Abstract
Objectives
Oral squamous cell carcinoma (OSCC) is a commonly reported cancer in men and is second only to breast cancer in women in Pakistan.. Investigations for identifying biomarkers of OSCC are essential for diagnostic, therapeutic, or prognostic significance. This study aims to examine the miR-31 expression in the pre- and post-operative OSCC patients and correlate this expression with clinicopathological characteristics.
Methods
Patients with histopathologically confirmed OSCC who had undergone surgical resections of tumours were recruited. A total of 40 saliva samples (pre- and post-operative) were collected from 19 patients and two healthy individuals. Levels of salivary miR-31 expressions were examined through quantitative reverse transcription polymerase chain reaction.
Results
The salivary miR-31 expression was significantly higher in the preoperative patients than in postoperative patients (p < 0.001). However, no significant correlation had been found between the salivary miR-31 expression and clinicopathological characteristics (p > 0.05).
Conclusion
Our data suggest that miR-31 can be used as an adjunct non-invasive marker to monitor surgery outcomes during postoperative follow-up in patients with OSCC.
Keywords: Circulating microRNA, miR-31, Saliva, Oral squamous cell carcinoma, Biomarkers
الملخص
أهداف البحث
سرطان الخلايا الحرشفية في الفم هو سرطان شائع لدى الرجال. والفحوصات لتحديد المؤشرات الحيوية لـسرطان الخلايا الحرشفية في الفم من أجل أهمية التشخيص أو العلاج أو التكهن ضرورية. تهدف هذه الدراسة إلى تحري تعبير miR-31 في مرضى سرطان الخلايا الحرشفية في الفم قبل الجراحة وما بعدها وربط التعبير بالخصائص السريرية.
طرق البحث
ضمت الدراسة المرضى الذين تأكد وجود سرطان الخلايا الحرشفية في الفم لديهم بعد الخضوع لاستئصال جراحي للأورام. وتم جمع ما مجموعه ٤٠ عينة من اللعاب (قبل الجراحة وبعدها) من ١٩ مريضا واثنين من الأفراد الأصحاء. تم التحقيق في مستويات تعبيرات miR-31 اللعابية باستخدام تفاعل البوليميراز المتسلسل للنسخ العكسي الكمي.
النتائج
كان التعبير اللعابي miR-31 مرتفعا بشكل ملحوظ في فترة ما قبل الجراحة مقارنة بالمرضى بعد الجراحة. ومع ذلك، لم يتم العثور على ارتباط كبير بين التعبير اللعابي ل miR-31 والخصائص السريرية المرضية.
الاستنتاجات
تشير بياناتنا إلى الاستخدام المحتمل لـ miR-31 كعلامة غير تداخلية لمراقبة نتائج الجراحة أثناء متابعة ما بعد الجراحة في المرضى الذين يعانون من سرطان الخلايا الحرشفية في الفم.
الكلمات المفتاحية: اللعاب, سرطان الخلايا الحرشفية في الفم, المؤشرات الحيوية
Introduction
Oral squamous cell carcinoma (OSCC) is ranked amongst the predominant malignancies worldwide.1 In Pakistan, it is the most common cancer in men and is second only to breast cancer in women.2 Despite progress in treatment modalities, the prognosis of OSCC remains poor. A majority of patients in Pakistan are diagnosed at an advanced stage.3 The delay in diagnosis is primarily due to the patients’ ignorance about the disease symptoms and delayed investigations or referral by general practitioners.3 To date, tissue biopsy, the gold standard, is the most suitable approach for definitive diagnosis of OSCC. However, it is invasive, costly, time consuming, and technique sensitive. Various minimally/non-invasive methods have been developed for oral cancer detection: brush or exfoliative cytology, toluidine blue, methylene blue, and photodynamic detection. Nevertheless, these methods have limited use as the first-line investigation for diagnostic purposes. Moreover, tissue/cytological methods preclude prognosis in follow-up patients and disease recurrence.4 To address these concerns, non-invasive markers should be identified which could improve detection and prognosis of OSCC. Saliva liquid biopsy is more advantageous for oral lesions than tissues and blood as it is non-invasive, convenient, cost-effective, reproducible, and painless, and can be obtained without the risk of needle-stick injury. Moreover, its direct contact with tumour tissue makes it a preferred diagnostic and prognostic tool in oral cancer research.5,6
Recent research has focused on microRNAs’ (miRNAs) potential utility for diagnosis, prognosis, and monitoring treatment response in cancer.7 MicroRNAs are short-non coding RNA (19–25 nucleotides in length) that regulate gene expression at transcription level. MicroRNA genes are transcribed into primary miRNA (>200 nucleotides) by RNA polymerase II. Primary miRNA is converted into precursor miRNA or pre-miRNA (70–100 nucleotides) through Drosha and DiGeorge Syndrome Critical Region 8 (DGCR8). Pre-miRNA is then transported to cytoplasm via exportin 5 where pre-miRNA it is cleaved into miRNA duplex (19–25 nucleotides) by Dicer. Subsequently, mature miRNA binds with Argonaute protein to form RNA-induced silencing complex (RISC), which in turn binds with the untranslated region (UTR) of mRNA, thereby leading to translational repression or degradation of mRNA.8
Interestingly, miRNAs are not only found in tissues, but also circulate in body fluids such as blood, saliva, urine, and cerebrospinal fluid.9 Recently, miRNAs have emerged as an excellent source for liquid biopsy biomarkers due to their stability and resistance to degradation in biofluids.10 Salivary miRNAs are secreted in the oral cavity via different sources, including the salivary glands, crevicular fluid, and desquamated cells. Mostly, miRNAs are present in saliva in partly degraded forms. The partly degraded miRNAs bind with macromolecules and thus sustain stability in saliva.11 Dysregulated salivary miRNAs have been detected in various cancers including OSCC.6,12 The expression of dysregulated miRNAs may help differentiate patients with oral squamous cell carcinoma from healthy individuals and may be utilized as biomarkers for diagnostic and prognostic purposes.11
MicroRNA-31 (miR-31) shows dysregulated levels of expression in various cancers. The miR-31 gene is positioned on chromosome 9p21.3, near the locus of cyclin dependent kinase inhibitor (CDKN2A and CDKN2B), a tumour suppressor gene that encodes p15 and p16 (cell-cycle inhibitor protein).13 Due to their proximity, co-deletion of miR-31 and CDKN2A gene loci or hypermethylation results in low expressions of miR-31 in gastric, liver, breast, ovarian, and prostate cancers. In contrast, oncogenic ability of miR-31 is regulated by mutated KRAS stimulating the promoter of miR-31 and increasing its expressions in lung, colorectal, esophageal, and oral squamous cell carcinoma.13 Increased expressions of miR-31 in tissue and liquid biopsies have been investigated in healthy individuals and OSCC patients.14,15 However, little is known about salivary miR-31 expression in OSCC patients with postoperative follow-up. The study aims to investigate salivary miR-31 expression in pre- and post-operative OSCC patients and correlate its expression with clinicopathological characteristics.
Materials and Methods
The study was conducted from June 2018 to November 2018 at Dr. Ruth K.M. Pfau Civil Hospital and Dow Research Institute of Biotechnology and Biomedical Sciences after approval from Institutional Review Board (IRB-1002/DUHS/Approval/2018/40) of Dow University of Health Sciences. Written, informed consent was obtained from patients after the objectives and nature of procedure were fully explained.
Openepi software was used for sample size calculation with 99% confidence interval and 95% power of test. Further data were used from a similar study: means ± standard deviations (6.56 ± 1.41 in preoperative group and 8.69 ± 1.41 in control group).16 The sample included 19 (preoperative group), 19 (postoperative group), and 2 controls.
Patients with histopathologically proven oral squamous cell carcinoma who were admitted to the ENT ward and underwent surgical resection of tumour were included via convenience sampling. Patients receiving chemoradiotherapy with recurrent OSCC and history of any other cancer were excluded from the study. Saliva samples from 19 preoperative and 19 postoperative patients (n = 38) were collected before and six weeks after surgery. Both preoperative and postoperative saliva samples were taken from the same patient. Saliva samples from healthy individuals (n = 2) without history of cancer were used as calibrator samples for quantitative polymerase chain reaction.
Saliva collection
Fasting and unstimulated saliva was collected via PureSAL ™ device (catalog no. PRSAL - 401, Oasis Diagnostics, Vancouver USA) following manufacturer's instructions. The device was positioned under the side of the tongue to absorb the pooled saliva. The sample was then stored at −80 °C until further processing.
RNA extraction
Four hundred microlitres of saliva were used for the miRNA extraction through mirVANA PARIS™ and native protein extraction kit (catalog No. AM1556, Invitrogen, Thermofisher Scientific, USA) following manufacturer's instruction. The final concentration and purity of isolated miRNA was measured on Nanodrop™ Lite Spectrophotometer using 2 μl of sample following manufacturer's instruction (Thermofisher Scientific, USA). A ratio of 1.8–2.0 (A260/A280) was accepted as pure for extracted RNA.
Complementary DNA (cDNA) synthesis
Complementary DNA (cDNA) for miR-31 (target gene) and miR-16 (internal control) was synthesized using the Taqman™ MicroRNA-Reverse-Transcription Kit (catalog no.4366596, Thermofisher Scientific, USA) following manufacturer guidelines. We used miR-16 for normalization of miRNAs as it is the most used housekeeping gene for saliva analysis.15 To prepare 15 μl final solution, the reverse transcriptase master mix was made using 0.15 μl dNTPs with dTTP (100 mM), 1 μl Multiscribe ™ reverse transcriptase (50 U/μl), 1.50 μl reverse transcription buffer (10X), 0.19 μl RNase inhibitor (20 U/μl), 10 ng of 5 μl RNA sample, 4.16 μl nuclease free water, and 3 μl reverse transcriptase primer. The 15 μl reaction was placed in SimpliAmp™ Thermal Cycler (catalog no. A24811, Applied Biosystems ™) in a 96-well plate and was run according to manufacturer guidelines as follows: incubation at 16 °C and 42 °C each for 30 min and then 85 °C and 4 °C each for 5 min. Following incubation, the synthesized cDNA products of miR-31 and miR-16 were stored at −20 °C until further processing.
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
Reverse transcription quantitative polymerase chain reaction (RT-qPCR) was conducted in Quant studio 7 flex for the quantitative expression of miR-31 (Applied Biosystems, USA) using PCR reaction mix, TaqMan MicroRNA assays kit (catalog no: 4427975, Thermofisher Scientific, USA), and TaqMan fast advanced master mix (catalog no: 4444557). The TaqMan microRNA Assay kit contains one tube of reverse transcriptase primers and one tube of TaqMan-Assay with separate preformed primers (forward and reverse) and MGB probe for miR-31 and miR-16. To compare quality of miR-31 expression, the amplification of miR-16 (endogenous control) was used as proof.15 The 10 μl PCR volume for amplification includes 0.50 μl TaqMan RNA assay, 5 μl PCR master mix, 3.84 μl nuclease free water, and 0.67 μl cDNA. The PCR thermal profile was primarily set to 95 °C for 10 min then 95 °C for 30 s, 56 °C for 30 s (40 cycles), and subsequently 72 °C for 20 s, wherein fluorescence was achieved. The reaction was performed in triplicates for each sample along with negative control (nuclease free water).
RT-qPCR data analyses
To calculate the fold change between patients (pre-operative and post-operative) and controls, the results of RT-qPCR for target gene (miRNA-31) and reference gene (miRNA-16) were analysed by the relative quantification method (ΔΔCT). The 2−ΔΔCT is a simple method to calculate the relative fold gene expression of sample when performing quantitative real time PCR. The following formula was used to calculate the relative expression.
| Fold change = 2−ΔΔCT |
| 2−ΔΔCT = ΔCT (OSCC patients) − ΔCT (controls) |
| ΔCT (OSCC patients) = CT (miR-31) – CT (miR-16) |
| ΔCT (controls) = CT (miR-31) – CT (miR-16) |
Statistical analyses
Data were analysed using SPSS version 24. Descriptive statistics were expressed in terms of frequency and percentage for categorical variables, including age, habit, site of tumour, size of tumour, grade of tumour, and pathological tumour node metastasis (pTNM) staging of tumour. Continuous variables (preoperative miR-31 expression levels) were presented as mean with standard deviation. Shapiro–Wilk's test was used to analyse normality of data. Additionally, data were normally distributed (p > 0.05). Paired-samples t-test was employed to compare miR-31 expression in preoperative and postoperative OSCC patients. Spearman's correlation was used to measure possible correlations between expression of preoperative miR-31 and clinicopathological characteristics. A p-value of 0.05 was set for all tests.
Results
Of the 40 saliva samples (19 preoperative, 19 same postoperative, and 2 controls) investigated, 3 preoperative and 3 postoperative saliva samples of same patients were discarded due to low purity ratio of RNA. Therefore, a total of 34 saliva samples (16 preoperative, 16 postoperative, and 2 controls) were finally included for qPCR data analyses. Samples from healthy individuals (2 controls) were used as calibrator sample for PCR reaction.
Mean age of patients was 50.05 ± 13.40 with age range of 32–79 years. The clinicopathological characteristics of patients are shown in Table 1.
Table 1.
Clinicopathological characteristics of patients.
| Variables | Frequency (n = 19) |
|---|---|
| Age | |
| >40 | 15 (78.9%) |
| ≤40 | 4 (21.1%) |
| Gender | |
| Male | 16 (84.2%) |
| Female | 3 (15.8%) |
| Habits | |
| Chewed tobacco | 12 (63.1%) |
| Smoked tobacco | 6 (31.6%) |
| No habit | 1 (5.3%) |
| Tumour site | |
| Buccal mucosa | 8 (42.1%) |
| Mandible | 3 (15.8%) |
| Tongue | 3 (15.8%) |
| Retromolar trigone | 3 (15.8%) |
| Maxilla | 2 (10.5%) |
| Tumour size | |
| pT2 | 4 (21.1%) |
| pT3 | 7 (36.8%) |
| pT4a | 8 (42.1%) |
| Tumour grade | |
| Well differentiated | 3 (15.8%) |
| Moderately differentiated | 12 (63.2%) |
| Poorly differentiated | 4 (21.1%) |
| Nodal Status | |
| Yes | 12 (63.2%) |
| No | 7 (36.8%) |
| pTNM∗staging | |
| pT2, pN0, pMx | 2 (10.5%) |
| pT2, pN1, pMx | 2 (10.5%) |
| pT3, pN0, pMx | 1 (5.3%) |
| pT3, pN1, pMx | 1 (5.3%) |
| pT3, pN2a, pMx | 1 (5.3%) |
| pT3, pN2b, pMx | 4 (21.1%) |
| pT4a, pN0, pMx | 4 (21.1%) |
| pT4a, pN1, pMx | 2 (10.5%) |
| pT4a, pN2b, pMx | 2 (10.5%) |
pTNM = pathological tumour node metastasis staging College of American Pathologists (CAP) protocol for lip & oral cavity cancer).
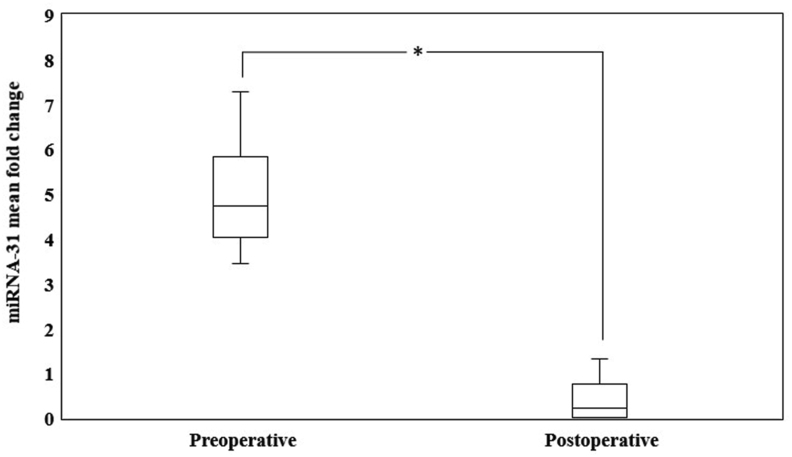
We compared expression levels of salivary miR-31 between preoperative and postoperative patients. As shown in Figure 1, increased miR-31 expression levels were observed in preoperative patients compared to the expression levels in postoperative patients, and this difference was statistically significant (p < 0.001). Mean fold change (2−ΔΔCT) for preoperative patients was 5.10 ± 1.21, whereas mean fold change (2−ΔΔCT) for postoperative patients was 0.31 ± 0.31. Next, we correlated miR-31 expression level and clinicopathological characteristics of OSCC patients and observed a weak–positive correlation between miR-31 and age, habits, tumour site, and size and pTNM staging, but this difference was not significant (p > 0.05). However, no correlation was found between miR-31 expression level and gender, nodal (regional) metastasis and grading (p > 0.05). Correlative analyses of miR-31 expression and clinicopathological characteristics are shown in Table 2.
Figure 1.
Significant fold change of salivary miR-31 expression levels between pre- and post-operative oral squamous cell carcinoma patients (∗p-value < 0.001).
Table 2.
Correlation between preoperative miR-31 expression and clinicopathological characteristics.
| Variables | R | p-value |
|---|---|---|
| Age | 0.43 | 0.09 |
| Gender | 0.08 | 0.76 |
| Habits | 0.41 | 0.10 |
| Tumour site | 0.38 | 0.14 |
| Tumour size | 0.32 | 0.21 |
| Tumour grade | 0.04 | 0.85 |
| Nodal status | −0.19 | 0.46 |
| pTNM staging | 0.38 | 0.14 |
R = correlation coefficient.
p < 0.05 significant.
Discussion
Researchers report that tobacco, alcohol, and human papilloma virus (HPV) are the known risk factors of OSCC that not only alter the function of tumour suppressor genes, proto-oncogene, and oncogenes but also produce aberrations in normal cellular mechanisms, such as chromosomal instability, copy number variation, loss of heterozygosity, telomere stability, DNA damage response, cell-cycle regulators, and Notch signaling pathways.17 The overall prognosis of OSCC patients depends on the stage at which the treatment is provided. Unfortunately, most patients in Pakistan are diagnosed at the advanced stage.3 Thus, novel biomarkers are urgently needed for patients with oral potentially malignant disorders (OPMD) and OSCC, as well as for postoperative follow-up. Saliva-liquid biopsy, due to its direct contact with oral mucosa, may provide a useful insight into the disease status of oral tissues. MicroRNAs are short non-coding, RNA molecules contributing to tumorigenesis. By targeting hundreds of mRNAs, a single miRNA potentially regulates cell proliferation, differentiation, migration, apoptosis, and signal transduction in tumorigenesis, thereby suggesting a possible utility of miRNAs for early detection and postoperative monitoring or follow-up in cancer patients.18,19
MiR-31 is overexpressed in OSCC, augmenting anchorage-independent growth and migration. Under normoxia, miR-31 targets 3´-UTR of factor-inhibiting hypoxia-inducible factor (FIH) and mutates hypoxia-inducible factor (HIF) pathway to promote tumorigenesis. Moreover, FIH works as an oxygen indicator. When oxygen levels are adequate, HIF1α is hydroxylated via asparagine. Subsequently, HIF1α represses the formation of vascular endothelial growth factor (VEGF). Under hypoxia, the suppressive activity of FIH on growth is decreased. When oxygen is above the FIH threshold, an inverse correlation ensues between FIH and VEGF. Thus, under normoxia, miR-31 contributes to OSCC development by mutating FIH and subsequently triggering VEGF.13 Additionally, epidermal growth factor receptors (EGFR) are overexpressed in OSCC tissues. Subsequent stimulation of EGF results in up-regulation of miR-31 through C/EBPβ signaling cascade.20 Recently, in vitro studies have suggested a potential role of miR-31 in regulating cell cycle and proliferation of OSCC tissues via Wnt and Hippo signalling pathways.21,22
The current study aimed to investigate miR-31 expression in preoperative and postoperative patients with oral squamous cell carcinoma. Our findings reveal significant difference in saliva miR-31 expression in preoperative and postoperative OSCC patients. The salivary miR-31 expression levels were high in the preoperative group, whereas the expression levels were reduced in the postoperative group after six weeks of surgery. These findings are in accordance with Liu et al.15 and Al-Malkey et al.23 who also reported increased expression of salivary miR-31 in preoperative patients and reduced expression in postoperative OSCC patients, suggesting that most of the upregulated miR-31 in preoperative patients was derived from OSCC tissues. Another study noted that miR-31 expression in plasma was reduced after surgery in OSCC patients, further establishing that miR-31 is tumor-associated.16,19 Although upregulation of salivary miR-31 in OSCC may be a contribution from multiple sources, in another study, Liu et al. reported increased expression of miR-31 in saliva compared to plasma in OSCC patients, implying a local release of miR-31 from tumour tissue to saliva.16 Moreover, at the molecular level, miR-31 targets EMP1 (epithelial membrane protein, a tumour suppressor gene) in squamous epithelium and suppresses its function, which may be a common phenomenon in various squamous cell carcinoma that explains the plausibility of overexpressed miR-31 in squamous cell carcinoma (SCC) tissues.24 Similarly, increased miR-31 expression in saliva was associated with OPMD and predicted the recurrence of OPMD and malignant transformation.25,26
Our results do not show a significant correlation between miR-31 expression and gender, habits, tumour site, tumour size, nodal status, grade of the tumour and pTNM staging. These findings are in alignment with previous studies.23,27 In contrast to our findings, previous studies showed significant associations of miR-31 with site, staging, and nodal metastasis.14,28 Moreover, another study revealed overexpression of miR-31 in non-metastatic samples of OSCC.29 However, no such correlation was found in our samples. The most plausible reason for these differences in our study may be due to the small sample size, comprising mainly advanced lesions, such as T3 and T4 lesions and positive lymph nodes. Our study shows no significant correlation between miR-31 and habit. To date, no study has compared the correlation of salivary miR-31 and habit in OSCC. However, in contrast to our findings, saliva miR-31 showed a significant association in smoked and smokeless tobacco in OPMD.30 Moreover, an in-vitro study reported altered expression of miR-31 in chewed tobacco and smoked tobacco.31 Despite cigarette, pan, and betel quid being the known risk factors of OPMD and subsequent OSCC development in our country,32 our findings showed a low positive correlation between saliva miR-31 and habit; however, the result was not statistically significant. The plausible reasons for this may be small size as well as lack of recording intensity and duration of habit.
Collectively, the above results indicate that increased saliva miR-31 expression can differentiate preoperative and postoperative OSCC patients. However, limitations of the study include small sample size, single-centre, one-time follow-up after six weeks of surgery, and only one-time postoperative sample. Since the follow-up was done only once after six months, long term follow-up studies are required to further validate this.
Conclusion
The saliva miR-31expression in preoperative patients of OSCC was significantly higher and its expression was subsequently reduced six weeks after surgery in postoperative patients. According to this observation, saliva miR-31 may be used as a non-invasive marker to monitor surgery outcomes during postoperative follow-up in oral squamous cell carcinoma patients.
Recommendations
Multi-centre studies with large sample size, chemoradiotherapy patients and postoperative saliva collection at different intervals may further explore the potential role of miR-31 in postoperative follow-up patients with OSCC. Salivary miR-31 can be used as an adjunct non-invasive marker for diagnosis of OSCC. MiR-31 seems to be a useful prognostic indicator for disease monitoring, recurrence, and survival of OSCC patients. Moreover, miR-31 may be used to measure surgical efficacy and develop a novel therapeutic target for OSCC patients.
Source of funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
The study was performed in line with the ethical regulation of the institutional research board and conformed with the 1964 Helsinki Declaration at Dr. Ruth K.M. Pfau Civil Hospital and Dow Research Institute of Biotechnology and Biomedical Sciences (DRIBBS) of Dow University of Health Sciences (DUHS) after obtaining ethical approval from the Institutional Review Board of the DUHS on 5th April 2018 (IRB-1002/DUHS/Approval/2018/40).
Consent
Patients were briefed about the objective of study. Written, informed consents were obtained from patients willing to participate in the study.
Authors’ contribution
PK performed the experiment, acquired data, and drafted the manuscript. SAS conceived and designed the study, analysed and interpreted data, and drafted the manuscript in correspondence with the journal. MW facilitated the implementation of the study and contributed towards manuscript drafting. MAQ implemented the study, analysed data, and drafted the manuscript. RK contributed towards sample collection and manuscript drafting. All authors have critically reviewed and approved the final draft and are responsible for the content and similarity index of the manuscript.
Acknowledgment
We acknowledge Dow University of Health Sciences for providing lab facilities.
Footnotes
Peer review under responsibility of Taibah University.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Qureshi M.A., Khan S., Sharafat S., Quraishy M.S. Common cancers in karachi, Pakistan: 2010-2019 cancer data from the Dow cancer registry. Pak J Med Sci. 2020;36(7):1572–1578. doi: 10.12669/pjms.36.7.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basharat S., Shaikh B.T., Rashid H.U., Rashid M. Correction to: Health seeking behaviour, delayed presentation and its impact among oral cancer patients in Pakistan: a retrospective qualitative study. BMC Health Serv Res. 2019;19(1):835. doi: 10.1186/s12913-019-4716-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chhabra N., Chhabra S., Sapra N. Diagnostic modalities for squamous cell carcinoma: an extensive review of literature-considering toluidine blue as a useful adjunct. J Maxillofac Oral Surg. 2015;14(2):188–200. doi: 10.1007/s12663-014-0660-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eguchi T. Organoids and liquid biopsy in oral cancer research. J Clin Med. 2020;9(11):3701. doi: 10.3390/jcm9113701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radhika T., Jeddy N., Nithya S., Muthumeenakshi R.M. Salivary biomarkers in oral squamous cell carcinoma - an insight. J Oral Biol Craniofac Res. 2016;6(Suppl 1):S51–S54. doi: 10.1016/j.jobcr.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bottani M., Banfi G., Lombardi G. Circulating miRNAs as diagnostic and prognostic biomarkers in common solid tumors: focus on lung, breast, prostate cancers, and osteosarcoma. J Clin Med. 2019;8(10):1661. doi: 10.3390/jcm8101661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang C., Li Y. Prospective applications of microRNAs in oral cancer. Oncol Lett. 2019;18(4):3974–3984. doi: 10.3892/ol.2019.10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weber J.A., Baxter D.H., Zhang S., Huang D.Y., Huang K.H., Lee M.J. The microRNA spectrum in 12 body fluids. Clin Chem. 2010;56(11):1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shigeyasu K., Toden S., Zumwalt T.J., Okugawa Y., Goel A. Emerging role of MicroRNAs as liquid biopsy biomarkers in gastrointestinal cancers. Clin Canc Res. 2017;23(10):2391–2399. doi: 10.1158/1078-0432.ccr-16-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Menini M., De Giovanni E., Bagnasco F., Delucchi F., Pera F., Baldi D. Salivary micro-RNA and oral squamous cell carcinoma: a systematic review. J Personalized Med. 2021;11(2):101. doi: 10.3390/jpm11020101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li F., Yoshizawa J.M., Kim K.M., Kanjanapangka J., Grogan T.R., Wang X. Discovery and validation of salivary extracellular RNA biomarkers for noninvasive detection of gastric cancer. Clin Chem. 2018;64(10):1513–1521. doi: 10.1373/clinchem.2018.290569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu T., Ma P., Wu D., Shu Y., Gao W. Functions and mechanisms of microRNA-31 in human cancers. Biomed Pharmacother. 2018;108:1162–1169. doi: 10.1016/j.biopha.2018.09.132. [DOI] [PubMed] [Google Scholar]
- 14.Wang L.L., Li H.X., Yang Y.Y., Su Y.L., Lian J.S., Li T. MiR-31 is a potential biomarker for diagnosis of head and neck squamous cell carcinoma. Int J Clin Exp Pathol. 2018;11(9):4339–4345. [PMC free article] [PubMed] [Google Scholar]
- 15.Liu C.J., Lin S.C., Yang C.C., Cheng H.W., Chang K.W. Exploiting salivary miR-31 as a clinical biomarker of oral squamous cell carcinoma. Head Neck. 2012;34(2):219–224. doi: 10.1002/hed.21713. [DOI] [PubMed] [Google Scholar]
- 16.Liu C.J., Kao S.Y., Tu H.F., Tsai M.M., Chang K.W., Lin S.C. Increase of microRNA miR-31 level in plasma could be a potential marker of oral cancer. Oral Dis. 2010;16(4):360–364. doi: 10.1111/j.1601-0825.2009.01646.x. [DOI] [PubMed] [Google Scholar]
- 17.Ali J., Sabiha B., Jan H.U., Haider S.A., Khan A.A., Ali S.S. Genetic etiology of oral cancer. Oral Oncol. 2017;70:23–28. doi: 10.1016/j.oraloncology.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Vannini I., Fanini F., Fabbri M. Emerging roles of microRNAs in cancer. Curr Opin Genet Dev. 2018;48:128–133. doi: 10.1016/j.gde.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu Z., He Q., Liang J., Li W., Su Q., Chen Z. miR-31-5p is a potential circulating biomarker and therapeutic target for oral cancer. Mol Ther Nucleic Acids. 2019;16:471–480. doi: 10.1016/j.omtn.2019.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu W.C., Kao S.Y., Yang C.C., Tu H.F., Wu C.H., Chang K.W. EGF up-regulates miR-31 through the C/EBPβ signal cascade in oral carcinoma. PloS One. 2014;9(9) doi: 10.1371/journal.pone.0108049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng Q.S., Cheng Y.N., Zhang W.B., Fan H., Mao Q.H., Xu P. circRNA_0000140 suppresses oral squamous cell carcinoma growth and metastasis by targeting miR-31 to inhibit Hippo signaling pathway. Cell Death Dis. 2020;11:112. doi: 10.1038/s41419-020-2273-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung J.E., Lee J.Y., Kim I.R., Park S.M., Kang J.W., Kim Y.H. MicroRNA-31 regulates expression of wntless in both Drosophila melanogaster and human oral cancer cells. Int J Mol Sci. 2020;21(19):7232. doi: 10.3390/ijms21197232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Malkey M.K., Abbas A.A., Khalaf N.F., Mubrak I.A., Jasim I.A. Expression analysis of salivary microrna-31 in oral cancer patients. Int J Curr Microbiol App Sci. 2015;4(12):375–382. [Google Scholar]
- 24.Zhang T., Wang Q., Zhao D., Cui Y., Cao B., Guo L. The oncogenetic role of microRNA-31 as a potential biomarker in oesophageal squamous cell carcinoma. Clin Sci. 2011;121(10):437–447. doi: 10.1042/CS20110207. [DOI] [PubMed] [Google Scholar]
- 25.Hung P.S., Tu H.F., Kao S.Y., Yang C.C., Liu C.J., Huang T.Y. miR-31 is upregulated in oral premalignant epithelium and contributes to the immortalization of normal oral keratinocytes. Carcinogenesis. 2014;35(5):1162–1171. doi: 10.1093/carcin/bgu024. [DOI] [PubMed] [Google Scholar]
- 26.Hung K.F., Liu C.J., Chiu P.C., Lin J.S., Chang K.W., Shih W.Y. MicroRNA-31 upregulation predicts increased risk of progression of oral potentially malignant disorder. Oral Oncol. 2016;53:42–47. doi: 10.1016/j.oraloncology.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 27.Ouyang S.B., Wang J., Huang Z.K., Liao L. Expression of microRNA-31 and its clinicopathologic significance in oral squamous cell carcinoma. Zhonghua Kou Qiang Yi Xue Za Zhi. 2013;48(8):481–484. [PubMed] [Google Scholar]
- 28.Siow M.Y., Ng L.P., Vincet-Chong V.K., Jamaludin M., Abraham M.T., Abdul Rahman Z.A. Dysregulation of miR-31 and miR-375 expression is associated with clinical outcomes in oral carcinoma. Oral Dis. 2014;20(4):345–351. doi: 10.1111/odi.12118. [DOI] [PubMed] [Google Scholar]
- 29.Severino P., Oliveira L.S., Andreghetto F.M., Torres N., Curioni O., Cury P.M. Small RNAs in metastatic and non-metastatic oral squamous cell carcinoma. BMC Med Genom. 2015;8:31. doi: 10.1186/s12920-015-0102-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uma Maheswari T.N., Nivedhitha M.S., Ramani P. Expression profile of salivary micro RNA-21 and 31 in oral potentially malignant disorders. Braz Oral Res. 2020;34 doi: 10.1590/1807-3107bor-2020.vol34.0002. [Internet] [cited 2020 Nov 12] [DOI] [PubMed] [Google Scholar]
- 31.Bhat M.Y., Advani J., Rajagopalan P., Patel K., Nanjappa V., Solanki H.S. Cigarette smoke and chewing tobacco alter expression of different sets of miRNAs in oral keratinocytes. Sci Rep. 2018;8:7040. doi: 10.1038/s41598-018-25498-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saira Ahmed R., Malik S., Khan M.F., Khattak M.R. Epidemiological and clinical correlates of oral squamous cell carcinoma in patients from north-west Pakistan. J Pakistan Med Assoc. 2019;69(8):1074–1078. [PubMed] [Google Scholar]