Abstract
After experiencing the COVID-19 pandemic, it is widely acknowledged that a rapid drug repurposing method is highly needed. A series of useful drug repurposing tools have been developed based on data-driven modeling and network pharmacology. Based on the disease module, we identified several hub proteins that play important roles in the onset and development of the COVID-19, which are potential targets for repositioning approved drugs. Moreover, different network distance metrics were applied to quantify the relationship between drug targets and COVID-19 disease targets in the protein–protein-interaction (PPI) network and predict COVID-19 therapeutic effects of bioactive herbal ingredients and chemicals. Furthermore, the tentative mechanisms of candidates were illustrated through molecular docking and gene enrichment analysis. We obtained 15 chemical and 15 herbal ingredient candidates and found that different drugs may play different roles in the process of virus invasion and the onset and development of the COVID-19 disease. Given pandemic outbreaks, our method has an undeniable immense advantage in the feasibility analysis of drug repurposing or drug screening, especially in the analysis of herbal ingredients.
Keywords: COVID-19, drug repurposing, disease module, network proximity, herbal ingredients
Introduction
In past 18 months, the rapid spread and destructiveness of the COVID-19 have presented significant challenges on public health and biomedical research. Combating COVID-19 requires rapid development of new drugs and vaccines [1–3]. The successful development of multiple vaccines gives people hope; however, the virus has madly evolved and mutated, and the pandemic large-scale outbreaks in India indicating potential huge risks still exist all over the world [4].Therefore, there is still an urgent need for therapeutic drugs. When an emergency pandemic breaks out, how to quickly and effectively obtain potential drugs or lead compound is a new challenge for drug discovery. Given the tight timescales, the methods relying on traditional development, experimental testing, clinical validation and approval of new compounds are not feasible. Repurposing clinically approved drugs whose mechanisms of action and safety profiles have been thoroughly investigated is an ideal alternative method.
In silico drug design, especially repurposing methodologies based on data-driven and network medicine techniques have been applied to develop a series of computational tools to identify drug repurposing candidates [5, 6]. Drug repurposing has enabled the identification of successful therapeutics for cancer [7] and Parkinson disease [8]. Drug repurposing can enhance the development of traditional drugs and accelerate the development of new treatments for patients with Alzheimer’s disease [9]. In addition, the data-driven repurposing strategies that can identify previously unknown relationships between drugs and diseases by integrating multiple sources of data using computational modeling and machine learning have transformed the current therapeutic method for rheumatic autoimmune inflammatory diseases [10]. A network-based drug-disease proximity method, shed light on the relationship between drug targets and disease targets [11], is a useful tool for effectively repurposing approved drugs to new indications and predicting potential side effects [12]. And network analysis method has been used in drug repurposing for the treatment of COVID-19 [13, 14]. Zhou used a state-of-the-art network proximity measure to quantify the relationship between host proteins from four known the human coronaviruses and drug targets in the human interactome by calculating the GSEA score to screen some candidates [13]. Their hypothesis focused on the selected host protein to explore the process of inhibiting virus invasion, without considering the complete occurrence and development of the COVID-19 disease. Gysi ranked all approved drugs based on their likely efficacy for COVID-19 patients, relying on network proximity, diffusion and AI-based metrics, to arrive at 81 chemical candidates [14]. Their ranking method is very meaningful and interested, but it only focused on the approved little molecule drugs or proteins (named ‘chemicals’ in this article), and it also gave few meaningful recommendations for the next step in ‘old’ drug use in clinical or drug development.
The role of traditional Chinese medicine (TCM) in successfully fighting the COVID-19 epidemic cannot be ignored [15]. The National Health Commission (NHC) of China proposed the ‘New Coronavirus Infection Pneumonia Diagnosis and Treatment Plan (Trial)’. It is clearly pointed out that there are some medications which could exert the antiviral effect by the combination of herbal ingredients and chemicals. For example, LianHuaQingWen [16], as a representative TCM for the respiratory system infections in public health events, had been recommended to be used during the COVID-19 medical observation period by the NHC. QingFeiPaiDu decotion is effective for COVID-19 patients at all stages according to the treatment guidelines, and the total effective rate is as high as 92.09% [17]. GanCao Mixture was on the list of the descriptions of all patients who are cured and discharged from the First People’s Hospital Jingmen City from January to April in 2020. Specifically, Dali previously used the patterns of hormetic responses to explain the applicability of these processes to herbal medicines of TCM [18]. The ‘regulating’ functions promote adaptive or preventive responses, while ‘curing’ treatments alleviate the clinical symptoms. Not only TCM but also chemicals play significant roles in treating the COVID-19 [19–21]. Hydroxychloroquine and chloroquine have shown antiviral activity against SARS-CoV-2 [22]. Lopinavir-ritonavir can significantly shorten the median time from positive to negative symptoms treating COVID-19 patients [20]. Both TCM and chemicals are useful to fight against the COVID-19, but the mechanism and effectiveness of these drugs are still unclear. The screening and mechanism analysis of TCM and chemicals were suggested to carry out in parallel.
Large data-driven methods involving a systematic analysis of multiple types of data [such as gene expression, chemical structure, genotype or proteomic data, electronic health records (EHRs) or clinical trials] enhance these computational approaches leading to the repurposing hypotheses [5, 23]. The principle of parsimony, known as ‘Occam’s razor’, resists being put on a rigorous footing, a difficulty that has become more pressing and topical with the ‘big-data’ explosion [24]. Hence, we selected molecular structures, genes related to the onset and development of diseases (disease module) and factors related to drugs and the human body or virus (drug target), genotype or proteomic data (human interactome), and clinical trials as effective and important information for analysis.
Therefore, this study applied network proximity and network diffusion to directly analyze the closeness between drug targets and effective disease targets without bias, convert it into the possibility of identifying new indications for old drugs. Possible mechanisms of the onset and development of disease after virus invasion were uncovered through the disease module. And the relationship between drug targets and disease targets is analyzed based on the network overlapping. Here, chemical drugs and TCMs are included in network analyses combining drug screening method to identify COVID-19 potential therapeutic targets and mechanisms. After chemicals and herbal ingredients were pick out and mentioned that different candidates treated disease by different pathways, our method could provide a simple fast tool for new use of ‘old’ drugs for major public pandemics or other urgent emerging diseases.
Materials and methods
Drug screening and targets collecting
In this study, drug screening includes chemicals screening and collecting, and herbal ingredients collecting. Chemicals are collected by two ways. Some chemicals are collected from some literatures of drug repurposing strategies based on previously approved for certain pathogens [25–34]. The other come from our results of virtual screening in 2454 approved drugs from the DrugBank database (https://go.drugbank.com/) by docking them to the 3CLpro, which is used the traditional repurposing strategies focus on drugs that target human proteins that bind to viral proteins. Herbal ingredients are mainly collected from classical herbal prescriptions that are currently reported to be effective to anti-COVID-19 [35–37]. Licorice Mixture, which is clinically effective to COVID-19 in Jinmen Hospital, Hubei province, is analyzed to obtain herbal ingredients through network pharmacology, too.
Target-collecting includes collecting disease targets and molecular targets. The COVID-19 disease targets are collected from the GeneCards database (https://www.genecards.org/, downloaded on 31 October 2020) using ‘novel coronavirus pneumonia’ or ‘novel coronavirus’. The targets of collected herbal ingredients are obtained from the TCMSP database, and those targets of chemicals are from the DrugBank database (downloaded on 2 November 2020). Then, all targets are converted into uniform gene names in UniProt (https://www.uniprot.org/).
The COVID-19 disease module constructing
The COVID-19 disease targets are distributed in the Menche’s protein–protein-interaction (PPI) network [38] instead of the human interactome to form a COVID-19 disease module. The module size S, which represents the LCC in this COVID-19 disease module, is defined. Both the distribution Prand(S), which test statistic could be calculated for the observed values of S, and the Z-Score to describe the size of S were defined and calculated by using the u-test [39]. To estimate whether the COVID-19 disease targets were significantly localized in the network than that by chance, S was also defined where the same number of proteins as the COVID-19 disease targets were randomly selected and repeated it 1000 times in the same network. Then, Prand(S), Z-Score with random was also calculated by Equation 1. The interconnected targets were defined as the core disease genes.
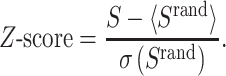 |
(1) |
Rank repurposing candidates
Network-based proximity analysis
The proximity between COVID-19 and a drug was evaluated using the closest distance measures that take into account the path lengths between drug targets and disease targets in the COVID-19 disease module [12]. The relative average shortest distance Zdc was used and calculated as follows. Given V (the set of collected COVID-19 disease targets), T (the set of drug targets including each herbal ingredient and chemical) and d(v, t), the shortest path length between nodes v (v∈V) and t (t∈T) in the human interactome [14], the closest distance measure (dc), which was the key parameter to describe the shortest path length for the node v and t in the menche PPI and represents the average shortest path length between the drug’s targets and the nearest disease protein, was defined in Equation 2.
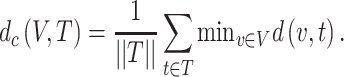 |
(2) |
To assess the significance of the distance between a drug and the COVID-19 disease (T, V), we determined a reference distance distribution corresponding to the expected distances between two randomly selected groups of proteins, matching the size and degrees of the original V and T sets. The reference distance distribution was generated by calculating the proximity between these two randomly selected groups, a procedure repeated 1000 times. To define the proximity measure and avoid repeatedly selecting the same high degree nodes, when the mean μd (V, T) and standard deviation σd (V, T) of the reference distribution were used to convert an observed distance to a normalized distance, the relative average shortest distance Zdc between T and V was calculated as follows.
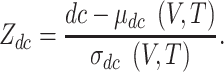 |
(3) |
If Zdc < 0, it means that the dc of the test group is significantly smaller than the reference, the drug with this test targets group is a potential candidate of anti-COVID-19. Whether it is an herbal ingredient or a chemical, the closest distance measure is calculated and estimated its network proximity.
Network-based diffusion analysis
Diffusion state distance (DSD) algorithm was used to rank drug candidates based on the network similarity of their targets to the COVID-19 targets [14]. In this study, the average minimum DSD of drug targets and disease targets in the PPI network of one candidate 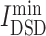 is calculated as follows. DSD was calculated as follows to measure the similarity that how a pair of proteins affect the rest global network in the human interactome. On an undirected graph G(V, E) with the vertex set V = {v1,v2,v3, …,vn} and |V| = n. Given k (k > 0), He{k}(i, j) represents the expected number of the times of the random walks from the node i to meet the node j after k steps. A node vi (vi∈V) has a He{k}(i, j) [denoted by He (vi, vj)] to any other node in the PPI network. An n-dimensional vector He(vi), ∀vi∈V was defined as follow:
is calculated as follows. DSD was calculated as follows to measure the similarity that how a pair of proteins affect the rest global network in the human interactome. On an undirected graph G(V, E) with the vertex set V = {v1,v2,v3, …,vn} and |V| = n. Given k (k > 0), He{k}(i, j) represents the expected number of the times of the random walks from the node i to meet the node j after k steps. A node vi (vi∈V) has a He{k}(i, j) [denoted by He (vi, vj)] to any other node in the PPI network. An n-dimensional vector He(vi), ∀vi∈V was defined as follow:
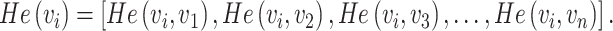 |
(4) |
He(vi) represents the influence of the node vi to all of the rest nodes in the global PPI network. Then, the DSD between two vertices i and j (∀i, j∈V) was defined as follow, which represents similarity through the sum over the absolute value of differences between the vertices of i and j.
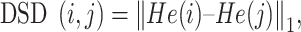 |
(5) |
where DSD is a non-zero (i ≠ j), positive definite and symmetric value for a given k, ||He(i) − He(j)||1 is the L1 norm (Manhattan distance) of the He vector of nodes i and j in the network. For the drug target t and the disease target v, the smaller DSD(t, v), the higher the similarity between t and v in the human interactome. Then, the average minimum DSD of drug targets and disease targets in the PPI network of one candidate,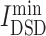 , is calculated to describe the impact of drug targets on the COVID-19 targets (Equation 6).
, is calculated to describe the impact of drug targets on the COVID-19 targets (Equation 6).
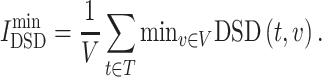 |
(6) |
Then, an herbal ingredient or a chemical with smaller 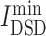 value is defined as a potential candidate to treat COVID-19 disease.
value is defined as a potential candidate to treat COVID-19 disease.
Aggregation ranking
An aggregation value was used to combine rankings returned by different methodologies into a single rank for each drug, which determined its anti-COVID-19 repurposing priority. The aggregation value was obtained for each candidate by setting the weight ratio of the values of network proximity and network diffusion similarity as 1:1 (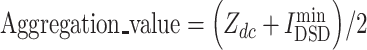 ). Then, the herbal ingredients and the chemicals were ranked by this aggregation value, respectively.
). Then, the herbal ingredients and the chemicals were ranked by this aggregation value, respectively.
Then, the drug repurposing feasibility of the candidate was confirmed by comparing the relative proximity Zdc of the candidate targets to its original therapeutic disease targets and the Zdc of the targets of this candidate to the COVID-19 disease targets in the PPI network.
Besides, to evaluate the predictive ability of the above-ranking methods, their power to recover the candidates in clinical trials as COVID-19 treatment were tested by computing the receiver operating characteristics (ROC) curves and the area under the curve (AUC) scores for measuring the quality of separation between positive and negative trials. Our herbal candidates were collected into a positive herbal set, and other herbal ingredients (retrieved from the TCMSP with degree = NA in the ‘herb-herbal ingredients-targets’ of the Licorice Mixture or OB<30, and DL<0.18) were selected into the negative set. In addition, when our chemical candidates were selected into the chemical positive set, other chemicals with lower LibDockScore listed on the bottom of our screening results of docking to 3CLpro were chosen as the negative set, or these chemicals had status of ‘Withdrawn’, ‘Terminated’ or ‘Suspended’ in ClinicalTrials.gov website (accessed on 01 March 2021) [40]. Then, both the true positive rate and the false positive rate of ROC and AUC were computed under the trapezoidal rule using the Scikit-learn package in Python.
Drug repurposing feasibility analysis of the candidates
Gene enrichment analysis
Gene enrichment is performed to explore the network vicinity of key targets in the COVID-19 disease and the potential pathways of drug treated COVID-19. The DAVID database (https://david.ncifcrf.gov/) was used for disease module functional annotations and network analysis. We examined enrichment in gene ontology for the following subsets of genes related to the COVID-19 disease module (P-value ≤ 0.05): (i) all risk genes, (ii) interconnected risk genes and (iii) risk genes in the LCC.
Key targets of the disease module
The key targets are pointed out by their appearing times in the results of top gene enrichment analysis on the COVID-19 disease module. Comparing ‘the greater the degree centrality (Degree) value of the network topology parameter node, the more important the node’ [41], the node with high degree centrality is selected as the effective target for COVID-19 treatment normally. All obtained COVID-19 disease targets chosen in the ‘Homo sapiens’ species with the minimum effect ‘high confidence’ to 0.7 were mapped into the STRING database by using the ‘NetworkAnalyzer’ module with Cytoscape 3.7.2 software to analyze its network topology parameters. Then the node with high degree centrality was selected as a control of key target.
Molecular docking
We chose several key targets to do molecular docking to evaluate drug-target interaction. Two key targets selected from the COVID-19 disease module are picked out according to above methods. The other two main targets (3CLpro and ACE2) are selected directly as effective targets for anti-COVID-19. Molecular docking of top 15 herbal ingredients and top 15 chemicals to 4 drug targets (http://www1.rcsb.org/) [42] are separately finished by Discovery Studio 2018.
Results
Drug screening and targets collecting
The 350 disease targets were obtained by using key words ‘novel coronavirus pneumonia’ or ‘novel coronavirus’ with MeSH method in the GeneCards database (Table S1A, see Supplementary Data available online at http://bib.oxfordjournals.org/). In chemicals drug screening, we collected the top 10 chemicals with highest LibDockScore screened 2454 approved drugs in DrugBank by docking to 3CLpro (Table S2A, see Supplementary Data available online at http://bib.oxfordjournals.org/) with Discovery Studio 2018 software. And those top chemical drugs with anti-COVID-19 effects screened by docking to 3CLpro [25–27], E protein [28], RDRP [29–31], S protein or ACE2 [43], respectively, were also collected (Table S2B, see Supplementary Data available online at http://bib.oxfordjournals.org/). After those chemicals without molecular targets in DrugBank were eliminated, 120 chemicals were chosen (Table S2E, see Supplementary Data available online at http://bib.oxfordjournals.org/). In herbal ingredients collecting, firstly, after the network pharmacology method was applied to screen the anti-COVID-19 ingredients of the Chinese medicine prescription called Gan-Cao Mixture (also named Licorice Mixture), eight ingredients with degree >5 in the overlap network are defined as key active ingredients for the treatment of diseases (Table S2C, see Supplementary Data available online at http://bib.oxfordjournals.org/, and Figure S1, see Supplementary Data available online at http://bib.oxfordjournals.org/). In addition, other 122 anti-COVID-19 herbal ingredients reported earlier [16, 35–37, 44], also screened by network pharmacology method, were collected from the 14 TCM prescriptions. After eliminating ingredients without known targets, 92 ingredients without duplicates were obtained (Table S2C, D and E, see Supplementary Data available online at http://bib.oxfordjournals.org/).
Construction of COVID-19 disease module
Among 350 collected COVID-19 disease targets, only 327 were significantly localized in the Menche human interactome [38, 39] (Z = 12.65, P < 0.0001, randomly sampling 1000 times) compared with the random gene sets, with an average of 1.19 edges between two disease targets, forming our COVID-19 disease module. There were 260 targets (S = 260, Z = 6.14, P = 4.07 × 10−10) formed the largest connected component (LCC) in the disease module (Figure S2, see Supplementary Data available online at http://bib.oxfordjournals.org/). Except for the LCC, the core disease module included only one interconnected COVID-19 disease target (Table S3, see Supplementary Data available online at http://bib.oxfordjournals.org/, and Figure S3, see Supplementary Data available online at http://bib.oxfordjournals.org/).
Prioritizing drug repurposing candidates for COVID-19
To identify drug repurposing candidates for COVID-19, collected chemicals and herbal ingredients were identified by network proximity and network diffusion repurposing strategies.
Network-based proximity analysis
After network-based proximity analysis, the relative average proximity distance between the drug targets and the COVID-19 disease targets distributed in the human interactome, Zdc, was calculated and listed in Table S4, see Supplementary Data available online at http://bib.oxfordjournals.org/. A negative Zdc value of a drug represents that the targets of this drug were close to the COVID-19-related targets. From Table S4A and B, see Supplementary Data available online at http://bib.oxfordjournals.org/, among chemicals, rescinnamine has the lowest Zdc value, −3.236, and the Zdc values of rilonacept, fingolimod, lypressin and nabumetone are following. Moreover, chloroquine has the Zdc value of −2.036, while the Zdc value of remdesivir is 0.071. Here, chloroquine, with Zdc < 0, is less proximal to COVID-19 targets comparing to remdesivir (with Zdc > 0), while chloroquine is a positive choice in antiviral drugs for COVID-19 [45] and remdesivir failed in clinical trial [46]. Among herbal ingredients, the Zdc value of quercetin is lowest, while the values of luteolin, apigenin, wogonin, rutin, rosmarinic acid and kaempferol are followed. Kaempferol have been proven effective [47], while quercetin and kaempferol are less proximal to COVID-19 targets causing with lower Zdc.
Network-based diffusion analysis
Network diffusion methodology was also used to probe the similarity of the collected the targets of drugs with COVID-19 disease targets. According the 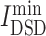 values show in Table S4, see Supplementary Data available online at http://bib.oxfordjournals.org/, all drugs were ranked. From Table S4C, see Supplementary Data available online at http://bib.oxfordjournals.org/, among chemicals, those antitumor drugs, such as baricitinib (
values show in Table S4, see Supplementary Data available online at http://bib.oxfordjournals.org/, all drugs were ranked. From Table S4C, see Supplementary Data available online at http://bib.oxfordjournals.org/, among chemicals, those antitumor drugs, such as baricitinib (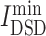 = 4.96), imatinib (
= 4.96), imatinib (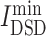 = 5.056) and ruxolitinib (
= 5.056) and ruxolitinib (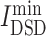 = 5.144), had lowest
= 5.144), had lowest 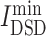 values, denoting that their targets had most similarity with COVID-19 targets. Baricitinib [48], for example, is a rheumatological drug in clinical trial for COVID-19, while its
values, denoting that their targets had most similarity with COVID-19 targets. Baricitinib [48], for example, is a rheumatological drug in clinical trial for COVID-19, while its 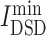 value is lowest among all chemicals. Furthermore, there are 11 anti-viral drugs, such as pibrentasvir, elbasvir, daclatasvir, ledipasvir, paritaprevir, glecaprevir, ombitasvir, velpatasvir, remdesivir, nelfinavir and tipranavir, which had relatively lower
value is lowest among all chemicals. Furthermore, there are 11 anti-viral drugs, such as pibrentasvir, elbasvir, daclatasvir, ledipasvir, paritaprevir, glecaprevir, ombitasvir, velpatasvir, remdesivir, nelfinavir and tipranavir, which had relatively lower 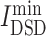 values. Here, remdesivir is described with the network similarity of its targets to COVID-19 targets. From Table S4D, see Supplementary Data available online at http://bib.oxfordjournals.org/, five herbal ingredients, such as acacetin, luteolin,quercetin, isorhamnetin and kaempferol, had the relatively low
values. Here, remdesivir is described with the network similarity of its targets to COVID-19 targets. From Table S4D, see Supplementary Data available online at http://bib.oxfordjournals.org/, five herbal ingredients, such as acacetin, luteolin,quercetin, isorhamnetin and kaempferol, had the relatively low 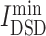 scores, indicating that their targets had most similarity with COVID-19 targets among all collected herbal ingredients.
scores, indicating that their targets had most similarity with COVID-19 targets among all collected herbal ingredients.
Rank
Combining two network-based methods, aggregation values of all candidates were calculated (Table S4, see Supplementary Data available online at http://bib.oxfordjournals.org/) and the top 15 herb ingredients and the top 15 chemicals with the lowest aggregation value were picked out and listed in Table S5, see Supplementary Data available online at http://bib.oxfordjournals.org/. Their structures are shown in Figure 1. All 15 chemicals are molecular diversity. Among chemicals, fingolimod is the best candidates with lowest aggregation value. Most anti-viral drugs with lowest 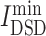 values were not included here. And chloroquine is listed as a positive candidate but remdesivir is not, which indicates that chloroquine may be better choice than remdesivir, which was confirmed by previous anti-COVID-19 reports [45, 46]. Observed top 15 herbal ingredients, quercetin, luteolin, acacetin and kaempferol, are candidates with high hit rate discussed in Chinese medicine clinical trial [35]. From Figure 1, there were 14 herbal ingredients with flavonoid structures (purple circle on herbal ingredient No. 6) indicating that most herbal candidates are similar in molecular structure.
values were not included here. And chloroquine is listed as a positive candidate but remdesivir is not, which indicates that chloroquine may be better choice than remdesivir, which was confirmed by previous anti-COVID-19 reports [45, 46]. Observed top 15 herbal ingredients, quercetin, luteolin, acacetin and kaempferol, are candidates with high hit rate discussed in Chinese medicine clinical trial [35]. From Figure 1, there were 14 herbal ingredients with flavonoid structures (purple circle on herbal ingredient No. 6) indicating that most herbal candidates are similar in molecular structure.
Figure 1.
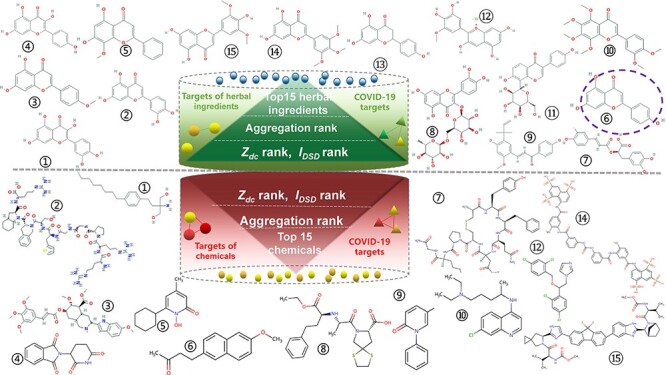
The molecular structures of the selected candidates (Chemicals No.11 and 13 as proteins are absent here).
Then, fingolimod and rosmarinic acid were randomly picked as positive candidates to discuss the possibility of correctness and the feasibility of treatment mechanism by comparing the relative proximity Zdc of the drug targets to its original therapeutic disease targets and the Zdc of the candidate targets to the COVID-19 disease targets, and the results were plotted in Figure 2. Fingolimod had a significant anti-human immunodeficiency virus (HIV) effect [49]. Rosmarinic acid has strong anti-inflammatory activity and even has anti-HIV effect [50]. Therefore, HIV was considered as an original therapeutic disease for both fingolimod and rosmarinic acid. To demonstrate the utility of the relative proximity, Figure 2A shows the shortest paths between drug targets and disease proteins for two known drug-disease associations: fingolimod-HIV and rosmarinic acid-HIV, and for two candidate-disease associations: fingolimod-COVID-19 and rosmarinic acid-COVID-19. In Figure 2B, the low Zdc value represents that both fingolimod and rosmarinic acid are expected drugs for the COVID-19 and HIV. The lower Zdc value is, the more potential the drug is.
Figure 2.
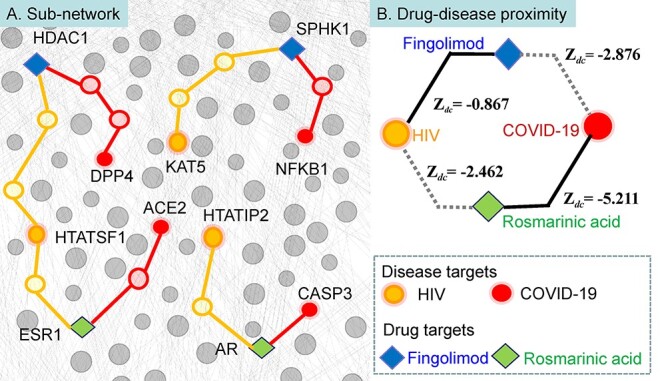
Network-based drug-disease proximity.
Fingolimod binds to sphingosine kinase 1(SPHK1) and histone deacetylase 1(HDAC1) and inhibits virus entry. Dipeptidyl peptidase 4 (DPP4) is a key receptor for the SARS-Cov-2 [51, 52], and there is at least one protein associated with COVID-19 targets within three steps of HDAC1 as neighborhood corresponding to an average distance of dc = 1.2 between fingolimod and the disease using the closest measure. The relative proximity between fingolimod and COVID-19 is Zdc = −2.876, suggesting that the targets of fingolimod are closer to the COVID-19 targets. Similarly, the relative proximity of rosmarinic acid, inhibiting the estrogen receptor (ESR1) and androgen receptor (AR), to COVID-19 is Zdc = −5.211, offering network-based support for the therapeutic effect of rosmarinic acid in COVID-19. The HIV proteins and drug targets are also closer than expected for randomly selected protein sets (Zdc = −0.867 and Zdc = −2.462, respectively), but farther than COVID-19 targets sets, respectively, suggesting that fingolimod and rosmarinic acid are nearer to COVID-19 disease module, they are specific to the COVID-19 disease module.
To evaluate the predictive ability of the above-ranking methods, and to identify candidate drugs with similar characteristics to known drugs used in a disease, by computing the ROC curves and the AUC scores for measuring the quality of separation between positive and negative trials, the AUC of the target proximity was calculated based on our positive candidates as a positive set and other reported negative herbal ingredients as a negative set. For herb, we collected our 86 herbal candidates into a positive set, while 44 herbal ingredients [retrieved from the TCMSP with degree = NA (no application) in the ‘herb-herbal ingredients-targets’ of the Licorice Mixture or OB < 30,DL < 0.18] were selected into a negative set. In addition, for chemicals, our positive chemical candidates were regarded as a positive set. And 31 chemicals that had status of ‘Withdrawn’, ‘Terminated’ or ‘Suspended’ (Table S6, see Supplementary Data available online at http://bib.oxfordjournals.org/) were chosen as a negative set. We evaluated the potential of the candidates for the COVID-19 by using three methods: (i) network proximity, (ii) network diffusion and (iii) aggregation ranking (Figure 3). From Figure 3A, the values of AUC on six sets were bigger than the value of a random set and bigger than 0.5, indicated our predicting results offering better candidates for the COVID-19 than random according to the proximity is a good proxy of therapeutic effect [12]. The AUC of two subsets by aggregation ranking methods was bigger than 0.8, which indicated that our final screened positive herbal and chemical candidates were reliable possible drugs for COVID-19.
Figure 3.
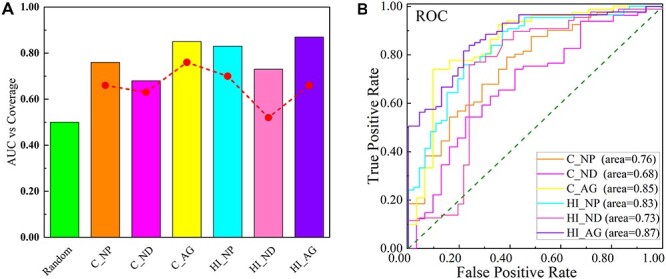
Validating drug-disease proximity. (The plot shows AUC and coverage values for network-based measures. Coverage is defined as the percentage of drug-disease associations for which the method can make predictions. C: chemicals; HI: herbal ingredients; NP: network proximity method; ND: network diffusion method; AG: aggregation rank).
Potential drug targets and mechanisms analysis
Gene enrichment analysis
After Gene enrichment analyses for (i) all risk genes (N = 350), (ii) interconnected risk genes (N = 261) and (iii) LCC risk genes (N = 260) were performed, all significant results (P < 0.05, corrected) were presented in Table S7, see Supplementary Data available online at http://bib.oxfordjournals.org/. Checked the top 10 gene enrichment results of the disease module, those pathways of ‘Influenza A’, ‘Measles’ and ‘Hepatitis B’, and some important biological processes (BP) including ‘inflammatory response’, ‘type I interferon signaling pathway’ and ‘defense response to the virus’ were some potential non-specific and significant pathogeneses in the COVID-19 disease development (Table S7A, see Supplementary Data available online at http://bib.oxfordjournals.org/). Normally, gene enriched pathway analyses on the COVID-19 disease module from the human interactome treated all COVID-19 disease targets equally and some targets located in the same loci would have some links with others [53]. Then IL-6 and TNF-α (or TNF) previously approved for general inflammation pathogens [54] were further identified as hub targets because they were found in these important BPs and pathways of the COVID-19 disease module with the highest hit (Table S7D, see Supplementary Data available online at http://bib.oxfordjournals.org/, both IL-6 and TNF appeared eight times in the top 10 pathways and also appeared eight times in BPs). Especially, by the analyses to genes in (ii) and (iii), the core genes of the COVID-19 disease module, it could be identified that enrichment items could not be identified while treating all risk genes equally [55], including the pathways of ‘adherens junction’ and ‘dilated cardiomyopathy’ and the BPs of ‘circadian rhythm’,‘thymus development’,‘regulation of cell adhesion’, etc. (See more details in Table S7F, see Supplementary Data available online at http://bib.oxfordjournals.org/). These items may be concluded as some specific indirect regulatory gene ontologies.
Gene enrichment on the overlapping network of the targets of the selected candidate and all disease module genes were systemically analyzed to discover important gene ontology of the candidate. Five candidates (including two herbal ingredients, ‘kaempferol’ and ‘rosmarinic acid’, two chemicals ‘fingolimod’ and ‘chloroquine’, and one chemical ranked as worse candidate, remdesivir) were analyzed their targets by overlapping to 350 disease targets, respectively (Figure S4, see Supplementary Data available online at http://bib.oxfordjournals.org/). As their targets with ds ≤ 1 show in Table S8, see Supplementary Data available online at http://bib.oxfordjournals.org/, kaempferol had 140, rosmarinic acid had 75, fingolimod had 29, chloroquine had 33 and remdesivir had only 6 targets. Then, we listed the results of gene enrichment analysis of their targets with ds ≤ 1 to the disease targets in Table S9, see Supplementary Data available online at http://bib.oxfordjournals.org/. From Table S9F, see Supplementary Data available online at http://bib.oxfordjournals.org/, four positive candidates had some similar significant pathways focused on the virus infection, including ‘Hepatitis B’ and ‘Influenza A’, but remdesivir had no. When four candidates had similar BPs on ‘negative regulation of apoptotic process’ and ‘lipopolysaccharide-mediated signaling pathway’, remdesivir still had none. It indicates that remdesivir is not a good choice, while other four candidates could be positive for anti-COVID-19 by different mechanism. As four candidates also had similar molecular functions (MFs, ‘protein binding’,‘identical protein binding’ and ‘enzyme binding’) and cellular components (CCs, ‘cytosol’ and ‘extracellular exosome’), remdesivir had ‘protein binding’, and similar CCs (cytosol and extracellular exosome) too. It indicates that remdesivir is easily to misunderstand as a positive candidate for anti-COVID-19 without systemic mechanism analysis.
Key targets of the COVID-19 disease
According to the gene enrichment analysis above, TNF and IL-6 were suggested to be two key targets for new medications, which is consistent with that previous experiment pointed out that the high IL-6 and TNF levels in the serum of COVID-19 patients are a strongly forward-looking predictor when host was infected by SARS-Cov-2 and even was taken as the predictor when the COVID-19 disease becoming extremely terrible [56]. Previous study identified that inflammatory monocytes, T cells secrete large amounts of IL-6 that can incur inflammatory storm for the COVID-19 [54, 57] and TNF is a crucial driver for multiple inflammatory macrophage phenotypes [58]. Comparing to analyzing the degree of 350 disease targets in PPI network from STRING database, IL-6 and TNF were also picked out as important potential drug-activated targets because their degree values were the highest when IL-6 had 65 and TNF had 66 (Table S10, see Supplementary Data available online at http://bib.oxfordjournals.org/). Degree centrality is the most convenient available centrality way to analyze potential drug targets [59]. But, here, it is only coincidence. The main reason of IL-6 and TNF chosen is according to the gene enrichment analysis results.
Molecular docking
After 30 candidates were docked to 3CLpro (PDB ID: 6 LU7), ACE2 (6 M17), TNF (2AZ5) and IL-6 (1ALU), separately, the best combinations were chosen based on -CDOCKER ENERGY scores. At the same time, the ligand N3 as the control of a binding site was docked to 3CLpro and tartaric acid was also docked to IL-6, separately. The docking results were listed in Table 1. In Table 1, some herb ingredients, such as rosmarinic acid, kaempferol and apigenin with the higher values, while the values of several chemicals, such as fingolimod, chloroquine, spirapril and miconazole, were relative higher, indicated that these compounds had good binding ability to 3CLpro. Observed the ACE2 complex, several drugs had greater values, such as rosmarinic acid, kaempferol, apigenin and luteolin as herbal ingredients and fingolimod as chemical. Except for nobiletin and rutin with negative values docking to TNF and IL-6, the rest 13 selected herbal ingredients had positive values docking to TNF and IL-6. Especially, four herbal ingredients (rosmarinic acid, kaempferol, apigenin and quercetin) had higher values docked to both TNF and IL-6. Among chemicals, fingolimod, spirapril and icatibant had the greater values docking to IL-6, even higher than tartaric acid. In addition, icatibant, lypressin, fingolimod and spirapril had the higher values, which meant that they had prominently interaction with TNF. Icatibant seemly had the prominently inhabitation for the TNF and IL-6, and interfered related the BP of inflammation and immunity, involved and interrupted the inflammation feedback loop and improve symptoms of inflammation clinically [60]. Lypressin displayed notable interaction with TNF, suggesting that it can inhibit relative targets and pathways to play the anti-inflammation effect.
Table 1.
The results of the candidates docking to 3CLpro, ACE2, IL-6 and TNF
| Molecules | - CDOCKER ENERGY | ||||
|---|---|---|---|---|---|
| 3CLpro | ACE2 | IL6 | TNF | ||
| Herbal ingredients | Quercetin | 36.407 | 4.201 | 43.300 | 25.354 |
| Luteolin | 31.357 | 18.330 | 27.322 | 18.687 | |
| Acacetin | 27.231 | 0.993 | 18.338 | 14.805 | |
| Kaempferol | 41.206 | 21.401 | 59.233 | 23.093 | |
| Wogonin | 23.072 | 10.181 | 20.031 | 13.073 | |
| Apigenin | 39.477 | 26.308 | 52.288 | 19.789 | |
| Rosmarinic acid | 43.409 | 34.407 | 51.048 | 28.870 | |
| Rutin | 10.244 | / | −15.917 | −14.400 | |
| Licochalcone a | 26.498 | −20.441 | 30.645 | 11.037 | |
| Nobiletin | 11.367 | / | −2.453 | −6.200 | |
| Puerarin | 28.539 | −10.652 | 41.049 | 13.833 | |
| Petunidin | 16.760 | −38.396 | 8.552 | 2.879 | |
| Naringenin | 28.325 | 7.524 | 22.635 | 15.269 | |
| 5,7-Dihydroxy-3,4,5- TrimethoxyFlavon | 22.382 | −70.834 | 10.330 | 7.869 | |
| Tricin | 34.672 | −32.983 | 39.708 | 13.734 | |
| Chemicals | Fingolimod | 43.926 | 21.114 | 31.440 | 28.750 |
| Icatibant | / | / | 41.574 | 92.676 | |
| Rescinnamine | −2.989 | / | −10.755 | −3.126 | |
| Thalidomide | 14.491 | / | 7.950 | 3.837 | |
| Ciclopirox | 2.862 | −96.214 | −3.363 | −4.608 | |
| Nabumetone | 18.360 | 6.696 | 11.717 | 8.498 | |
| Lypressin | / | / | / | 86.941 | |
| Spirapril | 33.588 | / | 30.796 | 21.097 | |
| Pirfenido ne | 8.261 | 3.705 | 2.128 | −1.544 | |
| Chloroquine | 35.518 | −21.148 | 12.146 | 12.035 | |
| Miconazole | 31.916 | / | 18.950 | 19.038 | |
| Suramin | −35.387 | −54.038 | −36.270 | −22.930 | |
| Ledipasvir | / | −92.034 | −13.859 | −1.371 | |
| Control ligands | N3 | 80.039 | / | / | / |
| Tartaric acid | / | / | 21.984 | / | |
Similar to some infectious diseases, such as hepatitis B and HIV, although there is still considerable value in optimizing access to virus-suppressing regimens, the researchers have embarked on a concerted journey to identify new antiviral drugs and immune interventions aimed at curing the virus infection [61]. The SARS-CoV-2 virus is not cytopathic. Hence, the treatment of COVID-19 should focus on the inhibition of virus replication and transfection in body and the treatment of immune symptoms and inflammatory reactions, not only on the inhibition of virus invasion. Comparing the results of herbal candidates docking to four targets, rosmarinic acid was identified the best inhibitor for the four targets, indicating that rosmarinic acid may have better potential to antiviral and regulate immune system by interacting with 3CLpro, ACE2, IL-6 and TNF. Previous study pointed out that rosmarinic acid can fight with immunodeficiency viruses, herpessimplex and influenza virus [62]. Chung found that magnesium lithospermate B combined rosmarinic acid can inhibit EV71 absorption stage during viral infection and rosmarinic acid blocked EV71 entering the cells to further hinder RNA expression and VP1 protein translation of the viral [62, 63]. Rosmarinic acid also can hinder the phosphorylation of TNF and reduce the production of chemokines, CCL11 and CCR3 [64], in line with that IL-6 and TNF in the COVID-19 disease module belong to chemokines, such as CXCL10, CXCL2 and CXCL8, precisely participating in the chemokine-mediated biology processes and triggering a series of cascade inflammation response [65]. Not only that, rosmarinic acid reduced more inflammatory cytokines and mediators likely TNF, IL-6, IL-1β and COX-2 to improve the symptoms of inflammation [66].
Observed the structures of herbal candidates, except rosmarinic acid, the other 14 molecules have C6-C3-C6 flavonoids framework. These flavonoids were suitable for embedding in the 3CLpro, IL-6 and TNF cavity (the results of 14 flavonoids docking to 3CLpro, IL-6 and TNF in Figure S5, see Supplementary Data available online at http://bib.oxfordjournals.org/), exerting the inhibition effects of virus proliferation and inflammation, especially kaempferol, apigenin and quercetin. Kaempferol showed strong inhibition with 3CLpro and ACE2, which is consistent with that it blocked the process of the proteolysis and SARS-CoV-2 cell entry [67]. Apigenin, luteolin and quercetin had the reliable IC50 values for the 3CLpro [68]. Quercetin can prevent the expression of inducible NO (Nitric oxide) synthase and IL-6 by interfering with the significantly enriched Lipopolysaccharide biology process [69].
Moreover, observed docking results, fingolimod showed us the best combination to four targets. Fingolimod is treated with multiple sclerosis (MM)—a CNS disease to inhibit ACE2 to hinder SARS-CoV-2 entry into nervous system preventing neurogenesis happening [70]. It can modulate a sphingosine-1-phosphate receptor to treat the neurogenesis and inflammation that are COVID-19 complication. Fingolimod could reduce the T-cell response when varicella-zoster virus infecting the host who suffered from multiple sclerosis disease [71], and influence virus-specific T cells proliferation and migration when viral triggering the acute-induced encephalomyelitis [72]. Hence, we conclude that fingolimod plays a role in anti-SARS-CoV-2 by inhibiting the proteins of 3CLpro and ACE2, while fingolimod also decreases T cells activity by interacting with TNF and IL-6, respectively, to evoke the antiviral and immunomodulatory.
In addition, combining the network-overlapping analysis (Table S9, see Supplementary Data available online at http://bib.oxfordjournals.org/) and docking results, we can find that four different positive candidates have different effects and regulate mechanisms in the occurrence and development of the COVID-19 disease. They have interactions with 3CLpro or ACE2 result in interference the SARS-Cov-2 duplication, transcription processes and intrusion process, while they interact with TNF or IL-6 to block some immunomodulatory and anti-inflammatory BPs and pathways or other antiviral ontology including influenza A, hepatitis B and hepatitis C to play anti-inflammatory and antiviral effects. The gene enrichment results of targets (ds = 1) of four candidates were together shown in Figure 4 and each was separately plotted on the same network attached in Figure S6, see Supplementary Data available online at http://bib.oxfordjournals.org/. Basically, the BPs of four candidates are different and there are fewer overlapping items with the BPs of the COVID-19 disease module (Table S9, see Supplementary Data available online at http://bib.oxfordjournals.org/), means that they have few effect on BPs. For kaempferol, rosmarinic acid and fingolimod (Figure S6, see Supplementary Data available online at http://bib.oxfordjournals.org/), there are focused on several similar viral infection pathways (such as Hepatitis B, Influenza A and Herpes simplex infection) to influence the onset and the development of the COVID-19 disease. Consistent with the good docking results, they three have a certain impact on the process of virus replication and transfection after virus invasion and the inflammatory and immune development in the COVID-19 disease. Moreover, kaempferol and rosmarinic acid, as classical TCM, are more similar in MF and CC to regulate the development of the COVID-19 disease. To chemicals, the overlapping pathways of chloroquine on the disease module are Hepatitis B and Influenza A, and its MFs and CCs are also similar on protein binding to the disease module. Combined with docking results, the combination with 3CLpro can indeed bind significantly, indicating that chloroquine has a significant effect in the process of the replication and transfection of the virus after it invaded. However, it may have not a significant role in inhibiting virus invasion, inflammation and immunity. The overlapping pathways of fingolimod are more similar to the disease module, the same as its MFs and its CCs, too. The combination with four receptors of fingolimod can indeed bind significantly, indicating that fingolimod has a significant effect on virus invasion, and it may have a good effect in the process of the replication and transfection of the virus, inflammation and immunity. Similar to treat with the hepatitis B, it is recommended to consider the above-mentioned aspects for subsequent experimental verification as a positive anti-COVID-19 candidate.
Figure 4.
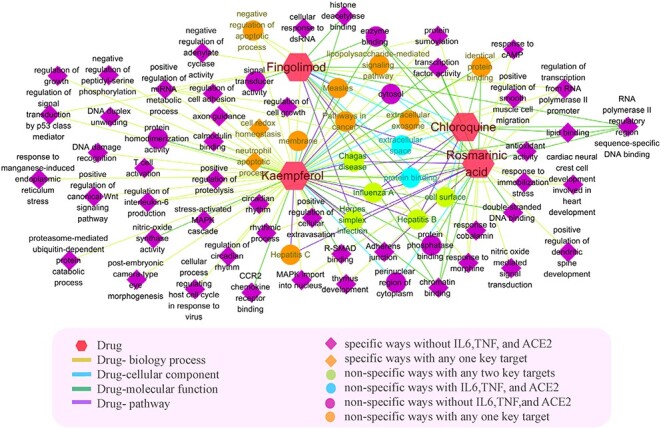
The gene enrichment analysis results of the targets with ds ≤ 1 of 4 candidates.
Furthermore, we focus on some specific and non-specific pathways in our results to explore different mechanisms. Through gene analysis, we found that compared with chemical, kaempferol and rosmarinic acid are significantly enriched in a specific pathway of adherens junction (Figure 5A). Researchers have reported that the death of ICU patients with COVID-19 is also related to cell adhesion and innate host reactions [73]. Normally adherens junction is an important pathway for the host immune response caused by virus invasion. One aspect, in adherens junction, CSNK2A2, an extracellular kinase, can phosphorylate various extracellular proteins involved in the life cycle of viral, coincidentally, which should be phosphorylated during viral infection [74]. Another aspect, TJP1 is a membrane-associated scaffold protein that assembles and locates a continuous network of tightly connected supramolecules on adherens junction, and plays a key role in cell adhesion [75]. TJP1 is a connected neighbor of Epidermal growth factor receptor (EGFR, a key pathway protein in adherens junction), which is closely related to COVID-19, especially related to the pathological effects of the inflammatory process in the brain [76, 77]. Previous studies have shown that SARS-CoV-2 can act on complex tip tight junctions, such as the E and 3a proteins of SARS-CoV-2, which bind to TJP1 protein and affect the entire cell connection and cell polarity [78]. In Figure 5A, AR, as a target of rosmarinic acid and kamepferol, directly regulates EGFR to affect TJP1; both ESR1 (a target of rosmarinic acid) and four targets of kamepferol (PGR, RELA, AKT1 and JUN) interact with CSNK2A2; ESR1 and two targets of kaempferol (STAT1 and ICAM1) interact with EGFR to indirectly regulate TJP1. Therefore, rosmarinic acid mainly regulates EGFR, SMAD3, CREBBP and CSNK2A2 by AR and ESR1. Kaempferol involves more high-end targets such as AR, STAT1, RELA, JUN, etc. to regulate multiple adhesion pathway targets comprehensively.
Figure 5.
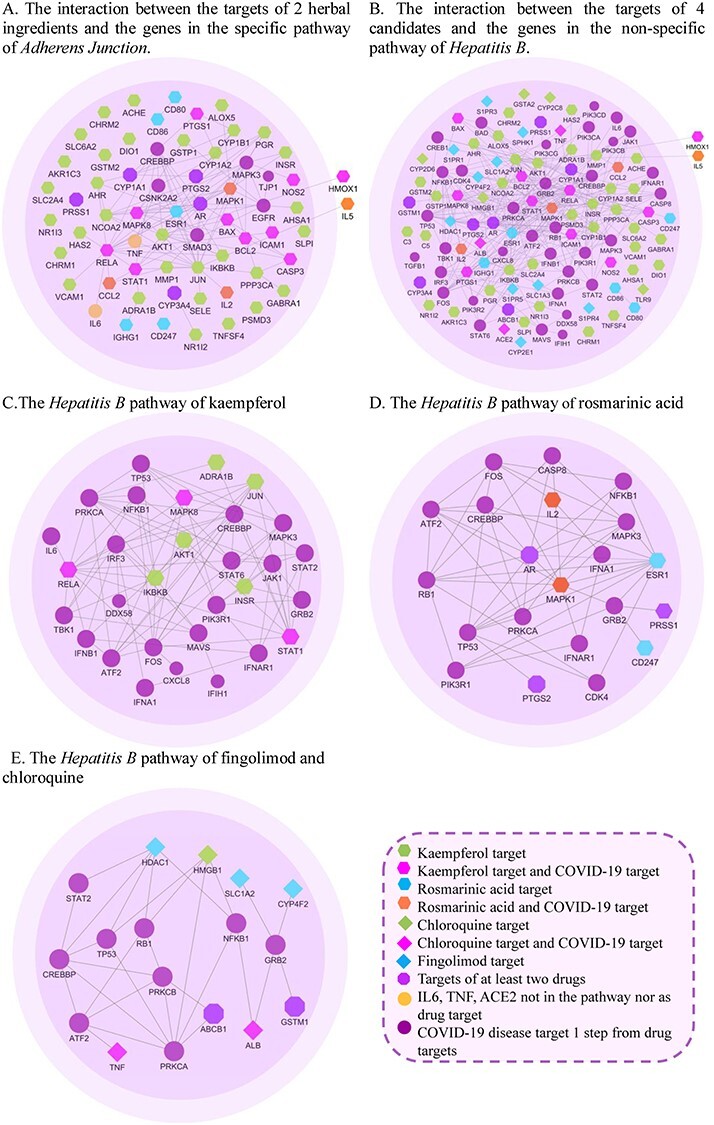
The relationship between drug targets and disease genes in adherens junction and hepatitis B.
In overlap analysis, four candidates have a regulatory effect on the Hepatitis B pathway (named non-specific pathway, shown in Figure 5B). From Figure 5C (the diagram of regulating in Hepatitis B by the targets of kaempferol), Kaempferol is related to NFκB1, TP53, PRKCA and other pathway targets regulating the production of interferon. For example, the target of kaempferol, RELA regulates the production of IFN-α by acting on IRF3 and FOS genes and affects IFNB1, too. IKBKB and RELA can establish an antiviral state by regulating TBK1. TBK1 phosphorylates IRF3 and dissociates from the adaptor protein to form a dimer, which then enters the nucleus to induce the expression of interferon [78]. RELA regulates the transcription of IL-6 to reduce the host inflammatory response caused by viral infection. In addition, STAT1 (as a kaempferol target) has an effect on JAK, STAT2, JUN, MAPK3, FOS and other targets. Previous studies have suggested that the abnormal STAT pathway is at the core of the treatment of COVID-19 [79]. Therefore, kaempferol affects the production of interferon and the expression of cytokines by interacting with these targets, resulting a therapeutic effect on viral infections [80, 81].
From Figure 5D, AR, ESR1 and MAPK1 (the targets of rosmarinic acid) with large node degrees can simultaneously adjust with multiple pathway targets, such as TP53, NFκB, MAPK3 and ATF2 (ds = 1). The targets of rosmarinic acid, MAPK1 and IL2 have a regulatory relationship with pathway targets IFNA1 and IFNAR1 (interferon, IFN, the cores of antiviral immunity). Virus recognition triggers the production of IFN, which in turn triggers gene transcription to produce various antiviral cytokines, thereby producing antiviral effects [82]. For example, IFN-γ and TNF are the main cytokines against hepatitis B virus [83]. Hadjadj et al. [84] showed that the peripheral blood immune cells of severe and critical COVID-19 patients reduced type I interferon, enhanced the response stimulated by pro-inflammatory IL-6 and TNF, and proposed that type I IFN deficiency is a sign of severe COVID-19.
From Figure 5E,in the interaction network between the targets of fingolimod and chloroquine and the targets of the hepatitis B pathway, ABCB1 and HMGB1 existing in fingolimod and chloroquine act on the pathway targets (such as TP53, NFκB1, RB1, PRKCB, PRKCA, etc.). Among them, HIV-1 viral infection factor can interact with TP53 to induce G2 cell cycle arrest and positively regulate virus replication [85]. TP53 is significantly related to the expression of TMPRSS2, a key protein of SARS-Cov-2 invasion [86]. NFκB with genetic polymorphisms helps to clear the virus [87]; NFκB also participates in T cell activation and enhances HIV-1 gene expression [88]. PRKCB and NFκB closely related to human T-cell leukemia virus type 1 (HTLV-1) [89]. Besides TP53, NFκB1 and RB1, the target of fingolimod, HDAC, also has connections with STAT2 [90]. There is also an interaction between chloroquine-acting COVID-19 pro-inflammatory factor TNF and hepatitis B virus pathway protein ATF2.
Discussion
We have studied a combined method of various data-driven computational approaches to identify drug repurposing candidates in clinical emergencies, such as COVID-19. In this method, we constructed a network-based framework approach to rank drug repurposing candidates based on the COVID-19 disease module. We proceeded with the top gene enrichment analysis of overlapping of disease genes and targets of candidates to identify effective disease targets and probe the anti-COVID-19 mechanisms of those chemical and herbal candidates. Furthermore, we applied molecular docking to key targets to verify some specific therapeutic hypotheses. We found that different drugs may play different roles in the process of virus invasion and the onset and development of the COVID-19 disease.
The COVID-19 disease phenotypes are typically determined by the defects of multiple genes whose concurrent and abnormal activity is significant for the onset and the development of the COVID-19 disease. The disease genes are not randomly distributed but clustered in the COVID-19 disease module corresponding to well-defined neighborhoods of the human interactome with the human interactome. We used a computational framework to quantify the relationship between disease and candidate using the distance measures that capture the network-based proximity of targets of a drug to all significant disease genes. The distribution characteristics and the relationships of disease genes in the COVID-19 disease module can be used to analyze the mechanism of disease onset and development. The enrichment analysis of disease genes in different parts of the COVID-19 disease module can distinguish specific and non-specific regulations for key targets that trigger the onset and development of the disease.
The network proximity of drug targets to the COVID-19 disease genes provides special insights into the candidate mechanism of action, uncovering the pathobiological components targeted by drug candidates, and improves the feasibility and interpretability for drug repurposing. The systematic analysis of the proximity of drug targets to the COVID-19 and to their original treatment diseases shows that drugs target the COVID-19 disease module effectively than aim at an original treatment disease of this drug. If a drug is proximal to the COVID-19 disease, it is more likely to be effective than a distant drug. The proximity can also be used to define similarity between two drug candidates and covered a great number of drug-disease associations.
Given the limitations of the current human interactome, from incompleteness to investigative bias, we have explored how the number and centrality of drug targets and disease proteins affect their network-based proximity. It was found that the proximity is unbiased with regard to both the number of targets of a candidate and their degrees in the network. Though some important targets may have bigger numbers and higher centralities of drug targets (such as TNF and IL-6), the proximity demonstrates the mechanism and effects of drug candidates much clearly than by docking to few key targets. Therefore, the use of proximity corrects a common pitfall in existing research that does not account for the elevated number of interactions of drug targets.
Conclusion
In summary, this research highlights probing the effective targets and drugs repurposing for the COVID-19, which may provide a quick way to find drug candidates. Basing on the COVID-19 disease module, different network distance strategies to quantify the relationship between drug targets and disease genes distribution in the PPI were adopted to predict drug candidates on herbal ingredients and chemicals, respectively. Integrating molecular docking and gene enrichment analysis of key targets offer the possible anti-disease mechanisms of candidates. However, disease modules and molecular targets still need to be improved due to the relationships of human interactome unexplored currently. Sure, we need to further verify the calculation results through experiments comprehensively. Despite the limitations in this study, for an outbreak of pandemic disease, it still has an undeniable immense advantage in the feasibility analysis of drug repurposing or drug screening, especially on herbal ingredients. Currently, the emergence of highly contagious variants of SARS-Cov-2 in many countries has brought huge challenges to the world’s public health. We will continue to study drug repurposing methods developed on the onset and development of diseases, verify their effectiveness for rapidly evolving and mutating viruses and further verify the calculation results through experiments.
Key Points
Based on the formed COVID-19 disease module, we identified several hub proteins that play important roles in the onset and development of the COVID-19, which are potential targets for repositioning approved drugs. The COVID-19 disease module revealed that the onset and development of COVID-19 mainly involve antiviral, inflammatory and immune responses, as two proinflammatory factors, TNF and IL-6, play vital role as hub targets in the development of the COVID-19.
Network strategies on different network distance metrics were successfully applied to quantify the relationship between drug targets and COVID-19 disease targets in the PPI network and predict COVID-19 therapeutic effects of bioactive herbal ingredients and chemicals. Our method has an undeniable immense advantage in the feasibility analysis of drug repurposing or drug screening, especially in the analysis of herbal ingredients.
The tentative mechanisms of candidates were illustrated through molecular docking and gene enrichment analysis. For example, rosmarinic acid and fingolimod could be multi-target inhibitors, not only restrain TNF, IL-6 activity to influence the inflammation and immunity relevant process but also inhibit 3CLpro and ACE2 to hinder the SARS-Cov-2’s reproduction and replication process.
Supplementary Material
Acknowledgements
The authors thank Professor Ailong Huang, Junqing Yang and Jingyuan Wan (Chongqing Medical University) for supporting some clinical and pharmacological advice and useful discussions.
Yonghong Zhang, Ph.D, Associate Professor, College of Pharmacy, Chongqing Medical University; Vice President, Committee, Chongqing Bioinformatics Society, Chongqing, China. She is also doing research in Medical Data Science Academy, Chongqing Medical University, Chongqing, 400016, China, and in Chongqing Engineering Research Center for Clinical Big-data and Drug Evaluation, Chongqing Medical University, Chongqing, 401331, China. Research interests are computer life science, computational aided drug analysis and evaluation.
Hong Wang is a master candidate in the College of Pharmacy, Chongqing Medical University, Chongqing, 400016, China. She is also doing research in Medical Data Science Academy, Chongqing Medical University, Chongqing, 400016, China, and in Chongqing Engineering Research Center for Clinical Big-data and Drug Evaluation, Chongqing Medical University, Chongqing, 401331, China. Her research interests include bioinformatics, drug evaluation and computer aided drug design.
Jingqing Zhang is a Professor in the College of Pharmacy, Chongqing Medical University, Chongqing, 400016, China. Her research interests are drug design, preparation, analysis and evaluation.
Zhigang Lu is currently an Associate Professor in Department of Neurology, the First People's Hospital of Jingmen affiliated to Hubei Minzhu University, China. His research interests are Pharmacology of Traditional Chinese Medicine, Basic and Clinical Study of Cerebrovascular Diseases.
Weina Dai is a master student in the College of Pharmacy, Chongqing Medical University. Her research interests are bioinformatics, computer aided drug analysis and drug design.
Chuanjiang Ma is an undergraduate student in College of Pharmacy, Chongqing Medical University. His research interests include bioinformatics, drug evaluation and computer aided drug design.
Yun Xiang is the deputy chief physician of the Obstetrics and Gynecology department of Guangzhou Women and Children's Medical Center. Her research interest is the analysis and evaluation of clinical drug use for children and pregnant women.
Contributor Information
Hong Wang, Chongqing Key Research Laboratory for Drug Metabolism, College of Pharmacy, Chongqing Medical University, Chongqing, 400016, China; Medical Data Science Academy, Chongqing Medical University, Chongqing, 400016, China; Chongqing Engineering Research Center for Clinical Big-data and Drug Evaluation, Chongqing Medical University, Chongqing, 401331, China.
Jingqing Zhang, Chongqing Key Research Laboratory for Drug Metabolism, College of Pharmacy, Chongqing Medical University, Chongqing, 400016, China.
Zhigang Lu, Department of Neurology, The First People's Hospital of Jingmen affiliated to Hubei Minzu University, Jingmen, 448000, China.
Weina Dai, Chongqing Key Research Laboratory for Drug Metabolism, College of Pharmacy, Chongqing Medical University, Chongqing, 400016, China.
Chuanjiang Ma, Chongqing Key Research Laboratory for Drug Metabolism, College of Pharmacy, Chongqing Medical University, Chongqing, 400016, China.
Yun Xiang, Gynaecology and Obstetrics, Guangzhou Women and Children's Medical Center, Guangzhou, 510623, China.
Yonghong Zhang, Chongqing Key Research Laboratory for Drug Metabolism, College of Pharmacy, Chongqing Medical University, Chongqing, 400016, China; Medical Data Science Academy, Chongqing Medical University, Chongqing, 400016, China; Chongqing Engineering Research Center for Clinical Big-data and Drug Evaluation, Chongqing Medical University, Chongqing, 401331, China.
Funding
National Natural Science Foundation of China (Grand numbers: 22176020, 32072262); Chongqing Natural Science Foundation (cstc2020jcyj-msxmX0150); The Science and Technology Research Program of Chongqing Municipal Education Commission of China (KJQN201900440); The Intelligent Medicine Research Project of Chongqing Medical University (ZHYX202020); Natural Science Foundation of Hubei Province of China (2020CFB722); Health Commission of Hubei Province of China (WJ2021M075); Science and Technology Planning Project of Jingmen City of China (2020YFZD025).
References
- 1. Dai L, Gao GF. Viral targets for vaccines against COVID-19. Nat Rev Immunol 2021;21:73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Walsh EE, Frenck RW, Jr, Falsey AR, et al. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N Engl J Med 2020;383:2439–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee SW, Ha EK, Yeniova AO, et al. Severe clinical outcomes of COVID-19 associated with proton pump inhibitors: a nationwide cohort study with propensity score matching. Gut 2021;70:76–84. [DOI] [PubMed] [Google Scholar]
- 4. Lancet T. India under COVID-19 lockdown. Lancet 2020;395:1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pushpakom S, Iorio F, Eyers PA, et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov 2019;18:41–58. [DOI] [PubMed] [Google Scholar]
- 6. Wang X, Yang Y, Li K, et al. BioERP: biomedical heterogeneous network-based self-supervised representation learning approach for entity relationship predictions. Bioinformatics 2021;btab565. doi: 10.1093/bioinformatics/btab565. [DOI] [PubMed] [Google Scholar]
- 7. Stewart AK. Medicine. How thalidomide works against cancer. Science 2014;343:256–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hubsher G, Haider M, Okun MS. Amantadine: the journey from fighting flu to treating Parkinson disease. Neurology 2012;78:1096–9. [DOI] [PubMed] [Google Scholar]
- 9. Ballard C, Aarsland D, Cummings J, et al. Drug repositioning and repurposing for Alzheimer disease. Nat Rev Neurol 2020;16:661–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kingsmore KM, Grammer AC, Lipsky PE. Drug repurposing to improve treatment of rheumatic autoimmune inflammatory diseases. Nat Rev Rheumatol 2020;16:32–52. [DOI] [PubMed] [Google Scholar]
- 11. Yildirim MA, Goh KI, Cusick ME, et al. Drug-target network. Nat Biotechnol 2007;25:1119–26. [DOI] [PubMed] [Google Scholar]
- 12. Guney E, Menche J, Vidal M, et al. Network-based in silico drug efficacy screening. Nat Commun 2016;7:10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou Y, Hou Y, Shen J, et al. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov 2020;6:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Morselli GD, do Valle Í, Zitnik M, et al. Network medicine framework for identifying drug-repurposing opportunities for COVID-19. Proc Natl Acad Sci U S A 2021;118:e2025581118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cao P, Wu S, Wu T, et al. The important role of polysaccharides from a traditional Chinese medicine-lung cleansing and detoxifying decoction against the COVID-19 pandemic. Carbohydr Polym 2020;240:116346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xian Y, Zhang J, Bian Z, et al. Bioactive natural compounds against human coronaviruses: a review and perspective. Acta Pharm Sin B 2020;10:1163–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang R, Liu H, Bai C, et al. Chemical composition and pharmacological mechanism of Qingfei Paidu Decoction and Ma Xing Shi Gan Decoction against Coronavirus Disease 2019 (COVID-19): in silico and experimental study. Pharmacol Res 2020;157:104820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang D, Calabrese EJ, Lian B, et al. Hormesis as a mechanistic approach to understanding herbal treatments in traditional Chinese medicine. Pharmacol Ther 2018;184:42–50. [DOI] [PubMed] [Google Scholar]
- 19. Yazdany J, Kim AHJ. Use of hydroxychloroquine and chloroquine during the COVID-19 pandemic: what every clinician should know. Ann Intern Med 2020;172:754–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hung IF-N, Lung K-C, Tso EY-K, et al. Triple combination of interferon beta-1b, lopinavir–ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet 2020;395:1695–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bronte V, Ugel S, Tinazzi E, et al. Baricitinib restrains the immune dysregulation in patients with severe COVID-19. J Clin Invest 2020;130:6409–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lei Z-N, Wu Z-X, Dong S, et al. Chloroquine and hydroxychloroquine in the treatment of malaria and repurposing in treating COVID-19. Pharmacol Ther 2020;216:107672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang S, Amahong K, Sun X, et al. The miRNA: a small but powerful RNA for COVID-19. Brief Bioinform 2021;22:1137–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Van Den Berg HA. Occam's razor: from Ockham's via moderna to modern data science. Sci Prog 2018;101:261–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fintelman-Rodrigues N, Sacramento CQ, Ribeiro Lima C, et al. Atazanavir, alone or in combination with ritonavir, inhibits SARS-CoV-2 replication and proinflammatory cytokine production. Antimicrob Agents Chemother 2020;64:e00825–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tripathi PK, Upadhyay S, Singh M, et al. Screening and evaluation of approved drugs as inhibitors of main protease of SARS-CoV-2. Int J Biol Macromol 2020;164:2622–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu X, Liu C, Liu G, et al. COVID-19: Progress in diagnostics, therapy and vaccination. Theranostics 2020;10:7821–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Das G, Das T, Chowdhury N, et al. Repurposed drugs and nutraceuticals targeting envelope protein: a possible therapeutic strategy against COVID-19. Genomics 2020;113:1129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Iftikhar H, Ali HN, Farooq S, et al. Identification of potential inhibitors of three key enzymes of SARS-CoV2 using computational approach. Comput Biol Med 2020;122:103848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bibi N, Gul S, Ali J, et al. Viroinformatics approach to explore the inhibitory mechanism of existing drugs repurposed to fight against COVID-19. Eur J Pharmacol 2020;885:173496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Indu P, Rameshkumar MR, Arunagirinathan N, et al. Raltegravir, Indinavir, Tipranavir, Dolutegravir, and Etravirine against main protease and RNA-dependent RNA polymerase of SARS-CoV-2: a molecular docking and drug repurposing approach. J Infect Public Health 2020;13:1856–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sencanski M, Perovic V, Pajovic SB, et al. Drug repurposing for candidate SARS-CoV-2 main protease inhibitors by a novel in silico method. Molecules 2020;25:3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shekhar N, Sarma P, Prajapat M, et al. In silico structure-based repositioning of approved drugs for spike glycoprotein S2 domain fusion peptide of SARS-CoV-2: rationale from molecular dynamics and binding free energy calculations. mSystems 2020;5:e00382–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Han Y, Duan X, Yang L, et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 2021;589:270–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen X, Wu Y, Chen C, et al. Identifying potential anti-COVID-19 pharmacological components of traditional Chinese medicine Lianhuaqingwen capsule based on human exposure and ACE2 biochromatography screening. Acta Pharm Sin B 2020;11:222–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao J, Tian S, Lu D, et al. Systems pharmacological study illustrates the immune regulation, anti-infection, anti-inflammation, and multi-organ protection mechanism of Qing-Fei-Pai-Du decoction in the treatment of COVID-19. Phytomedicine 2020;85:153315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tao Q, Du J, Li X, et al. Network pharmacology and molecular docking analysis on molecular targets and mechanisms of Huashi Baidu formula in the treatment of COVID-19. Drug Dev Ind Pharm 2020;46:1345–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Menche J, Sharma A, Kitsak M, et al. Uncovering disease-disease relationships through the incomplete interactome. Science 2015;347:1257601–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dai W, Tang T, Dai Z, et al. Probing the mechanism of hepatotoxicity of hexabromocyclododecanes through toxicological network analysis. Environ Sci Technol 2020;54:15235–45. [DOI] [PubMed] [Google Scholar]
- 40. Wang Y, Li F, Zhang Y, et al. Databases for the targeted COVID-19 therapeutics. Br J Pharmacol 2020;177:4999–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Luo E, Zhang D, Luo H, et al. Treatment efficacy analysis of traditional Chinese medicine for novel coronavirus pneumonia (COVID-19): an empirical study from Wuhan, Hubei Province. China Chin Med 2020;15:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. ww PDBc . Protein Data Bank: the single global archive for 3D macromolecular structure data. Nucleic Acids Res 2019;47:D520–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang WF, Stephen P, Theriault JF, et al. Novel coronavirus polymerase and nucleotidyl-transferase structures: potential to target new outbreaks. J Phys Chem Lett 2020;11:4430–5. [DOI] [PubMed] [Google Scholar]
- 44. Xin S, Cheng X, Zhu B, et al. Clinical retrospective study on the efficacy of Qingfei Paidu decoction combined with Western medicine for COVID-19 treatment. Biomed Pharmacother 2020;129:110500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Consortium WHOST, Pan H, Peto R, et al. Repurposed antiviral drugs for Covid-19 - Interim WHO solidarity trial results. N Engl J Med 2021;384:497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wang Y, Zhang D, Du G, et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet 2020;395:1569–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Khan A, Heng W, Wang Y, et al. In silico and in vitro evaluation of kaempferol as a potential inhibitor of the SARS-CoV-2 main protease (3CLpro). Phytother Res 2021;35:2841–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hoang TN, Pino M, Boddapati AK, et al. Baricitinib treatment resolves lower-airway macrophage inflammation and neutrophil recruitment in SARS-CoV-2-infected rhesus macaques. Cell 2021;184:460–75 e421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Resop RS, Fromentin R, Newman D, et al. Fingolimod inhibits multiple stages of the HIV-1 life cycle. PLoS Pathog 2020;16:e1008679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dubois M, Bailly F, Mbemba G, et al. Reaction of rosmarinic acid with nitrite ions in acidic conditions: discovery of nitro- and dinitrorosmarinic acids as new anti-HIV-1 agents. J Med Chem 2008;51:2575–9. [DOI] [PubMed] [Google Scholar]
- 51. Raj VS, Mou H, Smits SL, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013;495:251–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 2020;5:562–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sheahan TP, Sims AC, Leist SR, et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat Commun 2020;11:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang X, Xin B, Tan W, et al. DeepR2cov: deep representation learning on heterogeneous drug networks to discover anti-inflammatory agents for COVID-19. Brief Bioinform 2021;bbab226. doi: 10.1093/bib/bbab226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kauppi K, Rosenthal SB, Lo M-T, et al. Revisiting antipsychotic drug actions through gene networks associated with Schizophrenia. Am J Psychiatry 2018;175:674–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Del Valle DM, Kim-Schulze S, Huang HH, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med 2020;26:1636–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xu X, Han M, Li T, et al. Effective treatment of severe COVID-19 patients with tocilizumab. Proc Natl Acad Sci U S A 2020;117:10970–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Zhang F, Mears JR, Shakib L, et al. IFN-γ and TNF-α drive a CXCL10+ CCL2+ macrophage phenotype expanded in severe COVID-19 lungs and inflammatory diseases with tissue inflammation. Genome Med 2021;13:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sadegh S, Matschinske J, Blumenthal DB, et al. Exploring the SARS-CoV-2 virus-host-drug interactome for drug repurposing. Nat Commun 2020;11:3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Roche JA, Roche R. A hypothesized role for dysregulated bradykinin signaling in COVID-19 respiratory complications. FASEB J 2020;34:7265–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fanning GC, Zoulim F, Hou J, et al. Therapeutic strategies for hepatitis B virus infection: towards a cure. Nat Rev Drug Discov 2019;18:827–44. [DOI] [PubMed] [Google Scholar]
- 62. Chung YC, Hsieh FC, Lin YJ, et al. Magnesium lithospermate B and rosmarinic acid, two compounds present in Salvia miltiorrhiza, have potent antiviral activity against enterovirus 71 infections. Eur J Pharmacol 2015;755:127–33. [DOI] [PubMed] [Google Scholar]
- 63. Hsieh CF, Jheng JR, Lin GH, et al. Rosmarinic acid exhibits broad anti-enterovirus A71 activity by inhibiting the interaction between the five-fold axis of capsid VP1 and cognate sulfated receptors. Emerg Microbes Infect 2020;9:1194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lee J, Jung E, Kim Y, et al. Rosmarinic acid as a downstream inhibitor of IKK-beta in TNF-alpha-induced upregulation of CCL11 and CCR3. Br J Pharmacol 2006;148:366–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Catanzaro M, Fagiani F, Racchi M, et al. Immune response in COVID-19: addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Transduct Target Ther 2020;5:84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wei Y, Chen J, Hu Y, et al. Rosmarinic acid mitigates lipopolysaccharide-induced neuroinflammatory responses through the inhibition of TLR4 and CD14 expression and NF-kappaB and NLRP3 inflammasome activation. Inflammation 2018;41:732–40. [DOI] [PubMed] [Google Scholar]
- 67. Arokiyaraj S, Stalin A, Kannan BS, et al. Geranii herba as a potential inhibitor of SARS-CoV-2 Main 3CL(pro), spike RBD, and regulation of unfolded protein response: an in silico approach. Antibiotics (Basel) 2020;9:863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ryu YB, Jeong HJ, Kim JH, et al. Biflavonoids from Torreya nucifera displaying SARS-CoV 3CL(pro) inhibition. Bioorg Med Chem 2010;18:7940–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lee HN, Shin SA, Choo GS, et al. Antiinflammatory effect of quercetin and galangin in LPSstimulated RAW264.7 macrophages and DNCBinduced atopic dermatitis animal models. Int J Mol Med 2018;41:888–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Meacci E, Garcia-Gil M, Pierucci F. SARS-CoV-2 infection: a role for S1P/S1P receptor signaling in the nervous system? Int J Mol Med 2018;41:888–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yousefi H, Mashouri L, Okpechi SC, et al. Repurposing existing drugs for the treatment of COVID-19/SARS-CoV-2 infection: a review describing drug mechanisms of action. Biochem Pharmacol 2021;183:114296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Blanc C, Rosen H, Lane T. FTY720 (fingolimod) modulates the severity of viral-induced encephalomyelitis and demyelination. J Neuroinflammation 2014;11:138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. de Bruin S, Bos LD, van Roon MA, et al. Clinical features and prognostic factors in Covid-19: a prospective cohort study. EBioMedicine 2021;67:103378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gebauer N, Kunstner A, Ketzer J, et al. Genomic insights into the pathogenesis of Epstein-Barr virus-associated diffuse large B-cell lymphoma by whole-genome and targeted amplicon sequencing. Blood Cancer J 2021;11:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Beutel O, Maraspini R, Pombo-Garcia K, et al. Phase separation of zonula occludens proteins drives formation of tight junctions. Cell 2019;179:923–36 e911. [DOI] [PubMed] [Google Scholar]
- 76. Camara AB, Brandao IA. The main receptors involved with COVID-19: a systematic review and meta-analysis. Curr Med Chem 2021. doi: 10.2174/0929867328666210405113253. [DOI] [PubMed] [Google Scholar]
- 77. Horndler L, Delgado P, Abia D, et al. Flow cytometry multiplexed method for the detection of neutralizing human antibodies to the native SARS-CoV-2 spike protein. EMBO Mol Med 2021;13:e13549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Caillet-Saguy C, Durbesson F, Rezelj VV, et al. Host PDZ-containing proteins targeted by SARS-CoV-2. FEBS J 2021. doi: 10.1111/febs.15881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Matsuyama T, Kubli SP, Yoshinaga SK, et al. An aberrant STAT pathway is central to COVID-19. Cell Death Differ 2020;27:3209–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zheng HY, Xu M, Yang CX, et al. Longitudinal transcriptome analyses show robust T cell immunity during recovery from COVID-19. Signal Transduct Target Ther 2020;5:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Luo W, Li YX, Jiang LJ, et al. Targeting JAK-STAT signaling to control cytokine release syndrome in COVID-19. Trends Pharmacol Sci 2020;41:531–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Irving AT, Zhang Q, Kong PS, et al. Interferon regulatory factors IRF1 and IRF7 directly regulate gene expression in bats in response to viral infection. Cell Rep 2020;33:108345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Calabrese LH, Zein NN, Vassilopoulos D. Hepatitis B virus (HBV) reactivation with immunosuppressive therapy in rheumatic diseases: assessment and preventive strategies. Ann Rheum Dis 2006;65:983–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Hadjadj J, Yatim N, Barnabei L, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020;369:718–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Izumi T, Io K, Matsui M, et al. HIV-1 viral infectivity factor interacts with TP53 to induce G2 cell cycle arrest and positively regulate viral replication. Proc Natl Acad Sci U S A 2010;107:20798–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Sacconi A, Donzelli S, Pulito C, et al. TMPRSS2, a SARS-CoV-2 internalization protease is downregulated in head and neck cancer patients. J Exp Clin Cancer Res 2020;39:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhang Q, Ji X. XM. H, et al. Effect of functional nuclear factor-kappaB genetic polymorphisms on hepatitis B virus persistence and their interactions with viral mutations on the risk of hepatocellular carcinoma. Ann Oncol 2014;25:2413–9. [DOI] [PubMed] [Google Scholar]
- 88. O'Donnell SM, Hansberger MW, Connolly JL, et al. Organ-specific roles for transcription factor NF-kappaB in reovirus-induced apoptosis and disease. J Clin Invest 2005;115:2341–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Watanabe T. Adult T-cell leukemia: molecular basis for clonal expansion and transformation of HTLV-1-infected T cells. Blood 2017;129:1071–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Boudewijns R, Thibaut HJ, Kaptein SJF, et al. STAT2 signaling restricts viral dissemination but drives severe pneumonia in SARS-CoV-2 infected hamsters. Nat Commun 2020;11:5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


