Abstract
The nucleotide sequences of the internal transcribed spacer (ITS) 1 and 2 regions in the rRNA gene were determined by directly sequencing PCR-amplified fragments for all of the species (17 species and five varieties) in the genus Trichosporon. Comparative sequence analysis suggests that six medically relevant species, T. asahii, T. asteroides, T. cutaneum, T. inkin, T. mucoides, and T. ovoides, can be readily identified by their ITS sequences. In addition, the sequence analysis showed that conspecific strains have fewer than 1% nucleotide differences in the ITS 1 and 2 regions overall. Molecular phylogenetic trees are also presented.
Trichosporon Behrend is a medically important genus that includes the causative agents of both deep-seated, mucosa-associated infections and superficial infections, including white piedra (8, 28, 51). The majority of leukemia and lymphoma patients with fatal disseminated fungemia are in a profound neutropenic state when their infections develop. Recently, the number of patients with illness caused by Trichosporon species has been increasing (10, 14, 43, 44, 48, 50). Deep-seated trichosporonosis has a high mortality rate, and the prognosis for patients is very poor. Trichosporon species are also responsible for summer-type hypersensitivity pneumonitis (1, 2, 33, 49).
In 1992, the taxonomy of the genus Trichosporon was significantly revised by Guého et al. (5). Subsequently, Sugita et al. (37, 42) proposed a new classification that included new species, and 17 species and five varieties are presently accepted in the genus. Recent taxonomic studies indicate that trichosporonosis is caused by six species: T. asahii, T. asteroides, T. cutaneum, T. inkin, T. mucoides, and T. ovoides (4, 7, 40, 41). Moreover, it has been suggested that the major causative agents of trichosporonosis differ in each type of infection. T. asahii and T. mucoides are involved in deep-seated infection. T. asteroides and T. cutaneum are associated with superficial infection. T. ovoides and T. inkin are involved in white piedra of the head and genital area, respectively. T. pullulans is not a major causative agent of trichosporonosis and is rarely isolated from fungemia patients (16, 17).
On the other hand, at least four different serological types (I, II, III, and I-III) of Trichosporon have been identified in species by Ikeda et al. (9) and Nishiura et al. (30). The six medically relevant species are serotype I or II. Serotypes III and I-III do not seem to be responsible for infection.
Although we have developed a PCR-based identification system with genus-specific and T. asahii species-specific primers (38, 39), rapid identification of all Trichosporon species is not yet possible.
In this study, we sequenced the internal transcribed spacer (ITS) regions for all of the species in the genus Trichosporon and developed an identification system for all of the species.
MATERIALS AND METHODS
Strains used.
The strains used in this study are shown in Table 1. They included stock strains and clinical and environmental isolates. Trichosporon sp. strain M 9481 was isolated from the soil in Tokyo, Japan. Candida albicans, Candida famata, Debaryomyces hansenii, and Saccharomyces cerevisiae were also used.
TABLE 1.
Strains used and accession numbers of ITS sequences
| Species | Strain | Sourcea | Accession no. |
|---|---|---|---|
| Trichosporon aquatile | M 9317T | CBS 5973 | AB018011 (this study) |
| Trichosporon aquatile | M 9321 | CBS 5988 | AB018012 (this study) |
| Trichosporon aquatile | M 9472 | Environmental isolate | AB018012 (this study) |
| Trichosporon aquatile | M 9473 | Environmental isolate | AB018011 (this study) |
| Trichosporon asahii var. asahii | M 9306T | CBS 2479 | AB018013 (this study) |
| Trichosporon asahii var. asahii | M 9311 | CBS 2530 | AB018014 (this study) |
| Trichosporon asahii var. asahii | M 9470 | Clinical isolate | AB018013 (this study) |
| Trichosporon asahii var. asahii | M 9474 | Clinical isolate | AB018014 (this study) |
| Trichosporon asahii var. asahii | M 9475 | Clinical isolate | AB018013 (this study) |
| Trichosporon asahii var. asahii | M 9476 | Environmental isolate | AB018013 (this study) |
| Trichosporon asahii var. asahii | M 9477 | Environmental isolate | AB018013 (this study) |
| Trichosporon asahii var. coremiformis | M 9309T | CBS 2482 | AB018015 (this study) |
| Trichosporon asahii var. faecalis | M 9312T | CBS 4828 | AB018016 (this study) |
| Trichosporon asteroides | M 9308T | CBS 2481 | AB018017 (this study) |
| Trichosporon asteroides | M 9329 | CBS 7623 | AB018018 (this study) |
| Trichosporon asteroides | M 9330 | CBS 7624 | AB018017 (this study) |
| Trichosporon brassicae | M 9322T | CBS 6382 | AB018019 (this study) |
| Trichosporon cutaneum | M 9304T | CBS 2466 | AB018020 (this study) |
| Trichosporon cutaneum | M 9307 | CBS 2480 | AB018020 (this study) |
| Trichosporon domesticum | M 9401 | Environmental isolate (42) | AB018021 (this study) |
| Trichosporon domesticum | M 9412 | Clinical isolate | AB018021 (this study) |
| Trichosporon dulcitum | M 9337T | CBS 8257 | AB018022 (this study) |
| Trichosporon dulcitum | M 9318 | CBS 5785 | AB018022 (this study) |
| Trichosporon gracile | M 9334T | CBS 8189 | AB018023 (this study) |
| Trichosporon gracile | M 9335 | CBS 8193 | AB018023 (this study) |
| Trichosporon inkin | M 9316T | CBS 5585 | AB018024 (this study) |
| Trichosporon inkin | M 9333 | CBS 7629 | AB018024 (this study) |
| Trichosporon jirovecii | M 9325T | CBS 6864 | AB018025 (this study) |
| Trichosporon jirovecii | M 9326 | CBS 6950 | AB018026 (this study) |
| Trichosporon loubieri var. loubieri | M 9327T | CBS 7065 | AB018027 (this study) |
| Trichosporon loubieri var. laibachii | M 9319T | CBS 5790 | AB018028 (this study) |
| Trichosporon moniliiforme | M 9305T | CBS 2467 | AB018029 (this study) |
| Trichosporon montevideense | M 9323T | CBS 6721 | AB018021 (this study) |
| Trichosporon montevideense | M 9338 | CBS 8261 | AB018021 (this study) |
| Trichosporon mucoides | M 9331T | CBS 7625 | AB018030 (this study) |
| Trichosporon mucoides | M 9478 | Environmental isolate | AB018031 (this study) |
| Trichosporon mucoides | M 9479 | Environmental isolate | AB018031 (this study) |
| Trichosporon mucoides | M 9480 | Environmental isolate | AB018031 (this study) |
| Trichosporon mucoides | M 9422 | Clinical isolate (41) | AB018030 (this study) |
| Trichosporon ovoides | M 9315 | CBS 5580 | AB018033 (this study) |
| Trichosporon ovoides | M 9328T | CBS 7556 | AB018032 (this study) |
| Trichosporon ovoides | M 9458 | Environmental isolate (36) | AB018032 (this study) |
| Trichosporon pullulans | M 9339T | CBS 2532 | AB018034 (this study) |
| Trichosporon sporotrichoides | M 9336T | CBS 8245 | AB018035 (this study) |
| Trichosporon sp. | M 9481 | Environmental isolate | AB018036 (this study) |
| Candida albicans | M 1001T | CBS 562 | AB018037 (this study) |
| Candida albicans | M 1016 | ATCC 10264 | AB018038 (this study) |
| Candida albicans | M 1445 | NIHB 792 | AB018037 (this study) |
| Candida albicans | M 1447 | NIHA 207 | AB018037 (this study) |
| Candida albicans | M 1601 | CBS 1905 | AB018038 (this study) |
| Candida albicans | M 1602 | CBS 1918 | AB018038 (this study) |
| Candida albicans | M 2088 | IFO 1061 | AB018037 (this study) |
| Candida albicans | M 2089 | IFO 1389 | AB018037 (this study) |
| Candida albicans | M 2091 | IFO 0583 | AB018037 (this study) |
| Candida albicans | M 2093 | IFO 0579 | AB018037 (this study) |
| Candida famata var. famata | M 5033T | JCM 1521 | AB018039 (this study) |
| Candida famata var. flarei | M 5024T | JCM 2166 | AB018040 (this study) |
| Debaryomyces hansenii var. hansenii | M 5012T | JCM 1990 | AB018041 (this study) |
| Debaryomyces hansenii var. hansenii | M 5112 | Clinical isolate (29) | AB018041 (this study) |
| Debaryomyces hansenii var. fabryi | M 5011T | JCM 2104 | AB018042 (this study) |
| Debaryomyces hansenii var. fabryi | M 5102 | Clinical isolate (29) | AB018042 (this study) |
| Saccharomyces cerevisiae | M 6013T | CBS 1171 | AB018043 (this study) |
| Candida parapsilosis | MCO 429 | U10989 | |
| Candida parapsilosis | MCO 448 | U10989 | |
| Saccharomyces bayanus | CBS 380T | D89887 | |
| Saccharomyces bayanus | CBS 395 | Z95946 | |
| Saccharomyces bayanus | CBS 425 | Z95944 | |
| Saccharomyces bayanus | CBS 1546 | Z95948 | |
| Saccharomyces cerevisiae | CBS 382 | Z95936 | |
| Saccharomyces cerevisiae | CBS 400 | Z95939 | |
| Saccharomyces cerevisiae | CBS 423 | Z95932 | |
| Saccharomyces cerevisiae | CBS 2247 | Z95937 | |
| Saccharomyces cerevisiae | CBS 3081 | Z95941 | |
| Saccharomyces cerevisiae | CBS 3093 | Z95943 | |
| Saccharomyces cerevisiae | CBS 4903 | Z95940 | |
| Saccharomyces cerevisiae | CBS 5378 | Z95929 | |
| Saccharomyces cerevisiae | CBS 5635 | Z95942 | |
| Saccharomyces pastorianus | CBS 1513 | Z95950 | |
| Saccharomyces pastorianus | CBS 1538T | Z95949 | |
| Williopsis saturnus var. saturnus | CBS 5761T | Z93875 | |
| Williopsis saturnus var. saturnus | CBS 5761T | Z93882 | |
| Williopsis saturnus var. mrakii | NCYC 500T | Y11320 | |
| Williopsis saturnus var. mrakii | NCYC 500T | Y11319 | |
| Williopsis saturnus var. sargentensis | CBS 6342T | Z93879 | |
| Williopsis saturnus var. sargentensis | CBS 6342T | Z93886 | |
| Williopsis saturnus var. subsufficiens | CBS 5763T | Z93881 | |
| Williopsis saturnus var. subsufficiens | CBS 5763T | Z93888 | |
| Zygosaccharomyces cidri | CBS 4575T | Z48347 | |
| Zygosaccharomyces cidri | CBS 4575T | Z48361 | |
| Zygosaccharomyces fermentati | CBS 707T | Z48358 | |
| Zygosaccharomyces fermentati | CBS 707T | Z48362 |
ATCC, American Type Culture Collection, Manassas, Va.; CBS, Centraalbureau voor Schimmelcultures, Delft, The Netherlands; IFO, Institute of Fermentation, Osaka, Japan; JCM, Japan Collection of Microorganisms, Saitama, Japan; M, Meiji Pharmaceutical University, Tokyo, Japan; MCO, Medical College of Ohio, Toledo, Ohio; NCYC, National Collection of Yeast Cultures, Norwich, United Kingdom; NIH, National Institutes of Health, Rockville, Md.
Direct DNA sequencing.
Nuclear DNA was extracted by the method of Makimura et al. (22). ITSs 1 and 2 and the intervening 5.8S ribosomal DNA (rDNA) region were amplified with primers pITS-F (GTCGTAACAAGGTTAACCTGCGG) and pITS-R (TCCTCCGCTTATTGATATGC), which were designed from conserved regions of the 18S and 28S rRNA genes, respectively. The reactions were performed in a final reaction mixture (50 μl) containing 10 pmol of each primer; 200 mM (each) dATP, dTTP, dGTP, and dCTP; 2.5 mM MgCl2; 0.5 U of Ex Taq polymerase (Takara, Shiga, Japan); and 5 μl of 10× reaction buffer (Takara). The amplification reactions were performed in a GeneAmp PCR System 9700 (Perkin-Elmer Applied Biosystems, Foster City, Calif.) with the following cycling parameters: 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 57°C for 30 s, and 72°C for 45 s, with a final extension at 72°C for 10 min. The amplified products were purified with a NucleoSpin DNA purification kit (Macherey-Nagel GmbH, Duren, Germany) according to the manufacturer’s instructions. Direct sequencing of the PCR product was performed with a PRISM Cycle sequencing kit (Perkin-Elmer Applied Biosystems). Two external primers, pITS-F and pITS-R, were used to determine the sequences.
Nucleotide sequence similarity.
The similarities of the sequences were compared by using the nuclear DNA relatedness values taken from the literature (11–13, 18, 19, 24–27, 29, 36, 37, 40–42) and the similarity of the ITS 1 and 2 sequences separately. Sequence similarity was visually determined from pairwise alignments. The nucleotide sequences of species other than those in the genus Trichosporon included in this study were obtained from GenBank, and their accession numbers are cited in Table 1.
Molecular phylogenetic analysis.
The sequences were aligned with the computer program CLUSTAL W (45). For the neighbor-joining analysis (32), the distances between the sequences were calculated by using Kimura’s two-parameter model (15). Sites where gaps existed in any of the sequences were excluded. The program DNAML in PHYLIP version 3.5c was used for the maximum-likelihood analysis (3). T. pullulans was used as the outgroup.
Identification of ubiquinone.
The ubiquinone study was carried out only with the Trichosporon sp. strain M 9481 environmental isolate. The extraction of ubiquinone was performed following a procedure of Yamada and Kondo (47) with a slight modification. Ubiquinone was isolated by thin-layer chromatography (50 by 200 mm) (PK6F silica gel; Whatman, Clifton City, N.J.) in hexane-diethyl ether (85:15 [vol/vol]) and detected with UV light (254 nm). The type of ubiquinone was determined by high-performance liquid chromatography under the following conditions: reverse-phase column, Zorbax ODS (150 by 4.6 mm; Shimadzu, Kyoto, Japan); mobile phase, ethanol-water (97:3 [vol/vol]); column temperature, 53°C; flow rate, 1 ml/min; detection, 275 nm. Standard ubiquinones were used as references.
Serotyping.
Serotyping was carried out only with the Trichosporon sp. strain M 9481 environmental isolate. The serotype of the isolate was determined by the cell slide agglutination test with specific factor sera as described by Ikeda et al. (9).
Nucleotide sequence accession numbers.
The nucleotide sequences discussed in this paper have been deposited in the DNA Data Bank of Japan (DDBJ), and their accession numbers are given in Table 1.
RESULTS
Sequences of the ITS regions.
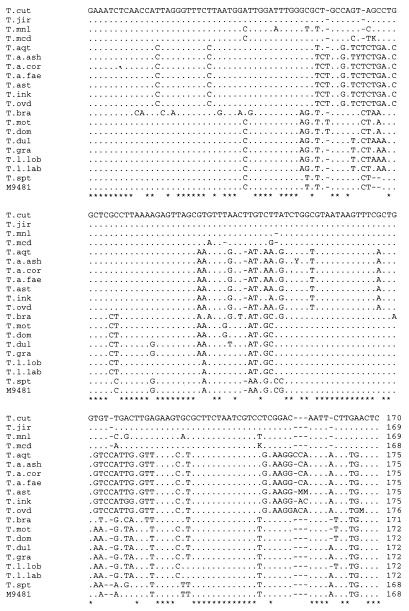
Figures 1 and 2 show the nucleotide sequences of ITS 1, including the 3′ end of 18S rDNA, and ITS 2 of all the species in the genus Trichosporon. With the exceptions of T. asahii var. coremiformis, T. asahii var. faecalis, T. brassicae, T. moniliiforme, and T. sporotrichoides, these are consensus sequences built from the sequences of more than two strains of each species. ITS 1 was from 115 to 123 bp long, while ITS 2 was from 168 to 176 bp long. The within-species length variation in either ITS 1 or ITS 2 was not remarkable.
FIG. 1.
Alignment of the Trichosporon ITS 1 sequences, including the 3′ end of 18S rDNA. Periods and asterisks are used when the nucleotide at a particular position is identical to that in T. cutaneum. Dashes represent deletions necessary for alignment. T. cut, T. cutaneum; T. jir, T. jirovecii; T. mnl, T. moniliiforme; T. mcd, T. mucoides; T. aqt, T. aquatile; T. a. ash, T. asahii var. asahii; T. a. cor, T. asahii var. coremiformis; T. a. fae, T. asahii var. faecalis; T. ast, T. asteroides; T. ink, T. inkin; T. ovd, T. ovoides; T. bra, T. brassicae; T. mot, T. montevideense; T. dom, T. domesticum; T. dul, T. dulcitum; T. gra, T. gracile; T. l. lob, T. loubieri var. loubieri; T. l. lab, T. loubieri var. laibachii; T. spt, T. sporotrichoides; M 9481, Trichosporon sp. strain M 9481.
FIG. 2.
Alignment of the Trichosporon ITS 2 sequences. For symbols and abbreviations, see the legend to Fig. 1.
Sequence similarity between strains of a single species.
Multiple strains of T. asahii, T. mucoides, C. albicans, and S. cerevisiae were examined to assess intraspecific variation (Table 2). In nine strains of T. asahii, <1- and 2-base differences were found in ITS 1 and 2, respectively, with overall similarities of 99.3 to 100% for both ITS 1 and 2. Five strains of T. mucoides had overall similarities of 98.9 to 100% in both the ITS 1 and 2 regions. For C. albicans, the similarities of the sequences were 99.7 to 100%. Although there were six different bases in the S. cerevisiae sequences in ITSs 1 and 2, the overall sequence similarity was 99.0%. Although strains in the same species do not necessarily have identical sequences, the overall sequence similarity of both ITSs 1 and 2 was 99% or more.
TABLE 2.
Number of nucleotide differences in ITSs 1 and 2 within a single species
| Species and strain | No. of differences
|
|||
|---|---|---|---|---|
| ITS 1 | ITS 2 | ITS 1 + 2 | % similarity | |
| Trichosporon asahii | ||||
| M 9306 | ||||
| M 9309 | 1 | 0 | 1 | 100 |
| M 9311 | 0 | 1 | 1 | 99.7 |
| M 9312 | 0 | 0 | 0 | 100 |
| M 9470 | 0 | 0 | 0 | 100 |
| M 9474 | 0 | 1 | 1 | 99.7 |
| M 9475 | 0 | 2 | 2 | 99.3 |
| M 9476 | 0 | 0 | 0 | 100 |
| M 9477 | 0 | 0 | 0 | 100 |
| Trichosporon mucoides | ||||
| M 9422 | ||||
| M 9331 | 2 | 1 | 3 | 98.9 |
| M 9478 | 0 | 0 | 0 | 100 |
| M 9479 | 0 | 0 | 0 | 100 |
| M 9480 | 0 | 0 | 0 | 100 |
| Candida albicans | ||||
| M 1001 | ||||
| M 1016 | 0 | 1 | 1 | 99.7 |
| M 1445 | 0 | 0 | 0 | 100 |
| M 1447 | 0 | 0 | 0 | 100 |
| M 1601 | 0 | 1 | 1 | 99.7 |
| M 1602 | 0 | 1 | 1 | 99.7 |
| M 2088 | 0 | 0 | 0 | 100 |
| M 2089 | 0 | 0 | 0 | 100 |
| M 2091 | 0 | 0 | 0 | 100 |
| M 2093 | 0 | 0 | 0 | 100 |
| Saccharomyces cerevisiae | ||||
| CBS 1171 | ||||
| CBS 382 | 1 | 0 | 1 | 99.8 |
| CBS 400 | 3 | 3 | 6 | 99.0 |
| CBS 423 | 1 | 2 | 3 | 99.5 |
| CBS 2247 | 1 | 0 | 1 | 99.8 |
| CBS 3081 | 0 | 0 | 0 | 100 |
| CBS 3093 | 0 | 0 | 0 | 100 |
| CBS 4903 | 1 | 0 | 1 | 99.8 |
| CBS 5378 | 0 | 0 | 0 | 100 |
| CBS 5635 | 3 | 0 | 3 | 99.5 |
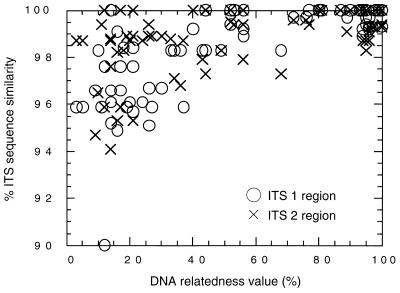
Relationship between the nuclear DNA relatedness value and ITS sequence similarity.
Table 3 shows the matrices of sequence similarity for ITSs 1 and 2 for all of the species in the genus Trichosporon. The matrix for the overall ITS sequence is shown in Table 4. Fig. 3 shows the relationship between the nuclear DNA relatedness value and the sequences’ ITS 1 and 2 similarities. This figure is based on the results for 74 pairs. A species concept has been defined on the basis of the nuclear DNA relatedness value, which corresponds well with biological relatedness (31). Within the same species (high-relatedness group), the value is approximately 80% or more. Species with values of approximately 40 to 80% are varieties of the same species or sibling species (intermediate-relatedness group). The value is less than 40% in different species (low-relatedness group). In the high-relatedness group, the sequence similarities of ITSs 1 and 2 were more than 98.9 and 98.8%, respectively. In the intermediate-relatedness group, the sequence similarities of ITSs 1 and 2 were more than 98.3 and 97.8%, respectively. In the low-relatedness group, data for ITS sequences with similarities of less than 90% were excluded from Figs. 3 and 4. The ITS similarities of the low-relatedness group were lower than the values for the high and intermediate groups. Exceptions were seen in T. asahii and T. asteroides. Their ITS 2 sequences were identical, but they had low nuclear DNA relatedness values of between 12 and 30% (37).
TABLE 3.
Matrix of ITS 1 and 2 similarities for Trichosporon speciesa
| Species | T. cutaneum | T. jirovecii | T. moniliiforme | T. mucoides | T. asahii var. asahii | T. asahii var. coremiformis | T. asahii var. faecalis | T. aquatile | T. asteroides | T. ovoides | T. inkin | T. brassicae | T. montevideense | T. domesticum | T. dulcitum | T. gracile | T. loubieri var. laibachii | T. loubieri var. loubieri | T. sporotrichoides | Trichosporon sp. strain M 9481 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T. cutaneum | 95.9 | 94.1 | 95.3 | |||||||||||||||||
| T. jirovecii | 96.6 | 94.7 | 98.2 | |||||||||||||||||
| T. moniliiforme | 96.6 | 95.7 | 92.9 | |||||||||||||||||
| T. mucoides | 95.7 | 94.9 | 99.1 | |||||||||||||||||
| T. asahii var. asahii | 100 | 100 | 98.9 | 100 | 99.4 | 98.3 | ||||||||||||||
| T. asahii var. coremiformis | 99.2 | 100 | 98.9 | 100 | 99.4 | 97.7 | ||||||||||||||
| T. asahii var. faecalis | 100 | 99.2 | 98.9 | 100 | 99.4 | 98.3 | ||||||||||||||
| T. aquatile | 96.7 | 95.9 | 96.7 | 98.9 | 98.9 | 97.7 | ||||||||||||||
| T. asteroides | 97.6 | 98.4 | 97.6 | 97.6 | 99.4 | 98.9 | ||||||||||||||
| T. ovoides | 95.9 | 95.1 | 95.9 | 97.6 | 94.3 | 98.3 | ||||||||||||||
| T. inkin | 95.1 | 95.1 | 97.6 | 93.5 | 92.7 | 98.4 | ||||||||||||||
| T. brassicae | ||||||||||||||||||||
| T. montevideense | 92.5 | 100 | ||||||||||||||||||
| T. domesticum | 92.5 | 100 | ||||||||||||||||||
| T. dulcitum | 98.8 | 97.7 | 96.5 | |||||||||||||||||
| T. gracile | 100 | 98.8 | 97.1 | |||||||||||||||||
| T. loubieri var. laibachii | 98.3 | 98.3 | 98.3 | |||||||||||||||||
| T. loubieri var. loubieri | 98.3 | 98.3 | 96.6 | |||||||||||||||||
| T. sporotrichoides | 98.2 | |||||||||||||||||||
| Trichosporon sp. strain M 9481 | 90.6 |
Similarities less than 90% are not indicated. Data in the upper right portion of the table refer to ITS 2 similarity, and data in the lower left portion refer to ITS 1 similarity.
TABLE 4.
Matrix of overall ITS similarity for Trichosporon speciesa
| Species | T. cutaneum | T. jirovecii | T. moniliiforme | T. mucoides | T. asahii var. asahii | T. asahii var. coremiformis | T. asahii var. faecalis | T. aquatile | T. asteroides | T. ovoides | T. inkin | T. brassicae | T. montevideense | T. domesticum | T. dulcitum | T. gracile | T. loubieri var. laibachii | T. loubieri var. loubieri | T. sporotrichoides | Trichosporon sp. strain M 9481 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T. cutaneum | ||||||||||||||||||||
| T. jirovecii | 96.1 | |||||||||||||||||||
| T. moniliiforme | 95.1 | 95.1 | ||||||||||||||||||
| T. mucoides | 95.4 | 96.8 | 95.8 | |||||||||||||||||
| T. asahii var. asahii | ||||||||||||||||||||
| T. asahii var. coremiformis | 99.7 | |||||||||||||||||||
| T. asahii var. faecalis | 100 | 99.7 | ||||||||||||||||||
| T. aquatile | 98.0 | 97.6 | 98.0 | |||||||||||||||||
| T. asteroides | 99.0 | 99.3 | 98.0 | 98.3 | ||||||||||||||||
| T. ovoides | 98.0 | 97.6 | 98.0 | 98.3 | 97.3 | |||||||||||||||
| T. inkin | 96.9 | 96.6 | 98.0 | 95.9 | 96.3 | 98.3 | ||||||||||||||
| T. brassicae | ||||||||||||||||||||
| T. montevideense | ||||||||||||||||||||
| T. domesticum | 100 | |||||||||||||||||||
| T. dulcitum | ||||||||||||||||||||
| T. gracile | 99.0 | |||||||||||||||||||
| T. loubieri var. laibachii | 97.9 | 98.6 | ||||||||||||||||||
| T. loubieri var. loubieri | 97.2 | 97.6 | 97.6 | |||||||||||||||||
| T. sporotrichoides | ||||||||||||||||||||
| Trichosporon sp. strain M 9481 | 95.1 |
Similarities of less than 90% are not indicated.
FIG. 3.
Relationship between nuclear DNA relatedness value and similarities of ITS 1 and 2 sequences considered separately.
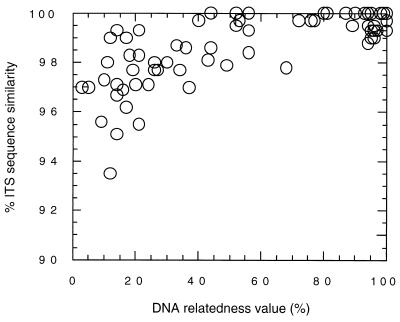
FIG. 4.
Relationship between nuclear DNA relatedness value and similarity of combined ITS 1 and 2 sequences.
The relationship between the nuclear DNA relatedness value and the ITS sequences is shown in Fig. 4. The high-relatedness group had more than 99.0% similarity, while the intermediate-relatedness group had more than 97.9% similarity. In the intermediate group, 100% sequence similarities were found between T. domesticum and T. montevideense and between the two varieties of C. famata. The low-relatedness group showed less than 99.3% sequence similarity. No identical sequences were found in the low-relatedness group, but the similarities between T. asteroides and the three varieties of T. asahii and between T. dulcitum and T. gracile exceeded 99%.
Species-specific sequences.
Table 5 shows the species-specific sequence used to distinguish each species. These sequences do not depend on the strain. The three varieties of T. asahii have identical sequences, so the specific sequences do not differentiate among them. T. domesticum and T. montevideense also have identical sequences and cannot be distinguished by ITS sequences.
TABLE 5.
Species-specific sequences in the ITS regions
| Species | Species-specific sequence | Position of nucleotide |
|---|---|---|
| Medically relevant species | ||
| T. asahii | TTTATAGGCTTAT | 10–22a |
| T. asteroides | TTAATTGGCTTAT | 10–22a |
| T. cutaneum | TCGGTCAATTGAT | 60–72a |
| T. inkin | TTTACAGGCTTAA | 10–22a |
| T. mucoides | TCGGTCGATTACT | 61–73a |
| T. ovoides | TTTATAGGCTTAA | 10–22a |
| Non-medically relevant species | ||
| T. aquatile | CATTGGCTTAAAA | 12–24a |
| T. brassicae | CGATTCAATTTTA | 64–76a |
| T. domesticum (=T. montevideense) | CGGATTCGATTTT | 64–76a |
| T. dulcitum | AAAGGAGTTAGCAAGTTTTACTAT | 69–92b |
| T. gracile | AAAGGAGTTAGCAAGTTTAACTAT | 69–92b |
| T. jirovecii | CCGGTCAATTACT | 60–72a |
| T. moniliiforme | TCGGTCAATTACT | 61–73a |
| T. loubieri var. loubieri | GATCATAACAAGA | 103–115a |
| T. loubieri var. laibachii | GATCATAACTAA | 102–103a |
| T. sporotrichoides | CCTCTGGGCTTAA | 8–20a |
ITS 1.
ITS 2.
Serotype and ubiquinone of Trichosporon sp. strain M 9481.
The slide agglutination test with specific factor sera demonstrated that environmental isolate M 9481 was serotype II. This isolate had Q9 as the major ubiquinone.
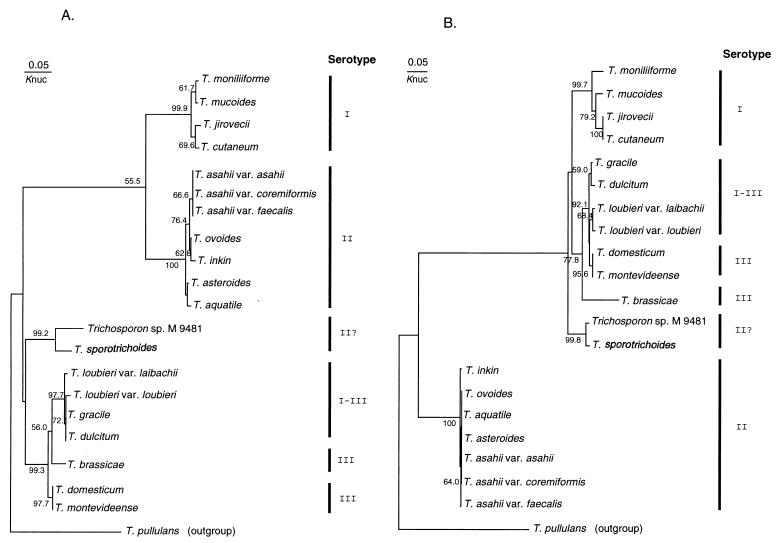
Molecular phylogenetic analysis based on the ITS sequences.
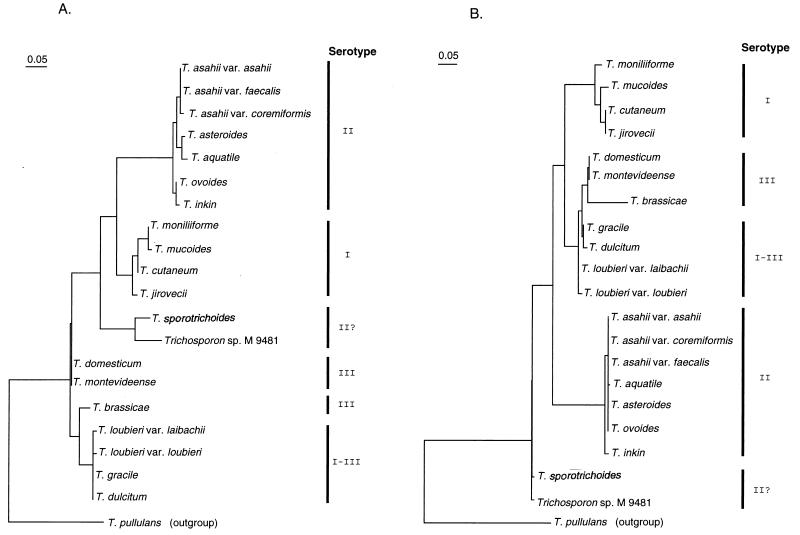
Figures 5 and 6 show the molecular phylogenetic trees based on the ITS 1 and 2 sequences constructed by the neighbor-joining and maximum-likelihood methods, respectively. The serotypes of the Trichosporon species are also shown in Fig. 5 and 6. The overall topology of the neighbor-joining tree is slightly different from that constructed by the maximum-likelihood method. This seems to be due to bootstrap values and differences in the method of analysis. However, the serotype correlated well with molecular phylogenetic trees, regardless of the ITS region or method. In the T. sporotrichoides clade, the strain M 9481 has serotype II, while T. sporotrichoides does not react with any factor sera.
FIG. 5.
Molecular phylogenetic trees based on the Trichosporon ITS 1 (A) and 2 (B) sequences. The trees were constructed by the neighbor-joining method. The numerals represent the confidence level from 1,000 replicate bootstrap samplings (frequencies less than 50% are not indicated). Knuc, Kimura’s parameter (15).
FIG. 6.
Molecular phylogenetic trees based on the Trichosporon ITS 1 (A) and 2 (B) sequences. The trees were constructed by the maximum-likelihood method. The scale marker indicates the distance in relative units that equals 5% of the total branch length depicted.
DISCUSSION
To detect pathogenic fungi rapidly, oligonucleotide primers for PCR have been designed, based on the sequences of 18S or 26S rDNA (6, 46). These sequences have been determined for most pathogenic yeasts. The relatively high levels of sequence similarity observed among some species appears to limit the value of 18S or 26S rDNA for differentiating species that are phylogenetically closely related. The ITS region is located between the 18S and 26S rRNA genes and is subdivided into the ITS 1 region, which separates the 18S and 5.8S rRNA genes, and the ITS 2 region, which is found between the 5.8S and 26S rRNA genes. It is generally thought that the ITS regions have higher rates of divergence than the 18S, 5.8S, or 26S rRNA genes. The lengths of the 18S and 26S rRNA genes are essentially identical in all species, while the lengths of the ITS regions depend on the species. For example, the ITS 2 regions of Candida glabrata (U70498), Candida kefyr (U70502), and S. cerevisiae (Z75722) are approximately 230 to 240 bp long; those of Candida guilliermondii (U70499) and C. famata (U70500) are approximately 190 bp long; those of C. albicans (L07796), Candida tropicalis (L11349), Candida parapsilosis (L11352), and Candida viswanathii (U70510) are approximately 130 to 140 bp long; and those of Candida lusitaniae (U70503) and Candida rugosa (U70506) are only 70 to 90 bp long (21). The clades in the molecular phylogenetic tree for these species correspond well to the ITS sequence length. ITSs 1 and 2 in the Trichosporon species are essentially the same size. With the exception of T. pullulans, the genus Trichosporon is monophyletic (35). T. pullulans was phylogenetically distinct from the other taxa in the genus, suggesting that this species does not belong in the genus. The lengths of the ITS 1 and 2 regions of T. pullulans are 40 and 50 bp longer, respectively, than those of the other Trichosporon species. As mentioned above, we believe that highly species-specific sequences can be found in the ITS sequences. To design highly specific oligonucleotide primers for PCR, sequence data for both the pathogenic and nonpathogenic species are required. Mannarelli and Kurtzman (23) developed a PCR-based identification system for the 14 Candida species that are human pathogens based on a ca. 600-nucleotide variable region (D1/D2) at the 5′-end of the 26S rDNA. Prior to this research work, their group determined the sequences of 204 Candida and related species, including nonpathogenic species (20). Their PCR system can clearly distinguish pathogenic species from species that are phylogenetically closely related.
DNA sequence can be determined quite rapidly with an automated DNA sequencer. Starting from DNA extraction from yeast cells, the ITS sequence can be determined within a working day, or 24 h at the most. Once an ITS sequence database has been constructed, rapid identification can be made. Since this identification method is not based on physiological characteristics, such as the carbon assimilation pattern, the chance of misidentification is reduced.
In this study, we found that conspecific species have less than a 1% overall nucleotide difference in both the ITS 1 and 2 regions. The species that have an intermediate DNA relatedness value have ITS sequences with 99% or more similarity, such as the three varieties of T. asahii and the two varieties of C. famata. Although T. domesticum and T. montevideense are distinct biological species, they have the same ITS sequences. They are considered to share an intermediate DNA relatedness value (42). With a few exceptions, the ITS sequence similarity between different species is less than 99.0%. The ITS sequence of T. asahii had two or three different bases from that of T. asteroides (99.0 to 99.3% similarity). It is very difficult to distinguish between these two species by using physiological characteristics (4), but the former is responsible for deep-seated infection while the latter is associated with superficial infection (4, 7).
Strain M 9481, which was isolated from a soil sample, was serotype II and had Q9 as the major ubiquinone. Although this strain is positioned in the T. sporotrichoides clade on the phylogenetic tree, its sequence similarity with the clade is only 95.1%. Strain M 9481 is considered a new species in the genus Trichosporon. At present, according to our data and a literature survey, the overall ITS sequence similarity of identical species is more than 99.0%. The accuracy of this value should be clarified as further data are accumulated. Kurtzman and Robnett (20) found a similar result in their analysis of the D1/D2 region of the 26S rDNA: the same species had fewer than 1% sequence differences.
The molecular phylogenetic trees of the genus Trichosporon constructed from the 18S and 26S rDNA sequences have already been reported (34, 35). The topology of these trees differs from that of the trees constructed from the ITS 1 and 2 sequences. However, the correlation between these trees depends on the molecular sequence and the molecular phylogenetic method. Although the factor sera we made are not species-specific, the serological groups correspond to the molecular phylogeny. Serological typing gives useful information to tentatively identify an isolate.
In conclusion, we constructed a readily used and accurate identification system for all of the species in the genus Trichosporon, including the six medically relevant species, based on comparative sequence analysis of the ITS regions. We expect that a sequence database will be constructed for other pathogenic fungi and related species.
ACKNOWLEDGMENTS
We thank Tomoe Ichikawa and Hiromi Shinohara for their technical skills in analyzing the ubiquinone.
REFERENCES
- 1.Ando M, Arima K, Yoneda R, Tamura M. Japanese summer-type hypersensitivity pneumonitis: geographic distribution, home environment, and clinical characteristics of 621 cases. Am Rev Respir Dis. 1991;144:765–769. doi: 10.1164/ajrccm/144.4.765. [DOI] [PubMed] [Google Scholar]
- 2.Ando M, Sakata T, Yoshida K. Serotype-related antigen of Trichosporon cutaneum in the induction of summer-type hypersensitivity pneumonitis: correlation between serotype of inhalation challenge-positive antigen and that of the isolates from patients’ homes. J Allergy Clin Immunol. 1990;85:36–44. doi: 10.1016/0091-6749(90)90218-s. [DOI] [PubMed] [Google Scholar]
- 3.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 4.Guého E, Improvisi L, de Hoog G S, Dupont B. Trichosporon on humans: a practical account. Mycoses. 1994;37:3–10. doi: 10.1111/j.1439-0507.1994.tb00277.x. [DOI] [PubMed] [Google Scholar]
- 5.Guého E, Smith M T, de Hoog G S, Grand G B, Christen R, Batenburg-van der Vegte W H. Contributions to a revision of the genus Trichosporon. Antonie Leeuwenhoek. 1992;61:289–316. doi: 10.1007/BF00713938. [DOI] [PubMed] [Google Scholar]
- 6.Haynes K A, Westerneng T J, Fell J W, Moens W. Rapid detection and identification of pathogenic fungi by polymerase chain reaction amplification of large subunit ribosomal DNA. J Med Vet Mycol. 1995;33:319–325. doi: 10.1080/02681219580000641. [DOI] [PubMed] [Google Scholar]
- 7.Herbrecht R, Koening H, Waller K, Liu L, Guého E. Trichosporon infections: clinical manifestations and treatment. J Mycol Med. 1993;3:129–136. [Google Scholar]
- 8.Hoy J, Hsu K C, Rolston K, Hopfer R L, Luna M, Bodey G P. Trichosporon beigelii infection: a review. Rev Infect Dis. 1986;8:959–967. [PubMed] [Google Scholar]
- 9.Ikeda R, Yokota M, Shinoda T. Serological characterization of Trichosporon cutaneum and related species. Microbiol Immunol. 1996;40:813–819. doi: 10.1111/j.1348-0421.1996.tb01146.x. [DOI] [PubMed] [Google Scholar]
- 10.Itoh T, Hosokawa H, Kohdera U, Toyazaki N, Asada Y. Disseminated infection with Trichosporon asahii. Mycoses. 1996;39:195–199. doi: 10.1111/j.1439-0507.1996.tb00124.x. [DOI] [PubMed] [Google Scholar]
- 11.James S A, Collins M D, Roberts I N. Use of an rRNA internal transcribed spacer region to distinguish phylogenetically closely related species of the genera Zygosaccharomyces and Torulaspora. Int J Syst Bacteriol. 1996;46:189–194. doi: 10.1099/00207713-46-1-189. [DOI] [PubMed] [Google Scholar]
- 12.James S A, Roberts I N, Collins M D. Phylogenetic heterogeneity of the genus Williopsis as revealed by 18S rRNA gene sequences. Int J Syst Bacteriol. 1998;48:591–596. doi: 10.1099/00207713-48-2-591. [DOI] [PubMed] [Google Scholar]
- 13.Kamiyama A, Niimi M, Tokunaga M, Nakayama H. DNA homology between Candida albicans strains: evidence to justify the synonymous status of C. stellatoidea. Mycopathologia. 1989;107:3–7. doi: 10.1007/BF00437584. [DOI] [PubMed] [Google Scholar]
- 14.Kataoka-Nishimura S, Akiyama H, Saku K, Kashiwa M, Mori S, Tanikawa S, Sakamaki H, Onozawa Y. Invasive infection due to Trichosporon cutaneum in patients with haematologic malignancies. Cancer. 1998;82:484–487. [PubMed] [Google Scholar]
- 15.Kimura M. A simple method for estimation of evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 16.Kunova A, Godal J, Sufliarsky J, Spanik S, Kollar T, Krcmery V., Jr Fatal Trichosporon pullulans breakthrough fungemia in cancer patients: report of three patients who failed on prophylaxis with itraconazole. Infection. 1996;24:273–274. doi: 10.1007/BF01781117. [DOI] [PubMed] [Google Scholar]
- 17.Kunova A, Sorkovska D, Sufliarsky J, Krcmery V., Jr First report of catheter associated Trichosporon pullulans breakthrough fungemia in a cancer patient. J Infect. 1996;32:70–71. doi: 10.1016/s0163-4453(96)80015-6. [DOI] [PubMed] [Google Scholar]
- 18.Kurtzman C P. DNA relatedness among species of the genus Zygosaccharomyces. Yeast. 1990;6:213–219. doi: 10.1002/yea.320060306. [DOI] [PubMed] [Google Scholar]
- 19.Kurtzman C P. DNA relatedness among saturn-spored yeasts assigned to the genera Williopsis and Pichia. Antonie Leeuwenhoek. 1991;60:13–19. doi: 10.1007/BF00580436. [DOI] [PubMed] [Google Scholar]
- 20.Kurtzman C P, Robnett C J. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol. 1997;35:1216–1223. doi: 10.1128/jcm.35.5.1216-1223.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lott T J, Burns B M, Zancope-Oliveira R, Elie C M, Reiss E. Sequence analysis of the internal transcribed spacer 2 (ITS2) from yeast species within the genus Candida. Curr Microbiol. 1998;36:63–69. doi: 10.1007/s002849900280. [DOI] [PubMed] [Google Scholar]
- 22.Makimura K, Murayama Y S, Yamaguchi H. Detection of a wide range of medically important fungal species by polymerase chain reaction (PCR) J Med Microbiol. 1994;40:358–364. doi: 10.1099/00222615-40-5-358. [DOI] [PubMed] [Google Scholar]
- 23.Mannarelli B M, Kurtzman C P. Rapid identification of Candida albicans and other human pathogenic yeasts by using short oligonucleotides in a PCR. J Clin Microbiol. 1998;36:1634–1641. doi: 10.1128/jcm.36.6.1634-1641.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martini A V. Saccharomyces paradoxus comb. nov., a newly separated species of the Saccharomyces sensu stricto complex based upon nDNA/nDNA homologies. Syst Appl Microbiol. 1989;12:179–182. [Google Scholar]
- 25.Martini A V, Kurtzman C P. Deoxyribonucleic acid relatedness among species of the genus Saccharomyces sensu stricto. Int J Syst Bacteriol. 1985;35:508–511. [Google Scholar]
- 26.Martini A V, Martini A. Three newly delimited species of Saccharomyces sensu stricto. Antonie Leeuwenhoek. 1987;53:77–84. doi: 10.1007/BF00419503. [DOI] [PubMed] [Google Scholar]
- 27.Montrocher R M, Verner C, Briolay J, Gautier C, Marmeisse R. Phylogenetic analysis of the Saccharomyces cerevisiae group based on polymorphisms of rDNA spacer sequences. Int J Syst Bacteriol. 1998;48:295–303. doi: 10.1099/00207713-48-1-295. [DOI] [PubMed] [Google Scholar]
- 28.Nahass G T, Rosenberg S P, Leonardi C L, Penneys N S. Disseminated infection with Trichosporon beigelii. Arch Dermatol. 1993;129:1020–1023. doi: 10.1001/archderm.129.8.1020. [DOI] [PubMed] [Google Scholar]
- 29.Nishikawa A, Tomomatsu H, Sugita T, Ikeda R, Shinoda T. Taxonomic position of clinical isolates of Candida famata. J Med Vet Mycol. 1996;34:411–419. doi: 10.1080/02681219680000731. [DOI] [PubMed] [Google Scholar]
- 30.Nishiura Y, Nakagawa-Yoshida K, Suga M, Shinoda T, Guého E, Ando M. Assignment and serotyping of Trichosporon species: the causative agents of summer-type hypersensitivity pneumonitis. J Med Vet Mycol. 1997;35:45–52. doi: 10.1080/02681219780000861. [DOI] [PubMed] [Google Scholar]
- 31.Price C W, Fuson G B, Phaff H J. Genome comparison in yeast systematics: delimitation of species within the genera Schwanniomyces, Saccharomyces, Debaryomyces, and Pichia. Microbiol Rev. 1978;42:161–193. doi: 10.1128/mr.42.1.161-193.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito N, Nei M. Neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 33.Shimazu K, Ando M, Sakata T, Yoshida K, Araki S. Hypersensitivity pneumonitis induced by Trichosporon cutaneum. Am Rev Respir Dis. 1984;130:407–411. doi: 10.1164/arrd.1984.130.3.407. [DOI] [PubMed] [Google Scholar]
- 34.Sugita T, Makimura K, Nishikawa A, Uchida K, Yamaguchi H, Shinoda T. Partial sequences of large subunit ribosomal DNA of a new yeast species, Trichosporon domesticum and related species. Microbiol Immunol. 1997;41:571–573. doi: 10.1111/j.1348-0421.1997.tb01893.x. [DOI] [PubMed] [Google Scholar]
- 35.Sugita T, Nakase T. Molecular phylogenetic study of the basidiomycetous anamorphic yeast genus Trichosporon and related taxa based on small subunit ribosomal DNA sequences. Mycoscience. 1998;39:7–13. [Google Scholar]
- 36.Sugita T, Nishikawa A, Ikeda R, Shinoda T, Sakashita H, Sakai Y, Yoshizawa Y. First report of Trichosporon ovoides isolated from the home of a summer-type hypersensitivity pneumonitis patient. Microbiol Immunol. 1998;42:475–478. doi: 10.1111/j.1348-0421.1998.tb02312.x. [DOI] [PubMed] [Google Scholar]
- 37.Sugita T, Nishikawa A, Shinoda T. Reclassification of Trichosporon cutaneum by DNA relatedness by using the spectrophotometric method and the chemiluminometric method. J Gen Appl Microbiol. 1994;40:397–408. [Google Scholar]
- 38.Sugita T, Nishikawa A, Shinoda T. Rapid detection of species of the opportunistic yeast Trichosporon by PCR. J Clin Microbiol. 1998;36:1458–1460. doi: 10.1128/jcm.36.5.1458-1460.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugita T, Nishikawa A, Shinoda T. Identification of Trichosporon asahii by PCR based on sequences of the internal transcribed spacer regions. J Clin Microbiol. 1998;42:475–478. doi: 10.1128/jcm.36.9.2742-2744.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sugita T, Nishikawa A, Shinoda T, Kume H. Taxonomic position of deep-seated, mucosa-associated, and superficial isolates of Trichosporon cutaneum from trichosporonosis patients. J Clin Microbiol. 1995;33:1368–1370. doi: 10.1128/jcm.33.5.1368-1370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugita T, Nishikawa A, Shinoda T, Kusunoki T. Taxonomic studies on clinical isolates from superficial trichosporonosis patients by DNA relatedness. Jpn J Med Mycol. 1996;37:107–110. [Google Scholar]
- 42.Sugita T, Nishikawa A, Shinoda T, Yoshida K, Ando M. A new species, Trichosporon domesticum, isolated from the house of a summer-type hypersensitivity pneumonitis patient in Japan. J Gen Appl Microbiol. 1995;41:429–436. [Google Scholar]
- 43.Tashiro T, Nagai H, Kamberi P, Goto Y, Kikuchi H, Nasu M, Akizuki S. Disseminated Trichosporon beigelii infection in patients with malignant diseases: immunohistochemical study and review. Eur J Clin Microbiol Infect Dis. 1994;13:218–224. doi: 10.1007/BF01974540. [DOI] [PubMed] [Google Scholar]
- 44.Tashiro T, Nagai H, Yamasaki T, Goto Y, Akizuki S, Nasu M. Disseminated Trichosporon beigelii infection: report of nine cases and review. Jpn J Infect. 1993;67:704–711. doi: 10.11150/kansenshogakuzasshi1970.67.704. [DOI] [PubMed] [Google Scholar]
- 45.Thompson J, Hompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Deventer A J M, Goessens W H F, van Belkum A, van Vliet H J, van Etten E W M, Verbrugh H A. Improved detection of Candida albicans by PCR in blood of neutropenic mice with systemic candidiasis. J Clin Microbiol. 1995;33:625–628. doi: 10.1128/jcm.33.3.625-628.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamada Y, Kondo K. Coenzyme Q system in the classification of the yeast genera Rhodotorula and Cryptococcus, and the yeast like genera Sporobolomyces and Rhodosporidium. J Gen Appl Microbiol. 1973;19:59–77. [Google Scholar]
- 48.Yamakami Y, Tashiro T, Tokimatsu I, Nagai H, Nagaoka H, Hashimoto A, Goto Y, Nasu M, Yamasaki T, Ito M. Microbiological and clinical study of fungemia between 1981 and 1992. Kansenshogaku Zasshi. 1995;69:890–894. doi: 10.11150/kansenshogakuzasshi1970.69.890. [DOI] [PubMed] [Google Scholar]
- 49.Yoshida K, Ando M, Sakata T, Araki S. Environmental mycological studies on the causative agent of summer-type hypersensitivity pneumonitis. J Allergy Clin Immunol. 1988;81:475–483. doi: 10.1016/0091-6749(88)90920-7. [DOI] [PubMed] [Google Scholar]
- 50.Yoss B S, Sautter R L, Brenker H J. Trichosporon beigelii, a new neonatal pathogen. Am J Perinatol. 1997;14:113–117. doi: 10.1055/s-2007-994109. [DOI] [PubMed] [Google Scholar]
- 51.Walsh T J. Trichosporonosis. Infect Dis Clin N Am. 1989;3:43–52. [PubMed] [Google Scholar]