Abstract
I performed metaviromic analysis of publicly available RNA-seq data from reptiles to understand the diversity of filoviruses (family Filoviridae). I identified a coding-complete sequence of a filovirus from the common lancehead (Bothrops atrox (Linnaeus, 1758)), tentatively named Tapajós virus (TAPV). Although the genome organization of TAPV is similar to mammalian filoviruses, our phylogenetic analysis showed that TAPV forms a cluster with a fish filovirus. However, TAPV is still distantly related to all the known filoviruses, suggesting that TAPV can be assigned as a species of a novel genus in Filoviridae. To our knowledge, this is the first report identifying a filovirus in reptiles, and thus contributes to a deeper understanding of the diversity and evolution of filoviruses.
Keywords: Bothrops atrox, common lancehead, Filoviridae, filovirus, virome
Filoviruses (family Filoviridae) are non-segmented negative-strand RNA viruses that belong to the order Mononegavirales [13]. Filoviruses were first discovered in 1967 as causative agents of hemorrhagic fever at laboratories in Germany and Yugoslavia [16]. In the following 50 years, several filoviruses, including Ebola viruses, have been detected. However, those filoviruses were detected in mammals exclusively [13]. In 2018, a large-scale viromic study identified distantly related filoviruses in fishes [9, 15]. Although the discovery of fish filoviruses expanded our knowledge on the diversity and evolution of filoviruses, there still exist large phylogenetic gaps among the fish and mammalian filoviruses, suggesting that filoviruses filling such phylogenetic gaps are present in vertebrates. Therefore, more extensive search for viruses would lead to the identification of novel filoviruses, leading to a better understanding of the diversity and evolution of filoviruses. In this study, I explored novel filoviruses using publicly available RNA-seq data obtained from reptiles.
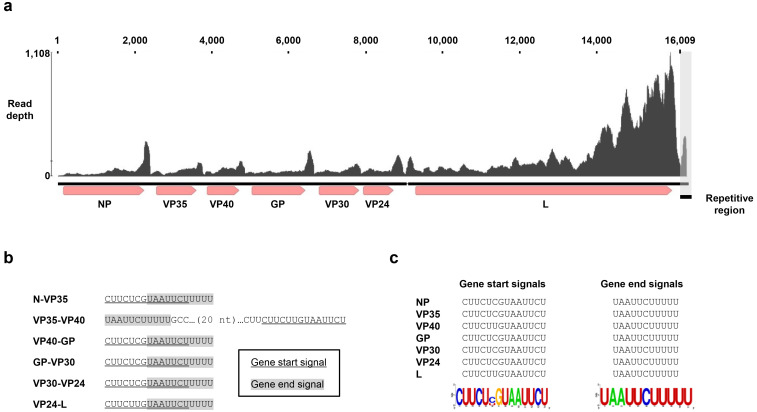
I first searched for filovirus-like sequences in the contigs that we previously assembled from paired end RNA-seq data obtained from reptiles [10]. Using BLASTx searches [4], I detected a 16,388-nucleotides (nt) long filovirus-like contig assembled from reads in SRR1953011, showing 39.87% amino acid identity to the L protein of Lloviu virus (LLOV; Filoviridae: Cuevavirus; YP_004928143). This RNA-seq data were obtained from the venom gland of a common lancehead (Bothrops atrox (Linnaeus, 1758)) sampled at Tapajós National Forest, Brazil [8]. To validate the quality of the contig, I mapped the original short reads to the contig. The contig sequence was well covered with short reads (Fig. 1a). The end of the filovirus-like contig contained an approximately 330-nt simple repeat-like sequence (the sequence is available in Supplementary information 1). The entire contig, except for the first two nucleotides, was covered with at least five reads of high quality, meeting the minimum quality for viral contigs previously suggested [14]. Nonetheless, I could not exclude the possibility that the repetitive sequence is derived from artifacts (data not shown). Therefore, I deleted the repetitive sequence present downstream of the gene end (GE) signal of the L gene (see below). The resultant viral contig consisted of 16,009 nt (Accession number BR001752). I named this virus Tapajós virus (TAPV). It should be noted that I cannot exclude the possibility that the contig contains nucleotides that have undergone post-transcriptional modifications, such as RNA-editing.
Fig. 1.
Genome organization of Tapajós virus (TAPV). (a) A schematic diagram of the TAPV genome and mapped reads. ORFs and gene start (GS) and end (GE) signals are shown. The upper graph indicates the mapped reads in SRR1953011. The gray box indicates the repetitive region observed in the contig (see text). (b and c) GS and GE signals for each gene of TAPV are shown. Sequence logos were based on the alignment of transcription initiation or termination signals by WebLogo [7]. Note that all the signals except for those between VP35 and VP40 are overlapping.
I next performed gene annotation for the TAPV contig. I searched for open reading frames (ORFs) more than 300 nt, identifying seven ORFs on the contig (Fig. 1a). I performed BLASTp searches against the filoviral sequences (taxid:11266) in the NCBI nr database [6] using the deduced amino acid sequences of identified ORFs as queries (Supplementary Table 1). Based on the results of BLASTp, I revealed that the seven ORFs correspond to the canonical filoviral genes, NP, VP35, VP40, GP, VP30, VP24, or L (Fig. 1a) [13]. Similar to marburgviruses and dianloviruses, the GP gene of TAPV contains a single ORF.
I further analyzed gene start (GS) and end (GE) signals on the contig. I extracted the nucleotide sequences of inter ORF regions and analyzed them by MEME [1]. The MEME searches identified putative GS and GE signals for each of the genes. The putative GS and GE signals are CUUCU (C/U) GUAAUUCU and UAAUUCUUUUU, respectively (Fig. 1b and c). As reported in other filoviruses, the first nucleotide of the TAPV GS signals is a cytidine residue. The 12-nucleotide stretch, which is well conserved in the GS signals of most of filoviruses, is also well conserved in the TAPV GS signals. The GE signals of TAPV are almost identical to those of other filoviruses. All the GS and GE signals except for those between the VP35 and VP40 genes are overlapping (Figs. 1b and 2).
Fig. 2.
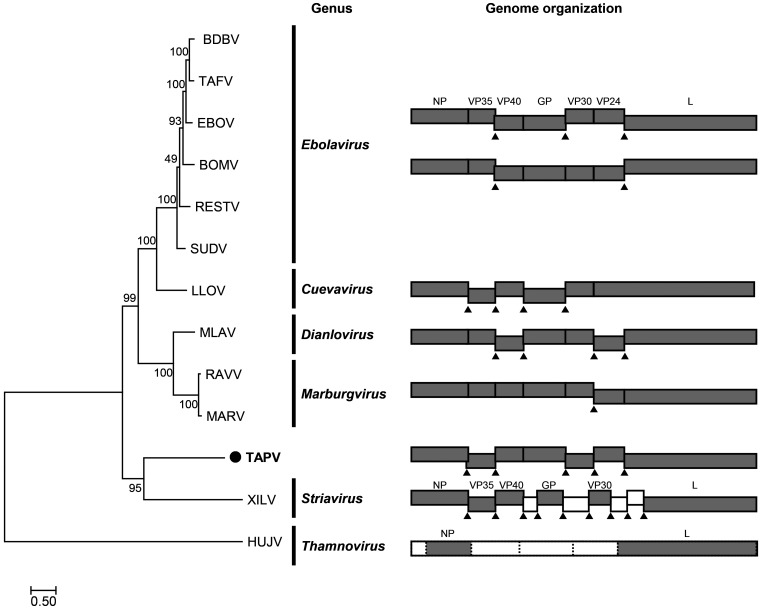
Phylogenetic relationship and comparison of gene structures of filoviruses. A phylogenetic tree was constructed by the maximum likelihood method using the deduced amino acid sequences of the L proteins of filoviruses. The scale bar indicates the number of amino acid substitutions per site. Tapajós virus (TAPV) is indicated with the black circle. The right panel shows the genome organizations of filoviruses. Each gene is indicated by a box. Gray boxes are the genes that show homology to known filoviral genes. White boxes are genes for which similarity to known filoviral genes could not be detected. Triangles show gene overlapping regions. Unclear gene start and end signals are depicted by the dashed lines.
To infer the evolutionary relationship between the TAPV and other filoviruses, I performed phylogenetic analysis using the sequences of L genes. I aligned the deduced amino acid sequences of L genes by MAFFT 7.455 using the L-INS-i algorithm [11], trimmed the ambiguously aligned regions by trimAl [5], and then constructed a phylogenetic tree by the maximum likelihood method with the resultant multiple alignments (available in Supplementary information 2) using RAxML-NG 0.9.0 [12]. The tree showed that TAPV formed a strongly supported cluster with Xīlǎng virus (XILV; Filoviridae: Striavirus) (Fig. 2). However, TAPV is still distantly related to XILV, and the gene organization of TAPV shares higher similarity with mammalian filoviruses than XILV (Fig. 2). These data suggest that TAPV should be assigned to a novel genus in the Filoviridae. To assess this possibility, I performed a pairwise sequence comparison (PASC) analysis [2] with known filovirus sequences. The highest identity to the TAPV contig was Bombali virus (BOMV; Filoviridae: Ebolavirus), showing 28.66% nucleotide identity (Supplementary Fig. 1). This suggests that TAPV can be a type virus of a novel genus in the Filoviridae family.
Finally, I analyzed whether TAPV is also detectable in other SRA data to understand the prevalence, distribution, and target tissues for the virus. I mapped short reads in 3480 RNA-seq data obtained from reptiles to the TAPV contig. I found that eight SRAs, except for the SRA in which TAPV was initially identified, contained reads mappable to the TAPV genome (Table 1). Especially, one of the eight SRAs (SRR1953010) contained many reads mappable to the TAPV sequence, reaching 669 reads per million (RPM). However, the detailed metadata are unavailable, and thus it is not clear whether SRR1953010 was obtained from a different individual or not. Further analyses are needed to understand the prevalence of TAPV in the animal population.
Table 1. Detection of sequence reads mapped to Tapajós virus.
| SRA accession | Reads per million mapped reads |
|---|---|
| SRR1953002 | 0.02 |
| SRR1953003 | 0.03 |
| SRR1953004 | 0.04 |
| SRR1953007 | 0.05 |
| SRR1953008 | 0.04 |
| SRR1953010 | 669.50 |
| SRR1953012 | 0.90 |
| SRR1953013 | 0.17 |
In this study, I identified a novel filovirus, TAPV, in the common lancehead. To the best of our knowledge, this is the first report showing the presence of a reptile filovirus. I further showed that TAPV could partially fill the phylogenetic gaps of filoviruses (Fig. 2). Based on the genus criteria of the family Filoviridae, where the genome sequences of filoviruses of different genera differ by more than 55% [3], I propose that TAPV can be a type virus of a novel genus in the family Filoviridae. Therefore, this study expands our knowledge of the diversity of filoviruses. The presence of diverse filoviruses in vertebrates, as shown here and in other studies, suggests that novel unidentified filoviruses still exist in vertebrate animals. Further studies are needed to understand the diversity and evolution of filoviruses.
CONFLICTS OF INTEREST
The author declares that they have no conflicts of interest.
Supplementary Materials
Acknowledgments
The supercomputing resources were provided by Human Genome Center, the Institute of Medical Science, the University of Tokyo and the NIG supercomputer at ROIS National Institute of Genetics. This study was supported by the Hakubi project at Kyoto University; Grant-in-Aid for Scientific Research on Innovative Areas from the Ministry of Education, Culture, Science, Sports, and Technology (MEXT) of Japan, Grant Numbers JP17H05821, JP19H04833.
REFERENCES
- 1.Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., Ren J., Li W. W., Noble W. S.2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37: W202-8. doi: 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao Y., Chetvernin V., Tatusova T.2014. Improvements to pairwise sequence comparison (PASC): a genome-based web tool for virus classification. Arch. Virol. 159: 3293–3304. doi: 10.1007/s00705-014-2197-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bào Y., Amarasinghe G. K., Basler C. F., Bavari S., Bukreyev A., Chandran K., Dolnik O., Dye J. M., Ebihara H., Formenty P., Hewson R., Kobinger G. P., Leroy E. M., Mühlberger E., Netesov S. V., Patterson J. L., Paweska J. T., Smither S. J., Takada A., Towner J. S., Volchkov V. E., Wahl-Jensen V., Kuhn J. H.2017. Implementation of objective PASC-derived taxon demarcation criteria for official classification of filoviruses. Viruses 9: 9. doi: 10.3390/v9050106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T. L.2009. BLAST+: architecture and applications. BMC Bioinformatics 10: 421. doi: 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T.2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. doi: 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coordinators N. R.NCBI Resource Coordinators. 2018. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 46 D1: D8–D13. doi: 10.1093/nar/gkx1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E.2004. WebLogo: a sequence logo generator. Genome Res. 14: 1188–1190. doi: 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freitas-de-Sousa L. A., Amazonas D. R., Sousa L. F., Sant’Anna S. S., Nishiyama M. Y., Jr., Serrano S. M., Junqueira-de-Azevedo I. L., Chalkidis H. M., Moura-da-Silva A. M., Mourão R. H.2015. Comparison of venoms from wild and long-term captive Bothrops atrox snakes and characterization of Batroxrhagin, the predominant class PIII metalloproteinase from the venom of this species. Biochimie 118: 60–70. doi: 10.1016/j.biochi.2015.08.006 [DOI] [PubMed] [Google Scholar]
- 9.Geoghegan J. L., Di Giallonardo F., Wille M., Ortiz-Baez A. S., Costa V. A., Ghaly T., Mifsud J. C. O., Turnbull O. M. H., Bellwood D. R., Williamson J. E., Holmes E. C.2021. Virome composition in marine fish revealed by meta-transcriptomics. Virus Evol. 7: veab005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horie M., Akashi H., Kawata M., Tomonaga K.2020. Identification of a reptile lyssavirus in Anolis allogus provided novel insights into lyssavirus evolution. Virus Genes. 57: 40–49. [DOI] [PubMed] [Google Scholar]
- 11.Katoh K., Standley D. M.2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30: 772–780. doi: 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozlov A. M., Darriba D., Flouri T., Morel B., Stamatakis A.2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35: 4453–4455. doi: 10.1093/bioinformatics/btz305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhn J., Amarasinghe G., Perry D., Howley P., Knipe D., Whelan S.2020. Fields virology: emerging viruses. pp. 449–503. In: Wolters Kluwer, Lippincott Williams & Wilkins, Philadelphia. [Google Scholar]
- 14.Ladner J. T., Beitzel B., Chain P. S., Davenport M. G., Donaldson E. F., Frieman M., Kugelman J. R., Kuhn J. H., O’Rear J., Sabeti P. C., Wentworth D. E., Wiley M. R., Yu G. Y., Sozhamannan S., Bradburne C., Palacios G., Threat Characterization Consortium. 2014. Standards for sequencing viral genomes in the era of high-throughput sequencing. MBio 5: e01360–e14. doi: 10.1128/mBio.01360-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi M., Lin X. D., Chen X., Tian J. H., Chen L. J., Li K., Wang W., Eden J. S., Shen J. J., Liu L., Holmes E. C., Zhang Y. Z.2018. The evolutionary history of vertebrate RNA viruses. Nature 556: 197–202. doi: 10.1038/s41586-018-0012-7 [DOI] [PubMed] [Google Scholar]
- 16.Siegert R., Shu H. L., Slenczka W., Peters D., Müller G.1967. On the etiology of an unknown human infection originating from monkeys. Dtsch. Med. Wochenschr. 92: 2341–2343. (in German) doi: 10.1055/s-0028-1106144 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.