Abstract
Polyploidy generally provides an advantage in phenotypic variation and growth vigor. However, the underlying mechanisms remain poorly understood. The tetraploid Liriodendron sino-americanum (Liriodendron × sinoamericanum P.C Yieh ex C.B. Shang & Zhang R.Wang) exhibits altered morphology compared with its diploid counterpart, including larger, thicker and deeper green leaves, bigger stomata, thicker stems and increased tree height. Such characteristics can be useful in ornamental and industrial applications. To elucidate the molecular mechanisms behind this variation, we performed a comparative transcriptome and proteome analysis. Our transcriptome data indicated that some photosynthesis genes and pathways were differentially altered and enriched in tetraploid L. sino-americanum, mainly related to F-type ATPase, the cytochrome b6/f complex, photosynthetic electron transport, the light harvesting chlorophyll protein complexes, and photosystem I and II. Most of the differentially expressed proteins we could identify are also involved in photosynthesis. Our physiological results showed that tetraploids have an enhanced photosynthetic capacity, concomitant with great levels of sugar and starch in leaves. This suggests that tetraploid L. sino-americanum might experience comprehensive transcriptome reprogramming of genes related to photosynthesis. This study has especially emphasized molecular changes involved in photosynthesis that accompany polyploidy, and provides a possible explanation for the altered phenotype of polyploidy plants in comparison with their diploid form.
Keywords: Liriodendron sino-americanum, photosynthetic, polyploid, proteomics, transcriptomics
Introduction
Many plant species have experienced, and are currently still undergoing, genome duplication, leading to an increase in chromosome number (Vyas et al. 2007). Polyploidy can be induced through two pathways: autoploidy, resulting from doubling a single genome (Cameron 1990), and alloploidy, resulting from chromosome doubling after interspecific hybridization or through unreduced gametes (Paterson 2005, Koehler et al. 2010). It has been estimated that only 15% of angiosperms have undergone polyploidization in the past (Eng and Ho 2019), which suggests that the extent of naturally occurring polyploidy is limited. In addition to natural polyploidy, polyploidy can be achieved through artificial induction using colchicine, trifluralin, pronamide and oryzalin (Chu et al. 2014). In addition, environmental factors such as wounding, water and temperature may also induce undiminished gametes (Ramsey and Schemske 1998).
Polyploids often exhibit increased growth vigor (Wendel 2000), and this remarkable superiority is mainly the result of larger plant morphology, higher growth rates, increased resistance to stress and an elevated metabolism in comparison with diploids—all traits that have been pursued by many plant breeders (Rubuluza et al. 2007, Yang et al. 2007, Q. Zhang et al. 2010, Zhang et al. 2019, Kim et al. 2014). Another often-seen consequence of polyploidy is a reduction in fertility, which can lead to seedless fruits, such as grapes (Vitis vinifera L.) (Yang et al. 2007) and watermelon (Citrullus lanatus (Thunb.) Matsum. et Nakai) (Miguel et al. 2001). Tetraploids can produce seeds and offspring, but pollen is not as effective as diploids, resulting in reduced fertility. But for triploid watermelons, which are grown from a seed of a cross between a natural diploid watermelon and a mutated tetraploid watermelon, the triploid plants cannot form normal germ cells because of the disordered chromosomal association during meiosis. The main reason is that plants need to undergo meiosis when they form gametes, and there will be syngamy during meiosis. The more the chromosome number, the more likely the syngamy will be disordered, and the more difficult the formation of gametes will be. It is worth mentioning that polyploidy is widely used in crops. For example, the fruits of decaploid strawberries (Fragaria×ananassa Duch.) become larger and more nutritious, and the plants have a higher resistance to cold (Luo et al. 2019). In muskmelon (Cucumis melo L.), the vegetative organs and fruits of a tetraploid plant increase in size, with the latter becoming sweeter as well (W. Zhang et al. 2010). Tetraploid hybrid rice (indica × japonica), a major food crop, can overcome the obstacle of low fertility and displays high heterosis (Guo et al. 2017). Thus, the application of polyploidy allowed the development of adapted cultivars and increased overall productivity for plant breeders (Haider 2013).
Among the many alterations caused by polyploidization, increased vegetative growth, which is closely related to increased effectiveness of photosynthetic capacity, has the potential to increase plant biomass (Warner et al. 1987, Vyas et al. 2007, Greer et al. 2017, Robinson et al. 2018). Photosynthesis rates are often influenced by leaf area, fresh weight and chlorophyll content (Warner et al. 1987, Warner and Edwards 1993, Dreyer et al. 2001). For example, in triploid poplar (Populus pseudosimonii × P. nigra) and tetraploid lemon (Citrus volkameriana Tan. and Pasq.), photosynthesis is increased, leading to higher accumulation of photosynthetic products (Du et al. 2019, Khalid et al. 2020). This indicates that the induction of polyploid plants should be a promising breeding approach for biomass production. Collectively, even though the enhanced photosynthesis capacity and biomass accumulation of polyploid plants have been recognized, many aspects of their fundamental molecular mechanisms underlying these traits remain unclear. In particular, it is worth mentioning that few studies have combined both physiological and molecular analysis to gain mechanistic insight.
Liriodendron is the only genus in the Magnoliaceae subfamily. At present, there are only two wild species: one naturally occurring in eastern Asia, namely, Liriodendron chinense (Hemsl.) Sargent, and the other occurring in north-eastern America, namely, the Liriodendron tulipifera Linn. Interbreeding is possible between the two species, and in the 1970s, Professor Peizhong Ye of Nanjing Forestry University successfully crossbred the two, giving rise to the hybrid Liriodendron sino-americanum. The growth and resistance of L. sino-americanum is significantly improved compared with both parents due to heterosis. The remarkable characteristics of this species include a straight and aesthetically pleasing tree shape, peculiar shaped leaves and tulip-like flowers (Zhou et al. 2016). It is one of the most important landscaping and industrial timber species (Qi and Rong 2000, Yao et al. 2016, Cheng and Li 2018, Chen et al. 2019). In addition to its ornamental value, it also provides key resources for furniture, paper, pulp and biomaterials. As a result of many years of research, great progress has been made in somatic embryogenesis and genetic improvement of this species (Li et al. 2012, Chen et al. 2013, 2019, Xia et al. 2013, Tan et al. 2015, Zhen et al. 2015). However, it is necessary to create new germplasm resources for this tree species. In L. sino-americanum, there have been no reports on the existence of natural polyploidy. We obtained a small number of tetraploid L. sino-americanum plants, which arose due to somatic variation, that showed significant phenotypic changes. To better understand the molecular process of the phenotypic differences between diploids and tetraploids, in the present study, we performed RNA-seq transcriptomic analysis coupled with a comparative proteomic analysis using iTRAQ of diploid and tetraploid mature leaves. Our analysis represents a vital step toward the clarification of gene and protein regulatory networks that underlie the polyploid phenotype and will contribute to a better understanding of L. sino-americanum polyploidy.
Materials and methods
Plant materials
All plants used in this study were from the same initial origin and regenerated by somatic embryogenesis. After 2 years in the greenhouse, we planted all plants in the field under the same conditions. According to our continuous observation, some regenerated plants showed significant phenotypic changes that were very similar to those found in polyploid plants. Therefore, after five growing seasons in the field, we performed a series of measurements. Currently, we have a total of 51 tetraploids.
Ploidy analysis
Young leaves (1--2 cm2) were finely chopped with a sharp razor blade into a plastic Petri dish containing 500-μl extraction buffer. After 30--60 s of incubation, 2-ml staining buffer was added. Samples were then passed through a 50-μm nylon filter. After incubation at room temperature for at least 30 min, the filtered solution with stained nuclei was analyzed using a flow cytometer (FCM, Sysmex, CyFlow Cube 6, Germany) (Dolezel et al. 2003), using diploids as a control. Bud tip meristems of both diploid and tetraploid plants were used for chromosome counting (Banyai et al. 2010) and chromosomes were observed using an optical microscope (ZEISS Axio Vert.A1, Oberkochen, Germany) at ×63 magnification. All experiments were carried out no less than three times.
Morphological and microscopic observation
Tree height and ground diameter were measured consecutively for 2 years after growth ceased in the autumn. Tree height was measured from the bottom to the highest of the canopy. The ground diameter (the trunk diameter at ground level) was measured using a Vernier caliper. The stem volume was estimated roughly according the formula (tree height × ground diameter2). Bud size from five diploids and five tetraploids was compared, using at least five replicates per plant. Leaf area was measured using software Win FOLIA. For examination of the leaf epidermis, adaxial epidermal cells were peeled off using nail enamel, then placed on a glass slide for examination and photographed with a light microscope (Zhou et al. 2017). The stomatal density, guard cell length and width were determined following a previously published method (Majdi et al. 2010). Morphological characteristics of the stomata were photographed using an optical microscope (ZEISS Axio Vert.A1). Experiments were conducted in triplicates (at least five views per replicate).
Paraffin sections were made as previously described (Chen et al. 2015), with a minor modification. The leaf materials were fixed for 24 h in formalin-acetoalcohol (FAA; a 1:1:18 ratio of formaldehyde, glacial acetic acid and ethanol by volume) and stored in 70% EtOH. The specimens were then dehydrated using a graded ethanol series (70, 85, 95, 100%). The ethanol was gradually replaced with xylene, and specimens were infiltrated with paraffin at 60 °C. Samples were embedded in paraffin, serially sectioned at 8 μm, then stained with a safranin and fast green counterstain. Slides were mounted with synthetic resin and photographed with an optical microscope (ZEISS Axio Vert.A1).
Transcriptome sequencing and analysis
For transcriptome analysis, three diploid and three tetraploid plants were chosen, from which we collected expanded mature leaf samples at the middle position carefully, after which they were immediately frozen in liquid nitrogen and stored at −80 °C until use. Additionally, we selected a diploid and tetraploid plant from which we collected the expanded mature leaves at three positions (see Top, Middle, Basal, Figure 2A) for the assay.
Figure 2.
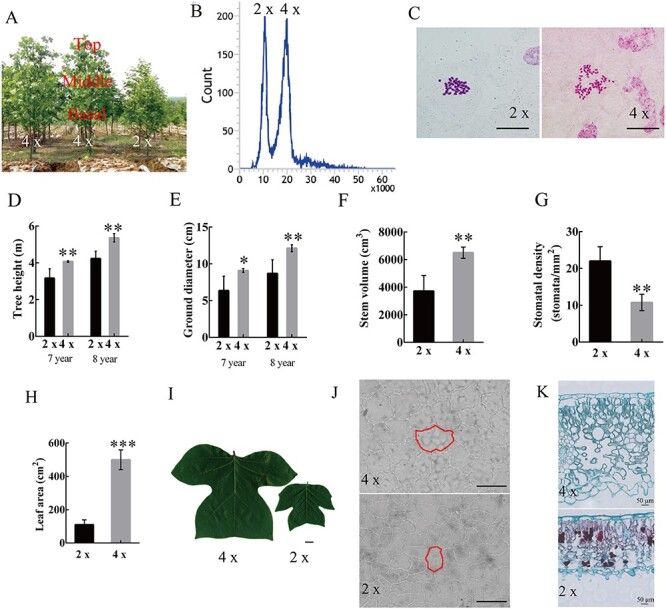
Ploidy analysis and morphological comparison between tetraploids and diploids. (A) Plant morphology and sample position, (B) ploidy level of diploid and tetraploid plants, as determined by FCM (flow cytometry), (C) ploidy level of diploid and tetraploid plants, as determined by chromosome counting, bar =10 μm, (D) tree height (n = 5), (E) ground diameter (n = 5), (F) stem volume (n = 5), (G) stomatal density, (H) leaf area (n = 15), (I) leaf morphology, bar = 1 cm, (J) light microscope micrographs of adaxial epidermal cells in diploid and tetraploid leaves with the red circle representing a single epidermal cell, bar =100 μm, (K) light microscope micrographs depicting cross sections of diploid and tetraploid leaves. Error bars indicate SE. Asterisks indicate significant differences between diploid and tetraploid (**P < 0.01, ***P < 0.001). 4×: tetraploid plants; 2×: diploid plants.
RNA was isolated using the CTAB method and quality-checked using Agilent. The extracted RNA sample was treated with DNase to remove contaminating DNA. Full-length transcripts and a quantitative analysis of gene expression were obtained using PacBio and HiSeq X-Ten, which were both performed at BGI China. The raw data were first processed using the PacBio official software package SMRTlink 5.0, then the full-length sequence was sorted. Clustering of full-length transcripts and PacBio calibration were performed using SMRTlink 5.0. The Illumina data were corrected using proofread (V2.12). Representative sequences were obtained from corrected transcripts by cd-hit-est (V4.7) and used for gene differential expression analysis. Trimmomatic (V0.36) was used to remove adapters from Illumina sequencing data, which was then aligned to transcript sequences, using Rsem (V1.3.0), to obtain the number of reads per transcript (Unigene). Gene differential expression analysis was performed using DEseq2 (V 3.6). The significance threshold was set at |Log2 FC| > 2 and Q-value <0.01. Gene Ontology (GO) functional classifications were performed using R scripts. Functional enrichment of signaling and metabolic pathways data was obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG;http://www.genome.jp/kegg/pathway.html). The heat map was displayed using the software Heml.
Protein identification and quantitation
Protein was extracted from six samples (the same samples that were used for RNA-seq of the middle leaf position). The protein concentration was determined according to the Bradford method (Hammond and Kruger 1988); 30-μg protein solution was taken for each sample, mixed with loading buffer, after which protein purity was determined by SDS-PAGE. For protein digestion, 100-μg protein solution was taken from each sample. iTRAQ labeling and mass spectrometry analysis were performed at BGI China. Raw MS/MS data were converted into MGF format using the thermo scientific tool Proteome Discoverer, and exported MGF files were searched using Mascot (v2.3.02) against our RNA-seq transcriptomic database. Quantitative analysis was performed using IQuant (Bo et al. 2015) software. The results were filtered at 1% false discovery rate (FDR) at the peptide level, with each identified protein having a minimum of one unique peptide. These data were used to obtain mass spectra and a peptide list of identified proteins.
Relation between transcriptome and proteome expression
To determine how consistent the transcriptomic and proteomic changes are between diploid and tetraploid L. sino-americanum, Venny (2.1) (https://bioinfogp.cnb.csic.es/tools/venny/index.html) was used to compare genes identified from our expression and functional enrichment analysis, as well as differentially expressed proteins (DEPs). First, the differentially expressed genes (DEGs) were classified into up-regulated and down-regulated groups, as were the DEPs. It was identified that the differentially expressed gene (protein) sets of transcriptome and proteome were not completely consistent, many genes differentially expressed in the transcriptome but not in the proteome. Similarly, many DEPs were not matched with DEGs. Then, the DEGs and DEPs were sub-divided into four combinations: (i) increased transcript level induces protein expression down-regulation (rUP-pDN), (ii) increased transcript level induces protein expression up-regulation (rUP-pUP), (iii) decreased transcript level induces protein expression up-regulation (rDN-pUP) and (iv) decreased transcript level induces protein expression down-regulation (rDN-pDN). In comparison data, all numbers were represented by an intersection rather than a union. Finally, we focused on DEG/DEP pairs with correlated expression changes (rUP-pUP and rDN-pDN) between diploids and tetraploids and classified these genes by performing KEGG enrichment to explore their functions.
Physiological measurements
We collected fully expanded mature leaves, cutting 1 cm2 of the leaf tissue into small pieces. Leaf filaments were then placed in a tube containing 5 ml of 80% acetone and extracted in the dark until the filaments turned white completely. The tube was shaken three to five times during the extraction process, and the extraction time was about 15 h. The solution was then gently poured into a cuvette and read at 663 nm and 645 nm using a UV-4802H spectrophotometer. The absorbance of 80% acetone was measured in the same cuvette as a reference for the test substance measurements.
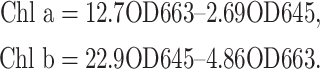 |
The leaves used for our photosynthetic parameter analysis grew on a similar location on the tree with comparable sun exposure; we chose leaves both on the sunny side and in the middle of the tree, taking leaves of identical maturity. Photosynthetic parameters were determined on leaves between 9 a.m. and 11 a.m. using a Portable Photosynthesis System (CIRAS-3, USA). The Pn (net photosynthetic rates), Gs (stomatal conductance) and Tr (transpiration rate) were detected. All photosynthetic parameters were obtained under a photosynthetically active radiation of 1500 mmol m−2 s−1, at 80–90% air relative humidity and with the CO2 concentration held at 390 mmol mol−1. The leaf temperature was kept at 27 ± 1.5 °C ensuring a maximum instantaneous Pn. The data were collected with at least three replicates (Ma et al. 2017). In addition, significant chlorophyll fluorescence parameters, including the photosynthetic electron transport rate (ETR), the maximal photochemical efficiency of PSII in the dark (Fv/Fm), the photochemical quantum yield of PSII (Y(II)) and the photochemical quenching coefficient (qP), were measured using DUAL-PAM-100 (Liao et al. 2016).
For the determination of leaf biomass, the leaf fresh weight of diploids and tetraploids was measured, using approximately 10 leaves for each ploidy level. For the determination of special leaf weight (SLW), the surface of the leaf was rinsed and placed in an oven at 105 °C for 24 h, then weighed with a balance. SLW = leaf dry weight (mg) leaf area (cm2)-1. To quantify total starch content and soluble sugars, fresh mature leaves were used and analyzed using assay kits (Nanjing Jiancheng Bioengineering Institute, Nanjing, China); 0.1-g fresh leaves were weighed for each sample, 1-ml ddH2O was added after which they were grounded into a homogenate. The homogenate was then bathed in boiling water for 10 min. After cooling, the samples were centrifuged at 4000 r.p.m. for 10 min. The supernatant was then absorbed and diluted 10-fold for later use. The reagent in the kit was used for subsequent reaction and the absorbance at a wavelength of 620 nm was determined.
Determination of endogenous hormone content
The content of leaf endogenous hormones was determined using ultra-performance liquid chromatography (UPLC). Chromatographic conditions: separation was performed on an Acquity UPLC BEH C18 column (1.7 μm, 2.1 mm × 50 mm) equipped with a VanGuard pre-column (BEHC18, 1.7 μm, 2.1 × 5 mm; Waters) and the column temperature was kept at 25 °C. The volume of injection was 5 μl. A solvent system of 0.01% formic acid-water (A) and 0.01% formic acid-methanol (B) was used to elute at a flow rate of 0.1 ml min-1. Mass spectrometry was evaluated using electrospray ionization, positive ion scanning and the multiple reaction monitoring scanning method. Ion source spray voltage was set to 5500 V and the atomizing temperature of the ion source to 500 °C, while the atomizing gas was set to a pressure of 25 p.s.i.
Real-time quantitative reverse transcription PCR analysis
A set of five DEGs from our RNA-seq analysis was selected for RT-qPCR. RNA samples were denatured, and first-strand cDNA of all the samples was synthesized using the HiScript II 1st Strand cDNA Synthesis Kit (Vazyme, China). Primers were designed using Oligo 7 (see Table S1 available as Supplementary data at Tree Physiology Online). To investigate relative gene expression, qRT-PCR was performed employing a LightCycler 480 II Real-Time PCR System with the AceQ qPCR SYBR Green Master mix (Vazyme). The relative expression levels of the genes were calculated using the 2−△△Ct method. All samples were examined in (technical) triplicate, using three biological replicates; 18S rRNA was used as a reference gene to normalize samples.
Statistical analysis
All data were analyzed using GraphPad Prism software (version 6.0). Statistical analysis of the data, shown as means ± SE, was conducted using one-way analysis of variance and t-tests, taking *P < 0.05, **P < 0.01 and ***P < 0.001 as levels of significance.
Results
Enlarged L. sino-americanum somatic embryos perform well in the field
While generating L. sino-americanum somatic embryos, we found that some regenerated plants induced from the same genotype grew significantly larger than others (Figure 1A), with enlarged and thickened leaves and well-developed roots (Figure 1B). We decided, after 2 years in the greenhouse, to transfer all plants to the field under the same conditions and observe them. According to our observations, the field survival rate of these plants was no difference from other plants, with all of them clearly thriving (Figure 1C). After 5 years of growth in the field, we propagated by taking cuttings from the maternal tree and found that the grafted surviving plants maintained the same growth trend as the maternal trees (Figure 1D). In addition, we observed their phenological period and found that the bud break of these large plants was 2 weeks later in the spring (Figure 1E) and similarly shed their leaves ~2 weeks later in the autumn (Figure 1F). There was also a significant difference in leaf endogenous hormone content between spring and autumn (see Figure S1 available as Supplementary data at Tree Physiology Online).
Figure 1.
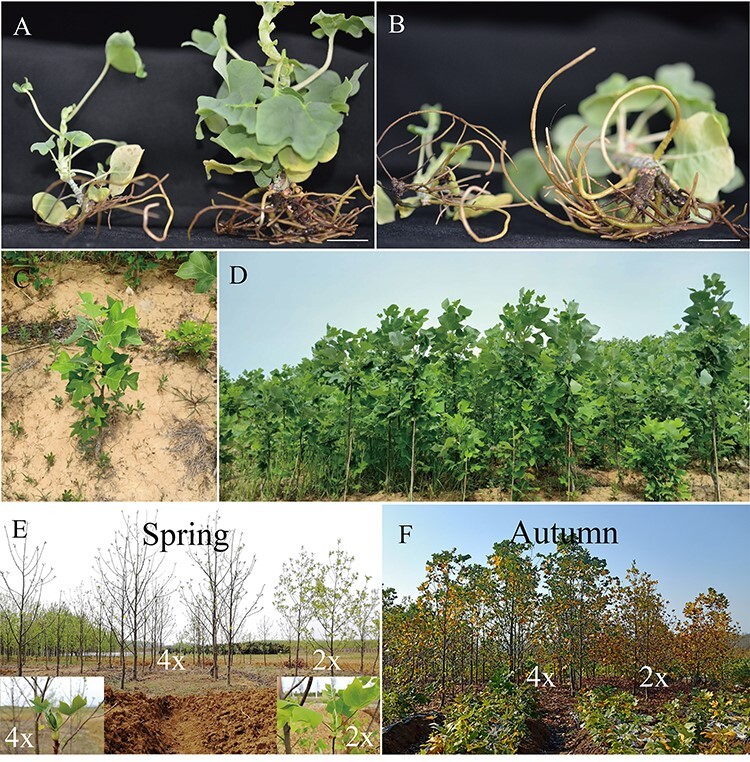
Field growth and phenological phase. (A) Phenotype and (B) root of 6-month-old plants regenerated from somatic embryo, bar = 1 cm, (C) plants regenerated from somatic embryos grew well in the field for 2 years, (D) grafted plants showed the same strong growth as their maternal plants, (E) bud break in spring, (F) yellowing of leaves in autumn. 4×: tetraploid plants; 2×: diploid plants.
Enlarged L. sino-americanum plants are tetraploids and show morphological alterations
We found that the significant phenotypic changes observed in our enlarged regenerated plants (Figure 2A) were very similar to the phenotypic alterations found in polyploid plants. Therefore, we speculated that somatic mutations occurred during somatic embryogenesis, leading to changes in plant ploidy levels. To determine plant ploidy, we combined FCM detection and chromosome counting. The FCM analysis indicated that the fluorescence intensity of the potential polyploids peaked at 20, a value twofold that of the control diploids (Figure 2B). Furthermore, the chromosome number of the diploid control was 2n = 38, while the potential polyploid displayed a chromosome number of 4n = 76 (Figure 2C). As a result, FCM and chromosome counting indicated that these putative polyploids were indeed tetraploids and belonged to the homologous tetraploids, because they were derived from soma-clonal variation.
Using the same growing conditions in the field, tetraploids grew faster than diploids, resulting in significantly higher trees and an increased ground diameter. We measured the tree height and ground diameter for two consecutive years and calculated their annual growth. The annual increase in tree height for tetraploids was 1.29 m, while for diploids, this was only 1.07 m per year (Figure 2D). In terms of ground diameter, the annual growth of tetraploids was 3.05 cm, while that of diploid was only 2.32 cm (Figure 2E). We made a rough estimate of the stem volume and it was found that the stem volume of the 8-year-old tetraploids was about 1.75 times that of the diploids (Figure 2F). Tetraploid buds were longer, wider and thicker (see Figure S2A and B available as Supplementary data at Tree Physiology Online). Furthermore, tetraploids showed approximately twofold lower stomatal density (yet an increased stomatal cell size: on average, by 53.7% in length and 25.4% in width) (Figure 2G, Figure S2C and D available as Supplementary data at Tree Physiology Online), and approximately fivefold larger leaves (Figure 2H and I). Comparing the upper epidermal cell number, we found that tetraploids have 1.4 times more cells than diploids (Figure 2J red circle, Figure S2E available as Supplementary data at Tree Physiology Online). Finally, their leaves showed differences at the tissue structure level: tetraploids have thicker palisade and especially spongy tissue (Figure 2K, Figure S2F available as Supplementary data at Tree Physiology Online), while no difference could be seen in transverse sections of epidermis. In general, these results demonstrate that tetraploids show morphological and anatomical differences from their diploid progenitors.
No significant transcriptomic difference between three tree positions of diploids and tetraploids
To understand the molecular mechanism that might underlie the phenotypic changes found in tetraploid L. sino-americanum, we first analyzed transcriptome from three tree positions of diploids and tetraploids (Figure 2A). The transcriptomic libraries of both diploid and tetraploid plants were assembled into unigenes and produced 146,591 unigenes altogether (see Table S2 available as Supplementary data at Tree Physiology Online). The size of these unigenes ranged from 282 to 11,808 bp, with a mean length of 2328.95 bp. The read number of each group is shown in Table S3, available as Supplementary data at Tree Physiology Online. Furthermore, ~53% of the unigenes were annotated to public databases (Swiss-Prot, KEGG and GO) (see Dataset S1 available as Supplementary data at Tree Physiology Online). The DEGs were screened from seven comparative transcriptome libraries (4× Top vs 2× Top, 4× Middle vs 2× Middle, 4× Basal vs 2× Basal, 4× Top vs 4× Basal, 4× Middle vs 4× Basal, 2× Top vs 2× Basal, 2× Middle vs 2× Basal), with the conditions|Log2 FC| > 2 and Q-value <0.01. As shown in Figure S3A, available as Supplementary data at Tree Physiology Online, a total of 3830, 2807 and 4519 DEGs from transcriptomes taken from the top, middle and basal sections of tetraploid and diploid tree, respectively, with each comparison giving rise to more up-regulated than down-regulated genes. In general, we found a large number of DEGs between analogous tree sections of tetraploid and diploid plants, while the number of DEGs between transcriptomes along different positions of a single variety was significantly smaller. We performed an initial evaluation of the effect of tetraploidization on gene expression using GO enrichment and found that diploids and tetraploids differed in kinase activity, photosynthetic system abundance, as well as the synthesis and metabolism of a large number of substances (see Figure S3B available as Supplementary data at Tree Physiology Online).
Polyploidization in L. sino-americanum is accompanied by transcriptomic alterations
We subsequently turned to compare the transcriptomes of L. sino-americanum leaves of different ploidies. RNA-seq was performed on samples from three diploid and three tetraploid plants and repeated in triplicate, thus sequencing 18 samples in total (see Dataset S2 available as Supplementary data at Tree Physiology Online). After read filtering, we mapped clean reads to the L. chinense reference genome (NCBI, Bio Project, PRJNA418360). On average, 86.75% reads were mapped (see Table S4 available as Supplementary data at Tree Physiology Online). We then determined the Pearson correlation between all samples, which is shown in Figure S4, available as Supplementary data at Tree Physiology Online. Additionally, gene expression levels based on FPKM value were evaluated, and the gene expression distribution of each sample was measured (see Figure S5A available as Supplementary data at Tree Physiology Online). After taking together all DEGs of each comparison group, we conducted clustering analysis and found that there were significant differences in gene expression trends between diploids and tetraploids (see Figure S5B available as Supplementary data at Tree Physiology Online). We also compared the expressed genes between three individual diploids and tetraploids and found that plants with the same ploidy show significantly fewer DEGs. When diploids and tetraploids were compared, tetraploids had more differential genes (see Figure S5C available as Supplementary data at Tree Physiology Online). In general, the replicate samples correlated strongly, and the gene expression difference between tetraploids and diploids was obvious.
Having identified a high number of DEGs between tetraploid and diploid L. sino-americanum, we asked what their main function is and how they may contribute to the phenotypic effects that we have observed. Through a GO-term analysis, we found that DEGs between diploids and tetraploids were categorized according to three classifications: ‘Biological Process (BP)’, ‘Cellular Component (CC)’ and ‘Molecular Function (MF)’ (Figure 3 (up-regulated), Figure S6 (down-regulated) available as Supplementary data at Tree Physiology Online). For the BP category, organonitrogen compound biosynthetic process, carbohydrate metabolic and catabolic process, ATP metabolic and biosynthetic process, plant hormone signal transduction, photosynthesis and electron transport chain were overrepresented, while for the CC, the largest group of genes corresponded to the cytoplasmic localization, followed by the photosystem, photosynthetic membrane, photosystem I and photosystem II and the oxygen evolving complex. For the MF category, the most prominent was ATP binding and tetrapyrrole binding (Figure 3). We also found methyltransferase and RNA methyltransferase activity. Furthermore, KEGG pathway analysis was used to further identify the biological behavior in which the DEGs were involved (Figure 4). The pathway that differed most significantly between tetraploids and diploids was that of the photosynthesis, photosynthesis-antenna proteins and carbon fixation in photosynthetic organisms, which was consistent with the GO analysis.
Figure 3.
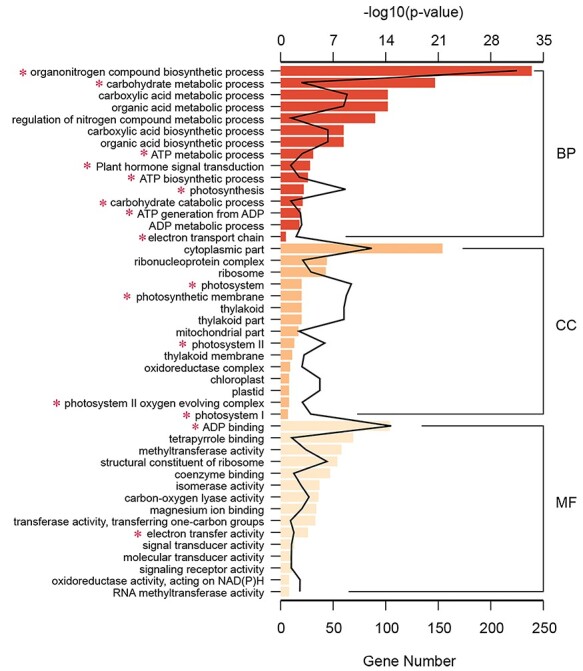
The GO enrichment analysis of up-regulated DEGs. The BP, CC and MF of identified DEGs in both diploids and tetraploids. The numbers above each column indicate the –log10 (P-value) in each group, and the numbers under the columns indicate the up-regulated DEGs of each group. *biological functions associated with photosynthesis.
Figure 4.
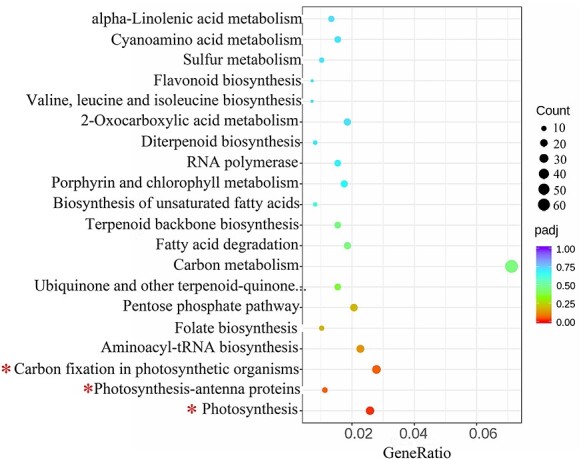
The KEGG pathway analysis of DEGs. The top 20 enriched KEGG pathways in which DEGs function, based on the tetraploids vs diploids comparison. The x-axis indicates the gene ratio. The dot color and size indicate the P-value and gene number as shown on the right. *biological functions associated with photosynthesis.
From our GO and KEGG classifications, we further identified 46 key photosynthesis-related genes and 20 key plant hormone signal transduction genes (Figure 5A), mainly enriched for F-type ATPase, cytochrome b6/f complex, photosynthetic electron transport, photosystem I, photosystem II, light-harvesting chlorophyll protein complex (LHC), auxin and cytokinin signal transduction. We reasoned that these 46 DEGs might play a key role in regulating photosynthesis. The 46 genes included five genes with known functions in photosynthesis, such as ATPF1G, psbW, LHCA2, LHCA3 and LHCB3. Expression levels of these five genes (Figure 5B), evaluated by qRT-PCR, were largely consistent with the RNA-seq data. According to the results of endogenous hormone determination, there was no significant difference between IAA and ZeaTin, while the content of 6-BA was significantly different (Figure 5C–E). All these findings suggested that genes related to photosynthesis and accumulation of photosynthetic organisms changed greatly in tetraploids.
Figure 5.
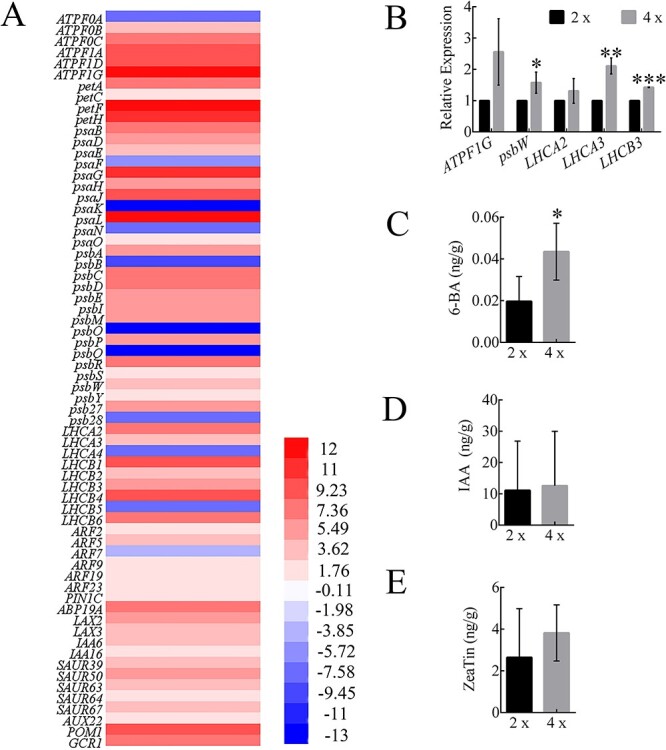
Expression level of DEGs and plant hormones. (A) A heatmap showing changes in expression levels of DEGs related to photosynthesis and plant hormone signal transduction of tetraploids vs diploids. The gradient color barcode at the right indicates log2 (fold change) values with up-regulated genes represented by positive values and down-regulated genes represented by negative values. (B) Quantitation of expression of several key photosynthesis genes in the leaves of tetraploids vs diploids, RT-qPCR data were means ± SE (n = 3). (C–E) Determination of 6-BA (C), IAA (D) and ZeaTin (E). 4×: tetraploid plants; 2×: diploid plants.
Polyploidy leads to changes in protein expression levels
We also examined proteomic changes between diploids and tetraploids. We generated a total of 289,226 mass spectra, identifying 14,542 peptides and 4018 proteins with a 1% FDR (see Table S5 available as Supplementary data at Tree Physiology Online). The protein mass, unique peptide number, protein coverage and protein functional annotation are provided in Datasets S3 and S4, available as Supplementary data at Tree Physiology Online, within the Supporting Information.
Proteins with |Log2 FC| > 1.2 and P-value < 0.05 are defined as DEPs. A total of 884 proteins were significantly differentially expressed between diploid and tetraploid trees. To estimate the expression pattern of DEPs, we performed a GO enrichment analysis of 884 DEPs (Figure 6). These 884 DEPs were functionally classified into 44 classes; the largest protein groups corresponded to catalytic activity, binding, cell, cell part, metabolic process and cellular process. We then used KEGG pathway analysis to further identify the BPs in which DEPs are involved (Figure 7A). The pathways differing most significantly between tetraploids and diploids were carbon fixation in photosynthetic organisms, photosynthesis and photosynthesis-antenna proteins, which is mostly consistent with our transcriptomic analysis.
Figure 6.
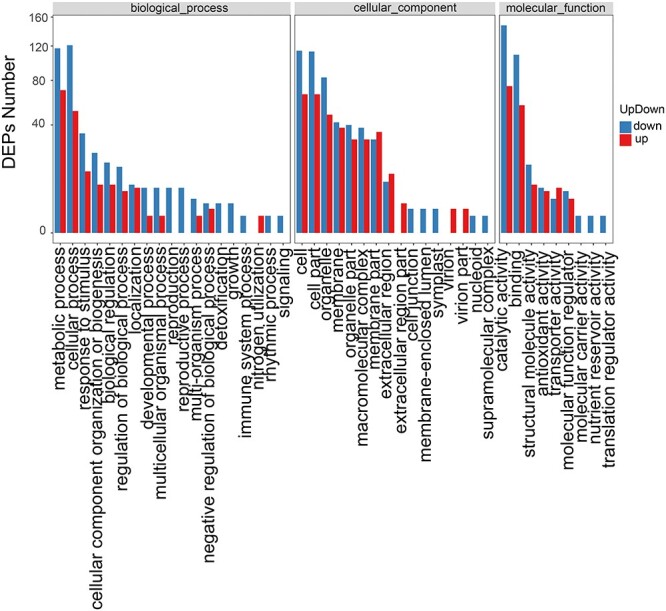
Gene Ontology enrichment analysis of DEPs; GO enrichment analysis of DEPs based on comparison of tetraploids vs diploids.
Figure 7.
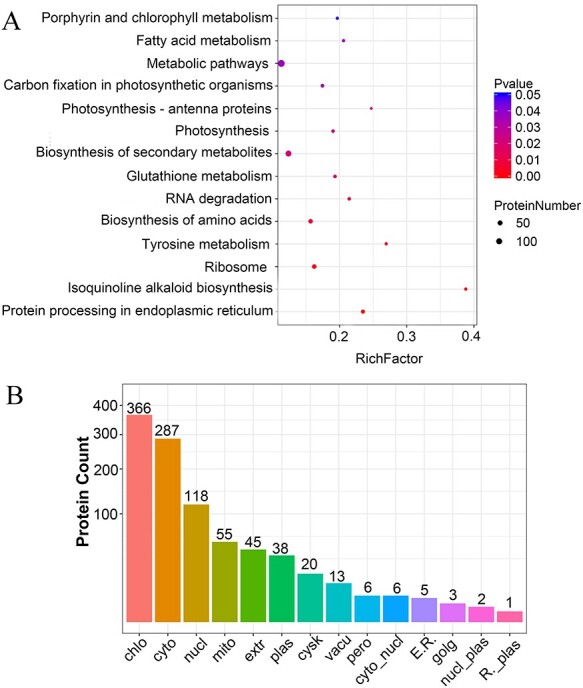
Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis and subcellular localization prediction of DEPs. (A) The top 14 enriched pathways in which DEPs were placed, based on the tetraploids vs diploids comparison. The x-axis indicates the enrichment factor. The dot color and size indicate the P-value and protein number as shown on the right, (B) subcellular localization prediction of DEPs. The x-axis displays subcellular structure; the y-axis displays protein count.
Proteins are synthesized in ribosomes and then transported to specific organelles guided by protein sorting signals. Proteins can also be secreted into the extracellular space or stay in the cytoplasm (C. S. Yu et al. 2010). Subcellular localization of proteins is an important part of protein functionality. We found that the predicted subcellular localization of the DEPs we identified was mainly enriched for the chloroplast and cytoplasm (Figure 7B). The main function of chloroplasts is to perform photosynthesis, absorb light energy and synthesize organic compounds. The cytoplasm is an important structural component of cells where a large number of intermediate metabolic processes, protein synthesis and fatty acid synthesis are carried out. The cytoplasmic matrix plays an important role in protein modification and selective degradation of proteins.
Comparison of the transcriptome and proteome suggests enhanced photosynthesis in tetraploid L. sino-americanum
Genes are carriers of genetic information, while proteins execute the gene function (Cadman et al. 2010). A series of expression regulatory mechanisms are involved in the processes of gene expression and protein synthesis. Therefore, to comprehensively explore the complex activities of plants, it is necessary to compare simultaneously detect mRNA and protein expression. Hence, transcriptomic analysis of samples simultaneously assayed by iTRAQ was performed, making it possible to compare transcript and protein expression levels between diploids and tetraploids. Subsequent analysis of expression changes in transcripts and proteins was compared at two levels, with the number of identified genes or proteins and their differential expression level. The parameters used for comparison analysis and the number of genes as well as proteins involved are listed in Dataset S5 available as Supplementary data at Tree Physiology Online.
There exist four possible combinations for the relation between DEGs and DEPs (Figure 8A). We focused on DEGs with correlated expression changes (rUP-pUP and rDN-pDN, being both transcript and protein level up or down, respectively) and identified 98 DEGs (with 98 proteins) satisfying these conditions between diploids and tetraploids. Biological pathways responsive to photosynthesis-antenna proteins, other glycan degradation and sulfur metabolism were enriched significantly among this rUP-pUP set. A large number of other BPs, such as phagosome, oxidative phosphorylation, protein processing in the endoplasmic reticulum and ribosome, also were enriched within this set (Figure 8B). For the rDN-pDN set, glutathione metabolism, cyanoamino acid metabolism, phenylpropanoid biosynthesis as well as starch and sucrose metabolism were enriched processes (Figure 8C). These results suggest that polyploidization may regulate growth by improving photosynthesis-responsive genes and protein expression. We proceeded to map these DEGs and DEPs onto the photosynthetic functional pathway and found that most genes within the pathway are involved (Figures 9 and 10).
Figure 8.
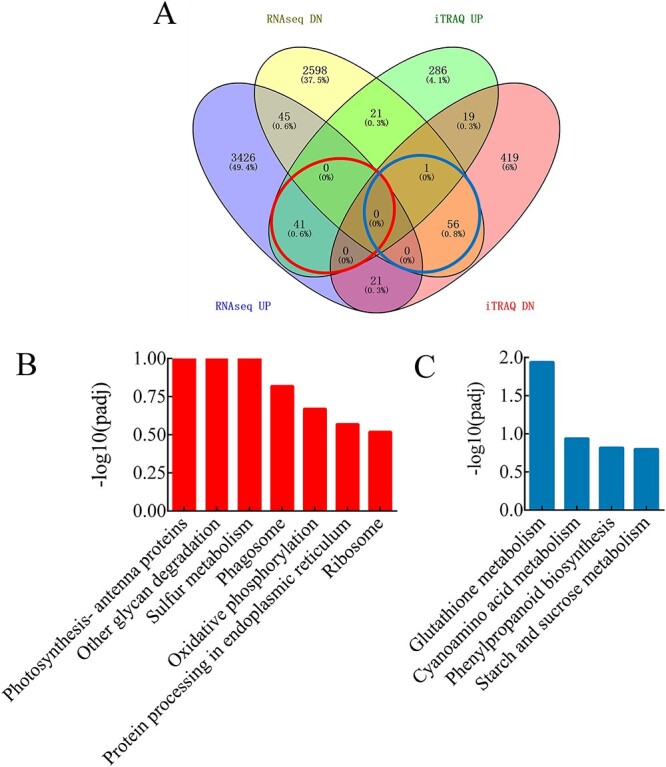
Comparison of the relation between transcriptome and proteome. (A) Venn diagram showing the number and proportion of DEGs and DEPs within each of the four categories resulting from a relation analysis. The DEGs and DEPs were divided into four combinations: (i) increased transcript level induces protein expression up-regulation (rUP-pUP) (encircled in red), (ii) decreased transcript level induces protein expression up-regulation (rDN-pUP), (iii) increased transcript level induces protein expression down-regulation (rUP-pDN) and (iv) decreased transcript level induces protein expression down-regulation (rDN-pDN) (encircled in blue). We focused on DEGs with correlated expression changes (rUP-pUP (i) and rDN-pDN (iv)), being both transcript and protein level up or down, respectively. (B, C) Kyoto Encyclopedia of Genes and Genomes enrichment of up-regulated (B) and down-regulated (C) genes and proteins.
Figure 9.
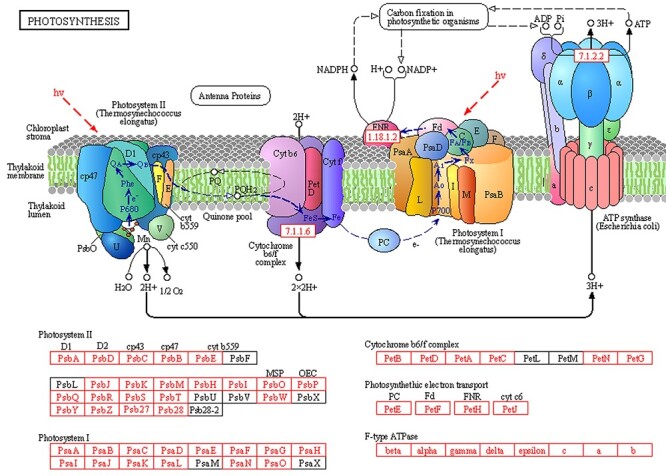
The KEGG pathway image of photosynthesis. Schematic representation of large protein complexes associated with photosynthesis, along with a list of proteins that make up these complexes. Protein complex components that we identified as DEGs (DEPs) are highlighted in red for photoreaction complexes. Images are from the KEGG website.
Figure 10.
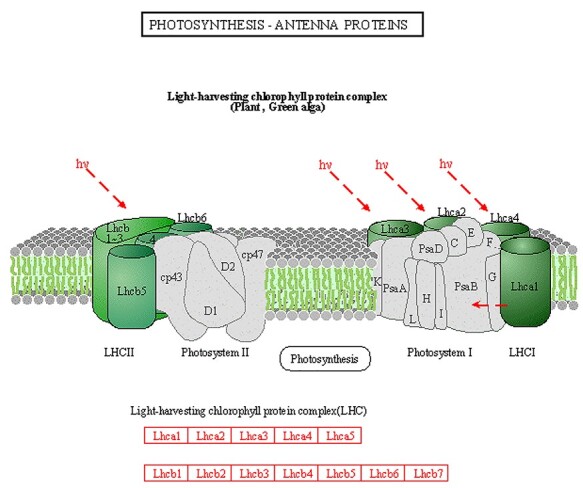
Kyoto Encyclopedia of Genes and Genomes pathway image of photosynthesis-antenna proteins. Schematic representation of photosynthesis-antenna protein complexes, DEGs (DEPs) are highlighted in red. Image is from the KEGG website.
Tetraploids exhibit enhanced photosynthesis capacity and biomass accumulation
As mentioned above, photosynthesis-related genes and proteins are all up-regulated in tetraploids. To confirm these results, we compared the photosynthetic capacity and chlorophyll content of diploids and tetraploids. As shown in Figure 11, the total chlorophyll content in tetraploids was ~2.7-fold higher than diploids, with chlorophyll a increasing more than chlorophyll b (Figure 11A). This result suggests that the biosynthesis of photosynthetic pigments may be sensitive to polyploidy. Additionally, three typical chlorophyll fluorescence traits (qP, Y(II) and ETR, Figure 11B and C) show significant differences (P < 0.05) between diploids and tetraploids. However, photosynthetic traits showed no significant correlation to the Fv/Fm ratio (Figure 11B), which illustrates that Fv/Fm was less sensitive than other traits under normal growth conditions. The net photosynthetic rate (Figure 11D), stomatal conductance levels (Figure 11E) and transpiration rates (Figure 11F) showed highly significant differences and increased ~1.6-fold, ~2.2-fold and ~1.6-fold in tetraploids, respectively. In general, these results collectively demonstrate that tetraploids have an improved photosynthetic capacity.
Figure 11.
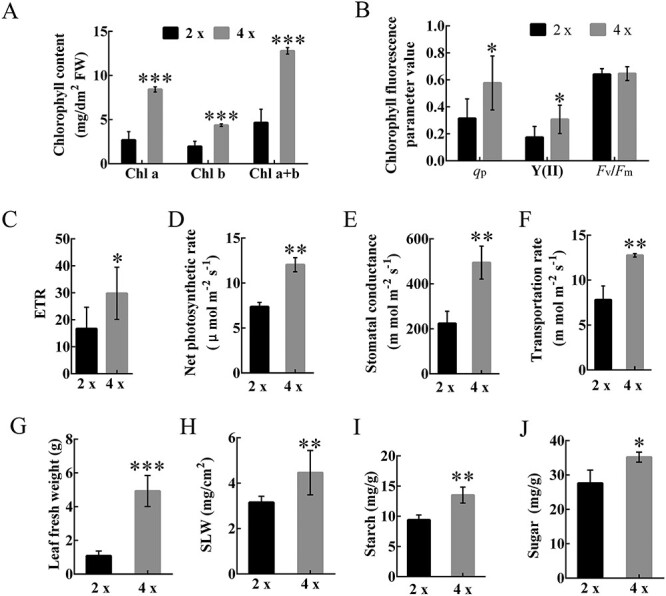
Enhanced photosynthetic capacity and biomass accumulation of tetraploids. (A–J) Bar graphs plotting (A) chlorophyll content (Chl a: chlorophyll a, Chl b: chlorophyll b and Chl a + b: total chlorophyll content), (B) chlorophyll fluorescence parameters (the photochemical quenching coefficient (qp), the photochemical quantum yield of PSII (Y(II)) and the maximal photochemical efficiency of PSII in the dark (Fv/Fm)), (C) ETR, (D) net photosynthetic rate, (E) stomatal conductance, (F) transportation rate, (G) leaf fresh weight, (H) mg/cm2, (I) starch content and (J) sugar content. Error bars indicate SE. Asterisks indicate significant differences between diploids and tetraploids (*P < 0.05; **P < 0.01; ***P < 0.001). 4×: tetraploid plants; 2×: diploid plants.
We also found that leaf biomass (represented as leaf fresh weight) and SLW (represented as SLW, leaf dry weight (mg) leaf area (cm2)-1) were significantly increased in tetraploids (Figure 11G and H). In addition, tetraploids accumulated more starch (Figure 11I) and sugar (Figure 11J) than diploids in leaves as measured using quantitative assays. In tetraploids, leaf starch and sugar contents were increased by 44 and 27%, respectively, suggesting high rates of sugar and starch accumulation and indicating the rapid transport and metabolism of sugar. Altogether, leaf biomass, SLW, sugar and starch contents were consistently higher in tetraploids.
Discussion
In this study, we found that tetraploid L. sino-americanum shows a series of phenotypic, physiological and biochemical differences, as well as an increased organic accumulation and wood yield, compared with the original diploid state (Figures 1, 2 and 11, Figure S2 available as Supplementary data at Tree Physiology Online). These changes reported here have also been observed in other tetraploid plants (Banyai et al. 2010, Majdi et al. 2010, Mu et al. 2012), which would indicate that the underlying molecular mechanism causative to these changes in polyploids might be similar. However, it has been shown that the autotetraploid apple (Malus × domestica) and Rangpur lime (Citrus limonia) are reduced in size compared with their diploid relatives (Allario et al. 2011, Ma et al. 2016). Hence, the effects of polyploidization as a whole are more contradictory and may vary depending on the species. Liriodendron sino-americanum is an important landscaping and industrial timber species for which a mature somatic embryogenesis system has been developed. In our study, tetraploid L. sino-americanum plants were obtained by somatic variation that occurred during somatic embryogenesis. As a matter of fact, induction of soma-clonal variation can be used as an approach to cultivate polyploid plants, and polyploidization is the most common form of soma-clonal variation (Yang et al. 2006). Tetraploids of many species have been successfully obtained via this approach (Sattler et al. 2016). Few mature and stable somatic embryogenesis systems exist in woody plants. Within our system, we can induce a large number of polyploid L. sino-americanum plants through soma-clonal variation during somatic embryogenesis, which provides a method for the systematic study of the potential regulatory changes associated with woody plant polyploidy. Our results also confirmed that this is an effective method for exploring polyploid plants.
Polyploid plants, as a special germplasm resource, have attracted much more attention in recent years because of their huge organs and biomass accumulation. According to our phenotypic observations and physiological measurements, the phenotypic variation of tetraploid L. sino-americanum can be summarized as a result of two factors; one is cellular expansion (Figure 2J, Figure S2C–E available as Supplementary data at Tree Physiology Online), and the other is the accumulation of more cells and cellular content (Figures 2K, 11A, I and J). Plant growth is closely associated with the accumulation of energy through photosynthesis (Dai et al. 2015). Any changes in photosynthetic activity are likely due to changes in the capacity for light harvest, energy transport or energy conversion (Corneillie et al. 2019). The leaves act as the main photosynthesis organ, allowing the accumulation of biomass (Zhang et al. 2019). For tetraploid L. sino-americanum, one of the reasons for the increased photosynthetic ability is that the tetraploid plants have larger leaves, which increases the available area for photosynthetic activity, allowing for the accumulation of more photosynthetic products (Figure 2H and I). In addition, the higher chlorophyll content in the leaves also increases the light-harvesting ability (Figure 11A). It is also worth mentioning that the tetraploid has a longer photosynthetic period according to our observation of its growing rhythm (Figure 1E and F). Even though many earlier studies have examined and characterized the huge organs and biomass accumulation of polyploid plants, advances in our understanding of how their traits are mechanistically established have been limited. It is imaginable that elucidating the molecular mechanisms underlying these traits has great theoretical and practical significance.
In order to reveal the molecular mechanisms that may lead to polyploid variation and enhanced photosynthesis, in the present work, we performed a transcriptomics and proteomics analysis of diploid and tetraploid L. sino-americanum. We found a large number of genes to be differentially expressed between plants of different ploidy levels with both analysis (DEGs and DEPs). We found a large relationship between DEGs and DEPs that were either co-up-regulated or co-down-regulated corroborating the validity of the data sets (Figure 8A). The up-regulated DEGs and DEPs are mostly involved in photosynthesis, including F-type ATPase, the cytochrome b6/f complex, photosynthetic electron transport, the light harvesting chlorophyll protein complex, photosystem I and II, as well as auxin and cytokinin signaling (Figures 3–7). Our findings resound with other studies on polyploidy, suggesting that the increase in photosynthetic rate is related to an increased dosage of relevant genes (Warner and Edwards 1989, Warner and Edwards 1993, Z. Yu et al. 2010, Zhou et al. 2015). Our verification of gene expression and endogenous hormone content confirmed this viewpoint (Figure 5C–E). From our analysis, we found that there was a good relation between the transcriptome and proteome, and the co-up-regulated expression pathway was also related to photosynthetic activity (Figure 8B). The DEGs and DEPs we identified will lay a foundation for further research to explore how photosynthesis affects plant vegetative growth and will shed light on the complexity of polyploidization. In general, chromosome doubling results in changes in gene expression levels involved in plant hormone signalings, organics accumulation and photosynthesis.
Many recent studies have verified that the impact of polyploidization on genome regulation is associated with epigenetic modification (Osborn et al. 2003, Chen 2007, Renny-Byfield and Wendel 2014). Epigenetic changes may alter gene expression without a change in DNA sequence and can generate dramatic phenotypic effects (Soltis et al. 2004). The altered expression of a few regulatory genes may causes a cascade of changes in downstream genes and physiological pathways, ultimately affecting plant growth and development (Adams and Wendel 2005, Roulin et al. 2013) and providing a mechanism for growth vigor and increased biomass. Some methyltransferases were found in the GO enrichment of our transcriptome (Figure 3). Whether the great phenotypic changes of polyploids and enhancement of photosynthesis are related to epigenetic regulation is a key question worthy of further study.
In conclusion, in this study, we obtained autotetraploid L. sino-americanum from soma-clonal variation resulting from somatic embryogenesis. Using these tetraploid plants, we investigated morphological differences, physiological changes, transcriptome and proteome profiles, and compared these with diploid L. sino-americanum in detail. Our omics data analysis allowed us to explain the potential mechanism underlying changes in the photosynthetic process in tetraploids and advances our understanding of how they accumulate more biomass. Taken together, information generated in this study provides the impetus for additional research to understand the foundation of many of these desirable traits resulting from polyploidization.
Supplementary Material
Funding
This research was supported by the Jiangsu Specially Appointed Professors Program, the Key research and development plan of Jiangsu Province (BE2017376), the Foundation of Jiangsu forestry bureau (LYKJ [2017]42), the Nature Science Foundation of China (31770715), the Qinglan project of Jiangsu province, the Distinguished Professor Project of Jiangsu province and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Contributor Information
Tingting Chen, Key Laboratory of Forest Genetics and Biotechnology of Ministry of Education of China, Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, 159 Longpan Rd, Xuanwu, Nanjing 210037, China.
Yu Sheng, Key Laboratory of Forest Genetics and Biotechnology of Ministry of Education of China, Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, 159 Longpan Rd, Xuanwu, Nanjing 210037, China.
Zhaodong Hao, Key Laboratory of Forest Genetics and Biotechnology of Ministry of Education of China, Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, 159 Longpan Rd, Xuanwu, Nanjing 210037, China.
Xiaofei Long, Key Laboratory of Forest Genetics and Biotechnology of Ministry of Education of China, Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, 159 Longpan Rd, Xuanwu, Nanjing 210037, China.
Fangfang Fu, College of Forestry, Nanjing Forestry University, 159 Longpan Rd, Xuanwu, Nanjing 210037, China.
Yang Liu, School of Forestry, Northeast Forestry University, 26 Hexing Rd, Xiangfang District, Harbin 150040, China.
Zhonghua Tang, College of Chemistry, Chemical Engineer and Resource Utilization, Northeast Forestry University, 26 Hexing Rd, Xiangfang District, Harbin 150040, China.
Asif Ali, Key Laboratory of Forest Genetics and Biotechnology of Ministry of Education of China, Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, 159 Longpan Rd, Xuanwu, Nanjing 210037, China.
Ye Peng, College of Biology and the Environment, Nanjing Forestry University, 159 Longpan Rd, Xuanwu, Nanjing 210037, China.
Yang Liu, Key Laboratory of Forest Genetics and Biotechnology of Ministry of Education of China, Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, 159 Longpan Rd, Xuanwu, Nanjing 210037, China.
Lu Lu, Key Laboratory of Forest Genetics and Biotechnology of Ministry of Education of China, Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, 159 Longpan Rd, Xuanwu, Nanjing 210037, China.
Xiangyang Hu, Shanghai Key Laboratory of Bio-Energy Crops, School of Life Sciences, Shanghai University, 266 Jufeng Rd, Baoshan, Shanghai 201900, China.
Jisen Shi, Key Laboratory of Forest Genetics and Biotechnology of Ministry of Education of China, Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, 159 Longpan Rd, Xuanwu, Nanjing 210037, China.
Jinhui Chen, Key Laboratory of Forest Genetics and Biotechnology of Ministry of Education of China, Co-Innovation Center for Sustainable Forestry in Southern China, Nanjing Forestry University, 159 Longpan Rd, Xuanwu, Nanjing 210037, China.
Conflicts of interest
The authors declare no conflict of interest.
Availability of data and materials
All data generated or analyzed during this study are included in this article and its Supplementary data files. The data are available from the corresponding author on reasonable request.
Authors’ contributions
C.J.H. and S.J.S. conceived and designed the study; C.T.T., S.Y., L.X.F., L.Y., T.Z.H., L.L., A.A. and P.Y. performed the experiments; C.T.T., S.Y., H.Z.D., F.F.F., L.Y. and H.X.Y. carried out the statistical analysis; C.T.T. wrote the manuscript. All authors contributed to manuscript revision, and read and approved the submitted version.
References
- Adams KL, Wendel JF (2005) Polyploidy and genome evolution in plants. Curr Opin Plant Biol 8:135–141. [DOI] [PubMed] [Google Scholar]
- Allario T, Brumos J, Manuel Colmenero-Flores J, Tadeo F, Froelicher Y, Talon M, Navarro L, Ollitrault P, Morillon R (2011) Large changes in anatomy and physiology between diploid Rangpur lime (Citrus limonia) and its autotetraploid are not associated with large changes in leaf gene expression. J Exp Bot 62:2507–2519. [DOI] [PubMed] [Google Scholar]
- Banyai W, Sangthong R, Karaket N, Inthima P, Mii M, Supaibulwatana K (2010) Overproduction of artemisinin in tetraploid Artemisia annua L. Plant Biotechnol 27:427–433. [Google Scholar]
- Bo W, Ruo Z, Qiang F, Quanhui W, Jun W, Siqi L (2015) IQuant: an automated pipeline for quantitative proteomics based upon isobaric tags. Proteomics 14:2280–2285. [DOI] [PubMed] [Google Scholar]
- Cadman CSC, Toorop PE, Hilhorst HWM, Finch-Savage WE (2010) Gene expression profiles of Arabidopsis Cvi seeds during dormancy cycling indicate a common underlying dormancy control mechanism. Plant J 46:805–822. [DOI] [PubMed] [Google Scholar]
- Cameron AD (1990) Autotetraploid plants from callus cultures of Betula pendula Roth. Tree Physiol 6:229–234. [DOI] [PubMed] [Google Scholar]
- Chen B, Wang C, Tian Y, Chu Q, Hu C (2015) Anatomical characteristics of young stems and mature leaves of dwarf pear. Sci Hortic 186:172–179. [Google Scholar]
- Chen J, Yang G, Ding Q, Xia T, She J, Jia A (2013) In vitro tumor cytotoxic activities of extracts from three Liriodendron plants. Pak J Pharm Sci 26:233–237. [PubMed] [Google Scholar]
- Chen J, Hao Z, Guang X et al. (2019) Liriodendron genome sheds light on angiosperm phylogeny and species-pair differentiation. Nat Plants 5:328–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ (2007) Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol 58:377–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Li H (2018) Interspecies evolutionary divergence in Liriodendron, evidence from the nucleotide variations of LcDHN-like gene. BMC Evol Biol 18:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu LH, Peng JJ, Wang Z, Zhao K, Yuan SX, Ming J, Liu C (2014) Studies on polyploid induction in Anthurium andraeanum using oryzalin, trifluralin and colchicine. Acta Hort Sin 41:2275–2280. [Google Scholar]
- Corneillie S, De Storme N, Van Acker R et al. (2019) Polyploidy affects plant growth and alters cell wall composition. Plant Physiol 179:74–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai F, Zhenjiang W, Guoqing L, Cuiming T (2015) Phenotypic and transcriptomic analyses of autotetraploid and diploid mulberry (Morus alba L.). Int J Mol Sci 16:22938–22956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolezel J, Bartos J, Voglmayr H, Greilhuber J (2003) Nuclear DNA content and genome size of trout and human. Cytometry A 51A:127–128. [DOI] [PubMed] [Google Scholar]
- Dreyer E, Roux XL, Montpied P, Daudet FA, Masson FA (2001) Temperature response of leaf photosynthetic capacity in seedlings from seven temperate tree species. Tree Physiol 21:223–232. [DOI] [PubMed] [Google Scholar]
- Du K, Han Q, Zhang Y, Kang X (2019) Differential expression of genes related to the formation of giant leaves in triploid poplar. Forests 10:920. [Google Scholar]
- Eng W, Ho W (2019) Polyploidization using colchicine in horticultural plants: a review. Sci Hortic 246:604–617. [Google Scholar]
- Greer BT, Still C, Cullinan GL, Brooks JR, Meinzer FC (2017) Polyploidy influences plant–environment interactions in quaking aspen (Populus tremuloides Michx.). Tree Physiol 38:630–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Mendrikahy JN, Xie L, Deng J, Lu Z, Wu J, Li X, Shahid M, Liu X (2017) Transcriptome analysis of neo-tetraploid rice reveals specific differential gene expressions associated with fertility and heterosis. Sci Rep 7:40139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haider N (2013) The origin of the B-genome of bread wheat (Triticum aestivum L.). Russ J Genet 49:263–274. [DOI] [PubMed] [Google Scholar]
- Hammond JBW, Kruger NJ (1988) The Bradford method for protein quantitation. Methods Mol Biol 32:9–15. [DOI] [PubMed] [Google Scholar]
- Khalid MF, Hussain S, Anjum MA, Ahmad S, Ali MA, Ejaz S, Morillon R (2020) Better salinity tolerance in tetraploid vs diploid volkamer lemon seedlings is associated with robust antioxidant and osmotic adjustment mechanisms. J Plant Physiol 244:153071–153071. [DOI] [PubMed] [Google Scholar]
- Kim CK, Chung M, Kim CY, Min JS, Lee D, Naing AH, Chung JD (2014) In vitro induction of tetraploids in an interspecific hybrid of Calanthe (Calanthe discolor x Calanthe sieboldii) through colchicine and oryzalin treatments. Plant Biotechnol Rep 8:251–257. [Google Scholar]
- Koehler C, Scheid OM, Erilova A (2010) The impact of the triploid block on the origin and evolution of polyploid plants. Trends Genet 26:142–148. [DOI] [PubMed] [Google Scholar]
- Li T, Chen J, Qiu S, Zhang Y, Wang P, Yang L, Lu Y, Shi J (2012) Deep sequencing and microarray hybridization identify conserved and species-specific MicroRNAs during somatic embryogenesis in hybrid yellow poplar. Plos One 7(8):e43451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao T, Cheng S, Zhu X, Min Y, Kang X (2016) Effects of triploid status on growth, photosynthesis, and leaf area in Populus. Trees Struct Funct 30:1137–1147. [Google Scholar]
- Luo G, Xue L, Xu W, Zhao J, Wang J, Ding Y, Kun L, Lei J (2019) Breeding decaploid strawberry with improved cold resistance and fruit quality. Sci Hortic 251:1–8. [Google Scholar]
- Ma N, Hu C, Wan L, Hu Q, Xiong J, Zhang C (2017) Strigolactones improve plant growth, photosynthesis, and alleviate oxidative stress under salinity in rapeseed (Brassica napus L.) by regulating gene expression. Front Plant Sci 8:1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Xue H, Zhang L, Zhang F, Ou C, Wang F, Zhang Z (2016) Involvement of auxin and brassinosteroid in dwarfism of autotetraploid apple (Malus × domestica). Sci Rep 6:26719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdi M, Karimzadeh G, Malboobi MA, Omidbaigi R, Mirzaghaderi G (2010) Induction of tetraploidy to feverfew (Tanacetum parthenium Schulz-Bip.): morphological, physiological, cytological, and phytochemical changes. Hortscience 45:16–21. [Google Scholar]
- Miguel A, Maroto JV, López-Galarza S (2001) Triploid seedless watermelon production without pollinators. Effect of the number of sprayed flowers on fruit size. Acta Hortic 142:135–138. [Google Scholar]
- Mu H, Liu Z, Lin L, Li H, Jiang J, Liu G (2012) Transcriptomic analysis of phenotypic changes in birch (Betula platyphylla) autotetraploids. Int J Mol Sci 13:13012–13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn TC, Pires JC, Birchler JA et al. (2003) Understanding mechanisms of novel gene expression in polyploids. Trends Genet 19:141–147. [DOI] [PubMed] [Google Scholar]
- Paterson AH (2005) Polyploidy, evolutionary opportunity, and crop adaptation. Genetica 123:191–196. [DOI] [PubMed] [Google Scholar]
- Qi LZ, Rong WZ (2000) Studies on the technique of cross breeding in liriodendron. J Nanjing Forest Univ 04:21–25. [Google Scholar]
- Ramsey J, Schemske DW (1998) Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu Rev Ecol Syst 29:467–501. [Google Scholar]
- Renny-Byfield S, Wendel JF (2014) Doubling down on genomes: polyploidy and crop plants. Am J Bot 101:1711–1725. [DOI] [PubMed] [Google Scholar]
- Robinson DO, Coate JE, Singh A, Hong L, Bush M, Doyle JJ, Roeder AHK (2018) Ploidy and size at multiple scales in the Arabidopsis sepal. Plant Cell 30:2308–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roulin A, Auer PL, Libault M, Schlueter J, Farmer A, May G, Stacey G, Doerge RW, Jackson SA (2013) The fate of duplicated genes in a polyploid plant genome. Plant J 73:143–153. [DOI] [PubMed] [Google Scholar]
- Rubuluza T, Nikolova RV, Smith MT, Hannweg K (2007) In vitro induction of tetraploids in Colophospermum mopane by colchicine. S Afr J Bot 73:259–261. [Google Scholar]
- Sattler MC, Carvalho CR, Clarindo WR (2016) The polyploidy and its key role in plant breeding. Planta 243:281–296. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Soltis PS, Tate JA (2004) Advances in the study of polyploidy since plant speciation. New Phytol 161:173–191. [Google Scholar]
- Tan X, Yang D, Yang G, Chen J, Dong W, Shi J, Jia A (2015) The investigation of inhibiting quorum sensing and methicillin-resistant Staphylococcus aureus biofilm formation from liriodendron hybrid. Pak J Pharm Sci 28:903–908. [PubMed] [Google Scholar]
- Vyas P, Bisht MS, Miyazawa S, Yano S, Noguchi K, Terashima I, Funayama-Noguchi S (2007) Effects of polyploidy on photosynthetic properties and anatomy in leaves of Phlox drummondii. Funct Plant Biol 34:673–682. [DOI] [PubMed] [Google Scholar]
- Warner DA, Edwards GE (1989) Effects of polyploidy on photosynthetic rates, photosynthetic enzymes, contents of DNA, chlorophyll, and sizes and numbers of photosynthetic cells in the C4 dicot Atriplex confertifolia. Plant Physiol 91:1143–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner DA, Edwards GE (1993) Effects of polyploidy on photosynthesis. Photosynth Res 35:135–147. [DOI] [PubMed] [Google Scholar]
- Warner DA, Ku MS, Edwards GE (1987) Photosynthesis, leaf anatomy, and cellular constituents in the polyploid C4 grass Panicum virgatum. Plant Physiol 84:461–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF (2000) Genome evolution in polyploids. Plant Mol Biol 42:225–249. [PubMed] [Google Scholar]
- Xia B, Dong C, Zhang W, Lu Y, Chen J, Shi J (2013) Highly efficient uptake of ultrafine mesoporous silica nanoparticles with excellent biocompatibility by liriodendron hybrid suspension cells. Sci China Life Sci 56:82–89. [DOI] [PubMed] [Google Scholar]
- Yang D, Li W, Li S, Yang X, Wu J, Cao Z (2007) In vitro embryo rescue culture of F-1 progenies from crosses between diploid and tetraploid grape varieties. Plant Growth Regul 51:63–71. [Google Scholar]
- Yang XM, Cao ZY, An LZ, Wang YM, Fang XW (2006) In vitro tetraploid induction via colchicine treatment from diploid somatic embryos in grapevine (Vitis vinifera L.). Euphytica 152:217–224. [Google Scholar]
- Yao J, Li H, Ye J, Shi L (2016) Relationship between parental genetic distance and offspring's heterosis for early growth traits in liriodendron: implication for parent pair selection in cross breeding. New Forests 47:163–177. [Google Scholar]
- Yu CS, Chen YC, Hwang JK (2010) Prediction of protein subcellular localization. Proteins-structure function. Bioinformatics 64:643–651. [DOI] [PubMed] [Google Scholar]
- Yu Z, Haberer G, Matthes M, Rattei T, Mayer KFX, Gierl A, Torres-Ruiz RA (2010) Impact of natural genetic variation on the transcriptome of autotetraploid Arabidopsis thaliana. Proc Natl Acad Sci USA 107:17809–17814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Luo F, Liu L, Guo F (2010) In vitro induction of tetraploids in crape myrtle (Lagerstroemia indica L.). Plant Cell Tiss Org Cult 101:41–47. [Google Scholar]
- Zhang W, Hao H, Ma L, Zhao C, Yu X (2010) Tetraploid muskmelon alters morphological characteristics and improves fruit quality. Sci Hortic 125:396–400. [Google Scholar]
- Zhang Y, Wang B, Qi S, Dong M, Wang Z, Li Y, Chen S, Li B, Zhang J (2019) Ploidy and hybridity effects on leaf size, cell size and related genes expression in triploids, diploids and their parents in Populus. Planta 249:635–646. [DOI] [PubMed] [Google Scholar]
- Zhen Y, Chen J, Chen Q, Shi J (2015) Elemental analyses of calli and developing somatic embryo of hybrid liriodendron. Pak J Bot 47:189–196. [Google Scholar]
- Zhou H, Zeng W, Yan H (2017) In vitro induction of tetraploids in cassava variety 'Xinxuan 048′ using colchicine. Plant Cell Tiss Org Cult 128:723–729. [Google Scholar]
- Zhou Y, Lei K, Shiying L, Qi P, Xianhong G, Zaiyun L, Jin-Song Z (2015) Transcriptomic analysis reveals differential gene expressions for cell growth and functional secondary metabolites in induced autotetraploid of Chinese Woad (Isatis indigotica Fort.). Plos One 10:e0116392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Li M, Zhao F, Zha H, Yang L, Lu Y, Wang G, Shi J, Chen J (2016) Floral nectary morphology and proteomic analysis of nectar of Liriodendron tulipifera Linn. Front Plant Sci 7:826. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this article and its Supplementary data files. The data are available from the corresponding author on reasonable request.


