Abstract
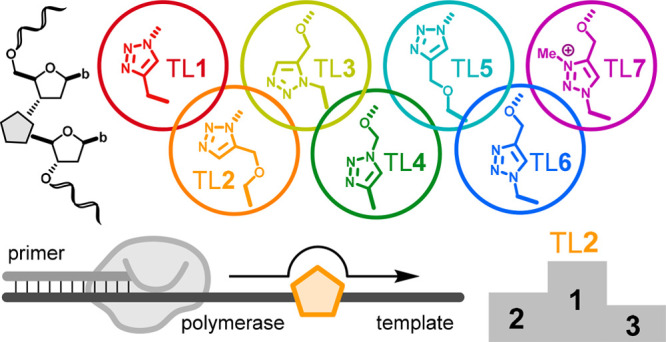
Triazole linkages (TLs) are mimics of the phosphodiester bond in oligonucleotides with applications in synthetic biology and biotechnology. Here we report the RuAAC-catalyzed synthesis of a novel 1,5-disubstituted triazole (TL2) dinucleoside phosphoramidite as well as its incorporation into oligonucleotides and compare its DNA polymerase replication competency with other TL analogues. We demonstrate that TL2 has superior replication kinetics to these analogues and is accurately replicated by polymerases. Derived structure–biocompatibility relationships show that linker length and the orientation of a hydrogen bond acceptor are critical and provide further guidance for the rational design of artificial biocompatible nucleic acid backbones.
Introduction
The replacement of natural phosphodiester (PO) bonds in DNA or RNA by artificial internucleoside linkages can generate remarkable biomimetic oligonucleotides (ONs) with applications as therapeutics,1 xenobiotic genetic polymers,2−4 aptamers,3,5−7 and synthetic genes.8−10 Biological integrity and favorable biophysical properties are critical, and good hybridization properties, mismatch discrimination, and compatibility with certain enzymes (e.g., RNase H) play crucial roles for antisense oligonucleotides.11−15 Other applications of backbone-modified, bioactive oligonucleotides include modified CRISPR-Cas9 systems,16 and compatibility with the cellular gene replication and expression machinery.
In general, replication-competent artificial DNA backbones can be used for gene synthesis,8,9 sequencing,17 or nucleic acid detection.18 A detailed study revealed several molecular characteristics of artificial backbones that are required for compatibility with DNA polymerases during replication.19 Such polymerase compatible artificial backbones comprise 5′-S-phosphorothioesters,20 phosphorothioates,21 disulfides,22 boranophosphates,23 phosphoramidates,10,24,25 amides,19,26 ureas,18 squaramides,18 and triazoles.8,9,19,27−29 Among these, the triazole linkage (TL) represents a powerful and versatile chemical moiety that can be readily formed by the CuI-catalyzed azide–alkyne cycloaddition (CuAAC) reaction, resulting in 1,4-disubstituted 1,2,3-triazoles.30 Examples include TL1,31 TL4,27 TL5,32 and TL629 (Figure 1).33,34 Further modification can be achieved by alkylation of the triazole as exemplified by cationic TL7,19 which is made by methylation of TL6. Many applications of such triazole backbones exist. For example, the CuAAC click ligation of 3′-azido ONs with 5′-alkyne adaptor ONs to form TL1 was recently described for the preparation of next-generation sequencing libraries.17 However, the authors reported low replication efficiencies by several polymerases through TL1. Furthermore, click ligation has been used to assemble long DNA templates with isolated TL6 modifications which can be replicated9,29 or transcribed8,9 by polymerases in bacterial9 or mammalian8 cells while retaining high fidelity read-through.8,9,19,29 However, TL6 suffers from reduced binding affinity to a complementary DNA or RNA target35−38 and induces a TL6-dependent slowdown in PCR replications.9 In search of the ideal nucleic acid triazole linkage, we recently developed a 1,5-disubstituted 1,2,3-triazole internucleoside linkage which was prepared by RuII-catalyzed azide–alkyne cycloaddition (RuAAC) (TL3; Figure 1).38 Altogether, a diverse toolbox of TLs has been reported over the past two decades, all having distinct triazole orientations and linker lengths as indicated in Figure 1. However, whether the more recent triazole isoforms, such as the 1,5-disubstituted TL3, retain compatibility for replication by DNA polymerases had not yet been determined.
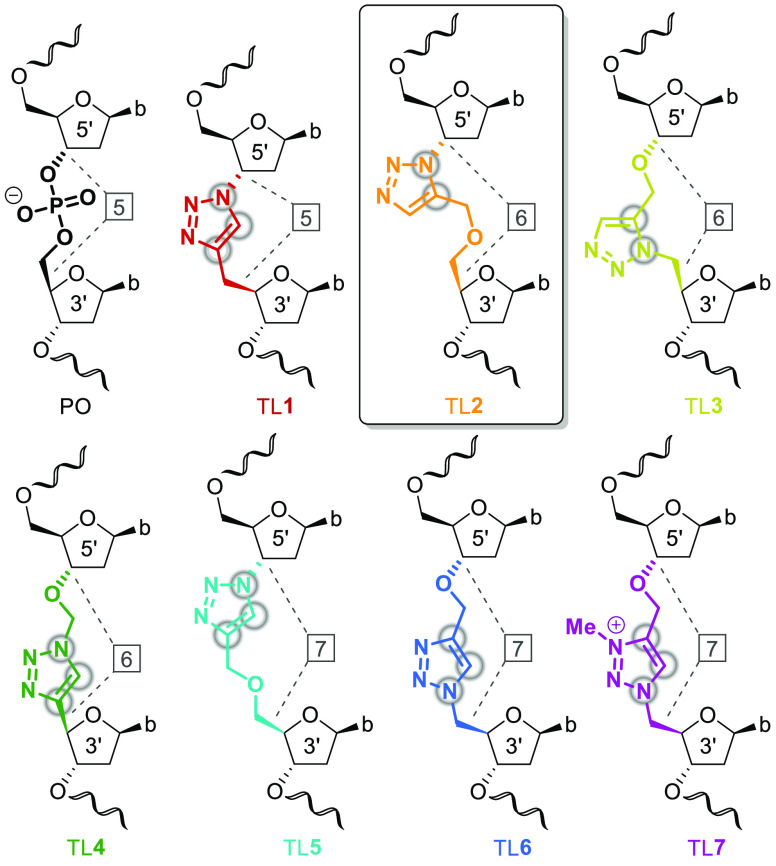
Figure 1.
Summary of TLs connecting the 5′- and the 3′-furanose rings through multiple bonds as indicated by the boxed numbers. Trigonal planar orbital geometries (sp2) along the linkages are highlighted with gray circles. The structure of the natural PO is shown for reference. The TL linkage (TL2) reported here is highlighted. Abbreviations: b, base.
Here we report the synthesis of a novel, 1,5-disubstituted TL connecting two nucleosides through 6 bonds (TL2; Figure 1). We directly compare the effect of RuAAC vs CuAAC to form TLs from the same precursors (TL2 vs TL5), and we study the ability of oligonucleotides containing these TLs to form duplexes with a DNA or RNA target in comparison to reported triazoles TL1, TL3, TL4, and TL6.38 Moreover, we test how efficiently polymerases are able to read through TLs 1–7 in replication templates. For the first time, we assess replication through a 1,5-disubstituted 1,2,3-triazole (TL2 and TL3). Evaluation of the molecular characteristics of the TLs with respect to conferred biocompatibility provides guidance for further biomimetic nucleic acid linkage designs.
Results and Discussion
Synthesis of TL-Linked Dinucleosides
While the 1,5-substitution of a TL (TL3) dinucleotide has been recently reported,38 the corresponding RuAAC ligation of azide- and alkyne-modified ONs to form such triazole isoforms has not yet been developed. Thus, we opted for a dinucleoside strategy to incorporate TL2 into ONs. A suitable dinucleoside phosphoramidite building block containing the 1,5-disubstituted triazole results from RuAAC between alkyne 2 and azide 3(39) (Scheme 1A). A previously described method to selectively alkylate the 5′-OH of thymidine in a single step40 was unsuccessful and resulted in alkylation of the thymine nucleobase instead of the desired 5′-OH (Figure S1). Thus, silyl-protected thymidine 1(41) served as a substrate for propargylation of the 5′-OH to give alkyne 2. RuAAC with azide 3(39) gave dimer 4 with the correct 1,5-substitution (Figure S2). TBAF-mediated deprotection of 4 gave alcohol 5, which was phosphitylated to dimer phosphoramidite 6 for ON synthesis.
Scheme 1. Synthesis of 1,5-TL Phosphoramidite 6 (A) and 1,4-TL Phosphoramidite 9 (B).
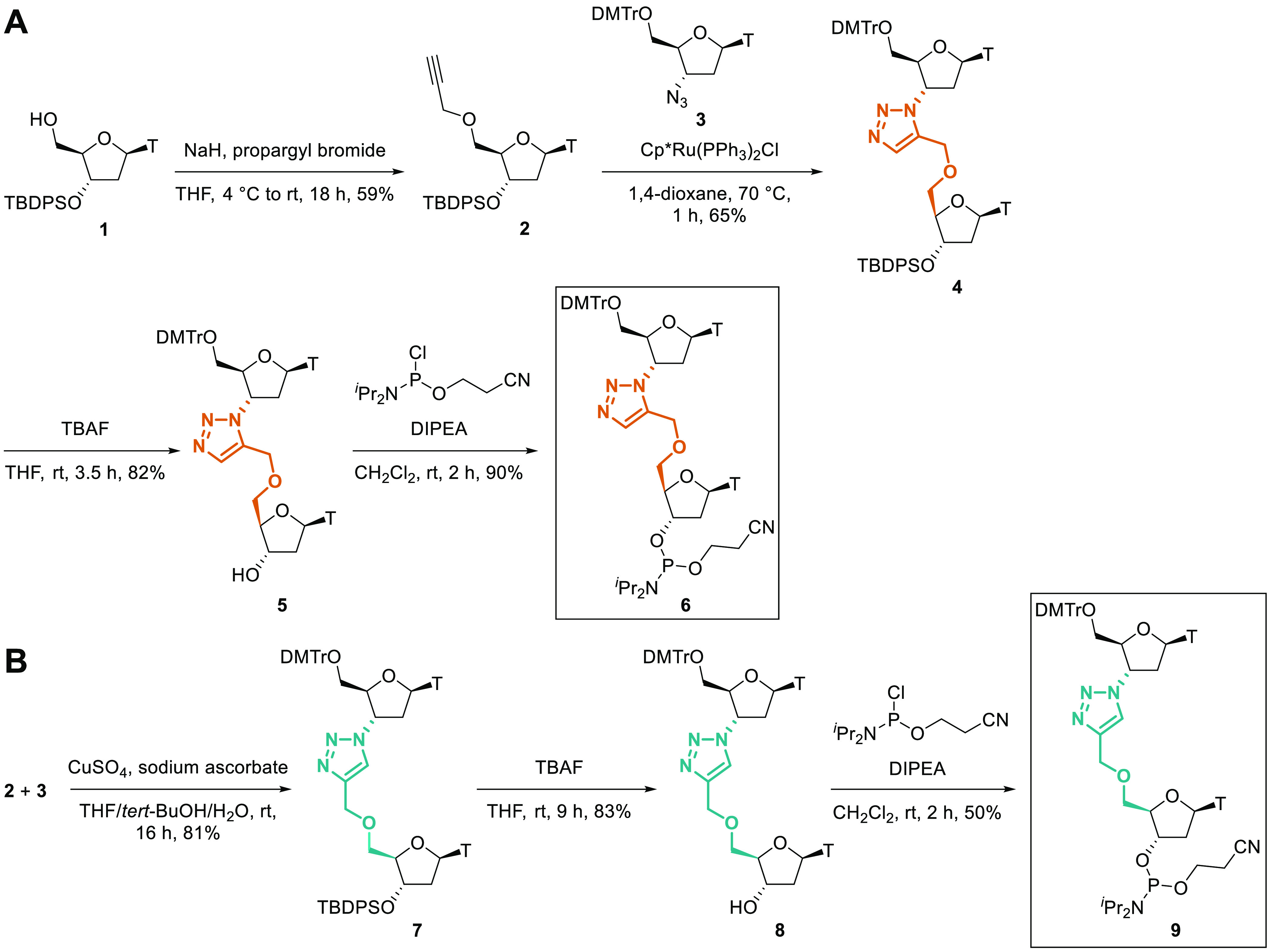
Abbreviations: DIPEA, N,N-diisopropylethylamine; DMTr, 4,4′-dimethoxytrityl; T, thymine; TBAF, tetra-n-butylammonium fluoride; TBDPS, tert-butyldiphenylsilyl.
In parallel, CuAAC of alkyne 2 and azide 3 resulted in the formation of 1,4-disubstituted dimer 7 (Scheme 1B). Desilylation gave alcohol 8, and phosphitylation using standard conditions yielded phosphoramidite 9. The 1,4-substitution of dimer 7—as opposed to the 1,5-substitution of dimer 4—was confirmed by 2D NMR characterization (Figure S3).
Hybridization Studies
Duplex Stability
Phosphoramidites 6 and 9 were coupled during standard ON synthesis to introduce TL2 and TL5 into ON2 and ON5, respectively (Table 1). The ON sequence and position of the TLs are identical to previously reported ONs with TL1, TL3, TL4, and TL6 (ON1, ON3, ON4, and ON6)38 to facilitate accurate comparisons among the TLs in UV melting experiments (Table 1). In comparison to unmodified ON7, introduction of TL2 results in destabilization of the duplex formed with DNA or RNA by −5.1 °C and −3.4 °C, respectively (Table 1). TL5 in ON5 destabilizes the duplexes with DNA and RNA by −8.6 °C, confirming previously reported destabilization by the TL5 modification in a different sequence context.32 Destabilizing effects have also been reported for TL1, TL3, TL4, and TL6 (Table 1).38 All 6-bond TLs (TLs 2–4) generally show more favorable duplex stability properties than their shorter 5-bond (TL1) or longer 7-bond (TL5 and TL6) counterparts. Rigid TL1 is the most destabilizing among the TLs. It is noteworthy that TL4 introduces minimal duplex destabilization (only −0.8 °C) when hybridized to an RNA target. To summarize, the relative duplex stabilities of TL-modified ONs with a DNA target are TL3 > TL4 > TL2 > TL6 > TL5 > TL1, and with an RNA target, TL4 > TL3 ≥ TL2 > TL6 > TL5 > TL1.
Table 1. Melting Temperatures (Tm)a of TL-Modified ONs with a DNAb and RNAc Target.
| ON | Sequence (5′ → 3′)d | DNAb target Tm (ΔTm)/°C | RNAc target Tm (ΔTm)/°C |
|---|---|---|---|
| ON1 | CGACGTTL1TTGCAGC | (−11.2)38 | (−13.6)38 |
| ON2 | CGACGTTL2TTGCAGC | 57.0 (−5.1) | 55.5 (−3.4) |
| ON3 | CGACGTTL3TTGCAGC | (−2.9)38 | (−3.3)38 |
| ON4 | CGACGTTL4TTGCAGC | (−3.2)38 | (−0.8)38 |
| ON5 | CGACGTTL5TTGCAGC | 53.5 (−8.6) | 50.3 (−8.6) |
| ON6 | CGACGTTL6TTGCAGC | (−7.4)38 | (−5.3)38 |
| ON7 | CGACGTTTGCAGC | 62.1 | 58.9 |
Values were obtained from the maxima dA260/dT vs T for 3 μM of each ON in 10 mM phosphate buffer, 200 mM NaCl, pH 7.0. The graphs of dA260/dT vs T are shown in Figure S4. ΔTm are relative to the unmodified control ON7.
Sequence of DNA target = 5′-GCTGCAAACGTCG-3′.
Sequence of RNA target = 5′-GCUGCAAACGUCG-3′.
TL indicates the site where a PO is replaced by TLs 1–6.
Duplex Structure
Despite some destabilizing effects, TL1, TL3, TL4, and TL6 are known to retain global B- or A-form duplex structures with the DNA or RNA target, respectively.38 In the current study, duplexes formed by TL2- and TL5-modified ON2 and ON5 with complementary DNA or RNA were analyzed by circular dichroism (CD) spectroscopy (Figure 2). The duplexes of ON2 with DNA and RNA show almost identical spectra to the equivalent duplexes formed by unmodified ON7. Duplexes of ON5 with DNA or RNA also retain global B- or A-form helical structures, respectively.
Figure 2.
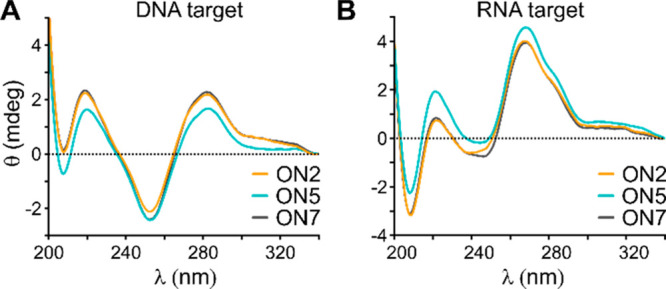
CD spectra of ON2, ON5 and ON7 in duplex with (A) DNA and (B) RNA containing 3 μM of ON and 3 μM of the target in 10 mM phosphate buffer, 200 mM NaCl, pH 7.0. Data points were taken as an average of four scans at rt. DNA target = 5′-GCTGCAAACGTCG-3′, RNA target = 5′-GCUGCAAACGUCG-3′.
Enzymatic Read-through Studies
The ability of TL-modified ONs (TLs 1–6) to form duplexes with DNA and RNA targets while maintaining global structural integrity suggests that these TLs could be promising biocompatible DNA backbone mimics. Indeed, TL1, TL6, and methylated TL7 have been extensively studied as modifications in templates for replication by DNA and RNA polymerases.8,9,16,19,29,42 Moreover, a previous study reported encouraging results for TL4 within modified primers in qPCR.27 However, no read-through compatibilities of 1,4-disubstituted TL5 or 1,5-disubstituted TLs (TL2 and TL3) have been reported. Thus, we compared TLs 1–7 in primer extension studies (Figure 3 and Figure 4). We synthesized unmodified templates and TL-modified templates, each containing a single TL (TLs 1–7), using phosphoramidite 6 (TL2), 9 (TL5), or previously reported phosphoramidites (TL1,19 TL3,38 TL4,27 TL6,43 and TL719). We then assessed the polymerase compatibilities of TLs 1–7 by enzymatic extension of fluorescein (FL)-labeled primers followed by analysis of the FL-labeled products after gel electrophoresis. The sequences of the templates and primers can be found in Table S1.
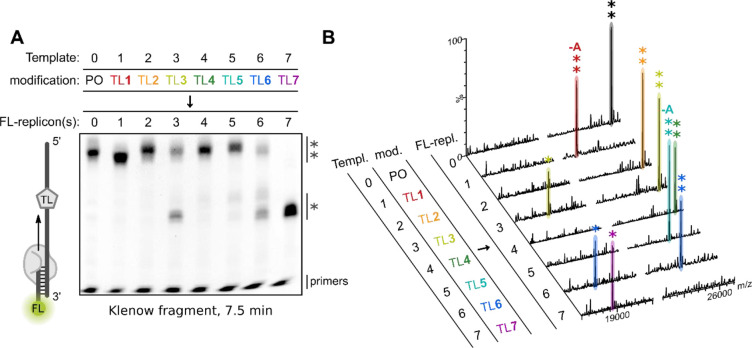
Figure 3.
Klenow fragment-mediated extension of FL-tagged primers. (A) Polyacrylamide gel electrophoresis (PAGE) of FL-labeled extension products after incubation with Klenow fragment at 37 °C for 7.5 min using either unmodified Template 0 (PO) or templates containing a single TL modification: Template 1 (TL1), Template 2 (TL2), Template 3 (TL3), Template 4 (TL4), Template 5 (TL5), Template 6 (TL6), or Template 7 (TL7). The FL-label was visualized (λex = 460 nm, λem = 516–600 nm). Read-through and truncated replicons are marked with (**) and (*), respectively. (B) MS analysis of the primer extension experiments using Klenow fragment at 37 °C for 7.5 min. Templates (Templ.) and their modifications (mod.) are identical to (A). Identified peaks for read-through and truncated FL-labeled replicons (FL-repl.) are marked with (**) and (*), respectively. –A denotes an identified A-deletion. For sequences of templates and FL-primers see Table S1. For full mass spectra and peak assignments see Figures S5–S7 and Table S2.
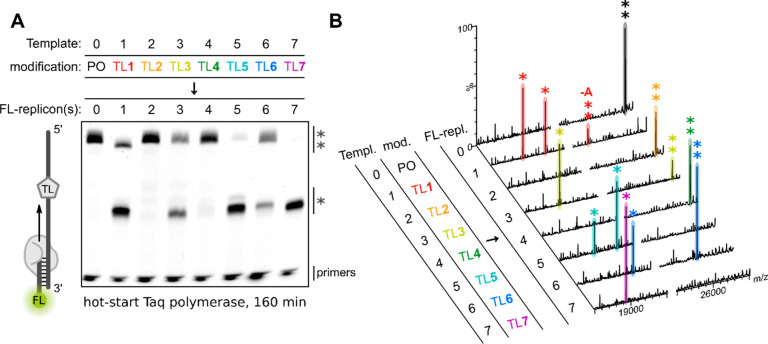
Figure 4.
Hot-start Taq-mediated extension of FL-tagged primers. (A) PAGE of FL-labeled extension products after incubation with hot-start Taq at 60 °Cfor 160 min using either unmodified Template 0 (PO) or templates containing a single TL modification: Template 1 (TL1), Template 2 (TL2), Template 3 (TL3), Template 4 (TL4), Template 5 (TL5), Template 6 (TL6), or Template 7 (TL7). The FL-label was visualized (λex = 460 nm, λem = 516–600 nm). (B) MS analysis of the primer extension experiments using hot-start Taq polymerase at 60 °C for 160 min. Templates (Templ.) and their modifications (mod.) are identical to (A). Identified peaks for read-through and truncated FL-labeled replicons (FL-repl.) are marked with (**) and (*), respectively. −A denotes an identified A-deletion. For sequences of templates and FL-primers see Table S1. For full mass spectra and peak assignments see Figures S8–S10 and Table S3.
Klenow Fragment
Extension of an FL-labeled primer by the large fragment of DNA polymerase I (Klenow fragment) using unmodified Template 0 (PO) resulted in formation of the expected full-length product, which was confirmed by mass spectrometry (MS) analysis (FL-replicon 0**; Figure 3A,B). Primer extension through the TL1-modified Template 1 gave a band that migrated slightly faster than FL-replicon 0 and a mass corresponding to a replicon with an A-deletion (FL-replicon 1**; Figure 3A,B). The fact that read-through of TL1 by Klenow fragment results in an A-deletion mutation which is situated next to the TL1 was previously shown by sequencing of the replicons.19 Templates bearing 6-bond TLs 2–4 resulted in the formation of the expected full-length products as confirmed by MS (FL-replicons 2**, 3**, and 4**; Figure 3A,B). However, replication of the TL3 template also produced a truncated product, shown by MS analysis to be a consequence of premature extension termination directly before the triazole (FL-replicon 3*; Figure 3A,B). The longer 7-bond TL5 in Template 5 was replicated but resulted in a product with an A-deletion (FL-replicon 5**; Figure 3A,B), and additional faint truncation bands were visible on the gel. Extension through TL6 produced the expected full-length product (FL-replicon 6**; Figure 3A,B) and truncated products, one of which results from termination directly before TL6 (FL-replicon 6*; Figure 3A,B). The presence of cationic methylated TL7 in Template 7 is catastrophic to the primer extension and exclusively results in a truncated product from extension termination directly before the methylated triazole (FL-replicon 7*; Figure 3A,B). This confirms previous reports for this TL.19
Hot-Start Taq Polymerase
It is known that replication by Taq polymerase is slowed down at high DNA concentrations such as those used in the primer extension assays.44 Moreover, a previous study showed that the replication of an unmodified template can be significantly slowed down under these conditions (up to 120 min).19 Thus, primer extensions along the TL-modified templates were performed with hot-start Taq polymerase at 60 °C for an extended time of 160 min (Figure 4). Replication of unmodified Template 0 (PO) resulted in the exclusive formation of the expected full-length product containing a Taq-characteristic 3′-A overhang as determined by MS (FL-replicon 0**; Figure 4A,B). Replication of the TL1-modified template resulted in minor amounts of a product for which MS suggested deletion of an A-nucleotide (FL-replicon 1**; Figure 4A,B). The major products from TL1 were truncated ONs where replication stopped directly before or after the triazole (FL-replicons 1*; Figure 4A,B). This supports the mutagenic effect of TL1 in causing deletions around the artificial linkage with a Taq polymerase.19 Remarkably, replication through TL2 yielded a clean full-length product including the expected Taq-characteristic 3′-A overhang (FL-replicon 2**; Figure 4A,B). No truncated products were detected on the gel or by MS. In contrast, a significant amount of truncated product was observed for the TL3 template where extension terminated directly before the triazole (FL-replicon 3*; Figure 4A,B). Extension through TL4 resulted in the expected full-length product without truncation (FL-replicon 4**; Figure 4A,B). In contrast to the previous primer extension experiment with Klenow fragment, replication through TL5 using hot-start Taq polymerase mainly resulted in truncated products from termination directly before or after the triazole (FL-replicons 5*; Figure 4A,B). Replication of the TL6 template produced the correct full-length construct (FL-replicon 6**; Figure 4A,B) and a truncated product (FL-replicon 6*; Figure 4A,B). As seen for Klenow fragment, methylated TL7 completely blocks extension (FL-replicon 7*; Figure 4A,B). Across the Klenow and hot-start Taq primer extension assays, TL2 and TL4 stand out as being the only TLs resulting in the clean formation of the expected full-length product without any detectable mutations or truncated fragments observed on gel or by MS. The excellent read-through compatibility of TL2 by Klenow fragment and hot-start Taq polymerase was confirmed with an alternative sequence (Template 9; Figures S11, S12 and Table S4).
qPCR Kinetics
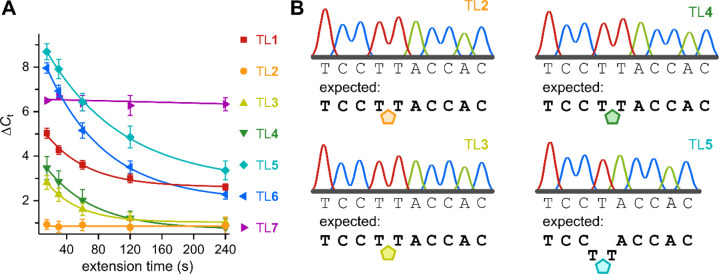
To assess the time dependencies of TLs 1–7, the modified templates were used in qPCR experiments with various extension times. Impaired read-through of a given template results in an increase of the threshold cycle (Ct)—the number of qPCR cycles after which a signal overcomes a set threshold. If impairment can be overcome with time, the resulting Ct value will decrease with longer extension times if within the range of the time dependency. Amplification of unmodified templates can be considered as being time independent under the conditions of the study, as all internucleoside linkages are natural POs which are formed rapidly. Ct values of qPCR reactions with extension times ranging from 15–240 s were determined for templates with TLs 1–7 using hot-start Taq polymerase (Figures S13–S18). The differences between the determined Ct values of the modified templates to the Ct values of the respective isosequential unmodified PO templates (ΔCt) are plotted for each extension time in Figure 5A. Except for TL2 and TL7, all TLs show time dependency for the read-through of the artificial linkage. The apparent time independency of TL7 has been previously explained by a thermodynamic barrier created by TL7 that can only be overcome when transitioning from extension (60 °C) to denaturation (95 °C) in each cycle.19 Most notably, the shorter 5- and 6-bond TLs 1–4 are less time dependent than the longer 7-bond TL5 and TL6. Indeed, ΔCt’s of TL5 and TL6 continued to decrease up to 240 s, the highest extension time tested in this study. The ΔCt’s of TL1, TL3, and TL4 decrease with longer extension times, but the fitted curves flatten toward an extension time of 240 s. In contrast, the ΔCt of TL2 showed no time dependency within the tested conditions. Together with the superior results from the primer extension experiments, this suggests that the observed time independency of TL2 does not originate from transitions between the cycles as seen for TL7. Remarkably, there is no significant kinetic barrier imposed by TL2 for read-through of hot-start Taq polymerase under the tested conditions.
Figure 5.
qPCR kinetics and PCR product sequencing. (A) Differences of threshold cycles (ΔCts) of TL-modified templates to their respective isosequential unmodified control template from qPCRs with hot-start Taq polymerase as a function of extension time. Unmodified templates: Template 0 and Templates 10–12. TL-modified templates: Template 1 (TL1), Template 2 (TL2), Template 3(TL3), Template 4 (TL4), Template 5 (TL5), Template 6 (TL6), and Template 7 (TL7). ΔCt = Ct(TL-modified) – Ct(unmodified) for each extension time. Data are presented as the average from triplicates ± standard error of the mean. For Ct’s see Figure S22. (B) Representative Sanger sequencing results for hot-start Taq PCR amplicons cloned into a vector. Amplicons result from PCR amplifications of Template 2 (TL2), Template 3 (TL3), Template 4 (TL4), and Template 5 (TL5) using hot-start Taq polymerase with an extension time of 30 s each cycle. Corresponding positions of the TLs in the templates are indicated as pentagons. For full template sequences see Table S1, Figure S23, and Figure S24.
DNA Sequencing
To verify that read-through of the TL-modified templates results in correct replication, PCR products from hot-start Taq polymerase-mediated amplification were sequenced. While TL1, TL6, and TL7 have been previously studied for replication fidelity,19 the question whether TLs 2–5 can be replicated correctly in PCR remains unanswered. Thus, PCR amplicons generated using hot-start Taq polymerase and templates containing TL2, TL3, TL4, or TL5 were cloned into a vector and transformed into Escherichia coli. Several colonies were randomly picked, and the recovered vectors were analyzed by Sanger sequencing (Figure 5B). The 6-bond triazole backbones (TLs 2–4) resulted in the correct read-through producing the expected sequence for all colonies picked (n = 4). In contrast, the longer 7-bond TL5 resulted in a deletion mutation around the artificial linkage for which the TTL5T motif has been read as a single T in four out of five colonies picked.
Structure–Biocompatibility Relationship
Analysis of the molecular properties of TLs 1–7 provides insight into the structure–biocompatibility relationship among the TLs for replication (Table 2). Most noticeable are the differences in TL backbone lengths. For 5-bond TL1, whose number of bonds is closest to the natural PO, inefficient read-through by hot-start Taq was observed confirming previous reports.17,19 Moreover, TL1 is mutagenic and results in single point deletions around the triazole when read by Klenow fragment or Taq polymerases.19 The read-through efficiencies of 6-bond TLs 2–4 gave different results in the primer extension assay. However, all 6-bond TLs imposed the lowest kinetic barriers on polymerase read-through and all amplicons from PCRs with hot-start Taq polymerase had the correct sequences. The longer 7-bond TL5 expressed poor read-through efficiency in the primer extension assay with hot-start Taq polymerase and resulted in deletion mutations in PCR amplicons. In contrast, 7-bond TL6 is known to be read-through correctly resulting in the expected amplicon sequences.19 Previous studies suggested that flexibility of the artificial backbone is required for read-through.19 There is an overall increase of flexibility along the TL backbone of 1,5-disubstituted TLs (TL2 and TL3: 2 × sp2; Table 2 and Figure 1) compared to 1,4-disubstituted TLs (TL1 and TLs 4–6: 3 × sp2; Table 2 and Figure 1). Moreover, the poor biocompatibility of 5-bond TL1 might be attributed to the combination of increased rigidity and highest duplex destabilization, defeating the positive contributions from being closest to the natural PO linker length. However, the flexibility model does not explain the lower replication efficiency of 6-bond TL3 (2 × sp2; Table 2 and Figure 1) compared to 6-bond TL4 (3 × sp2; Table 2 and Figure 1) and 6-bond TL2 (2 × sp2; Table 2 and Figure 1) in the primer extension studies (TL2, TL3 and TL4; Figure 3A and Figure 4A). Taq polymerase mainly interacts with the DNA through hydrogen bonding with the phosphates,45 and a crucial factor in read-through of artificial backbones is the ability to donate an electron pair as a hydrogen bond acceptor.19 As such, undesirable orientation of the hydrogen bond acceptor in TL3 is a plausible reason for its hampered read-through compatibility. Moreover, N3-methylation in TL7 is catastrophic, further suggesting that a hydrogen acceptor is important for replication. However, it is likely that the additional charge inversion and steric hindrance contribute to the replication incompatibility of TL7, too.
Table 2. Summary of the Structural Characteristics and Read-through Compatibilities of TLs 1–7.
| TL | linker lengtha | No. sp2 centersb | kinetic barrier (Taq) | Correct read-through (Taq)c |
|---|---|---|---|---|
| PO | 5 | 0 | – | – |
| TL1 | 5 | 3 | medium/high | no, deletion19 |
| TL2 | 6 | 2 | not significant | yes |
| TL3 | 6 | 2 | medium | yes |
| TL4 | 6 | 3 | medium | yes |
| TL5 | 7 | 3 | High | no, deletion |
| TL6 | 7 | 3 | High | yes19 |
| TL7 | 7 | 3 | very high | no, deletions19 |
Defined as the minimal number of bonds connecting the 5′- and the 3′-furanose rings.
Along the internucleoside linkage.
Defined as having the correct sequence after read-through has occurred.
Taking these observations together, we hypothesize that the ideal TL results from a combination of linker length, availability of a hydrogen bond acceptor, and overall flexibility. Extending this to any artificial DNA backbone, we propose that the artificial backbone should be 5–6 bonds in length and provide an accessible hydrogen bond acceptor, while maintaining an overall high degree of flexibility. The new 1,5-disubstituted TL2 combines these molecular requirements and possesses outstanding biocompatibility. The proposed molecular requirements also agree with our previously described model, which was derived from read-through compatibility tests of a range of backbone linkages including amides, phosphoro(di)thioates, phosphoramidates, and squaramides.18,19,24,25 It is important to emphasize that the observed read-through compatibilities are dependent on the polymerase and similar polymerase dependencies were reported for other PO mimics.18,19
Conclusions
In conclusion, triazole linkages are interesting phosphodiester mimics for applications in synthetic biology, biotechnology, and other areas. A profound understanding of the structure–biocompatibility relationship among different TLs is crucial for their successful application in biochemical and biological systems. Here we report the synthesis of novel 1,5-disubstituted TL2 by RuAAC and directly compare this to its 1,4-disubstituted equivalent TL5, which is formed by CuAAC. In duplex melting experiments, TL2-modified ON2 forms more stable duplexes with DNA and RNA targets than TL5-modified ON5. Moreover, the read-through kinetics and amplification of TL2 by Klenow fragment and Taq DNA polymerase exceed all other TLs tested. To the best of our knowledge, TL2 has the fastest read-through kinetics and the highest efficiency of full-length product formation by Taq polymerase among the TLs reported to date. Further studies will be necessary to determine if the excellent read-through compatibility of TL2 by Taq can be translated into a cellular environment and be accommodated by RNA polymerases as previously demonstrated for TL6.8,9,29 Moreover, direct comparison between TL2 and TL5 revealed a remarkable difference in read-through accuracy. While TL2 was read correctly by hot-start Taq polymerase, TL5 resulted in a deletion mutation around the triazole linkage. We have discussed the structure–biocompatibility relationship of several TLs, and we provide a rationale to assist future designs of artificial backbone mimics in replication templates. Importantly, the structure–biocompatibility relationship presented in this study is derived from ONs with TLs between two thymidines, providing a rather challenging sequence context for polymerase read-through. Efficiencies can vary with the nucleobase sequence and generally improve with greater duplex stability (a higher GC-content) around the triazole. Indeed, enhanced read-through compatibility was previously observed for a CTL6C motif.19,29 Hence, we anticipate that further investigations using different sequences will provide additional validation of the proposed model. The application of RuAAC to form TL2 exemplifies a powerful strategy to increase TL flexibility through formation of 1,5-disubstituted 1,2,3-triazoles. In contrast to the widely adopted CuAAC reaction, the feasibility of the RuAAC reaction to efficiently ligate azide- and alkyne-modified ONs in high yield, and without inducing DNA damage, remains elusive. However, in light of recent advances to obtain 1,5-disubstituted 1,2,3-triazoles via water and air compatible Ni-catalyzed click chemistry,46 TL2 represents a promising backbone for use in a plethora of applications across the life sciences.
Acknowledgments
S.E. is grateful to the EPSRC Centre for Doctoral Training in Synthesis for Biology and Medicine (EP/L015838/1) for a studentship, generously supported by AstraZeneca, Diamond Light Source, Defence Science and Technology Laboratory, Evotec, GlaxoSmithKline, Janssen, Novartis, Pfizer, Syngenta, Takeda, UCB, and Vertex. A.H.E.-S. is supported by BBSRC Grant BB/R008655/1, in partnership with ATDBio Ltd., and Y.R.B. is supported by BBSRC Grant BB/S018794/1. L.T. is supported by EPSRC Grant EP/S019944/1. P.W. thanks The National Science Centre (Poland, UMO-2018/28/T/ST5/00109) for a travel grant.
Glossary
Abbreviations
- Cas9
CRISPR associated protein 9
- CRISPR
clustered regularly interspaced short palindromic repeats
- DIPEA
N,N-diisopropylamine
- DMTr
4,4-dimethoxy trityl
- TL
triazole linkage
- qPCR
quantitative polymerase chain reaction
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.1c08057.
Experimental section, characterization data and NMRs of new compounds, additional primer extension experiments, Tm curves, oligonucleotide HPLC traces, oligonucleotide mass spectra, qPCR and sequencing data (PDF)
Author Present Address
§ Department of Chemistry, Ludwig Maximilians University, Munich, Germany
Author Present Address
# Faculty of Physics, University of Warsaw, L. Pasteura 5, 02-093, Warsaw, Poland.
The authors declare no competing financial interest.
Supplementary Material
References
- Wan W. B.; Seth P. P. The Medicinal Chemistry of Therapeutic Oligonucleotides. J. Med. Chem. 2016, 59 (21), 9645–9667. 10.1021/acs.jmedchem.6b00551. [DOI] [PubMed] [Google Scholar]
- Liu C.; Cozens C.; Jaziri F.; Rozenski J.; Maréchal A.; Dumbre S.; Pezo V.; Marlière P.; Pinheiro V. B.; Groaz E.; Herdewijn P. Phosphonomethyl Oligonucleotides as Backbone-Modified Artificial Genetic Polymers. J. Am. Chem. Soc. 2018, 140 (21), 6690–6699. 10.1021/jacs.8b03447. [DOI] [PubMed] [Google Scholar]
- Arangundy-Franklin S.; Taylor A. I.; Porebski B. T.; Genna V.; Peak-Chew S.; Vaisman A.; Woodgate R.; Orozco M.; Holliger P. A Synthetic Genetic Polymer with an Uncharged Backbone Chemistry Based on Alkyl Phosphonate Nucleic Acids. Nat. Chem. 2019, 11 (6), 533–542. 10.1038/s41557-019-0255-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eremeeva E.; Herdewijn P. Reprint of: Non Canonical Genetic Material. Curr. Opin. Biotechnol. 2019, 60, 259–267. 10.1016/j.copbio.2019.11.009. [DOI] [PubMed] [Google Scholar]
- Kang J.; Lee M. S.; Copland J. A.; Luxon B. A.; Gorenstein D. G. Combinatorial Selection of a Single Stranded DNA Thioaptamer Targeting TGF-B1 Protein. Bioorg. Med. Chem. Lett. 2008, 18 (6), 1835–1839. 10.1016/j.bmcl.2008.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashimoto Y.; Matsui T.; Nishino Y.; Taira J.; Inoue H.; Takeuchi M.; Yamagishi S. Blockade by Phosphorothioate Aptamers of Advanced Glycation End Products-Induced Damage in Cultured Pericytes and Endothelial Cells. Microvasc. Res. 2013, 90, 64–70. 10.1016/j.mvr.2013.08.010. [DOI] [PubMed] [Google Scholar]
- Varizhuk A. M.; Tsvetkov V. B.; Tatarinova O. N.; Kaluzhny D. N.; Florentiev V. L.; Timofeev E. N.; Shchyolkina A. K.; Borisova O. F.; Smirnov I. P.; Grokhovsky S. L.; Aseychev A. V.; Pozmogova G. E. Synthesis, Characterization and in Vitro Activity of Thrombin-Binding DNA Aptamers with Triazole Internucleotide Linkages. Eur. J. Med. Chem. 2013, 67, 90–97. 10.1016/j.ejmech.2013.06.034. [DOI] [PubMed] [Google Scholar]
- Birts C. N.; Sanzone A. P.; El-Sagheer A. H.; Blaydes J. P.; Brown T.; Tavassoli A. Transcription of Click-Linked DNA in Human Cells. Angew. Chem., Int. Ed. 2014, 53 (9), 2362–2365. 10.1002/anie.201308691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukwikila M.; Gale N.; El-Sagheer A. H.; Brown T.; Tavassoli A. Assembly of a Biocompatible Triazole-Linked Gene by One-Pot Click-DNA Ligation. Nat. Chem. 2017, 9 (11), 1089–1098. 10.1038/nchem.2850. [DOI] [PubMed] [Google Scholar]
- Nguyen H.; Abramov M.; Eremeeva E.; Herdewijn P. In Vivo Expression of Genetic Information from Phosphoramidate–DNA. ChemBioChem 2020, 21 (1–2), 272–278. 10.1002/cbic.201900712. [DOI] [PubMed] [Google Scholar]
- Iwamoto N.; Butler D. C. D.; Svrzikapa N.; Mohapatra S.; Zlatev I.; Sah Di. W. Y.; Standley S. M.; Lu G.; Apponi L. H.; Frank-Kamenetsky M.; Zhang J. J.; Vargeese C.; Verdine G. L. Control of Phosphorothioate Stereochemistry Substantially Increases the Efficacy of Antisense Oligonucleotides. Nat. Biotechnol. 2017, 35 (9), 845–851. 10.1038/nbt.3948. [DOI] [PubMed] [Google Scholar]
- Rait V. K.; Shaw B. R. Boranophosphates Support the RNase H Cleavage of Polyribonucleotides. Antisense Nucleic Acid Drug Dev. 1999, 9 (1), 53–60. 10.1089/oli.1.1999.9.53. [DOI] [PubMed] [Google Scholar]
- Šǐpová H.; Špringer T.; Rejman D.; Šimák O.; Petrová M.; Novák P.; Rosenbergová Š.; Páv O.; Liboska R.; Barvík I.; Štěpánek J.; Rosenberg I.; Homola J.; Šípová H.; Špringer T.; Rejman D.; Šimák O.; Petrová M.; Novák P.; Rosenbergová Š.; Páv O.; Liboska R.; Barvík I.; Štěpánek J.; Rosenberg I.; Homola J. 5′-O-Methylphosphonate Nucleic Acids—New Modified DNAs That Increase the Escherichia Coli RNase H Cleavage Rate of Hybrid Duplexes. Nucleic Acids Res. 2014, 42 (8), 5378–5389. 10.1093/nar/gku125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miroshnichenko S. K.; Patutina O. A.; Burakova E. A.; Chelobanov B. P.; Fokina A. A.; Vlassov V. V.; Altman S.; Zenkova M. A.; Stetsenko D. A. Mesyl Phosphoramidate Antisense Oligonucleotides as an Alternative to Phosphorothioates with Improved Biochemical and Biological Properties. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (4), 1229–1234. 10.1073/pnas.1813376116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan D.; Lunstad B.; Yamada C. M.; Stell B. G.; Caruthers M. H.; Dellinger D. J. Biochemical Properties of Phosphonoacetate and Thiophosphonoacetate Oligodeoxyribonucleotides. Nucleic Acids Res. 2003, 31 (14), 4109–4118. 10.1093/nar/gkg439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taemaitree L.; Shivalingam A.; El-Sagheer A. H.; Brown T. An Artificial Triazole Backbone Linkage Provides a Split-and-Click Strategy to Bioactive Chemically Modified CRISPR SgRNA. Nat. Commun. 2019, 10 (1), 1610. 10.1038/s41467-019-09600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura F.; Fujino T.; Kogashi K.; Shibata Y.; Miura M.; Isobe H.; Ito T. Triazole Linking for Preparation of a Next-Generation Sequencing Library from Single-Stranded DNA. Nucleic Acids Res. 2018, 46 (16), e95 10.1093/nar/gky452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivalingam A.; Taemaitree L.; El-Sagheer A. H.; Brown T. Squaramides and Ureas: A Flexible Approach to Polymerase-Compatible Nucleic Acid Assembly. Angew. Chem., Int. Ed. 2020, 59 (28), 11416–11422. 10.1002/anie.202000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivalingam A.; Tyburn A. E. S.; El-Sagheer A. H.; Brown T. Molecular Requirements of High-Fidelity Replication-Competent DNA Backbones for Orthogonal Chemical Ligation. J. Am. Chem. Soc. 2017, 139 (4), 1575–1583. 10.1021/jacs.6b11530. [DOI] [PubMed] [Google Scholar]
- Xu Y.; Kool E. T. Chemical and Enzymatic Properties of Bridging 5′-S-Phosphorothioester Linkages in DNA. Nucleic Acids Res. 1998, 26 (13), 3159–3164. 10.1093/nar/26.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreola M. L.; Calmels C.; Michel J.; Toulmé J. J.; Litvak S. Towards the Selection of Phosphorothioate Aptamers: Optimizing in Vitro Selection Steps with Phosphorothioate Nucleotides. Eur. J. Biochem. 2000, 267 (16), 5032–5040. 10.1046/j.1432-1327.2000.01557.x. [DOI] [PubMed] [Google Scholar]
- Hansen D. J.; Manuguerra I.; Kjelstrup M. B.; Gothelf K. V. Synthesis, Dynamic Combinatorial Chemistry, and PCR Amplification of 3′-5′ and 3′-6’ Disulfide-Linked Oligonucleotides. Angew. Chem., Int. Ed. 2014, 53 (52), 14415–14418. 10.1002/anie.201405761. [DOI] [PubMed] [Google Scholar]
- Porter K. W.; Briley J. D.; Shaw B. R. Direct PCR Sequencing with Boronated Nucleotides. Nucleic Acids Res. 1997, 25 (8), 1611–1617. 10.1093/nar/25.8.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sagheer A. H.; Brown T. Single Tube Gene Synthesis by Phosphoramidate Chemical Ligation. Chem. Commun. 2017, 53 (77), 10700–10702. 10.1039/C7CC00858A. [DOI] [PubMed] [Google Scholar]
- Chen J.; Baker Y. R.; Brown A.; El-Sagheer A. H.; Brown T. Enzyme-Free Synthesis of Cyclic Single-Stranded DNA Constructs Containing a Single Triazole, Amide or Phosphoramidate Backbone Linkage and Their Use as Templates for Rolling Circle Amplification and Nanoflower Formation. Chem. Sci. 2018, 9 (42), 8110–8120. 10.1039/C8SC02952K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara M.; Takeshima H.; Nagashima J.; Minezaki S.; Ozaki H.; Sawai H. Transcription and Reverse Transcription of Artificial Nucleic Acids Involving Backbone Modification by Template-Directed DNA Polymerase Reactions. Bioorg. Med. Chem. 2009, 17 (11), 3782–3788. 10.1016/j.bmc.2009.04.045. [DOI] [PubMed] [Google Scholar]
- Varizhuk A. M.; Kaluzhny D. N.; Novikov R. A.; Chizhov A. O.; Smirnov I. P.; Chuvilin A. N.; Tatarinova O. N.; Fisunov G. Y.; Pozmogova G. E.; Florentiev V. L. Synthesis of Triazole-Linked Oligonucleotides with High Affinity to DNA Complements and an Analysis of Their Compatibility with Biosystems. J. Org. Chem. 2013, 78 (12), 5964–5969. 10.1021/jo400651k. [DOI] [PubMed] [Google Scholar]
- El-Sagheer A. H.; Brown T. Efficient RNA Synthesis by in Vitro Transcription of a Triazole-Modified DNA Template. Chem. Commun. 2011, 47 (44), 12057–12058. 10.1039/c1cc14316f. [DOI] [PubMed] [Google Scholar]
- El-Sagheer A. H.; Sanzone A. P.; Gao R.; Tavassoli A.; Brown T. Biocompatible Artificial DNA Linker That Is Read through by DNA Polymerases and Is Functional in Escherichia Coli. Proc. Natl. Acad. Sci. U. S. A. 2011, 108 (28), 11338–11343. 10.1073/pnas.1101519108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantoni N. Z.; El-Sagheer A. H.; Brown T. A Hitchhiker’s Guide to Click-Chemistry with Nucleic Acids. Chem. Rev. 2021, 121 (12), 7122–7154. 10.1021/acs.chemrev.0c00928. [DOI] [PubMed] [Google Scholar]
- Isobe H.; Fujino T.; Yamazaki N.; Guillot-Nieckowski M.; Nakamura E. Triazole-Linked Analogue of Deoxyribonucleic Acid (TLDNA): Design, Synthesis, and Double-Strand Formation with Natural DNA. Org. Lett. 2008, 10 (17), 3729–3732. 10.1021/ol801230k. [DOI] [PubMed] [Google Scholar]
- Madhuri V.; Kumar V. A. Design and Synthesis of Dephosphono DNA Analogues Containing 1,2,3-Triazole Linker and Their UV-Melting Studies with DNA/RNA. Nucleosides, Nucleotides Nucleic Acids 2012, 31 (2), 97–111. 10.1080/15257770.2011.644100. [DOI] [PubMed] [Google Scholar]
- Rostovtsev V. V.; Green L. G.; Fokin V. V.; Sharpless K. B. A Stepwise Huisgen Cycloaddition Process: Copper(I)-Catalyzed Regioselective “Ligation” of Azides and Terminal Alkynes. Angew. Chem., Int. Ed. 2002, 41 (14), 2596–2599. . [DOI] [PubMed] [Google Scholar]
- Tornøe C. W.; Christensen C.; Meldal M. Peptidotriazoles on Solid Phase:[1, 2, 3]-Triazoles by Regiospecific Copper (I)-Catalyzed 1, 3-Dipolar Cycloadditions of Terminal Alkynes to Azides. J. Org. Chem. 2002, 67 (9), 3057–3064. 10.1021/jo011148j. [DOI] [PubMed] [Google Scholar]
- Dallmann A.; El-Sagheer A. H.; Dehmel L.; Mügge C.; Griesinger C.; Ernsting N. P.; Brown T. Structure and Dynamics of Triazole-Linked DNA: Biocompatibility Explained. Chem. - Eur. J. 2011, 17 (52), 14714–14717. 10.1002/chem.201102979. [DOI] [PubMed] [Google Scholar]
- El-Sagheer A. H.; Brown T. Combined Nucleobase and Backbone Modifications Enhance DNA Duplex Stability and Preserve Biocompatibility. Chem. Sci. 2014, 5 (1), 253–259. 10.1039/C3SC51753E. [DOI] [Google Scholar]
- Sharma V. K.; Singh S. K.; Krishnamurthy P. M.; Alterman J. F.; Haraszti R. A.; Khvorova A.; Prasad A. K.; Watts J. K. Synthesis and Biological Properties of Triazole-Linked Locked Nucleic Acid. Chem. Commun. 2017, 53 (63), 8906–8909. 10.1039/C7CC04092J. [DOI] [PubMed] [Google Scholar]
- Baker Y. R.; Traoré D.; Wanat P.; Tyburn A.; El-Sagheer A. H.; Brown T. Searching for the Ideal Triazole: Investigating the 1,5-Triazole as a Charge Neutral DNA Backbone Mimic. Tetrahedron 2020, 76 (7), 130914. 10.1016/j.tet.2019.130914. [DOI] [Google Scholar]
- Eisenhuth R.; Richert C. Convenient Syntheses of 3′-Amino-2′,3′-Dideoxynucleosides, Their 5′-Monophosphates, and 3′-Aminoterminal Oligodeoxynucleotide Primers. J. Org. Chem. 2009, 74 (1), 26–37. 10.1021/jo8018889. [DOI] [PubMed] [Google Scholar]
- Kawagoe N.; Kasori Y.; Hasegawa T. Highly C6-Selective and Quantitative Modification of Cellulose: Nucleoside-Appended Celluloses to Solubilize Single Walled Carbon Nanotubes. Cellulose 2011, 18 (1), 83–93. 10.1007/s10570-010-9468-9. [DOI] [Google Scholar]
- Szabó T.; Stawiński J. Synthesis and Some Conformational Features of the 5′-Deoxy-5′-Methylphosphonate Linked Dimer, 5′-Deoxy-5′-C-(Phosphonomethyl)Thymidin-3′-Yl (Thymidin-5′-Yl)Methylphosphonate [p(Ch2Tp(Ch2T]. Tetrahedron 1995, 51 (14), 4145–4160. 10.1016/0040-4020(95)00131-Q. [DOI] [Google Scholar]
- El-Sagheer A. H.; Brown T. Synthesis and Polymerase Chain Reaction Amplification of DNA Strands Containing an Unnatural Triazole Linkage. J. Am. Chem. Soc. 2009, 131 (11), 3958–3964. 10.1021/ja8065896. [DOI] [PubMed] [Google Scholar]
- Palframan M. J.; Alharthy R. D.; Powalowska P. K.; Hayes C. J. Synthesis of Triazole-Linked Morpholino Oligonucleotides via CuI Catalysed Cycloaddition. Org. Biomol. Chem. 2016, 14 (11), 3112–3119. 10.1039/C6OB00007J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainz P. The PCR Plateau Phase – towards an Understanding of Its Limitations. Biochim. Biophys. Acta, Gene Struct. Expression 2000, 1494 (1), 23–27. 10.1016/S0167-4781(00)00200-1. [DOI] [PubMed] [Google Scholar]
- Li Y.; Korolev S.; Waksman G. Crystal Structures of Open and Closed Forms of Binary and Ternary Complexes of the Large Fragment of Thermus Aquaticus DNA Polymerase I: Structural Basis for Nucleotide Incorporation. EMBO J. 1998, 17 (24), 7514–7525. 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W. G.; Kang M. E.; Lee J. B.; Jeon M. H.; Lee S.; Lee J.; Choi B.; Cal P. M. S. D.; Kang S.; Kee J. M.; Bernardes G. J. L.; Rohde J. U.; Choe W.; Hong S. Y. Nickel-Catalyzed Azide-Alkyne Cycloaddition to Access 1,5-Disubstituted 1,2,3-Triazoles in Air and Water. J. Am. Chem. Soc. 2017, 139 (35), 12121–12124. 10.1021/jacs.7b06338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.