Abstract
Telomeres are nucleoprotein complexes that protect the ends of eukaryotic linear chromosomes from degradation and fusions. Telomere dysfunction leads to cell growth arrest, oncogenesis, and premature aging. Telomeric RNAs have been found in all studied species; however, their functions and biogenesis are not clearly understood. We studied the mechanisms of development disorders observed upon overexpression of telomeric repeats in Drosophila. In somatic cells, overexpression of telomeric retrotransposon HeT-A is cytotoxic and leads to the accumulation of HeT-A Gag near centrosomes. We found that RNA and RNA-binding protein Gag encoded by the telomeric retrotransposon HeT-A interact with Polo and Cdk1 mitotic kinases, which are conserved regulators of centrosome biogenesis and cell cycle. The depletion of proteins Spindle E, Ccr4 or Ars2 resulting in HeT-A overexpression in the germline was accompanied by mislocalization of Polo as well as its abnormal stabilization during oogenesis and severe deregulation of centrosome biogenesis leading to maternal-effect embryonic lethality. These data suggest a mechanistic link between telomeric HeT-A ribonucleoproteins and cell cycle regulators that ensures the cell response to telomere dysfunction.
Introduction
The ends of linear chromosomes are protected from fusions and degradation by specialized nucleoprotein structures called telomeres. Telomeres promote genome stability by regulating DNA metabolism at chromosome ends. Telomere-dependent control of cellular proliferation ensures limited expansion of cells harboring chromosomal abnormalities [1–3]. This implies that there are mechanisms that link damaged telomeres to cell cycle checkpoints. The mechanisms of “telomeric signaling” appear to be based on the modulations of the levels of telomeric proteins that depend on the telomere state. Indeed, high levels of free telomeric proteins are a signal of shortened telomeres [4, 5]. Emerging evidence suggests that telomeric factors can control cellular targets elsewhere, thus acting as mediators of telomeric abnormalities [6]. For example, the release of Rap1 from shortened yeast telomeres and its ectopic binding to the non-telomeric targets results in transcriptional changes of non-telomeric genes [4]. Components of shelterin, the mammalian telomere protection protein complex, can also occupy non-telomeric sites and act as transcription factors [7, 8].
Telomeres can be transcribed into long telomeric repeat-containing RNAs (TERRA) in all taxa from yeast to mammals, suggesting a conserved role for telomeric RNAs [9]. Telomere transcription is highly dynamic, being regulated during the cell cycle and upon telomere dysfunction. Therefore, telomeric transcripts can be considered as possible candidates for telomeric signaling molecules. TERRA transcripts localize at telomeres in yeast and human cells [10, 11]. TERRA associates with the protein components of shelterin and telomeric chromatin, supporting its structural role in the recruitment of telomeric factors to telomeres [12]. Emerging evidence indicates that TERRA can localize outside of telomeres and regulate gene expression [13–15]. Because of the preferential nuclear localization of TERRA [16], investigation of TERRA partners has been limited to nuclear RNA and protein fractions. A large network of TERRA-interacting partners—including shelterin and chromatin proteins, telomerase components, transcription factors, DNA replication proteins, RNA-binding factors, and nuclear heterogenous ribonucleoproteins (RNPs)—has been discovered in the nuclei of mammalian cells [12, 15, 17–19].
TERRA has been found mainly in the nucleus, although cytoplasmic localization of shelterin proteins has been reported [20–22]. Interestingly, extranuclear TERRA foci have been detected in human cancer cell line after knockdown of retrotransposon Long Interspersed Nuclear Element 1 (LINE1) [23]. By studying Drosophila telomeres, we have found compelling evidence that Drosophila telomeric transcripts and their interacting proteins form RNP particles in the cytoplasm upon telomere dysfunction during oogenesis and early development [24], suggesting undiscovered functional implications of the cytoplasmic localization of telomeric factors.
The Drosophila genus and some other Diptera species have lost telomerase activity during evolution; their telomeres are maintained by transpositions of specialized telomeric retrotransposons (reviewed in [25–27]). Many functional and structural features of telomere homeostasis are conserved among species encoding telomerase or using telomeric retrotransposons. Such similarity could be explained by the general and pivotal role of telomeres in the maintenance of linear chromosomes and by the retrotransposon origin of telomerase reverse transcriptase [28, 29]. In both cases, telomeres are elongated by a reverse transcriptase. The protective complex terminin, a functional analogue of human shelterin, protects Drosophila chromosome ends from the DNA repair machinery and telomere fusions [30, 31]. Terminin consists of Drosophila-specific proteins binding telomeric DNA and single stranded overhang similar to the shelterin proteins. Studies of telomeric chromatin organization and regulation of telomeric repeat transcription in Drosophila and species possessing telomerase revealed many common mechanisms despite the structural differences [27, 32].
Drosophila melanogaster telomeres are elongated via the retrotransposition of three non-long terminal repeat (LTR) retrotransposons, HeT-A, TART, and TAHRE; non-autonomous HeT-A is the most abundant telomeric element [25]. Transcription of Drosophila telomeric retrotransposons was reported a decade before the discovery of TERRA [33]. HeT-A-encoded RNA-binding Gag protein performs typical telomeric functions: It localizes at telomeres, mediates specific targeting of the telomeric transcripts to chromosome ends, and prevents telomere fusions [24, 34, 35]. In wild type flies, the transcription of telomeric repeats is shut down primarily through the assembly of telomeric heterochromatin [36, 37]. In the Drosophila germline, the deposition and maintenance of repressive histone 3 mark, H3K9me3, and heterochromatin protein 1 (HP1) at telomeres is largely mediated by small Piwi-interacting RNAs (piRNAs). Accordingly, depletions of genes related to the heterochromatin assembly and piRNA pathway lead to the overexpression of telomeric elements and excessive telomere elongation [37–40]. We have also identified other factors related to different functional groups that downregulate the levels of HeT-A RNAs in the germline [41]. Among them the conserved RNA-binding arsenite-resistance protein 2 (Ars2) which acts also as a negative regulator of TERRA in human cells [17]. The deadenylase Ccr4–Not complex acts co-transcriptionally and mediates the degradation of Drosophila telomeric transcripts in telomere-associated bodies [42], whereas in human cells, deadenylases Ccr4 and Caf1 regulate telomerase RNA biogenesis in nuclear Cajal bodies [43]. These examples emphasize the similarity of regulatory processes despite the structural differences between the two types of telomeres.
It is believed that a balance between silencing mechanisms of Drosophila telomeric retroelements and the maintenance of appropriate levels of telomeric RNA template provides a proper telomere elongation and normal development [44]. The disruption of telomeric silencing in the germline leads to the accumulation of telomeric retrotransposon HeT-A RNPs in the cytoplasm of the germ cells and early embryos [24]. RNA-binding protein Egalitarian (Egl) [45, 46] was the first identified protein interacting with the cytoplasmic HeT-A RNP aggregates accumulated in the ovaries of the piRNA pathway mutants [24]. Egl mediates the proper localization of maternal mRNAs, and its ectopic localization within HeT-A Gag granules likely contributes to abnormalities of the embryonic axis specification, the phenomenon typical for the piRNA pathway mutants [47–49].
Maintaining telomere homeostasis during oogenesis is essential for genome stability during gametogenesis and early development, and telomere dysfunction is considered a potential biomarker for infertility in humans [50]. Depletion of any of the factors downregulating Drosophila HeT-A expression results in a highly similar phenotype characterized by mitotic defects during early development and embryonic lethality. This phenotype is accompanied by the accumulation of HeT-A RNPs consisting of HeT-A RNA and HeT-A Gag, an RNA binding protein, near centrosomes, which play the major role in mitotic spindle assembly in syncytial preblastoderm embryos [24, 41]. These observations inspired us to hypothesize about an unexpected role of cytoplasmic HeT-A RNPs in the embryonic lethality observed upon telomere dysfunction. Here, we have demonstrated the cytotoxicity of telomeric repeat HeT-A hyperexpression and revealed essential mitotic factors associated with cytoplasmic HeT-A RNA and Gag. Moreover, the depletion of Spindle E (SpnE), Ccr4 or Ars2 leads to accumulation of abundant HeT-A RNPs in the germline and is accompanied by impaired dynamics of Polo kinase, centrosome defects and aborted embryonic development. Our data suggest a mechanistic link between HeT-A mRNA, HeT-A Gag and cell cycle components, which could explain the severe developmental consequences of telomere dysfunction.
Results
Overexpression of telomeric retrotransposon HeT-A is cytotoxic and leads to the accumulation of HeT-A Gag near centrosomes
In our previous studies, we made a striking observation that the depletion of unrelated non-telomeric factors leading to HeT-A overexpression in the Drosophila ovaries, exhibited phenocopies during early development characterized by chromosome fusions, centrosome amplification and accumulation of HeT-A RNPs nearby centrosomes [24, 41]. However, this phenotype could be explained by the misregulation of various cellular mechanisms controlled by the studied factors rather than by telomere dysfunction. Therefore, we tested the cytotoxic effect of HeT-A overexpression in a normal cell line and in vivo in the wild type genetic background. For these experiments, we used pUAST-HeT-A-HA construct expressing HeT-A Gag protein tagged with HA and FLAG epitopes [24, 51].
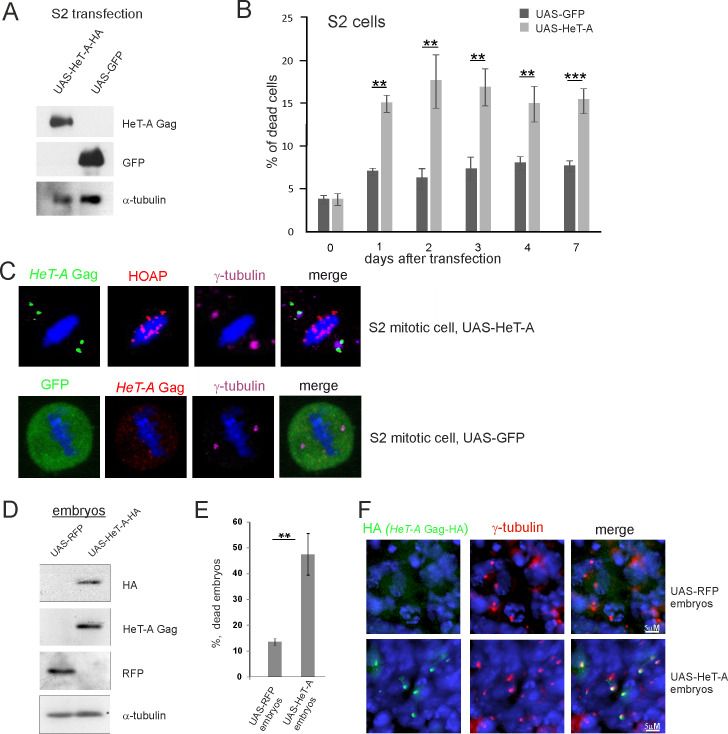
Drosophila embryonic Schneider 2 (S2) cultured cells were transiently co-transfected with the pUAST-HeT-A-HA and pAct-GAL4 driver constructs to induce HeT-A overexpression. A similar experiment with the pUAST-GFP construct was performed as a control. The increased levels of HeT-A Gag and green fluorescent protein (GFP) proteins were validated by western blotting (Fig 1A). Death curves were analyzed for week after transfection. Increased cell death was observed starting from the second day after transfection with the HeT-A- but not the GFP-expressing constructs (Fig 1B). Immunostaining revealed a specific pattern of HeT-A Gag localization near centrosomes stained for γ-tubulin, a pericentriolar matrix (PCM) component, in all studied mitotic S2 cells (n = 22) transfected with the HeT-A construct; however, HeT-A Gag was not found at telomeres stained for the telomere-specific HOAP protein (Fig 1C). GFP was evenly dispersed in the cells (Fig 1C).
Fig 1. Induced overexpression of HeT-A in somatic cells is cytotoxic.
(A) The samples of transfected S2 cells were separated by sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by western blotting using the antibodies indicated on the right. The constructs used for transfection are indicated above the lanes. Co-transfection with pAct-GAL4 driver was used in both cases. (B) The proportion of dead cells present in the S2 cell suspension after transfection by constructs expressing HeT-A Gag or GFP. (C) HeT-A Gag (green), HOAP (red), and γ-tubulin (magenta) immunostaining was performed on S2 cells transfected with the construct expressing full-length HeT-A (upper panel). GFP (green), HeT-A Gag (red), and γ-tubulin (magenta) immunostaining was performed on S2 cells transfected with the construct expressing GFP (lower panel). Mitotic cells at metaphase are shown. DNA is stained with DAPI (blue). (D) Western blot analysis of embryonic extracts from strains expressing UAS-RFP or UAS-HeT-A-HA under the ubiquitous daGAL4 driver. The antibodies used for western blotting are indicated on the right. (E) The percentage of dead embryos laid by transgenic females expressing HeT-A Gag or RFP. (B and E) The SEM for four independent experiments was calculated. Asterisks indicate statistically significant differences (* P < 0.05 to 0.01, ** P < 0.01 to 0.001, *** P < 0.001, unpaired t-test). (F) HeT-A Gag-HA (green) and γ-tubulin (red) immunostaining was performed on 3–5-h-old embryos expressing HeT-A Gag-HA or RFP. DNA is stained with DAPI (blue).
Next, we determined how overexpression of transgenic HeT-A in Drosophila embryos affects development. Expression of transgenic pUAST-HeT-A-HA was induced by GAL4 driven by the ubiquitous daughterless (da) promoter. Overexpression of the transgenic UAS-RedStinger driven by daGAL4 served as a control. There were high levels of HeT-A Gag-HA and red fluorescent protein (RFP) in 2–5-h-old embryos (Fig 1D). We next measured embryonic lethality. The survival of embryos with overexpression of HeT-A Gag-HA was significantly reduced compared with control embryos (Fig 1E). Immunostaining of 3–5-h-old embryos revealed centrosomal localization of HeT-A Gag-HA, whereas HeT-A Gag-HA was not detected in the embryos with RFP overexpression (Fig 1F). Similarly, expression of transgenic pUAST-HeT-A-HA induced by GAL4 driven by the neuronal elav promoter induced the accumulation of HeT-A Gag-HA around centrosomes of dividing neuroblasts in D. melanogaster larvae (S1 Fig).
We conclude that transgenic HeT-A overexpression in cultured cells and in vivo in the normal genetic background is cytotoxic and leads to cell death and embryonic lethality; this is accompanied by accumulation of HeT-A Gag-HA around centrosomes. Based on these observations, we suggested that centrosomal localization of HeT-A Gag may have a functional role, and HeT-A Gag may interfere with mitotic apparatus causing the arrest of cell divisions.
HeT-A Gag associates with mitotic regulators in early Drosophila embryos
Due to the activity of the piRNA pathway, expression of transgenic HeT-A is strongly repressed in the germline in contrast to the somatic tissues, making the experiment with transgenic HeT-A overexpression in the wild type germline impossible [24]. Therefore, to study the effects of HeT-A overexpression in the germline, we further used mutations or germline knockdowns (GLKD) of factors that negatively regulate HeT-A expression: SpnE, a component of the piRNA pathway, Ccr4 deadenylase, encoded by the gene twin, and RNA-binding protein Ars2 (S2 Fig). During its lifecycle in the germline, HeT-A RNA forms RNP complexes with HeT-A Gag [24]; therefore, we used HeT-A Gag immunostaining or HeT-A RNA fluorescence in situ hybridization (FISH) to trace the distribution of HeT-A RNP.
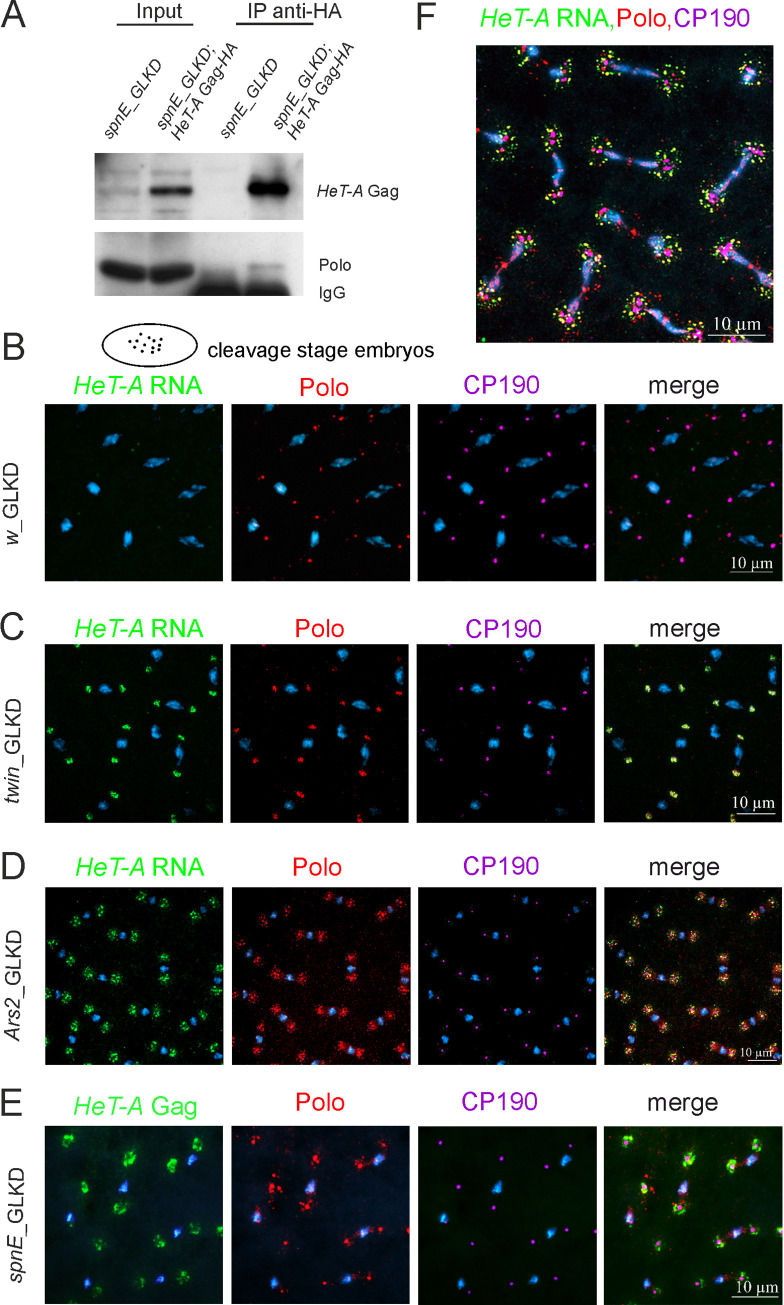
First, we studied the interaction partners of HeT-A RNPs in syncytial preblastoderm Drosophila embryos depleted of the piRNA pathway component spnE. In such embryos, the nuclei synchronously divide in a common cytoplasm, and abundant HeT-A RNPs surround the centrosomes [24, 41]. We performed affinity purification of the HeT-A Gag protein complex from the lysate of 0–2-h-old spnE_GLKD embryos expressing HeT-A Gag-HA using anti-hemagglutinin (HA) antibody–conjugated magnetic beads. In the control experiment, spnE_GLKD embryos did not carry the UAST-HeT-A-HA transgene. Purified proteins were separated by gel electrophoresis (S3 Fig) and analyzed by liquid chromatography coupled to mass spectrometry (LC-MS) (sample preparation and data analysis are described in the S2 File). The proteins involved in the regulation of cell division and centrosome maintenance were identified among potential partners of HeT-A Gag-HA (S1 Table). In this study, we have focused on Polo kinase, a conserved regulator of the cell cycle [52], which we have detected among HeT-A Gag-HA partners. We validated the association of Polo with HeT-A Gag-HA by co-immunoprecipitation (co-IP) performed using 0–2-h-old embryos of a transgenic strain with HA-tagged Gag HeT-A overexpression on the background of spnE_GLKD. The HeT-A Gag-HA protein complex was affinity purified using an anti-HA antibody and analyzed by western blotting. Polo was reproducibly co-purified with HeT-A Gag-HA (Fig 2A, S4A Fig), corroborating the MS data.
Fig 2. HeT-A ribonucleoproteins interact with Polo kinase in early Drosophila embryos.
(A) Coimmunoprecipitation experiments were performed on extracts from 0–2-h-old spnE_GLKD embryos using an anti-HA antibody. The samples were separated by SDS-PAGE and analyzed by western blotting using the antibodies indicated on the right. The starting lysate (input) and immunoprecipitated (IP) probes are indicated; the antibodies used for co-IP are indicated above the IP lanes. (B-E) The images demonstrate the localization of HeT-A Gag or HeT-A RNA (green), Polo (phosphorylated form, red), and CP190 (magenta) in the control (B), twin (C), Ars2 (D) and spnE (E) GLKD at metaphase in syncytial embryos. A cartoon of cleavage stage embryo is shown above the images. (F) The localization of HeT-A RNA (green), Polo (red), and CP190 (magenta) at anaphase in the Ars2_GLKD syncytial embryos. The blue color indicates DNA.
Polo is a highly dynamic enzyme, and the proper spatiotemporal control of its activity is crucial for correct cell divisions [53]. Polo colocalizes with centrosomal components throughout the entire embryonic mitotic cycle; in addition, Polo associates with the centromeres, mitotic spindle, and midbody [54]. Next, we asked whether the interaction of HeT-A Gag with Polo affects Polo localization during early embryogenesis. To this end, we performed Polo immunostaining in the syncytial embryos with spnE, Ars2 and twin GLKD causing HeT-A overexpression and revealed substantial differences in Polo localization compared with wild type embryos.
As expected, Polo associated with centrosomes stained with centrosomal protein 190 (CP190) in the control embryos with the germline knockdown of the white gene (w_GLKD) encoding adult eye pigment (Fig 2B). In spnE, Ars2, and twin GLKD embryos, Polo was also detected at centrosomes, and there were also Polo aggregates randomly dispersed around the centrosomes and colocalized with endogenous HeT-A RNA and Gag-HA during syncytial cleavages (Fig 2C–2E). In anaphase of Ars2_GLKD, Polo/HeT-A RNA aggregates become more dispersed moving away from the centrosomes (Fig 2F). HeT-A RNA and Gag-HA also colocalized with Cdk1 in syncytial embryos with spnE, Ars2, and twin GLKD (S5 Fig). In the cortex of spnE_GLKD embryos, Cdk1 and HeT-A Gag-HA aggregates associate with free centrosomes (S5 Fig), the accumulation of which indicates mitotic defects in the syncytial embryo [55]. Western blotting did not reveal the changes in the Polo and Cdk1 protein levels in spnE_GLKD early embryos (S6 Fig).
In embryos overexpressing HeT-A we observe similar localization pattern between HeT-A RNA and HeT-A Gag and conclude that HeT-A RNPs interact with Polo and Cdk1, which probably leads to mislocalization of these mitotic kinases during early development.
HeT-A RNA and Gag interact with mitotic kinases and centrosomal components during oogenesis
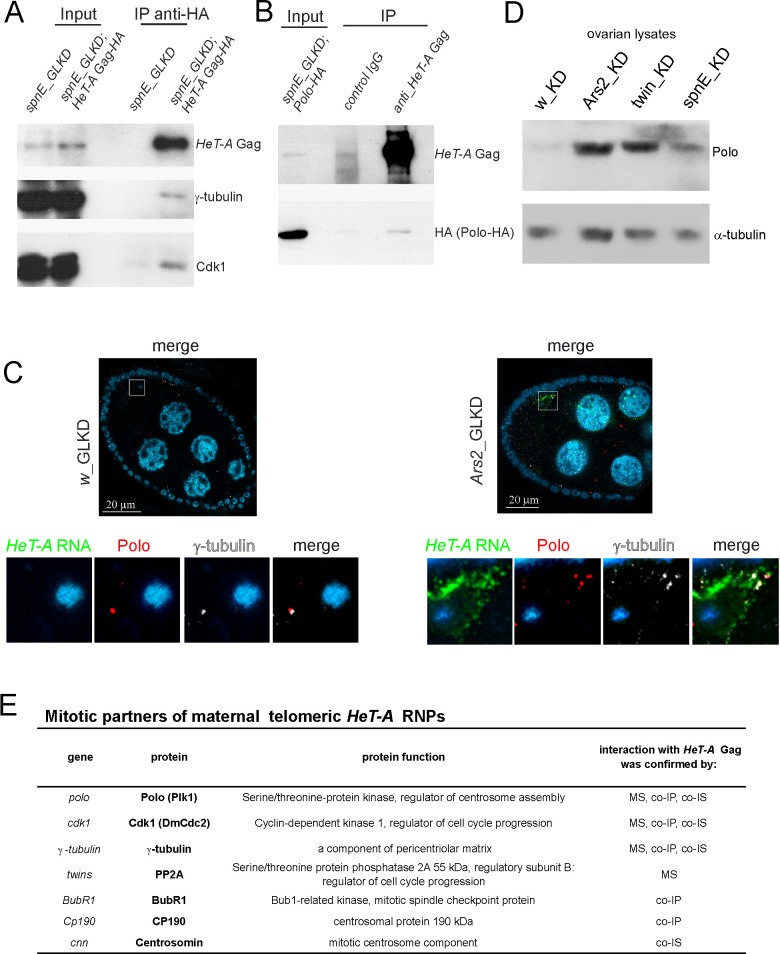
Based on the observed interactions of HeT-A Gag-HA in syncytial embryos, we examined the mitotic partners of HeT-A Gag during oogenesis upon HeT-A overexpression. Co-IP was performed using ovary extracts of the transgenic strain expressing HA-tagged Gag HeT-A with spnE_GLKD. The HeT-A Gag-HA protein complex was affinity purified using an anti-HA antibody and analyzed by western blotting. Cdk1 kinase and γ-tubulin were co-purified with HeT-A Gag-HA (Fig 3A). The HeT-A Gag–Polo interaction in ovaries was confirmed by the co-IP experiment with anti-HeT-A Gag antibodies using spnE_GLKD flies expressing Polo-HA in the germline (Fig 3B). In addition, CP190 and the mitotic kinase BubR1 [56] were revealed in the ovarian HeT-A Gag-HA complex (S4B Fig).
Fig 3. HeT-A ribonucleoprotein particles interact with Polo during oogenesis.
(A) A coimmunoprecipitation experiment was performed on ovarian extracts from spnE_GLKD flies expressing HeT-A Gag-HA. (B) A co-IP experiment was performed on ovarian extracts from spnE_GLKD flies expressing Polo-HA. The samples were separated by SDS-PAGE and analyzed by western blotting using the antibodies indicated on the right. The starting lysate (input) and immunoprecipitated (IP) probes are indicated; the antibodies used for co-IP are indicated above the IP lanes. (C) HeT-A RNA FISH (green) combined with immunostaining of Polo (phosphorylated form, red) and γ-tubulin (grey) in the control (left panel) and Ars2_GLKD ovaries (right panel). Egg chambers at 9–10 stages of oogenesis are shown. The lower panels show the enlarged areas highlighted by squares. The blue color indicates DNA. (D) Western blot analysis of ovary lysates probed with anti-Polo antibody; α-tubulin was used as a loading control. (E) Mitotic partners of maternal HeT-A RNPs revealed by different methods. MS, mass spectrometry; co-IP, co-immunoprecipitation; co-IS, co-immunostaining.
Thus, abundant HeT-A RNPs interact with Polo kinase and other cell cycle regulators during oogenesis and early development (Fig 3E).
Polo is localized to the centrosomes throughout the mid-stages of Drosophila oogenesis, and its level sharply decreases at later stages of normal oogenesis [57]. At stages 9–10 of normal oogenesis, there was weak Polo staining barely detected in the vicinity of the oocyte nucleus (Fig 3C). In contrast to the normally reduced Polo levels at later stages, Polo was dispersed in the cytoplasm of the oocyte and co-localized with HeT-A RNA in Ars2 and twin GLKD (Fig 3C, S7 Fig). The germline overexpression of UASg-Polo-HA transgene in spnE_GLKD resulted in the formation of Polo-HA aggregates co-localized with HeT-A RNA in the oocyte (S7 Fig). Accumulation of endogenous Polo in the ovaries of spnE, twin, and Ars2 GLKD compared with control w_GLKD ovaries was validated by western blotting (Fig 3D). There was a more pronounced effect obtained by western blot analysis of 12–14 stage egg chambers of spnE_GLKD (S7 Fig).
Polo kinase activity is required for centrosome maturation and separation [58–60]. polo depletion causes a failure in the recruitment of centrosomin (CNN), γ-tubulin and CP190 to the centrosome [60, 61]. Given the crucial role for Polo in centriole biogenesis, we hypothesized that HeT-A RNP interactions with Polo and other regulatory cell cycle proteins lead to centrosome biogenesis defects during oogenesis.
HeT-A overexpression is accompanied by centrosome dysfunction during oogenesis
The centrosomal structure is highly dynamic during oogenesis and early development. At the early stages of oogenesis, the centrosomes of the nurse cells move into the oocyte and form the microtubule-organization center (MTOC) near the nucleus [62].
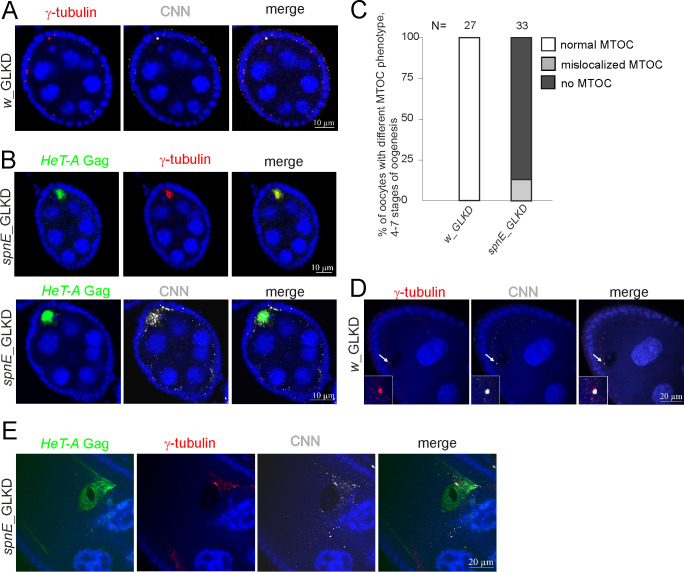
We studied the effect of HeT-A overexpression on MTOC formation during oogenesis. Immunostaining of CNN and γ-tubulin PCM was performed to monitor the state of the centrosome at different stages of oogenesis in the control and spnE_GLKD ovaries expressing HeT-A Gag-HA. In w_GLKD, the centrioles were clustered at a posterior pole of the oocyte, forming a strong focus of MTOC at stage 6 of oogenesis (Fig 4A). spnE_GLKD resulted in abnormal distributions of centrosomal proteins in the oocyte: CNN/γ-tubulin became dispersed in the oocyte at stage 6 of oogenesis and showed co-localization with abundant HeT-A Gag-HA (Fig 4B). Quantification of oocytes with a discrete MTOC revealed only a few of them in spnE_GLKD ovaries (Fig 4C). Even in these cases, the MTOC was unusually large and abnormally positioned (S8A Fig).
Fig 4. HeT-A overexpression caused by spnE knockdown is accompanied by centrosome dysfunction during oogenesis.
(A) Co-immunostaining demonstrates colocalization of CNN (grey) and γ-tubulin (red) in the control ovaries (w_GLKD). (B) Co-immunostaining demonstrates the localization of HeT-A Gag (green), CNN (grey), or γ-tubulin (red) in spnE_GLKD ovaries. Stage 6 of oogenesis is shown (A, B). (C) Bar diagrams show the MTOC phenotype (%) observed at 4–6 stages of oogenesis in spnE_GLKD. (D) Co-immunostaining of CNN (grey) and γ-tubulin (red) shows the centrosome reduction at stage 9 of oogenesis in w_GLKD ovaries. (E) Immunostaining reveals the accumulation of HeT-A Gag (green) and mislocalized CNN (grey) and γ-tubulin (red) at stage 10 of oogenesis in spnE_GLKD. The blue color indicates DNA.
In many species, including Drosophila, centrioles disappear during oogenesis [63]. In D. melanogaster, the zygotic centriole is provided paternally, whereas maternal centrioles are eliminated during late stages of oogenesis. At these stages, Polo levels decrease sharply, resulting in a reduction and the eventual elimination of centrosomes [57, 64]. In w_GLKD, we observed a normal reduction of centrosomes at stage 9 of oogenesis (Fig 4D). In spnE_GLKD, centriolar protein staining persisted until stage 10 of oogenesis at the anterior pole in the vicinity of the nucleus and was partially co-localized with HeT-A Gag-HA (Fig 4E). Multiple enlarged and abnormally positioned MTOCs were frequently observed at stage 9 of oogenesis in twin_GLKD (S8B Fig). In Ars2_GLKD, centrosomes were abnormally amplified as shown by Polo–γ-tubulin immunostaining (Fig 3C).
Thus, we discovered an impaired centrosome biogenesis to be a common result of depletions of such functionally different factors as SpnE, Ccr4, or Ars2, all of which are characterized by the HeT-A overexpression phenotype. Moreover, HeT-A transcripts and Gag were associated with abnormally localized centrosomal proteins in the oocyte.
HeT-A overexpression is accompanied by centriole dysfunction during late stages of oogenesis and early zygotic development
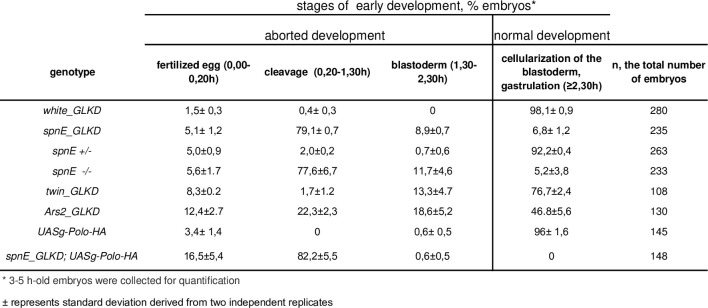
Next, we addressed how depletion of three factors leading to HeT-A overexpression in the germline can affect early embryogenesis. We determined the stage of developmental arrest in spnE_GLKD, spnE mutants, twin_GLKD, and Ars2_GLKD embryos. To this end, we quantified the percentage of 3–5-h-old embryos arrested at different stages of embryonic development. We found that spnE GLKD or spnE mutations caused strong maternal-effect embryonic lethality (Fig 5). In those embryos that overcame the first zygotic divisions, there was developmental arrest at the early syncytial cleavage stage, resulting in > 80% early embryonic lethality. Of note, 10% and 35% of twin_GLKD and Ars2_GLKD embryos, respectively, were arrested before blastoderm formation.
Fig 5. Maternal-effect embryonic lethality accompanying HeT-A overexpression.
It has been reported that the presence of maternal centrosomes caused by artificial Polo tethering to the centriole in the oocyte can lead to meiotic defects and developmental arrest during the initial zygotic divisions [57]. We observed that HeT-A overexpression was accompanied by abnormal accumulation of Polo until the late stages of oogenesis (Fig 3D). We therefore suggest that accumulation of Polo and centrosomal proteins during late stages of oogenesis could explain early developmental disorders in spnE, twin and Ars2 GLKD flies characterized by HeT-A overexpression.
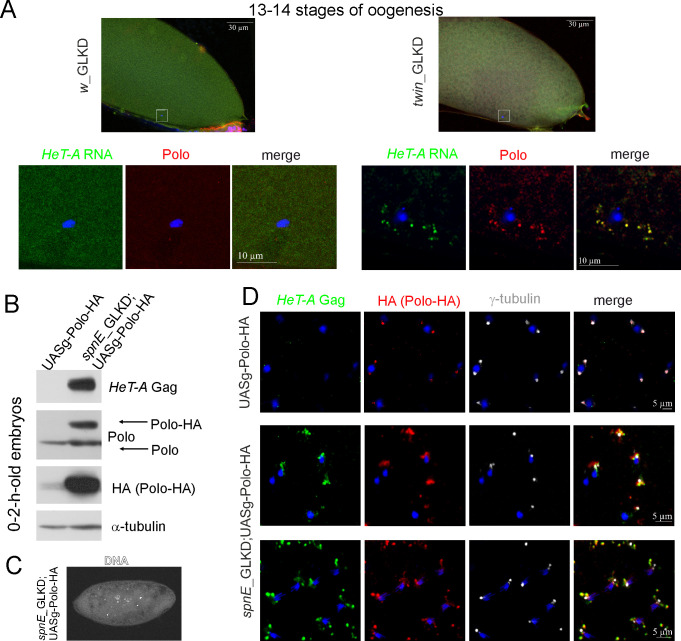
We performed HeT-A RNA FISH combined with Polo and CNN immunostaining at stages 13–14 of oogenesis, which are normally characterized by centriole elimination. In contrast to the control oocytes (w_GLKD), which did not contain centrioles, we found Polo and CNN foci to be co-localized with HeT-A RNA around the oocyte nucleus in twin_GLKD (Fig 6A, S9 Fig).
Fig 6. Retention of maternal Polo is mediated by its interaction with abundant telomeric HeT-A ribonucleoprotein particles.
(A) HeT-A RNA FISH (green) combined with Polo (phosphorylated form, red) immunostaining in the control (w_GLKD) and twin_GLKD egg chambers at 13–14 stages. The upper images demonstrate the general view; the lower panels show the enlarged area around the oocyte nucleus obtained with a 63× objective lens. (B) Western blot analysis of 0–2-h-old embryo lysates shows the overexpression and accumulation of Polo-HA in spnE_GLKD embryos. The antibodies are indicated on the right. (C) DAPI staining of spnE_GLKD;UASg-Polo-HA embryos reveals developmental arrest at early syncytial cleavage stage. (D) Co-localization of HeT-A Gag (green) and Polo-HA (red) in the spnE_GLKD;UASg-Polo-HA syncytial embryos. γ-tubulin appears in grey, and DNA appears in blue.
We then asked whether overexpressing Polo kinase in the germline can affect the early embryonic lethality in the progeny of spnE_GLKD females. Using an embryonic developmental arrest test, we observed that the germline expression of UASg-Polo-HA enhanced the maternal-effect embryonic lethality of spnE_GLKD embryos (Fig 5). Embryonic lethality in spnE_GLKD;UASg-Polo-HA was predominantly observed during the initial cleavage divisions (Fig 6C). Western blotting demonstrated endogenous HeT-A Gag overexpression and strong Polo-HA accumulation in the 0–2-h-old embryos in spnE_GLKD;UASg-Polo-HA but not in UASg-Polo-HA embryos (Fig 6B). Immunostaining revealed abundant Polo-HA aggregates associated with endogenous HeT-A Gag dispersed around the centrosomes during the early embryonic cleavages (Fig 6D). We suggest that interaction of HeT-A Gag with abundant Polo-HA followed by ectopic retention of Polo-HA in the proximity of the centrosomes could be the main reason for centrosome dysfunction and the increased early developmental arrest rate observed in spnE-depleted embryos overexpressing Polo-HA.
Thus, the depletion of spnE, twin and Ars2 characterized by HeT-A overexpression leads to Polo stabilization during oogenesis, a phenomenon that causes maternal centrosome dysfunction and severe mitotic defects during early development.
Discussion
Using a Drosophila model, we have deepened our understanding of the mechanisms underlying the link between telomere dysfunction and defects in gametogenesis and early development. We have addressed a mechanism of cytotoxicity of abundant HeT-A RNPs accumulated in cells that overexpress telomeric repeats. Using different approaches, we have demonstrated that maternal telomeric RNPs encoded by the telomeric retrotransposon HeT-A interact with the key regulators of the cell cycle during oogenesis and early development (Fig 3E).
In the wild type ovarian germline, the expression of HeT-A is strictly downregulated by various factors, including the piRNA pathway [39, 40], HP1 chromodomain protein [37, 65], Woc and Trf2 transcription factors, Ars2 RNA-binding protein [41], and the Ccr4–Not deadenylase complex [42]. Recent findings have revealed the role of histone-modifying enzymes in the HeT-A repression in the germline [66, 67]. Depletions of each of these factors increase HeT-A RNA levels up to 1000 fold in the ovaries and cause severe ovary degeneration, patterning defects and aborted development [41, 47, 49, 68, 69]. Despite the fact that the factors listed above regulate numerous targets beyond HeT-A, we wondered whether HeT-A overexpression could be directly related to the observed phenotype. In this study, we have addressed the mechanism of developmental arrest upon HeT-A overexpression caused by the depletions of SpnE, a component of the piRNA pathway, Ars2, a key factor in RNA nuclear metabolism [70], and Ccr4 deadenylase, which plays an essential role in the control of maternal mRNA stability during oogenesis and early development [71]. Noteworthy, HeT-A is the only common transposon target of these three pathways. Indeed, SpnE, a component of the piRNA pathway, controls many transposon families [72], and Ccr4 deadenylase co-transcriptionally regulates HeT-A and actively transcribed transposons [42]. However, RNA-binding protein Ars2 downregulates the expression of telomeric but not other transposons [41, 73]. We cannot attribute mitotic defects solely to HeT-A overexpression and exclude the involvement of other common targets of SpnE, Ccr4 and Ars2. Nevertheless, our results strongly support an essential role of telomeres in impaired centrosome biogenesis in the ovaries of spnE, twin and Ars2 GLKD. One may suggest that the observed high sensitivity of telomere signaling ensures a prompt cellular response to a wide range of genetic abnormalities.
Centrosomes are the key players in gametogenesis and the syncytial stage of embryogenesis. Their dynamics is regulated by mitotic kinases, and Polo plays a major role in centrosome maturation and separation [58]. We discovered the physical interactions between HeT-A Gag and the cell cycle kinases Polo and Cdk1 suggesting a mechanistic explanation for the role of HeT-A RNPs in the mitotic failure observed during early development after HeT-A overexpression [41]. Given the function of Polo in γ-tubulin and CNN recruitment to the centrosome, we suggested that the HeT-A Gag–Polo interaction may cause centrosome dysfunction. Supporting this idea, we observed severe defects in MTOC assembly during oogenesis: γ-tubulin and CNN were dispersed in the cytoplasm of oocytes instead of concentrating in the MTOC. However, because γ-tubulin, CNN, and CP190 also interact with HeT-A Gag and are found in Gag aggregates, the mechanism of centrosome dysfunction seems to be more complex. HeT-A Gag may directly or indirectly, through the assistance of Polo, interact with mitotic kinases and centrosome components, which collectively strongly affect centrosome dynamics. Abnormal accumulation of centrosomal proteins trapped in the HeT-A Gag complexes in the mature oocyte may explain the maternally determined embryonic lethality observed upon HeT-A overexpression. It seems that HeT-A RNP interactions with mitotic proteins do not play a role under normal conditions due to the extremely low levels of HeT-A expression [24], but they become a threat for development when HeT-A RNA levels exceed a certain threshold under telomere dysfunction conditions.
Analysis of maternal HeT-A Gag interactors identified a number of cell cycle proteins (Fig 3). Musaro et al. have reported a mechanistic link between Drosophila telomeres and components of anaphase checkpoint control [74]. In that work, studying the mechanism of metaphase arrest in the brain cells caused by unprotected Drosophila telomeres revealed that uncapped telomeres as well as the unattached kinetochores recruit the BubR1 kinase, which could inhibit anaphase onset. We found HeT-A Gag aggregates to be associated with Polo and Cdk1 in syncytial embryos around mitotic spindle poles, where components critical for proper mitotic progression are normally organized and orchestrated [75]. Frequently, HeT-A RNP and Polo/Cdk1 aggregates associate with free centrosomes in early embryos [41] (Fig 6D, S5E Fig). At the same time, detachment of γ-tubulin-positive centrosomes accompanied by elimination of the nuclei have been associated with compromised Polo function [76, 77]. These facts suggest the direct role of HeT-A RNP–Polo interactions in centrosome detachments during early development.
The mechanism of selective interaction of mitotic kinases with HeT-A Gag cytoplasmic granules is unclear. HeT-A Gag is phosphorylated, which is important for the protein stability [78]. It has been reported that Cdk1 activity produces the docking sites for Polo recruitment to its targets [79–81]. One may suggest that phosphorylation of HeT-A Gag by Cdk1 creates Polo docking sites leading to Polo targeting to the HeT-A Gag. Future studies will elucidate whether HeT-A Gag is a substrate of Polo and Cdk1 mitotic kinases.
What could be the evolutionary origin of the HeT-A Gag targeting to the centrosomes? Several viruses—including retroviruses and oncogenic viruses—have been shown to target the centrosome; moreover, viral proteins can directly interact with centrosomal proteins and alter their activity [82–86]. The evidence for cell cycle dysfunction promoted by viral proteins has been reported for the human immunodeficiency virus type 1 (HIV-1). HIV-1 Tat protein has been shown to induce apoptosis by interacting with the cell cycle proteins and suppressing Polo-like kinase 1 (Plk1) activity [87]. HIV-1 viral protein R (Vpr) localizes to the centrosome and modulates the activity of the ubiquitin ligase EDDDYRK2-DDB1DCAF1 or inhibits the Cdc25C phosphatase activity [88–90]. HIV-1 viral infectivity factor (Vif) causes dysfunction of Cdk1 and Cyclin B1 leading to cell cycle arrest [91]. Lastly, HIV-1 genomic RNA and Gag polyprotein colocalization near the centrosome is essential for viral particle assembly [92]. Centrosome targeting of retroviral GAG proteins has been described for mouse mammary tumor virus (MMTV), Mason-Pfizer monkey virus (M-PMV), and human foamy virus (HFV) [84, 93, 94]. It is believed that by targeting the centrosomal proteins, some viruses hijack their functions leading eventually to cell death or transformation. It is possible that HeT-A, as a retroelement, has inherited similar properties that have been adapted during evolution to the telomere signaling functions. In future studies, it will be important to determine the peptide sequences of HeT-A Gag responsible for the centrosomal targeting and interactions with the cell cycle and centrosomal proteins.
Interestingly, shelterin components being not of retrotransposon origin also localize at the centrosomes in human cells. Telomere repeat factor 1 (TRF1), a telomeric DNA-binding protein, localizes at mitotic spindles in human cancer cells [95] and mediates mitotic abnormalities via the Aurora-A kinase [96]. The telomeric factor tankyrase also localizes to both telomeres and centrosomes in HeLa cells [22]. Note that the association between telomeric proteins and the mitotic apparatus has been observed in cancer human cells for which telomere dysfunction is a typical hallmark. The molecular mechanism of the centrosome targeting of shelterin proteins is unclear. The retrotransposon origin of telomeres suggests that mechanisms of transposon control could be adopted for telomere regulation [26], and recent publications support this idea. Retrotransposon LINE1 knockdown leads to telomere dysfunction in human cancer cells [23, 97]. Moreover, LINE1 encoded proteins bind to telomeric ends and to TERRA [23]. One may suggest that the centrosome targeting of shelterin components could be mediated by retrotransposon proteins. It is tempting to speculate that interaction of the telomeric RNP with cell cycle regulators is an evolutionary conserved pathway present in both Drosophila and species possessing telomerase, and this process coupling telomere dysfunction and the cell cycle collapse can be considered as a telomeric checkpoint.
Materials and methods
D. melanogaster strains
The transgenic strain expressing HeT-A Gag protein tagged with HA and FLAG epitopes was described previously [24, 51]. As a control for the cell line experiments, the GFP open-reading frame (ORF) cloned into the pUASp-attB vector was used. Strain w[1118]; P{w[+mC] = UAS-RedStinger}6 (#8547, the Bloomington Drosophila Stock Center [BDSC]) was used as a control for the measurement of embryonic lethality. GLKD flies were F1 progeny of the genetic cross of a strain bearing a construct with short hairpin (sh)RNA (spnE_sh, #103913, the Vienna Drosophila Resource Center [VDRC]; twin_sh, #32490, BDSC; Ars2_sh, #106344, VDRC; w_sh, # 30033, VDRC, and a driver strain P{UAS-Dcr-2.D}. v1, w1118, P{GAL4-nos.NGT}40, #25751, BDSC). To induce tissue-specific expression of transgenic pUAST-HeT-A-HA, we used crosses with driver strains P{GAL4-nos.NGT}40 (germline), P{GAL4-da.G32}UH1 (ubiquitous) and P{GawB}elav[C155] (neuronal). The transgenic strain expressing poloORF-3xHA was from FlyORF (FO01229). The spnE mutant alleles were spnE1 and spnEhls3987.
Drosophila cell culture experiments
D. melanogaster embryonic S2 cells were transfected using the FuGENE® HD Transfection Reagent (Promega) according to the manufacturer’s instructions. Co-transfection of pUAST-HeT-A-HA plasmid [51] or pUAST-GFP along with the driver plasmid pAct-GAL4 was performed to induce HeT-A or GFP expression. Twenty-four hours post-transfection, the cells were resuspended and seeded on coverslips for 1 h. Immunostaining was performed as described previously [98], except that the samples were incubated with secondary antibodies 1 h at room temperature. The number of viable cells in culture was determined by the trypan blue exclusion test [99] every 24 h for 7 days after transfection.
Immunostaining and immunoprecipitation
Immunostaining and RNA FISH combined with immunostaining were carried out according to the previously described procedure [24]. A digoxigenin (DIG)-labeled antisense HeT-A riboprobe containing a fragment of the ORF (nucleotides 4330–4690 of GenBank sequence DMU06920) was used. Images were captured using a Zeiss LSM 900 confocal microscope and analyzed using ImageJ and Adobe Photoshop. Co-IP of HeT-A Gag-HA from embryo and ovary extracts was performed as described [24]. For co-IP of HeT-A Gag from the ovary extract, a Guinea pig anti-Gag antibody (kindly provided by Y. Rong) and Dynabeads Protein G magnetic beads (Invitrogen) were used (25 μL per sample).
Antibodies
The following primary antibodies were used: rabbit and mouse anti-γ-tubulin (Sigma); rabbit anti-Cdk1/Cdc2 (Sigma); rabbit anti-CNN (kindly provided by T.L. Megraw; [100]); rat anti-CP190 (kindly provided by A. Golovnin; [101]); rabbit anti-Plk1 (phosphor-Thr210, LSBio, used for immunostaining and western blot analysis); mouse anti-Polo (monoclonal antibody MA81 used for western blot analysis was kindly provided by C. Sunkel; [102]); rabbit anti-BubR1 (kindly provided by C. Sunkel; [56]); rabbit anti-DsRed (Clontech); rabbit and mouse anti-HA (Cell Signaling Technology); mouse anti-GFP (Abcam); guinea pig anti-HOAP (polyclonal antibodies against full-length HOAP protein generated by PrimeBioMed, Skolkovo, Russia); guinea pig anti-Gag HeT-A (kindly provided by Y. Rong); rabbit anti-Gag HeT-A (kindly provided by E. Casacuberta; [103]); and mouse polyclonal anti-Gag HeT-A (generated in the Immunochemistry Laboratory, Branch of IBC RAS, Pushchino, Russia, using recombinant protein corresponding to the 1–375 amino acids of HeT-A Gag). Alexa Fluor–conjugated secondary antibodies with minimal cross-reactivity to IgG from non-target species (Jackson ImmunoResearch) were used (dilution 1:500).
Quantification of embryonic phenotypes
Three hundred females were allowed to lay eggs on agar plates for 2 h. The flies were then removed, and the eggs were collected from the plates after 3 h. The 3–5-h-old embryos were fixed with methanol as described previously [104], incubated in phosphate-buffered saline (PBS) containing 0.5 μg/ml DAPI (4’,6-diamidino-2-phenylindole), and mounted in Vectashield Antifade Mounting Medium (Vector Laboratories). DNA staining of whole-mount embryos was used to visualize the stages of embryonic development according to [104].
Supporting information
(XLSX)
(RAR)
Mass spectrometry sample preparation and data analysis.
(PDF)
HeT-A Gag-HA (red) and α-tubulin (green) immunostaining was performed on larvae brain of the D. melanogaster strains expressing RFP (upper panel, control) or HeT-A Gag-HA (lower panel). DNA is stained with DAPI (blue).
(TIF)
(A) Analysis of HeT-A RNA levels in the ovaries of indicated strains. RT-qPCR analysis of the HeT-A RNA levels normalized to RNA levels of rp49 housekeeping gene. Bar diagrams show fold changes in steady-state HeT-A RNA levels in the ovaries of flies with the indicated genotypes relative to the control (w_GLKD). Error bars indicate SD (standard deviation) for three biological replicates. spnE mutants were spnE1/spnEhls3987. Heterozygous spnE/+ flies were a mix of spnE1/+ and spnEhls3987/+. (B) Efficiency of the germline knockdowns of SpnE, Ccr4 and Ars2. Western blot analysis of ovary extracts probed with indicated antibodies. α-tubulin staining was used as a loading control.
(TIF)
Coomassie brilliant blue staining of proteins copurified with HeT-A Gag-HA from 0-2-h-old spnE_GLKD embryos containing transgene expressing HeT-A Gag-HA. As a control, spnE_GLKD embryos without HeT-A transgene were used.
(TIF)
(A) Co-IP experiment performed on extracts from 0-2-h-old spnE_GLKD embryos expressing HeT-A Gag-HA reveals that Polo is co-purified with HeT-A Gag-HA. (B) Co-IP experiment performed on extracts from spnE_GLKD ovaries expressing HeT-A Gag-HA reveals that BubR1and CP190 are co-purified with HeT-A Gag-HA. Post-extraction insoluble pellet was loaded between Input and IP probes. The antibodies used for co-IP and western blotting are indicated on the right.
(TIF)
HeT-A RNA FISH (green) and coimmunostaining of Cdk1 (red) and CP190 (magenta) in the control (A), Ars2_GLKD (B) and twin_GLKD (C) early syncytial embryos. Lower panels in (C) show the enlarged area around the centrosome in twin_GLKD embryo. (D, E) Coimmunostaining of HeT-A Gag-HA (green), Cdk1 (red) and CP190 (magenta) in the spnE_GLKD is shown. Free centrosomes in the embryo cortex are shown (E). Blue, DNA.
(TIF)
Western blotting of 0–2 h old embryo lysates shows the overexpression of HeT-A Gag and unchanged Polo and Cdk1 levels in spnE_GLKD embryos compared with the control (w_GLKD). Antibodies are indicated on the right. Anti-Plk1 antibodies (pThr210, LSBio) used here recognize phosphorylated form of Polo.
(TIF)
(A) Colocalization of HeT-A RNA (green), Polo (phosphorylated form, red) and γ-tubulin (grey) in the oocyte of twin_GLKD at stage 9 is shown. Blue, DNA. (B) Accumulation of Polo-HA (red) and HeT-A Gag (green) in the oocyte of spnE_GLKD expressing Polo-HA at stage 6 of oogenesis is shown. Blue, DNA. (C) Western blotting confirms accumulation of phosphorylated Polo in spnE_GLKD egg chambers at 12–14 stages (rep1 and rep2, two biological replicates) compared with w_GLKD. The antibodies used are indicated on the right.
(TIF)
(A) Coimmunostaining demonstrating colocalization of HeT-A Gag (green) and γ-tubulin (red) in spnE_GLKD ovaries at stage 5 of oogenesis is shown. (B) Immunostaining reveals accumulation of HeT-A Gag (green) and multiple γ-tubulin (red) and CNN (grey) foci at stage 9 of oogenesis in twin_GLKD. Blue, DNA.
(TIF)
HeT-A RNA FISH (green) combined with CNN (gray) immunostaining in twin_GLKD at stage 13–14 egg chamber. The lower panel is a blow up of the area around the oocyte nucleus (the upper panel). DNA, blue.
(TIF)
Acknowledgments
We are grateful to Ivan Olovnikov for critical reading of this manuscript. We thank Y. Rong for the anti-HOAP and anti-Gag HeT-A antibodies; E. Casacuberta for the anti-Gag HeT-A antibody; T.L. Megraw for the anti-CNN antibody; A. Golovnin for the anti-CP190 antibody; and C. Sunkel for the anti-Polo and anti-BubR1 antibodies. We thank BDSC, VDRC, and FlyORF for fly strains. We thank the User Centre of the Institute of Molecular Genetics of National Research Centre «Kurchatov Institute» for the opportunity to use microscopy equipment.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
A.K. 19-04-00254 Russian Foundation for Basic Researches https://www.rfbr.ru/rffi/eng The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Anderson R, Lagnado A, Maggiorani D, Walaszczyk A, Dookun E, Chapman J, et al. Length-independent telomere damage drives post-mitotic cardiomyocyte senescence. EMBO J. 2019;38(5). doi: 10.15252/embj.2018100492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fumagalli M, Rossiello F, Clerici M, Barozzi S, Cittaro D, Kaplunov JM, et al. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat Cell Biol. 2012;14(4):355–65. doi: 10.1038/ncb2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hewitt G, Jurk D, Marques FD, Correia-Melo C, Hardy T, Gackowska A, et al. Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat Commun. 2012;3:708. doi: 10.1038/ncomms1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Platt JM, Ryvkin P, Wanat JJ, Donahue G, Ricketts MD, Barrett SP, et al. Rap1 relocalization contributes to the chromatin-mediated gene expression profile and pace of cell senescence. Genes Dev. 2013;27(12):1406–20. doi: 10.1101/gad.218776.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Straatman KR, Louis EJ. Localization of telomeres and telomere-associated proteins in telomerase-negative Saccharomyces cerevisiae. Chromosome Res. 2007;15(8):1033–50. doi: 10.1007/s10577-007-1178-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ye J, Renault VM, Jamet K, Gilson E. Transcriptional outcome of telomere signalling. Nat Rev Genet. 2014;15(7):491–503. doi: 10.1038/nrg3743 [DOI] [PubMed] [Google Scholar]
- 7.Martinez P, Thanasoula M, Carlos AR, Gomez-Lopez G, Tejera AM, Schoeftner S, et al. Mammalian Rap1 controls telomere function and gene expression through binding to telomeric and extratelomeric sites. Nat Cell Biol. 2010;12(8):768–80. doi: 10.1038/ncb2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang D, Xiong Y, Kim H, He Q, Li Y, Chen R, et al. Human telomeric proteins occupy selective interstitial sites. Cell Res. 2011;21(7):1013–27. doi: 10.1038/cr.2011.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azzalin CM, Lingner J. Telomere functions grounding on TERRA firma. Trends Cell Biol. 2015;25(1):29–36. doi: 10.1016/j.tcb.2014.08.007 [DOI] [PubMed] [Google Scholar]
- 10.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318(5851):798–801. doi: 10.1126/science.1147182 [DOI] [PubMed] [Google Scholar]
- 11.Cusanelli E, Romero CA, Chartrand P. Telomeric noncoding RNA TERRA is induced by telomere shortening to nucleate telomerase molecules at short telomeres. Mol Cell. 2013;51(6):780–91. doi: 10.1016/j.molcel.2013.08.029 [DOI] [PubMed] [Google Scholar]
- 12.Deng Z, Norseen J, Wiedmer A, Riethman H, Lieberman PM. TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell. 2009;35(4):403–13. doi: 10.1016/j.molcel.2009.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat Cell Biol. 2008;10(2):228–36. doi: 10.1038/ncb1685 [DOI] [PubMed] [Google Scholar]
- 14.Deng Z, Wang Z, Xiang C, Molczan A, Baubet V, Conejo-Garcia J, et al. Formation of telomeric repeat-containing RNA (TERRA) foci in highly proliferating mouse cerebellar neuronal progenitors and medulloblastoma. J Cell Sci. 2012;125(Pt 18):4383–94. doi: 10.1242/jcs.108118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chu HP, Cifuentes-Rojas C, Kesner B, Aeby E, Lee HG, Wei C, et al. TERRA RNA Antagonizes ATRX and Protects Telomeres. Cell. 2017;170(1):86–101 e16. doi: 10.1016/j.cell.2017.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porro A, Feuerhahn S, Reichenbach P, Lingner J. Molecular dissection of telomeric repeat-containing RNA biogenesis unveils the presence of distinct and multiple regulatory pathways. Mol Cell Biol. 2010;30(20):4808–17. doi: 10.1128/MCB.00460-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scheibe M, Arnoult N, Kappei D, Buchholz F, Decottignies A, Butter F, et al. Quantitative interaction screen of telomeric repeat-containing RNA reveals novel TERRA regulators. Genome Res. 2013;23(12):2149–57. doi: 10.1101/gr.151878.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lopez de Silanes I, Stagno d’Alcontres M, Blasco MA. TERRA transcripts are bound by a complex array of RNA-binding proteins. Nat Commun. 2010;1:33. doi: 10.1038/ncomms1032 [DOI] [PubMed] [Google Scholar]
- 19.Redon S, Reichenbach P, Lingner J. The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 2010;38(17):5797–806. doi: 10.1093/nar/gkq296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura M, Zhou XZ, Kishi S, Kosugi I, Tsutsui Y, Lu KP. A specific interaction between the telomeric protein Pin2/TRF1 and the mitotic spindle. Curr Biol. 2001;11(19):1512–6. doi: 10.1016/s0960-9822(01)00456-0 [DOI] [PubMed] [Google Scholar]
- 21.Lan J, Zhu Y, Xu L, Yu H, Yu J, Liu X, et al. The 68-kDa telomeric repeat binding factor 1 (TRF1)-associated protein (TAP68) interacts with and recruits TRF1 to the spindle pole during mitosis. J Biol Chem. 2014;289(20):14145–56. doi: 10.1074/jbc.M113.526244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith S, de Lange T. Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J Cell Sci. 1999;112 (Pt 21):3649–56. [DOI] [PubMed] [Google Scholar]
- 23.Aschacher T, Wolf B, Aschacher O, Enzmann F, Laszlo V, Messner B, et al. Long interspersed element-1 ribonucleoprotein particles protect telomeric ends in alternative lengthening of telomeres dependent cells. Neoplasia. 2020;22(2):61–75. doi: 10.1016/j.neo.2019.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kordyukova M, Morgunova V, Olovnikov I, Komarov PA, Mironova A, Olenkina OM, et al. Subcellular localization and Egl-mediated transport of telomeric retrotransposon HeT-A ribonucleoprotein particles in the Drosophila germline and early embryogenesis. PLoS One. 2018;13(8):e0201787. doi: 10.1371/journal.pone.0201787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casacuberta E. Drosophila: Retrotransposons Making up Telomeres. Viruses. 2017;9(7):192. doi: 10.3390/v9070192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kordyukova M, Olovnikov I, Kalmykova A. Transposon control mechanisms in telomere biology. Curr Opin Genet Dev. 2018;49:56–62. doi: 10.1016/j.gde.2018.03.002 [DOI] [PubMed] [Google Scholar]
- 27.Cacchione S, Cenci G, Raffa GD. Silence at the End: How Drosophila Regulates Expression and Transposition of Telomeric Retroelements. J Mol Biol. 2020;432(15):4305–21. doi: 10.1016/j.jmb.2020.06.004 [DOI] [PubMed] [Google Scholar]
- 28.Eickbush TH. Telomerase and retrotransposons: which came first? Science. 1997;277(5328):911–2. doi: 10.1126/science.277.5328.911 [DOI] [PubMed] [Google Scholar]
- 29.Servant G, Deininger PL. Insertion of Retrotransposons at Chromosome Ends: Adaptive Response to Chromosome Maintenance. Front Genet. 2015;6:358. doi: 10.3389/fgene.2015.00358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raffa GD, Cenci G, Ciapponi L, Gatti M. Organization and Evolution of Drosophila Terminin: Similarities and Differences between Drosophila and Human Telomeres. Front Oncol. 2013;3:112. doi: 10.3389/fonc.2013.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Zhang L, Tang X, Bhardwaj SR, Ji J, Rong YS. MTV, an ssDNA Protecting Complex Essential for Transposon-Based Telomere Maintenance in Drosophila. PLoS Genet. 2016;12(11):e1006435. doi: 10.1371/journal.pgen.1006435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoeftner S, Blasco MA. A ’higher order’ of telomere regulation: telomere heterochromatin and telomeric RNAs. EMBO J. 2009;28(16):2323–36. doi: 10.1038/emboj.2009.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Danilevskaya ON, Arkhipova IR, Traverse KL, Pardue ML. Promoting in tandem: the promoter for telomere transposon HeT-A and implications for the evolution of retroviral LTRs. Cell. 1997;88(5):647–55. doi: 10.1016/s0092-8674(00)81907-8 [DOI] [PubMed] [Google Scholar]
- 34.Silva-Sousa R, Lopez-Panades E, Pineyro D, Casacuberta E. The chromosomal proteins JIL-1 and Z4/Putzig regulate the telomeric chromatin in Drosophila melanogaster. PLoS Genet. 2012;8(12):e1003153. doi: 10.1371/journal.pgen.1003153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rashkova S, Karam SE, Kellum R, Pardue ML. Gag proteins of the two Drosophila telomeric retrotransposons are targeted to chromosome ends. J Cell Biol. 2002;159(3):397–402. doi: 10.1083/jcb.200205039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perrini B, Piacentini L, Fanti L, Altieri F, Chichiarelli S, Berloco M, et al. HP1 controls telomere capping, telomere elongation, and telomere silencing by two different mechanisms in Drosophila. Mol Cell. 2004;15(3):467–76. doi: 10.1016/j.molcel.2004.06.036 [DOI] [PubMed] [Google Scholar]
- 37.Savitsky M, Kravchuk O, Melnikova L, Georgiev P. Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster. Mol Cell Biol. 2002;22(9):3204–18. doi: 10.1128/MCB.22.9.3204-3218.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Radion E, Morgunova V, Ryazansky S, Akulenko N, Lavrov S, Abramov Y, et al. Key role of piRNAs in telomeric chromatin maintenance and telomere nuclear positioning in Drosophila germline. Epigenetics Chromatin. 2018;11(1):40. doi: 10.1186/s13072-018-0210-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rozhkov NV, Hammell M, Hannon GJ. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 2013;27(4):400–12. doi: 10.1101/gad.209767.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Savitsky M, Kwon D, Georgiev P, Kalmykova A, Gvozdev V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006;20(3):345–54. doi: 10.1101/gad.370206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morgunova V, Akulenko N, Radion E, Olovnikov I, Abramov Y, Olenina LV, et al. Telomeric repeat silencing in germ cells is essential for early development in Drosophila. Nucleic Acids Res. 2015;43(18):8762–73. doi: 10.1093/nar/gkv775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kordyukova M, Sokolova O, Morgunova V, Ryazansky S, Akulenko N, Glukhov S, et al. Nuclear Ccr4-Not mediates the degradation of telomeric and transposon transcripts at chromatin in the Drosophila germline. Nucleic Acids Res. 2020;48(1):141–56. doi: 10.1093/nar/gkz1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner E, Clement SL, Lykke-Andersen J. An unconventional human Ccr4-Caf1 deadenylase complex in nuclear cajal bodies. Mol Cell Biol. 2007;27(5):1686–95. doi: 10.1128/MCB.01483-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryazansky S, Radion E, Mironova A, Akulenko N, Abramov Y, Morgunova V, et al. Natural variation of piRNA expression affects immunity to transposable elements. PLoS Genet. 2017;13(4):e1006731. doi: 10.1371/journal.pgen.1006731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dienstbier M, Boehl F, Li X, Bullock SL. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 2009;23(13):1546–58. doi: 10.1101/gad.531009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mach JM, Lehmann R. An Egalitarian-BicaudalD complex is essential for oocyte specification and axis determination in Drosophila. Genes Dev. 1997;11(4):423–35. doi: 10.1101/gad.11.4.423 [DOI] [PubMed] [Google Scholar]
- 47.Klattenhoff C, Bratu DP, McGinnis-Schultz N, Koppetsch BS, Cook HA, Theurkauf WE. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell. 2007;12(1):45–55. doi: 10.1016/j.devcel.2006.12.001 [DOI] [PubMed] [Google Scholar]
- 48.Megosh HB, Cox DN, Campbell C, Lin H. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol. 2006;16(19):1884–94. doi: 10.1016/j.cub.2006.08.051 [DOI] [PubMed] [Google Scholar]
- 49.Rouget C, Papin C, Boureux A, Meunier AC, Franco B, Robine N, et al. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010;467(7319):1128–32. doi: 10.1038/nature09465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reig-Viader R, Garcia-Caldes M, Ruiz-Herrera A. Telomere homeostasis in mammalian germ cells: a review. Chromosoma. 2016;125(2):337–51. doi: 10.1007/s00412-015-0555-4 [DOI] [PubMed] [Google Scholar]
- 51.Olovnikov IA, Morgunova VV, Mironova AA, Kordyukova MY, Radion EI, Olenkina OM, et al. Interaction of Telomeric Retroelement HeT-A Transcripts and Their Protein Product Gag in Early Embryogenesis of Drosophila. Biochemistry (Mosc). 2016;81(9):1023–30. doi: 10.1134/S000629791609011X [DOI] [PubMed] [Google Scholar]
- 52.Joukov V, De Nicolo A. Aurora-PLK1 cascades as key signaling modules in the regulation of mitosis. Sci Signal. 2018;11(543). doi: 10.1126/scisignal.aar4195 [DOI] [PubMed] [Google Scholar]
- 53.Petronczki M, Lenart P, Peters JM. Polo on the Rise-from Mitotic Entry to Cytokinesis with Plk1. Dev Cell. 2008;14(5):646–59. doi: 10.1016/j.devcel.2008.04.014 [DOI] [PubMed] [Google Scholar]
- 54.Moutinho-Santos T, Sampaio P, Amorim I, Costa M, Sunkel CE. In vivo localisation of the mitotic POLO kinase shows a highly dynamic association with the mitotic apparatus during early embryogenesis in Drosophila. Biol Cell. 1999;91(8):585–96. [PubMed] [Google Scholar]
- 55.Archambault V, Pinson X. Free centrosomes: where do they all come from? Fly (Austin). 2010;4(2):172–7. [DOI] [PubMed] [Google Scholar]
- 56.Logarinho E, Bousbaa H, Dias JM, Lopes C, Amorim I, Antunes-Martins A, et al. Different spindle checkpoint proteins monitor microtubule attachment and tension at kinetochores in Drosophila cells. J Cell Sci. 2004;117(Pt 9):1757–71. doi: 10.1242/jcs.01033 [DOI] [PubMed] [Google Scholar]
- 57.Pimenta-Marques A, Bento I, Lopes CA, Duarte P, Jana SC, Bettencourt-Dias M. A mechanism for the elimination of the female gamete centrosome in Drosophila melanogaster. Science. 2016;353(6294):aaf4866. doi: 10.1126/science.aaf4866 [DOI] [PubMed] [Google Scholar]
- 58.Glover DM. Polo kinase and progression through M phase in Drosophila: a perspective from the spindle poles. Oncogene. 2005;24(2):230–7. doi: 10.1038/sj.onc.1208279 [DOI] [PubMed] [Google Scholar]
- 59.Bruinsma W, Raaijmakers JA, Medema RH. Switching Polo-like kinase-1 on and off in time and space. Trends Biochem Sci. 2012;37(12):534–42. doi: 10.1016/j.tibs.2012.09.005 [DOI] [PubMed] [Google Scholar]
- 60.Dobbelaere J, Josue F, Suijkerbuijk S, Baum B, Tapon N, Raff J. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 2008;6(9):e224. doi: 10.1371/journal.pbio.0060224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Donaldson MM, Tavares AA, Ohkura H, Deak P, Glover DM. Metaphase arrest with centromere separation in polo mutants of Drosophila. J Cell Biol. 2001;153(4):663–76. doi: 10.1083/jcb.153.4.663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tillery MML, Blake-Hedges C, Zheng Y, Buchwalter RA, Megraw TL. Centrosomal and Non-Centrosomal Microtubule-Organizing Centers (MTOCs) in Drosophila melanogaster. Cells. 2018;7(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cunha-Ferreira I, Bento I, Bettencourt-Dias M. From zero to many: control of centriole number in development and disease. Traffic. 2009;10(5):482–98. doi: 10.1111/j.1600-0854.2009.00905.x [DOI] [PubMed] [Google Scholar]
- 64.Manandhar G, Schatten H, Sutovsky P. Centrosome reduction during gametogenesis and its significance. Biol Reprod. 2005;72(1):2–13. doi: 10.1095/biolreprod.104.031245 [DOI] [PubMed] [Google Scholar]
- 65.Wang SH, Elgin SC. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc Natl Acad Sci U S A. 2011;108(52):21164–9. doi: 10.1073/pnas.1107892109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang F, Quan Z, Huang H, He M, Liu X, Cai T, et al. Ovaries absent links dLsd1 to HP1a for local H3K4 demethylation required for heterochromatic gene silencing. Elife. 2019;8. doi: 10.7554/eLife.40806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsai SY, Huang F. Acetyltransferase Enok regulates transposon silencing and piRNA cluster transcription. PLoS Genet. 2021;17(2):e1009349. doi: 10.1371/journal.pgen.1009349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cook HA, Koppetsch BS, Wu J, Theurkauf WE. The Drosophila SDE3 homolog armitage is required for oskar mRNA silencing and embryonic axis specification. Cell. 2004;116(6):817–29. doi: 10.1016/s0092-8674(04)00250-8 [DOI] [PubMed] [Google Scholar]
- 69.Morris JZ, Hong A, Lilly MA, Lehmann R. twin, a CCR4 homolog, regulates cyclin poly(A) tail length to permit Drosophila oogenesis. Development. 2005;132(6):1165–74. doi: 10.1242/dev.01672 [DOI] [PubMed] [Google Scholar]
- 70.Schulze WM, Stein F, Rettel M, Nanao M, Cusack S. Structural analysis of human ARS2 as a platform for co-transcriptional RNA sorting. Nat Commun. 2018;9(1):1701. doi: 10.1038/s41467-018-04142-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barckmann B, Simonelig M. Control of maternal mRNA stability in germ cells and early embryos. Biochim Biophys Acta. 2013;1829(6–7):714–24. doi: 10.1016/j.bbagrm.2012.12.011 [DOI] [PubMed] [Google Scholar]
- 72.Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, et al. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell. 2009;137(3):522–35. doi: 10.1016/j.cell.2009.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Czech B, Preall JB, McGinn J, Hannon GJ. A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway. Mol Cell. 2013;50(5):749–61. doi: 10.1016/j.molcel.2013.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Musaro M, Ciapponi L, Fasulo B, Gatti M, Cenci G. Unprotected Drosophila melanogaster telomeres activate the spindle assembly checkpoint. Nat Genet. 2008;40(3):362–6. doi: 10.1038/ng.2007.64 [DOI] [PubMed] [Google Scholar]
- 75.Ban KH, Torres JZ, Miller JJ, Mikhailov A, Nachury MV, Tung JJ, et al. The END network couples spindle pole assembly to inhibition of the anaphase-promoting complex/cyclosome in early mitosis. Dev Cell. 2007;13(1):29–42. doi: 10.1016/j.devcel.2007.04.017 [DOI] [PubMed] [Google Scholar]
- 76.Archambault V D ’Avino PP, Deery MJ, Lilley KS, Glover DM. Sequestration of Polo kinase to microtubules by phosphopriming-independent binding to Map205 is relieved by phosphorylation at a CDK site in mitosis. Genes Dev. 2008;22(19):2707–20. doi: 10.1101/gad.486808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Archambault V, Zhao X, White-Cooper H, Carpenter AT, Glover DM. Mutations in Drosophila Greatwall/Scant reveal its roles in mitosis and meiosis and interdependence with Polo kinase. PLoS Genet. 2007;3(11):e200. doi: 10.1371/journal.pgen.0030200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brar SS, Petrovich RM, Williams JG, Mason JM. Phosphorylation at serines 216 and 221 is important for Drosophila HeT-A Gag protein stability. PLoS One. 2013;8(9):e75381. doi: 10.1371/journal.pone.0075381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Neef R, Gruneberg U, Kopajtich R, Li X, Nigg EA, Sillje H, et al. Choice of Plk1 docking partners during mitosis and cytokinesis is controlled by the activation state of Cdk1. Nat Cell Biol. 2007;9(4):436–44. doi: 10.1038/ncb1557 [DOI] [PubMed] [Google Scholar]
- 80.Qi W, Tang Z, Yu H. Phosphorylation- and polo-box-dependent binding of Plk1 to Bub1 is required for the kinetochore localization of Plk1. Mol Biol Cell. 2006;17(8):3705–16. doi: 10.1091/mbc.e06-03-0240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Novak ZA, Wainman A, Gartenmann L, Raff JW. Cdk1 Phosphorylates Drosophila Sas-4 to Recruit Polo to Daughter Centrioles and Convert Them to Centrosomes. Dev Cell. 2016;37(6):545–57. doi: 10.1016/j.devcel.2016.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Afonso PV, Zamborlini A, Saib A, Mahieux R. Centrosome and retroviruses: the dangerous liaisons. Retrovirology. 2007;4:27. doi: 10.1186/1742-4690-4-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shumilov A, Tsai MH, Schlosser YT, Kratz AS, Bernhardt K, Fink S, et al. Epstein-Barr virus particles induce centrosome amplification and chromosomal instability. Nat Commun. 2017;8:14257. doi: 10.1038/ncomms14257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang G, Sharon D, Jovel J, Liu L, Wine E, Tahbaz N, et al. Pericentriolar Targeting of the Mouse Mammary Tumor Virus GAG Protein. PLoS One. 2015;10(6):e0131515. doi: 10.1371/journal.pone.0131515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wen Y, Golubkov VS, Strongin AY, Jiang W, Reed JC. Interaction of hepatitis B viral oncoprotein with cellular target HBXIP dysregulates centrosome dynamics and mitotic spindle formation. J Biol Chem. 2008;283(5):2793–803. doi: 10.1074/jbc.M708419200 [DOI] [PubMed] [Google Scholar]
- 86.Duensing A, Spardy N, Chatterjee P, Zheng L, Parry J, Cuevas R, et al. Centrosome overduplication, chromosomal instability, and human papillomavirus oncoproteins. Environ Mol Mutagen. 2009;50(8):741–7. doi: 10.1002/em.20478 [DOI] [PubMed] [Google Scholar]
- 87.Zhang SM, Song M, Yang TY, Fan R, Liu XD, Zhou PK. HIV-1 Tat impairs cell cycle control by targeting the Tip60, Plk1 and cyclin B1 ternary complex. Cell Cycle. 2012;11(6):1217–34. doi: 10.4161/cc.11.6.19664 [DOI] [PubMed] [Google Scholar]
- 88.Hossain D, Ferreira Barbosa JA, Cohen EA, Tsang WY. HIV-1 Vpr hijacks EDD-DYRK2-DDB1(DCAF1) to disrupt centrosome homeostasis. J Biol Chem. 2018;293(24):9448–60. doi: 10.1074/jbc.RA117.001444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chang F, Re F, Sebastian S, Sazer S, Luban J. HIV-1 Vpr induces defects in mitosis, cytokinesis, nuclear structure, and centrosomes. Mol Biol Cell. 2004;15(4):1793–801. doi: 10.1091/mbc.e03-09-0691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goh WC, Manel N, Emerman M. The human immunodeficiency virus Vpr protein binds Cdc25C: implications for G2 arrest. Virology. 2004;318(1):337–49. doi: 10.1016/j.virol.2003.10.007 [DOI] [PubMed] [Google Scholar]
- 91.Sakai K, Barnitz RA, Chaigne-Delalande B, Bidere N, Lenardo MJ. Human immunodeficiency virus type 1 Vif causes dysfunction of Cdk1 and CyclinB1: implications for cell cycle arrest. Virol J. 2011;8:219. doi: 10.1186/1743-422X-8-219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Poole E, Strappe P, Mok HP, Hicks R, Lever AM. HIV-1 Gag-RNA interaction occurs at a perinuclear/centrosomal site; analysis by confocal microscopy and FRET. Traffic. 2005;6(9):741–55. doi: 10.1111/j.1600-0854.2005.00312.x [DOI] [PubMed] [Google Scholar]
- 93.Sfakianos JN, LaCasse RA, Hunter E. The M-PMV cytoplasmic targeting-retention signal directs nascent Gag polypeptides to a pericentriolar region of the cell. Traffic. 2003;4(10):660–70. doi: 10.1034/j.1600-0854.2003.00125.x [DOI] [PubMed] [Google Scholar]
- 94.Yu SF, Eastman SW, Linial ML. Foamy virus capsid assembly occurs at a pericentriolar region through a cytoplasmic targeting/retention signal in Gag. Traffic. 2006;7(8):966–77. doi: 10.1111/j.1600-0854.2006.00448.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nakamura M, Zhou XZ, Kishi S, Lu KP. Involvement of the telomeric protein Pin2/TRF1 in the regulation of the mitotic spindle. FEBS Lett. 2002;514(2–3):193–8. doi: 10.1016/s0014-5793(02)02363-3 [DOI] [PubMed] [Google Scholar]
- 96.Ohishi T, Hirota T, Tsuruo T, Seimiya H. TRF1 mediates mitotic abnormalities induced by Aurora-A overexpression. Cancer Res. 2010;70(5):2041–52. doi: 10.1158/0008-5472.CAN-09-2008 [DOI] [PubMed] [Google Scholar]
- 97.Aschacher T, Wolf B, Enzmann F, Kienzl P, Messner B, Sampl S, et al. LINE-1 induces hTERT and ensures telomere maintenance in tumour cell lines. Oncogene. 2016;35(1):94–104. doi: 10.1038/onc.2015.65 [DOI] [PubMed] [Google Scholar]
- 98.Ilyin AA, Ryazansky SS, Doronin SA, Olenkina OM, Mikhaleva EA, Yakushev EY, et al. Piwi interacts with chromatin at nuclear pores and promiscuously binds nuclear transcripts in Drosophila ovarian somatic cells. Nucleic Acids Res. 2017;45(13):7666–80. doi: 10.1093/nar/gkx355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Strober W. Trypan Blue Exclusion Test of Cell Viability. Curr Protoc Immunol. 2015;111:A3 B 1–A3 B doi: 10.1002/0471142735.ima03bs111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Megraw TL, Kilaru S, Turner FR, Kaufman TC. The centrosome is a dynamic structure that ejects PCM flares. J Cell Sci. 2002;115(Pt 23):4707–18. doi: 10.1242/jcs.00134 [DOI] [PubMed] [Google Scholar]
- 101.Golovnin A, Volkov I, Georgiev P. SUMO conjugation is required for the assembly of Drosophila Su(Hw) and Mod(mdg4) into insulator bodies that facilitate insulator complex formation. J Cell Sci. 2012;125(Pt 8):2064–74. doi: 10.1242/jcs.100172 [DOI] [PubMed] [Google Scholar]
- 102.Llamazares S, Moreira A, Tavares A, Girdham C, Spruce BA, Gonzalez C, et al. polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev. 1991;5(12A):2153–65. doi: 10.1101/gad.5.12a.2153 [DOI] [PubMed] [Google Scholar]
- 103.Lopez-Panades E, Gavis ER, Casacuberta E. Specific Localization of the Drosophila Telomere Transposon Proteins and RNAs, Give Insight in Their Behavior, Control and Telomere Biology in This Organism. PLoS ONE. 2015;10(6):e0128573. doi: 10.1371/journal.pone.0128573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rothwell WF, Sullivan W. Fluorescent analysis of Drosophila embryos. In: Sullivan W, Ashburner M, Hawley RS, editors. Drosophilia protocols: CSHL Press; 2000. p. 148–52. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(RAR)
Mass spectrometry sample preparation and data analysis.
(PDF)
HeT-A Gag-HA (red) and α-tubulin (green) immunostaining was performed on larvae brain of the D. melanogaster strains expressing RFP (upper panel, control) or HeT-A Gag-HA (lower panel). DNA is stained with DAPI (blue).
(TIF)
(A) Analysis of HeT-A RNA levels in the ovaries of indicated strains. RT-qPCR analysis of the HeT-A RNA levels normalized to RNA levels of rp49 housekeeping gene. Bar diagrams show fold changes in steady-state HeT-A RNA levels in the ovaries of flies with the indicated genotypes relative to the control (w_GLKD). Error bars indicate SD (standard deviation) for three biological replicates. spnE mutants were spnE1/spnEhls3987. Heterozygous spnE/+ flies were a mix of spnE1/+ and spnEhls3987/+. (B) Efficiency of the germline knockdowns of SpnE, Ccr4 and Ars2. Western blot analysis of ovary extracts probed with indicated antibodies. α-tubulin staining was used as a loading control.
(TIF)
Coomassie brilliant blue staining of proteins copurified with HeT-A Gag-HA from 0-2-h-old spnE_GLKD embryos containing transgene expressing HeT-A Gag-HA. As a control, spnE_GLKD embryos without HeT-A transgene were used.
(TIF)
(A) Co-IP experiment performed on extracts from 0-2-h-old spnE_GLKD embryos expressing HeT-A Gag-HA reveals that Polo is co-purified with HeT-A Gag-HA. (B) Co-IP experiment performed on extracts from spnE_GLKD ovaries expressing HeT-A Gag-HA reveals that BubR1and CP190 are co-purified with HeT-A Gag-HA. Post-extraction insoluble pellet was loaded between Input and IP probes. The antibodies used for co-IP and western blotting are indicated on the right.
(TIF)
HeT-A RNA FISH (green) and coimmunostaining of Cdk1 (red) and CP190 (magenta) in the control (A), Ars2_GLKD (B) and twin_GLKD (C) early syncytial embryos. Lower panels in (C) show the enlarged area around the centrosome in twin_GLKD embryo. (D, E) Coimmunostaining of HeT-A Gag-HA (green), Cdk1 (red) and CP190 (magenta) in the spnE_GLKD is shown. Free centrosomes in the embryo cortex are shown (E). Blue, DNA.
(TIF)
Western blotting of 0–2 h old embryo lysates shows the overexpression of HeT-A Gag and unchanged Polo and Cdk1 levels in spnE_GLKD embryos compared with the control (w_GLKD). Antibodies are indicated on the right. Anti-Plk1 antibodies (pThr210, LSBio) used here recognize phosphorylated form of Polo.
(TIF)
(A) Colocalization of HeT-A RNA (green), Polo (phosphorylated form, red) and γ-tubulin (grey) in the oocyte of twin_GLKD at stage 9 is shown. Blue, DNA. (B) Accumulation of Polo-HA (red) and HeT-A Gag (green) in the oocyte of spnE_GLKD expressing Polo-HA at stage 6 of oogenesis is shown. Blue, DNA. (C) Western blotting confirms accumulation of phosphorylated Polo in spnE_GLKD egg chambers at 12–14 stages (rep1 and rep2, two biological replicates) compared with w_GLKD. The antibodies used are indicated on the right.
(TIF)
(A) Coimmunostaining demonstrating colocalization of HeT-A Gag (green) and γ-tubulin (red) in spnE_GLKD ovaries at stage 5 of oogenesis is shown. (B) Immunostaining reveals accumulation of HeT-A Gag (green) and multiple γ-tubulin (red) and CNN (grey) foci at stage 9 of oogenesis in twin_GLKD. Blue, DNA.
(TIF)
HeT-A RNA FISH (green) combined with CNN (gray) immunostaining in twin_GLKD at stage 13–14 egg chamber. The lower panel is a blow up of the area around the oocyte nucleus (the upper panel). DNA, blue.
(TIF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.