Abstract
The need to address the challenge of vagueness across several domains of applicability of ontology is gaining research attention. The presence of vagueness in knowledge represented with description logic impairs automating reasoning and inference making. The importance of reducing this vagueness in the formalization of medical knowledge representation is rising, considering the vulnerability of this domain to the expression of vague concepts or terms. This vagueness may be addressed from the perspective of ontology modeling language application such as ontology web language (OWL). Although several attempts have been made to tackle this problem in other disease prognoses such as diabetes and cardiovascular diseases, a similar effort is missing for breast cancer. Minimizing vagueness in breast cancer ontology is necessary to enhance automated reasoning and handle knowledge representation problems. This study proposes a framework for reducing vagueness in breast cancer ontology. The approach obtained breast cancer crisp ontology and applied fuzzy ontology elements based on the Fuzzy OWL2 model to formulate breast cancer fuzzy ontology. This was achieved by extending the elements of OWL2 (a more expressive version of OWL) with annotation properties to fuzzify the breast cancer crisp ontology. Results obtained showed a significant reduction of vagueness in the domain, yielding 0.38 for vagueness spread and 1.0 for vagueness explicitness. In addition, ontology metrics such as completeness, consistency, correctness and accuracy were also evaluated, and we obtained impressive performance. The implication of this result is the reduction of vagueness in breast cancer ontology, which provides increased computational reasoning support to applications using the ontology.
Keywords: Breast cancer, Ontology, Semantic web, Fuzzy logic, Fuzzy ontology, Description logic and reasoning
Introduction
Cancer is a chronic disease caused by uncontrolled cell growth. Female breast cancer is a leading cause of cancer death in women, with a record high of 48,530 cases in the USA in 2020. New cases of cancer itself are surging worldwide and are expected to rise to 15.5 million in 2030 with distribution according to 20–34 (0.9%), 35–44 (6.0%), 45–54 (15.0%), 55–64 (20.8%), 65–74 (19.7%), 75–84 (22.6%) and above 85 (15.1%) years [1–4]. As a result, studies approached the problem of detection and diagnosis of breast cancer using computational methods such as computer-aided diagnosis (CAD) and artificial intelligence [5–8]. But not many efforts are channelled toward knowledge representation to aid knowledge-based systems in detecting breast cancer. We observed that most CAD-based solutions that apply a computational approach to tackling the problem depend on a kind of knowledge conceptualization (explicitly or implicitly) and representation to improve performance, accuracy, acceptance, and confident value. Recently, the use of logic such as fuzzy and description logics (DL) has gained significant research interest in supporting this knowledge formalization or representation of domain knowledge. Ontology, an outcome of the application of DL, has proven to be useful for this task.
Ontologies are a pillar of the Semantic Web considering their roles in supporting the formalization and representation of domain knowledge by providing constructs for formal relations. They allow for the organization of biomedical concepts to describe domains such as anatomy, environment, genetics and others. They have been widely used in a machine-processable way for enhancing reasoning with domain knowledge [9]. Specifically, domain ontology consists of a hierarchical description of concepts and the description of the properties of each concept's instances in the domain. Domain ontology is constructed using ontology languages like Web Ontology Language (OWL), consisting of OWL1 and OWL2 versions is a family of knowledge representation languages and have variants such as OWL Lite, OWL full, and OWL DL. Although OWL DL provides a richer construct, it is limited where the knowledge in a domain consists of vague definition or expression as typical with medical sciences. Hence, encoding domain knowledge using the language often produce a knowledge-based system full of vagueness and imprecision. Vagueness and uncertainty related to domain knowledge arise from peculiarities of the domain, context-dependency or differences in experts’ conceptualization of a domain [10].
Vagueness is formally defined as a semantic phenomenon that is exhibited through predicates that supports borderline cases [11] and may appear as either degree or combinatory vagueness. A study describes degree vagueness as a lack of precise boundaries, while combinatory vagueness may be identifiable by lack of sharp discrimination between combinatorial conditions [12]. We acknowledge that vagueness needs to be disambiguated from similar notions such as uncertainty, inexactness, and ambiguity. Although, both vagueness and uncertainty remain a challenge that has motivated research gap interests in knowledge representation [13]. To demonstrate the existence of vagueness in the domain, consider the following: the outcome of Positron Emission Tomography (PET) scans may be reported as normal, abnormal Lymph Nodes, advanced PET Scan; urokinase plasminogen activator and its inhibitor are reported as higher (2.81–5.7), borderline (0.41–2.8) and normal (0.0–0.4) levels; carcinoma is defined as well differentiated, moderately differentiated, or poorly differentiated; results for estrogen receptor (ER) and progesterone receptor (PR) are as negative, weakly positive, positive, percent positive, and whether the staining is weak, moderate, or strong; all these are strong indications of the existence of a high level of vagueness in the domain.
Attempts have been made using fuzzy logic and probability [14] to address this gap in the literature. Fuzzy logic has gained research focus in tackling borderline cases of vagueness and cases of imprecision. The limitation of natural language in addressing the challenge of vagueness using a function that assigns fuzzy truth value has made it possible for the use of fuzzy logic [15–18]. Fuzzy ontologies are an extension of crisp ontologies through the addition of fuzzy elements like fuzzy concept, fuzzy object property, fuzzy data property, and fuzzy datatype [19]. The use of ontology annotation with fuzzy logic is applied to combine both crisp ontology (CO), crisp ontology instances (COI), fuzzy ontology (FO), fuzzy ontology instances (FOI) into a fuzzified ontology FO = (CO, FO, COI, and FOI) [20]. Traditionally, plugins such as Fuzzy OWL2, Fuzzy Protégé, and Extension of Protégé for automatic fuzzy (EPAF) ontology building may be used in developing fuzzified ontologies [20–22]. Fuzzy OWL2 allows for better expressivity of concepts in ontology to reduce vagueness. It supports the use of fuzzy membership function as a mechanism for modeling concepts with vagueness. These member function assigns to every presumably vague concept x a degree of truth, measured as a value in a truth space. Examples are left-shoulder, right-shoulder, trapezoidal, and triangular.
Although crisp ontology of breast cancer has been earlier developed in [23], however since the issue of vagueness was not addressed, we found that the computational reasoning process was impaired. In addition, whereas studies exist using fuzzy logic for breast cancer-based CADs [4, 24–27], but none is reported on fuzzification of breast cancer ontology. As a result, this study found it relevant to consider fuzzy logic in addressing the reduction of vagueness from domain knowledge used in knowledge representation for breast cancer. This study handles both degree and combinatory vagueness through the use of fuzzy ontology. This study aims to improve computational reasoning by reducing vagueness in the domain by fuzzification of crisp ontology. This is achieved using OWL2, which provides annotations on ontologies, axioms, and entities, which are then used to fuzzify the crisp ontology. We leveraged Fuzzy OWL2, FuzzyDL, Hermit/Pellet reasoners, and Protégé editor to develop crisp and then the fuzzification of OWL2 ontology.
The following are the contributions of the paper:
-
i
Development of a Fuzzy OWL-2 framework to improve computational reasoning on crisp ontology by reducing vagueness in the domain of breast cancer by the fuzzification of crisp ontology.
-
ii
Identification and implementation of a fuzzy concept, fuzzy data property (attribute), fuzzy relation, datatype, and modifiers to achieve a novel breast cancer fuzzy ontology using Fuzzy OWL2
The remaining part of this paper is organized as follows: Sect. 2 presents a review of related studies; Sect. 3 discusses the proposed framework; implementation and querying of the fuzzy ontology are presented in Sect. 4. We evaluated the quality of the resulting ontology and presented results and findings in Sect. 5; finally, the conclusion of the study is discussed in Sect. 6.
Related works
This section presents studies focused on the development of both fuzzy and non-fuzzy ontologies. The aim is to allow readers to understand research efforts channeled toward the reduction of vagueness in ontology and the negligence of such vagueness in other studies. We emphasize their approaches, contributions and limitation. In addition, the presentation of the studies is done in chronological order.
Dhingra and Bhatia, in their study, presented a strong argument for the need for the development of the ontology, related tools, query languages for knowledge formalism and representation. The authors proposed three ontologies for the domain of laptop usage. Their aim for this is to motivate effective mechanisms to curb problems associated with information retrieval. The study applied the use of DL query languages to validate the ontologies created [28]. Although the development of ontologies was advocated in the study, no consideration for tackling vagueness was reported. In a related work focused on ontology development alone, Chen and colleagues developed an ontology for diabetic patients to aid doctors in reaching an efficient diagnostic decision. They combined the ontology with Semantic Web Rule Language (SWRL) to populate the knowledge base with 766 cases, of which only 269 records had diabetes [29]. Whereas this study supports inference making through rule execution, we argue that the solution does not address the challenge of vagueness in their ontology. In an attempt to propose a CAD-based system driven by the case-based reasoning (CBR) paradigm, Oyelade & Ezugwu developed a COVID-19 ontology for formalizing and representing domain knowledge and case-level representation. The ontology assists the CBR reasoning framework for effective decision-making. The ontology was instantiated with a record of 71 cases [30]. While these contributions are impressive considering the case-level modeling using ontology, the crisp formalism reveals the omission to mitigate the issue of vagueness in the domain.
Considering the limitation of studies in [28–30] in addressing the challenge of vagueness, we, however, found some studies which made some attempts. For instance, in a study, Parry emphasized the need to handle vagueness through the postulation of fuzzy ontology. The author revealed that when the value for the degree of membership is added to every vague term, the ontology would have attained fuzzification, hence eliminating vagueness. The study applied its fuzzified ontology to the problem of corpus analysis and the use of a relevance feedback mechanism [31]. In a related study, the same author handled document structure identification using fuzzy ontology for knowledge representation in medical sciences. The fuzzified ontology was applied to an intelligent system that allows a group of users to identify the information they find useful [32]. While the two studies appeared to handle vagueness in ontology, we note that the method is inefficient. Gu and colleagues fuzzified a crisp ontology using the fuzzy instance relation, fuzzy concept relation, and fuzzy concept base relation fuzzy concepts element. The resulting fuzzy ontology was applied to reduce uncertainty in domain knowledge with the hope to improve domain reasoning on the knowledge base [33]. The approach of the study is interesting and presents the potential for reduction of vagueness in the ontology.
Alexopoulos et al. developed an enterprise-based fuzzy which modeled knowledge about the operation of a consulting firm. The authors claimed that the fuzzification of the ontology tackled vague knowledge in the domain, thereby aiding computational reasoning on when to write or not write a proposal for a tender call [11]. We found the presentation of the method used in the study to be unclear. Asma and Zizette fuzzified ontology to represent and classify individuals. The fuzzied ontology aims to allow for a reasoning mechanism capable of grouping individuals in their respective hierarchies [34]. Also, Fouda et al. proposed fuzzy ontology, modeled after the Disease Ontology (DO), for the cardiac arrhythmias disease to reduce the obvious level of real and uncertainty cases as seen in the domain. The authors achieved this using the combination of OWL2 and FuzzyOWL2 plugin in Protégé [35]. The approach used in the study is standard and proof capable of addressing the problem. A similar approach was applied to reducing vagueness in a study on the development of ontology on diabetes. The authors proposed a fuzzy case-based OWL2 ontology for use in a CBR framework. To demonstrate the usability of the ontology in a CBR framework, the fuzzy ontology was populated with 60 real diabetic cases [36].
Furthermore, Elhefny and colleagues developed a fuzzy ontology to represent obesity-related cancer. Using OWL2, the fuzzy ontology tackled the overlapping or vague concepts in the domain. The study was also able to cope with the linguistic domain variables, which were impossible using crisp ontology and query executed through the fuzzy DL reasoner [37]. Lastly, a study related to Fouda et al. proposed a novel case-base fuzzy OWL2 ontology and claimed to be the first fuzzy case-base ontology in the medical domain. The fuzzy ontology was based on a case-based fuzzy Extended Entity Relation (EER) data model. Fuzzified elements consist of fuzzy concepts/classes, fuzzy object properties, fuzzy data, and fuzzy data. They applied SPARQL-DL for query operation after instantiating the ontology with 60 cases [38]. Studies in both [37, 38] present an interesting approach for developing a medical ontology, which supports computational reasoning for the diagnostic process. In addition, they attempted to reduce the vagueness in their ontology using fuzzy logic.
Although we found studies addressing the need to reduce vagueness in ontologies on diabetes, obesity, and cardiac arrhythmias in medical sciences from 2004 to 2020, no report presented on breast cancer. In addition to the need to reduce vagueness in breast cancer ontology, a method capable of lowering the vagueness to the minimum is needed. This study is aimed at addressing this research gap.
Methods
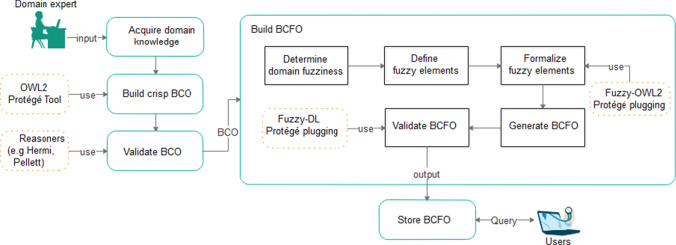
The representation of the proposed breast cancer ontology (BCO) and the improved breast cancer fuzzy ontology (BCFO) are presented in this section. We first describe the framework designed to guide the proposed work and then present the structure of the BCFO. In Fig. 1, the proposed framework showing the flow processes for the development of the proposed BCFO is presented. This study acquired the existing BCO ontology as presented in [39, 40]. However, this study found that the acquired BCO requires updating and improvement to allow omitted concepts and relations. The resulting BCO became input into the methodologies for the development of the BCFO.
Fig. 1.
Proposed framework for developing BCFO knowledge from BCO using Fuzzy OWL-2 plugging and Protégé
The BCFO follows the methodologies described in [11] to achieve its objective as described in the containing box on the right-hand side of Fig. 1. The outcome of the framework is the desired fuzzy ontology that may be queried using a semantic web-based query language. In addition, the resulting ontology may be applied to intelligent systems developed to aid the diagnosis of breast cancer.
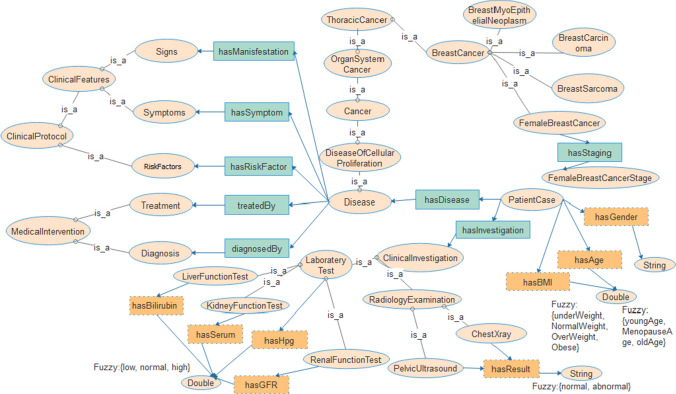
Following the disease ontology (DO) [41], we generated a structure for formalizing the proposed breast cancer fuzzy ontology. Figure 2 shows the major classes, the object and datatype properties and the use of the fuzzy OWL-2 plugging to add the fuzzy fragment of the breast cancer fuzzy ontology. The figure presents a wider view of the crisp ontology and a fragment of the fuzzy ontology. The figure aims to give an overview of the basic skeletal representation of the BCFO rooted in its source, the BCO. We identified some terms or concepts which may need fuzzification, especially those with numeric values and other datatype properties and even concepts. These fuzzified elements are detailed in Sect. 3.3. For brevity, we have excluded some other concepts, object and data properties and present details of the two fragments of the ontology in Sect. 4.
Fig. 2.
The structure of the BCO and BCFO as an addendum
The crisp breast cancer ontology
The crisp breast cancer ontology followed the structure of the DO and was corroborated with the provision in breast cancer-based clinical practice guidelines (CPG) in [42, 43]. This crisp ontology became the foundation for the proposed BCFO, making it a special case of a fuzzy ontology. The BCO does not allow for modeling vagueness; hence, all representations such as classes, object and datatype properties, and datatype belong to their membership class to a degree equal to 1. Therefore, we exploited this limitation to develop it further by identifying crisp elements that can be fuzzified to achieve BCFO.
Need for the fuzziness of breast cancer ontology
The need for fuzzification of the proposed BCO became necessary in an attempt to tackle the problem of vagueness resulting from the domain of consideration. In our previous work [39, 40], we realized that the BCO ontology is mostly limited and will be inefficient when vague descriptions are presented to the underlying intelligent system been supported. As a result, this study is focused on modeling vagueness without provision for handling uncertainty. Uncertainty often includes cases of probability, ambiguity, or inexactness, which we will leave for applying other techniques in solving them.
Definition of fuzzy ontology elements
This section presents fuzzy elements grouped into fuzzy concepts and fuzzy properties/relations, which allows for modeling both degree vagueness and combinatorial vagueness [10]. Our focus for the crisp ontology fuzzification is on object properties, data properties and data types contained in it. Therefore, we shall demonstrate the application of fuzzy elements such as fuzzy data types, weighted sum concepts, weighted concepts, fuzzy nominals, and Zadeh logic.
Representation of fuzzy concepts
Fuzzy concepts are so named because they are expected to contain at least fuzzy property. The need for the fuzzification of crisp concepts allows for the specification and representation of concepts or classes that belong to another class (superclass) to some degree. For instance, we may want to say that a patient case is diagnosed with breast cancer to a certain degree. We have the PatientCase class and the subclass SuspectiblePatientCase, which belongs to the parent class to a certain degree. The preceding describes concepts whose instances may be its members to a certain degree. As another example, consider classifying patients based on menopause status such as YoungPatientCase, MenopausePatientCase, OldPatientCase. Certainly, we know that these classes will be allocated vaguely due to different opinions held about young or old. Considering the preceding, a concept may have attribute values with no vagueness, such as “age” as a linguistic variable taking values “young,” “middle-aged,” or “old” defined by means of fuzzy numbers.
In addition to the provision for modeling fuzzy concepts, we can also achieve the fuzzification of concepts that have already been declared fuzzy but having more properties such as weight, modified with adjectives, and summation of concepts with weights. These categories of fuzzy concepts are the weighted concepts and the weighted sum. Whereas the former describes a fuzzy concept that is being weighted, the latter is created through the formation of a union from the former. The two cases are referred to as Weight Concepts and Weighted Sum Concepts. The need for weighted concepts may arise when we assign a weight to an already defined fuzzy concept. For instance, consider the fuzzy concepts: FemaleBreastCancerSignAssumed, FemaleBreastCancerRiskFactorAssumed, and FemaleBreastCancerSymptomAssumed, which may be assigned weights 0.3, 0.2, and 0.5, respectively, all summing to 1.0. Each of the weighted concepts has an impact on the overall diagnosis of patients with breast cancer. However, their weighted value on the overall decision is being regulated by their individual weights.
Representation of fuzzy relations and data types
Two types of relations are considered here, namely the fuzzy object property and the fuzzy datatype property. These relations exist between entities in ontology and may associate more than one paired entity. In our proposed BCFO, an object relation or property may associate two or more classes with different degrees of strength. For instance, a fuzzy object property is useful when there is a need to state that an instance of a class belongs to that class to a particular degree. As an example, patiencase01 (an instance of PatientCase, a crisp class) hasStage FemaleBreastCancerStage at a degree of 0.6, and another case where patiencase01 hasStage FemaleBreastTumorStage at a degree of 0.4. We see that the hasStage object relation associates two different entities at different degrees. Hence, we can classify hasStage as a fuzzy object relation.
The second fuzzy property is the fuzzy datatype property or attribute. Whereas the fuzzy object property links instances of two classes, the fuzzy datatype property links the instance of a class to literal at a given degree. It also associates instances of a class to fuzzy data types. An example for the former is patientcase01 hasOccupation ‘Trader’ at a degree of 0.8, while an example for the latter is patientcase01 has-Fuzzy-Age oldAge.
Another scenario requiring the use of fuzzy datatype or concrete role is the representation of crisp numerical values with linguistic values. For example, in Fig. 2 a PatientCase may have age using the hasAge data property. But we know that the age term may also assume a fuzzy definition ranging from {youngAge, menopauseAge, oldAge}. Hence, a concrete role otherwise known as fuzzy datatype property will be created for relating each of the numeric values to their linguistic values. As another example, we consider the BMI representation such that four fuzzy datatype data are created: hasUnderweight, hasNormalweight, hasOverweight, and hasObese. Using pre-defined fuzzy data types (to be defined shortly), underweight, normalweight, overweight, and obese. The fuzzy datatype properties will use the fuzzy data types as a range, while the class BMI is used as a domain. An example of usage is BMI hasUnderweight underweight. Another example is the age declaration which may have some form of vagueness where patience’s age may be declared as falling into this range: youngAge, menopauseAge, oldAge. Three fuzzy object properties are applicable here which are called hasYoungAge, hasMenopauseAge, and hasOldAge. Assuming the creation of a crisp class Age, we then write the axiom Age hasYoungAge youngAge.
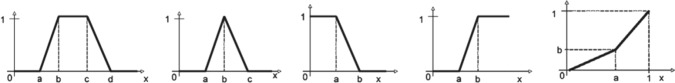
In the case of fuzzy data types, we have chosen to represent vague terms by using linguistic terms. We define fuzzy predicate using the functions in Fig. 3 over the interval [k1, k2], where k1, k2 are positive integers representing both minimum and maximum inclusive values. The trapezoidal function, triangular function, left-shoulder function, right-shoulder function, linear, and mod are illustrated in Fig. 3. In addition to k1, k2, we also need to state values for a, b, c, d.
Fig. 3.
Fuzzy functions used for representing fuzzy datatype are as follows: trapezoidal function; triangular function; left-shoulder function; right-shoulder function; linear function, respectively
The need to represent fuzzy data types became necessary due to the availability of numerical values in ranges. For example, in crisp ontology, patient blood pressure for the systolic and diastolic are written as 90–120 and 60–80, respectively. The cases of fuzzy terms as used in Fig. 2 such as {normal, abnormal} for a chest x-ray and abdominal pelvic ultrasound, {low, normal, high} for renal, liver and kidney tests, {underweight, normalweight, overweight, obese} for BMI and lastly {youngAge, menopauseAge, oldAge} for Age require the use of fuzzy data types for encoding. In the following paragraphs, we shall be defining the range, member function, equation and shape of items needing representation using fuzzy data types. After considering input from domain experts and consultation from domain-based clinical practice guidelines (CPG), range values were carefully chosen. Member function graphs were plotted using Python skfuzzy [44] library.
BMI: People having BMI < 19 are classified as underweight; 19.5–24.0 are classified as normal weight; 25–29 are classified as overweight, and > 30 are classified as obese; Considering the BMI linguistic variable, we have four linguistic values listed below and the range of their values. Taking the minimum BMI any person can have to be 0 and maximum as 250 for k1 and k2, then the following:
- underweight
= { (18.5,1), (19.5,0)} = > hasUnderweight (left shoulder (0, 250, 18.5, 19.5)).
- normal
= { (18.5,0), (19.5,1), (24,1), (25,0)} = > hasNormal (trapizoidal (0, 250, 18.5, 19.5, 19.5, 24.5)).
- overweight
= { (24,0), (25,1), (29,1), (30,0)} = > hasOverweight(triangle (0, 250, 24.0, 25.0, 29.0, 30.0)),
- obese
= { (29,0), (30,1)} = > hasObese (right shoulder (0, 250, 29.0, 30.0)).
The BMI linguistic variable, which is expected to have numerical features, has four concrete roles defined for each of its linguistic values hasUnderweight, hasNormal, hasOverweight, and hasObese, so that BMI is the domain and the four linguistic values are the ranges. Henceforth, the fuzzification function for numeric values in the ontology will be model using Eqs. (1), (2) and (3) based on the values of a and b.
| 1 |
when c is given:
| 2 |
when d is given:
| 3 |
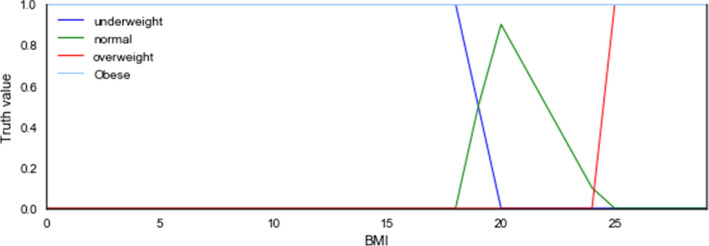
This study model each numeric attribute or linguistic variable to have datatype properties of the form has-Fuzzy-linguistic variable, e.g. has-Fuzzy-BMI, so that the numeric value of BMI is assigned to it. An example of an axiom using datatype property is patientcase01 has-Fuzzy-BMI 22. Since BMI has numeric value 22, it is modeled as funderweight(22) = 0, fnormal(22) = 0.5, foverweight(22) = 0, and fobese(22) = 0. Since fnormal(22) has the highest value, BMI representative linguistic results to normal, hence has-Fuzzy-BMI = normal. Therefore, the fuzzification of the numeric value is as follows:
The fuzzy member function is shownin Fig. 4 as follows:
Fig. 4.
Fuzzy member functions for BMI numerical attribute fuzzification
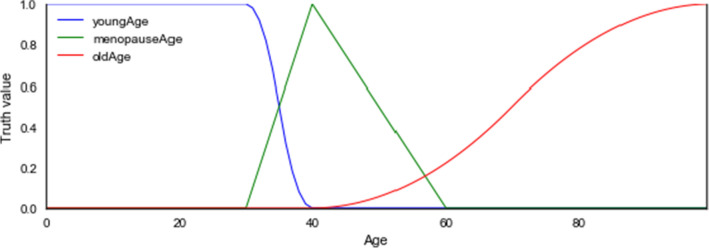
Age: We assumed that patients under the age of 40 are classified as young; those within 40–60 are classified as menopausal age; 60 and above are classified as old. Considering the Age linguistic variable, a numeric attribute, we have four linguistic values listed below and the range of their values. Taking the minimum Age to be 0 and maximum as 120 for k1 and k2, respectively, the following hold:
- youngAge
= { (30.0,1), (40.0,0)} = > hasYoungAge (left(0, 120, 30.0, 40.0)).
- menopauseAge
= { (30.0,0), (40.0,1), (60,0)} = > hasMenopauseAge (triangle(0, 120, 30.0, 40.0, 60.0)).
- oldAge
= { (60,1), (100,0)} = > hasOldAge (right(0, 120, 60.0, 120.0)).
The Age linguistic variable, which is expected to have numeric feature, has three concrete roles defined for each of its linguistic values: hasYoungAge, hasMenopauseAge, and hasOldAge. Age is the domain, and the three linguistic values or data types are the ranges.
The fuzzification of the numerical value that a patient case may present is as follows:
and the fuzzy member function is shown in Fig. 5 as follows:
Fig. 5.
Fuzzy member functions for Age numerical attribute fuzzification
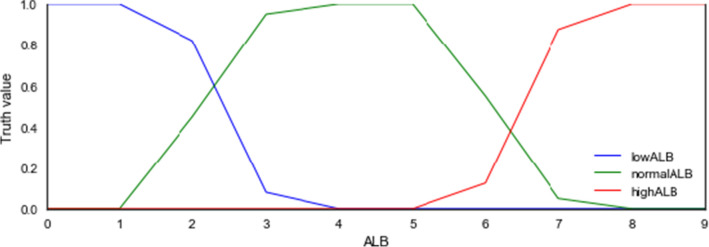
Liver Function Tests (LFTs): Consists of a set of tests carried out to check the liver's health through blood samples. These tests include alkaline phosphatase (ALP), prothrombin time/international normalized ratio (PT/INR), albumin, total protein, Gamma-glutamyl transferase (GGT), bilirubin, alkaline phosphatase (ALP), alanine aminotransferase (ALT) [45]. For instance, the range of values considered are low, normal and high for albumin (ALB) test are 1.4-3.4 g/dL, 3.4–5.4 g/dL and 5.5–7.4 g/dL respectively [46]. Taking the minimum of ALB as 0 and maximum as 10 for k1 and k2, then the following holds:
- lowALB
= { (1.1,1), (3.4,0)} = > hasLowALB (left(0, 10, 1.4, 3.4)).
- normalALB
= { (1.1,0), (3.1,1), (5.1,1), (7.1,0)} = > hasNormalALB (triangle (0, 10, 1.1, 3.1, 5.1)).
- highALB
= { (5.5,1), (7.5,0)} = > hasHighALB (triangle(0, 10, 3.1, 5.1, 7.1)).
The ALB linguistic variable which is expected to have numeric feature has three concrete roles defined for each of its linguistic values lowALB, normalALB, and highALB, so that ALB is the domain and the three linguistic values or data types are the ranges.
The fuzzification of the numeric value is as follows:
and the fuzzy member function is shown in Fig. 6 as follows:
Fig. 6.
Fuzzy member functions for ALB numerical attribute fuzzification
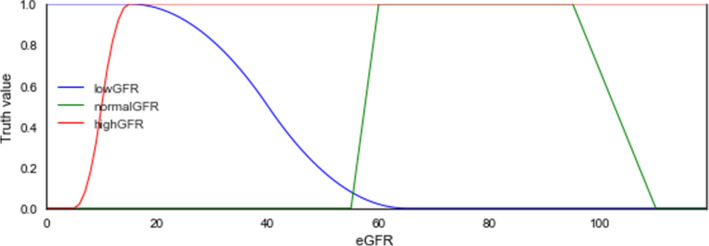
Kidney Function Tests (KFTs): Test for the performance state of the kidney is usually done by estimated glomerular filtration rate (eGFR), which is computed by the measurement of serum creatinine levels in combination with age, sex, weight or race [47]. However, measurement of eGFR is not sufficient alone to determine the kidney's health status, hence the need for other kidney function tests. These tests include urinalysis, serum creatinine test, blood urea nitrogen (BUN). A normal eGFR should be > = 60, 20–60 means low, and < = 15 is high [2, 48]. Considering the values of 0 and 120 for minimum and maximum as it relates with k1 and k2, then these follow:
- lowGFR
= { (15.0,1), (65.0,0)} = > hasLowGFR (left(0, 120, 15.0, 65.0)).
- normalGFR
= { (1.1,0), (60.0,1), (95,1), (7.1,0)} = > hasNormalGFR (trapizoidal (0, 120, 55.0, 60.0, 95.0, 110.0)).
- highGFR
= { (5.0,1), (15.0,0)} = > hasHighGFR (right(0, 120, 5.0, 15.0)).
The eGFR linguistic variable which is expected to have numerical feature has three concrete roles defined for each of its linguistic values lowGFR, normalGFR, and highGFR. This allows for eGFR to be used as the domain and the three linguistic values or data types are the ranges.
The fuzzification of the numeric value is as follows:
and the fuzzy membership function is shown in Fig. 7 as follows:
Fig. 7.
Fuzzy member functions for eGFR numerical attribute fuzzification
There are several other numerical values modeled in the ontology. These include blood tests such as creatinine levels, blood urea nitrogen, electrolytes, complete blood count, and rate of coagulation. Others are urinalysis, which measures the level of proteinuria; tumor markers, quantifying substances produced by cancer cells blood or urine, includes Alpha-fetoprotein (AFP), Lactate dehydrogenase (LDH), Prostate-specific antigen (PSA), CA 125, CA 19–9, CA 15–3, CA 27–29, Carcinoembryonic antigen (CEA), and Neuron-specific enolase (NSE) [49]. For brevity, the formalism given above will suffice to illustrate these numerical values are encoded using corresponding fuzzy values.
Representation and application of fuzzy modifier to concepts, data types and relations
The cardinal objective of introducing the OWL2 is to improve the web ontology language's expressivity in modeling de-vagued ontology. Fuzzy OWL2 plugin [50] made provision for construct allowing for the use of modifier. In addition to enhancing ontology expressiveness, modifiers also improve semantic queries. The fuzzy modifier is denoted using a mod function that changes a fuzzy set's membership to triangular or linear. Fuzzy modifiers are used to add semantics to existing constructs like data types.
Considering the domain of the proposed BCFO, some reoccurring terms suitable for representation as a modifier were obtained, namely severe, very, mild, advance, which are defined using fuzzy modifiers as linear (0.9), linear (0.8), linear (0.3), and linear (0.7), respectively. For instance, consider some of these fuzzy modifiers as they are applied to fuzzy concepts, fuzzy data types, and fuzzy relations already created in Sects. 3.3.1–3.3.2. The representations in Code Listings 1, 2, 3, and 4 describe the creation of the modifiers severe, very, mild, and advance, respectively.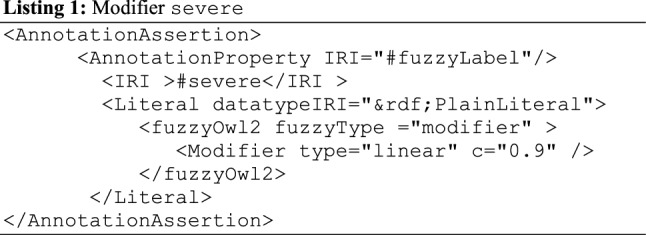



Modified concept: We consider an example of advance in the use of stating an advance BreastCancerPatientCase which is a subclass of PatientCase. This is represented in Listing 5.
We could choose to describe the case of a patient whose BMI value is very normal:
very(PatientCase П hasBMI.Normal)
-
b.
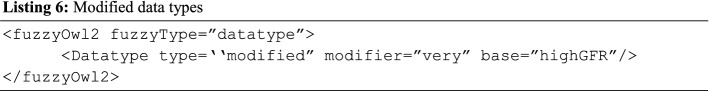
Modified data types: We earlier mentioned that eGFR lower than 15 indicates a high tendency of kidney failure. But assume a value of 7 will require using the modifier very to represent it in the BCFO. Hence, Listing 6 details the representation.
-
c.
Modified relations: to represent modified fuzzy relations like datatype properties, a modifier, say severe, can be used to note that a patient case ALB is severely low. This is formalized, as shown in Listing 7.
There are more cases of applying such modifiers in the fuzzy ontology. However, the few examples given above may suffice to describe the representation of modifiers in the ontology.
Representation of fuzzy logic used in breast cancer fuzzy ontology
As mentioned in the introduction of this section, this study applies its fuzzification task through Zadeh fuzzy logic. The motivation for this selection is based on the foundational nature of Zadeh over others as an example, and the Łukasiewicz many-valued logics are derived from Zadeh algebra [51], designed initially for modeling vague notions [52]. Fuzzy logic used in the proposed ontology is modeled using [20] Listing 8.
Where options for the replacement of fuzzy logic may assume any of these:
<FUZZY_LOGIC>:= "lukasiewicz" | "zadeh"
And the fuzzy logic representation as in Listing 9:
Validation of the breast cancer fuzzy ontology
Using the fuzzy ontology reasoners Fuzzy DL [53], similar to DeLorean [54], we tested the consistency of the ontology (BCFO). This is necessary to ensure that the knowledge of the domain is not wrongly represented even though correctly elicited from the domain expert. Similarly, we had previously applied OWL-based reasoners such as Pellet [55], Fact ++ [56], and Hermit [57] for validation of the crisp ontology. The result of the validation for the two cases shows that our ontology is consistent. To demonstrate ontology inconsistency, we allow for misrepresentation, thereby making the reasoner detect inconsistency. Once the inconsistency was removed, the reasoner confirmed its consistency.
Implementation and experimentation on the fuzzified ontology
The implementation and experimentation for ontology development and query were achieved using Protégé editor version 4.3.0, build 304. This editor uses the Java platform JVM 1.6.0_26-b03 and is pre-shipped with other plugins such as Cajun Visualization Plugin, Dlquery, OWLViz, SPARQL Plugin, HermiT Reasoner 1.3.8, the OWL API 3.4.2, and Factplusplus Plug-in 1.6.2. We added two plugins, namely FuzzyOWL 0.1.0 and Fuzzy DL, to provide support for the fuzzification process. The host system has a configuration of Intel (R) Core i5-4210U CPU at 1.70 GHz, 2.40 GHz; RAM size of 8 Gbytes on a Windows 10 OS version.
Implementation of the crisp and fuzzy ontology with protégé
The BCFO proposed in this study is built on the disease ontology (DO) [41]representing human disease information. The DO, which semantically integrates with other standard ontologies such as MeSH, ICD, NCI’s thesaurus, SNOMED and OMIM to enrich its disease and medical vocabularies, has proven useful in the biomedical community and other studies [38, 58]. The motivation to build the BCFO on DO is generated from its standards and provision for consistency and sustainable descriptions of human disease terms.
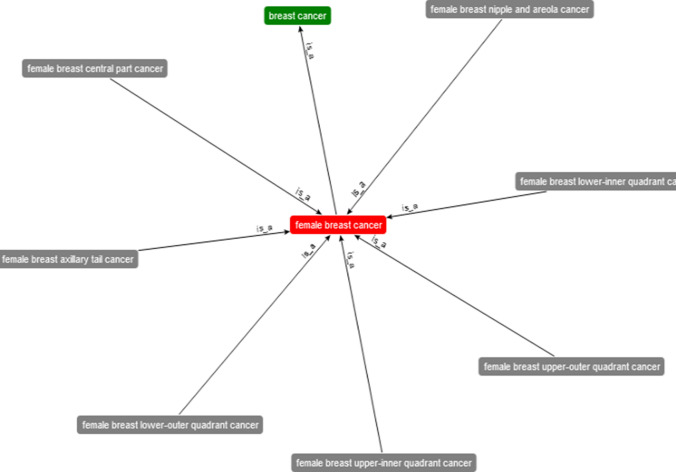
The root class in the DO is the disease concept that subclasses disease by an infectious agent, disease of anatomical entity, mental health disease, metabolism, cellular proliferation, and others. The domain of consideration in this study is rooted in the disease of cellular proliferation class. Cancer is one of the subclasses, and it relates with other subclasses such as organ system cancer. Furthermore, we found breast cancer as one of the subclasses, which in turn subclass female breast cancer. For brevity, we present a visualization of subclasses associated with the domain of interest, as shown in Fig. 8. Interestingly, we found that even the female breast cancer class has different types (subclasses)–providing our proposed BCFO with a rich, crisp ontology.
Fig. 8.
Crisp ontology of domain-based knowledge representation of breast cancer
Crisp ontology
The ontology development process for the crisp ontology acquired an existing ontology for improvement and enrichment. This study embarked on this enrichment considering two observations made on the existing ontology: (a) that the existing ontology was initially built as a lexicon of terms with no provision for associations of concepts with properties (object and datatype), (b) there was some observed structural deficiency in the existing ontology which may yield a less optimal solution to any underlying expert system being supported and (c) a further consideration of related CPG revealed that not the existing ontology captured sufficient domain knowledge.
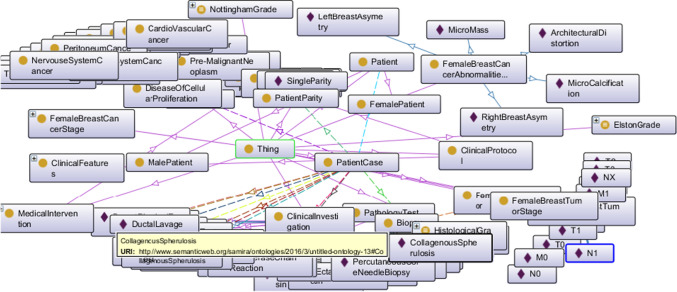
Efforts were therefore made in this study to tackle the limitations of the acquired breast cancer ontology. A well-structured crisp ontology was actualized as shown in Fig. 9, which shows a detailed listing of the classes in a coordinated hierarchical structure.
Fig. 9.
Proposed crisp ontology of breast cancer with a detailed listing of classes and the subclasses in addition to a visualization display of the complete ontology showing classes and some of their instances
The top-level classes consist of Patient, PatientCase (the latter describes patients case as modeled by patient’s hospital file while the former describes the patient), Disease (detailing the structure acquired from the DO mentioned in Sect. 3), MedicalIntervention (a class aimed for the description of the two major interventions patients often receive namely treatment and diagnosis), ClinicalInvestigation and ClinicalProtocol classes differ in the sense that one aim at collecting some categories of investigations such as LaboratoryTest, BloodMakerTest, GeneticTest, MolecularTest, PathologyTest, and other tests or investigations, the other models well-known domain knowledge such as Symptom, RiskFactor, and Sign/Manifestation. We note that most of these classes were further redefined to adhere to the domain of consideration in this study. For instance, the Symptom, RiskFactor, and Sign or Manifestation have subclasses such as FemaleBreastCancerSymptom, FemaleBreastCancerRiskFactor, and FemaleBreastCancerManifestation. This became necessary because concepts may inter-relate with other diseases like female breast cancer, though some are strictly used in the field of interest.
In Table 1, a detailed listing of the object and data properties are shown. Note that these properties were initially denoted by associating lines among classes in Fig. 9
Table 1.
A listing of object properties and data properties of the crisp ontology
| ObjectProperty | DataProperty | ||
|---|---|---|---|
|
diagnosedBy hasAgeVariable hasBMIVariable hasBiopsyTest hasBloodMakerTest hasBreastTumor hasBreastTumorStage hasDaignosis hasDisease hasE-cadherinTest hasFuzzyVariable hasGeneTest hasHeightVariable hasInvestigation hasKidney hasLiverTest hasManifestation |
hasMolecularTest hasParity hasPathologicalGrade hasPatientCase hasRadiolgyExamination hasRenalTest hasRiskFactors hasStage hasStaging hasSymptom hasTNM suspectibleHasStage treatedBy |
ageAtFirstChild ageAtMenopause hasALBCrisp hasAdenosis hasAgeCrisp hasApocrineMetaplasiaCysts hasAtypicalDuctalHyperplasia hasAtypicalLobularHyperplasia hasAverageHeight hasBCIPredictive hasBCIPrognostic hasBMI hasBMICrisp hasBloodSugarLevel hasCalciumLevel hasChlorideLevel hasColumnarCellChange hasComplexSclerosingLesion hasCrispValue |
hasDuctalCarcinomaInSitu hasEstrogenReceptor hasFuzzyDataProperty hasGFRCrisp hasGender hasGlandFormation hasHER2Level hasHeightCrisp hasHighALB hasHighGFR hasIn-SituLobularCarcinoma hasIntraductalCarcinoma hasKi-67 hasLiverEnzymesLevel hasLobularCarcinomaInSitu hasLowALB hasLowGFR hasLymphNodeFeature hasM |
Fuzzy ontology
Clearly, classical ontologies developed for different domains are mainly crisp and inefficient in handling difficult expressions with imprecision and vagueness. A fuzzy ontology, like the one proposed in this study, will exploit the concept and theories of fuzzy logic to overcome these limitations. To implement such fuzzy ontology for the female breast cancer situation, this study approached this by creating a fuzzy OWL ontology and fuzzy OWL2 representation. Existing annotation properties in OWL2 are backwardCompatibleWith, comment, deprecated, incompatibleWith, isDefinedBy, label, priorVersion, seeAlso and versionInfo. The fuzzy OWL2 representation provides additional annotation property named fuzzyLabel to encode its fuzzification. This is done in such a manner as to allow wrapper annotation tags around elements in the ontology.
Considering a wide range of crisp ontology elements that require fuzzification, each has been fuzzified turn as detailed in Sect. 3.3. The guiding principle for the fuzzification process is to recognize fuzzifiable terms such as the linguistic variable and their corresponding values. For instance, linguistic variables consist of items like Age, BMI, TotalProtein, PT/INR, GGT, ALB, albumin, Height, ALT, ALP, GFR, and several others. Also, some linguistic values fuzzified are overweight, underweight, obese, normal, oldage, youngage, menopauseage, nromalGFR, highGFR, lowGFR, normalALB, highALB, lowALB, normalALP, shortHeight, tallHeight, averageHeight, highALP, lowALP, normalALT, lowALT, hightALT, and others. We fuzzified datatype properties or fuzzy attributes such as severeHasLowALB. After completing the fuzzification process, we realized our BCFO has 55 fuzzy datatype properties, 34 fuzzy datatypes, and modified fuzzy data types, datatype properties, and existing classes, object properties and datatype properties in the crisp ontology.
Now that the fuzzy ontology structure is constructed, developed, and certified to be consistent, we approach the next phase of our development, populating the BCFO with instances.
Instantiation of the fuzzy ontology
The approach used for the population of the BCFO ontology with patient cases differs from the approach used in [38, 40, 58]. The instances of the cases and the instances of its describing features of their fuzzy ontologies were populated using either manual method or relational database. This study proposes a novel approach using the hybridization of data structures. The justification of this approach, which differs from the relational database approach, is to avoid data duplication in two different data representation formats. We argue that while the approach of [36] presents a good mapping of patient case data from a relational database to ontology, it suffers from the computational cost of data translation. The hybridization of using data structures to store patient cases before populating the BCFO is efficient, providing a good mapping between the data structure and the ontology with a reduced computational cost. The outcome of this approach is to store both the taxonomy and instances in the ontology structure so that the traditional TBox and ABox compartment are maintained. Whereas Sects. 4.11 and 4.1.2 describes the taxonomy box (TBox) of the BCFO, the instantiation, known as ABox (assertion box), for both crisp and fuzzy elements is presented in this subsection.
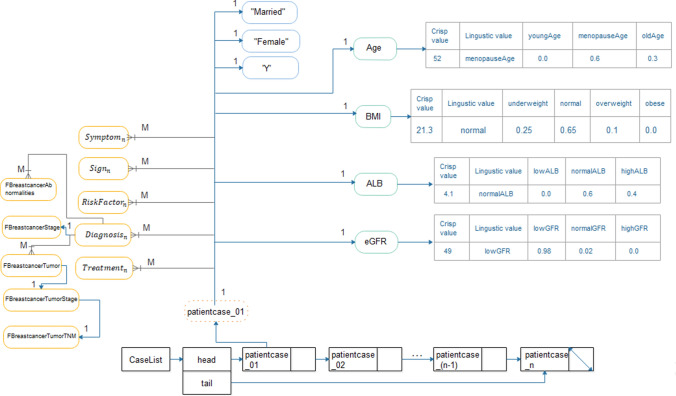
Instances of patient cases were first stored in a singly linked list. Generally, a linked structure is applied to overcome the limitation of an array to allow for easy insertion and deletion. This provides our approach with efficient in-memory insertion, editing and deletion before populating the BCFO. Linked lists have an organized collection of nodes that store data items and address them to other nodes so that each node has a data field and a reference field. The first node is called the head, and the last node is called the tail. Each element of the linked list contains a patient case, representing a container of other data structures. Let’s consider a patient case patientcasen that is expected to have numeric features such as age, BMI, GFR, ALB and several others and its no-fuzzifiable features, instances of classes associated through object properties. Our patientcasen can be modeled using a hybrid of data structures, as shown in Fig. 10.
Fig. 10.
An illustration of a representation of patient cases for ontology population using a composite of data structures
As shown in Fig. 10, each instance of patientcasen has a 1-M or 1–1 mapping to its data. Using an association pattern where an ordered pair of objects are paired together using key and value for the first and second objects, we associate the caseID of a patientcasen with the value (patientcasenValue). So, items having a 1-M relationship with patientcasen are aligned accordingly using this structure. Secondly, items, having a 1–1 relationship with patientcasen, have a simple solution using a simple data structure as an array while those with fuzzified features like Age, BMI, GFR and several others use the multi-way tree (a generic version of B-tree). This approach is convenient for two reasons: since the values are all expected to be numeric and that they maintain some order, say 0.0 (lowLB), 0.6 (normalALB) and 0.4 (highALB). Multi-way tree of order m ensures that each node has at-most m subtrees, at least 1 subtree, and m-1 distinct keys, where the subtrees may be empty and keys in each node are sorted. It also enjoys the requirement that the leaf nodes need not be at the same level.
Querying the fuzzy ontology
The scope of this study is limited to fuzzification of breast cancer ontology and no provision for applying it to any expert system. The outcome of this study will support studies on medical reasoning based on expert or knowledge-based systems. Notwithstanding, we attempted to demonstrate querying the fuzzified ontology to provide a query mechanism that supports expert systems or knowledge-based systems. Semantic web-based query languages such as SQWRL, f-SPARQL [59] and SPARQL-DL could be explored for querying the ontology. In addition to that, we applied the Fuzzy DL [60] plugin and the Gurobi optimization tools [60, 61], accompanied by the Fuzzy OWL2 plugin, to check the consistency of the fuzzy ontology and to also query it. The FuzzyDL reasoner is a DL-based reasoner that supports reasoning in both fuzzy logic and fuzzy rough set. The query is submitted to FuzzyDL, which is the underlying inference engine. Listing 10 show an example SPARQL-DL query applied to the implementation of the BCFO.
Also, a FuzzyDL-based query was executed through the Fuzzy OWL2 tab to output its result for confirmation of the consistency of the BCFO. A sample query executed is as follows:
(?min-instance PatientCase1 BreastCancerPatientCase) , (all-instances? BreastCancerPatientCase)
The query will retrieve the best entailment degree of an assertion that PatientCase1 belongs to BreastCancerPatientCase, and all instances of the BreastCancerPatientCase class and their degree of membership, respectively.
In this section, we have successfully implemented and experimented with fuzzified ontology. The implemented ontology was populated and then queried accordingly. In the next section, we shall evaluate the efficiency of the fuzzy ontology.
Results and discussion
In this section, we present the evaluation of the result on the ontology and make insightful discussions around it. We investigated the richness of the BCFO ontology, analyzed its semantic and content depth and richness. Furthermore, an evaluation of the coverage of the domain and the quality of information accessible from the associated concepts and data types is carried out.
Evaluation of the crisp ontology
A number of standard ontology metrics were considered for the evaluation of the ontology designed in this study. Each metric presents its value, as shown in the figure. For instance, it is clear that the ontology is rich in axiom and logical axiom, which are statements about concepts and relations in the TBox. Other notable metrics are the class assertion count, class count, SubclassOf axiom count, ObjectProperty assertion axiom count, DataProperty assertion axiom count, domain and range data property counts, domain and range of object property counts, object and data property counts, individual counts and others.
A description of these metrics and their count values are summarized in Table 2. This clarification of the atomic metrics became necessary to allow for their use in the computation of the composite ontology metrics described later in the section.
Table 2.
A listing of metrics and their respective counts in the BCFO ontology
| Metrics | Number of items | Description |
|---|---|---|
| Classes | 114 | Sets, collections, concepts, types of objects or things |
| Class axiom | 104 | Class-based statements that are asserted to be true in the domain being described: e.g. Subclass, Equivalent class, Disjoint class |
| Individuals | 194 | An instance of a class |
| Individuals (Object property assertion) | 59 | Statements made using object properties and individuals |
| Individuals (Data property assertion) | 37 | Statements made using data properties and individuals |
| Individuals (Class assertion) | 291 | Statements of individuals assigned to a class |
| Object property axiom (Object property domain/range) | 31/28 | Statements made by stating the domain and range of an object property |
| Date property axiom (Object property domain/range) | 71/71 | Statements made by stating the domain and range of an object property |
| Declarative axioms | 442 | A count of axioms declared or asserted |
| Maximum depth | 9 | The farthest route from the Thing class to a leaf class |
| Disjoint classes | 9 | Classes that cannot share an instance |
| Maximum number of children | 36 | |
| Average number of Children | 4.38 | Computational value of finding the average number of children immediate subclasses of the Thing class |
| Classes with a single child | 14 | |
| Classes with more than ~ 10 children | 3 | |
| Annotation | 87 | Comments on entities in an ontology |
| Axiom | 1115 | Statements asserted as a priori knowledge |
| Logical axiom count | 648 | Axioms that form the logical definition of terms |
| Object Property | 31 | Properties used to characterize classes |
| Data Property | 77 | Properties used to characterize the relationship between classes and data-values |
| Subclass of | 92 | A subdivision of a class |
| Subpropterty of | 4 | A subdivision of a property |
Considering the listing in Table 2, the following definitions hold:
-
i.
Class Complexity: Average number of paths to reach a class from the Thing class
-
ii.
Property Complexity
-
iii.
: Average number of semantic relations for object properties per class
-
iv.
Abstraction: Average depth of the ontology
-
v.
Cohesion: Average number of connected classes
-
vi.
Semantic richness: Ratio of the total number of semantic relations mapped to classes by all ontology relations consisting of object properties and subsumption relations.
-
vii.
Inheritance richness: Average number of subclasses in a class or computation of the broadness or depth of ontology graph
-
viii.
Attribute richness: Ratio of the total number of datatype properties by the number of classes. It also shows the average number of attributes defined per class within the ontology
-
ix.
Relationship richness—reflects the diversity of relations by comparing the number of non-subsumption relations to the number of subsumption relations
-
x.
Comprehension of properties (object and datatype): Percentage of annotation of the features in the ontology
-
xi.
Comprehension of classes: Percentage of annotation of the classes in ontology
The composite metrics listed above are used to generate the values listed in Table 3. For atomic metrics such as Axiom, Classes, Properties, and Instances, we obtained 1115, 114, 101 and 194, respectively. These figures demonstrate and confirm the richness of the ontology. Similarly, for the richness of conceptualization of the domain knowledge in terms of semantic relations, data value relations, and level of sub-classing (inheritance), our ontology obtained 2.0, 2.70, and 5.2, respectively. The ontology's complexity for the class and properties described above yielded 13.14 and 2.0, respectively. Lastly, we computed the average depth of the ontology and the average number of connected classes and obtained 4.38 and 191, respectively.
Table 3.
An evaluation of some related ontologies as compared with the fuzzy ontology proposed in this study
| Metrics | Ontology metrics | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Complexity | Conceptualization richness | Comprehension | Atomic metrics | ||||||||||
| Studies (Year) [Ref] | Class | Properties | Abstraction | Cohesion | Semantic | Data property | Inheritance | Classes (%) | Property (%) | Axiom | Classes | Properties | Instances |
| This study | 13.14 | 2.0 | 4.38 | 191 | 2.0 | 2.70 | 5.2 | 76 | 30 | 1115 | 114 | 101 | 194 |
| [30] | 4 | 11 | 4 | 181 | 1.041 | 0.12 | 8.2 | 10 | 10 | 3731 | 196 | 19 | 134 |
| [38] | 5 | 1.4 | 2 | 63 | 0.495 | 2.26 | 5.0 | 88.71 | 2.04% | 1316 | 62 | 196 | 2640 |
| [62] | 3 | 1.3 | 2 | 27 | 0.62 | 0.92 | 2.875 | 0.0 | 0.0 | 446 | 26 | 62 | 0 |
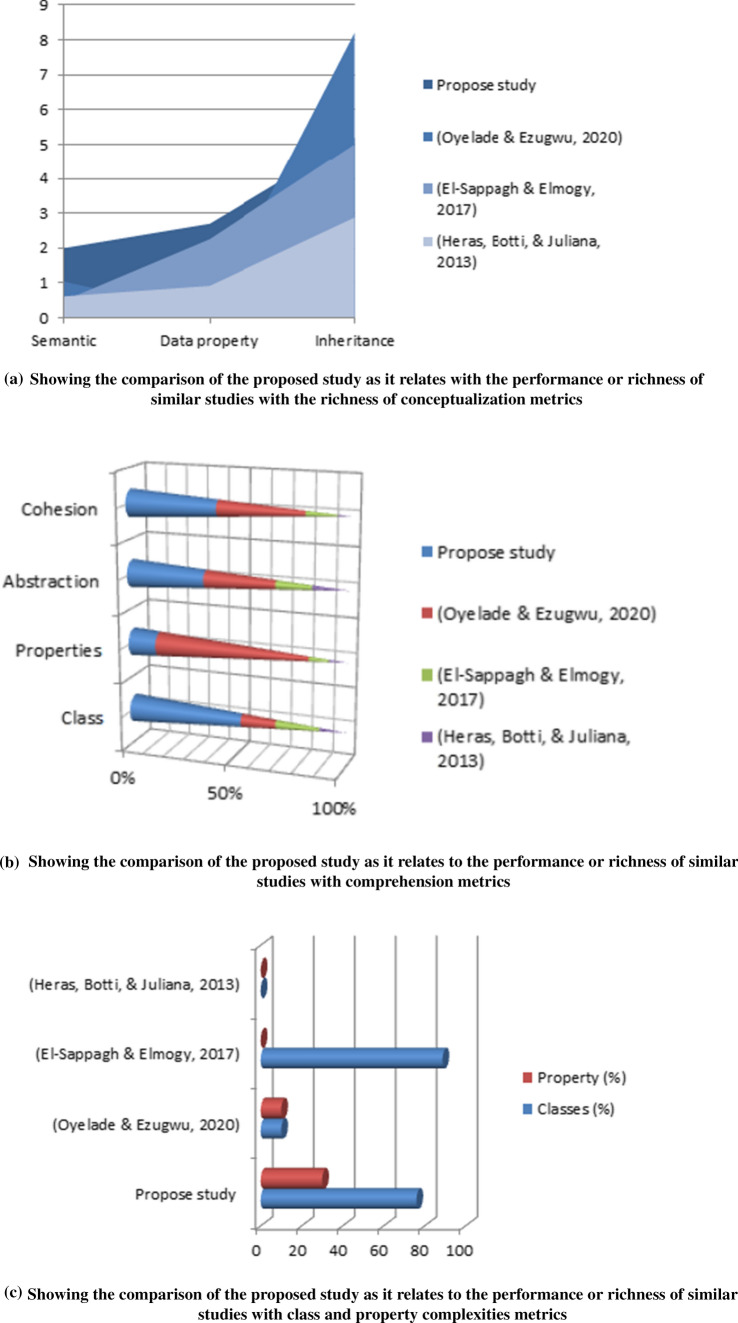
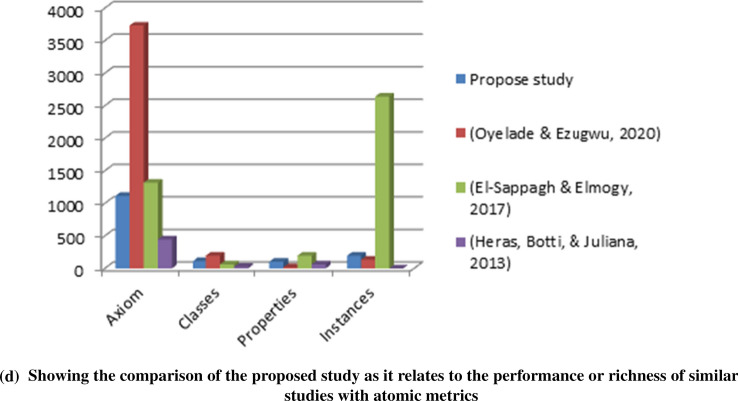
The performance of the BCFO ontology, as earlier described, was compared and graphed with other similar studies, which have also developed both crisp and fuzzy ontology. We found the performance of ontology proposed in this study to have yielded outperforming results. Figure 11a–d present a comparison of the performance of this study with related studies. The results of Table 3 show the richness of the axioms, properties (object and datatype) and instances of the proposed ontology used in this study. In the next section, we will concentrate on metrics demonstrating the level of fuzzification of the ontology.
Fig. 11.
a Showing the comparison of the proposed study as it relates with the performance or richness of similar studies with the richness of conceptualization metrics. b Showing the comparison of the proposed study as it relates to the performance or richness of similar studies with comprehension metrics. c Showing the comparison of the proposed study as it relates to the performance or richness of similar studies with class and property complexities metrics. d Showing the comparison of the proposed study as it relates to the performance or richness of similar studies with atomic metrics
The Class complexity and property complexity metrics measured the average number of paths to reach a class from the Thing class and the average number of semantic relations for object properties per class. These two metrics are supportive of demonstrating the semantic richness of the ontology. Table 3 shows that this study achieved an impressive level of class complexity. Figure 11b shows that though Oyelade and Ezugwu (2020) achieved a high level of property complexity, our domain ontology achieved the highest level of class complexity. The implication is that the BCFO proposed in this study presents a rich and detailed representation of domain entities.
Another category of metric evaluated is the measure of the abstraction and cohesion of the ontology. The measure of cohesiveness represented in the ontology demonstrates how the formalization sufficiently captures the intercommunication existing between entities in the domain. Medical knowledge often corroborates terms and entities together to deduce knowledge. Hence, the relevance of the ontology to the domain will determine the measure of cohesion seen in the ontology. The cohesion level shown in Table 3 for the proposed BCFO is considerable high. This indicates that this study demonstrates a good grasp of the interconnectivity exiting in the domain. Similarly, the abstraction metric measures the depth of the ontology, meaning it finds out the level at which the knowledge engineer understood the detailed definition of domain entities. Again, the BCFO proposed in this study achieved a significant level with regard to ontology abstraction. The result of this evaluation is illustrated in Fig. 11b with the class and property complexities.
Ontology comprehension metrics were also applied to evaluate the ontology proposed in this study. The level of comprehension of properties (object and datatype) classes was evaluated to determine the percentage of annotation achieved. Figure 11c illustrates the comparison of the measure of class complexity, property complexity, cohesion and abstraction attained in the proposed ontology compared with what similar studies achieved. Although El-Sappagh & Elmogy (2017) work achieved a slightly increased annotation level, in this study, the annotation level is not a deficiency to its richness since it significantly annotates its class and property, as seen in Table 3.
The richness of the conceptualization of the domain knowledge is of great importance when evaluating ontology. This is because this category of metrics measures semantic richness, inheritance richness, attribute richness and relationship richness. The semantic richness allows for reasoners to exploit ontology for allowing machine understandability of the domain. This metric is computed by finding the total number of semantic relations mapped to classes by all ontology relations consisting of object properties and subsumption relations. Inheritance richness reveals how the engineering process considers each concept and extrapolates its definition to determine other concepts. On the other hand, attribute richness measures the wideness of the attributes contained in each class. Table 3 shows that this study achieved a value of 2.7 compared with other studies. A significant result was obtained regarding the semantic richness compared with related studies. Figure 11a shows this comparative analysis of the conceptualization richness of the ontology as compared with other ontologies in medical sciences.
The ontology was also evaluated against some atomic metrics. These are the axiom, classproperty, and instance counts. The values returned by these metrics have a direct impact on the composite metrics. The evaluation is direct counts of the items associated with the definition of each metric. For instance, from Table 3, 1115, 114, 101, and 194 values were obtained for the axiom, class, property, and instance count, respectively. These are pointers to the depth of knowledge search made in the domain by the elicitation process before the engineering process. In Fig. 11d, we graphed these values with those obtained from other domains and showed an improvement compared with the level obtained in other domains.
Evaluating the depth of vagueness in the fuzzy ontology
We consider a number of fuzzy ontology-based metrics for evaluating the breast cancer fuzzy ontology proposed in this study. The following are the metrics proposed in the literature:
Since the reduction of vagueness to the barest minimum is what is conceived in the creation of fuzzy ontology, authors in [63] computed the metric vagueness explicitness (VE) described in Eq. 3, vagueness spread (VS) in Eq. 4, and vagueness intensity (VI). VS computes the measure of vagueness achieved in a fuzzy ontology, and VE, which computes the ontology's fuzzy relevance, are simple. VE is calculated by dividing the number of vague ontological elements that are explicitly represented (EVOE) by (VOE); VS is computed by dividing the number of vague ontology elements (VOE) which are classes, relations, and data types, divided by the total number of elements (OE). VI measures the level at which users of the fuzzy ontology can accept the definition of the instances of the fuzzy ontology. Since VI does not have a clear definition for computing its values, we ignore it in our evaluation.
| 4 |
| 5 |
The definition of Eq. 4 describes comprehensibility, shareability, and the level of reduction of vagueness achieved in a fuzzy ontology. In the fuzzy ontology proposed in this study, there are 114 classes, 31 object properties, 34 fuzzy datatypes, 55 fuzzy datatype properties, and fuzzy object properties are 1. We compute VS and VE as follows:
The numerator and denominator of VE are the same because we ensured that every fuzzy element we defined in the proposed ontology is defined explicitly. Therefore, obtaining the value of 1 for the VE signifies that the relevance of the proposed breast cancer fuzzy ontology is high since the value of VE is high. Similarly, we obtained the value of 0.38 for the VS. The implication of this demonstrates the level of vague elements this study was able to identify and fuzzify.
-
b.
In another study, Alexopoulos et al. suggested four fuzzy ontology-based metrics: accuracy, correctness, consistency, and completeness [11]. The following describes their applicability to fuzzy ontology:
-
c.
Correctness: A fuzzy ontology is correct when all its fuzzy elements convey a vague meaning in the given domain. According to the detailed presentation in Sect. 3.3, we identified all the vague elements as contemplated in the domain.
-
d.
Accuracy: A fuzzy ontology is accurate when the degrees of its fuzzy elements approximate the vagueness in an intuitively precise way for the given domain.
-
e.
Completeness: A fuzzy ontology is complete when all the vagueness in the domain has been well absorbed within the ontology. We might not have exhausted all the vague terms in the domain of consideration. However, a significant achievement was made.
-
f.
Consistency: A fuzzy ontology is consistent when it does not contain controversial information about the domain’s vagueness. This study has repeatedly emphasized the use of a domain expert and CPG in building the ontology. Hence, we assume the consistency of the breast cancer fuzzy ontology.
We found these metrics very relevant, and evaluating the fuzzy-based breast cancer ontology presented in this study showed that it performed well.
-
b.
Lastly, Ivanova proposed another metric for fuzzy ontology [64], which is defined in Eq. 5. This metric aims to apply to situations where a learning algorithm automates the extraction and representation of vague terms in the fuzzy ontology.
| 6 |
The author explained that the variables pi measures the probability of how correctly a fuzzy element is located in the ontology; n represents the total number of terms in the ontology; n1 denotes the number of valuable domain terms represented. A higher value of K shows a good learning process. We found that there is ambiguity in the definition of how pi is computed, hence the difficulty in applying the metric.
Evaluating the impact of fuzzy annotation
Similar studies that have developed fuzzy ontology have investigated the impact of the addition of annotations during the process of fuzzification. This impact is based on computational reasoning and speed during parsing and translation by the Fuzzy DL. This section presents the computational parsing time of the proposed ontology and the cumulative effect as the ontology gets bigger. To demonstrate the latter, we present the result in [20].
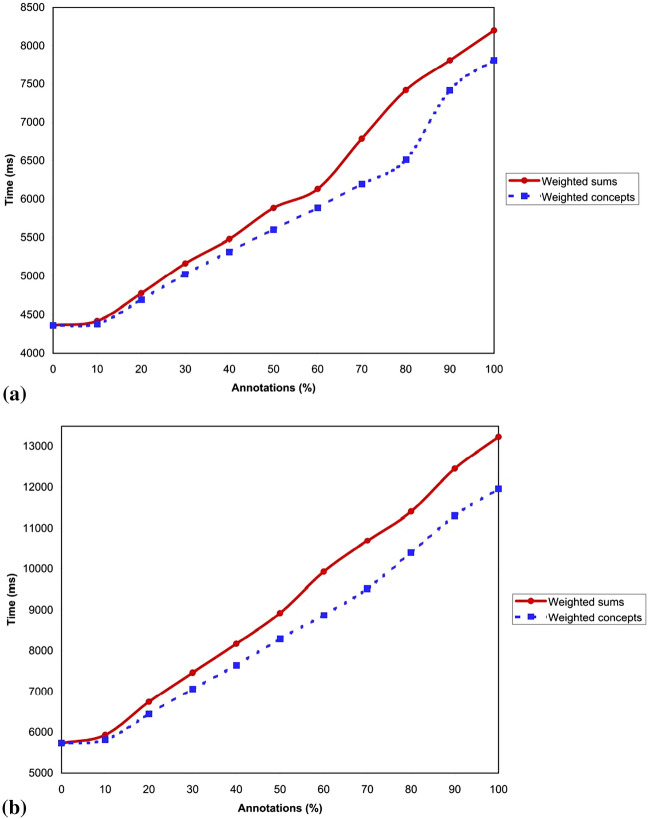
The proposed breast cancer fuzzy ontology has 41 annotations: 34 fuzzy data types, 55 fuzzy datatype properties, and 1 fuzzy concept. The parsing time obtained for crisp breast cancer ontology is 6.0 ms, whereas the fuzzified breast cancer ontology parsing time is 16.0 ms. Performance with a larger fuzzy ontology such as Galen ontology is examined. Authors showed the experimental result on the number of annotated elements as it affects parsing time (PT) in Fig. 12a; In Table 4, we show the numbers of annotated elements as it affects in the translation time (TT) Fig. 12b; and the influence of the percentage of annotations (%) in both PT and TT when translating into fuzzyDL syntax.
Fig. 12.
a Experimental evaluation results for the numbers of annotated elements influence in the PT. b Experimental evaluation results for the numbers of annotated elements influence in the TT
Table 4.
Listing of the parsing times for WSs and WCs
| % | Concepts | GCIs | RIAs | PT WCs (ms) | PT WSs (ms) | TT WCs (ms) | TT WSs (ms) |
|---|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 4364.1 | 4363.9 | 5731.7 | 5726.1 |
| 10 | 2385 | 2604 | 88 | 4420.3 | 4382.5 | 5932.8 | 5812.6 |
| 20 | 4590 | 5151 | 177 | 4773.6 | 4692 | 6746.8 | 6443.9 |
| 30 | 6990 | 7675 | 276 | 5166.8 | 5025.2 | 7465.5 | 7059.4 |
| 40 | 9312 | 10,152 | 383 | 5481.4 | 5320.3 | 8173.5 | 7648.1 |
| 50 | 11,588 | 12,760 | 462 | 5884.5 | 5603.4 | 8925.2 | 8295.4 |
| 60 | 13,888 | 15,260 | 569 | 6131.6 | 5889 | 9928.1 | 8875 |
| 70 | 16,216 | 17,764 | 672 | 6785.7 | 6193.9 | 10,690.5 | 9521.6 |
| 80 | 18,475 | 20,363 | 785 | 7418.6 | 6509.4 | 11,403.1 | 10,402.8 |
| 90 | 20,805 | 22,906 | 875 | 7809.2 | 7418.8 | 12,451.7 | 11,303.3 |
| 100 | 23,141 | 25,563 | 958 | 8201.6 | 7813.8 | 13,228.3 | 11,962.6 |
Parsing and translation times are computed on fuzzy ontology to know to what extent the fuzzification operation impacts the speed of reasoners. Reasoners are expected to parse the ontology for consistency and inference making (or reasoning) operations. There are different levels of fuzzification achieved in this study. For instance, fuzzification of fuzzy atomic concepts is much simpler. It requires little tagging compared to fuzzification of complex fuzzy concepts such as weighted concepts (WCs) and weighted sum concepts (WSs). Hence, we evaluated the performance of the reasoner when parsing and translating WCs and WSs.
Considering WCs and WSs' description, Fig. 12a–b show the computational time used to pars and translate WCs and WSs in the Galen ontology–the biggest fuzzified ontology. This computational time is constant across fuzzy ontologies representing WCs and WSs in the ontology. The figures revealed that as the percentage of the number of WCs and WSs increases in the ontology, the computational time for parsing and translating them using the FuzzyDL reasoner also increases. As graphed in the figure, the implication of this is that since WSs consists of two or more WCs, it is reasonable to have the computational time of WSs to be more than that of WCs, though the difference is not so significant. The benefit of using WSs in fuzzified ontology is achieving a rich or higher-level expression. Hence, this computational time is a tradeoff that allows for increased expressivity at the expense of time. Recall that expressivity, also synonymous with reduction of vagueness, is the major aim of ontology fuzzification.
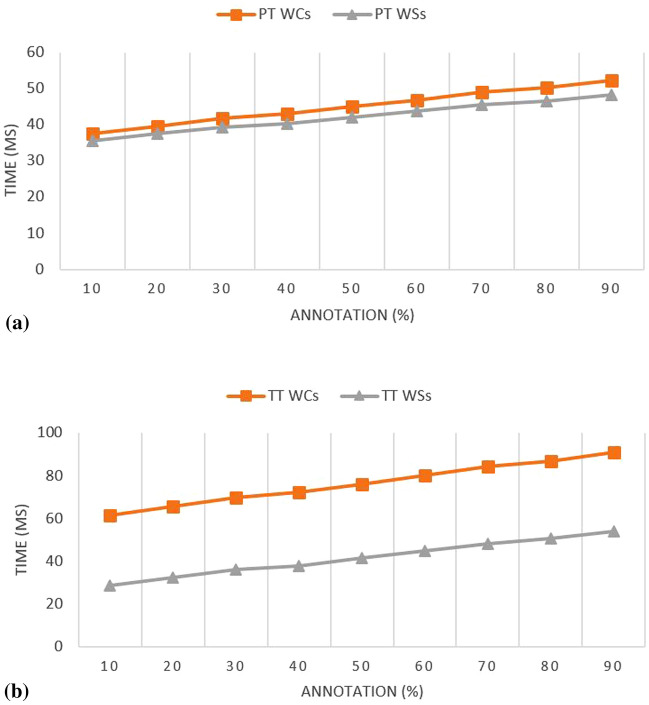
The parsing time and the translation time is shown in Table 5 for the fuzzified breast cancer ontology with emphasis on weighted sums (WSs) and weighted concepts (WCs). These were used for the time computation because of the increased number of nested tags required to represent them in the Fuzzy OWL2 syntax. It is assumed that using the WS and WC will capture a significant timing which will be required by other low-tag demanding constructs like a modifier, datatype and relations. Results obtained in Table 5 for the PT WCs, PT WSs, TT WCs, and TT WSs showed that there is a computational cost attached to the reduction of vagueness in the fuzzified ontology. This time cost also indicates that fuzzy elements contribute significant expressivity to the fuzzified ontology. As seen in Fig. 13a, b, the plot of the result showed that the difference between the parsing time (PT) and the translation time (TT) for both WCs and WSs are competing.
Table 5.
Listing of concepts, concept assertion, role assertion in computing the parsing time and in the translation time into fuzzyDL syntax for WSs and WCs in BCFO
| % | Concepts | CIs | RIAs | PT WCs (ms) | PT WSs (ms) | TT WCs (ms) | TT WSs (ms) |
|---|---|---|---|---|---|---|---|
| 10 | 12 | 33 | 17 | 35.40 | 33.76 | 57.16 | 25.19 |
| 20 | 25 | 65 | 33 | 37.48 | 35.59 | 61.40 | 28.82 |
| 30 | 38 | 98 | 49 | 39.56 | 37.42 | 65.64 | 32.45 |
| 40 | 51 | 131 | 65 | 41.64 | 39.25 | 69.88 | 36.08 |
| 50 | 58 | 163 | 84 | 42.92 | 40.23 | 72.16 | 38.04 |
| 60 | 70 | 196 | 101 | 44.85 | 41.92 | 76.07 | 41.39 |
| 70 | 82 | 228 | 116 | 46.78 | 43.61 | 79.98 | 44.74 |
| 80 | 95 | 261 | 133 | 48.87 | 45.44 | 84.22 | 48.37 |
| 90 | 103 | 294 | 150 | 50.15 | 46.56 | 86.83 | 50.61 |
| 100 | 115 | 325 | 167 | 52.08 | 48.25 | 90.74 | 53.96 |
Fig. 13.
a Experimental evaluation results for the numbers of annotated elements influence in the PT for the fuzzified ontology. bExperimental evaluation results for the numbers of annotated elements influence in the TT for the fuzzified ontology
Ontology evaluation is the task of measuring the quality of ontology, and it allows for the acceptance and adoption of a given ontology in its domain. The results obtained from the evaluation of the BCFO is demonstrating that it is a candidate ontology for use by domain experts and developers of expert systems or knowledge-based systems. The ontology possesses the quality for both acceptance and adoption. The result of this study reveals that fuzzification of an ontology is feasible and results in the elimination of the limitations of corresponding crisp ontology. In addition, the outcome of this study showed that reducing vagueness with the hope of improving computational reasoning in ontology is effective using the proposed approach. We have demonstrated that vagueness for the domain of female breast cancer can be reduced by impressively achieving a record high performance in reducing the vagueness of breast cancer ontology through the fuzzification of elements identified as vague.
Furthermore, the evaluation and result obtained from this study guide future research seeking to apply or improve on the proposed BCFO. Meanwhile, this will also serve as a guide to reducing vagueness in ontology for other domains. No study has been able to achieve the fuzzification and evaluation of fuzzy ontology related to female breast cancer, as shown in this study. Hence, this work is a rich contribution toward the body of knowledge advancing the reduction vagueness in formalization of knowledge in breast cancer diagnosis and knowledge representation.
Conclusion
In this study, the application of fuzzy logic to the challenge of reducing vagueness in crisp breast cancer ontology was proposed. The approach adopted is the use of IKARUS-Onto for fuzzy identification elements and Fuzzy OWL2 for the representation of the fuzzy elements. The study's outcome is a rich contribution toward the body of knowledge advancing the reduction vagueness in formalization of knowledge in breast cancer diagnosis and knowledge representation. We presented an elegant flow for the translation and conversion of a crisp ontology to fuzzy ontology using a novel framework. It follows the novel IKARUS-Onto methodology for fuzzification of the crisp ontology. The resulting fuzzified ontology was evaluated, and the result showed that a significant amount of vagueness had been removed from the ontology. Therefore, obtaining the value of 1 for the VE signifies that the relevance of the proposed breast cancer fuzzy ontology is high since VE's value is high.
Similarly, we obtained the value of 0.38 for the VS. The implication demonstrates the level of vague elements identified and fuzzified. We acknowledge that the proposed framework is limited by its handling of vagueness with no mechanism for tackling imprecision. As future work, we recommend that the framework be modified to accommodate the reduction of imprecision. Also, we suggest that the resulting BCFO be used to support knowledge-based systems that may be using reasoning algorithms such as Select and Test (ST) [6]or case-based reasoning (CBR) paradigm [30]. Considering the need for continuous engineering of ontologies, starting from a benchmark version obtained from this study, this fuzzification level can be further improved in future studies.
Declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Olaide N. Oyelade, Email: Oyeladeo@ukzn.ac.za
Absalom E. Ezugwu, Email: ezugwua@ukzn.ac.za
Sunday A. Adewuyi, Email: sadewuyi@abu.edu.ng
References
- 1.Dheebaa J, Albert Singh NN, Tamil Selvi S. Computer-aided detection of breast cancer on mammograms: a swarm intelligence optimized wavelet neural network approach. J Biomed Informa. 2014 doi: 10.1016/j.jbi.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 2.Silva VTDCE, Costalonga EC, Coelho FO, Caires RA, Burdmann AEA. Assessment of kidney function in patients with cancer. Adv Chron Kidney Dis. 2018;25(1):49–56. doi: 10.1053/j.ackd.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 3.Costa A, Oliveira H, Catani J, de Barros N, Melo C, Vieira M (2020) Data augmentation for detection of architectural distortion in digital mammography using deep learning approach.. Comput Vis Pattern Recognit. (2018). arXiv:1807.031672020
- 4.Zadeh HG, Pakdelazar O, Haddadnia J, Rezai-Rad G, Mohammad-Zadeh M. Diagnosing breast cancer with the aid of fuzzy logic based on data mining of a genetic algorithm in infrared Images. Middle East J Cancer. 2012;3(4):119–129. [Google Scholar]
- 5.Hadjiiski L, Sahiner B, Chan H-P. Advances in CAD for diagnosis of breast cancer. Curr Opin Obstet Gynecol. 2006;18(1):64–70. doi: 10.1097/01.gco.0000192965.29449.da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oyelade O, Obiniyi A, Junaidu S, Adewuyi S. ST-ONCODIAG: a semantic rule-base approach to diagnosing breast cancer base on Wisconsin datasets. Inform Med Unlocked. 2018;10:117–125. doi: 10.1016/j.imu.2017.12.008. [DOI] [Google Scholar]
- 7.Olaide Oyelade N, Absalom Ezugwu E. A deep learning model using data augmentation for detection of architectural distortion in whole and patches of images. Biomed Signal Processing Control. 2020;65(2020):1–17. [Google Scholar]
- 8.Ramadan SZ. Methods used in computer-aided diagnosis for breast cancer detection using mammograms: a review. J Healthc Eng. 2020;2020:1–21. doi: 10.1155/2020/9162464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vrandecic D (2010) Ontology evaluation, A Doctoral dissertation submitted to des Karlsruher Instituts fur Technologie (KIT) pp 1–235
- 10.Samani ZR, Shamsfard M (2017) The State of the Art in Developing Fuzzy Ontologies: A Survey, Faculty of Computer Science and Engineering, Shahid Beheshti University pp 1–46
- 11.Alexopoulos P, Wallace M, Kafentzis K, Askounis D. IKARUS-Onto: a methodology to develop fuzzy ontologies from crisp ones. Knowl Inf Syst. 2012;32(3):667–695. doi: 10.1007/s10115-011-0457-6. [DOI] [Google Scholar]
- 12.Hyde D (2008) Agueness, logic and ontology, shgate new critical thinking in philosophy
- 13.Ramos-Soto A, Pereira-Fari˜na M (2017) On modeling vagueness and uncertainty in data-to-text systems through fuzzy sets. Inf Sci pp 1–32
- 14.Lassiter D. Vagueness as probabilistic linguistic knowledge. In: Nouwen R, van Rooij R, Sauerland U, Schmitz HC, editors. Vagueness in communication. Berlin: Springer; 2011. pp. 1–21. [Google Scholar]
- 15.Dinis B (2017) Old and new approaches to the sorites paradox, arXiv:1704.00450v1 [math.HO] pp 1–17
- 16.Smith NJ (2006) Fuzzy logic and higher-order vagueness, Department of Philosophy, Main Quadrangle, The University of Sydney, pp 1-19
- 17.Gerla G. Vagueness and formal fuzzy logic: some criticisms. Logic Log Philo. 2017 doi: 10.12775/LLP.2017.031. [DOI] [Google Scholar]
- 18.Fermuller CG, Ajek PH (2011) A conversation about fuzzy logic and vagueness, A conversation pp 4015–416, accessible from https://publik.tuwien.ac.at/files/PubDat_206476.pdf
- 19.Ghorbel H, Bahri A, Bouaziz R (2010) Fuzzy ontologies building method: fuzzy ontomethodology, Conference Paper pp 1–9
- 20.Bobilloa F, Straccia U. Fuzzy ontology representation using OWL 2. Int J Approx Reason. 2011;52:1073–1094. doi: 10.1016/j.ijar.2011.05.003. [DOI] [Google Scholar]
- 21.Amira A, Amel GT. An extension of protégé for an automatic fuzzy-ontology building using clustering and FCA. Int J Comput Sci Inf Technol (IJCSIT) 2015;7(2):13–20. [Google Scholar]
- 22.Agrebi HG, Bahri A, Bouaziz R (2009) Fuzzy protégé for fuzzy ontology models, Conference: IPC'09
- 23.Oyelade O, Obiniyi A, Junaidu S, Adewuyi S. Patient symptoms elicitation process for breast cancer medical expert systems: a semanti web and natural language parsing approach. Future Comput Inform J. 2017;3(1):72–81. doi: 10.1016/j.fcij.2017.11.003. [DOI] [Google Scholar]
- 24.Balanică V, Dumitrache I, Caramihai M, Rae W, Herbst C (2011) Evaluation of breast cancer risk by using fuzzy logic. U.P.B Sci Bull, Series C vol 73(1): pp 53–64
- 25.Muhic I. Fuzzy analysis of breast cancer disease using fuzzy c-means and pattern recognition. Southeast Eur J Soft Comput. 2013;2(1):50–55. [Google Scholar]
- 26.Sen S, Patra K, Mondal SK. Fuzzy risk analysis in familial breast cancer using a similarity measure of interval-valued fuzzy numbers. Pac Sci Rev A Nat Sci Eng. 2016;18(2016):203–221. [Google Scholar]
- 27.Ameur STB, Sellami D, Wendling L, Cloppet F. Breast cancer diagnosis system based on semantic analysis and choquet integral feature selection for high risk subjects. Big Data Cogn Comput. 2019;3(41):1–24. [Google Scholar]
- 28.Dhingra V, Bhatia KK (2015) Development of ontology in laptop domain for knowledge representation. In: Procedia Computer Science: International conference on information and communication technologies (ICICT 2014), vol 46 (2015): pp 249 – 256
- 29.Chen L, Lu D, Zhu M, Muzammal M, Samuel OW, Huang G, Li W, Wu H. OMDP: an ontology-based model for diagnosis and treatment of diabetes patients in remote healthcare systems. Int J Distrib Sens Netw. 2019;15(5):1–15. [Google Scholar]
- 30.Oyelade ON, Ezugwu AE. A case-based reasoning framework for early detection and diagnosis of novel coronavirus. Inform Med Unlocked. 2020;20:1–41. doi: 10.1016/j.imu.2020.100395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parry D (2004) A fuzzy ontology for medical document retrieval, The Australasian Workshop on DataMining and Web Intelligence (DMWI2004), Dunedin pp 1–6
- 32.Parry DT (2005) Fuzzy ontology and intelligent systems for discovery of useful medical information, A Doctoral Thesis Submitted to Auckland University of Technology pp 1–294
- 33.Gu H, Lv H, Gao J, Shi J. Towards a general fuzzy ontology and its construction, college of computer science. Hangzhou: Zhejiang University; 2007. pp. 1–6. [Google Scholar]
- 34.Asma D, Zizette B. Fuzzy ontology evolution: classification of a new individual. J Emerg Technol Web Intell. 2014;6(1):9–14. [Google Scholar]
- 35.Heba F, Mohammed E, Ahmed A, Raziq MA (2015) Constructing fuzzy ontology for cardiac arrhythmias, Conference IEEE pp 402–409
- 36.Shaker ES, Mohammed E, Riad A. A fuzzy-ontology oriented case-based reasoning framework for semantic diabetes diagnosis. Artif Intell Med. 2015;65(3):179–208. doi: 10.1016/j.artmed.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Elhefny MA, Elmogy M, Elfetouh AA. Developing a fuzzy OWL ontology for obesity related cancer domain. Int J Med Eng Inform. 2017;9(2):162–187. [Google Scholar]
- 38.El-Sappagh S, Elmogy M. A fuzzy ontology modeling for case base knowledge in diabetes mellitus domain. Eng Sci Technol Int J. 2017;20(3):1025–1040. [Google Scholar]
- 39.Oyelade ON, Kana AFD. "OWL formalization of cases: an improved case-based reasoning in diagnosing and treatment of breast cancer. Inte J Inf Secur Priv Digit Forensics (IJIS) 2019;3(2):92–105. [Google Scholar]
- 40.Oyelade ON, Obiniyi AA, Junaidu SB, Adewuyi SA. Patient symptoms elicitation process for breast cancer medical expert systems: a semanti web and natural language parsing approach. Future Comput Inform J. 2017;3(1):72–81. doi: 10.1016/j.fcij.2017.11.003. [DOI] [Google Scholar]
- 41.Disease ontology, "Disease ontology, [Online]. Available: https://disease-ontology.org/. [Accessed 20 January 2021]
- 42.Association of Directors of Anatomic and Surgical Pathology (2020) Understanding Your Pathology Report: Breast Cancer," 6 August 2020. [Online]. Available: https://www.cancer.org/treatment/understanding-your-diagnosis/tests/understanding-your-pathology-report/breast-pathology/breast-cancer-pathology.html. [Accessed 21 January 2021]
- 43.Webster LR, Bilous AM, Willis L, Byth K, Burgemeister FC, Salisbury EL, Clarke CL, Balleine RL. Histopathologic indicators of breast cancer biology: insights from population mammographic screening. Br J Cancer. 2005;92(28):1366–1371. doi: 10.1038/sj.bjc.6602501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skfuzzy2.0 (2021) skfuzzy 0.2 docs. [Online]. Available: https://pythonhosted.org/scikit-fuzzy/. [Accessed 19 January 2021]
- 45.Team O (2020) Liver Function Test (LFT), 22 July 2020. [Online]. Available: https://www.oncolink.org/cancer-treatment/procedures-diagnostic-tests/blood-tests-tumor-diagnostic-tests/liver-function-test-lft. [Accessed 20 January 2021]
- 46.Haldeman-Englert C, Trevino H, Turley K (2021) Albumin (Blood), [Online]. Available: https://www.urmc.rochester.edu/encyclopedia/content.aspx?contenttypeid=167&contentid=albumin_blood. [Accessed 20 January 2021]
- 47.Shahinian VB, Bahl A, Niepel D, Lorusso V. Considering renal risk while managing cancer. Cancer Manag Res. 2017;9:167–178. doi: 10.2147/CMAR.S125864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.National Kidney Foundation (2021) Tests to measure kidney function, Damage and Detect Abnormalities," 21 April 2017. [Online]. Available: https://www.kidney.org/atoz/content/kidneytests. [Accessed 20 January 2021]
- 49.Stump-Sutliff M, Cunningham L, Gersten T (2021) Lab Tests for Cancer," [Online]. Available: https://www.urmc.rochester.edu/encyclopedia/content.aspx?contenttypeid=85&contentid=p07248. [Accessed 20 January 2021]
- 50.Bobillo F (2021) Fuzzy ontology representation using OWL 2," [Online]. Available: http://www.umbertostraccia.it/cs/software/FuzzyOWL/. [Accessed 20 Janaury 2021].
- 51.Mattila JK (2012) Zadeh algebra as the basis of Łukasiewicz logics. In: Conference: fuzzy information processing society (NAFIPS), 2012 Annual Meeting of the North American
- 52.Godo L, Gottwald S. Fuzzy sets and formal logics. Fuzzy Sets Syst. 2016;281:44–60. doi: 10.1016/j.fss.2015.06.021. [DOI] [Google Scholar]
- 53.Bobillo F (2021) Fuzzy DL, [Online]. Available: http://www.umbertostraccia.it/cs/software/fuzzyDL/fuzzyDL.html. [Accessed 20 Janauary 2021].
- 54.Bobillo F, Calvo-Flores MD, Gómez-Romero J. DeLorean: a reasoner for fuzzy OWL 2. Expert Syst Appl. 2012;39(1):258–272. doi: 10.1016/j.eswa.2011.07.016. [DOI] [Google Scholar]
- 55.Sirin E, Parsia B, Grau BC, Kalyanpur A, Katz Y. Pellet: a practical OWL-DL reasoner. Journal Web Semant. 2007;5(2):51–53. doi: 10.1016/j.websem.2007.03.004. [DOI] [Google Scholar]
- 56.Tsarkov D, Horrocks I (2006) FaCT++ description logic reasoner: system description. In: Furbach U,Shankar N (Eds) Automated reasoning. IJCAR 2006. Lecture notes in computer science, Springer, Berlin 4130
- 57.Shearer R, Motik B, Horrocks I (2008) HermiT: a highly-efficient OWL reasoner. In: Proceedings of the 5th international workshop on OWL: experiences and directions
- 58.Fouda H, Elmogy M, Aboelfetoh A, Maat AR (2015) Constructing Fuzzy ontology for cardiac arrhythmias, ICCES IEEE pp 402–409
- 59.Pan J, Stamou G, Stoilos G, Thomas E, Taylor S (2008) Scalable querying service over fuzzy ontologies. In: Proceedings of the 17th international world wide web conference (WWW 2008) pp 575–584
- 60.Bobillo F, Straccia U. The fuzzy ontology reasoner fuzzyDL. Knowl Based Syst. 2016;95:12–34. doi: 10.1016/j.knosys.2015.11.017. [DOI] [Google Scholar]
- 61.Gurobi Optimizer (2021) Gurobi_Optimizer," [Online]. Available: https://protegewiki.stanford.edu/wiki/Gurobi_Optimizer. [Accessed 23 January 2021]
- 62.Heras S, Botti V, Juliana V. A Knowledge representation formalism for case-based-reasoning. In: Chesñevar CI, Onaindia E, Ossowski S, Vouros G, editors. Agreement technol. Berlin: Springer; 2013. pp. 105–119. [Google Scholar]
- 63.Alexopoulos P, Mylonas P. Towards vagueness-oriented quality assessment of ontologies. Artif Intell Methods Appl. 2014;8445(2014):448–453. [Google Scholar]
- 64.Ivanova TI (2008) A metic and approach for fuzzy ontology evaluation. In Proceedings of international scientific conference computer science, 17th pp 822–827