Abstract
Since the outbreak of SARS-CoV-2, the etiologic agent of the COVID-19 pandemic, the viral genome has acquired numerous mutations with the potential to alter the viral infectivity and antigenicity. Part of mutations in SARS-CoV-2 spike protein has conferred virus the ability to spread more quickly and escape from the immune response caused by the monoclonal neutralizing antibody or vaccination. Herein, we summarize the spatiotemporal distribution of mutations in spike protein, and present recent efforts and progress in investigating the impacts of those mutations on viral infectivity and antigenicity. As mutations continue to emerge in SARS-CoV-2, we strive to provide systematic evaluation of mutations in spike protein, which is vitally important for the subsequent improvement of vaccine and therapeutic neutralizing antibody strategies.
Keywords: SARS-CoV-2, spike protein, mutation, infectivity, antigenicity
Introduction
The coronavirus disease 2019 (COVID-19), caused by a single-stranded RNA virus called severe acute respiratory syndrome-coronavirus 2 (SARS-CoV-2), has become a pandemic disease globally [1–4]. By 25 July 2021, nearly 194 million cases of COVID-19 have been reported. What’s more, the number of COVID-19 cases remains at the high level with over 3.8 million new weekly cases according to the World Health Organization (WHO) [5]. COVID-19 is a type of acute respiratory disease with varied manifestations such as mild infection, pneumonia, lung failure and even death [2]. As early as 30 January 2020, the WHO declared COVID-19 as the sixth public health emergency of international concern. Therefore, the epidemic status of COVID-19 requires global cooperation to prevent its spread.
SARS-COV-2 is a new RNA virus strain belonging to the coronaviridae family, its whole genome consists of 29 903 nucleotides and shares a higher nucleotide sequence identity to a bat coronavirus RaTG13 (96.2%) rather than SARS-CoV (79.6%) [1, 6]. The genome of SARS-CoV-2 encodes four structural proteins including spike (S), envelope (E), membrane (M) and nucleocapsid (N) [7]. Among those proteins, the spike protein is a type I fusion protein that forms trimers on the surface of the virus and mediates the process of coronavirus entering into host cells [8]. It can be cleaved by the protein convertase furin at the S1/S2 site and the transmembrane serine protease 2 (TMPRSS2) at the S2′ site into S1 and S2 functional subunits [9]. S1 subunit contains multiple domains including N-terminal domain (NTD), intermediary domain (IND), C-terminal domain (CTD) and receptor-binding domain (RBD) [10], and it is responsible for binding to the host cell receptor (including ACE2 [8, 11], neuropilin-1 [12] and CD-147 [13]) or cathepsin L/B in the endosome pathway. The S2 subunit is responsible for the fusion of the viral and cellular membranes [11]. As the spike protein is surface-exposed and mediates the process of coronavirus entering into host cells, it is the main target of neutralizing antibodies (Abs) and the most promising source of antigens for vaccine design [8]. Several monoclonal antibodies (mAbs) [14–18] and vaccines [19–21] have been developed according to the sequence of initial SARS-CoV-2 strain, which have been proved to inhibit viral replication and alleviate severe clinical symptoms effectively. Most of the monoclonal antibodies and neutralizing antibodies induced by infection or vaccination inhibit the viral by blocking the binding of spike protein to its receptor [22–25].
As SARS-CoV-2 uses an intrinsically error-prone RNA polymerase for replication, it has a relatively higher mutation rate than DNA viruses [7]. SARS-CoV-2 has accumulated a considerable amount of mutations in spike protein during the spread of COVID-19 [26]. The great majority of those mutations are either ineffective or detrimental to some aspect of virus function and removed by natural selection [27]; however, there are still some mutations in spike protein that have been reported to significantly alter virus infectivity and antigenicity [7, 28]. The mutations in spike may promote the viral fitness by increasing the binding affinity to the hACE2 or conferring virus resistance to immune response induced by mAbs and vaccination (Figure 1). Consequently, these mutations bring great challenges to the prevention of COVID-19 and the development of mAbs and vaccines. Here, we summarize the spatiotemporal distribution of mutations in SARS-CoV-2 spike protein, and present recent efforts and progress in investigating the impact of mutations on viral infectivity and antigenicity.
Figure 1.
The schematic of the impact of mutations on viral fitness, including enhancement of infectivity (top), escape from neutralizing antibodies (middle) and reduction of vaccine protection (bottom).
The spatiotemporal distribution of mutations in SARS-COV-2 spike protein
As of 1 June 2021, a total of 1 750 995 SARS-CoV-2 spike protein sequences were available in GISAID, which were used to characterize the spatiotemporal distribution of mutations in spike protein. As is shown in Figure 2A, the mutations leading to an amino acid change in spike protein show a steady increase with each passing month. The mutations in spike protein were uncommon at the beginning of the pandemic. By contrast, the spike protein harbored an average of 10.24 ± 1.58 mutations according to the new cases reported in May 2021. There were as high as 6166 mutations in spike protein and their occurrence is in long-tail distribution (Figure 2B), which means most mutations are specific existed in few SARS-CoV-2 strains and a few mutations tend to occur frequently during the epidemic. It coincides with the fact most mutations happened to appear and are little to no impact on the virus’ properties. However, a few mutations with relatively high mutation frequency may promote the virus’ properties, such as infectivity, disease severity or the performance of mAbs, convalescent plasma and vaccines.
Figure 2.
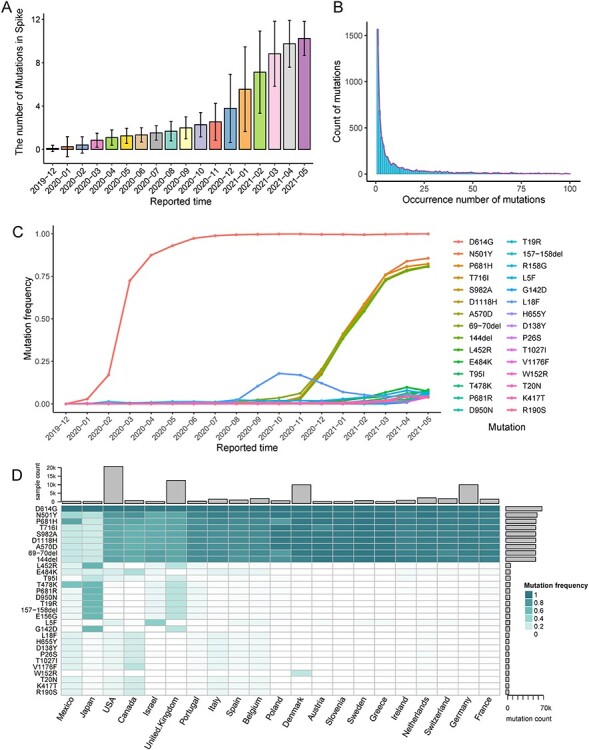
The spatiotemporal evolution of common mutations in SARS-COV-2 spike protein. (A) The average number of mutations in S protein since the beginning of the SARS-COV-2. (B) The distribution of occurrence number of mutations. (C) The mutation frequencies of current 30 most common mutations with the evolution of SARS-CoV-2. (D) The frequencies of mutations among different countries during May 2021.
To uncover the evolution of mutations along with virus spread, we delineated the mutation frequencies of the current 30 most common mutations (May 2021). As Figure 2C (Supplementary Table S1) shows, D614G is the first mutation that attracted worldwide attention and has become the most prevalent mutation since its appearance. In addition to D614G, both N501Y, P681H, T716I, S982A, D1118H, A570D, 70del and 144del show higher mutation frequency than other mutations. Both of them are the characteristic mutations of B.1.1.7 lineage [29], and the increase of their frequency is consistent with the outbreak of B.1.1.7 lineage. According to mutation data, B.1.1.7 has become the most dominant lineage of SARS-CoV-2 by far. What’s more notable is that the frequency of some mutations (including L453R, E484K, T95I, etc.) continue to rise in recent months, which is associated with the spread of other lineages (especially B.1.617.2) [30]. The L18F maintains a higher frequency during the latter half of 2020; however, its frequency starts to descend with the pandemic of B.1.1.7 lineage. To further characterize the present situation and spatial distribution of mutations, we calculated the frequencies of mutations among different countries with more than 100 samples during May 2021 (Figure 2D and Supplementary Table S2). B.1.1.7 characteristic mutations are widely distributed in all countries. On the contrary, the distribution of some mutations shows a degree of region specific. For example, B.1.617.2 characteristic mutations show a relatively high frequency in UK and Japan, P.1 characteristic mutations are mainly distributed in Canada, Mexico, the USA and Italy. W152R is still in part of Denmark strains.
The method of evaluating the mutation impact
Molecular dynamics simulation
Molecular dynamics (MD) is a computer method to analyze the equilibrium and transport properties of a classical many-body system by simulating real experiments [31, 32]. At the early stage of covid-19 epidemics, molecular dynamic simulations revealed that SARS-CoV-2 spike protein mediates a higher receptor affinity compared with SARS-CoV [33, 34]. Along with the emergence of mutation, molecular dynamics simulations have been utilized to elucidate its infectivity and antigenicity changes by calculating the binding free energy between spike protein and ACE2 or mAbs [35–37]. For example, the MD simulations of spike-ACE2 complexes revealed that five mutations (A348T, V367F, G476S, V483A and S494P) in the receptor binding domain of spike protein alter the binding affinity of RBD with ACE2 [36]. Meanwhile, the MD simulations of spike-mAbs complexes have been used to explore the structural mechanisms under the neutralization activity change of mAbs [37].
Those MD findings are in congruence with the experimental results and provide insights into the infection and neutralization processes at the molecular level [35]. Significantly, as the SARS-CoV-2 virus is expected to continue evolving in populations, molecular dynamics simulation is characteristic of high speed and low cost, which is especially suitable for high throughput screening of high-risk mutations in spike protein.
Pseudotyped virus system
SARS-CoV-2 is highly pathogenic in humans and has been classified as a biosafety level 3 (BSL3) pathogen according to WHO guidelines, impeding the basic research on live viruses [38]. The pseudotyped virus system has been developed to overcome this limitation; it is a chimeric virus consisting of a surrogate virus core surrounded by surface spike protein of SARS-CoV-2 [39, 40]. This single-cycle pseudovirus is much safer due to the absence of virulent viral components [40].
By utilizing the pseudotyped virus system, a study has investigated the biological significance of 80 natural mutations and found that most mutations show less infectious than wild-type [7]. However, there are still some mutations conferring virus resistance against several neutralizing antibodies and enhanced binding with ACE2 [7, 41–46], which bring serious challenges to current prevention, antibody therapies and vaccine protection. The SARS-CoV-2 pseudotyped virus system can mimic the infectious and neutralization process of the live virus [39, 47], making it an ideal serological tool to study the impact of mutations in vitro.
Cryo-electron microscopy technology
Cryo-electron microscopy (cryo-EM) is a mainstream tool for the structural visualization of biological macromolecular complexes and has been used to shed light on the infection and neutralization mechanisms of SARS-COV-2 [48, 49]. For example, although D614G seems to reduce affinity for ACE2 due to its faster dissociation rate, the crystal structure of spike protein trimer shows that D614G can induce more open conformation of its RBD region and increase the infectivity on human lung cells [10], which is consistent with the current high frequency of D614G. Moreover, K417N can decrease the neutralization activity of mAbs by reducing the polar contacts with complementarity determining regions [50]. Therefore, crystal structural studies, particularly on the conformational state of the mutant spike protein, are paramount for the mechanism studies of mutations on viral infectivity and antigenicity.
The impact of mutations on SARS-CoV-2 infectivity and antigenicity
Four lineages (including B.1.1.7, B.1.351, P.1 and B.1.617.2 lineages) have been officially classified as variants of concern (VOCs) by the WHO because of their risk to global public health [30, 51]. Those four lineages harbor a considerable amount of mutations in spike protein and show different levels of transmissibility, clinical presentation and severity changes. Thus, we mainly focus on the mutations in those lineages and review their impact on SARS-CoV-2 infectivity and antigenicity.
Mutations in B.1.1.7 lineage
The B.1.1.7 lineage, which is also known as 501Y.V1, was discovered in the UK as early as September 2020 [52]. It appears to be better at spreading between people and accounts for an increasing proportion of cases in parts of England (about 26% of cases in mid-November [53]). Besides D614G, which has already been found, the genomic of B.1.1.7 lineage has acquired other 17 mutations (14 non-synonymous mutations and 3 deletions) all at once. Even more worrying, nine mutations are located in spike protein (Table 1). Among those mutations, D614G appears earlier than B.1.1.7 lineage, and its frequency has increased from 10 to 67% on March 2020 and become the most prevalent mutation in the global pandemic (Figure 2C) [54]. More importantly, D614G can enhance viral replication [55, 56], induce more open conformation [10] and increase the infectivity of virus. Fortunately, the D614G mutation does not decrease the neutralizing activity of the antibodies induced by current vaccines [56–58] and the monoclonal antibodies [56, 57]. The N501Y, located in key residues of the ACE2-RBD interface, increases the binding affinity of the SARS-CoV-2 spike to ACE2 by inserting into a cavity at the binding interface [59, 60]. Most antibodies induced by vaccines and monoclonal antibodies retain the ability to neutralize the N501Y mutant [45, 61–63], but there are still some vaccines and monoclonal antibodies that show reduced neutralization to N501Y [45, 63]. The 69-70del and 144del mutations lead to the deletion of amino acids in the NTD and may allosterically change the S1 conformation of spike protein, which show a decreased susceptibility to convalescent plasma and vaccines [41, 64].
Table 1.
The impact of mutations on viral infectivity and antigenicity in different SARS-CoV-2 lineages
| SARS-CoV-2 lineages | First detected region | First detected time | Characteristic mutations in spike |
|---|---|---|---|
| B.1.1.7 (501Y.V1, Alpha) | UK | Sep 2020 | 69-70dela, 144dela, N501Ya,b, A570D, D614Gb, P681H, T716I, S982A, D1118H |
| B.1.351 (501Y.V2, Beta) | South Africa | Aug 2020 | L18F, D80A, D215G, 242-244dela, R246I, K417Na,b, E484Ka,b, N501Ya,b, D614Gb, and A701V |
| P.1 (501Y.V3, Gamma) | Northern Brazil | Dec 2020 | L18F, T20N, P26S, D138Y, R190S, K417Ta, E484Ka,b, N501Ya,b, D614Gb, H655Y, T1027I, and V1176F |
| B.1.617.2 (478 K.V1, Delta) | India | Dec 2020 | T19R, G142D, 157-158del, L452Ra, T478K, D614Gb, P681R, and D950N |
| Other lineages | — | — | W152 substitutionsa, and N439Ka,b |
aThe mutations decrease the susceptibility to mAbs, convalescent plasma or vaccines.
bThe mutations increase infectivity of the SARS-CoV-2.
Mutations in B.1.351 lineage
The B.1.351 lineage (also termed 501Y.V2) emerged in early August 2020 from South Africa. It spread rapidly and has become the dominant lineage in some provinces within weeks [65]. B.1.351 contains 10 mutations in the spike protein including the deletion of three amino acids in the NTD (242-244del), three substitutions (K417N, E484K and N501Y) in the RBD region and a substitution (A701V) near the furin cleavage site (Table 1) [44]. Recent studies find single N501Y mutation increases affinity for ACE 2.7-fold, and the co-mutation of K417N, E484K and N501Y further increase the affinity for ACE 19-fold [66, 67]. Even more striking, B.1.351 is particularly resistant to monoclonal antibodies, convalescent plasma and vaccines [41, 44, 68–70]. The 242-244del mutation shows resistance to neutralization of most monoclonal antibodies targeting the NTD. Meanwhile, three substitutions in the RBD region, especially E484K, confer SARS-Cov-2 resistance to monoclonal antibodies targeting the RBD region [41, 44]. The B.1.351 lineage potentially increases the risk of infection in immunized individuals as it is less sensitive or even insensitive to antibodies.
Mutations in P.1 lineage
The P.1 lineage (also termed 501Y.V3) was first reported in Northern Brazil and counted for about 42% located COVID-19 patients in December 2020 [71], and it exhibited more transmissible and led to a large wave of infection in Brazil [72]. The P.1 lineage contains 12 mutations spread throughout spike protein and shows a similar RBD mutation profile (K417T/N, E484K and N501Y) with the B.1.351 lineage (Table 1) [66], which are of the main concern because of their potential to alter viral infectivity and antigenicity [73]. Studies have indicated that the P.1 lineage increases affinity for ACE2 as that observed in B.1.351. E484K improves the electrostatic complementarity and N501Y induces a favorable ring stacking interaction with ACE2 [66]. Fortunately, the P.1 lineage shows lower resistance to monoclonal antibodies, convalescent plasma, and vaccines than the B.1.351 lineage. K417T, E484K, and N501Y are key mutations in conferring the P.1 lineage resistance to antibodies targeting the RBD region [41]. Although P.1 does not harbor deletion (69-70del, 144del or 242-244del), it also shows resistance to some mAbs targeting the NTD, which reveals that multiple substitutions in the NTD region may also disrupt its epitope [41, 66, 67].
Mutations in B.1.617.2 lineage
The B.1.617.2 lineage (also called 478 K.V1), a sub-lineage of B.1.617, was first detected in India in December 2020 [74]. B.1.617 appears to have a higher rate of transmission and has been spread across the globe [51]. In particular, the B.1.617.2 lineage shows a clear competitive advantage compared with B.1.1.7, B.1.351 and P.1, and includes a notable increase in the UK recently [75]. The B.1.617.2 possesses multiple characteristic mutations in its spike protein including T19R, Δ157–158, L452R, T478K, D614G, P681R and D950N (Table 1) [51]. Although some studies suggested that B.1.617.2 shows some degree of resistance to convalescent plasma, vaccines and part of mAbs [76–79], two doses of vaccine can still provide protection against the B.1.617 lineage [74, 79–81]. The mechanisms under high transmission and antibody resistance remain unknown. Multiple mutations in the NTD region may be responsible for the reduced neutralizing activity of mAbs targeting the NTD region [79]. L452R and T478K may have an impact on mAbs and vaccines directing towards the RBD region [7]. P681R is at the S1/S2 cleavage site may result in higher transmissibility [51]. Further robust studies are also required to validate the efficacy of the currently available mAbs and vaccines against B.1.617.2 lineage and understand the phenotypic impacts of these mutations.
Mutations in other lineages
In addition to the above-mentioned mutations in SARS-CoV-2 variants of concern, other mutations have been proved to alter the viral infectivity and antigenicity. For example, the mutations in W152 (including W152L, W152R, and W152C) are independently recruited numerous times across diverse geographical locations. The substitutions of W152, a residue present in the NTD of spike protein, promote virus immune escape by removing an important interaction point for multiple potent neutralizing antibodies [82]. N439K is most present in late 2019 and early 2020 (accounts for 3.37% of global SARS-CoV-2 in Dec 2020) and has been found across multiple countries. N439K, located in the spike receptor binding motif region, has been reported to enhance binding affinity to the hACE2 receptor and confer resistance against several mAbs [83, 84]. Fortunately, our data show that the percentage of N439K has fallen to 1‰. Here, we summarized the impact of common mutations on viral infectivity and antigenicity, however, there will be more mutations with the potential to alter viral infectivity and antigenicity as the COVID-19 pandemic.
Conclusion
The COVID-19 pandemic has become a main threat to human health and prosperity; the mutations in the SARS-COV-2 spike are making matters worse by altering viral infectivity and antigenicity. We summarize the spatiotemporal distribution of mutations and point that mutations in spike protein change gradually over time. The current viruses have accumulated more than 10 mutations on average by far. Although there are numerous mutations located in spike protein, only a few mutations tend to occur frequently which should be paid more attention to.
Considering the increasing proportion of mutant lineages, mutations in spike protein have posed a challenge to the existing mAbs and vaccines. Since the emergence of D614G, the scientific community has begun to investigate the impact of mutations on viral fitness and their underlying mechanisms using various bioinformatics and experimental methods. Many mutations in spike protein have been reported to promote the virus’ properties by increasing affinity to ACE2 or providing resistance to the immune response induced by mAbs, natural infection or vaccination. These findings are crucial for the prevention of SARS-CoV-2 and the next period of vaccine and therapeutic antibody development.
The recent emergence of B.1.617.2 lineage has been detected across the globe and shows a clear competitive advantage. To prevent a further spread of B.1.617.2 lineage, more studies are needed to confirm the effectiveness of current vaccines and uncover the mechanisms under its high transmission and antibody resistance. Fortunately, vaccines had not been demonstrated to lose their potency against most SARS-CoV-2 strains, and it still needs to maximize vaccination with two doses among populations. New mutations will continue to emerge as the SARS-CoV-2 persists; although most changes have little impact on viral fitness, there are still some mutations that can promote the virus’s properties and bring serious challenges to current prevention, antibody therapies and vaccine protection. So, there is an urgent need for continued monitoring of the frequency shifts of mutations in spike at regional and global levels and identifying potential risk mutations.
Key Points
SARS-CoV-2 evolution has been characterized by the emergence of sets of mutations, which bring great challenges to the prevention of COVID-19.
The bioinformatics and experimental methods are summarized to evaluate the impact of mutations in the SARS-CoV-2 spike protein.
Systematic review of the most common mutations in current SARS-CoV-2 lineages (e.g. B.1.1.7, B.1.351, P.1, B.1.617.2 and so on) and their impact on viral infectivity and antigenicity.
Supplementary Material
Wenyang Zhou is a Ph.D. student of biomedicine engineering at Harbin Institute of Technology, China.
Chang Xu is a Ph.D. student of biomedicine engineering at Harbin Institute of Technology, China.
Pingping Wang is a Ph.D. student of biomedicine engineering at Harbin Institute of Technology, China.
Anastasia A. Anashkina is a senior researcher at Engelhardt Institute of Molecular Biology, Russia.
Qinghua Jiang is a professor at Harbin Institute of Technology, China, focusing on immune mechanisms of complex diseases based on multi-omics data.
Contributor Information
Wenyang Zhou, School of Life Science and Technology, Harbin Institute of Technology, Harbin 150000, China.
Chang Xu, School of Life Science and Technology, Harbin Institute of Technology, Harbin 150000, China.
Pingping Wang, School of Life Science and Technology, Harbin Institute of Technology, Harbin 150000, China.
Anastasia A Anashkina, Engelhardt Institute of Molecular Biology RAS, Vavilova, 32, Moscow 119991, Russia.
Qinghua Jiang, School of Life Science and Technology, Harbin Institute of Technology, Harbin 150000, China.
Data Availability Statement
All presented data are free to available online. The SARS-CoV-2 spike protein sequences are available in GISAID database (https://www.gisaid.org/).
Funding
The National Natural Science Foundation of China (61822108, 62032007 and 62041102 to Q.J.); and the Emergency Research Project for COVID-19 of Harbin Institute of Technology (2020-001 to Q.J.)
References
- 1. Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020;579(7798):270–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lai CC, Shih TP, Ko WC, et al. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int J Antimicrob Agents 2020;55(3):105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 2020;382(8):727–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jin X, Zhou W, Luo M, et al. Global characterization of B cell receptor repertoire in COVID-19 patients by single-cell V(D)J sequencing. Brief Bioinform 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. WHO . Coronavirus Disease (COVID-19): Weekly Epidemiological Update (27 July 2021).
- 6. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature 2020;579(7798):265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Q, Wu J, Nie J, et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell 2020;182(5):1284–94 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Walls AC, Park YJ, Tortorici MA, et al. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020;181(2):281–92 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bestle D, Heindl MR, Limburg H, et al. TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance 2020;3(9):e202000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yurkovetskiy L, Wang X, Pascal KE, et al. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell 2020;183(3):739–51 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020;181(2):271–80 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cantuti-Castelvetri L, Ojha R, Pedro LD, et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 2020;370(6518):856–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bittmann S. COVID-19: ACE-2 receptor, TMPRSS2, Cathepsin-L/B and CD-147 receptor. J Regen Biol Med 2020. [Google Scholar]
- 14. Kim C, Ryu DK, Lee J, et al. A therapeutic neutralizing antibody targeting receptor binding domain of SARS-CoV-2 spike protein. Nat Commun 2021;12(1):288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lv Z, Deng YQ, Ye Q, et al. Structural basis for neutralization of SARS-CoV-2 and SARS-CoV by a potent therapeutic antibody. Science 2020;369(6510):1505–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shi R, Shan C, Duan X, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 2020;584(7819):120–4. [DOI] [PubMed] [Google Scholar]
- 17. Wu Y, Wang F, Shen C, et al. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 2020;368(6496):1274–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li F, Luo M, Zhou W, et al. Single cell RNA and immune repertoire profiling of COVID-19 patients reveal novel neutralizing antibody. Protein Cell 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. WHO . Draft landscape and tracker of COVID-19 candidate vaccines. 2021. 1 June. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines.
- 20. Yu J, Tostanoski L H, Peter L, et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 2020;369(6505):806–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Smith TRF, Patel A, Ramos S, et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat Commun 2020;11(1):2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Awadasseid A, Wu Y, Tanaka Y, et al. Current advances in the development of SARS-CoV-2 vaccines. Int J Biol Sci 2021;17(1):8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang S, Hillyer C, Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol 2020;41(5):355–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yadav T, Srivastava N, Mishra G, et al. Recombinant vaccines for COVID-19. Hum Vaccin Immunother 2020;16(12):2905–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang P, Xu Z, Zhou W, et al. Identification of potential vaccine targets for COVID-19 by combining single-cell and bulk TCR sequencing. Clin Transl Med 2021;11(5):e430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li R, Liu J, Zhang H. The challenge of emerging SARS-CoV-2 mutants to vaccine development. J Genet Genomics 2021;48(2):102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harvey WT, Carabelli AM, Jackson B, et al. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol 2021;19(7):409–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu A, Wang L, Zhou HY, et al. One year of SARS-CoV-2 evolution. Cell Host Microbe 2021;29(4):503–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. WHO . Coronavirus Disease (COVID-19): Weekly Epidemiological Update (04 May 2021), 2021.
- 30. WHO . Coronavirus Disease (COVID-19): Weekly Epidemiological Update (15 Jun 2021), 2021.
- 31. Pluhackova K, Wassenaar TA, Bockmann RA. Molecular dynamics simulations of membrane proteins. Methods Mol Biol 2013;1033:85–101. [DOI] [PubMed] [Google Scholar]
- 32. Wedekind J, Strey R, Reguera D. New method to analyze simulations of activated processes. J Chem Phys 2007;126(13):21502. [DOI] [PubMed] [Google Scholar]
- 33. Wang Y, Liu M, Gao J. Enhanced receptor binding of SARS-CoV-2 through networks of hydrogen-bonding and hydrophobic interactions. Proc Natl Acad Sci USA 2020;117(25):13967–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. He J, Tao H, Yan Y, et al. Molecular mechanism of evolution and human infection with SARS-CoV-2. Viruses 2020;12(4):428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dehury B, Raina V, Misra N, et al. Effect of mutation on structure, function and dynamics of receptor binding domain of human SARS-CoV-2 with host cell receptor ACE2: a molecular dynamics simulations study. J Biomol Struct Dyn 2020;1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chakraborty S. Evolutionary and structural analysis elucidates mutations on SARS-CoV2 spike protein with altered human ACE2 binding affinity. Biochem Biophys Res Commun 2021;534:374–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kwarteng A, Asiedu E, Sylverken AA, et al. Molecular characterization of interactions between the D614G variant of SARS-CoV-2 S-protein and neutralizing antibodies: a computational approach. Infect Genet Evol 2021;91:104815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ju X, Zhu Y, Wang Y, et al. A novel cell culture system modeling the SARS-CoV-2 life cycle. PLoS Pathog 2021;17(3):e1009439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carnell GW, Ferrara F, Grehan K, et al. Pseudotype-based neutralization assays for influenza: a systematic analysis. Front Immunol 2015;6:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nie J, Li Q, Wu J, et al. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg Microbes Infect 2020;9(1):680–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang R, Zhang Q, Ge J, et al. Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity 2021;54(7):1611–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Volz E, Hill V, McCrone JT, et al. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 2021;184(1):64–75 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. The CITIID-NIHR BioResource COVID-19 Collaboration, The COVID-19 Genomics UK (COG-UK) Consortium, Collier DA, et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature 2021;593(7857):136–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang P, Nair MS, Liu L, et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 2021;593(7857):130–5. [DOI] [PubMed] [Google Scholar]
- 45. Wang Z, Schmidt F, Weisblum Y, et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature 2021;592(7855):616–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tada T, Dcosta BM, Samanovic MI, et al. Convalescent-phase sera and vaccine-elicited antibodies largely maintain neutralizing titer against global SARS-CoV-2 variant spikes. MBio 2021;12(3):e0069621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ou X, Liu Y, Lei X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun 2020;11(1):1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020;367(6483):1260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cao Y, Zhu Q, Yu P, et al. Structurally resolved SARS-CoV-2 antibody shows high efficacy in severely infected hamsters and provides a potent cocktail pairing strategy—ScienceDirect. Cell 2020;183:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cao Y, Yisimayi A, Bai Y, et al. Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines. Cell Res 2021;31:732–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. WHO . Coronavirus Disease (COVID-19): Weekly Epidemiological Update (11 May 2021). 2021. https://reliefweb.int/report/world/coronavirus-disease-covid-19-weekly-epidemiological-update-11-may-2021.
- 52. Loman ARN. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Virological 2020. [Google Scholar]
- 53. Kupferschmidt K. Mutant coronavirus in the United Kingdom sets off alarms, but its importance remains unclear. Science 2020. [Google Scholar]
- 54. Korber B, Fischer WM, Gnanakaran S, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell 2020;182(4):812–27 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shi AC, Xie X. Making sense of spike D614G in SARS-CoV-2 transmission. Sci China Life Sci 2021;64(7): 1062–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hou YJ, Chiba S, Halfmann P, et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 2020;370(6523):1464–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Plante JA, Liu Y, Liu J, et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021;592(7852):116–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Weissman D, Alameh MG, de Silva T, et al. D614G spike mutation increases SARS CoV-2 susceptibility to neutralization. Cell Host Microbe 2021;29(1):23–31 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhu X, Mannar D, Srivastava SS, et al. Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies. PLoS Biol 2021;19(4):e3001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Starr TN, Greaney AJ, Hilton SK, et al. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell 2020;182(5):1295–310 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xie X, Liu Y, Liu J, et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nat Med 2021;27(4):620–1. [DOI] [PubMed] [Google Scholar]
- 62. Gu H, Chen Q, Yang G, et al. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science 2020;369(6511):1603–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cheng L, Song S, Zhou B, et al. Impact of the N501Y substitution of SARS-CoV-2 Spike on neutralizing monoclonal antibodies targeting diverse epitopes. Virol J 2021;18(1):87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. The CITIID-NIHR BioResource COVID-19 Collaboration, The COVID-19 Genomics UK (COG-UK) Consortium, Kemp SA, et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature 2021;592(7853):277–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Tegally H, Wilkinson E, Giovanetti M, et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv 2020. p. 2020.12.21.20248640.
- 66. Dejnirattisai W, Zhou D, Supasa P, et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell 2021;184(11):2939–54 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhou D, Dejnirattisai W, Supasa P, et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell 2021;184(9):2348–61 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hoffmann M, Arora P, Groß R, et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell 2021;184(9):2384–93 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Planas D, Bruel T, Grzelak L, et al. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med 2021;27(5):917–24. [DOI] [PubMed] [Google Scholar]
- 70. Abu-Raddad LJ, Chemaitelly H, Butt AA, et al. Effectiveness of the BNT162b2 Covid-19 vaccine against the B.1.1.7 and B.1.351 variants. N Engl J Med 2021;385(2):187–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Claro NRFIM. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. Virological 2020. [Google Scholar]
- 72. Faria NR, Mellan TA, Whittaker C, et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 2021;372(6544):815–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dejnirattisai W, Zhou D, Ginn HM, et al. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell 2021;184(8):2183–200 e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Liu J, Liu Y, Xia H, et al. BNT162b2-elicited neutralization of B.1.617 and other SARS-CoV-2 variants. Nature 2021;596(7871):273–5. [DOI] [PubMed] [Google Scholar]
- 75. Archer FCB. Increased transmissibility and global spread of SARSCoV-2 variants of concern as at June 2021. Eurosurveillance 2021;26(24):2100509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wall E C, Wu M, Harvey R, et al. Neutralising antibody activity against SARS-CoV-2 VOCs B.1.617.2 and B.1.351 by BNT162b2 vaccination. Lancet 2021;397(10292):2331–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pragya D. Yadav, Gajanan N. Sapkal, Raches Ella, et al. Neutralization against B.1.351 and B.1.617.2 with sera of COVID-19 recovered cases and vaccinees of BBV152. bioRxiv 2021; 2021.06.05.447177. [Google Scholar]
- 78. Planas D, Veyer D, Baidaliuk A, et al. Reduced sensitivity of infectious SARS-CoV-2 variant B.1.617.2 to monoclonal antibodies and sera from convalescent and vaccinated individuals. Nature 2021;596(7871):276–80. [DOI] [PubMed] [Google Scholar]
- 79. Liu C, Ginn HM, Dejnirattisai W, et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell 2021;184(16):4220–36.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of COVID-19 vaccines against the B.1.617.2 variant. medRxiv 2021;385(7):585–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Lopez Bernal J, Andrews N, Gower C, et al. Effectiveness of Covid-19 vaccines against the B.1.617.2 (Delta) variant. N Engl J Med 2021;385(7):585–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kubik S, Arrigo N, Bonet J, et al. Mutational hotspot in the SARS-CoV-2 spike protein N-terminal domain conferring immune escape potential. bioRxiv 2021; p. 2021.05.28.446137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Thomson EC, Rosen LE, Shepherd JG, et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell 2021;184(5):1171–87 e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Zhou W, Xu C, Wang P, et al. N439K variant in spike protein alter the infection efficiency and antigenicity of SARS-CoV-2 based on molecular dynamics simulation. Front Cell Dev Biol 2021;9(2071):697035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All presented data are free to available online. The SARS-CoV-2 spike protein sequences are available in GISAID database (https://www.gisaid.org/).


