Abstract
Healthcare sensors represent a valid and non-invasive instrument to capture and analyse physiological data. Several vital signals, such as voice signals, can be acquired anytime and anywhere, achieved with the least possible discomfort to the patient thanks to the development of increasingly advanced devices. The integration of sensors with artificial intelligence techniques contributes to the realization of faster and easier solutions aimed at improving early diagnosis, personalized treatment, remote patient monitoring and better decision making, all tasks vital in a critical situation such as that of the COVID-19 pandemic. This paper presents a study about the possibility to support the early and non-invasive detection of COVID-19 through the analysis of voice signals by means of the main machine learning algorithms. If demonstrated, this detection capacity could be embedded in a powerful mobile screening application. To perform this important study, the Coswara dataset is considered. The aim of this investigation is not only to evaluate which machine learning technique best distinguishes a healthy voice from a pathological one, but also to identify which vowel sound is most seriously affected by COVID-19 and is, therefore, most reliable in detecting the pathology. The results show that Random Forest is the technique that classifies most accurately healthy and pathological voices. Moreover, the evaluation of the vowel /e/ allows the detection of the effects of COVID-19 on voice quality with a better accuracy than the other vowels.
Keywords: Healthcare sensors, Machine learning techniques, COVID-19 detection, Voice analysis, Vowel sounds
Introduction
The ageing of the population, the diffusion of chronic conditions and the outbreaks of infectious diseases and new pandemics, such as COVID-19 which has been affecting the world since last year, all represent major challenges in our present-day society [22]. The acquisition, processing and analysis of health information constitute significant tasks in relation to the early detection and treatment of major diseases. The use of unobtrusive sensing and wearable devices constitutes a valuable support in the acquisition of health data anywhere and anytime. Sensors can be integrated into clothing, accessories and the living environment. Advanced electronic devices can provide detailed health information, monitoring continuously and in real-time biochemical and physiological parameters during the daily life of the patient [20, 21, 34, 54].
These sensors are particularly useful in the monitoring of the health condition of the millions of people who have been afflicted by COVID-19. This pandemic has so far affected more than 80 million people and at present its diffusion shows no sign of reaching a conclusion [53]. While the application of increasingly advanced, easy-to-use and wearable technologies has helped to improve the processes of patient care through the continuous real-time monitoring of vital parameters and the definition of personalized treatment, on the other hand this has contributed to the accumulation of a large amount of data from such devices and the consequent need for its accurate analysis and processing. The successful application of these sensors, in fact, hinges on the ability to extract, interpret and elaborate the considerable volume of heterogeneous data they generate in a consistent and constructive way. The complexity and variety of these data require the provision of new models, solutions and technologies able to process and analyse them reliably and easily. This objective can be achieved by using Machine Learning (ML) algorithms. These techniques, in fact, represent an important tool capable of providing different possibilities to transform such data into valid insights to be used for the construction of reliable decision-making models necessary to provide accurate healthcare to patients. In addition, their use in health care contributes to the optimization of resources through the improvement of processes and services with a consequent reduction in costs [10, 32].
In this study, we explore the opportunity to support the early assessment and detection of COVID-19 symptoms through the evaluation of voice sounds. COVID-19 patients, including asymptomatic subjects, have, in fact, reported difficulties in voice production as well as abnormalities in vocal folds oscillation [24, 40]. In detail, we have analysed and studied the sounds of three vowels, /a/, /e/ and /o/. These were selected from the Coswara database, an available crowd-sourced database [46]. The aim has been to evaluate the possibility to support the early and non-invasive detection of COVID-19 by analysing the vocalization of a vowel through the use of the most appropriate ML algorithm. In particular, on one hand, we have investigated among all the three vowels sound present in the dataset considered, which vowel reflects more accurately the effects of COVID-19 on voice production, so which vowel is more appropriate to detect the coronavirus disease. On the other hand, we have performed an evaluation of which, among an ample number, ML technique is more performing in terms of correct classification. The possibility to detect COVID-19 by easily analysing short vowel sounds could be significant in terms of the realization of a mobile health (m-health) solution able to acquire the vocalization sound, analyse it and distinguish between healthy and pathological subjects through the most reliable ML technique. Such an m-health system could constitute a valid instrument for a fast and easy screening, reducing the time required to map the spread of the infection, as well as the costs involved in its detection.
The remaining sections of the manuscript are organized as follows. The main works relating to the diagnosis of COVID-19 existing in literature are discussed in Sect. 2. The voice samples, features and ML models evaluated in this study are, instead, presented in Sect. 3. Finally, the results obtained are discussed in Sect. 4, while our conclusions are presented in Sect. 5.
Related Works
Due to the recent diffusion of the COVID-19 pandemic and the continuous mutations of the virus with the formation of new variants, very few studies exist in the literature about the detection of COVID-19 symptoms through the analysis of voice signals. Respiratory signals [5, 18, 48] or coughing sounds [4, 7, 23, 27, 36, 37, 47] constitute the main vocal signals for the detection of the effects of COVID-19. Other studies, instead, have identified pathomorphological modifications caused by the pandemic in the patient’s chest by analysing Computed Tomography (CT) images [35, 55] or chest radiographic [1, 52].
However, limited and, often, non-accessible datasets, have been used to perform these preliminary studies, reducing the possibility for further development of reliable classification approaches on standardized datasets for the research community. A small dataset composed of only 9 healthy and 10 pathological subjects, for example, was used in [42] to evaluate the accuracy of the proposed Support Vector Machine (SVM) model. Mel filter bank features represent the inputs to this model, achieving an F1-score and an accuracy, respectively, of 77.0% and 70.5%.
A SVM algorithm was, also, used in [17] to detect pandemic symptoms by evaluating voice samples. The authors proposed a system able to analyse the severity of the disease by evaluating the vocalization of five sentences. Voice samples were collected from 52 pathological subjects in two hospitals in Wuhan, China. The Geneva Minimalistic Acoustic Parameter and Computational Paralinguistics Challenge sets were estimated and used as inputs of the SVM. An accuracy of 69% was achieved.
A Convolutional Neural Network (CNN) model capable of detecting the anomalies in the dynamics of the glottal flow waveform (GFW) during voice production was, instead, proposed in [11]. A private database containing recordings of the vowels /a/, /i/ and /u/ voiced by a limited sample of only 9 pathological and 10 healthy subjects was analysed. The performance is presented in terms of the Receiver Operating Characteristic (ROC-AUC) and its standard deviation, respectively, equal to 0.900 and 0.062.
In [18], a deep learning approach was proposed. The features extracted from the vocal, breathing and coughing sounds were processed by a Long Short-Term Memory (LSTM) architecture. The dataset evaluated consists of 80 subjects (60 healthy and 20 pathological). As features, the Spectral Roll-off (SR), Spectral Centroid (SC), Mel-Frequency Cepstral Coefficients (MFCC), the first and second derivates of MFCC and the Zero-Crossing rate (ZCR) were considered. The LSTM model, relating to the voice samples, achieved an accuracy and F1-score, of 88.2% and 92.5%, respectively.
A new feature, called the COVID-19 Coefficient (C-19CC), was, instead, proposed by Dash et al. [8] to detect the presence of COVID-19 symptoms through an analysis of opportune sounds selected from the Coswara database. The reliability of these cepstral features to distinguish correctly between pathological and healthy subjects was evaluated by classifying the sample with a SVM algorithm. The best performance was obtained through an analysis of the coughing sounds, with an accuracy of about 85%.
To the best of our knowledge, our study is the first and the only study that presents a reproducible analysis by using a freely available dataset and an exhaustive overview of how different vowels sounds and ML techniques impact on the COVID-19 detection.
Materials and Methods
Voice samples were selected from the Coswara database, a readily available database [9]. The Indian Institute of Science (IISc) Bangalore realized this database and it contains coughing, breathing and voice sounds of healthy and pathological subjects. In this preliminary study, we analysed voice samples from 166 subjects, 83 healthy and 83 COVID-19 positive. 46 female and 120 male voices were selected with a mean age of 33 years. More details about the number, gender and age of the subjects involved in this study are shown in Table 1.
Table 1.
Details about the subjects involved in this study. For the age, we report the mean and standard deviation (SD)
| Category | Gender | No | Age |
|---|---|---|---|
| Mean SD | |||
| Healthy | Female | 21 | 29.6 ± 10.1 |
| Male | 62 | 36.4 ± 13.1 | |
| Total | 83 | 34.7 ± 12.7 | |
| Covid-positive | Female | 25 | 32.08 ± 11.4 |
| Male | 58 | 31.2 ± 11.5 | |
| Total | 83 | 31.5 ± 11.4 | |
| Total | Female | 46 | 30.9 ± 10.7 |
| Male | 120 | 33.9 ± 12.6 | |
| Total | 166 | 33.1 ± 12.1 |
Bold italics indicate the total obtained for each category (healthy, covid-positive and all subjects involved in this study)
In this preliminary study voice samples with an adequate quality, not particularly corrupted by noise, were selected, although all the sounds were filtered by using an opportune filter to reduce the effect of this noise [33]. It is important to note that Coswara is a crowd-sourced database, with all samples being recorded by volunteers, and therefore it is necessary to control the quality of the voice signals. Although the database adopted, Coswara, provides voice, coughing and breathing sounds for each subject, we decided to evaluate the effects of COVID-19 by using only the vowels sounds. This choice has been made because in the medical practice, accordingly with the medical guidelines, experts analyse the vowels characteristics in order to estimate any pneumo-phono-articulatory apparatus disorders [30, 38]. Scientific studies confirmed that sustained vowels are rated significantly more than continuous speech [29]. Based on this scientific evidence, we have also consulted a medical team of the Department of Otorhinolaryngology, of the University Hospital (Policlinico) Federico II of Naples (Italy), that confirmed to us that the analysis of vowels sounds allows us to exhaustively extract the most relevant features useful to identify specific changes in vowel articulation and to quantify any pneumo-phono-articulatory apparatus alterations.
The sounds of three vowels, /a/, /e/ and /o/, were processed for each subject to extract the features that constitute the inputs of the considered ML algorithms. Due to the recent diffusion of the COVID-19 pandemic and the consequent scarcity of studies about the effects of this infection on voice quality, the choice of the features to be extracted from the voice sounds and to be used as inputs of ML algorithms cannot be performed in accordance with a specific medical protocol. Therefore, we decided to use as features the acoustic parameters indicated in medical protocol [28] to evaluate voice quality, such as the Fundamental Frequency (F), shimmer, jitter and Harmonic to Noise Ratio (HNR), as well as other parameters used in literature for the voice classification when using ML algorithms [16, 39, 44, 51], such as Mel-Frequency Cepstral Coefficients (MFCC) or Spectral Centroid or Roll-Off.
The F is useful for an assessment of the correct functioning of the larynx as it shows the rate of oscillation of the vocal folds. The instabilities of these oscillations in amplitude and frequency are represented, respectively, by the shimmer and jitter. The incorrect closure of the vocal folds due to a pathology is, instead, represented by the noise in the voice signals. This noise is evaluated by the HNR parameter. These acoustic parameters were calculated by adopting the Java Programming Language through the use of Eclipse IDE (version 4.6.3) according to the procedures indicated in [13, 45, 50]. Other parameters, such as the MFCC coefficients represent the voice signal as the linear cosine transform of a log power spectrum on a non-linear mel scale of frequency. The dynamic behaviour of the voice signal is represented by the first and second derivatives of the cepstral coefficients. Finally, the spectral centroid (SC) and Spectral Roll-off (SR) were considered. The former is useful for an evaluation of the modifications of the signal frequency over time, while the latter is used to distinguish between unvoiced and voiced sounds. These were evaluated by using Matlab, version R2020a with the function audioFeatureExtractor being adopted [31].
These features have been used as inputs of the main ML techniques used in the literature for voice classification, as described in the following subsection.
Machine Learning Classifiers
Currently, many biomedical applications use appropriate methodologies based on machine learning (ML) techniques to support the early and reliable diagnosis of specific pathologies [43]. These techniques are, in fact, able to distinguish between a pathological and healthy subject through the processing and analysis of specific data. The evaluation of the set of data allows the construction of a model. This model approximates the so-called features, namely the values assumed by independent variables corresponding to the measurable characteristics of each sample. ML techniques are capable of learning from the observed data and adapting their structure to optimize the classification.
In this study, several ML techniques were used to distinguish between a pathological and a healthy voice. The Waikato Environment for Knowledge Analysis project (WEKA) [15] tool, version 3.8.4, was used to perform the analyses. All the experiments were performed on a machine with an 8 GB memory and Intel(R) Core(TM) i5-6200U CPU with 2.40 GHz. The ML algorithms are subdivided into several categories. In this work for a better readability, only the performances of the best techniques are reported for each category. In detail, the performances of the following ML classifiers were evaluated:
- Bayes: the classification, for these algorithms, is based on a probabilistic model where the nodes and strings represent, respectively, a set of random variables and their conditional dependencies. This category is based on the Bayes theorem:
This allows you to find the probability of event A, given event B. As shown in Eq. 1, this is estimated by means of the relationship between a priori probability of A (P(A)) and a posteriori probability of B (P(B)), i.e. the probability of event A after evidence is seen, as well as the probability of event B, given the event A (P(B|A)). The BayesNet (BN) and Naive Bayes (NB) algorithms were used in this study. This latter assumes that predictions are independent, namely that the presence of an appropriate feature in a class is unconnected to the presence of other features. In BayesNet, instead, the conditional probability is estimated on each node by building a Bayesian Network. More details are provided in [25].1 Functions: the operation of the classifiers of this category can be interpreted as a mathematical equation. The classification performances of Stochastic Gradient Descent (SGD) and Support Vector Machine (SVM) were evaluated. SVM is a supervised machine learning algorithm. Its aim is to find the optimal classification function that distinguishes between the samples of the classes. The optimal hyperplane that is searched, shown in Fig. 1, is the one equally distant from the support vectors of the classes [6]. SGD, instead, implements stochastic gradient descent learning of the linear models. The true gradient is approximated by considering one training sample at a time. It is an iterative algorithm, its parameters being updated for each sample analysed [3].
Lazy: the k nearest neighbours were evaluated by means of an Instance-based Learning approach. This evaluation is necessary to decide the class to which a sample belongs. A group of k objects of the training set that has the closest proximity to the test set was localized. A label derived from the prevalence of a class in the closest proximity was assigned. The Locally Weighted Learning (LWL) and k-nearest neighbour (Ibk) algorithms were used in this work. The LWL algorithm, is a non-parametric method, where a local model for each point of interest was used to achieve the prediction. This model is based on the neighbouring data of the classifiers analysed [14]. The Ibk model, instead, represents the simplest lazy learner. The nearest neighbours can be found with a variety of different search algorithms. The predictions from more than one neighbour are weighted based on their distance from the test instance [2].
Meta: the classification of this category was achieved combining multiple ML models to improve the performance but with a consequent increment of computational time and network complexity [12]. The performances of the Adaboost and Bagging techniques were estimated in this study. Baggings bags the classifier. The averaging probability estimates generate the predictions. Adaboost, instead, was designed so that subsequent models try to correct the prediction errors made by previous models. The weights of each instance of the training set are, in fact, updated based on the accuracy of the model. In detail, Adaboost is a boosting algorithm constitute from n number of decision trees. The records incorrectly classified during the first model are priority. These records are sent as inputs for the second model and this process continues until the indicated number of base learners as shown in Fig. 2.
Rules: the voice classification for these approaches is governed by rules. One-R and Decision Table (DT) have been evaluated. DT is a decision table classifier, where the features subsets are evaluated using best-first search [26]. The One-R algorithm, instead, uses minimum-error attributes for the prediction. The rules are based on the most informative attribute. The ranking of the attributes is estimated based on the error rate [19].
Trees: the classification is based on data attributes, hierarchical models composed of decision nodes and terminal leaves, while the branches are labelled with the discrete outcomes of the function that each decision node implements. The Random Forest and C4.5 decision tree (J48) algorithms constitute the algorithms belonging to this category considered in this study. The Random Forest algorithm consists of a various number of decision trees, as shown in Fig. 3. Each tree makes a class prediction using as inputs from samples of the initial dataset. The features extracted from these samples are randomly selected and process from the tree to predict the class. The class with the most votes constitute the model’s prediction. The C4.5 algorithm is based on the theories of Shannon. In particular, the entropy of Shannon measures the disorder of the data and defines the amount of information provided by the event [41]. Random Forest is an ensemble of trees with each tree building via bagging with replacement (bootstrap) and with a random selection of features at each tree node [49].
Fig. 1.
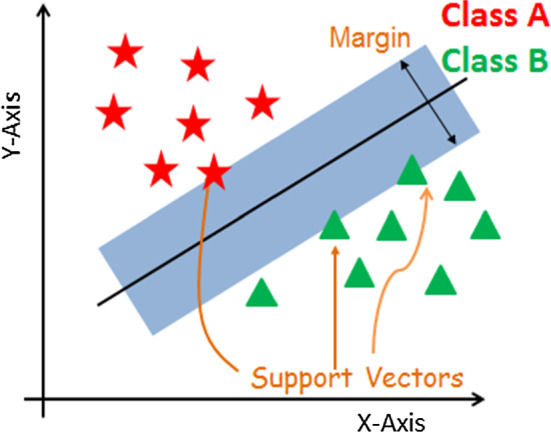
An overview of Support Vector Machine algorithm
Fig. 2.
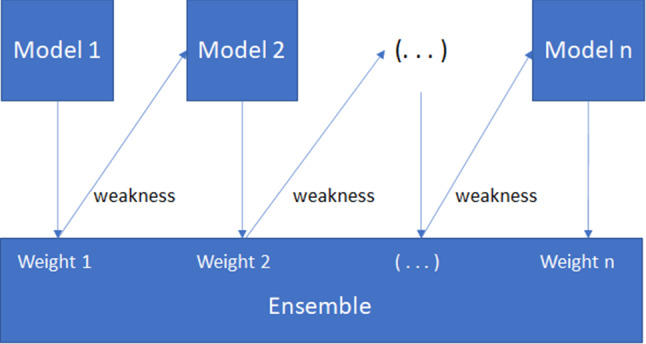
An overview of Adaboost algorithm
Fig. 3.
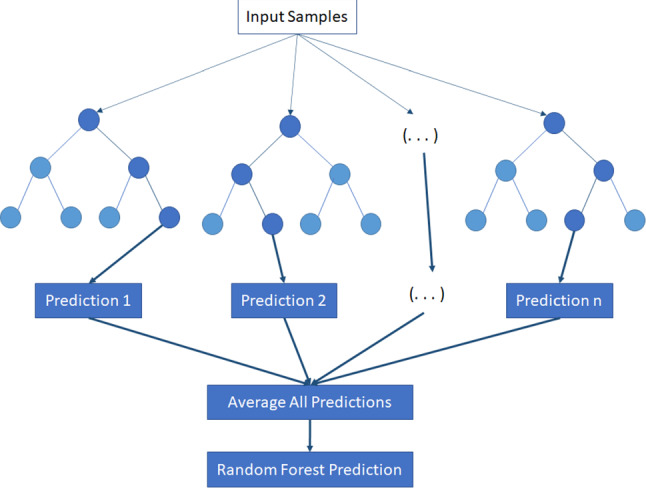
An overview of Random Forest algorithm
Results and Discussion
In order to evaluate the classification reliability of the ML approaches considered, the accuracy, sensitivity, specificity, F1-score, recall and Receiver Operating Characteristic (ROC) area were estimated. The accuracy is defined as the number of correct predictions out of all the samples, estimated according to Eq. 2:
| 2 |
where True Positives (TP) and True Negatives (TN) are defined as the number of samples correctly classified, respectively, as pathological or healthy, while False Positives (FP) and False Negatives (FN) represent the number of samples incorrectly classified, respectively, as pathological and healthy. The sensitivity represents the number of pathological cases the classifier correctly classifies, out of all the pathological cases in the dataset. The specificity, instead, measures the number of healthy correct predictions made, while the precision represents the measurement of how many of the pathological predictions made are correct. These measures are calculated by using the following equations:
| 3 |
| 4 |
| 5 |
The harmonic mean of the precision and sensitivity represents the F1-score, estimated by Eq. 6:
| 6 |
Finally, the performance of the classifiers is evaluated considering the area under the ROC curve (AUC). This is useful for an evaluation of the goodness of the classifier. When the AUC is, in fact, the minimum (AUC=0), the ML technique incorrectly classifies all the data, while, when the AUC is equal to 1 and so is the maximum, the algorithm distinguishes perfectly between the pathological and healthy samples.
The voice samples were divided randomly into training (80% of the samples) and testing (20% of the samples) sets. In detail, the sounds of three vowels (/a/, /e/ and /o/) of 132 subjects (66 healthy and 66 COVID-19 positive) constitute the training set, while the remaining recordings of 34 subjects (17 healthy and 17 COVID-19 positive) compose the testing set. The Coswara database is unbalanced, in that it contains more healthy voices than pathological voices. In this preliminary study, we have adopted a balanced dataset, selecting an equal number of healthy and pathological voice samples. However, it is important to note that the data collection is still in progress and that, in future studies, the increase in data may improve the analyses.
Table 4.
Results achieved on the training set for the vowel /a/
| Algorithm | Sensibility (%) | Specificity (%) | Accuracy (%) | Precision (%) | F1-score (%) | AUC |
|---|---|---|---|---|---|---|
| Naive Bayes [25] | 92.42 | 28.79 | 60.61 | 56.48 | 70.11 | 0.801 |
| Bayes Net [25] | 78.79 | 68.18 | 73.48 | 71.23 | 74.82 | 0.775 |
| SVM [6] | 95.45 | 93.94 | 94.70 | 94.03 | 94.74 | 0.947 |
| SGD [3] | 95.45 | 100.00 | 97.73 | 100.00 | 97.67 | 0.977 |
| Ibk [2] | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 1.000 |
| LWL [14] | 80.30 | 65.15 | 72.73 | 69.74 | 74.65 | 0.908 |
| Adaboost [12] | 78.79 | 93.94 | 86.36 | 92.86 | 85.25 | 0.952 |
| Bagging [12] | 89.39 | 87.88 | 88.64 | 88.06 | 88.72 | 0.958 |
| OneR [19] | 75.00 | 86.36 | 81.15 | 82.35 | 78.50 | 0.750 |
| Decision Table [26] | 75.76 | 71.21 | 73.48 | 72.46 | 74.07 | 0.753 |
| J48 [41] | 98.48 | 96.97 | 97.73 | 97.01 | 97.74 | 0.993 |
| Random Forest [49] | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 1.000 |
Table 6.
Results achieved on the training set for the vowel /e/
| Algorithm | Sensibility (%) | Specificity (%) | Accuracy (%) | Precision (%) | F1-score (%) | AUC |
|---|---|---|---|---|---|---|
| Naive Bayes [25] | 66.67 | 84.85 | 75.76 | 81.48 | 73.33 | 0.861 |
| Bayes Net [25] | 60.61 | 91.30 | 76.30 | 86.96 | 71.43 | 0.880 |
| SVM [6] | 96.97 | 95.45 | 96.21 | 95.52 | 96.24 | 0.960 |
| SGD [3] | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 1.000 |
| Ibk [2] | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 1.000 |
| LWL [14] | 78.79 | 98.48 | 88.64 | 98.11 | 87.39 | 0.941 |
| Adaboost [12] | 80.30 | 96.97 | 88.64 | 96.36 | 87.60 | 0.965 |
| Bagging [12] | 89.39 | 87.88 | 88.64 | 88.06 | 88.72 | 0.961 |
| OneR [19] | 86.36 | 71.21 | 78.79 | 75.00 | 80.28 | 0.788 |
| Decision Table [26] | 89.16 | 80.30 | 86.64 | 91.93 | 90.52 | 0.800 |
| J48 [41] | 96.97 | 98.48 | 97.73 | 98.46 | 97.71 | 0.991 |
| Random Forest [49] | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 1.000 |
Table 8.
Results achieved on the training set for the vowel /o/
| Algorithm | Sensibility (%) | Specificity (%) | Accuracy (%) | Precision (%) | F1-score (%) | AUC |
|---|---|---|---|---|---|---|
| Naive Bayes [25] | 96.97 | 28.79 | 62.88 | 57.66 | 72.32 | 0.647 |
| Bayes Net [25] | 95.45 | 72.73 | 84.09 | 77.78 | 85.71 | 0.900 |
| SVM [6] | 89.39 | 86.36 | 87.88 | 86.76 | 88.06 | 0.879 |
| SGD [3] | 96.97 | 95.45 | 96.21 | 95.52 | 96.24 | 0.962 |
| Ibk [2] | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 1.000 |
| LWL [14] | 83.33 | 69.70 | 76.52 | 73.33 | 78.01 | 0.919 |
| Adaboost [12] | 89.39 | 80.30 | 84.85 | 81.94 | 85.51 | 0.923 |
| Bagging [12] | 93.94 | 91.18 | 92.54 | 91.18 | 92.54 | 0.993 |
| OneR [19] | 83.33 | 72.73 | 78.03 | 75.34 | 79.14 | 0.780 |
| Decision Table [26] | 95.45 | 54.55 | 75.00 | 67.74 | 79.25 | 0.750 |
| J48 [41] | 100.00 | 98.48 | 99.24 | 98.51 | 99.25 | 1.000 |
| Random Forest [49] | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 1.000 |
Tables 2 and 3 report the results, respectively, for the training set and testing set, of several of the ML algorithms for each sample, analysing for each subject the sound of all three vowels, /a/, /e/ and /o/. These show that the best-performing ML algorithms are Random Forest, Adaboost and SVM. Among these three algorithms, the best performance in the testing set was obtained by Random Forest, achieving a classification accuracy and F1-score, respectively, of about 82% and 84%. An accuracy of about 74% is, instead, obtained by Adaboost and SVM algorithms. Meanwhile, observing the sensitivity and specificity values, we can affirm that both Random Forest and SVM are able to accurately distinguish between the voices of people suffering from COVID-19 and those of healthy ones. This is confirmed by the sensitivity value obtained (about 94%), while the sensitivity achieved by the Adaboost algorithm stops at about 71%. Nevertheless, Adaboost obtains the best specificity (76.47% vs. 52.94% of SVM and 70.59% of Random Forest) among the three classifiers. This means that Adaboost is better able to identify healthy voices from pathological ones.
Table 2.
Results achieved on the training set for the vowels /a/, /e/ and /o/
| Algorithm | Sensibility (%) | Specificity (%) | Accuracy (%) | Precision (%) | F1-score (%) | AUC |
|---|---|---|---|---|---|---|
| Naive Bayes [25] | 96.97 | 53.03 | 75.00 | 67.37 | 79.50 | 0.799 |
| Bayes Net [25] | 93.94 | 78.79 | 86.36 | 81.58 | 87.32 | 0.953 |
| SVM [6] | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 1.000 |
| SGD [3] | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 1.000 |
| Ibk [2] | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 1.000 |
| LWL [14] | 68.18 | 83.33 | 75.76 | 80.36 | 73.77 | 0.941 |
| Adaboost [12] | 92.42 | 89.39 | 90.91 | 89.71 | 91.04 | 0.966 |
| Bagging [12] | 98.48 | 92.42 | 95.45 | 92.86 | 95.59 | 0.988 |
| OneR [19] | 71.21 | 86.36 | 78.79 | 83.93 | 77.05 | 0.788 |
| Decision Table [26] | 86.36 | 78.79 | 82.58 | 80.28 | 83.21 | 0.847 |
| J48 [41] | 98.48 | 100.00 | 99.24 | 100.00 | 99.24 | 1.000 |
| Random Forest [49] | 100.00 | 100.00 | 100.00 | 100.00 | 100.00 | 1.000 |
Table 3.
Results achieved on the testing set for the vowels /a/, /e/ and /o/
| Algorithm | Sensibility (%) | Specificity (%) | Accuracy (%) | Precision (%) | F1-score (%) | AUC |
|---|---|---|---|---|---|---|
| Naive Bayes [25] | 100.00 | 35.29 | 67.65 | 60.71 | 75.56 | 0.706 |
| Bayes Net [25] | 88.24 | 29.41 | 58.82 | 55.56 | 68.18 | 0.671 |
| SVM [6] | 94.12 | 52.94 | 73.53 | 66.67 | 78.05 | 0.735 |
| SGD [3] | 94.12 | 47.06 | 70.59 | 64.00 | 76.19 | 0.706 |
| Ibk [2] | 88.24 | 47.06 | 67.65 | 62.50 | 73.17 | 0.676 |
| LWL [14] | 58.82 | 52.94 | 55.88 | 55.56 | 57.14 | 0.647 |
| Adaboost [12] | 70.59 | 76.47 | 73.53 | 75.00 | 72.73 | 0.785 |
| Bagging [12] | 76.47 | 64.71 | 70.59 | 68.42 | 72.22 | 0.747 |
| OneR [19] | 29.41 | 76.47 | 52.94 | 55.56 | 38.46 | 0.529 |
| Decision Table [26] | 64.71 | 47.06 | 55.88 | 55.00 | 59.46 | 0.554 |
| J48 [41] | 64.71 | 52.94 | 58.82 | 57.89 | 61.11 | 0.588 |
| Random Forest [49] | 94.12 | 70.59 | 82.35 | 76.19 | 84.21 | 0.901 |
Considering the performance of each vowel, we can observe that the recordings of the vowels /e/ identify the presence of COVID-19 better than those of the vowels /a/ and /o/. An accuracy and F1-score of about 85% and 84%, respectively, were obtained when using the features extracted from the vowel /e/ as inputs of the Random Forest algorithm in the testing set, as indicated in Table 7. When analysing the sound of the vowels /a/ and /o/, instead, the classification accuracies and F1-scores were lower than those achieved for the only vowel /e/, as shown in Tables 5 and 9.
Table 7.
Results achieved on the testing set for the vowel /e/
| Algorithm | Sensibility (%) | Specificity (%) | Accuracy (%) | Precision (%) | F1-score (%) | AUC |
|---|---|---|---|---|---|---|
| Naive Bayes [25] | 70.59 | 70.59 | 70.59 | 70.59 | 70.59 | 0.754 |
| Bayes Net [25] | 52.94 | 82.35 | 67.65 | 75.00 | 62.07 | 0.749 |
| SVM [6] | 84.62 | 71.43 | 76.47 | 64.71 | 73.33 | 0.765 |
| SGD [3] | 64.71 | 76.47 | 70.59 | 73.33 | 68.75 | 0.706 |
| Ibk [2] | 58.82 | 52.94 | 55.88 | 55.56 | 57.14 | 0.559 |
| LWL [14] | 35.29 | 100.00 | 67.65 | 100.00 | 52.17 | 0.709 |
| Adaboost [12] | 64.71 | 76.47 | 70.59 | 73.33 | 68.75 | 0.763 |
| Bagging [12] | 82.35 | 70.59 | 76.47 | 73.68 | 77.78 | 0.820 |
| OneR [19] | 76.47 | 29.41 | 52.94 | 52.00 | 61.90 | 0.529 |
| Decision Table [26] | 58.82 | 64.71 | 61.76 | 62.50 | 60.61 | 0.606 |
| J48 [41] | 58.82 | 70.59 | 64.71 | 66.67 | 62.50 | 0.652 |
| Random Forest [49] | 76.47 | 94.12 | 85.29 | 92.86 | 83.87 | 0.867 |
Table 5.
Results achieved on the testing set for the vowel /a/
| Algorithm | Sensibility (%) | Specificity (%) | Accuracy (%) | Precision (%) | F1-score (%) | AUC |
|---|---|---|---|---|---|---|
| Naive Bayes [25] | 82.35 | 23.53 | 52.94 | 51.85 | 63.64 | 0.713 |
| Bayes Net [25] | 47.06 | 70.59 | 58.82 | 61.54 | 53.33 | 0.616 |
| SVM [6] | 52.94 | 82.35 | 67.65 | 75.00 | 62.07 | 0.676 |
| SGD [3] | 47.06 | 94.12 | 70.59 | 88.89 | 61.54 | 0.706 |
| Ibk [2] | 52.94 | 70.59 | 61.76 | 64.29 | 58.06 | 0.618 |
| LWL [14] | 47.06 | 58.82 | 52.94 | 53.33 | 50.00 | 0.626 |
| Adaboost [12] | 41.18 | 70.59 | 55.88 | 58.33 | 48.28 | 0.683 |
| Bagging [12] | 70.59 | 70.59 | 70.59 | 70.59 | 70.59 | 0.744 |
| OneR [19] | 47.06 | 58.82 | 52.94 | 53.33 | 50.00 | 0.529 |
| Decision Table [26] | 52.94 | 58.82 | 55.88 | 56.25 | 54.55 | 0.578 |
| J48 [41] | 58.82 | 76.47 | 67.65 | 71.43 | 64.52 | 0.713 |
| Random Forest [49] | 47.06 | 76.47 | 61.76 | 66.67 | 55.17 | 0.739 |
Table 9.
Results achieved on the testing set for the vowel /o/
| Algorithm | Sensibility (%) | Specificity (%) | Accuracy (%) | Precision (%) | F1-score (%) | AUC |
|---|---|---|---|---|---|---|
| Naive Bayes [25] | 94.12 | 11.76 | 52.94 | 51.61 | 66.67 | 0.585 |
| Bayes Net [25] | 64.71 | 35.29 | 50.00 | 50.00 | 56.41 | 0.533 |
| SVM [6] | 70.59 | 58.82 | 64.71 | 63.16 | 66.67 | 0.647 |
| SGD [3] | 70.59 | 35.29 | 52.94 | 52.17 | 60.00 | 0.529 |
| Ibk [2] | 64.71 | 70.59 | 67.65 | 68.75 | 66.67 | 0.676 |
| LWL [14] | 76.47 | 58.82 | 67.65 | 65.00 | 70.27 | 0.775 |
| Adaboost [12] | 64.71 | 41.18 | 52.94 | 52.38 | 57.89 | 0.576 |
| Bagging [12] | 64.71 | 35.29 | 50.00 | 50.00 | 56.41 | 0.474 |
| OneR [19] | 47.06 | 35.29 | 41.18 | 42.11 | 44.44 | 0.412 |
| Decision Table [26] | 76.47 | 41.18 | 58.82 | 56.52 | 65.00 | 0.592 |
| J48 [41] | 58.82 | 41.18 | 50.00 | 50.00 | 54.05 | 0.517 |
| Random Forest [49] | 70.59 | 52.94 | 61.76 | 60.00 | 64.86 | 0.666 |
It is important to note that the data collected for the considered database is still in progress. This means that the number of data changes constantly, making the task of building a common database for the entire scientific community difficult until data collection is finished, not allowing in this way the comparison of the results obtained by different studies existing in literature.
Conclusions
Nowadays, wearable sensors contribute to disease detection and patient monitoring and rehabilitation. Physiological signals coming from these sensors constitute, in fact, a fundamental resource to support specific healthcare applications. The increasingly common practice of the collection of healthcare data and the rapid development of artificial intelligence algorithms have contributed to promoting an increase in the successful application of these techniques in the healthcare sector. Artificial intelligence methods offer an opportunity to process and interpret healthcare data in a fast and reliable way, detecting, at times, clinically relevant information hidden in a vast amount of data, thereby assisting medical decision making.
In this paper we have presented an overview of the artificial intelligence algorithms most frequently used for voice signal analysis in relation to the early detection of disorders caused by COVID-19. The performance of several ML techniques is presented. The aim is to identify the most reliable ML technique to be used and the most accurate vowel sound to be applied in order to distinguish between healthy and COVID-19 positive subjects. The objective is to embed this ML technique within a mobile health solution, capable of acquiring a vowel sound, analysing it and classifying it as healthy or COVID-19 positive. The utility of such a mobile health solution is indisputable in that it would support the early detection of COVID-19 in a faster, cheaper and more reliable way. The analyses have shown that the Random Forest algorithm achieves the best performance, obtaining an accuracy of about 82% in the analysis of the sound of three vowels (/a/, /e/ and /o/) for each subject. This performance improves when only the sound of the vowel /e/ is analysed (the accuracy is equal of 85%).
It is important to note that all the experimental tests were performed on a dataset selected from a crowd-sourced database. Currently, all available voice databases containing samples from subjects suffering from COVID-19 are crowd-sourced, volunteers independently recording all the samples contained in this database without any control from an expert. Therefore, in order to validate an approach able to support the early detection of the pandemic, the currently available recordings need to be improved. It is fundamental to obtain samples labelled by a medical specialist during a controlled clinical trial. Additionally, it is necessary to enhance the quality of the collected data by reducing the effects of noise added during the recording, as well as by ensuring a correct execution of the vocalization. Moreover, in this preliminary study, only the effects of COVID-19 were evaluated. Due to the recent and rapid diffusion of COVID-19, there is still very little information about the causes and development of the pandemic, as well as the association with patient data. In our future research, it will be interesting to analyse data about the etiopathogenesis of the pandemic, clinical data such as age and data about pre-existing pathologies. It is important to consider the effects of these factors on voice quality, combining this information with that extracted from an analysis of the voice parameters. It would be expedient to consider the data already obtained as well as data obtained from an analysis of the other two sounds provided by the database, the coughing and respiratory samples.
Acknowledgements
The authors thank for medical support the specialists of the Department of Otorhinolaryngology, of the University Hospital (Policlinico) Federico II of Naples (Italy).
Author Contributions
Author 1 and Author 3 contributed to the conception and the design of this study and to carrying out the analysis. The first draft of the manuscript was written by Author 1, and all the other authors commented on the previous versions of this manuscript. All the authors read and approved the final manuscript.
Funding
No sources of funding were used in the performing of this study.
Availability of Data and Material
The Coswara database used in this study is a freely available crow sourced database.
Declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Code Availability
The data processing and the feature extraction phases have been carried out by using Matlab version R2020a. Instead, the classification analysis has been performed by using Weka version 3.8.4 (available at https://waikato.github.io/weka-wiki/downloading_ weka/).
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent to Publication
Not applicable.
References
- 1.Afshar, P.; Heidarian, S.; Naderkhani, F.; Oikonomou, A.; Plataniotis, K.N.; Mohammadi, A.: Covid-caps: a capsule network-based framework for identification of COVID-19 cases from x-ray images. arXiv:2004.02696 (2020) [DOI] [PMC free article] [PubMed]
- 2.Aha DW, Kibler D, Albert MK. Instance-based learning algorithms. Mach. Learn. 1991;6(1):37–66. [Google Scholar]
- 3.Alaoui SS, Labsiv Y, Aksasse B. Classification algorithms in data mining. Int. J. Tomogr. Simul. 2018;31:34–44. [Google Scholar]
- 4.Andreu-Perez, J.; Pérez-Espinos, H.; Timone, E.; Girón-Pérez, M.I.; Kiani, M.; Benitez-Trinidad, A.B.; Jarchi, D.; Rosales-Pérez, A.; Ali, Z.; Gatzoulis, N.: A novel deep learning based recognition method and web-app for COVID-19 infection test from cough sounds with a clinically validated dataset (2020)
- 5.Brown, C.; Chauhan, J.; Grammenos, A.; Han, J.; Hasthanasombat, A.; Spathis, D.; Xia, T.; Cicuta, P.; Mascolo, C.: Exploring automatic diagnosis of COVID-19 from crowdsourced respiratory sound data. In: Proceedings of the 26th ACM SIGKDD International Conference on Knowledge Discovery & Data Mining. pp. 3474–3484 (2020)
- 6.Burges, C.J.; Smola, A.J.: Advances in kernel methods. Support Vector Learning (1999)
- 7.Chaudhari, G.; Jiang, X.; Fakhry, A.; Han, A.; Xiao, J.; Shen, S.; Khanzada, A.: Virufy: global applicability of crowdsourced and clinical datasets for ai detection of COVID-19 from cough. arXiv:2011.13320 (2020)
- 8.Dash, T.K.; Mishra, S.; Panda, G.; Satapathy, S.C.: Detection of COVID-19 from speech signal using bio-inspired based cepstral features. Pattern Recognit. 117, 107999 (2021) [DOI] [PMC free article] [PubMed]
- 9.Database, C.: Coswara-Data. https://github.com/iiscleap/Coswara-Data/ (2020), [Online; accessed 11-Junuary-2021]
- 10.Davenport T, Kalakota R. The potential for artificial intelligence in healthcare. Future Healthc. J. 2019;6(2):94. doi: 10.7861/futurehosp.6-2-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deshmukh, S.; Ismail, M.A.; Singh, R.: Interpreting glottal flow dynamics for detecting COVID-19 from voice. arXiv:2010.16318 (2020)
- 12.Dietterich, T.G.: Ensemble methods in machine learning. In: International workshop on multiple classifier systems. pp. 1–15. Springer (2000)
- 13.Farrús, M.; Hernando, J.; Ejarque, P.: Jitter and shimmer measurements for speaker recognition. In: Eighth Annual Conference of the International Speech Communication Association (2007)
- 14.Frank, E.; Hall, M.; Pfahringer, B.: Locally weighted Naive Bayes. In: 19th Conference in Uncertainty in Artificial Intelligence. pp. 249–256. Morgan Kaufmann (2003)
- 15.Garner SR. Weka: the waikato environment for knowledge analysis. Proc. New Z. Comput. Sci. Res. Stud. Conf. 1995;1995:57–64. [Google Scholar]
- 16.Gupta, V.: Voice disorder detection using long short term memory (lstm) model. arXiv:1812.01779 (2018)
- 17.Han, J.; Qian, K.; Song, M.; Yang, Z.; Ren, Z.; Liu, S.; Liu, J.; Zheng, H.; Ji, W.; Koike, T.: An early study on intelligent analysis of speech under COVID-19: severity, sleep quality, fatigue, and anxiety. arXiv:2005.00096 (2020)
- 18.Hassan, A.; Shahin, I.; Alsabek, M.B.: COVID-19 detection system using recurrent neural networks. In: 2020 International Conference on Communications, Computing, Cybersecurity, and Informatics (CCCI). pp. 1–5. IEEE (2020)
- 19.Holte R. Very simple classification rules perform well on most commonly used datasets. Mach. Learn. 1993;11:63–91. doi: 10.1023/A:1022631118932. [DOI] [Google Scholar]
- 20.Hossain MS. Cloud-supported cyber-physical localization framework for patients monitoring. IEEE Syst. J. 2017;11(1):118–127. doi: 10.1109/JSYST.2015.2470644. [DOI] [Google Scholar]
- 21.Hossain MS, Muhammad G, Alamri A. Smart healthcare monitoring: a voice pathology detection paradigm for smart cities. Multimed. Syst. 2019;25(5):565–575. doi: 10.1007/s00530-017-0561-x. [DOI] [Google Scholar]
- 22.Hossain MS, Muhammad G, Guizani N. Explainable AI and mass surveillance system-based healthcare framework to combat COVID-i9 like pandemics. IEEE Netw. 2020;34(4):126–132. doi: 10.1109/MNET.011.2000458. [DOI] [Google Scholar]
- 23.Imran, A.; Posokhova, I.; Qureshi, H.N.; Masood, U.; Riaz, S.; Ali, K.; John, C.N.; Nabeel, M.: Ai4covid-19: Ai enabled preliminary diagnosis for COVID-19 from cough samples via an app. arXiv:2004.01275 (2020) [DOI] [PMC free article] [PubMed]
- 24.Ismail, M.A.; Deshmukh, S.; Singh, R.: Detection of COVID-19 through the analysis of vocal fold oscillations. arXiv:2010.10707 (2020)
- 25.John, G.H.; Langley, P.: Estimating continuous distributions in bayesian classifiers. In: Eleventh Conference on Uncertainty in Artificial Intelligence. pp. 338–345. Morgan Kaufmann, San Mateo (1995)
- 26.Kohavi, R.: The power of decision tables. In: 8th European Conference on Machine Learning. pp. 174–189. Springer (1995)
- 27.Laguarta, J.; Hueto, F.; Subirana, B.: COVID-19 artificial intelligence diagnosis using only cough recordings. IEEE Open J. Eng. Med. Biol. 1, 275–281 (2020) [DOI] [PMC free article] [PubMed]
- 28.Maccarini, A.R.; Lucchini, E.: La valutazione soggettiva ed oggettiva della disfonia. il protocollo sifel. Acta Phoniatrica Latina 24(1/2), 13–42 (2002)
- 29.Maryn Y, Roy N. Sustained vowels and continuous speech in the auditory-perceptual evaluation of dysphonia severity. Jornal da Sociedade Brasileira de Fonoaudiologia. 2012;24:107–112. doi: 10.1590/S2179-64912012000200003. [DOI] [PubMed] [Google Scholar]
- 30.Maryn Y, Roy N, De Bodt M, Van Cauwenberge P, Corthals P. Acoustic measurement of overall voice quality: a meta-analysis. J. Acoust. Soc. Am. 2009;126(5):2619–2634. doi: 10.1121/1.3224706. [DOI] [PubMed] [Google Scholar]
- 31.Matlab: audioFeatureExtractor Function. https://it.mathworks.com/help/audio/ref/audiofeatureextractor.html/ (2020), [Online; accessed 25-Junuary-2021]
- 32.Mehta N, Pandit A, Shukla S. Transforming healthcare with big data analytics and artificial intelligence: a systematic mapping study. J. Biomed. Informatics. 2019;100:103311. doi: 10.1016/j.jbi.2019.103311. [DOI] [PubMed] [Google Scholar]
- 33.Meiniar, W.; Afrida, F.A.; Irmasari, A.; Mukti, A.; Astharini, D.: Human voice filtering with band-stop filter design in matlab. In: 2017 International Conference on Broadband Communication, Wireless Sensors and Powering (BCWSP). pp. 1–4. IEEE (2017)
- 34.Muhammad G, Hossain MS, Kumar N. Eeg-based pathology detection for home health monitoring. IEEE J. Sel. Areas Commun. 2021;39(2):603–610. doi: 10.1109/JSAC.2020.3020654. [DOI] [Google Scholar]
- 35.Ni, Q.; Sun, Z.Y.; Qi, L.; Chen, W.; Yang, Y.; Wang, L.; Zhang, X.; Yang, L.; Fang, Y.; Xing, Z.: A deep learning approach to characterize 2019 coronavirus disease (COVID-19) pneumonia in chest ct images. Eur. Radiol. 30(12), 6517–6527 (2020) [DOI] [PMC free article] [PubMed]
- 36.Orlandic, L.; Teijeiro, T.; Atienza, D.: The coughvid crowdsourcing dataset: a corpus for the study of large-scale cough analysis algorithms. arXiv:2009.11644 (2020) [DOI] [PMC free article] [PubMed]
- 37.Pahar, M.; Klopper, M.; Warren, R.; Niesler, T.: COVID-19 cough classification using machine learning and global smartphone recordings. arXiv:2012.01926 (2020) [DOI] [PMC free article] [PubMed]
- 38.Parsa, V.; Jamieson, D.G.: Acoustic discrimination of pathological voice. J. Speech Lang. Hear. Res. 44(2), 327–339 (2001) [DOI] [PubMed]
- 39.Pishgar, M.; Karim, F.; Majumdar, S.; Darabi, H.: Pathological voice classification using mel-cepstrum vectors and support vector machine. arXiv:1812.07729 (2018)
- 40.Qian, K.; Schuller, B.W.; Yamamoto, Y.: Recent advances in computer audition for diagnosing COVID-19: an overview. arXiv:2012.04650 (2020)
- 41.Quinlan R. C4.5: programs for machine learning. San Mateo: Morgan Kaufmann Publishers; 1993. [Google Scholar]
- 42.Ritwik, K.V.S.; Kalluri, S.B.; Vijayasenan, D.: COVID-19 patient detection from telephone quality speech data. arXiv:2011.04299 (2020)
- 43.Sajda P. Machine learning for detection and diagnosis of disease. Annu. Rev. Biomed. Eng. 2006;8:537–565. doi: 10.1146/annurev.bioeng.8.061505.095802. [DOI] [PubMed] [Google Scholar]
- 44.Saldanha JC, Ananthakrishna T, Pinto R. Vocal fold pathology assessment using mel-frequency cepstral coefficients and linear predictive cepstral coefficients features. J. Med. Imag. Health Informatics. 2014;4(2):168–173. doi: 10.1166/jmihi.2014.1253. [DOI] [Google Scholar]
- 45.Severin, F.; Bozkurt, B.; Dutoit, T.: Hnr extraction in voiced speech, oriented towards voice quality analysis. In: 2005 13th European Signal Processing Conference. pp. 1–4. IEEE (2005)
- 46.Sharma, N.; Krishnan, P.; Kumar, R.; Ramoji, S.; Chetupalli, S.R.; Ghosh, P.K.; Ganapathy, S.: Coswara–a database of breathing, cough, and voice sounds for COVID-19 diagnosis. arXiv:2005.10548 (2020)
- 47.Subirana, B.; Hueto, F.; Rajasekaran, P.; Laguarta, J.; Puig, S.; Malvehy, J.; Mitja, O.; Trilla, A.; Moreno, C.I.; Valle, J.F.M.: Hi sigma, do i have the coronavirus?: Call for a new artificial intelligence approach to support health care professionals dealing with the COVID-19 pandemic. arXiv:2004.06510 (2020)
- 48.Trivedy, S.; Goyal, M.; Mohapatra, P.R.; Mukherjee, A.: Design and development ofsmartphone-enabled spirometer with a disease classification system using convolutional neural network. IEEE Trans. Instrum. Meas. 69(9), 7125–7135 (2020)
- 49.Venkatesan N, Priya G. A study of random forest algorithm with implementation using weka. Int. J. Innov. Res. Comput. Sci. Eng. 2015;1(6):156–162. [Google Scholar]
- 50.Verde L, De Pietro G, Sannino G. A methodology for voice classification based on the personalized fundamental frequency estimation. Biomed. Signal Process. Control. 2018;42:134–144. doi: 10.1016/j.bspc.2018.01.007. [DOI] [Google Scholar]
- 51.Verde L, De Pietro G, Sannino G. Voice disorder identification by using machine learning techniques. IEEE Access. 2018;6:16246–16255. doi: 10.1109/ACCESS.2018.2816338. [DOI] [Google Scholar]
- 52.Wang, L.; Lin, Z.Q.; Wong, A.: Covid-net: a tailored deep convolutional neural network design for detection of COVID-19 cases from chest x-ray images. Sci. Rep. 10(1), 1–12 (2020) [DOI] [PMC free article] [PubMed]
- 53.World Health Organization: WHO Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int/?gclid=EAIaIQobChMIht_qyL_K6gIVB-7tCh2AIgwMEAAYASAAEgLyX_D_BwE/ (2020), [Online; accessed 01-Junuary-2021]
- 54.Wu, M.; Luo, J.: Wearable technology applications in healthcare: a literature review. Online J. Nurs. Inform. 23(3) (2019). [online] Available at: https://www.himss.org/resources/wearabletechnology-applications-healthcare-literature-review
- 55.Xu, X.; Jiang, X.; Ma, C.; Du, P.; Li, X.; Lv, S.; Yu, L.; Ni, Q.; Chen, Y.; Su, J.: A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering (2020). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The Coswara database used in this study is a freely available crow sourced database.


