A stimulated proinflammatory feature of leukemic progenitors facilitates their survival and immune evasion after chemotherapy.
Abstract
Chemotherapy can effectively reduce the leukemic burden and restore immune cell production in most acute myeloid leukemia (AML) cases. Nevertheless, endogenous immunosurveillance usually fails to recover after chemotherapy, permitting relapse. The underlying mechanisms of this therapeutic failure have remained poorly understood. Here, we show that abnormal IL-36 production activated by NF-κB is an essential feature of mouse and human leukemic progenitor cells (LPs). Mechanistically, IL-36 directly activates inflammatory monocytes (IMs) in bone marrow, which then precludes clearance of leukemia mediated by CD8+ T cells and facilitates LP growth. While sparing IMs, common chemotherapeutic agents stimulate IL-36 production from residual LPs via caspase-1 activation, thereby enabling the persistence of this immunosuppressive IL-36–IM axis after chemotherapy. Furthermore, IM depletion by trabectedin, with chemotherapy and PD-1 blockade, can synergistically restrict AML progression and relapse. Collectively, these results suggest inhibition of the IL-36–IM axis as a potential strategy for improving AML treatment.
INTRODUCTION
Acute myeloid leukemia (AML) represents a group of common hematopoietic malignancy characterized with infiltration of hematopoietic tissues such as bone marrow (BM) by accumulated aberrant immature myeloid blasts, resulting in severely repressed normal hematopoiesis. Although most AML patients initially respond to the conventional therapies comprising chemotherapy and allogeneic BM transplantation (allo-BMT), many of them would relapse in a few months and acquired therapy resistance (1). Longitudinal studies showed that AML that relapsed after allo-BMT needed to escape graft-versus-leukemia immune rejection, while AML that relapsed after chemotherapy did not experience a selection pressure emanated from a possibly recovered endogenous immunosurveillance (2, 3). Also in line with the notion that AML largely belongs to a group of immune-cold tumors, most AML cases showed overwhelming de novo resistance to immune checkpoint blockade (4, 5), and only a portion of immune-infiltrated AML cases responded to flotetuzumab immunotherapy (6). On the other hand, the observation that the chronic immunosuppression after organ transplantation greatly increased AML incidence suggested the existence of endogenous immunosurveillance against AML, albeit its effects probably were at a restrained level (7). Thus, activating endogenous immunosurveillance against AML holds great potential to improve AML treatment by synergizing with other contemporary therapeutic modes.
It is reported that both leukemia stem/progenitor cells and their monocytic progeny are capable of suppressing and evading immunosurveillance (8, 9). Because chemotherapy largely spares leukemia stem/progenitor cells while effectively reducing bulk leukemia cells (10, 11), it is likely that residual leukemia stem/progenitor cells are capable of creating an immunosuppressive microenvironment that permits a possible relapse. Similarly, during the very early stage of leukemic propagation, a few leukemic progenitor cells (LPs) have to defy a possible local immunosurveillance (for example, in BM) to strive. However, the underlying mechanisms in these particular situations remain poorly defined.
Three functional paralogs of interleukin-36α (IL-36α), IL-36β, and IL-36γ, as well as an antagonist IL-36Ra, constitute the IL-36 family that share a common receptor complex composed of IL-36R and IL-1RAcP, and the abnormal activation of IL-36 signaling is involved in the pathogenesis of multiple types of autoimmunity disease (12–14). While IL-36γ artificially expressed in solid tumor was shown to promote type I antitumor immunity (15), its role in the regulation of immunosurveillance against AML is unreported. In this study, we show that proinflammatory LPs, as marked by nuclear factor κB (NF-κB) pathway activation, secrete IL-36, and this feature is sustained after chemotherapy via a chemotherapy-stimulated caspase-1–IL-1β–NF-κB autocrine signaling pathway. Mechanistically, IL-36 activates inflammatory monocytes (IMs) within BM, which not only nurses the survival and proliferation of AML cells, as indicated before (16), but also precludes a CD8+ T cell–mediated AML clearance. Collectively, our study suggests inhibition of the IL-36–IM axis as a potential strategy for improving AML treatment.
RESULTS
Leukemia cell–derived Il-36γ supports early disease progression in BM
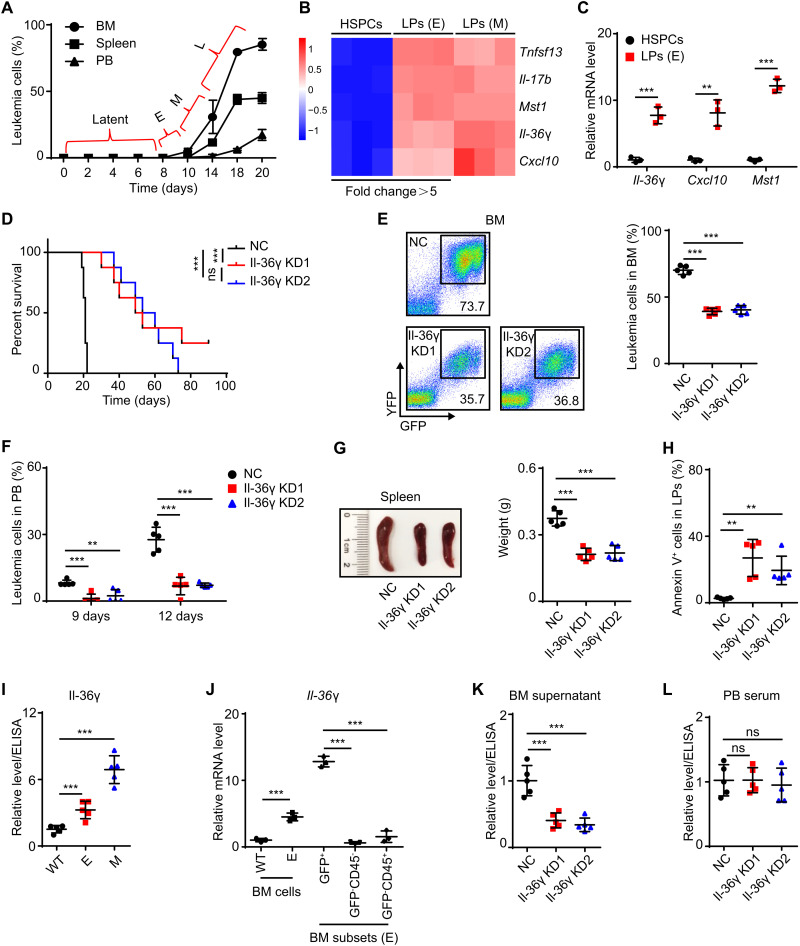
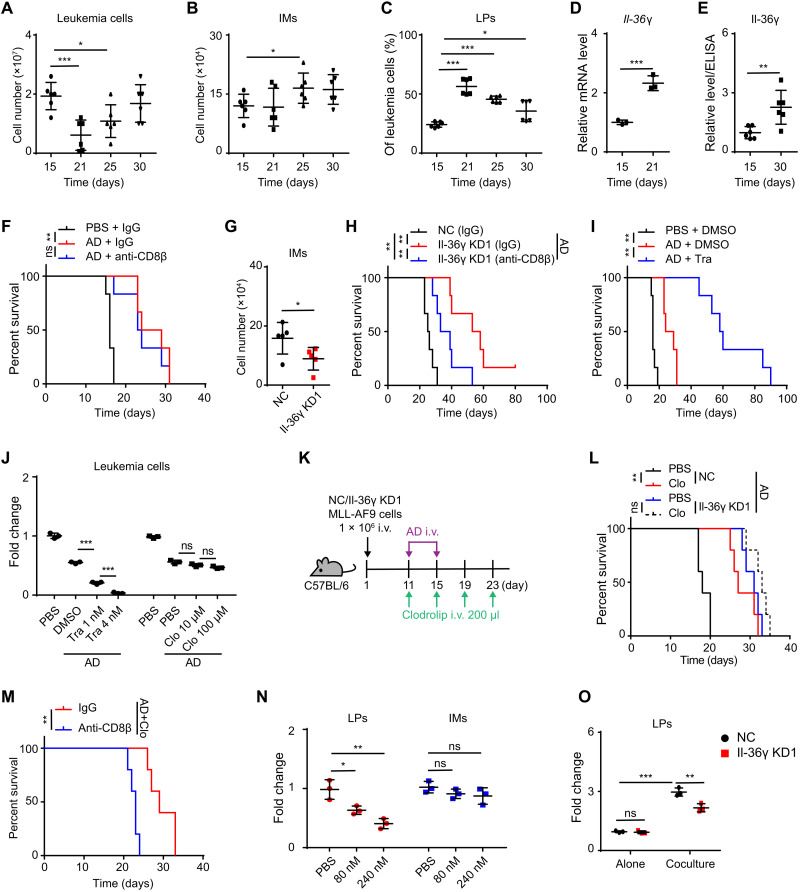
To investigate how leukemia cells remodel the BM microenvironment to propagate, we labeled the murine acute promyelocytic leukemia (APL) cells derived from BM of hMRP8-PML/RARα transgenic mice (FVB/NJ) (17) with green fluorescent protein (GFP)–expressing retroviral vector MigR1 and transplanted 1 × 104 GFP+ APL cells into nonirradiated syngeneic recipients (fig. S1A). This resulted in leukemic predominance within approximately 3 weeks (fig. S1B). The leukemic propagation spanned four phases: a relatively long latent phase followed by a transient early phase (E) wherein the proportion of leukemia cells in BM was between 1 and 5%, but was almost undetectable in spleen and peripheral blood (PB); leukemic propagation then intensified throughout the middle phase (M) (leukemic proportion was 5 to 50% in BM), followed by a late phase (L) with high leukemic proportion in BM (≥50%) (Fig. 1A).
Fig. 1. Leukemic Il-36γ promotes early disease progression in BM.
(A) APL mouse models were established as described in fig. S1A. The percentages of APL cells in recipients’ organs were determined at the indicated time points (n = 5 mice per time point). E (early), M (middle), and L (late). (B and C) Whole transcriptome RNA-seq (B) and qRT-PCR analysis (C) of proinflammatory cytokines were conducted in BM HSPCs (lineage−c-Kit+) sorted from WT FVB/NJ mice and BM LPs (c-Kit+) sorted from APL mice (n = 3). (D to H) NC/Il-36γ KD APL mouse models were established as described in fig. S1D. Dox (200 μg/ml) was supplied in drinking water beginning on day 10. (D) Survival curves were generated (n = 8 mice per group). (E to H) Mice were examined on day 9 of Dox treatment and sacrificed on day 12 (n = 5 mice per group). The percentage of APL cells in BM (E) and PB (F), spleen weights (G), and percentages of annexin V+ cells in BM LPs (H) are shown. Photo credit: He-Zhou Guo, Shanghai Institute of Hematology, Ruijin Hospital affiliated with Shanghai Jiao-Tong University School of Medicine. (I and J) WT FVB/NJ mice and APL mice in E or M phases were sacrificed. (I) Il-36γ relative protein levels in BM supernatant were measured using ELISA (n = 5 mice per group). (J) Relative mRNA levels of Il-36γ in discrete subsets of BM cells were measured using qRT-PCR (n = 3). (K and L) NC/Il-36γ KD APL mouse models were established as described in fig. S1D. Dox was started on day 10, and mice were sacrificed after 12 days of Dox treatment (n = 5 mice per group). Il-36γ relative protein levels in BM supernatant (K) and PB serum (L) are shown. Data are presented as means ± SD. **P < 0.01, ***P < 0.001. ns, not significant.
Given that complex changes occurred in BM during the M/L phases and that leukemia cells were sparse in the latent phase, we focused on leukemia cells in the E phase, most of which were LPs (c-Kit+) (fig. S1C), which can initiate leukemia (18). Because normal hematopoietic stem and progenitor cells (HSPCs) (lineage−c-Kit+) can regulate the BM microenvironment through cytokine secretion (19, 20), we then tested whether this regulation might be abnormal in LPs. Whole transcriptome RNA sequencing (RNA-seq) and quantitative reverse transcription polymerase chain reaction (qRT-PCR) assays revealed that the mRNA levels of several cytokines, including Il-36γ, Cxcl10, Mst1, Tnfsf13, and Il-17b, were substantially up-regulated (more than fivefold) in E-phase LPs compared to their expression in wild-type (WT) HSPCs (Fig. 1, B and C).
Previous reports have shown the oncogenic role of Tnfsf13 and Il-17b via autocrine signaling to facilitate the AML cell survival (21, 22). We therefore used a doxycycline (Dox)–inducible, leukemia cell–specific expression system to characterize the function of the remaining three cytokines in regulating leukemic progression (fig. S1D). We screened out two short hairpin RNAs (shRNAs) that effectively suppressed each cytokine and confirmed that they decreased target cytokine transcription in leukemia cells in vivo after Dox administration (fig. S1E). The inducible knockdown (KD) of leukemic Il-36γ starting from the E phase resulted in significantly longer survival of leukemic mice compared with controls, but not Mst1 and Cxcl10 (Fig. 1D and fig. S1F). Meanwhile, Il-36γ KD delayed leukemic progression and resulted in increased levels of apoptosis among LPs in BM (Fig. 1, E to H). Furthermore, enzyme-linked immunosorbent assay (ELISA) showed that the level of Il-36γ in BM supernatant increased during the E or M phases compared to that in normal BM (Fig. 1I). Because Il-36γ was preferentially expressed by leukemia cells compared with nonleukemia cells (Fig. 1J), Dox-induced leukemic Il-36γ KD significantly decreased Il-36γ in BM supernatant, although not in PB serum (Fig. 1, K and L). Together, these results suggested that Il-36γ locally promoted early leukemic progression within BM.
LP-derived Il-36γ supports their capacity to initiate leukemia
Notably, Il-36γ mRNA levels were higher in APL LPs compared to non-LPs (c-Kit−) (Fig. 2A). To further validate the relevance of this result in AML, we examined GFP+ murine MLL-AF9 AML cells (fig. S2A) and observed similar patterns of Il-36γ expression (Fig. 2B). Chromatin immunoprecipitation (ChIP)–qPCR assays to quantify H3K4me3 and H3K27me3 levels revealed that the 5′ regulatory element of the Il-36γ locus was in an activated state in LPs compared with that in non-LPs and HSPCs in both APL and MLL-AF9 models (fig. S2, B to D), further supporting the above findings that Il-36γ transcription was highly activated within the LPs.
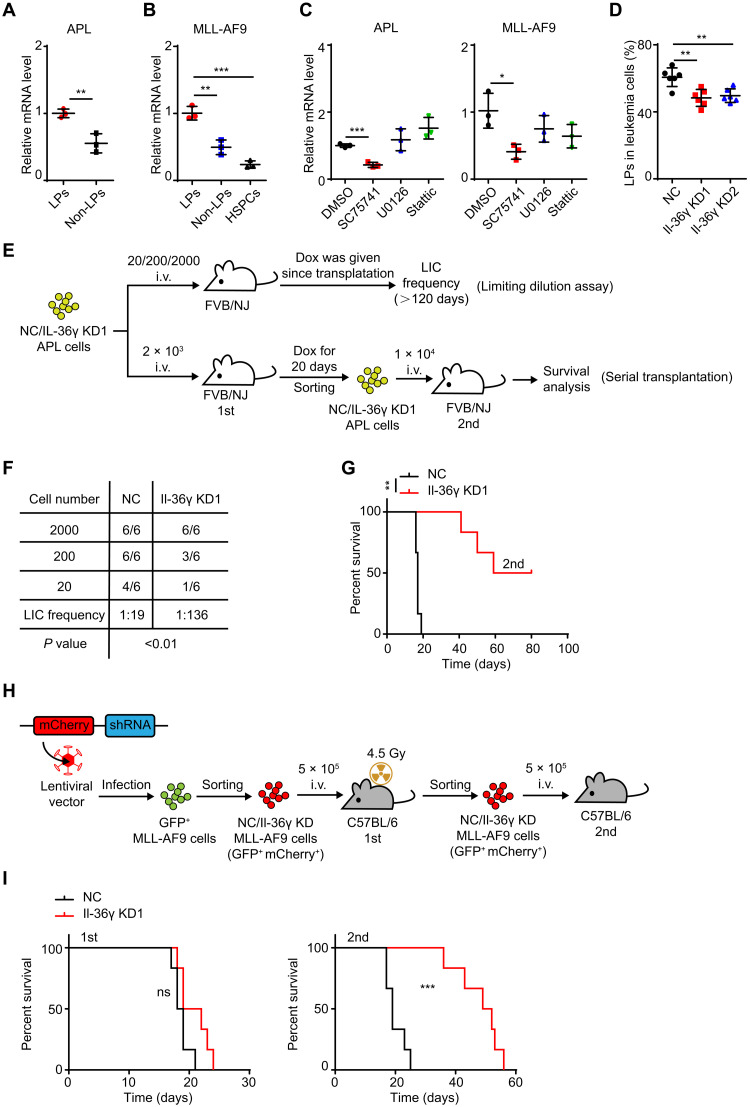
Fig. 2. LP-derived Il-36γ enhances leukemia initiation.
(A to C) qRT-PCR analysis of Il-36γ mRNA in (A) BM LPs and non-LPs from E-phase APL mice; (B) BM HSPCs from WT C57BL/6, BM LPs, and non-LPs from MLL-AF9 mice (see fig. S2A); and (C) LPs treated with inhibitor of NF-κB (SC75741, 5 μM), Erk (U0126, 10 μM), or Stat3 (Stattic, 1 μM) for 24 hours in vitro (n = 3). (D) NC/Il-36γ KD APL mice (see fig. S1D) were treated with Dox beginning on day 10 and sacrificed after 12 days of Dox treatment. The percentage of LPs in BM APL cells were measured (n = 6 mice per group). (E to G) Flow chart (E) of the limiting dilution assay (F) and serial transplantation (G). i.v., intravenously. (F) BM NC/Il-36γ KD APL cells were serially diluted to 20, 2 × 102, and 2 × 103 total cells and transplanted into nonirradiated FVB/NJ mice to calculate LIC frequency (n = 6 mice per dilution). Dox was given since transplantation and continually for >120 days. (G) BM NC/Il-36γ KD APL cells (2 × 103) were transplanted into nonirradiated FVB/NJ mice. Dox was given since transplantation and continually for 20 days. Then, 1 × 104 BM NC/Il-36γ KD APL cells from moribund leukemic mice in the first transplantation experiment were transplanted into nonirradiated FVB/NJ mice for survival analysis without Dox treatment (n = 6 mice per group). (H and I) BM NC/Il-36γ KD MLL-AF9 cells (5 × 105) were transplanted into sublethally irradiated C57BL/6 mice for survival analysis. Then, 5 × 105 BM NC/Il-36γ KD MLL-AF9 cells from the moribund leukemic mice in the first transplantation assay were transplanted into nonirradiated C57BL/6 mice for survival analysis (n = 6 mice per group). Data are presented as means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
As Il-36γ is an inflammatory cytokine, and inflammatory pathway NF-κB was previously shown to promote leukemic progression in multiple subtypes of AML (23–25), we speculated that NF-κB could induce Il-36γ expression in LPs. Analysis of published ChIP sequencing (ChIP-seq) data revealed a strong NF-κB p65 subunit binding site in the Il-36γ promoter region in kdo2-lipidA (KLA)-treated macrophages (fig. S2E). The results of ChIP-qPCR confirmed that NF-κB p65 occupied this site in the Il-36γ promoter in MLL-AF9 LPs (fig. S2F). Furthermore, Gene Set Enrichment Analysis (GSEA) indicated higher NF-κB activity, but not extracellular signal–regulated kinase (ERK) and signal transducer and activator of transcription 3 (Stat3) pathways, in APL LPs compared with HSPCs (fig. S2G), in agreement with Western blots that showed that NF-κB p-p65 levels were higher in the LPs of both AML models than in their non-LPs and HSPCs (fig. S2H). As expected, NF-κB DNA binding inhibitor, but not Stat3 or Erk inhibitor, reduced Il-36γ mRNA levels in the LPs of both AML models (Fig. 2C).
Furthermore, Il-36γ KD reduced the percentage of APL LPs among the leukemic population in BM (Fig. 2D). This finding prompted us to test whether AML LP–derived Il-36γ enhanced their leukemogenic potential using limiting dilution assay and serial transplantation (Fig. 2E). Limiting dilution assay showed that Il-36γ KD resulted in reduced frequency of functional leukemia-initiating cells compared with the negative control (NC) counterparts (1 in 136 versus 1 in 19) (Fig. 2F), and Il-36γ KD prolonged the survival of the secondary APL recipients (Fig. 2G). We also knocked down Il-36γ in GFP+ MLL-AF9 cells and performed serial transplantation (Fig. 2H). Similarly, Il-36γ KD prolonged the survival of the secondary leukemic recipients (Fig. 2I). Collectively, these results strongly suggested that Il-36γ expressed by LPs supported their potential to initiate leukemia.
Il-36γ thwarts the CD8+ T cell response against AML
On the basis of the above findings, we next sought to determine the mechanism by which Il-36γ promoted LP leukemogenic potential. Il-36γ KD caused no observable inhibition of LP growth in vitro (fig. S3A). However, the prolonged survival conferred by Il-36γ KD in NOG recipients [lacking T, B, and natural killer (NK) cells] was substantially limited compared to that observed in syngeneic immunocompetent recipients (Fig. 3A). These observations implied that Il-36γ–induced promotion of AML involved other microenvironmental factors, such as immunosurveillance components.
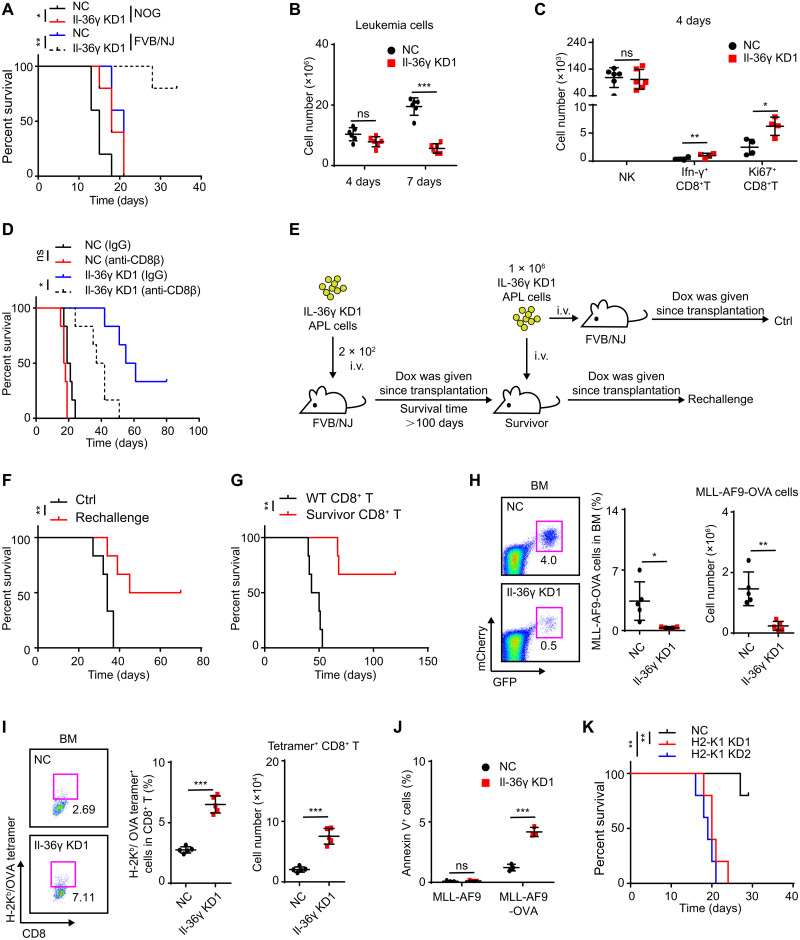
Fig. 3. Il-36γ thwarts a CD8+ T cell response against AML.
(A) NC/Il-36γ KD APL cells (1 × 104) were transplanted into nonirradiated FVB/NJ or NOG mice for survival analysis (n = 5 mice per group). Dox was given since transplantation. (B and C) NC/Il-36γ KD APL mice (see fig. S1D) were sacrificed after 4 or 7 days of Dox treatment (Dox was started on day 10). The numbers of APL cells (n = 6 mice per group) (B), NK cells (n = 6 mice per group), and CD8+ T cells (n = 4 mice per group) (C) in BM were determined. (D) Survival curves were generated for NC/Il-36γ KD APL mice (n = 6 mice per group) after anti-CD8β treatment (see fig. S3D). (E to G) Experimental schematic (E) for evaluation of immune memory. Il-36γ KD APL recipients that survived more than 100 days were identified as survivors. (F) Il-36γ KD APL cells were transplanted into survivors (Rechallenge) or WT FVB/NJ mice (Ctrl) for survival analysis (n = 6 mice per group). (G) Spleen CD8+ T cells (1 × 106) from survivors or WT FVB/NJ mice were cotransplanted with 1 × 105 Il-36γ KD APL cells into nonirradiated FVB/NJ mice for survival analysis (n = 6 mice per group). Dox was given since transplantation. (H to J) MLL-AF9-OVA cells (H) and tetramer+ CD8+ T (I) in BM of MLL-AF9-OVA model (see fig. S3H) on day 12 after AML inoculation were examined (n = 5 mice per group). (J) T cell killing assay was conducted using activated spleen CD8+ T cells from NC/Il-36γ KD mice as effector cells. MLL-AF9-OVA or MLL-AF9 cells were used as targets (n = 3). (K) Survival curves were generated for H2-K1 KD MLL-AF9 mice (see fig. S3K) (n = 5 mice per group). Data are presented as means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
To identify the participating components, we next examined the abundance of CD8+ T and NK cells because these cells serve as the main killers of AML cells (8, 26). We found that BM CD8+ T cell counts decreased in the E phase, while NK cells remained stable (fig. S3, B and C). Conversely, at 4 days after Il-36γ KD, when the leukemic burden and NK cell counts were unchanged in BM, the number of functional CD8+ T cells was significantly increased (Fig. 3, B and C). Accordingly, depletion of CD8+ T cells, but not NK cells, mediated by antibodies led to significantly shorter survival of Il-36γ KD APL recipient mice (Fig. 3D and fig. S3, D to G). Moreover, the recipients that survived through primary transplantation with Il-36γ KD APL cells were conferred protection against rechallenge with APL cells (Fig. 3, E and F), while CD8+ T cells isolated from the surviving recipients showed a stronger protective effect against APL establishment than that provided by unprimed CD8+ T cells (Fig. 3G). These results indicated an immunosuppressive role of leukemic Il-36γ on the APL-responsive cytotoxic T lymphocytes.
To further explore this effect, MLL-AF9 cells expressing ovalbumin (OVA) as a model tumor antigen were established (fig. S3H). We first verified that MLL-AF9 cells with puromycin resistance presented OVA-derived SIINFEKL peptide on H-2Kb (fig. S3I) and that MLL-AF9-OVA cells, but not MLL-AF9 cells, were killed in vitro by activated OVA-specific CD8+ T cells derived from splenocytes of OT-1 transgenic mice (fig. S3J). Subsequently, we found that Il-36γ KD decreased the percentage and number of MLL-AF9-OVA cells while increasing the abundance of CD8+ T cells in BM that specifically bound H-2Kb/OVA tetramer (Fig. 3, H and I). In addition, the CD8+ T cells isolated from the Il-36γ KD leukemic mice exhibited enhanced cytotoxicity toward MLL-AF9-OVA cells but not MLL-AF9 cells in vitro as compared to the CD8+ T cells isolated from the NC leukemic mice (Fig. 3J). To test whether killing by CD8+ T cells required recognition of major histocompatibility complex (MHC) class I–restricted antigen, we generated a second KD of H2-K1 in Il-36γ KD MLL-AF9 cells and found that loss of H2-K1 led to shortened survival of AML recipients (Fig. 3K and fig. S3, K and L). Together, these results strongly suggested that leukemic Il-36γ attenuated a classic cytotoxic T lymphocyte (CTL) response induced by AML tumor antigens.
To test whether Il-36γ can directly promote the suppression of CD8+ T cells by AML cells, we cultured APL cells together with CD8+ T cells in vitro. The results showed that APL cells moderately promoted, rather than attenuated, CD8+ T cell activation in vitro and that this effect was not affected by Il-36γ KD (fig. S3M), thus excluding a direct suppressive effect of Il-36γ on CD8+ T cells in AML. Moreover, in contrast with Il-36γ elevation in BM during APL progression, levels of interferon-γ (Ifn-γ) declined (fig. S3N), while Il-12 (p70) and Il-2 that synergized with Il-36γ to activate CD8+ T cells were not increased (fig. S3N) (27). These results suggested that Il-36γ did not result in the activation of type I immunity in AML, in contradiction with published reports of the effects of Il-36γ overexpression in a solid tumor model (15).
Il-36γ activates IMs to increase leukemic propagation by inactivating CD8+ T cells and promoting LP growth
We next searched for missing factors that could mediate Il-36γ–induced inactivation of CD8+ T cells in vivo. Cell counts indicated that IMs (CD3−B220−Ter119−Gr-1+CD115+) increased in the E-phase BM, whereas other potential CD8+ T cell–inactivating cells such as neutrophils (Neu) (CD3−B220−Ter119−Gr-1+CD115−), macrophages (Mac) (CD3−B220−Ter119−Gr-1−CD115−F4/80+SSCLow), and regulatory T cells (Tregs) (CD3+CD4+FOXP3+) did not (Fig. 4, A and B). Conversely, IMs decreased after 4 days of Dox-induced Il-36γ KD, while other cell types did not (Fig. 4C). Given that the IM subtype of myeloid-derived suppressor cell (MDSC) can facilitate evasion of tumor immunity (28), we next used trabectedin, which eliminates IMs and their progeny (29, 30), to examine whether Il-36γ–stimulated IMs may contribute to leukemic progression and CD8+ T cell inactivation. After a single trabectedin injection, IMs in BM were decreased and trabectedin exhibited inhibition effects on leukemic progression of the treated APL mice (fig. S4, A to C).
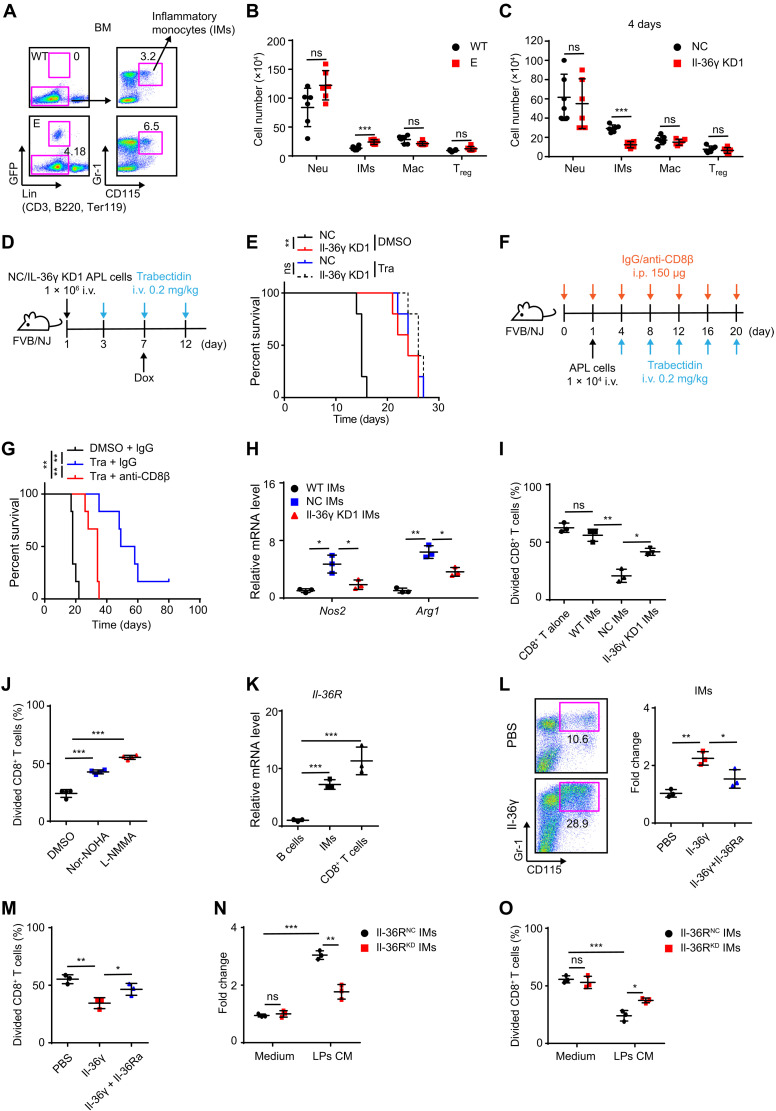
Fig. 4. Il-36γ–activated IMs suppress CD8+ T cells.
(A and B) Representative flow cytometric analysis of IMs (A) and the number of indicated cells in BM of WT FVB/NJ and E-phase APL mice (B) (n = 6 mice per group). (C) NC/Il-36γ KD APL mice (see fig. S1D) were sacrificed after 4 days of Dox treatment (Dox was started on day 10). The number of indicated cells in BM was measured (n = 6 mice per group). (D and E) Experimental schematic (D) of trabectedin treatment for NC/Il-36γ KD APL mice and corresponding survival curves (E) (n = 5 mice per group). (F and G) Schematic (F) for trabectedin and/or anti-CD8β treatment for APL mice and corresponding survival curves (G) (n = 5 mice per group). (H to J) NC/Il-36γ KD APL mice (see fig. S1D) were sacrificed after 7 days of Dox treatment (Dox was started on day 10). Then, BM IMs in NC recipients (NC IMs), Il-36γ KD recipients (Il-36γ KD1 IMs), or WT FVB/NJ mice (WT IMs) were sorted to measure Nos2 and Arg1 mRNA levels (n = 3) (H) and T cell proliferation (I and J). DMSO, nor-NOHA (Arg1 inhibitor, 0.5 mM), or L-NMMA (Nos2 inhibitor, 0.5 mM) was used to treat NC IMs-CD8+ T cell cocultures (n = 3) (J). (K) qRT-PCR assay on Il-36R in BM subsets from APL mice (n = 3). (L and M) BM cells of APL mice were cultured with Il-36γ (200 ng/ml) and/or Il-36Ra (2 μg/ml) for 4 days (n = 3). Then, IMs were counted (L) and sorted for T cell proliferation assay (M). (N and O) Il-36RNC/Il-36RKD BM cells were cultured in control medium or APL LPs CM for 4 days (n = 3). Then, IMs were counted (N) and sorted for T cell proliferation assay (O). Data are presented as means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
To test whether targeting of IMs plays a major role in the antitumor activity of trabectedin, we also used clodrolip, which is only toxic to phagocytic cells and can be used to deplete IMs (31, 32). Although clodrolip and trabectedin exerted no direct toxicity on APL cells in vitro (fig. S4D), IMs and APL propagation were decreased by clodrolip in BM (fig. S4, E to G). Nevertheless, trabectedin showed no additional antileukemia effects in clodrolip-treated APL mice (fig. S4, E to G). We further performed adoptive transfer of IMs after trabectedin treatment and found that IMs promoted the leukemic progression in the NC group but not in the Il-36γ KD group (fig. S4, H and I). Accordingly, the inhibitory effects on leukemic progression conferred by Il-36γ KD were abolished by depletion of IMs (Fig. 4, D and E). These results suggested that the increased abundance of IMs was required for the promotion of leukemia downstream of the LP-derived Il-36γ.
Notably, multiple applications of trabectedin led to increased survival time of APL mice, while CD8+ T cell depletion attenuated the protective effect (Fig. 4, F and G). These findings suggested that elevated IM levels contributed to inactivation of CD8+ T cells with AML-clearing potential. To examine potential mechanisms controlling the effect, we checked Nos2 and Arg1 expression, two functional markers of MDSCs, and found that they were up-regulated in leukemic IMs but were attenuated by leukemic Il-36γ KD (Fig. 4H). In addition, exposure to leukemic IMs led to inhibited CD8+ T cell proliferation in vitro, but WT IMs did not, while both Il-36γ KD and treatment with Nos2 or Arg1 inhibitor attenuated this ability of IMs to inhibit the proliferation of CD8+ T cells (Fig. 4, I and J). Cumulatively, these results indicated that elevated Il-36γ levels not only increased IM abundance but also enhanced their MDSC activity.
Because Il-36R was highly expressed in the BM IMs (Fig. 4K), Il-36RNC or Il-36RKD hematopoietic chimeric mice were generated by inoculating lethally irradiated syngeneic recipients with BM HSPCs carrying an shRNA for targeted suppression of Il-36R (fig. S4, J and K). Compared to Il-36RNC IMs, transplantation of Il-36RKD IMs that were sorted from the chimeric mice exhibited a decreased ability to enhance leukemic propagation (fig. S4L), thus suggesting that Il-36γ could directly activate IMs. In support of this possibility, we observed that exposure to recombinant Il-36γ stimulated the proliferation of IMs and their suppressive activity, which could be subsequently blocked by addition of Il-36Ra, a natural antagonist of Il-36γ (Fig. 4, L and M). Likewise, compared to control medium, LP-conditioned medium (CM) enhanced the proliferation of Il-36RNC IMs and their ability to suppress CD8+ T cells (Fig. 4, N and O), and Il-36RKD IMs displayed lower activation by LP-CM in the proliferation and inhibition of CD8+ T cell (Fig. 4, N and O).
It warrants mention here that these separate lines of evidence pointed to an additional T cell–independent leukemia-promoting effect conferred by the Il-36γ–IM axis (Figs. 3, A and D, and 4G). We suspected that this effect could be related to a previously reported nursery role by activated IMs in facilitating AML cell growth (16). Thus, we cultured NC or Il-36γ KD LPs alone or together with IMs in vitro. The results showed that exposure to IMs promoted the LP growth in a manner dependent on Il-36γ signaling (fig. S4M). Together, these results demonstrated the essential contribution of Il-36γ–stimulated IMs in creating a BM microenvironment conducive to LP growth while circumventing CD8+ T cell–mediated immunosurveillance.
Stimulated residual LPs can sustain the Il-36γ–IM axis after chemotherapy
We then used the MLL-AF9 murine AML model to examine a plausible involvement of the Il-36γ–IM axis in the phenomenon that immunosurveillance against AML was not activated by chemotherapy (2, 3). After establishing an overt leukemic phenotype, we performed intravenous administration of cytarabine (Ara-C) plus doxorubicin (AD), a common first-line chemotherapeutic regimen for human AML (fig. S5A). Treatment with AD for five consecutive days significantly diminished the leukemic burden in BM, and we observed leukemic regrowth within 5 days after AD withdrawal (Fig. 5A). The number of IMs was not reduced, and LPs were enriched by AD treatment (Fig. 5, B and C). Furthermore, AD treatment led to higher levels of Il-36γ mRNA in the residual LPs, leading to elevated levels of Il-36γ in BM supernatant after chemotherapy (Fig. 5, D and E). We also found that CD8+ T cell depletion did not compromise the therapeutic effects of AD (Fig. 5F), suggesting that recovery of immunosurveillance functions could be blocked by the persistence of the Il-36γ–IM axis. As expected, KD of leukemia cell–derived Il-36γ resulted in lower IM numbers in BM after chemotherapy (Fig. 5G). In this scenario, CD8+ T cell depletion could partially block the protective effects of Il-36γ KD and chemotherapy on host survival (Fig. 5H).
Fig. 5. LPs sustain Il-36γ–IMs axis after chemotherapy.
(A to E) MLL-AF9 mice were treated with AD (see fig. S5A). The numbers of leukemia cells (A) and IMs (B) in BM, LP percentage in BM leukemia cells (C), Il-36γ in BM supernatant (E) (n = 6 mice per group), and Il-36γ mRNA level in BM LPs (n = 3) (D) were measured. (F) MLL-AF9 mice treated with PBS or AD (see fig. S5A) were injected with IgG or anti-CD8β on days 15, 16, 18, and 20 for survival analysis (n = 6 mice per group). (G and H) NC/Il-36γ KD MLL-AF9 mice treated with AD (see fig. S5A) were sacrificed on day 25. The number of BM IMs was measured (n = 5 mice per group) (G). (H) The mice were also injected with IgG or anti-CD8β on days 15, 16, 18, and 20 for survival analysis (n = 6 mice per group). (I) MLL-AF9 mice treated with PBS or AD (see fig. S5A) were injected with DMSO or trabectadin on days 24, 27, 30, 33, and 36 for survival analysis (n = 6 mice per group). (J) Relative numbers of MLL-AF9 cells after treatment with trabectedin or clodrolip combined with AD (Ara-C, 80 nM; Doxo, 0.1 μg/ml) for 3 days (n = 3). (K to M) Survival curves were generated for NC/Il-36γ KD MLL-AF9 mice after AD and clodrolip treatment (n = 6 mice per group) (K and L). (M) NC MLL-AF9 mice treated with AD and clodrolip were also injected with IgG or anti-CD8β on days 11, 15, 19, and 23 for survival analysis (n = 5 mice per group). (N) Relative numbers of MLL-AF9 LPs or IMs after treatment with Ara-C for 3 days (n = 3). (O) Relative numbers of NC/Il-36γ KD MLL-AF9 LPs after culture alone or with IMs at a 1:10 ratio in the presence of Ara-C (200 nM) for 2 days (n = 3). Data are presented as means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
We then treated AML mice with AD alone or AD combined with trabectedin and found that the combination therapy significantly improved the survival of AML recipients (Fig. 5I). However, because this therapeutic benefit was potentially attributable to the synergistic cytotoxicity of AD and trabectedin against MLL-AF9 cells, we then tested the role of IMs in relapse by using clodrolip, which has no additive toxicity on leukemic cells in vitro (Fig. 5J). The results showed that IM depletion increased the survival time of AML recipients but did not enhance the effects induced by Il-36γ KD (Fig. 5, K and L). This anti-AML effect was compromised by CD8+ T cell depletion (Fig. 5M). To examine whether Il-36γ–activated IMs could also directly protect LPs from chemo-cytotoxicity, we then treated the LP-only cultures, IM-only cultures, and LP-IM cocultures with Ara-C. IMs were refractory to Ara-C–mediated cytotoxicity (Fig. 5N), and the presence of IMs reduced the chemo-cytotoxicity toward LPs, which could be attenuated by Il-36γ KD (Fig. 5O). These results demonstrated the protective effects of Il-36γ–activated IMs on chemotherapy-treated LPs.
Next, we investigated how chemotherapeutic reagents stimulated Il-36γ expression in MLL-AF9 LPs. An autocrine Il-1β–NF-κB signaling pathway has been shown to be central to maintain the malignant stemness of MLL-AF9 LPs (33). In addition, chemotherapy can reportedly activate Nlrp3 inflammasomes in myeloid cells to trigger caspase-1 activation, which, in turn, promotes Il-1β release (34). These observations led us to test whether an activated caspase-1–Il-1β–NF-κB axis functioned in AML LPs after chemotherapy. We found that activation of caspase-1 in residual LPs was enhanced by AD treatment (fig. S5B), and double staining with annexin V and FLICA-1 confirmed that chemotherapy elevated the levels of active caspase-1 activity within the surviving LPs (fig. S5C), with higher concentrations of LP-derived Il-1β detected after AD treatment (fig. S5D). As expected, chemotherapy further increased the level of p-p65 in the surviving LPs (fig. S5E). Similarly, treatment with low Ara-C concentrations (80 nM), which did not induce LP cell death, elevated the activated caspase-1 in LPs in vitro (fig. S5, F and G). Moreover, Ara-C also enhanced the expression of Il-1β, p-p65, and Il-36γ (fig. S5, H and I), whereas caspase-1 KD attenuated these effects induced by Ara-C (fig. S5, J to L), thereby verifying a functional role of chemotherapy-induced caspase-1. Furthermore, the effects of Il-1β–neutralizing antibody on reducing Il-36γ secretion and endogenous p-p65 levels in LPs suggested that Il-1β autocrine signaling likely contributed to enhancing Il-36γ production (fig. S5, M and N).
Human LP–derived IL-36α stimulates IMs to inactivate CD8+ T cells
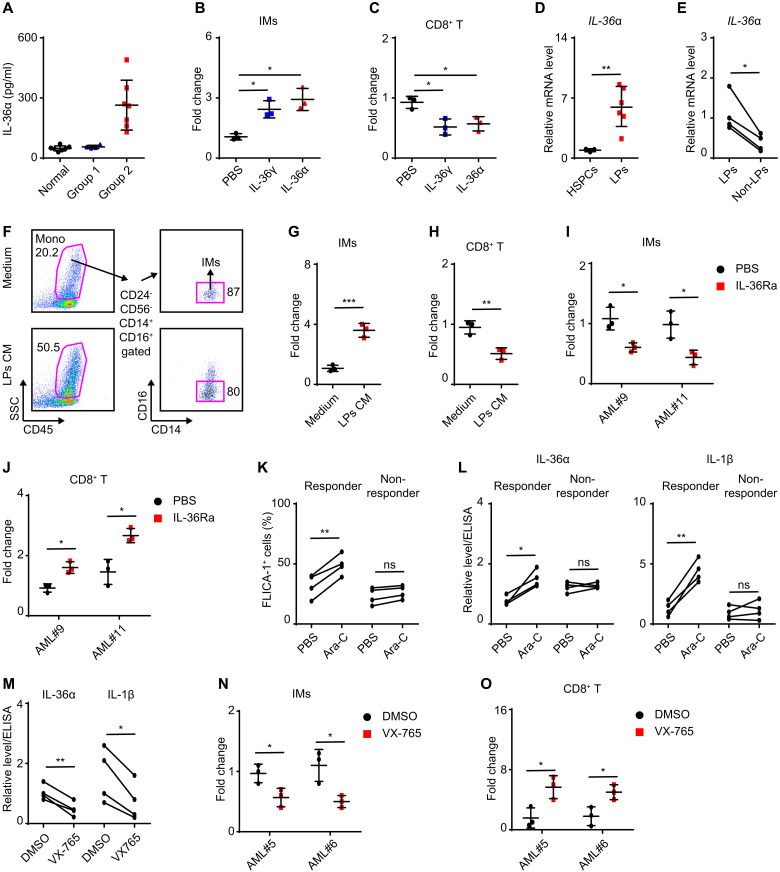
We then investigated whether this IL-36–IM axis is conserved in human AML. To this end, we checked inflammatory factor expression in human AML cell lines and found that some of these, such as MLL-AF9+ MV4-11 and THP-1, expressed IL-36α rather than IL-36β and IL-36γ concurrently with elevated NF-κB p-p65 level (fig. S6, A and B). Subsequent analysis of published ChIP-seq data revealed that NF-κB can directly bind IL-36α promoter region in IL-1β–stimulated human aortic endothelial cells (fig. S6C), while ChIP-qPCR assays verified that Il-36α was a direct target of NF-κB (fig. S6D). These results indicated that NF-κB preferentially activated IL-36α expression, rather than IL-36γ, in human AML. In line with this finding, we determined that IL-36α was higher in 7 of 11 primary AML samples (group 2) compared with that in normal BM samples, while IL-36γ was undetectable (Fig. 6A and table S1). We cultured healthy BM mononuclear cells (BMMCs) with recombinant IL-36γ or IL-36α, and these could directly promote the proliferation of human IMs (SSCHighCD45+CD24−CD56−CD16−CD14+) and their CD8+ T cell–suppressive activity (Fig. 6, B and C). Therefore, IL-36α could serve as a functional substitute for Il-36γ in the pathogenicity of human AML.
Fig. 6. Human AML LP–derived IL-36α stimulates IMs to suppress CD8+ T cells.
(A) ELISA assay on IL-36α level in BM supernatant of healthy donors (n = 7) and AML patients (group 1, n = 4; group 2, n = 7). (B and C) Healthy BMMCs (n = 3) were treated with IL-36γ or IL-36α (200 ng/ml) for 4 days. Then, IMs were counted (B) and sorted for T cell proliferation assay (C). (D) qRT-PCR assay on IL-36α level in CD34+ cord blood cells (n = 3) and AML LPs (CD33+CD34+) (n = 6). (E) qRT-PCR assay on IL-36α level in LPs and non-LPs (CD33+CD34−) from four AML blastic BM samples. (F to H) Healthy BMMCs were cultured with normal medium or LP-derived CM from three patients for 4 days. (F) Representative flow cytometric analyses of IMs after culture. The IMs were further counted (G) and sorted for T cell proliferation assay (H). (I and J) Healthy BMMCs were cultured in LP-derived CM from two patients and treated with PBS or IL-36Ra (2 μg/ml) for 4 days. Then, IMs were counted (n = 3) (I) and sorted for T cell proliferation assay (n = 3) (J). (K and L) LPs from eight AML patients were treated with PBS or Ara-C (80 nM) for 30 hours. The percentage of FLICA-1+ LPs (K) and the relative levels of IL-36α and IL-1β in medium (L) are shown. (M to O) LPs from four responders were treated with Ara-C (80 nM) in the presence of DMSO or VX-765 (100 μM) for 30 hours to collect CM. (M) Relative levels of IL-36α and IL-1β in CM. Furthermore, healthy BMMCs were cultured in CM from two patients for 4 days. Then, IMs were counted (N) and sorted for T cell proliferation assay (O). Data are presented as means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Using the published AML BM blastic complementary DNA (cDNA) microarray data (35, 36), we performed unsupervised clustering analysis with 39-gene MDSC signatures, which categorized 494 patients into three subtypes: MDSC-high, MDSC-medium, and MDSC-low (fig. S6E) (37). This analysis showed that IL-36α mRNA was significantly up-regulated in the MDSC-high group compared to the MDSC-medium and MDSC-low groups, as well as that detected in normal BM samples (fig. S6F). Among individual samples, IL-36α mRNA levels were positively correlated with the expression of genes related to IM recruitment and activation (such as CCL2 and CSF2) as well as MDSC function (such as NOS2 and PD-L1) (fig. S6G). Likewise, GSEA indicated that NF-κB pathway activity was higher in the MDSC-high group compared to MDSC-medium and MDSC-low groups (fig. S6H). Results drawn from the HemaExplorer database showed that IL-36α mRNA levels were significantly higher in multiple types of AML blastic BM samples compared to those of normal hematopoietic stem cells (HSCs) and myeloid subsets at various stages of differentiation (fig. S6I). Together, these results suggested a possible link between leukemic blast–derived IL-36α and IM activation.
According to a published dataset of human CD34+CD38− AML leukemia stem cell (LSC) expression profiles, IL-36α was one proinflammatory cytokine among 128 LSC signature genes (38), which we were able to verify in cord blood and AML BM samples (Fig. 6, D and E, and table S1). Next, we collected LP-CM to stimulate healthy BMMCs and found that the CM exerted a strong stimulatory effect on the proliferation of IMs and their CD8+ T cell–suppressive activity, while control medium did not (Fig. 6, F to H, and table S1). As expected, addition of Il-36Ra attenuated IM proliferation and activation induced by LP-CM (Fig. 6, I and J, and table S1), thus verifying a pathogenic role for human AML LP–derived IL-36α in the stimulation of IM proliferation and subsequent CD8+ T cell inactivation.
Moreover, Ara-C–activated caspase-1 was able to stimulate IL-1β production and increase the levels of p-p65 and IL-36α transcription in caspase-1–expressing THP-1 cells, but not in U937 cells that did not express caspase-1 (fig. S6, J to M) (39), while IL-36α transcription was decreased by NF-κB or caspase-1 inhibitors (fig. S6N). These findings indicated the participation of chemotherapy-stimulated IL-1β autocrine signaling in driving IL-36α expression. Similarly, Ara-C was shown to stimulate caspase-1 activation and promote IL-1β and IL-36α secretion in four of eight AML blastic samples, which were defined as responders (Fig. 6, K and L, and table S1). Among these responders, caspase-1 inhibitor decreased the levels of IL-1β and IL-36α as stimulated by Ara-C (Fig. 6M and table S1). In addition, we treated primary LPs sorted from two responders with Ara-C in the presence or absence of the caspase-1 inhibitor VX-765 and then collected the conditioned medium to test its effects on healthy IM–containing BMMCs. The results showed that VX-765 decreased the ability of Ara-C–treated LPs to stimulate IM proliferation and immunosuppressive activity (Fig. 6, N and O, and table S1). Together, these results suggested that chemotherapeutic agents could stimulate a portion of human AML blasts to produce IL-36α that, in turn, activated immunosuppressive IMs.
Il-36γ–IM axis disruption sensitizes AML to immune checkpoint blockade
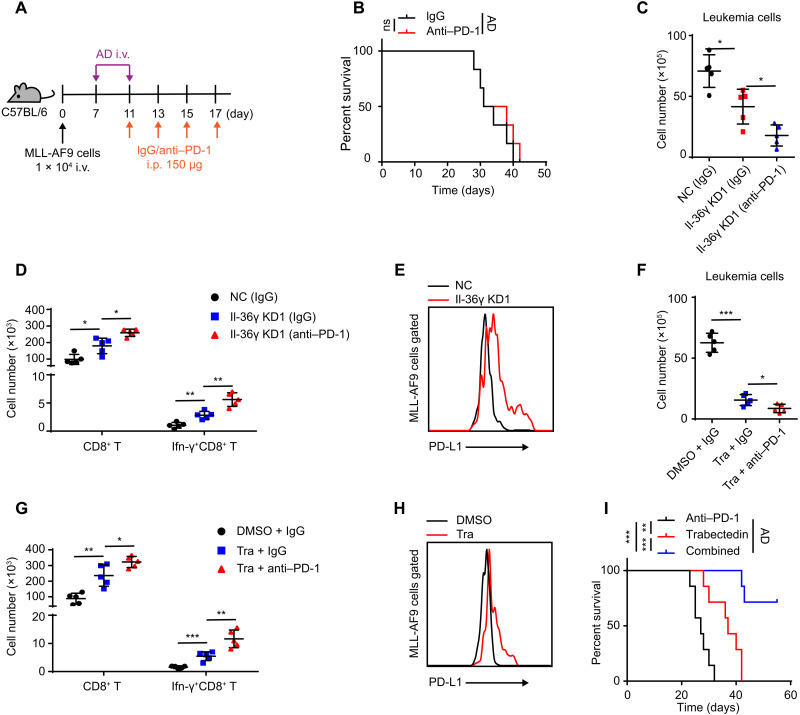
To date, programmed cell death protein 1 (PD-1) blockade has shown limited success as a monotherapy or in combination with chemotherapy in AML treatment (4, 5). We found that MLL-AF9 murine AML model showed no response to anti–PD-1 therapy whether it was combined with AD treatment or not (Fig. 7, A and B, and fig. S7, A and B). Because MDSCs are known to perform major roles in establishing de novo resistance to immune checkpoint blockade (40), we tested whether the refractory response of AML to anti–PD-1 therapy could be due, at least in part, to the Il-36γ–IM axis. The results showed that Il-36γ KD sensitized AML to anti–PD-1 therapy, thus resulting in a lower leukemic burden and increased accumulation of CD8+ T cells in BM (fig. S7, C to E). Similarly, combination of anti–PD-1 antibody with trabectedin led to a potent, synergistic, antileukemia effect and further enhanced the accumulation of CD8+ T cells in BM (fig. S7, F to H).
Fig. 7. Il-36γ–IM axis disruption sensitizes AML to anti–PD-1 therapy.
(A and B) The MLL-AF9 mice model was treated with IgG or anti–PD-1 [150 μg, intraperitoneally (i.p.)] on indicated days. The AD regimen was conducted from days 7 to 11 (A). (B) Survival curves were generated (n = 6 mice per group). (C to E) NC/Il-36γ KD MLL-AF9 cells (1 × 104) were inoculated into nonirradiated C57BL/6 and treated as depicted in (A). The mice were sacrificed on day 23. The numbers of leukemia cells (C) and T cells (D) in BM were counted (n = 5 mice per group). (E) Representative flow cytometric analysis of PD-L1 in BM MLL-AF9 cells. (F to I) MLL-AF9 cells (1 × 106) were inoculated into nonirradiated C57BL/6 and treated as described in (A). Meanwhile, DMSO or trabectadin (0.2 mg/kg, intravenously) was injected on days 13, 15, and 17. (F to H) The mice were sacrificed on day 23. The numbers of leukemia cells (F) and T cells (G) in BM were counted (n = 5 mice per group). (H) Representative flow cytometric analysis of PD-L1 in BM MLL-AF9 cells. (I) Survival curves were generated (n = 7 mice per group). Data are presented as means ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Il-36γ KD also improved the therapeutic efficacy of anti–PD-1 treatment following AD administration, which was accompanied by further accumulation of functional Ifn-γ+CD8+ T cells in BM (Fig. 7, C and D). In accordance with the possibility that type I immunity was activated, PD-L1 expression was induced in AML cells by Il-36γ KD (Fig. 7E), and we observed similar results under trabectedin treatment (Fig. 7, F to H). Last, we found that combined regimen of chemotherapy, anti–PD-1, and trabectedin could obviously improve survival of AML mice (Fig. 7I). Collectively, these results indicated a new combinatorial therapeutic regimen capable of sustained eradication of AML.
DISCUSSION
Inflammation is frequently detectable in tumorigenic microenvironment and involves tumorigenesis (41). Proinflammatory IL-1 signaling pathway elevated NF-κB activity and supported AML progression in an autocrine manner (16, 33). Here, we show that BM-infiltrating AML LPs were highly proinflammatory compared to normal HSPCs and that the marked IL-36 overexpression, as regulated by NF-κB activation, represented an essential feature of the AML LPs. Moreover, we noticed that the chemotherapeutic agents further stimulated IL-1β signaling pathways, thus contributing to an enhanced IL-36 expression in residual LPs and persistence of the immunosuppressive IL-36–IM axis. In this study, we also describe a new scenario in which IL-36 production protects the AML LPs from CTL killing and chemo-cytotoxicity by activating BM-resident IMs, expanding our understanding about how proinflammatory features of the AML LPs contribute to the persistence of AML malignancy.
We propose two mechanisms to explain why the LP-derived Il-36γ and solid tumor cell–derived Il-36γ exert opposite roles in regulating T cell activation. First, the immune microenvironment in AML BM is quite different from that in solid tumor. For example, IMs are derived from BM hematopoiesis and thus are more enriched in BM than in solid tumor. Second, the level of two cytokines—Il-2 and Il-12—that act synergistically with Il-36γ to activate the type I immunity did not increase during AML progression.
The abundance of MDSCs was correlated with the pathogenesis and poor therapeutic response of AML (42), while their activation pathways remain to be further explored. It was reported that the monocytic progenies of AML LPs act as MDSCs (9). Alternatively, AML blasts also showed to activate MDSCs through secreting exomes and/or soluble molecules (43, 44). After excluding that AML LPs were able to directly inactivate CD8+ T cells, our work specifies an involvement of IMs that, as activated by LP-derived IL-36 in paracrine manner, not only act as a subtype of monocytic MDSCs but also protect the LPs from chemo-induced cytotoxicity.
The persistence of the Il-36γ–IMs axis, before or after chemotherapy, dampened the response of AML toward PD-1 blockade therapy, implicating it as an important immunosuppressive pathway in AML. Trabectedin, a U.S. Food and Drug Administration–approved drug that is known to deplete IMs (29), combined with chemotherapy and immune checkpoint blockade, markedly improves the survival of MLL-AF9 AML mice, suggesting the potential of a new and available combinational therapeutic regimen in improving the treatment of human AML.
MATERIALS AND METHODS
Mice
FVB/NJ mice, C57BL/6 mice, and NOD.Cg-PrkdcscidIl2rgtm1Sug/JicCrl (NOG) mice were purchased from Beijing Vital River Laboratory Animal Technology. OT-1 transgenic mice were purchased from Shanghai Model Organisms. For each experiment, the same sex- and age-matched (4 to 8 weeks) mice were used and randomly allocated to each group. The number of mice in each group treatment or strain of mice for each experiment is indicated in the figures or figure legends. All animal experiments were carried out in accordance with the guidelines provided by the Laboratory Animal Resource Center of Shanghai Jiao Tong University School of Medicine.
Human and mouse samples
Human samples were obtained after receiving informed consents from AML patients and healthy donors admitted to the Department of Hematology in Ruijin Hospital, Shanghai Ninth People’s Hospital and Children’s Hospital of Soochow University according to the principles of the Declaration of Helsinki (see also table S1). The Human Ethics Committee of Ruijin Hospital approved the protocol for the recruitment of AML patients and healthy volunteers. Human primary AML samples, BMMCs, and PBMCs of healthy donors were isolated by Ficoll-Hypaque density gradient centrifugation. Human IMs were sorted from BMMCs of healthy donors by using the human Classical Monocyte Isolation Kit (130-117-337, Miltenyi Biotec). Human CD34+ cells were isolated using EasySep Human Cord Blood CD34 Positive Selection Kit III (17897, STEMCELLTechnologies). Human T cells were isolated from PBMCs of healthy donors by using the human Pan T Cell Isolation Kit (130-096-535, Miltenyi Biotec). Murine APL cells were originally isolated from hMRP8-PML-RARα transgenic mice generated in the laboratory of J. M. Bishop (University of California, Los Angeles). Mouse CD8+ T cells were isolated by using the mouse CD8a+ T Cell Isolation Kit (130-104-075, Miltenyi Biotec). Mouse IMs were fluorescence-activated cell sorting (FACS)–sorted from the enriched BM monocytes by using a mouse Monocyte Isolation kit (BM) (130-100-629, Miltenyi Biotec).
Cell culture
Human leukemia cell lines (THP-1, U937, MV4-11, and HL-60) were obtained from the cell bank of Shanghai Institute of Hematology (Shanghai, China) and cultured in RPMI 1640 medium (A1049101, Gibco) containing 10% fetal bovine serum (FBS; 10099, Gibco). Human primary leukemia cells and BMMCs were cultured with Iscove’s modified Dulbecco’s medium (IMDM) supplemented with 20% BIT (09500, STEMCELL Technologies). Mouse BM cells were cultured in IMDM (12440061, Gibco) containing 10% FBS. Mouse leukemia cells and IMs were cultured alone or cocultured in IMDM containing 10% FBS, mouse stem cell factor (mSCF) (50 ng/ml; 455-MC-050, R&D Systems), mouse IL-6 (mIL-6) (10 ng/ml; 406-ML-200, R&D Systems), and mouse IL-3 (mIL-3) (6 ng/ml; 403-ML-050, R&D Systems). Plate-E cells and 293T cells were cultured with Dulbecco’s modified Eagle’s medium (11965092, Gibco) supplemented with 10% FBS.
GFP labeling and genetic incorporation of a Dox-inducible shRNA expression system into APL cells
For GFP labeling, the retroviral plasmid MigR1-GFP and the packaging plasmid Ecopack were cotransfected into the plate-E cells to produce retrovirus. Then, c-Kit+CD11b− BM cells derived from hMRP8-PML-RARα transgenic mice were incubated with the retroviral supernatant in RPMI 1640 medium containing 10% FBS, mSCF (50 ng/ml), mIL-6 (10 ng/ml), mIL-3 (6 ng/ml), and polybrene (4 μg/ml; Sigma-Aldrich) for 48 hours. Then, the GFP+ cells were sorted and transplanted into sublethally irradiated (4.5 Gy) FVB/NJ mice to allow GFP-labeled cells to repopulate in vivo. After 3 to 4 weeks, the recipients were sacrificed when they became moribund, and the GFP+ APL cells were sorted and inoculated into nonirradiated FVB/NJ mice to establish APL model. For the integration of Dox-inducible shRNA expression systems, MigR1-TET3G-IRES-YFP retroviral plasmid was used to produce retrovirus 1. The shRNAs of Il-36γ, Mst1, or Cxcl10 were designed at website (http://sirna.wi.mit.edu/) and cloned into Dox-inducible retroviral plasmid coexpressing GFP to produce retrovirus 2. Then, c-Kit+CD11b− BM cells derived from the hMRP8-PML-RARα transgenic mice were infected with retrovirus 1 and transplanted into sublethally irradiated (4.5 Gy) FVB/NJ mice to allow the yellow fluorescent protein (YFP)–labeled cells to repopulate in vivo. Furthermore, YFP+ APL cells were infected with retrovirus 2. Last, YFP+GFP+ APL cells were inoculated into nonirradiated FVB/NJ mice to establish APL model that were responsive to Dox. The shRNA target sequences are as follows: IL-36γ KD1, 5′-GAGCTGGGCTATTTGTATCTTC-3′; IL-36γ KD2, 5′-AAGTGATGTTGAGATGGAACAC-3′; Mst1 KD1, 5′-CAAGGACCTTCGTGAGAATTTC-3′; Mst1 KD2, 5′-GAGAGTTGACAAGAGTAATAAA-3′. Cxcl10 KD1, 5′-CATAGGGAAGCTTGAAATCATC-3′; Cxcl10 KD2, 5′-AATCTAAGACCATCAAGAATTT-3′.
MLL-AF9 mice model and RNA KD
MLL-AF9 cDNA was cloned into retroviral plasmid MigR1-GFP to produce retrovirus. Lineage− (Ter119−, CD3−, B220−, CD11b−, Gr-1−) BM cells derived from C57BL/6 mice were incubated with the retroviral supernatant in RPMI 1640 medium containing 10% FBS, mSCF (50 ng/ml), mIL-6 (10 ng/ml), mIL-3 (6 ng/ml), and polybrene (4 μg/ml) for 48 hours. The cells were harvested and sorted for the GFP+ subpopulation, which was then transplanted into lethally irradiated (9 Gy) C57BL/6 mice. After 4 to 5 weeks, the recipients were sacrificed when they became moribund, and the labeled GFP+ MLL-AF9 cells were sorted and inoculated into nonirradiated C57BL/6 mice to establish MLL-AF9 mice model. To knock down Il-36γ and caspase-1, shRNAs were cloned into lentiviral plasmid that coexpress mCherry and transfected into 293T cells with package plasmid Vsvg and Pspax2 to produce lentivirus. GFP+ MLL-AF9 cells were infected with these lentiviruses for 48 hours. Then, GFP+mCherry+ MLL-AF9 cells were sorted for experiments. To knock down H2-K1, shRNAs were cloned into lentiviral plasmid that coexpress Puro to produce lentivirus. GFP+mCherry+ Il-36γ KD MLL-AF9 cells were infected with these lentiviruses for 48 hours. After infection, cells were treated with puromycin (2 μg/ml; Sigma-Aldrich) for 2 days. Then, cells were used for experiments. The shRNA target sequences are as follows: caspase-1 KD1, 5′-CAGCTGAAACATTTGTTGCTA-3′; caspase-1 KD2, 5′-GGGCAAAGAGGAAGCAATTTA-3′; H2-K1 KD1, 5′-GAATGTAACCTTGATTGTTAT-3′; H2-K1 KD2, 5′-CAGATACCTGAAGAACGGGAA-3′.
MLL-AF9-OVA mice model
OVA cDNAs were subcloned into lentiviral plasmid coexpressing Puro to produce lentivirus. GFP+ MLL-AF9 cells were infected with the lentivirus for 2 days. After infection, cells were treated with puromycin (2 μg/ml) for 2 days. Then, cells were transplanted into sublethally irradiated (4.5 Gy) C57BL/6 mice to allow GFP+ MLL-AF9-OVA cells to repopulate in vivo. Then, GFP+ MLL-AF9-OVA cells were isolated from these recipients and infected with control or Il-36γ KD lentivirus for 48 hours. After infection, 5 × 105 GFP+mCherry+ control or Il-36γ KD MLL-AF9-OVA cells were transplanted into nonirradiated C57BL/6 mice to establish the model.
T cell killing assay
Splenocytes from the OT-1 transgenic mice or leukemic mice were cultured in RPMI 1640 medium containing 10% FBS, mouse Il-2 (65 IU/ml; 402-ML-020, R&D Systems), and SIINFEKL peptide (7.5 μg/ml; Nanjing Peptide Biotech) for 5 days. Then, CD8+ T cells were purified and then cocultured with leukemia cells at 8:1 ratio for 4 hours. After that, leukemia cells were assessed for annexin V staining.
T cell proliferation assay
Mouse CD8+ T cells were stained with carboxyfluorescein diacetate succinimidyl ester (CFSE; 423801, BioLegend) according to the manufacturer’s instruction. Then, CD8+ T cells (1 × 105 per well) were cocultured with leukemia cells at a 16:1 ratio for 3 days or IMs at a 1:1 ratio for 5 days in 96-well plates coated with anti-CD3 (354720, Corning) in RPMI 1640 medium containing 10% FBS. The percentages of proliferating CD8+ T cells were determined by CFSE dilution. In some experiments, Dox (1 μg/ml); a nitric oxide synthase inhibitor, L-NMMA (0.5 mM); or an arginase inhibitor, nor-NOHA (0.5 mM) was added (see Fig. 4). Human T cells (1 × 105 per well) were stimulated with anti-CD3/CD28–coated beads (130-091-441, Miltenyi Biotec) and cocultured with human IMs sorted from the same donors at a 1:1 ratio in 96-well plates for 3 days. Then, the harvested cells were analyzed by flow cytometry.
In vivo treatment
To knock down the genes of interest in APL cells in vivo, Dox (200 μg/ml) was administrated in the drinking water on the indicated day and water was changed every 3 days. To deplete IMs in leukemic mice, trabectedin (0.2 mg/kg) or clodronate liposomes (200 μl) were administrated intravenously on the indicated day. Control mice were given vehicle [2% dimethyl sulfoxide (DMSO)] or control liposomes [phosphate-buffered saline (PBS)]. To deplete CD8+ T cells or NK cells in leukemic mice, anti-CD8β, anti-NK1.1, or immunoglobulin G (IgG) antibodies were injected intraperitoneally (150 μg per mouse) on the indicated days. To conduct immunotherapy, anti–PD-1 or IgG antibodies were injected intraperitoneally (150 μg per mouse) on the indicated days. To conduct chemotherapy, leukemic mice were administered intravenously by injecting Ara-C (100 mg/kg) for 5 days and doxorubicin (Doxo) (3 mg/kg) for 3 days. Ara-C was co-delivered with Doxo on the first 3 days and then alone for 2 days, mimicking the 7 + 3 regimen used in treating human AML patients.
Statistical analyses
Leukemia-initiating cell (LIC) frequencies and P values were calculated at website (http://bioinf.wehi.edu.au/software/elda/). Survival curves use two-sided Mantel-Cox log-rank test. Normal distribution and similar variation between experimental groups were examined for appropriateness before statistical tests were conducted. Comparisons between two groups were performed by unpaired, two-tailed t test. Comparisons between more than two groups were performed by one-way ANOVA using Prism 6.0 software (GraphPad Software). Statistical tests were done with biological replicates. Data are presented as means ± SD. P < 0.05 was considered statistically significant.
Acknowledgments
Funding: This work was supported by grants from the National Major Scientific and Technological Special Project (2018ZX09101001), the National Natural Science Foundation of China (NSFC) (81430002, 81770206, 81800099, 81900146, and 81900148), Shanghai Collaborative Innovation Program on Regenerative Medicine and Stem Cell Research (2019CXJQ01), the National Key Research and Development Program of China (2018YFA0107802), Samuel Waxman Cancer Research Foundation, Novo Nordisk Hemophilia Research Fund in China, and Shanghai Rising-Star Program (19QA1407800 and 20QC1400100). Author contributions: H.-Z.G. and J.Z. designed the study. H.-Z.G., Z.-H.G., L.-T.N., W.-T.Q., and M.-M.H. performed the experiments and interpreted the data. J.C., S.-H.Y., H.Y., and X.-Q.W. provided technical support. Y.-Y.T., Y.Z., J.S., S.-H.Y., S.-Y.H., and W.Z. were involved in providing experimental materials and interpreting data. H.-Z.G. and J.Z. prepared the manuscript. J.Z. supervised the entire project. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The accession number of whole transcriptome RNA-seq data is GSE120479.
Supplementary Materials
This PDF file includes:
Supplementary Materials and Methods
Figs. S1 to S7
Table S1
References
REFERENCES AND NOTES
- 1.Short N. J., Zhou S., Fu C., Berry D. A., Walter R. B., Freeman S. D., Hourigan C. S., Huang X., Nogueras Gonzalez G., Hwang H., Qi X., Kantarjian H., Ravandi F., Association of measurable residual disease with survival outcomes in patients with acute myeloid leukemia: A systematic review and meta-analysis. JAMA Oncol. 6, 1890–1899 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christopher M. J., Petti A. A., Rettig M. P., Miller C. A., Chendamarai E., Duncavage E. J., Klco J. M., Helton N. M., O’Laughlin M., Fronick C. C., Fulton R. S., Wilson R. K., Wartman L. D., Welch J. S., Heath S. E., Baty J. D., Payton J. E., Graubert T. A., Link D. C., Walter M. J., Westervelt P., Ley T. J., DiPersio J. F., Immune escape of relapsed AML cells after allogeneic transplantation. N. Engl. J. Med. 379, 2330–2341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Toffalori C., Zito L., Gambacorta V., Riba M., Oliveira G., Bucci G., Barcella M., Spinelli O., Greco R., Crucitti L., Cieri N., Noviello M., Manfredi F., Montaldo E., Ostuni R., Naldini M. M., Gentner B., Waterhouse M., Zeiser R., Finke J., Hanoun M., Beelen D. W., Gojo I., Luznik L., Onozawa M., Teshima T., Devillier R., Blaise D., Halkes C. J. M., Griffioen M., Carrabba M. G., Bernardi M., Peccatori J., Barlassina C., Stupka E., Lazarevic D., Tonon G., Rambaldi A., Cittaro D., Bonini C., Fleischhauer K., Ciceri F., Vago L., Immune signature drives leukemia escape and relapse after hematopoietic cell transplantation. Nat. Med. 25, 603–611 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Daver N., Garcia-Manero G., Basu S., Boddu P. C., Alfayez M., Cortes J. E., Konopleva M., Ravandi-Kashani F., Jabbour E., Kadia T., Nogueras-Gonzalez G. M., Ning J., Pemmaraju N., DiNardo C. D., Andreeff M., Pierce S. A., Gordon T., Kornblau S. M., Flores W., Alhamal Z., Bueso-Ramos C., Jorgensen J. L., Patel K. P., Blando J., Allison J. P., Sharma P., Kantarjian H., Efficacy, safety, and biomarkers of response to azacitidine and nivolumab in relapsed/refractory acute myeloid leukemia: A nonrandomized, open-label, phase II study. Cancer Discov. 9, 370–383 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lichtenegger F. S., Krupka C., Haubner S., Kohnke T., Subklewe M., Recent developments in immunotherapy of acute myeloid leukemia. J. Hematol. Oncol. 10, 142 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vadakekolathu J., Minden M. D., Hood T., Church S. E., Reeder S., Altmann H., Sullivan A. H., Viboch E. J., Patel T., Ibrahimova N., Warren S. E., Arruda A., Liang Y., Smith T. H., Foulds G. A., Bailey M. D., Gowen-MacDonald J., Muth J., Schmitz M., Cesano A., Pockley A. G., Valk P. J. M., Löwenberg B., Bornhäuser M., Tasian S. K., Rettig M. P., Davidson-Moncada J. K., DiPersio J. F., Rutella S., Immune landscapes predict chemotherapy resistance and immunotherapy response in acute myeloid leukemia. Sci. Transl. Med. 12, eaaz0463 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gale R. P., Opelz G., Commentary: Does immune suppression increase risk of developing acute myeloid leukemia? Leukemia 26, 422–423 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Paczulla A. M., Rothfelder K., Raffel S., Konantz M., Steinbacher J., Wang H., Tandler C., Mbarga M., Schaefer T., Falcone M., Nievergall E., Dörfel D., Hanns P., Passweg J. R., Lutz C., Schwaller J., Zeiser R., Blazar B. R., Caligiuri M. A., Dirnhofer S., Lundberg P., Kanz L., Quintanilla-Martinez L., Steinle A., Trumpp A., Salih H. R., Lengerke C., Absence of NKG2D ligands defines leukaemia stem cells and mediates their immune evasion. Nature 572, 254–259 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Galen P., Hovestadt V., Wadsworth Ii M. H., Hughes T. K., Griffin G. K., Battaglia S., Verga J. A., Stephansky J., Pastika T. J., Story J. L., Pinkus G. S., Pozdnyakova O., Galinsky I., Stone R. M., Graubert T. A., Shalek A. K., Aster J. C., Lane A. A., Bernstein B. E., Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell 176, 1265–1281.e24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shlush L. I., Mitchell A., Heisler L., Abelson S., Ng S. W. K., Trotman-Grant A., Medeiros J. J. F., Rao-Bhatia A., Jaciw-Zurakowsky I., Marke R., McLeod J. L., Doedens M., Bader G., Voisin V., Xu C. J., McPherson J. D., Hudson T. J., Wang J. C. Y., Minden M. D., Dick J. E., Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature 547, 104–108 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Thomas D., Majeti R., Biology and relevance of human acute myeloid leukemia stem cells. Blood 129, 1577–1585 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garlanda C., Dinarello C. A., Mantovani A., The interleukin-1 family: Back to the future. Immunity 39, 1003–1018 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gresnigt M. S., van de Veerdonk F. L., Biology of IL-36 cytokines and their role in disease. Semin. Immunol. 25, 458–465 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Bassoy E. Y., Towne J. E., Gabay C., Regulation and function of interleukin-36 cytokines. Immunol. Rev. 281, 169–178 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Wang X., Zhao X., Feng C., Weinstein A., Xia R., Wen W., Lv Q., Zuo S., Tang P., Yang X., Chen X., Wang H., Zang S., Stollings L., Denning T. L., Jiang J., Fan J., Zhang G., Zhang X., Zhu Y., Storkus W., Lu B., IL-36γ transforms the tumor microenvironment and promotes type 1 lymphocyte-mediated antitumor immune responses. Cancer Cell 28, 296–306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carey A., Edwards D. K. V, Eide C. A., Newell L., Traer E., Medeiros B. C., Pollyea D. A., Deininger M. W., Collins R. H., Tyner J. W., Druker B. J., Bagby G. C., McWeeney S. K., Agarwal A., Identification of interleukin-1 by functional screening as a key mediator of cellular expansion and disease progression in acute myeloid leukemia. Cell Rep. 18, 3204–3218 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brown D., Kogan S., Lagasse E., Weissman I., Alcalay M., Pelicci P. G., Atwater S., Bishop J. M., A PMLRARα transgene initiates murine acute promyelocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 94, 2551–2556 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu X., Chen J., Yu S., Yan L., Guo H., Dai J., Zhang W., Zhu J., All-trans retinoic acid and arsenic trioxide fail to derepress the monocytic differentiation driver Irf8 in acute promyelocytic leukemia cells. Cell Death Dis. 8, e2782 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou B. O., Ding L., Morrison S. J., Hematopoietic stem and progenitor cells regulate the regeneration of their niche by secreting Angiopoietin-1. eLife 4, e05521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J. L., Ma C., O’Connell R. M., Mehta A., DiLoreto R., Heath J. R., Baltimore D., Conversion of danger signals into cytokine signals by hematopoietic stem and progenitor cells for regulation of stress-induced hematopoiesis. Cell Stem Cell 14, 445–459 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chapellier M., Peña-Martínez P., Ramakrishnan R., Eriksson M., Talkhoncheh M. S., Orsmark-Pietras C., Lilljebjörn H., Högberg C., Hagström-Andersson A., Fioretos T., Larsson J., Järås M., Arrayed molecular barcoding identifies TNFSF13 as a positive regulator of acute myeloid leukemia-initiating cells. Haematologica 104, 2006–2016 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo H. Z., Niu L.-T., Qiang W.-T., Chen J., Wang J., Yang H., Zhang W., Zhu J., Yu S.-H., Leukemic IL-17RB signaling regulates leukemic survival and chemoresistance. FASEB J. 33, 9565–9576 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Guzman M. L., Neering S. J., Upchurch D., Grimes B., Howard D. S., Rizzieri D. A., Luger S. M., Jordan C. T., Nuclear factor-κB is constitutively activated in primitive human acute myelogenous leukemia cells. Blood 98, 2301–2307 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Kuo H.-P., Wang Z., Lee D.-F., Iwasaki M., Duque-Afonso J., Wong S. H. K., Lin C.-H., Figueroa M. E., Su J., Lemischka I. R., Cleary M. L., Epigenetic roles of MLL oncoproteins are dependent on NF-κB. Cancer Cell 24, 423–437 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kagoya Y., Yoshimi A., Kataoka K., Nakagawa M., Kumano K., Arai S., Kobayashi H., Saito T., Iwakura Y., Kurokawa M., Positive feedback between NF-κB and TNF-α promotes leukemia-initiating cell capacity. J. Clin. Invest. 124, 528–542 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng M., Gui X., Kim J., Xie L., Chen W., Li Z., He L., Chen Y., Chen H., Luo W., Lu Z., Xie J., Churchill H., Xu Y., Zhou Z., Wu G., Yu C., John S., Hirayasu K., Nguyen N., Liu X., Huang F., Li L., Deng H., Tang H., Sadek A. H., Zhang L., Huang T., Zou Y., Chen B., Zhu H., Arase H., Xia N., Jiang Y., Collins R., You M. J., Homsi J., Unni N., Lewis C., Chen G.-Q., Fu Y.-X., Liao X. C., An Z., Zheng J., Zhang N., Zhang C. C., LILRB4 signalling in leukaemia cells mediates T cell suppression and tumour infiltration. Nature 562, 605–609 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsurutani N., Mittal P., St. Rose M. C., Ngoi S. M., Svedova J., Menoret A., Treadway F. B., Laubenbacher R., Suárez-Ramírez J. E., Cauley L. S., Adler A. J., Vella A. T., Costimulation endows immunotherapeutic CD8 T cells with IL-36 responsiveness during aerobic glycolysis. J. Immunol. 196, 124–134 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veglia F., Perego M., Gabrilovich D., Myeloid-derived suppressor cells coming of age. Nat. Immunol. 19, 108–119 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Germano G., Frapolli R., Belgiovine C., Anselmo A., Pesce S., Liguori M., Erba E., Uboldi S., Zucchetti M., Pasqualini F., Nebuloni M., van Rooijen N., Mortarini R., Beltrame L., Marchini S., Fuso Nerini I., Sanfilippo R., Casali P. G., Pilotti S., Galmarini C. M., Anichini A., Mantovani A., D’Incalci M., Allavena P., Role of macrophage targeting in the antitumor activity of trabectedin. Cancer Cell 23, 249–262 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Galletti G., Caligaris-Cappio F., Bertilaccio M. T. S., B cells and macrophages pursue a common path toward the development and progression of chronic lymphocytic leukemia. Leukemia 30, 2293–2301 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Conrady C. D., Zheng M., Mandal N. A., van Rooijen N., Carr D. J. J., IFN-α-driven CCL2 production recruits inflammatory monocytes to infection site in mice. Mucosal Immunol. 6, 45–55 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanna B. S., McClanahan F., Yazdanparast H., Zaborsky N., Kalter V., Rößner P. M., Benner A., Dürr C., Egle A., Gribben J. G., Lichter P., Seiffert M., Depletion of CLL-associated patrolling monocytes and macrophages controls disease development and repairs immune dysfunction in vivo. Leukemia 30, 570–579 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Liang K., Volk A. G., Haug J. S., Marshall S. A., Woodfin A. R., Bartom E. T., Gilmore J. M., Florens L., Washburn M. P., Sullivan K. D., Espinosa J. M., Cannova J., Zhang J., Smith E. R., Crispino J. D., Shilatifard A., Therapeutic targeting of MLL degradation pathways in MLL-rearranged leukemia. Cell 168, 59–72.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruchard M., Mignot G., Derangère V., Chalmin F., Chevriaux A., Végran F., Boireau W., Simon B., Ryffel B., Connat J. L., Kanellopoulos J., Martin F., Rébé C., Apetoh L., Ghiringhelli F., Chemotherapy-triggered cathepsin B release in myeloid-derived suppressor cells activates the Nlrp3 inflammasome and promotes tumor growth. Nat. Med. 19, 57–64 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Haferlach T., Kohlmann A., Wieczorek L., Basso G., Kronnie G. T., Béné M. C., De Vos J., Hernández J. M., Hofmann W.-K., Mills K. I., Gilkes A., Chiaretti S., Shurtleff S. A., Kipps T. J., Rassenti L. Z., Yeoh A. E., Papenhausen P. R., Liu W.-M., Williams P. M., Foà R., Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: Report from the International Microarray Innovations in Leukemia Study Group. J. Clin. Oncol. 28, 2529–2537 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohlmann A., Kipps T. J., Rassenti L. Z., Downing J. R., Shurtleff S. A., Mills K. I., Gilkes A. F., Hofmann W.-K., Basso G., Dell’Orto M. C., Foà R., Chiaretti S., De Vos J., Rauhut S., Papenhausen P. R., Hernández J. M., Lumbreras E., Yeoh A. E., Koay E. S., Li R., Liu W.-M., Williams P. M., Wieczorek L., Haferlach T., An international standardization programme towards the application of gene expression profiling in routine leukaemia diagnostics: The Microarray Innovations in LEukemia study prephase. Br. J. Haematol. 142, 802–807 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang G., Lu X., Dey P., Deng P., Wu C. C., Jiang S., Fang Z., Zhao K., Konaparthi R., Hua S., Zhang J., Li-Ning-Tapia E. M., Kapoor A., Wu C. J., Patel N. B., Guo Z., Ramamoorthy V., Tieu T. N., Heffernan T., Zhao D., Shang X., Khadka S., Hou P., Hu B., Jin E. J., Yao W., Pan X., Ding Z., Shi Y., Li L., Chang Q., Troncoso P., Logothetis C. J., McArthur M. J., Chin L., Wang Y. A., DePinho R. A., Targeting YAP-dependent MDSC infiltration impairs tumor progression. Cancer Discov. 6, 80–95 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gal H., Amariglio N., Trakhtenbrot L., Jacob-Hirsh J., Margalit O., Avigdor A., Nagler A., Tavor S., Ein-Dor L., Lapidot T., Domany E., Rechavi G., Givol D., Gene expression profiles of AML derived stem cells; similarity to hematopoietic stem cells. Leukemia 20, 2147–2154 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Johnson D. C., Taabazuing C. Y., Okondo M. C., Chui A. J., Rao S. D., Brown F. C., Reed C., Peguero E., de Stanchina E., Kentsis A., Bachovchin D. A., DPP8/DPP9 inhibitor-induced pyroptosis for treatment of acute myeloid leukemia. Nat. Med. 24, 1151–1156 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu X., Horner J. W., Paul E., Shang X., Troncoso P., Deng P., Jiang S., Chang Q., Spring D. J., Sharma P., Zebala J. A., Maeda D. Y., Wang Y. A., DePinho R. A., Effective combinatorial immunotherapy for castration-resistant prostate cancer. Nature 543, 728–732 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crusz S. M., Balkwill F. R., Inflammation and cancer: Advances and new agents. Nat. Rev. Clin. Oncol. 12, 584–596 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Wang L., Jia B., Claxton D. F., Ehmann W. C., Rybka W. B., Mineishi S., Naik S., Khawaja M. R., Sivik J., Han J., Hohl R. J., Zheng H., VISTA is highly expressed on MDSCs and mediates an inhibition of T cell response in patients with AML. Onco. Targets Ther. 7, e1469594 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pyzer A. R., Stroopinsky D., Rajabi H., Washington A., Tagde A., Coll M., Fung J., Bryant M. P., Cole L., Palmer K., Somaiya P., Karp Leaf R., Nahas M., Apel A., Jain S., McMasters M., Mendez L., Levine J., Joyce R., Arnason J., Pandolfi P. P., Kufe D., Rosenblatt J., Avigan D., MUC1-mediated induction of myeloid-derived suppressor cells in patients with acute myeloid leukemia. Blood 129, 1791–1801 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trabanelli S., Chevalier M. F., Martinez-Usatorre A., Gomez-Cadena A., Salomé B., Lecciso M., Salvestrini V., Verdeil G., Racle J., Papayannidis C., Morita H., Pizzitola I., Grandclément C., Bohner P., Bruni E., Girotra M., Pallavi R., Falvo P., Leibundgut E. O., Baerlocher G. M., Carlo-Stella C., Taurino D., Santoro A., Spinelli O., Rambaldi A., Giarin E., Basso G., Tresoldi C., Ciceri F., Gfeller D., Akdis C. A., Mazzarella L., Minucci S., Pelicci P. G., Marcenaro E., McKenzie A. N. J., Vanhecke D., Coukos G., Mavilio D., Curti A., Derré L., Jandus C., Tumour-derived PGD2 and NKp30-B7H6 engagement drives an immunosuppressive ILC2-MDSC axis. Nat. Commun. 8, 593 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu S.-H., Zhu K.-Y., Chen J., Liu X.-Z., Xu P.-F., Zhang W., Yan L., Guo H.-Z., Zhu J., JMJD3 facilitates C/EBPβ-centered transcriptional program to exert oncorepressor activity in AML. Nat. Commun. 9, 3369 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bagger F. O., Rapin N., Theilgaard-Mönch K., Kaczkowski B., Jendholm J., Winther O., Porse B., HemaExplorer: A Web server for easy and fast visualization of gene expression in normal and malignant hematopoiesis. Blood 119, 6394–6395 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Bagger F. O., Rapin N., Theilgaard-Mönch K., Kaczkowski B., Thoren L. A., Jendholm J., Winther O., Porse B. T., HemaExplorer: A database of mRNA expression profiles in normal and malignant haematopoiesis. Nucleic Acids Res. 41, D1034–D1039 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heinz S., Romanoski C. E., Benner C., Allison K. A., Kaikkonen M. U., Orozco L. D., Glass C. K., Effect of natural genetic variation on enhancer selection and function. Nature 503, 487–492 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hogan N. T., Whalen M. B., Stolze L. K., Hadeli N. K., Lam M. T., Springstead J. R., Glass C. K., Romanoski C. E., Transcriptional networks specifying homeostatic and inflammatory programs of gene expression in human aortic endothelial cells. eLife 6, e22536 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mei S., Qin Q., Wu Q., Sun H., Zheng R., Zang C., Zhu M., Wu J., Shi X., Taing L., Liu T., Brown M., Meyer C. A., Liu X. S., Cistrome Data Browser: A data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 45, D658–D662 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Figs. S1 to S7
Table S1
References