Abstract
Objectives
To analytically and clinically evaluate the semiquantitative Elecsys anti-severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein antibody (S-Ab) assay on the Roche cobas e602 analyzer.
Methods
The S-Ab assay is a 1-step, double-antigen sandwich electrochemiluminescent immunoassay that semiquantitatively measures total IgG, IgM, and IgA antibodies specific for the receptor binding domain of SARS-CoV-2 spike protein in serum or plasma. The S-Ab assay was evaluated for precision, linearity, interference (by hemoglobin, bilirubin, triglycerides, and biotin), cross-reactivity, and clinical performance, and was compared to the qualitative Elecsys anti-nucleocapsid (N-Ab) immunoassay, a lateral flow device that qualitatively detects S-Ab and N-Ab, and an anti-spike enzyme-linked immunosorbent assay (ELISA).
Results
S-Ab assay is precise, exhibits linearity from 0.4 to 250 U/mL, is unaffected by significant cross-reactivity or interferences, and qualitatively demonstrates greater than 90% concordance with N-Ab assay and lateral flow device. Readouts of S-Ab assay correlate with ELISA, which in turn correlates strongly with SARS-CoV-2 virus neutralization assay, and exhibit 100% sensitivity and specificity for COVID-19 patient samples obtained at or more than 14 days after PCR positivity.
Conclusions
The S-Ab assay is a robust clinical test for qualitative and semiquantitative detection of seropositivity following SARS-CoV-2 infection or spike-encoding mRNA COVID-19 vaccination.
Keywords: SAR-CoV-2, COVID-19, Coronavirus, Antibodies, Immunoassay, Serology, Diagnostics, Semiquantitative, Spike protein, Anti-spike antibodies
Key Points.
The S-Ab assay is a precise, semiquantitative test that measures total serum or plasma IgG, IgM, and IgA antibodies with an analytical measurement range of 0.4 to 250 U/mL.
The antibody concentrations determined by the S-Ab assay correlate reasonably well with an anti-spike ELISA and a SARS-CoV-2 virus neutralization assay by proxy.
The S-Ab assay demonstrates excellent clinical performance with 100% sensitivity and 100% specificity for COVID-19 patient samples obtained at or more than 14 days after PCR positivity.
Introduction
The etiologic agent of the coronavirus disease of 2019 (COVID-19) is severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a pathogenic RNA virus that—in the span of a year—has led to over 80 million confirmed cases and nearly 2 million deaths worldwide since its emergence in late 2019 from Wuhan, China.1-3 Although case counts continue to rise across the globe, the issuance of emergency use authorization (EUA) of the novel Pfizer/BioNTech and Moderna mRNA vaccines on December 11, 2020, and December 18, 2020, respectively, and of the Janssen (Johnson & Johnson) COVID-19 vaccine more recently on February 27, 2021, by the US Food and Drug Administration (FDA) have provided cause for much optimism.4-6 However, the challenges of mass vaccine distribution are enormous and inevitably require significant resources to implement.7
Since the onset of the pandemic, the diagnostic approach toward COVID-19 has remained largely unchanged.8,9 Real-time reverse transcription polymerase chain reaction (RT-PCR) remains the laboratory test of choice for the direct detection of viral genetic material and thus evidence of active infection by SARS-CoV-2. Antigen tests, which are based on the direct detection of distinct structural components of the viral particle, have become increasingly available. Both molecular and antigen tests have gained further popularity through their implementation as point-of-care tests, aimed towards providing results more rapidly than their conventional formats.8 Saliva tests and home collection kits have further increased access to testing. However, nonmolecular-based tests remain generally less sensitive than RT-PCR.10,11
In contrast to direct viral detection methods, antibody (serological) assays indirectly test for prior infection or exposure to SARS-CoV-2 by detecting evidence of a humoral response to components of the virus. To date, numerous anti–SARS-CoV-2 antibody tests are available for clinical use through EUA by the FDA12; however, their use has been limited largely by concerns that a complete understanding of the kinetics of SARS-CoV-2 seroconversion remains lacking.13 In fact, the US Centers for Disease Control and Prevention continues to recommend against routine antibody testing following mRNA COVID-19 vaccination, citing the currently unclear clinical utility of postvaccination antibody testing.14 Variability in the performance of antibody assays, particularly as they first became available, also led to concerns regarding their overall usefulness.15 Many of the first-generation antibody tests are qualitative—or binary—in nature, which further limited their utility for monitoring the kinetics of seroconversion.
In this study, we present a comprehensive analytical and clinical evaluation of the semiquantitative Elecsys anti–SARS-CoV-2, anti-spike antibody (S-Ab) assay on the Roche cobas e602 analyzer. The S-Ab assay is designed to detect total immunoglobulin G (IgG), IgM, and IgA antibodies against the receptor binding domain (RBD) of the SARS-CoV-2 spike protein. Our study shows that the S-Ab assay is precise, unaffected by relatively high concentrations of several common interferents (hemoglobin, bilirubin, triglycerides, and biotin), and demonstrates excellent clinical performance. We compared the S-Ab assay with other previously validated anti–SARS-CoV-2 immunoassays,16,17 as well as an anti-spike enzyme-linked immunosorbent assay (ELISA) that has previously been shown to correlate strongly with a SARS-CoV-2 virus neutralization assay.18,19 Our study demonstrates that the semiquantitative S-Ab assay is a useful clinical test for both monitoring the humoral component of the adaptive immune response against both SARS-CoV-2 infection and spike-encoding mRNA COVID-19 vaccination.
Materials and Methods
Study Samples
Leftover patient samples collected in lithium heparin gel tubes for clinical testing were used for all validation studies, whereas donor range studies were conducted using volunteer samples collected in K2-EDTA tubes. Plasma samples derived from these tubes were stored temporarily at 4°C for testing conducted typically within 2 weeks of collection and were frozen at −80°C for longer-term storage. All samples were obtained via a quality assurance protocol, which qualified for an institutional review board waiver, and no patient identifiers were used.
Analyzer Systems and Test Device
The Roche Elecsys S-Ab assay and its predecessor, the Roche Elecsys anti-nucleocapsid antibody (N-Ab) assay,16 are 1-step, double-antigen sandwich electrochemiluminescent immunoassays that detect total IgG, IgM, and IgA antibodies against the SARS-CoV-2 spike (S) and nucleocapsid (N) proteins, respectively. The S-Ab test is a semiquantitative assay with an analytical measurement range (AMR) claim of 0.4 to 250 U/mL; positive results are defined as concentrations at or greater than 0.8 U/mL.20 In contrast, the N-Ab assay is a qualitative assay that utilizes a calibrator-based cutoff index (COI) at or greater than 1.0 as the definition of positivity/reactivity. Acceptable specimen types for the S-Ab assay include serum and Li-heparin, K2-EDTA, K3-EDTA, and sodium citrate plasma. In our laboratory, both S-Ab and N-Ab assays are performed in the cobas e602 module of the Roche cobas 8000 total automation system. The S-Ab and N-Ab assays received EUA from the US FDA on November 25, 2020, and May 2, 2020, respectively.
The Truvian Easy Check COVID-19 IgM/IgG device is a standalone lateral flow device, manufactured by Access Bio, for the detection of IgM and IgG antibodies against both SARS-CoV-2 S (S1 subunit, RBD) and N proteins. The Easy Check is a qualitative test that has been authorized for use with plasma, serum, or whole blood samples. The analytical and clinical evaluation of the Easy Check device has been previously reported.17 Access Bio received FDA EUA for this device on July 24, 2020.
Precision Studies
A within-run precision study was performed with 20 replicates of low (approximately 1 U/mL) and high (approximately 9 U/mL) quality control materials in 1 complete run. Between-day precision was assessed using the same quality control materials assayed daily over 23 days. The mean, standard deviation (SD), and coefficient of variation (CV, %), defined as (SD × 100)/mean, for each precision series were determined.
Analytical Measurement Range
A positive sample pool of approximately 240 U/mL was serially diluted with pooled negative plasma (<0.4 U/mL) to several dilutions between 242 U/mL and 0.5 U/mL. All dilutions were tested in replicates. A linear regression of the mean observed vs target anti-spike levels was performed to verify the 0.5 to 250 U/mL AMR of the S-Ab assay. We consider the claimed AMR to be validated if the measuring limit is within 20% of the lower and upper targets.
Dilution and Diluent Studies
The Elecsys S-Ab assay will perform a 10-fold autodilution with an onboard universal diluent for values greater than 250 U/mL. Thus, 2 dilution series comprising a positive sample pool of approximately 240 U/mL diluted by 10, 20, 40, 50, and 100-fold using either Elecsys universal diluent or S-Ab–negative plasma were assessed for matrix effects based on the calculated recovery for each fold dilution. All dilution samples were prepared directly from the positive pool stock solution and were measured in duplicates. In a similar manner, the autodilution function of the e602 analyzer for the S-Ab assay was also validated by comparisons with 10-fold manual dilutions using Elecsys universal diluent.
Interference Studies
Positive and Negative Plasma Pools
Leftover antibody-positive plasma samples were combined to generate an antibody-positive plasma pool with an S-Ab concentration of 2 U/mL, whereas an antibody-negative plasma pool was prepared by diluting antibody-positive samples with antibody-negative plasma to yield a pool with a S-Ab concentration of 0.6 U/mL. The antibody-positive and antibody-negative pools were spiked with increasing concentration of the following interferents: hemoglobin (0-1,740 mg/dL), bilirubin (0-60 mg/dL), triglycerides (0-4,000 mg/dL), and biotin (0-1,000 ng/mL).
Hemoglobin Stock Solution
Type O, rhesus negative RBCs were packed by centrifugation at 13,000 rpm at room temperature and resuspended in an isotonic saline solution. Following a total of 3 isotonic saline washes, the RBCs were lysed by vigorous vortexing in deionized water and subsequent overnight freezing at −20°C. The RBC debris was removed by centrifugation at 13,000 rpm the following day. The total hemoglobin concentration of the hemolysate stock solution was measured on a GEM5000 co-oximeter (Instrumentation Laboratory) to be 17,400 mg/dL.
Bilirubin Stock Solution
Lyophilized conjugated bilirubin (Sigma-Aldrich) was dissolved with normal saline to obtain a bilirubin stock solution. The total bilirubin concentration of the stock solution was determined on the cobas 702 analyzer (Roche Diagnostics) to be approximately 600 mg/dL.
Triglycerides Stock Solution
An intralipid solution (20% fat emulsion; Fresenius Kabi) was used as the stock lipid solution. The triglycerides concentration of this stock solution was determined to be approximately 40,000 mg/dL based on that of a 10-fold diluted sample measured on the cobas 702 analyzer.
Biotin Stock Solution
A total of 5 mg of biotin (Sigma-Aldrich) was dissolved in 50 mL of deionized water and further diluted 10-fold to yield a stock biotin solution of concentration of 10,000 ng/mL.
Preparation of Interference Sets
Each interferent stock solution was diluted between 10- and 60-fold (ie, 10, 15, 20, 30, and 60) with either antibody-positive or antibody-negative pooled plasma to generate the antibody-positive or antibody-negative interference sets. For each particular sample, the S-Ab assay was performed in duplicates and the recovery (%) was determined as the ratio of the measured S-Ab concentration relative to the S-Ab concentration of the untreated (ie, no interferent) sample. Hemolysis, icteric, and lipemic indices were additionally measured on the cobas 702 analyzer.
Cross-Reactivity Studies
Anonymized lithium heparinized samples from 47 patients admitted in 2020 who concurrently tested negative by RT-PCR for SARS-CoV-2 via the BioFire Respiratory 2.1 Panel on the FilmArray Torch System at our institution’s Microbiology and Immunology Laboratory were used to test for potential cross-reactivity with the S-Ab assay.
Method Comparisons
A comparison of the semiquantitative S-Ab assay vs the qualitative N-Ab assay was made using 53 prepandemic (ie, COVID-19–negative) and 112 PCR-confirmed, COVID-19–positive samples. Of these 165 samples, a subset consisting of 120 samples (41 negatives and 79 positives) were used for a qualitative 3-way comparison using the S-Ab assay, N-Ab assay, and the Truvian Easy Check lateral flow immunoassay (LFIA). The overall concordance was calculated for each of the comparisons.
SARS-CoV-2 Anti-Spike Protein ELISA
The laboratory-developed SARS-CoV-2 ELISA has been described and applied previously.18,19,21 In brief, Nunc 96-well microplate (Thermo Scientific) were coated with SARS-CoV-2 spike ectodomain, which complexes with SARS-CoV-2 antibodies in samples or controls. Recombinant anti–SARS-CoV-2 spike monoclonal antibody CR3022 (Abcam) was used as a positive titer control. Secondary antibody detection of S-Ab in patient plasma or serum was achieved using peroxidase-conjugated anti-human IgG Fc antibodies (Invitrogen) and visualized using 3,3′,5,5′-tetramethylbenzidine substrate (Thermo Scientific). The optical density (OD) for each sample and control was then measured at a wavelength of 450 nm using a plate spectrophotometer. For each sample dilution, the measured OD is normalized and compared against predefined cutoffs and thus the positive well with the greatest dilution was defined as the end titer for the sample.
Clinical Performance
The clinical performance of the S-Ab assay was assessed using 112 leftover lithium heparinized samples from deidentified inpatients with PCR-confirmed, COVID-19 active infection collected between 0 and 6 days (60 samples), 7 and 13 days (32 samples), and 14 or more days (20 samples) since initial PCR positivity, and 53 prepandemic samples (41 banked samples from a reference range study prior to 2018 and 12 from a banked respiratory viral panel study from early 2019). Calculations of sensitivity, specificity, and positive and negative predictive values were performed using the SciStat online software,22 assuming a 10% disease prevalence.
Serial Daily S-Ab Concentrations After Initial PCR Positivity
Anonymized lithium heparinized samples from 12 deidentified inpatients were retrieved consecutively on a daily basis within the first month of initial SARS-CoV-2 PCR positivity (typically on admission) for a serial time-course study of an infected patient population.
Vaccinated Donor Study
Blood samples offered by self-reportedly healthy volunteers were obtained mostly 2 to 5 weeks after mRNA COVID-19 vaccination. In several cases, a prevaccination sample and a between-dose sample were available for testing by both the semiquantitative S-Ab and qualitative N-Ab assays. The Mann-Whitney U test was used to determine statistical difference between the means of the individuals at 3 weeks after the first dose vs individuals at 2 to 5 weeks after the second dose of the vaccine.
RESULTS
Precision Studies
The within-run CVs for the low-positive and positive samples were 1.5% and 0.91%, respectively, whereas the between-day CVs for the low-positive and positive samples were 3.2% and 2.4%, respectively Table 1 . Overall, the S-Ab assay showed excellent precision and was well within the claims established for the cobas e602 module.
Table 1.
Precision Studies of the Elecsys Anti–SARS-CoV-2 S-Ab Assaya
| SARS-CoV-2 Controls | Within Run (n = 20) | Between Day (n = 23) | ||||
|---|---|---|---|---|---|---|
| Mean, U/mL | SD, U/mL | CV, % | Mean, U/mL | SD, U/mL | CV, % | |
| Low positive control | 0.96 | 0.014 | 1.50 | 1.02 | 0.03 | 3.2 |
| High positive control | 8.95 | 0.081 | 0.91 | 8.98 | 0.22 | 2.4 |
CV, coefficient of variation; S-Ab, spike protein antibody.
aA positive S-Ab assay result was defined by a measured S-Ab concentration ≥0.8 U/mL. CV (%) = (SD/mean) × 100.
Analytical Measurement Range
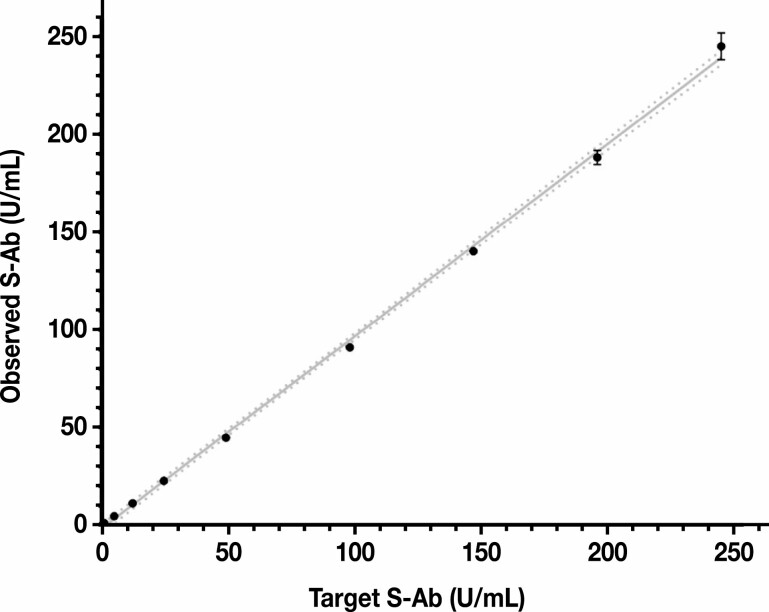
The AMR of the S-Ab assay was validated effectively as 0.4 to 250 U/mL. Antibody concentrations below 0.8 U/mL are considered to be negative and thus reported as less than 0.8 U/mL. Specimens testing above 250 U/mL trigger an automatic 10-fold dilution performed using the onboard Elecsys Diluent Universal. As shown in Figure 1 , linearity was demonstrated within the AMR of the S-Ab assay by serial dilution of a positive plasma pool (242 U/mL) with a negative plasma pool (<0.4 U/mL). A linear regression model resulted in an equation of y = 0.98x − 1.59 (r2 = 0.999).
Figure 1.
Analytic measurement range of the Elecsys anti–SARS-CoV-2 spike protein antibody (S-Ab) assay. All S-Ab measurements were performed in replicates (error bars shown) and the 95% confidence interval of the linear regression slope is 0.96 to 1.00; y = 0.98x – 1.59; r2 = 0.999. S-Ab, spike protein antibody. The best fit line is shown as a solid line and the 95% confidence bands of the best fit line are represented by the dotted lines.
Dilution and Diluent Studies
A comparison was made between the use of Elecsys universal diluent and negative plasma for manual sample dilutions (Supplementary Figure 1; all supplemental materials can be found at AmericanJournal of Clinical Pathology online). We verified up to 40-fold manual dilutions, whereas there was an overrecovery greater than 15% with manual dilutions 50-fold and beyond using either diluent. The use of universal diluent or negative plasma yielded clinically equivalent results (Supplementary Table 1). Additionally, the autodilution function of e602 analyzer for the S-Ab assay was validated by direct comparisons with 10-fold manual dilutions performed using Elecsys universal diluent (Supplementary Table 2).
Interference Studies
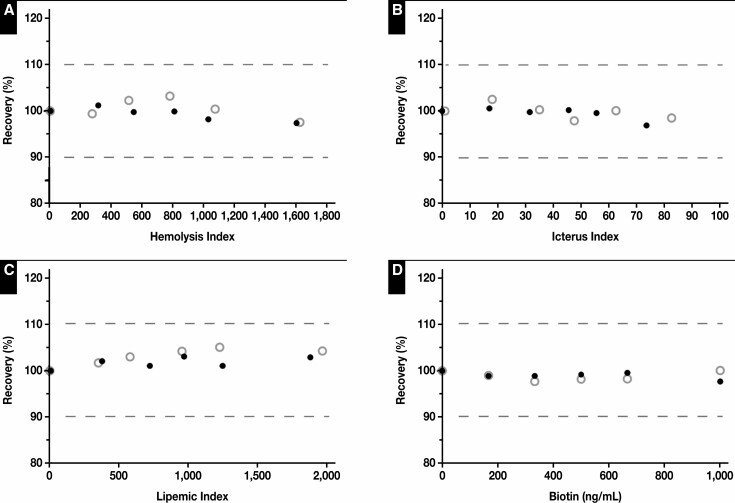
Overall, the S-Ab assay performed robustly over a relatively wide concentration range of several interferents. In particular, the S-Ab readings for the 2 representative negative and positive sample pools were not significantly affected (defined as a recovery of 100% ± 10%) by up to hemolysis index of 1,600, icteric index of 80, lipemic index of 1,800, and 1,000 ng/mL of biotin Figure 2 .
Figure 2.
Interference studies of Elecsys anti–SARS-CoV-2 spike protein antibody assay. The recovery (%) was assessed vs the hemolysis index (A), icterus index (B), lipemic index (C), and biotin concentration (D). The dotted lines denote a ±10% threshold relative to a recovery of 100%. Both negative (open circles) and positive (closed circles) sample pools were unaffected by up to 1,740 mg/dL of hemoglobin (hemolysis index of approximately 1,600), 60 mg/dL of bilirubin (icteric index of approximately 80), 4,000 mg/dL of triglycerides (lipemic index of approximately 1,800), and 1,000 ng/mL of biotin.
Cross-Reactivity Studies
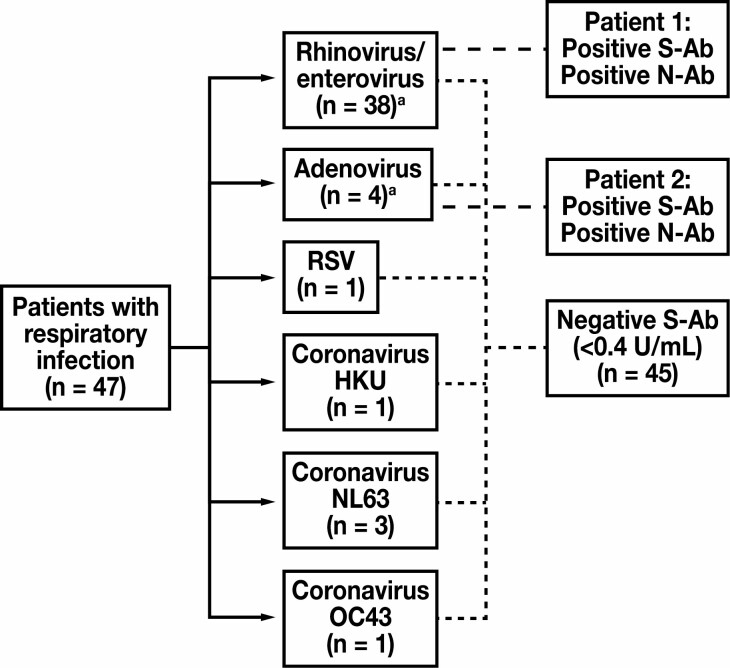
Possible cross-reactivity of the S-Ab assay was examined using sera obtained from patients who tested negative for SARS-CoV-2 but positive for another, non-SARS-CoV-2 respiratory virus by RT-PCR performed concurrently on a nasal or nasopharyngeal swab specimen (via the BioFire Respiratory 2.1 Panel). Samples obtained from 47 patients were tested in this manner, and of these 4 samples from 2 patients tested positive with the S-Ab assay—1 patient tested positive for adenovirus and the other tested positive for rhinovirus/enterovirus by PCR Figure 3 . Sera from both patients also concurrently tested positive using the N-Ab assay. Therefore, these 2 patients were likely previously infected with SARS-CoV-2 and, in which case, their positive S-Ab assay results represent true positives. On the other hand, all 8 samples obtained from 5 patients infected with PCR-confirmed coronavirus HKU, NL63, or OC43 tested negative by both the S-Ab assay (all samples < 0.4 U/mL) and the N-Ab assay, showing no cross-reactivity for these samples.
Figure 3.
Cross-reactivity with samples derived from patients with a non-COVID-19 respiratory infection. Samples were obtained from 47 patients who tested concurrently positive for a non-SARS-CoV-2 respiratory infection and negative for SARS-CoV-2 by reverse transcription polymerase chain reaction (via the BioFire Respiratory 2.1 Panel). Among these patients, 2 tested positive for spike protein antibody (S-Ab). Given that they also tested positive via the qualitative nucleocapsid antibody (N-Ab) assay, these patients likely had a true SARS-CoV-2 infection that elicited antibody formation against both S and N antigens, as opposed to representing false positives of S-Ab and N-Ab assays due to cross-reactivity. aOne patient tested positive for rhinovirus/enterovirus and adenovirus, and therefore was counted toward the total for both categories. RSV, respiratory syncytial virus.
Comparisons With Qualitative N-Ab and a Lateral Flow Immunoassay
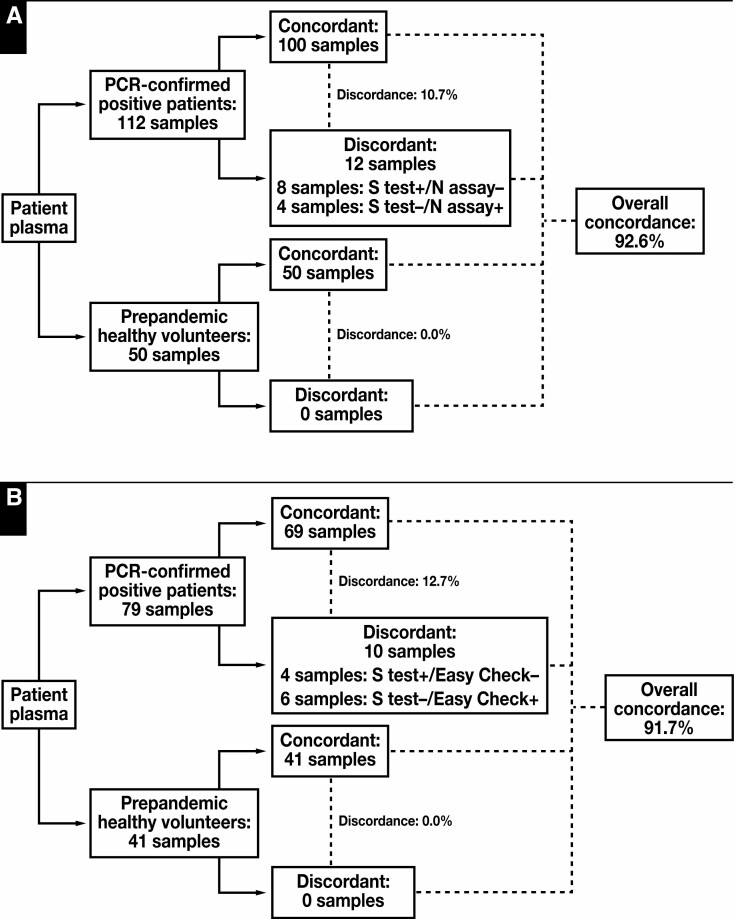
A comparison made between the S-Ab assay and the N-Ab assay, which detects total IgG, IgM, and IgA antibodies against the SARS-CoV-2 nucleocapsid protein, showed an overall concordance of 92.7% Figure 4 . Of the 12 PCR-confirmed, COVID-19–positive specimens that made up the 7.3% discordance, 8 samples were S-Ab positive/N-Ab negative, whereas 4 samples were S-Ab negative/N-Ab positive. These different patterns of discordance suggest that the precise kinetics of seroconversion differ depending on what type of antibodies are present at that time point.
Figure 4.
Method comparison of the Elecsys anti–SARS-CoV-2 S-Ab semiquantitative assay vs the Elecsys anti–SARS-CoV-2 nucleocapsid antibody (N-Ab) qualitative assay (A) and the Easy Check lateral flow device (B). The overall concordance between the spike protein (S-Ab) assay and the N-Ab assay or the Easy Check device are greater than 90%. Discordant cases are comprised of both positive and negative S-Ab assay results, which likely reflect differences in test principle and/or the specific analytes assayed between the 3 methods. PCR, polymerase chain reaction.
Similarly, a comparison between the S-Ab assay and the LFIA that qualitatively detects IgG and IgM antibodies specific for the SARS-CoV-2 nucleocapsid and spike proteins resulted in an overall concordance of 91.7%. Of the 10 PCR-confirmed, COVID-19–positive specimens that led to the 8.3% discordance, 4 samples were S-Ab positive/LFIA-Ab negative, whereas 6 samples were S-Ab negative/LFIA-Ab positive. Although it is difficult to pinpoint the precise causes of the discordant cases, some degree of discordance likely arises as a result of differences between the test principles. Even so, the overall concordance rate remains relatively high.
Regarding the discordant positive samples, of the 12 Figure 4A and 10 discordant samples Figure 4B , 4 were the same samples, while the other 8 and 6, respectively, were different samples.
Comparisons With an Anti-Spike ELISA
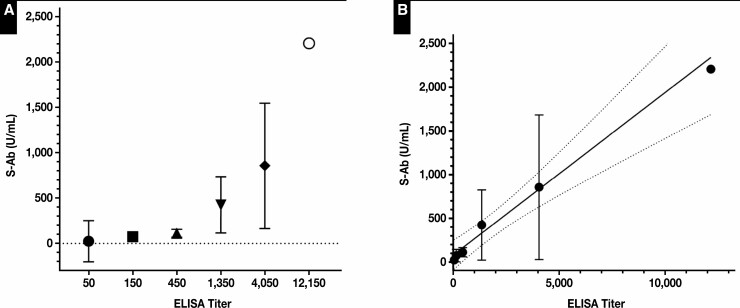
As shown in Figure 5 , S-Ab concentrations determined by the S-Ab assay show a direct correlation with increasing titers measured using an anti-spike ELISA. However, there is a high degree of S-Ab variability within each ELISA titer category. Each ELISA titer group differs by 3-fold, and there is a ±1 titer imprecision in the ELISA, leading to a large overlap of S-Ab assay results between adjacent ELISA titer groups. Overall, 22 of 26 (85%) S-Ab assay results of greater than 80 U/mL correspond to an ELISA titer at or greater than 1:450; 9 of 12 (75%) S-Ab assay results of less than 80 U/mL correspond to an ELISA titer ≤ 1:150. In the 2 highest titer groups (1:4,050 and 1:12,150), all but 1 S-Ab assay result (89%) was greater than 200 U/mL. A linear regression of the mean S-Ab concentration vs ELISA titer, when treated as a quasicontinuous variable, resulted in an equation of y = 0.19x + 80 (r2 = 0.52, P < .0001). Moreover, it has been shown that titers obtained using this ELISA correlate strongly with a SARS-CoV-2 virus neutralization assay. Therefore, by extension, high S-Ab concentrations determined by the S-Ab assay should also correlate to a greater degree of humoral protection against SARS-CoV-2.
Figure 5.
Spike protein antibody (S-Ab) concentration vs an anti-spike ELISA titer. A, Comparison between S-Ab concentrations and anti-spike ELISA titers shown for 38 samples grouped by increasing ELISA titers. The mean S-Ab concentration and the corresponding 95% confidence interval for each ELISA titer group are shown. The sample size of each ELISA titer group are as follows: N(50) = 2, N(150) = 10, N(450) = 8, N(1,350) = 9, N(4,050) = 8, N(12,150) = 1. B, A linear regression of S-Ab concentrations vs ELISA titers plotted on a linear scale shows moderate correlation (r2 = 0.52) despite significant variability seen in S-Ab concentrations. y = 0.19x + 80 (depicted as a solid line and the 95% confidence bands of the best fit line are represented by the dotted lines).
Clinical Performance of the S-Ab Assay
As shown in Table 2 and Table 3 , the S-Ab assay exhibited excellent clinical performance for patient samples obtained at or more than 14 days after PCR positivity with a 100% sensitivity (95% confidence interval [CI], 83.2%-100.0%). In contrast, the sensitivities of the S-Ab assay 0 to 6 days and 7 to 13 days after PCR positivity were 60.0% (95% CI, 46.5%-72.4%) and 78.1% (95% CI, 60.0%-90.7%), respectively. Based on 53 prepandemic samples, the specificity of the S-Ab assay was 100% (95% CI, 93.3%-100.0%).
Table 2.
Clinical Performance of the Elecsys Anti–SARS-CoV-2 S-Ab Assaya
| Semiquantitative S-Ab Assay | Prepandemic | PCR-Positive 0-6 Days | PCR-Positive 7-13 Days | PCR-Positive ≥14 Days |
|---|---|---|---|---|
| No Disease | Disease | Disease | Disease | |
| Positive, ≥0.8 U/mL | 0 | 36 | 25 | 20 |
| Negative, <0.8 U/mL | 53 | 24 | 7 | 0 |
| Total | 53 | 60 | 32 | 20 |
S-Ab, spike protein antibody.
aData are number of samples. A total of 112 COVID-19 patient samples were collected 0 to 6 days (60 samples), 7 to 13 days (32 samples), or at or more than 14 days (20 samples) after initial confirmation of positivity by reverse transcription quantitative polymerase chain reaction (PCR). The 53 prepandemic samples were collected prior to mid-2019 and were therefore COVID-19 negative by definition.
Table 3.
Test Characteristics of the Elecsys Anti–SARS-CoV-2 S-Ab Assay Clinical Performancea
| S-Ab Assay Statistics | PCR Positive 0-6 Days (95% CI) | PCR Positive 7-13 Days (95% CI) | PCR Positive ≥14 Days (95% CI) |
|---|---|---|---|
| Sensitivity, % | 60.0 (46.5-72.4) | 78.1 (60.0-90.7) | 100 (83.2-100.0) |
| Specificity, % | 100 (93.3-100.0) | 100 (93.3-100.0) | 100 (93.3-100.0) |
| Disease prevalence, % | 10 | 10 | 10 |
| PPV, % | 100 | 100 | 100 |
| NPV, % | 95.7 (94.3-96.8) | 97.6 (95.5-98.8) | 100 |
| Accuracy, % | 96.0 (90.6-98.8) | 97.8 (92.0-99.8) | 100 (95.1-100.0) |
CI, confidence interval; NPV, negative predictive value; PPV, positive predictive value; S-Ab; spike protein antibody.
aA 10% disease prevalence was assumed for PPV, NPV, and accuracy calculations for samples grouped by collection time: 0 to 6 days, 7 to 13 days, and at or more than 14 days after initial confirmation of positivity by reverse transcription quantitative polymerase chain reaction (PCR).
The median S-Ab concentrations 0 to 6 days, 7 to 13 days, and at or more than 14 days after PCR positivity were 7.3 U/mL, 56.1 U/mL, and 166.0 U/mL, respectively. Although there was significant variability and thus a wide range of S-Ab concentrations for samples obtained across these different time frames, the median S-Ab concentration at or more than 14 days after PCR positivity was significantly greater (P < .015) than the median S-Ab concentration of samples less than 14 days after PCR positivity (Supplementary Figure 2).
Assuming a 10% prevalence of COVID-19, the calculated positive predictive value is 100% and the negative predictive value (NPV) is greater than 96%, with an overall accuracy of greater than 96% for samples collected 0 to 13 days and 100% with an overall accuracy of 100% (95% CI, 95.1%-100.0%) for samples collected 14 or more days after PCR positivity Table 3 .
Serial Daily S-Ab Assaying After Initial PCR Positivity
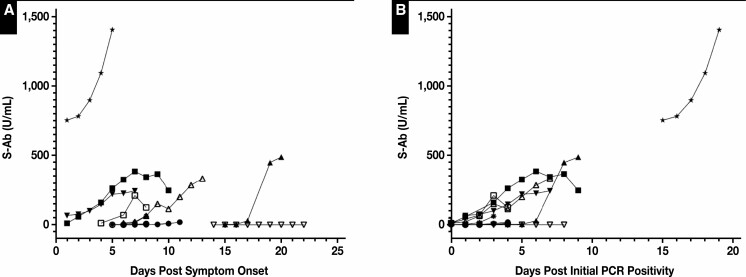
Significant but highly variable daily increases in S-Ab concentrations were observed for hospitalized patients infected with SARS-CoV-2 Figure 6 . Despite notable interpersonal variations, S-Ab concentrations typically undergo an exponential increase during the first several days after initial PCR positivity for most individuals that develop a robust humoral response Figure 6B , whereas this trend becomes much less obvious when S-Ab concentrations are plotted against the days after symptom onset Figure 6A . In 2 cases, the antibody levels appeared to have peaked approximately 1 week after symptom onset while in another case, the S-Ab concentrations remained undetectable (<0.4 U/mL) over the course of 9 days.
Figure 6.
Daily spike protein antibody (S-Ab) time course of hospitalized patients with COVID-19. The S-Ab assay was performed on leftover plasma samples collected daily from a total of 11 hospitalized patients infected with COVID-19 during their admission. Each patient is represented by a unique set of symbols. The measured S-Ab concentrations were plotted against the number of days after reported symptom onset (A) and the number of days after initial polymerase chain reaction positivity (B). Of note, S-Ab concentrations were undetectable (<0.4 U/mL) over the course of 9 days for 1 patient (downward facing open triangle).
Vaccinated Donor Study
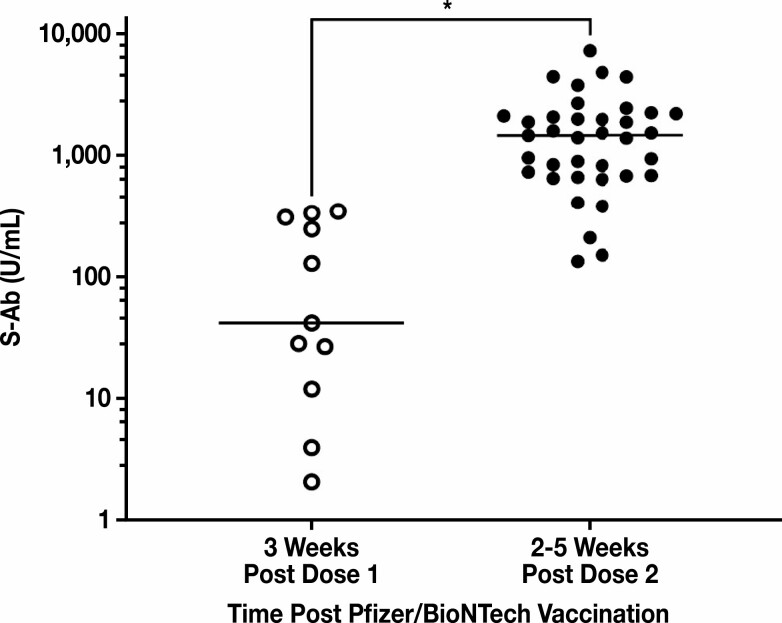
An S-Ab donor range was examined using samples obtained mostly 2 to 5 weeks after mRNA COVID-19 vaccination from self-reportedly healthy volunteers. The general pattern that was observed among all tested individuals was that there was a robust and exponentially increasing antibody response within the first month after vaccination, even though there was significant variability in the actual antibody concentrations. We observed a range of S-Ab values from 2 to 351 U/mL (median of 42.23 U/mL) for individuals after approximately 3 weeks of receiving the first dose, and a range of 135 to 7,326 U/mL (median of 1,482 U/mL) for individuals after approximately 2 to 5 weeks of receiving the second dose. There was a statistically significant difference (P < .0001) between the median of the 2 groups Figure 7 .
Figure 7.
Healthy donor range of spike protein antibody (S-Ab) concentrations postvaccination. S-Ab concentrations were measured in 11 healthy donors approximately 3 weeks after administration of the first dose of the Pfizer/BioNTech mRNA COVID-19 vaccine and in 37 healthy donors between 2 and 5 weeks after administration of the second dose of the Pfizer/BioNTech vaccine. All donors received both doses of the Pfizer/BioNTech vaccine and within the recommended timeframe. The median of the first group was 42.23 U/mL, while the median of the second group was 1,482 U/mL. Using the Mann-Whitney U test, the difference in median S-Ab concentrations measured for the 2 groups was statistically significant (* P < .0001).
Discussion
The COVID-19 pandemic remains a global threat and public health crisis since the emergence of SARS-CoV-2 in late 2019.23 While RT-PCR remains the primary testing modality for the detection of active infection in exposed individuals, numerous immunoassays have been developed over the past year intended for detecting antibodies formed against various structural components of SARS-CoV-2 as part of an individual’s adaptive immune response. One of the more recent iterations of immunoassays are the semiquantitative assays, which allow for measurements of actual antibody concentrations and therefore the potential for seroconversion kinetics to be tracked numerically over time. In the present study, we performed a complete analytical and clinical evaluation of the semiquantitative Elecsys anti–SARS-CoV-2 S-Ab assay on the Roche cobas e602 analyzer and thereby demonstrate its potential utility for seroprevalence surveillance, antibody time-course studies, and postvaccination-related monitoring.
We demonstrate that the S-Ab assay is precise, accurate, and can detect changes in S-Ab concentrations elicited by natural infection or by vaccination in a quantitative manner. Comparisons with other serological assays—namely, the Roche Elecsys N-Ab assay and the Truvian Easy Check lateral flow device—reveal overall a high degree of concordance between the 3 methods with discordant cases likely arising from inherent differences between the tests: the Elecsys S-Ab and N-Ab assays detect total IgG, IgM, and IgA against the SARS-CoV-2 spike and nucleocapsid proteins, respectively, whereas the Easy Check device is designed to detect IgG and IgM against both SARS-CoV-2 spike and nucleocapsid proteins. The S-Ab assay demonstrates excellent clinical performance with 100% sensitivity and 100% specificity for COVID-19 patient samples obtained at or more than 14 days after PCR positivity, and, moreover, the antibody concentrations determined by the S-Ab assay correlate reasonably well with an anti-spike ELISA and thereby a SARS-CoV-2 virus neutralization assay by proxy.
A major criticism of quantitative or semiquantitative anti–SARS-CoV-2 assays, such as the S-Ab assay, is that positive test results do not necessarily indicate humoral immunity or protection from the virus.24 A comparison between the S-Ab assay and an anti-spike ELISA results for antibody-positive samples obtained from inpatients infected with SARS-CoV-2 demonstrates reasonable correlation between both test readouts. Moreover, the anti-spike ELISA in turn correlates strongly with a SARS-CoV-2 virus neutralization assay.19 Because the S-Ab assay measures total IgG, IgM, and IgA antibodies against the SARS-CoV-2 spike protein RBD in a semiquantitative fashion, we posit that S-Ab concentrations provided by the S-Ab assay do indeed reflect some degree of humoral immunity against SARS-CoV-2.25
As part of our S-Ab assay evaluation, a post–Pfizer-BioNTech mRNA COVID-19 vaccination donor range was determined using samples obtained from apparently healthy volunteers. Although there was significant variation in the measured S-Ab concentrations, our overall finding was that apparently healthy individuals do generally show a robust humoral response (several thousand-fold increase in S-Ab concentrations) to mRNA COVID-19 vaccination several weeks following the second dose. Interestingly, all of the vaccinated individuals with positive S-Ab showed negative N-Ab results, indicating the high specificity of the N-Ab assay with zero cross-reactivity to the S-Ab assay. Therefore, a potential use of N-Ab assay is to differentiate between individuals with natural infection vs individuals vaccinated with spike proteins-based vaccines. Given the greater than 90% efficacy of the mRNA COVID-19 vaccines was based on early clinical trial data,26,27 it is not unreasonable to infer that positive antibody concentrations as determined by the S-Ab assay do reflect some degree of protection against SARS-CoV-2. At minimum, the S-Ab assay provides a means to ascertain whether an individual has developed the expected humoral response postvaccination.28,29 For patients infected with SARS-CoV-2, our findings suggest that efforts to monitor seroconversion kinetics in infected individuals might benefit more from tracking antibody concentrations relative to initial PCR positivity rather than the time after symptom onset.
It has been over a year since the emergence of SARS-CoV-2, but the kinetics of SARS-CoV-2 seroconversion still require elucidation and the clinical utility of antibody level monitoring remains to be established.13,30,31 Such endeavor would require a reliable and robust quantitative serological assay such as the S-Ab assay, which would allow for concentrations to be trended over time.32 Importantly, quantitation would allow for more detailed examination of interpersonal variability and, ultimately, the ability to objectively determine the potential timing of further vaccine booster doses should there be a need. For example, for immunocompromised patients or patients receiving immunosuppressive drug regimens, the measurement of S-Ab concentration postvaccination might help determine if an individual requires further vaccine boosters. On the other hand, serological testing remains an important tool for convalescent plasma so as long as passive immunotherapy is used for treating COVID-19.33
Although serological assays do not test for cell-mediated immunity, antibody levels provide an important assessment of humoral immunity, which constitutes a key component of the adaptive immune response of an otherwise healthy individual to infection or vaccination. There is a continued need for further elucidation of the temporal characteristics of antibody development, particularly as it relates to individuals receiving certain drug therapies and of different age groups, sex, and health status. Any serious efforts to achieve this aim would require a fully evaluated, quantitative serological assay. Given the tremendous ongoing public health initiative to vaccinate the general population to achieve herd immunity, it would be sensible to concurrently implement seroprevalence surveillance programs to help organize mass vaccine distribution. What awaits to be seen is whether the available COVID-19 vaccines—and, by extension, laboratory testing including serological assays—are effective against the novel strains of SARS-CoV-2, and recent data so far suggest that they are for at least some strains.34-36
Supplementary Material
Acknowledgment
The authors thank Truvian for providing the Easy Check devices for this project.
Disclosure: Dr Yeo is a member of the Scientific Advisory Board of Truvian and has received honoraria and equities from Truvian.
References
- 1. Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20:533-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hu B, Guo H, Zhou P, et al. . Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2021;19:141-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. V’kovski P, Kratzel A, Steiner S, et al. . Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19:155-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Food and Drug Administration. Pfizer-BioNTech COVID-19 vaccine.2021. https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/pfizer-biontech-covid-19-vaccine. Accessed March 08, 2021.
- 5. Food and Drug Administration. Moderna COVID-19 vaccine.2021. https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/moderna-covid-19-vaccine. Accessed March 08, 2021.
- 6. Food and Drug Administration. Janssen COVID-19 vaccine. 2021. https://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/janssen-covid-19-vaccine. Accessed March 08, 2021.
- 7. Goralnick E, Kaufmann C, Gawande AA. Mass-vaccination sites—an essential innovation to curb the Covid-19 pandemic. N Engl J Med. 2021;384:e67. [DOI] [PubMed] [Google Scholar]
- 8. Cheng MP, Papenburg J, Desjardins M, et al. . Diagnostic testing for severe acute respiratory syndrome-related coronavirus 2: a narrative review. Ann Intern Med. 2020;172:726-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kevadiya BD, Machhi J, Herskovitz J, et al. . Diagnostics for SARS-CoV-2 infections. Nat Mater. 2021;20:593-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vandenberg O, Martiny D, Rochas O, et al. . Considerations for diagnostic COVID-19 tests. Nat Rev Microbiol. 2021;19:171-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Centers for Disease Control and Prevention. Interim guidance for antigen testing for SARS-CoV-2.2021. https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html. Accessed March 16, 2021.
- 12. Food and Drug Administration. EUA authorized serology test performance.2021. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/eua-authorized-serology-test-performance. Accessed March 16, 2021.
- 13. Huang AT, Garcia-Carreras B, Hitchings MDT, et al. . A systematic review of antibody mediated immunity to coronaviruses: kinetics, correlates of protection, and association with severity. Nat Commun. 2020;11:4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Centers for Disease Control and Prevention. Interim clinical considerations for use of COVID-19 vaccines currently authorized in the United States.2021. https://www.cdc.gov/vaccines/covid-19/info-by-product/clinical-considerations.html. Accessed March 08, 2021.
- 15. Whitman JD, Hiatt J, Mowery CT, et al. . Evaluation of SARS-CoV-2 serology assays reveals a range of test performance. Nat Biotechnol. 2020;38:1174-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chan CW, Parker K, Tesic V, et al. . Analytical and clinical evaluation of the automated Elecsys anti-SARS-CoV-2 antibody assay on the Roche cobas e602 analyzer. Am J Clin Pathol. 2020;154:620-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chan CW, Shahul S, Coleman C, et al. . Evaluation of the Truvian easy check COVID-19 IgM/IgG lateral flow device for rapid anti-SARS-CoV-2 antibody detection. Am J Clin Pathol. 2021;155:286-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Salazar E, Christensen PA, Graviss EA, et al. . Treatment of coronavirus disease 2019 patients with convalescent plasma reveals a signal of significantly decreased mortality. Am J Pathol. 2020;190:2290-2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Salazar E, Kuchipudi SV, Christensen PA, et al. . Convalescent plasma anti-SARS-CoV-2 spike protein ectodomain and receptor-binding domain IgG correlate with virus neutralization. J Clin Invest. 2020;130:6728-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Roche. Elecsys® anti-SARS-CoV-2 S.2021. https://diagnostics.roche.com/us/en/products/params/elecsys-anti-sars-cov-2-s.html#productSpecs. Accessed March 18, 2021.
- 21. Salazar E, Perez KK, Ashraf M, et al. . Treatment of coronavirus disease 2019 (COVID-19) patients with convalescent plasma. Am J Pathol. 2020;190:1680-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. MedCalc Software. SciStat. https://www.scistat.com/statisticaltests/diagnostic_test.php. Accessed June 14, 2021.
- 23. Commissioners of the Lancet COVID-19 Commission; Task Force Chairs and Members of the Lancet COVID-19 Commission; Commission Secretariat and Staff of the Lancet COVID-19 Commission. Priorities for the COVID-19 pandemic at the start of 2021: statement of the Lancet COVID-19 commission. Lancet. 2021;397:947-950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Giri B, Pandey S, Shrestha R, et al. . Review of analytical performance of COVID-19 detection methods. Anal Bioanal Chem. 2021;413:35-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lumley SF, O’Donnell D, Stoesser NE, et al. ; Oxford University Hospitals Staff Testing Group . Antibody status and incidence of SARS-CoV-2 infection in health care workers. N Engl J Med. 2021;384:533-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baden LR, El Sahly HM, Essink B, et al. ; COVE Study Group . Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N Engl J Med. 2021;384:403-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Polack FP, Thomas SJ, Kitchin N, et al. ; C4591001 Clinical Trial Group . Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383:2603-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krammer F, Simon V. Serology assays to manage COVID-19. Science. 2020;368:1060-1061. [DOI] [PubMed] [Google Scholar]
- 29. Krammer F, Srivastava K, Alshammary H, et al. . Antibody responses in seropositive persons after a single dose of SARS-CoV-2 mRNA vaccine. N Engl J Med. 2021;384:1372-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sakhi H, Dahmane D, Attias P, et al. . Kinetics of anti-SARS-CoV-2 IgG antibodies in hemodialysis patients six months after infection. J Am Soc Nephrol. 2021;32:1033-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pflüger LS, Bannasch JH, Brehm TT, et al. . Clinical evaluation of five different automated SARS-CoV-2 serology assays in a cohort of hospitalized COVID-19 patients. J Clin Virol. 2020;130:104549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Føns S, Krogfelt KA. How can we interpret SARS-CoV-2 antibody test results? Pathog Dis. 2021;79:ftaa069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. DomBourian MG, Annen K, Huey L, et al. . Analysis of COVID-19 convalescent plasma for SARS-CoV-2 IgG using two commercial immunoassays. J Immunol Methods. 2020;486:112837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang Z, Schmidt F, Weisblum Y, et al. . mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Edara VV, Floyd K, Lai L, et al. . Infection and mRNA-1273 vaccine antibodies neutralize SARS-CoV-2 UK variant. medRxiv, doi: 10.1101/2021.02.02.21250799, 5. February 2021, preprint: not peer reviewed. [DOI] [Google Scholar]
- 36. Fontanet A, Autran B, Lina B, et al. . SARS-CoV-2 variants and ending the COVID-19 pandemic. Lancet. 2021;397:952-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.