Abstract
In humans, the telomere consists of tandem 5′TTAGGG3′ DNA repeats on both ends of all 46 chromosomes. Telomere shortening has been linked to aging and age-related diseases. Similarly, telomere length changes have been associated with chemical exposure, molecular-level DNA damage, and tumor development. Telomere elongation has been associated to tumor development, caused due to chemical exposure and molecular-level DNA damage. The methods used to study these effects mostly rely on average telomere length as a biomarker. The mechanisms regulating subtelomere-specific and haplotype-specific telomere lengths in humans remain understudied and poorly understood, primarily because of technical limitations in obtaining these data for all chromosomes. Recent studies have shown that it is the short telomeres that are crucial in preserving chromosome stability. The identity and frequency of specific critically short telomeres potentially is a useful biomarker for studying aging, age-related diseases, and cancer. Here, we will briefly review the role of telomere length, its measurement, and our recent single-molecule telomere length measurement assay. With this assay, one can measure individual telomere lengths as well as identify their physically linked subtelomeric DNA. This assay can also positively detect telomere loss, characterize novel subtelomeric variants, haploiypes, and previously uncharacterized recombined subtelomeres. We will also discuss its applications in aging cells and cancer cells, highlighting the utility of the single molecule telomere length assay.
Keywords: Telomere length, Optical mapping, Telomere dysfunction
1. Background
1.1. The human telomere system and telomere length regulation
Telomeres are made up of a G-rich dsDNA with (TTAGGG)n repeats and a specialized six member protein complex called Shelterin; functional telomeres comprised of this nucleoprotein complex prevent aberrant DNA repair and fusion of the telomeric DNA ends by preventing recognition of these linear DNA ends by cellular DNA Damage and Repair pathways. Telomeres contribute to genomic stability by protecting the exposed chromosome ends from agents that cause genomic DNA to undergo nucleolytic degradation, aberrant recombination, repair, or fusion (Blackburn et al., 2015; Shammas, 2011; Wise et al., 2009). Human diploid cells have 92 telomeres of heterogenous lengths (0.5–15 kbp), one at each chromosome end. In somatic cells, telomeres shorten with every cell division and when they become critically short, their structure is disrupted resulting in telomere dysfunction, which eventually triggers replicative senescence or cell apoptosis (Aubert et al., 2012; Kaul et al., 2012; Meier et al., 2007; Samassekou et al., 2010). Telomerase adds telomeric repeats at chromosome ends to lengthen telomeres in germ cells. Unregulated telomerase activity and compromised tumor suppressor pathways can destabilize chromosomes, resulting in telomeric fusions (Capper et al., 2007), genomic instability and formation of tumors (Coppe et al., 2010; Davalos et al., 2010; Jaskelioff et al., 2011; Sabatier et al., 2005). In some cell types lacking telomerase, a variety of DNA repair and recombination-based pathways, collectively called Alternative Lengthening of Telomeres (ALT) pathways are involved in maintaining telomere lengths (Lazzerini-Denchi and Sfeir, 2016). ALT is a telomerase independent mechanism that is exhibited in ~15 % of cancers; in osteosarcoma and glioblastoma ALT tumors are usually associated with poor prognosis (Blasco, 2007; Cesare and Reddel, 2010; Xu and Blackburn, 2007).
1.2. Telomere and disease
In the last two decades, numerous studies associating shortening of telomere length to conditions like type 2 diabetes (Zhao et al., 2013), cancers (bladder, esophageal, gastric, head, breast, neck, ovarian, renal) (Blackburn et al., 2015; Wentzensen et al., 2011), Alzheimer’s (Zhan et al., 2015) have been published. Genetically transmitted telomere shortening-related disorders present characteristic diseased states like loss of immune function, deficits in bone marrow stem cells, elevated cancer levels, pulmonary fibrosis, gastrointestinal disorders, liver cirrhosis, and neuropsychiatric symptoms. Patients are often co-symptomatic for diabetes, myocardial infarction, graying hair and skin pigmentation (Armanios and Blackburn, 2012). Critically short telomere lengths are thought to be responsible for telomere dysfunction, instead of average telomere length, and are a better indication of chromosome instability (Hemann et al., 2001; Kaul et al., 2012; Samassekou et al., 2010; Vera and Blasco, 2012). They may be used as a biomarker for disease states caused by mechanisms leading to telomere dysfunction and determining cellular phenotypes (Hemann et al., 2001; Kaul et al., 2012; McEachern et al., 1996; Teixeira et al., 2004). In addition to increasing age, processes like nuclease action, oxidative damage, and/or DNA replication stress, chemical-induced damage, environmental stressors, and genetic alterations in shelterin member proteins also contribute to telomere shortening. Identifying the frequency and lengths of critically short telomeres could be a useful assaying tool in predicting disease risk factors and pathology of aging-related diseases or cancer.
1.3. Telomere length as a biomarker
Telomere length has a hereditary component but is also significantly impacted by lifestyle, health, and disease state (Lin et al., 2012). Therefore, there is considerable interest in using telomere length as a reliable clinical biomarker for disease risk, especially for cardiovascular diseases, cancers, viral respiratory infections and autoimmune disorders (Cohen et al., 2013; Fasching, 2018). Almost all existing epidemiological studies using telomere length as a biomarker have relied on measurements of average telomere lengths in samples. However, it is clear that the shortest telomere(s) in a cell can trigger biologically relevant effects such as senescence, apoptosis, and chromosome instability. Since there are clear arm-specific and haplotype-dependent single-telomere length biases in cells, methods to acquire these types of datasets should increase the resolution and sensitivity of telomere lengths as biomarkers. Examples of these biases and trends can be found in McCaffrey et al. (2017), including striking examples of haplotype-specific differences at in (TTAGGG)n lengths at single telomeres that are presumably allele-specific cis-effects of subtelomere sequences upon telomere length regulation. Bulk telomere length measuring methods are limited in acquiring both single telomere and haplotype-resolved data. The distal 500kbp of each chromosome, subtelomeres, known to have repeat elements and large structural variations, contain both coding and non-coding regions. They actively participate in regulating telomere length and stability (Britt-Compton et al., 2006). In addition, genes located in subtelomeric DNA are subject to regulation by telomere length; for example, expression of three subtelomeric genes interferon stimulated gene 15 kb (ISG15 in 1p subtelomeric region), Desmoplakin (DSP in 6p subtelomeric region), and C1S (complement component 1 s subcomplement (CIS in 12p subtelomeric region) were specifically associated with telomere length changes (Robin et al., 2014). Further, subtelomere regions are hard to identify because of high degree of sequence similarities within telomeres (Stong et al., 2014). Lack of good reference sequences, high segmental duplication content, and extensive levels of structural variation have prevented some subtelomeric regions from being well characterized. Therefore, single molecule methods capable of measuring telomere length connecting to specific subtelomeres as well as identifying haplotypes are well suited for studying telomere regulation and dynamics of telomeres with complex and variable telomere-adjacent regions (McCaffrey et al., 2017).
1.4. Telomere length as a biomarker in toxicology
In addition to aging, lifestyle and psychological stress have also been known to affect telomere lengths (Lin et al., 2012). In recent years, exposure to environmental toxins is being linked to premature aging and decreased lifespan (Chen et al., 2013). Based on the environmental stressor, telomeres may either shorten or lengthen, depending on the cellular and molecular mechanisms triggered by the particaular stressor, such as oxidative stress, telomerase inhibition, DNA damage, etc. (Romano et al., 2013). For example, chronic exposure to cadmium is thought to cause telomere length shortening by way of oxidative stress and DNA damage (Huang et al., 2013). Other metals, such as lead and tungsten have similar effects as well. Organic chemicals, such as polycyclic aromatic hydrocarbons and pesticides can also cause telomere length shortening (Yuan et al., 2018). Polychlorinated biphenyls, on the other hand, have been linked to increased telomere length in leukocytes (Mitro et al., 2016). It has been suggested that such telomere elongation could be preceding tumor development in cancer (Joyce and Hou, 2015). Prolonged exposure to particulate matter was also shown to be correlated to elongated telomeres. However, telomerase expression and méthylation levels were not different, leaving the mechanism unknown (Dioni et al., 2011).
The concentration of the pollutant too seems to decide whether the telomeres lengthen or shorten. For example, chronic exposure to arsenic, via drinking water, at low concentrations is thought to enhance the function of telomerase resulting in telomere lengthening. At high arsenic concentrations (>1 μM), this resulted in oxidative stress and cell apoptosis. This telomeric lengthening is thought to be the mode of action in cancer caused due to arsenic, by way of upregulation in WRN, a gene involved in both normal telomere replication and maintenance as well as the ALT mechanism of telomere lengthening (Gao et al., 2015). In one study, telomere lengths of coal workers were measured to observe the effect of exposure to coal and associated chemicals. Shortening of telomere and excessive DNA methylation were observed in the exposed individuals. However, the telomere shortening did not correlate with premature aging or oxidative stress in the workers (de Souza et al., 2018).
Telomere length is unique type of DNA-based biomarker for testing the mutagenic effects of environmental toxins. From the above exam pies, it is implicit that a closer inspection of the telomere is necessary in understanding the short-term and long-term effects of exposure. Single molecule sensitivity to telomeres and a genome-wide approach will likely contribute to future studies.
1.5. Telomere length measurement techniques
Several methods have been developed to measure telomere length, each having advantages and disadvantages that differ from each other. The most common methods, Terminal Restriction Fragment (TRF) and Quantitative-PCR (qPCR), can only yield an average telomere length value of all the telomeres present in the cell population. TRF uses a set of frequently cutting restriction enzymes (RE) to digest non telomeric DNA within the genomic DNA sample. This includes the adjacent subtelomeric regions that contain the restriction sites. After digestion, the DNA sample contains substantially intact and longer telomeric fragments that are then visualized via a Southern Blot, using probes that target the TTAGGG repeats. The resolution of this method is 1 kbp and requires at least 1.5 μg of genomic DNA. This method is not sensitive to short telomeres that are < 2 kbp, which are known to be of greater significance in affecting senescence. Due to the inclusion of subtelomeric DNA fragments, the average telomere length value could exceed the true value by several kbp. There is also a significant variation introduced due to use of different set of REs for the digestion (Aubert et al., 2012; Counter et al., 1992; Levy et al., 1992). The assay time for a widely used commercial TRF kit, TeloTAGGG (Roche), is approximately 18 h, starting from genomic DNA digestion to chemiluminescent detection (Altshuler et al., 2005). In qPCR method, telomere length is evaluated as a ratio between the relative ratio of telomere copies in experimental over reference DNA (T) and relative ratio of single gene copies in experimental over reference DNA (S). Thus, at T/S = 1, the experimental DNA sample is identical to the reference DNA sample (Cawthon, 2002). In comparison to TRF, the DNA amount and the assay time are much lower, 50 ng and a few hours, respectively. Since T/S is a relative measure and does not quantify telomere length in kbp, calibration against a gold standard method (TRF) becomes necessary to interpret T/S in kbp. Telomere length from qPCR is known to vary between samples and within (Cawthon, 2009; Gadalla et al., 2010). Due to this, qPCR data across different labs and studies is susceptible to misinterpretation due to inter-lab variability and dissimilar calibration curves, highlighting the need for rigorous standardization (Lin et al., 2019; Wang et al., 2018). Although qPCR is cost-effective and is high throughput, the inability to directly determine the telomere length and the associated variability limit its application. Due to average telomere length being insufficient in detailed telomere investigations, other techniques like Single Telomere Length Analysis (STELA), Universal STELA, Telomere Shortest Length Assay (TeSLA), Single Telomere Absolute length Rapid (STAR), Quantitative Fluorescence In Situ Hybridization (Q-FISH), Flow-FISH and Telomere length Combing Assay (TCA) were developed to detect and measure telomeres at specific chromosome ends. STELA (Baird et al., 2003) is based on PCR with one primer targeting the telomeric end (the G-rich 3′ overhang) and the other targeting a unique subtelomeric region (for chromosome-specific targeting). Since most chromosomes have their subtelomeric region enriched with duplications and finding a unique priming site for a given chromosome is challenging, Telomere-specific STELA is limited to interrogating a small subset of all chromosome ends with unique subtelomeric priming sites (comprising XpYp, 2p, 11q, 12q, and 17p (Aubert et al., 2012; Britt-Compton et al., 2006; Montpetit et al., 2014; Samassekou et al., 2010). This technique can accurately measure single-telomere specific telomere lengths and requires low amount of starting material compared to other techniques. However, due to the low throughput of analysis, it is labor intensive. Allelic variations in the subtelomeric region may also be targeted using this technique. STELA has a resolution of 0.1 kbp and is sensitive to detection of short telomeres and the maximum measurable telomere length is about 20 kbp (Baird et al., 2003).
To overcome the limitations of STELA, Universal STELA (U-STELA), a variation of STELA. U-STELA was primarily developed to detect all short telomeres, which were found to trigger cellular senescence (di Fagagna et al., 2003). Using a frequent cutting enzyme to digest non-telomeric DNA, and a telomere primer akin to that used in STELA, PCR was performed to determine the sizes of short telomeres. Unlike in STELA, U-STELA can interrogate telomeres on all chromosomes but detection of telomeres longer than 8 kbp was found to be inefficient and was hence, deemed to be unsuitable for mean telomere length measurement(Lai et al., 2017). Even so, U-STELA is an accurate technique for measuring the load of short telomeres and requires low amounts of starting material. However, the low throughput analysis makes this technique laborious (Bendix et al., 2010).
To overcome the limitations of STELA and U-STELA and measure telomeres of all lengths, Telomere Shortest Length Assay (TeSLA) was developed (Lai et al., 2017). TeSLA is also PCR based but employs an alternative background suppression mechanism to make it a more accurate and sensitive method. While the analysis is automated, with throughput and resolution improved over the previous versions, TeSLA is also labor intensive, and it is not possible to acquire data about ITS loci, haplotypes, or subtelomere-linkages.
Most recently, Luo et. al developed single telomere absolute-length rapid (STAR) assay, digital-PCR based approach to quantify telomere repeats based on end-point florescence. The florescence intensity is compared to that generated using synthetic copy-number standards and length of telomere in each molecule is estimated. The resolution of this method is said to be 0.2kbp - 320 kbp. While this assay is a definite improvement over other PCR-based assays, it only provides the average telomere length across all chromosomes and it is not possible to acquire subtelomere-linked and haplotype-resolved data (Luo et al., 2020).
Q-FISH (Quantitative fluorescence in situ hybridization) is a technique that uses fluorescent peptide nucleic acid (PNA) probes to detect the telomere repeat sequence, TTAGGG. The number of probes hybridized (total fluorescence intensity) corresponds to the total number of telomeric repeats and by extension, the length of the telomere. Only 15–20 cells are required per sample to visualize and measure all the telomeres at the same time. Q-FISH has a resolution of 0.3 kbp, it can detect ultra-short telomeres as small as 0.1 kbp as well as fused telomeres (Aubert et al., 2012; Lansdorp et al., 1996). Q-FISH can be applied to cells isolated from tissue, cultured cells, FFPE tissue and agar-embedded cells, but a major disadvantage is that it is limited to metaphase cells and it cannot be used to measure telomere length in terminally senescent cells (Montpetit et al., 2014).
In Flow FISH (Baerlocher and Lansdorp, 2003), Q-FISH (Lansdorp et al., 1996) has been combined with flow cytometry to analyze mean TL across various cell types within a single individual sample. Telomeres of cells in a suspension are labeled with fluorescent PNA probes. The mean fluorescence intensity of all the telomeres from a single cell type is measured following cell sorting of the distinct cell types in the sample and compared against a mean intensity value obtained from TRF measurements of a control cell population to estimate the average telomere lengths of each cell type. Flow-FISH has been improved and automated to make reproducible telomere length measurements (Baerlocher and Lansdorp, 2003; Baerlocher et al., 2006) and demonstrated for fresh blood samples that do not contain fixative reagents with a resolution of 0.2–0.3 kbp, (Aubert et al., 2012). This method is the first for telomere length measurement approved as a clinical diagnostics test.
Kahl et al. developed telomere length combing assay (TCA) by leveraging molecular combing technique to stretch DNA fibers on a functionalized glass surface (Allemand et al., 1997) and probing telomere sequences by hybridizing florescent PNA probes onto the stretched and immobilized DNA. They have demonstrated length measurements from approximately 1 kbp - 145 kbp using this technique (Kahl et al., 2020). Even though it is difficult to identify the chromosome here, combing enables high throughput telomere length measurements and is a cost-effective technique, especially for large population studies. However, it just provides the average telomere length of all chromosomes.
Using the above telomere length measurement methods, it is not feasible to efficiently acquire global subtelomere and haplotype resolved telomere length data. To address this need, we have developed a method that measures individual telomere lengths and identifies their physically linked subtelomeric DNA at the same time (discussed in detail below) (McCaffrey et al., 2015, 2017). This method combines nanochannel array linearization of DNA with CRISPR-Cas9 labeling of telomeric and subtelomeric repeats. The range of measurable telomere length extends from 0.1–30 kbp (McCaffrey et al., 2017). Complete or near total telomere loss can be ascertained with this method unlike with other methods which are not definitive about telomere loss. Using this method, previously uncharacterized subtelomere regions can be characterized. It can also detect novel subtelomeric structural variants (SVs) and haplotypes and associate them to the specific telomere lengths in a single assay.
In studies that involve large number of individual samples, a cost-effective high-throughput method is most desired, while in studies that require close observation of telomere length dynamics in a cell population at different population doublings (PDs), a high-resolution and highly sensitive telomere length assay is desired. When studying exposure to environmental toxins and its effect on telomeres, given limited information and variabilities associated with commonly used methods compared to the high resolution and accuracy offered by the single molecule method, the choice of telomere length measurement method plays a crucial role.
2. High throughput single molecule telomere length assay in nanochannel array
We have recently developed a novel method which enables global subtelomere and haplotype-resolved analysis of telomere lengths at the single-molecule level (McCaffrey et al., 2015, 2017) using an optical mapping in nano-channel technology (Das et al., 2010; Lam et al., 2012).
Optical mapping in nanochannel arrays is a commercial technology by Bionano Genomics. DNA sample loaded on to their chips via the sample inlet port can be controlled via electrophoresis and is made to enter wide microchannels region followed by a micropillar region, where the long molecules are uncoiled and are then made to enter a series of narrowing nanoehannels(100 nm and 45 nm). Once the molecules enter the confinements of nanochannels, the electrophoresis is tinned off. Here, the molecules are linearized and stretched. They are imaged at 60X magnification with a high-resolution florescent microscope. Red and Green labels are first excited with 637 nm and 532 nm lasers, respectively. Then, YOYO-1 stained DNA backbone is excited with a 473 nm laser. Semi-automated iterations of this loading and imaging are performed to image several thousands of molecules across hundreds of nanochannels in a high throughput format. Single molecule raw images are analyzed and assembled de no into consensus maps using Bionano Genomics Software tools, Refaligner and Assembler (Mak et al., 2016). These consensus maps are refined further and merged to output a final set of consensus maps which can be used for subsequent analyses.
An overview of the single molecule telomere length assay is shown in Fig. 1. First, high molecular weight DNA molecules are purified from cells that were embedded into agarose-gel plugs using commercial kits (BioRad no. 170–3592) as per the manufacturer’s specifications. Typically, 10 μg DNA can be obtained with 1 million embedded cells. To detect the telomeres and characterize their linked-subtelomeres at the same time, a sequence-specific fluorescent co-labeling strategy of long DNA molecules was then developed. 300 ng genomic DNA was globally labelled at specific short sequence motifs that occur throughout the human genome to serve in the identification of the subtelomeric regions. Nt.BspQI, a common nickase enzyme with GCTCTTC motifs was used for this global labeling purpose (Lam et al., 2012; Mak et al., 2016; Xiao et al., 2007). The second set of labels were specifically tagged at the telomeric repeats, (TTAGGG)n of above Nt.BspqI labeled DNA molecules to measure the individual telomere lengths based on the idea that number of detected fluorescent labels are proportional to the number of repeats. A CRISPR-Cas9 nickase (Cas9n) based labeling method directed to the telomeric (TTAGGG)n tracts by a synthetic guide RNA containing UUAGGGUUAGGGUUAGGGUU as its recognition sequence, was employed.
Fig. 1.
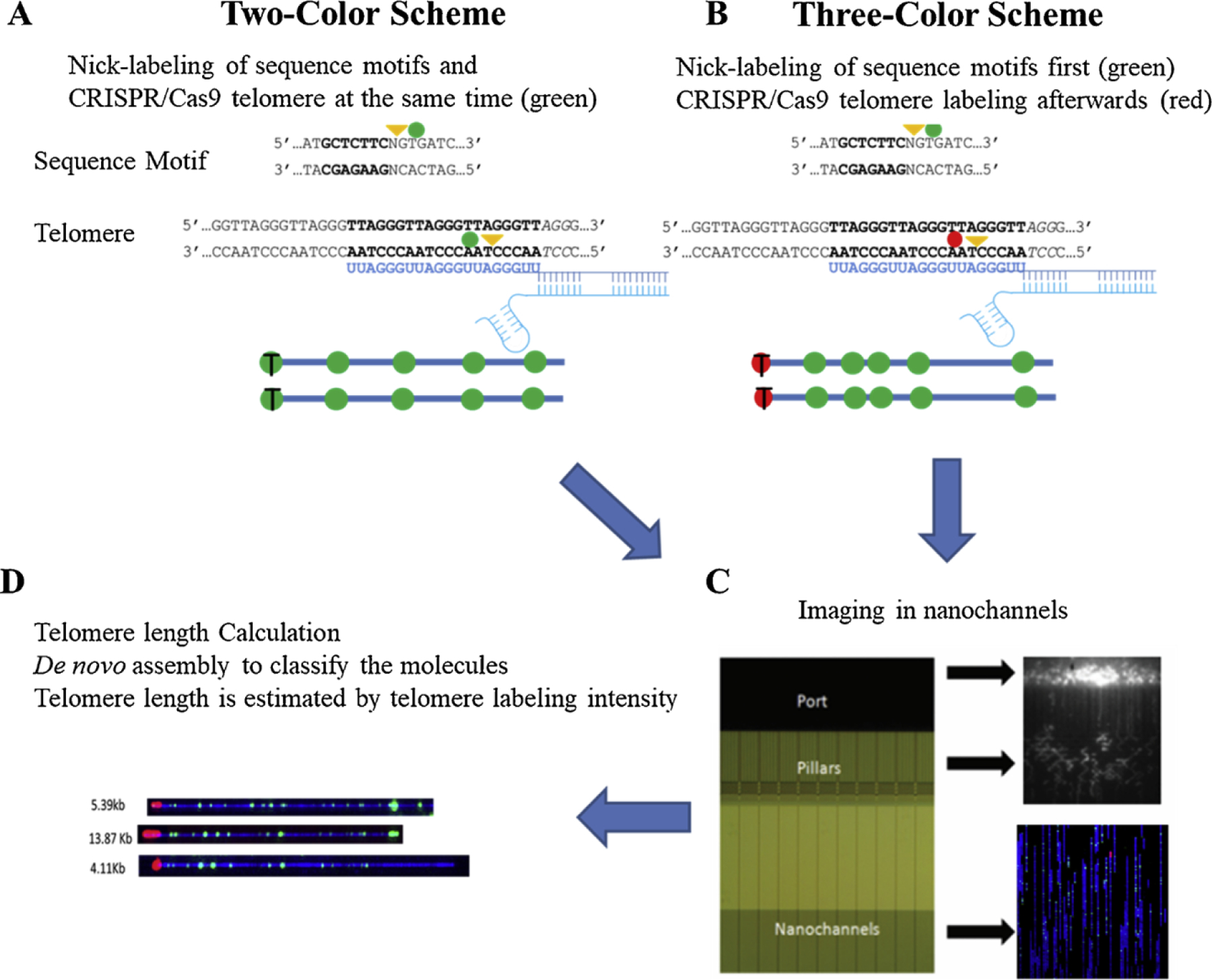
Schematic overview of single molecule telomere length assay (by McCaffrey et al. is licensed under CC BY 4.0). A) Two-color scheme shows simultaneous labeling at Cas9n-telomere gRNA directed nicks and Nt.BspQI global nicks. All labeled sites are shown as green spots, telomere ends aie marked with ‘T’. B) Three-color scheme showing staggered labeling of Nt.BspQI global nicks (shown as green spots) followed by Cas9n-telomere gRNA directed oicks (show as red spots). C) Labeled DNA from both schemes are loaded onto chips containing nanochannels and imaged in green, red and blue modules. D) Image shows examples of raw microscope images of molecules containing red (telomere ends) and green (Nt.BspQI) labels on blue DNA backbone. Consensus maps of such labeled molecules are generated using IrysSolve (bionano genomics) and compared to hg38 reference genome. Telomere length is estimated by measuring telomere label intensities.
The labeled DNA sample is then loaded onto a nanochannel array via electrophoresis. The single DNA molecules are uncoiled, linearized, and confined into nanochannels, and are optically imaged using a high-speed scanning microscope. Using the Nt.BspQI (GCTCTTC) label patterns, de novo assembly of the large DNA fragments is performed and the resulting assemblies are mapped to the subtelomeric regions of the hg38 human genome reference. The individual DNA molecules containing the telomere and the subtelomere region are used to simultaneously calculate the telomere length and identify the chromosome end. The telomere length can be estimated based on total Cas9n labeling intensity. The basis of this approach is that with longer telomeres, Cas9n will nick more frequently and the total number of fluorophores incorporated into the telomere will scale up accordingly, making it reasonable to use total intensity for telomere length estimation. Because even a single point emitter lights up several pixels due to photo scattering, the total fluorescence intensity method can be used to quantitate telomere length in these cases, improving resolution for very short telomeres.
We measured the telomere lengths based on the labeling intensity. As a baseline control, we used fosmids with known telomere lengths (800bp, 100bp) to characterize the system.
In our optical set up, the laser was expended 28.3 times to cover the entire CMOS camera sensor. Due to the exponential decay of laser power from the center of focus, the total raw intensity of telomere labels from individual fosmid molecules is calculated based on its distance relative to the imaging frame center.
After normalization, the measured length of the 800bp fosmid telomere repeats was estimated to be 814bp ± 195bp. Similarly, we tested the system with another fosmid with 100bp telomere repeats where the estimated length was 153bp ± 97bp. Even though the shorter telomere length measurement had higher variation, we could still detect and measure telomeric repeats as short as 100bp (McCaffrey et al., 2017).
When analyzing telomeres in a new sample, we first mapped the sample using Nt. BspQI without any telomere labels and defined the consensus contigs. Next, we performed the two or three-color labeling and classified any additional labeling found on the single molecules as telomeres. Events like loss of end telomere signal in some molecules, recombined molecules (displaced Nt. BspQI labels with or without retention of internal telomere like labels) were mostly observed in the subtelomeric regions of ALT cells and were chromosome-specific and sample-specific.
Two-color and three-color co-labeling schemes are designed for the single molecule telomere length assay (Fig. 1). In the two color-scheme, the Cas9n and Nt.BspQI nick sites are labeled with the same color (green) fluorescent labeled nucleotides using a Taq DNA polymerase. In the three-color scheme, Nt.BspQI nick sites are first labeled with green fluorescent nucleotides with Taq DNA polymerase, following which the unincorporated green fluorescent nucleotides are removed with Shrimp Alkaline Phosphatase (SAP) enzyme. This DNA solution is then nicked with Cas9n and labeled with red fluorescent nucleotides using Taq DNA polymerase.
Fig. 2 shows data from the two co-labeling schemes. The three-color scheme was applied to characterizing the telomeres of human DNA, GM11832. The telomeres, 3p (top) and 12p (middle) are shown in Fig. 2. In this case, the telomeres are labeled red and the global labeling is in green. Although it is much easier to distinguish the telomere from the subtelomere due to different colors, the three-color scheme is tedious and time-consuming. The additional steps (pipetting and chemical modifications) of intermediate fluorescent nucleotide removal and separate nick-labeling reaction tend to break the DNA molecules. This results in fewer long DNA molecules, which are necessary to identify the subtelomeric regions accurately and efficiently. The two-color scheme obviates this issue by employing a single nick-labeling reaction. The data from the two-color scheme, generated from the IMR90 cell line, is shown in Fig. 2 (bottom). The microscope images of single DNA molecules from the chromosome arm, 2q, the consensus maps of the two parental haplotypes, and the hg38 reference are all presented. The 2-color scheme to perform telomere characterization is simpler, faster (savings in imaging time), and provides more information of the subtelomere (longer terminal DNA molecules).
Fig. 2.
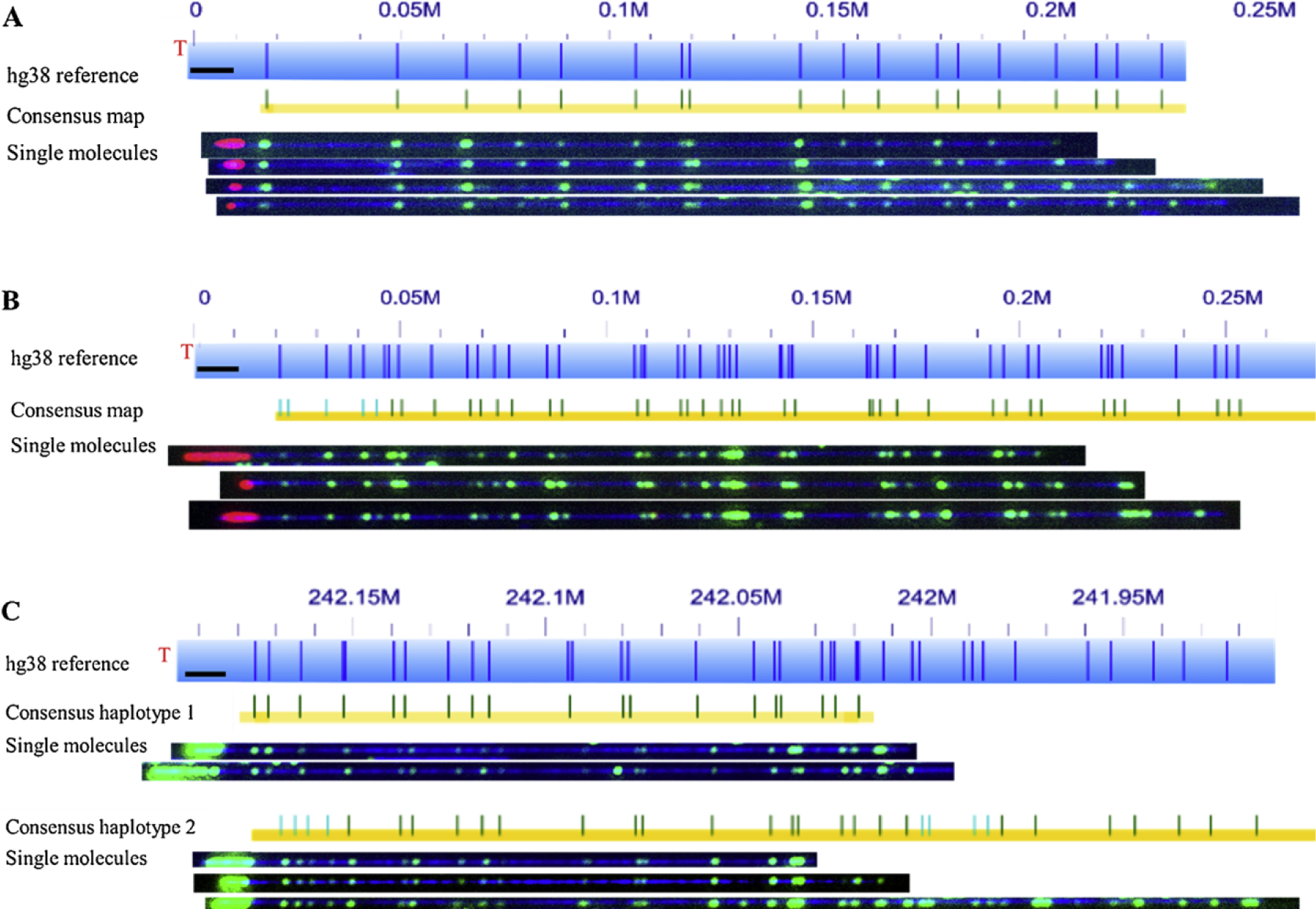
Microscope images of molecules from both labeling schemes (by McCaffrey et al. is licensed tinder CC BY 4.0. A and B) Co-labeling of telomere (TTAGGG)n tracts using gRNA-directed CRISPR/Cas9 nickase and subtelomeres using global motif-dependent nickase Nt.BsPQI. The three-color scheme was applied to characterize telomeres of human DNA. In the image, telomeres appear as red labels and the Nt. BspQI nick-label sites appear green. Reference genome hg38 appears as light blue bar with dark blue marks where the Nt. BspQI nick-label sites occur. Blue numbers above the reference indicate the location in million base pair scale. For example, 0.1 M = 0.1 × 10^6 bp from the chromosome end as indicated in hg38 reference. De novo assembled consensus maps are shown as yellow lines with vertical blue and green marks. Green marks show the Nt. BspQI sites that align to the reference and light blue marks show non-aligned sites A shows data of chromosome arm 3p and B shows data of chromosome arm 12p. C) Data from two-color labeling scheme as applied to IMR90-PD17, chromosome arm 2q. Image shows consensus map and single molecules grouped together into two separate haplotypes.
The assay can be used to identify and characterize telomere length distributions of 30–35 discrete telomeres simultaneously and accurately, for most chromosome arms. Five acrocentric short arms of 13p, 14p, 15p, 21p, and 22p lack reference sequences, and thus the associated telomere could not be identified and measured. The telomeres of XpYp were also not measured due to many gaps in the hg38 reference and a large highly variable region containing minisatellite repeats near this telomere (Inglehearn and Cooke, 1990). Lastly, 16p, 17p, 19q, and 22q failed the assembly due to the two closely spaced nicking enzymes sites on opposite strands sequences, which causes double-strand breaks (DSB) in molecules to be mapped.
This assay is unique in providing haplotype-resolved telomere information. The method is very sensitive to short telomeres, which are biologically most relevant, detecting telomeres as short as 0.1 kbp. Also, telomere loss can be detected unambiguously, a unique feature of the assay. The assay is also not limited to metaphase cells and can be adapted to cells from various tissue samples. With high throughput and resolution, and robust procedures, the single molecule telomere length assay will find significant utility in future telomere studies. Using this method, telomeres were characterized extensively in aging cells (IMR90), telomerase positive cells (UMUC3, LNCaP) as well as ALT-positive cells (U2OS, SK-MEL-2, and Saos-2). In the following sections, we will review the above results and describe for each type, characteristic features of telomeres, observed trends and relevant insights enabled by the technology.
2.1. Telomere shortening in aging cells
We applied our single telomere measurement assay technique to obtain haplotype resolved telomere length data of cellular aging model (human fetal lung fibroblasts IMR90) cell line at early passage, late passage, and senescence as characterized by number of population doublings (PD) in the cell culture. Early passage was chosen at PD17, late and senescence were chosen to be at PD45 and PD61, respectively. Typical raw imaging results used for determining the average single-telomere lengths, in this case for chromosome arm 8q, are shown at the top of Fig. 3A. The consensus maps from this cell line aligned well to hg38 reference genome. The early passage had the highest intensity telomere labels, indicating that telomeres were long. Among senescent stage telomeres, a fraction of the 8q were very short and a fraction was still relatively long. This could be explained by different doubling rates of mixed clonal subpopulations of IMR90 cells and mixed clone-specific telomere attrition rates (Britt Compton et al., 2006). Fig. 3B shows a plot comparison of early, late, and senescent telomere lengths. Each dot represents a single TL measurement. Mean of the distribution is represented by the horizontal bar in each cluster. The coefficients of variation (CV) calculated for average single-telomere lengths measured using our method was 14.6 %, which agrees with similar CV values published when using QFISH, Flow-FISH, and qPCR (Gutierrez-Rodrigues et al., 2014; Martin-Ruiz er al., 2015; O’Sullivan et al., 2004).
Fig. 3.
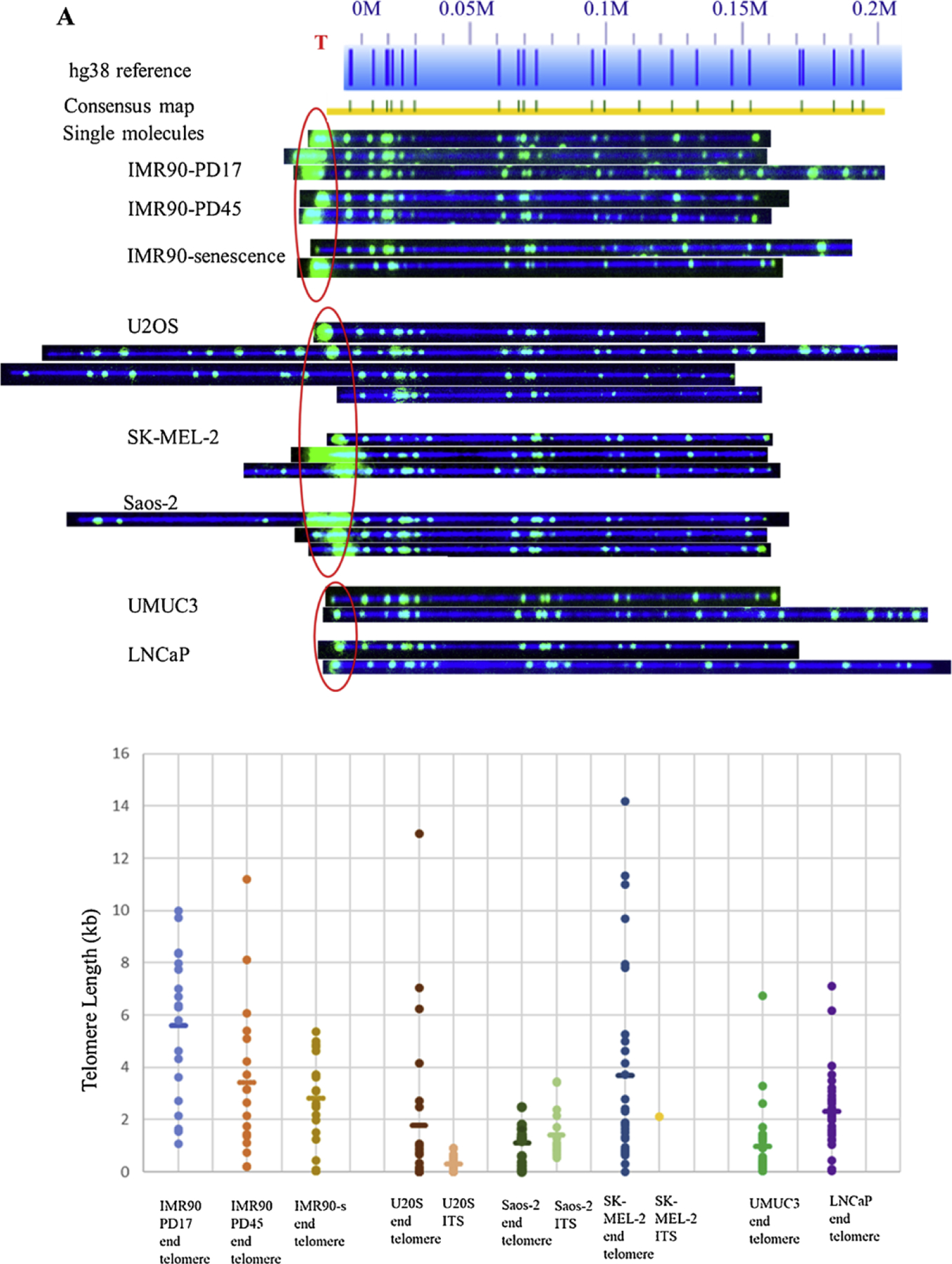
Comparison of 8q molecules across various cell lines. A) Raw microscope images of 8q molecules across among aging (IMR90 fibroblasts), ALT-positive (U2OS, SK-MEL-2, Saos-2), and telomerase-positive (UMUC3 and LNCaP) cell lines. Reference genome hg38 appears as light blue bar with dark blue marks where the Nt, BspQI nick-label sites occur. Blue numbers above the reference indicate the location in million base pair scale. For example, 0.1 M = 0.1 × 10^6 bp from the chromosome end as indicated in hg38 reference. De novo assembled consensus maps are shown as yellow lines with vertical blue and green marks. Green marks show the Nt. BspQI sites that align to the reference and light blue marks show non-aligned sites. Telomere labels are shown in red circle. Long telomere lengths were observed in early passages and decreased with increasing passage number. ALT-positive cells had a heterogeneous distribution of telomere lengths. Also, an example of recombinant molecule with ITS can be seen in Saos-2 group. Very short telomere ends consistent with ‘t-stumps’ observed in telomerase-positive cells, B) Plot showing the distributions of telomere length I measured at 8q molecules across among IMR90 aging cells (light blue, orange, mustard dots representing early, late and senescent passages respectively), U2OS (brown), Saos-2 (dark green), SK-MEL-2(dark blue) ALT-positive, and UMUC3 (bright green), LNCaP (purple) telomerase-positive cell lines. Each dot represents a single telomere length measurement, and the average length is marked as the short horizontal bar. The lengths of ITS sequences observed in each of the ALT-positive cell lines are represented as pink (U2OS), light green (Saos-2) and yellow (SK-MEL-2) dots.
We observed many interesting trends and features from measured telomere lengths of IMR90 samples. For instance, the average telomere lengths in the fibroblasts reduced from 7.2kbp in PD17 to 4.5kbp in PD45, to 4.0 kbp at senescence. Another trend was, the differences between the lengths of longest and shortest telomeres also decreased as the cell culture progressed toward cellular senescence (13.2 kb vs 3.6 kb at PD17, 9.3 kb vs 2.4 kb at PD45, and 5.5 kb vs 2.4 kb at senescence). This decreasing trend in average telomere length was found to be highly variable dependent on specific chromosome arms. Most chromosome arms (including 1p, 3p, 4p, 5p, 6p-1, 6p-2, 8p, 11p, 12p, 1q, 5q, 12q, 14q, 15q, 21q) exhibited 50 % decrease from PD17 to PD45, others do not decrease significantly from PD 17 to PD45 (2p, 8q, 9q and 11q) and the average telomere lengths of 9p, 20p, 2q-1 and 17q remained same.
Very short telomere lengths (<500bp) started to appear in late passage. The frequency of very short telomeres, approaching length < 100bp, increased fivefold from PD45 to senescence. Despite the fivefold increase in very short telomeres (<100bp), the average telomere lengths of PD61 do not differ statistically from those of PD45. It is worth mentioning that a third of very short telomeres in senescent IMR90 were found on 8q and one-eighth were on 14q suggesting subtelomere-speciflc vulnerabilities.
The overall average telomere length we measured from our single-telomere length data for senescent IMR90 cells (4.0 kb, Table 1) was very close to the average of the four STELA-measured single telomeres measured in bulk IMR90 senescent cultures previously (4.1 kb)(Britt Compton et al., 2006). Multiple trends of telomere length reduction with population doublings were found to be consistent between both methods (Britt-Compton et al., 2006; McCaffrey et al., 2017).
Table 1.
Longest and shortest average single-telomere lengths of end telomeres (terminal) and telomere loss.
| Cell line used in telomere characterization | Longest single telomere lengtha | Shortest single telomere lengtha | Overall Average Telomere Length | Total Telomere loss | Telomeres <500bp (% total) |
|---|---|---|---|---|---|
| IMR90_PD17 | (1p) 13.2 ± 5.4 (24) | (9p) 3.6 ± 3.0 (34) | 7.2 ± 1.7 (802) | 0 | 0(0%) |
| IMR90_PD17 | (3p) 8.9 ± 4.4 (31) | (3q) 3.9 ± 3.0 (23) | |||
| IMR90_PD17 | (8p) 8.9 ± 6.8 (33) | (16q1) 5.3 ± 4.5 (14) | |||
| IMR90_PD45 | (2q1) 9.3 ± 4.5 (13) | (6p2) 1.6 ± 1.4 (20) | 4.5 ± 1.7 (742) | 0 | 22(3.0 %) |
| IMR90_PD45 | (2q2) 7.4 ± 6.1 (16) | (8p) 2.1 ± 1.7 (9) | |||
| IMR90_PD45 | (1p) 6.7 ± 4.5 (10) | (6p1) 2.1 ± 2.0 (14) | |||
| IMR90_Senescence | (2q1) 5.5 ± 4.8(24) | (8p) 2.4 ± 2.1 (31) | 4.0 ± 1.0(851) | 0 | 74(8.7 %) |
| IMR90_Senescence | (20p) 5.4 ± 3.7 (23) | (8q) 2.6 ± 2.5(31) | |||
| IMR90_Senescence | (7q) 5.1 ± 3.0(30) | (19p) 3.0 ± 2.1 (31) | |||
| UMUC3 | (10q) 6.0 ± 3.4(23) | (8q) 1.0 ± 0.5(31) | 3.1 ± 1.3 (871) | 0 | 65(7.4 %) |
| UMUC3 | (12q) 5.8 ± 3.8(31) | (6p2) 1.2 ± 0.8 (12) | |||
| UMUC3 | (3p) 5.4 ± 3.5 (31) | (20p) 1.6 ± 1.1 (23) | |||
| LNCaP | (5q1) 4.5 ± 1.9(27) | (20p) 1.8 ± 0.9 (12) | 3.0 ± 0.7 (959) | 0 | 20(2.1 %) |
| LNCaP | (6p1) 4.2 ± 2.0 (10) | (10q) 2.0 ± 0.8(26) | |||
| LNCaP | (5q2) 4.1 ± 1.8(22) | (8q) 2.2 ± 1.6(31) | |||
| U20S | (20p)7.8 ± 12.2 (20) | (18q)0.5 ± 1.2 (5) | 3.1 ± 3.5 (906) b | 165(57,108) c | 158(6593)(17 %) d |
| U20S | (6q)7.8 ± 7.9 (21) | (8q)1.8 ± 3.5 (23) | |||
| U20S | (19p)7.0 ± 8.1 (29) | (2p)2.1 ± 2.5 (12) | |||
| SK-MEL-2 | (19q)5.1 ± 5.4 (31) | (XqYq)1.2 ± 1.9 (16) | 3.4 ± 3.5 (862) b | 43(38,5) c | 92(85,7) (11 %) d |
| SK-MEL-2 | (12q)4.9 ± 4.3 (31) | (9q)1.3 ± 2.0 (7) | |||
| SK-MEL-2 | (15q)4.8 ± 3.5 (30) | (16q)1.3 ± 1.1 (20) | |||
| Saos-2 | (6q)8.9 ± 10.0 (27) | (8q)1.1 ± 0.7 (15) | 4.3 ± 4.2 (862) b | 49(46,3) c | 53(42,10) (7.8 %) d |
| Saos-2 | (6p)7.7 ± 7.6 (10) | (17q)1.6 ± 1.9 (10) | |||
| Saos-2 | (3p)7.5 ± 5.5 (30) | (7q)1.9 ± 1.4 (27) |
Average Single Telomere Lengths in kb: Mean ± Std (number of telomeres).
End telomere loss: Events where some molecules in a specific sample have demonstrated lack of telomere-like signal at the end of chromosome. Some of these events manifest as additional internal telomere like sequences with blight label while some did not have any ITS.
ITS loss: Events where ITS signal loss is observed.
Total Telomere loss: This is defined as the total number of molecules where either loss of end telomeres signal is observed or loss of ITS signal is observed.
End telomere length.
Includes both ends and recombined ends.
Total telomere loss (end telomere loss, ITS loss).
Total telomere less than 500bp (end telomere<500bp, ITS<500bp) (% of total telomeres).
2.2. Abundant short telomeres detected in telomerase positive cancer cells
The telomerase positive bladder cancer UMUC3 and the prostate cancer LNCaP cell lines were investigated using our single molecule telomere length measurement assay and we found that the telomere lengths of both cell lines tend to be shorter with more uniform distributions compared to IMR90 cell lines. UMUC3 exhibited a relatively large number of very short telomeres (average length <500bp) at the same level of IMR90 senescence cell, while LNCaP cell line has less abundant shorter telomeres (Table 1). These shorter telomeres tend to concentrate on a few chromosome arms of UMUC3 and LNCaP and IMR90 senescence cell lines (8q and 9p of UMUC3; 8q and 14q of IMR90 senescence cell; 8q of LNCaP) (Table 2). These results are consistent with the previously published STELA results for this cell line which designated these short telomeres “t-stumps” (Xu and Blackburn, 2007). As the chromosome arm 8q is common between both cell lines with highest frequency of short telomeres, we show the raw images of 8q molecules and their telomere length distributions in Fig. 3.
Table 2.
Veiy short telomeres and telomere loss in senescent and cancer cell lines.
| Cell line used in telomere characterization | Characterized Chromosome arm | Average length | <500bpe | Telomere lossf |
|---|---|---|---|---|
| IMR90-senescence | Total (Bulk) | 4.0 ± 1.0 (851) | 49(5.8 %) | 0 |
| UMUC3 | Total (Bulk) | 3.1 ± 1.3 (871) | 57(6.5 %) | 0 |
| LNCaP | Total (Bulk) | 3.0 ± 0.7 (959) | 16(1.7 %) | 0 |
| U20S | Total (Bulk) | 3.1 ± 3.5 (906) f | 158(17 %) | 165 |
| SK-MEL-2 | Total (Bulk) | 3.4 ± 3.5 (862) f | 92(11 %) | 43 |
| Saos-2 | Total (Bulk) | 4.3 ± 4.2 (862) f | 53(7.8 %) | 49 |
| IMR90-senescence | 9p | 4.4 ± 2.2 (32) | 3(9.4 %) | 0 |
| IMR90-senescence | 8q | 2.6 ± 2.5 (31) | 8(25.8 %) | 0 |
| IMR90-senescence | 14q | 3.0 ± 1.9 (26) | 7(26.9 %) | 0 |
| UMUC3 | 9p | 1.7 ± 1.5 (30) | 12(40 %) | 0 |
| UMUC3 | 8q | 1.0 ± 0.5 (31) | 15(48.4 %) | 0 |
| UMUC3 | 14q | 1.9 ± 1.4 (10) | 1(10 %) | 0 |
| LNCaP | 9p | 3.4 ± 2.9 (32) | 0 | 0 |
| LNCaP | 8q | 2.2 ± 1.6 (31) | 5(16.1 %) | 0 |
| LNCaP | 14q | 3.9 ± 2.0 (14) | 0 | 0 |
| U20S | 3q | 0 | 0 | 23(13.9 %) |
| U20S | 8q | 1.2 ± 2.1 (42)g | 15(9.5 %) | 13(7.9 %) |
| U20S | 21q | 3.2 ± 4.4 (25)g | 9(5.7 %) | 18(10.9 %) |
| SK-MEL-2 | 8p | 3.1 ± 32.8 (32)g | 5(5.4 % %) | 5(11.6 %) |
| SK-MEL-2 | 14q | 1.7 ± 3.0 (23)g | 8(8.6 % %) | 4(9.3 %) |
| SK-MEL-2 | 20q | 1.4 ± 2.3 (29)g | 12(13.3 %) | 4(9.3 %) |
| Saos-2 | 1P | 2.8 ± 4.6 (37)g | 5(10.2 %) | 6(11.5 %) |
| Saos-2 | 19q | 4.8 ± 5.3 (31)g | 1(2.0 %) | 12(23.1 %) |
End telomere loss: Events where some molecules in a specific sample have demonstrated lack of telomere-like signal at the end of chromosome. Some of these events manifest as additional internal telomere like sequences with blight label while some did not have any ITS.
ITS loss: Events where ITS signal loss is observed.
Total Telomere loss: This is defined as the total number of molecules where either loss of end telomeres signal is observed 01 loss of ITS signal is observed.
Telomere loss not included. Percentage is calculated out of the total telomeres<500bp.
Total telomere loss (end telomere loss, ITS loss). Percentage is calculated out of the total telomere losses.
Average of both terminal telomere ends and recombinant ends.
2.3. Highly variable telomere-associated structures and telomere lengths detected in ALT cancer cells
Using our single molecule telomere measurement method, we characterized the telomere lengths and telomere-associated structures in ALT positive U2OS, SK-MEL-2, and Saos-2 cell lines (Abid et al., 2020). For all the subtelomeres analyzed, specific examples of linked terminal (TTAGGG)n tract end fragments as well as recombinant end fragments were found. Average subtelomere-specific terminal (TTAGGG)n tract lengths, the ratio of terminal end fragments to recombined end fragments, as well as other telomere-associated structural features varied widely depending upon the specific subtelomere. These general global observations were anticipated from prior studies of ALT cells (Cesare and Reddel, 2010). We observed that most of the subtelomeres have more terminal (TTAGGG)n ends than recombinant ends in this three ALT cell lines. Examples of recombinant molecules that often result in internal telomere-like (TTAGGG)n sequence tracts (ITSs) are also observed among the arms with primarily end telomeres (Fig. 3A). Among all telomeres, U2OS, Saos-2 and SK-MEL-2 had an average of 24 %, 19 % and 11 % of recombinant telomeres, respectively. Chromosome arms 1q, 3q, 7p, 8q, 11p 18q, 19q and 21q of U2OS; 3q and 20q of SK-MEL-2; and 1p, 3q, 8q, 17q and 21q of Saos-2, each had over 50 % recombinant telomeres. The recombination partner DNA fragment for most of these subtelomeres typically shows a defined stable pattern. The ITSs associated with recombinant telomeres are generally short. Very short ITSs (< 500bp) and ITS loss at recombinant telomeres are observed mainly on above chromosome arms (Tables 1,2). Overall, the terminal telomere lengths of the ALT positive cells U2OS, SK-MEL-2, and Saos2 cell lines varied from undetectable to extremely long, and more heterogenous than in aging or telomerase positive cancer cell lines. As an example, the 8q telomere tracts lengths (Fig. 3A) were shorter than in other chromosome arms but were similar across all ALT-positive cells with higher heterogeneity (Fig. 3B). Unlike in aging or telomerase positive cells, a very high level of signal-free ends (no detectable (TTAGGG)n tracts) were also observed in these ALT-positive cell lines: 8 % in U2OS cells and SK-MEL-2, and 5% in Saos-2 cell lines. Among these, over half were associated with just a few chromosome arms for U2OS and SK-MEL-2 cell lines, including 8q (Fig. 3A). We also observed very large linear extrachromosomal telomere repeat (ECTR) DNA molecules in all three cell lines. Two-thirds of chromosome ends and 89 % of all long extrachromosomal DNA fragments in all three ALT-positive cell lines exhibited punctated (TTAGGG)n labels interspersed with non-telomeric sequences. While the association of short ECTR fragments including c-circles and t-loops, with DNA repair and ALT-maintenance mechanisms has been reported before,(Fasching, 2018; Sobinoff and Pickett, 2017) the potential role of large linear ECTRs (like template molecules for break-induced repair in generating new telomeric DNA) with the telomere length maintenance was a new insight (Abid et al., 2020).
3. Conclusion
Telomere length is a unique type of clinical biomarker for testing not only disease states but monitoring mutagenic effects caused by environmental toxins. Although the telomere length trait is partially heritable, it can also be impacted significantly by lifestyle, health state, and exposure to environmental toxins and stress factors. Telomere length is commonly measured as an average value of all telomeres. The various methods involved in measuring telomere lengths have been summarized with respect to their advantages and disadvantages. Although the average telomere length is useful, it does not illuminate on the heterogeneity of the telomeres. It has been found that the shortest telomere or a small number of short telomeres in cells rather than the average telomere length that is critical for chromosome stability and cell viability. Hence, it is crucial to obtain chromosome-specific telomere length information. Additionally, haplotype-specific telomere length could be useful to gaining deeper insight into effects on telomere length.
One of the challenges to obtaining chromosome-specific telomere length is the identification of the individual telomeres. This mostly arises out of the complicated structure of the subtelomere. Our new single molecule method addresses this need. The high-throughput single molecule telomere length assay combines telomere-specific labeling using CRISPR-Cas9 and nanochannel-based linearization of long DNA molecules to achieve this. The assay comprises fluorescent labeling of single genomic DNA molecules at repeat sequences specific to the telomeres and at specific motifs that occur all throughout the genome. Following imaging, one can measure individual telomere lengths as well as identify their physically linked subtelomeric DNA.
In a single assay, we can accurately measure individual chromosome and haplotype-specific telomere lengths, distinguish absolute telomere loss from telomeres as short as 100 bp, analyze previously uncharacterized subtelomeric regions, and detect large subtelomeric structural variants, all at single molecule resolution.
Results from extensive characterization of aging cells (IMR90), telomerase positive cells (UMUC3, LNCaP) and ALT-positive cells (U2OS, SK-MEL-2, and Saos-2), using this method are summarized. Multiple subtelomere-specific telomere attrition trends have been observed. We observed a biased distribution of very short telomeres likely to be dysfunctional, suggesting subtelomere specific vulnerabilities to telomere dysfunction. In the case of IMR90, 8q and 14q were more frequently found to be short C<500 bp) when compared to other chromosome arms. This trend was also observed in specific chromosome arms of telomerase-positive cancer cell lines (8q and 9p of UMUC3; 8q of LNCaP). Markedly distinct telomeric and subtelomeric features were observed in the ALT-positive cancer cells. Here, the presence of unexpectedly high signal-free ends and surprisingly large linear ECTR DNA molecules were observed. The frequency and size of retained ITSs at recombinant telomere fusion junctions also varied according to the subtelomere. These results provide molecular level insight into the mechanisms of telomere damage, repair, and recombination in ALT cells. We believe our tractable assay, with high throughput and robust procedures, will find significant utility in telomere length association studies with large sample number.
Footnotes
Declaration of Competing Interest
The author declares that there is no conflict of interest.
References
- Abid HZ, McCaffrey J, Raseley K, Young E, Lassahn K, Varapula D, Riethman H, Xiao M, 2020. Single-molecule analysis of subtelomeres and telomeres in Alternative Lengthening of Telomeres (ALT) cells. BMC Genomics 21, 485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allemand J, Bensimon D, Jullien L, Bensimon A, Croquette V, 1997. pH-dependent specific binding and combing of DNA. Biophys. J 73, 2064–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P, Gibbs R, Belmont J, Boudreau A, Leal SJN, 2005. A haplotype map of the human genome. Nature 437, 1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armanios M, Blackburn G, E.H.J.N.R., 2012. The telomere syndromes. Nat. Rev. Genet 13, 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert G, Hills M, Lansdorp PM, 2012, Telomere length measurement-caveats and a critical assessment of the available technologies and tools. Mutat. Res 730, 59–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baerlocher GM, Lansdorp PM, 2003. Telomere length measurements in leukocyte subsets by automated multicolor flow-FISH. Cytometry Part A 55, 1–6. [DOI] [PubMed] [Google Scholar]
- Baerlocher GM, Vulto L, De Jong G, Lansdorp PM, 2006. Flow cytometry and FISH to measure the average length of telomeres (flow FISH). Nat. Protoc 1, 2365. [DOI] [PubMed] [Google Scholar]
- Baird DM, Rowson J, Wynford-Thomas D, Kipling D, 2003. Extensive allelic variation and ultrashort telomeres in senescent human cells. Nat. Genet 33, 203–207. [DOI] [PubMed] [Google Scholar]
- Bendix L, Horn PB, Jensen UB, Rubelj I, Kolvraa S, 2010. The load of short telomeres, estimated by a new method, Universal STELA, correlates with number of senescent cells. Aging Cell 9, 383–397. [DOI] [PubMed] [Google Scholar]
- Blackburn EH, Epel ES, Lin J, 2015. Human telomere biology: a contributory and interactive factor in aging, disease risks, and protection. Science 350, 1193–1198. [DOI] [PubMed] [Google Scholar]
- Blasco MA, 2007. Telomere length, stem cells and aging. Nat. Chem. Biol 3, 640–649. [DOI] [PubMed] [Google Scholar]
- Britt-Compton B, Rowson J, Locke M, Mackenzie I, Kipling D, Baird DM, 2006. Structural stability and chromosome-specific telomere length is governed by cis-acting determinants in humans, a Hum. Mol. Genet 15, 725–733. [DOI] [PubMed] [Google Scholar]
- Capper R, Britt-Compton B, Tankimanova M, Rowson J, Letsolo B, Man S, Haughton M, Baird DM, 2007. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes Dev. 21, 2495–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon RM, 2002. Telomere measurement by quantitative PCR. Nucleic Acids Res. 30 e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon RM, 2009. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 37 e21–e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesare AJ, Reddel RR, 2010. Alternative lengthening of telomeres: models, mechanisms and implications. Nat. Rev. Genet 11, 319–330. [DOI] [PubMed] [Google Scholar]
- Chen Y, Ebenstein A, Greenstone M, Li H, 2013. Evidence on the impact of sustained exposure to air pollution on life expectancy from China’s Huai River policy. Proc. Natl. Acad. Sci 110, 12936–12941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Janicki-Deverts D, Turner RB, Casselbrant ML, Li-Korotky H-S, Epel ES, Doyle WJ, 2013. Association between telomere length and experimentally induced upper respiratory viral infection in healthy adults. Jama 309, 699–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppe JP, Desprez PY, Krtolica A, Campisi J, 2010. The senescence-associated secretory phenotype: the dark side of tumor suppression. Annu. Rev. Pathol 5, 99–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Counter CM, Avilion AA, LeFeuvre GE, Stewart NG, Greider CW, Harley GB, Bacchetti S, 1992. Telomere shortening associated with chromosome instability is arrested in immortal cells which express telomerase activity. EMBO J. 11, 1921–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SK, Austin MD, Akana MC, Deshpande P, Cao H, Xiao M, 2010. Single molecule linear analysis of DNA in nano-channel labeled with sequence specific fluorescent probes. Nucleic Acids Res. 38 e177–e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davalos AR, Coppe JP, Campisi J, Desprez PY, 2010. Senescent cells as a source of inflammatory factors for tumor progression. Cancer Metastasis Rev. 29, 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza MR, Kahl VFS, Rohr P, Kvitko K, Cappetta M, Lopes WM, da Silva J, 2018, Shorter telomere length and DNA hypermethylation in peripheral blood cells of coal workers. Mutat. Res. Genet. Toxicol, Environ. Mutagen 836, 36–41. [DOI] [PubMed] [Google Scholar]
- di Fagagna Fd.A., Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP, 2003. A DNA damage checkpoint response in telomere-initiated senescence. Nature 426, 194–198. [DOI] [PubMed] [Google Scholar]
- Dioni L, Hoxha M, Nordio F, Bonzini M, Tarantini L, Albetti B, Savarese A, Schwartz J, Bertazzi PA, Apostoli P, Hou L, Baccarelli A, 2011. Effects of short-term exposure to inhalable particulate matter on telomere length, telomerase expression, and telomerase methylation in steel workers. Environ. Health Perspect, 119, 622–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasching CL, 2018. Telomere length measurement as a clinical biomarker of aging and disease. Crit. Rev. Clin. Lab. Sci 55, 443–465. [DOI] [PubMed] [Google Scholar]
- Gadalla SM, Cawthon R, Giri N, Alter BP, Savage SA, 2010. Telomere length in blood, buccal cells, and fibroblasts from patients with inherited bone marrow failure syndromes. Aging (Albany NY) 2, 867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Roy S, Tong L, Argos M, Jasmine F, Rahaman R, Rakibuz-Zaman M, Parvez F, Ahmed A, Hore SK, Sarwar G, Slavkovich V, Yunus M, Rahman M, Baron JA, Graziano JH, Ahsan H, Pierce BL, 2015. Arsenic exposure, telomere length, and expression of telomere-related genes among Bangladeshi individuals. Environ, Res. 136, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Rodrigues F, Santana-Lemos BA, Scheucher PS, Alves-Paiva RM, Calado RT, 2014. Direct comparison of flow-FISH and qPCR as diagnostic tests for telomere length measurement in humans. PLoS One 9, e113747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemann MT, Strong MA, Hao L-Y, Greider CWJC, 2001. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell 107, 67–77. [DOI] [PubMed] [Google Scholar]
- Huang J, Okuka M, Lu W, Tsibris JCM, McLean MP, Keefe DL, Liu L, 2013. Telomere shortening and DNA damage of embryonic stem cells induced by cigarette smoke, Reprod. Toxicol. 35, 89–95. [DOI] [PubMed] [Google Scholar]
- Inglehearn C, Cooke H, 1990. A VNTR immediately adjacent to the human pseudoautosomal telomere. Nucleic Acids Res. 18, 471–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaskelioff M, Muller FL, Paik JH, Thomas E, Jiang S, Adams AC, Sahin E, Kost-Alimova M, Protopopov A, Cadinanos J, Homer JW, Maratos-Flier E, Depinho RA, 2011. Telomerase reactivation reverses tissue degeneration in aged telomerase-deficient mice. Nature 469, 102–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce BT, Hou L, 2015. Organic pollutants and telomere length: a new facet of carcinogenesis. EBioMedicine 2, 1854–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahl VFS, Allen JAM, Nelson CB, Sobinoff AP, Lee M, Kilo T, Vasireddy RS, Pickett HA, 2020. Telomere length measurement by molecular combing. Front. Cell Dev. Biol 8, 493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul Z, Cesare AJ, Huschtscha LI, Neumann AA, Reddel RR, 2012. Five dysfunctional telomeres predict onset of senescence in human cells. EMBO Rep. 13, 52–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai T-P, Zhang N, Noh J, Mender I, Tedone E, Huang E, Wright WE, Danuser G, Shay JW, 2017. A method for measuring the distribution of the shortest telomeres in cells and tissues. Nat. Commun 8, 1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam ET, Hastie A, Lin C, Ehrlich D, Das SK, Austin MD, Deshpande P, Cao H, Nagarajan N, Xiao M, Kwok P-Y, 2012. Genome mapping on nanochannel arrays for structural variation analysis and sequence assembly. Nat. Biotechnol 30, 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansdorp PM, Verwoerd NP, van de Rijke FM, Dragowska V, Little M-T, Dirks RW, Raap AK, Tanke HJ, 1996. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet 5, 685–691. [DOI] [PubMed] [Google Scholar]
- Lazzerini-Denchi E, Sfeir A, 2016. Stop pulling my strings — what telomeres taught us about the DNA damage response. Nat. Rev. Mol. Cell Biol 17, 364–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy MZ, Allsopp RC, Futcher AB, Greider CW, Harley CB, 1992. Telomere end-replication problem and cell aging. J. Mol. Biol 225, 951–960. [DOI] [PubMed] [Google Scholar]
- Lin J, Epel E, Blackburn E, 2012. Telomeres and lifestyle factors: roles in cellular aging. Mutat. Res. Mol. Mech. Mutagen 730, 85–89. [DOI] [PubMed] [Google Scholar]
- Lin J, Smith DL, Esteves K, Drury S, 2019. Telomere length measurement by qPCR - Summary of critical factors and recommendations for assay design. Psychoneuroendocrinology 99, 271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y, Viswanathan R, Hande MP, Loh AHP, Cheow LF, 2020. Massively parallel single-molecule telomere length measurement with digital real-time PCR. Sci. Adv 6 eabb7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak AC, Lai YY, Lam ET, Kwok TP, Leung AK, Poon A, Mostovoy Y, Hastie AR, Stedman W, Anantharaman T, Andrews W, Zhou X, Pang AW, Dai H, Chu C, Lin C, Wu JJ, Li CM, Li JW, Yim AK, Chan S, Sibert J, Dzakula Z, Cao H, Yiu SM, Chan TF, Yip KY, Xiao M, Kwok PY, 2016. Genome-wide structural variation detection by genome mapping on nanochannel arrays. Genetics 202, 351–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Ruiz CM, Baird D, Roger L, Boukamp P, Krunic D, Cawthon R, Dokter MM, van der Harst P, Bekaert S, de Meyer T, Roos G, Svenson U, Codd V, Samani NJ, McGlynn L, Shiels PG, Pooley KA, Dunning AM, Cooper R, Wong A, Kingston A, von Zglinicki T, 2015. Reproducibility of telomere length assessment - an international collaborative study (voi 44, pg 1673, 2015). Int. J. Epidemiol 44, 1749–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey J, Sibert J, Zhang B, Zhang Y, Hu W, Riethman H, Xiao M, 2015. CRISPR-CAS9 D10A nickase target-specific fluorescent labeling of double strand DNA for whole genome mapping and structural variation analysis. Nucleic Acids Res. 44 e11–e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey J, Young E, Lassahn K, Sibert J, Pastor S, Riethman H, Xiao M, 2017. High-throughput single-molecule telomere characterization. Genome Res. 27, 1904–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Blackburn EHJG, Development, 1996. Cap-Prevented Recombination between Terminal Telomeric Repeat Arrays (Telomere CPR) Maintains Telomeres in Kluyveromyces Lactis Lacking Telomerase, Vol. 10, pp. 1822–1834. [DOI] [PubMed] [Google Scholar]
- Meier A, Fiegler H, Muñoz P, Ellis P, Rigler D, Langford C, Blasco MA, Carter N, Jackson SP, 2007. Spreading of mammalian DNA-damage response factors studied by ChlP-chip at damaged telomeres. EMBO J. 26, 2707–2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitro SD, Birnbaum LS, Needham BL, Zota AR, 2016. Cross-sectional associations between exposure to persistent organic pollutants and leukocyte telomere length among U.S. Adults in NHANES, 2001–2002. Environ. Health Perspect 124,651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montpetit AJ, Alhareeri AA, Montpetit M, Starkweather AR, Elmore LW, Filler K, Mohanraj L, Burton CW, Menzies VS, Lyon DE, Jackson-Cook CK, 2014. Telomere length: a review of methods for measurement. Nurs. Res 63, 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Sullivan JN, Finley JC, Risques Ra., Shen WT, Gollahon KA, Moskovitz AH, Gryaznov S, Harley CB, Rabinovitch PS, 2004. Telomere length assessment in tissue sections by quantitative FISH: image analysis algorithms. Cytometry Part A 58, 120–131. [DOI] [PubMed] [Google Scholar]
- Robin JD, Ludlow AT, Batten K, Magdinier F, Stadler G, Wagner KR, Shay JW, Wright WE, 2014. Telomere position effect: regulation of gene expression with progressive telomere shortening over long distances. Genes Dev. 28, 2464–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano GH, Harari Y, Yehuda T, Podhorzer A, Rubinstein L, Shamir R, Gottlieb A, Silberberg Y, Pe’er D, Ruppin E, Sharan R, Kupiec M, 2013. Environmental stresses disrupt telomere length homeostasis. PLoS Genet. 9, e1003721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatier L, Ricoul M, Pottier G, Murnane JP, 2005. The loss of a single telomere can result in instability of multiple chromosomes in a human tumor cell line. Mol. Cancer Res 3, 139–150. [DOI] [PubMed] [Google Scholar]
- Samassekou O, Gadji M, Drouin R, Yan J, 2010. Sizing the ends: normal length of human telomeres. Ann. Anat 192, 284–291. [DOI] [PubMed] [Google Scholar]
- Shammas MA, 2011. Telomeres, lifestyle, cancer, and aging. Curr. Opin. Clin. Nutr. Metab. Care 14, 28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobinoff AP, Pickett H.A.J.Ti.G., 2017. Alternative lengthening of telomeres: DNA repair pathways converge. Trends Genet. 33, 921–932. [DOI] [PubMed] [Google Scholar]
- Stong N, Deng Z, Gupta R, Hu S, Paul S, Weiner AK, Eichler EE, Graves T, Fronick CC, Courtney L, 2014. Subtelomeric CTCF and cohesin binding site organization using improved subtelomere assemblies and a novel annotation pipeline. Genome Res. 24, 1039–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MT, Arneric M, Sperisen P, Lingner JJC, 2004. Telomere length homeostasis is achieved via a switch between telomerase-extendible and- nonextendible states. Cell 117, 323–335. [DOI] [PubMed] [Google Scholar]
- Vera E, Blasco MA, 2012. Beyond average: potential for measurement of short telomeres. Aging (Albany NY) 4, 379–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Savage SA, Alsaggaf R, Aubert G, Dagnall CL, Spellman SR, Lee SJ, Hicks B, Jones K, Katki HA, Gadalla SM, 2018. Telomere length calibration from qPCR measurement: limitations of current method. Cells 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzensen IM, Mirabello L, Pfeiffer RM, Savage SA, 2011. The association of telomere length and cancer: a meta-analysis. Cancer Epidemiol. Prevent. Biomark 20, 1238–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise JL, Crout RJ, McNeil DW, Weyant RJ, Marazita ML, Wenger SL, 2009, Human telomere length correlates to the size of the associated chromosome arm. PLoS One 4, e6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Phong A, Ha C, Chan TF, Cai D, Leung L, Wan E, Kistler AL, DeRisi JL, Selvin PR, Kwok P-Y, 2007. Rapid DNA mapping by fluorescent single molecule detection. Nucleic Acids Res. 35 e16–e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Blackburn EH, 2007. Human cancer cells harbor T-stumps, a distinct class of extremely short telomeres. Mol. Cell 28, 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Liu Y, Wang J, Zhao Y, Li K, Jing Y, Zhang X, Liu Q, Geng X, Li G, Wang F, 2018, Long-term persistent organic pollutants exposure induced telomere dysfunction and senescence-associated secretary phenotype. J. Gerontol, A Biol. Sci. Med. Sci 73, 1027–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Y, Song C, Karlsson R, Tillander A, Reynolds CA, Pedersen NL, Hägg S, 2015. Telomere length shortening and Alzheimer disease—a Mendelian randomization study, JAMA Neurol. 72, 1202–1203. [DOI] [PubMed] [Google Scholar]
- Zhao J, Miao K, Wang H, Ding H, Wang DW, 2013. Association between telomere length and type 2 diabetes mellitus: a meta-analysis. PLoS One 8, e79993. [DOI] [PMC free article] [PubMed] [Google Scholar]


