Abstract
Artificial intelligence (AI) has given the electrocardiogram (ECG) and clinicians reading them super-human diagnostic abilities. Trained without hard-coded rules by finding often subclinical patterns in huge datasets, AI transforms the ECG, a ubiquitous, non-invasive cardiac test that is integrated into practice workflows, into a screening tool and predictor of cardiac and non-cardiac diseases, often in asymptomatic individuals. This review describes the mathematical background behind supervised AI algorithms, and discusses selected AI ECG cardiac screening algorithms including those for the detection of left ventricular dysfunction, episodic atrial fibrillation from a tracing recorded during normal sinus rhythm, and other structural and valvular diseases. The ability to learn from big data sets, without the need to understand the biological mechanism, has created opportunities for detecting non-cardiac diseases as COVID-19 and introduced challenges with regards to data privacy. Like all medical tests, the AI ECG must be carefully vetted and validated in real-world clinical environments. Finally, with mobile form factors that allow acquisition of medical-grade ECGs from smartphones and wearables, the use of AI may enable massive scalability to democratize healthcare.
Keywords: Artificial intelligence, Machine learning, Electrocardiograms, Digital health
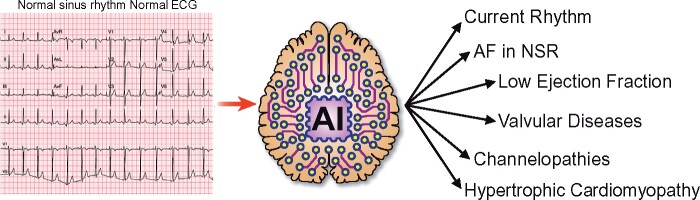
Graphical Abstract
Graphical Abstract.
The application of artificial intelligence to the standard electrocardiogram enables it to diagnose conditions not previously identifiable by an electrocardiogram, or to do so with a greater performance than previously possible. This includes identification of the current rhythm, identification of episodic atrial fibrillation from an ECG acquired during sinus rhythm, the presence of ventricular dysfunction (low ejection fraction), the presence of valvular heart disease, channelopathies (even when electrocardiographically ‘concealed’), and the presence of hypertrophic cardiomyopathy.
Despite the fact that the electrocardiogram (ECG) has been in use for over 100 years and is a central tool in clinical medicine, we are only now beginning to unleash its full potential with the application of artificial intelligence (AI). The ECG is the cumulative recording at a distance (the body surface) of the action potentials of millions of individual cardiomyocytes (Figure 1). Traditionally, clinicians have been trained to identify specific features such as ST-segment elevation for acute myocardial infarction, T-wave changes to suggest potassium abnormalities, and other gross deviations to identify specific clinical entities. By their very nature, the magnitude of the changes must be substantial in order to significantly alter a named ECG feature to result in a clinical diagnosis. With the application of convolutional neural networks to an otherwise standard ECG, multiple non-linear potentially interrelated variations can be recognized in an ECG. Thus, neural networks have been used to: identify a person’s sex with startling precision [area under the curve (AUC) 0.97]; recognize the presence of left ventricular dysfunction; uncover the presence of silent arrhythmia not present at the time of the recording; as well as identify the presence of non-cardiac conditions such as cirrhosis.1–3 Many biological phenomena, each of which can leave its imprint on cardiomyocytes electrical function in a unique manner, lead to multiple, subtle, non-linear, subclinical ECG changes. Although ECGs are filtered between 0.05 and 100 Hertz to augment capture of cardiac signals, they likely also are influenced directly and indirectly by nerve activity, myopotentials, as well as anatomic considerations such as cardiac rotation, size, and surrounding body habitus. With large datasets to train a network as to the multiple and varied influences of each of these conditions, powerful diagnostic tools can be developed. In this review, we will offer an overview of machine learning (ML), show specific examples of conditions not previously diagnosed with an ECG that are now recognized, and provide an update of the application and practice and future directions of the AI processed ECG (AI ECG).
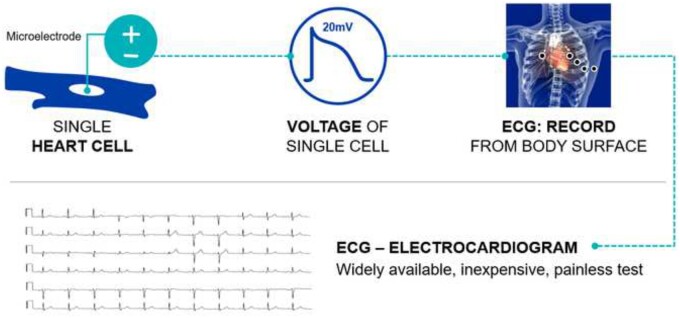
Figure 1.
Microelectrodes in a single myocyte (top left) record an action potential (depicted middle panel). Ionic currents and their propagation are sensitive to cardiac and non-cardiac conditions and structural changes. When the aggregated action potentials are recorded at the body surface (top right), the insuring tracing is the electrocardiogram (bottom). ECG, electrocardiogram.
Broadly speaking, AI can be applied in two ways to the ECG.4 In one, currently performed human skills, such as determining arrhythmias or acute infarction, are performed in an automated manner making those skills massively scalable. The second utilization is to extract information from an ECG beyond which a human can typically perform. In this review, we will focus on the latter.
Machine learning introduction
The term machine learning was coined more than 70 years ago to describe the ability of man-made machines to learn how to perform complex tasks, and to improve task performance based on additional experience. Practically, it refers to replacing algorithms that define the relationship between inputs and outputs using man-made rules with statistical tools that identify (learn) the most probable relationship between input and output based on repeated exposure to exemplar data elements. As with other revolutionary ideas, when first introduced, it was ahead of its time, and the data and computing power required to enable this revolution did not exist. Today, with exponentially growing digital datasets and a significant increase in the computational power available to train networks, ML, sometimes referred to as AI, performs highly complex tasks. In supervised ML, the task is defined as an optimization problem, which seeks to find the optimal solution using labelled data. By defining a ‘loss function’ or a matrix of how well the machine performs a task and minimizing the error and maximizing the success matrix, a complex task is transformed into a mathematical problem.
Most ML models are parametric and define a function between an input space and an output space. In neural networks, designed to mimic human visual cortex, each ‘neuron’ is a simple mathematical equation with parameters that are adjusting during network training. When the neurons are connected in many layers, it is referred to as a deep network. The function applies parameters to the input in linear and non-linear ways to derive an estimated output (Figure 2). During the learning phase, both the inputs and the outputs are shown to the function, and the parameters (sometimes called ‘weights’) are adjusted in an iterative manner, to minimize the difference between the estimated output and the known outputs. This learning or training phase often requires large data sets and robust computing power. The most common way to adjust these weights is by applying a gradient-based optimization method, which adjusts each parameter weight based on its effect on the error, with the network weights iteratively adjusted until an error minimum is found.
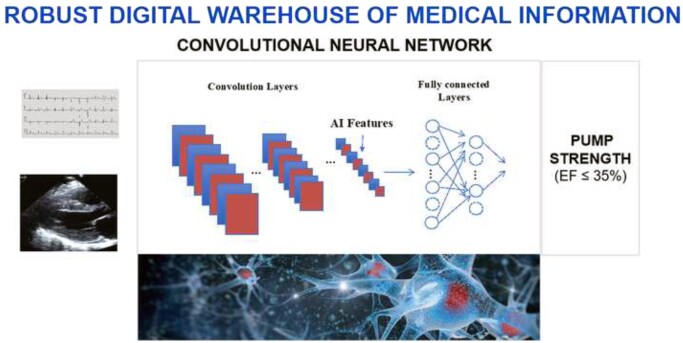
Figure 2.
A convolutional neural network is trained by feeding in labelled data (in this case voltage time waveforms), and through repetition it identifies the patterns in the data that are associated with the data labels (in this example, heart pump strength, or ejection fraction). The network has two components, convolution layers that extract image components to create the artificial intelligence features, and the fully connected layers that comprise the model, that leads to the network output. While large data sets and robust computing are required to train networks, once trained, the computation requirements are substantially reduced, permitting smartphone application. AI, artificial intelligence; EF, ejection fraction.
Once the learning phase is complete, the parameters are set, and the function becomes a simple deterministic algorithm that can be applied to any unseen input. The computing power required to deploy a trained network is modest and can often be performed from a smartphone. A neural network is composed of different nodes (representing neurons) connected to each other in a directed way, where each node calculates a function of its input based on an embedded changeable parameter (weight) followed by a non-linear function, and the inputs flow from one set of neurons (called a layer) to the next. Each layer calculates an intermediate function of the inputs (features), and the final layer is responsible to calculate the final output. In some architectures, the neurons are connected to each other to enable determination of temporal patterns (recurrent neural networks), used when the input is a time series; others, such as convolutional neural networks, originally developed for computer vision tasks, have also been used for natural language processing, video analysis, and time series.
As these models are mathematical functions, the inputs are comprised of real numbers created by sampling physical signals. For example, an ECG is converted to a time series sampled at a consistent sampling frequency in which each sample represents the signal amplitude for a given time point. For a 10-s 12 lead ECG sampled at 500 Hz, the digital representation of the input is a matrix with 5000 samples (500 Hz × 10 s) per lead. With 12 leads, the final input will be a set of 60 000 numbers (5000 × 12).
For binary models in which the output is ‘yes/no’, such as the presence of silent atrial fibrillation (AF) determined from an ECG acquired in sinus rhythm, the input numbers (i.e. the digital ECG) are used by the network to calculate the probability of silent AF. The output will be a single number, ranging from 0 (no silent AF) to 1 (silent AF present). To dichotomize the output, a threshold value determines whether the model output is positive or negative. By adjusting the threshold, the test can be made more sensitive, with more samples considered positive and fewer missed events, but at the cost of a higher number of false positives; or alternatively, more specific, with fewer false positives but more missed events. Since the training of supervised ML models requires only labelled data [e.g. ECGs and associated ejection fraction (EF) values], and no explicit rules, machines learn to solve tasks that humans don’t know how to solve, giving the machines (and humans who use them) what appear to be super-human abilities.
Artificial intelligence model explainability, robustness, and uncertainty
Explainability
Neural networks are often described as ‘black boxes’ since the signal features a network selects to generate an output and the network’s intermediate layers typically are not comprehensible to humans. Explainability refers to uncovering the underlying rules that a model finds during training, Explainability may help humans understand what gives a model its super-human abilities, enhance user trust of AI tools, uncover novel pathophysiologic mechanisms by identifying relationships between input signals and outputs, and reveal network vulnerabilities. One method of understanding a model is by looking at specific examples and highlighting the parts of the input that contribute most to the final output. In the Grad-CAM method,5 for example, network gradients are used to produce a coarse localization of drivers of the output. While the method was developed for images, it works with ECGs as well. In published valvular disease detection algorithms, for example,6,7 the authors use saliency-based methods to highlight the portions of the ECG that contributed to the model’s output in selected samples. While this is a first step towards developing explainable AI, current methods explain the results for a particular example and do not reveal the general rules used by the models. This remains an open research question.
Uncertainty
Unless a data quality check is performed before using a model, the model will generate a result for any given input, including inputs outside of the distribution of data used to train the model. If a model is fed data outside the range of its training domain, whether due to deficits in the training set or shifts in the data over time, the model may generate results that do not accurately classify the input. Other factors may also lead to uncertainty. Methods to measure uncertainty have been described.8–10 To minimize this error, models are evaluated for their consistent performance with inputs that are anticipated within the input distribution that should not affect the outputs.
Robustness
This refers to the ability of a model to accurately classify inputs that are synthetically modified in and adversarial way in order to favour misclassification. For example, in Han et al.,11 the authors show that by using almost-invisible perturbations to an ECG, a model with very high accuracy for the detection of AF can be fooled into thinking an normal sinus rhythm (NSR) recording is actually AF with high certainty, and vice versa, even though to the human reviewer the ECGs looks unchanged.
Metrics for assessing neural networks
Most neural networks generate an output that is a continuous probability. To create binary outputs a threshold value is selected, with probabilities above the threshold considered positive. The selection of the threshold impacts model sensitivity and specificity, with an increase in one at the cost of the other. The receiver operating characteristic (ROC) curve represents all the sensitivity and specificity pairs for a model, and the AUC (AUC of the ROC), measures how well the model separates two classes, and is often used to represent model function. For example, a model with random outputs will have complete overlap of scores for positive and negative input samples, yielding an AUC of 0.5, and a perfect model that gives all positive inputs scores above the threshold, and all negative samples scores below that threshold (hence perfectly separating the classes) will have an AUC of 1 (Figure 3). Once a threshold is selected, a confusion matrix can be calculated, indicating the true and false positive and negative values, allowing real-world calculation of the sensitivity (also called recall), specificity, accuracy, weighted accuracy (important when the classes are imbalanced, due to low prevalence of a disease, for example), positive predictive value (precision), negative predictive value (NPV), and more specialized scores as the F1 accuracy.
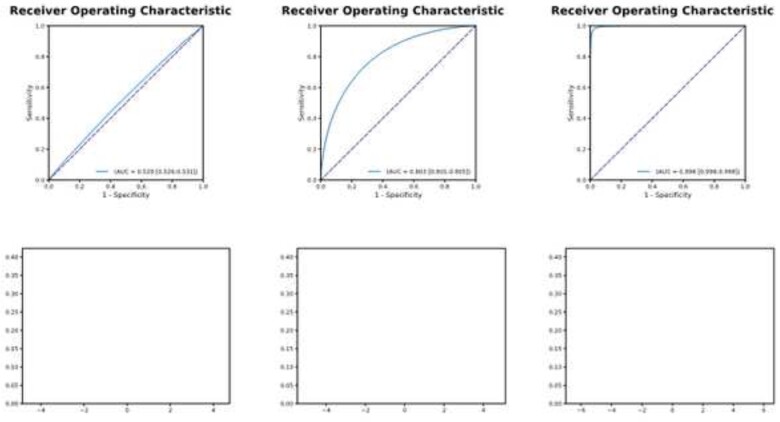
Figure 3.
The receiver operating characteristic curve and model performance. Left panel: A test with an area under the curve of 0.529 (top) results in very poor separation of the classes (bottom left). As the area under the curve increases (0.803 middle panel, top and 0.998 right panel, top) the separation of the classes and utility of the test improves (bottom panels). This results in improved sensitivity and specificity. See text for additional details.
In cases with imbalanced datasets, some measurements will appear optimistic. For example, in detecting a disease with a 1% prevalence, a model that always generates a score of 0 will be right 99% of the times, yielding an accuracy of 99%, but a sensitivity of 0%. In less extreme cases, an imbalanced dataset might still appear optimistic with some measurements.12 With the AUC of the ROC, for example, one of the axes of the curve (the false positive rate) is calculated as false positive/(false positive + true positive) and is less sensitive to changes in false positives when the negative class grows relative to the positive samples. While with unbalanced data sets, it may be an optimistic measurement, and a different metric might be more accurate (the area under the recall-precision curve, that takes prevalence into account as part of the precision, for example), the AUC of the ROC is often used to compare models, particularly with regards to medical tests. In this review article, we present the statistics reported in the original work (predominantly AUC values).
Using machine learning and deep learning to accomplish tasks a human cannot
Left ventricular systolic dysfunction (LVSD), AF, and hypertrophic cardiomyopathy (HCM) share three characteristics: they are frequently under-diagnosed; they are associated with significant morbidity; and once detected, effective, evidence-based therapies are available.2,3,13,14 Routine screening strategies are not currently recommended due to the absence of effective screening tools.15–17 The ECG is a rapid, cost-effective, point-of-care test that requires no blood and no reagents, that is massively scalable with smartphone technology to medical and non-medical environments. The AI ECG provides information beyond what is visible to the eyes of an experienced clinician with manual ECG interpretation today. Specific use cases are detailed below. Many of AI models have been developed in parallel by different research groups. In Table 1, we summarize these and include key model performance and characteristic information.
Table 1.
Summary of artificial intelligence electrocardiogram algorithms and their performance and characteristics
| Model | Author/Group | Test geography hospital vs. development | Prospective or retrospective | Number of patients tested | Disease prevalence (%) | Description of controls | Hardware specification (12 lead vs. other; specify manufactures/performance of 12 lead) | Bias analysis: population reporting (age, sex, race, other) | AUC | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| LVSD/HF |
Attia et al.2 Mayo |
All Mayo Clinic Sites | Retrospective | 52 870 | 7.8 | Low EF confirmed by TTE | 12 Lead ECG (GE-Marquette) | No formal analysis | 0.93 | 86.3 | 85.7 |
| LVSD/HF |
Attia et al.18 Mayo |
All Mayo Clinic Sites | Prospective | 3874 | 7.0 | Low EF confirmed by TTE or HF prediction by NT-proBNP | 12 Lead ECG (GE-Marquette) | No formal analysis | 0.918 | 82.5 | 86.8 |
| LVSD/HF |
Adedinsewo et al.19 Mayo |
All Mayo Clinic Sites | Retrospective | 1606 | 10.2 | Low EF confirmed by TTE | 12 Lead ECG (GE-Marquette) | Age, Sex | 0.89 | 73.8 | 87.3 |
| LVSD/HF |
Noseworthy et al.20 Mayo |
All Mayo Clinic Sites | Retrospective | 52 870 | 7.8 | Low EF confirmed by TTE | 12 Lead ECG (GE-Marquette) | Race | >0.93 in all groups tested | — | — |
| LVSD/HF |
Attia et al.21 Mayo |
Mayo Rochester | Prospective | 100 | 7 | Low EF confirmed by TTE | AI-enhanced ECG-enabled stethoscope (Eko); single lead | No formal analysis | 0.906 | — | — |
| LVSD/HF |
Attia et al.22 Mayo/Multi-Institution |
Know Your Heart Sites (Russia) |
Retrospective | 4277 | 0.6 | Low EF confirmed by TTE | 12 Lead ECG (Cardiax; IMED Ltd, Hungary) | Age, sex | 0.82 | 26.9 | 97.4 |
| LVSD/HF |
Cho et al.23 Sejong/Korea |
Mediplex/Sejong (Korea) | Retrospective |
IV-2908 EV-4176 |
6.8 | Low EF confirmed by echo | 12 Lead ECG (Page Writer Cardiograph; Philips, Netherlands) | Age, sex, obesity |
IV-0.913 EV-0.961 |
IV-90.5 EV-91.5 |
IV-75.6 EV-91.1 |
| LVSD/HF |
Cho et al.23 Sejong/Korea |
Mediplex/Sejong (Korea) | Retrospective |
IV-2908 EV-4176 |
6.8 | Low EF confirmed by echo | Single lead (LI) from 12 lead ECG (Page Writer Cardiograph; Philips, Netherlands) | Performance of all single leads |
IV-0.874 EV-0.929 |
IV-93.2 EV-92.1 |
IV-63.2 EV-82.1 |
| LVSD/HF |
Kwon et al.24 Sejong/Korea |
Mediplex/Sejong (Korea) | Retrospective |
IV-3378 EV-5901 |
IV-9.7 EV-4.2 |
Low EF confirmed by echo | 12 Lead ECG (Page Writer Cardiograph; Philips, Netherlands) | No formal analysis |
IV-0.843 EV-0.889 |
IV-n/a EV-90 |
IV-n/a EV-60.4 |
| HCM |
Ko et al.13 Mayo |
All Mayo Clinic Sites | Retrospective | 13 400 | 4.6 | Sex/age matched | 12 Lead ECG (GE-Marquette) | Age, sex, ECG finding | 0.96 | 87 | 90 |
| HCM |
Rahman et al.25 Hopkins Queens (CA) |
Hopkins Baltimore |
Retrospective | 762 | 29.0 | Patients with ICD and CM diagnosis | 12 Lead ECG (unspecified) | No formal analysis |
RF-0.94 SVM-0.94 |
RF-87 SVM-0.91 |
RF-92 SVF-0.91 |
| Hyperkalaemia |
Galloway et al.26 Mayo |
All Mayo Clinic Sites | Retrospective |
MN-50 099 AZ-5855 FL-6011 |
MN-2.6 AZ-4.6 FL-4.8 |
Confirmation by serum potassium | 12 Lead ECG (GE-Marquette); 2 Lead evaluation LI/LII | No formal analysis |
MN-0.883 AZ-0.853 FL-0.860 |
MN-90.2 AZ-88.9 FL-91.3 |
MN-54.7 AZ-55.0 FL-54.7 |
| Sex and age >40 years |
Attia et al.1 Mayo |
All Mayo Clinic Sites | Retrospective | 275 056 | n/a | Confirmed age/sex in medical record | 12 Lead ECG (GE-Marquette) | Co-morbidity impact on ECG age |
Sex-0.968 Age-0.94 |
Sex-n/a Age-87.8 |
Sex-n/a Age-86.8 |
| Afib |
Attia et al.3 Mayo |
All Mayo Clinic Sites | Retrospective | 36 280 | 8.4 | Patients without Afib on prior EKG | 12 Lead ECG (GE-Marquette) | Analysis with ‘window of interest’ | 0.87 | 79.0 | 79.5 |
| Afib |
Tison et al.27 UCSF |
Remote study; UCSF | Prospective | 9750 | 3.4 | 12 lead EKG diagnosis of Afib | Apple Watch photoplethysmography (Apple Inc.) | No formal analysis | 0.97 | 98.0 | 90.2 |
| Afib |
Hill et al.28 UK-Multi-institution |
UK | Retrospective | 2 994 837 | 3.2 | CHARGE-AF score | Time-varying neural network; based on clinic data and risk scores | No formal analysis | 0.827 | 75.0 | 74.9 |
| Afib |
Jo et al.29 Sejong/Korea |
Multiple sites (Korea) | Retrospective |
IV-6287 EV-38 018 |
IV-13 EV-6.0 |
Patients without afib | 12 lead, 6 lead, and single lead ECG (unspecified) | No formal analysis | IV/EV for 12, 6, single lead all >0.95 | All >98% | All >99% |
| Afib |
Poh et al.30 Boston |
Hong Kong | Retrospective | 1013 | 2.8 | Patients without afib | Photoplethysmographic pulse waveform | No formal analysis | 0.997 | 97.6 | 96.5 |
| Afib | Raghunath et al.31 | Geisinger Clinic, PA, USA | Retrospective | 1.6M | Patients without afib | 12 lead ECG | Age, sex, race analysed | 0.85 | 69 | 81 | |
| Long QT (>500 ms) |
Giudicessi et al.32 Mayo |
Mayo Clinic Rochester | Both; prospective data reported | 686 | 3.6 | QT expert/lab over-read of 12 lead ECGs | 6 lead smartphone-enabled ECG (AliveCor Kardia Mobile 6L) | No formal analysis | 0.97 | 80.0 | 94.4 |
| Long QT |
Bos et al.33 Mayo |
Mayo Clinic Rochester | Retrospective | 2059 | 47 | Patients without LQTS | 12 Lead ECG (GE-Marquette) | LQTS genotype subgroup analysis | 0.900 | 83.7 | 80.6 |
| Multiple Pathologies |
Tison et al.34 UCSF |
UCSF | Retrospective | 36 816 (ECGs) |
HCM-27.4 PAH-29.8 Amyloid-28.3 MVP-21.0 |
Individual pathologies determined by standard care (i.e. echo, biopsy) | 12 Lead ECG (GE-Marquette) | No formal analysis |
HCM-0.91 PAH-0.94 Amyloid-0.86 MVP-0.77 |
— | — |
| Mod-Sev AS |
Cohen-Shelly et al.6 Mayo |
All Mayo Clinic Sites | Retrospective | 102 926 | 3.7 | Mod-Sev AS confirmed by TTE | 12 Lead ECG (GE-Marquette) | Age, sex | 0.85 | 78 | 74 |
| Significant AS |
Kwon et al.7 Sejong/Korea |
Mediplex/Sejong (Korea) | Retrospective |
IV-6453 EV-10 865 |
IV-3.8 EV-1.7 |
Significant AS confirmed by echo | 12 Lead ECG (Unspecified) | No formal analysis |
IV-0.884 EV-0.861 |
IV-80.0 EV-80.0 |
IV-81.4 EV-78.3 |
| Significant AS |
Kwon et al.7 Sejong/Korea |
Mediplex/Sejong (Korea) | Retrospective |
IV-6453 EV-10 865 |
IV-3.8 EV-1.7 |
Significant AS confirmed by echo | Single lead (L2) from 12 lead ECG (unspecified) | No formal analysis |
IV-0.845 EV-0.821 |
— | — |
| Mod-Sev MR |
Kwon et al.35 Sejong/Korea |
Mediplex/Sejong (Korea) | Retrospective |
IV-3174 EV-10 865 |
IV-n/a EV-3.9 |
Mod-Sev MR confirmed by echo | 12 Lead ECG (Unspecified) | No formal analysis |
IV 0.816 EV 0.877 |
IV 0.900 EV 0.901 |
IV 0.533 EV 0.699 |
| Mod-Sev MR |
Kwon et al.35 Sejong/Korea |
Mediplex/Sejong (Korea) | Retrospective |
IV-3174 EV-10 865 |
IV-n/a EV 3.9 |
Mod-Sev MR confirmed by echo | Single lead (aVR) from 12 lead ECG (unspecified) | No formal analysis |
IV 0.758 EV 0.850 |
IV 0.900 EV 0.901 |
IV 0.408 EV 0.560 |
Afib, atrial fibrillation; AI, artificial intelligence; AUC, area under the curve; AZ, Arizona; CA, Canada; ECG, electrocardiogram; echo, echocardiography; EV, external validation; FL, Florida; HCM, hypertrophic cardiomyopathy; HF, heart failure; ICD, implantable cardiac defibrillator; IV, internal validation; LVSD, left ventricular systolic dysfunction; LQTS, long QT syndrome; MN, Minnesota; mod-sev AS, moderate to severe aortic stenosis; mod-sev MR, moderate to severe mitral regurgitation; MVP, mitral valve prolapse; NT-proBNP, N-terminus of brain natriuretic peptide; PAH, pulmonary arterial hypertension; RF, random forest classifier; SVM, support vector machine classifier; TTE, transthoracic echocardiogram.
Artificial intelligence to screen for left ventricular systolic dysfunction
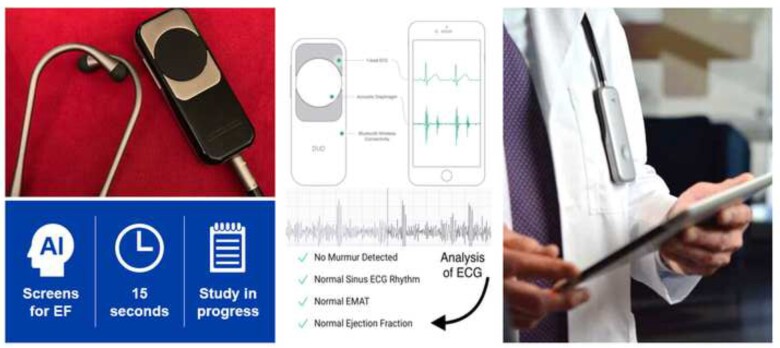
Several research groups have used ECG-based deep learning networks to detect LVSD.2,4,19,23,24,36 We engineered a neural network using 50 000 ECG-echocardiogram pairs for training that was able to discriminate low EF (≤35%) from EF >35% with a high accuracy (AUC 0.93) in a testing population not previously seen by the network (Figure 2).2 In the emergency room setting, LVSD is identified with similar accuracy (85.9%; AUC 0.89) in patients with symptoms of an acute HF exacerbation (i.e. dyspnoea on exertion, shortness of breath).19 At the time of this writing, the algorithm has received US Food and Drug Administration (FDA) breakthrough technology designation and, during the pandemic, emergency use authorization. The algorithm and algorithms analogous to ours have been adapted for use with a single lead, and our own algorithm has been embedded in an electrode-equipped digital stethoscope that identifies ventricular dysfunction in 15 s (Figure 4).21,23,37
Figure 4.
Embedding of an artificial intelligence electrocardiogram into a stethoscope. Electrodes on the stethoscope acquire an electrocardiogram during normal auscultation, permitting the identification of ventricular dysfunction with 15 s of skin contact time. This workflow provides immediate notification to the healthcare provider via a smartphone connection. ECG, electrocardiogram; EF, ejection fraction.
These algorithms have been tested in diverse populations and found to function well across race and ethnicity.15,16,20
Artificial intelligence algorithms may experience dataset shift errors when applied to previously untested environments. These errors occur when the new populations differ from those used to train the network in a substantive manner so that the network has not been exposed to key data characteristics required for accurate output. Artificial intelligence ECG networks for the detection of LVSD, developed by our team and Cho et al., have demonstrated performance stability and robustness with regards to sex, age, and body mass index in internal and external validation sets,18,22,23 supporting wide applicability. Prospective application of this AI ECG algorithm in various clinical settings is essential to establish the accuracy of LVSD diagnosis in a real-world setting and the impact on clinical decision-making, and is discussed further below.36
Artificial intelligence electrocardiogram to detect silent atrial fibrillation
Atrial fibrillation is often paroxysmal, asymptomatic, and elusive. It is associated with stroke, heart failure, and mortality.14,38 In patients with embolic stroke of uncertain source, the choice of anti-platelet vs. anticoagulant therapy depends on the absence or presence of AF. Holter monitors and 14- to 30-day mobile cardiac outpatient telemetry have a low yield, leading to the use of implanted loops recorders, which find AF less in <15% of patients at 1 year.38,39 Clinical risk scores and electronic medical record-based ML tools have had limited power to predict AF.3,28
Since neural networks can detect multiple, subtle, non-linear-related patterns in an ECG, we hypothesized that they may be able to detect the presence of intermittent AF from an NSR ECG recorded before or after an AF episode, as patients with AF may have subclinical ECG changes associated with fibrosis or transient physiologic changes. To test this hypothesis, we utilized ∼1 million ECGs from patients with no AF (controls) and patients with episodic AF (cases). The network was never shown ECGs with AF, but only NSR ECGs from patients with episodic AF and from controls. After training, the AI ECG network accurately detected paroxysmal AF from an ECG recorded during NSR (accuracy of 79.4%; AUC 0.87).3 When an ECG from the patients’ ‘window of interest’ (31-day period prior to first ECG showing AF) was evaluated, the accuracy of the AI ECG algorithm improved (83.3%; AUC 0.9).3 As noted in a letter to the editor, there were differences in the AF vs. no AF populations.40 This algorithm was subsequently tested as a predictor of AF compared to the CHARGE-AF (cohorts for ageing and research in genomic epidemiology-AF) score.41 The incidence of AF predicted by each model—AI ECG AF probability and CARGE-AF score—was assessed in a quartile analysis over time. The cumulative incidence of AF was greatest in the highest quartile for each method at 10 years (AI ECG AF 36.1% when AF probability >12.4%; CHARGE-AF 31.4% when score >14.7).41 Both methods revealed a c-statistic of 0.69 independently (c-statistic 0.72 when combined) indicating that the AI ECG model can provide a simple means for assessing AF risk without clinical data abstraction (i.e. CHARGE-AF).41 Importantly, individuals with AI ECG AF model output of >0.5 at baseline had cumulative incidence of AF 21.5% at 2 years and 52.2% at 10 years, identifying a high-risk subset.
This work has subsequently been independently confirmed by others. Raghunath et al.31 used 1.6 M 12 lead ECGs to train a network that could predict new-onset AF in patients with no AF history (AUC ROC 0.85, AUC precision-recall curve 0.22). Additionally, multiple groups have used deep learning algorithms to detect atrial fibrillation present during recording (non-‘silent’ AF) using lead, single lead, and photoplethysmographic devices.27,29,30
Artificial intelligence to screen for hypertrophic cardiomyopathy
Hypertrophic cardiomyopathy can result in symptoms or sudden cardiac death in young athletes. Various ECG criteria have been proposed for diagnosis, but none have shown consistent diagnostic performance.42–44 Similarly, previous attempts to detect HCM with AI application have focused on high-risk patient characteristics,45 specific ECG criteria, or beat-to-beat morphologic ECG features.25 Noteworthy, up to 10% of HCM patients may exhibit a ‘normal’ ECG rendering diagnostic criteria and algorithms useless.42–44
The AI ECG is a powerful tool for the detection of HCM, with high accuracy found from multiple groups.25,34,46 The Mayo Clinic algorithm maintained its robust accuracy when the testing group was narrowed to patients with left ventricular hypertrophy (LVH) ECG criteria (AUC 0.95) and ‘normal ECG’ by manual interpretation (AUC 0.95), as found by others.25,34 This implies that the AI ECG algorithm does not the typical ECG characteristics associated with HCM for diagnosis.13,47 Importantly, the AI ECG effectively differentiates ‘normal ECG’ findings, from LVH and benign LVH-like ECG features related to athletic training.
Lastly, we identified the potential for cost-effective HCM screening as the NPV of the model remained high at all probability thresholds (NPV 98–99%).13 Appropriate deployment of this test could provide reassurance to patients, prevent unnecessary, expensive diagnostic workup associated with manual interpretation, and the clinical conundrum of the athletic heart vs. HCM, resulting in improved utilization of healthcare resources.
Artificial intelligence for arrhythmia syndromes
As with other cardiac genetic disorders, arrhythmia syndromes such as long QT syndrome (LQTS) and Brugada syndrome exhibit incomplete penetrance: disease expression and severity in individuals even from the same family and carrying the same mutation can be very different. As diagnosis is based primarily upon the ECG phenotype, the potential for human misinterpretation and mismeasurement is significant.
Artificial intelligence therefore offers the opportunity for more reliable measurement of diagnostic markers and then the ability to recognize the diagnosis when expression is incomplete or even absent. This may result from the ability of neural networks to classify subtle morphologic ECG changes. The best exemplar is measurement of the QT interval and diagnosis of the LQTS. A deep neural network (DNN) was trained on ECGs from 250 767 patients, tested on a further 107 920 and then validated on a further 179 513, using the institutional ECG repository with the cardiologist over-read QTc as the gold standard. There was strong agreement in the test set between the gold standard and the DNN predicted value. Furthermore, when tested against a prospective genetic heart disease clinic population including LQTS patients, the DNN performed just as robustly. The DNN relied upon a two lead ECG input and was therefore also tested against an input from a mobile ECG device with excellent concordance. For a cut-off of QTc >500 ms, a strong diagnostic and risk marker for the likelihood of LQTS, the AUC was 0.97, with sensitivity and specificity of 80.0%, and 94.4%, respectively, indicating strong utility as a screening method.
Even when the QTc is normal, concealed LQTS has been detectable by computerized analysis of T-wave morphology. A DNN has been applied to the whole ECG from all patients from the genetic heart disease clinic (N = 2059) with a diagnosis of LQTS and those who were seen and then discharged without a diagnosis. The QTc interval alone identified patients with LQTS and a QTc <450 ms compared to patients without LQTS with an AUC of 0.741. The AI algorithm increased the AUC to 0.863 and also distinguished genetic subtypes of LQTS, offering potential as a tool to guide evaluation in the clinic.
Other opportunities include the diagnosis of concealed Brugada syndrome, which is made using sodium channel blocker challenge. Artificial intelligence could potentially avoid the process of drug challenge. Furthermore, risk stratification is heavily dependent on and limited by ECG markers that are only visually interpretable. Artificial intelligence algorithms may identify markers from the ECG that would otherwise go undetected.
Artificial intelligence valvular heart disease
Early diagnosis of valvular diseases such as aortic stenosis (AS) and mitral regurgitation (MR) permits prevention of irreversible damage. A growing body of data supports early treatment, including in asymptomatic patients.48 Current screening methods rely on expert examination and auscultation that prompt echocardiography. With the adoption of echocardiography in practice, auscultation is in decline,49 resulting in a need for improved screening. The AI ECG detects moderate–severe AS, as reported by two independent groups, with AUC of 0.87–0.9.6,7 Using echocardiography as the ground truth, Cohen-Shelly et al.6 found that the AI ECG also predicted future development of severe AS, prior to clinical manifestation. Kwon et al.35 have demonstrated a similar yield in screening for moderate to severe MR (AUC: 0.88). These algorithms, developed independently using two geographically and racially diverse populations, highlight potential for the AI ECG to detect valvular diseases at earlier stages, even when tested using a single ECG lead and in asymptomatic patients. The use of mobile form factors to acquire ECGs that can be augmented by AI makes the test available at point of care and massively scalable; in combination with percutaneous and novel treatments for valvular lesions, a significant potential exists to improve patient outcomes in a scalable and cost-efficient way.
Validation in practice
Artificial intelligence in medicine has been predominantly developed and tested in silico, using large aggregates of medical records containing analysable medical information. However, in order for AI to have a meaningful impact on human health, it must be applied and integrated in medical workflows, and used to treat patients. Differences between the training population and applied population, lack of use of AI output by clinicians who may not trust, understand, or easily access the information impact real-world effectiveness of AI. Several studies have explicitly tested this. The EAGLE study50 and Yao et al.36 were a pragmatic clustered trial study that randomized 120 primary care teams including 358 clinicians to an intervention group (access to AI screening results, 181 clinicians) or control (usual care, 177 clinicians), across 48 clinics/hospitals. A total of 22 641 adult patients who had an ECG performed for any indication between August 5, 2019, and March 31, 2020, without prior heart failure were included. The primary endpoint was a new diagnosis of an EF of ≤50% within 3 months. The proportion of patients with a positive result was similar between groups (6.0% vs. 6.0%). Among patients with a positive screening result, a high percentage of intervention patients received echocardiograms (38.1% for control and 49.6% for intervention, P < 0.001). The AI screening tool also increase the diagnosis of low EF from 1.6% in the control group to 2.1% in the intervention group, odds ratio 1.32, P = 0.007. This study highlighted a number of important points. The AI ECG integrated into primary care improved the diagnosis of low EF. This study found that the environment in which the algorithm was used impacted its performance, with a higher yield whose ECGs were obtained in outpatient clinics compared to those who were hospitalized. This may potentially reflect a higher probability of undiagnosed low EF patients in outpatient settings. Another important finding is that the performance of the intervention was highly dependent on clinician adoption of the recommendation. Among patients with a positive screening result, the intervention increased the percentage of patients receiving an echocardiogram from 38.1% to 49.6%, indicating that a large number of clinicians did not respond to the AI recommendation. Many of the decisions to forgo echocardiography were based on logical clinical reasoning such as a patient undergoing palliative care in whom additional diagnostic testing was not needed. Nonetheless, the study highlighted the impact of AI both in its capability of identifying undiagnosed disease and the importance of the use of clinical judgement, as with any tool. Importantly, this trial of over 20 000 patients was executed rapidly and inexpensively, highlighting the ability to rigorously and effectively assess AI tools developed in a pragmatic manner due to the tool’s software foundation, allowing for rapid design development and iteration.
The BEAGLE study (ClinicalTrials.gov Identifier: NCT04208971) is applying a silent atrial fibrillation algorithm3 to identify patients previously seen at Mayo Clinic to determine whether undetected atrial fibrillation is present. If found by the application of AI to stored ECGs (acquired at a previous visit to the clinic), a separate natural language processing AI algorithm screens the patient record to determine anticoagulation eligibility based on the CHADS-VASc score and bleeding risk. Those individuals found to have silent atrial fibrillation by the AI ECG screen who would benefit from anticoagulation are invited to enrol in a study via an electronic portal message. Study participants are monitored for 30 days using a wearable monitor. This trial is ongoing, and the results are pending. However, instances of individuals previously not known to have atrial fibrillation (despite many NSR ECGs) have been identified, clinical atrial fibrillation has been subsequently recorded with prospective ambulatory monitoring, and following consultation, anticoagulation initiated. This trial seeks to demonstrate how the AI ECG may improve our systematic clinical capabilities, eliminate variability resulting from the random chance of capturing a time-limited arrhythmia with standard ECG recordings and variations in clinical knowledge among practitioners to comprehensively find individuals who may benefit from evidence-based stroke prevention therapies.
New opportunities and implications
High school athlete and large-scale screening
As alluded to above, the AI ECG has the potential to measure and detect ECG markers for cardiomyopathy and arrhythmia syndromes, automatically and more effectively than humans, including the use of mobile ECG devices. This may have specific utility in the screening of young people to exclude risk prior to participation in sports. Objections to employing the ECG in the screening algorithm include issues of cost, false positives and negatives, and the lack of experts to read ECGs to minimize false positives.51 Artificial intelligence may be able to address these concerns but requires a large dataset with linkage to outcomes to test the hypothesis.
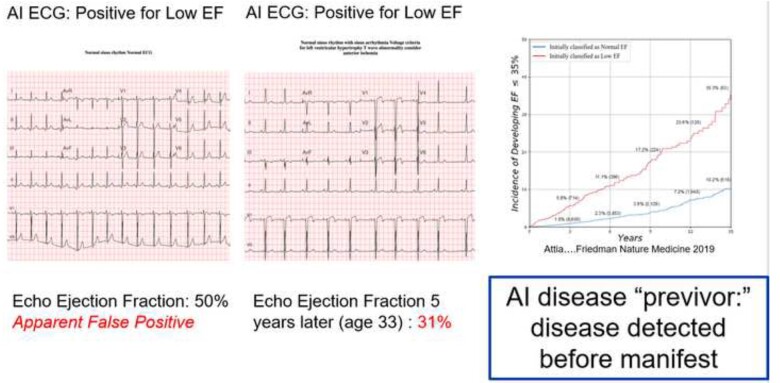
Concept of artificial intelligence disease ‘previvors’
The AI ECG identifies LVSD in some individuals with normal imaging findings and no manifest disease. However, those individuals with apparent false positive AI ECG findings have a five-fold increased risk of developing ventricular dysfunction over the ensuing 5 years.2 This raises the possibilities that with sufficient data AI tools may predict who will develop a disease, leading to the concept of disease ‘previvors’—individuals who are healthy but have a markedly elevated predisposition to develop a disease (Figure 5). This concept was similarly evident with other AI ECG models.41 This raises issues of preventive interventions and their potential risks, patient anxiety, and insurance coverage, data privacy and larger societal issues that must be considered as increasingly powerful AI tools are developed.
Figure 5.
Artificial intelligence disease previvor. Left panel—an apparently normal electrocardiogram is identified by artificial intelligence as being associated with a low ejection fraction. A contemporaneous echocardiogram depicts normal ventricular function (ejection fraction 50%). Middle panel—5 years later, at age 33, additional electrocardiograms changes are now visible to the human eye, and repeat echocardiography shows depressed ventricular function (ejection fraction 31%). Right panel—risk of developing ejection fraction <35% with a positive artificial intelligence electrocardiogram (red line) vs. with a negative artificial intelligence electrocardiogram for low ejection fraction (blue line). Further details in the text. AI, artificial intelligence; ECG, electrocardiogram; EF, ejection fraction.
Data sharing and privacy
Data availability is the driving force behind the AI revolution, as deep neural networks and other AI models require large and high-quality datasets.52,53 In some cases, there is not a single institution with sufficient data to train an accurate AI model, and data from multiple institutions is required. Sharing data among institutions introduces a risk of disclosing including protected health information. Sharing identified health information without proper approval is unethical,54 risks eroding trust in health care institutions, and violates national and international laws, including HIPAA in the USA55 and GDPR in Europe56 To avoid infringing on patient privacy and to comply with regulations, data are often de-identified. While this reduces the risk of re-identification, recent research showed that data that appear de-identified to humans often continues to embed patient information, which can be extracted and reconstituted. Packhauser et al. have shown that in an open dataset containing supposably de-identified chest X-ray images, using an AI similarity model, images from the same patient could be identified even when acquired 10 years or more from one another (AUC of 0.994). While their method required additional medical data to re-identify patients, this is not always the case. Schwarz et al. were able to reconstruct subject faces using an anonymized magnetic resonance imaging dataset from the three-dimensional data and to then match the faces to subject photographs, with an accuracy of 84%. While these methods require access to multiple data sources, they highlight data exposure risks. Therefore, we envision that data sharing will be replaced with privacy preserving methods that reduce the probability of re-identification significantly in the very near future.
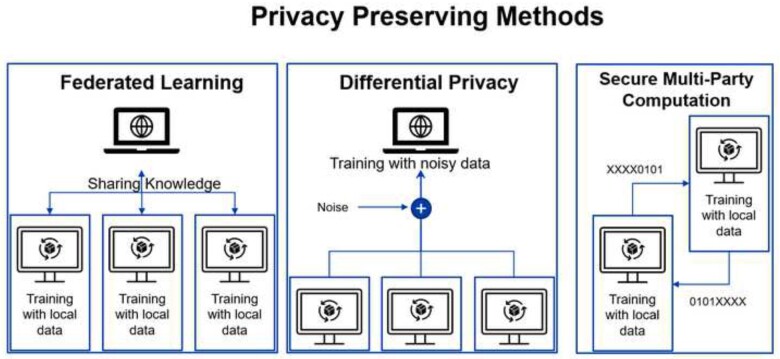
The most common technique to allow training of AI models without sharing data is federated learning,57 an approach that trains a single model in a de-centralized fashion (Figure 6). Rather than sending data to a single location, each site with private data uses a local training node that only sees its own data. The output from each local node—which is no longer interpretable—is then sent to a main node that aggregates the knowledge and consolidates it to a single model without having access to the original raw data. A second method that allows training of a model with privacy in mind is differential privacy (DP).58 Instead of using raw data directly, the data are shared with added noise, allowing the data contributor to blend in a sea of data contributors. To prevent noise averaging out with the addition of data from a given individual, the amount of information contributed from one person is limited (privacy budget). A common use case for DP is smartphone keyboard suggestions. By adding noise and limiting the number of contributions from a single user, smartphones can predict of the next word or emoji a user will type, but without disclosing all the user’s previous conversations.59 A third privacy preserving method mostly used for inference using a pre-trained model is secure multi party computation. In this method, only an encrypted portion of each data sample is shared, each part of the data is analysed separately, and the results are combined in a secured way, allowing data analysis without sending any complete samples.
Figure 6.
Privacy preserving methods. Details in the text.
Conclusion
The ECG is rich in physiologic information that is unique, identifying, and encodes many health conditions. Since ionic currents are frequently affected very early in many disease processes, the addition of AI to a standard ECG—a ubiquitous, inexpensive, test that requires no body fluids or reagents—transforms it into a powerful diagnostic screening tool that may also permit monitoring and assessment of response to therapy. When coupled to smartphones, it enables a massively scalable point of care test. Like any test, clinicians will need to understand when and how to use it in order to best care for patients.
Conflict of interest: Z.I.A. and P.A.F. are coinventors of several of the AI algorithms described (including screen for low EF, hypertrophic cardiomyopathy, QT tool, atrial fibrillation detection during NSR). These have been licensed to Anumana, AliveCor, and Eko. Mayo Clinic and Z.I.A. and P.A.F. may receive benefit from their commercialization. All other authors declared no conflict of interest.
Data availability
No new data were generated or analysed in support of this research.
References
- 1. Attia ZI, Friedman PA, Noseworthy PA, Lopez-Jimenez F, Ladewig DJ, Satam G, Pellikka PA, Munger TM, Asirvatham SJ, Scott CG, Carter RE, Kapa S. Age and sex estimation using artificial intelligence from standard 12-lead ECGs. Circ Arrhythm Electrophysiol 2019;12:e007284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Attia ZI, Kapa S, Lopez-Jimenez F, McKie PM, Ladewig DJ, Satam G, Pellikka PA, Enriquez-Sarano M, Noseworthy PA, Munger TM, Asirvatham SJ, Scott CG, Carter RE, Friedman PA. Screening for cardiac contractile dysfunction using an artificial intelligence-enabled electrocardiogram. Nat Med 2019;25:70–74. [DOI] [PubMed] [Google Scholar]
- 3. Attia ZI, Noseworthy PA, Lopez-Jimenez F, Asirvatham SJ, Deshmukh AJ, Gersh BJ, Carter RE, Yao X, Rabinstein AA, Erickson BJ, Kapa S, Friedman PA. An artificial intelligence-enabled ECG algorithm for the identification of patients with atrial fibrillation during sinus rhythm: a retrospective analysis of outcome prediction. Lancet 2019;394:861–867. [DOI] [PubMed] [Google Scholar]
- 4. Siontis KC, Noseworthy PA, Attia ZI, Friedman PA. Artificial intelligence-enhanced electrocardiography in cardiovascular disease management. Nat Rev Cardiol 2021;18:465–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Selvaraju RR, Cm Das A, Vedantam R, Parikh D, Batra D. Grad-CAM: visual explanations from deep networks via gradient-based localization. IEEE Int Conf Comput Vis (ICCV) 2017;618–626. [Google Scholar]
- 6. Cohen-Shelly M, Attia ZI, Friedman PA, Ito S, Essayagh BA, Ko WY, Murphree DH, Michelena HI, Enriquez-Sarano M, Carter RE, Johnson PW, Noseworthy PA, Lopez-Jimenez F, Oh JK. Electrocardiogram screening for aortic valve stenosis using artificial intelligence. Eur Heart J 2021;42:2885–2896. [DOI] [PubMed] [Google Scholar]
- 7. Kwon JM, Lee SY, Jeon KH, Lee Y, Kim KH, Park J, Oh BH, Lee MM. Deep learning-based algorithm for detecting aortic stenosis using electrocardiography. J Am Heart Assoc 2020;9:e014717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alizadehsani R, Roshanzamir M, Hussain S, Khosravi A, Koohestani A, Zangooei MH, Abdar M, Beykikhoshk A, Shoeibi A, Zare A, Panahiazar M, Nahavandi S, Srinivasan D, Atiya AF, Acharya UR. Handling of uncertainty in medical data using machine learning and probability theory techniques: a review of 30 years (1991-2020). Ann Oper Res 2021;1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gal Y, Ghahramani Z. Dropout as a Bayesian Approximation: Representing Model Uncertainty in Deep Learning. Proceedings of the 33rd International Conference on Machine Learning 2016;48:1050–1059. [Google Scholar]
- 10. Vranken JF, van de Leur RR, Gupta DK, Juarez Orozco LE, Hassink RJ, van der Harst P, Doevendans PA, Gulshad S, van Es R. Uncertainty estimation for deep learning-based automated analysis of 12-lead electrocardiograms. Eur Heart J Dig Health 2021;ztab045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Han X, Hu Y, Foschini L, Chinitz L, Jankelson L, Ranganath R. Deep learning models for electrocardiograms are susceptible to adversarial attack. Nat Med 2020;26:360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson JM, Khoshgoftaar TM. Survey on deep learning with class imbalance. J Big Data 2019;6:27. [Google Scholar]
- 13. Ko WY, Siontis KC, Attia ZI, Carter RE, Kapa S, Ommen SR, Demuth SJ, Ackerman MJ, Gersh BJ, Arruda-Olson AM, Geske JB, Asirvatham SJ, Lopez-Jimenez F, Nishimura RA, Friedman PA, Noseworthy PA. Detection of hypertrophic cardiomyopathy using a convolutional neural network-enabled electrocardiogram. J Am Coll Cardiol 2020;75:722–733. [DOI] [PubMed] [Google Scholar]
- 14. Hindricks G, Potpara T, Dagres N, Arbelo E, Bax JJ, Blomström-Lundqvist C, Boriani G, Castella M, Dan GA, Dilaveris PE, Fauchier L, Filippatos G, Kalman JM, La Meir M, Lane DA, Lebeau JP, Lettino M, Lip GYH, Pinto FJ, Thomas GN, Valgimigli M, Van Gelder IC, Van Putte BP, Watkins CL; ESC Scientific Document Group. 2020 ESC Guidelines for the diagnosis and management of atrial fibrillation developed in collaboration with the European Association for Cardio-Thoracic Surgery (EACTS): the Task Force for the diagnosis and management of atrial fibrillation of the European Society of Cardiology (ESC). Developed with the special contribution of the European Heart Rhythm Association (EHRA) of the ESC. Eur Heart J 2021;42:373–498. [DOI] [PubMed] [Google Scholar]
- 15. Betti I, Castelli G, Barchielli A, Beligni C, Boscherini V, De Luca L, Messeri G, Gheorghiade M, Maisel A, Zuppiroli A. The role of N-terminal PRO-brain natriuretic peptide and echocardiography for screening asymptomatic left ventricular dysfunction in a population at high risk for heart failure. The PROBE-HF study. J Card Fail 2009;15:377–384. [DOI] [PubMed] [Google Scholar]
- 16. Redfield MM, Rodeheffer RJ, Jacobsen SJ, Mahoney DW, Bailey KR, Burnett JC Jr. Plasma brain natriuretic peptide to detect preclinical ventricular systolic or diastolic dysfunction: a community-based study. Circulation 2004;109:3176–3181. [DOI] [PubMed] [Google Scholar]
- 17. McDonagh TA, McDonald K, Maisel AS. Screening for asymptomatic left ventricular dysfunction using B-type natriuretic Peptide. Congest Heart Fail 2008;14:5–8. [DOI] [PubMed] [Google Scholar]
- 18. Attia ZI, Kapa S, Yao X, Lopez-Jimenez F, Mohan TL, Pellikka PA, Carter RE, Shah ND, Friedman PA, Noseworthy PA. Prospective validation of a deep learning electrocardiogram algorithm for the detection of left ventricular systolic dysfunction. J Cardiovasc Electrophysiol 2019;30:668–674. [DOI] [PubMed] [Google Scholar]
- 19. Adedinsewo D, Carter RE, Attia Z, Johnson P, Kashou AH, Dugan JL, Albus M, Sheele JM, Bellolio F, Friedman PA, Lopez-Jimenez F, Noseworthy PA. Artificial intelligence-enabled ECG algorithm to identify patients with left ventricular systolic dysfunction presenting to the emergency department with dyspnea. Circ Arrhythm Electrophysiol 2020;13:e008437. [DOI] [PubMed] [Google Scholar]
- 20. Noseworthy PA, Attia ZI, Brewer LC, Hayes SN, Yao X, Kapa S, Friedman PA, Lopez-Jimenez F. Assessing and mitigating bias in medical artificial intelligence: the effects of race and ethnicity on a deep learning model for ECG analysis. Circ Arrhythm Electrophysiol 2020;13:e007988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Attia IZ, Dugan J, Rideout A, Lopez-Jimenez F, Noseworthy PA, Asirvatham S, Pellikka PA, Ladewig DJ, Satam G, Pham S, Venkatraman S, Friedman PA, Kapa S. Prospective analysis of utility signals from an ECG-enabled stethoscope to automatically detect a low ejection fraction using neural network techniques trained from the standard 12-lead ECG. Circulation 2019;140:A13447. [Google Scholar]
- 22. Attia IZ, Tseng AS, Benavente ED, Medina-Inojosa JR, Clark TG, Malyutina S, Kapa S, Schirmer H, Kudryavtsev AV, Noseworthy PA, Carter RE, Ryabikov A, Perel P, Friedman PA, Leon DA, Lopez-Jimenez F. External validation of a deep learning electrocardiogram algorithm to detect ventricular dysfunction. Int J Cardiol 2021;329:130–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cho J, Lee BT, Kwon J-M, Lee Y, Park H, Oh B-H, Jeon K-H, Park J, Kim K-H. Artificial intelligence algorithm for screening heart failure with reduced ejection fraction using electrocardiography. ASAIO J 2021;67:314–321. [DOI] [PubMed] [Google Scholar]
- 24. Kwon JM, Kim KH, Jeon KH, Kim HM, Kim MJ, Lim SM, Song PS, Park J, Choi RK, Oh BH. Development and validation of deep-learning algorithm for electrocardiography-based heart failure identification. Korean Circ J 2019;49:629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rahman QA, Tereshchenko LG, Kongkatong M, Abraham T, Abraham MR, Shatkay H. Utilizing ECG-based heartbeat classification for hypertrophic cardiomyopathy identification. IEEE Trans Nanobiosci 2015;14:505–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Galloway CD, Valys AV, Shreibati JB, Treiman DL, Petterson FL, Gundotra VP, Albert DE, Attia ZI, Carter RE, Asirvatham SJ, Ackerman MJ, Noseworthy PA, Dillon JJ, Friedman PA. Development and validation of a deep-learning model to screen for hyperkalemia from the electrocardiogram. JAMA Cardiol 2019;4:428–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tison GH, Sanchez JM, Ballinger B, Singh A, Olgin JE, Pletcher MJ, Vittinghoff E, Lee ES, Fan SM, Gladstone RA, Mikell C, Sohoni N, Hsieh J, Marcus GM. Passive detection of atrial fibrillation using a commercially available smartwatch. JAMA Cardiol 2018;3:409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hill NR, Ayoubkhani D, McEwan P, Sugrue DM, Farooqui U, Lister S, Lumley M, Bakhai A, Cohen AT, O'Neill M, Clifton D, Gordon J. Predicting atrial fibrillation in primary care using machine learning. PLoS One 2019;14:e0224582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jo YY, Cho Y, Lee SY, Kwon JM, Kim KH, Jeon KH, Cho S, Park J, Oh BH. Explainable artificial intelligence to detect atrial fibrillation using electrocardiogram. Int J Cardiol 2021;328:104–110. [DOI] [PubMed] [Google Scholar]
- 30. Poh MZ, Poh YC, Chan PH, Wong CK, Pun L, Leung WW, Wong YF, Wong MM, Chu DW, Siu CW. Diagnostic assessment of a deep learning system for detecting atrial fibrillation in pulse waveforms. Heart 2018;104:1921–1928. [DOI] [PubMed] [Google Scholar]
- 31. Raghunath S, Pfeifer JM, Ulloa-Cerna AE, Nemani A, Carbonati T, Jing L, vanMaanen DP, Hartzel DN, Ruhl JA, Lagerman BF, Rocha DB, Stoudt NJ, Schneider G, Johnson KW, Zimmerman N, Leader JB, Kirchner HL, Griessenauer CJ, Hafez A, Good CW, Fornwalt BK, Haggerty CM. Deep neural networks can predict new-onset atrial fibrillation from the 12-lead ECG and help identify those at risk of atrial fibrillation-related stroke. Circulation 2021;143:1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Giudicessi JR, Schram M, Bos JM, Galloway CD, Shreibati JB, Johnson PW, Carter RE, Disrud LW, Kleiman R, Attia ZI, Noseworthy PA, Friedman PA, Albert DE, Ackerman MJ. Artificial intelligence-enabled assessment of the heart rate corrected QT interval using a mobile electrocardiogram device. Circulation 2021;143:1274–1286. [DOI] [PubMed] [Google Scholar]
- 33. Bos JM, Attia ZI, Albert DE, Noseworthy PA, Friedman PA, Ackerman MJ. Use of artificial intelligence and deep neural networks in evaluation of patients with electrocardiographically concealed long QT syndrome from the surface 12-lead electrocardiogram. JAMA Cardiol 2021;6:532–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tison GH, Zhang J, Delling FN, Deo RC. Automated and interpretable patient ECG profiles for disease detection, tracking, and discovery. Circ Cardiovasc Qual Outcomes 2019;12:e005289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kwon JM, Kim KH, Akkus Z, Jeon KH, Park J, Oh BH. Artificial intelligence for detecting mitral regurgitation using electrocardiography. J Electrocardiol 2020;59:151–157. [DOI] [PubMed] [Google Scholar]
- 36. Yao X, McCoy RG, Friedman PA, Shah ND, Barry BA, Behnken EM, Inselman JW, Attia ZI, Noseworthy PA. ECG AI-Guided Screening for Low Ejection Fraction (EAGLE): rationale and design of a pragmatic cluster randomized trial. Am Heart J 2020;219:31–36. [DOI] [PubMed] [Google Scholar]
- 37. Attia ZI, Kapa S, Noseworthy PA, Lopez-Jimenez F, Friedman PA. Artificial intelligence ECG to detect left ventricular dysfunction in COVID-19: a case series. Mayo Clin Proc 2020;95:2464–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sanna T, Diener HC, Passman RS, Di Lazzaro V, Bernstein RA, Morillo CA, Rymer MM, Thijs V, Rogers T, Beckers F, Lindborg K, Brachmann J. Cryptogenic stroke and underlying atrial fibrillation. N Engl J Med 2014;370:2478–2486. [DOI] [PubMed] [Google Scholar]
- 39. Seet RC, Friedman PA, Rabinstein AA. Prolonged rhythm monitoring for the detection of occult paroxysmal atrial fibrillation in ischemic stroke of unknown cause. Circulation 2011;124:477–486. [DOI] [PubMed] [Google Scholar]
- 40. Freedman B. An AI-ECG algorithm for atrial fibrillation risk: steps towards clinical implementation. Lancet 2020;396:236. [DOI] [PubMed] [Google Scholar]
- 41. Christopoulos G, Graff-Radford J, Lopez CL, Yao X, Attia ZI, Rabinstein AA, Petersen RC, Knopman DS, Mielke MM, Kremers W, Vemuri P, Siontis KC, Friedman PA, Noseworthy PA. Artificial intelligence-electrocardiography to predict incident atrial fibrillation: a population-based study. Circ Arrhythm Electrophysiol 2020;13:e009355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Drezner JA, Ackerman MJ, Anderson J, Ashley E, Asplund CA, Baggish AL, Börjesson M, Cannon BC, Corrado D, DiFiori JP, Fischbach P, Froelicher V, Harmon KG, Heidbuchel H, Marek J, Owens DS, Paul S, Pelliccia A, Prutkin JM, Salerno JC, Schmied CM, Sharma S, Stein R, Vetter VL, Wilson MG. Electrocardiographic interpretation in athletes: the ‘Seattle criteria’. Br J Sports Med 2013;47:122–124. [DOI] [PubMed] [Google Scholar]
- 43. Pickham D, Zarafshar S, Sani D, Kumar N, Froelicher V. Comparison of three ECG criteria for athlete pre-participation screening. J Electrocardiol 2014;47:769–774. [DOI] [PubMed] [Google Scholar]
- 44. Sheikh N, Papadakis M, Ghani S, Zaidi A, Gati S, Adami PE, Carré F, Schnell F, Wilson M, Avila P, McKenna W, Sharma S. Comparison of electrocardiographic criteria for the detection of cardiac abnormalities in elite black and white athletes. Circulation 2014;129:1637–1649. [DOI] [PubMed] [Google Scholar]
- 45. Bhattacharya M, Lu DY, Kudchadkar SM, Greenland GV, Lingamaneni P, Corona-Villalobos CP, Guan Y, Marine JE, Olgin JE, Zimmerman S, Abraham TP, Shatkay H, Abraham MR. Identifying ventricular arrhythmias and their predictors by applying machine learning methods to electronic health records in patients with hypertrophic cardiomyopathy (HCM-VAr-Risk Model). Am J Cardiol 2019;123:1681–1689. [DOI] [PubMed] [Google Scholar]
- 46. Dai H, Hwang HG, Tseng VS. Convolutional neural network based automatic screening tool for cardiovascular diseases using different intervals of ECG signals. Comput Methods Programs Biomed 2021;203:106035. [DOI] [PubMed] [Google Scholar]
- 47. Engel TR. Diagnosis of hypertrophic cardiomyopathy: who is in charge here-the physician or the computer? J Am Coll Cardiol 2020;75:734–735. [DOI] [PubMed] [Google Scholar]
- 48. Kang DH, Park SJ, Lee SA, Lee S, Kim DH, Kim HK, Yun SC, Hong GR, Song JM, Chung CH, Song JK, Lee JW, Park SW. Early surgery or conservative care for asymptomatic aortic stenosis. N Engl J Med 2020;382:111–119. [DOI] [PubMed] [Google Scholar]
- 49. Alam UA, Khan SQ, Hayat S, Malik RA. Cardiac auscultation: an essential clinical skill in decline. Br J Cardiol 2010;17:8–10. [Google Scholar]
- 50. Yao X, Rushlow DR, Inselman JW, McCoy RG, Thacher TD, Behnken EM, Bernard ME, Rosas SL, Akfaly A, Misra A, Molling PE, Krien JS, Foss RM, Barry BA, Siontis KC, Kapa S, Pellikka PA, Lopez-Jimenez F, Attia ZI, Shah ND, Friedman PA, Noseworthy PA. Artificial intelligence–enabled electrocardiograms for identification of patients with low ejection fraction: a pragmatic, randomized clinical trial. Nat Med 2021;27:815–819. [DOI] [PubMed] [Google Scholar]
- 51. Van Brabandt H, Desomer A, Gerkens S, Neyt M. Harms and benefits of screening young people to prevent sudden cardiac death. BMJ 2016;353:i1156. [DOI] [PubMed] [Google Scholar]
- 52. Cho J, Lee K, Shin E, Choy G, Do S. How much data is needed to train a medical image deep learning system to achieve necessary high accuracy? 2015. arXiv:1511.06348.
- 53. Hlynsson H, Escalante BA, Wiskott L. Measuring the Data Efficiency of Deep Learning Methods. 2019. arXiv: 1907.02549
- 54. Larson DB, Magnus DC, Lungren MP, Shah NH, Langlotz CP. Ethics of using and sharing clinical imaging data for artificial intelligence: a proposed framework. Radiology 2020;295:675–682. [DOI] [PubMed] [Google Scholar]
- 55. Services USDoHH. Health Information Privacy. https://www.hhs.gov/hipaa/index.html (Date accessed 23 July 2021).
- 56. European Commission. EU Data protection rules. https://ec.europa.eu/info/law/law-topic/data-protection/eu-data-protection-rules_en (Date accessed 23 July 2021).
- 57. Bonawitz K, Eichner H, Grieskamp W, Huba D, Ingerman A, Ivanov V, Kiddon C, Konecny J, Mazzocchi S, McMahan HB, Van Overveldt T, Petrou D, Ramage D, Roselander J. Towards Federated Learning at Scale: System Design. 2019. arXiv: 1902.01046.
- 58. Dwork C. Differential Privacy: A Survey of Results. In Agrawal M, Du D, Duan Z, Li A, eds. Theory and Applications of Models of Computation from Lecture Notes in Computer Science Vol. 4978. Berlin/Heidelberg: Springer; 2008. pp. 1–19. [Google Scholar]
- 59. Apple. Differential Privacy Technical Overview. https://www.apple.com/privacy/docs/Differential_Privacy_Overview.pdf (Date accessed 23 July 2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analysed in support of this research.