Abstract
The “gold standard” for identifying Yersinia pestis-infected fleas has been inoculation of mice with pooled flea material. Inoculated mice are monitored for 21 days, and those that die are further analyzed for Y. pestis infection by fluorescent-antibody assay and/or culture. PCR may provide a more rapid and sensitive alternative for identifying Y. pestis in fleas. To compare these assays, samples were prepared from 381 field-collected fleas. Each flea was analyzed individually by both PCR and mouse inoculation. Sixty of the 381 flea samples were positive for Y. pestis by PCR; 48 of these PCR-positive samples caused death in mice (80.0% agreement). None of the 321 PCR-negative samples caused death. Among the 12 mice that survived inoculation with PCR-positive samples, 10 were later demonstrated by serology or culture to have been infected with Y. pestis. This suggests that death of inoculated mice is less reliable than PCR as an indicator of the presence of Y. pestis in flea samples. Mouse inoculation assays produce results that are comparable to PCR only when surviving as well as dead mice are analyzed for infection. The rapidity and sensitivity (10 to 100 CFU of Y. pestis) of PCR suggest that it could serve as a useful alternative to mouse inoculation for routine plague surveillance and outbreak investigations.
Yersinia pestis, the etiological agent of plague, is typically transmitted between rodent hosts and other mammals by the bites of infectious fleas. The risks of human infection are highest during periods of epizootic activity in local rodent populations (9). These risks are reduced by public education and the implementation of measures to limit the spread of plague, including the use of insecticides and rodent management techniques. Timely application of preventative measures is made possible by effective surveillance programs that are capable of rapidly identifying epizootics. For surveillance and outbreak investigation purposes, fleas are often easier and safer to collect and handle than animals potentially infected with Y. pestis or other pathogens (5). Data on which host and flea species are involved in a given epizootic also can provide valuable information for designing locally appropriate control programs.
The “gold standard” for testing fleas for Y. pestis infection has been the inoculation of mice with ground flea suspensions (14, 15). Typically, inoculated mice are monitored for 21 days, and tissues from mice that die are tested by fluorescent-antibody analysis for evidence of Y. pestis infection (8). Surviving mice are not routinely tested because mice are extremely susceptible to most wild-type strains of Y. pestis (50% lethal dose, 1 to 100 organisms) (14), and considerable time and personnel are required to process these additional samples. Mouse assays usually require at least 3 days, and often more, until infected mice succumb to Y. pestis infection; this creates a considerable delay in obtaining results from Y. pestis-infected flea samples. Also, false-negative results can occur when death is used as the assay endpoint, because some mice occasionally survive infection with Y. pestis (4).
Recently, molecular techniques have been proposed as a means of more rapidly identifying plague bacteria in fleas. DNA hybridization probes were developed but were unable to reliably detect fewer than 105 plague bacteria (13, 17). More recently, several PCR assays have been developed for plague diagnosis (1, 7, 11). These assays provide much greater sensitivity than the above DNA probe techniques. Identification of Y. pestis in fleas by PCR has been described by Hinnebusch and Schwan (7). Although this assay decreases the time necessary to detect bacteria in fleas, it has not been compared to the standard mouse inoculation assay. We report here a modification of the PCR assay described by Hinnebusch and Schwan (7) and a comparison of this assay with mouse inoculation.
MATERIALS AND METHODS
Y. pestis strains.
To test the sensitivity of the PCR assay, the virulent strain CO96-3188 was used. The avirulent A1122 strain was used as a positive control for all PCR assays involving field-collected fleas. Y. pestis strains were grown in brain heart infusion (BHI) broth at 28°C and harvested during log-phase growth. Serial 10-fold dilutions were then spread on blood-agar plates to determine the number of CFU, according to standard methods.
Flea samples.
For comparison of mouse inoculation and PCR assays, 381 fleas of 12 different species were collected at various locations in Colorado and New Mexico from rodent burrows, domestic animals, captured animals, or animal carcasses (Table 1). Following collection, fleas were stored individually at −70°C or in pools in 2% saline held at 4°C. Epizootic plague activity was identified or suspected at each of the locations where fleas were collected. Fleas for the sensitivity assays were from a colony of Oropsylla montana which was derived from fleas originally collected in Bernalillo County, New Mexico, in 1992.
TABLE 1.
Location, collection type, species, and number of fleas used in comparison between PCR and mouse inoculation
| Location and collection type | Species | No. tested |
|---|---|---|
| Applewood Estates, Colo. | ||
| Rodent burrow | Oropsylla hirsuta | 113 |
| Lighthouse Church, Colo. | ||
| Rodent burrow | Oropsylla hirsuta | 27 |
| Rodent burrow | Oropsylla tuberculata cynomuris | 21 |
| Tribly Creek, Colo. | ||
| Rodent burrow | Oropsylla hirsuta | 29 |
| Rodent burrow | Oropsylla tuberculata cynomuris | 7 |
| Devil’s Backbone, Colo. | ||
| Rodent burrow | Oropsylla hirsuta | 43 |
| Cedar Grove, N. Mex. | ||
| Rodent burrow | Oropsylla montana | 24 |
| Rodent burrow | Hoplopsyllus anomalus | 6 |
| Domestic animal | Hoplopsyllus anomalus | 1 |
| Domestic animal | Cediopsylla inaequalis | 4 |
| Captured animal | Aetheca wagneri | 1 |
| Captured animal | Orchopeas leucopus | 1 |
| Abiquiu, N. Mex. | ||
| Captured animal | Aetheca wagneri | 2 |
| Captured animal | Orchopeas leucopus | 2 |
| Captured animal | Orchopeas sexdentatus | 2 |
| Captured animal | Malaraeus sinomus | 1 |
| Carson, N. F., N. Mex. | ||
| Captured animal | Aetheca wagneri | 51 |
| Captured animal | Cediopsylla inaequalis | 4 |
| Captured animal | Orchopeas leucopus | 3 |
| Captured animal | Malaraeus sinomus | 1 |
| Captured animal | Euhoplopsyllus glacialis | 1 |
| Captured animal | Epitedia sp. | 1 |
| C.F. Prairie, Colo. | ||
| Animal carcass | Oropsylla hirsuta | 6 |
| Animal carcass | Oropsylla tuberculata cynomuris | 30 |
Preparation of fleas for PCR and mouse inoculation.
Fleas for comparison of PCR and mouse inoculation assays were prepared by triturating individual fleas in 100 μl of BHI broth in 1.5-ml microcentrifuge tubes with sterile sea sand and disposable pestles. Fleas pooled and stored in saline at the time of collection were individually washed in sterile saline before trituration. Fifty microliters of the triturated flea-BHI infusion was pipetted into a separate 1.5-ml microcentrifuge tube to which 350 μl of sterile 0.85% (physiological) saline was added for mouse inoculation. The remaining 50 μl of the ground flea suspension was heated to 95°C for 10 min and then immediately centrifuged for 10 s at maximum speed (15,600 × g) to pellet flea tissue and sand. The supernatants were assayed by pla PCR within 10 min. Leftover supernatants were stored at −70°C.
Fleas used for testing the sensitivity of the PCR assay were triturated in six separate tubes containing 100 μl of BHI broth spiked with 106, 105, 104, 103, 102, and 101 CFU of Y. pestis CO96-3188. The triturated flea material was then heated and pelleted as described above. Pools of 0, 1, 5, 10, 20, and 25 fleas were used per sample.
PCR amplification targets.
Primer sequences were the same as those described by Hinnebusch and Schwan (7) and were derived from the published sequence data for the plasminogen activator gene (pla) (16). These primers, Yp1 (5′-ATCTTACTTTCCGTGAGAA-3′) and Yp2 (5′-CTTGGATGTTGAGCTTCCTA-3′), correspond to nucleotides 971 to 990 and 1431 to 1450, respectively, and produce a 478-bp amplification product. All PCR tests with the pla primers will hereafter be referred to as pla PCR assays. A confirmatory PCR assay used primers targeting the caf1 gene of Y. pestis, as described by Chu et al. (2). These primers correspond to nucleotides 1 to 18 and 496 to 513 of the caf1 gene, which encodes the structural region of the F1 antigen (6), and produce a 513-bp amplification product. All PCR tests with the caf1 primers will hereafter be referred to as caf1 PCR assays.
PCR.
The pla PCR protocol used in this study is a modification of the protocol described by Hinnebusch and Schwan (7). Briefly, for each PCR assay, 2.5 μl of the flea-BHI preparation was combined with 0.25 μl of each primer (Yp1 and Yp2; 30 pmol of primer/μl), 50 mM MgCl2, and 21.75 μl of deionized, distilled water in a 0.65-ml tube containing a Ready-To-Go PCR bead (Amersham Pharmacia Biotech, Piscataway, N.J.). The PCR bead contained 1.5 U of Taq DNA polymerase, 10 mM Tris HCl, 1.5 mM MgCl2, 200 μM (each) deoxynucleoside triphosphate (dNTP) and stabilizers, including bovine serum albumin. Positive controls were prepared by adding 2.5 μl of Y. pestis A1122 to the above reagents, and negative controls were prepared by adding 2.5 μl of sterile BHI to the above reagents. Two drops of sterile mineral oil were then added to the tubes before they were sealed and placed in a thermocycler (Minicycler; MJ Research, Watertown, Mass.) with the following amplification program: initial denaturation at 95°C for 5 min, followed by 35 cycles of denaturing at 95°C for 1 min, annealing at 51°C for 1 min, and primer extension at 72°C for 2 min. After the last cycle, primer extension was continued at 72°C for 10 min. The preparation of the caf1 PCR samples for the confirmatory PCR, with caf1 primers, was the same as described above, but the denaturation, annealing, and extension steps of the thermocycler program were done according to Chu et al. (2). Samples were analyzed by electrophoresis on 2% agarose gels, according to standard methods (12).
Mouse inoculation and sample collection.
Three hundred microliters of each 400-μl flea-saline suspension was inoculated subcutaneously in the lower abdomen of individual white laboratory (Swiss Webster) mice (one mouse/flea suspension). Following inoculation, mice were labeled and held in cages and supplied with food and water ad libitum. Mice were checked twice daily for signs of morbidity or mortality. Mice that died within 21 days after inoculation were stored at −20°C prior to a postmortem examination, which included macroscopic observations of the appearance of the lymph nodes, liver, and spleen, along with collection of portions of these organs or nodes for bacterial examination. Surviving mice were held for 21 days before tests were canceled. Postmortem tissue samples were taken from all mice that were inoculated with pla PCR-positive fleas and survived to day 21. Blood samples also were collected from these mice and from a random sampling of 30 mice inoculated with PCR-negative fleas. All experiments with mice were done according to a protocol approved by the Division of Vector Borne Infectious Disease’s Animal Care and Use Committee (AUC no. 97-09-008-AM).
PHA.
The passive hemagglutination assay (PHA) was used to examine mouse serum samples for anti-F1 antibodies. Samples with titers greater than 1:10 were considered to be positive. This methodology, which is described elsewhere (18), was selected because it is the standard serological assay used by the World Health Organization Collaborating Center on Plague at the Centers for Disease Control and Prevention.
FA testing.
Impression smears of mouse tissues were analyzed by fluorescent-antibody (FA) assays as a presumptive test for the presence of Y. pestis cells. The FA tests were conducted as described elsewhere (3).
Bacterial culture.
To obtain bacterial isolates from mouse tissue, small pieces of spleen, liver, and lymph node tissues were streaked on sheep blood (6%) agar plates. To obtain bacterial isolates from flea suspensions, 50 μl of the flea-saline suspension was also streaked on blood agar plates. All plates were incubated at 37°C for at least 48 h and examined for characteristic colonial morphology consistent with Y. pestis growth. Confirmation of identity of the isolate was by a positive FA test, biochemical profiles, and by evidence of lysis with Y. pestis-specific bacteriophage at both 25°C and 37°C (3). Bacteriophage lysis assays were done at both temperatures to eliminate false-positive reactions with Yersinia pseudotuberculosis, which can occur at 37°C but not at 25°C.
RESULTS
Sensitivity and pooling.
Our modification of the pla PCR assay appeared to have sensitivity comparable to that of the original assay described by Hinnebusch and Schwan (7) and was able to detect as few as 10 CFU per sample, as determined by testing 10-fold serial dilutions of Y. pestis suspended in BHI broth containing ground, uninfected flea material (data not shown). The sensitivity of this assay did not noticeably change when up to 20 fleas were suspended in each serial dilution. Consistent results were not obtained, however, when 25 fleas per pool were used.
PCR-mouse inoculation comparison.
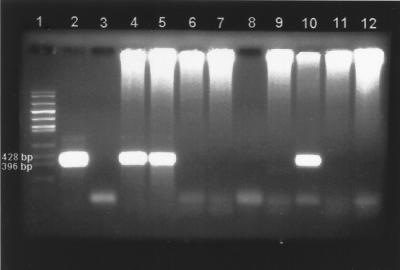
Of the 381 field-collected flea samples tested by pla PCR, 60 were positive (i.e., contained a predicted 478-bp fragment that appeared identical in size to the one observed in the positive controls) (Table 2). The pla PCR-positive specimens included 28 of 208 (13.5%) Oropsylla hirsuta specimens and 32 of 58 (55.2%) O. tuberculata cynomuris specimens tested. None of the other flea species in our study were pla PCR positive. Figure 1 shows an example of the pla PCR assay results for flea samples collected in northern Colorado.
TABLE 2.
Collection type, test results, and percent agreement between PCR and mouse inoculation assays
| Collection type | No. of fleas tested | No. of PCR-positive fleas (%) | No. of PCR-positive fleas that caused death in mice (% agreement) | No. of PCR-positive fleas that caused infection in mice (% agreement) |
|---|---|---|---|---|
| Rodent burrow | 270 | 24 (8.8) | 20 (83.3) | 24 (100.0) |
| Domestic animal | 5 | 0 | 0 | 0 |
| Captured animal | 70 | 0 | 0 | 0 |
| Animal carcass | 36 | 36 (100.0) | 28 (77.8) | 34 (94.4) |
| Total | 381 | 60 (15.7) | 48 (80.0) | 58 (96.7) |
FIG. 1.
Ethidium bromide-stained agarose gel showing pla PCR results for samples of individual O. hirsuta collected from Applewood Estates, Colo. Large bands represent a 478-bp fragment amplified from the Y. pestis pla gene. Lane 1, molecular size marker; lane 2, positive control (A1122); lanes 3 and 8, negative controls; lanes 4 to 7 and 9 to 12, flea samples (4, 5, and 10 are positive; 6, 7, 9, 11, and 12 are negative). Note: The high-molecular-weight DNA at the top of the gel is probably flea DNA in that it appears only in the lanes containing PCR samples from flea suspensions.
Of the 60 pla PCR-positive flea suspensions, 48 caused death in mice; all dead mice were positive by FA for Y. pestis infection. This resulted in 80% agreement between PCR and mouse inoculation when death was used as an endpoint for the assay. Of the 12 mice that survived inoculation with pla PCR-positive flea suspensions, 10 had positive PHA serology titers (range, 1:32 to 1:8,192) at the end of the 21-day test period. When killed on day 21, 5 of these 10 mice also were determined by FA assay to have Y. pestis in the liver, spleen, or lymph nodes. Additionally, Y. pestis was isolated from the tissues of four of these five FA-positive mice. Overall, inoculation of mice with 58 of the 60 pla PCR-positive flea suspensions produced a positive FA assay, PHA serology, or Y. pestis isolate from mouse tissues (96.7% agreement). Both of the pla PCR-positive flea suspensions that did not produce a detectable infection in mice were positive when retested by caf1 PCR. Y. pestis also was isolated directly from one of these two flea suspensions. All PCR-negative flea suspensions were negative by mouse inoculation (100% agreement) when death of mouse was used as the endpoint. Additionally, each of 30 randomly selected serum samples from mice inoculated with pla PCR-negative flea suspensions was negative by PHA.
DISCUSSION
We have shown that our modification of the pla PCR assay has many advantages over mouse inoculation for identifying Y. pestis in fleas, including its sensitivity and rapidity. Hinnebusch and Schwan (7) demonstrated that the pla primers used in our study were suitable for amplifying target DNA from Y. pestis strains collected in Asia, Africa, and the Americas, suggesting that PCR assays based on these primers should have wide applicability for surveillance purposes and epidemiological investigations. These authors also indicated that the pla primers did not produce false positives when tested with samples of Y. pseudotuberculosis, Yersinia enterocolitica, and Rickettsia typhi. The translated coding region of the pla gene has some degree of homology with the ompT gene of Escherichia coli (16), but the pla primers used in this study target the 3′ noncoding region of the plasminogen activator gene. A comparison of the pla primer sequences used in this study showed no homology with the ompT gene sequence (GenBank accession no. X06903).
The sensitivity of the pla PCR assay (10 to 100 CFU of Y. pestis) did not appear to diminish noticeably when assays were done with pools of as many as 20 fleas, which is only slightly less than the maximum of 25 fleas per pool usually recommended for the mouse inoculation assay (15). Testing to date has indicated that our pla PCR results are less reliable when the pool size is increased to 25 fleas. Although positives were occasionally obtained with samples that contained 25 fleas and were spiked with as few as 10 CFU, the 100 μl of BHI appeared to be insufficient to suspend that amount of flea material. This difficulty in suspending flea material may partially explain the inconsistent results. Consistent results may be obtainable by further modifying the procedures used to prepare the suspensions of flea material, but this type of modification was not evaluated during our study.
Our results suggest that the mouse inoculation assay can produce false-negative results when death is used as an endpoint, as indicated by the fact that only 80% of the mice that were inoculated with pla PCR-positive flea suspensions actually died. Among the 12 mice that survived inoculation with pla PCR-positive flea suspensions, 10 were later demonstrated by serology or bacterial isolation to have been infected with Y. pestis. These 10 flea suspensions might have contained insufficient Y. pestis to produce a fatal infection in mice or might have contained bacteria of lower virulence than typical wild-type strains. Regardless of why some mice failed to die when inoculated with Y. pestis-infected flea suspensions, pla PCR reliably detected Y. pestis in these samples. The results of pla PCR and mouse inoculation, however, agreed closely (96.7%) when all mice, including those that survived to 21 days, were analyzed for evidence of Y. pestis infection. It is important that all the PCR-positive fleas were collected either from prairie dog burrows in areas of suspected plague activity or from a dead prairie dog known to have died from plague. Although Y. pestis can be isolated occasionally from fleas collected from apparently healthy animals, all such fleas examined during our study were PCR negative.
Additional testing of the two pla PCR-positive mouse inoculation-negative flea suspensions clearly demonstrated or strongly suggested that these suspensions did indeed contain Y. pestis. Both flea suspensions were also positive by caf1 PCR. Although culturing Y. pestis directly from flea material is often ineffective because of contamination (15), we were able to isolate Y. pestis from one of the pla and caf1 PCR-positive, mouse inoculation-negative flea suspensions, providing clear proof that this flea was infected with Y. pestis. We were unable to isolate Y. pestis from the remaining pla and caf1 PCR-positive flea suspension. Although, as with all PCR assays, the positive pla and caf1 PCR results obtained from the culture-negative sample could have resulted from an external source of contamination, it is possible that this flea suspension contained dead bacteria with sufficient amounts of intact Y. pestis DNA to be amplified by PCR. However, it is unlikely that the positive result for this last flea suspension was due to amplicon contamination, because we obtained positive results with two different primer sets (pla and caf1), including one (caf1) not previously used in our laboratory.
In addition to standard precautions taken to minimize possible sources of contamination in PCR assays (10), we took further steps to ensure sample quality. For example, field-collected fleas are often stored under various conditions, including on dry ice (−70°C), in 2% saline with Tween 80 detergent, or in 70% ethyl alcohol. Prior to this study, we found that the 2% saline-Tween 80 solutions used to store infected fleas occasionally tested positive for Y. pestis by PCR and mouse inoculation (data not shown). Contamination of the 2% saline, therefore, is likely to result in the contamination of other fleas stored in the same container. These findings led us to wash fleas stored in saline solutions in fresh, sterile, 2% saline before triturating these samples for PCR, which appeared to eliminate the false-positive results occasionally obtained with unwashed flea material. Other methods, however, may prove superior for removing foreign DNA from the outside of fleas stored under this condition. Obviously, fleas stored in 70% alcohol cannot be tested by mouse inoculation but are suitable for PCR. It seems reasonable that fleas stored in alcohol also should be rinsed thoroughly prior to analysis by PCR to minimize cross-contamination.
Storage of fleas in alcohol is likely to be advantageous under certain circumstances, including those situations in which fleas are to be held without freezing for long periods (more than a few days) prior to analysis by PCR. We obtained good results when fleas were held dry on dry ice and at −70°C in an ultra-low freezer. Immediate freezing of samples reduces handling of the specimens, thereby reducing the risk of contamination. Our modified PCR protocol provided a simpler assay that required fewer steps to complete than the one described initially by Hinnebusch and Schwan (7). The PCR beads used with the mineral oil appeared to result in an assay whose sensitivity was comparable to that of these authors’ “hot start” method. When commercially prepared PCR beads are used, the likelihood of PCR reagent contamination is minimized, and the potential for contamination is further reduced because the number of times the tubes need to be opened is minimized. Sample preparation times also were decreased, by decreasing the number of preparation steps, without a noticeable change in performance.
On average, the sample preparation, PCR amplification, and electrophoresis of reaction products take about 4 to 5 h to complete. This is much faster than the mouse inoculation assay, which usually requires a minimum of 3 days to identify a positive sample and 21 days to confirm a negative one, not including the additional time required to perform serological or cultural assays on serum samples or tissues, respectively. We have not calculated a cost comparison, but the price of PCR reagents is likely to be offset by the additional person-hours needed to care for the mice and to conduct the bacteriological and serological testing involved with mouse inoculation assays. More importantly, the rapid turnaround time for samples ensures that surveillance data and results from epidemiological investigations will be available within hours after samples are received. This information can be invaluable in determining what control and prevention methods should be implemented to reduce risks posed by individual rodent and flea species found within the affected area.
Although this PCR assay should be adequate for most surveillance purposes requiring rapid identification of Y. pestis in fleas, it cannot substitute for bacterial culture methods when additional data are needed, such as those pertaining to virulence, plasmid profile analysis, or antibiotic susceptibility. Because the PCR assay is unable to distinguish between Y. pestis strains with various degrees of virulence or antibiotic susceptibility, we recommend that representative samples of the triturated flea material be kept at −70°C for future analysis.
Our results show that the death of inoculated mice is a less reliable indicator than PCR of the presence of Y. pestis in flea samples. The results of the modified pla PCR assay and the mouse inoculation assay were similar only when all surviving mice were tested for evidence of Y. pestis infection at the end of the 21-day assay period. This is important, because under the current protocols, surviving mice are rarely tested after 21 days because of time and personnel constraints. It should be noted, however, that even follow-up testing of the surviving mice in our study failed to identify two Y. pestis-infected flea pools. In conclusion, the rapidity of the pla PCR test (∼4 h), along with its sensitivity, makes it a suitable alternative to mouse inoculation assay for identifying infected fleas during surveillance activities and outbreak investigations.
ACKNOWLEDGMENTS
We thank Pam Reynolds and Ted Brown of the New Mexico Division of Health and Richard Grossman of the Larimer County Health Department (Colo.) for their assistance in collecting flea samples. We also thank Brook M. Yockey, Zenda L. Berrada, and Todd S. Deppe for their assistance in performing diagnostic serology tests and culture identification and William Black and three anonymous reviewers for their comments on the manuscript.
REFERENCES
- 1.Campbell J, Lowe J, Walz S, Ezzel J. Rapid and specific identification of Yersinia pestis by using a nested polymerase chain reaction procedure. J Clin Microbiol. 1993;31:758–759. doi: 10.1128/jcm.31.3.758-759.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu, M. C., J. E. Bracher, K. J. Moss, and B. M. Yockey. 1998. Unpublished data.
- 3.Chu, M. C., B. A. Wilmoth, L. G. Carter, and T. A. Quan. Laboratory manual of plague diagnostic tests, in press. World Health Organization, Geneva, Switzerland.
- 4.Englesberg E, Chen T H, Levy J B, Foster L E, Meyer K F. Virulence in Pasteurella pestis. Science. 1954;119:413–414. doi: 10.1126/science.119.3091.413. [DOI] [PubMed] [Google Scholar]
- 5.Gage K L. Plague. In: Collier L, editor. Topley and Wilson’s microbiology and microbial infections. 9th ed. New York, N.Y: Oxford University Press, Inc.; 1998. pp. 885–903. [Google Scholar]
- 6.Galyov E E, Smirnov O Y, Karlishev A V, Volkovoy K I, Denesyuk A I, Nazimov I V, Rubtsov K S, Abramov V M, Dalvadyanz S M, Zavyalov V P. Nucleotide sequence of the Yersinia pestis gene encoding F1 antigen and the primary structure of the protein. FEBS Lett. 1990;277:230–232. doi: 10.1016/0014-5793(90)80852-a. [DOI] [PubMed] [Google Scholar]
- 7.Hinnebusch J, Schwan T G. New method for plague surveillance using polymerase chain reaction to detect Yersinia pestis in fleas. J Clin Microbiol. 1993;31:1511–1514. doi: 10.1128/jcm.31.6.1511-1514.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hudson B W, Kartman L, Prince F M. Pasteurella pestis detection in fleas by fluorescent antibody staining. Bull W H O. 1966;34:709–714. [PMC free article] [PubMed] [Google Scholar]
- 9.Kartman L, Prince F M, Quan S F, Stark H E. New knowledge on the ecology of sylvatic plague. Ann N Y Acad Sci. 1958;70:668–711. doi: 10.1111/j.1749-6632.1958.tb35421.x. [DOI] [PubMed] [Google Scholar]
- 10.Kwok S. Procedures to minimize PCR-product carry-over. In: Innis M A, Gelfand D G, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 142–145. [Google Scholar]
- 11.Leal N C, Abath F G C, Alves L C, de Almedia A M P. A simple PCR-based procedure for plague diagnosis. Rev Inst Med Trop Sao Paulo. 1996;38:371–373. doi: 10.1590/s0036-46651996000500009. [DOI] [PubMed] [Google Scholar]
- 12.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 13.McDonough K A, Schwan T G, Thomas R E, Falkow S. Identification of a Yersinia pestis-specific DNA probe with potential for use in plague surveillance. J Clin Microbiol. 1988;25:2515–2519. doi: 10.1128/jcm.26.12.2515-2519.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poland J D, Barnes A M. Plague. In: Steele J H, editor. CRC handbook series in zoonoses. Boca Raton, Fla: CRC Press, Inc.; 1979. pp. 515–597. [Google Scholar]
- 15.Quan T J, Barnes A M, Polland J D. Yersiniosis. In: Barlows A, Hausler W J Jr, editors. Diagnostic procedures for bacterial, mycotic and parasitic infections. 6th ed. Washington, D.C: APHA, Inc.; 1981. pp. 723–745. [Google Scholar]
- 16.Sodeinde O A, Goguen J D. Nucleotide sequence of the plasminogen activator gene of Yersinia pestis: relationship to ompT of Escherichia coli and gene E of Salmonella typhimurium. Infect Immun. 1989;57:1517–1523. doi: 10.1128/iai.57.5.1517-1523.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomas R E, McDonough K A, Schwan T G. Use of DNA hybridization probes for detection of the plague bacillus (Yersinia pestis) in fleas (Siphonoptera: Pulicidae and Ceratophyillidae) J Med Entomol. 1989;26:342–348. doi: 10.1093/jmedent/26.4.342. [DOI] [PubMed] [Google Scholar]
- 18.World Health Organization. Passive hemagglutination test. WHO Tech Rep Ser. 1970;447:23–25. [Google Scholar]