INTRODUCTION:
To report the prevalence and outcomes of unselected pancreatic cancer (PC) patients with pathogenic/likely pathogenic germline variants (PGVs) detected using a universal testing approach.
METHODS:
We undertook a prospective, multisite study of germline sequencing using a >80 gene next-generation sequencing platform among 250 patients with PC (not selected for age or family history of cancer) between April 1, 2018, and March 31, 2020. Demographic, tumor characteristics, and clinical outcomes were compared between PGV carriers and noncarriers.
RESULTS:
Of 250 patients, the mean age was 65 years (SD 8.7), 56% was male, 83.6% was White, and 65.6% had advanced disease (stages III and IV). PGVs were found in 15.2% (N = 38) of patients, and 2 patients had more than 1 PGV. Variants of uncertain significance were found in 44.4% (N = 111). Family history of cancer (odds ratio: 2.36, 95% confidence interval: 1.14–5.19, P = 0.025) was associated with a higher risk of PGV. In a median follow-up of 16.5 months, the median overall survival was 16.8 months in PGV carriers compared with 16.5 months in noncarriers (hazard ratio: 0.51, 95% confidence interval: 0.25–1.01, P = 0.05). Higher levels of carbohydrate antigen 19-9 and advanced disease stages (III and IV) were associated with worse outcomes in both groups. Overall, 68% of PGV carriers had mutations in homologous recombination repair genes, including BRCA1, BRCA2, PALB2, ATM, CHEK2, NBN, and RAD51C.
DISCUSSION:
Universal multigene panel testing in PC reveals that 1 in 6 patients are carriers of PGV. Multigene germline testing should be used to aid in treatment selection, prognostication, and familial cancer counseling.
INTRODUCTION
Pancreatic cancer (PC) is a lethal disease; 5-year relative survival rate for all stages of the disease combined during 2009 through 2015 was just 9% (1). It is estimated 57,600 new cases of PC were diagnosed in the United States in 2020, with 47,050 resultant deaths (1). Approximately 5%–10% of overall cases occur in patients with a family history of the disease (2–4).
Prevalence of mismatch repair deficiency in PC is rare, ranging between 1% and 2% in recent studies (5–7). Genomic alterations in genes related to homologous recombination repair (HRR) are of extreme interest because strategies exploiting these molecular features have shown promising results in randomized trials with platinum-based therapies and PARP inhibitors (8,9). The prevalence of pathogenic/likely pathogenic germline variants (PGVs) in PC can vary between 3% and 20%, causally related to the type of multigene panel test performed and population studied (10–13). Most of the actionable PGVs in PC are from HRR genes (14–17).
Data from prospective cohorts evaluating disease-specific outcomes in patients with PC harboring HRR defective tumors beyond BRCA are limited (11,17). Some studies have reported improved survival in patients with PC and mutations in the DNA damage response pathway treated with platinum-based regiments and PARP inhibitors (8,9,11,17).
In this article, we report clinical characteristics and outcomes of a multicenter, prospective cohort of patients with PC who underwent germline testing with next-generation sequencing (NGS) using a >80 genes platform. Patients were not selected for stage of disease, family history of cancer, ethnicity, or age.
METHODS
Patient selection
From April 1, 2018, through March 31, 2020, a total of 2,984 unselected adult (aged 18 years or older) patients with a new or active diagnosis of cancer were recruited from medical oncology, radiation oncology, dermatology, and surgical oncology clinics at any of the 4 Mayo Clinic destination Cancer Centers in Phoenix, AZ, Jacksonville, FL, or Rochester, MN, and a community oncology practice in Eau Claire, WI. This represents the prospective Interrogating Cancer Etiology using Proactive Genetic Testing cohort study of patients with solid tumor undergoing germline genetic testing as previously described (18). Patients were recruited using central lists of daily oncology clinic visits by research coordinators at each site. Participants were offered germline sequencing using an NGS panel of 83 genes (84 genes as of July 2019) on the Invitae Multicancer panel at no cost and had disclosure of results. This panel included all cancer predisposing genes identified in the American College of Medical Genetics and Genomics guidelines. Patients were unselected for cancer type, stage of disease, family history of cancer, ethnicity, age at diagnosis, multifocal tumor, or personal history of multiple malignancies. This cohort included 250 patients with a diagnosis of pancreatic adenocarcinoma who underwent genetic testing and comprised the patients analyzed in this study.
Before undergoing genetic testing, all patients viewed a standard pretest education video and were offered additional pretest genetic counseling if they desired. All test results were reviewed by a certified genetic counselor or physicians with expertise in cancer genetics. Individuals with PGVs were invited for genetic counseling. Free family cascade testing was offered. Clinical, demographic, family history, treatment types, and clinical outcome information was collected on all patients in this study either from medical records or self-administered electronic questionnaires for family pedigree information.
This study was approved by the Mayo Clinic Institutional Review Board (18-000326). All patients provided written informed consent. Data were deidentified except to investigators of the study. Neither patients nor the public were involved in the design, conduct, reporting, or dissemination plans of our research.
Sequencing, variant calling, and result reporting
All patients underwent NGS germline genetic testing with a multigene cancer panel of 83 genes (84 genes as of July 2019) on the Invitae Multicancer panel. Full gene sequencing, deletion/duplication analysis, and variant interpretation were performed at Invitae (San Francisco, CA), as previously described (18–20). All patients had their variant findings source verified and confirmed by independent review of the test results by a medical geneticist. PGVs were classified as high (relative risk [RR] > 4), intermediate (RR 2–4) or low (RR < 2) penetrant, recessive, or of a variant of uncertain clinical actionability (variant of uncertain significance [VUS]) based on disease risks and previous modeling.
Statistical analysis
Descriptive statistics for demographic, clinical, and treatment-related characteristics of the cohort were examined. Tests for group differences by site of enrollment and germline testing results were conducted using the Pearson χ2 tests for differences in rates of categorical variables. Analysis of variance F tests to compare distributions of continuous variables. Univariate logistic regression models were used to predict PGVs with patients' age, sex, cancer stage, localization of primary tumor, and family history of cancer as predictors. P values <0.05 were considered statistically significant. Kaplan-Meier analysis of overall survival (OS) and recurrence-free survival was conducted, and survival curves were produced to look for differences in rates of genetic variants (PGV vs VUS vs negative). Rates of detection of clinically actionable findings using 2018 and 2020 National Comprehensive Cancer Network (NCCN) guidelines were calculated. Rates of uptake of family variant testing (FVT) and mutation rates of tested family members were examined.
RESULTS
Cohort characteristics
The distribution of sex, age, comorbidities, stage, and location of primary tumor is summarized in Table 1, stratified by site of enrollment: Southwest (Phoenix, AZ), Midwest (Rochester, MN and Eau Claire, WI), and Southeast (Jacksonville, FL). The median age was 66.5 years at diagnosis, and 56% was male. Fifty-six percentage of patients was never smokers, 13.3% had a body mass index more than 30 kg/m2, and 28% had type 2 diabetes. The proportions with stage I, II, III, and IV disease at the time of enrollment were 14.4%, 20%, 24.8%, and 40.8%, respectively. Race and ethnicity distributions included 6.8% Hispanic/Latino, 5.2% Black/African American, 1.6% Asian, 1.6% American Indian/Alaskan Native, with 83.6% being White. The primary location of the pancreatic tumor was in the head (66.8%), pancreatic body in (17.6%), and pancreatic tail (15.6%). Detailed family history information was available for 124 patients (49.6%), of whom 17 patients (13.7%) had a family history of PC in a first-degree relative.
Table 1.
Clinical and demographic characteristics of included patients
| Southwest (N = 132) | Midwest (N = 54) | Southeast (N = 64) | Total (N = 250) | |
| Sex | ||||
| Male, n (%) | 78 (59.1) | 28 (51.9) | 34 (53.1) | 140 (56.0) |
| Female | 54 (40.9) | 26 (48.1) | 30 (46.9) | 110 (44.0) |
| Age | ||||
| Mean (SD) | 64.9 (8.8) | 65.6 (8.1) | 64.6 (9.2) | 65.0 (8.7) |
| Median | 67.0 | 66.5 | 66.0 | 66.5 |
| Range | 38.0–80.0 | 44.0–79.0 | 40.0–80.0 | 38.0–80.0 |
| Race (grouped) | ||||
| White | 107 (81.1%) | 53 (98.1%) | 49 (76.6%) | 209 (83.6%) |
| Hispanic/Latino | 13 (9.8%) | 0 (0.0%) | 4 (6.2%) | 17 (6.8%) |
| Black/African American | 4 (3.0%) | 1 (1.9%) | 8 (12.5%) | 13 (5.2%) |
| Asian | 3 (2.3%) | 0 (0.0%) | 1 (1.6%) | 4 (1.6%) |
| American Indian/Alaskan Native | 4 (3.0%) | 0 (0.0%) | 0 (0.0%) | 4 (1.6%) |
| Native Hawaiian/Pacific Islander | 1 (0.8%) | 0 (0.0%) | 1 (1.6%) | 2 (0.8%) |
| Other | 0 (0.0%) | 0 (0.0%) | 1 (1.6%) | 1 (0.4%) |
| Ethnicity (dichotomized) | ||||
| Hispanic/Latino | 13 (9.8%) | 0 (0.0%) | 4 (6.2%) | 17 (6.8%) |
| Non-Hispanic | 119 (90.2%) | 54 (100.0%) | 60 (93.8%) | 233 (93.2%) |
| Smoking | ||||
| Yes | 59 (44.7%) | 27 (50.0%) | 25 (39.1%) | 111 (44.4%) |
| No | 73 (55.3%) | 27 (50.0%) | 39 (60.9%) | 139 (55.6%) |
| BMI > 30 kg/m2 | ||||
| Yes | 14 (10.6%) | 5 (9.4%) | 14 (21.9%) | 33 (13.3%) |
| No | 118 (89.4%) | 49 (90.6%) | 50 (78.1%) | 217 (86.7%) |
| Diabetes mellitus | ||||
| Yes | 38 (28.8%) | 9 (16.7%) | 23 (35.9%) | 70 (28.0%) |
| No | 94 (71.2%) | 45 (83.3%) | 41 (64.1%) | 180 (72.0%) |
| Hypertension | ||||
| Yes | 47 (35.6%) | 20 (37.0%) | 30 (46.9%) | 97 (38.8%) |
| No | 85 (64.4%) | 34 (63.0%) | 34 (53.1%) | 153 (61.2%) |
| Germline result | ||||
| Positive | 19 (14.4%) | 6 (11.1%) | 13 (20.3%) | 38 (15.2%) |
| Negative | 55 (41.7%) | 24 (44.4%) | 22 (34.4%) | 101 (40.4%) |
| VUS | 58 (43.9%) | 24 (44.4%) | 29 (45.3%) | 111 (44.4%) |
| Result (dichotomized) | ||||
| Positive | 19 (14.4%) | 6 (11.1%) | 13 (20.3%) | 38 (15.2%) |
| VUS/negative | 113 (85.6%) | 48 (88.9%) | 51 (79.7%) | 212 (84.8%) |
| Pedigree complete | ||||
| Yes | 67 (50.8%) | 24 (44.4%) | 33 (51.6%) | 124 (49.6%) |
| No | 65 (49.2%) | 30 (55.6%) | 31 (48.4%) | 126 (50.4%) |
| Family history of cancer (any) in first-degree relatives | ||||
| Yes | 47 (35.6%) | 21 (38.9%) | 23 (35.9%) | 91 (36.4%) |
| No | 20 (15.2%) | 3 (5.6%) | 10 (15.6%) | 33 (13.2%) |
| Family history of pancreatic cancer in first-degree relative | ||||
| Yes | 10 (14.9%) | 5 (20.8%) | 2 (6.1%) | 17 (13.7%) |
| No | 57 (85.1%) | 19 (79.2%) | 31 (93.9%) | 107 (86.3%) |
| Staging AJCC 8th edition at diagnosis (clinical stage) | ||||
| 1 | 18 (13.6%) | 13 (24.1%) | 5 (7.8%) | 36 (14.4%) |
| 2 | 27 (20.5%) | 10 (18.5%) | 13 (20.3%) | 50 (20.0%) |
| 3 | 33 (25.0%) | 9 (16.7%) | 20 (31.2%) | 62 (24.8%) |
| 4 | 54 (40.9%) | 22 (40.7%) | 26 (40.6%) | 102 (40.8%) |
| Staging AJCC 8th edition (early vs late) | ||||
| Early stage (0–2) | 45 (34.1%) | 23 (42.6%) | 18 (28.1%) | 86 (34.4%) |
| Late stage (3–4) | 87 (65.9%) | 31 (57.4%) | 46 (71.9%) | 164 (65.6%) |
| Location | ||||
| Head | 87 (65.9%) | 35 (64.8%) | 45 (70.3%) | 167 (66.8%) |
| Body | 29 (22.0%) | 9 (16.7%) | 6 (9.4%) | 44 (17.6%) |
| Tail | 16 (12.1%) | 10 (18.5%) | 13 (20.3%) | 39 (15.6%) |
| CA 19-9 | ||||
| >37 | 90 (73.2%) | 40 (75.5%) | 45 (70.3%) | 175 (72.9%) |
| ≤37 | 33 (26.8%) | 13 (24.5%) | 19 (29.7%) | 65 (27.1%) |
| Missing | 9 | 1 | 0 | 10 |
| CA 19-9 | ||||
| >300 | 52 (42.3%) | 23 (43.4%) | 31 (48.4%) | 106 (44.2%) |
| ≤300 | 71 (57.7%) | 30 (56.6%) | 33 (51.6%) | 134 (55.8%) |
| Missing | 9 | 1 | 0 | 10 |
| Deceased | ||||
| Yes | 58 (43.9%) | 11 (20.4%) | 23 (35.9%) | 92 (36.8%) |
| No | 74 (56.1%) | 43 (79.6%) | 41 (64.1%) | 158 (63.2%) |
AJCC, American Joint Committee on Cancer; BMI, body mass index; CA 19-9, carbohydrate antigen; VUS, variant of uncertain significance.
Variants detection and clinical outcomes
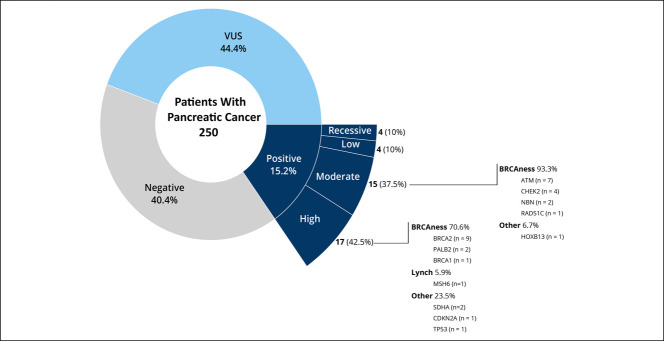
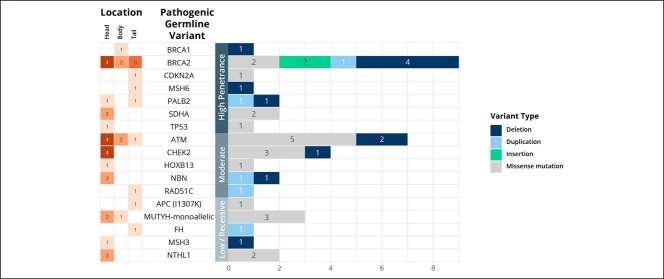
Of the 250 patients undergoing germline analysis, 38 patients (15.2%) harbored 40 PGVs conferring cancer predisposition (Figure 1). Two patients had more than 1 PGV detected, involving PALB2 and MSH3 in 1 patient and TP53 and CHEK2 in the other. PGVs could be stratified into those with high (n = 17), moderate (n = 14), or low (n = 4) penetrance, and 3 patients were carriers of variants associated with recessive syndromes (see Supplementary Table 1, Supplementary Digital Content 1, http://links.lww.com/CTG/A701). The most common of the 40 PGVs were found in BRCA2 (22.5%), ATM (17.5%), and CHEK2 (10%). A molecular diagnosis of Lynch syndrome (PGV in MSH6) was confirmed in 1 patient, and 26 (10.4%) patients had a mutation in a gene associated with HRR pathway (ATM, BRCA1, BRCA2, CHEK2, NBN, PALB2, and RAD51C). Figure 2 shows the distribution of PGVs by gene and pancreatic tumor site. Most of the PGVs were missense mutations (52.5%), followed by deletion (30%), duplication (12.5%), and insertion (5%). Table 2 summarizes the distribution of PGV carriers stratified by site of enrollment, sex, age at diagnosis, race/ethnicity, comorbidities, stage of disease, and tumor location. Having a family history of cancer in a first-degree relative was the only significant predictor of PGV (odds ratio: 2.36, 95% confidence interval [CI]: 1.14–5.19, P = 0.025), whereas age, sex, stage, tumor location, and carbohydrate antigen (CA) 19-9 were not predictive of PGV (see Supplementary Table 2, Supplementary Digital Content 1, http://links.lww.com/CTG/A701). Patients with early-stage PC (0–2) had a higher prevalence of high penetrance genes compared with patients with late-stage (3–4) disease, 46.2% vs 40.7% respectively; however, this was not statistically significant (P = 0.523).
Figure 1.
Distribution of germline testing results.
Figure 2.
Distribution of pathogenic germline variants and pancreatic location of primary tumor.
Table 2.
Clinical and demographic characteristics of pathogenic germline variant carriers
| Positive (N = 38) | VUS/Negative (N = 212) | Total (N = 250) | P value | |
| Enrollment region | 0.356a | |||
| Southwest | 19 (14.4%) | 113 (85.6%) | 132 (100.0%) | |
| Midwest | 6 (11.1%) | 48 (88.9%) | 54 (100.0%) | |
| Southeast | 13 (20.3%) | 51 (79.7%) | 64 (100.0%) | |
| Sex | 0.334a | |||
| Male | 24 (17.1%) | 116 (82.9%) | 140 (100.0%) | |
| Female | 14 (12.7%) | 96 (87.3%) | 110 (100.0%) | |
| Age | 0.206b | |||
| Mean (SD) | 63.3 (8.5) | 65.3 (8.8) | 65.0 (8.7) | |
| Median | 66.0 | 67.0 | 66.5 | |
| Range | 44.0–78.0 | 38.0–80.0 | 38.0–80.0 | |
| Age group, yrs | 0.438a | |||
| Younger than 60 | 11 (28.9%) | 49 (23.1%) | 60 (24.0%) | |
| Aged 60 yrs or older | 27 (71.1%) | 163 (76.9%) | 190 (76.0%) | |
| Race | 0.453a | |||
| White | 32 (15.3%) | 177 (84.7%) | 209 (100.0%) | |
| Hispanic/Latino | 1 (5.9%) | 16 (94.1%) | 17 (100.0%) | |
| Black/African American | 2 (15.4%) | 11 (84.6%) | 13 (100.0%) | |
| Asian | 2 (50.0%) | 2 (50.0%) | 4 (100.0%) | |
| American Indian/Alaskan Native | 1 (25.0%) | 3 (75.0%) | 4 (100.0%) | |
| Native Hawaiian/Pacific Islander | 0 (0.0%) | 2 (100.0%) | 2 (100.0%) | |
| Other | 0 (0.0%) | 1 (100.0%) | 1 (100.0%) | |
| Smoking | 0.143a | |||
| Yes | 21 (18.9%) | 90 (81.1%) | 111 (100.0%) | |
| No | 17 (12.2%) | 122 (87.8%) | 139 (100.0%) | |
| BMI > 30 kg/m2 | 0.590a | |||
| Yes | 4 (12.1%) | 29 (87.9%) | 33 (100.0%) | |
| No | 34 (15.7%) | 183 (84.3%) | 217 (100.0%) | |
| Diabetes mellitus | 0.802a | |||
| Yes | 10 (14.3%) | 60 (85.7%) | 70 (100.0%) | |
| No | 28 (15.6%) | 152 (84.4%) | 180 (100.0%) | |
| Hypertension | 0.528a | |||
| Yes | 13 (13.4%) | 84 (86.6%) | 97 (100.0%) | |
| No | 25 (16.3%) | 128 (83.7%) | 153 (100.0%) | |
| Pedigree complete | 0.685a | |||
| Yes | 20 (16.1%) | 104 (83.9%) | 124 (100.0%) | |
| No | 18 (14.3%) | 108 (85.7%) | 126 (100.0%) | |
| Family history of cancer (any) in first-degree relatives | 0.587a | |||
| Yes | 13 (14.3%) | 78 (85.7%) | 91 (100.0%) | |
| No | 7 (21.2%) | 26 (78.8%) | 33 (100.0%) | |
| Family history of pancreatic cancer in first-degree relative | 0.598a | |||
| Yes | 2 (11.8%) | 15 (88.2%) | 17 (100.0%) | |
| No | 18 (16.8%) | 89 (83.2%) | 107 (100.0%) | |
| Proband/first-degree relative cancer match | 0.372a | |||
| Yes | 4 (11.4%) | 31 (88.6%) | 35 (100.0%) | |
| No | 16 (18.0%) | 73 (82.0%) | 89 (100.0%) | |
| Missing | 18 | 108 | 126 | |
| Staging AJCC 8th edition at diagnosis (clinical stage) | 0.473a | |||
| 1 | 6 (16.7%) | 30 (83.3%) | 36 (100.0%) | |
| 2 | 7 (14.0%) | 43 (86.0%) | 50 (100.0%) | |
| 3 | 6 (9.7%) | 56 (90.3%) | 62 (100.0%) | |
| 4 | 19 (18.6%) | 83 (81.4%) | 102 (100.0%) | |
| Location | 0.141a | |||
| Head | 22 (13.2%) | 145 (86.8%) | 167 (100.0%) | |
| Body | 6 (13.6%) | 38 (86.4%) | 44 (100.0%) | |
| Tail | 10 (25.6%) | 29 (74.4%) | 39 (100.0%) | |
| Deceased | 0.069a | |||
| Yes | 9 (23.7%) | 83 (39.2%) | 92 (36.8%) | |
| No | 29 (76.3%) | 129 (60.8%) | 158 (63.2%) |
AJCC, American Joint Committee on Cancer; BMI, body mass index; VUS, variant of uncertain significance.
The Pearson χ2 test.
Linear model analysis of variance.
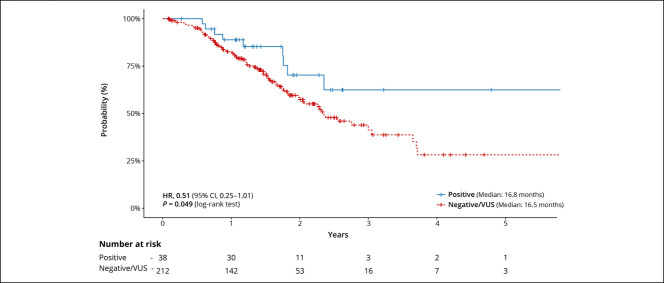
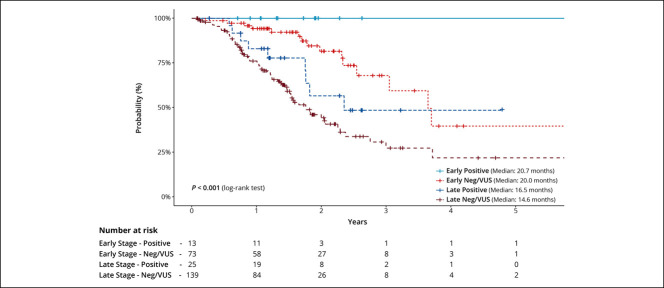
The median OS in PGV carriers was 16.8 months compared with 16.5 months in noncarriers (hazard ratio [HR]: 0.51, 95% CI: 0.25–1.01, P = 0.05, Figure 3). A similar trend (P = 0.054) was observed when comparing PC patients with a PGV in the HRR genes vs noncarriers (see Supplementary Figure 1, Supplementary Digital Content 1, http://links.lww.com/CTG/A701). After stratifying PGV carriers and VUS/negative into subgroups of early-stage (I and II) disease and late-stage (III and IV) disease, PGV carriers had better survival when compared with VUS/negative patients within each stage group (P < 0.0001) (Figure 4). No differences in recurrence-free survival were observed by cancer stage or among those with or without a PGV. Among the entire PC cohort, both late-stage disease (vs early stage, P < 0.001) and higher levels of CA 19-9 (>300 vs ≤ 300, P < 0.001) at diagnosis were associated with worse survival (see Supplementary Figures 2 and 3, Supplementary Digital Content 1, http://links.lww.com/CTG/A701). HR for survival based on PGV status, CA 19-9 level, disease stage, and tumor location is summarized in Supplementary Table 3 (Supplementary Digital Content 1, http://links.lww.com/CTG/A701).
Figure 3.
Kaplan-Meier curves for overall survival of pathogenic germline variants carriers and noncarriers. CI, confidence interval; HR, hazard ratio; VUS, variant of uncertain significance.
Figure 4.
Kaplan-Meier curves for overall survival stratified by stage of pathogenic germline variants carriers and noncarriers. VUS, variant of uncertain significance.
Platinum-based therapy was used in 65% of HRR gene carriers during systemic treatment. Four patients with PGVs were treated with targeted-directed therapy, including PARP inhibitors and immune checkpoint inhibitors, and complete data of systemic treatment in PGV carriers are summarized in Supplementary Tables 4 and 5 (Supplementary Digital Content 1, http://links.lww.com/CTG/A701). The response rate (partial response and complete response) in patients submitted to neoadjuvant treatment or metastatic first-line treatment was higher in PGV carriers compared with noncarriers, 90.9% vs 77.1%, respectively, in neoadjuvant treatment and 68.2% vs 54.5%, respectively, in metastatic first-line treatment (see Supplementary Table 5, Supplementary Digital Content 1, http://links.lww.com/CTG/A701).
Application of clinical genetic referral criteria
Twelve cases (31.5% of PGV) had incremental clinically actionable findings that would not have been detected by phenotype or family history–based testing criteria using the 2018 NCCN guidelines; however, all cases would be detected using the 2020 NCCN guidelines that broadened testing for PC (see Supplementary Table 6, Supplementary Digital Content 1, http://links.lww.com/CTG/A701) (21). Of the 12 patients with incremental findings using the 2018 NCCN guidelines, 1 (8.3%), 7 (58.3%), 2 (16.7%), and 2 (16.7%) patients carried high-penetrance, moderate-penetrance, or low-penetrance mutations and recessive alleles, respectively.
Family variant cascade testing
No-cost FVT was offered to all blood relatives of affected participants. Only 7 (18.4%) patients with PGV had family members undergo FVT within a 3-month window of their test result. On average, only 1 family member was tested (mean = 1.3).
DISCUSSION
In this multisite, prospective study of unselected PC patients, universal multigene panel testing was able to identify 15.2% PGV carriers, equating to nearly 1 in 6 patients with PC harboring a germline predisposition to cancer. Family history of cancer was associated with a greater than 2-fold elevated risk of carrying a PGV. No associations with sex, age, or disease stage were found. Most of the PGVs were in HRR genes, comprising 10% of overall patients included in the cohort. A trend in better OS was observed in PGV carriers vs noncarriers.
The prevalence of PGVs in PC in the literature has shown a large variation between 3% and 20% (3,10–13,22,23). This variation in prevalence can be due to the populations studied and size of gene panel used. Previous prevalence studies of germline variants in PC were often conducted in registry and referral-based (high-risk) populations with targeted gene panels (10,13,22,23). A case-control study with more than 3,000 adults enrolled in a Mayo Clinic registry between 2000 and 2016, using a 21-gene panel, identified PGV in 5.5% of all patients with PC, including 7.9% of patients with a family history of PC (13). Another study in a large group of patients performing expanded next generating sequencing with at least 76 genes associated with cancer susceptibility genes identified 19.8% of PGV carriers, most in HRR genes pathway (11). However, this cohort included a larger proportion of patients with Ashkenazi Jewish heritage (18%), justifying higher PGV when compared with other cohorts (11). Our results are concordant with a recent metanalysis with 60 studies and a combined sample of more than 20,000 patients that found similar rates of prevalence of homologous recombination deficiency in PC (24).
We report a trend in better OS for PGV carriers compared with noncarriers, primarily due to the HRR genes and independent of cancer stage at diagnosis. However, in one of the largest cohorts of unselected patients with pancreatic adenocarcinoma submitted to germline testing, no statistical differences in OS were observed among PGV carriers and noncarriers (48.4 vs 50.8 months) (11). In an analysis of a retrospective biorepository of patients with PC from Mayo Clinic (recruited from 2000 to 2017, before any patients recruited for the current prospective study) found that carriers of PGV in 8 HRR genes had longer OS when compared with noncarriers (HR: 0.83, 95% CI: 0.70–0.97, P = 0.02) (10). A higher response rate is observed in patients with PGV who received platinum-based therapy in comparison with noncarriers, with the best responders being patients with BRCA1 and BRCA2 mutations, although clinical outcomes were not obtained for all patients included in the cohort (11). In our cohort, 65% of PGV carriers was treated with platinum-based therapy at any time in their treatment course, including neoadjuvant treatment, adjuvant treatment, or metastatic disease. Among patients where a response rate could be evaluated (neoadjuvant treatment and metastatic disease), a higher response rate (71% vs 54%) was seen in PGV carriers. In the study by Yadav et al. (25), analysis restricted to PGV carriers with metastatic and nonresectable disease showed that the group treated with platinum-based chemotherapy regimens had better OS compared with carriers not treated with platinum-based therapies. The observation of better response exploiting platinum-based chemotherapy regimens in HRR-defective PC support the hypothesis that this group of patients have greater benefit from platinum-based chemotherapy strategies (9,17). It is unclear what the real impact of incorporation of germline-based targeted therapies is in localized PC, but the findings reported in our manuscript and by others indicate that broader germline testing has clinically meaningful value in the identification of targetable PGV in unselected pancreatic adenocarcinoma. These higher responses rates to chemotherapy could partly explain the better outcomes observed in PGV carriers.
The most common PGV in our study was in BRCA2; 40% of PGV carriers in our report had mutations in non-BRCA HRR genes, with some patients harboring more than 1 PGV. This raises the possibility of expanded treatment options. Olaparib, a PARP inhibitor, improved progression-free survival in a randomized phase 3 trial compared with placebo in germline BRCA1 or BRCA2 mutated metastatic PC after platinum-based therapy for advanced disease; however, OS was not different among groups (8). Veliparib plus upfront cisplatin and gemcitabine in advanced disease does not improve survival when compared with chemotherapy in a randomized phase II trial with BRCA and PALB2 germline patients with PC. However, impressive results with the platinum doublet were achieved (9). In our cohort, 2 patients were treated with PARP inhibitors. These results should highlight the real applicability of PARP inhibitors in maintenance therapy in this highly selective group of advanced PC who have not progressed after platinum-based first-line therapy. The prevalence of microsatellite instability is rare in PC, approximately 1% (7). Just 1 patient was diagnosed with Lynch syndrome (MSH6) in our study and was treated with pembrolizumab on second-line therapy for advanced disease with successful stable disease.
Overall, family cascade testing remained low (<20%) even when available for no cost, suggesting the importance of nonfinancial barriers in this process. This represents a missed opportunity for preventive intervention in family members identified to be at increased risk. PC screening is recommended for family members at increased risk due to PGV in BRCA1, BRCA2, MSH6, ATM, CDKN2A, PALB2, and TP53, among others, which takes the form of annual contrast-enhanced magnetic resonance imaging/magnetic resonance cholangiopancreatography and/or endoscopic ultrasound, with consideration of shorter screening intervals (26). Extra-PC preventive intervention is recommended for family members at increased risk due to PGV in BRCA1, BRCA2, MSH6, ATM, CHEK2, PALB2, RAD51C, and TP53, among others. This can include annual mammography or breast magnetic resonance imaging starting at age 40 years, colonoscopy starting at age 20 years, risk-reducing salpingo-oophorectomy, and risk-reducing mastectomy for early detection and prevention of malignancy in at-risk family members (26,27). Using the 2018 NCCN guidelines, 12 patients with PGV would have been missed. These results reinforce the need to fully realize germline testing for all patients with PC as advocated in the current NCCN guidelines.
Strengths of the study include its prospective, multicenter design, including recruitment from a community oncology clinic, broad stage distribution (35% earlier stage), inclusion of both resected and unresectable cases, and use of a large gene panel. Some limitations of our results include that the demographic characteristics of patients may not completely reflect those in other regions of the country or other countries and the relatively short duration of follow-up. Further long-term follow-up will be necessary to address implications of PGV status and treatment selection.
Universal multigene panel testing in PC reveals that nearly 1 in 6 patients are carriers of a germline cancer predisposition, with most of the mutations being in HRR genes, including BRCA2. PGVs in pancreatic adenocarcinoma are associated with better OS. Multigene germline testing should be offered to all patients with PC (regardless of stage, family history, or age at onset) the findings of which can be used for disease prognosis, treatment selection, and familial cancer counseling.
CONFLICTS OF INTEREST
Guarantor of the article: N. Jewel Samadder, MD, MSc, FRCPC and Katie Kunze, PhD.
Specific author contributions: P.L.S.U., N.J.S., A.K.S., and T.B.-S: study concept and design. P.L.S.U., N.J.S., A.K.S., D.R.J., L.B., D.A., M.B.S., D.O.F., N.F., R.P., K.K., M.G., M.K., E.D.E., R.L.N., and T.B.-S.: acquisition, analysis, and interpretation of data. P.L.S.U., N.J.S., D.R.J., L.B., M.J.B., D.A., M.B.S., D.O.F., N.F., R.P., K.K., M.G., E.D.E., R.L.N., and T.B.-S.: drafting of the manuscript and critical revision of the manuscript for important intellectual content. K.K. and M.G.: statistical analysis. N.J.S. and A.K.S.: obtained funding.
Financial support: Support for this project was provided by Mayo Transform the Practice Grant, Mayo Clinic Center for Individualized Medicine, Desert Mountain Members' CARE Foundation, David and Twila Woods Foundation, and a Faculty Career Development Award from the Gerstner Foundation (N.J.S.).
Potential competing interests: N.J.S. is a consultant for Jansen Research and Development and Cancer Prevention Pharmaceuticals. E.D.E. and R.L.N. are employees and stockholders of Invitae. No other authors have a conflict of interest to disclose.
Study Highlights.
WHAT IS KNOWN
✓ Approximately 5%–10% of pancreatic cancer (PC) cases occur in patients with family history of disease.
✓ Multiple genes are associated with hereditary PC beyond BRCA 1/2, and 5%–10% of pathogenic germline alterations are therapeutically targetable. However, prognostic implications of germline positive testing are still not clear.
WHAT IS NEW HERE
✓ Universal multigene panel testing in PC reveals that nearly 1 in 6 patients with PC harbors a pathogenic germline alteration.
✓ Inherited genetic predisposition plays a critical role in PC including survival probabilities.
✓ Family cascade testing remained low (<20%) even when available for no cost to participants.
Supplementary Material
ACKNOWLEDGMENTS
We thank the following persons for their assistance with this project—Sydney Welp, Jessie Fox, Plush Guiterrez, Sara Hernandez, Sharon Levy, Eric Nelson, Anne Bofferding, Arta Palaj, Lorelei Bandel, Megan Mulcahy, and David Upjohn.
Footnotes
SUPPLEMENTARY MATERIAL accompanies this paper at http://links.lww.com/CTG/A701
Pedro L. S. Uson and N. Jewel Samadder contributed equally to this article.
Contributor Information
Pedro L. S. Uson, Jr, Email: pedroluiz_uson@hotmail.com.
Douglas Riegert-Johnson, Email: riegertjohnson.douglas@mayo.edu.
Lisa Boardman, Email: Boardman.Lisa@mayo.edu.
Mitesh J. Borad, Email: borad.mitesh@mayo.edu.
Daniel Ahn, Email: Ahn.Daniel@mayo.edu.
Mohamad B. Sonbol, Email: Sonbol.Mohamad@mayo.edu.
Douglas O. Faigel, Email: faigel.douglas@mayo.edu.
Norio Fukami, Email: fukami.norio@mayo.edu.
Rahul Pannala, Email: pannala.rahul@mayo.edu.
Katie Kunze, Email: Kunze.Katie@mayo.edu.
Michael Golafshar, Email: Golafshar.Michael@mayo.edu.
Edward D. Esplin, Email: ed.esplin@invitae.com.
Robert L. Nussbaum, Email: robert.nussbaum@invitae.com.
A. Keith Stewart, Email: stewart.keith@mayo.edu.
Tanios Bekaii-Saab, Email: Bekaii-Saab.Tanios@mayo.edu.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7–30. [DOI] [PubMed] [Google Scholar]
- 2.Grover S, Syngal S. Hereditary pancreatic cancer. Gastroenterology 2010;139(4):1076–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salo‐Mullen EE, O'Reilly EM, Kelsen DP, et al. Identification of germline genetic mutations in patients with pancreatic cancer. Cancer 2015;121(24):4382–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brand RE, Lerch MM, Rubinstein WS, et al. Advances in counselling and surveillance of patients at risk for pancreatic cancer. Gut 2007;56(10):1460–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Humphris JL, Patch AM, Nones K, et al. Hypermutation in pancreatic cancer. Gastroenterology 2017;152(1):68–74. [DOI] [PubMed] [Google Scholar]
- 6.Connor AA, Denroche RE, Jang GH, et al. Association of distinct mutational signatures with correlates of increased immune activity in pancreatic ductal adenocarcinoma. JAMA Oncol 2017;3(6):774–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu ZI, Shia J, Stadler ZK, et al. Evaluating mismatch repair deficiency in pancreatic adenocarcinoma: Challenges and recommendations. Clin Cancer Res 2018;24(6):1326–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Golan T, Hammel P, Reni M, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med 2019;381(4):317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Reilly EM, Lee JW, Zalupski M, et al. Randomized, multicenter, phase II trial of gemcitabine and cisplatin with or without veliparib in patients with pancreas adenocarcinoma and a germline BRCA/PALB2 mutation. J Clin Oncol 2020;38(13):1378–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shindo K, Yu J, Suenaga M, et al. Deleterious germline mutations in patients with apparently sporadic pancreatic adenocarcinoma. J Clin Oncol 2017;35(30):3382–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lowery MA, Wong W, Jordan EJ. Prospective evaluation of germline alterations in patients with exocrine pancreatic neoplasms. J Nat Cancer Inst 2018;110(10):1067–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yurgelun MB, Chittenden AB, Morales-Oyarvide V, et al. Germline cancer susceptibility gene variants, somatic second hits, and survival outcomes in patients with resected pancreatic cancer. Gen Med 2019;21(1):213–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu C, Hart SN, Polley EC, et al. Association between inherited germline mutations in cancer predisposition genes and risk of pancreatic cancer. JAMA 2018;319(23):2401–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Witkiewicz AK, McMillan EA, Balaji U, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Comm 2015;6(1):6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47–52. [DOI] [PubMed] [Google Scholar]
- 16.Lowery MA, Jordan EJ, Basturk O, et al. Real-time genomic profiling of pancreatic ductal adenocarcinoma: Potential actionability and correlation with clinical phenotype. Clin Cancer Res 2017;23:6094–100. [DOI] [PubMed] [Google Scholar]
- 17.Pishvaian MJ, Blais EM, Brody JR, et al. Overall survival in patients with pancreatic cancer receiving matched therapies following molecular profiling: A retrospective analysis of the Know Your Tumor registry trial. Lancet Oncol 2020;21(4):508–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samadder NJ, Riegert-Johnson D, Boardman L, et al. Comparison of universal genetic testing vs guideline-directed targeted testing for patients with hereditary cancer syndrome. JAMA Oncol 2021;7(2):230-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurian AW, Hare EE, Mills MA, et al. Clinical evaluation of a multiple-gene sequencing panel for hereditary cancer risk assessment. J Clin Oncol 2014;32(19):2001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nykamp K, Anderson M, Powers M, et al. Sherloc: A comprehensive refinement of the ACMG–AMP variant classification criteria. Genet Med 2017;19(10):1105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.https://www.nccn.org/professionals/physician_gls/pdf/pancreatic.pdf. Accessed September 15, 2020.
- 22.Hu C, Hart SN, Bamlet WR, et al. Prevalence of pathogenic mutations in cancer predisposition genes among pancreatic cancer patients. Cancer Epidemiol Prev Biomarkers 2016;25(1):207–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holter S, Borgida A, Dodd A, et al. Germline BRCA mutations in a large clinic-based cohort of patients with pancreatic adenocarcinoma. J Clin Oncol 2015;33(28):3124–9. [DOI] [PubMed] [Google Scholar]
- 24.Casolino R, Paiella S, Azzolina D, et al. Homologous recombination deficiency in pancreatic cancer: A systematic review and prevalence meta-analysis. J Clin Oncol 2021;39(23):2617-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yadav S, Kasi PM, Bamlet WR, et al. Effect of germline mutations in homologous recombination repair genes on overall survival of patients with pancreatic adenocarcinoma. Clin Cancer Res 2020;26(24):6505-6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daly MB, Pal T, Berry MP, et al. Genetic/familial high-risk assessment: Breast, ovarian, and pancreatic, version 2.2021, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw 2021;19(1):77–102. [DOI] [PubMed] [Google Scholar]
- 27.Gupta S, Provenzale D, Llor X, et al. NCCN guidelines insights: Genetic/familial high-risk assessment: Colorectal, version 2.2019: Featured updates to the NCCN guidelines. J Natl Compr Canc Netw 2019;17(9):1032–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.