Abstract
Monoclonal antibodies (mAbs) are emerging as safe and effective therapeutics against SARS-CoV-2. However, variant strains of SARS-CoV-2 have evolved, with early studies showing that some mAbs may not sustain their efficacy in the face of escape mutants. Also, from the onset of the COVID-19 pandemic, concern has been raised about the potential for Fcγ receptor-mediated antibody-dependent enhancement (ADE) of infection. In this study, plaque reduction neutralization assays demonstrated that mAb 1741-LALA neutralizes SARS-CoV-2 strains B.1.351, D614 and D614G. MAbs S1D2-hIgG1 and S1D2-LALA mutant (STI-1499-LALA) did not neutralize B.1.351, but did neutralize SARS-CoV-2 strains D614 and D614G. LALA mutations did not result in substantial differences in neutralizing abilities between clones S1D2-hIgG1 vs STI-1499-LALA. S1D2-hIgG1, STI-1499-LALA, and convalescent plasma showed minimal ability to induce ADE in human blood monocyte-derived macrophages. Further, no differences in pharmacokinetic clearance of S1D2-hIgG1 vs STI-1499-LALA were observed in mice expressing human FcRn. These findings confirm that SARS-CoV-2 has already escaped some mAbs, and identify a mAb candidate that may neutralize multiple SARS-CoV-2 variants. They also suggest that risk of ADE in macrophages may be low with SARS-CoV-2 D614, and LALA Fc change impacts neither viral neutralization nor Ab clearance.
Keywords: SARS-CoV-2, COVID-19, Antibody therapy, Antibody dependent enhancement
1. Introduction
In just over one year's time, COVID-19 has caused over 4 million deaths worldwide in the SARS-CoV-2 pandemic. At the time of this writing it is estimated that 41.5% of the world's population has received at least one dose of SARS-CoV-2 vaccine (Our World in Data), and the breadth and durability of immunity induced by the various vaccine products being deployed has yet to be determined. Concurrently, SARS-CoV-2 variants with enhanced infectivity have begun to emerge and already contributed to additional waves of the pandemic on multiple continents. While only one small molecule antiviral, an adenosine nucleotide triphosphate analog, has been approved by the US Food and Drug Administration for treatment of COVID-19 (McCreary and Angus, 2020), monoclonal antibodies (mAbs) appear to be emerging as specific, safe, and effective therapies against SARS-CoV-2 in the early stages of infection (Pallotta et al., 2021). Yet there is already evidence showing variant resistance to convalescent plasma (Hoffmann et al., 2021; Planas et al., 2021) and mAbs targeting the original pandemic SARS-CoV-2 strain D614 (Tada et al., 2021; Wang et al., 2021). Antibody-dependent enhancement (ADE) of macrophage infection has been demonstrated with other viruses including dengue virus (Fowler et al., 2018; Halstead et al., 2010), Zika virus (Brown et al., 2019; Dejnirattisai et al., 2016), and mutant feline enteric coronavirus (Olsen et al., 1992), and concerns about SARS-CoV-2 ADE were raised at the onset of the pandemic (Arvin et al., 2020) as public health agencies and pharmaceutical companies raced to trial vaccines and mAbs at unprecedented speeds.
In the present study we examined the neutralizing abilities and potential for ADE of three mAbs and convalescent plasma. In two of the clones (1741-LALA and S1D2 LALA mutant [STI-1499-LALA]), positions 234 and 235 have been engineered with leucine to alanine (LALA) mutations, which silence Fc binding to Fcγ receptors (FcγRs) and uptake by FcγR-expressing cells (Magnani et al., 2017). Phase 1 clinical safety studies have been carried out by Sorrento Therapeutics using STI-1499-LALA under the name COVI-GUARD™, and there were no safety concerns in normal volunteers at each COVI-GUARD dose tested (30 mg, 100 mg, and 200 mg) (clinicaltrials.gov). To examine the influence of FcγR binding on pharmacokinetic clearance, we compared STI-1499-LALA to its unsilenced twin S1D2-hIgG1 in vivo.
Using a human blood monocyte-derived macrophage model of ADE, we show that wild-type (WT) Fc mAbs, LALA Fc mAbs, and convalescent plasma show minimal ability to induce SARS-CoV-2 D614 ADE. Further, no differences in pharmacokinetic clearance of S1D2-hIgG1 vs STI-1499-LALA were observed in mice expressing human FcRn, the receptor responsible for the half-life of IgG (Roopenian and Akilesh, 2007). Our plaque reduction neutralization assays demonstrate that clone 1741-LALA neutralizes SARS-CoV-2 strains B.1.351, D614, and D614G. Clones STI-1499-LALA and S1D2-hIgG1 did not neutralize B.1.351, but did neutralize strains D614 and D614G.
2. Materials and methods
2.1. Viruses and cell lines
Vero E6 and Vero-CCL81 (ATCC, CCL-81) were used in SARS-CoV-2 propagation. Both Vero E6 and Vero-CCL81 were maintained in Dulbecco's Modification of Eagle's Medium (DMEM) (Corning) supplemented with 10% fetal bovine serum (FBS), 100 U/mL of penicillin–streptomycin, 10 mM HEPES buffer and 1% of non-essential amino acid. Three SARS-CoV-2 strains (D614, D614G, and B.1.351) were used in this study. SARS-CoV-2 strain D614 was kindly provided by Dr. Sumit Chanda, Sanford Burnham Prebys Medical Discovery Institute. SARS-CoV-2 strain D614G was obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate Germany/BavPat1/2020, NR-52370. SARS-CoV-2 strain B.1.351 was obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate hCoV-19/South Africa/KRISP-K005325/2020, NR-54009. SARS-CoV-2 strain D614 was propagated in Vero E6 cells; strains D614G and B.1.351K were propagated in Vero-CCL81 cells. Viruses were collected at 3 dpi and viral titers were determined by plaque assay on Vero E6 cells (Case et al., 2020). Briefly, viral supernatant was subjected to a 1:10 serial dilution prior to infection of confluent Vero E6 cells in 24 well plate for 2 h at 37 °C. The plates were gently rocked every 10–15 min within the incubation time. The inoculum was further removed and 0.8% low melting agarose medium or 1% carboxymethylcellulose medium was added to each well. After 3 days incubation, the cells were fixed with 10% formaldehyde in PBS for 1h at room temperature (RT), washed once with PBS before stained with 0.1% crystal violet solution in 20% methanol for 20 min at RT. All the assays were performed in a biosafety level 3 facility. All the viral preparations were deep-sequenced by the La Jolla Institute for Immunology Sequencing Core or the Chanda lab at Sanford Burnham Prebys Medical Discovery Institute.
2.2. Human cells
Human blood was collected from healthy blood donors under the La Jolla Institute for Immunology Protocol VD-057-0217. All donors were tested negative for HIV, hepatitis B, and hepatitis C virus. Peripheral blood mononuclear cells (PBMCs) were separated using Histopaque 1077. Monocytes were negatively selected with pan monocyte isolation kits (Miltenyi Biotec). Monocytes were plated in a 24-well plate at the concentration of 4 x 105 cells/400 μl/well with macrophage serum-free medium (Gibco) containing 1% penicillin/streptomycin, 1% Nutridoma-SP (Roche), and supplemented freshly with cytokines macrophage colony-stimulating factor (rhM-CSF, 100 ng/mL) (PeproTech). Cells were cultured at 37 °C with 5% CO2. The media was changed every 3–4 days for a period of 7 days to differentiate monocytes into monocyte-derived macrophages.
2.3. Mice and pharmacokinetic assay
Six to eight-week-old human FcRn-expressing mice (Ifnar1 −/− x Fcgrt −/ − x Tg32 hFcRn+/+ C57BL/6 mice) were bred and housed in a specific pathogen-free facility at the La Jolla Institute for Immunology. All procedures of the present study were in accordance with the guidelines set by the La Jolla Institute for Immunology Animal Care and Use Committee. For in vivo pharmacokinetics, mice were weighed 1d prior to mAb administration. S1D2-hIgG1 or STI-1499-LALA were prepared in PBS for 5 mg/kg per mice given in 0.1 mL of total volume retro-orbitally (n = 3–4 mice per time point). Two blood samples were harvested from each mouse, first collection via submandibular vein, and then terminal blood sample was harvested at 7 days or more after first blood draw. Serum samples were collected after 30 min, 1h, 8h, 7d, 14d, 21d and 28d of mAb administration to detect hIgG by ELISA (Bethyl Laboratories) according to manufacturer's instructions.
2.4. ELISA
Antibodies S1D2-hIgG1, STI-1499-LALA, or 1741-LALA were individually coated at 5 μg/mL in PBS onto wells of microtiter plates. After washing and blocking, Serially-diluted SARS-CoV-2 Spike S1 subunit antigens (3-fold serial dilution starting at concentration of 3 μg/mL) (2019-nCoV S1: ACRO, Cat No. SIN–C52H3, Spike S1 (B.1.1.7 lineage mut): SARS-CoV-2 (2019-nCoV) Spike S1 (HV69-70 deletion, Y144 deletion, N501Y, A570D, D614G, P681H)-His: Sino, Cat: 40,591-V08H12, Spike S1 (B.1.351 lineage mut): SARS-CoV-2 (2019-nCoV) Spike S1 (K417N, E484K, N501Y, D614G)-His: Sino, Cat: 40,591-V08H10) and Spike S1 (B.1.617.2 lineage mut): SARS-CoV-2 (2019-nCoV) Spike S1 (T19R, G142D, E156G, 157–158 deletion, L452R, T478K, D614G, P681R)-His: Sino, Cat: 40,591-V08H23) were added to the corresponding wells for a 1h RT incubation. The plate was washed three times and incubated with 1:5000 dilution of HRP-conjugated anti-HIS antibody. Color development was performed with 3,3′,5,5′-Tetramethylbenzidine (TMB). Absorbance was read at 450 nm.
2.5. Surface plasmon resonance
Kinetic interactions between the antibodies and His-tagged Spike S1 proteins were measured at 25 °C using BIAcore T200 surface plasmon resonance (SPR) (GE Healthcare). Anti-human fragment crystallizable region (Fc region) antibody was immobilized on a CM5 sensor chip to approximately 8000 resonance units (RU) using standard N hydroxysuccinimide/N Ethyl-N′-(3-dimethylaminopropyl) carbodiimide hydrochloride (NHS/EDC) coupling methodology. 1741-LALA (1–2 μg/mL) was captured for 60 s at a flow rate of 10 μL/min. The SARS-CoV-2 Spike S1, SARS-CoV-2 (2019-nCoV) Spike S1– B.1.1.7 lineage mut (HV69-70 deletion, Y144 deletion, N501Y, A570D, D614G, P681H)-His, SARS-CoV-2 (2019-nCoV) Spike S1– B.1.351 lineage mut (K417N, E484K, N501Y, D614G)-His and Spike S1– B.1.617.2 lineage mut (T19R, G142D, E156G, 157–158 deletion, L452R, T478K, D614G, P681R)-His proteins were prepared at six different dilutions (range 1.56 nM–50 nM) in a running buffer of 0.01 M HEPES pH 7.4, 0.15 M NaCl, 3 mM EDTA, 0.05% v/v Surfactant P20 (HBS EP+). Blank flow cells were used for correction of the binding response. All measurements were conducted in HBS-EP + buffer with a flow rate of 30 μl/min. A 1:1 binding model and was used to fit the data.
2.6. COVID-19 patient convalescent plasma and anti SARS-CoV-2 monoclonal antibodies (mAbs)
Anti-SARS-CoV-2 mAbs S1D2-hIgG1, S1D2-LALA mutant (STI-1499-LALA) and 1741-LALA were generated at Sorrento Therapeutics, Inc. These three mAbs were each expressed in Chinese hamster ovary (CHO) cells and purified by Protein A affinity chromatography. Plasma samples from convalescent COVID-19 patients were tested in this study. Two males and one female (aged 23 to 65-year-old), six to eight weeks from being tested positive for SARS-CoV-2 were enrolled from Scripps Clinic Bio-Repository & Bio-Informatics Core under La Jolla Institute for Immunology protocol IB-233-0820.
2.7. Plaque reduction neutralization assay (PRNT)
Vero E6 cells were plated at 8 x 104 cells/well in a 24-well plate one day prior to infection. COVID-19 patient serum or anti SARS-CoV-2 mAbs were serially diluted and then incubated with 30–40 plaque forming units (PFU) of virus for 1 h at 37 °C. The antibody/virus mixture was further transferred onto Vero E6 cells for 1 h at 37 °C. The infectious media was removed afterward and 1% carboxymethylcellulose medium was overlaid onto each well. After 3 days incubation, the cells were fixed with 10% formaldehyde for 1h at RT and then stained with 0.1% crystal violet solution. All the conditions were performed in duplication. Antibody titers (PRNT50) were defined as the highest serum dilution that reduce 50% of virus plaques.
2.8. Antibody dependent enhancement (ADE) assay
Serial diluted COVID-19 patient serum or anti SARS-CoV-2 mAb was incubated with SARS-CoV-2 D614 at multiplicities of infection (MOI) of 0.25 and 1 for 1 h at 37 °C. The antibody/virus mixture was further transferred onto 7 day-differentiated human monocyte-derived macrophages for 1 h at 37 °C. The infectious media was removed, and the cells were washed once with PBS before adding fresh pre-warmed complete macrophage medium. After 2 dpi, culture supernatant was collected for plaque assays to determine the level of infectious viral particles as described previously, and infected cells were lysed with RLT lysis buffer (Qiagen) supplemented freshly with 1% of 2-Mercaptoethanol (Gibco) for 1h at RT. RNA from lysed cells was isolated (Qiagen) and SARS-CoV-2 genomic RNA for E gene was detected by qRT-PCR.
2.9. Quantification and statistical analysis
Statistics and graphs were prepared using Prism software version 9.1.0 (GraphPad Software) and are expressed as the mean ± standard deviation (SD). Statistically differences between ADE groups were determined using one-way ANOVA. The pharmacokinetic difference of serum human IgG between S1D2-hIgG1 and LALA mutant was evaluated with non-parametric Mann–Whitney test. p < 0.05 was considered significant. Further details on statistical analysis are listed in the figure legends.
3. Results
3.1. Generation of LALA mAbs against SARS-CoV-2
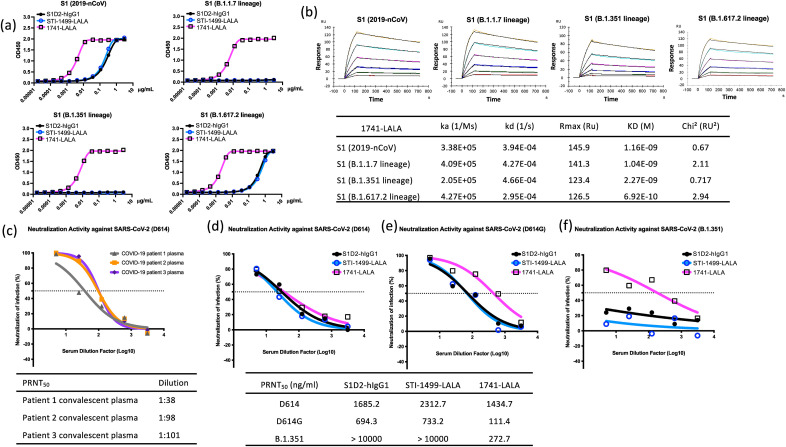
MAb clones S1D2-hIgG1, S1D2-LALA Fc (STI-1499-LALA) and 1741-LALA originated as a single chain variable fragment (scFv) isolated from the Sorrento G-MAB library, a phage-display library with an estimated coding complexity of approximately 1010 unique antibody sequences derived from over 600 healthy individuals, as screened (Cao et al., 2020). The library was panned using the Spike protein S1 fragment derived from the SARS-CoV-2/human/USA/WA-CDC-WA1/2020 (USA-WA1/2020, GenBank: MN985325.1). ScFv fragments with potent Spike S1 binding properties were further confirmed with the capacity to block interactions between Spike S1 and a recombinant ACE2 ectodomain by competition ELISA. Candidates are sequenced, reformatted with CHO–S cells to a full-length human IgG1 or a human IgG1 with LALA mutations in the hinge region and purified by affinity chromatography. Candidates were further evaluated in a USA-WA1/2020 virus neutralization assay and mAb clones S1D2-hIgG1, STI-1499-LALA, and 1741-LALA were selected as among the most potent neutralizing antibodies of the candidates isolated from the original library screen. Binding affinity of clones S1D2-hIgG1, STI-1499-LALA, and 1741-LALA to Spike S1 derived from 2019-nCoV was further confirmed by ELISA (Fig. 1 a). Clone S1D2-hIgG1, STI-1499-LALA also showed binding affinity to Spike S1 derived from B.1.617.2 lineage with 50% binding inhibition (IC50) at 411.2 and 561.1 ng/mL. Additionally, clone 1741-LALA revealed strong binding affinity to Spike S1 harboring SARS-CoV-2 B.1.1.7, B.1.351, and B.1.617.2 lineage mutation with IC50 at 5.211, 6.155, and 1.78 ng/mL, respectively (Fig. 1a). Furthermore, the binding kinetics of clone 1741-LALA against S1 fragment of 2019-nCoV, B.1.1.7-, B.1.351-, and B.1.617.2- lineage mutation determined by BiaCore surface plasmon resonance showed high binding affinity and low dissociation rate with equilibrium dissociation constants (KD) of 1.16 x 10−9, 1.04 x 10−9, 2.27 x 10−9, and 6.92 x 10−10 M, respectively (Fig. 1b).
Fig. 1.
Kinetic interaction between anti-SARS-CoV-2 Spike mAbs and variant spike protein, neutralization abilities of convalescent plasma and anti-SARS-CoV-2 spike mAbs against SARS-CoV-2 strains B.1.351, D614, and D614G. (a) Binding responses of anti-SARS-CoV-2 S protein mAbs, S1D2-hIgG1, STI-1499-LALA and 1741-LALA to the S1 fragment of the S protein from SARS-CoV-2 variants, including 2019-nCoV, B.1.1.7 (HV69-70 deletion, Y144 deletion, N501Y, A570D, D614G, P681H)-, B.1.351 (K417N, E484K, N501Y, D614G)-, and B.1.617.2 (T19R, G142D, E156G, 157–158 deletion, L452R, T478K, D614G, P681R)- lineage. (b) Panels indicate the association and dissociation of 1741-LALA at various concentrations binding to S1 of 2019-nCoV, B.1.1.7, B.1.351 and B.1.617.2 lineage. The table shows the association rate (ka), dissociation rate (kd), and dissociation constant (KD). Neutralization titers of (c) convalescent plasma from three different COVID-19 patients against SARS-CoV-2 D614 and PRNT50 curves of anti-SARS-CoV-2 spike mAbs against (d) D614 (e) D614G, and (f) B.1.351. Tables indicate the titers of each plasma sample or mAb concentrations in neutralizing 50% of input virus.
3.2. SARS-CoV-2 B.1.351 is neutralized by mAb 1741-LALA but not by other clones
Emerging evidence shows SARS-CoV-2 variant resistance to convalescent plasma (Hoffmann et al., 2021; Planas et al., 2021) and mAbs targeting the original pandemic SARS-CoV-2 strain D614 (Tada et al., 2021; Wang et al., 2021). To determine the neutralizing abilities of mAbs 1741-LALA, S1D2-hIgG1, and STI-1499-LALA, we used the 50% plaque reduction neutralization test (PRNT50) to assess the capacities of these mAbs to neutralize SARS-CoV-2 strains B.1.351, D614, and D614G. We compared the mAb neutralizing abilities to convalescent plasma samples from three COVID-19 patients who had previously tested positive for SARS-CoV-2. Plasma samples neutralized SARS-CoV-2 D614 as expected at dilutions ranging from 1:101–1:38 (Fig. 1c). MAbs S1D2-hIgG1 and STI-1499-LALA neutralized SARS-CoV-2 strains D614 and D614G but not B.1.351 (Fig. 1d and e). In contrast, 1741-LALA neutralized B.1.351 at a PRNT50 concentration of 272.7 ng/mL (Fig. 1f). MAb 11741-LALA also neutralized SARS-CoV-2 D614 and D614G at 1434.7 and 111.4 ng/mL, respectively (Fig. 1d and e). Thus among the three mAbs tested, mAb 1741-LALA demonstrated the broadest spectrum of neutralization at the lowest concentration.
LALA mutated antibodies are designed to prevent mAbs from binding FcγR and prevent adverse consequences such as ADE, while not impacting neutralizing ability (Magnani et al., 2017). For SARS-CoV-2 strain D614, the PRNT50 concentrations of S1D2-hIgG1 and STI-1499-LALA were comparable at 1685 ng/mL and 2312.7 ng/mL, respectively (Fig. 1d). For strain D614G the PRNT50 concentrations of S1D2-hIgG1 and STI-1499-LALA were also comparable at 694.3 ng/mL and 733.2 ng/mL, respectively (Fig. 1e). Thus no substantial difference in neutralizing ability was seen between the WT Ab and the LALA mutant.
3.3. Monocyte-derived macrophages do not show evidence of SARS-CoV-2 D614 antibody-dependent enhancement
At the onset of the pandemic, researchers cautioned against hasty vaccine and therapeutic antibody deployment (Arvin et al., 2020) due to the potential for ADE of infection, as has been documented with other viruses including dengue virus (Zellweger et al., 2010), Zika virus (Brown et al., 2019; Dejnirattisai et al., 2016), and mutant feline enteric coronavirus (Olsen et al., 1992). Briefly, ADE results when poorly neutralizing Abs or subneutralizing concentrations of neutralizing Abs bind virions and allow these immune-complexed virions to be uptaken into FcγR-expressing cells, where viral replication ensues (Ngono and Shresta, 2018). Because macrophages are recognized as being susceptible to ADE and have also been implicated in cytokine storm, we tested the mAbs in this study for SARS-CoV-2 D614 ADE in human blood monocyte-derived macrophages as previously described for Zika virus (Carlin et al., 2018).
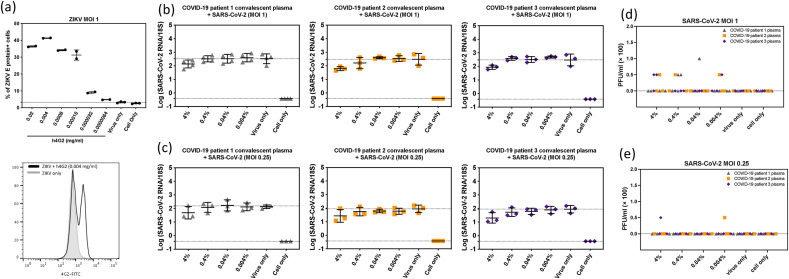
To assess the risk of ADE mediated by a diverse/polyclonal pool of Abs, we serially diluted convalescent plasma samples from 3 donors who had recovered from SARS-CoV-2 between March to May in 2020 and incubated these dilutions with SARS-CoV-2 D614 at multiplicities of infection (MOIs) of 0.25 and 1. These immune-complex solutions were then inoculated onto monocyte-derived macrophages from 3 to 4 donors. At 2 days post-infection, supernatants were evaluated by plaque forming assay on Vero E6 cells and infected cells were lysed and tested for viral genomic RNA for E gene by RT-qPCR (Corman et al., 2020). At all four plasma dilutions ranging from 0.004% to 4%, minimal increase in viral RNA (Fig. 2 b and c) and supernatant virus (Fig. 2d and e) was seen for any of the plasma-macrophage donor pairs. This contrasted with our control experiment in which a higher percentage of Zika virus E protein-expressing cells were detected in macrophage cultures incubated with pan-flavivirus humanized mAb 4G2 at concentrations of 0.000032 mg/mL and higher (Fig. 2a).
Fig. 2.
Antibody-dependent enhancement (ADE) effect of COVID-19 convalescent plasma in primary human monocyte derived macrophages. (a) Representative figure and histogram plot of intracellular ZIKV E protein showing ADE induced by humanized 4G2 antibody. This assay served as a positive control in these experiments. Intracellular viral E protein was increased in human macrophages when ZIKV was incubated with various concentrations of humanized 4G2 antibody prior to inoculation onto macrophages. ADE of SARS-CoV-2 was evaluated by changes in intracellular viral genomic RNA in macrophages and extracellular infectious particles released in culture supernatant. Viral RNA (b, c) and infectious viral particles (d, e) at 2 days post infection with SARS-CoV-2 pre-incubated with serial diluted COVID-19 patient convalescent plasma at MOI 1 (b, d) and MOI 0.25 (c, e). These data are shown as the mean with SD (n = 3–4). Each data point represents the average of a duplicate of each testing condition with individual donor macrophages. One-Way ANOVA was used to test the difference of viral titers between various concentration of patient plasma and virus only group.
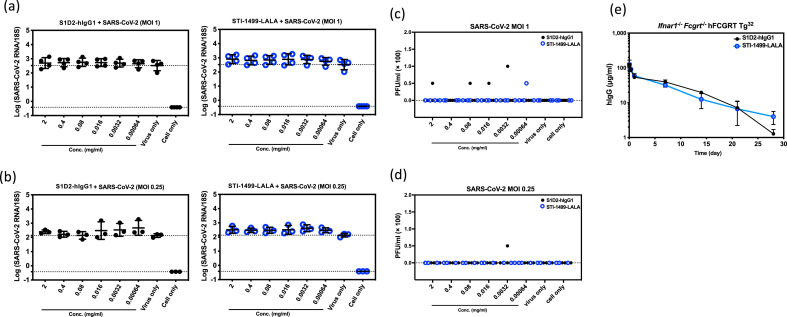
Next, to assess the risk of ADE mediated by the mAbs in this study, clones S1D2-hIgG1, and STI-1499-LALA were serially diluted from 2 to 0.00064 mg/mL and examined under the same incubations and macrophage culture inoculations. At 2 days post-infection, for all 6 mAb dilutions, limited increases in intracellular viral genomic RNA (Fig. 3 a and b) and infectious virus level in the supernatant (Fig. 3c and d) were seen for any of the mAb-macrophage donor pairs. Thus no evidence of SARS-CoV-2 D614 ADE was seen with WT Fc mAbs, LALA Fc mAbs, or convalescent plasma, and together these data suggest the risk of ADE in macrophages may be low with D614.
Fig. 3.
Negligible enhancement of SARS-CoV-2 infection with anti-SARS-CoV-2 spike mAbs on primary human monocyte derived macrophages. Serial diluted anti-SARS-CoV-2 spike mAbs S1D2-hIgG1 and STI-1499-LALA were incubated with SARS-CoV-2 at MOI 1 (a, c) or MOI 0.25 (b, d) for 1 h prior to inoculation onto macrophages. Viral RNA (a, b) and infectious viral particles (c, d) at 2 days post infection were evaluated. Data are presented as the mean with SD (n = 3–4). Each data point represents the average of a duplicate from each testing condition with macrophages derived from different donors. The differences of ADE conditions between various concentration of mAbs and virus only group were tested with one-way ANOVA. (e) Mean serum human IgG concentration-time profiles of anti-SARS-CoV-2 spike mAbs S1D2-hIgG1 and STI-1499-LALA in Ifnar1−/−Fcgrt−/− hFCGRT Tg32 mice. Each data point indicates a mean serum concentration from 3 to 4 mice. Error bars show the SD from mean. Mann-Whitney test was used to test the difference of serum hIgG concentration between S1D2-hIgG1 and STI-1499-LALA harvested at various time points.
3.4. No difference in mAb clearance between wildtype and LALA Fc in vivo
IgG plasma half-life is largely determined by binding affinity of the neonatal Fc receptor at acidic pH (FcRn; encoded by the FCGRT gene) to its binding site at an Ab's Fc region, and binding is then further modulated by pH (Roopenian and Akilesh, 2007). Previous antibody studies have indicated that LALA mutation does not affect the FcRn binding site (Schlothauer et al., 2016). To determine whether LALA mutation influences the pharmacokinetic clearance of anti-SARS-CoV-2 mAbs, we employed a humanized mouse which expresses human FcRn and is knocked out for murine FcRn. Murine FcRn binds human IgG with supraphysiologic affinity, resulting in prolonged clearance times and half-lives which are suboptimally representative of human pharmacokinetics (Roopenian and Akilesh, 2007). For use as a general virus susceptibility model, this mouse is also knocked out for the interferon-α/β receptor (Ifnar1). Mice were injected with a single 5 mg/kg dose of S1D2-hIgG1 or STI-1499-LALA. Sera were then harvested after 30 min, 1h, 8h, 1d, 7d, 14d, 21d and 28d to measure human IgG by ELISA. No significant difference was seen between the clearance curves of S1D2-hIgG1 vs STI-1499-LALA (Fig. 3e), indicating that the pharmacokinetics of anti-SARS-CoV-2 mAb S1D2 are not altered by LALA mutation.
4. Discussion
The COVID-19 pandemic remains a rapidly evolving situation for the pathogen, therapeutics, and vaccines. Emerging evidence showing variant resistance to convalescent plasma (Hoffmann et al., 2021; Planas et al., 2021) is particularly alarming as immune plasma would ideally represent a diverse, multitargeted, polyclonal defense against virus variants. The inabilities of mAbs S1D2-hIgG1 and STI-1499-LALA to neutralize SARS-CoV-2 B.1.351 in this study support studies showing variant resistance to convalescent plasma (Hoffmann et al., 2021; Planas et al., 2021) and mAbs (LY-CoV555 and Regeneron cocktails) currently in clinical trials targeting the original pandemic SARS-CoV-2 strain D614 (Hoffmann et al., 2021; Tada et al., 2021). LY-CoV555 (Jones et al., 2021) and Regeneron 10933 (Zhou et al., 2021) harbored neutralizing activity against original SARS-CoV-2 D614 strain at a NT50 concentration of 20 and 4 ng/mL, respectively, but neutralizing abilities were completely or markedly abolished against B.1.351 (Wang et al., 2021; Zhou et al., 2021). Further evidence shows receptor-binding domain (RBD) mutations E484K (Chen et al., 2021; Wang et al., 2021; Zhou et al., 2021) and combined K417N and N501Y mutations (Zhou et al., 2021) of SARS-CoV-2 B.1.351 and P.1 variants as vulnerabilities to multiple SARS-CoV-2 neutralizing mAbs engaging the RBD domain, and polyclonal Abs from convalescent sera or vaccine-induced immune sera. However, our demonstration of B.1.351 neutralization by clone 1741-LALA suggests that monoclonal therapy or single target vaccines against all SARS-CoV-2 strains may remain possible. SARS-CoV-2 is already proving to be a formidable immunologic foe, and in all likelihood a multimodal approach including humoral and cellular immunity will have to be employed to control the virus’ spread and clinical consequences.
Although macrophages are the archetype permissive cell for Ab-enhanced infection of viruses, only negligible evidence of SARS-CoV-2 D614 ADE was seen in our macrophage model with convalescent plasma and both WT hIgG and LALA mutant mAbs. These findings suggest that ADE may be less of a concern with SARS-CoV-2 D614 as compared to a virus such as dengue. Similarly, both positive- and negative-stranded RNA can be detected in SARS-CoV infected human monocyte-derived macrophages, but productive replication of the virus is aborted in the presence of anti-spike serum (Yip et al., 2014) or without serum (Cheung et al., 2005). Recent studies have also detected intracellular SARS-CoV-2 viral RNA (Grant et al., 2021) and Spike protein (Rendeiro et al., 2021) in patient alveolar macrophages, and suggested that viral replication in these macrophages is a key event in the pathogenesis of severe COVID. However, it remains unclear whether this RNA and S protein represent replicating vs phagocytosed virus. ADE is a multifactorial and dynamic process (Halstead et al., 2010), and one model cannot account for all the complexities in pathogen mutations, hosts, vaccines and therapies that could combine to produce ADE. Although our system demonstrated ADE in the Zika virus control, macrophages are highly plastic and heterogenous in phenotypes, and our cultured macrophages may not represent all characteristics of patient alveolar macrophages.
Previous studies have revealed LALA variants were able to abrogate ADE of dengue and Zika virus infection while retaining the same neutralizing abilities as unmodified parental mAbs (Beltramello et al., 2010; Khandia et al., 2018; Xu et al., 2017). However, Fc-effector function has been shown to decrease viral load by 6-14-fold in SARS-CoV-2 infected mouse lungs (Winkler et al., 2021). Finally, no differences in pharmacokinetic clearance of S1D2-hIgG1 vs STI-1499-LALA were observed in mice expressing human FcRn. These findings indicate that either WT Fc or LALA mutant Abs may be deployed as SARS-CoV-2 therapeutics without concern for altering clearance or resulting efficacy.
5. Conclusion
The rapidly spreading SARS-CoV-2 variants exhibit reduced susceptibility in vitro to immune sera and therapeutic mAbs. In this study, clone 1741-LALA showed a broad neutralizing activity against SARS-CoV-2 D614G and B.1.351 variants, suggesting this mAb may protect against other SARS-CoV-2 variants. In addition, our data showed that anti-SARS-CoV-2 mAbs and convalescent plasma from COVID-19 patients did not facilitate SARS-CoV-2 D614 infection in human blood monocyte-derived macrophages that readily promote Zika virus ADE, providing no evidence of SARS-CoV-2 D614 ADE.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by Sorrento Therapeutics, Inc. The following reagent, SARS-CoV-2 strain B.1.351, was obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate hCoV-19/South Africa/KRISP-K005325/2020, NR-54009, contributed by Alex Sigal and Tulio de Oliveira. The following reagent, SARS-CoV-2 strain D614G, was obtained through BEI Resources, NIAID, NIH: SARS-Related Coronavirus 2, Isolate Germany/BavPat1/2020, NR-52370.
References
- Arvin A.M., Fink K., Schmid M.A., Cathcart A., Spreafico R., Havenar-Daughton C., Lanzavecchia A., Corti D., Virgin H.W. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584:353–363. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- Beltramello M., Williams K.L., Simmons C.P., Macagno A., Simonelli L., Quyen N.T., Sukupolvi-Petty S., Navarro-Sanchez E., Young P.R., de Silva A.M., Rey F.A., Varani L., Whitehead S.S., Diamond M.S., Harris E., Lanzavecchia A., Sallusto F. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe. 2010;8:271–283. doi: 10.1016/j.chom.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.A., Singh G., Acklin J.A., Lee S., Duehr J.E., Chokola A.N., Frere J.J., Hoffman K.W., Foster G.A., Krysztof D., Cadagan R., Jacobs A.R., Stramer S.L., Krammer F., García-Sastre A., Lim J.K. Dengue virus immunity increases zika virus-induced damage during pregnancy. Immunity. 2019;50:751–762. doi: 10.1016/j.immuni.2019.01.005. e755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X., Maruyama J., Zhou H., Kerwin L., Sattler R., Manning J.T., Johnson S., Richards S., Li Y., Shen W., Blair B., Du N., Morais K., Lawrence K., Lu L., Pai C.-I., Li D., Brunswick M., Zhang Y., Ji H., Paessler S., Allen R.D. Discovery and development of human SARS-CoV-2 neutralizing antibodies using an unbiased phage display library approach. bioRxiv. 2020 doi: 10.1101/2020.09.27.316174. [DOI] [Google Scholar]
- Carlin A.F., Vizcarra E.A., Branche E., Viramontes K.M., Suarez-Amaran L., Ley K., Heinz S., Benner C., Shresta S., Glass C.K. Deconvolution of pro- and antiviral genomic responses in Zika virus-infected and bystander macrophages. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E9172–E9181. doi: 10.1073/pnas.1807690115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Bailey A.L., Kim A.S., Chen R.E., Diamond M.S. Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology. 2020;548:39–48. doi: 10.1016/j.virol.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., Gilchuk P., Zost S.J., Tahan S., Droit L., Turner J.S., Kim W., Schmitz A.J., Thapa M., Wang D., Boon A.C.M., Presti R.M., O'Halloran J.A., Kim A.H.J., Deepak P., Pinto D., Fremont D.H., Crowe J.E., Jr., Corti D., Virgin H.W., Ellebedy A.H., Shi P.Y., Diamond M.S. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C.Y., Poon L.L., Ng I.H., Luk W., Sia S.F., Wu M.H., Chan K.H., Yuen K.Y., Gordon S., Guan Y., Peiris J.S. Cytokine responses in severe acute respiratory syndrome coronavirus-infected macrophages in vitro: possible relevance to pathogenesis. J. Virol. 2005;79:7819–7826. doi: 10.1128/JVI.79.12.7819-7826.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brunink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejnirattisai W., Supasa P., Wongwiwat W., Rouvinski A., Barba-Spaeth G., Duangchinda T., Sakuntabhai A., Cao-Lormeau V.M., Malasit P., Rey F.A., Mongkolsapaya J., Screaton G.R. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with zika virus. Nat. Immunol. 2016;17:1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler A.M., Tang W.W., Young M.P., Mamidi A., Viramontes K.M., McCauley M.D., Carlin A.F., Schooley R.T., Swanstrom J., Baric R.S., Govero J., Diamond M.S., Shresta S. Maternally acquired zika antibodies enhance dengue disease severity in mice. Cell Host Microbe. 2018;24:743–750. doi: 10.1016/j.chom.2018.09.015. e745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant R.A., Morales-Nebreda L., Markov N.S., Swaminathan S., Querrey M., Guzman E.R., Abbott D.A., Donnelly H.K., Donayre A., Goldberg I.A., Klug Z.M., Borkowski N., Lu Z., Kihshen H., Politanska Y., Sichizya L., Kang M., Shilatifard A., Qi C., Lomasney J.W., Argento A.C., Kruser J.M., Malsin E.S., Pickens C.O., Smith S.B., Walter J.M., Pawlowski A.E., Schneider D., Nannapaneni P., Abdala-Valencia H., Bharat A., Gottardi C.J., Budinger G.R.S., Misharin A.V., Singer B.D., Wunderink R.G., Investigators N.S.S. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature. 2021;590:635–641. doi: 10.1038/s41586-020-03148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead S.B., Mahalingam S., Marovich M.A., Ubol S., Mosser D.M. Intrinsic antibody-dependent enhancement of microbial infection in macrophages: disease regulation by immune complexes. The Lancet. Infectious diseases. 2010;10:712–722. doi: 10.1016/S1473-3099(10)70166-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., Krüger N., Graichen L., Hofmann-Winkler H., Kempf A., Winkler M.S., Schulz S., Jäck H.M., Jahrsdörfer B., Schrezenmeier H., Müller M., Kleger A., Münch J., Pöhlmann S. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., Higgs R.E., Balasubramaniam D., Wang L., Zhang Y., Yang E.S., Bidshahri R., Kraft L., Hwang Y., Zentelis S., Jepson K.R., Goya R., Smith M.A., Collins D.W., Hinshaw S.J., Tycho S.A., Pellacani D., Xiang P., Muthuraman K., Sobhanifar S., Piper M.H., Triana F.J., Hendle J., Pustilnik A., Adams A.C., Berens S.J., Baric R.S., Martinez D.R., Cross R.W., Geisbert T.W., Borisevich V., Abiona O., Belli H.M., de Vries M., Mohamed A., Dittmann M., Samanovic M.I., Mulligan M.J., Goldsmith J.A., Hsieh C.L., Johnson N.V., Wrapp D., McLellan J.S., Barnhart B.C., Graham B.S., Mascola J.R., Hansen C.L., Falconer E. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci Transl Med. 2021;13 doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandia R., Munjal A., Dhama K., Karthik K., Tiwari R., Malik Y.S., Singh R.K., Chaicumpa W. Modulation of dengue/zika virus pathogenicity by antibody-dependent enhancement and strategies to protect against enhancement in zika virus infection. Front. Immunol. 2018;9:597. doi: 10.3389/fimmu.2018.00597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani D.M., Ricciardi M.J., Bailey V.K., Gutman M.J., Pedreño-Lopez N., Silveira C.G.T., Maxwell H.S., Domingues A., Gonzalez-Nieto L., Su Q., Newman R.M., Pack M., Martins M.A., Martinez-Navio J.M., Fuchs S.P., Rakasz E.G., Allen T.M., Whitehead S.S., Burton D.R., Gao G., Desrosiers R.C., Kallas E.G., Watkins D.I. Dengue virus evades AAV-mediated neutralizing antibody prophylaxis in rhesus monkeys. Molecular therapy. the journal of the American Society of Gene Therapy. 2017;25:2323–2331. doi: 10.1016/j.ymthe.2017.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCreary E.K., Angus D.C. Efficacy of remdesivir in COVID-19. Jama. 2020;324:1041–1042. doi: 10.1001/jama.2020.16337. [DOI] [PubMed] [Google Scholar]
- Ngono A.E., Shresta S. Immune response to dengue and zika. Annu. Rev. Immunol. 2018;36:279–308. doi: 10.1146/annurev-immunol-042617-053142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen C.W., Corapi W.V., Ngichabe C.K., Baines J.D., Scott F.W. Monoclonal antibodies to the spike protein of feline infectious peritonitis virus mediate antibody-dependent enhancement of infection of feline macrophages. J. Virol. 1992;66:956–965. doi: 10.1128/jvi.66.2.956-965.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Our World in Data. https://ourworldindata.org/covid-vaccinations?country=OWID_WRL.
- Pallotta A.M., Kim C., Gordon S.M., Kim A. Monoclonal antibodies for treating COVID-19. Cleve. Clin. J. Med. 2021 doi: 10.3949/ccjm.88a.ccc074. [DOI] [PubMed] [Google Scholar]
- Planas D., Bruel T., Grzelak L., Guivel-Benhassine F., Staropoli I., Porrot F., Planchais C., Buchrieser J., Rajah M.M., Bishop E., Albert M., Donati F., Prot M., Behillil S., Enouf V., Maquart M., Smati-Lafarge M., Varon E., Schortgen F., Yahyaoui L., Gonzalez M., De Seze J., Pere H., Veyer D., Seve A., Simon-Loriere E., Fafi-Kremer S., Stefic K., Mouquet H., Hocqueloux L., van der Werf S., Prazuck T., Schwartz O. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat. Med. 2021;27:917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- Rendeiro A.F., Ravichandran H., Bram Y., Chandar V., Kim J., Meydan C., Park J., Foox J., Hether T., Warren S., Kim Y., Reeves J., Salvatore S., Mason C.E., Swanson E.C., Borczuk A.C., Elemento O., Schwartz R.E. The spatial landscape of lung pathology during COVID-19 progression. Nature. 2021;593:564–569. doi: 10.1038/s41586-021-03475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roopenian D.C., Akilesh S. FcRn: the neonatal Fc receptor comes of age. Nature reviews. Immunology. 2007;7:715–725. doi: 10.1038/nri2155. [DOI] [PubMed] [Google Scholar]
- Schlothauer T., Herter S., Koller C.F., Grau-Richards S., Steinhart V., Spick C., Kubbies M., Klein C., Umaña P., Mössner E. Novel human IgG1 and IgG4 Fc-engineered antibodies with completely abolished immune effector functions. Protein engineering, design & selection. PEDS. 2016;29:457–466. doi: 10.1093/protein/gzw040. [DOI] [PubMed] [Google Scholar]
- Tada T., Dcosta B.M., Zhou H., Vaill A., Kazmierski W., Landau N.R. Decreased neutralization of SARS-CoV-2 global variants by therapeutic anti-spike protein monoclonal antibodies. bioRxiv. 2021 doi: 10.1101/2021.02.18.431897. [DOI] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S., Mascola J.R., Chang J.Y., Yin M.T., Sobieszczyk M., Kyratsous C.A., Shapiro L., Sheng Z., Huang Y., Ho D.D. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Winkler E.S., Gilchuk P., Yu J., Bailey A.L., Chen R.E., Chong Z., Zost S.J., Jang H., Huang Y., Allen J.D., Case J.B., Sutton R.E., Carnahan R.H., Darling T.L., Boon A.C.M., Mack M., Head R.D., Ross T.M., Crowe J.E., Jr., Diamond M.S. Human neutralizing antibodies against SARS-CoV-2 require intact Fc effector functions for optimal therapeutic protection. Cell. 2021;184:1804–1820. doi: 10.1016/j.cell.2021.02.026. e1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Zuest R., Velumani S., Tukijan F., Toh Y.X., Appanna R., Tan E.Y., Cerny D., MacAry P., Wang C.I., Fink K. A potent neutralizing antibody with therapeutic potential against all four serotypes of dengue virus. NPJ Vaccines. 2017;2:2. doi: 10.1038/s41541-016-0003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip M.S., Leung N.H., Cheung C.Y., Li P.H., Lee H.H., Daeron M., Peiris J.S., Bruzzone R., Jaume M. Antibody-dependent infection of human macrophages by severe acute respiratory syndrome coronavirus. Virol. J. 2014;11:82. doi: 10.1186/1743-422X-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellweger R.M., Prestwood T.R., Shresta S. Enhanced infection of liver sinusoidal endothelial cells in a mouse model of antibody-induced severe dengue disease. Cell Host Microbe. 2010;7:128–139. doi: 10.1016/j.chom.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Dejnirattisai W., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., Wang B., Paesen G.C., Lopez-Camacho C., Slon-Campos J., Hallis B., Coombes N., Bewley K., Charlton S., Walter T.S., Skelly D., Lumley S.F., Dold C., Levin R., Dong T., Pollard A.J., Knight J.C., Crook D., Lambe T., Clutterbuck E., Bibi S., Flaxman A., Bittaye M., Belij-Rammerstorfer S., Gilbert S., James W., Carroll M.W., Klenerman P., Barnes E., Dunachie S.J., Fry E.E., Mongkolsapaya J., Ren J., Stuart D.I., Screaton G.R. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361. doi: 10.1016/j.cell.2021.02.037. e2346. [DOI] [PMC free article] [PubMed] [Google Scholar]