To the Editor:
Nodular lymphocyte-predominant Hodgkin lymphoma (NLPHL) is a rare subtype of lymphoma that usually has an indolent clinical behavior. Similar to classical Hodgkin lymphoma (cHL), the tumor cells in NLPHL represent a minority of the cells in the tumor and, therefore, have been a challenge to study because of insufficient material for analysis. Transformation of NLPHL (tNLPHL) to diffuse large B-cell lymphoma (DLBCL) is seen in up to 30% of the cases [1] and can have varying morphologic patterns. However, two main patterns are seen, T-cell/histiocyte-rich large B-cell lymphoma (THRLBCL) and more typical DLBCL with sheets of large cells. Because the tumor cells are also sparse in THRLBCL, it has also been difficult to study these cases. However, with laser capture micro-dissection, a limited number of cases of tNLPHL have been investigated by next-generation sequencing [2, 3] using limited gene panels.
In this study, we evaluated tNLPHL cases that have the sheet-like growth pattern typical of de novo DLBCL. We characterized the genomic landscape of such cases using next-generation sequencing and copy number analysis (CNA) to gain a better understanding of the pathogenesis of these tumors.
We identified 19 cases of tNLPHL from three institutions (National Cancer Institute, University of Chicago, and City of Hope National Medical Center). Cases that had a pattern of THRLBCL were excluded to ensure that only cases with typical DLBCL growth pattern were studied. A diagnosis of NLPHL preceded the development of DLBCL in 5 patients (average interval, 4.7 years), whereas both NLPHL and DLBCL were seen concurrently in 11 patients (see Table S1). Three patients developed NLPHL after the diagnosis of DLBCL (1, 7, and 12 years after). Cases with concurrent NLPHL and DLBCL were macrodissected to obtain NLPHL areas with only a few tumor cells (<5%) (n = 8). These NLPHL areas were sequenced and used as matched germline DNA for the corresponding DLBCL. This study was approved by the local institutional review board and was conducted in accordance with the Declaration of Helsinki. DNA was extracted from formalin-fixed, paraffin-embedded tissue blocks and targeted sequencing was performed using a 356 gene panel containing the mutations most frequently seen in B-cell lymphoma and Hodgkin lymphoma (see Supplemental Methods and Tables S2, S3, and S4).
There were 14 males and 5 females with tNLPHL included in this study (M:F ratio of 2.8:1). The median age of the patients was 37 years (range, 15–68 years). The majority of cases of DLBCL had nodal involvement (68%), but other sites included the spleen, submandibular gland, and gastrointestinal tract (Table S1).
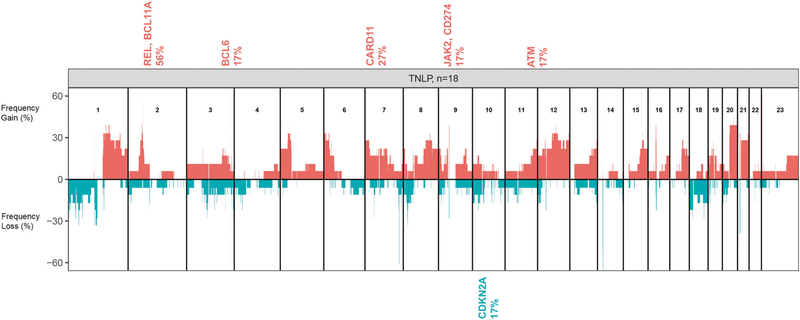
We performed CNA on 19 cases of tNLPHL, but one case failed quality control metrics and was excluded. In the remaining 18 cases, we observed that the number of genomic aberrations in these cases was comparable to that seen in de novo DLBCL cases, and almost all chromosomes showed aberrations. Using GISTIC, we found copy number (CN) gains of several oncogenes (REL, BCL11A, BCL6, CARD11, JAK2) and CN loss of the tumor suppressor gene CDKN2A (Q-bound < 0.05 and G-score < 1.0) in tNLPHL (Fig. 1).
Fig. 1. Copy number analysis of 18 cases of transformed nodular lymphocyte-predominant Hodgkin lymphoma.
We found copy number gains of several oncogenes (REL, BCL11A, BCL6, CARD11, JAK2) and copy number loss of the tumor suppressor gene CDKN2A as significant targets within the aberrant locus (Q-bound <0.05 and G-score <1.0) in tNLPHL using GISTIC.
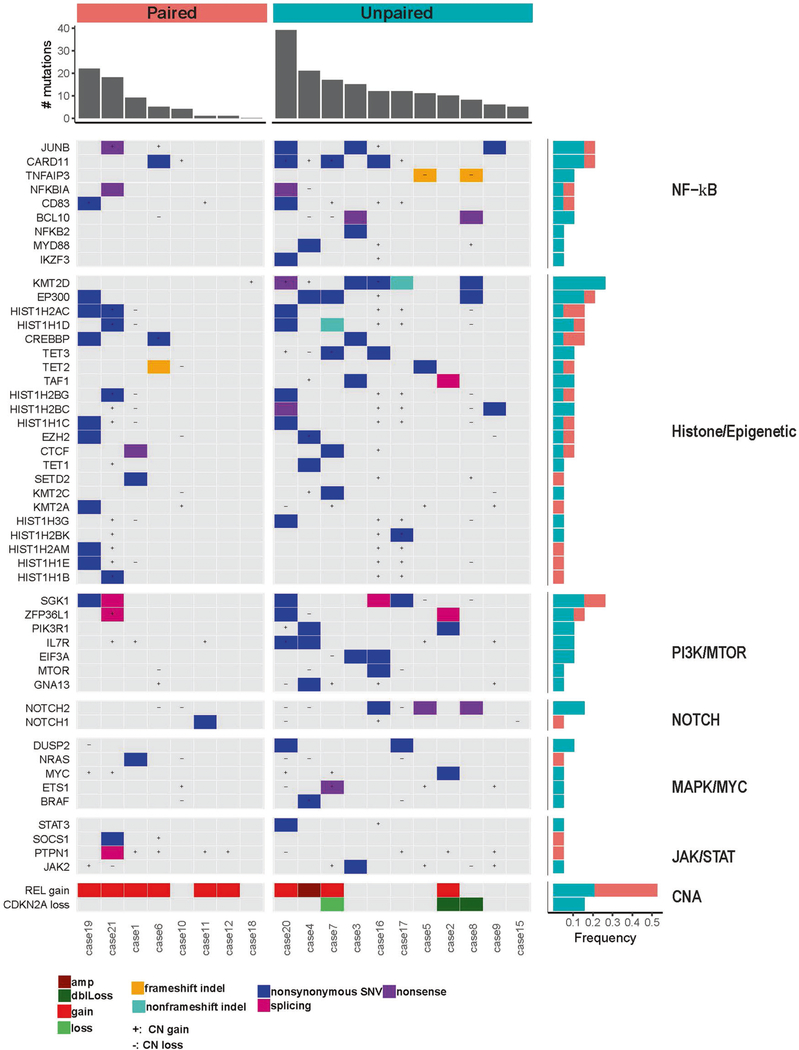
Although a number of pathways are identified, we found that genomic abnormalities in tNLPHL were most often associated with the PI3K (SGK1) and NF-kB (CARD11, JUNB) pathways, and chromatin modification regulators (Fig. 2). This mutational spectrum of tNLPHL resembles the DLBCL Cluster 4 of Chapuy et al. [4], which are primarily germinal center B-cell (GCB) type DLBCL. Transformed NLPHL also had frequent mutations in genes not often seen in de novo DLBCL (TET2, JUNB, and NOTCH2) and this provides new insights into the pathogenesis of these rare tumors.
Fig. 2. Select copy number abnormalities and mutation analysis of 19 cases (8 paired and 11 unpaired) of tNLPHL.
Mutations and copy number abnormalities involved key pathways such as NF-kB, PI3K, NOTCH, MAPK, and JAK/STAT. Histone modifiers were also commonly mutated. The REL gene was commonly gained as well as loss of CDKN2A. +, copy number gain; −, copy number loss; amp, amplification; dblLoss, double loss. Paired cases (salmon colored) are ones with corresponding germline DNA used to remove germline variants. Unpaired cases (teal colored) are those without corresponding germline DNA. Frequency of these mutations and copy number abnormalities are seen on the left by bar graphs.
Mutations in the PI3K pathway (SGK1, ZFP36L1, EIF3A, IL7R, and PIK3R1) were common in tNLPHL. Mutations of SGK1 (serum and glucocorticoid-regulated kinase 1; 26% of cases mutated in our study) have been reported in NLPHL [2] as well as de novo DLBCL, particularly those of germinal center origin [5]. The role of SGK1 in B-cell lymphoma is still unclear, but some data suggest that the mutation acts as an oncogene rather than a tumor suppressor gene in lymphoma [2]. Our cases, as well as the two cases seen by Hartmann et al. [2], showed SGK1 mutations particularly between positions 20 and 200 (Fig. S1a). We compared the location and frequency of SGK1 mutations in the three large de novo DLBCL studies [4–6], which found a predisposition for mutations in this region as well. Some studies suggest this region is important for localization of the protein into the mitochondria and nucleus [7]. Mutations and splice mutations in these regions, as seen in our tNLPHL cases, may alter SGK1 localization and change its normal regulatory functions [8].
Mutations in the NF-kB pathway were common in tNLPHL including CARD11, JUNB, BCL10, NFKBIA, and TNFAIP3. JUNB mutations (Fig. S1b) are rarely reported in de novo DLBCL but were frequently detected in our series of tNLPHL (21%), as well as other studies of NLPHL [2, 3]. JunB is a member of the activator protein-1 (AP-1) family of transcription factors, which collectively regulate genes important for proliferation, apoptosis, differentiation, and the immune response. A study by Szremska et al. [9] using a mouse model showed that JunB inhibits proliferation and transformation of B-cells and, thus, one can hypothesize that inactivation or lack of JunB makes B-cells more susceptible to transformation.
We observed CN gains of REL in 56% of our cases of tNLPHL, supporting the notion that this gene is important in the pathogenesis of tNLPHL. Heise et al. [10] found that c-REL-deficient germinal center B-cells failed to upregulate genes involved in metabolic functions that allow the cell to meet the energy demands required by rapid proliferation. Genomic gains in REL, as seen in tNLPHL, may contribute to lymphomagenesis by dysregulating cellular metabolism and promoting survival and proliferation [10].
We also observed frequent genetic abnormalities affecting DNA methylation and histone and chromatin modification pathways such as mutations of EP300 (21% of our cases), CREBBP (16%) and TET2 (11%). TET2, is infrequently mutated in DLBCL (6% of DLBCL, 7% of GCB DLBCL), but was mutated in 11% of our cases. Recent work has shown that TET2 should be considered a tumor suppressor gene and loss or mutation results in altered DNA 5hmC and 5mC modifications [11], which may disrupt the germinal center reaction or deregulate B-cell receptor signaling [12]. We also found mutations in other epigenetic modifiers such as EZH2 and KMT2D (mutated in 11 and 26% of cases, respectively) in tNLPHL.
There are some similarities of tNLPHL to transformed follicular lymphoma (tFL). Both FL and NLPHL have a rich tumor microenvironment (TME) but in transformation to DLBCL, much of the TME is lost and, thus, the dependence on this interaction is altered. Genes more frequently mutated in tFL compared with FL include TP53, EZH2, MYC, CCND3, KMT2C, and CARD11 [13]. Importantly, TP53 alterations do not appear to be common in tNLPHL, as we saw mutation in only one case and no deletions were detected by CNA. Another notable difference is the low frequency of mutations affecting genes associated with immune surveillance (B2M, MHC class I and II, CD58) in tNLPHL. However, four cases had gains in the region of PD-L1 and PD-L2 by CNA which has been seen in NLPHL as well cHL. Unfortunately, we were unable to enrich for LP cells in our cases of NLPHL and, thus, we could not confirm this finding on our cases without transformation.
A limitation to this study is that we did not perform sequencing of the NLPHL cells and, therefore, we cannot determine if these mutations were acquired or were already present at the NLPHL stage of disease. Prior studies have sequenced the LP cells of NLPHL but this was done with a very limited panel [2, 3] as compared with our mutation panel that covers the majority of genes implicated in lymphoma. In regard to the unpaired samples, some mutations may represent germline variants (Figs. S2 and S3) since our unpaired cases showed a higher mutation load, but recurrent genes mutated in B-cell lymphoma were selected (Fig. 2).
In summary, this is the largest series of tNLPHL with a conventional DLBCL growth pattern analyzed by CNA and mutational analysis. These lesions are rare but are very informative since they are highly enriched in tumor cells. We found that tNLPHL has a mutational spectrum simulating the DLBCL Cluster 4 of Chapuy et al. [4] with frequent mutations affecting the PI3K pathway (SGK1, ZFP36L1), the NF-kB pathway (CARD11, TNFAIP3, NFKBIA), and linker histones. On the other hand, BRAF/JAK/STAT3 mutations were uncommon in our cases, whereas there were frequent CN gains affecting REL and CN losses involving CDKN2A. Transformed NLPHLs are also distinctive in enrichment of mutations in genes uncommonly altered in sporadic DLBCL, including TET2, JUNB and NOTCH2. Interestingly, distinct from tFL, TP53 abnormalities and mutations affecting immune surveillance genes are uncommonly observed. This information provides new insights into the biology of tNLPHL and may highlight potential targets for therapy in the future.
Supplementary Material
Footnotes
Compliance with ethical standards
Conflict of interest The authors declare that they have no conflict of interest.
Supplementary information The online version of this article (https://doi.org/10.1038/s41375-020-0739-7) contains supplementary material, which is available to authorized users.
References
- 1.Al-Mansour M, Connors JM, Gascoyne RD, Skinnider B, Savage KJ. Transformation to aggressive lymphoma in nodular lymphocyte-predominant Hodgkin’s lymphoma. J Clin Oncol. 2010;28:793–9. [DOI] [PubMed] [Google Scholar]
- 2.Hartmann S, Schuhmacher B, Rausch T, Fuller L, Doring C, Weniger M, et al. Highly recurrent mutations of SGK1, DUSP2 and JUNB in nodular lymphocyte predominant Hodgkin lymphoma. Leukemia. 2016;30:844–53. [DOI] [PubMed] [Google Scholar]
- 3.Schuhmacher B, Bein J, Rausch T, Benes V, Tousseyn T, Vornanen M, et al. JUNB, DUSP2, SGK1, SOCS1 and CREBBP are frequently mutated in T-cell/histiocyte-rich large B-cell lymphoma. Haematologica. 2019;104:330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24:679–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N. Engl J Med. 2018;378:1396–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171:481–494 e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelsberg A, Kobelt F, Kuhl D. The N-terminus of the serum- and glucocorticoid-inducible kinase Sgk1 specifies mitochondrial localization and rapid turnover. Biochemical J. 2006;399: 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordas E, Naray-Fejes-Toth A, Fejes-Toth G. Subcellular location of serum- and glucocorticoid-induced kinase-1 in renal and mammary epithelial cells. Am J Physiol Cell Physiol. 2007;292: C1971–1981. [DOI] [PubMed] [Google Scholar]
- 9.Szremska AP, Kenner L, Weisz E, Ott RG, Passegué E, Artwohl M, et al. JunB inhibits proliferation and transformation in B-lymphoid cells. Blood. 2003;102:4159–65. [DOI] [PubMed] [Google Scholar]
- 10.Heise N, De Silva NS, Silva K, Carette A, Simonetti G, Pasparakis M, et al. Germinal center B cell maintenance and differentiation are controlled by distinct NF-kappaB transcription factor subunits. J Exp Med. 2014;211:2103–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dominguez PM, Ghamlouch H, Rosikiewicz W, Kumar P, Béguelin W, Fontán L, et al. TET2 deficiency causes germinal center hyperplasia, impairs plasma cell differentiation, and promotes B-cell lymphomagenesis. Cancer Discov. 2018;8: 1632–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mouly E, Ghamlouch H, Della-Valle V, Scourzic L, Quivoron C, Roos-Weil D, et al. B-cell tumor development in Tet2-deficient mice. Blood Adv. 2018;2:703–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bouska A, Zhang W, Gong Q, Iqbal J, Scuto A, Vose J, et al. Combined copy number and mutation analysis identifies oncogenic pathways associated with transformation of follicular lymphoma. Leukemia. 2016;31:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.