacts are shown in cyan, pale turquoise, and blue, respectively. For non-RBM antibodies, heavy chain, light chain, and shared contacts are shown in magenta, pale violet, and fuchsia, respectively. SARS-CoV RBD is shown at the bottom, with substitutions relative to SARS-CoV-2 RBD highlighted in green. SARS-CoV-2 RBD contacts with ACE2 are identified by stars at the bottom of the alignment, based on Lan et al. (2020). Escape mutants contacts are shown above the alignment as cyan circles for 2B04 and magenta triangles for 2H04, with the N343 glycan demarcated by a magenta diamond. Secondary structure annotation is based on the locally refined 2B04/RBD model.
For 2H04, two antibodies were identified that overlapped its footprint: the SARS-CoV cross-reactive mAb S309 and SARS-CoV-2-specific C135 (Figure 3B, magenta group) (Barnes et al., 2020b; Pinto et al., 2020). Notably, the orientation of the heavy and light chain was inverted between 2H04 and S309. All three antibodies contact the core fucose of the glycan at N343. Despite their overlap, each antibody also forms unique contacts with the RBD (Figure 3C). Epitope alignment shows that S309 makes more extensive contacts with the N terminus of the RBD as well as the β1 strand (residues 353–358), whereas 2H04 makes unique contacts with N448, N450, and P499 (Figure 3C). In contrast to both 2H04 and S309, C135 displays fewer contacts overall to the RBD, but makes a number of unique contacts: W436, N437, and S438 (Barnes et al., 2020b). Twelve of 19 and 9 of 20 contact residues are strictly conserved between SARS-CoV-2 and SARS-CoV spike for 2H04 and 2B04, respectively, suggesting these mAbs might cross-react to SARS-CoV. Quantitative binding analysis by BLI showed weak binding to SARS-CoV by 2H04, with a KD of 1417 nM and half-life of 0.43 min; 2B04 did not bind to SARS-CoV RBD at the tested concentrations (Figures S5C and S5D) (Alsoussi et al., 2020).
Proximal to the 2H04/S309 epitope are four additional groups of antibodies. Immediately adjacent and partially overlapping with the 2H04/S309 epitope is REGN-10987 (Figure 3B, blue group) (Hansen et al., 2020). Further rotated away from 2H04 and REGN-10987 are three groups targeting similar yet distinct epitopes, bound by H014 (Figure 3B, light blue group) (Lv et al., 2020), COVA1-16 (Figure 3B, dark green group) (Liu et al., 2020a), and CR3022 or EY6A (Figure 3B, light green group) (Yuan et al., 2020a; Zhou et al., 2020a), respectively.
Overall, the RBD appears to be highly immunogenic with a multitude of epitopes that cluster roughly into three distinct groups. Antibodies targeting the RBM ridge and loop are differentiated by their orientation of binding on the ridge, but share a common mechanism of action and tend to display potent neutralization. Antibodies targeting RBM-adjacent (i.e., 2H04, S309, REGN-10987, H014, and C135) or RBM-distal (i.e., COVA1-16, CR3022, and EY6A) epitopes are differentiated by their rotation about the core RBD beta sheet and show less potent neutralization than RBM antibodies, typically via non-ACE2-competitive mechanisms.
SARS-CoV-2 can escape neutralization by both 2B04 and 2H04
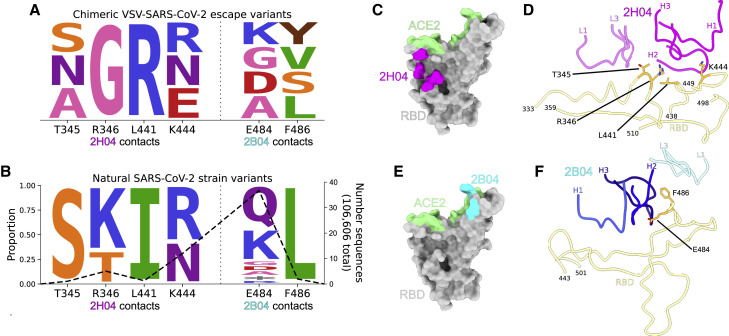
Escape from antibody-mediated neutralization by mutation of key binding residues is a feature of RNA viruses that hinders therapeutic development. A wide variety of amino acid substitutions in the spike protein of SARS-CoV-2 have been observed in human infections, suggesting that SARS-CoV-2 might be able to escape antibody-mediated neutralization. Escape mutations generated using a SARS-CoV-2 chimeric vesicular stomatitis virus (VSV-SARS-CoV-2) (Case et al., 2020) under selection pressure from 2B04 and 2H04 revealed mutations at T345, R346, L441, K444, E484, and F486 (Figure 4 A) (Liu et al., 2021). Interrogation of human-derived SARS-CoV-2 sequences in the global initiative on sharing all influenza data (GISAID) database via COV-GLUE revealed naturally occurring variants with substitutions at identical residues (Figure 4B; Table 1 ) (Shu and McCauley, 2017; Singer et al., 2020). T345, R346, L441, and K444 lie outside the binding site for ACE2 (Figure 4C) and are all contained within the epitope for 2H04 (Figure 4D), whereas E484 and F486 lie in the 2B04 footprint, with F486 also acting as an ACE2 contact residue (Figures 4E and 4F). Although these variants currently are present at low frequency in human populations (Figure 4B), these results suggest that SARS-CoV-2 can generate resistance to 2B04, 2H04, and other antibodies when used as monotherapies, which could result in expansion of resistant strains under prolonged selection pressure.
Figure 4.
SARS-CoV-2 can escape neutralization by 2B04 and 2H04
(A) Logo plot depicting escape mutants generated by selection pressure from 2B04 or 2H04 using a chimeric VSV expressing SARS-CoV-2 spike.
(B) Identity and frequency of mutations seen in clinical isolates of SARS-CoV-2 at identical positions as the VSV-SARS-CoV-2 escape mutants. The right axis shows the number of total sequences with mutations at each residue position, out of a total 106,606 sequences. Logo plots were generated using dmslogo (Bloom lab).
(C) RBD with ACE2 contacts colored in green, and 2H04 escape mutants colored in magenta.
(D) Ribbon diagram showing escape mutants at the binding interface of 2H04 on the RBD.
(E) RBD with ACE2 contacts colored in green, and 2B04 escape mutants colored in cyan.
(F) Ribbon diagram showing escape mutants at the binding interface of 2B04 on the RBD.
Table 1.
SARS-CoV-2 isolates with mutations at 2B04 and 2H04 escape residues
| Viral protein | Replacement | No. of sequences | Virus name | GISAID ID | Collection date |
|---|---|---|---|---|---|
| Spike | T345S | 1 | hCoV-19/USA/WA-S1049/2020 | EPI_ISL_463539 | 03/05/2020 |
| R346K | 4 | hCoV-19/USA/MI-MDHHS-SC21980/2020 | EPI_ISL_566030 | 06/11/20 | |
| hCoV-19/Spain/AN-IBV-98001706/2020 | EPI_ISL_538134 | 04/11/20 | |||
| hCoV-19/USA/MI-MDHHS-SC20812/2020 | EPI_ISL_471833 | 05/20/20 | |||
| hCoV-19/Netherlands/Gelderland_35/2020 | EPI_ISL_460935 | 04/29/20 | |||
| R346T | 2 | hCoV-19/India/GJ-GBRC-380b/2020 | EPI_ISL_524740 | 06/18/20 | |
| hCoV-19/India/GJ-GBRC333/2020 | EPI_ISL_512069 | 07/07/20 | |||
| L441I | 1 | hCoV-19/USA/FL-BPHL-0297/2020 | EPI_ISL_480948 | 05/16/2020 | |
| K444N | 5 | hCoV-19/USA/OR-OHSU-0911/2020 | EPI_ISL_525925 | 06/01/2020 | |
| hCoV-19/Australia/VIC4515/2020 | EPI_ISL_518926 | 07/16/2020 | |||
| hCoV-19/USA/OR-OHSU-0419/2020 | EPI_ISL_509133 | 05/24/2020 | |||
| hCoV-19/England/BRIS-12FB45/2020 | EPI_ISL_488423 | 04/02/3020 | |||
| hCoV-19/England/201060040/2020 | EPI_ISL_464376 | 04/03/2020 | |||
| K444R | 7 | hCoV-19/England/NOTT-112F58/2020 | EPI_ISL_526429 | 08/14/2020 | |
| hCoV-19/England/NOTT-112ED3/2020 | EPI_ISL_514453 | 08/07/2020 | |||
| hCoV-19/England/NOTT-112EB5/2020 | EPI_ISL_514452 | 08/04/2020 | |||
| hCoV-19/England/NOTT-112E79/2020 | EPI_ISL_512382 | 07/31/2020 | |||
| hCoV-19/England/NOTT-112E5B/2020 | EPI_ISL_512381 | 07/30/2020 | |||
| hCoV-19/England/BRIS-12CE21/2020 | EPI_ISL_481886 | 04/24/2020 | |||
| hCoV-19/Spain/Barcelona_VH7773/2020 | EPI_ISL_444976 | 04/06/2020 | |||
| E484∗ | 2 | hCoV-19/England/NORW-EA01B/2020 | EPI_ISL_448386 | 04/30/2020 | |
| hCoV-19/Australia/VIC1221/2020 | EPI_ISL_430518 | 04/01/2020 | |||
| E484A | 2 | hCoV-19/Northern Ireland/NIRE-106625/2020 | EPI_ISL_501014 | 05/05/2020 | |
| hCoV-19/Spain/Valencia578/2020 | EPI_ISL_447508 | 03/27/2020 | |||
| E484D | 2 | hCoV-19/Germany/BY-MVP-0253/2020 | EPI_ISL_466906 | 05/04/2020 | |
| hCoV-19/Thailand/Trang_5008/2020 | EPI_ISL_455588 | 03/23/2020 | |||
| E484G | 2 | hCoV-19/Switzerland/ZH-230059-753-G04/2020 | EPI_ISL_516595 | 08/05/2020 | |
| hCoV-19/England/CAMB-1AB7D4/2020 | EPI_ISL_470242 | 04/20/2020 | |||
| E484K | 12 | hCoV-19/Switzerland/BL-UHB-42201557/2020 | EPI_ISL_528300 | 03/21/2020 | |
| hCoV-19/USA/CA-ALSR-1928/2020 | EPI_ISL_512239 | 07/08/2020 | |||
| hCoV-19/Sweden/20-51736/2020 | EPI_ISL_510839 | 05/19/2020 | |||
| hCoV-19/Spain/EX-004992/2020 | EPI_ISL_510307 | 05/09/2020 | |||
| hCoV-19/USA/IL-UW-379/2020 | EPI_ISL_480361 | 05/22/2020 | |||
| hCoV-19/Wales/PHWC-1652D5/2020 | EPI_ISL_479447 | 05/25/2020 | |||
| hCoV-19/Sweden/20-51636/2020 | EPI_ISL_476136 | 05/22/2020 | |||
| hCoV-19/England/NOTT-111DB9/2020 | EPI_ISL_472405 | 06/08/2020 | |||
| hCoV-19/England/BRIS-12B8F7/2020 | EPI_ISL_469369 | 05/18/2020 | |||
| hCoV-19/Spain/GA-002758/2020 | EPI_ISL_467270 | 04/07/2020 | |||
| hCoV-19/England/20188152504/2020 | EPI_ISL_465520 | 04/30/2020 | |||
| hCoV-19/England/NOTT-1115C0/2020 | EPI_ISL_461895 | 05/18/2020 | |||
| E484Q | 17 | hCoV-19/South Africa/KRISP-K001324/2020 | EPI_ISL_515809 | 07/18/2020 | |
| hCoV-19/India/GJ-GBRC278b/2020 | EPI_ISL_495015 | 06/18/2020 | |||
| hCoV-19/India/GJ-GBRC278a/2020 | EPI_ISL_495014 | 06/18/2020 | |||
| hCoV-19/USA/CA-ALSR-1462/2020 | EPI_ISL_494618 | 03/20/2020 | |||
| hCoV-19/Wales/PHWC-168CC5/2020 | EPI_ISL_494352 | 05/20/2020 | |||
| hCoV-19/Wales/PHWC-168C5C/2020 | EPI_ISL_494348 | 05/20/2020 | |||
| hCoV-19/Wales/PHWC-168B5F/2020 | EPI_ISL_494335 | 05/20/2020 | |||
| hCoV-19/Wales/PHWC-168991/2020 | EPI_ISL_494311 | 05/21/2020 | |||
| hCoV-19/Wales/PHWC-16873D/2020 | EPI_ISL_494279 | 05/20/2020 | |||
| hCoV-19/Wales/PHWC-16859D/2020 | EPI_ISL_494256 | 05/21/2020 | |||
| hCoV-19/Wales/PHWC-168418/2020 | EPI_ISL_494240 | 05/20/2020 | |||
| hCoV-19/Wales/PHWC-16806F/2020 | EPI_ISL_494194 | 05/21/2020 | |||
| hCoV-19/Wales/PHWC-167EB1/2020 | EPI_ISL_494170 | 05/20/2020 | |||
| hCoV-19/Wales/PHWC-162A85/2020 | EPI_ISL_473071 | 05/20/2020 | |||
| hCoV-19/Wales/PHWC-16296A/2020 | EPI_ISL_473042 | 05/20/2020 | |||
| hCoV-19/Wales/PHWC-15FCEF/2020 | EPI_ISL_472846 | 05/21/2020 | |||
| hCoV-19/India/MH-NIV-4271/2020 | EPI_ISL_454530 | 03/22/2020 | |||
| E484R | 1 | hCoV-19/England/20342000104/2020 | EPI_ISL_528438 | 08/12/2020 | |
| F486L | 2 | hCoV-19/Netherlands/NB-EMC-277/2020 | EPI_ISL_523399 | 07/12/2020 | |
| hCoV-19/Netherlands/NB-EMC-266/2020 | EPI_ISL_523389 | 07/12/2020 |
Citations for GISAID sequence data can be found in Table S2 (generated 09/16/2020).
Neutralization of SARS-CoV-2 variants by 2B04 and 2H04
Several novel strains of SARS-CoV-2 bearing mutations in the RBD have emerged since late 2020, some of which contain substitutions at E484, suggesting that these strains may not be as sensitive to neutralization by 2B04 and 2H04 (Figure 5 B). To address this, we tested neutralization of these variant strains by 2B04 and 2H04. In line with results from aforementioned escape mutant studies, variants containing substitutions at E484 (B.1.351, B.1.1.28, B.1.617.1, and B.1.526 E484K) displayed significantly decreased neutralization by 2B04 (Figure 5A). 2H04 also displayed decreased neutralization potency against certain variants, particularly those bearing the N501Y substitution (B.1.1.7, B.1.351, and B.1.1.28), but to a lesser extent than 2B04 (Figure 5A). Additionally, 2H04 was less able to neutralize B.1.1.298 and B.1.222, bearing the Y453R and N439K mutations, respectively. A cocktail of 2B04 and 2H04 was generally able to rescue neutralization against variants that displayed resistance to either 2B04 or 2H04 individually, although variants containing both E484 and N501 substitutions (most notably B.1.1.28) still displayed significantly reduced neutralization compared with wild-type (WT) strains.
Figure 5.
Neutralization of SARS-CoV-2 variants by 2B04 and 2H04
(A) Neutralization curves of SARS-CoV-2 variants of concern by 2B04 or 2H04 individually and in combination. Data are mean of 2 independent experiments. Inset table summarizes chimeric viruses and variants of concern used.
(B) Structural depiction of the SARS-CoV-2 RBD shown as a silver surface with 2B04 and 2H04 shown as ribbon diagrams and colored cyan/pale turquoise and magenta/violet for heavy and light chains, respectively. Residues substituted in variants of concern are shaded orange, with lineages for each variant listed below the substitution and colored as in (A).
Mechanism of SARS-CoV-2 neutralization by 2B04 and 2H04
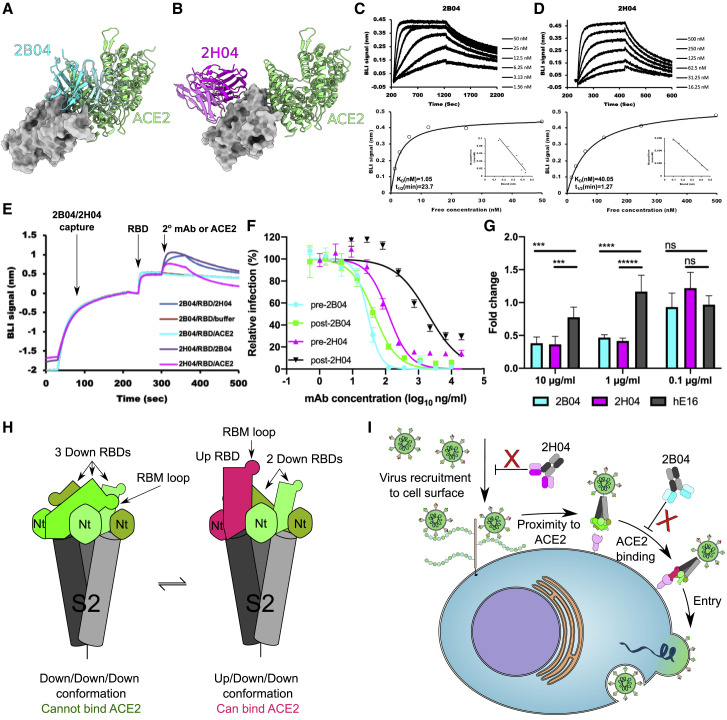
Based on our structural analysis and previous reports, 2B04 potently neutralizes SARS-CoV-2 by blocking ACE2 binding, while 2H04 neutralizes less potently without blocking ACE2 (Figures 6A and 6B). To begin to understand why 2H04 is less efficient at neutralizing SARS2-CoV-2 than 2B04, we measured the binding affinities of these two mAbs to recombinant RBD expressed in Escherichia coli by biolayer interferometry (BLI). 2B04 tightly bound RBD with a kinetic KD value of 1.05 nM and a relatively slow dissociation rate (half-life of 23.70 min) (Figure 6C). While 2H04 bound RBD strongly, it displayed weaker affinity (KD of 40.1 nM) and a faster off rate (half-life of 1.27 min) than 2B04 (Figure 6D). Since 2H04 recognizes the glycan of residue N343 (Figure S4), we also assessed the binding of these two mAbs to RBD expressed in mammalian cells (Figures S5A and S5B). Both mAbs showed comparable affinities and kinetics to recombinant RBDs derived from either Expi293 cells (mammalian, (+)-glycosylation) or E. coli (bacterial, no glycosylation), suggesting that the glycans on the RBD do not contribute to the binding of 2H04 or 2B04.
Figure 6.
Mechanism of SARS-CoV-2 neutralization by 2B04 and 2H04
(A) Structural alignment of 2B04/RBD with ACE2/RBD complex (PDB: 6M0J). RBD is displayed as a surface model colored in gray. 2B04 is colored in cyan, and ACE2 is colored in green.
(B) Structural alignment of 2H04/RBD with ACE2/RBD complex (PDB: 6M0J). RBD is colored in gray, 2H04 is colored in magenta, and ACE2 is colored green.
(C and D) Binding affinity of 2B04 (C) and 2H04 (D) for SARS-CoV-2 RBD to bacterially derived SARS-CoV-2 RBD. The kinetic values were fitted to a 1:1 Langmuir binding model (KD, kinetic). Steady-state analysis is shown on bottom (KD, equilibrium), with inset Scatchard plots. The data were analyzed using Biaevaluation 3.1 (GE Healthcare) with a single representative sensogram of two independent experiments shown for each mAb.
(E) BLI traces of competitive binding of 2B04, 2H04, and ACE2 against SARS-CoV-2 RBD. mAbs were loaded onto anti-human IgG Fc biosensors, and recombinant RBD was captured, followed by either 2H04 Fab, 2B04 Fab, or monomeric human ACE2 (hACE2).
(F) Pre- and post-attachment inhibition assays. Serial dilutions of mAbs were either pre-incubated with GFP expressing VSV-SARS-CoV-2 followed by addition of mAb-virus mixture to Vero E6-TMPRSS2 cells (pre-attachment), or the mAbs were added to the cells after viral attachment (post-attachment). GFP+ infected cells were measured by flow cytometry 8 h post-infection. Experiments were performed two independent times in triplicate. Error bars indicate mean ± SEM.
(G) Attachment blockade assay with authentic SARS-CoV-2. The fold change of viral RNA on cell surfaces was measured by quantitative real-time PCR and compared with control cells infected with untreated virus. hE16 (anti-West Nile virus) was used as an isotype control.
(H) Cartoon depicting different SARS-CoV-2 spike configurations. Spike trimers with three RBDs in the “down” configuration cannot bind ACE2, whereas spike configurations with at least one RBD in the “up” configuration can bind ACE2.
(I) Cartoon model of SARS-CoV-2 neutralization by 2B04 and 2H04. 2H04 binds to SARS-CoV-2 virions and inhibits viral accumulation at cellular surfaces, possibly by preventing heparan sulfate or other attachment factors from interacting with the RBD. 2B04 neutralizes by binding to the RBM, directly preventing RBD binding to ACE2, which is required for cellular entry. Data in (G) are the mean of three independent experiments performed in duplicate. Error bars indicate SD. Statistical significance was determined by two-way ANOVA. ns, not significant; ∗p < 0.05; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
To investigate the neutralization mechanisms of 2B04 and 2H04, we performed a BLI-based competition binding assay. When 2B04 was immobilized, the binding of ACE2 to captured RBD was abrogated (Figure 6E), consistent with our structural predictions (Figure 6A). By contrast, 2H04 failed to block ACE2 binding, suggesting it might neutralize SARS-CoV-2 in a non-ACE2-competitive manner, also in agreement with our structural predictions (Figure 6B). Additionally, 2B04 and 2H04 were able to bind RBD simultaneously, supporting our structural observations that 2B04 and 2H04 recognize spatially distinct epitopes (Figures 2E and 6A and 6B).
We next performed pre- and post-attachment neutralization assays using a GFP-expressing replication-competent VSV-SARS-CoV-2 chimeric virus on Vero E6-TMPRSS2 cells (Case et al., 2020). 2B04 and 2H04 were incubated with VSV-SARS-CoV-2 before or after virus adsorption to the cell surface, and infection was monitored 8 h post-infection by flow cytometry. 2B04 efficiently inhibited VSV-SARS-CoV-2 infection when mixed with the virus before or after cell attachment (Figure 6F). 2H04, which did not compete with ACE2 for the binding of RBD (Figure 6E), neutralized infection more efficiently when added before virus attachment to cells. Thus, 2H04 likely affects viral attachment to these target cells (Figure 6F).
To further explore how 2H04 neutralizes SARS-CoV-2, we used quantitative real-time PCR to perform a cellular attachment inhibition assay with authentic SARS-CoV-2 on Vero E6-TMPRSS2 cells. As expected, 2B04 prevented SARS-CoV-2 attachment compared with the control mAb (hE16), which agrees with our ACE2 competition binding results and indicates that 2B04 interferes with viral engagement to cellular ACE2. Unexpectedly, 2H04 showed comparable attachment inhibition ability as 2B04 (Figure 6G), suggesting that 2H04 limits viral infection by blocking interaction with non-ACE2 attachment factors on cell surfaces, or possibly by sterically blocking the virion from close contact with host cells, thus inhibiting ACE2 engagement.
Recent studies have suggested that some neutralizing antibodies inhibit SARS-CoV-2 infection more potently than their monovalent Fab derivatives (Barnes et al., 2020b; Hansen et al., 2020; Liu et al., 2020a). To understand whether 2B04 or 2H04 also utilize avidity to neutralize SARS-CoV-2, we tested their potency in both bivalent and monovalent formats against chimeric VSV-SARS-CoV-2. Both 2B04 and 2H04 Fabs still showed inhibitory activity against VSV-SARS-CoV-2, with an ∼50- and ∼560-fold decrease in neutralization potency compared with intact 2B04 and 2H04 immunoglobulin Gs (IgGs), respectively (Figure S6). These data suggest that bivalent 2B04 and 2H04 IgG inhibit virus infection more effectively than their Fab derivatives.
Discussion
We have shown that two potently neutralizing mouse antibodies target distinct epitopes on the SARS-CoV-2 RBD, with one engaging the RBM and another binding nearby. Notably, both 2B04 and 2H04 target epitopes shared by human mAbs (i.e., 2-4 for 2B04 and S309 for 2H04). This suggests that these epitopes are immunogenic in both humans and mice and highlights the possibility of raising therapeutic antibodies in animals.
It was noted recently that certain antibodies targeting the RBM epitope, such as CC12.1 and B38, preferentially utilize the human IgHV3-53 heavy-chain gene and display relatively little somatic hypermutation (Yuan et al., 2020b). CC12.1 showed only 4 somatic mutations, whereas CC12.3 showed 3 substitutions and a deletion compared with its germline precursor (Yuan et al., 2020b). IgBLAST analysis of 2B04 shows a similarly low number of somatic mutations, with only 3 amino acid substitutions in the heavy chain and 2 in the light chain (Figures S7A and S7B). By contrast, 2H04 showed 6 heavy-chain mutations and 5 light-chain mutations (Figures S7C and S7D). Interestingly, despite bearing half as many somatic mutations as 2H04, 2B04 displays approximately 40-fold higher binding affinity for SARS-CoV-2 RBD (Figures 6C and 6D). Also notable is the conservation of paratope contact residues in 2B04. Only 2 of 15 contact residues in the heavy chain were mutated, with one of the two bearing a functionally conservative S31N mutation. Seven of these contact residues also were identical in the human IgHV3-53 gene, with a further 6 bearing conservative substitutions. Only two residues in 2B04 were completely non-conserved compared with human IgHV3-53, I30, and N58 (S30 and Y58 in IgHV3-53). Like IgHV3-53-derived antibodies in humans, the epitope targeted by 2B04 also appears to be relatively immunodominant in mice, as clonally and functionally similar antibodies have been identified (Alsoussi et al., 2020; Hassan et al., 2020).
One notable feature of 2B04 is its ability to bind RBD subunits in both the “up” and “down” conformations (Figures 1A and 1B and 6H). mAb 2-4 was only reported to bind “down” RBDs, whereas antibodies targeting the opposite flank of the RBM, such as CC12.1, only bind to RBD in the “up” conformation (Barnes et al., 2020a; Liu et al., 2020b; Wu et al., 2020a; Yuan et al., 2020b). Notably, the antibody BD23 was observed to bind only one “down” RBD per trimer (Cao et al., 2020), suggesting that binding orientation on the RBM ridge modulates antibody-binding stoichiometry to SARS-CoV-2 spike trimers. Because antibodies in these groups tend to display low ng/mL IC50 values, more studies are needed to determine whether differential binding stoichiometry has functional consequence. It is worth noting, however, that 2B04 appears to be the most potently neutralizing antibody covered in our analysis (IC50 of 1.47ng/mL against authentic virus). Additionally, a few other RBM antibodies that have been shown to bind both “up” and “down” RBDs, such as COVA2-15 and C144, display similarly potent neutralization (IC50 of 9 ng/mL against authentic virus for COVA2-15; IC50 of 2.55 ng/mL against authentic virus for C144) (Brouwer et al., 2020; Robbiani et al., 2020).
Several potently neutralizing mAbs inhibit SARS-CoV-2 infection through unknown mechanisms. Non-RBD mAbs 4A8 and COVA1-21 potently neutralize SARS-CoV-2 infection in vitro (Brouwer et al., 2020; Chi et al., 2020). S309 and 47D11 are mAbs that bind RBD, but cannot block spike binding to ACE2 (Pinto et al., 2020; Wang et al., 2020). We have shown that although 2H04 does not block ACE2 binding to the isolated RBD, it still impedes virus attachment as efficiently as the ACE2-blocking mAb 2B04, and unlike 2B04, loses much of its potency against virions already attached to host cells. These data indicate there may be alternative attachment receptor(s) in addition to ACE2 that 2H04 is blocking. Indeed, heparan sulfates and C-type lectins (DC-SIGN or L-SIGN) may have roles in SARS-CoV-2 infection and transmission (Amraie et al., 2020; Bermejo-Jambrina et al., 2020; Clausen et al., 2020; Partridge et al., 2020). Thus, we speculate that 2H04 might neutralize SARS-CoV-2 by inhibiting viral binding to target cell surfaces via unknown receptors or attachment factors. Importantly, these experiments do not exclude the possibility that 2H04 can block the trimeric spike binding ACE2 by selecting a particular spike conformation or otherwise sterically preventing dimeric ACE2 engagement (Barnes et al., 2020b; Huo et al., 2020; Liu et al., 2020a). Further studies are required to delineate which of these possibilities plays a dominant role in neutralization.
An antibody identified in a SARS-CoV patient from 2003, mAb S309, was shown to cross-neutralize SARS-CoV-2 (Pinto et al., 2020). Both S309 and 2H04 target an overlapping RBM-adjacent epitope with a relatively high degree of conservation of contact residues between SARS-CoV and SARS-CoV-2. Both antibodies, as well as the SARS-CoV-2-specific C135 (Barnes et al., 2020b), also neutralize SARS-CoV-2 without blocking ACE2 binding. As 2H04 neutralizes SARS-CoV-2 by reducing cellular attachment without blocking ACE2 engagement, we predict that S309 and C135 also inhibit SARS-CoV-2 infection through a similar mechanism. Although 2H04 exhibits relatively low-affinity binding to SARS-CoV RBD, it may be possible to enhance the affinity of 2H04 for SARS-CoV RBD by structure-guided mutagenesis, thus enhancing cross-neutralization of 2H04, enabling its potential use as a dual therapeutic option for both SARS-CoV and SARS-CoV-2.
The facts that VSV expressing the SARS-CoV-2 spike can escape antibody-mediated neutralization and that mutations at identical residues have been observed in naturally occurring SARS-CoV-2 variants suggest that SARS-CoV-2 might escape antibody-mediated neutralization when used as monotherapy. In line with these predictions, variants of SARS-CoV-2 containing RBD mutations that disrupt neutralization by RBM-targeting antibodies, including 2B04 and 2H04, and convalescent and vaccinee serum-derived polyclonal antibodies, have recently emerged (Figure 5) (Chen et al., 2021; Garcia-Beltran et al., 2021; Wang et al., 2021a, 2021b). K417, E484, and N501 are substituted in multiple variants of concern and are contact residues for a variety of previously described RBM-targeted antibodies, particularly those utilizing the IgHV3-53 germline sequence (Figures 3C, 5, and S7). Notably, the epitopes targeted by REGN-10933 and REGN-10987, which comprise the REGN-COV2 therapeutic, encompass contacts that are subtituted in these variants (K417, E484, and N501 for REGN-10933; N501 for REGN-10987) (Figure 3C). Although REGN-10933 displays less neutralizing potency against B.1.351, in combination with REGN-10987 it retains most of its neutralizing efficacy (Tada et al., 2021; Wang et al., 2021a). The emergence and spread of neutralization-resistant variants highlights the ongoing threat of resistance, underscoring the need for continued development of next generation therapeutic antibody cocktails.
Overall, we report high-resolution cryo-EM structures of two anti-SARS-CoV-2 neutralizing antibodies that bind to the RBD portion of the spike protein. These antibodies target distinct epitopes and act through disparate mechanisms of action (Figure 6I). These findings advance our understanding of the epitopes targeted by multiple species of vertebrate when challenged with SARS-CoV-2 and highlight two candidate antibodies that can be used together, and in conjunction with other antibodies, for the prevention or treatment of SARS-CoV-2 infection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| 2B04, anti-SARS-CoV-2 mAb | Alsoussi et al., 2020 | N/A |
| 2H04, anti-SARS-CoV-2 mAb | Alsoussi et al., 2020 | N/A |
| Bacterial and virus strains | ||
| E. coli XL10 | Agilent | Cat # 200314 |
| E. coli BL21(DE3) | Agilent | Cat # 200131 |
| SARS-CoV-2 (strain 2019 n-CoV/USA_WA1/2020) | CDC | NR52281 |
| Chimeric VSV-SARS-CoV-2 virus | Case et al., 2020 | Spike mutated from MN908947.3 |
| SARS-CoV-2 D614G | Chen et al., 2021 | N/A |
| SARS-CoV-2 N501Y + D614G | Chen et al., 2021 | N/A |
| SARS-CoV-2 E484K + D614G | Chen et al., 2021 | N/A |
| SARS-CoV-2 B.1.1.7 | Chen et al., 2021 | N/A |
| SARS-CoV-2 Wash-B.1.351 | Chen et al., 2021 | N/A |
| SARS-CoV-2 Wash-B.1.1.28 | Chen et al., 2021 | N/A |
| SARS-CoV-2 B.1.429 | Chen et al., 2021 | N/A |
| SARS-CoV-2 B.1.617.1 | Chen et al., 2021 | N/A |
| SARS-CoV-2 B.1.526 (S477N) | Chen et al., 2021 | N/A |
| SARS-CoV-2 B.1.526 (E484K) | Chen et al., 2021 | N/A |
| SARS-CoV-2 B.1.1.298 | VanBlargan et al., 2021 | N/A |
| SARS-CoV-2 B.1.222 | VanBlargan et al., 2021 | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant SARS-CoV-2 spike protein (Mammalian cell) | This paper | GenBank: MN908947.3 |
| Recombinant SARS-CoV-2 RBD protein (Mammalian cell) | This paper | GenBank: MN908947.3 |
| Recombinant SARS-CoV-2 RBD protein (E. coli) | This paper | GenBank: MN908947.3 |
| Human ACE2-Fc | This paper | GenBank: BAB40370.1 & AAC82527.1 |
| Deposited data | ||
| 2B04 bound to SARS-CoV-2 spike model | This paper | PDB: 7K9H |
| 2B04 bound to SARS-CoV-2 spike (local refinement) model | This paper | PDB: 7K9I |
| 2H04 bound to SARS-CoV-2 spike model | This paper | PDB: 7K9J |
| 2H04 bound to SARS-CoV-2 spike (local refinement) model | This paper | PDB: 7K9K |
| 2B04 bound to SARS-CoV-2 spike U/D/D map | This paper | EMDB: EMD-22748 |
| 2B04 bound to SARS-CoV-2 spike (local refinement) map | This paper | EMDB: EMD-22749 |
| 2B04 bound to SARS-CoV-2 spike U/U/D map | This paper | EMDB: EMD-22752 |
| 2H04 bound to SARS-CoV-2 spike D/D/D map | This paper | EMDB: EMD-22750 |
| 2H04 bound to SARS-CoV-2 spike (local refinement) map | This paper | EMDB: EMD-22751 |
| 2H04 bound to SARS-CoV-2 spike U/D/D map | This paper | EMDB: EMD-22753 |
| Experimental models: Cell lines | ||
| Expi293F cells | Expi293F | Cat# A14527 |
| Vero E6-TMPRSS2 cells | Case et al., 2020 | N/A |
| Vero CCL81 | ATCC | Cat# CCL-81 |
| Oligonucleotides | ||
| SARS-CoV-2-Fwd, 5′-ATGCTGCAATCG TGCTACAA-3′ |
This paper | N/A |
| SARS-CoV-2-Rev, 5′-GACTGCCGCCT CTGCTC-3′ |
This paper | N/A |
| Probe SARS-CoV-2-P, /56-FAM/TCAAG GAACAACATTGCCAA/3BHQ_1/ |
This paper | N/A |
| GAPDH-Fwd, 59-TGTAGTTGA GGTCA ATGAAGGG-39 |
This paper | N/A |
| Probe GAPDH, 56-FAM/AAGGTCGGA/ZEN/GTCAAC GGATTTGGTC/3IABkFQ | This paper | N/A |
| Recombinant DNA | ||
| pFM1.2-hACE2-Fc | This paper | GenBank: AB046569.1 |
| pCAGGS-SARS-CoV-2-spike | This paper | GenBank: MN908947.3 |
| pET21a-SARS-CoV-2-RBD | This paper | GenBank: MN908947.3 |
| pCAGGS-SARS-CoV-2-RBD | This paper | GenBank: MN908947.3 |
| Software and algorithms | ||
| Prism 8.0 | GraphPad | v8 |
| FlowJo | FlowJo, LLC | v10 |
| BIAevaluation version 3.1 | GE Healthcare | v4.1 |
| Relion 3.1 | Zivanov et al., 2018 | v3.1 |
| MotionCor2 | Zheng et al., 2017 | v1.3.1 |
| GCTF | Zhang, 2016 | v1.06 |
| CryoSPARC v2.15 | Structura Biotechnology Inc. | v2.15 |
| Coot | Emsley et al., 2010 | v0.91 |
| Isolde | Croll, 2018 | v0.93 |
| UCSF ChimeraX | Goddard et al., 2018 | v0.9 |
| Other | ||
| Anti-human Fc Biosensor | ForteBio | Cat # 18-5063 |
| Protein A Agarose Resin | GoldBio | Cat # P-400-100 |
| Lacey carbon TEM grid | Ted Pella | Cat # 01895-F |
| Lacey carbon TEM grid with ultra-thin carbon film | Ted Pella | Cat # 01824-G |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Daved H. Fremont (fremont@wustl.edu).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Cells
Vero cells were cultured at 37°C in DMEM supplemented with 10% FBS. Expi293 cells were cultured in Expi293 serum-free media at 37°C in an 8% CO2 atmosphere.
Viruses
SARS-CoV-2 (2019 n-CoV/USA_WA1/2020) was obtained from the Centers for Disease Control and amplified in Vero CCL81 cells. Chimeric VSV-SARS-CoV-2 stocks were generated as previously described (Case et al., 2020). The chimeric virus was amplified on MA104 cells, and neutralization assays were performed on Vero-E6-TMPRSS2 cells. SARS-CoV-2 variants viruses were amplified on Vero-E6-TMPRSS2 cells as previously described (Chen et al., 2021; VanBlargan et al., 2021).
Recombinant proteins
Recombinant mammalian-derived SARS-CoV-2 spike protein RBD was produced in Expi293 cells grown in a 37°C 8% CO2 atmosphere humidified shaker incubator. Bacterially-derived SARS-CoV-2 RBD was produced in E. coli BL21(DE3) cells grown in liquid culture at 37°C in a shaker incubator.
Method details
Protein production and purification
Genes encoding SARS-CoV-2-spike (residues 1-1213, GenBank: MN908947.3) and RBD (residues 319-541) were cloned into a mammalian expression vector with a C-terminal hexahistidine tag. For spike, 986KV987 was mutated to 986PP987, S1/S2 furin cleavage sites were disrupted to stabilize the prefusion conformation, and a C-terminal foldon trimerization motif was incorporated. The vectors were transiently transfected into Expi293F cells with ExpiFectamine, and recombinant spike and RBD proteins were purified from culture supernatants using cobalt-charged resin (G-Biosciences). In some cases, size exclusion chromatography was used for additional purification (superose 6 increase for spike, and superdex 75 for RBD).
Untagged RBD was expressed in E. coli BL21(DE3) cells and oxidatively refolded from inclusion bodies as previously described (Oliphant et al., 2007). RBD variants were made using a Q5 Site-Directed Mutagenesis Kit (NEB), and were expressed and purified as described for WT RBD. Genes encoding human ACE2 (hACE2 residues 1-615) were synthesized (IDT) and placed into a mammalian expression vector with a C-terminal HRV-3C protease cleavage site and a human Fc fragment, as previously reported (Case et al., 2020). The vector was transfected into Expi293F cells using FectoPRO (Poly-plus) and hACE2-hFc was purified from culture supernatants 4 days post transfection by affinity chromatography using protein A resin (GoldBio). Monomeric hACE2 was generated by incubating hACE2-hFc with HRV-3C protease overnight at 4°C. hACE2 was subsequently purified by passage over a protein A column to remove cleaved Fc and further purified through size exclusion chromatography.
Cryo-EM sample preparation
For standard lacey carbon grid (Ted Pella #01895-F) datasets, trimeric SARS-CoV-2 spike at a concentration of 1 mg/mL in 20 mM HEPES pH 7.5, 150 mM NaCl was combined with 1 molar equivalent of a papain-cleaved Fab form of either 2B04 or 2H04 in 20 mM HEPES pH 7.5, 150 mM NaCl, and incubated for 10 minutes before flash-freezing on lacey carbon grids using a Vitrobot Mk IV (ThermoFisher Scientific). For lacey carbon grids with ultra-thin carbon film (Ted Pella #01824G), SARS-CoV-2 spike was diluted to 0.2 mg/mL prior to mixing with 1 molar equivalent of either 2B04 or 2H04 Fab and then vitrified on thin-film lacey carbon grids using a Vitrobot Mk IV (ThermoFisher Scientific). Both sets of grids were glow discharged prior to sample application in a GloQube (EMS) for at least 20 s under vacuum.
Cryo-EM data collection
Frozen grids were transferred to a Cs-corrected FEI Titan Krios 300KV microscope equipped with a Gatan K2 Summit detector mounted on a BioQuantum 968 energy filter operating in zero loss mode with a slit width of 20 eV. Movies were collected at a nominal magnification of 105,000X resulting in a pixel size of 1.1 Å/pixel, with 45 frames per movie at 200ms each with a dose of 1.49 e-/Å2/frame, resulting in a total dose of 66.9 e-/Å2/movie.
Cryo-EM data processing
Raw movies for both datasets were motion corrected using MotionCor2 v1.3.1 (Zheng et al., 2017). Micrograph contrast transfer function correction parameters were estimated using GCTF v1.06 (Zhang, 2016). Particles were picked on each dataset using CrYOLO v1.7.1 employing a general model (Wagner et al., 2019). Particles were first subjected to 2D classification in relion 3.1 (Scheres, 2012; Zivanov et al., 2018). Good 2D classes from both holey lacey carbon and thin-film lacey carbon particle sets were picked and merged for both 2B04 and 2H04. The merged particle datasets were then subjected to 3D classification using a 7.5 degree angular search for 25 iterations followed by 25 iterations of local searches at 1.8 degree sampling to separate spike conformational states, as previously described (Walls et al., 2020). Particles for each conformational state were put through CTF refinement and Bayesian polishing in relion 3.1 (Zivanov et al., 2019) prior to non-uniform refinement in cryoSPARC v2.15 to generate the final full-spike maps (Punjani et al., 2017). To enhance resolution and map quality at the Fab/RBD interface, masks encompassing either one FV and RBD of the U/D/D 2B04 dataset or all three FV and RBD positions for the D/D/D 2H04 dataset were used for focused classification on their respective particle sets in relion 3.1. Classes with well-resolved detail in the masked region were selected, and the particles expanded with C3 symmetry. Finally, the particles were subjected to local non-uniform refinement in cryoSPARC v2.15 using a mask encompassing one FV/RBD for both 2B04 and 2H04 to generate the locally refined maps.
Model building
For the locally refined maps, an initial model was generated by using protein BLAST with the Fab sequences to identify pre-existing Fab models that displayed high sequence similarity to 2B04 (PDB: 1GIG) and 2H04 (PDB: 6DG2 for heavy chain, PDB: 1K4C for light chain). The initial model for the RBD for 2B04 was taken from the crystal structure of Fab CR3022 bound to SARS-CoV-2 RBD (PDB 6W41), while for 2H04 it was derived from the cryo-EM structure of SARS-CoV-2 spike (PDB 6VXX). All starting model components were combined and rigid body fit into their respective maps. Model building and refinement were performed with Coot 0.91 (Emsley et al., 2010), Isolde v0.93 (Croll, 2018), and phenix (Adams et al., 2010). Initial models for the full spike/Fab complex structures were generated by combining the locally-refined structures of 2B04 and 2H04 with previously solved cryo-EM structures of SARS-CoV-2 spike proteins in the proper conformational configurations (PDB 6VYB for 2B04 U/D/D, PDB 6VXX for 2H04 D/D/D). These models were refined subsequently using Coot 0.91, Isolde v0.93, and Phenix. Contact residues at the interfaces of the locally refined models were identified using qtPISA to generate epitope footprints. Buried surface areas were calculated in UCSF ChimeraX with a probe radius of 1.4 Å. Structures were visualized using UCSF ChimeraX (Goddard et al., 2018).
Binding analysis via biolayer interferometry (BLI)
Biolayer interferometry (BLI) was used to investigate the binding capacity of mAbs to wild-type or mutant RBDs on an Octet-Red96 device (ForteBio). 10 μg/mL of 2B04, 2H04 or 20 μg/mL ACE2-hFc were immobilized onto anti-human IgG Fc biosensors (ForteBio) for 3 minutes. After a 30 s wash, the pins were dipped into running buffer (10 mM HEPES, 150 mM NaCl, 3 mM EDTA, and 0.005% P20 surfactant with 3% BSA) containing 500 nM recombinant RBDs (RBD343-346, RBD486-490, RBD500-505 and RBDWT, respectively) to measure association, followed by a dissociation step in running buffer alone. The BLI traces were recorded and the maximum binding signals were averaged at steady state and analyzed with Prism (Version 8.0).
Competition-binding analysis through BLI
Anti-human IgG Fc biosensors were loaded with 2B04 or 2H04 IgG (10 μg/mL), with a parallel control of buffer alone to monitor non-specific binding. After a 30 s wash, the sensors were immersed into RBD-containing wells for 1 minute to capture RBD molecules, followed by immersion into buffer containing either 2H04 Fab, 2B04 Fab, or monomeric ACE2 for 1 minute, with binding curves recorded in real time. The two tested subunits were considered competitive if no increase in BLI signal was observed.
Pre- and post- attachment neutralization assay
For the pre-attachment assay, serially diluted 2B04 or 2H04 were incubated with GFP expressing chimeric VSV-SARS-CoV-2 for 1 hour at 4°C. The virus-mAb mixture was subsequently added to pre-cooled Vero E6-TMPRSS2 cells and incubated for 1 hour. Cells were washed three times with cold DMEM to remove unbound virus, and the plates were transferred to a 37°C incubator with 5% CO2. 8 hours post-infection, the cells were trypsinized (Trypsin-EDTA) and fixed with 4% paraformaldehyde. Infection frequency was quantified by measuring the number of GFP-positive infected cells via flow cytometry. For the post-attachment assay, the virus was first incubated with chilled cells at 4°C. One hour later, cells were washed with cold media to remove unbound virus, followed by addition of serial dilutions of 2B04 or 2H04 and incubation for 1 hour at 4°C. Viral infection was quantified as described above. Relative infection was calculated by comparing cells infected with mAb-bound virus to cells infected with untreated virus. IC50 values were determined by non-linear regression with values constrained between 0 and 100. During this experiment, we observed a viral fraction that resisted neutralization by 2H04; similar findings have been reported for other viruses (i.e., alphaviruses and flaviviruses) (Earnest et al., 2019; Nelson et al., 2008)
Attachment blockade assay
Vero E6-TMPRSS2 cells (2 × 105) were seeded in 24-well plates and incubated at 37°C for 24 hours. Diluted mAbs (2B04 or 2H04) were premixed with SARS-CoV-2 (MOI of 0.01) and incubated for 1 hour at 4°C, followed by addition of the mAb-virus mixture to chilled Vero E6-TMPRSS2 cells for 1 hour at 4°C. Virus alone and a control antibody (humanized anti-West Nile virus mAb, hE16) (Oliphant et al., 2005) were included. Cells were then rinsed 4 times with chilled DMEM and once with PBS on ice before total cellular RNA extraction using a MagMAX mirVana Total RNA Isolation Kit (A27828). Viral RNA levels were measured by qRT-PCR on an ABI 7500 Real Time-PCR system (Applied Biosystems) and normalized to an internal GAPDH control. The following primers and probes were used: SARS-CoV-2-Fwd, 5′-ATGCTGCAATCGTGCTACAA-3′; SARS-CoV-2-Rev, 5′-GACTGCCGCCTCTGCTC-3′; Probe SARS-CoV-2-P, /56-FAM/TCAAGGAACAACATTGCCAA/3BHQ_1/; GAPDH-Fwd, 59-TGTAGTTGA GGTCAATGAAGGG-39; GAPDH-Rev, 59-ACATCGCTCAGACAC CATG-39; Probe GAPDH, 56-FAM/AAGGTCGGA/ZEN/GTCAAC GGATTTGGTC/3IABkFQ.
SARS-CoV-2 variant neutralization assays
Variant focus reduction neutralization assays were conducted as previously described (Chen et al., 2021). Briefly, Serial 10-fold dilutions of 2B04 and 2H04 were incubated with 100 FFUs of SARS-CoV-2 variant strains for 1 hour at 37°C. The antibody-virus complexes were then added to Vero-hACE2-TMPRSS2 cell monolayers and incubated for 1 hour at 37°C. The cells were then overlaid with 1% w/v methylcellulose in MEM supplemented with 2% FBS. After 24 hours, overlays were removed and cells were fixed with 4% PFA in PBS for 20 minutes at room temperature. Plates were washed and incubated with an oligoclonal pool of anti-SARS-CoV-2 spike antibodies followed by HRP-conjugated goat anti-mouse IgG (Sigma) in PBS with 0.1% saponin and 0.1% BSA. Plates were developed with TrueBlue peroxidase substrate and quantified using an ImmunoSpot microanalyzer.
Quantification and statistical analysis
No statistical methods were used to determine appropriate powering of experiments a priori. Details of statistical methods can be found in figure legends associated with the experiment upon which statistical methods were applied.
Acknowledgments
This work was supported by National Institute of Allergy and Infectious Diseases contract 75N93019C00062 (to M.S.D., A.H.E., and D.H.F.), contract HHSN272201700060C (to D.H.F.), and grant R01 AI157155 (to M.S.D.). We gratefully acknowledge Raul Andino, Charles Chiu, Mehul Suthar, and BEI for contributing SARS-CoV-2 variant strains, and members of the Fremont and Diamond laboratories for valuable discussions.
Author contributions
J.M.E., H.Z., Z.L., S.P.J.W., and D.H.F. designed the experiments and D.H.F. supervised the project. J.M.E., M.J.R., and J.A.J.F. collected cryo-EM data. J.M.E. processed cryo-EM data, built models, and performed structural analyses. H.Z. and M.M. expressed recombinant proteins. H.Z. performed the BLI experiments and pre- and post-attachment experiments. H.Z. and J.B.C. performed SARS-CoV-2 cellular binding inhibition studies. R.E.C. tested neutralization against variants; P.-Y.S. provided variant SARS-CoV-2 viruses; A.J.S. and A.H.E. generated and provided the recombinant mAbs; and J.M.E. generated Fabs. J.M.E., H.Z., Z.L., S.P.J.W., M.S.D., and D.H.F. analyzed the data. J.M.E., H.Z., and D.H.F. wrote the initial manuscript, with all authors providing editorial input.
Declaration of interests
D.H.F. is a founder of Courier Therapeutics. A.H.E. is a consultant for Inbios and Fimbrion Therapeutics. M.S.D. is a consultant for Inbios, Vir Biotechnology, and NGM Biopharmaceuticals and is on the Scientific Advisory Board of Moderna and Immunome. D.H.F., M.S.D., and A.H.E. have received unrelated funding support from Emergent BioSolutions. M.S.D. has sponsored research agreements from Moderna and Vir Biotechnology, A.H.E. has a sponsored research agreement from Abbvie, and D.H.F. has a sponsored research agreement from Mallinckrodt Pharmaceuticals.
Published: October 8, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109881.
Supplemental information
Data and code availability
-
•
Density maps used to build models were deposited in the EMDB with the following accession numbers: 2B04 U/D/D as EMDB: EMD-22748, 2B04/RBD locally refined as EMDB: EMD-22749, 2H04 D/D/D as EMDB: EMD-22750, and 2H04/RBD locally refined as EMDB: EMD-22751. Models built with these maps were deposited in the PDB with the following accession codes: 2B04 U/D/D as PDB: 7K9H, 2B04/RBD locally refined as PDB: 7K9I, 2H04 D/D/D as PDB: 7K9J, and 2H04/RBD locally refined as PDB: 7K9K. Additional maps of 2B04 U/U/D and 2H04 U/D/D were deposited in the EMDB with the following accession numbers: 2B04 U/U/D as EMDB: EMD-22752, and 2H04 U/D/D as EMDB: EMD-22753.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalayze the data reported in this paper is available from the lead contact upon request.
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.-W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alsoussi W.B., Turner J.S., Case J.B., Zhao H., Schmitz A.J., Zhou J.Q., Chen R.E., Lei T., Rizk A.A., McIntire K.M., et al. A Potently Neutralizing Antibody Protects Mice against SARS-CoV-2 Infection. J. Immunol. 2020;205:915–922. doi: 10.4049/jimmunol.2000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amraie R., Napoleon M.A., Yin W., Berrigan J., Suder E., Zhao G., Olejnik J., Gummuluru S., Muhlberger E., Chitalia V., et al. CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2 and are differentially expressed in lung and kidney epithelial and endothelial cells. BioRxiv. 2020 doi: 10.1101/2020.06.22.165803. [DOI] [Google Scholar]
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 2020;182:828–842.e16. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo-Jambrina M., Eder J., Kaptein T.M., Helgers L.C., Brouwer P.J.M., van Hamme J.L., Vlaar A.P.J., van Baarle F.E.H.P., de Bree G.J., Nijmeijer B.M., et al. SARS-CoV-2 Infection and Transmission Depends on Heparan Sulfates and Is Blocked by Low Molecular Weight Heparins. BioRxiv. 2020 2020.08.18.255810. [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G., et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C., et al. Potent Neutralizing Antibodies against SARS-CoV-2 Identified by High-Throughput Single-Cell Sequencing of Convalescent Patients’ B Cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Rothlauf P.W., Chen R.E., Liu Z., Zhao H., Kim A.S., Bloyet L.-M., Zeng Q., Tahan S., Droit L., et al. Neutralizing Antibody and Soluble ACE2 Inhibition of a Replication-Competent VSV-SARS-CoV-2 and a Clinical Isolate of SARS-CoV-2. Cell Host Microbe. 2020;28:475–485.e5. doi: 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N., et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell. 2020;183:1043–1057.e15. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll T.I. ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D Struct. Biol. 2018;74:519–530. doi: 10.1107/S2059798318002425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnest J.T., Basore K., Roy V., Bailey A.L., Wang D., Alter G., Fremont D.H., Diamond M.S. Neutralizing antibodies against Mayaro virus require Fc effector functions for protective activity. J. Exp. Med. 2019;216:2282–2301. doi: 10.1084/jem.20190736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., St. Denis K., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383.e9. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.J., Ni Z.Y., Hu Y., Liang W.H., Ou C.Q., He J.X., Liu L., Shan H., Lei C.L., Hui D.S.C., et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K., et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan A.O., Case J.B., Winkler E.S., Thackray L.B., Kafai N.M., Bailey A.L., McCune B.T., Fox J.M., Chen R.E., Alsoussi W.B., et al. A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell. 2020;182:744–753.e4. doi: 10.1016/j.cell.2020.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo J., Zhao Y., Ren J., Zhou D., Duyvesteyn H.M.E., Ginn H.M., Carrique L., Malinauskas T., Ruza R.R., Shah P.N.M., et al. Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike. Cell Host Microbe. 2020;28:445–454.e6. doi: 10.1016/j.chom.2020.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurlburt N.K., Seydoux E., Wan Y.-H., Edara V.V., Stuart A.B., Feng J., Suthar M.S., McGuire A.T., Stamatatos L., Pancera M. Structural basis for potent neutralization of SARS-CoV-2 and role of antibody affinity maturation. Nat. Commun. 2020;11:5413. doi: 10.1038/s41467-020-19231-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju B., Zhang Q., Ge J., Wang R., Sun J., Ge X., Yu J., Shan S., Zhou B., Song S., et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature. 2020;584:115–119. doi: 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B., et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- Liu H., Wu N.C., Yuan M., Bangaru S., Torres J.L., Caniels T.G., van Schooten J., Zhu X., Lee C.D., Brouwer P.J.M., et al. Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity. Immunity. 2020;53:1272–1280.e5. doi: 10.1016/j.immuni.2020.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F.-W., Sahi V., Figueroa A., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Liu Z., VanBlargan L.A., Bloyet L.-M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J., et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488.e4. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Z., Deng Y.-Q., Ye Q., Cao L., Sun C.-Y., Fan C., Huang W., Sun S., Sun Y., Zhu L., et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science. 2020;369:1505–1509. doi: 10.1126/science.abc5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson S., Jost C.A., Xu Q., Ess J., Martin J.E., Oliphant T., Whitehead S.S., Durbin A.P., Graham B.S., Diamond M.S., Pierson T.C. Maturation of West Nile virus modulates sensitivity to antibody-mediated neutralization. PLoS Pathog. 2008;4:e1000060. doi: 10.1371/journal.ppat.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant T., Engle M., Nybakken G.E., Doane C., Johnson S., Huang L., Gorlatov S., Mehlhop E., Marri A., Chung K.M., et al. Development of a humanized monoclonal antibody with therapeutic potential against West Nile virus. Nat. Med. 2005;11:522–530. doi: 10.1038/nm1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant T., Nybakken G.E., Austin S.K., Xu Q., Bramson J., Loeb M., Throsby M., Fremont D.H., Pierson T.C., Diamond M.S. Induction of epitope-specific neutralizing antibodies against West Nile virus. J. Virol. 2007;81:11828–11839. doi: 10.1128/JVI.00643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge L.J., Urwin L., Nicklin M.J.H., James D.C., Green L.R., Monk P.N. ACE2-independent interaction of SARS-CoV-2 spike protein to human epithelial cells can be inhibited by unfractionated heparin. BioRxiv. 2020 doi: 10.3390/cells10061419. 2020.05.21.107870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H.W. A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer J., Gifford R., Cotten M., Robertson D. CoV-GLUE: A Web Application for Tracking SARS-CoV-2 Genomic Variation. Preprints. 2020 doi: 10.20944/preprints202006.0225.v1. [DOI] [Google Scholar]
- Tada T., Dcosta B.M., Zhou H., Vaill A., Kazmierski W., Landau N.R. Decreased neutralization of SARS-CoV-2 global variants by therapeutic anti-spike protein monoclonal antibodies. BioRxiv. 2021 2021.02.18.431897. [Google Scholar]
- VanBlargan L.A., Adams L.J., Liu Z., Chen R.E., Gilchuk P., Raju S., Smith B.K., Zhao H., Case J.B., Winkler E.S., et al. A potently neutralizing SARS-CoV-2 antibody inhibits variants of concern by utilizing unique binding residues in a highly conserved epitope. Immunity. 2021 doi: 10.1016/j.immuni.2021.08.016. S1074-7613(21)00348-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T., Merino F., Stabrin M., Moriya T., Antoni C., Apelbaum A., Hagel P., Sitsel O., Raisch T., Prumbaum D., et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2019;2:218. doi: 10.1038/s42003-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Li W., Drabek D., Okba N.M.A., van Haperen R., Osterhaus A.D.M.E., van Kuppeveld F.J.M., Haagmans B.L., Grosveld F., Bosch B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N.C., Yuan M., Liu H., Lee C.D., Zhu X., Bangaru S., Torres J.L., Caniels T.G., Brouwer P.J.M., van Gils M.J., et al. An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain. Cell Rep. 2020;33:108274. doi: 10.1016/j.celrep.2020.108274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y., et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.-P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Duyvesteyn H.M.E., Chen C.-P., Huang C.-G., Chen T.-H., Shih S.-R., Lin Y.-C., Cheng C.-Y., Cheng S.-H., Huang Y.-C., et al. Structural basis for the neutralization of SARS-CoV-2 by an antibody from a convalescent patient. Nat. Struct. Mol. Biol. 2020;27:950–958. doi: 10.1038/s41594-020-0480-y. [DOI] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Forsberg B.O., Kimanius D., Hagen W.J., Lindahl E., Scheres S.H. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivanov J., Nakane T., Scheres S.H.W. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ. 2019;6:5–17. doi: 10.1107/S205225251801463X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S., et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Density maps used to build models were deposited in the EMDB with the following accession numbers: 2B04 U/D/D as EMDB: EMD-22748, 2B04/RBD locally refined as EMDB: EMD-22749, 2H04 D/D/D as EMDB: EMD-22750, and 2H04/RBD locally refined as EMDB: EMD-22751. Models built with these maps were deposited in the PDB with the following accession codes: 2B04 U/D/D as PDB: 7K9H, 2B04/RBD locally refined as PDB: 7K9I, 2H04 D/D/D as PDB: 7K9J, and 2H04/RBD locally refined as PDB: 7K9K. Additional maps of 2B04 U/U/D and 2H04 U/D/D were deposited in the EMDB with the following accession numbers: 2B04 U/U/D as EMDB: EMD-22752, and 2H04 U/D/D as EMDB: EMD-22753.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalayze the data reported in this paper is available from the lead contact upon request.