Abstract
The trillions of bacteria that constitutively colonize the human gut collectively generate thousands of unique small molecules. These microbial metabolites can accumulate both locally and systemically and potentially influence nearly all aspects of mammalian biology, including immunity, metabolism, and even mood and behavior. Here, we briefly summarize recent work identifying bioactive microbiota metabolites, the means through which they are synthesized, and their effects on host physiology. Rather than offering an exhaustive list of all known bioactive microbial small molecules, we select a few examples from each key class of metabolites to illustrate the diverse impacts of microbiota-derived compounds on the host. In addition, we attempt to address the microbial logic behind specific biotransformations. Finally, we outline current and emerging strategies for identifying previously undiscovered bioactive microbiota metabolites that may shape human health and disease.
Introduction
Recent interest in the microbiome field has shifted from identifying associations between microbial taxa and human health towards mechanistic studies of the specific impacts of defined microbes and microbial products. One prominent mechanism by which the microbiome influences both local and systemic physiology is through the production of thousands of unique bioactive small molecules. Indeed, up to 50% of all serum metabolites are either produced by or modulated by commensal microbes. In this review, we will provide an overview of recent studies illuminating the roles of diverse microbial metabolites in shaping mammalian biology, with a general focus on immunological phenotypes. We will not exhaustively detail all classes of microbial metabolites or all known effects on the immune system as these topics have been covered previously by others (1, 2). Instead, we hope to provide illustrative examples from throughout the field for three broad classes of microbial metabolites defined based on the origins of their core building blocks (diet vs. host vs. de novo synthesis) and to provide proof-of-concept examples for specific microbial metabolite-mediated impacts on immunity. Throughout this review, we will also attempt to tackle the less well-discussed subject of why bacteria initially evolved the capacity to perform specific biotransformations. In this vein, we will classify microbial biotransformations into four non-mutually exclusive categories: attainment of nutrients, signalling (either to other microbes or the host), detoxification, and competition (Figure 1). Finally, we will outline existing and emerging strategies for identifying new bioactive compounds that are hidden among the vast array of tens of thousands of uncharacterized microbiota metabolites.
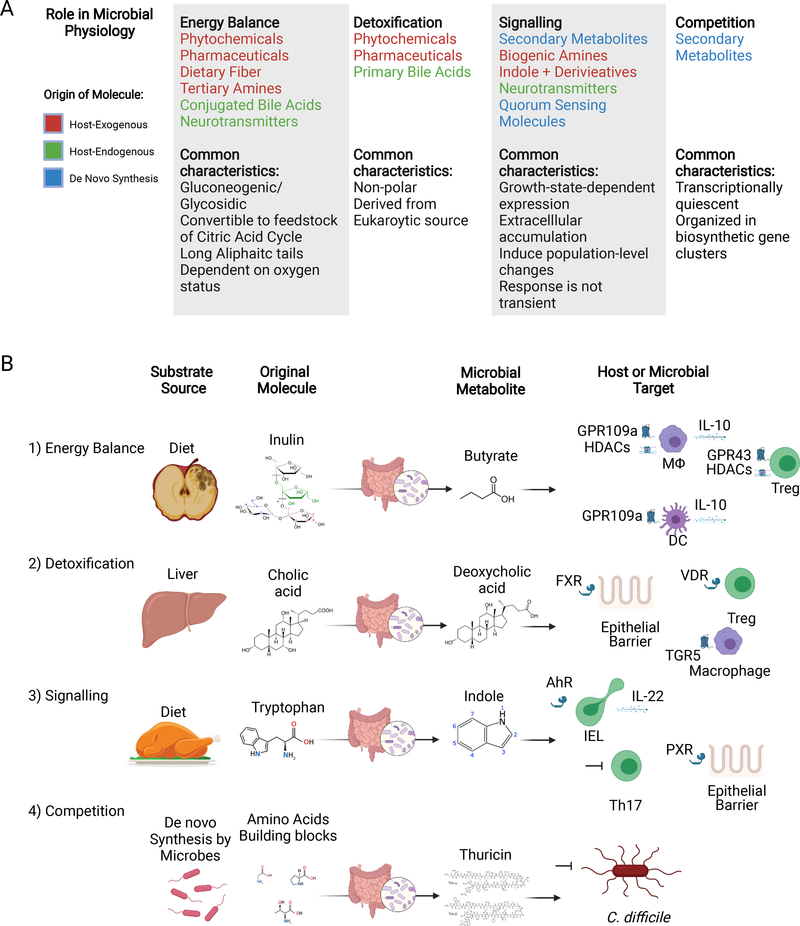
Figure 1. Examples of microbial metabolites that impact host immunity categorized by their role in microbial physiology.
(A) Classes of small molecules, arranged by their role in microbial physiology. Note: the text for each chemical is color-coded based on the origin of the building blocks for that compound.
(B) For each category, we provide one example of a microbial biotransformation relevant to mammalian biology, including the substrate source, molecular origins, microbial metabolite, and host or bacterial targets. (1) Microbial fermentation of the dietary fiber inulin results in production of the SCFA butyrate, which has diverse impacts on the immune system. For example, butyrate induces an anti-inflammatory state in macrophages and dendritic cells via HDAC inhibition and engagement of GPR109a, and enhances Treg differentiation via HDAC inhibition and engagement of GPR43. (2) Primary bile acids (e.g., cholic acid) are dehydroxylated by gut microbes to produce secondary bile acids (e.g., deoxycholic acid). Secondary bile acids such as deoxycholic acid can enhance gut barrier function via FXR in epithelial cells, enhance Treg differentiation via the Vitamin D receptor, and suppress macrophage activation by engaging TGR5. (3) Dietary tryptamine is metabolized by gut microbes to produce indole and indole derivatives, which can enhance gut barrier function via PXR in epithelial cells, induce IL-22 production by IELs via AhR, and inhibit Th17 differentiation via an unknown mechanism. (4) Thuricin, a RiPP is synthesized de novo by commensal microbes and specifically inhibits the growth of the opportunistic pathogen C. difficile. Created with BioRender.com.
Biotransformations of Exogenously Consumed Compounds
Dietary fiber and the Short Chain Fatty Acids
Dietary fiber refers to plant-derived carbohydrates that cannot be metabolized by host enzymes, including complex polysaccharides, lignans, and resistant starches. As dietary fiber cannot be absorbed or metabolized into absorbable components by host enzymes, it passes unaltered through the small intestine to the large intestine where it is fermented by resident microbes that encode specialized machinery to import and process dietary carbohydrates. In non-Westernized populations, members of the Prevotella genus are the major consumers of ingested fiber, whereas Faecalibacterium prausnitzii, Clostridium leptum, Eubacterium rectale, and Roseburia spp. are more predominant in the West (3, 4). Fermentation restores NAD+ consumed during glycolysis of monosaccharides liberated from fiber, and generally leads to the production of short chain fatty acids (SCFAs), which are the 2–4 carbon carboxylic acids: acetate, propionate, and butyrate. SCFA producers preferentially resort to fermentation despite their capacity for respiration, which may be partially explained by biogeography and mutualism. An appreciable portion of gut microbes have been observed to have respiratory machinery, but no biosynthetic capability for quinones (intermediary electron carriers). It is speculated that these microbes must rely on quinones synthesized by other microbes to carry out respiration (5). Accordingly, some SCFA-producers will switch from fermentation to respiration in oxygen rich environments (6, 7).
SCFAs can accumulate at up to 100 mM concentrations in the colonic lumen (8) and have been implicated in shaping diverse aspects of host biology. (See (9) and (10) for a comprehensive review of the many effects of SCFA on host immunity and beyond.) As just a few examples, colonocytes preferentially utilize butyrate as an energy source, and SCFA are critical regulators of both innate and adaptive immune responses through both receptor-mediated and non-receptor-mediated mechanisms. For instance, SCFA drive regulatory T cell differentiation and production of anti-inflammatory cytokines by macrophages through histone deacetylase inhibition (11–13) and regulate diverse immune responses through the engagement of G protein-coupled receptors (GPCRs) including GPR41, GPR43, and GPR109a (9, 14–17).
In addition to the SCFAs, the metabolic intermediate succinate is also produced during bacterial fermentation of dietary fibers (18–20). Microbiota-derived succinate can potentially modulate the immune response via diverse mechanisms, including engagement of the succinate receptor GPR91 (SUCNR1), which is expressed on multiple immune cell types (21). For example, succinate can directly activate tuft cells in the small intestine via GPR91 and elicits a multifaceted type 2 immune response through engagement of the tuft cell-IL-25-ILC2 circuit (22, 23).
TMAO
Longitudinal human cohort studies initially implicated trimethylamine N-oxide (TMAO) in the development of cardiovascular disease and follow-up mouse studies suggested a causal role for TMAO in these disorders (24, 25). TMAO is produced exclusively by the microbiota via metabolism of dietary choline and L-carnitine, which are abundant in red meat and shellfish (24, 26, 27). Both choline and L-carnitine are tertiary amines and microbial cleavage yields trimethylamine (TMA), which is oxidized to TMAO by flavin monooxygenases in the liver (28). The impetus for bacterial production of TMA is potentially multifactorial, but likely supports bacterial metabolism and growth. For example, catabolism of L-carnitine produces a 4-carbon intermediate that can enter the TCA cycle (29) and acetaldehyde, which is used in the biosynthesis of Acetyl-CoA (30). Carnitine can also serve as a terminal electron acceptor in anaerobic respiration, although this functionality is repressed by access to superior electron acceptors such as nitrate (31). When carnitine is used as an electron acceptor, gamma-butyrobetaine and not TMA is generated, which suggests that the nutritive status of a microbe may affect TMA levels. Despite the intense interest in TMAO as a therapeutic target, the precise molecular mechanisms by which it promotes cardiovascular disease remain largely unclear (32, 33). However, recent studies suggest that TMAO may contribute to cardiovascular pathology by directly promoting platelet hyperreactivity (34) or by facilitating chronic inflammation (35). For example, TMAO can trigger inflammasome activation in both macrophages and endothelial cells, leading to increased release of proinflammatory cytokines including IL-1β and IL-18 (36, 37).
Amino acid derivatives
Due to their ubiquity, amino acids are prime candidates for microbial energy balance and derivatization into signalling molecules. As any autotrophic diet contains proteins, diet-derived amino acid substrates are readily available in the mammalian gut lumen. In addition, many bacteria directly synthesize amino-acids from basic building blocks, providing an additional source of endogenous amino acid substrates in the distal gut. Host-derived proteins may provide an additional minor source of lumenal amino acids. Thus, although we have included amino acid-derived compounds under the heading of exogenously consumed compounds, microbiota-derived amino acid metabolites may also be derived from amino acids produced by other microbes, the host, or synthesized de novo by individual microbes themselves (38).
Biogenic amines
Amino acid-derivatives constitute many of the key mammalian signaling molecules, including the neurotransmitters histamine, dopamine, and serotonin, trace amines such as phenethylamine and tyramine, and hormones such as melatonin and epinephrine. While best known for their roles in regulating diverse physiological processes in mammals, nearly all of these compounds can also be produced by microbes, including select gut microbes (39–42). Given the availability of large amounts of amino acid substrates in the intestinal lumen, gut microbes can potentially produce high concentrations of amino acid-derivatives, raising the possibility that microbially-derived biogenic amines may influence local or systemic host physiology, including mood and behavior (39). For example, microbially-derived histamine can enhance gut motility (43) and modulate allergic asthma in the lung (44). Diverse immune cell types express myriad receptors for neurotransmitters, trace amines, and hormones, but our understanding of how engagement of these receptors by microbiota-derived metabolites impacts the immune response remains limited (45). In addition to production of neurotransmitters, microbial breakdown of amino acids can also lead to the production of polyamines, such as spermine, which have been shown to exhibit immunomodulatory properties, including regulation of inflammasome activation (46).
Branched Chain Amino Acids
The branched chain amino acids (BCAA), isoleucine, L-valine and leucine, are essential amino acids that are synthesized from the metabolic precursors pyruvate and threonine (47). Gut microbes can affect BCAA levels in the intestine by acting as both producers and consumers of BCAA (48). BCAA can impact various aspects of mammalian physiology, including the immune response, through their effects on protein synthesis, metabolism, and cell proliferation (49). For example, gut microbes such as Prevotella copri and Bacteroides vulgatus produce excess BCAA, and microbial BCAA-production is associated with increased systemic BCAA levels and insulin resistance in humans and mice (48).
Indole and Indole Derivatives
A variety of commensal microbes convert tryptophan into indole or related indole derivatives that can shape diverse immune processes by engaging the aryl hydrocarbon receptor (AhR) or pregnane X receptor (PXR). For example, indole 3-propionic acid can enhance intestinal barrier function through PXR (50), indole-3-aldehyde can induce intestinal IL-22 production and Lactobacillus-derived tryptophan derivatives can regulate intraepithelial lymphocyte differentiation through AhR (51, 52), and indole-3-lactic acid can inhibit Th17 differentiation and experimental autoimmune encephalomyelitis through an unknown mechanism (53). Microbial tryptophan metabolism was systematically dissected in Clostridium sporogenes, which can convert aromatic amino acids into their respective propionic acid derivatives (54). Targeted disruption of a key enzyme for these biotransformations (fldC) revealed critical roles for C. sporogenes-derived indole metabolites in shaping gut physiology and systemic immunity as gnotobiotic mice colonized with fldC-mutant C. sporogenes displayed increased gut permeability and increased mucosal and systemic C. sporogenes-specific antibody responses (54). Indole may also serve as a potential signalling molecule. Indeed, in many microbial species, indole and indole derivatives regulate biofilm formation and the expression of virulence factors (55, 56) and it has been speculated that indole may function as a quorum-sensing-like regulator (57).
Phytochemicals
The gut microbiota is constantly exposed to a diverse array of phytochemicals from the diet. As just one example, the lignans are a large group of polyphenols found in many plants and lignan consumption has been associated with lower cancer risk (58). Despite the plenitude of plant lignans, only two biologically active subtypes are prevalent in the mammalian gut, enterodiol and enterolactone (59), both of which are structurally similar to estrogen. The lignan pinoresinol (PINO) is biotransformed by the gut microbiota to enterolactone via sequential processing by multiple bacterial species and nicely illustrates the role of microbial metabolite exchanges in the production of bioactive metabolites. Select strains of E. lenta encode the enzyme ber, which catalyzes the first two reactions of the pathway, PINO to lariciresinol (LAR) to secoisolariciresinol (SECO). After these initial processing steps in E. lenta, three separate bacterial taxa, B. producta, Gordonibacter pamelaeae, and Lactonifactor longoviformis were found to sequentially convert SECO to its bioactive end product enterolactone (60).
The microbial benefit of lignan metabolism may be purely energetic in nature as some bacteria can grow on lignans as a sole carbon source (61). However, in vitro studies may confound this simple interpretation as supplementation of lignans to stool-derived mixed cultures led to an increase in Proteobacteria (62) even though the main species capable of metabolizing lignans belonged to the Actinobacteria and Firmicutes. Interestingly, lignans exhibit significant antimicrobial activity against select species and also exhibit strong antioxidant properties which could benefit strict anaerobes in the gut (63, 64). Nonetheless, given that enterolactone and enterodiol both have negligible free radical scavenging ability relative to their precursors (65), it seems likely that lignan biotransformation is mainly a means of detoxification, although additional studies are necessary to rigorously test this assumption.
Pharmaceuticals
Orally ingested drugs may encounter commensal microbes both prior to absorption and during enterohepatic circulation (66). Biotransformations of therapeutic small molecules by the microbiome can potentially 1) reduce the bioavailability of the active form of a drug via conversion to an inert intermediate, 2) convert prodrugs into their active form, or 3) lead to the generation of toxic drug metabolites. While pharmaceutical agents are likely to be entirely foreign to the standard biochemistry of any given gut microbe, microbial life has evolved to encode an arsenal of enzymes devoted to xenobiotic metabolism. Microbial xenobiotic metabolism in the gut generally involves either reduction or hydrolysis. The prevalence of reductive biochemistry may be a consequence of the anaerobic nature of the gut. Xenobiotics may thus serve as alternative substrates for anaerobic respiration, or reactants of oxidoreductases that reduce NADH (67). Hydrolysis may reflect the need to obtain substrates for growth. Recent pioneering studies have revealed the prevalence and diversity of microbial metabolism of medical drugs (68–70).
By systematically evaluating the biotransformations of 271 drugs by 76 bacterial species/strains from the human gut microbiome, Zimmerman et al. revealed a rich landscape of microbial modifications of common pharmaceuticals (68). They found that over two-thirds of assayed drugs were metabolized by at least one strain and that phylogenetically-related taxa often processed drugs with similar structural features. For instance drugs containing an ester or amide group were metabolized mainly by Bacteroidetes species, while nitro- or azo-group containing drugs were metabolized by members of all phyla except Proteobacteria (68). These results suggest that microbial metabolism of medical drugs is the rule and further highlight the possibility that interindividual variation in gut microbial communities may impact individual responses to medical drugs.
Identifying the ultimate fate of a pharmaceutical in the gut is complicated by microbial metabolite exchanges where the product of one microbial transformation is the substrate for a subsequent biotransformation by another bacterium. For example, levo-dopa, a drug for Parkinson’s disease that is converted into dopamine by aromatic L-amino acid decarboxylase after crossing the blood-brain barrier, can be converted by Eggerthella lenta into dopamine, which is further metabolized by Enterococcus faecalis species into tyramine (71). This pre-processing restricts the bioavailability of L-dopa to the brain and can affect responsiveness to L-dopa treatment. L-dopa is commonly co-administered with carbidopa, an inhibitor of mammalian aromatic L-amino acid decarboxylase, to prevent the premature processing of L-dopa into dopamine, which cannot cross the blood-brain barrier and can cause undesired side-effects in the periphery (72). However, carbidopa is a poor inhibitor of the microbial enzyme that catalyzes L-dopa processing in the gut. Instead, a different drug, (S)-ɑ-Fluoromethyltyronsine can selectively inhibit this microbial enzyme and, in gnotobiotic mice colonized with an L-dopa metabolizing microbiota, co-administration of (S)-ɑ-Fluoromethyltyronsine and L-dopa increased the serum concentration of L-dopa (71). Notably, in these studies, only E. lenta strains containing a specific SNP in the enzyme Dadh were capable of decarboxylating L-dopa, which underscores additional challenges in determining the metabolic potential of the microbiome based on marker-genes alone (71).
Biotransformations of Compounds Produced by the Host
Bile acids
Bile acids are amphipathic molecules synthesized from cholesterol in the liver and secreted into the duodenum to aid in the absorption of dietary fat. Primary bile acids synthesized by the host are conjugated with either glycine or taurine. Given their detergent-like nature, bile acids can disrupt the lipid bilayer of cellular membranes and are toxic to many microbes (73). Accordingly, bile acid supplementation significantly alters the gut microbiome (74) and recent studies suggest that bile acid secretion in the neonate may facilitate maturation of the microbiome in early life (75). Commensal microbes have evolved several means of biotransformation and detoxification of bile-acids including, epimerization, deconjugation by Bile Salt Hydrolase (removing the conjugated glycine or taurine), reduction/oxidation, hydroxylation, and dehydroxylation (a multistep pathway which is found almost exclusively in gut anaerobes) (76). Interestingly, microbiota-mediated bile acid metabolism is at least partially host species-specific as human microbiota are incapable of transforming select bile acids from rodents (77). Bacterially-modified bile acids are referred to collectively as secondary bile acids, though this term most commonly describes deconjugated bile acids. Many microbial bile acid modifications reduce their membrane-damaging effects by decreasing hydrophobicity and potential toxicity (78). However, somewhat paradoxically, deconjugation appears to increase the hydrophobicity of bile acids. Given that deconjugation releases free glycine and taurine, which can be catabolized to ammonia and carbon dioxide, bacterial metabolism of primary bile acids via this route may serve primarily as a source of energy and amino acid building blocks for select commensal microbes.
The consequences of bacterial modification of bile acids for the host stem partially from the autoregulation of primary biliary synthesis. Activation of Farnesoid X receptor (FXR), a nuclear receptor expressed in the gut and liver, by primary bile acids suppresses CYP7A1 expression, which reduces the conversion of cholesterol into primary bile acids. Thus, FXR provides autoregulatory control of the bile acid pool. Since secondary bile acids have differential affinity for the FXR receptor, microbial transformation of primary bile acids can alter FXR signalling and therefore bile acid pool size. Indeed,germ-free rodents exhibit increased bile acid pools relative to conventionalized animals due to microbial processing of the FXR-antagonist T-BetaMCA (a mouse-specific bile acid) (79). Bile acids also serve as ligands for multiple GPCRs, including Takeda GPCR 5 (TGR5). Activation of TGR5 in the intestine and pancreas induces secretion of glucagon-like peptide-1 and insulin, regulating circulating blood glucose (80). Fascinatingly, unconjugated bile acids can also cross the blood brain barrier and activation of TGR5 by these compounds in the hypothalamus reduces food intake and increases energy expenditure (80). TGR5 is also the predominant bile acid receptor of liver-resident macrophages, and activation in these cells inhibits inflammation (81). Finally, recent studies have revealed that specific secondary bile acids can regulate the differentiation of intestinal T cells towards Treg or Th17 phenotypes (82, 83). For example, microbial production of the secondary bile acid isoDCA enhanced peripheral Treg generation and abolition of secondary bile-acid production by individual commensals significantly decreased their ability to induce colonic T regs (82, 84). The impact of bile acids on T cells may also depend on the specific composition of the bile acid pool as distinct bile acid species derived from the same precursor control unique aspects of T cell differentiation via distinct molecular mechanisms (48).
Human Milk Oligosaccharides
Human Milk Oligosaccharides (HMOs) are prebiotic fibers in breastmilk that support the establishment of a healthy microbiome during early development (85). They are a specialized case of dietary fiber as they are the only source of complex carbohydrates available to nursing infants. We have classified HMOs here as ‘host-derived’, but they could also be classified as ‘exogenously consumed’ from the perspective of the infant. HMOs largely pass unaffected to the large intestine where they support the growth of Bifidobacterium species (i.e., the bifidogenic effect), which are early colonizers of the human gut and the dominant member of the microbiome in breast-feeding infants. Bifidobacteria are uniquely suited to metabolize HMOs via the bifid shunt pathway (86). Bifidobacteria support healthy immune system development through diverse mechanisms, including the production of acetate and lactate, and are critical mediators of colonization resistance in early life (87).
‘Neurotransmitters’ as a Microbial Food Source
The gut harbors more than 90% of the serotonin in the body. Enteroendocrine cells and enteric neurons are major sources of gut neurotransmitters, and recent studies have revealed that specific commensal microbes critically modulate intestinal neurotransmitter production (42). GF mice and mice colonized with altered Schaedler flora exhibit reduced concentrations of colonic serotonin. However, colonization of germ-free mice with spore forming microbes, predominantly Clostridia species, or administration of associated microbial metabolites restored normal gut serotonin levels by stimulating colonic enterochromaffin cells (88). Artificially increasing colonic serotonin, either through gavage or by genetic ablation of the enteric serotonin transporter SERT, also increased the abundance of spore-formers including Turicibacter sanguinis. T. sanguinis itself was found to encode a protein similar to human SERT, which conferred the ability to uptake serotonin, and treatment of T. sanguinis with serotonin induced the downregulation of genes related to cell differentiation, morphogenesis and sporulation, as well as the upregulation of transporters (89). Taken together, these data reveal a narrative whereby serotonin serves as a signal that the microbe is in an environment conducive to vegetative growth (i.e., the gut lumen). Having exited the spore, the microbe then induces further host production of serotonin presumably for use as an energy source. However, because this endogenous factor has its own signalling role in the host, microbial induction of this factor also leads to altered host physiology.
Serotonin is not the only neurotransmitter that can serve as a microbial growth factor. For example, bacteria are also known to consume γ-aminobutyric acid (GABA) (90), largely through the GABA shunt whereby GABA is converted to succinate which subsequently enters the TCA cycle (91). More recently, GABA was found to be an essential growth factor for a previously uncultivatable human gut microbe of a first-in genus member of the Ruminococcaceae family (92).
Small Molecules Synthesized De Novo
Quorum Sensing Molecules
Quorum sensing (QS) molecules enable microbes to regulate their behavior based on population size. QS molecules are often subject to autoregulation, whereby a QS compound induces increased QS biosynthesis. Among the classical autoinducers (AIs), AI-1, has generally eluded detection in the human gut (93), while both AI-2 and AI-3 have been detected (94). The dearth of AI-1 could potentially be explained by the expression of the paraoxonases, PON1/PON2/PON3 by gut epithelium (95), which can efficiently cleave Acyl homoserine lactone (AI-1) and disrupt QS in pathogens such as P. aeruginosa (96). Interestingly, the host may have also evolved the capacity to listen in on QS-mediated microbial chatter. For example, AI-2 induces IL-8 secretion in HCT-8 cells (97) and AI-3 analogs have been shown to increase macrophage IL-8 secretion (98). The synthetic process for AI-3 was unknown until it was recently discovered that “abortive” tRNA synthetase reactions rather than enzymatic transformations mediate AI-3 production (98). Quorum sensing can also alter the course of re-establishment of the gut microbiota following antibiotic treatment. In mice treated with streptomycin, the presence of an AI-2 producer will shift the composition of a recovered community towards a greater abundance of Firmicutes (99). Within the densely-packed multispecies context of the gut, quorum sensing may also mediate interspecies signalling in addition to intraspecies signalling. For example, Ruminococcus obeum, synthesizes AI-2 in response to V. cholera infection, which moderates expression of V. cholera colonization factors (100).
Secondary Metabolites
Aside from modifications to molecules ingested or produced by the host, microbes can produce a wide array of complex small molecules from smaller modular components. These secondary metabolites can be broadly classified as polyketides (PKs), non-ribosomally synthesized peptides (NRPs), ribosomally synthesized, posttranslationally modified peptides (RiPPs), terpenes, NRP synthetase-independent siderophores, and saccharides. The genes responsible for the biosynthesis of these compounds typically co-localize on the genome in what are termed biosynthetic gene clusters (BGCs). Notably, the addition of subunits to a nascent secondary metabolite encoded by a BGC generally proceeds in gene order (101–103).
RiPPs are the most widely distributed BGCs in the human microbiome, while PKs and NRPs are less prevalent and abundant. Many BGC-derived natural products from the microbiome exhibit narrow-spectrum antimicrobial activities and may mediate bacterial competition within complex host-associated microbial communities. For example, Lactocillin, a RiPP derived from Lactobacillus gasseri inhibits Gram-positive urogenital pathogens, with no commensurate activity against Gram-positive urogenital commensals (104). The NRP Lugdunin from Staphylococcus lugdunensis, specifically antagonizes S. aureus (105). And, the RiPP Thuricin specifically antagonizes C. difficile while sparing most other members of the gut microbiota (106). Although many previously-characterized secondary metabolites are antimicrobials, BGC-derived compounds may also exhibit immunomodulatory activities. For example, the RiPP commendamide activates GPR132/G2A, a GPCR implicated in autoimmune disease and cardiovascular disease (107) and a family of NRPs from the gut microbiome inhibits host cathepsins, potentially as a means of avoiding antigen processing and presentation (108).
Experimental approaches to identify bioactive microbiota metabolites
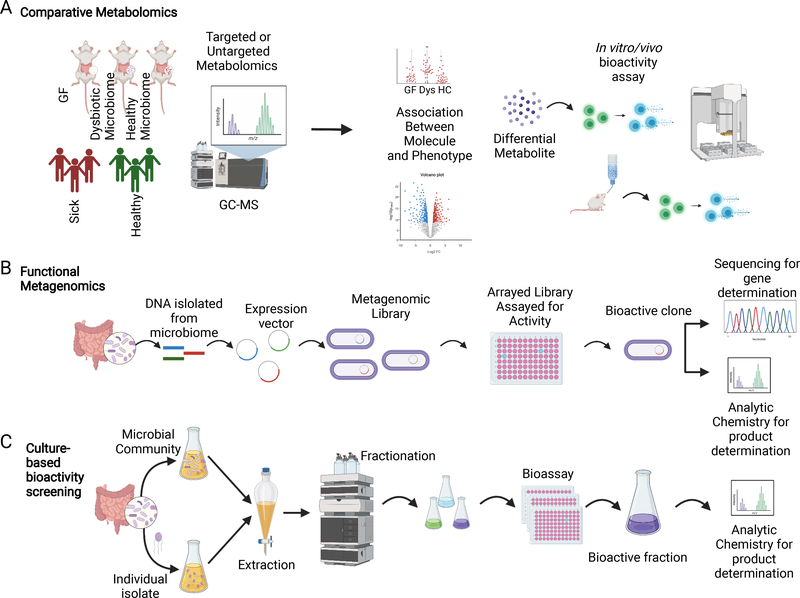
The enormous biochemical potential of the microbiome and the resulting complexity of the microbiota metabolome imply that the majority of bacterially-derived chemicals that shape human health remain to be discovered. Here, we will briefly outline existing and emerging strategies for discovering and characterizing novel bioactive microbiota metabolites (see also (2) and (109) for comprehensive reviews on this topic). These approaches can be categorized into three general frameworks outlined in Figure 2.
Figure 2. Experimental approaches for the discovery of novel bioactive microbiota metabolites.
(A) Comparative Metabolomics. In comparative metabolomics, metabolomes from groups of mice or humans with distinct phenotypes are assessed via targeted or untargeted metabolomics to generate a list of putative causal metabolites that correlate with a defined phenotype. The potential impacts of these specific metabolites on the host can then be assessed in vitro (e.g., using primary cells or reporter cells) or in vivo using mouse models. (B) Functional metagenomics. Metagenomic DNA from a microbiome of interest is fragmented, incorporated into expression vectors, and expressed in a facile recipient to produce large libraries of metagenomic clones, which are typically assayed for bioactivity via high-throughput screening. Bioactive clones are then sequenced to reveal the genes responsible for biosynthesis of active compounds and can be used to facilitate chemical identification. (C) Culture-based bioactivity profiling. Supernatants from cultures of individual commensal microbes or mixed communities of microbes are subjected to extraction and fractionation. Fractions are then assayed for a bioactivity of interest (e.g., GPCR activation). Bioactive fractions are further interrogated using bioassay-guided natural products discovery approaches to identify and characterize the compound of interest. Created with BioRender.com
The first pipeline is exemplified by the “correlation-first” approach whereby comparative metabolomics analyses of samples from humans or mice with divergent phenotypes is used to identify potentially causal metabolites of interest (Figure 2A). These metabolites can then be interrogated for relevant bioactivities either in vitro or in vivo (e.g., cytokine induction (107) or Th17 polarization (110)). Two prominent successes based on this approach are the discovery of the role of TMAO in cardiovascular disease (24) and SCFAs in Treg differentiation (12). However, a major limitation of correlation-based comparative metabolomics approaches is that they are typically restricted to the discovery of previously known metabolites as deconvolving and resolving unknown MS peaks remains a major challenge. Related approaches such as assessments of previously-characterized families of small molecule metabolites (e.g., secondary bile acids), have led to similar successes (83), but suffer from the same general limitation. Thus, additional approaches have been developed to enable the unbiased discovery of truly novel bioactive microbiota metabolites.
The most prominent approach for identifying previously undiscovered bioactive compounds is ‘functional metagenomics’ (Figure 2B). Here, sheared metagenomic DNA is cloned into a surrogate host (usually E. coli) and large libraries of clones are subjected to high-throughput bioactivity assays. This approach has been particularly successful for isolating natural products synthesized by cryptic BGCs (104, 107, 108, 111, 112) in part because the products of BGCs are often created using simple building blocks present in all microbes including E. coli. Two advantages of this approach are that it does not require cultivation of the microbial metabolite producer and that the clones of interest greatly facilitate both chemical identification and genetic dissection of the relevant BGC. The clustered nature and conserved motifs of BGCs makes them ideally suited for in silico discovery (113–116). From here, it is occasionally possible to proceed directly to chemical synthesis based on computational predictions of potential chemical structures, dispensing with genetics entirely (117, 118), or to leverage recent developments in synthetic biology to reconstruct complete biosynthetic pathways using cell-free systems (119).
Finally, high-throughput bioactivity screening of chemical extracts from in vitro cultures of commensal microbes has also led to the discovery of novel bioactive microbiota metabolites, as well as their specific microbial sources in the gut (Figure 2C). For example, recent efforts to screen microbial metabolomes against nearly all GPCRs have revealed a rich array of GPCR-active commensal microbes and metabolites (43, 120). In these studies, microbial monocultures were screened against individual receptors using engineered reporter assays. However, similar approaches can be applied to any phenotype of interest that can be assessed in vitro (from simplified reporter-assays to remodeling of tissue organoids) using microbial metabolomes of any complexity (from monocultures to complete gut microbial communities). Continued improvements in anaerobic culturomics, tissue organoids, and organ-on-a chip models will undoubtedly facilitate further insights into the bioactive microbiota metabolome in the coming years.
Conclusions
The past two decades of technological advances in microbiome science have ushered in a new golden era of molecular-level dissection of the reciprocal interactions between mammalian hosts and their associated microbiomes. However, many barriers still remain before we can realize the full fundamental and therapeutic potential of the microbiome. For example, the effects of specific metabolites on microbial physiology have been characterized almost exclusively in model organisms grown in axenic culture, and the impacts of specific microbes and metabolites on host biology are most often examined in exquisitely controlled, but admittedly highly artificial, gnotobiotic mouse models. Realizing the next era of microbiome science will require a movement beyond proof-of-concept studies and collections of anecdotes towards a broader and more generalizable understanding of the fundamental principles that dictate beneficial and detrimental interactions between indigenous microbes and their mammalian hosts.
Acknowledgments
The authors thank members of the Palm lab for useful discussions.
Abbreviations used in this article
- SCFA
Short Chain Fatty Acid
- HMO
Human Milk Oligosaccharide
- BCAA
Branched-chain Amino Acid
- GPCR
G protein-coupled receptors
- TMAO
Trimethylamine N-oxide
- AhR
Aryl hydrocarbon receptor
- PXR
Pregnane X Receptor
- FXR
Farnesoid X Receptor
- QS
Quorum sensing
- AI
Autoinducer
- BGC
Biosynthetic gene cluster
- GF
Germ Free
Footnotes
We gratefully acknowledge funding from the NIH (DK125119, AI137935, GM141649, AG068863), the Helmsley and Pew Charitable Trusts, Chan Zuckerberg Initiative, and Michael J. Fox, Kenneth Rainin, Mathers, Ludwig, and Smith Foundations.
N.W.P. is an advisor for and academic co-founder of Artizan Biosciences and Design Pharmaceuticals and has received research funding from Artizan Biosciences and Hoffman-LaRoche, Ltd.
References
- 1.Skelly AN, Sato Y, Kearney S, and Honda K. 2019. Mining the microbiota for microbial and metabolite-based immunotherapies. Nat. Rev. Immunol. 19: 305–323. [DOI] [PubMed] [Google Scholar]
- 2.Shine EE, and Crawford JM. 2021. Molecules from the Microbiome. Annu. Rev. Biochem. 789–815. [DOI] [PubMed] [Google Scholar]
- 3.Parada Venegas D, De la Fuente MK, Landskron G, González MJ, Quera R, Dijkstra G, Harmsen HJM, Faber KN, and Hermoso MA. 2019. Short Chain Fatty Acids (SCFAs)-Mediated Gut Epithelial and Immune Regulation and Its Relevance for Inflammatory Bowel Diseases. Front. Immunol. 10: 277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makki K, Deehan EC, Walter J, and Bäckhed F. 2018. The Impact of Dietary Fiber on Gut Microbiota in Host Health and Disease. Cell Host Microbe 23: 705–715. [DOI] [PubMed] [Google Scholar]
- 5.Ravcheev DA, and Thiele I. 2016. Genomic Analysis of the Human Gut Microbiome Suggests Novel Enzymes Involved in Quinone Biosynthesis. Front. Microbiol. 7: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khan MT, Duncan SH, Stams AJM, van Dijl JM, Flint HJ, and Harmsen HJM. 2012. The gut anaerobe Faecalibacterium prausnitzii uses an extracellular electron shuttle to grow at oxic-anoxic interphases. ISME J. 6: 1578–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tegtmeier D, Thompson CL, Schauer C, and Brune A. 2016. Oxygen Affects Gut Bacterial Colonization and Metabolic Activities in a Gnotobiotic Cockroach Model. Appl. Environ. Microbiol. 82: 1080–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cummings JH, Pomare EW, Branch WJ, Naylor CP, and Macfarlane GT. 1987. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28: 1221–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan J, McKenzie C, Potamitis M, Thorburn AN, Mackay CR, and Macia L. 2014. Chapter Three - The Role of Short-Chain Fatty Acids in Health and Disease. In Advances in Immunology vol. 121. Alt FW, ed. Academic Press. 91–119. [DOI] [PubMed] [Google Scholar]
- 10.van der Hee B, and Wells JM. 2021. Microbial Regulation of Host Physiology by Short-chain Fatty Acids. Trends Microbiol.. [DOI] [PubMed] [Google Scholar]
- 11.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, Liu H, Cross JR, Pfeffer K, Coffer PJ, and Rudensky AY. 2013. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504: 451–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, Nakanishi Y, Uetake C, Kato K, Kato T, Takahashi M, Fukuda NN, Murakami S, Miyauchi E, Hino S, Atarashi K, Onawa S, Fujimura Y, Lockett T, Clarke JM, Topping DL, Tomita M, Hori S, Ohara O, Morita T, Koseki H, Kikuchi J, Honda K, Hase K, and Ohno H. 2013. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504: 446–450. [DOI] [PubMed] [Google Scholar]
- 13.Chang PV, Hao L, Offermanns S, and Medzhitov R. 2014. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. U. S. A. 111: 2247–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang W, Yu T, Huang X, Bilotta AJ, Xu L, Lu Y, Sun J, Pan F, Zhou J, Zhang W, Yao S, Maynard CL, Singh N, Dann SM, Liu Z, and Cong Y. 2020. Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat. Commun. 11: 4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, Glickman JN, and Garrett WS. 2013. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341: 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, Lee JR, Offermanns S, and Ganapathy V. 2014. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40: 128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macia L, Tan J, Vieira AT, Leach K, Stanley D, Luong S, Maruya M, Ian McKenzie C, Hijikata A, Wong C, Binge L, Thorburn AN, Chevalier N, Ang C, Marino E, Robert R, Offermanns S, Teixeira MM, Moore RJ, Flavell RA, Fagarasan S, and Mackay CR. 2015. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 6: 6734. [DOI] [PubMed] [Google Scholar]
- 18.Louis P, and Flint HJ. 2017. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 19: 29–41. [DOI] [PubMed] [Google Scholar]
- 19.Strobel HJ 1992. Vitamin B12-dependent propionate production by the ruminal bacterium Prevotella ruminicola 23. Appl. Environ. Microbiol. 58: 2331–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Vadder F, Kovatcheva-Datchary P, Zitoun C, Duchampt A, Bäckhed F, and Mithieux G. 2016. Microbiota-Produced Succinate Improves Glucose Homeostasis via Intestinal Gluconeogenesis. Cell Metab. 24: 151–157. [DOI] [PubMed] [Google Scholar]
- 21.Caffaratti C, Plazy C, Mery G, Tidjani A-R, Fiorini F, Thiroux S, Toussaint B, Hannani D, and Le Gouellec A. 2021. What We Know So Far about the Metabolite-Mediated Microbiota-Intestinal Immunity Dialogue and How to Hear the Sound of This Crosstalk. Metabolites 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneider C, O’Leary CE, von Moltke J, Liang H-E, Ang QY, Turnbaugh PJ, Radhakrishnan S, Pellizzon M, Ma A, and Locksley RM. 2018. A Metabolite-Triggered Tuft Cell-ILC2 Circuit Drives Small Intestinal Remodeling. Cell 174: 271–284.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nadjsombati MS, McGinty JW, Lyons-Cohen MR, Jaffe JB, DiPeso L, Schneider C, Miller CN, Pollack JL, Nagana Gowda GA, Fontana MF, Erle DJ, Anderson MS, Locksley RM, Raftery D, and von Moltke J. 2018. Detection of Succinate by Intestinal Tuft Cells Triggers a Type 2 Innate Immune Circuit. Immunity 49: 33–41.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z, Klipfell E, Bennett BJ, Koeth R, Levison BS, Dugar B, Feldstein AE, Britt EB, Fu X, Chung Y-M, Wu Y, Schauer P, Smith JD, Allayee H, Tang WHW, DiDonato JA, Lusis AJ, and Hazen SL. 2011. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu W, Wang Z, Tang WHW, and Hazen SL. 2017. Gut Microbe-Generated Trimethylamine N-Oxide From Dietary Choline Is Prothrombotic in Subjects. Circulation 135: 1671–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang WHW, Wang Z, Levison BS, Koeth RA, Britt EB, Fu X, Wu Y, and Hazen SL. 2013. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 368: 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L, Smith JD, DiDonato JA, Chen J, Li H, Wu GD, Lewis JD, Warrier M, Brown JM, Krauss RM, Tang WHW, Bushman FD, Lusis AJ, and Hazen SL. 2013. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19: 576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warrier M, Shih DM, Burrows AC, Ferguson D, Gromovsky AD, Brown AL, Marshall S, McDaniel A, Schugar RC, Wang Z, Sacks J, Rong X, de T, Vallim A, Chou J, Ivanova PT, Myers DS, Brown HA, Lee RG, Crooke RM, Graham MJ, Liu X, Parini P, Tontonoz P, Lusis AJ, Hazen SL, Temel RE, and Brown JM. 2015. The TMAO-Generating Enzyme Flavin Monooxygenase 3 Is a Central Regulator of Cholesterol Balance. Cell Rep. 10: 326–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seim H, Löster H, Claus R, Kleber H-P, and Strack E. 1982. Splitting of the C-N bond in carnitine by an enzyme (trimethylamine forming) from membranes of Acinetobacter calcoaceticus. FEMS Microbiol. Lett. 15: 165–167. [Google Scholar]
- 30.Martínez-del Campo A, Bodea S, Hamer HA, Marks JA, Haiser HJ, Turnbaugh PJ, and Balskus EP. 2015. Characterization and detection of a widely distributed gene cluster that predicts anaerobic choline utilization by human gut bacteria. MBio 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jung K, Jung H, and Kleber HP. 1987. Regulation of L-carnitine metabolism in Escherichia coli. J. Basic Microbiol. 27: 131–137. [DOI] [PubMed] [Google Scholar]
- 32.Roberts AB, Gu X, Buffa JA, Hurd AG, Wang Z, Zhu W, Gupta N, Skye SM, Cody DB, Levison BS, Barrington WT, Russell MW, Reed JM, Duzan A, Lang JM, Fu X, Li L, Myers AJ, Rachakonda S, DiDonato JA, Brown JM, Gogonea V, Lusis AJ, Garcia-Garcia JC, and Hazen SL. 2018. Development of a gut microbe-targeted nonlethal therapeutic to inhibit thrombosis potential. Nat. Med. 24: 1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Witkowski M, Weeks TL, and Hazen SL. 2020. Gut Microbiota and Cardiovascular Disease. Circ. Res. 127: 553–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu W, Gregory JC, Org E, Buffa JA, Gupta N, Wang Z, Li L, Fu X, Wu Y, Mehrabian M, Sartor RB, McIntyre TM, Silverstein RL, Tang WHW, DiDonato JA, Brown JM, Lusis AJ, and Hazen SL. 2016. Gut Microbial Metabolite TMAO Enhances Platelet Hyperreactivity and Thrombosis Risk. Cell 165: 111–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, and Dai M. 2020. Trimethylamine N-Oxide Generated by the Gut Microbiota Is Associated with Vascular Inflammation: New Insights into Atherosclerosis. Mediators Inflamm. 2020: 4634172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu K, Yuan Y, Yu H, Dai X, Wang S, Sun Z, Wang F, Fei H, Lin Q, Jiang H, and Chen T. 2020. The gut microbial metabolite trimethylamine N-oxide aggravates GVHD by inducing M1 macrophage polarization in mice. Blood 136: 501–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boini KM, Hussain T, Li P-L, and Koka S. 2017. Trimethylamine-N-Oxide Instigates NLRP3 Inflammasome Activation and Endothelial Dysfunction. Cell. Physiol. Biochem. 44: 152–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang HT 1964. Microbial production of amino acids. Prog. Ind. Microbiol. 5: 55–92. [PubMed] [Google Scholar]
- 39.Bambury A, Sandhu K, Cryan JF, and Dinan TG. 2018. Finding the needle in the haystack: systematic identification of psychobiotics. Br. J. Pharmacol. 175: 4430–4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wall R, Cryan JF, Ross RP, Fitzgerald GF, Dinan TG, and Stanton C. 2014. Bacterial Neuroactive Compounds Produced by Psychobiotics. In Microbial Endocrinology: The Microbiota-Gut-Brain Axis in Health and Disease Lyte M, and Cryan JF, eds. Springer New York, New York, NY. 221–239. [DOI] [PubMed] [Google Scholar]
- 41.Clarke G, Stilling RM, Kennedy PJ, Stanton C, Cryan JF, and Dinan TG. 2014. Minireview: Gut microbiota: the neglected endocrine organ. Mol. Endocrinol. 28: 1221–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strandwitz P 2018. Neurotransmitter modulation by the gut microbiota. Brain Res. 1693: 128–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen H, Nwe P-K, Yang Y, Rosen CE, Bielecka AA, Kuchroo M, Cline GW, Kruse AC, Ring AM, Crawford JM, and Palm NW. 2019. A Forward Chemical Genetic Screen Reveals Gut Microbiota Metabolites That Modulate Host Physiology. Cell 177: 1217–1231.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barcik W, Pugin B, Brescó MS, Westermann P, Rinaldi A, Groeger D, Van Elst D, Sokolowska M, Krawczyk K, Frei R, Ferstl R, Wawrzyniak M, Altunbulakli C, Akdis CA, and O’Mahony L. 2019. Bacterial secretion of histamine within the gut influences immune responses within the lung. Allergy 74: 899–909. [DOI] [PubMed] [Google Scholar]
- 45.Shea-Donohue T, and Urban JF. 2017. Neuroimmune Modulation of Gut Function. In Gastrointestinal Pharmacology Greenwood-Van Meerveld B, ed. Springer International Publishing, Cham. 247–267. [DOI] [PubMed] [Google Scholar]
- 46.Levy M, Thaiss CA, Zeevi D, Dohnalová L, Zilberman-Schapira G, Mahdi JA, David E, Savidor A, Korem T, Herzig Y, Pevsner-Fischer M, Shapiro H, Christ A, Harmelin A, Halpern Z, Latz E, Flavell RA, Amit I, Segal E, and Elinav E. 2015. Microbiota-Modulated Metabolites Shape the Intestinal Microenvironment by Regulating NLRP6 Inflammasome Signaling. Cell 163: 1428–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Amorim Franco TM, and Blanchard JS. 2017. Bacterial Branched-Chain Amino Acid Biosynthesis: Structures, Mechanisms, and Drugability. Biochemistry 56: 5849–5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pedersen HK, Gudmundsdottir V, Nielsen HB, Hyotylainen T, Nielsen T, Jensen BAH, Forslund K, Hildebrand F, Prifti E, Falony G, Le Chatelier E, Levenez F, Doré J, Mattila I, Plichta DR, Pöhö P, Hellgren LI, Arumugam M, Sunagawa S, Vieira-Silva S, Jørgensen T, Holm JB, Trošt K, MetaHIT Consortium, Kristiansen K, Brix S, Raes J, Wang J, Hansen T, Bork P, Brunak S, Oresic M, Ehrlich SD, and Pedersen O. 2016. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 535: 376–381. [DOI] [PubMed] [Google Scholar]
- 49.Nie C, He T, Zhang W, Zhang G, and Ma X. 2018. Branched Chain Amino Acids: Beyond Nutrition Metabolism. Int. J. Mol. Sci. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, Qiu Z, Maher L, Redinbo MR, Phillips RS, Fleet JC, Kortagere S, Mukherjee P, Fasano A, Le Ven J, Nicholson JK, Dumas ME, Khanna KM, and Mani S. 2014. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 41: 296–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, Carvalho A, Puccetti P, and Romani L. 2013. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39: 372–385. [DOI] [PubMed] [Google Scholar]
- 52.Cervantes-Barragan L, Chai JN, Tianero MD, Di Luccia B, Ahern PP, Merriman J, Cortez VS, Caparon MG, Donia MS, Gilfillan S, Cella M, Gordon JI, Hsieh C-S, and Colonna M. 2017. Lactobacillus reuteri induces gut intraepithelial CD4+CD8αα+ T cells. Science 357: 806–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilck N, Matus MG, Kearney SM, Olesen SW, Forslund K, Bartolomaeus H, Haase S, Mähler A, Balogh A, Markó L, Vvedenskaya O, Kleiner FH, Tsvetkov D, Klug L, Costea PI, Sunagawa S, Maier L, Rakova N, Schatz V, Neubert P, Frätzer C, Krannich A, Gollasch M, Grohme DA, Côrte-Real BF, Gerlach RG, Basic M, Typas A, Wu C, Titze JM, Jantsch J, Boschmann M, Dechend R, Kleinewietfeld M, Kempa S, Bork P, Linker RA, Alm EJ, and Müller DN. 2017. Salt-responsive gut commensal modulates TH17 axis and disease. Nature 551: 585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dodd D, Spitzer MH, Van Treuren W, Merrill BD, Hryckowian AJ, Higginbottom SK, Le A, Cowan TM, Nolan GP, Fischbach MA, and Sonnenburg JL. 2017. A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551: 648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu M, Zhang C, Mu Y, Shen Q, and Feng Y. 2010. Indole affects biofilm formation in bacteria. Indian J. Microbiol. 50: 362–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee J, Attila C, Cirillo SLG, Cirillo JD, and Wood TK. 2009. Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microb. Biotechnol. 2: 75–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee J, Jayaraman A, and Wood TK. 2007. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 7: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Webb AL, and McCullough ML. 2005. Dietary lignans: potential role in cancer prevention. Nutr. Cancer 51: 117–131. [DOI] [PubMed] [Google Scholar]
- 59.Yoder SC, Lancaster SM, Hullar MAJ, and Lampe JW. 2015. Chapter 7 - Gut Microbial Metabolism of Plant Lignans: Influence on Human Health. In Diet-Microbe Interactions in the Gut Tuohy K, and Del Rio D, eds. Academic Press, San Diego. 103–117. [Google Scholar]
- 60.Bess EN, Bisanz JE, Yarza F, Bustion A, Rich BE, Li X, Kitamura S, Waligurski E, Ang QY, Alba DL, Spanogiannopoulos P, Nayfach S, Koliwad SK, Wolan DW, Franke AA, and Turnbaugh PJ. 2020. Genetic basis for the cooperative bioactivation of plant lignans by Eggerthella lenta and other human gut bacteria. Nat Microbiol 5: 56–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tabak HH, Chambers CW, and Kabler PW. 1959. Bacterial utilization of lignans. I. Metabolism of alpha-conidendrin. J. Bacteriol. 78: 469–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Corona G, Kreimes A, Barone M, Turroni S, Brigidi P, Keleszade E, and Costabile A. 2020. Impact of lignans in oilseed mix on gut microbiome composition and enterolignan production in younger healthy and premenopausal women: an in vitro pilot study. Microb. Cell Fact. 19: 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barbary OM, El-Sohaimy SA, El-Saadani MA, and Zeitoun AMA. 2010. Antioxidant, antimicrobial and anti-HCV activities of lignan extracted from flaxseed. Res. J. Agric. Biol. Sci. 6: 247–256. [Google Scholar]
- 64.Kyselka J, Rabiej D, Dragoun M, Kreps F, Burčová Z, Němečková I, Smolová J, Bjelková M, Szydłowska-Czerniak A, Schmidt Š, Šarman L, and Filip V. 2017. Antioxidant and antimicrobial activity of linseed lignans and phenolic acids. Eur. Food Res. Technol. 243: 1633–1644. [Google Scholar]
- 65.Hu C, Yuan YV, and Kitts DD. 2007. Antioxidant activities of the flaxseed lignan secoisolariciresinol diglucoside, its aglycone secoisolariciresinol and the mammalian lignans enterodiol and enterolactone in vitro. Food Chem. Toxicol. 45: 2219–2227. [DOI] [PubMed] [Google Scholar]
- 66.Lam KN, Alexander M, and Turnbaugh PJ. 2019. Precision Medicine Goes Microscopic: Engineering the Microbiome to Improve Drug Outcomes. Cell Host Microbe 26: 22–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koppel N, Maini Rekdal V, and Balskus EP. 2017. Chemical transformation of xenobiotics by the human gut microbiota. Science 356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, and Goodman AL. 2019. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 570: 462–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, and Goodman AL. 2019. Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Javdan B, Lopez JG, Chankhamjon P, Lee Y-CJ, Hull R, Wu Q, Wang X, Chatterjee S, and Donia MS. 2020. Personalized Mapping of Drug Metabolism by the Human Gut Microbiome. Cell 181: 1661–1679.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maini Rekdal V, Bess EN, Bisanz JE, Turnbaugh PJ, and Balskus EP. 2019. Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tolosa E, Martí MJ, Valldeoriola F, and Molinuevo JL. 1998. History of levodopa and dopamine agonists in Parkinson’s disease treatment. Neurology 50: S2–10; discussion S44–8. [DOI] [PubMed] [Google Scholar]
- 73.Begley M, Gahan CGM, and Hill C. 2005. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29: 625–651. [DOI] [PubMed] [Google Scholar]
- 74.Islam KBMS, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, Ogura Y, Hayashi T, and Yokota A. 2011. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 141: 1773–1781. [DOI] [PubMed] [Google Scholar]
- 75.Van Best N, Rolle-Kampczyk U, Schaap FG, Basic M, Damink SO, Bleich A, Savelkoul PHM, Von Bergen M, Penders J, and Hornef MW. 2020. Bile acids drive the newborn’s gut microbiota maturation. Nat. Commun. 11: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ridlon JM, Harris SC, Bhowmik S, Kang D-J, and Hylemon PB. 2016. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 7: 22–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sacquet EC, Gadelle DP, Riottot MJ, and Raibaud PM. 1984. Absence of transformation of beta-muricholic acid by human microflora implanted in the digestive tracts of germfree male rats. Appl. Environ. Microbiol. 47: 1167–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Heuman DM 1989. Quantitative estimation of the hydrophilic-hydrophobic balance of mixed bile salt solutions. J. Lipid Res. 30: 719–730. [PubMed] [Google Scholar]
- 79.Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall H-U, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, and Bäckhed F. 2013. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17: 225–235. [DOI] [PubMed] [Google Scholar]
- 80.Castellanos-Jankiewicz A, Guzmán-Quevedo O, Fénelon VS, Zizzari P, Quarta C, Bellocchio L, Tailleux A, Charton J, Fernandois D, Henricsson M, Piveteau C, Simon V, Allard C, Quemener S, Guinot V, Hennuyer N, Perino A, Duveau A, Maitre M, Leste-Lasserre T, Clark S, Dupuy N, Cannich A, Gonzales D, Deprez B, Mithieux G, Dombrowicz D, Bäckhed F, Prevot V, Marsicano G, Staels B, Schoonjans K, and Cota D. 2021. Hypothalamic bile acid-TGR5 signaling protects from obesity. Cell Metab.. [DOI] [PubMed] [Google Scholar]
- 81.Perino A, and Schoonjans K. 2015. TGR5 and Immunometabolism: Insights from Physiology and Pharmacology. Trends Pharmacol. Sci. 36: 847–857. [DOI] [PubMed] [Google Scholar]
- 82.Song X, Sun X, Oh SF, Wu M, Zhang Y, Zheng W, Geva-Zatorsky N, Jupp R, Mathis D, Benoist C, and Kasper DL. 2020. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature 577: 410–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hang S, Paik D, Yao L, Kim E, Trinath J, Lu J, Ha S, Nelson BN, Kelly SP, Wu L, Zheng Y, Longman RS, Rastinejad F, Devlin AS, Krout MR, Fischbach MA, Littman DR, and Huh JR. 2019. Bile acid metabolites control TH17 and Treg cell differentiation. Nature 576: 143–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Campbell C, McKenney PT, Konstantinovsky D, Isaeva OI, Schizas M, Verter J, Mai C, Jin W-B, Guo C-J, Violante S, Ramos RJ, Cross JR, Kadaveru K, Hambor J, and Rudensky AY. 2020. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 581: 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhang S, Li T, Xie J, Zhang D, Pi C, Zhou L, and Yang W. 2021. Gold standard for nutrition: a review of human milk oligosaccharide and its effects on infant gut microbiota. Microb. Cell Fact. 20: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Devika NT, and Raman K. 2019. Deciphering the metabolic capabilities of Bifidobacteria using genome-scale metabolic models. Sci. Rep. 9: 18222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alessandri G, Ossiprandi MC, MacSharry J, van Sinderen D, and Ventura M. 2019. Bifidobacterial Dialogue With Its Human Host and Consequent Modulation of the Immune System. Front. Immunol. 10: 2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, Nagler CR, Ismagilov RF, Mazmanian SK, and Hsiao EY. 2015. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 161: 264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fung TC, Vuong HE, Luna CDG, Pronovost GN, Aleksandrova AA, Riley NG, Vavilina A, McGinn J, Rendon T, Forrest LR, and Hsiao EY. 2019. Intestinal serotonin and fluoxetine exposure modulate bacterial colonization in the gut. Nat Microbiol 4: 2064–2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dover S, and Halpern YS. 1972. Utilization of -aminobutyric acid as the sole carbon and nitrogen source by Escherichia coli K-12 mutants. J. Bacteriol. 109: 835–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Feehily C, and Karatzas KAG. 2013. Role of glutamate metabolism in bacterial responses towards acid and other stresses. J. Appl. Microbiol. 114: 11–24. [DOI] [PubMed] [Google Scholar]
- 92.Strandwitz P, Kim KH, Terekhova D, Liu JK, Sharma A, Levering J, McDonald D, Dietrich D, Ramadhar TR, Lekbua A, Mroue N, Liston C, Stewart EJ, Dubin MJ, Zengler K, Knight R, Gilbert JA, Clardy J, and Lewis K. 2019. GABA-modulating bacteria of the human gut microbiota. Nat Microbiol 4: 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hughes DT, Terekhova DA, Liou L, Hovde CJ, Sahl JW, Patankar AV, Gonzalez JE, Edrington TS, Rasko DA, and Sperandio V. 2010. Chemical sensing in mammalian host-bacterial commensal associations. Proc. Natl. Acad. Sci. U. S. A. 107: 9831–9836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sperandio V, Torres AG, Jarvis B, Nataro JP, and Kaper JB. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. U. S. A. 100: 8951–8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shamir R, Hartman C, Karry R, Pavlotzky E, Eliakim R, Lachter J, Suissa A, and Aviram M. 2005. Paraoxonases (PONs) 1, 2, and 3 are expressed in human and mouse gastrointestinal tract and in Caco-2 cell line: selective secretion of PON1 and PON2. Free Radic. Biol. Med. 39: 336–344. [DOI] [PubMed] [Google Scholar]
- 96.Stoltz DA, Ozer EA, Ng CJ, Yu JM, Reddy ST, Lusis AJ, Bourquard N, Parsek MR, Zabner J, and Shih DM. 2007. Paraoxonase-2 deficiency enhances Pseudomonas aeruginosa quorum sensing in murine tracheal epithelia. Am. J. Physiol. Lung Cell. Mol. Physiol. 292: L852–60. [DOI] [PubMed] [Google Scholar]
- 97.Zargar A, Quan DN, Carter KK, Guo M, Sintim HO, Payne GF, and Bentley WE. 2015. Bacterial secretions of nonpathogenic Escherichia coli elicit inflammatory pathways: a closer investigation of interkingdom signaling. MBio 6: e00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim CS, Gatsios A, Cuesta S, Lam YC, Wei Z, Chen H, Russell RM, Shine EE, Wang R, Wyche TP, Piizzi G, Flavell RA, Palm NW, Sperandio V, and Crawford JM. 2020. Characterization of Autoinducer-3 Structure and Biosynthesis in E. coli. ACS Cent Sci 6: 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Thompson JA, Oliveira RA, Djukovic A, Ubeda C, and Xavier KB. 2015. Manipulation of the quorum sensing signal AI-2 affects the antibiotic-treated gut microbiota. Cell Rep. 10: 1861–1871. [DOI] [PubMed] [Google Scholar]
- 100.Hsiao A, Ahmed AMS, Subramanian S, Griffin NW, Drewry LL, Petri WA Jr, Haque R, Ahmed T, and Gordon JI. 2014. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature 515: 423–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Helfrich EJN, Lin G-M, Voigt CA, and Clardy J. 2019. Bacterial terpene biosynthesis: challenges and opportunities for pathway engineering. Beilstein J. Org. Chem. 15: 2889–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schmid J, Sieber V, and Rehm B. 2015. Bacterial exopolysaccharides: biosynthesis pathways and engineering strategies. Front. Microbiol. 6: 496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nivina A, Yuet KP, Hsu J, and Khosla C. 2019. Evolution and Diversity of Assembly-Line Polyketide Synthases. Chem. Rev. 119: 12524–12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Donia MS, Cimermancic P, Schulze CJ, Wieland Brown LC, Martin J, Mitreva M, Clardy J, Linington RG, and Fischbach MA. 2014. A systematic analysis of biosynthetic gene clusters in the human microbiome reveals a common family of antibiotics. Cell 158: 1402–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, Burian M, Schilling NA, Slavetinsky C, Marschal M, Willmann M, Kalbacher H, Schittek B, Brötz-Oesterhelt H, Grond S, Peschel A, and Krismer B. 2016. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535: 511–516. [DOI] [PubMed] [Google Scholar]
- 106.Rea MC, Sit CS, Clayton E, O’Connor PM, Whittal RM, Zheng J, Vederas JC, Ross RP, and Hill C. 2010. Thuricin CD, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile. Proc. Natl. Acad. Sci. U. S. A. 107: 9352–9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cohen LJ, Kang H-S, Chu J, Huang Y-H, Gordon EA, Reddy BVB, Ternei MA, Craig JW, and Brady SF. 2015. Functional metagenomic discovery of bacterial effectors in the human microbiome and isolation of commendamide, a GPCR G2A/132 agonist. Proc. Natl. Acad. Sci. U. S. A. 112: E4825–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Guo C-J, Chang F-Y, Wyche TP, Backus KM, Acker TM, Funabashi M, Taketani M, Donia MS, Nayfach S, Pollard KS, Craik CS, Cravatt BF, Clardy J, Voigt CA, and Fischbach MA. 2017. Discovery of Reactive Microbiota-Derived Metabolites that Inhibit Host Proteases. Cell 168: 517–526.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Milshteyn A, Colosimo DA, and Brady SF. 2018. Accessing Bioactive Natural Products from the Human Microbiome. Cell Host Microbe 23: 725–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chang Y-L, Rossetti M, Vlamakis H, Casero D, Sunga G, Harre N, Miller S, Humphries R, Stappenbeck T, Simpson KW, Sartor RB, Wu G, Lewis J, Bushman F, McGovern DPB, Salzman N, Borneman J, Xavier R, Huttenhower C, and Braun J. 2019. A screen of Crohn’s disease-associated microbial metabolites identifies ascorbate as a novel metabolic inhibitor of activated human T cells. Mucosal Immunol. 12: 457–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Seyedsayamdost MR 2014. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc. Natl. Acad. Sci. U. S. A. 111: 7266–7271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bushin LB, Covington BC, Rued BE, Federle MJ, and Seyedsayamdost MR. 2020. Discovery and Biosynthesis of Streptosactin, a Sactipeptide with an Alternative Topology Encoded by Commensal Bacteria in the Human Microbiome. J. Am. Chem. Soc. 142: 16265–16275. [DOI] [PubMed] [Google Scholar]
- 113.Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, Birren BW, Takano E, Sali A, Linington RG, and Fischbach MA. 2014. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158: 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Skinnider MA, Dejong CA, Rees PN, Johnston CW, Li H, Webster ALH, Wyatt MA, and Magarvey NA. 2015. Genomes to natural products PRediction Informatics for Secondary Metabolomes (PRISM). Nucleic Acids Res. 43: 9645–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach MA, Weber T, Takano E, and Breitling R. 2011. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 39: W339–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sugimoto Y, Camacho FR, Wang S, Chankhamjon P, Odabas A, Biswas A, Jeffrey PD, and Donia MS. 2019. A metagenomic strategy for harnessing the chemical repertoire of the human microbiome. Science 366. [DOI] [PubMed] [Google Scholar]
- 117.Chu J, Koirala B, Forelli N, Vila-Farres X, Ternei MA, Ali T, Colosimo DA, and Brady SF. 2020. Synthetic-Bioinformatic Natural Product Antibiotics with Diverse Modes of Action. J. Am. Chem. Soc. 142: 14158–14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vila-Farres X, Chu J, Inoyama D, Ternei MA, Lemetre C, Cohen LJ, Cho W, Reddy BVB, Zebroski HA, Freundlich JS, Perlin DS, and Brady SF. 2017. Antimicrobials Inspired by Nonribosomal Peptide Synthetase Gene Clusters. J. Am. Chem. Soc. 139: 1404–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Karim AS, and Jewett MC. 2016. A cell-free framework for rapid biosynthetic pathway prototyping and enzyme discovery. Metab. Eng. 36: 116–126. [DOI] [PubMed] [Google Scholar]
- 120.Colosimo DA, Kohn JA, Luo PM, Piscotta FJ, Han SM, Pickard AJ, Rao A, Cross JR, Cohen LJ, and Brady SF. 2019. Mapping Interactions of Microbial Metabolites with Human G-Protein-Coupled Receptors. Cell Host Microbe 26: 273–282.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]