Abstract
Background
Prostate cancer in African American (AA) men has a poor prognosis. This study aimed to identify potential genetic risk factors for prostate cancer in AA men.
Methods
We used prostate cancer tissue from 61 patients who underwent radical prostatectomy. We compared somatic gene expression in Caucasian (CA) and AA men using RNA sequencing.
Results
By comparing the RNA-seq data obtained from prostate cancer tissue between AA and CA men, this study showed a significant difference in expression levels of 45 genes. Pathway analysis of 45 genes using Kyoto Encyclopedia of Genes and Genomesenrichment analysis revealed a neuroactive ligand–receptor interaction signal. In addition, the results of the Ingenuity Pathway Analysis showed pathways involved sphingosine-1-phosphate signaling. Furthermore, validating 45 genes in the The Cancer Genome Atlas (TCGA) Provisional cohort, cholinergic receptor muscarinic 3 expression level was significantly lower in AA than in CA men, and the results showed a significantly higher rate of biochemical recurrence in patients with low expression.
Conclusions
We identified genetic differences of clinically localized prostate cancer in AAs and CAs by RNA sequencing.
Keywords: African American, Caucasian, Prostate cancer, RNA-seq
1. Introduction
Prostate cancer is the second most common cancer in men, with 164,690 estimated new cases diagnosed in the United States through 20181. In the United States, African American (AA) men are about 1.4 times more likely to have prostate cancer than Caucasian (CA) men and have about twice the mortality rate2. Furthermore, in AA men, prostate cancer has a poor prognosis and may have a worse prognosis even if patients are diagnosed with a low-risk prostate cancer. Brian et al. reported that AA men who chose active surveillance for a low-risk prostate cancer have a worse prognosis than non-AA men3. A number of studies have reported early upgrade of prostate cancer in AA men4,5. Therefore, AA men may need a different screening and treatment strategy than CA men6.
Differences in morbidity and mortality could be due to genetic predisposition. Epidemiological studies of men with similar genetic backgrounds give us the hypothesis that genetic factors are associated with high incidence and mortality in AA men. For example, men in Nigeria and Ghana have a high incidence of prostate cancer, and similar results have been observed in African descent in the Caribbean and the United Kingdom7. Genome-wide sequencing of high-risk prostate cancer in AA men showed race-specific genetic differences8. Thus, differences in somatic gene expression in AA prostate cancer are expected, suggesting that there may be biomarkers for predicting early progression. However, studies of gene expression levels for prostate cancer in AA men have been inadequate9.
The aim of this study was to compare somatic gene expression in CA and AA men using RNA sequences and to identify potential genetic risk factors for prostate cancer in AA men from these differences. The results obtained in this study were compared and validated using publicly available prostate cancer database. Then we searched for genes that are specific to AA men and may have prognostic impact.
2. Materials and Methods
2.1. Selection of patients
Sixty-one men who underwent radical prostatectomy at the Rutgers University Cancer Institute New Jersey (CINJ) between 2011 and 2017 were selected. Thirty-one were AA and 30 were CA men. Tumor collection was approved by the Institutional Review Board. All patients agreed to genetic testing of surgical explants. Prostate specific antigen (PSA) was measured before radical prostatectomy. Prostatectomy specimens were pathologically diagnosed at our institution after surgery, Gleason score was evaluated, and pathologic staging was determined. To determine whether there were significant differences in clinical backgrounds between AA and CA men, a correlation analysis was performed for each background factor.
2.2. Tissue preparation and sequencing
After prostatectomy, the prostate was fixed with formalin and paraffin. To adequately extract only prostate cancer from formalin-fixed paraffin-embedded tissue, a pathologist identified a site that was morphologically diagnosed with cancer. Library preparation for whole-transcriptome sequencing, with rRNA depletion, was performed using the NuGen Ovation Universal RNA-Seq system (Part#: 0343) according to manufacturer's protocol. The libraries were analyzed on Agilent 4200 TapeStation System using High Sensitivity D1000 ScreenTape Assay (Cat#: 5067-5584) and quantified using KAPA qPCR (Cat# KK4835). Libraries were then normalized to 10nM, and specific number of libraries were combined per pool to get about 100 million reads per sample. Each pooled library was then clustered and sequenced on Illumina NextSeq 550 instrument using NextSeq 500/550 High Output Kit v2 (300 cycles) in 2 × 150 bp paired end sequencing format.
2.3. Differential expression analysis
The RNA expression values in transcripts per million were determined using this kallisto package10 The kallisto results were grouped based upon AA and CA status and differential expression was calculated using the sleuth R package11. The threshold for significance in gene expression was a Benjamini-Hochberg corrected Pvalue (q-value) < 0.05. Enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Gene Ontologies were determined using DAVID12. Furthermore, we analyzed RNA signaling pathways with software, Ingenuity Pathway Analysis (IPA, http://www.ingenuity.com/).
2.4. Statistical analysis
To compare the patients' background, Fisher's exact test was used for a categorical variable and the Mann–Whitney's U test was used for continuous variables. Kaplan–Meier analysis was used to analyze the disease-free survival in the low- and high-RNA expression group (cutoff = 0.5), and log-rank test was used to detect the statistical significance. We analyzed only cases that had no missing values in the required data, when we found missing values in each analysis. All statistical analyses were performed with R Statistical Software (version 3.5.2.)13.
2.5. Validation data sets
Candidate genes obtained by comparing differences in gene expression between AA and CA men in our cohort were tested for gene expression levels and clinical outcomes in available public data to search for genes which are specific for AA men and likely to be involved in recurrence. The RNA expression levels between AA and CA men were compared again using TCGA Provisional database from the cBioPortal for Cancer Genomics (http://www.cbioportal.org/). Because TCGA Provisional contained the most ethnic information available in public databases, correlation analysis between gene expression and clinical outcome was performed using this dataset. RNA expressions were compared using the Mann–Whitney U test. We compared disease-free survival between the high- and low-expression groups using Kaplan–Meier analysis. Disease relapse was either biochemical recurrence or radiological tumor recurrence/metastasis. We were available RNA expression levels as a Z-score calculated by comparing to the expression distribution of each gene tumors that are diploid for these gene.
3. Results
3.1. Characteristics of the 31 AA and 30 CA men
The results of the comparison of patient background factors are shown in Table 1. There were no significant differences between AA and CA men in age, Gleason Score in surgical specimens, and pathological stage. The median preoperative PSA was 8.4 in AA and 5.9 in CA men, which were significantly higher in AA than in CA men (P = 0.021).
Table 1.
Characteristics of 61 patients of AAs and CAs
| Baseline characteristics | All patients (N = 61) | AAs (N = 31) | CAs (N = 30) | P |
|---|---|---|---|---|
| Age (years) median (range) | 63 (47–80) | 62 (47–78) | 64 (52–80) | 0.373 |
| Gleason sum at postoperative (6–7/8–10) N (%) | 48 (78.7%)/7 (11.5%) | 23 (74.2%)/4 (12.9%) | 25 (83.3%)/3 (10.0%) | 0.705 |
| Pathological tumor stage (T2/T3) N (%) | 36 (59.0%)/19 (31.1%) | 18 (58.1%)/9 (29.0%) | 18 (60.0%)/10 (33.3%) | 0.999 |
| PSA median (ng/mL) (range) | 6.9 (2.1–214) | 8.4 (3.7–214) | 5.9 (2.1–53.9) | 0.021 |
AAs, African Americans; CA, Caucasian; PSA, Prostate specific antigen.
3.2. mRNA expression
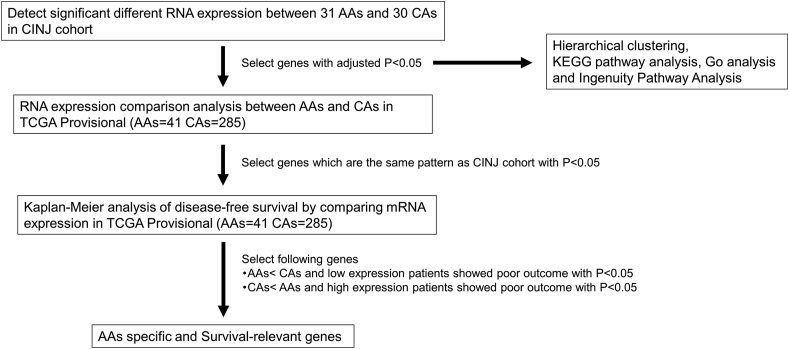
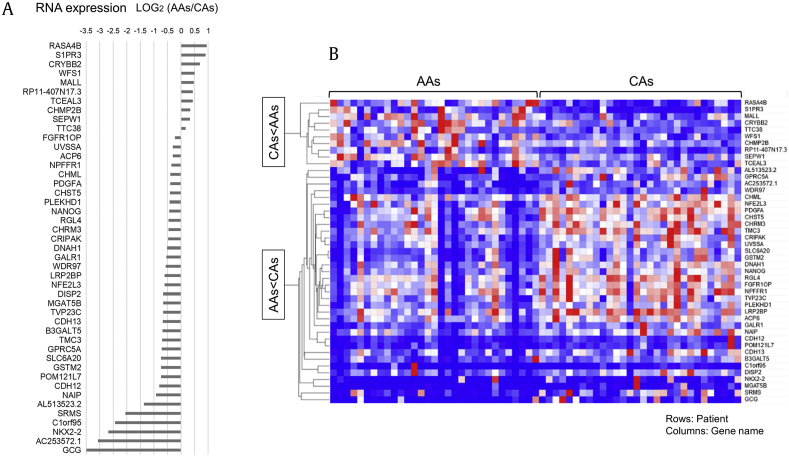
Fig. 1 showed a flowchart of this study. From whole-transcriptome RNA-seq in prostate cancers from AA and CA men, we found a significant difference in expression of 45 genes (adj. P < 0.05) (Fig. 2-a and Supplementary Table 1). Comparison of the log fold change RNA expression levels in AA and CA men (Log2 AAs/CAs) revealed that the genes specifically upregulated in CA men were AL513523.2, SRMS C1orf95 NKX2-2, AC25357.1, GCG (LOG2 (AAs/CAs) < −1.0). On the other hand, the top 3 most upregulated genes in AA men were RASA4B, S1PR3, and CRYBB2 (LOG2 (AAs/CAs) > 0.5). We divided the genes into two groups after hierarchical clustering by versatile matrix visualization and analysis software, Morpheus (https://software.broadinstitute. org/morpheus) (Fig. 2-b). CAs<AAs genes included increased expression levels in AA than in CA men and AAs<CAs genes included increased expression levels in CA than AA men. The results of Gene Ontologies analysis identified biological processes such as negative regulation of molecular function in AAs<CAs genes (Supplementary Table 2). On the other hand, no significant biological process was identified in CAs<AAs genes. Pathway analysis of 45 genes using KEGG enrichment analysis identified a neuroactive ligand–receptor interaction signal containing S1PR3, GALR1, cholinergic receptor muscarinic 3 (CHRM3), and NPFFR1 (Supplementary Table 3).
Fig. 1.
A flowchart for RNA sequence in CINJ cohort and validation analysis in TCGA Provisional. TCGA, The Cancer Genome Atlas
Fig. 2.
(A) Forty-five genes which have a significant difference in expression between AAs and CAs in CINJ cohort. RNA expressions were showed by log fold change in AAs and CAs (Log2 AAs/CAs). (B) Heat Map 45 RNA expression of AAs and CAs in CINJ cohort. Forty-five genes were classified into two groups (CAs<AAs and AAs<CAs) by hierarchical clustering in CINJ cohort. Blue represent downregulated, and red represents upregulated. AAs, African Americans; CA, Caucasian.
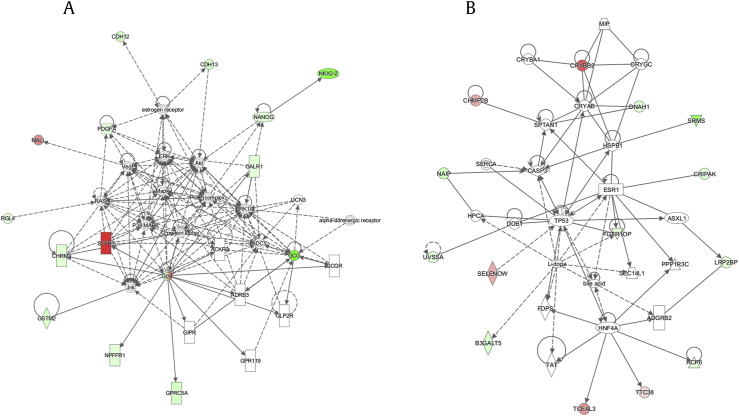
The analysis of 45 RNA expression in AAs by IPA identified four molecular pathways and two networks involving these genes. The network analyzed by IPA included a human embryonic stem (ES) cell pluripotency pathway involving NANOG, PDGFA, and S1PR3 (Table 2). In addition, CHRM3, S1PR3, NPFFR1, and GPRC5A formed a network centered on G protein–coupled receptors (Fig. 3-a). We also identified another network involving TP53 (Fig. 3-b).
Table 2.
The pathways involving 45 RNA expression of AAs in CINJ cohort by Ingenuity Pathway Analysis software
| Pathways detected by IPA | P | Genes |
|---|---|---|
| Human embryonic stem cell pluripotency | 0.002 | NANOG, PDGFA, S1PR3 |
| GPCR-mediated integration of enteroendocrine signaling exemplified by an L cell | 0.007 | GALR1, GCG |
| Sphingosine-1-phosphate signaling | 0.022 | PDGFA, S1PR3 |
| NAD phosphorylation and dephosphorylation | 0.023 | ACP6 |
AAs, African Americans.
Fig. 3.
The networks involving 45 RNA expression of AAs in CINJ cohort by Ingenuity Pathway Analysis (IPA) software. IPA identified two significant molecular pathways using 45 RNA expression of AAs in CINJ cohort. Red represents genes were the higher expression in AAs than CAs, and green represent genes were the lower expression in AAs than CAs. Solid lines represent the direct relationship, and dashed lines represent the indirect relationship, and arrows represent acts on. AAs, African Americans; CA, Caucasian.
3.3. Validation RNA-seq result by TCGA data set
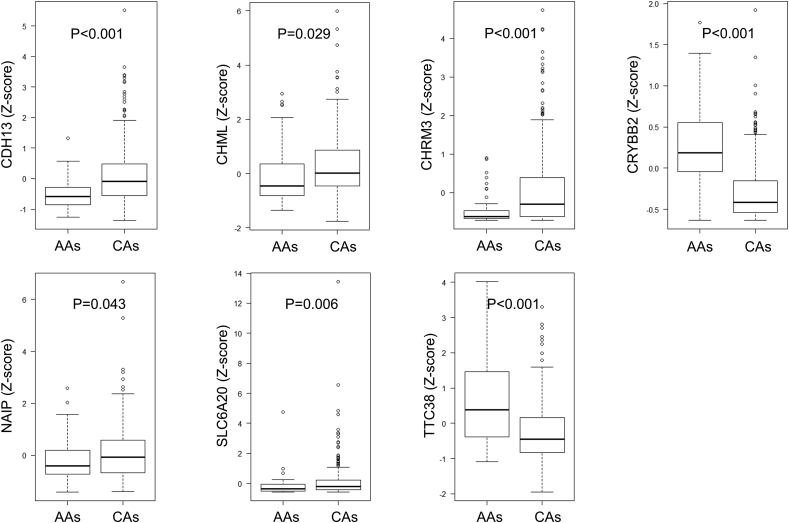
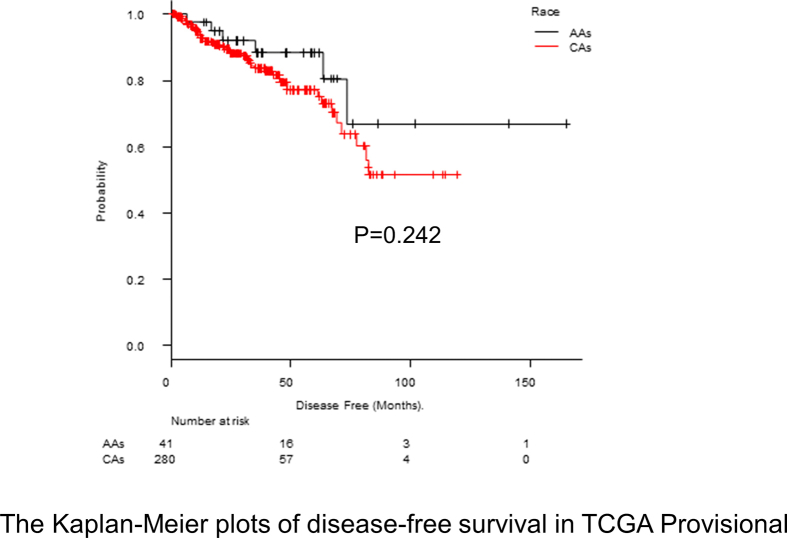
In TCGA Provisional, RNA sequence data were obtained from 326 patients, of which 285 were CA and 41 were AA men. Although there were significant differences between AA and CA men in diagnosis age (P = 0.004) and prostatectomy Gleason sum (P < 0.001) (Table 3), there was no significant differences in disease-free survival (median survival, 73.3 vs. 77.3 months; P = 0.242) (Supplementary Figure 1). Regarding genes with significant differences in our RNA sequence results, we also examined whether there were differences in RNA expression levels between AA and CA men in TCGA Provisional. The results showed a significant difference in the expression levels of 7 genes, which exhibited a similar pattern to our results with respect to whether AA or CA men expression increased levels (Fig. 4). In other words, the expression level of RNA in genes CRYBB2 (median Z score, 0.187 vs −0.414; P < 0.001) and TTC38 (median Z score, 0.381 vs −0.449; P < 0.001) was significantly higher in AA men, whereas the expression level of RNA in genes CDH13 (median Z score, −0.583 vs −0.087; P < 0.001), CHML (median Z score, −0.448 vs 0.013; P = 0.029), CHRM3 (median Z score, −0.623 vs −0.306; P < 0.001), NAIP (median Z score, −0.406 vs −0.074; P = 0.043), and SLC6A20 (median Z score, −0.372 vs −0.211; P = 0.006) was significantly higher in CA men in both CINJ cohort and TCGA Provisional.
Table 3.
Characteristics of 326 patients of AAs and CAs in TCGA Provisional
| Baseline characteristics | All patients (N = 326) | AAs (N = 41) | CAs (N = 285) | P |
|---|---|---|---|---|
| Age (years) median (range) | 61 (44–76) | 57 (44–71) | 62 (44–76) | 0.004 |
| Prostatectomy Gleason sum (6-7/8-10) N (%) | 204 (62.6%)/122 (37.4%) | 35 (85.4%)/6 (14.6%) | 169 (59.3%)/116 (40.7%) | <0.001 |
| Tumor stage at diagnosis (T2/T3) N (%) | 212 (65.0%)/39 (12.0%) | 34 (82.9%)/3 (7.3%) | 178 (62.5%)/36 (12.6%) | 0.224 |
AAs, African Americans; CA, Caucasian.
Fig. 4.
Forty-five RNA expression between AAs and CAs in TCGA Provisional. Among 45 genes, seven genes showed a significant difference between AAs and CAs in TCGA Provisional with the same pattern as CINJ cohort. RNA expression levels are shown as a Z-score. AAs, African Americans; CA, Caucasian.
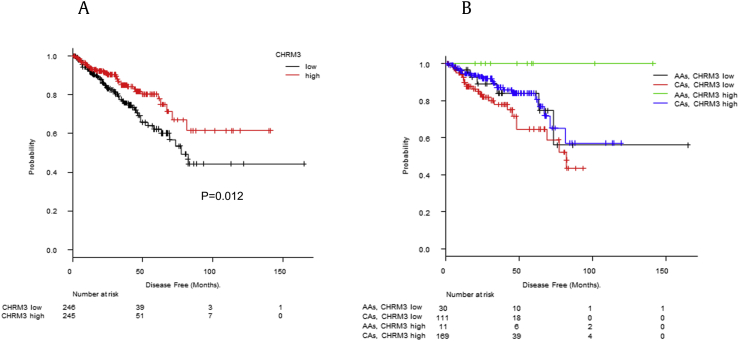
In addition, these gene expression levels and disease-free survival were compared using Kaplan–Meier analysis. There was significant difference in disease-free survival between the high- and low-expression groups only in CHRM3. High-expression group in CHRM3 had significantly longer disease-free survival (median survival, 77.3 vs. not reached months; P = 0.012) (Fig. 5-a). Furthermore, as a result of racial stratification, AA men with high expression of CHRM3 tended not to relapse after surgery compared with other patients (median survival, CHRM3 low in AA men; not reached, CHRM3 low in CA men; 82.2 months, CHRM3 high in AA men; not reached months, CHRM3 high in CA men; not reached) (Fig. 5-b).
Fig. 5.
The Kaplan–Meier plots of disease-free survival in TCGA Provisional. (A) CHRM3 expression and disease-free survival. Patients were divided into the low- and high- CHRM3 expression group (cutoff = 0.5). CHRM3 expression levels and disease-free survival were compared (median survival, 77.3 vs. not reached months; P = 0.012). (B) CHRM3 expression and disease-free survival with racial stratification. CHRM3 expression levels and disease-free survival were compared with racial stratification (median survival, CHRM3 low in AAs; not reached months, CHRM3 low in CAs; 82.2 months, CHRM3 high in AAs; not reached months, CHRM3 high in CAs; not reached months). AAs, African Americans; CA, Caucasian; CHRM3, cholinergic receptor muscarinic 3.
4. Discussion
By comparing the RNA-seq data obtained from prostate cancer tissue of AA and CA men, we found a significant difference in expression levels of 45 genes. The results of KEGG pathway analysis revealed four genes involved in neuroactive ligand–receptor interaction. In addition, the results of the IPA showed that there were two networks and five pathways in 45 genes. These pathways involved sphingosine-1-phosphate signaling. Furthermore, testing of 45 genes in the public database TCGA Provisional cohort showed that seven genes had significant differences in AA and CA men in the pattern similar to that of the present cohort. In particular, CHRM3 was significantly less expressed in AA than in CA men, and the results showed a significantly higher rate of biochemical recurrence in patients with low expression levels.
Several race-specific pathways in prostate cancer have been reported comparing AA with CA men14,15. Protein analysis by Myers et al.15 showed that neuroactive ligand–receptor interaction signaling was significantly associated with AA than with CA men in prostate cancer. They report that five proteins involved in neuroactive ligand–receptor interaction signaling were significantly higher in AA men. Neuroactive ligand–receptor interaction signaling is the assembly of receptors and ligands on the plasma membrane, which is associated with an intracellular and extracellular signaling pathway composed of 272 genes. Neuroactive ligand–receptor interaction signaling has also been reported to be associated with disease progression in other carcinomas16. In this study, three genes, which were significantly higher in CA men, and one gene, which was significantly higher in AA men, were involved in neuroactive ligand–receptor interaction signaling. These results suggest that prostate cancer in both AA and CA men are associated with neuroactive ligand–receptor interaction signaling.
IPA revealed that the human embryonic stem cell pluripotency pathway including S1PR3, which was highly expressed in AA men, was involved in prostate cancer in AA men. Sphingosine-1-phosphate (S1P) has a potential role in the self-replicating process in human ES cells. S1P is involved in a wide variety of biological processes, including cell proliferation, differentiation, migration, and apoptosis, in many different cell types. Thus, S1P plays a role in the cell's self-renewal process by preventing apoptosis17,18. S1PR3 is a functional receptor for S1P and has been reported to have oncogenic effects by activating AKT19. Significantly elevated S1PR3 levels in AA suggest that AA men may have increased self-renewal capacity in prostate cancer cells.
Activation of muscarinic receptors has been reported to promote the proliferation of prostate cancer cells in vitro20. In addition, Wang et al.21 reported that darifenacin, a selective CHRM3 antagonist, inhibits prostate cancer growth and castration resistance. Contrary to the biological functions of CHRM3 inferred from previous reports, patients with elevated CHRM3 levels had a better prognosis in the TCGA Provisional database. Furthermore, all AA men with elevated CHRM3 levels were free from recurrence during the observation period. In endometrial cancer, Wang et al.22 reported that patients with high levels of CHRM3 have significantly lower survival rates, but there are no reports on the expression and prognosis of CHRM3 in prostate cancer, and the significance of CHRM3 expression in human prostate cancer is still controversial. Yin et al.23 reported that pirenzepine, an antagonist of the same muscarinic receptor, CHRM1, inhibited migration and infiltration of PC-3, LNCaP, and A549 cells, whereas the agonist carbachol promoted migration and infiltration of these three cell lines. However, they also reported higher expression of CHRM1 in stage Ⅰ-Ⅱ prostate cancer than in stage Ⅲ-Ⅳ prostate cancer, based on staining of tumor tissue obtained from human patients. The authors explained these paradoxical results by speculating that patients with elevated CHRM1 levels may have more effective denervation, tumor resection24. Because patients with TCGA Provisional have undergone radical prostatectomy, we may be able to make similar speculation.
This study has several limitations. First, our cohort's observation period was relatively short. Therefore, we were unable to compare the results of RNA sequencing with those of the patient's clinical outcome. Second, the different sample sizes of AA and CA men in the TCGA Provisional cohort may influence the statistical analysis. Therefore, we are planning to reanalyze the results obtained in this study once clinical outcomes in the CINJ cohort are available. Third, although low expression of CHRM3 was associated with poor prognosis in overall patients and CHRM3 was significantly less expressed in AA than in CA men, it is not clear whether low expression of CHRM3 in AA causes a poorer prognosis of prostate cancer.
4.1. Conclusion
We identified specific genetic features in AAs by comparing the levels of AA and CA men RNA expression in tumor samples obtained from radical prostatectomy. In particular, low expression of CHRM3 and human ES cell pluripotency pathway and neuroactive ligand–receptor interaction, which includes S1PR3, may be one of the mechanisms that characterize poor prognosis prostate cancer in AA.
Author contributions
All authors have significantly contributed to the study and are in agreement with the content of the manuscript. Each author's contribution is as follows: I.K. designed and directed the project. N.N., J.R., and G.L. processed the experimental data and performed the analysis. N.N. and J.R. wrote the manuscript, and I.K. revised it.
Funding
This work was supported by the United States Department of Defense Office of the Congressionally Directed Medical Research Program (W81XWH-10-1-0359), cancer center grant from the National Cancer Institute (Grant P30CA072720), and generous support from the Marion and Norman Tanzman Charitable Foundation and Mr. Malcolm Wernik.
Conflict of interest
Authors have no significant conflict of interest.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.prnil.2020.11.002.
Appendix A. Supplementary data
The following is/are the Supplementary data to this article:
Supplementary Figure 1.
The Kaplan–Meier plots of disease-free survival in TCGA Provisional. Disease-free survival is compared between AAs and CAs (median survival, 73.3 vs. 77.3 months; P = 0.242).
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. 2018. [DOI] [PubMed] [Google Scholar]
- 2.DeSantis C.E., Miller K.D., Goding Sauer A., Jemal A., Siegel R.L. Cancer statistics for African Americans. CA Cancer J Clin. 2019;69(3):211–233. doi: 10.3322/caac.21555. 2019. [DOI] [PubMed] [Google Scholar]
- 3.Odom B.D., Mir M.C., Hughes S., Senechal C., Santy A., Eyraud R. Active surveillance for low-risk prostate cancer in African American men: a multi-institutional experience. Urology. 2014;83(2):364–368. doi: 10.1016/j.urology.2013.09.038. [DOI] [PubMed] [Google Scholar]
- 4.Sundi D., Ross A.E., Humphreys E.B., Han M., Partin A.W., Carter H.B. African American men with very low-risk prostate cancer exhibit adverse oncologic outcomes after radical prostatectomy: should active surveillance still be an option for them? J Clin Oncol. 2013;31(24):2991–2997. doi: 10.1200/JCO.2012.47.0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jalloh M., Myers F., Cowan J.E., Carroll P.R., Cooperberg M.R. Racial variation in prostate cancer upgrading and upstaging among men with low-risk clinical characteristics. Eur Urol. 2015;67(3):451–457. doi: 10.1016/j.eururo.2014.03.026. [DOI] [PubMed] [Google Scholar]
- 6.Shenoy D., Packianathan S., Chen A.M., Vijayakumar S. Do African-American men need separate prostate cancer screening guidelines? BMC Urol. 2016;16(1):19. doi: 10.1186/s12894-016-0137-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Odedina F.T., Akinremi T.O., Chinegwundoh F., Roberts R., Yu D., Reams R.R. Prostate cancer disparities in Black men of African descent: a comparative literature review of prostate cancer burden among Black men in the United States, Caribbean, United Kingdom, and West Africa. Infect Agents Canc. 2009;4(Suppl 1):S2. doi: 10.1186/1750-9378-4-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindquist K.J., Paris P.L., Hoffmann T.J., Cardin N.J., Kazma R., Mefford J.A. Mutational landscape of aggressive prostate tumors in African American Men. Canc Res. 2016;76(7):1860–1868. doi: 10.1158/0008-5472.CAN-15-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.The molecular taxonomy of primary prostate cancer. Cell. 2015;163(4):1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 11.Pimentel H., Bray N.L., Puente S., Melsted P., Pachter L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods. 2017;14:687. doi: 10.1038/nmeth.4324. [DOI] [PubMed] [Google Scholar]
- 12.Jiao X., Sherman B.T., Huang D.W., Stephens R., Baseler M.W., Lane H.C. DAVID-WS: a stateful web service to facilitate gene/protein list analysis. Bioinformatics. 2012;28(13):1805–1806. doi: 10.1093/bioinformatics/bts251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanda Y. Investigation of the freely available easy-to-use software 'EZR' for medical statistics. Bone Marrow Transplant. 2013;48(3):452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang B.D., Yang Q., Ceniccola K., Bianco F., Andrawis R., Jarrett T. Androgen receptor-target genes in african american prostate cancer disparities. Prostate Cancer. 2013;2013:763569. doi: 10.1155/2013/763569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Myers J.S., von Lersner A.K., Sang Q.X. Proteomic upregulation of fatty acid synthase and fatty acid binding protein 5 and identification of cancer- and race-specific pathway associations in human prostate cancer tissues. J Canc. 2016;7(11):1452–1464. doi: 10.7150/jca.15860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang Z.Q., Zang W.D., Chen R., Ye B.W., Wang X.W., Yi S.H. Gene expression profile and enrichment pathways in different stages of bladder cancer. Genet Mol Res. 2013;12(2):1479–1489. doi: 10.4238/2013.May.6.1. [DOI] [PubMed] [Google Scholar]
- 17.Inniss K., Moore H. Mediation of apoptosis and proliferation of human embryonic stem cells by sphingosine-1-phosphate. Stem Cell Dev. 2006;15(6):789–796. doi: 10.1089/scd.2006.15.789. [DOI] [PubMed] [Google Scholar]
- 18.Pyne S., Pyne N.J. Sphingosine 1-phosphate signalling in mammalian cells. Biochem J. 2000;349(Pt 2):385–402. doi: 10.1042/0264-6021:3490385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee H.M., Lo K.W., Wei W., Tsao S.W., Chung G.T.Y., Ibrahim M.H. Oncogenic S1P signalling in EBV-associated nasopharyngeal carcinoma activates AKT and promotes cell migration through S1P receptor 3. J Pathol. 2017;242(1):62–72. doi: 10.1002/path.4879. [DOI] [PubMed] [Google Scholar]
- 20.Rayford W., Noble M.J., Austenfeld M.A., Weigel J., Mebust W.K., Shah G.V. Muscarinic cholinergic receptors promote growth of human prostate cancer cells. Prostate. 1997;30(3):160–166. doi: 10.1002/(sici)1097-0045(19970215)30:3<160::aid-pros3>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 21.Wang N., Yao M., Xu J., Quan Y., Zhang K., Yang R. Autocrine activation of CHRM3 promotes prostate cancer growth and castration resistance via CaM/CaMKK-mediated phosphorylation of Akt. Clin Canc Res. 2015;21(20):4676–4685. doi: 10.1158/1078-0432.CCR-14-3163. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y., Li J., Wen S., Yang X., Zhang Y., Wang Z. CHRM3 is a novel prognostic factor of poor prognosis in patients with endometrial carcinoma. Am J Tourism Res. 2015;7(5):902–911. [PMC free article] [PubMed] [Google Scholar]
- 23.Yin Q.Q., Xu L.H., Zhang M., Xu C. Muscarinic acetylcholine receptor M1 mediates prostate cancer cell migration and invasion through hedgehog signaling. Asian J Androl. 2018;20(6):608–614. doi: 10.4103/aja.aja_55_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saloman J.L., Albers K.M., Rhim A.D., Davis B.M. Can Stopping Nerves, Stop Cancer? Trends Neurosci. 2016;39(12):880–889. doi: 10.1016/j.tins.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.