Abstract
An immunodominant protein from Nocardia brasiliensis, P61, was subjected to amino-terminal and internal sequence analysis. Three sequences of 22, 17, and 38 residues, respectively, were obtained and compared with the protein database from GenBank by using the BLAST system. The sequences showed homology to some eukaryotic catalases and to a bromoperoxidase-catalase from Streptomyces violaceus. Its identity as a catalase was confirmed by analysis of its enzymatic activity on H2O2 and by a double-staining method on a nondenaturing polyacrylamide gel with 3,3′-diaminobenzidine and ferricyanide; the result showed only catalase activity, but no peroxidase. By using one of the internal amino acid sequences and a consensus catalase motif (VGNNTP), we were able to design a PCR assay that generated a 500-bp PCR product. The amplicon was analyzed, and the nucleotide sequence was compared to the GenBank database with the observation of high homology to other bacterial and eukaryotic catalases. A PCR assay based on this target sequence was performed with primers NB10 and NB11 to confirm the presence of the NB10-NB11 gene fragment in several N. brasiliensis strains isolated from mycetoma. The same assay was used to determine whether there were homologous sequences in several type strains from the genera Nocardia, Rhodococcus, Gordona, and Streptomyces. All of the N. brasiliensis strains presented a positive result but only some of the actinomycetes species tested were positive in the PCR assay. In order to confirm these findings, genomic DNA was subjected to Southern blot analysis. A 1.7-kbp band was observed in the N. brasiliensis strains, and bands of different molecular weight were observed in cross-reacting actinomycetes. Sequence analysis of the amplicons of selected actinomycetes showed high homology in this catalase fragment, thus demonstrating that this protein is highly conserved in this group of bacteria.
Mycetoma is a chronic, localized, subcutaneous disease caused by both fungi and actinomycetes (29). In Mexico about 98% of the cases are produced by actinomycetes, and Nocardia brasiliensis accounts for about 86.6% of the isolates (16). Although the mechanisms of defense against Nocardia are not completely known, some studies indicate that the cellular immune response is very important in resistance (8, 12, 20, 32). Conversely, the role of antibodies is not well established, and their production could even be considered a detrimental factor for the host during N. brasiliensis infection (20). Most immunological assays have been conducted by using complex mixtures of nocardial antigens. In order to determine the immunodominant antigens of N. brasiliensis recognized by the patient’s immune system, we analyzed by Western blot a crude extract from N. brasiliensis with a panel of sera from patients with mycetoma (23). In this study we also analyzed the cross-reactivity with other actinomycetes by testing sera from patients with tuberculosis and leprosy. In these assays, we observed that mycetoma patients developed antibodies that more frequently recognized three proteins of 61, 26, and 24 kDa that were designated as P61, P26, and P24, respectively. The sera from patients with mycetoma identified other proteins in the molecular mass range of 35 to 45 kDa, but sera from patients with tuberculosis and leprosy also recognized these bands.
We have isolated the P61 and P24 proteins (26), and the latter (P24) has been found to be useful in the detection of antinocardial antibodies (24). In order to determine the identity of these proteins, it is important to determine their N-terminal amino-acid sequences and to clone the genes. In this work we subjected one of them, P61, to amino acid sequence analysis and were able to obtain a partial nucleotide sequence of this gene. By comparison to the GenBank database as well as by studying its enzymatic activity on H2O2, we conclude that it is an N. brasiliensis catalase. We also determined the presence of this sequence or similar sequences in other actinomycetes. For this and following studies, we have designated the gene coding for the N. brasiliensis catalase as katN (for nocardial catalase).
MATERIALS AND METHODS
Purification of the N. brasiliensis HUJEG-1 P61.
The technique used to purify P61 has been published previously (26). Briefly, a batch culture (7 to 10 liters) of N. brasiliensis HUJEG-1 was prepared in brain heart infusion (Difco) and incubated for 7 days at 37°C. The cells were harvested, washed with distilled water, and defatted with ethanol-ethyl ether. A crude cellular extract was obtained by sonication of the bacterial mass in a Biosonik apparatus (Bronwill Scientific, Rochester, N.Y.) at a 60-probe intensity for 30 min in an ice bath. The suspension was centrifuged at 3,000 × g for 15 min to remove fragments and unbroken cells, and the soluble fraction was obtained by centrifugation at 144,000 × g for 3 h at 4°C in an L8-70M ultracentrifuge (Beckman, Palo Alto, Calif.). P61 was isolated by precipitation from the supernatant by using ammonium sulfate at a 50% saturation. After the pellet was separated by centrifugation at 600 × g, it was dialyzed and subjected to electrophoresis in a nondenaturing polyacrylamide gel electrophoresis (PAGE) system with a 5% stacking gel and a 10% running gel. The protein was characterized by a greenish color, which facilitated its detection in the 3-mm-thick gel. The band was excised and electroeluted, and the protein was quantified by the Bradford technique.
Amino acid sequence analysis.
A 30-μg sample of the pure protein was electrophoresed in a sodium dodecyl sulfate (SDS)–12% PAGE gel system in a Protean IIxi cell (Bio-Rad Laboratories, Richmond, Calif.). The protein was transferred at 100 mA for 18 h to polyvinylidine difluoride (PVDF) membranes (0.2-μm pore size; Bio-Rad) by using 10 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS) with 1% methanol (pH 11.0) as a transfer buffer. The filter was stained with Coomassie brilliant blue R-250 and destained with 50% methanol. A broad-staining band with minor contaminant bands of lower molecular weight was observed. P61 was excised from the paper, and sequence analysis was performed directly on the protein bound to the PVDF membrane (1).
P61 was also subjected to hydrolysis by using CNBr in order to obtain peptide fragments for sequencing. The digested protein was analyzed by reverse-phase high-pressure liquid chromatography with a Microbore C8-C18 column, and several absorbance peaks were observed. Peaks with retention times of 60 and 68 min were selected for the sequence analysis.
Staining method for the detection of catalase activity in PAGE gels.
In order to detect the catalase and peroxidase activity of P61, 3 μg of the pure protein was run in a 7.5% nondenaturing polyacrylamide gel prepared according to the method of Laemmli (15). We used 30 μg of N. brasiliensis crude extract, as well as 30 μg of a crude extract of N. farcinica, as controls. The gels were processed according to the method described previously by Wayne and Diaz (28). Briefly, after the electrophoresis, the gel was washed three times for 10 min with 0.01 M phosphate-buffered saline (PBS). The gel was then incubated with 3,3′-diaminobenzidine (DAB) tetrahydrochloride (0.05% in PBS) for 20 min at room temperature. The DAB reagent was poured off, and the gel was washed with H2O. The gel was then suspended in a solution containing 10 μl of 30% H2O2 in 100 ml of H2O and agitated continuously for 20 min. The hydrogen peroxide solution was discarded, and the gel was rinsed in water. The gel was then put in a pan containing 30 ml each of 2% ferric chloride and 2% potassium ferricyanide. After a green color began to appear, the ferricyanide reagent was discarded and replaced with water. Dark bands indicated peroxidase activity, and catalase activity was observed as clear areas over a green background.
Determination of P61 native molecular weight by gel filtration.
The molecular weight of the native P61 protein was estimated by gel filtration chromatography with Sephadex G-200 (Sigma) in a 90-cm-by-1.6-cm column. A flow rate of 20 ml/min was maintained, and the following standards were applied to the column separately in 5- to 10-mg quantities: apoferritin, β-amylase, carbonic anhydrase, and alcohol dehydrogenase.
Production and sequencing analysis of the NB2-NB3 katN gene fragment.
In order to determine the presence of consensus amino acid sequences in catalases related to P61, we aligned a group of catalase sequences of eukaryotic and prokaryotic origin that included the following organisms: Bacteroides fragilis, Pseudomonas aeruginosa, Brucella abortus, Haemophilus influenzae, Campylobacter jejuni, Neisseria gonorrhoeae, Proteus vulgaris, Rhizobium meliloti, Schizosaccharomyces pombe, Oryza sativa, Onchocerca volvulus, and Mus musculus. By this process, we detected the presence of several common sequences, such as IPER, RGFA, and VGNNTP, located in positions 75, 136, and 152 of the P. aeruginosa catalase. Based on this analysis we designed a PCR assay with degenerate oligonucleotide primers that utilized part of SeqnNB2 (FDLTQV) and VGNNTP (see Table 4). The PCR assay was carried out by using the three-step program in a PTC-200 DNA Engine (MJ Research, Watertown, Mass.), but with an annealing temperature of 50°C to generate a 500-bp product. For the sequence analysis of the PCR product, a 200-μl reaction was used, and the product was gel purified with the Wizard PCR Prep system of Promega (Madison, Wis.). The sequence of the purified PCR product was determined with the Prism Dye Terminator sequence kit (Applied Biosystems, Foster City, Calif.) in a 377 automatic sequencer.
TABLE 4.
Internal amino acid sequences of peptides obtained by disruption of P61 with cyanogen bromidea
| Sequence and organism | Amino acid residues |
|---|---|
| Nocardia brasiliensis SeqNB2 | XVAEAENY_FR_FDLTQVV |
| Streptomyces coelicolor | A AENY F FD LT V |
| Streptomyces violaceus | A A Y F FDLT V |
| Schizosaccharomyces pombe | EAE Y FDLT V |
| Saccharomyces cerevisiae | EAE Y FDLT V |
| Mus musculus | EAE F FDLT V |
| Nocardia brasiliensis SeqNB3 | VRAGYIEHAEDGDFTQPGTLVREV(N)DAQRDRLVSNVVG |
| Streptomyces violaceous | HAED DFTQ G L R RL N G |
| Streptomyces coelicolor | D F QPG L R Q L N |
| Pseudomonas putida | G L R D Q D L SN G |
BLAST sequence alignment analysis was used to indicate homology to other catalase amino acid sequences.
Detection of the NB2-NB3 katN fragment in N. brasiliensis clinical isolates and other actinomycetales.
In order to confirm the distribution of this gene fragment in N. brasiliensis strains, genomic DNA from a group of N. brasiliensis (Table 1) strains isolated from mycetoma cases in Monterrey, Mexico, was subjected to PCR but with primers NB10 and NB11 derived from the sequence of the fragment NB2-NB3 (Table 2). Since a subtaxon of N. brasiliensis, N. pseudobrasiliensis, has been recently described (22), we took care to include only N. brasiliensis sensu stricto strains in this study. The identification of these strains was made by using the conventional biochemical tests and confirmed by DNA sequencing of a region located between nucleotides 70 and 334 of the N. brasiliensis 16S RNA gene (sequence accession number Z36935). This fragment was amplified with the primers NOC-3 and NOC-4 that were located in conserved areas, although some internal regions allowed us to differentiate most of the Nocardia species by DNA sequencing.
TABLE 1.
Actinomycete strains utilized in this study
| Strain |
|---|
| Nocardia spp. |
| N. asteroides NCTC 6761 |
| N. asteroides “complex” LCDC 92431 |
| N. asteroides “complex” LCDC 95-0355 |
| N. asteroides “complex” LCDC 91-005 |
| N. asteroides “complex” LCDC 96-0153 |
| N. asteroides “complex” LCDC 93-0637 |
| N. asteroides “complex” LCDC 91-352 |
| N. asteroides “complex” LCDC 94-0238 |
| N. brasiliensis HUJEG-1 |
| N. brasiliensis NCTC 10300 |
| N. brasiliensis HUJEGH-217 |
| N. brasiliensis HUJEGH-489 |
| N. brasiliensis HUJEGC-542 |
| N. brasiliensis HUJEGD-980 |
| N. brasiliensis HUJEGD-915 |
| N. brasiliensis HUJEGD-572 |
| N. brasiliensis HUJEGD-403 |
| N. brasiliensis HUJEGI-923 |
| N. brasiliensis HUJEGJ-93 |
| N. brasiliensis HUJEGH-743 |
| N. brasiliensis HUJEGL-1114 |
| N. brasiliensis HUJEGI-531 |
| N. brasiliensis HUJEGJ-209 |
| N. brasiliensis HUJEGL-912 |
| N. brevicatena ATCC 15333 |
| N. carnea ATCC 06847 |
| N. farcinica ATCC 03318 |
| N. nova ATCC 33726 |
| N. otitidis-caviarum ATCC 14629 |
| N. transvalensis ATCC 06865 |
| Other actinomycetes |
| Actinomadure madurae ATCC 19425 |
| Actinomadure pelletieri ATCC 33385 |
| Gordona bronchialis ATCC 25592 |
| Gordona rubropertinctus ATCC 14352 |
| Gordona sputi ATCC 33610 |
| Gordona terrae ATCC 25594 |
| Rhodococcus chubuensis ATCC 33609 |
| Rhodococcus equi ATCC 06939 |
| Rhodococcus erythropolis ATCC 04277 |
| Rhodococcus rhodochrous ATCC 13808 |
| Streptomyces somaliensis ATCC 19437 |
| Streptomyces somaliensis ATCC 33201 |
TABLE 2.
Oligonucleotide primers used in this study for the PCR and sequence analysis
| Name | Sequence |
|---|---|
| NB2 | 5′-SAC SAC CTT SGT SAG GTC GAA-3′ |
| NB3 | 5′-GTS GGX AAC AAC ACS CCS-3′ |
| NB10 | 5′-CCA CAA CAT GCA GTG GGA CTT-3′ |
| NB11 | 5′-CAC AGG TCT TTG CGG TGG TAG T-3′ |
| NOC3 | 5′-ACG GGT GAG TAA CAC GTG-3′ |
| NOC4 | 5′-AGT CTG GGC CGT GTC TCA GTC-3′ |
The DNA was extracted by using the previously reported CTAB-NaCl method (30); 100 ng of DNA of each strain was used for the PCR assay. We also tested DNA from other species of actinomycetes that belong to the genera Nocardia, Rhodococcus, Streptomyces, and Gordona (Table 1). As an internal PCR control we utilized primers NOC-3 and NOC-4.
Southern blot analysis.
In order to confirm the presence of positive results in the PCR assays, we carried out Southern blot analysis with genomic DNA of the actinomycetes mentioned above by utilizing BamHI to cut the DNA and the PCR fragment NB10-NB11 as a probe. Briefly, 4 μg of DNA were digested with 5 U of BamHI (Stratagene, La Jolla, Calif.) for 4 h at 37°C. The samples were loaded on a 0.8% agarose gel and run overnight at 60 V. As molecular weight standards, we used a PvuII-digested supercoiled ladder DNA and HaeIII-digested φx174 DNA. After electrophoresis, the DNA samples were transferred to Nytran plus nylon membranes (Schleicher & Schuell, Keene, N.H.) by using the turboblotter system according to the manufacturer’s directions. The blot was prehybridized and then incubated overnight with the peroxidase-labeled probe (NB10-NB11 fragment) at 42°C prepared with the enhanced chemiluminescence kit (Amersham, Arlington Heights, Ill.). Hybridization, washings, and development of the blots were all performed according to the manufacturer’s instructions.
RESULTS
Amino acid sequence analysis.
Twenty-nine cycles were performed on the blotted protein by using an Applied Biosystems model 473A amino acid sequencer that resulted in a 22-residue sequence (Table 3). This was compared with the sequences of other proteins in the GenBank database by using the internet BLAST system. The N-terminal N. brasiliensis P61 protein sequence showed similarity to catalases from Oryza sativa (68%), Hordeum vulgare (68%), Secale cereale (68%), and Zea mays (63%) (Table 3). By aligning these sequences with the P61 N-terminal sequence we observed a consensus sequence (T_TTTN_G_PV_DNE_LT_G [underscores indicate variability]) in all of them. As anticipated, a higher homology was observed among the N-terminal sequences of the cereal catalases than with the N. brasiliensis catalase sequence.
TABLE 3.
Similarity of N. brasiliensis N-terminal P61 sequence to some eukaryotic catalases
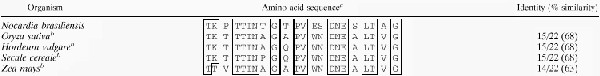 |
The consensus amino acid motifs are boxed.
Residues 14 to 35 are shown.
We were also able to obtain two amino acid internal sequences by a chemical breakage of P61 with CNBr: a 17-amino-acid residue from the 60-min (SeqNB2) peak and a 38-residue sequence from the 68-min peak (SeqNB3) (Table 4). These sequences were also analyzed by using the BLAST system, and the results are shown in Table 4. According to the homology analysis, SeqNB2 showed similarity to catalases from Streptomyces coelicolor, S. violaceus, Schizosaccharomyces pombe, Saccharomyces cerevisiae, and catalases from rodents such as the mouse (Mus musculus) and rat (Rattus norvegicus, not shown). Among these sequences there was a conserved motif, FDLT_V. Another sequence, SeqNB3, showed the highest similarity to S. violaceus bromoperoxidase-catalase (48% in a stretch of 30 amino acids) and a lower percentage of homology to S. coelicolor (38%) and Pseudomonas putida (36%) catalases.
Sequencing of the NB2-NB3 fragment of P61.
By using a PCR assay with degenerate primers derived from the sequences FDLTQV and VGNNTP, we were able to obtain a 500-bp amplicon that was subjected to sequence analysis. The sequence was parsed to the National Center for Biotechnology Information network BLAST server to identify database homologies. The highest similarity observed was of 86% to the bromoperoxidase-catalase (bca) gene of Streptomyces violaceus in a stretch of 151 nucleotides (from nucleotides 1159 to 1310 of the bca gene). The NB2-NB3 fragment also showed similarity, although to a lesser extent, to catalases from Drosophila melanogaster, Streptomyces coelicolor, P. aeruginosa, and Methylobacterium extorquens.
PCR test.
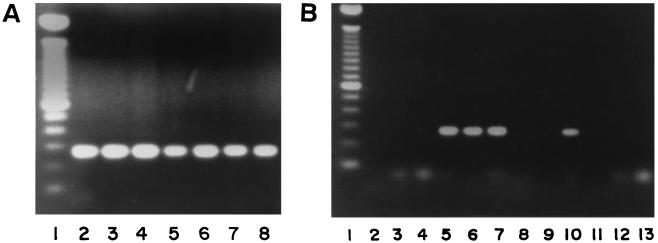
In order to determine the presence of the NB2-NB3 sequence or other homologous sequences in N. brasiliensis strains as well as in some other actinomycetes, we utilized a PCR assay based on the internal primers NB10 and NB11 that amplify a 250-bp fragment of the NB2-NB3 sequence (Fig. 1). All N. brasiliensis strains were positive for this gene. Of the other Nocardia species tested, only N. nova was positive. A group of N. asteroides complex strains from the Special Pathogens Section of the Laboratory Centre for Disease Control was also tested, and only one strain, later confirmed to be N. nova by sequencing of its 16S RNA gene, presented a positive PCR test. Of the other actinomycetes tested, positive reactions were observed with R. equi, R. erythropolis, R. chubuensis, and G. sputi.
FIG. 1.
PCR assay of genomic DNA from actinomycete species with primers NB10 and NB11. (A) Lanes: 1, 100-bp ladder; 2, N. brasiliensis NCTC 10300; lanes 3 to 8, N. brasiliensis clinical isolates. (B) Lanes: 1, markers; 2, S. somaliensis ATCC 33201; 3, S. somaliensis ATCC 19437; 4, R. rhodochrous ATCC 13808; 5, R. chubuensis ATCC 33609; 6, R. erythropolis ATCC 04277; 7, R. equi ATCC 06939; 8, G. rubropertinctus ATCC 14352; 9, G. terrae ATCC 25594; 10, G. sputi ATCC 33610; 11, G. bronchialis ATCC 25592; 12, A. pelletieri ATCC 33385; 13, N. otitidiscaviarum ATCC 14629.
Southern blot analysis.
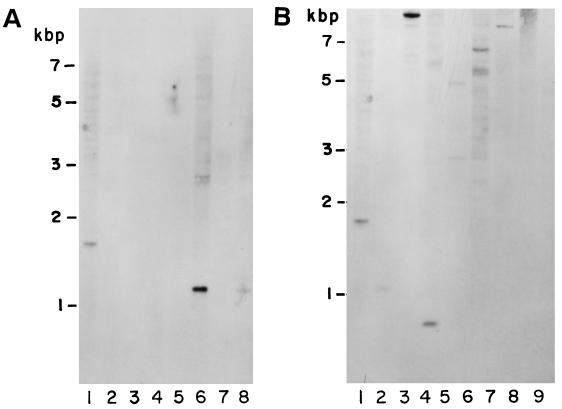
In order to determine the presence of this gene or similar sequences present in cross-reacting actinomycetes in the PCR assay, genomic DNA from a series of actinomycetes were subjected to Southern blot analysis by utilizing the restriction endonuclease BamHI. As shown in Fig. 2, the N. brasiliensis NCTC 10300 showed a band of approximately 1.7 kbp (lane 1 in both panels A and B). The N. brasiliensis HUJEG-1, as well as the other N. brasiliensis clinical isolates, also produced the same size band. N. otitidiscaviarum, N. brevicatena, N. transvalensis, N. asteroides (Fig. 2A, lanes 3, 4, 5, and 7, respectively) and N. carnea (not shown) type strains utilized in this study were negative in this assay. However, N. nova showed a band of about 1.150 kbp, as well as a lighter band of approximately 3 kbp (Fig. 2A, lane 6). N. farcinica presented a band of approximately 1.5 kbp (data not shown). No cross-reaction was observed with S. somaliensis (two strains), S. lavendulae (not shown), Actinomadura pelletieri, and A. madurae (not shown). Other microorganisms belonging to the nocardioform group that were positive included G. bronchialis, G. sputi, G. terrae, R. equi, R. erythropolis, and R. chubuensis (Fig. 2B, lanes 2, 3, 4, 5, 6, and 7 respectively). These microorganisms presented one or several bands that cross-hybridized with the NB10-NB11 probe but were a different size than that presented by the N. brasiliensis strains. Table 5 summarizes the PCR and Southern blot results.
FIG. 2.
Southern blot analysis of genomic DNA probed with the NB10-NB11 fragment of N. brasiliensis katN. (A) Lanes: 1, N. brasiliensis NCTC 10300; 2, A. pelletieri ATCC 33385; 3, N. otitidiscaviarum ATCC 14629; 4, N. brevicatena ATCC 15333; 5, N. transvalensis ATCC 06865; 6, N. nova ATCC 33726; 7, N. asteroides NCTC 6761; 8, N. asteroides LCDC 940238. (B) Lanes: 1, N. brasiliensis NCTC 10300; 2, G. bronchialis ATCC 25592; 3, G. sputi ATCC 33610; 4, G. terrae ATCC 25594; 5, R. equi ATCC 06939; 6, R. erythropolis ATCC 04277; 7, R. chubuensis ATCC 33609; 8, S. somaliensis ATCC 19437; 9, S. somaliensis ATCC 33201.
TABLE 5.
Results of the PCR and Southern blot assays with genomic DNA from the actinomycetes utilized in this studya
| Strain(s) | PCR | Southern blot | Approximate length of the band(s) in the Southern blot (kbp) |
|---|---|---|---|
| N. brasiliensis strains | Pos. | Pos. | 1.7 |
| N. asteroides NCTC 6761 | Neg. | Neg. | |
| N. farcinica ATCC 03318 | Neg. | Pos. | 1.5 |
| N. nova ATCC 33726 | Pos. | Pos. | 1.15, 3.0 |
| N. carnea ATCC 06847 | Neg. | Neg. | |
| N. transvalensis ATCC 06865 | Neg. | Neg. | |
| N. brevicatena ATCC 15333 | Neg. | Neg. | |
| N. otitidis-caviarum ATCC 14629 | Neg. | Neg. | |
| A. pelletieri ATCC 33385 | Neg. | Neg. | |
| A. madurae ATCC 19425 | Neg. | Neg. | |
| S. somaliensis ATCC 19437 | Neg. | Neg. | |
| S. somaliensis ATCC 33201 | Neg. | Neg. | |
| R. equi ATCC 06939 | Pos. | Pos. | 2.7, 5.0 |
| R. erythropolis ATCC 04277 | Pos. | Pos. | 5.3, 5.5, 7.0 |
| R. chubuensis ATCC 33609 | Pos. | Pos. | 9.0 |
| R. rhodochrous ATCC 13808 | Neg. | Neg. | |
| G. bronchialis ATCC 25592 | Neg. | Pos. | 1.1 |
| G. sputi ATCC 33610 | Pos. | Pos. | 10 |
| G. terrae ATCC 25594 | Neg. | Pos. | 0.7 |
| G. rubropertinctus ATCC 14352 | Neg. | Neg. |
Primers NB10 and NB11 were utilized in the PCR assay. Amplicon NB10-NB11 was used as a probe in the Southern blot assays. Positive (Pos.) and negative (Neg.) assay results are as indicated.
Sequence analysis of NB2-NB3 fragments of R. erythropolis and G. sputi.
In order to sequence the PCR products of cross-reacting bacteria, genomic DNA from R. erythropolis and G. sputi were subject to preparative PCR. Fragments of 500 bp were obtained, and the sequence was analyzed as described above. The resulting sequences were aligned (Fig. 3), and a high similarity was observed. The G. sputi and R. erythropolis NB2-NB3 sequences presented 78 and 81% homologies, respectively, to the N. brasiliensis NB2-NB3 fragment.
FIG. 3.
Alignment of the NB2-NB3 sequences of N. brasiliensis HUJEG-1 (catn_nb), G. sputi ATCC 33610 (cats_gs), and R. erythropolis ATCC 04277 (catr_re). Asterisks identify nucleotides common to all three sequences. Hyphens indicate gaps introduced to increase similarity.
DISCUSSION
Little is known about the immunogenic components of pathogenic Nocardia species that is useful in studying the host-parasite relationship in this infection. In this work we purified and determined the N-terminal and internal amino acid sequences of P61, an immunodominant protein of N. brasiliensis. These sequences showed a high degree of similarity to catalases of eukaryotic and prokaryotic origin. Although other microorganisms have several catalases and/or peroxidases (17), P61 seems to be the only enzyme of this kind in N. brasiliensis, since we observed only one band with the ferricyanide staining method.
P61 appears to be related to a group of catalases that comprise those from Comamonas compransoris, Klebsiella pneumoniae, S. violaceus, B. subtilis, Bacteroides fragilis, and Lactobacillus sakee (11, 18, 21). These catalases share amino acid sequences and are arranged in dimers or tetramers formed with subunits of 61 kDa. According to our gel filtration data, the native molecular mass of P61 is about 180 kDa, a size which can probably be attributed to the formation of a trimer.
N. brasiliensis is classified according to the ninth edition of the Bergey’s Manual of Determinative Bacteriology as part of group 22, the nocardioform actinomycetes, in subgroup 1, the mycolic-acid-containing bacteria, which also includes the genera Gordona, Nocardia, Rhodococcus, and Tsukamurella (13). The genus Nocardia comprises many species, the most commonly associated with human disease being N. brasiliensis, N. asteroides, N. farcinica, N. otitidiscaviarum, N. transvalensis, and N. nova. Of these the most extensively studied species is N. asteroides, for which some immunodominant antigens have been described (2, 3, 5, 10, 14). A 54-kDa protein of N. asteroides has been shown to be useful in detecting antibodies in patients with nocardiosis, although this protein is also present in N. brasiliensis and N. otitidiscaviarum (2, 25). Neither a complete biochemical characterization of this antigen nor the cloning of the gene encoding this protein have yet been reported.
N. asteroides is a heterogeneous taxon with many possible subvarieties (4); recently, it has been subgrouped in three species: N. asteroides sensu stricto, N. nova, and N. farcinica. Although these species can be differentiated by testing their abilities to utilize several carbon sources, by their growth at 45°C, as well as by their different mycolic acid or antimicrobial sensitivity patterns, these methods are time-consuming and often not definitive (27). In our experiments we observed that the P61 fragment NB10-NB11 is distributed differently in the N. asteroides complex strains we tested. N. asteroides sensu stricto tested negative in the PCR and in the Southern blot assay; N. nova was positive in both tests, and N. farcinica was positive only in the Southern blot assay. These findings correlate perfectly with the enzymatic profile of those species (6), where it has been reported that N. asteroides is catalase negative, whereas N. farcinica and N. nova are positive. The Southern blot results reflect differences at the nucleotide level between the N. farcinica and N. nova catalases, differences which can be exploited in the future to develop a genetic test to differentiate the N. asteroides complex species. These differences can also explain the negativity of N. farcinica in the PCR test, a result which is probably due to significant differences at the annealing sites of the oligonucleotides that did not permit the formation of an amplicon. A larger number of strains would be required to determine if the differences in the catalase genes of the N. asteroides complex bacteria can be useful for differentiating its members.
As mentioned above, N. brasiliensis is classified as part of the subgroup of mycolic-acid-containing bacteria which includes other genera, such as Nocardia, Rhodococcus, Tsukamurella, and Gordona (13). This group of bacteria share many biological, biochemical, and genetic characteristics. Therefore, we considered it important to study the distribution of the NB10-NB11 katN fragment or related sequences in these bacteria. In the Southern blot analysis, it was observed that some of the species tested cross-react with the NB10-NB11 probe, although bands of a different molecular weight were observed, reflecting variations in nucleotide sequence. This similarity was confirmed after sequence analysis of the NB10-NB11 amplicons of R. erythropolis and G. sputi, where a high degree of homology was observed. This finding was anticipated since other catalase genes similar to katN, such as the bca gene of S. violaceus or the catA gene of S. coelicolor, present a conserved or homologous region in positions 500 to 1500 of the bca gene open reading frame, while lower homology percentages were observed in the 5′ and 3′ ends of the ORF (data not shown). The cloning of the entire katN gene and/or related catalases from other actinomycetes will help to develop rapid genetic techniques for identifying and differentiating these species of bacteria. These data may also be useful for phylogenetic analysis in catalase-positive actinomycetes, as has been seen with other proteins (19).
Although we isolated and purified P61 because it is an immunodominant antigen in patients and experimental animals, we did not expect it to have catalase activity. Catalases have been claimed to play an important role in the survival and protection of microorganisms from the lysosomal oxygen-dependent microbicidal agents. This protective effect is carried out by nullifying toxic derivatives of oxygen produced by the respiratory burst in phagocytic cells. It has been demonstrated that in M. tuberculosis infection the loss of catalase activity correlates to isoniazid resistance and to a marked decrease in virulence for guinea pigs compared to infection with M. tuberculosis strains that have a complete katG gene (7, 9, 33). Wilson et al. has corroborated this finding more recently (31); they observed that the integration of a functional katG gene into an isoniazid-resistant M. bovis strain fully restores its virulence in experimental animals. N. brasiliensis virulence factors are presently unknown. The complete genetic and biochemical characterization of this catalase will facilitate the study of the role of this N. brasiliensis protein as a potential virulence factor.
ACKNOWLEDGMENTS
We are grateful for the valuable help of S. Tyler, R. Vogrig, and E. Torres for performing the gel filtration assays. We also thank K. Bernard for kindly providing the actinomycete type strains used in this study.
This research was supported in part by the Consejo Nacional de Ciencia y Tecnología (Conacyt) grants 25650-M and M92011F-123.
REFERENCES
- 1.Andrews D W, Girardi M, Mark J. Chemistry and sequencing of covalently immobilized proteins and peptides. Technical literature MG 345. Marlborough, Mass: Millipore Corp.; 1990. [Google Scholar]
- 2.Angeles A M, Sugar A M. Identification of a common immunodominant protein in culture filtrates of three Nocardia species and use in etiologic diagnosis of mycetoma. J Clin Microbiol. 1987;25:2278–2280. doi: 10.1128/jcm.25.12.2278-2280.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angeles A M, Sugar A M. Rapid diagnosis of nocardiosis with an enzyme immunoassay. J Infect Dis. 1987;155:292–296. doi: 10.1093/infdis/155.2.292. [DOI] [PubMed] [Google Scholar]
- 4.Beaman B L, Beaman L. Nocardia species: host-parasite relationships. Clin Microbiol Rev. 1994;7:213–264. doi: 10.1128/cmr.7.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boiron P, Provost F. Use of partially purified 54-kilodalton antigen for diagnosis of nocardiosis by Western blot (immunoblot) assay. J Clin Microbiol. 1990;28:328–331. doi: 10.1128/jcm.28.2.328-331.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boiron P, Provost F, Dupont B. Méthodes de laboratoire pour le diagnostic de la nocardiose. Paris, France: Institut Pasteur; 1993. pp. 24–32. [Google Scholar]
- 7.Cohn M L, Kovitz C, Oda V, Middlebrook G. Studies on isoniazid and tubercle bacilli. II. The growth requirements, catalase activities and pathogenic properties of INH-resistant mutants. Am Rev Tuberc Pulm Dis. 1954;70:641–650. doi: 10.1164/art.1954.70.4.641. [DOI] [PubMed] [Google Scholar]
- 8.Deem R L, Beaman B L, Gershwin M E. Adoptive transfer of immunity to Nocardia asteroides in nude mice. Infect Immun. 1982;38:914–920. doi: 10.1128/iai.38.3.914-920.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz G A, Wayne L G. Isolation and characterization of catalase produced by Mycobacterium tuberculosis. Am Rev Respir Dis. 1974;110:312–319. doi: 10.1164/arrd.1974.110.3.312. [DOI] [PubMed] [Google Scholar]
- 10.El-Zaatari F A, Reiss E, Yakrus M A, Bragg S L, Kaufman L. Monoclonal antibodies against ioselectrically focused Nocardia asteroides proteins characterized by the enzyme-linked immunoelectro-transfer blot method. Diagn Immunol. 1986;4:97–106. [PubMed] [Google Scholar]
- 11.Facey S J, Gross F, Vining L C, Yang K, van Pée K H. Cloning, sequencing and disruption of a bromoperoxidase-catalase gene in Streptomyces venezuelae: evidence that is not required for chlorination in chloramphenicol biosynthesis. Microbiology. 1996;142:657–665. doi: 10.1099/13500872-142-3-657. [DOI] [PubMed] [Google Scholar]
- 12.Folb P I, Timme A, Horowitz A. Nocardia infections in congenitally athymic (nude) mice and in other inbred mouse strains. Infect Immun. 1977;18:459–466. doi: 10.1128/iai.18.2.459-466.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holt J G, Krieg N R, Sneath P H A, Staley J T, Williams S T. Group 22, nocardioform actinomycetes. In: Holt J G, et al., editors. Bergey’s manual of determinative bacteriology. 9th ed. Baltimore, Md: Williams & Wilkins; 1994. pp. 625–650. [Google Scholar]
- 14.Kjelstrom J A, Beaman B L. Development of a serologic panel for the recognition of nocardial infections in a murine model. Diagn Microbiol Infect Dis. 1993;16:291–301. doi: 10.1016/0732-8893(93)90079-m. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Martinez R, Mendez-Tovar L J, Lavalle P, Welsh O, Saul A, Macotela-Ruiz E. Epidemiología del micetoma en México: estudio de 2105 casos. Gac Med Mex. 1992;128:477–481. [PubMed] [Google Scholar]
- 17.Menéndez M C, Ainsa J A, Martin C, García M J. katGI and katGII encode two different catalases-peroxidases in Mycobacterium fortuitum. J Bacteriol. 1997;179:6880–6886. doi: 10.1128/jb.179.22.6880-6886.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nies D, Schlegel H G. Catalase from Comamonas compransoris. J Gen Appl Microbiol. 1982;28:311–319. [Google Scholar]
- 19.Ochi K. Phylogenetic analysis of mycolic acid-containing wall-chemotype in actinomycetes and allied taxa by partial sequences of ribosomal protein AT-L30. Int J Syst Bacteriol. 1995;45:653–660. doi: 10.1099/00207713-45-4-653. [DOI] [PubMed] [Google Scholar]
- 20.Rico G, Ochoa R, Oliva A, González-Mendoza A, Walker S M, Ortiz-Ortiz L. Enhanced resistance to Nocardia brasiliensis infection in mice depleted of antigen-specific B cells. J Immunol. 1982;129:1688–1693. [PubMed] [Google Scholar]
- 21.Rocha E R, Smith C J. Biochemical and genetic analysis of a catalase from the anaerobic bacterium Bacteroides fragilis. J Bacteriol. 1995;177:3111–3119. doi: 10.1128/jb.177.11.3111-3119.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruimy R, Riegel P, Carlotti A, Boiron P, Bernardin G, Monteil H, Wallace R J, Christen R. Nocardia pseudobrasiliensis sp. nov., a new species of Nocardia which groups bacterial strains previously identified as Nocardia brasiliensis and associated with invasive diseases. Int J Syst Bacteriol. 1996;46:259–264. doi: 10.1099/00207713-46-1-259. [DOI] [PubMed] [Google Scholar]
- 23.Salinas-Carmona M C, Vera L, Welsh O, Rodriguez M A. Antibody response to Nocardia brasiliensis in man. Zentbl Bakteriol. 1992;276:390–397. doi: 10.1016/s0934-8840(11)80546-3. [DOI] [PubMed] [Google Scholar]
- 24.Salinas-Carmona M C, Welsh O, Casillas S M. Enzyme-linked immunosorbent assay for serological diagnosis of Nocardia brasiliensis and clinical correlation with mycetoma infections. J Clin Microbiol. 1993;31:2901–2906. doi: 10.1128/jcm.31.11.2901-2906.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sugar A M, Schoolnik G K, Stevens D A. Antibody response in human nocardiosis: identification of two immunodominant culture filtrate antigens derived from Nocardia asteroides. J Infect Dis. 1985;151:895–901. doi: 10.1093/infdis/151.5.895. [DOI] [PubMed] [Google Scholar]
- 26.Vera-Cabrera L, Salinas-Carmona M C, Welsh O, Rodriguez M A. Isolation and purification of two immunodominant antigens from Nocardia brasiliensis. J Clin Microbiol. 1992;30:1183–1188. doi: 10.1128/jcm.30.5.1183-1188.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Workman M R, Philpott-Howard J, Yates M, Beighton D, Casewell M W. Identification and antibiotic susceptibility of Nocardia farcinica and N. nova in the UK. J Med Microbiol. 1998;47:85–90. doi: 10.1099/00222615-47-1-85. [DOI] [PubMed] [Google Scholar]
- 28.Wayne L G, Diaz G A. A double staining method for differentiating between two classes of mycobacterial catalase in polyacrylamide electrophoresis gels. Anal Biochem. 1986;157:89–92. doi: 10.1016/0003-2697(86)90200-9. [DOI] [PubMed] [Google Scholar]
- 29.Welsh O, Salinas M C, Rodriguez M A. Mycetoma. In: Hoeprich P D, Jordan M-C, Ronald A R, editors. Infectious disease. 5th ed. Philadelphia, Pa: J. B. Lippincott Co.; 1994. p. 1405. [Google Scholar]
- 30.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 1. New York, N.Y: Wiley Interscience; 1990. pp. 2.4.1–2.4.2. [Google Scholar]
- 31.Wilson T M, de Lisle G W, Collins D M. Effect of inhA and katG on isoniazid resistance and virulence of Mycobacterium bovis. Mol Microbiol. 1995;15:1001–1015. doi: 10.1111/j.1365-2958.1995.tb02276.x. [DOI] [PubMed] [Google Scholar]
- 32.Ximenez C, Melendro E I, Gonzalez-Mendoza A, Garcia A M, Martinez A, Ortiz-Ortiz L. Resistance to Nocardia brasiliensis infection in mice immunized with either Nocardia or BCG. Mycopathologia. 1980;70:117–122. doi: 10.1007/BF00443077. [DOI] [PubMed] [Google Scholar]
- 33.Zhongming L, Kelley C, Collins F, Rouse D, Morris S. Expression of katG in Mycobacterium tuberculosis is associated with its growth and persistance in mice and guinea pigs. J Infect Dis. 1998;177:1030–1035. doi: 10.1086/515254. [DOI] [PubMed] [Google Scholar]