Abstract
The highly infectious disease COVID-19 is induced by SARS-coronavirus 2 (SARS-CoV-2), which has spread rapidly around the globe and was announced as a pandemic by the World Health Organization (WHO) in March 2020. SARS-CoV-2 binds to the host cell’s angiotensin converting enzyme 2 (ACE2) receptor through the viral surface spike glycoprotein (S-protein). ACE2 is expressed in the oral mucosa and can therefore constitute an essential route for entry of SARS-CoV-2 into hosts through the tongue and lung epithelial cells. At present, no effective treatments for SARS-CoV-2 are yet in place. Blocking entry of the virus by inhibiting ACE2 is more advantageous than inhibiting the subsequent stages of the SARS-CoV-2 life cycle. Based on current published evidence, we have summarized the different in silico based studies and repurposing of anti-viral drugs to target ACE2, SARS-CoV-2 S-Protein: ACE2 and SARS-CoV-2 S-RBD: ACE2. This review will be useful to researchers looking to effectively recognize and deal with SARS-CoV-2, and in the development of repurposed ACE2 inhibitors against COVID-19.
Keywords: Coronavirus, SARS-CoV-2, Repurposed, Angiotensin Converting Enzyme-2 (ACE2), COVID-19
Introduction
Coronaviruses (CoVs) are single-stranded RNA (+ ssRNA) non-segmented positive-sense viruses with four groups: alpha and beta from rodents and bats; gamma and delta from aerial species [1]. Since the beginning of the twenty-first century, coronaviruses have triggered large outbreaks of lethal human pneumonia [2]. In 2003, Severe Acute Coronaviral Syndrome (SARS-CoV / SARS-CoV 1) broke out and spread to five continents, with a mortality rate of 10% [3, 4]. In 2012, the Middle East Coronavirus Respiratory Syndrome (MERS-CoV) erupted in Saudi Arabia with a mortality rate of 35% [5–7]. SARS-CoV and MERS-CoV are both bat/covetous and dromedary camel host’s zoonotic viruses [8, 9]. No effective therapeutic drug or vaccine has been approved to date for the treatment of human coronavirus. CoVs are thus viewed as viruses, the spread of which poses a major threat to humans. In December 2019, Chinese researchers detected a new virus in China called Wuhan coronavirus or 2019 novel coronavirus (2019-nCov) [10–13]. The virus shared a SARS-CoV-1 genetic homology of 79.5% and thus the International Committee on Taxonomy of Viruses (ICTV) classified the virus as SARS-CoV-2; this virus has been responsible for the 2019 Coronavirus Disease (COVID-19) pandemic [14, 15]. The genome of virus has been fully sequenced and appears to be most similar to the strain in bats, indicating that it may have evolved from bats [16]. The genome of SARS-CoV-2 (30 kb in size) covers a large non-structural polyprotein, an open read frame (ORF) of 1a/b that is further cleaved into 15/16 proteins, five accessory proteins (ORF3a, ORF6 and ORF7), and four structural proteins (S, M, E, and N) which are required for assembly and infection [17].
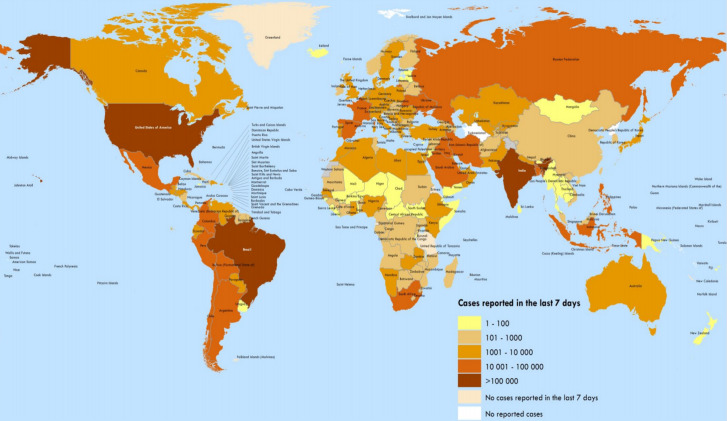
SARS-CoV-2 uses the same receptor as SARS-CoV for entry into human cells: the angiotensin converting enzyme 2 (ACE2) receptor [18]. A person with COVID-19 has typical signs and symptoms such as nasal congestion (4.8% per person), nausea or vomiting (5.0%), chills (11.4%), headache (13.6%), sore throat (13.9%), myalgia or artralgia (14.8%), respiratory shortness (18.6%), production of sputum (33%), fatigue (38.1%), dry cough (67.7%) and fever (87.9%) [19]. SARS-COV-2 is the third extremely virulent human coronavirus of the twenty-first century, preceded by SARS-COV and MERS-COV [20]. There is evidence that SAR SCoV-2 is more highly contagious/transmissible than SARS-CoV [21]. Although the COVID-19 epicenter was located in Wuhan, China, in December 2019, the disease has spread to over 100 countries (Fig. 1). A recent World Health Organization (WHO) report (10–16 August 2020) indicates that it has severely affected the United States (US), Brazil and India, with more than 100,000 confirmed cases. As per the COVID-19 situation report-109 of WHO (30 December 2019–23 September 2020) 31,425,029 confirmed cases and 967,164 deaths were reported worldwide, mostly affecting the US, Europe and South East Asia (Fig. 2). Millions of people have also been affected by obligatory isolation/quarantine [22]. There are currently no successful therapies that target SARS-CoV-2 and it takes months and years for these drugs to develop. Researchers are working to identify promising treatments to save lives and develop potential preventive vaccines [22]. Targeting virus entry by inhibiting ACE2 has a greater advantage than inhibiting subsequent steps of the viral life cycle [23]. Based on current published data, we have summarized the different in silico based studies and repurposing of anti-viral drugs targeting ACE2, SARS-CoV-2 S-Protein-ACE2 and SARS-CoV-2 S-RBD: ACE2 interface. Such efforts may lead to the development of efficient therapeutic and preventive agents for COVID-19.
Fig. 1.
Worldwide confirmed 2019 Coronavirus Disease (COVID-19) cases reported from 10 August 2020 to 16 August 2020 as per the COVID-19 situation report-209 of the World Health Organization (WHO).
Reproduced with permission from WHO [24]
Fig. 2.
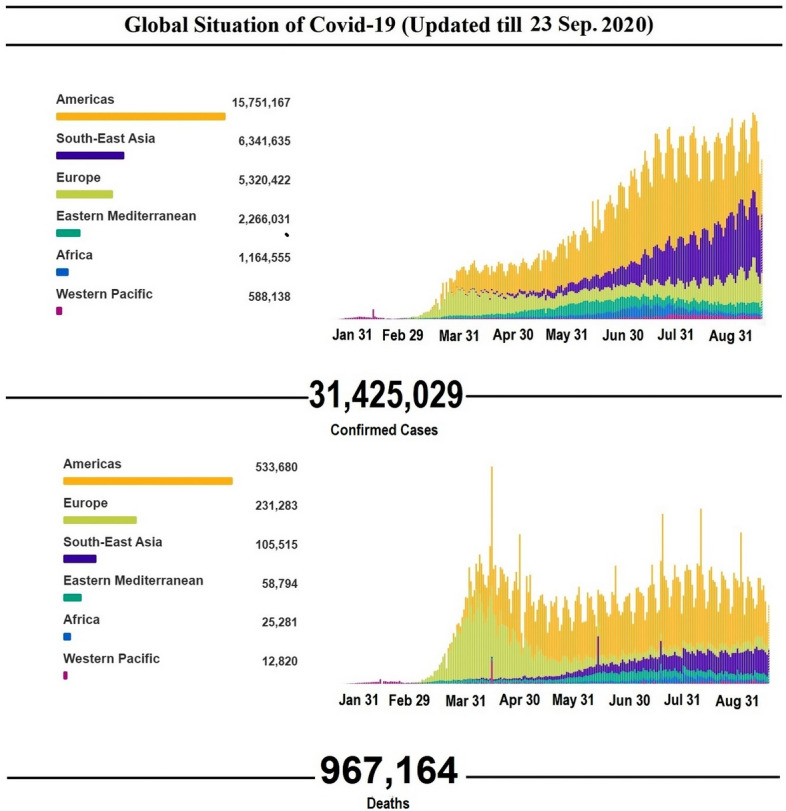
Number of confirmed COVID-19 cases and deaths from 30 December 2019 to 23 September 2020 as per the WHO COVID-19 dashboard.
Reproduced with permission from WHO [24]
ACE2 and SARS CoV-2 Interaction
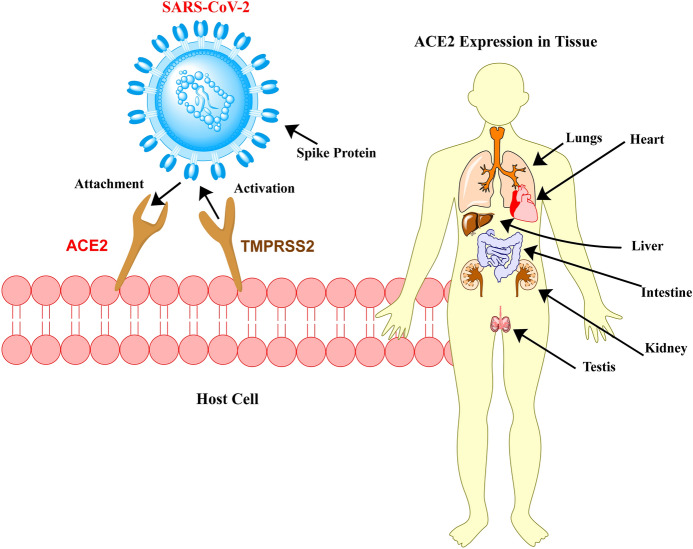
Current studies have shown that SARS-CoV-2 resembles other beta-coronaviruses, being composed of a 5′ untranslated region (UTR), a spike protein (S) gene, a replicase complex (ORF1ab) encoding non-structural proteins (nsps), envelope protein (E) gene, a nucleocapsid protein (N) gene and a membrane protein (M) gene [23]. ACE2 is a type I transmembrane-metallocarboxypeptidase enzyme with ACE homology. This is a key player enzyme in the renin-angiotensin system (RAS) and a target for the treatment of hypertension [25]. ACE2 is a functional receptor that acts as a point of entry for SARS-CoV-2 into human lung cells (binds through its spike (S) protein) [26–29]. S-protein consists of two subunits, S1 and S2; S1 as the receptor-binding domain (RBD), while the S2 subunit is responsible for viral membrane fusion [30, 31]. The S2 subunit of SARS-CoV-2 is highly conserved with a similarity of ~ 99%, while the S1 subunit is 70% identical to other SARS-CoV in bats and human SARS-CoV. The core RBD domain is extremely conserved amongst them [32]. A study found that ACE2 is expressed in the oral mucosa and enriched in the lungs, intestine and tongue epithelial cells, and thus constitutes an essential route for SARS-CoV-2 to reach hosts [33].
The SARS-CoV-2 spike S1 protein binds to the ACE-2 receptor. This complex is proteolytically handled via type-2 transmembrane cellular serine protease (TMPRSS2) enzyme, leading to cleavage of ACE2 and activation of the spike protein, thus allowing viral admittance into the target cell (Fig. 3). Cells containing ACE2 and TMPRSS2 are more prone to SARS-CoV-2 entry [34–38]. Binding to ACE2 allows the virus to penetrate cells inside the epithelial oropharyngeal. Infection normally begins with respiratory mucosal cells and then spreads to alveolar epithelial cells in the lungs, leading to acute respiratory distress syndrome (ARDS) in patients [36, 39–43].
Fig. 3.
Binding mechanism of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) to angiotensin converting enzyme 2 (ACE2) receptors on the surface of the target cell
Binding Site Analysis of ACE2 and SARS-CoV-2 S-RBD: ACE2
The crystal structure of the SARS-CoV-2 S protein in complex with human ACE2 (SARS-CoV-2 S-RBD-ACE2) offers insights into the mechanisms of binding of the virus and its structural differences from SARS [39]. SARS-CoV and SARS-CoV-2 receptor binding regions were structurally examined by Veeramachanenia’s group [39] by comparing the protein structures of SARS-CoV and SARS-CoV-2 from the protein database (PDB Id: 3SCI and 6M0J, respectively). In both sequences, the F-chain relates to the virus S protein RBD, and the A-chain depicts the ACE2 receptor [36, 39]. Many structural changes have been identified in the spike protein SARS-CoV-2 RBD due to mutations over time compared with SARS-CoV. The three-dimensional (3D) structure of the spike protein RBD is made up of most of the loop structure with two helices and five sheets in SARS-CoV [39, 40]. The key structural changes were noted in residues 306–382 of SARS-CoV in the loop area, while the corresponding residues of SARS-CoV-2, i.e., 333–395, were incorporated into three sheets and two helices. In the corresponding region of SARS-CoV-2 (residues 439–442), the helix found in the SARS-CoV protein (residues 426–429) was absent and these residues were located in the loop area. Additional sheets of SARS-CoV-2 proteins between Tyr 473 and Gln 474, Cys 488 and Tyr 489 and Val 524 and Cys 525 were not present in SARS-CoV and these residues were formed into loops [39, 41]. It has been observed that, in the SARS-CoV-2 spike protein RBD, there were more ACE2 interacting residues compared with SARS-CoV due to substantial mutations present in the SARS-CoV-2-sequence (specifically in the RBD domain) [36, 40, 41]. This binding site analysis will help in the design of the better ACE2 inhibitors for SARS-CoV-2 (Fig. 4).
Fig. 4.
Ribbon structure of ACE2 enzyme (retrieved from wikimedia.org under the Creative Commons CC BY-SA 3.0) and SARS-CoV-2 S-RBD: ACE2 complex (retrieved from wikimedia.org under the “Creative Commons CC0 1.0 Universal Public Domain”) [44, 45]
ACE2 Binding Site Analysis (Active Residue to be Targeted)
Wan et al. [42] stated that Lys31 and Lys353 on ACE2 are known to be binding hotspot residues that are more sensitive to S-protein binding. In human receptor ACE2, hotspot 31 (Lys31) consists of the salt bridge between Lys31 and Glu35, and hotspot 353 consists of another salt bridge between Lys353 and Asp38, covered by a hydrophobic residue.
SARS-CoV-2 S-RBD: ACE2 Binding Site Analysis (Active Residue to be Targeted)
The RBD of the S1 subunit of the spike protein plays an indispensable role in the binding of SARS-CoV-2 to the ACE2. In vitro binding studies suggest good binding affinity at low nanomolar range between ACE2 and the RBD (residues 331–524 of spike protein) of the SARS-CoV-2 spike protein [43].
The recently released crystal structure PDB Id: 6M0J, deposited by Lan and co-workers [46], revealed the structural interactions of SARS-CoV-2 spike protein RBD with ACE2. They observed that the interfaces of RBD/ACE2 have hydrophilic networks. At the SARS-CoV-2 RBD: ACE2 interface, there are 13 hydrogen bonds and two salt bridges as shown in Fig. 5 [46]. The second important feature is that the various tyrosine residues form hydrogen bonding interactions with hydroxyl groups [46]. The key tyrosine residues from the SARS-CoV-2 RBD are Tyr449, Tyr489, and Tyr505. The third key feature is the involvement of the Asn90-associated glycans of ACE2 in binding different RBDs [46–48].
Fig. 5.
The SARS-CoV-2 RBD: ACE2 interaction
Apart from the above mentioned studies, other in silico studies have also been reported in the literature, where authors discussed the crucial residues for targeting SARS-CoV-2 RBD. Jani et al. [43] reported that, within the RBD domain, the receptor binding motif (RBM) (residue range 438–506) interacts directly with ACE2. The interacting residues on the surface of the RBD domain of the spike protein are Tyr505, Gly502, Asn501, Thr500, Gln498, Gln493, Tyr489, Asn487, Phe486, Ala475, Phe456, Leu455, Tyr453, Tyr449, Gly446 and Lys417 [36, 49]. The interacting residues belonging to the ACE2 receptor are Arg393, Arg357, Asp355, Gly354, Lys353, Asn330, Tyr83, Met82, Leu79, Gln42, Tyr41, Asp38, Glu37, Lys35, His34, Lys31, Asp30, Phe28, Thr27, and Gln24. These residues form various interactions between the ACE2 receptor and the RBD of the spike that include 13 hydrogen bonds and 2 salt bridges between the ACE2 receptor and the RBD-spike interface residues [43].
Wan et al. [42] stated that the RBD of SARS-CoV-2 recognizes the Gln493 and Leu455 residues of human ACE2, which are suggested to have favorable molecular associations with the 31-hotspot residue, thus enhancing human ACE2 viral binding—approximately 10 times higher than the RBD of SARS-CoV. Han et al. [50] reported the ACE2 amino acid sequence that was crucial for drug development (Glu57, Glu56, Asp38, Glu37, Glu35, His34, Lys31, Asp30, Lys26, Glu22 and Glu23). Li et al. [51] identified essential amino acids between the RBD of the S-Protein of SARS-CoV-2 with ACE2 (Gly354, Lys353, Asn330, Glu329, Gln325, Asn90, Tyr83, Met82, Leu79, Leu45, Gln42, Tyr41, Asp38, Glu37, His34, Lys31, Thr27, Gln24 in ACE2). Yan et al. [52] published the crystallographic structure of the SARS-CoV-2-RBD-ACE2 interaction. They reported Tyr505, Ser494, Phe497, Gly496, Tyr495, Tyr453, Lys403, Arg393, Phe390, Pro389, Gln388, Ala387, Lys353, Asp38, Glu37, His34, and Asn33 as the vital residues involved in binding of viral S-protein with the human ACE2 receptor.
ACE2 and Target Organs at Risk
It has been observed that cells in which ACE2 and TMPRSS2 are present are more prone to SARS-CoV-2 entry (Fig. 6). These organs are thus at higher risk of COVID-19.
Fig. 6.
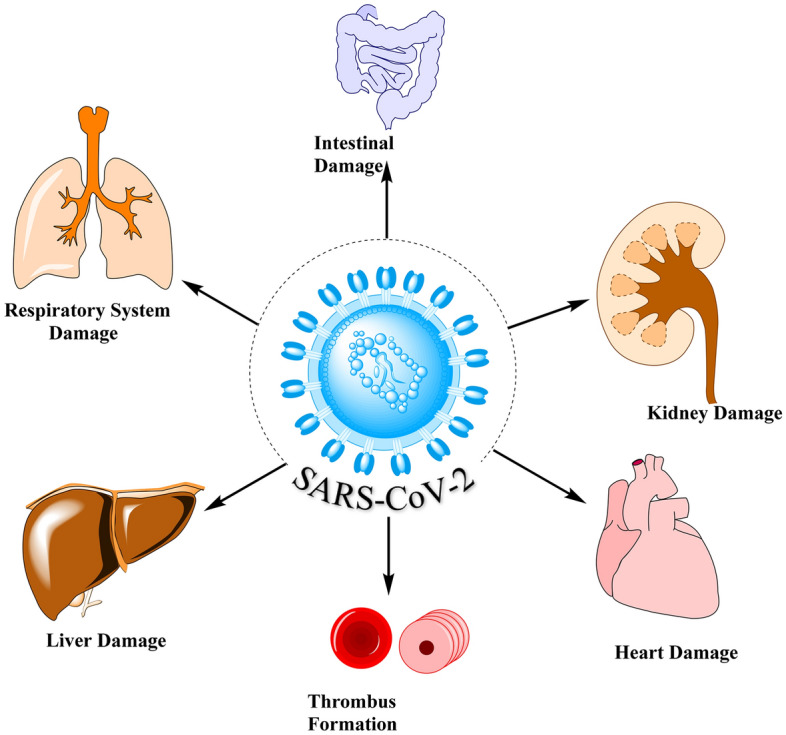
SARS CoV-2 and organs at high risk
Damage Risk to the Respiratory System
The rapid occurrence of respiratory symptoms is common among patients as the lung is the main target organ for infection with COVID-19. ACE2 is not only a COVID-19 gateway in the lung but also likely to be involved in lung injury development [53]. Research by Kubal et al. and Imai et al. revealed that blocking the signal pathway for renin-angiotensin can reduce extreme acute SARS-CoV-2 spike protein injury. This suggests that RAS, consisting of the ACE, angiotensin II and 1a angiotensin II receptors (AT1a), facilitates disease pathogenesis, causing lung edema and adverse pulmonary function [54, 55]. As the RAS counter regulator, ACE2 thus plays a role in improving extreme pulmonary edema and acute lung failure. Finding the downstream mechanisms of ACE2 responsible for the inflammatory storm would resolve the problem [53–55].
Damage Risk to the Heart
ACE2 is significantly found in the heart and its receptors allow the virus to penetrate the heart [53]. Oudience et al. [56] reported that SARS-CoV-2-infected mice had myocardial infection, and expression of ACE2 has notably decreased, suggesting that ACE2 has a major role to play in mediating SARS-CoV-2 cardiac infection. SARS CoV-2 is also linked with a substantial decrease in expression of ACE2 protein in the heart. This would lead to an increase in Ang II levels. Increased Ang II levels are further responsible for cardiovascular disease pathophysiology through regulation of growth cardiomyocytes. Intercellular and intracellular signaling pathways are impaired, intercellular communication is decreased, body immunity is hindered, lipid peroxidation is triggered and insulin resistance occurs.
Hence, SARS-CoV-2 is also implicated in myocardial inflammation and in down-regulation-related damage to the myocardial ACE2 system. Antagonizing the SARS-CoV-2 on ACE2 in myocardial cells is the key to improving cardiac function. If the number of cases of COVID-19 pneumonia increases, the number of patients suffering from heart failure cannot be overlooked, and it needs to be identified and treated in time. Changes in pathways linked to ACE2 and myocardial histopathology will help our understanding of the basic mechanism of myocardial damage due to COVID-19 and lead to appropriate clinical treatment [53].
Damage Risk to the Intestine
Other common symptoms of COVID-19 infection other than fever and cough include gastrointestinal tract (GIT)-related symptoms, like nausea, diarrhoea and vomiting. These gastrointestinal (GI) symptoms are more serious in COVID-19 infections than in SARS-CoV and MERS-CoV. In addition to recurrent fever and dry cough, a number of scholars in the US reported diarrhea and abdominal pain during their hospitalizations. It should be noted that COVID-19 has also been found in stool samples of diarrhea [53, 57]. Zhang et al. [58] reported that SARS-CoV-2 genetic analysis showed high levels of COVID-19 receptor ACE2 expression in esophageal stratified epithelial cells and indicated a possible transmission route via the GI system. GIT symptoms of COVID-19 infection can therefore be associated with an invasion of intestinal epithelial cells that express ACE2. The intestine is a potential target organ for COVID-19 infection, but much more research is needed to determine whether the digestive system is a transmission path [58].
Abnormal Coagulation Risk
Studies have shown that certain patients with a new form of coronavirus have impaired coagulation activity, and coagulation dysfunction exists in almost all critically ill patients [53, 59, 60]. There may be several factors that endorse thrombosis in patients with novel coronavirus. The acute inflammatory reaction caused by serious infection or sepsis may have several effects on coagulation and the fibrinolytic system. Furthermore, there is some connection between ACE2 and coagulation. Fraga-Silva et al. [61] assessed ACE2 and ACE thrombus activity by inducing thrombosis in the vena cava of spontaneously hypertensive (SHR) and Wistar Kyoto (WKY) rats. They observed that thrombus weight increased by 30% by ACE2 inhibition by giving DX600 0.1 mmol L−1 kg−1 and reduced thrombus formation in SHR by 30% by giving 10 mg kg−1 of XNT (ACE2 activator). XNT also decreased platelet adherence to damaged blood vessels, decreased thrombus size and extended the time to complete blood vessel occlusion in mice [53]. Consequently, a reduction in thrombotic ACE2 activity is linked with a rise in SHR thrombosis. Furthermore, the activation of ACE2 will minimize thrombus formation and platelet adhesion to blood vessels [53].
Kidney Damage Risk
Renal tubular cells are involved in reabsorption and excretion, and play a vital role in metabolite excretion, maintenance of body fluid balance, and acid–base balance [53]. COVID-19 can reach tubular renal cells by binding to ACE2, which can cause cytotoxicity and impaired renal function. Neo-coronavirus-infected patients should undergo renal function tests and follow up to diagnose renal dysfunction in a timely fashion to deal with it as early as possible [53]. A study found that the expression of ACE2 is almost highest in the human testis [61].
COVID-19 is therefore likely to affect and damage patient’s testicular tissue by binding to ACE2-positive cells. A research study analyzed data from 59 patients in several hospitals, including 28 serious cases, to examine the condition of their renal function from 21 January to 7 February 2020. Among these 66 patients, 63% (32/51) had proteinuria, indicating renal insufficiency. Plasma urea nitrogen and creatinine were raised in 27% and 19% of patients, respectively. A computed tomography (CT) scan report showed abnormal kidney damage among all the infected patients [53, 62].
Liver Damage Risk
A retrospective analysis of 99 Wuhan COVID-19 patients showed that 43 had varying degrees of irregular liver biochemical tests, of which 1 found substantial increases in serum aminotransferase [63]. There was a rise in serum lactate dehydrogenase in 75 patients and a rise in serum creatine kinase in 13 patients. The paper did not discuss whether such enzymatic changes were attributable to either COVID-19 infection or liver damage triggered by the drugs used, nor did it identify any underlying liver condition of the patient. Researchers analyzed cell types expressing ACE2 in healthy hepatic tissue and identified higher ACE2 levels in bile duct cells compared with hepatocytes [64]. These findings suggest that either virus binding to the ACE2-positive bile duct cells causes bile duct dysfunction or toxic side effects caused by therapeutic drugs could be responsible for the observed liver damage [53, 64].
Repurposed Inhibitors
Researchers have so far targeted three crucial enzymes of the ACE2 family, i.e., ACE2, SARS-CoV-2 S-Protein-ACE2 and SARS-CoV-2 S-RBD: ACE2 as understanding about the disease has progressed since December 2019.
Repurposed ACE2 Inhibitors
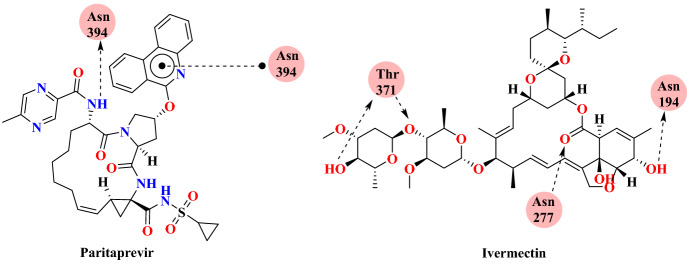
Oliveira and colleagues [65] performed molecular docking with the ACE2 receptor in complex with S-glycoprotein SARS-CoV-2 (PDB ID: 6M0J) using 60 drugs, which were published in the literature against SARS-CoV-2. The docking result showed that paritaprevir and ivermectin have highest binding affinity to the ACE2 receptor with interaction energies of −11.8 kcal mol−1 and −11.6 kcal mol−1, respectively (Fig. 7). The strongest binding affinity with the ACE2 receptor was demonstrated by paritaprevir, principally due to two factors: the hydrogen bonding interaction with residue Asn394 and the hydrophobic interaction of aminophenanthrene with Trp349 residue, as shown in Fig. 8. Paritaprevir and ivermectin are used for the treatment of hepatitis C virus and parasitic infections, respectively. In silico results indicated that both candidates could be used for the treatment of COVID-19. Remdesivir and azithromycin are in the second group of promising drugs. Compared with other medications, the repurposed medications hydroxychloroquine and chloroquine were not effective as monotherapies against SARS-CoV-2 infection.
Fig. 7.
Chemical structures of Paritaprevir and Ivermectin
Fig. 8.
Binding interaction of Paritaprevir and Ivermectin interaction with the ACE2 receptor
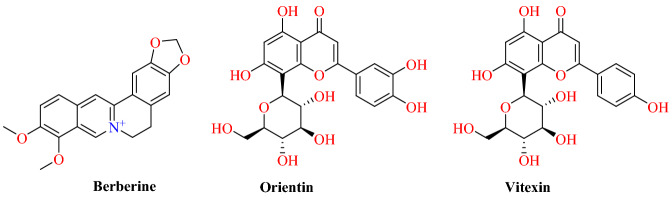
Lakshmi and colleagues [66] documented the interaction of bioactive compounds from 10 different Indian medicinal plants employed widely to treat cold and respiratory disorders through molecular docking research. Totally, 47 bioactive compounds were identified and investigated against the structural target of the human ACE2 receptor (PDB ID: 1R42) from medicinal plants (Fig. 9).
Fig. 9.
Chemical structures of Berberine, Orientin, and Vitexin
Top leads were selected based on the number of hydrogen bonds, interaction energies and other parameters that describe their potency in inhibiting ACE2. The docking results showed significant interactions between 2-monolinolenin, berberine, orientine and vitexin and the human ACE2 receptor (Fig. 10). Among them, 2-monolinolenin was approved by the US Food and Drug Administration (FDA) and can therefore be used for preclinical studies to treat COVID-19 infection. Another best compound is Berberine, which has a docking score of −95.33 and forms 2 hydrogen bond interactions with residues Lys26 and Thr27. Orientin is a C-glycosyl compound that forms 5 hydrogen bond interactions with 3 residues: Glu22, Lys26 and Asn90. Vitexin, which is an apigenin flavone glycoside, has a docking score of −85.24; the strong affinity can be explained due to the two hydrogen bonds with Glu22 and Asn90 (Fig. 10).
Fig. 10.
Binding interaction of Berberine, Orientin and Vitexin with ACE2
Abdelli and coworkers [67] reported virtual screening of drugs capable of blocking the action of ACE2 using natural compounds (γ-terpinene, p-cymene, limonene, thymol and isothymol) obtained from the anti-viral plant Ammoides verticillata (Desf.) Briq. This aromatic plant is commonly used in western Algeria for dietary and therapeutic purposes. They assessed the active principles of A. verticillata (γ-terpinene, p-cymene, limonene, thymol and isothymol) against the enzyme ACE2 and conducted a molecular docking analysis using regular parameter settings in the MOE-2013 software package against the ACE2 enzyme containing β-d-mannose co-crystallized ligand (PDB ID: 6vw1). The docking result analysis showed that docked isothymol has the best docking score compared with the co-crystallized ligand β-d-mannose.
They also evaluated the drug likeness, PASS prediction, ADME/T, the P450 site of metabolism, pharmacophore mapping and molecular dynamics (MD) simulation of the best ligand. The result indicated that the top ligand isothymol (51.2%) in Ammoides verticillata has a significant binding score and a stable interaction with the target ACE2. Isothymol will block the ACE2 receptor, making it harder for coronavirus to enter cells (Fig. 11).
Fig. 11.
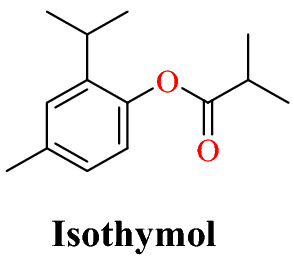
Interaction of isothymol with ACE2
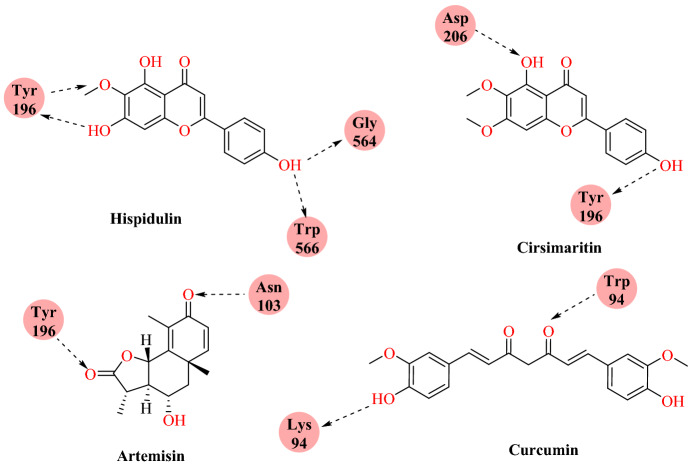
Omar and his group studied the in silico interaction between the active site of ACE2 with natural compounds (eugenol, hispidulin, quercetin, curcuma, hydroxy-chloroquine, artemisin, thymoquinone and cirsimaritin) [68].
Molecular docking was performed using Auto Dock Vina with ACE2 protein (PDB ID: 1r42). In silico results revealed that hispidulin (−7.8 kcal mol−1), cirsimaritin (−7.6 kcal mol−1), artemisin (−7.2 kcal mol−1) and curcuma (−7.2 kcal mol−1) displayed the best potential inhibitor activity against ACE2 (Fig. 12). Hispidulin displayed the highest affinity at the active site of ACE2; it formed hydrogen bonds with Tyr196, Gly564 and Trp566 residues (Fig. 13). Artemisin, hispidulin, and cirsimaritin are flavonoids isolated from Artemesia herba Alba, which displays wide range of biological activities, including nephroprotective, antihyperlipidemic, antidiabetic, cardioprotective, anticancer, antioxidant, antiprotozoal, gastroprotective and antibacterial effects.
Fig. 12.
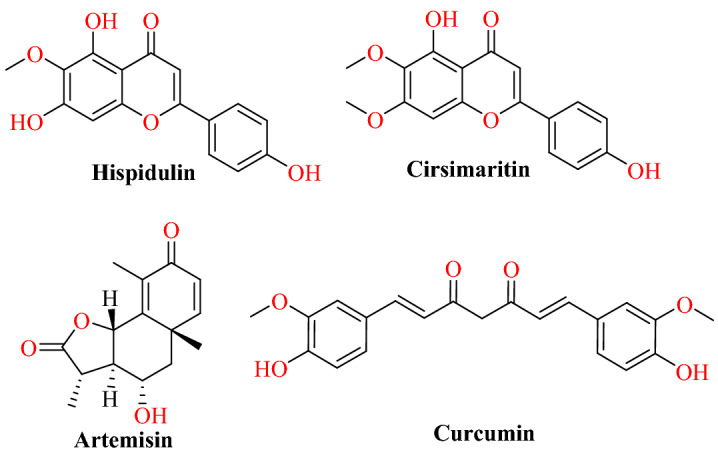
Chemical structures of hispidulin, artemisin cirsimaritin, and curcumin
Fig. 13.
Binding interaction of hispidulin, cirsimaritin, artemisin and curcumin with ACE2
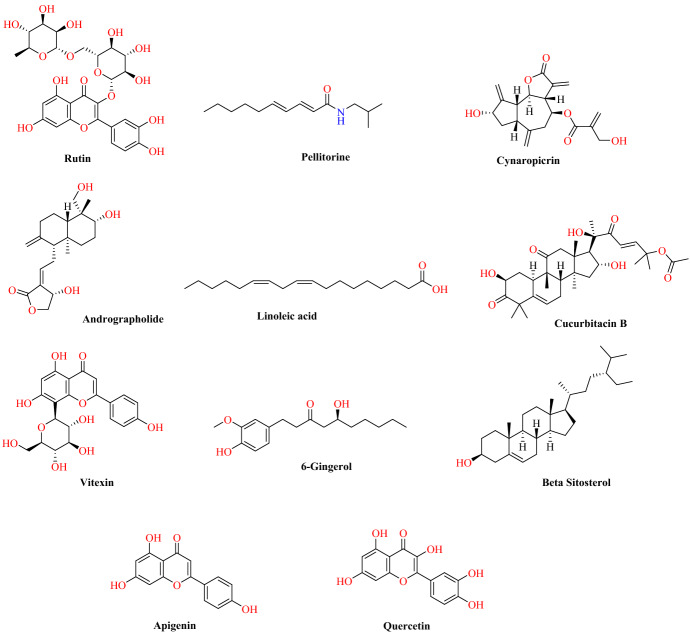
Dhanasekaran and colleagues examined ACE2 lead molecules from well-known herbs (Nilavembu Kudineer and Kaba Sura Kudineer) [69]. These compounds underwent a molecular docking study targeting the ACE2 receptor and their ability to inhibit the host–viral interface was assessed. These two traditional Siddha medicinal products (Nilavembu Kudineer and Kaba Sura Kudineer), recommended by Indian medicine practitioners for the treatment of viral infections, are officially suggested by government authorities for the following reasons. The rationale behind these Indian medicines is that they are believed to be useful in the treatment of epidemic viral infections. A total of 28 bioactive leads (vitexin, thymol, eugenol, cynaropicrin, vetiverol, vasicine, spathulenol, santalic acid, rutin, pellitorine, linoleic acid, gallic acid, cucurbitacin-B, costunolide, cissamine, carvacrol, bharangin, andrographolide, alpha-bisabolol, beta-pinene, chlorogenic acid, quercetin, piperine, apigenin, piperidine, beta-sitosterol, 6-gingerol and 6-shogaol), which were present in both traditional medicines, were obtained for in silico investigation (Fig. 14). A molecular modelling study was performed using Auto Dock against the ACE2 with PDB ID of 2JAF. Lys 31 and Lys 353 are the two hot-spot residues (active binding sites) associated with the ACE2 receptor [70–72]. Amino acid sequence (Leu455, Phe486, Ser494), which is located on the primary binding SARS-CoV-2 motif, mediates the virus’ binding paradigm with hotspot residues Lys 31 and Lys 353 of the ACE2.
Fig. 14.
Chemical structures of rutin, andrographolide, vitexin, pellitorine, apigenin, linoleic acid, 6-gingerol, quercetin, cynaropicrin, beta sitosterol, cucurbitacin B
Hierarchically, the active site residue Lys 31 bridged between the amino acids Lys 31 and Glu 35 and residue Lys 353 was bridging between Asp 38 and Lys 353. Results of computational analysis indicated that, among the 28 ligands, 11 bioactive lead molecules, including cucurbitacin-B, rutin, andrographolide, linoleic acid, pellitorin, cynaropicrin, vitexin, 6-gingerol, apigenin, beta-sitosterol, and quercetin, exhibited potential binding affinity to the target amino acid residues (Lys 31 and Lys 353) (Fig. 15).
Fig. 15.
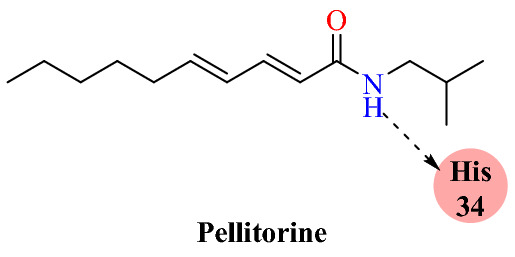
Binding interaction of pellitorine with ACE2
Wang and colleagues have documented ACE2 modulation with Cannabis sativa extracts, which could be a reliable strategy for reducing disease susceptibility [73]. C. sativa is the cannabinoid cannabidiol (CBD), which has been suggested to modulate gene expression and inflammation and possess anti-inflammatory and anti-cancer properties. In order to analyze the effects of C. sativa on ACE2, extracts of 22 novel C. sativa lines have been used to treat artificial human 3D tissue models of oral epithelium, intestinal tissues and airways. Using the pro-inflammatory cytokines tumor necrosis factor alpha (TNF-α) and interferon gamma (IFN-γ) (TNF-IFN), inflammation is induced in these 3D tissue models. Results indicated that 13 high extracts of CBD C. sativa could modulate gene expression of ACE2 and the levels of ACE2 proteins. It was also noted that some C. sativa extracts down-regulated gene expression of TMPRSS2, which is another essential protein for SARS-CoV2 entry into host cells, in epi-oral and epi-intestinal tissues.
Cheng and colleagues reported flavonoids from Citrus fruits as a promising candidate in the prevention and treatment of SARS-CoV-2 [74]. They determined six flavonoid compounds (naringenin, naringin, hesperetin, hesperidin, neohesperidin and nobiletin) from three citrus species by using the LC–MS technique. These flavonoid compounds were collected from three major species of mandarin (Citrus reticulata), pummelo (Citrus maxima) and sweet orange (Citrus sinensis). In the most severe COVID-19 patients, cytokine storm was observed along with increased plasma concentrations of TNF-α, interleukin (IL)-1β, IL-10 and IFNγ.
Corticosteroids are used frequently for severe cases of treatments to reduce inflammatory-induced lung injury. Therefore, treatment with an anti-inflammatory approach is critical to alleviating clinical symptoms related to COVID-19. These cytokine storms can be induced by the bacterial gram-negative endotoxin lipopolysaccharide, which increases cytokines such as IL-1β, TNF-α, IFNγ, IL-6 and MCP-1. They determined the anti-inflammatory effect of citrus Naringin (extracted from Citrus wilsonii Tanaka) on inflammatory cytokines, COX-2, iNOS, IL-1β and IL-6 at mRNA levels in LPS-induced RAW macrophages. The results revealed that the iNOS, IL-1β, IL-6 mRNA and COX-2 expression levels in LPS-treated macrophage cells were increased compared to the control group. Application of naringin (10, 20, 40 μg ml−1) significantly diminished the effects of lipopolysaccharide induced iNOS, IL-1β, IL-6 mRNA and COX-2 expression.
It was further demonstrated that naringin could restrain cytokine storm to a certain extent through inhibiting high mobility group box 1 (HMGB1) expression, which acts as a pro-inflammatory cytokine and regulates the cytokine storm. They also found that naringin pretreatment could attenuate levels of p-α38 MAPK, which plays an important role in HMGB1-mediated production of proinflammatory cytokines. Their docking study with ACE enzyme (PDB code: 6ACG) showed that naringin had significant binding activity to the ACE2 enzyme with a docking score of −6.85 kcal mol−1, with potential binding sites at Glu-398, Tyr-515, Glu-402 and Asn394 (Fig. 16). The results suggested that naringin could have the potential to prevent COVID-19 cytokine storms. The findings indicated that naringin may have the ability to prevent cytokine storms in COVID-19.
Fig. 16.
Binding interaction of Naringin with ACE2 enzyme (PDB code: 6ACG)
In a docking study, Yong-Ming’s group investigated 38 Chinese patent drugs (CPDs) that are used commonly in the respiratory diseases for SARS-CoV-2 [75]. Their ligand data set contains total of 38 marketed CPDs covering 93 herbs used for the treatment of respiratory diseases. These CPDs were selected and docked into the ACE2 receptor. According to the docking result analysis, the top 19 compounds (hesperidin, saikosaponin A, mulberroside A, rutin, bilirubin, verbascoside, vincetoxicoside B, baicalin, prim-O-glucosylcimifugin, corosolic acid, cynaroside, orientin, corynoline, astragaloside A, protostemonine, ilexgenin A, amygdalin, paeoniflorin, and ursolic acid) bind directly to the ACE2 receptor with high affinity. Among them, hesperidin has highest docking score (−11.4 kcal mol−1) and showed key binding interaction with Cys344, His345, Asp368, Arg514, Tyr515 and Arg518 residue of ACE2. Another best hit is saikosaponin A, which has a docking score of −11 kcal mol−1 and binding interaction with Ala348, Glu402, Arg514, Tyr515 and Arg518 (Fig. 17).
Fig. 17.
Chemical structure of hespiridine and saikosapnin A
Repurposed SARS-CoV-2 S-Protein-ACE2 Inhibitors
Smith et al. [76] reported combined restricted “temperature replica-exchange molecular dynamics (T-REMD) simulations with high-throughput virtual screening in an ensemble docking protocol” using the world’s most efficient supercomputer, SUMMIT, to find well-characterized drugs that bind either the S-protein:ACE2 receptor interface or the S-protein receptor recognition region.
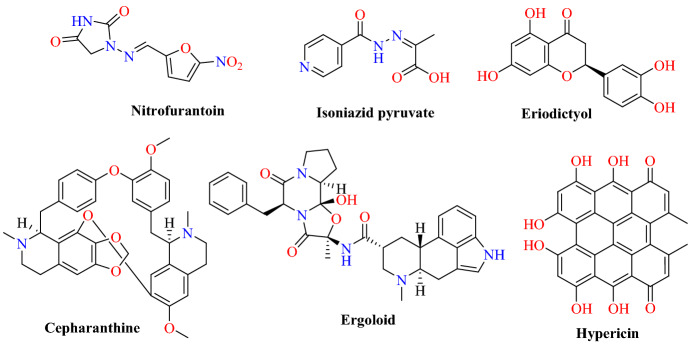
These small molecules can interrupt the host recognition and infection pathway of SARS-CoV-2 by binding the host viral interface and/or the isolated host recognition region of S-proteins. They ranked more than 8000 drugs, natural products, and metabolites against the SARS-CoV-2 S-protein and the S-protein:ACE2 receptor by implementing the molecular docking and molecular simulation approaches. They filtered the top 6 molecules, among which nitrofurantoin, isoniazid pyruvate, and eriodictyol had higher affinity for the ACE2 receptor portion of the S-protein:ACE2 receptor interface (Figs. 18 and 19). On the other hand, cepharanthine, ergoloid, and hypericin were identified as compounds of interest against the isolated S-protein’s receptor recognition region. The outcome of their study indicated that these six compounds would be suitable candidates for experimental investigations into COVID-19.
Fig. 18.
Chemical structures of nitrofurantoin, cepharanthine, isoniazid pyruvate, ergoloid, eriodictyol and hypericin
Fig. 19.
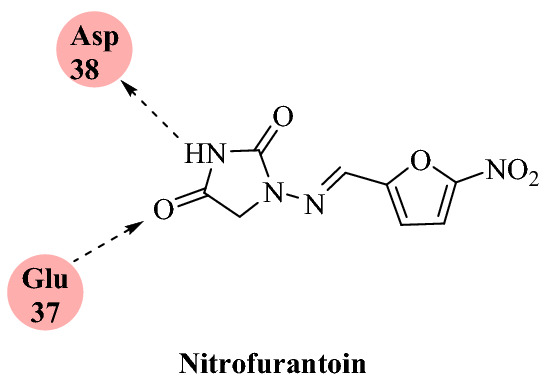
Interaction of nitrofurantoin with SARS-CoV-2 S protein-ACE2
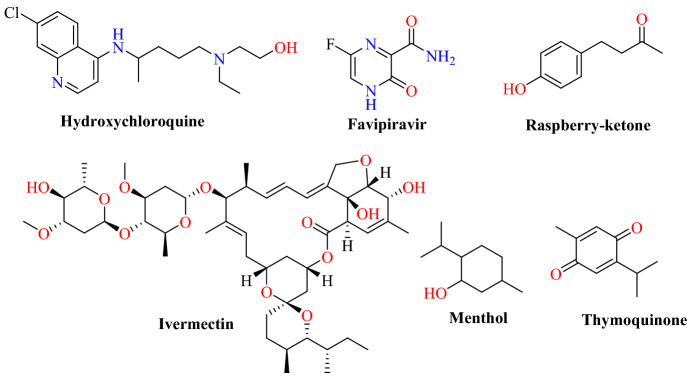
Abdel-Mottaleb et al. [77] exploited the electronic characteristics of nutraceuticals and antiviral drugs for the treatment of COVID-19. The electron distribution surface of the molecules and the frontier orbital energies [highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO)] of the interacting species were defined by electrostatic potential energy maps (ESP-maps), and manual docking was carried out. Ligands containing seven natural antivirals (raspberry ketone, quercetin, menthol, curcumin, eriodictyol, thymoquinone and resveratrol) and four synthesized antivirals (ivermectin, hydroxychloroquine, nicolosamide, and favipiravir) were selected and their bindings were determined for unique peptide sequences at the ACE2 interface. Results of this study showed that hydroxychloroquine, ivermectin and favipiravir have significant drug binding with the both S protein and ACE2, while raspberry ketones, menthol, eriodictyol, and thymoquinone have affinity for the viral S protein (Figs. 20 and 21). Raspberry ketone has equivalent binding strength to the known antiviral agents that are currently being clinically tested. The natural drugs had no affinity with S protein or ACE2. However, they interfere with the direct S-protein–ACE2 interaction by displaying the ability to receive electrons (electron sink) from S-protein in the presence of ACE2 into its disturbed LUMO.
Fig. 20.
Chemical structures of hydroxychloroquine, favipiravir, raspberry-ketone, ivermectin, menthol and thymoquinone
Fig. 21.
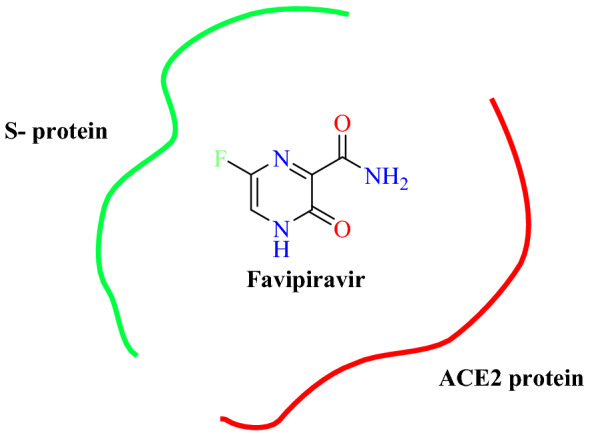
Favipiravir prevents the interaction of the S protein with the ACE2 enzyme and functions as the electron sink of the S protein nucleophile
Repurposed SARS-CoV-2 RBD: ACE2 Inhibitors
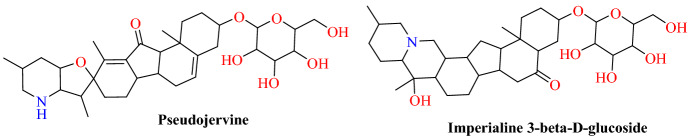
Cheng and colleagues [78] identified active chemical constituents from the traditional Mongolian medicine Agsirga to inhibit ACE2. Mongolian medicine is an integral part of Chinese traditional medicine. It has a very significant clinical impact on the treatment and prevention of infectious diseases like influenza, tuberculosis, H7N9, SARS and H1N1. Alkaloids and stilbenes are the key components of the Mongolian medicine Agsirga. A total of 96 major alkaloids and stilbenes, including 31 alkaloids of type cevanin, 8 of type Jervine, 10 of type veratramine, 7 of type secosolanidin and 18 stilbenes, were screened. The molecular modelling study was carried out using Auto Dock.
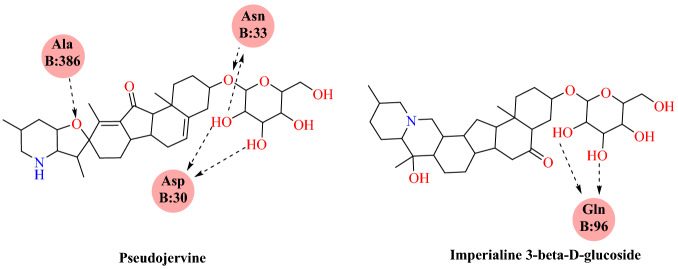
The findings of molecular docking have shown that the key binding site between alkaloids and human ACE2 protein is located in the middle bridge (residue His34 and Asp30). The docking results showed that imperialine-3-β-d-glucoside (−7.1 kcal mol−1) and pseudojervine (−6.8 kcal mol−1) significantly inhibited the SARS-CoV-2-RBD:ACE2 interaction at His34 and Asp30 (Figs. 22 and 23). These findings suggest that the combination of imperialin-3-β-d-glucoside and pseudojervine will possibly block the binding of S-protein and ACE2.
Fig. 22.
Chemical structures of pseudojervine and imperialine-3-β-d-glucoside
Fig. 23.
Binding interaction of pseudojervine and imperialine-3-β-d-glucoside SARS-CoV-2 RBD: ACE2
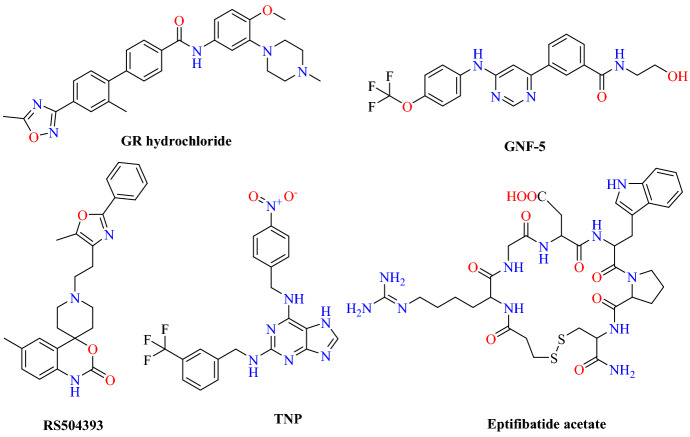
Using virtual structure screening, Choudhary et al. [79] repurposed the clinically approved drug against SARS-CoV-2-RBD: ACE2 and a newly published SARS-CoV-2 genome sequence. They found small molecules targeting the hotspot residues 31 and 353 of the ACE2, which are considered to provide a binding site for S-protein. They used 1280 molecules from the LOPAC drug library to identify possible antiviral drugs.
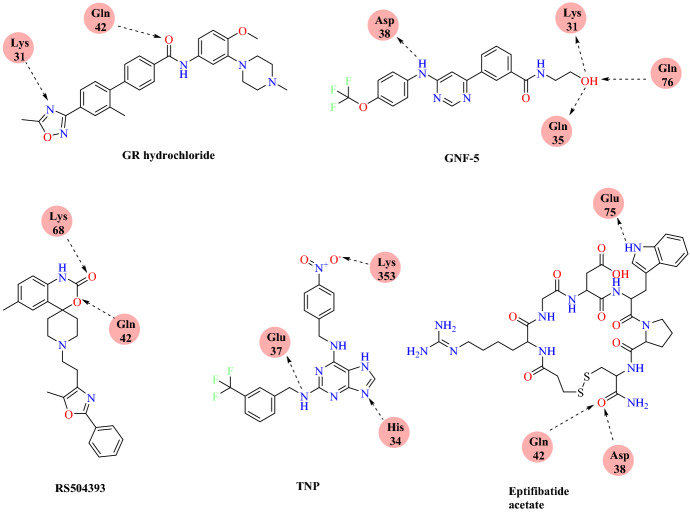
By generating the grid around the hotspot residues 31 and 353 of ACE 2, high throughput virtual screening (HTVS) was performed using PyRx and Auto Dock Vina. The results of the docking showed that GR127935 hydrochloride hydrate forms two H-bonds with Glu37 and Asp350 ACE2 receptors and hydrophobic interactions with Lys353 hotspot residues, clearly indicating its ability to bind to hotspot residues 353. Along with GR hydrochloride, GNF-5 showed that binding to hotspot 353 could potentially inhibit virus entry. Docked conformations of ligand RS504393, TNP and Eptifibatide acetate indicate that these ligands exhibit affinity to hotspot 31 residues, and, to some degree, even to hotspot 353. Among the screened compounds, the best hits were subjected to MD simulation to evaluate the stability of the protein–ligand complex using GROMACS 5.4.1.
On the basis of MD simulation results, Eptifibatide acetate (−6.05 kcal mol−1), TNP (−7.42 kcal mol−1), GNF-5 (−7.57 kcal mol−1), RS504393 (−8.32 kcal mol−1) and GR 127,935 hydrochloride hydrate (−11.23 kcal mol−1) were the best scoring hits against the ACE2 host–virus interface. This result provides proof that these repurposed molecules are potential drugs for subsequent studies (Figs. 24 and 25).
Fig. 24.
Chemical structures of GR hydrochloride, RS504393, GNF-5, TNP and eptifibatide acetate
Fig. 25.
Binding interaction of GR hydrochloride, GNF-5, RS504393, TNP and eptifibatide acetate with SARS-CoV-2 RBD: ACE2
Durdagi and colleagues [80] reported a virtual drug repurposing study, where they screened clinically approved and investigational drugs as potential inhibitors of spike receptor-binding domain (SBD) bound to ACE2.
They used the standard precision (SP) protocol of Maestro Glide docking module to dock the 7922 FDA approved clinical drugs, which were obtained from both the Pharmaceutical Collection (NPC) and NIH Chemical Genomics Center (NCGC) databases, into spike protein/ACE2.
Docking scores do not necessarily lead to identification of suitable compounds but they provide a preliminary insight into ligand–protein interactions. To understand ligand–protein interactions, they chose the top 100 compounds from each docking study and ran MD simulations with Desmond for 10-ns. The binding energies of these complexes were calculated by the MM/GBSA using the 1000 trajectories, which were collected throughout the MD simulations. Based on the MM/GBSA score, selected hits were further run for 100–500 ns in MD simulations.
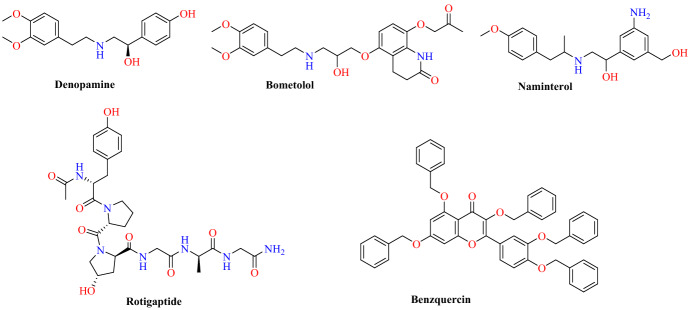
Finally, they identified five compounds (benzquercin, rotigaptide, naminterol, bometolol, denopamine) as SARS-CoV-2-RBD: ACE2 inhibitors (Fig. 26). Among them, in the 100 ns simulation study, denopamine was identified as the highest MM/GBSA scorer (−79.63 kcal mol−1) in the spike protein/ACE2 binding region. The secondary amine group of denopamine interacts primarily with Glu37 and the terminal methoxy group with Gly496 through hydrogen bonding, respectively. These residues interact with denopamine persistently throughout the 100 ns simulation (Fig. 27). These compounds could be tested clinically against COVID-19 and used for the treatment of COVID-19.
Fig. 26.
Chemical structures of denopamine, rotigaptide, bometolol, benzquercin and naminterol
Fig. 27.

Ligand interactions diagram of denopamine with SARS-CoV-2 RBD: ACE2
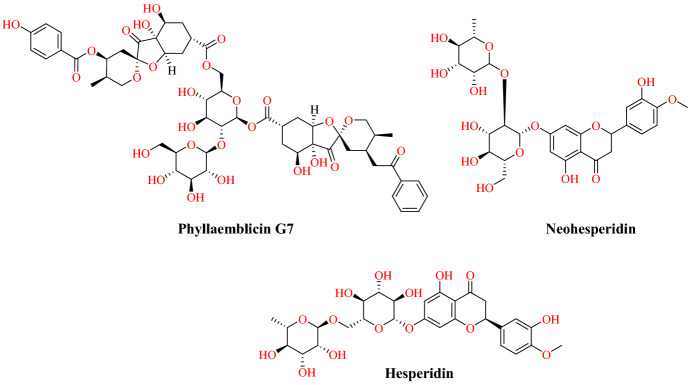
Wu and collaborators [7] reported target-based virtual ligand screening using the ZINC drug database (including 78 widely used antiviral drugs) and natural product databases against 21 targets (SARS-CoV-2). They developed 20 homology structures of SARS-CoV-2 and 1 human protein (human ACE2 structure). The ICM-Pro soft special double scoring system was used to determine the performance of the docking. The result obtained from virtual screening of ACE2 protein showed that troglitazone (mfScore −130.636059), losartan (mfScore −122.963538), ergotamine (mfScore −169.307566), cefmenoxime (mfScore −113.497767) and silybin (mfScore −115.502335) had higher binding affinity with ACE2. SARS CoV-2 mainly affects the respiratory system, so compounds affecting systems other than the respiratory system were excluded. Among the natural products, phyllaemblicin G7 (mfScore −262.834528) from Phyllanthus emblica, neohesperidin (mfScore −198.729505) and hesperidin (mfScore −151.82086) from Citrus aurantium have significant binding affinity to ACE2 (Figs. 28 and 29). A docking study found that hesperidine disrupts the interaction between ACE2 and RBD by forming hydrogen bonds with Tyr440 of the SARS-CoV-2 S-RBD: ACE2 complex (Fig. 29).
Fig. 28.
Chemical structure of phyllaemblicin G7, neohesperidin, hesperidin
Fig. 29.
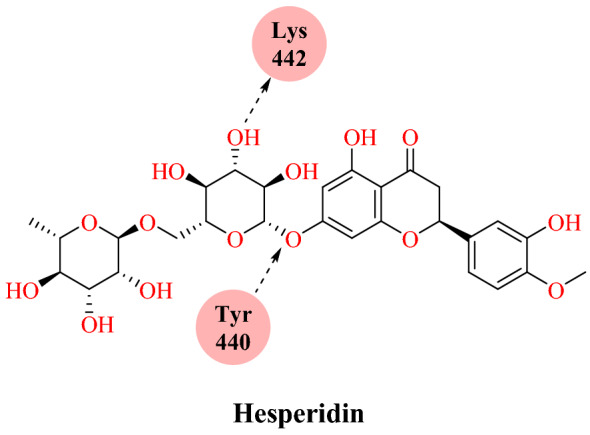
Blockage of the ACE2 and spike RBD interface by hesperidin
Patel’s group [81] reported the virtual screening of curcumin and its derivatives against the SARS-CoV-2 RBD: ACE2 complex. Curcumin has diverse pharmacological properties with over 100 cellular objectives targets, including cytokines, proteins, transcription factors and receptors. Earlier research identified curcumin as a possible cure for influenza virus infection by modulating the immune response to avert lung tissue injury. It also has anti-neuraminidase (NA) activity against the NA protein of the influenza virus. Inspired by these previous reports, Patel and colleagues screened curcumin and 24 analogues against the SARS-CoV-2 RBD: ACE2 complex using Auto Dock 4.2. According to docking result analysis, all the compounds displayed one or more hydrogen bond interactions with the SARS-CoV-2 RBD: ACE2 complex except 3-5-di-tert-butyl-4-hydroxybenzaldehyde curcumin.
All the curcumin compounds showed hydrogen bond, hydrophobic and van der Waals (VdW) interaction with Phe515, Leu517, Ser514, Glu516, Phe464, Phe429, Gly431, Thr430, Asp428, Pro426, Tyr396 and Arg355 amino acids. Using pkCSM online prediction platforms, they also predicted the drug-likeness based on Lipinski’s rule of five. All the curcumin derivatives obeyed the rules defined for bioavailability except compound-16, 4-methoxy-1-naphthaldehyde curcumin (MNC), syringaldehyde curcumin (SYC), and 5-di-tert-butyl-4-hydroxybenzaldehyde curcumin (BHBC). Based on the molecular docking method and in silico ADMET prediction, they concluded that bis-demethoxycurcumin was a suitable candidate for SARS-CoV-2 RBD: ACE2 inhibition (Fig. 30).
Fig. 30.
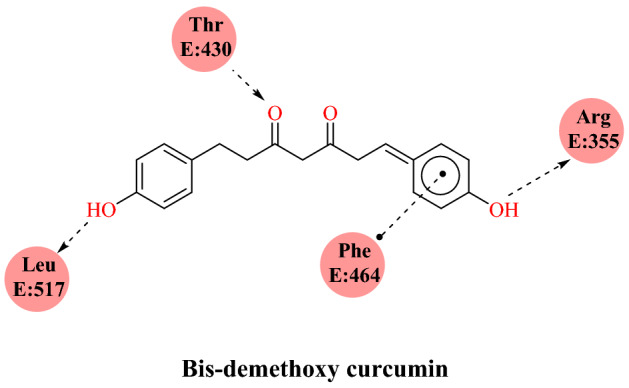
Binding approach of bis-demethoxy curcumin with SARS-CoV-2 RBD: ACE2 complex
Omotuyi et al. [82] reported that darunavir disturbs the critical nodes in the metastable conformation of SARS-CoV-2 RBD: ACE2 complex using computational methods. In their in silico study by applying cut-off criteria of greater than 250 and less than 1800 g mol−1, approximately 800 compounds were selected for docking into the SARS-CoV-2 RBD: ACE2 complex from the FDA library. Among them, two compounds—darunavir and ubrogepant—were selected based on the ligand efficiency index. Both compounds havea molecular weight of less than 550 g mol−1 and a docking score of around −11.0 kcal mol−1. Free energy by MMPBSA was calculated for darunavir in complex with SARS-CoV-2 RBD and SARS-CoV-2 RBD: ACE2, respectively. The result showed that darunavir preferably interacts with SARS-CoV-2 RBD: ACE2 (−174.36 kJ mol−1) compared with the SARS-CoV-2 RBD (−110.06 kJ mol−1). Thus, darunavir was discovered as a candidate of high ligand affinity capable of disrupting contact between the SARS-CoV-2 RBD and ACE2. Darunavir, known as an anti-HIV protease inhibitor, is now repurposeable for the treatment of COVID-19 disease by disrupting cell recognition, binding and invasion (Fig. 31).
Fig. 31.
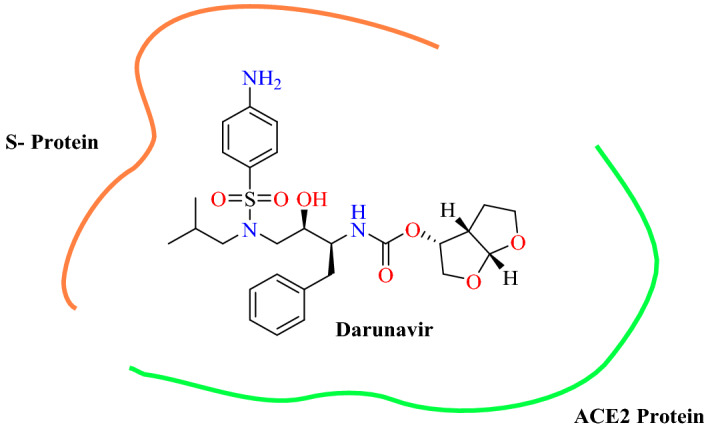
Binding of darunavir between the SARS-CoV-2 RBD and ACE2 complex
Using high-throughput screening, Zamai [83] identified MLN-4760 as a human ACE2 carboxypeptidase inhibitor with an IC50 value of 0.44 nM (Fig. 32). They reported that MLN-4760 is the best candidate to treat patients with severe SARS, but it can neither prevent COVID-19 nor treat patients with mild symptoms of COVID-19. MLN-4760 is a small synthetic molecule also known as C16, GL1001 or ORE1001. Its binding to ACE2 does not disturb the S-protein-binding region of ACE2, suggesting that ACE2 activity has been inhibited regardless of the S-protein-mediated binding of SARS-CoV-2 to ACE2. Interestingly, no adverse effects on its administration were described in in vivo rodent experiments or in human phase I clinical trials.
Fig. 32.
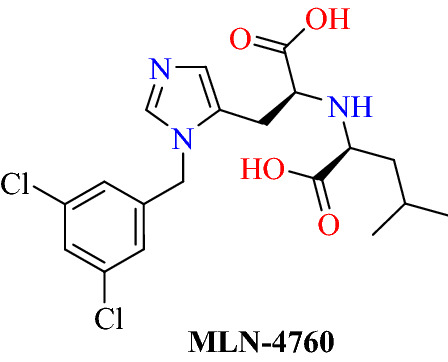
Chemical structure of MLN-4760
Nami and co-workers [84] studied the interaction between MLN-4760 (potent ACE2 inhibitor) and SARS-CoV-2-RBD: ACE2 by MD simulation for 100 ns using GROMACS. Their MD simulation study showed that MLN-4760 binds to ACE2 at the enzymatic active site with stronger affinity, which changes the ACE2 protein conformation and alters the binding site and the residues of hydrogen and hydrophobic binding involved in SARS-CoV-2 S-RBD: ACE2. However, the binding affinity of the interaction between RBD and ACE2 was not significantly affected. Binding of RBD to ACE2, coupled with MLN-4760, reversed the inhibitory effect of MLN-4760 and reversed the conformation of the ACE2 enzyme site with dissociation of the MLN-4760 enzyme site into the free native conformation. This observation indicates that MLN-4760 neither blocks nor increases the RBD spike binding of SARS-CoV-2 to human ACE2 and is unlikely to affect viral entry. Binding of the spike protein to ACE2 can therefore rescue ACE2’s enzymatic function from its inhibitor (Fig. 32). These in silico results are in alignment with the in vitro and in vivo studies of Zamai et al. [83], who found that binding of MLN-4760 to ACE2 does not disturb the S-protein-binding region of ACE2, suggesting that ACE2 activity has been inhibited regardless of the S-protein-mediated binding of SARS-CoV-2 to ACE2 (Fig. 32).
Wang’s group [84] published Arbidol (Umifenovir) an anti-influenza virus drug—an effective inhibitor of SARS-CoV-2 in vitro. They evaluated the six approved anti-influenza medications, including Zanamivir, Peramivir, Oseltamivir, Laninamivir, Baloxavir, and Arbidol against SARS-CoV-2. Among these 6 anti-influenza drugs, only Arbidol was found to efficient inhibit SARS-CoV-2 infection. Arbidol had EC50 and IC50 (cytotoxicity) values of 4.11 and 31.79 μM, respectively, and a selectivity index of 7.73. Arbidol, an indole-analog, has been approved to combat influenza in Russia and China for decades. Inspired by the work of Wang et al. [85] and Padhi et al. [86], they performed the MD simulation of Arbidol with SARS-CoV-2-RBD: ACE2 complex to gain molecular insights into the interaction. Arbidol monotherapy was considered to be superior to Lopinavir, Favipiravir or Ritonavir in the treatment of COVID-19. Their MD simulation study demonstrated that Arbidol has a stronger intermolecular interaction with the RBD domain than ACE2 in the SARS-CoV-2-RBD: ACE2 complex. MD simulation findings indicated that the binding of Arbidol induced structural rigidity in the glycoprotein of the virus, which interferes with virus attachment and entry (Fig. 33).
Fig. 33.
Binding interaction of the Arbidol with the SARS-CoV-2-RBD: ACE2 complex
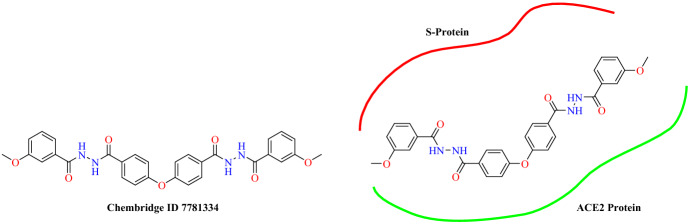
Benítez-Cardoza and co-workers [87] reported potential interaction inhibitors between the SARS-CoV-2 RBD and ACE2 Spike protein using an in silico virtual screening approach. Chembridge Corp.’s small molecule library’s express-pick array Stock was used for the virtual screening. For docking studies, the crystal structure of ACE2 with RBD complex (PDB: 6M17) and Molecular Operating Environment (MOE) software were used. Benítez-Cardoza and colleagues performed virtual screening using the reported amino acids of SARS-CoV-2-RBD: ACE2 interface (Asp30, Gln24, His34, Met82, Gln42, Lys353, Tyr41 and Arg357 in ACE2). According to ΔG binding values (−5.87 to −5.50 kcal mol−1) they selected 20 compounds, which interact with Arg393, Pro389, Ala387, Ala386, Arg357, Lys353, Asn330, Gln325, Gln96, Tyr41, Asp38, Glu37, His34 and Lys26 and thus interfere in the interaction with the SARS-CoV-2-RBD: ACE2. Moreover, these 20 compounds are likely to be safe in humans as they were confirmed by the ProTox-II and PreADMET servers. Among these virtually screened compounds, Chembridge ID 7781334 has a docking score of −5.87 kcal mol−1 and predicted lethal dose (LD50) 575 mg kg−1, which indicates interaction with Lys353, Asn330, Lys68, Gln42, Tyr41, Asp38, Glu37, His34 and Asp30 (Fig. 34).
Fig. 34.
Chemical structure and binding mode of Chembridge ID 7781334 with SARS-CoV-2 RBD: ACE2
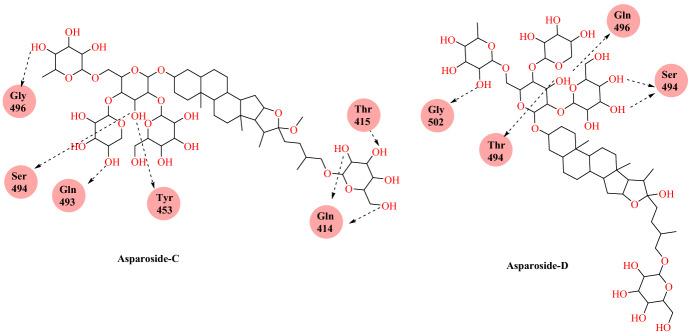
Chikhale et al. [88] reported Asparagus racemosus’ phytochemicals as a plausible antiviral agent for COVID-19 following an in silico study. A. racemosus (traditionally known as Shatavari) is a well-known medicinal plant cultivated in tropical and subtropical areas of India, where its therapeutic significance is well known in Indian and British Pharmacopoeia as well as in several traditional drug systems such as Ayurveda, Siddha and Unani.Chikhale et al. [88] conducted docking studies of 32 phytochemicals obtained from the A. racemosus on the SARS-CoV-2 S-RBD: ACE2 domain. The results revealed that asparoside-C, racemoside-A, shatavarin-X, shatavarin-I and asparoside-D were the most effective against the SARS-CoV-2 S-RBD: ACE2 domain among the 32 phytochemicals. Asparoside-C was correctly placed amongst the top five plant components into the SARS-CoV-2 S-RBD: ACE2 binding pocket and showed hydrogen binding interaction with Tyr 453, Thr 415, Ser 494, Gln 414 and Gly 496 with a docking score of −7.542 kcal mol−1 and free-binding energy −62.61 kcal mol−1. Similarly, Asparoside-D was showing hydrogen bonding interaction with Gln 498, Tyr 449, Asp 420, Lys, 417, Ser 494, Gly 502 with docking score of −7.069 kcal mol−1 and binding free energy of − 66.49 kcal mol−1 (Figs. 35, 36).
Fig. 35.
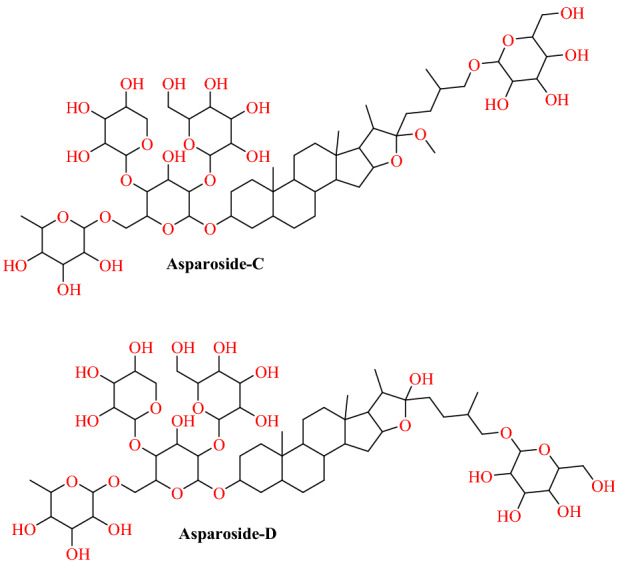
Chemical structure of Asparoside-C and Asparoside-D
Fig. 36.
Binding of Asparoside-C and Asparoside-D with SARS-CoV-2 RBD: ACE2
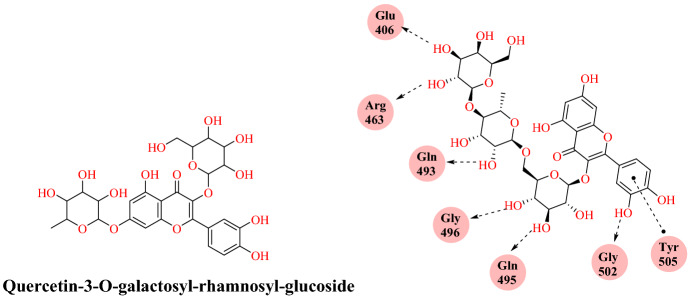
A molecular modelling study by Chikhale et al. [89] reported the SARS-CoV-2 host entry and replication inhibitors from the Indian ginseng (Withania somnifera). W. somnifera is a leading plant in Ayurvedic and Indian medicine from India. It is known as a healthy food and herbal tonic and used to treat different types of diseases and human illnesses, traditionally called ‘Ashwagandha.’ It was reported for its strong antiviral activity against distinct types of viruses, including bursal disease virus, human immunodeficiency virus (HIV), hepatitis, H1N1 influenza, coxsackie virus, herpes simplex type-1. The Government of India (Ministry of AYUSH) recently permitted Ashwagandha for clinical trials against SARS-CoV-2 along with the Council of Scientific and Industrial Research (CSIR) and the Indian Council for Medical Research (ICMR). Thus, taking into account the above view, they determined the effectiveness of 48 different phytoconstituents from W. somnifera against the SARS-CoV-2 S-RBD: ACE2. These 48 bioactive molecules were retrieved from the PubChem database and docked into the SARS-CoV-2 S-RBD: ACE2 crystal structure. Among them, quercetin-3-O-galactosyl-rhamnosyl-glucoside (QGRG) has the most favourable interaction with the SARS-CoV-2 S-RBD: ACE2 with a binding energy of −9.246 kcal mol−1. The oxane ring (2H-tetrahydropyran ring) of the QGRG formed hydrogen bond interactions with Gln 498, Glu 406, Gln 493, and Gly 496 and the OH functional of terminal phenyl ring showed an H-bonding interaction with Gly 502 (Fig. 37).
Fig. 37.
Binding interaction of quercetin-3-O-galactosyl-rhamnosyl-glucoside (QGRG) with SARS-CoV-2 RBD: ACE2
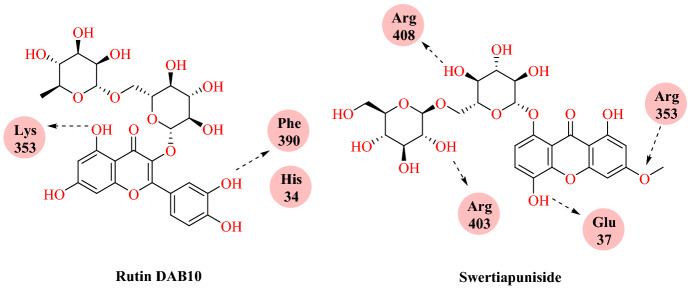
Jani and collaborators [43] reported a computational study of FDA-approved drugs and the phytochemicals from Indian medicinal plants against the ACE2-spike complex. They included only those phytochemicals from plants that are already used to treat respiratory disorders, namely Ocimum sanctum, Withania somnifera, Piper longum, Tinospora cordifolia, Curcuma longa, Terminalia arjuna, Andrographis paniculata, Swertia chirata, Azadirachta indica and Aloe barbadensis. This selection resulted in a database consisting of 250 phytochemicals that were downloaded from PubChem database. The top-ranked molecules were obtained from the direct molecular docking of the SARS-CoV-2 S-RBD: ACE2 (PDB ID: 6LZG) using DOCK6 software. According to the grid score, the five best hits from the FDA-approved database are rutin DAB10, fulvestrant, cefoperazone acid, pinaverium bromide, and abitrexate. These molecules are known to have diverse roles in terms of their therapeutic properties. Among these, rutin DAB10 is a flavonoid known to have a variety of biological activities, which showed the highest grid score (−52.58 kcal mol−1) and exhibited hydrogen bonding with Lys353 and Phe390. The docked pose of Rutin DAB10 with SARS-CoV-2 S-RBD: ACE2 complex showed hydrophobic interactions with Asn33 and Glu37 of ACE2 protein. Besides these, it showed a π-cation interaction with His34 of ACE2.
Grid scores obtained for all these phytochemicals were sorted and the top five ranked molecules were selected, namely swertiapuniside (Swertia chirayita), octadecanoate (Azadirachta indica), guineensine (Piper longum), oleic acid (Azadirachta indica) and 3-O-caffeoyl-d-quinic acid (Andrographis paniculata). These plants are also known to have anti-bacterial properties and help in increasing immunity. Among the selected hits, swertiapuniside, which is used as a bitter tonic in fevers and is also found useful in asthma and bronchitis, had the best grid score (−60.46 kcal mol−1). It formed hydrogen bonds with Glu37, Lys353, Ala387, Arg393 of the ACE2 and Arg403, Glu406, Arg408, and Lys417 of the spike. It also formed hydrophobic interactions with Glu37 of the ACE2, and Tyr453 of the spike protein (Figs. 38 and 39).
Fig. 38.

Chemical structure of rutin DAB10 and swertiapuniside
Fig. 39.
Binding interaction of Rutin DAB10 and Swertiapuniside with SARS-CoV-2 RBD: ACE2
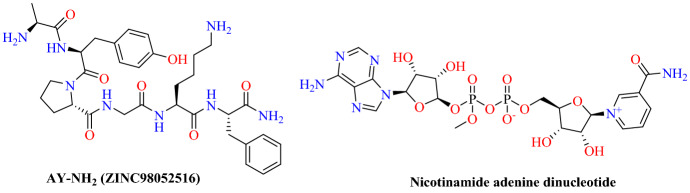
Smieszek and coworkers [90] reported small molecules and peptides that were capable of binding with SARS-CoV-2-RBD: ACE2 interface. They found that genetic variation in ACE2 possibly affects the propagation ability of the virus and variation in individual susceptibility. As SARS-CoV-2 is poorly expressed in pigs, dogs, ducks and chickens, while cats and ferrets are more contagious, they studied the amino acid sequences of ACE2 at the RBD-interacting domain by aligning them. They observed that His-34 is a crucial amino acid (an inter-species susceptibility amino acid). They hypothesized that His34 is necessary for successful viral entry, so they focused on small molecules and peptides that can bind effectively to His34 of the ACE2 protein and interferes in viral binding.
They conducted an in silico screening of approximately 11,000 compounds, including FDA-approved compounds retrieved from the Zinc database, against the SARS-CoV-2-RBD: ACE2 structure. Based on docking score, four potential small molecules were identified, namely: AY-NH2, nicotinamide adenine dinucleotide (NAD +), reproterol and thymopentin. Among these four small molecules, AY-NH2 (ZINC98052516) had the highest binding affinity with a −8.541 kcal mol−1 docking score that formed hydrogen bonding with the ACE2 Asp30, Ala387, Gln388 and Glu564 amino acids (Figs. 40 and 41).
Fig. 40.
Chemical structure of AY-NH2 and nicotinamide adenine dinucleotide
Fig. 41.

Binding interaction of AY-NH2 with SARS-CoV-2-RBD: ACE2
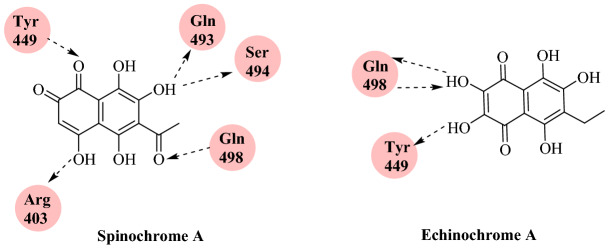
Barbieri’s group [91] reported sea urchin pigments against the ACE2-spike domain as potential therapeutic agents in in silico studies. Sea urchin pigments are a very interesting group of bioactive compounds that not only have anti-viral and anti-bacterial properties but also reduce reactive oxygen species (ROS) stress. 1,4-Naphtoquinones polihydroxilates (PHNQs) are one of the related families of sea urchin pigments and comprises spinochrome A (Spin A) and echinochrome A (Ech A). Ech A is the active compound of two Russian preparations, Gistochrome and Histochrome, used for glaucoma and cardiopathy diseases that reached the pharmaceutical market and passed all the regulatory requirements. Hence, given the urgent requirement for a specific antiviral drug, Barbieri’s group examined the in silico properties of Ech A and Spin A against SARS-CoV-2-RBD of the spike protein co-crystalized with ACE2 (PDB ID: 6M0J) using Auto Dock tools. The top ensemble docking pose of Ech A and Spin A exhibited binding affinities of −5.9 and −6.7 kcal mol−1, respectively. These Vina docking scores indicate the stability of the complex for both urchin pigment molecules. Ech A showed three hydrogen bonding with Tyr449 and Gln498 and 32 VdW contacts with Arg403, Ser494, Gln498, Q493, Tyr 449, Tyr 505, Gln496 and Tyr495. Similarly, Spin A showed five hydrogen bond interaction with Arg403, Tyr449, Gln498, Gln493 and Ser494 (Figs. 42 and 43).
Fig. 42.

Chemical structure of spinochrome A (Spin A) and echinochrome A (Ech A)
Fig. 43.
Binding interaction of Spin A and Ech A with SARS-CoV-2-RBD: ACE2
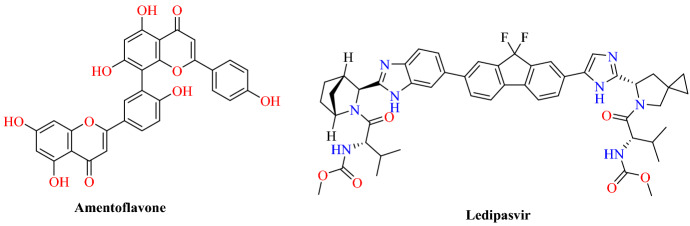
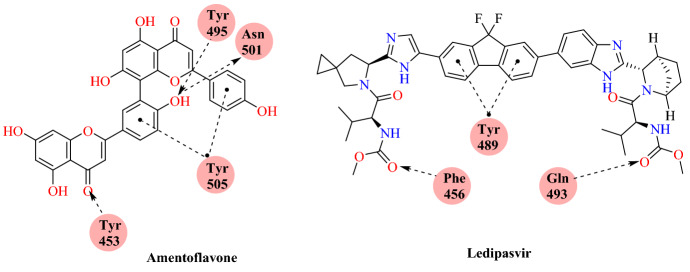
Miroshnychenko and colleagues [92] reported the repurposing of drugs against the spike glycoprotein of SARS-CoV-2 using the crystal structure of RBD/ACE2-B0AT1. Their ligand dataset includes 248 BLDPharm drugs. They identified the top 12 ligands based on the Vina Energy score of the AutoDock Vina 1.1.2. Among 12 top-ranked ligands, 9 are used as drugs for hepatitis C virus treatment. The first two ranked ligands, amentoflavone (−8.5 kcal mol−1) and ledipasvir (−8.4 kcal mol−1) have multiple binding sites on RBD, which makes their combined use possible. Their results suggest that amentoflavone and the hepatitis C virus treatment drug ledipasvir are potential candidates for COVID-19 (Figs. 44 and 45).
Fig. 44.
Chemical structure of amentoflavone and ledipasvir
Fig. 45.
Binding interaction of the amentoflavone and ledipasvir with SARS-CoV-2-RBD: ACE2
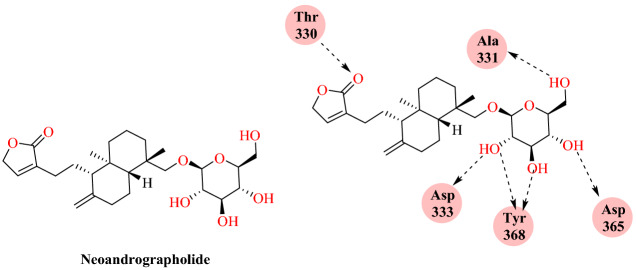
Murugan’s group reported in silico investigation of Andrographis paniculata plant phytochemicals to assess their efficacy against SARS-CoV-2 [93]. They included the four most active compounds of A. paniculata: andrographolide, eoandrographolide, 12-didehydro andrographolide, 14-deoxy andrographolide and 14-deoxy 11. They created a grid box on the S1 RBD bound to the mammalian ACE-2 receptor. Docking analysis showed that the phytochemical AGP3 (neoandrographolide) has a greater binding affinity to the SARS-CoV-2-RBD: ACE2 complex (−8.9 kcal mol−1). Neoandrographolide’s binding analysis showed that it was not binding in the interfacial regions; on the other hand, it indirectly influenced the binding of the spike protein to ACE-2 receptors (Fig. 46).
Fig. 46.
Binding of neoandrographolide with SARS-CoV-2-RBD: ACE2
Ahmad and collaborators [94] reported the bioactive constituents of Nigella sativa against COVID-19. Inspired by the broad-spectrum antiviral effect of N. sativa, they screened bioactive constituents of N. sativa against SARS-CoV-2 S protein: ACE2 receptor (PDB ID: 6VW1). They retrieved active ingredients of N. sativa, especially 4-terpineol, dithymoquinone, carvacrol, carvone, thymoquinone, p-cymene, thymohydroquinone thymol, and t-anethole.
The docking study disclosed that, among the screened compounds, dithymoquinone displayed a high binding affinity of −8.6 kcal mol−1 for the target as compared with chloroquine (−7.2 kcal mol−1). Binding analysis demonstrated that dithymoquinone had two hydrogen bonds and two ionic interactions with key residues Lys31 and Gln493 of ACE2. Additionally, dithymoquinone has good profile of drug likeness in terms of gut absorption and high solubility. In an aqueous dynamic environment, the predicted conformation of the compound is stable at SARS-CoV-2: ACE2 interface as confirmed by the binding free energy calculation. Based on these outcomes, they hypothesized that N. sativa compounds, especially dithymoquinone, interfere in the key residue interaction of the interface by disrupting the S-protein interaction with ACE2 (Fig. 47).
Fig. 47.
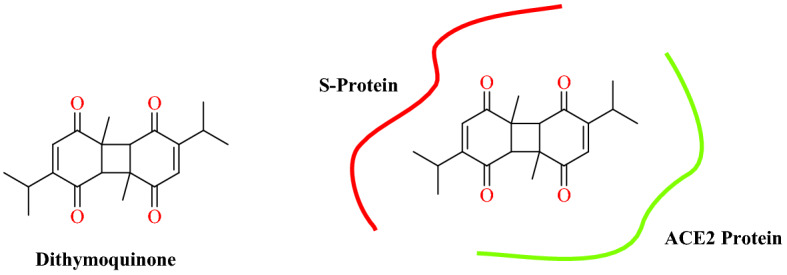
Binding interaction of dithymoquinone at the SARS-CoV-2-RBD: ACE2 interface
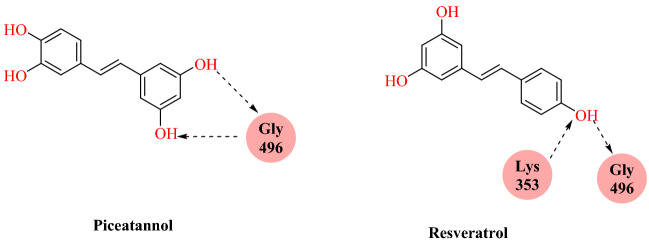
Wahedi’s group reported stilbene-based phyto-compounds as promising candidates against the SARS-CoV-2-RBD: ACE2 complex [95]. They collected four stilbenoids, namely piceatannol 581006, pinosylvin 4444110, cis-resveratrol 1265933, resveratrol 445154, pterostilbene 4445042 and trans-resveratrol 392875 from the Chemspider database based on their previous biological activities. Using autodock/vina docking study was carried out at the interface of the SARS-CoV-2 spike protein and human ACE2. Docking result analysis indicates that compounds form multiple hydrogen bond interactions with SARS-CoV-2 spike protein: ACE2 complex.
Among the screened compounds; resveratrol and piceatannol were chosen as top-class compounds based on binding affinity. Resveratrol and piceatannol were formed seven and six hydrogen bonds with the SARS-CoV-2-RBD: ACE2 complex, respectively. The 50 ns MD simulation and MM-PBSA binding free energy calculation, however, showed that resveratrol was more stable compared with piceatannol (Figs. 48 and 49).
Fig. 48.
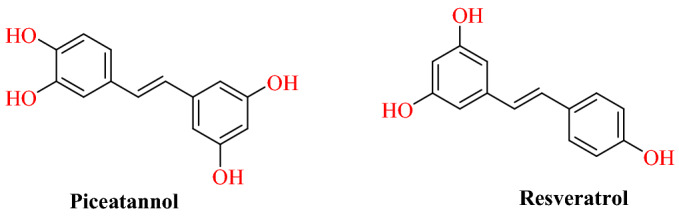
Chemical structure of piceatannol and resveratrol
Fig. 49.
Binding interaction of piceatannol and resveratrol with SARS-CoV-2 spike protein and ACE2 complex
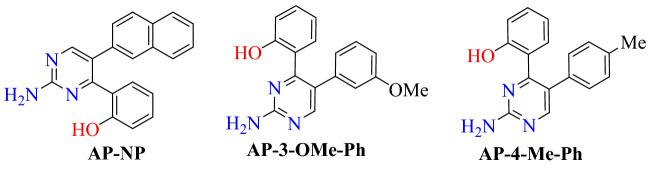
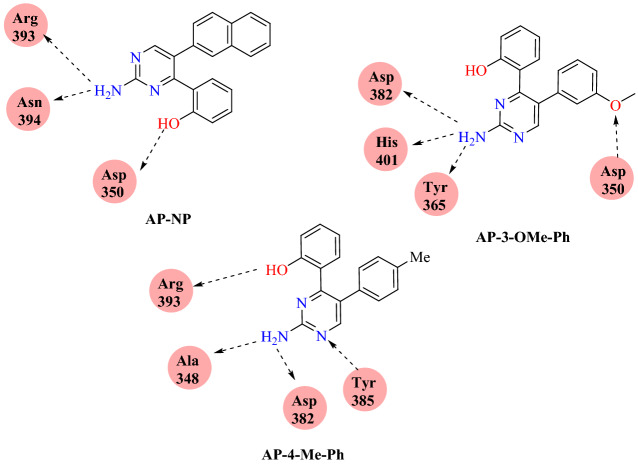
Rane and co-workers [96] reported a diaryl pyrimidine derivative as an effective inhibitor for spike protein and the human ACE2 receptor complex using an in silico approach. They performed computational analyses of diaryl pyrimidine derivative at the interface of the SARS-CoV-2-RBD: ACE2 complex (PDB ID: 6VW1) using AutoDock Vina software. Docking results showed that AP-4-Me-Ph, AP-3-OMe-Ph, AP-NP and with binding energies of −8.1, −8.1 and –8.95 kcal mol−1, respectively, showed strong binding affinity towards the ACE2 receptor. The compound AP-NP showed hydrogen bonding interaction with the polar amino acids Arg393, Asn394 and Asp350 and hydrophobic interaction with Phe40, Trp69, Leu73, Phe390 and Leu391 amino acids.
Compound AP-3-OMe-Ph forms hydrogen bond interactions with Gly352, Asp350, Arg393, Asp382, Ala348 and a hydrophobic interaction with Trp349, Phe390 and Phe40. AP-4-Me-Ph forms hydrophobic interactions with the Phe40, Ser44, Ser47, Trp349, Ala 348 and hydrogen bond interactions with His401, Asp382, Tyr385 and Asp350 (Figs. 50 and 51).
Fig. 50.
Chemical structure of AP-NP, AP-3-OMe-Ph and AP-4-Me-Ph
Fig. 51.
Binding interaction of of AP-NP, AP-3-OMe-Ph and AP-4-Me-Ph with SARS-CoV-2-RBD: ACE2
Srivastava’s group [97] reported the in silico identification of phytocompounds with activity towards the SARS-CoV-2-RBD: ACE2 complex. They conducted molecular docking studies using Auto Dock Vina software to analyze molecular interactions between various phytocompounds and ACE2 Receptor (PDB ID: 6LZG). Phytochemicals were obtained from ashwagandha, fennel, liquorice, tulsi, neem, moringa, elderberry and amla. The results of docking showed that the phytochemicals chebulagic acid, pedunculagin and nimbolide have the highest binding affinity for the SARS-CoV-2-RBD: ACE2 complex. These phytochemicals such as chebulagic acid (−18.4 kcal mol−1), Pedunculagin (−17.5 kcal mol−1), Azadirachtin (−12.4 kcal mol−1) and Nimbolide (−16.7 kcal mol−1) may become possible candidates for targeting the SARS-CoV-2-RBD: ACE2 complex (Fig. 52).
Fig. 52.
Chemical structure of chebulagic acid, azadirachtin, pedunculagin, and nimbolide
Discussion and Future Perspectives
The highly infectious COVID-19 is induced by SARS-CoV-2. It has been announced as a pandemic by the WHO. ACE2 is a functional receptor that acts as the point of entry for SARS-CoV-2 into human lung cells [binding through its spike (S) protein]. The S-protein consists of two subunits: S1 as the RBD and the S2 subunit responsible for viral membrane fusion. The S2 subunit of SARS-CoV-2 has ~ 99% similarity, while the S1 subunit is 70% identical to another SARS-CoV bat and human SARS-CoV, but the core RBD domain is highly preserved among them. It has been observed that cells in which ACE-2 and TMPRSS2 are present are more prone to SARS-CoV-2 entry. In SARS-CoV-2 spike protein RBD there were more ACE2-interacting residues compared with SARS-CoV due to substantial mutations present in the SARS-CoV-2-sequence, more specifically in the RBD domain.
Lys31 and Lys353 are the crucial amino acids of the human ACE2 receptor, and considered as virus-binding hotspot residues, more sensitive to S-protein binding. In human receptor ACE2, hotspot 31 (Lys31) consists of the salt bridge between lysine 31 and glutamic acid 35, and hotspot 353 (Lys353) consists of another salt bridge between lysine 353 and asparagine 38, covered by a hydrophobic residue. The RBD domain of the S1 subunit of the spike protein plays a crucial role in binding of SARS-CoV-2 to ACE2. In vitro binding studies revealed good binding affinity at low nanomolar range between ACE2 and the RBD. The recently released crystal structure PDB Id: 6M0J explains the structural interactions of SARS-CoV-2 S-RBD with ACE2. The RBD/ACE2 interface has a hydrophilic network, which consists of 13 hydrogen bonds and two salt bridges. ACE2 is expressed in respiratory system, heart, intestine, kidney and liver and hence these organs are at higher risk of infection. Researchers have so far targeted the three crucial forms of ACE2 enzymes, i.e., ACE2, SARS-CoV-2 S-Protein-ACE2 and SARS-CoV-2 S-RBD: ACE2 as the understanding about the disease processed since December 2019 as SARS-CoV-2 entry blockers (Fig. 53). Targeting the virus entry point by inhibiting ACE2 has a greater benefit than inhibiting later stages of the viral life cycle. Here, we have summarized the different in silico based studies and the repurposing of anti-viral drugs to target ACE2, SARS-CoV-2 S-Protein-ACE2 and SARS-CoV-2 S-RBD:ACE2.
Fig. 53.
Pardigm shifts in the discovery of SARS-CoV-2 entry blockers
It takes years to design a new drug and test its pharmacokinetics and pharmacodynamics properties. In repurposing studies, molecules have already passed through multiple stages and have well-defined profiles; they would not need prolonged pre-clinical studies and would thus be excellent candidates for emergencies or outbreaks. In the beginning of 2020, researchers targeted the ACE2 enzyme and reported the repurposing of the previously used drugs. They reported that ivermectin, rutin, hesperdin, naringin, paritaprevir, vitexin, orientin, berberine, artemisnin, curcumin, apigenin, crisimaritin, hispudulin, quercetin, 6-gingerol, isothymol, cucurbitacin B, beta-sitosterol, cynaropicrin, andrographolide and saikosaponin A could inhibit ACE2 enzyme based on their in silico studies (Fig. 54). Later, when it was clear that, via S-protein, SARS-CoV-2 was bound with ACE2 enzyme, researchers targeted the SARS-CoV-2-S protein:ACE2 complex. They reported nitrofurantoin, favipravir, hydroxychloroquine, cepharanthine, ergolide, menthol, isoniazid pyruvate, eriodictyol, raspberry ketone, ivermectin, hypericin, and thymoquinone as potential inhibitors of the SARS-CoV-2-S protein:ACE2 complex (Fig. 55). Moreover, SARS-CoV-2 S-RBD residues are highly conserved in bats, human and palm civet cat SARS-CoVs. The affinity between SARS-CoV-2 and ACE2 S-RBD is almost ten times higher than that of SARS-CoV RBD, suggesting that ACE2 is the unique receptor responsible for binding the virus to the host cell membrane. Hence, researchers tilted towards targeting the SARS-CoV-2-RBD:ACE2 complex. They reported pseudojervine, hesperidin, bis-demethoxy curcumin, asparoside C, quercetin-3-O-galatosyl-rhamnosyl-glucoside, rutin DAB10, AY-NH2, spinochrome A, amentoflvaone, neoandrographolide, dithymoquinone, piceatannol, AP-NP, chebulagic acid from natural sources and GR hydrochloride, denopamine, darunavir, MLN-4760, arbidol, chembridge ID 7781334 from synthetic source as SARS-CoV-2-RBD:ACE2 complex inhibitors (Fig. 56). These molecules could be used against COVID-19; however, further studies are required to validate these compounds in in vitro and in vivo studies. Thanks to repositioning studies, new uses for various compounds different from their original purposes were identified even though they had failed in their original purpose and/or had been withdrawn from the market. Computational methods such as virtual screening will minimize the time needed to find new targets for existing drug molecules, with the benefit of also being cost-effective, as shown in review papers.
Fig. 54.
Repurposed ACE2 inhibitors from natural and synthetic sources
Fig. 55.
Repurposed SARS-CoV-2 S Protein:ACE2 inhibitors from natural and synthetic sources
Fig. 56.
Repurposed SARS-CoV-2 S-RBD: ACE2 inhibitors from natural and synthetic sources
Acknowledgements
The authors would like to thank “Indian Council of Medical Research (ICMR)-New Delhi, Govt. of India” (Grant No. 2018-2953) for funding the Adhoc Research Grant.
Abbreviations
- CoV
Coronavirus
- + ssRNA
Single-stranded RNA
- ICTV
International Committee on Taxonomy of Viruses
- SARS
severe acute respiratory syndrome
- SARS-CoV 1
Severe acute respiratory syndrome coronavirus 1
- MERS-CoV
Middle East respiratory syndrome virus
- MERS
Middle East respiratory syndrome
- 2019-nCoV
2019 novel coronavirus
- WHO
World Health Organization
- RAS
Renin-angiotensin system
- ACE2
Angiotensin converting enzyme 2
- SARS-CoV 2
Virus responsible for the coronavirus disease 2019 pandemic
- COVID-19
Coronavirus disease 2019
- ORF
Open reading frame
- N Protein
Nucleocapsid protein
- UTR
Untranslated region
- NSP
Nonstructural protein
- RBD
Receptor binding domain
- ARDS
Acute respiratory distress syndrome
- TMPRSS2
Transmembrane protease serine 2
- RMSD
Root mean square deviation
- mfScore
Mean force score
Author Contributions
This article is dedicated to Mr. Sudhakar Chaudhari (Department of Pharmacognosy, R. C. Patel Institute of Pharmaceutical Education and Research, Shirpur) and Dr. Siraj Shaikh (Ali-Allana College of Pharmacy-Akkalkua), who left us due to COVID-19.
Footnotes
This article is part of the Topical Collection ‘‘Antiviral Drug Research’’; edited by Guangxin Liang, Zheng Yin.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Denison MR, Graham RL, Donaldson EF, Eckerle LD, Baric RS (2011) Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol 8(2):270–279 [DOI] [PMC free article] [PubMed]
- 2.Banerjee A, Kulcsar K, Misra V, Frieman M, Mossman K. Bats and coronaviruses. Viruses. 2019;11(1):41. doi: 10.3390/v11010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng VC, Lau SK, Woo PC, Yuen KY. Severe acute respiratory syndrome coronavirus as an agent of emerging and reemerging infection. Clin Microbiol Rev. 2007;20(4):660–694. doi: 10.1128/CMR.00023-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee N, Hui D, Wu A, Chan P, Cameron P, Joynt GM, Ahuja A, Yung MY, Leung CB, To KF, Lui SF. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 2003;348(20):1986–1994. doi: 10.1056/NEJMoa030685. [DOI] [PubMed] [Google Scholar]
- 5.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367(19):1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 6.de Groot RJ, Baker SC, Baric RS, Brown CS, Drosten C, Enjuanes L, Fouchier RA, Galiano M, Gorbalenya AE, Memish ZA, Perlman S. Commentary: Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J Virol. 2013;87(14):7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, Wang Q, Xu Y, Li M, Li X, Zheng M. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau SK, Woo PC, Li KS, Huang Y, Tsoi HW, Wong BH, Wong SS, Leung SY, Chan KH, Yuen KY. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102(39):14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reusken CB, Haagmans BL, Müller MA, Gutierrez C, Godeke GJ, Meyer B, Muth D, Raj VS, Smits-De Vries L, Corman VM, Drexler JF. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;13(10):859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KS, Lau EH, Wong JY, Xing X. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N Engl J Med. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lai CC, Shih TP, Ko WC, Tang HJ, Hsueh PR. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents. 2020;55(31):05924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.World Health Organization (2020) Laboratory testing for coronavirus disease 2019 (COVID-19) in suspected human cases: interim guidance, 2 March 2020. https://apps.who.int/iris/handle/10665/331329 (accessed 14 August 2020).
- 13.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang S, Peng W, Zhu Y, Lu S, Zhou M, Lin W, Wu W, Huang S, Jiang L, Luo X, Deng M. Recent progress in understanding 2019 Novel Coronavirus associated with human respiratory disease: detection, mechanism and treatment. Int J Antimicrob Agents. 2020;55(5):105950. doi: 10.1016/j.ijantimicag.2020.105950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y. Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau SK, Woo PC, Li KS, Huang Y, Wang M, Lam CS, Xu H, Guo R, Chan KH, Zheng BJ, Yuen KY. Complete genome sequence of bat coronavirus HKU2 from Chinese horseshoe bats revealed a much smaller spike gene with a different evolutionary lineage from the rest of the genome. Virology. 2007;367(2):428–439. doi: 10.1016/j.virol.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ortega JT, Serrano ML, Pujol FH, Rangel HR. Unrevealing sequence and structural features of novel coronavirus using in silico approaches: the main protease as molecular target. EXCLI J. 2020;19:400–409. doi: 10.17179/excli2020-1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rivellese F, Prediletto E. ACE2 at the center of COVID-19 from paucisymptomatic infections to severe pneumonia. Autoimmun Rev. 2020;19(6):102536. doi: 10.1016/j.autrev.2020.102536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Health Organization (2020) Report of the WHO-China joint mission on coronavirus disease 2019 (COVID-19) https://www.who.int/publications/i/item/report-of-the-who-china-joint-mission-on-coronavirus-disease-2019-(covid-19).
- 20.Biswas A, Bhattacharjee U, Chakrabarti AK, Tewari DN, Banu H, Dutta S. Emergence of novel coronavirus and COVID-19: whether to stay or die out? Crit Rev Microbiol. 2020;46(2):182–193. doi: 10.1080/1040841X.2020.1739001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang B, Bragazzi NL, Li Q, Tang S, Xiao Y, Wu J. An updated estimation of the risk of transmission of the novel coronavirus (2019-nCov) Infect Dis Model. 2020;5:248–255. doi: 10.1016/j.idm.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cynthia L, Qiongqiong Z, Yingzhu L, Garner LV, Watkins SP, Carter LJ, Smoot J, Gregg AC, Daniels AD, Jervey S, Albaiu D. Research and development on therapeutic agents and vaccines for COVID-19 and related human Coronavirus Diseases. ACS Cent Sci. 2020;6(3):315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boopathi S, Poma AB, Novel KP. coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J Biomol Struct Dyn. 2019;2020:1–10. doi: 10.1080/07391102.2020.1758788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.WHO Coronavirus (COVID-19) Dashboard. https://covid19.who.int/
- 25.Riordan JF. Angiotensin-I-converting enzyme and its relatives. Genome Biol. 2003;4(8):225. doi: 10.1186/gb-2003-4-8-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bosch BJ, van der Zee R, de Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77(16):8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581(7807):221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H, Penninger JM, Li Y, Zhong N, Slutsky AS. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46(4):586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bombardini T, Picano E. Angiotensin-converting enzyme 2 as the molecular bridge between epidemiologic and clinical features of COVID-19. Can J Cardiol. 2020;36(5):784.e1–784.e2. doi: 10.1016/j.cjca.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Malik YS, Sircar S, Bhat S, Sharun K, Dhama K, Dadar M, Tiwari R, Chaicumpa W. Emerging novel coronavirus (2019-nCoV)—current scenario, evolutionary perspective based on genome analysis and recent developments. Vet Q. 2020;40(1):68–76. doi: 10.1080/01652176.2020.1727993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan JFW, Kok KH, Zhu Z, Chu H, To KKW, Yuan S, Yuen KY. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 2020;27(4):671–680.e2. doi: 10.1016/j.chom.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu H, Zhong L, Deng J, Peng J, Dan H, Zeng X, Li T, Chen Q. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020;12(1):8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwata-Yoshikawa N, Okamura T, Shimizu Y, Hasegawa H, Takeda M, Nagata N. TMPRSS2 contributes to virus spread and immunopathology in the airways of murine models after Coronavirus infection. J Virol. 2019;93(6):e01815–e1818. doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleine-Weber H, Elzayat MT, Wang L, Graham BS, Müller MA, Drosten C, Pöhlmann S, Hoffmann M. Mutations in the spike protein of middle east respiratory syndrome Coronavirus transmitted in Korea increase resistance to antibody-mediated neutralization. J Virol. 2019;93(2):e01381–e1418. doi: 10.1128/JVI.01381-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 37.Luan J, Lu Y, Jin X, Zhang L. Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS-CoV-2 infection. Biochem Biophys Res Commun. 2020;526(1):165–169. doi: 10.1016/j.bbrc.2020.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.R&D Systems (2020) ACE-2: the receptor for SARS-CoV-2. https://www.rndsystems.com/resources/articles/ace-2-sars-receptor-identified. Assesed on 14 August 2020
- 39.Veeramachaneni GK, Thunuguntla VBSC, Bobbillapati J, Bondili JS. Structural and simulation analysis of hotspot residues interactions of SARS-CoV 2 with human ACE2 receptor. J Biomol Struct Dyn. 2021;39(11):4015–4025. doi: 10.1080/07391102.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sinha SK, Shakya A, Prasad SK, Singh S, Gurav NS, Prasad RS, Gurav SS. An in-silico evaluation of different Saikosaponins for their potency against SARS-CoV-2 using NSP15 and fusion spike glycoprotein as targets. J Biomol Struct Dyn. 2021;39(9):3244–3255. doi: 10.1080/07391102.2020.1762741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pant S, Singh M, Ravichandiran V, Murty USN, Srivastava HK. Peptide-like and small-molecule inhibitors against Covid-19. J Biomol Struct Dyn. 2021;39(8):2904–2913. doi: 10.1080/07391102.2020.1757510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol. 2020;94(7):e00127–e220. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jani V, Koulgi S, Uppuladinne VN, Sonavane U, Joshi R. Computational drug repurposing studies on the ACE2-Spike (RBD) interface of SARS-CoV-2. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12652832.v2. [DOI] [Google Scholar]
- 44.https://en.wikipedia.org/wiki/Angiotensin-converting_enzyme_2
- 45.https://ko.m.wikipedia.org/wiki/%ED%8C%8C%EC%9D%BC:RBD_in_complex_with_ACE2.png
- 46.Jun L, Jiwan G, Jinfang Y, Sisi S, Huan Z, Shilong F, Qi Z, Xuanling S, Qisheng W, Linqi Z, Xinquan W. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 47.Li F, Li W, Farzan M, Harrison SC. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 48.Wenhui L, Chengsheng Z, Jianhua S, Jens HK, Michael JM, Shiwen L, Sweem KW, I-Chueh H, Keming X, Natalya V, Akikazu M, Yaqing H, Wayne AM, Yi, Hyeryun C, Michael F, Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24(8):1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han DP, Penn-Nicholson A, Cho MW. Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology. 2006;350(1):15–25. doi: 10.1016/j.virol.2006.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309(5742):1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 52.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li SR, Tang ZJ, Li ZH, Liu X. Searching therapeutic strategy of new coronavirus pneumonia from angiotensin-converting enzyme 2: the target of COVID-19 and SARS-CoV. Eur J Clin Microbiol Infect Dis. 2020;39(6):1021–1026. doi: 10.1007/s10096-020-03883-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Keiji K, Yumiko I, Shuan R, Hong G, Feng G, Bin G, Yi H, Peng Y, Yanli Z, Wei D, Linlin B, Binlin Z, Guang L, Zhong W, Mark C, Yanxin L, Dexian Z, Andreas L, Teiji W, Arthur SS, Depei L, Chuan Q, Chengyu J, Josef MP. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat Med. 2005;11(8):875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yumiko I, Keiji K, Shuan R, Yi H, Feng G, Bin G, Peng Y, Renu S, Teiji W, Howard L, Michael AC, Akiyoshi F, Chi-Chung H, Lutz H, Stefan U, Arthur SS, Chengyu J, Josef MP. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436(7047):112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oudit GY, Kassiri Z, Jiang C, Liu PP, Poutanen SM, Penninger JM, Butany J. SARS-coronavirus modulation of myocardial ACE2 expression and inflammation in patients with SARS. Eur J Clin Invest. 2009;39(7):618–625. doi: 10.1111/j.1365-2362.2009.02153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, Diaz G. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382(10):929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang H, Kang Z, Gong H, Xu D, Wang J, Li Z, Cui X, Xiao J, Meng T, Zhou W, Liu J. The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes. Gut. 2020;69(6):1010–1018. [Google Scholar]
- 59.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z. Clinical features of patients infected with 2019 novel coronavirus in Wuhan China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Yu T. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fraga-Silva RA, Sorg BS, Wankhede M, deDeugd C, Jun JY, Baker MB, Li Y, Castellano RK, Katovich MJ, Raizada MK, Ferreira AJ. ACE2 activation promotes antithrombotic activity. Mol Med. 2010;16(5–6):210–215. doi: 10.2119/molmed.2009.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fan C, Li K, Ding Y, Lu WL, Wang J. ACE2 expression in kidney and testis may cause kidney and testis damage after 2019-nCoV infection. Front Med. 2021;7:1045. doi: 10.3389/fmed.2020.563893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen N, Zhou M, Dong X, Qu J, Gong F, Qiu Y, Han Y, Wang J, Liu Y, Wei Y, Yu T. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chai X, Hu L, Zhang Y, Han W, Lu Z, Ke A, Zhou J, Shi G, Fang N, Fan J, Cai J. Specific ACE2 expression in cholangiocytes may cause liver damage after 2019-nCoV infection. bioRxiv. 2020 doi: 10.1101/2020.02.03.931766. [DOI] [Google Scholar]
- 65.Micael DLO, Kelson MTO. Comparative docking of SARS-CoV-2 receptors antagonists from repurposing drugs. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12044538.v4. [DOI] [Google Scholar]
- 66.Alagu Lakshmi S, Shafreen RMB, Priya A, Shunmugiah KP. Ethnomedicines of Indian origin for combating COVID-19 infection by hampering the viral replication: using structure-based drug discovery approach. J Biomol Struct Dyn. 2020;39:1–16. doi: 10.1080/07391102.2020.1778537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abdelli I, Hassani F, Bekkel B, Ghalem S. In silico study the inhibition of angiotensin converting enzyme 2 receptor of COVID-19 by Ammoides verticillata components harvested from western Algeria. J Biomol Struct Dyn. 2021;39(9):3263–3276. doi: 10.1080/07391102.2020.1763199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Omar S, Bouziane I, Bouslama Z, Djemel A. In-silico identification of potent inhibitors of COVID-19 main protease (Mpro) and angiotensin converting enzyme 2 (ACE2) from natural products: quercetin, hispidulin, and cirsimaritin exhibited better potential inhibition than hydroxy-chloroquine against COVID-19 main protease active site and ACE2. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12181404.v1. [DOI] [Google Scholar]
- 69.Dhanasekaran S. Scope of phytotherapeutics in targeting ACE2 mediated host–viral interface of SARS-CoV2 that causes COVID-19. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12089730.v1. [DOI] [Google Scholar]
- 70.Andersen KG, Rambaut A, Lipkin WI, Holmes EC, Garry RF (2020) The proximal origin of SARSCoV-2. Nat Med 26:450–452. 10.1038/s41591-020-0820-9 [DOI] [PMC free article] [PubMed]
- 71.Chen Y, Liu Q, Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wan Y, Shang J, Graham R, Baric RS, Li F (2020) Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J Virol 94(7):e00127-20. 10.1128/jvi.00127-20 [DOI] [PMC free article] [PubMed]
- 73.Wang B, Kovalchuk A, Li D, Ilnytskyy Y, Kovalchuk I and Kovalchuk O (2020) In search of preventative strategies: novel anti-inflammatory high-CBD Cannabis sativa extracts modulate ACE2 expression in COVID-19 gateway tissues. Aging (Albany NY) 12(22):22425–22444 [DOI] [PMC free article] [PubMed]
- 74.Cheng L, Zheng W, Li M, Huang J, Bao S, Xu Q, Ma Z (2020) Citrus fruits are rich in flavonoids for immunoregulation and potential targeting ACE2. (https://www.preprints.org/manuscript/202002.0313/v1) [DOI] [PMC free article] [PubMed]
- 75.Yong-Ming Y, Shen X, Cao YK, Jiao-Jiao Z, Wang Y, Yong-Xian C. Discovery of anti-2019-nCoV agents from 38 Chinese patent drugs toward respiratory diseases via docking screening. Preprints. 2020 doi: 10.20944/preprints202002.0254.v2. [DOI] [Google Scholar]
- 76.Smith M, Smith JC. Repurposing therapeutics for COVID-19: supercomputer-based docking to the SARS-CoV-2 viral spike protein and viral spike protein-human ACE2 interface. ChemRxiv. 2020 doi: 10.26434/chemrxiv.11871402.v4. [DOI] [Google Scholar]
- 77.Abdel-Mottaleb MS, Abdel-Mottaleb Y. In Search for effective and safe drugs against SARS-CoV-2: Part I] Simulated interactions between selected nutraceuticals, ACE2 enzyme and S Protein simple peptide sequences. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12155235.v1. [DOI] [Google Scholar]
- 78.Cheng J, Tang Y, Bao B, Zhang P. Exploring the active compounds of traditional mongolian medicine agsirga in intervention of Novel Coronavirus (2019-nCoV) based on HPLC-Q-exactive-MS/MS and molecular docking method. ChemRxiv. 2020 doi: 10.26434/chemrxiv.11955273.v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Choudhary S, Malik YS, Tomar S. Identification of SARS-CoV-2 cell entry inhibitors by drug repurposing using in silico structure-based virtual screening approach. Front Immunol. 2020;11:1664. doi: 10.3389/fimmu.2020.01664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Durdagi S, Aksoydan B, Dogan B, Sahin K, Shahraki A, Birgül-İyison N. Screening of clinically approved and investigation drugs as potential inhibitors of SARS-CoV-2 main protease and spike receptor-binding domain bound with ACE2 COVID19 target proteins: a virtual drug repurposing study. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12032712.v2. [DOI] [Google Scholar]
- 81.Patel A, Rajendran M, Pakala SB, Shah A, Patel H, Karyala P (2021) Virtual screening of curcumin and its analogs against the spike surface glycoprotein of SARS-CoV-2 and SARS-CoV. J Biomol Struct Dyn. DOI: 10.1080/07391102.2020.1868338 [DOI] [PMC free article] [PubMed]
- 82.Omotuyi OI, Nash O, Ajiboye BO, Metibemu DS, Oyinloye BE, Adelakun NS, Hurdayal R, Aruleba RT, Kao AP (2020) Darunavir disrupts critical nodes in metastable 2019-nCoV-RBD/ACE-2 complex. https://www.preprints.org/manuscript/202003.0125/v1
- 83.Zamai L. The rational for administration of ACE2 pathway inhibitors in patients infected by SARS-CoV-2: devising an administration strategy. Cells. 2020;9:1704. [Google Scholar]
- 84.Nami B, Ghanaeian A, Ghanaeian K, Nami N, Ghasemi-Dizgah A, Caluseriu O. The interaction of the severe acute respiratory syndrome coronavirus 2 spike protein with drug-inhibited angiotensin converting enzyme 2 studied by molecular dynamics simulation. J Hypertens. 2021;39(8):1705–1716. doi: 10.1097/HJH.0000000000002829. [DOI] [PubMed] [Google Scholar]
- 85.Wang X, Cao R, Zhang H, Liu J, Xu M, Hu H, Li Y, Zhao L, Li W, Sun X, Yang X. The anti-influenza virus drug, arbidol is an efficient inhibitor of SARS-CoV-2 in vitro. Cell Discov. 2020;6(1):1–5. doi: 10.1038/s41421-020-0169-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Padhi A, Seal A, Tripathi T. How does Arbidol Inhibit the Novel Coronavirus SARS-CoV-2. Atomistic insights from molecular dynamics simulations. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12464576.v1. [DOI] [Google Scholar]
- 87.Benítez-Cardoza CG, Vique-Sánchez JL. Potential inhibitors of the interaction between ACE2 and SARS-CoV-2 (RBD), to develop a drug. Life Sci. 2020;256:117970. doi: 10.1016/j.lfs.2020.117970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chikhale RV, Sinha SK, Patil RB, Prasad SK, Shakya A, Gurav N, Prasad R, Dhaswadikar SR, Wanjari M, Gurav SS. In-silico investigation of phytochemicals from Asparagus racemosus as plausible antiviral agent in COVID-19. J Biomol Struct Dyn. 2020;39:1–15. doi: 10.1080/07391102.2020.1784289. [DOI] [PubMed] [Google Scholar]
- 89.Chikhale RV, Gurav SS, Patil RB, Sinha SK, Prasad SK, Shakya A, Shrivastava SK, Gurav NS, Prasad RS. SARS-CoV-2 host entry and replication inhibitors from Indian ginseng: an in-silico approach. J Biomol Struct Dyn. 2020;39:1–12. doi: 10.1080/07391102.2020.1778539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smieszek S, Przychodzen B, Polymeropoulos V, Polymeropoulos C, Polymeropoulos M. Direct ACE2-spike RBD binding disruption with small molecules: a strategy for COVID-19 treatment. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12159945.v1. [DOI] [Google Scholar]
- 91.Barbieri ES, Rubilar Panasiuk CT, Gázquez A, Avaro M, Seiler EN, Vera Piombo M, Gittardi Calderón AA, Chaar F, Fernandez JP, Sepúlveda LR. Sea urchin pigments as potential therapeutic agents against the spike protein of SARS-CoV-2 based on in silico analysis. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12568595.v1. [DOI] [Google Scholar]
- 92.Miroshnychenko K, Shestopalova AV. Combined use of amentoflavone and ledipasvir could interfere with binding of spike glycoprotein of SARS-CoV-2 to ACE2: the results of molecular docking study. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12377870.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Murugan NA, Pandian CJ, Jeyakanthan J. Computational investigation on Andrographis paniculata phytochemicals to evaluate their potency against SARS-CoV-2 in comparison to known antiviral compounds in drug trials. J Biomol Struct Dyn. 2020;39:1–12. doi: 10.1080/07391102.2020.1777901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ahmad S, Abbasi HW, Shahid S, Gul S, Abbasi SW. Molecular docking, simulation and MM-PBSA studies of Nigella sativa compounds: a computational quest to identify potential natural antiviral for COVID-19 treatment. J Biomol Struct Dyn. 2020;39:1–16. doi: 10.1080/07391102.2020.1775129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wahedi HM, Ahmad S, Abbasi SW. Stilbene-based natural compounds as promising drug candidates against COVID-19. J Biomol Struct Dyn. 2020;39:1–10. doi: 10.1080/07391102.2020.1762743. [DOI] [PubMed] [Google Scholar]
- 96.Rane JS, Pandey P, Chatterjee A, Khan R, Kumar A, Prakash A, Ray S. Targeting virus–host interaction by novel pyrimidine derivative: an in silico approach towards discovery of potential drug against COVID-19. J Biomol Struct Dyn. 2020;39:1–11. doi: 10.1080/07391102.2020.1794969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Srivastava AK, Guptab SK, Kumara U. Computational studies towards identification of lead herbal compounds of medicinal importance for development of nutraceutical against COVID-19. ChemRxiv. 2020 doi: 10.26434/chemrxiv.12581819.v1. [DOI] [Google Scholar]