PURPOSE
Clinical outcomes of patients with neuroblastoma range from spontaneous tumor regression to fatality. Hence, understanding the mechanisms that cause tumor progression is crucial for the treatment of patients. In this study, we show that FOXR2 activation identifies a subset of neuroblastoma tumors with unfavorable outcome and we investigate the mechanism how FOXR2 relates to poor outcome in patients.
MATERIALS AND METHODS
We analyzed three independent transcriptional data sets of in total 1030 primary neuroblastomas with full clinical annotation. We performed immunoprecipitation for FOXR2 and MYCN and silenced FOXR2 expression in two neuroblastoma cell lines to examine the effect on cellular processes, transcriptome, and MYCN protein levels. Tumor samples were analyzed for protein levels of FOXR2 and MYCN.
RESULTS
In three combined neuroblastoma data sets, 9% of tumors show expression of FOXR2 but have low levels of MYCN mRNA. FOXR2 expression identifies a group of patients with unfavorable outcome, showing 10-year overall survival rates of 53%-59%, and proves to be an independent prognostic factor compared with established risk factors. Transcriptionally, FOXR2-expressing tumors are very similar to MYCN-amplified tumors, suggesting that they might share a common mechanism of tumor initiation. FOXR2 knockdown in FOXR2-expressing neuroblastoma cell lines resulted in cell cycle arrest, reduced cell growth, cell death, and reduced MYCN protein levels, all indicating that FOXR2 is essential for these tumors. Finally, we show that FOXR2 binds and stabilizes MYCN protein and MYCN protein levels are highly increased in FOXR2-expressing tumors, in several cases comparable with MYCN-amplified samples.
CONCLUSION
The stabilization of MYCN by FOXR2 represents an alternative mechanism to MYCN amplification to increase MYCN protein levels. As such, FOXR2 expression identifies another subset of neuroblastoma patients with unfavorable clinical outcome.
INTRODUCTION
Neuroblastoma, derived of the sympathetic nervous system, represents the most common extra-cranial solid tumor in pediatric cancer.1,2 The clinical outcome of patients with neuroblastoma is highly variable ranging from spontaneous tumor regression to progression with fatal outcome.3 Low-risk neuroblastoma, primarily classified in International Neuroblastoma Staging System (INSS) 1 and 2, shows 5-year overall survival (OS) rates > 95%, whereas high-risk tumors, often diagnosed in patients above 18 months showing INSS4 status and/or harboring MYCN amplification, indicate a poor clinical outcome of around 40%-50% 5-year OS.4–7 Furthermore, TERT activation, ALK mutations, or alternative lengthening of telomeres (ALT) also identify subgroups of neuroblastoma with an unfavorable clinical outcome.8–11 Here, we report on another independent prognostic group of neuroblastoma with unfavorable outcome that is characterized by expression of Forkhead Box R2 (FOXR2). FOXR2 expression has previously been associated with tumorigenesis, aberrant cell growth, and poor prognosis in, for instance, breast cancer and endometrial adenocarcinoma.12–18 In pediatric tumors, FOXR2 activation by enhancer hijacking is the genetic hallmark of CNS neuroblastomas with FOXR2 activation (CNS NB-FOXR2), a novel distinct pediatric brain tumor entity.19 In addition, subsets or single cases of medulloblastoma, pineoblastoma, or glioblastoma also express elevated FOXR2 levels.16,20,21 First insights into the function of FOXR2 revealed that FOXR2 binds to MYC and MYCN proteins.15,22 The interaction between MYC and FOXR2 proteins seems to stabilize the short-lived MYC protein, which is broadly implicated in the oncogenesis of often aggressive, poorly differentiated tumors.15,22,23
CONTEXT
Key Objective
In this study, we report on a yet unidentified subset of neuroblastoma expressing FOXR2. What is the role of FOXR2 in neuroblastoma tumorigenesis and does it identify an independent prognostic risk factor?
Knowledge Generated
FOXR2 expression identifies a subset of neuroblastoma patients with a poor outcome and proves to be prognostic independent of other well-established risk factors. Furthermore, we found that FOXR2 stabilizes MYCN, thereby presenting an alternative mechanism to MYCN amplifications increasing MYCN protein levels.
Relevance
Identification of FOXR2 as independent prognostic factor in neuroblastoma may further improve the current risk stratification of patients with neuroblastoma. Revealing the mechanism of MYCN stabilization by FOXR2 may explain the correlation with poor outcome and generates important knowledge for therapeutic approaches.
Here, we have investigated the role of FOXR2 in neuroblastoma of the sympathetic nervous system. Using in vitro models and tumor samples, we investigated the mechanistic interaction of FOXR2 and MYCN to understand why patients with FOXR2-expressing tumors have a poor outcome.
MATERIALS AND METHODS
Neuroblastoma Cohorts
Three RNA-based data sets with full clinical annotation were analyzed using the R2 platform24:
Tumor-Neuroblastoma-SEQC-498-RPM-seqcnb1, containing RNA-seq data on 498 primary neuroblastoma25 (Gene Expression Omnibus (GEO):gse62564)
Tumor-Neuroblastoma-Primary-NRC-283-rma_sketch_(bc)-huex10t, containing microarray data on 283 primary neuroblastoma (GEO:gse85047)
Tumor-Neuroblastoma-TARGET-Asgharzadeh-249-custom-huex10t, containing primarily high-risk neuroblastoma26 (dbGaP_ID:phs000218.v22.p8).
Generation of Knockdown Cell Lines
FOXR2 knockdown cell lines were generated by lentiviral transduction of pLKO-TET-ON27,28 with inducible FOXR2 shRNA insert 5′-CTGGAAGAGCACCATTCATTA-3′ or control scrambled insert 5′-CCTAAGGTTAAGTCGCCCTCG-3′. Lentivirus was produced as described previously.29 Experiments were conducted under Biosafety Level 2 conditions in accord with the National Institutes of Health Guidelines.
Quantitative Real-Time Polymerase Chain Reaction
RNA was isolated with the Maxwell RSC using simplyRNA Tissue Kit. Quantitative real-time polymerase chain reaction was performed applying the Power SYBR Green RNA-to-CT 1-Step Kit (Applied Biosystems, Waltham, MA). The fold change was calculated using the 2−ΔΔct method. Primers are listed in the Data Supplement (online only).
Cell Viability
The viability of SK-N-AS was assessed using the CellTiter-Glo Luminescent Cell Viability Assay (Promega, Madison, WI). RealTime-Glo MT Cell Viability Assay (Promega) was used to quantify viability of SK-N-FI. RealTime-Glo dilution was added to the cells and incubated for 1 hour at 37°C before measurement.
Cell Cycle and Cell Death
Cell death was determined using the Annexin V-FITC Apoptosis Detection Kit (Sigma-Aldrich, St Louis, MO) and detected by flow cytometry using BD FACSCanto II. Cell cycle analysis was conducted by measurement of the cellular DNA content by PI staining and flow cytometry. Cells were fixed in 70% ethanol and incubated in PI staining solution (0.1% TritonX-100, 10 µg/mL PI, and 100 µg/mL DNase-free RNase) for 30 minutes at 37°C. Analysis was conducted using FlowJo, on the basis of at least 10,000 events.
Western Blot Analysis
Western blots were performed as described previously29 using FOXR2 (Sigma-Aldrich, HPA057358), MYCN (Sigma-Aldrich, MABE333), MYC (Abcam, Cambridge, United Kingdom, ab32072), HA-tag (Abcam, ab9110), and β-actin (Abcam, ab49900) antibodies and were quantified using ImageJ.
Cyclohexamide Assay
300 ng/µL cyclohexamide (CHX, Sigma-Aldrich) was added to the cells stopping translation. After specific time points, cells were analyzed by western blot.
Co-Immunoprecipitation
Cells were lysed in NETN buffer and precleared using 10 µl of magnetic A/G beads for 1 hour at 4°C. 2 µg of antibody was added to the precleared lysate for 1 hour at 4°C, before 25 µg of magnetic beads were added and incubated overnight at 4°C. Subsequently, target proteins were eluted and analyzed by western blot.
Gene Expression Analysis
Gene expression profiles of SK-N-AS and SK-N-FI with inducible FOXR2 knockdown were generated in triplicates on Affymetrix GeneChip Human U133 Plus2.0 arrays (GEO:GSE156092) and analyzed using the R2 platform. Pathway analyses were conducted through Gene Set Enrichment Analysis30,31 and Ingenuity Pathway Analysis (Qiagen, Hilden, Germany).
Statistical Analyses
FOXR2 levels between knockdown cell lines were assessed using two-tailed t-tests. Differentially expressed genes in SK-N-AS and SK-N-FI upon FOXR2 knockdown were determined using ANOVA P value < .05, corrected using false discovery rate. The value for minimal expression difference and the minimal expression value that should be met in at least one sample of the data set were set to 50 units. Chi-squared analyses were performed to determine the distribution of FOXR2-expressing patients with neuroblastoma between INSS stages and of TERT-positive patients with neuroblastoma over the expression-based groups. Differences in survival (months from initial diagnosis) between groups were analyzed using log-rank tests. Multivariable Cox regression analyses were conducted, evaluating FOXR2 expression in the context of established risk factors, using SPSS.
RESULTS
FOXR2 Is Expressed in a Subset of Neuroblastoma
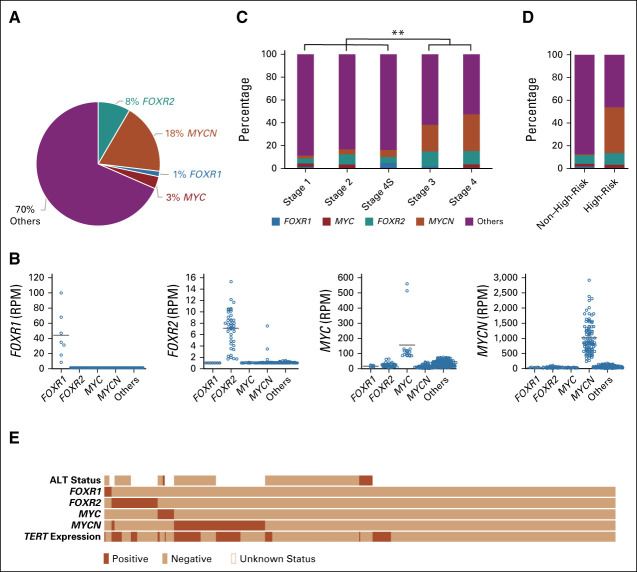
In healthy human tissue, FOXR2 mRNA is usually not expressed, except for male reproductive tissues.32 By contrast, FOXR2 levels are elevated in subsets of various tumors, including brain, breast, prostate, or colorectal cancer, but expression of FOXR2 in neuroblastoma has not been reported, yet.13,16,19–21,33,34 Investigating an RNA-Seq data set of 498 primary neuroblastomas, we identified 42 cases (8%) expressing FOXR2 (Fig 1A). FOXR1, a homologue of FOXR2 and previously identified to be expressed in a small subset of neuroblastomas,35 was expressed mutually exclusive from FOXR2 in seven cases (1%). Interestingly, elevated MYC and MYCN levels, in 90 of 92 (98%) cases driven by gene amplifications, were identified in 14 (3%) and 92 (18%) cases, respectively, and almost mutually exclusive from each other and from the FOXR1- and FOXR2-expressing cases (Figs 1A and 1B).36 All remaining cases (n = 341), here called others group, did not have elevated expression levels of any of these four genes.
FIG 1.
FOXR2 is expressed in a distinct subset of neuroblastomas. (A) Cohort overview of the primary neuroblastoma RNA-seq data set (N = 498). (B) Expression of FOXR1, FOXR2, MYC, and MYCN is in nearly all cases mutually exclusive from another. (C) Percentage of FOXR1-, FOXR2-, MYC-, and MYCN-expressing cases within the INSS stages 1, 2, 3, 4, and 4S. The FOXR2 group is significantly enriched in stages 3 and 4 when compared with stages 1, 2, and 4S. (D) Percentage of MYCN-, FOXR2-, FOXR1-, and MYC-expressing cases within the non–high-risk and the high-risk group (N = 1,016; legend of [C] applies). (E) Oncoplot of the RNA-seq data set (N = 498), showing the status of the FOXR1, FOXR2, MYC, MYCN, and TERT expression and the ALT phenotype. ALT, alternative lengthening of telomeres; INSS, International Neuroblastoma Staging System; RPM, Reads per million mapped reads. **P ≤ .005.
In two other independent neuroblastoma Affymetrix expression data sets, the Neuroblastoma Research Consortium (NRC) series (N = 283) and the TARGET series (N = 249), FOXR2 was present in 17 (6%) and 34 (14%) cases, respectively, and again almost mutually exclusive with subsets that express elevated levels of FOXR1, MYCN, or MYC (Data Supplement).
Investigating the proportion of the FOXR2 group (90 of 1025 cases, 9%) within the INSS stages in all three data sets, we found FOXR2 expression in neuroblastoma of all stages, but significantly enriched among stage 3 and stage 4 tumors (χ2-test, P value .001), similar to that for the MYCN group (Fig 1C). Subdividing by risk group, we observed 8% (39 of 509) FOXR2-expressing cases in the non–high-risk subset and 10% (52 of 507) in the high-risk subset within the three combined data sets (Fig 1D).
FOXR2-Expressing Cases Show Elevated TERT Expression But No ALT Phenotype
MYCN-amplified cases identify a subset of high-risk neuroblastoma and are usually mutually exclusive from cases with TERT rearrangements or ALT phenotype, two other markers associated with poor outcome.9,11,37,38 Analyzing cases with available ALT status (174 of 498 cases), assessed via C-Circle assay as part of another study by Hartlieb et al (submitted), indicated that FOXR2- or FOXR1-expressing neuroblastomas occur like MYCN amplifications mutually exclusively from an ALT phenotype in neuroblastoma (Fig 1E).
On the basis of a defined cutoff (lowest expression level of TERT- among TERT-rearranged cases), we found that TERT-activated cases are significantly enriched for the FOXR2 group (χ2-test, P < .001) and, as expected, the MYCN group (χ2 test, P < .001), whereas the residual groups showed less TERT-activated neuroblastomas (Fig 1E, Data Supplement). This is in line with our observation on higher TERT expression levels in FOXR2-activated cases compared with cases with low FOXR2 expression in an MYCN-nonamplified subset (n = 406; Data Supplement). Because of a low number of cases with status annotations on TERT rearrangement (62 of 498), no conclusions on distribution can be made for those cases. Altogether, FOXR2 expression identifies a subset of neuroblastomas distinct from MYCN-amplified tumors or cases with ALT phenotype, but they share the activated TERT expression with MYCN-amplified tumors.
FOXR2 Identifies Neuroblastoma Patients With Unfavorable Clinical Outcome
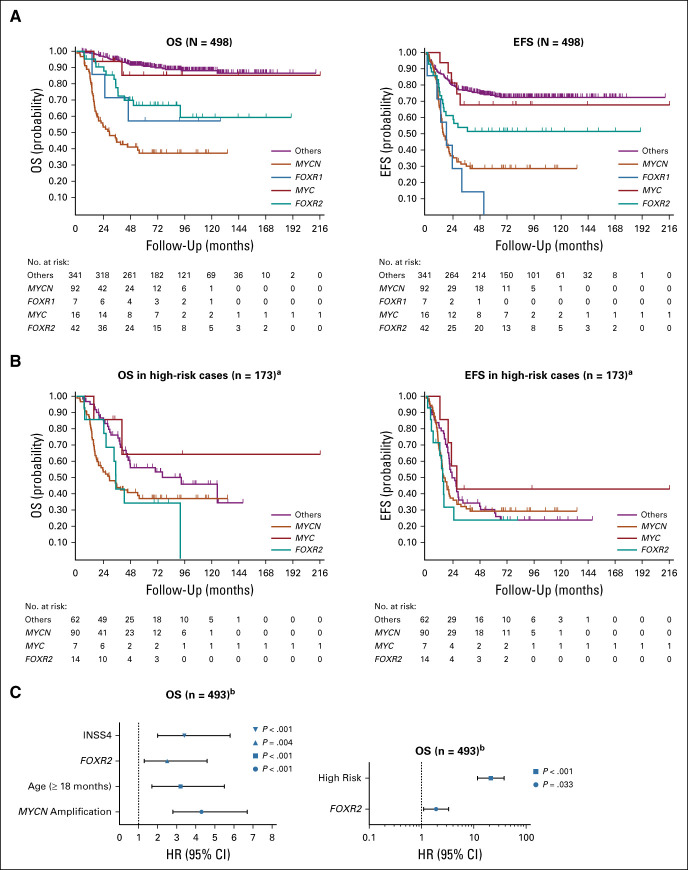
Survival analyses in the RNA-seq data set revealed that patients with FOXR2-expressing neuroblastoma have a significantly (P < .001) reduced survival, showing a 5- and 10-year OS of 67% (95% CI, 49 to 79) and 59% (95% CI, 38 to 75) and a consistent 5- and 10-year event-free survival (EFS) of 51% (95% CI, 35 to 65), whereas the others group shows a comparably good clinical outcome with a 5- and 10-year OS of 91% (95% CI, 87 to 93) and 88% (95% CI, 83 to 91) and a 5- and 10-year EFS of 74% (95% CI, 69 to 78) and 72% (95% CI, 67 to 77; Fig 2A). Only the outcome of the MYCN group was significantly (P = .002) worse than that of the FOXR2 group, showing the consistent 5- and 10-year OS and EFS of 37% (95% CI, 26 to 48) and 29% (95% CI, 19 to 39). Also in the independent NRC data set, FOXR2-expressing tumors have a significantly (P < .001) worse outcome, showing a 5- and 10-year OS of 53% (95% CI, 25 to 74) than the others group with a 5- and 10-year OS of 82% (95% CI, 74 to 87) and 80% (95% CI, 71 to 86), but a better outcome than the MYCN group (5- and 10-year OS of 26% [95% CI, 14 to 40]; Data Supplement). The TARGET series was due to its bias to high-risk tumors solely used as a reference for high-risk subsets.
FIG 2.
Survival data of distinct molecular groups. (A) OS and EFS from initial diagnosis of patients with neuroblastoma of the RNA-seq data set (N = 498) separated by the groups FOXR1, FOXR2, MYC, MYCN, and others. (B) OS and EFS of exclusively high-risk patients in the RNA-seq data set (n = 173). aThe FOXR1 group is excluded because of low number (n = 2). (C) Multivariable Cox regression analyses of the RNA-seq data set (N = 498) for OS taking into account FOXR2 and the established prognostic factors age above 18 months at diagnosis, MYCN amplification, INSS 4, and FOXR2 and risk group in a separate analysis. HRs with 95% CIs and P values are indicated. bThe MYCN amplification status is unavailable for five cases in the RNA-seq data set, and therefore, this multivariate analysis was conducted for 493 of 498 cases. EFS, event-free survival; HR, hazard ratio; OS, overall survival.
Investigating high-risk neuroblastoma only in the RNA-seq, NRC, and TARGET data sets, we observed the very poor 5-year OS of 34% (95% CI, 11 to 60), 0%, and 24% (95% CI, 11 to 40), respectively, for the patients with FOXR2-expressing tumors, which is significantly worse than the others group in NRC and TARGET (P < .001) and almost significant in the RNA-seq data set (P = .052; Fig 2B, Data Supplement). No significant difference in outcome was observed between the FOXR2 and MYCN groups in high-risk neuroblastoma in the RNA-seq (P = .971), NRC (P = .063), and TARGET (P = .172) data sets.
FOXR1-expressing tumors showed a comparable clinical outcome with the FOXR2 group (P = .715; 5- and 10-year OS of 57% [95% CI, 17 to 84]), whereas MYC-expressing tumors revealed a relatively good outcome (5- and 10-year OS of 85% [95% CI, 52 to 96]), but numbers were small in both groups (Fig 2A). Finally, we also compared the FOXR2 group with patients with an ALT phenotype in the ALT-annotated subset of the RNA-seq data set (n = 174), revealing the similar 5-year OS of 72% (95% CI, 41 to 88) and 61% (95% CI, 25 to 84) and the 5-year EFS of 47% (95% CI, 22 to 69) and 29% (95% CI, 9 to 53), respectively. Both the FOXR2 and ALT group showed a significantly worse overall outcome (P < .001 in both cases) than the remaining patients without ALT phenotype or elevated expression of FOXR1, FOXR2, MYC, and MYCN (5-year OS 99% [95% CI, 92 to 100] and an EFS of 74% [95% CI, 64 to 82]; Data Supplement).
FOXR2 Expression Is an Independent Prognostic Risk Factor
The clinical relevance of FOXR2 expression in neuroblastoma was further evaluated by multivariable Cox regression analyses, comparing FOXR2 expression with other well-established risk factors like INSS4 stage, MYCN amplification, and age above 18 months (Data Supplement). In both the RNA-seq and NRC data sets, FOXR2 expression is an independent and significant prognostic risk factor with hazard ratios of 2.5 and 3.5 (P = .004 and P = .003, respectively; Data Supplement). Also, when comparing FOXR2 expression with high-risk status (defined by INSS4 patients above 18 months and MYCN-amplified patients independent from age and INSS stage7) within the whole RNA-seq and NRC cohort, FOXR2 expression sustained in both data sets an independent and significant prognostic marker in multivariable analyses (Fig 2C, Data Supplement). In line with this, within the non–high-risk subset of the NRC and RNA-seq data sets, FOXR2-expressing patients show a significantly reduced survival (P = .012 and P < .001, respectively) compared with patients without or very low FOXR2 expression, indicating that FOXR2 expression identifies patients at higher risk of recurrence among the group of children who are not currently designated as having high-risk disease (Data Supplement). FOXR2 expression remained prognostic also when taking into account other prognostic markers like 11q status or segmental chromosome alterations in general (including 1p, 3p, and 11q deletion and 17q gain) for which data were available in a subset (n = 183) of the RNA-seq data set (Data Supplement). Because of its high-risk bias, the TARGET data set was excluded from these analyses.
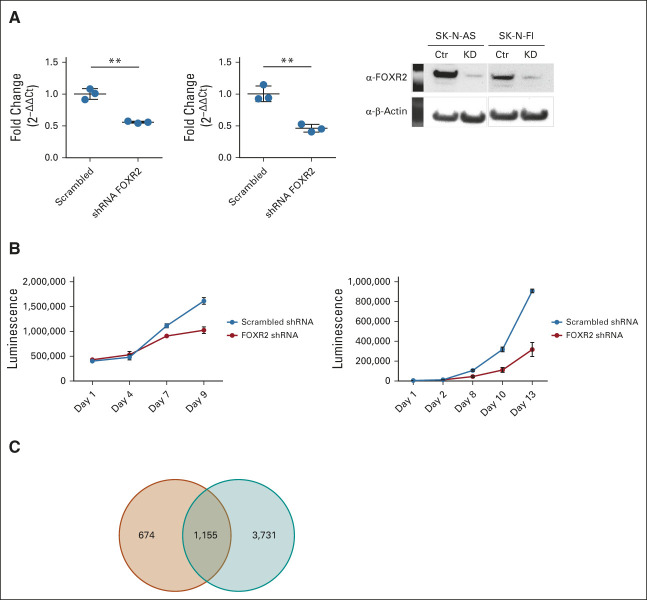
FOXR2 Knockdown Is Associated With Reduced Cellular Growth, Cell Cycle Arrest, and Cell Death in Neuroblastoma Cell Lines
To investigate the role of FOXR2 in neuroblastoma, we selected two neuroblastoma cell lines (SK-N-AS and SK-N-FI) with FOXR2 expression for shRNA-induced knockdown experiments (Fig 3A). Interestingly, in the SK-N-AS line, FOXR2 is activated by enhancer hijacking of an active gene KLHL13. How FOXR2 is activated in SK-N-FI is unclear as no fusions could be detected. Both cell lines do not have an MYCN amplification, but still express relatively low levels of either MYCN mRNA (SK-N-FI) or MYC mRNA, because of a t(4;8) rearrangement (SK-N-AS).39 FOXR2 knockdown resulted in a significantly reduced growth in both cell lines (Fig 3B). Expression analyses after FOXR2 knockdown in SK-N-AS and SK-N-FI identified 4886 and 1829 significantly differentially expressed genes (P value: .05, ANOVA), with 1155 mutually affected genes (Fig 3C). Pathway analyses illustrate the oncogenic role of FOXR2 (Data Supplement), which was further validated with in vitro experiments, showing that FOXR2 knockdown is associated with cell cycle arrest and increased cellular death (Data Supplement).
FIG 3.
FOXR2 silencing reduces proliferation. (A) FOXR2 KD in neuroblastoma cell lines SK-N-AS (left) and SK-N-FI (right) shown by quantitative real-time polymerase chain reaction and western blot. (B) Growth curve of SK-N-AS (left) and SK-N-FI (right) upon FOXR2 KD reveals reduced cell proliferation. (C) Venn diagram indicating the FOXR2 KD signature overlap of SK-N-FI (red) and SK-N-AS (blue). Ctr, control; KD, knockdown. **P ≤ .005.
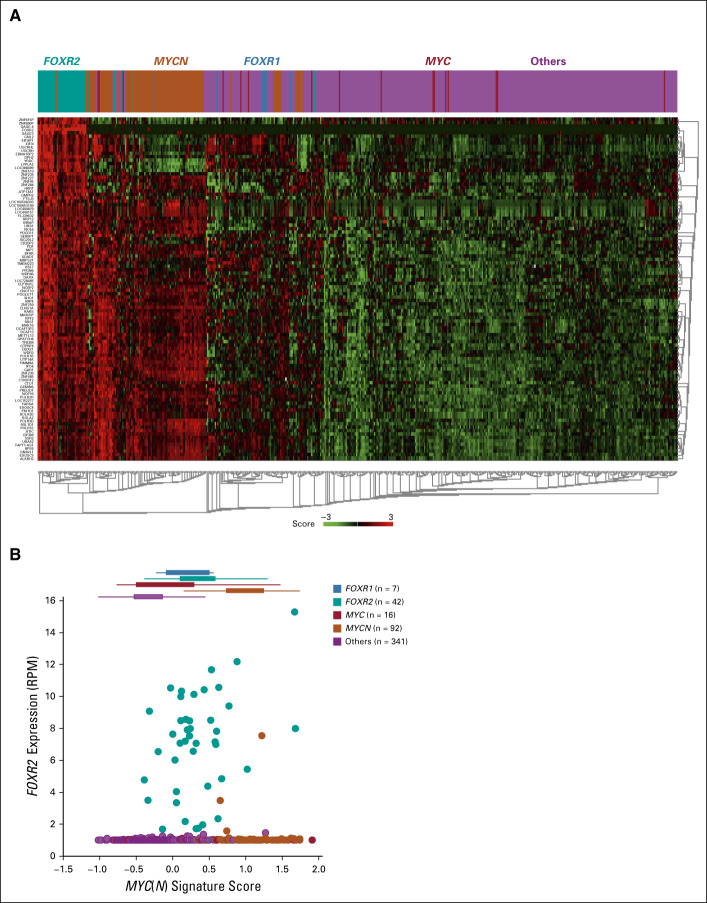
The FOXR2 Group Is Transcriptionally Similar to the MYCN Group
On the basis of transcriptomic data, we found that FOXR2-expressing cases do not form a separate distinct cluster when clustering all cases (Data Supplement), but when comparing FOXR2-expressing cases with the others group and using the differentially expressed genes (Data Supplement) as a gene signature to cluster all cases, we found that the MYCN-expressing cases shared most of the genes that are upregulated in the FOXR2 group (Fig 4A). Also, when using our previously developed MYC(N) signature36 to cluster all cases, we found that the FOXR2 and MYCN groups are transcriptionally related (Data Supplement). Finally, we used our MYC(N) signature to calculate the MYC(N) activity score for all cases. FOXR2-expressing tumors have a positive MYC(N) activity score, in many cases similar in magnitude to MYCN-amplified tumors, indicating that FOXR2-expressing neuroblastomas also activate MYCN target genes despite low MYCN mRNA expression levels (Fig 4B).
FIG 4.
FOXR2 and MYCN tumors are transcriptionally similar. (A) Applying the FOXR2 tumor signature on the RNA-seq data set (N = 498), the MYCN group resembles the FOXR2 group transcriptionally. (B) FOXR2 expression plotted against the MYC(N) signature score reveals a positive MYC(N) activity score for the FOXR2 group.
FOXR2 Binds and Stabilizes MYCN and MYC Protein Levels
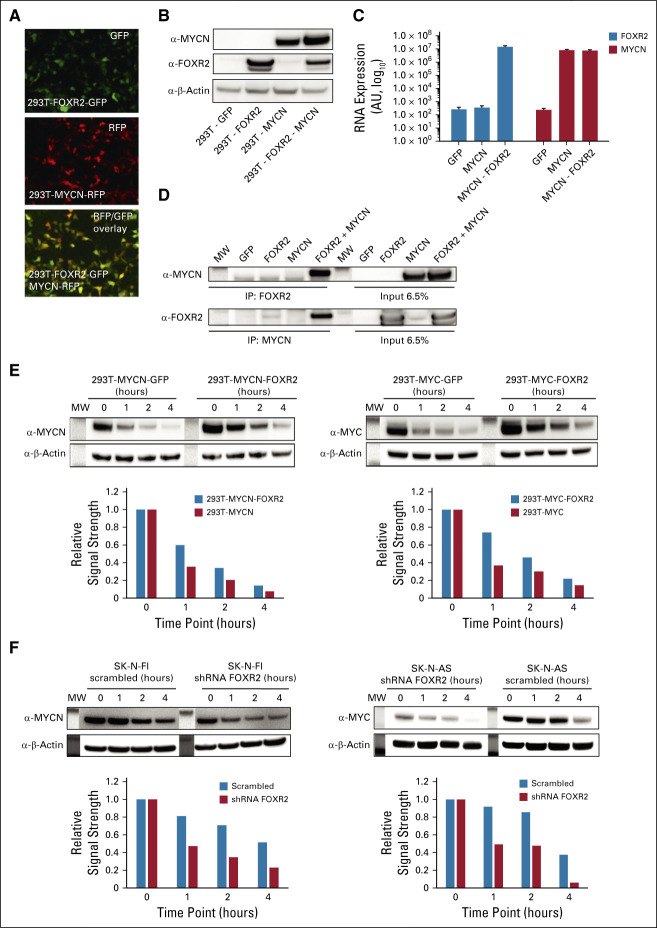
To investigate FOXR2 and MYCN mechanistically, we transfected 293T cells with constructs expressing HA-tagged FOXR2 co-expressed with GFP or MYCN co-expressed with RFP (Fig 5A). Interestingly, western blot analyses showed that when FOXR2 and MYCN were expressed together, MYCN protein levels were about 2-fold higher (Fig 5B), which could not be attributed to increased expression of MYCN RNA (Fig 5C). Co-immunoprecipitation analyses showed that FOXR2 binds to MYCN (Fig 5D) and is thereby stabilizing the short-lived protein, as demonstrated by cycloheximide chase assays, tracking MYCN, and also MYC protein turnover in the presence and the absence of FOXR2 (Figs 5E and 5F).
FIG 5.
FOXR2 protein binds to and stabilizes MYCN protein. FOXR2, MYCN, and a combination of both overexpressed in HEK 293T cells as shown by (A) fluorescence immunostainings, (B) on protein level, and (C) on RNA level. (D) Immunoprecipitation analysis revealed that FOXR2 binds to MYCN. (E) CHX assays of FOXR2- and MYC(N)-overexpressing HEK 293T cells show MYC and MYCN stabilization by FOXR2 on western blot (upper panels). Band intensities were quantified and signal intensities are shown for each time point, normalized to β-actin and the initial signal intensity at the time point 0 hour (lower panel). (F) CHX assays of the FOXR2 knockdown cell lines SK-N-AS and SK-N-FI indicate that MYCN and MYC degrade more rapidly upon FOXR2 knockdown (upper panel). Band intensities were quantified, and signal intensities are shown for each time point, normalized to β-actin and the initial signal intensity at the time point 0 hour (lower panel). CHX, cyclohexamide.
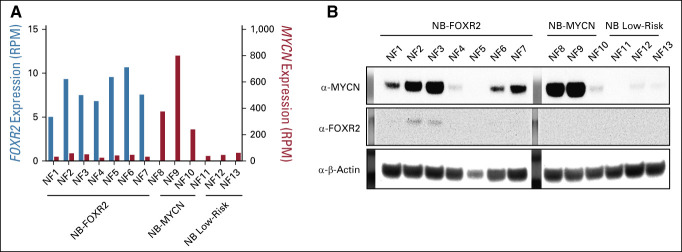
Finally, to confirm stabilization of MYCN protein by FOXR2 in neuroblastoma tissue, we compared FOXR2-activated tumors showing minimal MYCN expression with low-risk samples (without FOXR2 or low MYCN) and MYCN-amplified samples. Despite the minimal MYCN RNA expression, MYCN protein levels were highly increased in 5 of 7 of FOXR2-expressing samples compared with the low-risk tissues. In two of the FOXR2-expressing cases, we even observed MYCN to a level that is similar in the MYCN-amplified cases (Fig 6). FOXR2 protein could hardly be detected by western blot; nonetheless, the two samples that showed an FOXR2 signal were precisely the ones with the highest levels of MYCN protein (Fig 6). From these findings in vitro and in tumor tissue, we conclude that FOXR2 stabilizes MYCN in neuroblastoma on the protein level.
FIG 6.
MYCN protein levels are high in FOXR2-expressing neuroblastoma. (A) mRNA levels and (B) western blot of seven FOXR2-expressing neuroblastomas, three MYCN-amplified neuroblastomas, and three low-risk neuroblastomas reveal that MYCN protein levels are highly increased in most FOXR2 expressing neuroblastoma samples despite low MYCN expression.
DISCUSSION
We have identified FOXR2 expression as a new prognostic marker associated with unfavorable clinical outcome in neuroblastoma, independent of other previously well-established prognostic factors.12,13
Moreover, we have shown that FOXR2 is essential in neuroblastoma and stabilizes MYCN protein, indicating that FOXR2 forms an alternative mechanism to MYCN amplification.40,41 The mechanism of FOXR2 activation in peripheral neuroblastoma remains elusive since in contrast to CNS neuroblastoma with FOXR2 activation,19 gene fusions driving FOXR2 expression in peripheral neuroblastoma were rarely detected. Poor survival of the FOXR2 group was similarly observed within the separately analyzed NB97 and NB2004 treatment studies of the RNA-seq data set (data not shown). Since the response of high-risk patients to different induction42 or consolidation treatment arms was similar in the RNA-seq data set (data not shown), it is unlikely that the outcome of the FOXR2 groups is influenced by the different protocols. Interestingly, the almost identical 5- and 10-year EFS and progression-free survival of FOXR2-expressing neuroblastoma indicate that primarily, only early events occur in this group of patients.
Moreover, we observed enriched TERT expression in the FOXR2-expressing group, most likely a direct consequence of stabilized MYCN, which is a transcriptional activator of TERT.9,43,44
In our study, we observed that high MYC mRNA–expressing patients show a relatively good outcome, in contrast to previous studies.39,45 However, Zimmermann et al39 considered exclusively high-risk patients, whereas in the RNA-seq data set analyzed here, 8 of 16 MYC-expressing cases belong to the non–high-risk group, which might explain varying findings. Moreover, Wang et al45 analyzed MYC protein levels, which are not directly translatable to mRNA levels used in our study.
Limitations of this study, including its retrospective nature and potential confounding related to variability in treatment, underline the need for further prospective studies of uniformly treated patients who are otherwise considered non–high-risk. These requirements in combination with the relatively small number of FOXR2-expressing neuroblastomas would likely require an international collaboration. Additionally, our study was limited by the unavailable subclassification into low risk and intermediate risk and did not allow for further characterization of FOXR2 within these risk groups. The need for biomarkers to identify patients with poor outcome, especially in subsets such as patients age above 18 months, INSS 3, and unfavorable histology, where risk classification is not consistent between different international studies, underlines the importance of further characterization of FOXR2 within risk groups.
Altogether, our study identifies an FOXR2-activated subset in neuroblastoma that shows an unfavorable clinical outcome and we have identified FOXR2 expression as a new independent prognostic factor. Our mechanistic study reveals that MYCN is stabilized by FOXR2, which may explain the poor clinical outcome of patients and provides crucial knowledge for a targeted treatment of FOXR2-activated neuroblastoma.
ACKNOWLEDGMENT
We thank Young-Gyu Park for excellent technical assistance. Furthermore, we thank Sander Lambo, Aniello Federico, Sonja Krausert, and Annette Büllesbach for their support and constructive input. We thank the Microarray Unit of the Genomics and Proteomics Core Facility, German Cancer Research Center (DKFZ), for providing excellent Expression Profiling services.
Jens-Martin Hübner
Employment: InfectoPharm
Sabine Hartlieb
Research Funding: Bayer
Stefan M. Pfister
Research Funding: Lilly, Bayer, Roche, PharmaMar, Pfizer
Patents, Royalties, Other Intellectual Property: Patent on utilizing DNA methylation profiling for tumor classification
No other potential conflicts of interest were reported.
SUPPORT
Supported by funding from the TransOnc priority program of the German Cancer Aid within Grant #70112951 (ENABLE) to S.M.P., F.W., and M.K.
F.S.-H., S.v.R., F.W., and M.K. contributed equally to the work.
AUTHOR CONTRIBUTIONS
Conception and design: Frank Westermann, Marcel Kool
Financial support: Stefan M. Pfister, Frank Westermann, Marcel Kool
Administrative support: Stefan M. Pfister, Frank Westermann, Marcel Kool
Provision of study materials or patients: Kai-Oliver Henrich, Frank Westermann
Collection and assembly of data: Felix Schmitt-Hoffner, Sjoerd van Rijn, Umut H. Toprak, Monika Mauermann, Felix Rosemann, Anke Heit-Mondrzyk, Jens-Martin Hübner, Aylin Camgöz, Sabine Hartlieb, Kai-Oliver Henrich, Frank Westermann, Marcel Kool
Data analysis and interpretation: Felix Schmitt-Hoffner, Sjoerd van Rijn, Umut H. Toprak, Anke Heit-Mondrzyk, Jens-Martin Hübner, Stefan M. Pfister, Kai-Oliver Henrich, Frank Westermann, Marcel Kool
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
FOXR2 Stabilizes MYCN Protein and Identifies Non–MYCN-Amplified Neuroblastoma Patients With Unfavorable Outcome
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Jens-Martin Hübner
Employment: InfectoPharm
Sabine Hartlieb
Research Funding: Bayer
Stefan M. Pfister
Research Funding: Lilly, Bayer, Roche, PharmaMar, Pfizer
Patents, Royalties, Other Intellectual Property: Patent on utilizing DNA methylation profiling for tumor classification
No other potential conflicts of interest were reported.
REFERENCES
- 1.Van Arendonk KJ, Chung DH: Neuroblastoma: Tumor biology and its implications for staging and treatment. Children (Basel, Switzerland) 6:12, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maris JM, Matthay KK: Molecular biology of neuroblastoma. J Clin Oncol 17:2264-2279, 1999 [DOI] [PubMed] [Google Scholar]
- 3.Rosswog C, Schmidt R, Oberthuer A, et al. : Molecular classification substitutes for the prognostic variables stage, age, and MYCN status in neuroblastoma risk assessment. Neoplasia 19:982-990, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meany HJ: Non-high-risk neuroblastoma: Classification and achievements in therapy. Children (Basel) 6:5, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maris JM: Recent advances in neuroblastoma. N Engl J Med 362:2202-2211, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinto NR, Applebaum MA, Volchenboum SL, et al. : Advances in risk classification and treatment strategies for neuroblastoma. J Clin Oncol 33:3008-3017, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W, Yu Y, Hertwig F, et al. : Comparison of RNA-seq and microarray-based models for clinical endpoint prediction. Genome Biol 16:133, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mossé YP, Laudenslager M, Longo L, et al. : Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 455:930-935, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peifer M, Hertwig F, Roels F, et al. : Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature 526:700-704, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheung N-KV, Zhang J, Lu C, et al. : Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA 307:1062-1071, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Valentijn LJ, Koster J, Zwijnenburg DA, et al. : TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet 47:1411-1414, 2015 [DOI] [PubMed] [Google Scholar]
- 12.Deng X, Hou C, Liang Z, et al. : miR-202 suppresses cell proliferation by targeting FOXR2 in endometrial adenocarcinoma. Dis Markers 2017:2827435, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song H, He W, Huang X, et al. : High expression of FOXR2 in breast cancer correlates with poor prognosis. Tumour Biol 37:5991-5997, 2016 [DOI] [PubMed] [Google Scholar]
- 14.Rahrmann EP, Watson AL, Keng VW, et al. : Forward genetic screen for malignant peripheral nerve sheath tumor formation identifies new genes and pathways driving tumorigenesis. Nat Genet 45:756-766, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Wang W, Xi Y, et al. : FOXR2 interacts with MYC to promote its transcriptional activities and tumorigenesis. Cell Rep 16:487-497, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koso H, Tsuhako A, Lyons E, et al. : Identification of FoxR2 as an oncogene in medulloblastoma. Cancer Res 74:2351-2361, 2014 [DOI] [PubMed] [Google Scholar]
- 17.Wang X, He B, Gao Y, et al. : FOXR2 contributes to cell proliferation and malignancy in human hepatocellular carcinoma. Tumour Biol 37:10459-10467, 2016 [DOI] [PubMed] [Google Scholar]
- 18.Li B, Huang W, Cao N, et al. : Forkhead-box R2 promotes metastasis and growth by stimulating angiogenesis and activating hedgehog signaling pathway in ovarian cancer. J Cell Biochem 119:7780-7789, 2018 [DOI] [PubMed] [Google Scholar]
- 19.Sturm D, Orr BA, Toprak UH, et al. : New brain tumor entities emerge from molecular classification of CNS-PNETs. Cell 164:1060-1072, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu APY, Gudenas B, Lin T, et al. : Risk-adapted therapy and biological heterogeneity in pineoblastoma: Integrated clinico-pathological analysis from the prospective, multi-center SJMB03 and SJYC07 trials. Acta Neuropathol 139:259-271, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu X, Liu N, Yue C, et al. : FoxR2 promotes glioma proliferation by suppression of the p27 pathway. Oncotarget 8:56255-56266, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beckmann PJ, Larson JD, Larsson AT, et al. : Sleeping beauty insertional mutagenesis reveals important genetic drivers of central nervous system embryonal tumors. Cancer Res 79:905, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vita M, Henriksson M: The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol 16:318-330, 2006 [DOI] [PubMed] [Google Scholar]
- 24.R2: Genomics Analysis and Visualization Platform : http://r2.amc.nl
- 25.Su Z, Fang H, Hong H, et al. : An investigation of biomarkers derived from legacy microarray data for their utility in the RNA-seq era. Genome Biol 15:523, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pugh TJ, Morozova O, Attiyeh EF, et al. : The genetic landscape of high-risk neuroblastoma. Nat Genet 45:279-284, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wee S, Wiederschain D, Maira SM, et al. : PTEN-deficient cancers depend on PIK3CB. Proc Natl Acad Sci USA 105:13057-13062, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wiederschain D, Wee S, Chen L, et al. : Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle 8:498-504, 2009 [DOI] [PubMed] [Google Scholar]
- 29.Hübner J-M, Müller T, Papageorgiou DN, et al. : EZHIP/CXorf67 mimics K27M mutated oncohistones and functions as an intrinsic inhibitor of PRC2 function in aggressive posterior fossa ependymoma. Neuro Oncol 21:878-889, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subramanian A, Tamayo P, Mootha VK, et al. : Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102:15545-15550, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liberzon A, Subramanian A, Pinchback R, et al. : Molecular signatures database (MSigDB) 3.0. Bioinformatics 27:1739-1740, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.The Human Protein Atlas : https://www.proteinatlas.org/ENSG00000189299-FOXR2/tissue
- 33.Xu W, Chang J, Liu G, et al. : Knockdown of FOXR2 suppresses the tumorigenesis, growth and metastasis of prostate cancer. Biomed Pharmacother 87:471-475, 2017 [DOI] [PubMed] [Google Scholar]
- 34.Lu SQ, Qiu Y, Dai WJ, et al. : FOXR2 promotes the proliferation, invasion, and epithelial-mesenchymal transition in human colorectal cancer cells. Oncol Res 25:681-689, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santo EE, Ebus ME, Koster J, et al. : Oncogenic activation of FOXR1 by 11q23 intrachromosomal deletion-fusions in neuroblastoma. Oncogene 31:1571-1581, 2012 [DOI] [PubMed] [Google Scholar]
- 36.Westermann F, Muth D, Benner A, et al. : Distinct transcriptional MYCN/c-MYC activities are associated with spontaneous regression or malignant progression in neuroblastomas. Genome Biol 9:R150, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koneru B, Lopez G, Farooqi A, et al. : Telomere maintenance mechanisms define clinical outcome in high-risk neuroblastoma. Cancer Res 80:2663-2675, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeineldin M, Federico S, Chen X, et al. : MYCN amplification and ATRX mutations are incompatible in neuroblastoma. Nat Commun 11:1-20, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zimmerman MW, Liu Y, He S, et al. : MYC drives a subset of high-risk pediatric neuroblastomas and is activated through mechanisms including enhancer hijacking and focal enhancer amplification. Cancer Discov 8:320-335, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang JT, Weng ZH, Tsang KS, et al. : MycN is critical for the maintenance of human embryonic stem cell-derived neural crest stem cells. PLoS One 11:e0148062, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang M, Weiss WA: Neuroblastoma and MYCN. Cold Spring Harb Perspect Med 3:a014415, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berthold F, Faldum A, Ernst A, et al. : Extended induction chemotherapy does not improve the outcome for high-risk neuroblastoma patients: Results of the randomized open-label GPOH trial NB2004-HR. Ann Oncol 31:422-429, 2020 [DOI] [PubMed] [Google Scholar]
- 43.Mac SM, D'Cunha CA, Farnham PJ: Direct recruitment of N-myc to target gene promoters. Mol Carcinog 29:76-86, 2000 [PubMed] [Google Scholar]
- 44.Ackermann S, Cartolano M, Hero B, et al. : A mechanistic classification of clinical phenotypes in neuroblastoma. Science 362:1165-1170, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang LL, Teshiba R, Ikegaki N, et al. : Augmented expression of MYC and/or MYCN protein defines highly aggressive MYC-driven neuroblastoma: A Children’s Oncology Group study. Br J Cancer 113:57-63, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]