Abstract
We have developed a high-throughput, semiautomated, quantitative fluorescence-based PCR assay to detect and type herpes simplex virus (HSV) DNA in clinical samples. The detection assay, which uses primers to the type-common region of HSV glycoprotein B (gB), was linear from <10 to 108 copies of HSV DNA/20 μl of sample. Among duplicate samples in reproducibility runs, the assay showed less than 5% variability. We compared the fluorescence-based PCR assay with culture and gel-based liquid hybridization system with 335 genital tract specimens from HSV type 2 (HSV-2)-seropositive persons attending a research clinic and 380 consecutive cerebrospinal fluid (CSF) samples submitted to a diagnostic virology laboratory. Among the 162 culture-positive genital tract specimens, TaqMan PCR was positive for 157 (97%) specimens, whereas the quantitative-competitive PCR was positive for 144 (89%) specimens. Comparisons of the mean titer of HSV DNA detected by the two assays revealed that the mean titer detected by the gel-based system was slightly higher (median, 1 log). These differences in titers were in part related to the fivefold difference in the amount of HSV DNA used in the amplicon standards with the two assays. Among the 380 CSF samples, 42 were positive by both assays, 13 were positive only by the assay with the agarose gel, and 3 were positive only by the assay with the fluorescent probe. To define the subtype of HSV DNA detected in the screening assay, we also designed one set of primers which amplifies the gG regions of both types of HSV and probes which are specific to either HSV-1 (gG1) or HSV-2 (gG2). These probes were labeled with different fluorescent dyes (6-carboxyfluorescein for gG2 and 6-hexachlorofluorescein for gG1) to enable detection in a single PCR. In mixing experiments the probes discriminated the correct subtype in mixtures with up to a 7-log-higher concentration of the opposite subtype. The PCR typing results showed 100% concordance with the results obtained by assays with monoclonal antibodies against HSV-1 or HSV-2. Thus, while the real-time PCR is slightly less sensitive than the gel-based liquid hybridization system, the high throughput, the lack of contamination during processing, the better reproducibility, and the better ability to type the isolates rapidly make the real-time PCR a valuable tool for clinical investigation and diagnosis of HSV infection.
The rising seroprevalence of herpes simplex virus (HSV) type 2 (HSV-2) infections and the increasing reactivation of HSV among immunocompromised patients have made clinical management of HSV infections a common and increasing problem in clinical practice. Because of the varied clinical manifestations of HSV and the similarity with other mucocutaneous and central nervous system (CNS) infections, accurate diagnosis is a key to effective clinical management. Increasingly, HSV DNA detection in clinical samples is being used to define clinical infection with HSVs (1, 2, 4, 10, 11). PCR testing of cerebrospinal fluid (CSF) has become the method of choice for defining HSV infection of the CNS (1, 10, 11, 13). HSV DNA detection by PCR is more sensitive than viral isolation for defining the presence of HSV-1 or HSV-2 in genital lesions and for demonstrating the presence of subclinical shedding of HSV on mucosal surfaces in the male or female genital tract (2–5, 14, 16, 17). Quantitation of HSV DNA has been used as a means of evaluating the antiviral effects of candidate compounds and as a means of defining the threshold of infectivity of HSV in mucosal and other tissue sites (19). While current strategies for the quantitation of HSV DNA are accurate, they require considerable manipulation and require considerable expertise and time to perform (8).
High-throughput assays with a high degree of sensitivity for the detection of HSV DNA in clinical samples would continue to enhance the utility of this technology in investigative and clinical medicine. As such, we sought to develop a high-output, semiautomated, non-gel-based, quantitative PCR method for the detection of HSV in clinical samples. This report describes our results with a fluorescent dye-based quantitative detection assay that both accurately quantitates and subtypes HSV DNA in clinical samples.
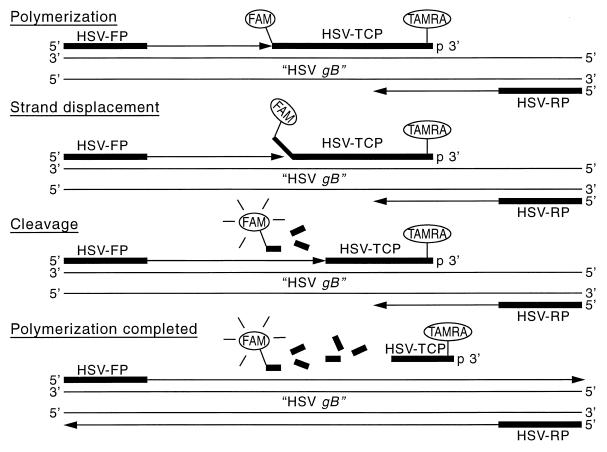
The assay is based upon the commercially available TaqMan PCR detection system used in combination with the Applied Biosystem 7700 analytical PCR system and sequence detector (6, 7). The TaqMan PCR detection system takes advantage of the 5′ exonuclease activity of Taq polymerase to digest an internal probe dually labeled with two fluorescent dyes (9). Due to the proximity of the fluorescent dyes, they undergo fluorescent resonance energy transfer (FRET) prior to cleavage (7). The internal fluorescently labeled probe is typically 20 to 30 bp in length and is added directly to the PCR amplification mixture. The fluorescent probe hybridizes to a region internal to the flanking PCR primers. Upon primer elongation, the probe is cleaved by Taq polymerase’s 5′ to 3′ exonuclease activity, which interrupts the FRET and allows a reporter dye to no longer be physically attached to the quencher dye on the internal probe. The result of the exponential amplification of the PCR target is the exonuclease digestion of the fluorescent probe, which causes the release of the reporter dye from the quencher for every cycle of the PCR (Fig. 1). The amount of reporter dye released is proportional to the amount of DNA being amplified by the PCR.
FIG. 1.
Construction of quantitative fluorescence-based assay for detection of HSV DNA (see text for the sequences of the HSV forward primer [primer HSV-FP], HSV reverse primer [HSV-RP], and HSV gB type-common probe [probe HSV-TCP]. The reporter dye was FAM, and the quencher dye was TAMRA. As described in the text, the proportional release of FAM that occurs during primer elongation results in the ability to monitor the amount of DNA amplified during the exponential phase of the PCR.
PCR products can be detected by determination of the increase in fluorescence intensity of the reporter dye. The increase in fluorescence during the PCR amplification can be monitored while the reaction is proceeding, and the fluorescence data can be analyzed continuously from each of the 96 wells. This approach has the advantage of quantitating the PCR in the exponential phase rather than the endpoint accumulation of PCR product or trying to capture the PCR in the exponential phase, as was done previously in many quantitative PCRs. The determination of copy number is done by comparison of the number of PCR cycles at which fluorescence was detected to the number of PCR cycles necessary for known amounts of DNA to cross a fluorescent threshold on a standard curve. The threshold is set at the beginning of the exponential phase for the amplifications being run.
MATERIALS AND METHODS
Sample collection and preparation.
Genital tract specimens were obtained from patients with suspected genital lesions who were seen at the University of Washington Virology Research Clinic (8, 19, 20). We assayed 335 samples from participants enrolled in studies of viral shedding from the genital area. Specimens were collected directly from genital skin, the cervix, or the perianal region with Dacron swabs, and the swabs were placed into vials containing 1 ml of filter-sterilized digestion buffer (100 mM KCl, 10 mM Tris [pH 8.0], 25 mM EDTA, 0.5% Nonidet P-40 (2, 19). In addition, we also assayed 380 consecutive CSF samples submitted to the University of Washington Diagnostic Virology Laboratory for diagnosis of viral CNS infections. All samples were stored at −20°C until they were ready for assay. The samples were then thawed and left at room temperature, and 200 μl of sample was mixed with 200 μl of AL lysis buffer (Qiagen, Inc., Santa Clarita, Calif.). The mixture was vortexed, 25 μl of proteinase was added, and the solution was vortexed again and placed in a 70°C heat block for 10 min. The tubes were then placed in a 95°C heating block for 15 min to inactivate the proteinase, 210 μl of 100% ethanol was added, and the ethanol-buffer-specimen mixture was then placed into a Qiagen centrifuge column (catalog no. 29163). The column was centrifuged at 6,000 × g for 1 min; 500 μl of AW wash buffer (Qiagen Inc.) was added, and the sample was then centrifuged at 8,000 rpm for 1 min and at 20,000 × g for 2 min. A total of 100 μl of preheated 10 mM Tris was then added, and the tube was placed in a 70°C dry heat block for 5 min and centrifuged at 6,000 × g for 1 min to elute the DNA; 10 μl of the DNA was used for each PCR.
PCR primers and probes.
The PCR primers are directed to the HSV glycoprotein B (gB) gene (2, 4, 8, 19). The forward primer (primer HSV-FP) was 5′-TCC CGG TAC GAA GAC CAG, and the reverse primer (primer HSV-RP) was 5′-AGC AGG CCG CTG TCC TTG. For the gel-based system, purified DNA was amplified with 833 nM concentrations of each primer for 35 cycles by using the conditions as described in a previous paper (2, 8), and PCR products were detected by liquid hybridization with 32P-labeled probe (probe HSV-2A), gel electrophoresis, and autoradiography (2, 8, 19). Because the fluorescent dye system requires a longer probe, we designed the following type-common gB probe (probe: HSV-TCP) 5′-TGG TCC TCC AGC ATG GTG ATG TTG/C AGG TCG-3′. The probe was labeled at the 5′ end with 6-carboxyfluorescein (FAM) and at the 3′ end with 6-carboxytetramethylrhodamine (TAMRA) (Synthegen, Houston, Tex.).
We designed separate primers and probes to distinguish between the two viral subtypes on the basis of the glycoprotein G (gG) gene of HSV (12, 15). The forward HSV typing primer (primer HSV-1 gG1-FP) was 5′-TCC TG/CG TTC CTA/C ACG/T GCC TCC C-3′, and the reverse HSV typing primer (primer HSV-1 gG1-RP) was 5′-GCA GIC AC/TA CGT AAC GCA CGC T-3′. The fluorescent HSV-1 typing probe (probe HSV-1 gG1-P) was 5′-CGT CTG GAC CAA CCG CCA CAC AGG T-3′; the probe was labeled at the 5′ end with 6-hexachlorofluorescein (HEX) and at the 3′ end with TAMRA. The sequence and labels of the HSV-2 gG2 probe (probe HSV-gG2-P) were 5′-FAM-CGA CCA GAC AAA CGA ACG CCG CCG T-3′-TAMRA.
TaqMan PCR.
Each 50 μl-PCR mixture contained 10 μl of purified DNA, 833 nM concentrations of each primer, and 100 nM probe. After 2 min of incubation at 50°C and 2 min of incubation at 95°C for denaturation, the sample was subjected to 45 cycles of PCR. Each cycle was 95°C for 20 s and 58°C for 1 min. The intensities of the fluorescent dyes in each reaction were read automatically during PCR cycling in a PE-Applied Biosystem Sequence Detector 7700 machine. To control for pipetting variability, an internal passive control consisting of the passive fluorescent dye 6-carboxy-X-rhodamine (ROX), which was conjugated to the 5′ end of 5′-GATTAG-3′, was included in the master mixture for each reaction. This reagent was used at a working concentration of 60 nM. The 7700 machine detects this dye and standardizes the quantity of dye in each sample to the quantity of this dye in each reaction mixture. The real-time data that are generated are analyzed with sequence detector software (version 1.6.3; Perkin Elmer, Inc., Foster City, Calif.). The threshold of detection is set at the point that is >10 standard deviations above the background and that occurs when the PCR enters the exponential phase. Each PCR run contained several negative controls, including two reaction mixtures without DNA as well as several specimens that were known to contain no HSV DNA, a positive amplicon control, and a standard dilution curve for amplicon DNA. Each specimen was run in duplicate; only those specimens for which the values in both replications were above the cutoff were considered positive. All positive specimens tested in these studies met these criteria. The sequence detector software determines the standard curve, which is then used to calculate the precise quantities of starting template molecules for the unknown sample. All of the titers in this report are reported as number of copies per 20 μl of specimen.
RESULTS
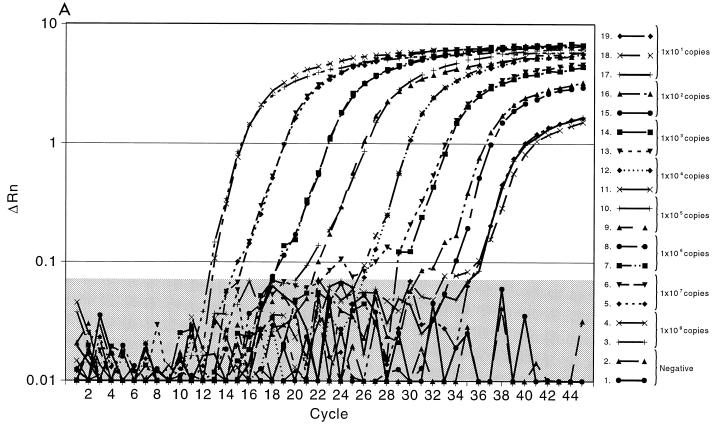
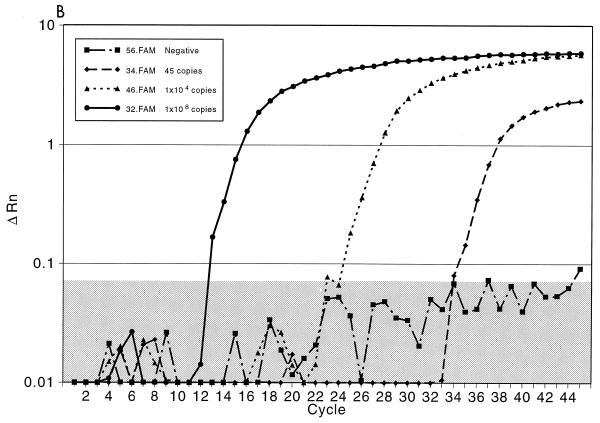
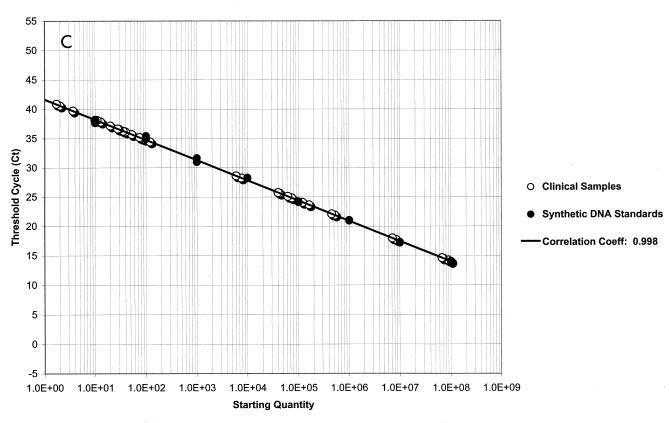
Initially, we evaluated the ability of the TaqMan PCR to detect purified HSV DNA using an HSV-2 gB amplicon (8). For these experiments, 10-fold dilutions of the amplicon were made. As shown in Fig. 2A, linear quantitation was achieved with each of the 1-log dilutions of sample. The standard deviation for each curve was less than 1% for each dilution. Excellent discrimination between the results for the positive samples and those for the negative samples, which contained all reagents except the HSV gB amplicon, was seen with all dilutions. Figure 2B demonstrates the curve for detection of HSV in three genital tract specimens by PCR; one specimen contained a high titer of HSV DNA (108 copies/20 μl of specimen) that was detected after 12 cycles, one contained a modest titer (104 copies/20 μl sample) that was detected after 24 cycles, and one contained a low titer (45 copies/20 μl of sample) that was detected at cycle 33. Again, marked discrimination between the results for the positive samples containing the probe and those for the negative control samples was noted. Figure 2C plots the results for the HSV-2 gB amplicon standards and replicates of 17 representative patient samples to illustrate the quantitative calculation of the titers compared to the titers of the standards. These experiments demonstrated that the linear range of the assay is from 101 to 108 copies of HSV DNA. Clear separation on a logarithmic scale between high- and low-titer patient specimens, along with the closeness of the results for the replicate samples for each specimen, indicates the precision and accuracy of the technique and the instrumentation.
FIG. 2.
Fluorescent probe assay for detection of HSV DNA. ΔRn corresponds to the increase in reporter dye intensity relative to the passive internal reference. (A) Detection of gB amplicon with the gB forward and gB reverse primers and the gB type-common primer. The results are for triple replicates of serial 10-fold dilutions of gB amplicon DNA. The negative control contains the PCR master mixture with unrelated DNA. The shaded area delineates the negative threshold. (B) Results for three representative genital tract samples. Specimen A contained 108 copies of HSV DNA, specimen B contained 104 copies of HSV DNA, and specimen C contained 45 copies of HSV DNA. (C) Results for 17 duplicate clinical samples compared with the results on a standard curve for HSV amplicon DNA. The standard curve is linear between 101 and 108 copies of amplicon DNA. The correlation coefficient between the 17 samples and the standard curve is 0.998. Each sample was run in duplicate, and the results for both replicates are depicted.
Next, we conducted a series of experiments to evaluate the quantitation of HSV DNA in genital samples using both the fluorescent probe and our previously described quantitative-competitive (QC) PCR method using agarose gels and a liquid hybridization detection system (8). For these experiments, purified DNA from the clinical samples was aliquoted and run in the two assays. The technicians who performed the two assays were blinded to the results of the cultures and PCRs. Table 1 presents the results of the two assays for 335 genital tract specimens. All specimens were obtained from persons known to be HSV-2 seropositive. HSV was isolated in tissue culture from 162 of these samples; 173 of the samples were culture negative. Among the 162 culture-positive samples, the PCR assay with the fluorescent probe detected HSV DNA in 157 (97%) of the samples, and the liquid hybridization system detected HSV DNA in 144 (89%) of the samples. While the detection frequency was slightly higher by the fluorescent probe assay, the median titer of HSV DNA per 20 μl of specimen was 105 for the liquid hybridization PCR assay, whereas it was 104 for the TaqMan PCR assay (Table 1). The greatest discrepancy in the two assays was for the 21 specimens in which the HSV DNA titers were 107 and 108 by the QC PCR system (Table 2). To investigate this further we compared the DNA “standards” used for comparison of the quantity of HSV DNA in the two assays, because the lengths of the amplicons used as the standards in the two assays differed by 82 bp. The titer of HSV DNA obtained with the amplicon used in the fluorescent probe system was 0.5 log higher than that obtained with the amplicon used in the gel-based system. Thus, some of the differences in titer appear to be related to the differences in the amplification efficiencies of the two standards. Among the 173 culture-negative specimens, HSV DNA was detected by liquid hybridization in 132 (76%) specimens, whereas it was detected by the TaqMan system in 104 (60%) specimens. The median titer in the culture-negative samples was higher by the QC PCR than with the TaqMan system, 102 versus 101, respectively (Table 2). Again, the differences were more marked for high-titer specimens (Table 2).
TABLE 1.
Comparison between TaqMan and QC PCR for detection of HSV DNA in culture-positive and culture-negative genital swab specimens
| No. of HSV copies/ 20 μl of specimen | No. of specimens with the following results:
|
|||
|---|---|---|---|---|
| Culture positive (n = 162)
|
Culture negative (n = 173)
|
|||
| QC PCR | TaqMan | QC PCR | TaqMan | |
| 0 | 18 | 5 | 41 | 68 |
| 101 | 3 | 1 | 24 | 23 |
| 102 | 1 | 16 | 24 | 44 |
| 103 | 10 | 38 | 32 | 22 |
| 104 | 31 | 57 | 29 | 14 |
| 105 | 32 | 42 | 13 | 2 |
| 106 | 46 | 3 | 7 | 0 |
| 107 | 17 | 0 | 3 | 0 |
| 108 | 4 | 0 | 0 | 0 |
TABLE 2.
Comparison of HSV DNA titer detected by TaqMan PCR versus that detected by QC PCR for culture-positive specimensa
| QC PCR titer (log10) | No. of specimens with the following TaqMan PCR titer (log10):
|
||||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | |
| 0 | 3 | 0 | 1 | 3 | 8 | 3 | 0 |
| 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 |
| 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 3 | 0 | 0 | 5 | 2 | 2 | 1 | 0 |
| 4 | 0 | 0 | 10 | 14 | 4 | 3 | 0 |
| 5 | 0 | 0 | 0 | 12 | 16 | 4 | 0 |
| 6 | 1 | 0 | 0 | 4 | 18 | 22 | 1 |
| 7 | 0 | 0 | 0 | 2 | 5 | 8 | 2 |
| 8 | 0 | 0 | 0 | 0 | 4 | 0 | 0 |
| Total | 5 | 1 | 16 | 38 | 57 | 42 | 3 |
Titers are given as number of HSV DNA copies per 20 μl of specimen. Boldface indicates samples with the same titer in both assays. A total of 162 specimens were tested.
We then evaluated the two assays with 380 consecutive CSF samples submitted to the University of Washington Diagnostic Virology Laboratory for HSV DNA detection (Table 3). HSV DNA was detected in 42 of these samples by both assays, in 13 samples by the agarose gel liquid hybridization assay only, and in 3 samples by the TaqMan PCR only. Seven of the 13 samples with negative TaqMan PCR results had titers of ≤10 copies in 20 μl of specimen. The three specimens which were negative by liquid hybridization but positive by the TaqMan PCR also had low copy numbers. These discrepancies probably relate to the inability to obtain reproducible results for PCR amplification due to the low amounts of target DNA in the sample.
TABLE 3.
Comparison between real-time TaqMan qualitative PCR and semiquantitative PCR with liquid hybridization for detection of HSV DNA in CSFa
| Liquid hybridization assay HSV DNA titer | No. of specimens with the following TaqMan PCR titer (log10):
|
|||||
|---|---|---|---|---|---|---|
| Negative | 101 | 102 | 103 | ≥104 | Total | |
| Negative | 322 | 3 | 0 | 0 | 0 | 325 |
| 101 | 7 | 9 | 0 | 0 | 0 | 16 |
| 102 | 6 | 3 | 10 | 3 | 0 | 22 |
| ≥103 | 0 | 1 | 3 | 6 | 7 | 17 |
| Total | 335 | 16 | 13 | 9 | 7 | 380 |
A total of 380 specimens were tested.
HSV DNA typing.
The primers and probes used in a previous study (4) had type-common sequences and detected both HSV-1 and HSV-2 DNAs (4). For the typing assays we amplified the HSV-1 and HSV-2 gG region and probed the samples with type-specific probes gG1p and gG2P. Figure 3 illustrates the fluorescent dye assay results both for prototype isolates and for HSV isolates previously typed with monoclonal antibodies. Complete concordance was achieved between the two assays. To date, 60 clinical samples previously typed with monoclonal antibodies as either HSV-1 or HSV-2 have been run in the typing assay; 100% concordance with the types obtained with monoclonal antibodies was noted in all cases.
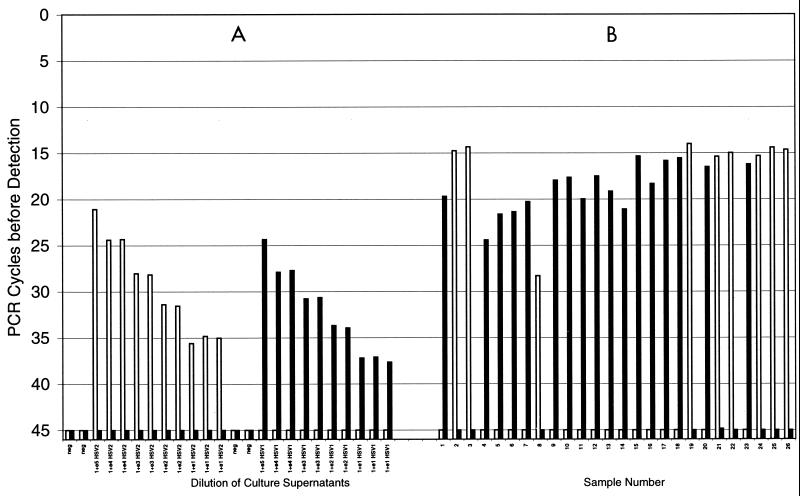
FIG. 3.
Typing of HSV DNA with a fluorescent probe system with the gG1 and gG2 probes. (A) Results obtained with serial dilutions of prototype clinical HSV-2 and HSV-1 isolates. Serial 10-fold dilutions of 105 to 101 of the culture supernatant from a prototype HSV-2 isolate and HSV-1 isolate were made. Note the excellent discrimination between the results obtained with the two probes, even with the large amounts of the heterologous virus type present in the reaction mixtures. (B) Typing results for 26 consecutive clinical isolates from the nasopharyngeal and genital region (17 HSV-1 isolates and 9 HSV-2 isolates). A 100% concordance with monoclonal antibody typing was present. □, HSV-2; ■, HSV-1.
DISCUSSION
We describe the development of a quantitative, easily automated HSV PCR system for the detection of HSV DNA in clinical samples. The assay was able to detect as few as 10 copies of HSV DNA. The linear range of the assay was from 10 to 108 copies; the assay variability was less than 3%. This non-gel-based technique has several advantages over our previous QC agarose gel-based technique (8). First, this system allows a large increase in throughput. The fluorescent probe assay is run in a 96-well format, and many of the steps in the assay are automated. Second, the assay is a closed system in which the tube is never opened postamplification. Third, it uses an automated detection system that quantitates and calculates the degree of fluorescence over that for the control at each cycle and, hence, accurately defines the cycle number and linear range for a positive result. Finally, the inclusion of the internal control dye (ROX) in every reaction mixture allows the PCR machine to normalize the difference in the volumes of the reaction mixtures due to pipetting variation during the PCR setup. The fluorescent probe PCR method missed only 5 of 162 culture-positive genital samples, whereas the gel-based system missed 18 of 162 culture-positive genital samples. However, with culture-negative specimens the TaqMan assay was slightly less sensitive than the QC PCR system for the detection of HSV DNA in either CSF or genital tract specimens. Whether this slightly reduced sensitivity for the culture-negative samples makes the TaqMan assay less useful clinically will require further studies. Little is known about the transmissibility and clinical importance of culture-negative specimens that contain HSV DNA.
One of the advantages of the fluorescent dye system is its reproducibility and precision, both of which were much better than those of our gel-based system. The one disadvantage of the fluorescent dye assay in its current format was that we did not have a way to confirm whether negative specimens were negative because of the presence of nonspecific inhibition of the reaction or due to the absence of viral DNA in the specimen. More recently, we have added an exogenous internal control into our type-common PCR assays to ensure that amplification has occurred; this provides assurance that a negative PCR result is not the result of inhibition of the reaction during the thermocycling procedure. The efficiency of the PCR reaction can be determined with this second probe and fosters confidence in the quantitation result obtained for the sample.
To distinguish between the HSV subtypes, we developed new primers and probes that rely on the differences between HSV-1 and HSV-2 in the gG region (12, 15, 18, 21). Two type-specific fluorescent probes homologous to the regions specific for HSV-1 or HSV-2 were designed. The probes used different reporter dyes that allowed for their simultaneous use in a single PCR tube. Distinction between the two types was readily apparent, even if high titers of heterotypic virus were present in the reaction tube. The model 7700 sequence detector easily differentiated the respective fluorescent emissions of the two viral types. This approach allows us to retest a sample of the original purified DNA to quantify as well as subtype the virus.
In summary, we have described a semiautomated, non-gel-based technique for the quantitation of HSV DNA in clinical samples. The technique is accurate and reproducible and has a large linear range. The use of this assay in clinical studies to evaluate the response to antiviral chemotherapy as well as to define the natural history of HSV infections in a variety of clinical settings is now possible.
ACKNOWLEDGMENTS
This study was supported in part by NIH grant AI-30731 and an unrestricted gift from Glaxo Wellcome Inc.
REFERENCES
- 1.Cinque P, Cleator G M, Weber T, Monteyne P, Sindic van Loon A M. The role of laboratory investigation in the diagnosis and management of patients with suspected herpes simplex encephalitis: a consensus report. J Neurol Neurosurg Psych. 1996;61:339–345. doi: 10.1136/jnnp.61.4.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cone R W, Hobson A C, Brown Z, Ashley R, Berry S, Ishak L, Winter C, Corey L. Frequent reactivation of genital herpes simplex viruses among pregnant women. JAMA. 1994;272:792–796. [PubMed] [Google Scholar]
- 3.Cone R W, Hobson A C, Huang M-L. Coamplified positive control detects inhibition of polymerase chain reactions. J Clin Microbiol. 1992;30:3185–3189. doi: 10.1128/jcm.30.12.3185-3189.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cone R W, Hobson A C, Palmer J, Remington M, Corey L. Extended duration of herpes simplex virus DNA in genital lesions detected by polymerase chain reaction. J Infect Dis. 1991;164:757–760. doi: 10.1093/infdis/164.4.757. [DOI] [PubMed] [Google Scholar]
- 5.Cullen A P, Long C D, Lorincz A T. Rapid detection and typing of herpes simplex virus DNA clinical specimens by the hybrid capture II signal amplification probe test. J Clin Microbiol. 1997;35:2275–2278. doi: 10.1128/jcm.35.9.2275-2278.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibson U, Heid C A, Williams P M. A novel method for real time quantitative RT-PCR. Genome Res. 1996;6:995–1001. doi: 10.1101/gr.6.10.995. [DOI] [PubMed] [Google Scholar]
- 7.Heid C A, Stevens J, Livak K J, Williams P M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 8.Hobson A, Wald A, Wright N, Corey L. Evaluation of a quantitative competitive PCR assay for measuring herpes simplex virus DNA content in genital tract secretions. J Clin Microbiol. 1997;35:548–552. doi: 10.1128/jcm.35.3.548-552.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holland P, Abramson R, Watson R, Gelfand D. Detection of specific polymerase chain reaction product by utilizing the 5′ → 3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimberlin D W, Lakeman F D, Arvin A M, Prober C G, Corey L, Powell D A, Burchett S K, Jacobs R F, Starr S E, Whitley R J the National Institute of Allergy and Infectious Diseases Collaborative Antiviral Study Group. Application of the polymerase chain reaction to the diagnosis and management of neonatal herpes simplex virus disease. J Infect Dis. 1996;174:1162–1167. doi: 10.1093/infdis/174.6.1162. [DOI] [PubMed] [Google Scholar]
- 11.Lakeman F D, Whitley R J. Diagnosis of herpes simplex encephalitis: application of polymerase chain reaction to cerebrospinal fluid from brain-biopsied patients and correlation with disease. J Infect Dis. 1995;171:857–863. doi: 10.1093/infdis/171.4.857. [DOI] [PubMed] [Google Scholar]
- 12.McGeoch D J, Moss H W M, McNab D, Frame M C. DNA sequence and genetic content of HindIII 1 region in the short unique component of the herpes simplex virus type 2 genome: identification of the gene encoding glycoprotein G, and evolutionary comparisons. J Gen Virol. 1987;68:19–38. doi: 10.1099/0022-1317-68-1-19. [DOI] [PubMed] [Google Scholar]
- 13.Mitchell P S, Espy M J, Smith T F, Toal D R, Rys P, Barbari E F, Osmon D R, Persing D H. Laboratory diagnosis of central nervous system infections with herpes simplex virus PCR performed with cerebrospinal fluid specimens. J Clin Microbiol. 1997;35:2873–2877. doi: 10.1128/jcm.35.11.2873-2877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Safrin S, Shaw H, Bolan G, Cuan J, Chiang C S. Comparison of virus culture and the polymerase chain reaction for diagnosis of mucocutaneous herpes simplex virus infection. Sex Transm Dis. 1997;24:176–280. doi: 10.1097/00007435-199703000-00010. [DOI] [PubMed] [Google Scholar]
- 15.Sanchez-Martinez D, Pellett P E. Expression of HSV-1 and HSV-2 glycoporotein G in insect cells by using a novel baculovirus vector. Virology. 1991;182:229–238. doi: 10.1016/0042-6822(91)90666-y. [DOI] [PubMed] [Google Scholar]
- 16.Scott D A, Coulter W A, Biagioni P A, O’Neill H O, Lamey P J. Detection of herpes simplex virus type 1 shedding in the cavity by polymerase chain reaction and enzyme-linked immunosorbent assay at the prodromal stage of recrudescent herpes labialis. J Oral Pathol Med. 1997;26:305–309. doi: 10.1111/j.1600-0714.1997.tb00220.x. [DOI] [PubMed] [Google Scholar]
- 17.Slomka M J, Emry L, Munday P E, Moulsdale M, Brown D W. A comparison of PCR with virus isolation and direct antigen detection for diagnosis and typing of genital herpes. J Med Virol. 1998;55:177–183. [PubMed] [Google Scholar]
- 18.Srinivas R V, Balachandran N, Alonso-Caplen F V, Copans R W. Expression of herpes simplex virus glycoproteins in polarized epithelial cells. J Virol. 1986;58:689–693. doi: 10.1128/jvi.58.2.689-693.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wald A, Corey L, Cone R, Hobson A, Davis G, Zeh J. Frequent genital herpes simplex virus 2 shedding in immunocompetent women: effect of acyclovir treatment. J Clin Invest. 1997;99:1092–1097. doi: 10.1172/JCI119237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wald A, Zeh J, Selke S, Ashley R L, Corey L. Virologic characteristics of subclinical and symptomatic genital herpes infections. N Engl J Med. 1995;333:770–775. doi: 10.1056/NEJM199509213331205. [DOI] [PubMed] [Google Scholar]
- 21.Weldon S K, Su H K, Fetherston J D, Courtney R J. In vitro synthesis and processing of herpes simplex 2 gG-2, using cell-free transcription and translation. J Virol. 1990;64:1357–1359. doi: 10.1128/jvi.64.3.1357-1359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]