Abstract
NF-Y is a CCAAT-binding trimer with two histonic subunits, NF-YB and NF-YC, resembling H2A-H2B. We previously showed that the short conserved domains of NF-Y efficiently bind to the major histocompatibility complex class II Ea Y box in DNA nucleosomized with purified chicken histones. Using wild-type NF-Y and recombinant histones, we find that NF-Y associates with H3-H4 early during nucleosome assembly, under conditions in which binding to naked DNA is not observed. In such assays, the NF-YB–NF-YC dimer forms complexes with H3-H4, for whose formation the CCAAT box is not required. We investigated whether they represent octamer-like structures, using DNase I, micrococcal nuclease, and exonuclease III, and found a highly positioned nucleosome on Ea, whose boundaries were mapped; addition of NF-YB–NF-YC does not lead to the formation of octameric structures, but changes in the digestion patterns are observed. NF-YA can bind to such preformed DNA complexes in a CCAAT-dependent way. In the absence of DNA, NF-YB–NF-YC subunits bind to H3-H4, but not to H2A-H2B, through the NF-YB histone fold. These results indicate that (i) the NF-Y histone fold dimer can efficiently associate DNA during nucleosome formation; (ii) it has an intrinsic affinity for H3-H4 but does not form octamers; and (iii) the interactions between NF-YA, NF-YB–NF-YC, and H3-H4 or nucleosomes are not mutually exclusive. Thus, NF-Y can intervene at different steps during nucleosome formation, and this scenario might be paradigmatic for other histone fold proteins involved in gene regulation.
Gene expression is controlled by gene-specific trans-acting factors and general transcription proteins recognizing discrete elements in promoters-enhancers and operating in the context of chromatin structures (reviewed in reference 45). The fundamental chromatin unit is the nucleosome, a DNA-protein complex formed by 146 bp of DNA that wraps around core histones, H2A, H2B, H3, and H4. Histones are among the most conserved proteins in evolution, having in their C-terminal regions a 65-amino-acid histone fold motif (HFM) with low sequence identity (14 to 18%) and high structural resemblance (2). It is minimally composed of three α helices, α1, α2, and α3. The long central α2 (28 amino acids) is flanked by two short ones separated by loop-strand regions; this structure enables histone-histone interactions and contacts with the DNA (2, 29). Histones also possess N-terminal tails that are acetylated at specific lysine residues. This process is highly regulated by several acetylases, including some transcriptional coactivators, and is thought to contribute to regulation of gene expression in multiple ways: facilitation of transcription factors binding (43) and of RNA polymerase progression (42) and chromatin solubility (38). Formation of nucleosomes is a stepwise phenomenon initiated by tetramerization of H3-H4 through H3-H3 interactions and binding of the tetramer to DNA; this nucleates wrapping of DNA, but formation of a stable complex requires subsequent association of two H2A-H2B dimers, mainly through H2B-H4 contacts (12, 16–18). H3-H4 tetramers, but not H2A-H2B dimers, dictate nucleosome positioning (12).
Several polypeptides involved in the process of transcriptional regulation were shown to contain HFM domains (3): (i) Drosophila TAFII60 (dTAFII60)/human TAFII80 (hTAFII80), dTAFII40/hTAFII31, hTAFII28, hTAFII18, and hTAFII20/dTAFII30, which are part of the general TFIID complex (crystallization of dTAFII60/dTAFII40 and hTAFII28/hTAFII18 dimers revealed their histone-like structures [6, 8]); (ii) the two subunits of NC2 (also called Dr1/DRAP1), which bind TATA-binding protein and repress transcription (15); (iii) an hTAFII80-like subunit (PAF65α) of the P/CAF histone acetylase complex (37); and (iv) two subunits (NF-YB and NF-YC) of NF-Y, a ubiquitous CCAAT-binding heterotrimeric complex (3).
The CCAAT box is present in 25% of eukaryotic promoters, with a strong position preference at −60 to −80 (33). In vivo footprinting of several promoters invariably found this element protected, and functional experiments indicate that it plays an important and sometimes essential role in transcription. NF-Y, originally identified as the protein binding to the major histocompatibility complex (MHC) class II Ea promoter Y box, has an almost absolute requirement for the five CCAAT nucleotides and has been implicated in the activation of most, if not all, CCAAT-containing promoters (reviewed in references 31 and 33). It is composed of three different subunits, NF-YA, NF-YB, NF-YC, each containing evolutionarily conserved domains. NF-YB and NF-YC belong to the H2A-H2B subfamily; their dimerization, elicited through strong HFM interactions, is required for NF-YA binding. For this function, a complex surface resulting from heterodimerization and comprising specific residues in NF-YC α1, in NF-YB α2, and at the C terminus of α3 is necessary (21, 40). Detailed mutational analysis of NF-YA and of the yeast homologue HAP2 identified a 56-amino-acid region that can be split into two short separable parts, responsible for contacting NF-YB–NF-YC and DNA (32, 46). Similarly, NF-YB and NF-YC histone folds contain DNA-binding subdomains (21, 40, 47).
We have started to investigate the relationships between NF-Y and higher-order structures on the MHC class II Ea promoter, using an in vitro chromatin reconstitution system from the brine shrimp Artemia franciscana and nucleosome assembly assays with purified chicken histones. We found that a small NF-Y formed by the homology domains can associate with preformed nucleosomes. Translational analysis indicated that CCAAT positioning at one end of the fragment leads to NF-Y binding with slightly higher affinity (36). Detailed DNA-binding studies with bending and phasing assays indicated that the small NF-Y associates the CCAAT box and distorts DNA in a way that is reminiscent of histones in the nucleosome (36). However, careful examination of a set of wild-type (wt) and mutant NF-Y subunit combinations indicated that important DNA-binding parameters, such as flexure angles and off rates, are remarkably influenced by regions outside the conserved parts (28). In this study, we pursued our characterization of NF-Y-nucleosome interactions by using wt NF-Y and recombinant histones, focusing in particular on the separated H2A-H2B and H3-H4 dimers.
MATERIALS AND METHODS
Expression and purification of recombinant histone and NF-Y proteins.
The vectors coding for Xenopus laevis histones (29) (kindly provided by K. Luger, ETH, Zurich, Switzerland) were used to transform Escherichia coli BL21(DE3) (LysS). Protein expression was induced at an A600 of 0.6 by addition of isopropyl-β-d-thiogalactopyranoside to a final concentration of 1 mM for 3 h for H2A, H2B, and H3 and 1.5 h for histone H4. Bacterial pellets were resuspended and sonicated in sonication buffer (150 mM KCl, 20 mM Tris-HCl [pH 7.8], 0.05% NP-40, 0.1 mM EDTA, 5 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride [PMSF; Sigma], protein inhibitors) and centrifuged at 23,000 × g in a Beckman SW27Ti rotor for 30 min at 4°C. The inclusion body pellet was resuspended in sonication buffer, sonicated, and centrifuged again. Three cycles of this procedure yield proteins that are >90% pure. Inclusion bodies were finally resuspended in 6 M GnCl–20 mM sodium acetate (pH 5.2)–5 mM 2-mercaptoethanol–1 mM PMSF, and unfolding was allowed to proceed for 1 h at room temperature on a rotating wheel. The four histones, or H3-H4 and H2A-H2B couples, were mixed to a final concentration of 1.6 mg/ml and dialyzed against a 100-fold excess of refolding buffer (30) in 3-kDa-cutoff dialysis bags; the glycerol concentration was adjusted to 20%, and proteins were stored at −80°C. The full-length NF-Y subunits were expressed and purified as described in reference 28; NF-YB4 and NF-YC5 purification was also described previously (5).
Nucleosome reconstitution and EMSA.
Labeled probes used for nucleosome reconstitutions were fragments 2, 2m, and 6 described in reference 36. One microgram of renatured core histones or 600 ng of H3-H4 or H2A-H2B dimers was incubated with 250 ng of competitor DNA (salmon sperm DNA sonicated to an average length of 200 bp) and 1 ng of labeled DNA (105 cpm) in a final volume of 10 μl in 1 M NaCl-10 mM Tris-HCl (pH 7.8)–500 ng of bovine serum albumin (BSA)/μl–1 mM 2-mercaptoethanol for 30 min at 20°C. The reaction mixture was serially diluted to 0.8, 0.67, 0.57, 0.5, and 0.1 M NaCl by addition of TE buffer (10 mM Tris-HCl [pH 7.6], 1 mM EDTA) every 15 min. NF-Y binding reactions and electrophoretic mobility shift assays (EMSAs) were performed as in reference 36.
For most of the experiments described, we used PCR-derived labeled fragment 2, containing positions −115 to +60 of the Ea promoter derived from the PE3 plasmid (36). For Fig. 3C, we used an identical fragment mutated in the Y box, generated by PCR (32, 36). For Fig. 4, we also used fragment 6, harboring a CCAAT box in a central position (36). Antibody challenge experiments were performed by adding 200 ng of anti-NF-Y (33), anti-Gata1 (Santa Cruz Biotechnology, Santa Cruz, Calif.), or antihistone (Boehringer Mannheim, Mannheim, Germany) antibodies to the binding reaction mixture and then incubating it on ice for further 30 min.
FIG. 3.
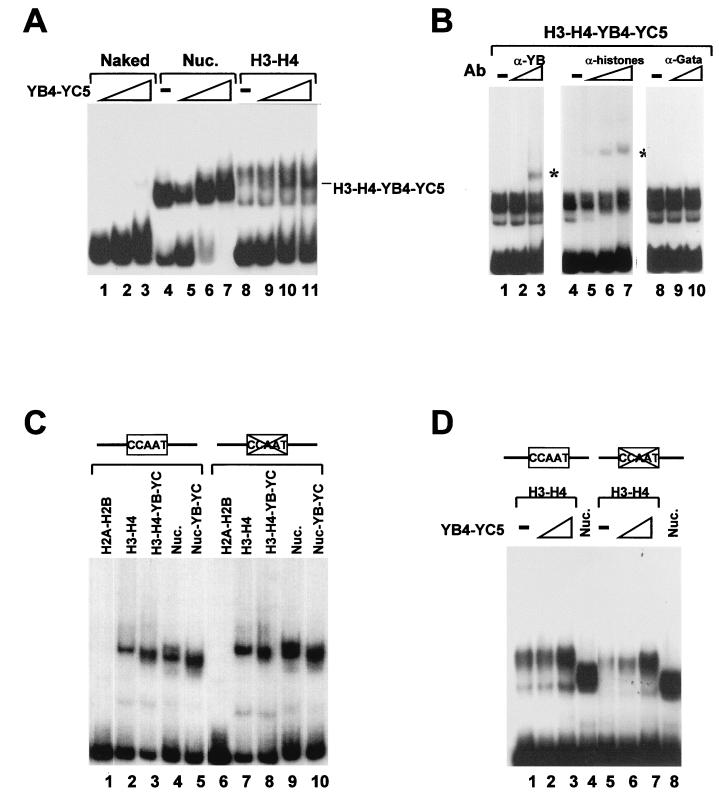
EMSA of a NF-YB–NF-YC–H3–H4 complex on the Ea promoter DNA. (A) Reconstitution of fragment 2 with increasing amounts of NF-YB4–NF-YC5 alone (200 ng, 400 ng, and 1 μg; lanes 1 to 3), with all core histones (lanes 5 to 7), or with H3-H4 only (lanes 9 to 11). Control reconstitutions with core histones and H3-H4 alone are in lanes 4 and 8. The hybrid complex is indicated. Nuc., nucleosomes. (B) Antibody challenge of the hybrid NF-YB–NF-YC–H3–H4 complex. The hybrid complex was incubated for 30 min at 4°C with increasing amounts of anti-NF-YB (100 and 500 ng; lanes 2 and 3), antihistone (10 ng, 100 ng, and 1 μg; lanes 5 to 7), or control anti-Gata1 (100 and 500 ng; lanes 9 and 10) antibodies (Ab). The supershifted complexes are indicated by asterisks. (C) The indicated combinations of proteins (H2A-H2B [lanes 1 and 6], H3-H4 [lanes 2 and 7], H3–H4–NF-YB–NF-YC [lanes 3 and 8], H2A-H2B-H3-H4 [lanes 4 and 9], and H2A-H2B-H3-H4-NF-YB-NF-YC [lanes 5 and 10]) were incubated with wt Ea DNA (lanes 1 to 5) or with a Y-box mutant (lanes 6 to 10). (D) Same as panel C except that H3-H4 tetramers were incubated alone (lanes 1 and 5) or with NF-YB4-NF-YC5 (200 ng [lanes 2 and 6] and 1 μg [lanes 3 and 7]). Nucleosomes are in lanes 4 and 8.
FIG. 4.
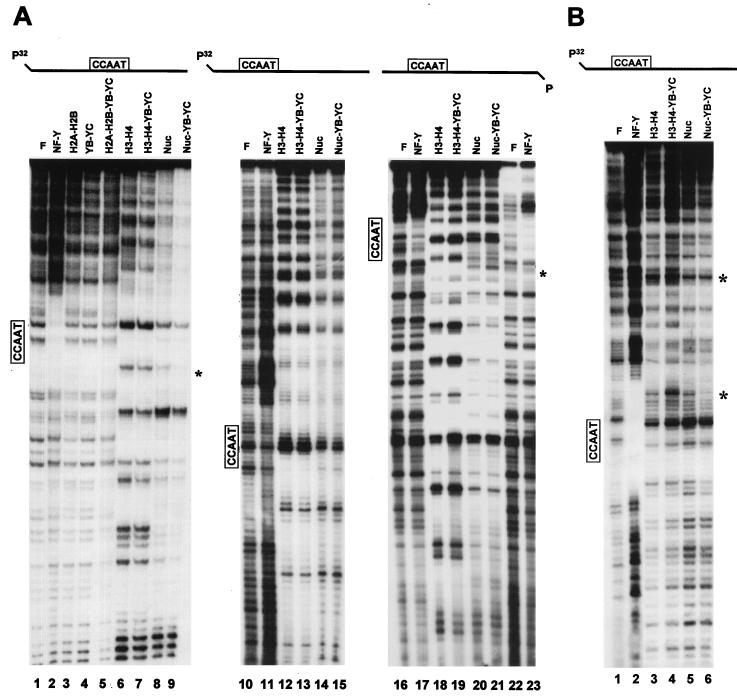
DNase I footprint of histone-NF-YB–NF-YC combinations. (A) The DNAs used were Ea fragment 2 (labeled on the top strand; lanes 1 to 9) and fragment 6 (−145 to +35 of Ea [36]; labeled on the top strand [lanes 10 to 15] and bottom strand [lanes 16 to 23]). After reconstitutions with stoichiometric amounts of the indicated HFM protein combinations, aliquots were digested with DNase I and analyzed in sequencing gels. Asterisks denote bands that were diminished (lanes 8 and 9) or increased (lanes 20 and 21) when NF-YB–NF-YC was added to reconstitutions. To locate the position of the NF-Y footprinted area, samples in lanes 2, 11, 17, and 23 contained only the NF-Y trimer, without histones. Asterisks indicate protections (lane 9) and hypersensitivities (lanes 19 and 21) upon addition of NF-YB–NF-YC to histones. F, Free DNA; Nuc, nucleosomes. (B) The indicated complexes were DNase I digested, purified from gels (see Materials and Methods), eluted, and run on sequencing gels. Lane 1, free DNA; lane 2, DNA and NF-Y; lanes 3 to 6, H3-H4, H3–H4–NF-YB–NF-YC, nucleosome, and nucleosome–NF-YB–NF-YC, respectively. Asterisks indicate protections (lane 4) and hypersensitivity (lane 6).
DNase I footprinting, MNase accessibility assay, and Exo III digestion.
For DNase I footprinting, micrococcal nuclease (MNase) accessibility assays, and exonuclease III (Exo III) digestions, we used twice the amount of recombinant histones as employed for EMSA. In this procedure, 20,000 cpm of reconstituted tetramer or octamer particles was digested with 0.1 U of DNase I (grade I; Boehringer Mannheim) supplemented with 5 mM MgCl2 and 10 mM CaCl2 at 37°C for 3 min, while free DNA was digested with 0.001 U of DNase I. The same amounts of reconstituted tetramers or octamers and free DNA were digested in 2 mM CaCl2 with 0.01 U of MNase (Sigma) at room temperature for 2 min. DNase I and MNase digestions were terminated by adding 2 volumes of DNase I stop buffer (1.5% sodium dodecyl sulfate [SDS], 30 mM EDTA, 450 mM sodium acetate [pH 5.2]). Then 50,000 cpm of reconstituted tetramers, octamers, or free DNA was digested with 100 U of Exo III (Boehringer Mannheim) with 66 mM Tris-HCl (pH 8)–2.5 mM MgCl2–1 mM 2-mercaptoethanol at 37°C, and aliquots of 28 μl were taken after 15, 30, 60, and 120 min and added to 56 μl of DNase I stop buffer. DNase I, MNase, and Exo III digestion products were phenol-chloroform extracted, ethanol precipitated, and analyzed on 7 M urea–8% polyacrylamide gels.
Polyacrylamide gel purification of DNase I-digested complexes.
Probe (105 cpm) was assembled with histones, histones and NF-YB–NF-YC, H3-H4, and H3–H4–NF-YB–NF-YC and digested with DNase I as described above. Reactions were stopped on ice in 5 mM EDTA and immediately loaded on a 4% polyacrylamide gel. Bands corresponding to the different complexes located by autoradiography of the wet gel were cut, crushed, and incubated with 500 μl of diffusion buffer (0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA [pH 8.0], 0.1% SDS) at 50°C for 30 min. Samples were centrifuged at 14,000 rpm for 5 min, and supernatants were passed through packed glass wool to eliminate any residual polyacrylamide. DNA was phenol extracted, ethanol precipitated, and analyzed on a sequencing gel as described above.
Protein-protein interactions.
Pure histone dimers and the His-tagged NF-YB4–NF-YC5 dimer (10 μg of each in 80 μl) were incubated together in BC2000 (2 M KCl, 20 mM Tris-HCl [pH 7.5], 1 mM β-mercaptoethanol, 0.05% NP-40, 100 μg of BSA/ml, 0.25 mM PMSF) and step diluted with BC100 (same as BC2000 but containing 100 mM KCl) over a period of 2 h, until a salt concentration of 0.35 M KCl was reached; 20 μl of nickel-nitrilotriacetic acid (NTA)-agarose resin (Qiagen, Hilden, Germany) was then added, and the samples were rocked for 1 h and washed twice with 1 ml of BC500. All procedures were performed at 4°C. Bound proteins were eluted by boiling samples in SDS buffer and analyzed in 17% gels stained with Coomassie blue. The NF-YB4 and NF-YB43 Sepharose columns were described earlier (4); the NF-YB43 mutant contain amino acids 51 to 117, lacking the C-terminal six amino acids of the HFM. Histones (10 μg) were incubated overnight at 4°C with 50 μl of either column in 200 μl of BC300 supplemented with 0.05% NP-40, 100 μg of BSA/ml, and 0.25 mM PMSF. Samples were then washed with the same buffer, eluted, and analyzed as described above.
RESULTS
Binding of wt NF-Y to nucleosomal DNA.
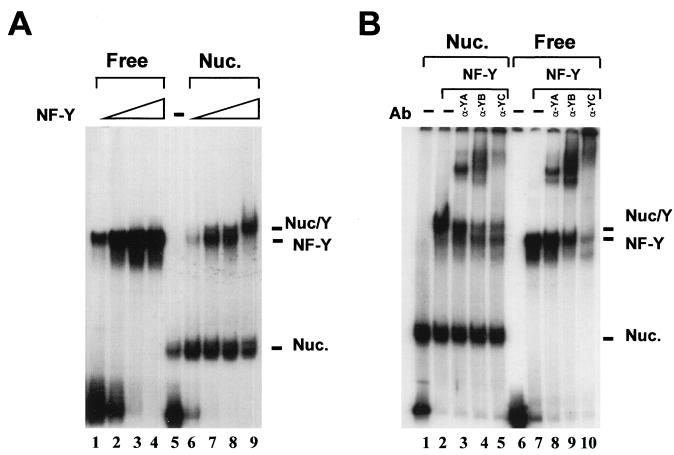
In a previous study, we used the short evolutionary conserved domains of NF-Y to show that the trimer can associate with DNA that was preassembled with purified chicken histones. By analyzing DNA binding, bending, and phasing of several mutant combinations, we became aware of two important facts: (i) the two large Q-rich regions of NF-YA and NF-YC influence angle amplitude and (ii) the presence of NF-YC αN and NF-YB αC, absent in our YB4 and YC5 mutants, alters some of the DNA-binding parameters, most notably shortening the off rate of the DNA complex (28). For these reasons, we felt important to assess the affinity of the wt NF-Y trimer for preassembled nucleosomal DNA. A fragment of the MHC class II Ea promoter containing the Y box in a semicentral position (fragment 2 [36]) was assembled with recombinant X. laevis histones, recently used for crystallographic studies (Fig. 1A, lane 5). Increasing amounts of wt NF-Y were added, either on naked DNA (lanes 1 to 4) or on 30% nucleosomized DNA (lanes 6 to 9); upper complexes of slower mobilities were readily seen at relatively low NF-Y concentrations (compare lanes 2 and 7). To ascertain whether these complexes contain all NF-Y subunits, we challenged them with anti-NF-Y antibodies (33). Figure 1B shows that antibodies directed against all three subunits supershift the upper complexes, as well as the NF-Y band on naked DNA (compare lanes 3 to 5 and 8 to 10). Note that in the latter experiments, the 70% nucleosomized fragment yielded, with comparable NF-Y concentrations, only nucleosome–NF-Y complexes (compare lanes 2 and 7); this behavior is similar to that observed with the small YA9-YB4-YC5 mutant previously used in these assays (36). Thus, like the short NF-Y mutant, wt NF-Y preferentially binds to a naked CCAAT box, but the amount of wt NF-Y required (1.3 ng) to bind DNA in a nucleosomal context is low compared to other transcription factors (see reference 36 and references therein).
FIG. 1.
wt NF-Y associates a nucleosome-bound Ea Y box. (A) EMSA of a dose response of wt NF-Y on mock-reconstituted (0.5, 1, 3, and 10 ng; lanes 1 to 4) or nucleosome (NUC.)-reconstituted (lanes 6 to 9) Ea fragment 2 (−115 to +60 of Ea [36]). Lane 5, no NF-Y added to nucleosomal DNA. (B) A 70% nucleosomized Ea fragment 2 was run without NF-Y (lane 1), with 5 ng of NF-Y (lane 2), and with the same amount of NF-Y incubated with the indicated anti-NF-Y antibody (Ab; 200 ng of purified antibody; lanes 3 to 5). The same was in lanes 7 to 10 except that mock-reconstituted DNA was used.
Association of NF-Y during nucleosome reconstitution.
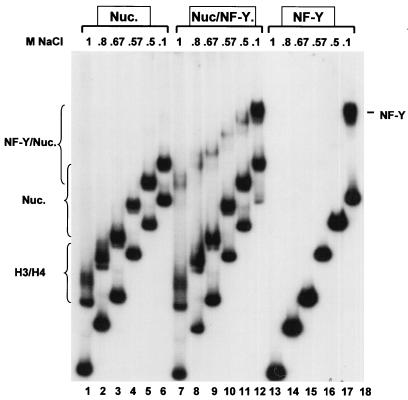
Our reconstitution assay can also be used to dissect NF-Y binding during histone-DNA association. We incubated histones (Fig. 2, lanes 1 to 6), NF-Y (lanes 13 to 18), or histones and NF-Y together (lanes 7 to 12) in the presence of cold competitor DNA and 1 M NaCl; the salt concentration was then progressively lowered over 15-min periods, and at each point an aliquot of the samples was loaded on a running EMSA. The resulting patterns gave indications as to the binding of the different complexes to DNA. During nucleosome assembly, two bands were observed at 1 M (lane 1); they have been observed in other studies (17, 18, 41) and most likely represent the H3-H4 tetramer and ditetramers species detailed in glycerol gradient experiments by Spangenberg et al. (41). In line with this interpretation, they are observed upon reconstitution with H3-H4 only (see below). At 0.8 M, the H3-H4 lower complex is still present, while an intermediate band becomes apparent and progressively predominant at lower salt concentrations (lanes 2 and 3 to 6); this represents H2A-H2B association and formation of a stable histone octamer. With NF-Y alone, no binding is observed until the NaCl concentration is lower than 0.5 M (compare lanes 13 to 17 with lane 18); this finding is in full agreement with previous results (19). When histones and NF-Y are incubated together, two slowly migrating complexes are observed at 1 M NaCl, together with the H3-H4 tetrameric complexes (compare lanes 1, 7, and 13). These complexes persist at lower concentrations, progressively merging into one major complex as H2A and H2B associate (compare lanes 7 to 11). These results indicate that in the presence of histones, NF-Y can bind DNA in nonpermissive salt conditions and suggest that this is accomplished through association with H3-H4.
FIG. 2.
Binding of NF-Y during nucleosome assembly. Stoichiometric amounts of histones were assembled in high salts with Ea fragment 2 and cold competitor DNA, in the absence (lanes 1 to 6) or presence (lanes 7 to 12) of 5 ng of wt NF-Y. In lanes 13 to 18, 5 ng of wt NF-Y was used as in lanes 7 to 12, in the absence of histones. Mixtures were progressively diluted, and at each NaCl concentration, aliquots were added to a running polyacrylamide gel. The NF-Y, H3-H4, nucleosome, and NF-Y–nucleosome bands are indicated.
Association of NF-Y HFM subunits with H3-H4 on the DNA.
The experiments described above suggest that the histone fold subunits of NF-Y can bind histones while assembly takes place, in the absence of NF-YA. To gain insight into the mechanism, we reconstituted nucleosomes by incubating increasing near-stoichiometric amounts of the NF-YB–NF-YC dimer alone, with the four core histones, or with H3-H4 in the presence of cold competitor DNA. Initially, we used the YB4-YC5 mutant containing the evolutionarily conserved domains; as expected, they had no DNA-binding capacity on their own (Fig. 3A, lanes 1 to 3) However, upon reconstitution with all core histones (lanes 4 to 7) and with H3-H4 (lanes 8 to 11), a distinct band was generated, with an electrophoretic mobility different from that of the nucleosome or of the H3-H4 tetramers (compare lanes 9 and 7 with lanes and 11). This complex was stable for several days at 4°C (not shown). To investigate the protein composition in such complexes, we challenged it with increasing amounts of purified anti-NF-YB (Fig. 3B, lanes 1 to 3) and antihistone antibodies (lanes 4 to 7). Both antibodies were able to specifically supershift the H3–H4–NF-YB–NF-YC complex, while an irrelevant anti-Gata1 antibody had no effect (lanes 8 to 10). These experiments suggest that a hybrid complex containing both H3-H4 and NF-YB–NF-YC dimers can be formed on DNA. We repeated the experiments with full-length NF-Y subunits; the different combinations gave results similar to those for YB4-YC5, as complexes of dissimilar electrophoretic mobility were observed (Fig. 3C, lanes 1 to 5). In parallel, we also checked whether the integrity of the CCAAT box was required for formation of the NF-YB–NF-YC-nucleosome and NF-YB–NF-YC–H3–H4 or YB4-YB5-H3-H4 complexes. For this, we used an Ea fragment of identical length containing in the Y box a 10-bp mutation that renders it unable to interact with NF-Y (36, 44). As shown in Fig. 3C and D, the patterns generated with the different combinations were essentially identical to that for the wt Ea fragment (Fig. 3C [compare lanes 1 to 5 with lanes 6 to 10] and D [compare lanes 2 and 3 with lanes 6 and 7]), indicating that association of NF-YB–NF-YC does not require the CCAAT box. Altogether, these results indicate that NF-YB–NF-YC dimers associate with H3-H4 and suggest the possibility that they might be incorporated with H3-H4 into hybrid nucleosomes or nucleosome-like complexes.
Analysis of histone–NF-YB–NF-YC complexes with DNase I, MNase, and Exo III assays.
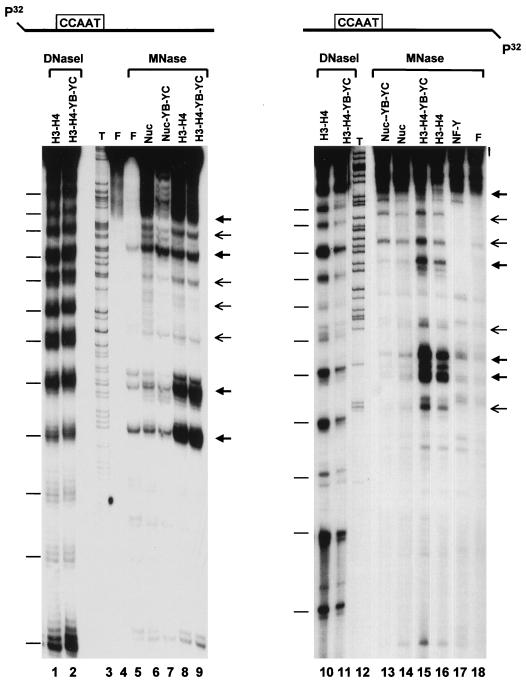
To verify this possibility, we performed analysis with DNase I, MNase, and Exo III. These enzymes, particularly MNase, have been widely used to assess the presence of nucleosomes and map their exact positions. Figure 4A shows DNase I footprints on fragment 6 (a fragment with the CCAAT box in the dyad symmetry) and fragment 2 (CCAAT in a semicentral position) on both strands (36). Reconstitution of fragment 2 with either H2A-H2B, wt NF-YB–NF-YC, or combinations of the four proteins resulted in no differences in cutting patterns with respect to the mock-reconstituted control (Fig. 4A; compare lanes 1 and 3 to 5); note that the same result was obtained with the small-homology-containing proteins (data not shown). H3-H4 and nucleosome reconstitutions generated regular DNase I patterns of 10-bp cuts very similar, albeit not identical, among each other (compare lane 1 with lanes 6 and 8). Addition of the wt NF-YB–NF-YC dimer did not modify substantially the H3-H4 pattern, failing to render it identical to the pattern of the nucleosome (compare lanes 6 to 8). However, addition of NF-YB–NF-YC to core histones provoked a decrease in the intensity of a band in the NF-Y footprinted region (compare lanes 8 and 9; see lane 2). Similar experiments were performed with fragment 2 labeled on both strands: when a labeled top strand was used, essentially identical patterns were obtained with the different combinations tested (compare lanes 10 and 12 to 15); on the bottom strand, more pronounced differences were seen between H3-H4 and nucleosomes (compare lanes 18 and 20). Addition of NF-YB–NF-YC failed to alter the patterns, with the exception of an increased accessibility of an area at the edge of the NF-Y footprinted region (compare lanes 20 and 21; see lanes 16, 17, 21, and 23). We also performed footprinting experiments following gel isolation of the complexes shown in Fig. 3. Nucleosomes, nucleosome–NF-YB–NF-YC, H3-H4, and H3–H4–NF-YB–NF-YC were reconstituted as usual and treated with DNase I; whole reconstitutions were loaded on a polyacrylamide gel; complexes were separated; corresponding bands were excised, eluted, and run on sequencing gels. Results of such experiment on fragment 2 labeled on the top strand are shown in Fig. 4B. The overall patterns are rather similar, with two prominent hypersensitive sites in H3–H4–NF-YB–NF-YC complexes compared to H3-H4 (Fig. 4B; compare lanes 3 and 4), while a clear protection is observed in nucleosome–NF-YB–NF-YC complexes compared to nucleosomes (compare lanes 5 and 6). Note that one of the hypersensitive/protection sites is located within the NF-Y footprinted area (lanes 1 and 2). In summary, modifications on H3-H4 and on nucleosomes induced by NF-YB–NF-YC addition are subtle in this assay.
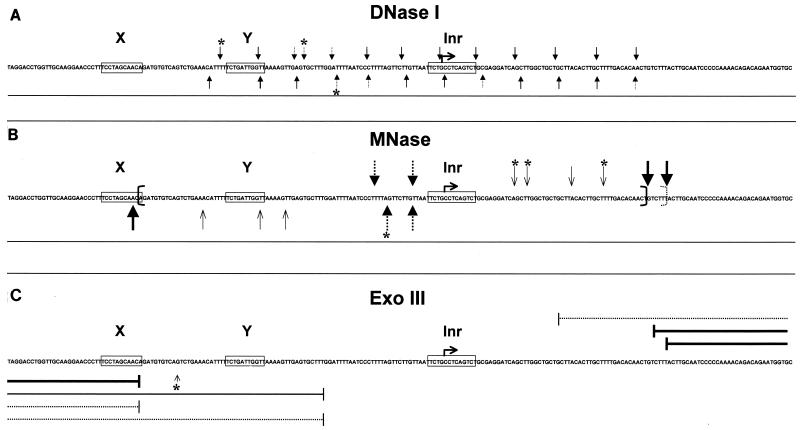
In DNase I assays, differences between the H3-H4 and nucleosome patterns are relatively subtle, as expected from the notion that H3-H4 tetramers are primarily responsible for the 10-bp cuts; thus, the differences effected by NF-YB–NF-YC could be largely missed. To obviate this possibility, we used MNase cleavage. This assay is based on the differential cuts generated after reconstitution of H3-H4 tetramers, which tend to protect a 75-bp fragment generated from the dyad symmetry, from those observed with nucleosomes, which protect a larger 150-bp fragment corresponding to the whole nucleosome. After reconstitution of nucleosomes on fragment 2, MNase was added and fragments of different lengths were indeed generated, as judged from the protein composition of the complexes (Fig. 5). On the top strand, H3-H4 yielded prevalent fragments of 95 and 106 bp (Fig. 5, lane 8); addition of NF-YB–NF-YC to H3-H4 generated a predominant H3-H4-like pattern but gave protections of intermediate bands at positions +10 to 15 (compare lanes 8 and 9). On the other hand, the nucleosome protected a larger fragment of 170 bp (lane 6); NF-YB–NF-YC induced a large protection in the region at the 3′ end of the nucleosome (compare lanes 6 and 7). In parallel, we performed DNase I footprints on the same H3-H4 and H3–H4–NF-YB–NF-YC reconstitutions and observed regular 10-bp cutting patterns, strongly suggesting that the MNase-resistant bands observed in these experiments are indeed generated by the presence of highly positioned H3-H4 tetramers (lanes 1 and 2). The same type of analysis was performed on the bottom strand of this fragment, also giving regular 10-bp cuts in parallel DNase I experiments (lanes 10 and 11). MNase cuts at positions −11 to −17 with H3-H4 and at position −84 with all four histones (lanes 14 and 16). Addition of NF-YB–NF-YC increased the intensities of the H3-H4-generated cuts and yielded shorter fragments with nucleosomes in the NF-Y protected region at −65 (compare lanes 13 and 14 with lanes 15 and 16). Again, NF-YB–NF-YC did not generate a nucleosome-like pattern. Overall, these MNase experiments are consistent with the idea that a nucleosome is highly positioned on the Ea promoter, with boundaries at −85 and +60 and a dyad symmetry at −10 with respect to the major start site (see Fig. 7 for a summary).
FIG. 5.
MNase accessibility assay of histone–NF-YB–NF-YC combinations. Stoichiometric amounts of the indicated combinations of HFM proteins were reconstituted with fragment 2 (labeled on the top strand [lanes 4 to 9] and bottom strand [lanes 13 to 18]), cut with MNase, and analyzed on sequencing gels. In lane 17, the NF-Y trimer was used to show the NF-Y footprinted area. F refers to free, mock-reconstituted DNA (lanes 4 and 5; uncut and cut with MNase, respectively). Arrows correspond to the major and minor hypersensitive sites. Part of the H3-H4 and H3–H4–NF-YB–NF-YC reconstitutions were cut with DNase I and run in parallel (lanes 1, 2, 10, and 11). Bars correspond to the 10-bp cutting patterns of DNase I. Sequencing reactions (T; lanes 3 and 12) were run in parallel to precisely map the sites of MNase cuts.
FIG. 7.
Recapitulation of the data obtained with DNase I, MNase, and Exo III assays. The X, Y, and Inr elements are boxed, and the major +1 start site of the Ea promoter is outlined. (A) DNase I. Arrows indicate the cuts in the nucleosomal region. (B) MNase. Thick arrows indicate the cuts with nucleosomes and dotted arrows with H3-H4 tetramers, thin arrows represent minor cutting sites, and brackets delimit the boundaries of the nucleosome. (C) Exo III. Thick and dotted lines indicate major stops with nucleosomes and H3-H4 tetramers, respectively. The small arrow indicates a major stop which is bypassed when NF-YB–NF-YC is reconstituted with nucleosomes. Asterisks refer to differences in the pattern observed when the NF-YB and NF-YC subunits are added to histones.
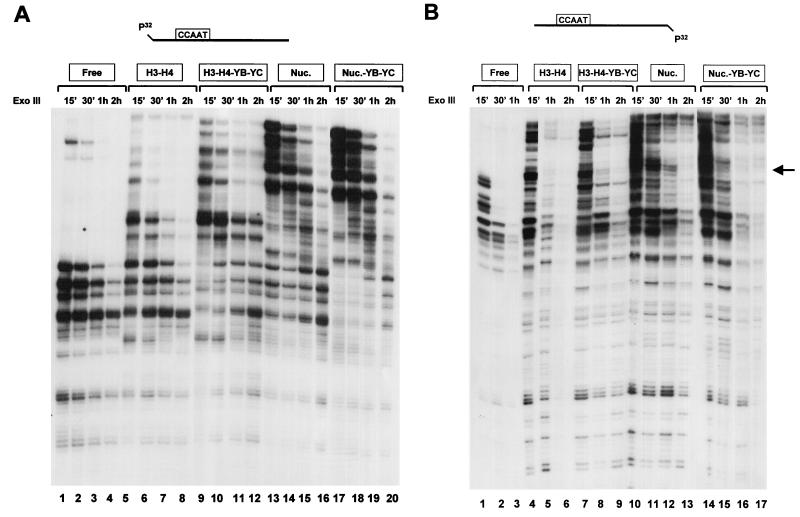
We also performed Exo III experiments on our reconstitutions (Fig. 6); this assay is based on the 3′-5′ nuclease activity of this processive enzyme, which is a measure of the stability of the protein-DNA complexes. As expected, Exo III digestions of nucleosomes generated protections larger than with tetramers, on both the top (Fig. 6A; compare lanes 1 to 4 with lanes 5 to 8 and 13 to 16) and bottom (Fig. 6B; compare lanes 1 to 3, 4 to 6, and 10 to 13) strands of fragment 2. Addition of NF-YB–NF-YC stabilized the H3-H4 tetramers (compare lanes 5 to 8 and 9 to 12 in Fig. 6A and lanes 4 to 6 and 7 to 9 in Fig. 6B) and nucleosomes (compare lanes 13 to 16 and 16 to 20 in Fig. 6A and lanes 10 to 13 and 14 to 17 in Fig. 6B). Moreover, NF-YB–NF-YC reduced some hypersensitive sites (Fig. 6B; compare lanes 11 and 15). However, clear differences between H3–H4–NF-YB–NF-YC and nucleosomal patterns were present (compare lanes 9 to 12 and 13 to 16 in Fig. 6A and lanes 7 to 9 and 10 to 13 in Fig. 6B). The results with Exo III, summarized in Fig. 7, are in agreement with the nucleosome position derived by MNase and are consistent with the hypothesis that the HFM subunits are associated to tetramers and nucleosomes but do not transform the formers in octamer-like structures. From this set of experiments, we conclude that reconstitutions of NF-YB–NF-YC with H3-H4 do not lead to the formation of bona fide nucleosomes, but addition of NF-YB–NF-YC does modify the tetramer (and octamer) patterns in ways that are consistent with association of the HFM subunits.
FIG. 6.
Exo III assay of histone–NF-YB–NF-YC combinations. Stoichiometric amounts of the indicated combinations of HFM proteins were reconstituted with fragment 2 (labeled on the top strand [A] and bottom strand [B]), cut with Exo III for the indicated length of time, and analyzed on sequencing gels. Nuc., nucleosome.
NF-YB–NF-YC dimers bind H3-H4 in solution.
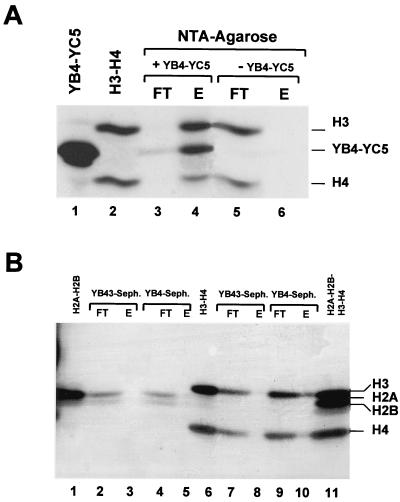
Because of the data obtained from the DNA reconstitutions with NF-YB–NF-YC and the evidence that HFM-containing TAFIIs are able to interact with core histones in solution (8), we wanted to test whether H3-H4 could bind directly to NF-YB–NF-YC in the absence of DNA. To do this, we incubated equimolar amounts of H3-H4 with homology domains containing His-tagged NF-YB–NF-YC in high-salt (2 M KCl) conditions and progressively diluted the sample to 0.35 M KCl. We then added the NTA-agarose resin, to which the recombinant NF-YB–NF-YC complexes bind; following an extensive wash with 0.5 M KCl buffers, we eluted bound proteins by boiling in SDS buffer and analyzed them in SDS-gels. The result of such experiment is shown in Fig. 8A. H3-H4 complexes were efficiently retained by the nickel-NTA column only in the presence of NF-YB–NF-YC (lanes 3 and 4), whereas they were in the unbound material when incubated alone on the column (lanes 5 and 6). In similar experiments, H2A-H2B were not bound to NTA columns, either in the presence or in the absence of NF-YB–NF-YC (not shown).
FIG. 8.
Protein-protein interactions between NF-YB–NF-YC and core histone dimers. (A) NTA-agarose columns. Lanes: 1 and 2, NF-YB4–NF-YC5 and H3-H4, respectively; 3 and 4, flowthrough and eluted material from NTA-agarose supplemented with His-tagged NF-YB4–NF-YC5; 5 and 6, control NTA-agarose columns run without NF-YB4–NF-YB5. (B) NF-YB4 and NF-YB43–Sepharose (Seph.) columns. Load, flowthrough, and eluates of pure H2A-H2B (lanes 1 to 5) and H3-H4 (lanes 6 to 10) are indicated. The four histones are run together in lane 11.
To confirm these data and verify whether the HFM of NF-YB is involved in interactions, as in the case of H2B-H4 (29), we used a different approach (4). The homology domains containing YB4 and NF-YB43 (a mutant lacking part of helix α3) were coupled to a Sepharose matrix, and the resulting columns were loaded with H3-H4 or H2A-H2B histone dimers. Bound material was recovered in SDS buffer and analyzed. As shown in Fig. 8B, H3-H4 were retained, albeit not completely, by the NF-YB4 column, but not by NF-YB43 (lanes 6 to 10), while H2A-H2B did not bind to either columns (lanes 1 to 5); the lower efficiency with respect to the NTA columns is most likely due to the fact that coupling of recombinant proteins to CnBr-activated Sepharose is a random process, involving active sites in the HFM of the short YB4 mutant. Taken together, these data prove that (i) NF-YB–NF-YC can efficiently bind to H3-H4 but not to H2A-H2B, (ii) NF-YB–NF-YC regions outside the homology domains are expendable for this activity, and (iii) the HFM of NF-YB is involved.
Association of NF-YA with the NF-YB–NF-YC dimer in a nucleosomal context.
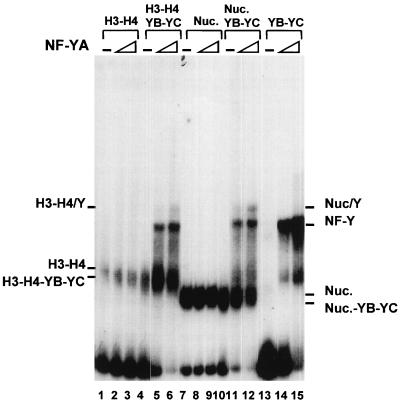
The experiments described so far suggest that the NF-YB–NF-YC dimer is able to interact with H3-H4, both in solution and during reconstitutions, in a way that is different from H2A-H2B association. Since NF-YB–NF-YC binding to H3-H4 is mediated by the histone folds, which are also required for trimer formation with NF-YA, an important issue was to establish whether such NF-YB–NF-YC–H3–H4 complexes are compatible with NF-YA association and CCAAT box binding or whether preengagement of HFM subunits with histones would preclude NF-YA binding. To address this point, we added increasing amounts of NF-YA to the reconstituted combinations detailed in Fig. 3 to 6. As expected, no effect was seen on nucleosomes and on H3-H4 tetramers (Fig. 9, lanes 1 to 3 and 7 to 9); an upper complex was observed when NF-YA was added to the NF-YB–NF-YC-containing reconstitutions, either with H3-H4 (lanes 5 and 6) or with nucleosomes (lanes 11 and 12). This upper complex has an electrophoretic mobility different from that of the band generated by the NF-Y trimer on naked DNA (compare lanes 5, 6, 11, and 12 with lanes 14 and 15). Moreover, the efficiencies of upper complex formation are similar whether a nucleosome or H3-H4 tetramers are present, further suggesting that H3-H4, and not H2A-H2B, tetramers are NF-Y docking spots. The protein composition in this upper band was checked with anti-NF-Y antibodies, and all three NF-Y subunits were found to be involved in this interaction (not shown). Thus, association of NF-YB–NF-YC with H3-H4 and with nucleosomes is not incompatible with subsequent binding of NF-YA.
FIG. 9.
NF-YA associates with histone-bound NF-YB–NF-YC. Increasing concentrations of wt NF-YA (1 ng [lanes 2, 5, 8, 11, and 14] and 5 ng [lanes 3, 6, 9, 12, and 15]) were incubated with mixed complexes previously reconstituted with the indicated combinations of HFM proteins: H3-H4 (lanes 1 to 3), H3–H4–NF-YB–NF-YC (lanes 4 to 6), H3-H4-H2A-H2B (lanes 7 to 9), H3–H4–H2A–H2B–NF-YB–NF-YC (lanes 10 to 12), and NF-YB–NF-YC (lanes 13 to 15). Nuc., nucleosome.
DISCUSSION
In this study, we pursued our investigation on the interactions between NF-Y and nucleosomal structures and detailed its remarkable interactions with core histones. We found evidence that NF-YB–NF-YC complexes bind to H3-H4, but not to H2A-H2B, both in solution and in reconstitution assays with DNA. The integrity of the NF-YB HFM is necessary for histone interactions, thus suggesting that the HFMs of histones, not their N-terminal tails, mediate association. Interestingly, a highly positioned nucleosome is bound to the Ea core promoter. DNase I and MNase digestions documented that addition of the NF-YB–NF-YC dimer to H3-H4 tetramers or nucleosomes generated differences consistent with NF-YB–NF-YC association but did not provide unambiguous proof of an hybrid octamer-like structures. Finally, the complexes retain remarkable NF-YA binding capacity.
NF-Y is thought to be an architectural protein playing a major role in promoter and/or enhancer organization. Its peculiar structure leads to the idea that part of its function stems from special connections with nucleosomes. Few studies have focused on the chromatin structure of NF-Y-dependent systems in vivo; in the Xenopus HSP70 promoter, NF-Y prevents the formation of repressing arrays of nucleosomes, facilitating the activity of the heat shock factor and of the general machinery (23). In the rat uteroglobin enhancer, NF-Y is unable to bind at the linker between two positioned nucleosomes unless progesterone, the inducer of enhancer activity, is added (39). Previous experiments of our lab started to dissect nucleosome–NF-Y interactions in vitro; an NF-Y mutant, containing only the evolutionarily conserved domains, showed specific and rather efficient interactions with nucleosomal DNA (36). Although the CCAAT specificity of this NF-Y mutant is identical to that of the wt trimer, bending and phasing studies of several combinations of NF-Y mutants on the double CCAAT of the γ-globin promoter yielded clear indications that two important parameters are considerably different from the wt protein: (i) the half-life of the small NF-Y-CCAAT complex is much longer, 4 h compared to 15 min; and (ii) the flexure angles are ampler, 80° versus 56° (28). Because of these findings, it was important to verify the histone–NF-Y–DNA interactions with a physiologically relevant trimer. One of the most interesting, and somewhat surprising, results of the present study is that NF-Y associates DNA during histone deposition under high-salt conditions which do not allow CCAAT binding on naked DNA (reference 19 and Fig. 2). The most likely explanation for this phenomenon is that NF-Y is recruited on DNA through hydrophobic interactions with histone dimers, mediated by HFM subunits. Because the NF-Y complexes are seen at 1 M NaCl, when only H3-H4 tetramers are bound to DNA, it seems logical to suppose that H3-H4 tetramers are responsible of this recruitment, especially since we have shown H3-H4 binding in protein-protein interactions in solution and de novo assembly without H2A-H2B (Fig. 3 and 8). Other factors are able to associate with H3-H4 tetramers: Tup1, a yeast global repressor, interacts with H3-H4 through the N terminus of H3 (13); the NF-1 P-rich activation domain also interacts with H3 and H3-H4 (1); NF1 and OTF1 were shown to bind tetramers but not nucleosomes on a reconstituted mouse mammary tumor virus promoter (41); similarly, H2A-H2B inhibits binding of TFIIIA to H3-H4 tetramers in X. borealis somatic 5S RNA gene (18). NF-Y is the first transcription factor for which histone fold association during nucleosome assembly has been documented.
Our in vitro observations might have important physiological consequences: many genes that are active early after replication require a DNA-bound NF-Y, and CCAAT-containing promoters of cell cycle regulated genes are constantly bound by NF-Y in vivo (reference 33 and references therein). Thus, early association of this protein during nucleosome deposition might be an essential signal of a soon to be active promoter and a pivotal step in building up of additional interactions with nearby DNA-binding activators and with the general transcriptional machinery or holoenzyme. The demonstration of a remarkably well positioned nucleosome on the Ea promoter, which overlaps the CCAAT and Inr elements and whose 5′ boundaries are adjacent to the crucial X-box element, will spur further investigation on the existence of such structures in vivo. Moreover, the recent recapitulation of the natural RFX complex, which binds the X box as a trimer and makes cooperative interactions with NF-Y (reference 35 and references therein), will allow studies aimed at clarifying the interactions between these two trimers in the well-characterized nucleosome context described here.
We feel that our results have wider implications pertaining the histone fold family, a growing group of proteins known to form complex interactions among them, as exemplified by HFM TAFIIs (6, 8). Results of TAFII-histone interactions showed that the H4-like hTAFII80 can bind to H3, the H3-like hTAFII31 binds to H4, and hTAFII20 binds to H2A and H2B (8). Our findings of interactions of NF-YB–NF-YC with H3-H4, but not with H2A-H2B, support the hypothesis that distinct subfamilies of HFM proteins have coevolved cross-dimer preferences. The H3-H4 tetramer interacts with H2A-H2B mainly via H2B-H4 association, elicited by hydrogen bonds of H4-H75 and H4-K93 with H2B-E90 and H2B-E73, respectively (29); in the corresponding positions, NF-YB also harbors acidic residues, D115 and E98, two of the relatively few amino acids that are absolutely conserved in 26 sequences from different species (33a), suggesting that indeed NF-YB contacts H4 and indicating a reason for the strong evolutionary pressure on these residues. On the other hand, the inability to interact with H2A-H2B is in accordance with our recent observation that NF-YB–NF-YC cannot cross-dimerize with the H2A-H2B-like subunits of NC2 despite their closer relatedness (47). Moreover, interactions between NF-Y and HFM TAFIIs have been recently documented in our lab (14a).
NF-YB–NF-YC, either the wt complex or short versions containing the evolutionary conserved parts, can form complexes in stoichiometric amounts with H3-H4 in our reconstitution assays, in which a large (250-fold) excess of cold sonicated salmon sperm DNA is present; neither NF-YA nor the CCAAT box is necessary. It was therefore of some importance to establish whether such complexes could be considered as nucleosome-like structures. DNase I, MNase, and Exo III assays were informative in this respect: in general, our data are not in favor of this hypothesis, especially since a clear difference exists between the patterns generated by MNase on nucleosomal DNA and those of NF-YB–NF-YC–H3–H4, which resemble those of H3-H4 tetramers. However, differences were found upon addition of NF-YB–NF-YC in all assays, with respect to both H3-H4 tetramers and nucleosomes. Our data imply that the NF-YB–NF-YC dimer is involved in contacts outside the octameric or tetrameric structures, by associating directly with H3-H4. The extended protections are an indication that additional sequences are contacted by the NF-YB–NF-YC dimer; this might have important consequences for the three-dimensional structure of the promoter by altering, disturbing, or even arresting nucleosome deposition or hampering association of linker histones. It should be remembered that H2A-H2B association is an essential step in formation of a transcriptionally repressive unit (16); histone folds interfering with this step might thus counteract repression.
What is the physiological basis for performing experiments with isolated NF-YB–NF-YC dimers? Studies on immortalized cell lines suggested that NF-Y was a constant, noninducible trimeric factor. Indeed, evaluation of HFM subunits expression in different systems indicated that they are ubiquitous (9, 14, 34). However, two types of evidence challenge this view. (i) HFM dimers were shown to engage in high-molecular-weight complexes in the absence of NF-YA, and evidence of association with TFIID has been presented (4). Recent experiments confirm these findings, as proteins involved in histone acetylation are found associated with NF-Y: human GCN5 binds to the NF-YB–NF-YC dimer (10), p300 binds to NF-YB (26), and P/CAF binds to NF-YA (20). It should be noted that P/CAF is part of a large complex containing more than 20 polypeptides, including hTAFII31, hTAFII20, and PAF65α, all proteins containing histone folds (37). Moreover, binding of p300 to Xenopus NF-YB results in acetylation of NF-YB, the functional consequences of which are unknown (26). Interestingly, activation assays with HFM subunits fused to GAL4 indicated that they are sufficient to activate transcription robustly, two- to fourfold better than the NF-Y trimer (11); thus, even in the absence of NF-YA, NF-YB–NF-YC could serve the dual function of being able to confer nucleosome binding, as shown here, and transcriptional activation potential to associated complexes. (ii) The expression of NF-YA in physiological cellular systems is sometimes limiting and highly regulated posttranscriptionally: NF-YA is dramatically down-modulated in IMR90 fibroblasts upon senescence and in terminally differentiated C2C12 myotubes (9, 14) but up-modulated in human peripheral monocytes following macrophage maturation (34). The latter process is accomplished without cell division and de novo chromatin deposition; it is possible that NF-YA can directly interact with the structures described here, activating, among others, genes of the antigen presentation pathway, such as MHC class II, all dependent on CCAAT boxes. Within this conceptual framework, we feel that the remarkable efficiency of NF-YA binding to a NF-YB–NF-YC dimer preengaged in histone interactions is an important finding.
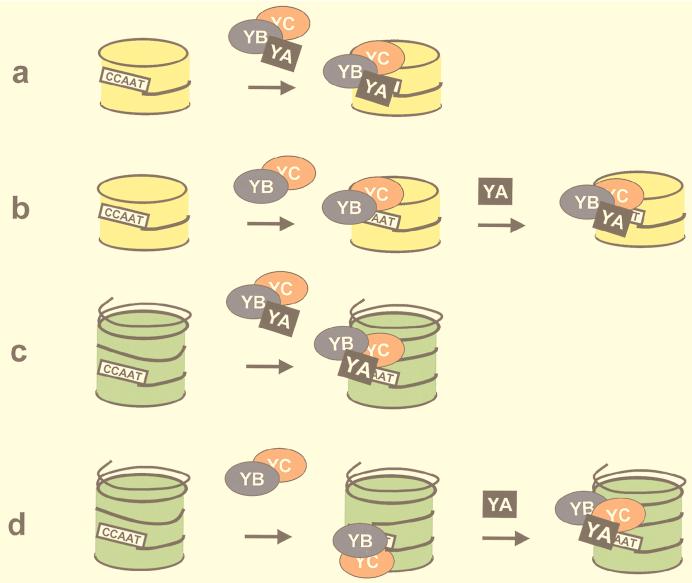
In summary, we think that NF-Y is a well-suited interface with basic chromatin structures that can employ multiple mechanisms to “open up” a promoter, as outlined in the scheme presented in Fig. 10. It can prevent promoters from being shut off by nucleosome deposition, and it can bind sites that need to be activated but are embedded in nucleosomes. NF-YB–NF-YC dimers, thanks to their histone-like structures, can associate DNA during nucleosome formation; further deposition of NF-YA will then lead to proper CCAAT box binding, local changes in the nucleosomal structure, and access of activators binding nearby. The assays used here will now be implemented in the study of facilitation of other activators binding to the Ea promoter, both upstream and downstream of the CCAAT box.
FIG. 10.
Models for NF-Y–histone association. The NF-Y trimer associates with H3-H4 tetramers (a) or nucleosomes (c). The NF-YB–NF-YC dimer associates with H3-H4 tetramers (b) and nucleosomes, presumably through H3-H4, and forms structures which can still be bound by NF-YA (d). H3-H4 tetramers are in yellow; nucleosomes are in green.
ACKNOWLEDGMENTS
We thank K. Luger and T. Richmond for the PET3 histones plasmids and G. F. Badaracco for encouragement and helpful discussions.
G.C. was a recipient of a Telethon fellowship. This work was supported by grants from MURST (PRIN-“Nucleic acid-proteins interactions”) and CNR to R.M. The contribution of Telethon grant E582 to R.M. is gratefully acknowledged.
REFERENCES
- 1.Alevizopoulos A, Dusserre Y, Tsai-Pflugfelder M, von der Weid T, Whali W, Mermod N. A proline-rich TGF-β-responsive transcriptional activator interacts with histones H3. Genes Dev. 1995;9:3051–3066. doi: 10.1101/gad.9.24.3051. [DOI] [PubMed] [Google Scholar]
- 2.Arents G, Moudrianakis E N. The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc Natl Acad Sci USA. 1995;92:11170–11174. doi: 10.1073/pnas.92.24.11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baxevanis A D, Arents G, Moudrianakis E N, Landsman D. A variety of DNA-binding and multimeric proteins contain the histone fold motif. Nucleic Acids Res. 1995;23:2685–2691. doi: 10.1093/nar/23.14.2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellorini M, Lee D K, Dantonel J C, Zemzoumi K, Roeder R G, Tora L, Mantovani R. CCAAT binding NF-Y-TBP interactions: NF-YB and NF-YC require short domains adjacent to their histone fold motifs for association with TBP basic residues. Nucleic Acids Res. 1997;25:2174–2181. doi: 10.1093/nar/25.11.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellorini M, Zemzoumi K, Farina A, Berthelsen J, Piaggio G, Mantovani R. Cloning and expression of human NF-YC. Gene. 1997;193:119–125. doi: 10.1016/s0378-1119(97)00109-1. [DOI] [PubMed] [Google Scholar]
- 6.Birck C, Poch O, Romier C, Ruff M, Mengus G, Lavigne A-C, Davidson I, Moras D. Human TAFII28 and TAFII18 interact through a histone fold encoded by the atypical evolutionary conserved motifs also found in SPT3 family. Cell. 1998;94:239–249. doi: 10.1016/s0092-8674(00)81423-3. [DOI] [PubMed] [Google Scholar]
- 7.Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 8.Burley S L, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1997;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 9.Chang Z F, Liu C-J. Human thymidine kinase CCAAT-binding factor is NF-Y, whose A subunit expression is serum-dependent in human IMR90 diploid fibroblasts. J Biol Chem. 1994;269:17893–17898. [PubMed] [Google Scholar]
- 10.Currie R A. NF-Y is associated with the histone acetyltransferases GCN5 and P/CAF. J Biol Chem. 1998;273:1430–1434. doi: 10.1074/jbc.273.3.1430. [DOI] [PubMed] [Google Scholar]
- 11.Di Silvio A, Imbriano C, Mantovani R. Dissection of the NF-Y transcriptional activation potential. Nucleic Acids Res. 1999;27:2578–2584. doi: 10.1093/nar/27.13.2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong F, van Holde K E. Nucleosome positioning is determined by the (H3-H4) tetramer. Proc Natl Acad Sci USA. 1991;88:10596–10600. doi: 10.1073/pnas.88.23.10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edmondson D G, Mitchell Smith M, Roth S Y. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 14.Farina A, Manni I, Fontemaggi G, Tiainen M, Cenciarelli C, Bellorini M, Mantovani R, Sacchi A, Piaggio G. Down regulation of cyclin B1 gene transcription in terminally differentiated skeletal muscle cells is associated with loss of functional CCAAT-binding NF-Y complex. Oncogene. 1999;18:2818–2827. doi: 10.1038/sj.onc.1202472. [DOI] [PubMed] [Google Scholar]
- 14a.Frontini, M., C. Imbriano, and R. Mantovani. Submitted for publication.
- 15.Goppeldt A, Steltzer G, Lottspeich F, Meisterernst M. A mechanism for repression of class II gene transcription through specific binding of NC2 to TBP-promoter complexes via heterodimeric histone fold domains. EMBO J. 1996;15:3105–3116. [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen C J, Wolffe A. A role for histones H2A/H2B in chromatin folding and transcriptional repression. Proc Natl Acad Sci USA. 1994;91:2339–2343. doi: 10.1073/pnas.91.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayes J J, Clark D J, Wolffe A. Histone contribution to the structure of DNA in the nucleosome. Proc Natl Acad Sci USA. 1991;88:6829–6833. doi: 10.1073/pnas.88.15.6829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayes J J, Wolffe A. Histones H2A/H2B inhibit the interaction of transcription factor IIIA with the Xenopus borealis somatic 5S RNA gene in a nucleosome. Proc Natl Acad Sci USA. 1992;89:1319–1233. doi: 10.1073/pnas.89.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hooft van Huijsduijnen R, Bollekens J, Dorn A, Benoist C, Mathis D. Properties of a CCAAT-binding protein. Nucleic Acids Res. 1987;15:7265–7272. doi: 10.1093/nar/15.18.7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin S, Scotto K W. Transcription regulation of the MDR1 gene by histone acetyltransferase and deacetylase is mediated by NF-Y. Mol Cell Biol. 1998;18:4377–4384. doi: 10.1128/mcb.18.7.4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim I-S, Sinha S, deCrombrugghe B, Maity S N. Determination of functional domains in the C subunit of the CCAAT-binding factor (CBF) necessary for formation of a CBF-DNA complex: CBF-B interacts simultaneously with both the CBF-A and CBF-C subunits to form a heterotrimeric CBF molecule. Mol Cell Biol. 1996;16:4003–4013. doi: 10.1128/mcb.16.8.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kouzarides T. Histone acetylases and deacetylases in cell proliferation. Curr Opin Genet Dev. 1999;9:40–48. doi: 10.1016/s0959-437x(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 23.Landsberger N, Wolffe A P. Role of chromatin and Xenopus laevis heat shock transcription factor in regulation of transcription from the X. laevis hsp70 promoter in vivo. Mol Cell Biol. 1995;15:6013–6024. doi: 10.1128/mcb.15.11.6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee D Y, Hayes J J, Pruss D, Wolffe A P. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 25.Lefebvre P, Mouchon A, Lefebvre B, Formstecher P. Binding of retinoic acid receptor heterodimers to DNA. J Biol Chem. 1998;273:12288–12295. doi: 10.1074/jbc.273.20.12288. [DOI] [PubMed] [Google Scholar]
- 26.Li Q, Herrler M, Landsberger N, Kaludov N, Ogryzko V V, Nakatani Y, Wolffe A P. Xenopus NF-Y pre-sets chromatin to potentiate p300 and acetylation-responsive transcription from the Xenopus hsp70 promoter in vivo. EMBO J. 1998;17:6300–6315. doi: 10.1093/emboj/17.21.6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liberati C, Ronchi A, Lievens P, Ottolenghi S, Mantovani R. NF-Y organizes the γ-globin CCAAT boxes region. J Biol Chem. 1998;273:16880–16889. doi: 10.1074/jbc.273.27.16880. [DOI] [PubMed] [Google Scholar]
- 28.Liberati C, diSilvio A, Ottolenghi S, Mantovani R. NF-Y binding to twin CCAAT boxes: role of Q-rich domains and histone fold helices. J Mol Biol. 1999;285:1441–1455. doi: 10.1006/jmbi.1998.2384. [DOI] [PubMed] [Google Scholar]
- 29.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 30.Luger K, Rechsteiner T J, Flaus A J, Waye M M Y, Richmond T J. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1998;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 31.Maity S N, deCrombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci. 1998;23:174–178. doi: 10.1016/s0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- 32.Mantovani R, Li X-Y, Pessara U, Hooft van Huijsduijnen R, Benoist C, Mathis D. Dominant negative analogs of NF-YA. J Biol Chem. 1994;269:20340–20346. [PubMed] [Google Scholar]
- 33.Mantovani R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998;26:1135–1143. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33a.Mantovani, R. Unpublished data.
- 34.Marziali G, Perrotti E, Ilari R, Coccia U E, Mantovani R, Testa U, Battistini A. The activity of the CCAAT-box binding factor NF-Y is modulated through the regulated expression of its A subunit during monocyte to macrophage differentiation: regulation of tissue-specific genes through a ubiquitous transcription factor. Blood. 1999;93:519–526. [PubMed] [Google Scholar]
- 35.Masternak K, Barras E, Zufferey M, Conrad B, Corthals G B, Aebersold R, Sanchez J C, Hochstrasser D F, Mach B, Reith W. A gene encoding a novel RFX-associated transactivator is mutated in the majority of the MHC class II deficiency patients. Nat Genet. 1998;3:273–277. doi: 10.1038/3081. [DOI] [PubMed] [Google Scholar]
- 36.Motta M C, Caretti G, Badaracco F, Mantovani R. Interactions of histone-fold containing CCAAT-binding trimer NF-Y with the nucleosome. J Biol Chem. 1999;274:1326–1333. doi: 10.1074/jbc.274.3.1326. [DOI] [PubMed] [Google Scholar]
- 37.Ogryzko V V, Kotani T, Zhang X, Schiltz R L, Howard T, Yang X-J, Howard B H, Qin J, Nakatani Y. Histone-like TAFs within the PCAF histone acetylase complex. Cell. 1998;94:35–44. doi: 10.1016/s0092-8674(00)81219-2. [DOI] [PubMed] [Google Scholar]
- 38.Perry M, Chalkley R. Histone acetylation increases the solubility of chromatin and occurs sequentially over most of the chromatin. J Biol Chem. 1982;257:7336–7347. [PubMed] [Google Scholar]
- 39.Scholz A, Truss M, Beato M. Hormone dependent recruitment of NF-Y to the uteroglobin gene enhancer associated with chromatin remodeling in rabbit endometrial epithelium. J Biol Chem. 1999;274:4017–4026. doi: 10.1074/jbc.274.7.4017. [DOI] [PubMed] [Google Scholar]
- 40.Sinha S, Kim I-S, Sohn K Y, deCrombrugghe B, Maity S N. Three classes of mutations in the A subunit of the CCAAT-binding factor CBF delineate functional domains involved in the three-step assembly of the CBF-DNA complex. Mol Cell Biol. 1996;16:328–337. doi: 10.1128/mcb.16.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spangenberg C, Eisfeld K, Stuckel W, Luger K, Flaus A, Richmond T J, Truss M, Beato M. The mouse mammary tumour virus promoter positioned on a tetramer of histone H3 and H4 binds nuclear factor 1 and OTF1. J Mol Biol. 1998;278:725–739. doi: 10.1006/jmbi.1998.1718. [DOI] [PubMed] [Google Scholar]
- 42.Tse C, Sera T, Wolffe A P, Hansen J C. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol Cell Biol. 1998;18:4629–4638. doi: 10.1128/mcb.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vettese-Dadey M, Grant P A, Hebbes T R, Crane-Robinson C, Allis C D, Workman J. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 44.Viville S, Jongeneel V, Koch W, Mantovani R, Benoist C, Mathis D. The Ea promoter: a linker-scanning analysis. J Immunol. 1991;146:3211–3217. [PubMed] [Google Scholar]
- 45.Workman J L, Kingston R E. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu Rev Biochem. 1998;67:745–779. doi: 10.1146/annurev.biochem.67.1.545. [DOI] [PubMed] [Google Scholar]
- 46.Xing Y, Fikes J D, Guarente L. Mutations in yeast HAP2/HAP3 define a hybrid CCAAT box binding domain. EMBO J. 1993;12:4647–4655. doi: 10.1002/j.1460-2075.1993.tb06153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zemzoumi K, Frontini M, Bellorini M, Mantovani R. NF-Y histone fold α1 helices help impart CCAAT specificity. J Mol Biol. 1999;286:327–337. doi: 10.1006/jmbi.1998.2496. [DOI] [PubMed] [Google Scholar]